Abstract
Background
Metabolic side effects of psychotropic medications are a major drawback to patients’ successful treatment. Using an epigenome-wide approach, we aimed to investigate DNA methylation changes occurring secondary to psychotropic treatment and evaluate associations between 1-month metabolic changes and both baseline and 1-month changes in DNA methylation levels. Seventy-nine patients starting a weight gain inducing psychotropic treatment were selected from the PsyMetab study cohort. Epigenome-wide DNA methylation was measured at baseline and after 1 month of treatment, using the Illumina Methylation EPIC BeadChip.
Results
A global methylation increase was noted after the first month of treatment, which was more pronounced (p < 2.2 × 10–16) in patients whose weight remained stable (< 2.5% weight increase). Epigenome-wide significant methylation changes (p < 9 × 10−8) were observed at 52 loci in the whole cohort. When restricting the analysis to patients who underwent important early weight gain (≥ 5% weight increase), one locus (cg12209987) showed a significant increase in methylation levels (p = 3.8 × 10–8), which was also associated with increased weight gain in the whole cohort (p = 0.004). Epigenome-wide association analyses failed to identify a significant link between metabolic changes and methylation data. Nevertheless, among the strongest associations, a potential causal effect of the baseline methylation level of cg11622362 on glycemia was revealed by a two-sample Mendelian randomization analysis (n = 3841 for instrument-exposure association; n = 314,916 for instrument-outcome association).
Conclusion
These findings provide new insights into the mechanisms of psychotropic drug-induced weight gain, revealing important epigenetic alterations upon treatment, some of which may play a mediatory role.
Supplementary Information
The online version contains supplementary material available at 10.1186/s13148-024-01648-4.
Keywords: DNA methylation, Metabolic side effects, Psychotropic drugs, EWAS
Background
Psychiatric disorders including schizophrenia, bipolar and major depression disorders are associated with a high prevalence of cardiovascular diseases (CVDs) leading to premature death [1]. This excessive cardiovascular risk results from a combination of factors including psychiatric disease-related aspects, shared genetic susceptibilities, unhealthy lifestyle and adverse effects of treatment. Psychotropic medications, such as antipsychotics (most atypical and some typical), mood stabilizers (i.e. lithium and valproate) and some antidepressants (i.e. mirtazapine) can indeed increase the risk of metabolic disorders including obesity, dyslipidemia, type 2 diabetes and hypertension [2, 3]. Although weight gain and obesity are major risk factors for the development of other metabolic abnormalities, dysregulations following psychotropic treatment have also been observed without or with only slight weight gain, suggesting the implication of other mechanisms. Psychotropic drug-induced disturbances in lipid and glucose levels may thus occur following an increase in adiposity and body weight, but also through independent pathways [2, 4, 5]. The multifactorial mechanisms underlying the development of these adverse effects are only partially understood, and epigenetic changes, driven by both environmental and genetic factors, may contribute to explaining their occurrence. Administration of psychotropic drugs may induce alterations in DNA methylation, profoundly influencing gene regulation and expression [6–9]. Although this molecular mechanism is currently extensively studied in relation to treatment response, the pharmacoepigenetic of psychotropic drug-induced metabolic side effects remains underexplored [10–14]. In the general population, some differentially methylated sites within the genome have already been reported to be causally linked to CVDs [15]. It is thus likely that psychotropic drugs also act through epigenetic mechanisms to increase CVDs risks.
Global methylation in relation to atypical antipsychotic treatment and metabolic parameters was assessed in three different studies [16–18]. While the first study yielded no conclusive results [16], the second study showed that atypical antipsychotic use and insulin resistance were both significantly associated with lower global methylation [17], and the last study highlighted a positive correlation between methylation levels and insulin resistance [18]. These mixed preliminary results point towards an effect of psychotropic drugs on global methylation levels leading to metabolic side effects.
In a candidate gene approach, epigenetic analyses focusing on genes or genetic pathways with a highly probable role in the development of metabolic syndrome (MetS) induced by psychotropic medications were performed. DNA methylation of the catechol-O-methyl transferase (COMT) gene [19] and insulin growth factor 2 (IGF2) gene [20] was measured, but no significant relationships between epigenetic variability and metabolic parameters were found. Nonetheless, an association between changes in the methylation level of the CREB-regulated transcription coactivator 1 (CRTC1) gene and early weight gain following psychotropic treatment initiation was shown [21], and a positive trend for increased methylation of protein kinase B (AKT2) associated with insulin resistance was observed in patients treated with atypical antipsychotics, while the opposite correlation was revealed in mood stabilizer users [22].
Hypothesis-driven studies may help reveal how modulation of genes leads to metabolic side effects, but given the mixed results obtained to date, they might fail to capture the complex effects of psychotropic drugs in targeting only specific sites. To overcome this limitation, epigenome-wide association studies (EWAS) may help to further investigate the role of epigenetics in psychotropic drug-induced metabolic side effects. The only studies that used this approach so far allowed to discover a differentially methylated site in the fatty acyl CoA reductase 2 (FAR2) gene that was associated with insulin resistance [23] and another site in the cadherin-like 22 (CDH22) gene that was associated with MetS [24].
Within the current study, we aimed to use the same hypothesis-free strategy to explore global methylation associated with metabolic alterations, in combination with a hypothesis-driven approach, addressing the relationship between site- or gene-specific DNA methylation patterns and metabolic side effects. Unlike previous cross-sectional studies that highlighted associations present in samples of patients at a given time, we wished to capture the effects of psychotropic drugs occurring with treatment onset and followed a longitudinal design, analysing samples collected at the start of treatment and after one month. We then sought to investigate whether baseline methylation or changes in methylation were associated with changes in weight and metabolic parameters.
Methods
Study design
Patients were recruited at the Department of Psychiatry of the Lausanne University Hospital as part of a large observational cohort study (PsyMetab) described elsewhere [25]. Briefly, patients were recruited in PsyMetab when starting a treatment with a psychotropic drug known to induce weight gain and metabolic alterations (including most antipsychotics, mood stabilizers and some antidepressants). They were then followed-up in compliance with a local clinical guideline to control the occurrence of side effects. From this cohort, 79 patients with informed consent, included between February 2008 and February 2016, were selected, as illustrated in the flow chart available in the Additional file 1: Fig. S1. Most patients were not drug naïve and had already received psychotropic medications before entering the study. Early weight gain is a good predictor of metabolic complications following treatment introduction [26, 27]. Patients with a ≥ 5% weight increase in one month show important long-term weight gain (≥ 15% after 3 months; ≥ 20% after 12 months), and the reasons for this rapid and consistent weight gain are only poorly understood. We thus chose to include patients with important early weight gain (n = 39) and patients with no or minimal weight gain (n = 40). A comparative table (Additional file 1: Table S1) with the main characteristics of selected vs unselected patients is provided to ensure that the sample is representative of the whole cohort.
When available, metabolic parameters including blood pressure, plasma levels of glucose, triglycerides, and cholesterol (total cholesterol, LDL cholesterol and HDL cholesterol), were retrieved from medical files. Metabolic syndrome status was assessed using the International Diabetes Federation (IDF) definition [28]. Information on diagnosis, age at medication onset, smoking status and sex was also extracted from medical records and/or specific questionnaires. Diagnostic groups were established according to ICD-10 classification, and psychotropic medications were categorized according to their propensity to induce weight gain in three groups, as already defined in previous analyses involving the PsyMetab cohort [21, 29–31], i.e., low-risk (e.g., amisulpride n = 3, aripiprazole n = 10); medium-risk (e.g., quetiapine n = 23, risperidone n = 8, lithium n = 8, and mirtazapine n = 3), and high-risk (e.g., valproate n = 1, olanzapine n = 19, and clozapine n = 4) [32–35].
DNA methylation
Blood samples were collected for each patient at the start of treatment (T0) and after one month (T1). Genomic DNA was obtained from whole blood as previously described [21]. DNA methylation was analysed at the iGE3 genomics platform of the University of Geneva (Home - iGE3 Genomics Platform - UNIGE) using the Illumina Infinium Methylation EPIC BeadChip, enabling the measurement of over 850,000 methylation sites (Illumina, San Diego, CA, USA).
Statistical analysis
Demographic and clinical characteristics of patients were described and compared between patients who gained 5% or more of their initial weight (considered cases) and patients whose weight remained stable (considered controls) using Wilcoxon Mann–Whitney rank-sum and Pearson χ2 tests for continuous and categorical variables, respectively.
All following statistical analyses were conducted using M-values to estimate methylation levels as this metric shows good statistical validity, and significant results were illustrated using β-values as it enables better interpretability [36]. Principal component analysis was performed on M-values, and the top three principal components were used as covariates in regression analyses to capture unmeasured confounding effects. Additional multivariable models adjusting for cell type composition were also conducted as sensitivity analyses, i.e., estimating the proportion of cell subtypes using the EpiDISH (Epigenetic Dissection of Intra-Sample Heterogeneity) algorithm [37] and alternatively using percent neutrophils measured in blood samples in a subset of patients with available data. All analyses were 2-sided with alpha = 0.05. Data preparation was conducted using Stata 16 (StataCorp; College Station, Texas), and analyses were performed using the R environment for statistical computing version 4.0.2.
Global and epigenome-wide methylation changes and their association with weight change
Global DNA methylation levels were estimated using the mean methylation level of all the analyzed sites, and differences between T0 and T1 were evaluated using paired Student’s t-tests, once in all participants and separately in cases and controls.
To identify loci with epigenome-wide significant T0–T1 changes in the entire cohort, and again, separately according to early weight gain groups, paired Student’s t-tests were performed, not adjusting on additional variables. The family-wise error rate (FWER) was controlled by the Bonferroni correction, and hence, nominal P values passing the 9 × 10–8 threshold (alternatively 4.5 × 10–8 for the stratified analyses) were considered statistically significant [38]. The association between the CpG sites with epigenome-wide significant T0-T1 changes and weight gain was evaluated using a linear model. Weight change was adjusted for baseline BMI, smoking status, sex, age, treatment propensity to induce weight gain (categorized as low, moderate, or high as described above) and the first three principal components of methylation data (or cellular composition, in sensitivity analyses) and normalized using an inverse normal quantile transformation (INQT).
Epigenome-wide association analyses (EWAS) with metabolic phenotypes in PsyMetab and investigation of causality by Mendelian randomization
To identify CpG sites with baseline methylation levels or changes in methylation levels (between T0 and T1) associated with increased body weight gain, linear regressions were performed, using the same linear model as described above. Similarly, to identify CpG sites associated with changes in glucose and lipid plasma levels (fewer patients with available data: glucose (n = 25), triglycerides (n = 39) and total- (n = 38), HDL- (n = 38) and LDL-cholesterol (n = 37)), linear regression models were run, adjusting for covariates of baseline phenotype, smoking status, sex, age, treatment propensity to induce weight gain and the first three principal components of methylation data as well as for the presence of a treatment for diabetes or dyslipidemia for glucose and lipid phenotypes, respectively. For all EWAS, phenotypic traits were normalized, using an INQT. FWER was again controlled by the Bonferroni correction, with nominal P values below 9 × 10–8 considered statistically significant [38].
We selected CpG sites among the top 10 most significant associations with each metabolic phenotype and estimated their causal effect by Mendelian randomization (MR) in independent study samples. MR methodology has been presented in depth elsewhere [39]. Briefly, the random distribution of single nucleotide polymorphisms (SNPs) at birth reduces the possibility of reverse causation or confounding as explanations for the association between the exposure and outcome in the same way that the allocation of an intervention in a randomized controlled trial minimizes this possibility. Methylation quantitative trait loci (cis-meQTLs), discovered by Bonder et al. in a cohort of 3841 European individuals [40], were used as instrumental variables and CpGs linked to a minimum of two SNPs were retained. The association of these genetic variants with the metabolic phenotypes were then derived from genome wide association studies (GWAS) performed in the UKBiobank (UKB), selecting British unrelated individuals (http://www.nealelab.is/uk-biobank, n ≈ 300,000, depending on the phenotype). The details of the UKB have been described elsewhere [41]. Briefly, UKB is a prospective cohort study including more than 500,000 individuals (40–69 years) recruited from the United Kingdom during 2006–2010. Two-sample MR analyses, using the inverse-variance weighted method, were eventually performed to estimate the causal relationships between CpGs and related metabolic phenotypes [42].
Validation of previous findings
The role of methylation patterns in the three genes that have already been associated with metabolic outcomes in psychotropic drug treated patients was further characterized with our data. Thus, the associations between the average methylation in the region of the candidate gene AKT2 (5 out of 22 CpG sites present in our data) [22] and 1-month glucose change were evaluated; the association between 1-month glucose change and the EWAS hit with insulin resistance [23], namely, cg10171063 located in FAR2, was assessed; and the association between the EWAS hit with MetS [24], namely, cg04640913 located in CDH22, and MetS was eventually tested. Linear and logistic models, adjusting for baseline phenotype, smoking status, sex, age, treatment propensity to induce weight gain and the first three principal components of methylation data, were used for these analyses.
Hypothesis-driven analyses
Three hypothesis-driven analyses were performed to select specific subgroups of CpG sites with a putative key role in weight gain or in psychotropic drug-induced metabolic side effects. The associations between baseline or T0-T1 change in methylation level with the metabolic phenotypes in the psychiatric cohort were assessed using the same models as described above. Associations with Bonferroni-corrected P values below 0.05 were considered statistically significant. These three analyses are further described in the Additional file 1.
Results
Population characteristics
The clinical and demographic parameters of the 79 included participants are presented in Table 1. The median age of the cohort was 37 years (range = 16–84) and men represented 50.6% of the patients. The proportion of smokers was 35.9% in patients with early weight gain, while it reached 65.0% in patients whose weight remained stable during the first month of treatment (p = 0.01). Patients suffered mainly from psychotic disorders (45.6%), followed by bipolar disorder (22.8%), schizoaffective disorder (12.7%) and depressive disorders (10.1%), with no difference in relation to early weight gain status (p = 0.33). They were treated primarily with psychotropic drugs carrying an intermediate propensity to induce metabolic side effects (53.2%), while 30.4% and 16.5% of participants received a treatment with a high and low risk, respectively. The median BMI at treatment initiation was 23.1 kg/m2 (range = 15.2–37.5) and it did not differ according to early weight gain status (p = 0.16). Consistent with the study design and participant selection, the weight gain difference across both patient groups during the study period was statistically significant (p < 10–4), and interestingly, a significant increase in MetS prevalence was noted among cases (p = 0.05).
Table 1.
Clinical and demographic parameters of the study sample, with global methylation change in participants stratified according to early weight gain status
| N | Total sample | Controlsa (n = 40) | Casesa (n = 39) | p-valueb | |
|---|---|---|---|---|---|
| Age, median (range), y | 79 | 37 (16–84) | 37 (17–84) | 39 (16–83) | 0.56 |
| Men, n (%) | 79 | 40 (50.6) | 20 (50.0) | 20 (51.3) | 0.9 |
| Smoking, n (%) | 79 | 40 (50.6) | 26 (65.0) | 14 (35.9) | 0.01 |
| Main diagnosis, n (%) | 79 | 0.33 | |||
| Psychotic disorders (F20-F24; F28-F29) | 36 (45.6) | 17 (42.5) | 19 (48.7) | ||
| Schizoaffective disorders (F25) | 10 (12.7) | 6 (15.0) | 4 (10.3) | ||
| Bipolar disorders (F30–F31) | 18 (22.8) | 12 (30.0) | 6 (15.4) | ||
| Depressive disorders (F32–F33) | 8 (10.1) | 2 (5.0) | 6 (15.4) | ||
| Other | 7 (8.9) | 3 (7.5) | 4 (10.3) | ||
| Psychotropic treatment group, n (%)c | 79 | 0.74 | |||
| Low risk of WG | 13 (16.5) | 6 (15.0) | 7 (18.0) | ||
| Medium risk of WG | 42 (53.2) | 23 (57.5) | 19 (48.7) | ||
| High risk of WG | 24 (30.4) | 11 (27.5) | 13 (33.3) | ||
| BMI, median (range), kg/m2 | 79 | ||||
| Baseline | 23.1 (15.2–37.5) | 23.5 (17.1–36.5) | 21.9 (15.2–37.5) | 0.16 | |
| First month | 23.9 (17.0–39.5) | 23.8 (17.1–36.5) | 24.1 (17.0–39.5) | 0.81 | |
| p-valued | < 10–4 | < 10–4 | < 10–4 | ||
| WG, median (range), % | 2.4 (0–23.0) | 0.5 (0–2.4) | 7 (5.2–23.0) | < 10–4 | |
| Metabolic syndrome prevalence, n (%)e | 51 | ||||
| Baseline | 3 (5.9) | 3 (11.1) | 0 (0.0) | 0.09 | |
| First month | 8 (15.7) | 4 (14.8) | 4 (16.7) | 0.86 | |
| p-valued | 0.23 | 0.32 | 0.05 | ||
| Global baseline (T0) methylation (β-value), mean (range), % | 79 | 61.78 (58.52–64.01) | 61.80 (58.52–64.01) | 61.77 (59.20–63.86) | 0.87 |
| Global methylation (β-value) increase (T1–T0), mean (95%CI), % | 79 | 0.187 (0.185–0.190) | 0.201 (0.198–0.204) | 0.174 (0.170–0.177) | < 2.2 × 10–16 |
BMI body mass index, WG weight gain
aPatients who gained 5% or more of their initial weight were considered cases, and patients whose weight remained stable were considered controls
bStatistical significance for differences between groups was tested using the Wilcoxon Mann–Whitney rank-sum test for continuous variables (except for the differences in methylation levels which were assessed using Student’s t-test) and Pearson χ2 test of independence for categorical variables. Significant p-values (< 0.05) are indicated in bold
cPsychotropic drugs are considered to confer a low, medium and high risk of metabolic side effects for amisulpride and aripiprazole; risperidone, quetiapine, mirtazapine and lithium; and valproate, clozapine and olanzapine, respectively
dStatistical significance for differences between baseline and 1-month values was tested using the Wilcoxon signed rank test for matched pairs for continuous variables and McNemar test for categorical variables. Significant p values (< 0.05) are indicated in bold
eMetabolic syndrome was evaluated according to the definition of the International Diabetes Federation [28]
Global and epigenome-wide methylation changes and their association with weight change
The global mean methylation level was higher one month after treatment initiation (Table 1), and this increase was greater in patients whose weight remained stable (p < 2.2 × 10–16).
A significant change between baseline and 1-month methylation levels (p < 9 × 10−8) was observed in 52 methylation sites in the entire cohort. The complete list of CpG sites and their methylation levels is available in Table 2, with information related to the genomic location of these sites. The change in methylation level at these 52 CpG sites was not associated with weight gain following treatment onset (nominal p > 0.09, data not shown).
Table 2.
Baseline and 1-month methylation levels of 52 methylation sites with significant changes following treatment initiation
| CpG site | Baseline methylation level (β-value), median (IQR), % | 1-month methylation level (β-value), median (IQR), % | p-valuea | CHR | CpG positionb | Reference genec | Location of CpG related to genec | Relation to CpG Islandd |
|---|---|---|---|---|---|---|---|---|
| cg10992198 | 67.7 (64.3–71.5) | 70.7 (66.2–73.1) | 0.00005 | 19 | 36,552,038 | WDR62* | Body | |
| cg12120973 | 68.3 (65.1–71.7) | 70.0 (67.1–74.3) | 0.00009 | 1 | 1,215,925 | SCNN1D* | 5'UTR | |
| cg05034501 | 84.6 (82.9–86.4) | 87.1 (85.1–89.5) | 0.0001 | 3 | 148,985,369 | |||
| cg05235884 | 81.3 (80.2–84.1) | 82.8 (80.9–84.8) | 0.0001 | 6 | 30,131,806 | TRIM15 | 1stExon | |
| cg11678481 | 75.6 (73.9–77.8) | 77.8 (75.6–78.9) | 0.0001 | 1 | 151,682,882 | CELF3* | Body | |
| cg13422535 | 61.9 (58.9–64.1) | 62.2 (60.2–65.6) | 0.0001 | 22 | 47,077,682 | S_Shelf | ||
| cg22329201 | 76.1 (73.1–77.4) | 77.1 (74.8–79.3) | 0.0002 | 4 | 3,569,189 | S_Shelf | ||
| cg20548564 | 89.4 (88.3–90.6) | 90.5 (89.3–91.3) | 0.0004 | 13 | 28,240,073 | POLR1D | Body | |
| cg20626144 | 74.0 (72.0–76.1) | 75.1 (73.9–77.3) | 0.0005 | 17 | 157,107 | RPH3AL | Body | Island |
| cg24357026 | 9.8 (7.7–11.4) | 8.3 (6.7–9.9) | 0.0005 | 19 | 36,705,589 | ZNF146* | 5'UTR | Island |
| cg02644728 | 74.1 (71.8–76.7) | 75.6 (73.7–77.9) | 0.0005 | 11 | 83,324,374 | DLG2* | Body | |
| cg02692850 | 65.5 (63.0–67.3) | 66.4 (64.3–67.9) | 0.0006 | 21 | 44,607,862 | |||
| cg05129295 | 56.4 (54.2–58.7) | 58.4 (55.7–60.4) | 0.0007 | 8 | 1,316,294 | |||
| cg05696006 | 79.0 (76.5–79.9) | 79.9 (77.9–81.4) | 0.0007 | 19 | 7,622,906 | PNPLA6* | Body | S_Shore |
| cg15223933 | 74.5 (72.9–76.1) | 75.5 (74.1–77.0) | 0.0008 | 17 | 39,916,203 | JUP* | Body | S_Shelf |
| cg13691093 | 87.4 (84.9–89.2) | 89.0 (86.9–91.0) | 0.002 | 8 | 2,031,672 | MYOM2 | Body | |
| cg27452651 | 7.1 (6.1–8.1) | 6.2 (5.4–6.9) | 0.002 | 22 | 50,312,357 | ALG12* | TSS1500 | Island |
| cg13982468 | 88.8 (87.3–89.6) | 89.8 (88.9–90.6) | 0.004 | 12 | 120,571,393 | GCN1 | Body | |
| cg22800959 | 94.6 (93.6–95.3) | 95.2 (94.4–95.9) | 0.004 | 6 | 32,020,477 | TNXB | Body | |
| cg11702503 | 82.2 (80.3–84.0) | 83.3 (81.6–85.0) | 0.004 | 19 | 6,215,254 | MLLT1 | Body | Island |
| cg20710898 | 69.8 (68.5–71.4) | 71.4 (69.7–73.2) | 0.004 | 10 | 96,996,833 | |||
| cg23628099 | 68.2 (64.7–72.0) | 70.0 (67.3–73.6) | 0.004 | 8 | 48,089,762 | |||
| cg25226092 | 63.2 (61.4–64.6) | 64.3 (62.4–66.3) | 0.005 | 3 | 39,508,863 | MOBP* | TSS1500 | |
| cg07769732 | 65.3 (61.9–67.7) | 67.0 (62.9–70.3) | 0.005 | 2 | 8,815,465 | N_Shore | ||
| cg10825881 | 77.7 (75.7–79.0) | 78.5 (77.4–80.0) | 0.005 | 15 | 52,202,509 | TMOD3 | 3'UTR | |
| cg17866025 | 62.4 (59.7–64.5) | 64.0 (61.6–65.5) | 0.006 | 1 | 39,051,682 | |||
| cg18930928 | 79.2 (77.7–81.2) | 80.2 (78.6–82.1) | 0.006 | 1 | 205,210,953 | TMCC2 | Body | S_Shore |
| cg02624558 | 5.2 (4.3–6.7) | 4.9 (3.6–5.8) | 0.008 | 1 | 202,777,611 | KDM5B | TSS200 | Island |
| cg05103574 | 80.3 (78.6–82.0) | 81.4 (79.9–83.2) | 0.008 | 1 | 29,527,219 | MECR* | Body | |
| cg08243790 | 64.1 (62.1–65.9) | 65.4 (63.8–67.3) | 0.008 | 8 | 127,485,353 | |||
| cg14885690 | 78.4 (76.2–80.3) | 79.9 (77.8–82.4) | 0.009 | 1 | 43,195,659 | |||
| cg27275821 | 75.5 (72.8–78.0) | 78.6 (76.0–80.5) | 0.009 | 1 | 89,144,331 | PKN2-AS1 | Body | |
| cg00661205 | 75.6 (74.4–77.6) | 77.0 (75.6–78.3) | 0.01 | 13 | 42,400,902 | KIAA0564* | Body | |
| cg08581040 | 83.1 (80.4–85.0) | 84.5 (82.5–86.6) | 0.01 | 4 | 39,172,577 | S_Shore | ||
| cg09518293 | 71.0 (68.6–73.1) | 72.2 (70.8–74.9) | 0.01 | 2 | 179,673,636 | TTN* | TSS1500 | |
| cg10231096 | 85.8 (82.7–89.2) | 87.5 (84.3–90.5) | 0.01 | 15 | 31,451,780 | TRPM1 | Body | |
| cg13003350 | 81.2 (78.4–83.7) | 83.2 (80.0–85.2) | 0.01 | 6 | 27,830,500 | N_Shelf | ||
| cg13091133 | 67.4 (65.9–70.1) | 69.4 (66.7–71.7) | 0.01 | 4 | 42,190,340 | |||
| cg16106297 | 75.2 (73.2–77.0) | 76.3 (74.5–77.8) | 0.01 | 19 | 58,556,609 | ZSCAN1 | Body | Island |
| cg24870895 | 76.0 (74.1–77.6) | 77.6 (75.3–78.9) | 0.01 | 15 | 40,768,542 | |||
| cg26032412 | 52.5 (51.0–54.1) | 53.8 (51.6–55.5) | 0.01 | 10 | 130,300,419 | S_Shore | ||
| cg01823005 | 79.0 (75.8–82.6) | 82.0 (78.3–84.1) | 0.02 | 10 | 90,691,868 | ACTA2-AS1 | TSS1500 | |
| cg03797660 | 89.6 (88.1–91.1) | 90.6 (89.7–91.9) | 0.02 | 16 | 89,035,228 | CBFA2T3 | Body | S_Shore |
| cg04043710 | 67.6 (64.0–70.4) | 69.4 (65.9–71.0) | 0.02 | 16 | 9,448,891 | |||
| cg05864168 | 89.7 (88.8–91.0) | 90.8 (89.6–91.8) | 0.02 | 1 | 185,110,088 | TRMT1L* | Body | |
| cg11867718 | 70.7 (68.5–72.9) | 72.1 (69.8–73.7) | 0.02 | 11 | 61,647,697 | FADS3 | Body | |
| cg19267144 | 76.5 (75.2–78.5) | 77.8 (76.2–79.6) | 0.02 | 17 | 40,956,947 | CNTD1 | Body | |
| cg20073412 | 95.5 (94.9–96.3) | 96.3 (95.6–97.0) | 0.02 | 11 | 131,560,558 | NTM | Body | Island |
| cg23313005 | 82.7 (81.6–84.2) | 84.0 (82.7–85.4) | 0.02 | 5 | 176,965,046 | FAM193B* | Body | |
| cg25470611 | 74.8 (71.4–78.0) | 76.3 (72.9–79.7) | 0.02 | 11 | 71,121,630 | FLJ42102 | Body | |
| cg26856604 | 74.4 (71.0–76.7) | 75.8 (73.6–77.8) | 0.02 | 1 | 10,370,540 | KIF1B | Body | |
| cg05058976 | 86.1 (84.5–87.3) | 87.1 (85.6–88.3) | 0.03 | 16 | 3,637,956 | BTBD12 | Body | N_Shore |
CHR chromosome, IQR interquartile range
aStatistical significance for differences between baseline and 1-month methylation was tested using paired Student’s t-test, without adjustment on additional factors. Bonferroni correction for epigenome-wide analyses was applied
bCpG positions refer to Genome Research Consortium human genome build 37 (GRCh37)/UCSC human genome 19 (hg19)
cReference genes for the methylation sites, and gene regions where the CpGs are located according to the UCSC database. Empty fields indicate an intergenic location
*Specifies there exists > 1 gene or gene transcript at this location. TSS200 = 0–200 bases upstream of the transcriptional start site (TSS); TSS1500 = 200–1500 bases upstream of the TSS; 5′UTR = within the 5′ untranslated region, between the TSS and the ATG start site; Body = Between the ATG and stop codon, irrespective of the presence of introns, exons, TSS, or promoters; 3′UTR = between the stop codon and poly A signal
dThe relation to a putative nearby CpG island, according to the UCSC database, is given. Shore = 0–2 kb from island; Shelf = 2–4 kb from island; N = upstream (5′) of CpG island; S = downstream (3′) of CpG island
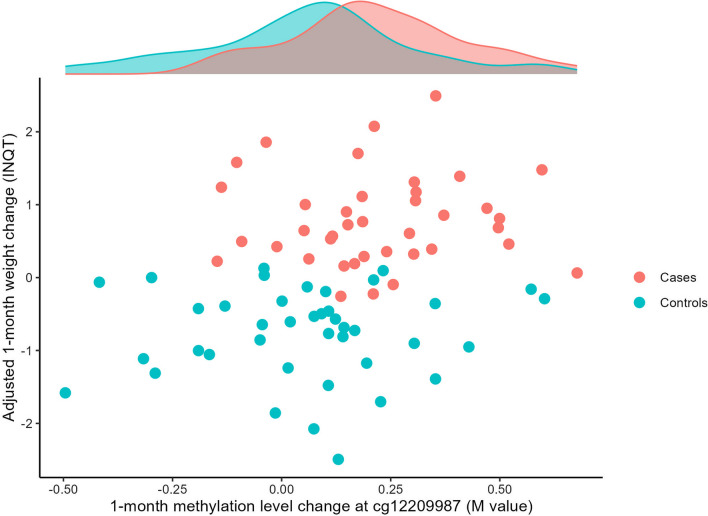
The stratified analyses, restricted to samples with early weight gain, revealed a significant change (between T0 and T1) in methylation at one specific locus, namely cg12209987. This methylation site is located on chromosome 5, in an intergenic region 60 kb upstream of the PSMC1P5 pseudogene. Its median methylation level increased from 69.7% (interquartile range [IQR] = 66.1–73.6) at baseline to 72.5% (IQR = 70.7–74.4) after a 1-month treatment (pcorr = 0.04). In the entire cohort, the change in its methylation level was found to be related to weight change, as shown in Fig. 1, although this association did not reach epigenome-wide statistical significance. The linear model predicted that a 2.8% difference in methylation level was associated with 0.85 kg of excess weight gain (95% CI 0.28–1.43, p = 0.004). Of note, this association remained significant when adjusting for white blood cell composition (0.78 kg of excess weight gain (95% CI 0.19–0.59, p = 0.01) when adjusting for estimated cell subtype variation and 0.86 kg of excess weight gain (95% CI 0.08–1.65, p = 0.03) when adjusting for measured neutrophil variation in the restricted sample).
Fig. 1.
Association between 1-month methylation level change at cg12209987 and weight change. Weight change was adjusted for baseline BMI, smoking status, sex, age, treatment propensity to induce weight gain and the first three principal components of methylation data, and normalized using an inverse normal quantile transformation (INQT). The methylation level change was assessed using the M-value metric. Patients who gained 5% or more of their initial weight were considered cases, and patients whose weight remained stable were considered controls
Epigenome-wide association analyses (EWAS) with metabolic phenotypes in PsyMetab and investigation of causality by Mendelian randomization
Associations between CpG methylation sites and changes in metabolic outcomes did not reach epigenome-wide significance. The Q–Q plots were checked and showed the absence of inflation, as shown in the Additional file 1: Fig. S2, for the EWAS investigating the association between the change in methylation levels (T1–T0) and the increase in body weight. Among the top 10 most significant associations of each EWAS (6 metabolic traits evaluated with baseline methylation and 1-month methylation change: 120 sites selected), only seven methylation loci were associated with a minimum of two meQTLs, enabling the estimation of a causal effect through MR (Table 3). One of the tested associations, cg11622362 (on chromosome 11), located close to the transcription start site of the APIP gene and opposite the PDHX gene body, revealed a significant effect. Namely, a higher methylation level was shown to causally relate to higher glucose levels (p = 10–4).
Table 3.
Mendelian randomization analysis results of causal CpG sites for metabolic traits, selected among the top ten EWAS most significant hits
| Exposure | MR resultsa | EWAS resultsb | CHR | CpG position in Genome Build 37 | Reference genec | Location of CpG related to genec | Relation to CpG Islandd | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Outcome | N SNPs | Beta | SE | p-value | CpG data | Beta | SE | p-value | ||||||
| cg11622362 | Glucose | 2 | 0.019 | 0.005 | 0.0001 | T0 | − 1.05 | 0.22 | 2.96 × 10–5 | 11 | 34,938,112 | APIP* | TSS200 | Island |
| cg09137125 | Glucose | 2 | − 0.003 | 0.007 | 0.63 | T0 | − 0.93 | 0.19 | 3.51 × 10–5 | 10 | 7,486,981 | |||
| cg26295559 | Glucose | 4 | 0.003 | 0.004 | 0.55 | T0-T1 | − 2.40 | 0.46 | 1.02 × 10–5 | 7 | 158,550,318 | ESYT2 | Body | |
| cg09694300 | HDL | 2 | − 0.025 | 0.019 | 0.18 | T0 | − 1.25 | 0.28 | 5.58 × 10–5 | 3 | 159,429,667 | SCHIP1 | Body | |
| cg23700124 | LDL | 2 | 0.001 | 0.005 | 0.86 | T0 | − 0.69 | 0.14 | 1.15 × 10–5 | 4 | 123,843,736 | SPATA5* | TSS1500 | Island |
| cg03803210 | TG | 3 | 0.001 | 0.005 | 0.87 | T0-T1 | 1.93 | 0.34 | 8.93 × 10–7 | 8 | 17,438,816 | PDGFRL | Body | S_Shelf |
| cg08387293 | TG | 2 | 0.002 | 0.008 | 0.82 | T0-T1 | − 3.22 | 0.58 | 1.13 × 10–6 | 1 | 984,383 | AGRN | Body | Island |
CHR chromosome, EWAS epigenome-wide association study, HDL high density lipoprotein cholesterol, LDL low-density lipoprotein cholesterol, MR Mendelian randomization, SE standard error, SNPs single nucleotide polymorphisms, TG triglycerides
aTwo-sample MR analyses using the inverse-variance weighted method were performed. Summary statistics for instrument-exposure association (n = 3,841) were retrieved from Bonder et al. [40], while instrument-outcome association statistics were derived from GWAS performed in the UKBiobank (http://www.nealelab.is/uk-biobank; n = 314′916 for glucose, n = 315′133 for HDL, n = 343′621 for LDL, n = 343,992 for TG). Significant p-values (< 0.05) are indicated in bold
bThe results of the EWAS using methylation M-values are displayed. For the first line, the magnitude of the effect in β-value is to be interpreted as follows: a difference of 1.4% in baseline methylation level (corresponding to baseline methylation IQR in PsyMetab sample) is associated with a 0.39 mM (SE = 0.07) smaller glucose increase. No result reached statistical significance
cReference genes for the methylation sites, and gene regions where the CpGs are located according to the UCSC database. Empty fields indicate an intergenic location
*Specifies there exists > 1 gene or gene transcript at this location. TSS200 = 0–200 bases upstream of the transcriptional start site (TSS); TSS1500 = 200–1500 bases upstream of the TSS; Body = between the ATG and stop codon, irrespective of the presence of introns, exons, TSS, or promoters
dThe relation to a putative nearby CpG island, according to the UCSC database, is given. S_Shelf = 2–4 kb downstream (3′) of CpG island
In the psychiatric cohort, its baseline methylation level (median = 3.8%, IQR = 3.0–4.4) was negatively associated with a 1-month glucose change. Indeed, the model estimated a difference of 1.4% (IQR) in baseline methylation level to be associated with a 0.39 mM smaller glucose increase (95% CI 0.24–0.55), although this association did not reach epigenome-wide significance (p = 2.96 × 10–5). Of note, this association remained similar when adjusting for white blood cell composition (0.40 mM smaller glucose increase (95% CI 0.24–0.57, p = 1.91 × 10–5) when adjusting for estimated cell subtype variation and 0.21 mM smaller glucose increase (95% CI − 0.21 to 0.63, p = 0.3) when adjusting for measured neutrophil variation in the restricted sample).
Validation of previous findings
Previous findings concerning AKT2 and FAR2 genes could be confirmed whereas results on CDH22 gene could not be validated (Additional file 1: Table S2).
An association between AKT2 methylation and glucose was indeed observed in our cohort. The median methylation level of the averaged 5 loci considered decreased from 3.4% (IQR = 2.9–3.8) at baseline to 3.3% (IQR = 2.3–4.2) after a 1-month treatment. The linear model predicted that a decrease of 0.1% in methylation level was associated with a 0.04 mM (SE = 0.02) reduction in glucose change (p = 0.02). However, no association between the baseline methylation level and glucose change was found.
The EWAS hit located in FAR2 was also associated with glucose in our sample. Its baseline methylation level (median = 3.0%, IQR = 2.0–4.3), but not the change in methylation, was negatively associated with a 1-month glucose change. Indeed, the linear model estimated a difference of 2.3% (IQR) in baseline methylation level to be associated with a 0.43 mM (SE = 0.16) smaller glucose increase (p = 0.01).
The EWAS hit located in CDH22 was not associated with MetS in our sample. Indeed, the logistic models did not reveal any significant association between the development of MetS and baseline methylation or change in methylation (p > 0.14 for all).
Hypothesis-driven analyses
The three hypothesis-driven analyses did not uncover significant associations and only some trends were revealed. The results are detailed in the Additional file 1: Tables S3, S4 and S5.
Discussion
Global methylation changes
This study provided insights into a putative effect of psychotropic treatment on methylation as a mechanism leading to metabolic dysregulations. It assessed rapid changes in methylation following treatment initiation, and a global increase in methylation level was observed. This univariate analysis further showed that patients who gained ≥ 5% of their initial weight experienced a smaller 1-month methylation change than patients whose weight remained stable. This last observation shows that the increase in methylation levels likely occurs secondary to treatment and is not (at least not exclusively) a consequence of weight increase.
Nevertheless, it is challenging to distinguish medication-specific effects on DNA methylation from effects mediated by weight gain. For this purpose, future studies should include control groups of psychiatric patients with and without weight gain, but free from antipsychotics and compare the difference in the change in methylation occurring within one month. Unfortunately, such control groups were not available in the present study, and it is thus not possible to delineate the effect of the treatment, of weight increase and that of the natural course of disease. Most studies integrating control groups to date performed cross-sectional analyses, and the few researchers who have already evaluated longitudinal methylation changes pre- and post- psychotropic drug therapy only included baseline comparisons with control groups [8, 14, 43].
Epigenome-wide methylation changes and association with weight change
Independent of weight increase, 52 methylation loci were shown to be significantly modified following the start of treatment. Most of these sites were located within a protein coding gene sequence and displayed an increase in methylation levels. The function of intragenic methylation is still largely unknown, but recent evidence supports a role of these methylation patterns in the regulation of alternative splicing [44–46]. In addition, a decrease in methylation was measured in three sites (cg24357026, cg27452651, cg02624558), found within CpG islands associated with promoters. As hypomethylation of CpG islands often correlates with active transcription of nearby genes [47, 48], the observed change in methylation may induce an increase in the expression of Zinc Finger Protein 146 (ZNF146), ALG12 Alpha-1,6-Mannosyltransferase (ALG12) and Lysine Demethylase 5B (KDM5B) genes. Interestingly, ZNF146 and KDM5B gene ontology annotations according to GeneCards [49], both include DNA-binding transcription factor activity, implying that their activation likely affects the regulation of many other genes.
Regarding changes in methylation related to metabolic side effects, we observed that individuals who underwent marked weight gain experienced a significant increase in methylation level at one specific site (cg12209987). Moreover, this methylation change remained associated with weight evolution in multivariate analysis performed in all patients. This methylation locus is located within an enhancer, according to the Ensembl database [50], 60 kb upstream of the pseudogene PSMC1P5. The increased methylation at this site has no currently known function but may contribute to the regulation of enhancer activity [51].
These last three cited methylation modifications, rather than having a direct phenotypic impact, likely influence downstream pathways. As this analysis was conducted shortly after treatment onset, only early changes were detected. The modifications observed might lead to the regulation of a broader set of genes. In future studies, it would be interesting to follow the evolution of the methylation profile over time, using additional blood samples and detect whether the observed changes remain stable over the course of treatment, continue to evolve in the same direction or return to baseline; and whether methylation of other genes would occur in a second step.
Epigenome-wide association analyses (EWAS) with metabolic phenotypes in PsyMetab and investigation of causality by Mendelian randomization
The statistically nonsignificant results we obtained in EWAS analyses possibly result from a lack of statistical power, considering the expected small effect sizes of single variations in DNA methylation. Besides, we used a conservative cutoff for FWER controlling, possibly leading to a certain rate of false negatives. Given the heterogeneity of the psychiatric population included—broad age range, diverse diagnoses, different current and past psychotropic drugs with various medication history—and the known impact these variables may have on metabolic side effects [52], a larger sample size would be required to identify hits. This would allow to further characterize the common involved mechanisms, and enable powerful stratified analyses, with more homogeneous groups with respect to specific factors.
Nevertheless, among the strongest associations observed, the causal role of the cg11622362 methylation level on glycemia could be established through an MR analysis. Importantly, the direction of the association was not concordant with the MR results, as high methylation at this locus was causally associated with elevated plasma glucose levels, while the longitudinal data showed an association of high methylation with a lower 1-month increase in plasma glucose. One thus needs to be cautious with causal assumptions because adiposity was shown to determine the alterations in methylation at the majority of the previously identified BMI-associated CpG sites [15, 53, 54]. Nevertheless, this finding deserves further research as the cg11622362 methylation site is located opposite the PDHX gene body. This gene encodes a subunit of the pyruvate dehydrogenase complex, which enables the conversion of pyruvate to acetyl coenzyme A, thereby linking glycolysis to the Krebs cycle. In addition, 3 SNPs located on this gene have been associated with type II diabetes mellitus, fasting blood glucose and insulin measurements, as well as with insulin resistance (HOMA-B) in recent GWAS [55, 56]. The functional and clinical relevance of this finding is thus notable, and methylation at cg11622362 may indeed have downstream effects influencing the change in plasma glucose levels following psychotropic treatment.
Validation of previous findings
Interestingly, we validated previous findings in this specific field. We indeed found a signal for the implication of AKT2 methylation in glucose homeostasis and confirmed an association between the previously identified EWAS hit in FAR2 and glucose. The direction of both analyses was concordant with the literature [22, 23]. The reason why these two associations did not reach genome-wide statistical significance can be explained by several factors. First, AKT2 was initially analysed in skeletal muscle and not blood samples. It is conceivable that the signal is attenuated in this marker tissue, but it is still very interesting to observe a convergent result. Additionally, the association was evaluated between an average of 22 methylation loci and insulin resistance, while we only had the data for 5 methylation sites in this genomic region, giving a less precise estimation of the methylation pattern. Last, the associations for both genes were estimated with glucose change, which is less stringent to indicate an abnormality in glucose homeostasis than insulin resistance. The relevance of the methylation patterns of AKT2 and FAR2 could be confirmed in a slightly different context, which gives a promising character to further research on these two genes.
Limitations
It is important to mention that methylation patterns are highly tissue—or even cell-type—specific [57, 58]. Although we controlled for cellular heterogeneity using principal components (with additional sensitivity analyses using estimated cell subtype proportions and cellular composition for a subset of patients), the putative effects of psychotropic medication on blood might not be identical to those occurring in target tissues involved in metabolic dysfunctions (i.e.: brain, adipose tissue, liver, etc.), as previously reported in in vitro models [59]. Whether methylation patterns observed in blood might be relevant in the biological process leading to metabolic side effects or whether they can only be used as biomarkers attesting to related changes in the less accessible tissues of interest still needs validation [60]. Besides, and as previously mentioned, our cohort was treated with a variety of psychotropic drugs. The results observed in this study reflect putative shared mechanisms leading to metabolic adverse effects, but there may also be individual drug-specific changes that cannot be revealed. In addition, there is no perfect way to categorize treatments in terms of propensity to induce weight gain. There indeed exists various meta-analysis ordering antipsychotics according to their risk of weight gain. While it is clear that olanzapine induces greater weight gain than aripiprazole, there is considerable overlap in the confidence intervals for the drugs in between, and depending on the methodologies and the articles considered, the drug order sometimes differs [2, 33, 35, 52, 61, 62]. Moreover, a lack of head-to-head comparisons with the other psychotropic drugs prevents to make indisputable drug categories. Last, we were not able to control for diet, somatic comorbidities, substance use, trauma and other factors known for influencing specific epigenetic sites as well as metabolic health. It is thus important to keep in mind that the changes observed after the introduction of a psychotropic drug may be modulated by the patient’s history.
Conclusion
In summary, appreciable changes in methylation levels were observed following the prescription of psychotropic treatments, but their role in the onset of metabolic side effects remains to be fully elucidated. With an improved understanding of the mechanisms behind such side effects, epigenetic biomarkers may contribute to precision medicine in the future.
Supplementary Information
Additional file 1. Appendix: Supplementary Methods and Results. Supplementary figures and tables.
Acknowledgements
The authors are grateful to A-C. Aubert, A. Vullioud, A. Pittet, C. Brogli, G. Viret, M. Brawand, M. Brocard, M. Delessert, N. Cochard, and S. Jaquet for sample analysis. The authors thank the nursing and medical staff who were involved in the metabolic monitoring program.
Author contributions
Study concept and design: CBE, ZK, CD. Acquisition of data: CD, AD, CG, NL, MP and AVG, PC and KJP. Analyses and interpretation: EP, CD, ZK and CBE. Drafting of the manuscript: CD. Critical revision of the manuscript for important intellectual content: all authors. Obtained funding: CBE, PC and KJP. Administrative, technical, or material support: KJP, AVG, and PC.
Funding
Open access funding provided by University of Lausanne. This work was funded in part by the Swiss National Science Foundation (CE and PC: 320030-120686, 324730-144064, and 320030-173211; CBE, PC and KJP: 320030-200602). The funding sources had no role in the writing of the manuscript or in the decision to submit it for publication.
Availability of data and materials
The datasets analysed during the current study are not publicly available due to the sensitivity of the human personal data involved, which requires specific precautions and limitations. Procedures and documents related to data and/or material sharing (research application form, data and / or the material transfer agreements) are available via the link: http://www.chuv.ch/cnp-psymetab. The datasets analysed during the current study and/or the biological material used are available on reasonable request via the centralized e-mail address: research.psymetab@chuv.ch.
Declarations
Ethical approval and consent to participate
This study was carried out in accordance with the Declaration of Helsinki, the Swiss law, and local requirements. The study protocol was approved by the Ethics Committee of Vaud (CER-VD) with written informed consent from all subjects.
Consent for publication
Not applicable.
Competing interests
CBE received honoraria for conferences or teaching CME courses from Janssen-Cilag, Lundbeck, Otsuka, Sandoz, Servier, Sunovion, Takeda, Vifor-Pharma, and Zeller in the past 3 years. All authors declare that they have no conflicts of interest in relation to the content of this paper.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Céline Dubath, Email: celine.dubath@chuv.ch.
Chin Bin Eap, Email: chin.eap@chuv.ch.
References
- 1.Correll CU, Solmi M, Veronese N, Bortolato B, Rosson S, Santonastaso P, et al. Prevalence, incidence and mortality from cardiovascular disease in patients with pooled and specific severe mental illness: a large-scale meta-analysis of 3,211,768 patients and 113,383,368 controls. World Psychiatry Offic J World Psychiatric Assoc. 2017;16(2):163–180. doi: 10.1002/wps.20420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Abosi O, Lopes S, Schmitz S, Fiedorowicz JG. Cardiometabolic effects of psychotropic medications. Hormone molecular biology and clinical investigation. 2018. [DOI] [PMC free article] [PubMed]
- 3.Barton BB, Segger F, Fischer K, Obermeier M, Musil R. Update on weight-gain caused by antipsychotics: a systematic review and meta-analysis. Expert Opin Drug Saf. 2020;19(3):295–314. doi: 10.1080/14740338.2020.1713091. [DOI] [PubMed] [Google Scholar]
- 4.Henderson DC, Vincenzi B, Andrea NV, Ulloa M, Copeland PM. Pathophysiological mechanisms of increased cardiometabolic risk in people with schizophrenia and other severe mental illnesses. Lancet Psychiatry. 2015;2(5):452–464. doi: 10.1016/S2215-0366(15)00115-7. [DOI] [PubMed] [Google Scholar]
- 5.Grajales D, Ferreira V, Valverde ÁM. Second-generation antipsychotics and dysregulation of glucose metabolism: beyond weight gain. Cells. 2019;8:11. doi: 10.3390/cells8111336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lockwood LE, Youssef NA. Systematic review of epigenetic effects of pharmacological agents for bipolar disorders. Brain Sci. 2017;7:11. doi: 10.3390/brainsci7110154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ovenden ES, McGregor NW, Emsley RA, Warnich L. DNA methylation and antipsychotic treatment mechanisms in schizophrenia: progress and future directions. Prog Neuropsychopharmacol Biol Psychiatry. 2018;81:38–49. doi: 10.1016/j.pnpbp.2017.10.004. [DOI] [PubMed] [Google Scholar]
- 8.Gardea-Resendez M, Kucuker MU, Blacker CJ, Ho AM-C, Croarkin PE, Frye MA, et al. Dissecting the epigenetic changes induced by non-antipsychotic mood stabilizers on schizophrenia and affective disorders: a systematic review. Front Pharmacol. 2020;11:467. doi: 10.3389/fphar.2020.00467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhou J, Li M, Wang X, He Y, Xia Y, Sweeney JA, et al. Drug response-related DNA methylation changes in schizophrenia, bipolar disorder, and major depressive disorder. Front Neurosci. 2021;15:674273. doi: 10.3389/fnins.2021.674273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Goud Alladi C, Etain B, Bellivier F, Marie-Claire C. DNA methylation as a biomarker of treatment response variability in serious mental illnesses: a systematic review focused on bipolar disorder, schizophrenia, and major depressive disorder. Int J Mol Sci. 2018;19(10):3026. doi: 10.3390/ijms19103026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Marie-Claire C, Lejeune FX, Mundwiller E, Ulveling D, Moszer I, Bellivier F, et al. A DNA methylation signature discriminates between excellent and non-response to lithium in patients with bipolar disorder type 1. Sci Rep. 2020;10(1):12239. doi: 10.1038/s41598-020-69073-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Webb LM, Phillips KE, Ho MC, Veldic M, Blacker CJ. The Relationship between DNA methylation and antidepressant medications: a systematic review. Int J Mol Sci. 2020;21(3):826. doi: 10.3390/ijms21030826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Swathy B, Saradalekshmi KR, Nair IV, Nair C, Banerjee M. Understanding the influence of antipsychotic drugs on global methylation events and its relevance in treatment response. Epigenomics. 2018;10(3):233–247. doi: 10.2217/epi-2017-0086. [DOI] [PubMed] [Google Scholar]
- 14.Burghardt KJ, Khoury AS, Msallaty Z, Yi Z, Seyoum B. Antipsychotic medications and DNA methylation in schizophrenia and bipolar disorder: a systematic review. Pharmacotherapy. 2020;40(4):331–342. doi: 10.1002/phar.2375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Huan T, Joehanes R, Song C, Peng F, Guo Y, Mendelson M, et al. Genome-wide identification of DNA methylation QTLs in whole blood highlights pathways for cardiovascular disease. Nat Commun. 2019;10(1):4267. doi: 10.1038/s41467-019-12228-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Burghardt KJ, Pilsner JR, Bly MJ, Ellingrod VL. DNA methylation in schizophrenia subjects: gender and MTHFR 677C/T genotype differences. Epigenomics. 2012;4(3):261–268. doi: 10.2217/epi.12.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Burghardt KJ, Goodrich JM, Dolinoy DC, Ellingrod VL. DNA methylation, insulin resistance and second-generation antipsychotics in bipolar disorder. Epigenomics. 2015;7(3):343–352. doi: 10.2217/epi.15.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Burghardt KJ, Howlett BH, Sanders E, Dass SE, Msallaty Z, Mallisho A, et al. Skeletal muscle DNA methylation modifications and psychopharmacologic treatment in bipolar disorder. Eur Neuropsychopharmacol J Eur Coll Neuropsychopharmacol. 2019;29(12):1365–1373. doi: 10.1016/j.euroneuro.2019.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lott SA, Burghardt PR, Burghardt KJ, Bly MJ, Grove TB, Ellingrod VL. The influence of metabolic syndrome, physical activity and genotype on catechol-O-methyl transferase promoter-region methylation in schizophrenia. Pharmacogenomics J. 2013;13(3):264–271. doi: 10.1038/tpj.2012.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Moons T, De Hert M, Kenis G, Viechtbauer W, van Os J, Gohlke H, et al. No association between genetic or epigenetic variation in insulin growth factors and antipsychotic-induced metabolic disturbances in a cross-sectional sample. Pharmacogenomics. 2014;15(7):951–962. doi: 10.2217/pgs.14.46. [DOI] [PubMed] [Google Scholar]
- 21.Delacrétaz A, Glatard A, Dubath C, Gholam-Rezaee M, Sanchez-Mut JV, Gräff J, et al. Psychotropic drug-induced genetic-epigenetic modulation of CRTC1 gene is associated with early weight gain in a prospective study of psychiatric patients. Clin Epigen. 2019;11(1):198. doi: 10.1186/s13148-019-0792-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Burghardt KJ, Seyoum B, Dass SE, Sanders E, Mallisho A, Yi Z. Association of protein kinase B (AKT) DNA hypermethylation with maintenance atypical antipsychotic treatment in patients with bipolar disorder. Pharmacotherapy. 2018;38(4):428–435. doi: 10.1002/phar.2097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Burghardt KJ, Goodrich JM, Dolinoy DC, Ellingrod VL. Gene-specific DNA methylation may mediate atypical antipsychotic-induced insulin resistance. Bipolar Disord. 2016;18(5):423–432. doi: 10.1111/bdi.12422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Burghardt KJ, Goodrich JM, Lines BN, Ellingrod VL. The influence of metabolic syndrome and sex on the DNA methylome in schizophrenia. Int J Genom. 2018;2018:8076397. doi: 10.1155/2018/8076397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dubath C, Delacretaz A, Glatard A, Vollenweider P, Preisig M, Richard-Lepouriel H, et al. Evaluation of cardiometabolic risk in a large psychiatric cohort and comparison with a population-based sample in Switzerland. J Clin Psychiatry. 2020;81:3. doi: 10.4088/JCP.19m12796. [DOI] [PubMed] [Google Scholar]
- 26.Vandenberghe F, Gholam-Rezaee M, Saigi-Morgui N, Delacretaz A, Choong E, Solida-Tozzi A, et al. Importance of early weight changes to predict long-term weight gain during psychotropic drug treatment. J Clin Psychiatry. 2015;76(11):e1417–e1423. doi: 10.4088/JCP.14m09358. [DOI] [PubMed] [Google Scholar]
- 27.El Asmar K, Feve B, Colle R, Trabado S, Verstuyft C, Gressier F, et al. Early weight gain predicts later metabolic syndrome in depressed patients treated with antidepressants: findings from the METADAP cohort. J Psychiatr Res. 2018;107:120–127. doi: 10.1016/j.jpsychires.2018.10.021. [DOI] [PubMed] [Google Scholar]
- 28.Alberti KG, Zimmet PZ, Shaw J, Grundy SM. IDF Consensus Worldwide Definition of the Metabolic Syndrome Brussels, Belgium: International Diabetes Federation; 2006 [updated 29.07.2020. Available from: https://www.idf.org/e-library/consensus-statements/60-idfconsensus-worldwide-definitionof-the-metabolic-syndrome.html.
- 29.Piras M, Ranjbar S, Laaboub N, Grosu C, Gamma F, Plessen KJ, et al. Evolutions of Metabolic Parameters Following Switches of Psychotropic Drugs: A Longitudinal Cohort Study. Schizophrenia Bull. 2022. [DOI] [PMC free article] [PubMed]
- 30.Dubath C, Gholam-Rezaee M, Sjaarda J, Levier A, Saigi-Morgui N, Delacrétaz A, et al. Socio-economic position as a moderator of cardiometabolic outcomes in patients receiving psychotropic treatment associated with weight gain: results from a prospective 12-month inception cohort study and a large population-based cohort. Transl Psychiatry. 2021;11(1):360. doi: 10.1038/s41398-021-01482-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Laaboub N, Dubath C, Ranjbar S, Sibailly G, Grosu C, Piras M, et al. Insomnia disorders are associated with increased cardiometabolic disturbances and death risks from cardiovascular diseases in psychiatric patients treated with weight-gain-inducing psychotropic drugs: results from a Swiss cohort. BMC Psychiatry. 2022;22(1):342. doi: 10.1186/s12888-022-03983-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Huhn M, Nikolakopoulou A, Schneider-Thoma J, Krause M, Samara M, Peter N, et al. Comparative efficacy and tolerability of 32 oral antipsychotics for the acute treatment of adults with multi-episode schizophrenia: a systematic review and network meta-analysis. Lancet. 2019. [DOI] [PMC free article] [PubMed]
- 33.Pillinger T, McCutcheon RA, Vano L, Mizuno Y, Arumuham A, Hindley G, et al. Comparative effects of 18 antipsychotics on metabolic function in patients with schizophrenia, predictors of metabolic dysregulation, and association with psychopathology: a systematic review and network meta-analysis. Lancet Psychiatry. 2020;7(1):64–77. doi: 10.1016/S2215-0366(19)30416-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Correll CU, Detraux J, De Lepeleire J, De Hert M. Effects of antipsychotics, antidepressants and mood stabilizers on risk for physical diseases in people with schizophrenia, depression and bipolar disorder. World Psychiatr Offic J World Psychiatr Assoc. 2015;14(2):119–136. doi: 10.1002/wps.20204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hasnain M, Vieweg WV. Weight considerations in psychotropic drug prescribing and switching. Postgrad Med. 2013;125(5):117–129. doi: 10.3810/pgm.2013.09.2706. [DOI] [PubMed] [Google Scholar]
- 36.Du P, Zhang X, Huang C-C, Jafari N, Kibbe WA, Hou L, et al. Comparison of beta-value and M-value methods for quantifying methylation levels by microarray analysis. BMC Bioinform. 2010;11(1):587. doi: 10.1186/1471-2105-11-587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Teschendorff AE, Breeze CE, Zheng SC, Beck S. A comparison of reference-based algorithms for correcting cell-type heterogeneity in epigenome-wide association studies. BMC Bioinform. 2017;18(1):105. doi: 10.1186/s12859-017-1511-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mansell G, Gorrie-Stone TJ, Bao Y, Kumari M, Schalkwyk LS, Mill J, et al. Guidance for DNA methylation studies: statistical insights from the Illumina EPIC array. BMC Genom. 2019;20(1):366. doi: 10.1186/s12864-019-5761-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Evans DM, Davey SG. Mendelian randomization: new applications in the coming age of hypothesis-free causality. Annu Rev Genom Hum Genet. 2015;16:327–350. doi: 10.1146/annurev-genom-090314-050016. [DOI] [PubMed] [Google Scholar]
- 40.Bonder MJ, Luijk R, Zhernakova DV, Moed M, Deelen P, Vermaat M, et al. Disease variants alter transcription factor levels and methylation of their binding sites. Nat Genet. 2017;49(1):131–138. doi: 10.1038/ng.3721. [DOI] [PubMed] [Google Scholar]
- 41.Bycroft C, Freeman C, Petkova D, Band G, Elliott LT, Sharp K, et al. The UK Biobank resource with deep phenotyping and genomic data. Nature. 2018;562(7726):203–209. doi: 10.1038/s41586-018-0579-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Burgess S, Butterworth A, Thompson SG. Mendelian randomization analysis with multiple genetic variants using summarized data. Genet Epidemiol. 2013;37(7):658–665. doi: 10.1002/gepi.21758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Xavier G, Santoro ML, Ota VK, Spindola LM, Oliveira G, Vieira T, et al. Blood gene expression changes after Risperidone treatment in an antipsychotic-naïve cohort of first episode of psychosis patients. Schizophr Res. 2020;220:285–286. doi: 10.1016/j.schres.2020.03.033. [DOI] [PubMed] [Google Scholar]
- 44.Zheng Z, Wei X, Hildebrandt A, Schmidt B. A computational method for studying the relation between alternative splicing and DNA methylation. Nucleic Acids Res. 2016;44(2):e19. doi: 10.1093/nar/gkv906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lev Maor G, Yearim A, Ast G. The alternative role of DNA methylation in splicing regulation. Trends Genet. 2015;31(5):274–280. doi: 10.1016/j.tig.2015.03.002. [DOI] [PubMed] [Google Scholar]
- 46.Shayevitch R, Askayo D, Keydar I, Ast G. The importance of DNA methylation of exons on alternative splicing. RNA. 2018;24(10):1351–1362. doi: 10.1261/rna.064865.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ma X, Kang S. Functional implications of DNA methylation in adipose biology. Diabetes. 2019;68(5):871. doi: 10.2337/dbi18-0057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Moore LD, Le T, Fan G. DNA methylation and its basic function. Neuropsychopharmacol Offic Pub Am Coll Neuropsychopharmacol. 2013;38(1):23–38. doi: 10.1038/npp.2012.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Stelzer G, Rosen N, Plaschkes I, Zimmerman S, Twik M, Fishilevich S, et al. The genecards suite: from gene data mining to disease genome sequence analyses. Curr Protocols Bioinform. 2016;54(1):130–113. doi: 10.1002/cpbi.5. [DOI] [PubMed] [Google Scholar]
- 50.Howe KL, Achuthan P, Allen J, Allen J, Alvarez-Jarreta J, Amode MR, et al. Ensembl 2021. Nucleic Acids Res. 2021;49(D1):D884–D891. doi: 10.1093/nar/gkaa942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Angeloni A, Bogdanovic O. Enhancer DNA methylation: implications for gene regulation. Essays Biochem. 2019;63(6):707–715. doi: 10.1042/EBC20190030. [DOI] [PubMed] [Google Scholar]
- 52.De Hert M, Detraux J, van Winkel R, Yu W, Correll CU. Metabolic and cardiovascular adverse effects associated with antipsychotic drugs. Nat Rev Endocrinol. 2011;8(2):114–126. doi: 10.1038/nrendo.2011.156. [DOI] [PubMed] [Google Scholar]
- 53.Wahl S, Drong A, Lehne B, Loh M, Scott WR, Kunze S, et al. Epigenome-wide association study of body mass index, and the adverse outcomes of adiposity. Nature. 2016;541:81. doi: 10.1038/nature20784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mendelson MM, Marioni RE, Joehanes R, Liu C, Hedman AK, Aslibekyan S, et al. Association of body mass index with DNA methylation and gene expression in blood cells and relations to cardiometabolic disease: a mendelian randomization approach. PLoS Med. 2017;14(1):e1002215. doi: 10.1371/journal.pmed.1002215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mahajan A, Taliun D, Thurner M, Robertson NR, Torres JM, Rayner NW, et al. Fine-mapping type 2 diabetes loci to single-variant resolution using high-density imputation and islet-specific epigenome maps. Nat Genet. 2018;50(11):1505–1513. doi: 10.1038/s41588-018-0241-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Masotti M, Guo B, Wu B. Pleiotropy informed adaptive association test of multiple traits using genome-wide association study summary data. Biometrics. 2019;75(4):1076–1085. doi: 10.1111/biom.13076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Farré P, Jones MJ, Meaney MJ, Emberly E, Turecki G, Kobor MS. Concordant and discordant DNA methylation signatures of aging in human blood and brain. Epigenet Chromatin. 2015;8(1):19. doi: 10.1186/s13072-015-0011-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ziller MJ, Gu H, Müller F, Donaghey J, Tsai LT, Kohlbacher O, et al. Charting a dynamic DNA methylation landscape of the human genome. Nature. 2013;500(7463):477–481. doi: 10.1038/nature12433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lamb J, Crawford ED, Peck D, Modell JW, Blat IC, Wrobel MJ, et al. The connectivity map: using gene-expression signatures to connect small molecules, genes, and disease. Science. 2006;313(5795):1929. doi: 10.1126/science.1132939. [DOI] [PubMed] [Google Scholar]
- 60.Campbell KA, Colacino JA, Park SK, Bakulski KM. Cell types in environmental epigenetic studies: biological and epidemiological frameworks. Curr Environ Health Rep. 2020;7(3):185–197. doi: 10.1007/s40572-020-00287-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bowden CL, Mosolov S, Hranov L, Chen E, Habil H, Kongsakon R, et al. Efficacy of valproate versus lithium in mania or mixed mania: a randomized, open 12-week trial. Int Clin Psychopharmacol. 2010;25(2):60–67. doi: 10.1097/YIC.0b013e328333ac1b. [DOI] [PubMed] [Google Scholar]
- 62.De Hert M, Correll Cu, Bobes J, Cetkovich-Bakmas M, Cohen D, Asai I, et al. Physical illness in patients with severe mental disorders: I—prevalence, impact of medications and disparities in health care. World Psychiatry Offic J World Psychiatr Assoc. 2011;10(1):52–77. doi: 10.1002/j.2051-5545.2011.tb00014.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1. Appendix: Supplementary Methods and Results. Supplementary figures and tables.
Data Availability Statement
The datasets analysed during the current study are not publicly available due to the sensitivity of the human personal data involved, which requires specific precautions and limitations. Procedures and documents related to data and/or material sharing (research application form, data and / or the material transfer agreements) are available via the link: http://www.chuv.ch/cnp-psymetab. The datasets analysed during the current study and/or the biological material used are available on reasonable request via the centralized e-mail address: research.psymetab@chuv.ch.