Abstract
The macrolide antibiotic rapamycin inhibits cellular proliferation by interfering with the highly conserved TOR (for target of rapamycin) signaling pathway. Growth arrest of budding yeast cells treated with rapamycin is followed by the program of molecular events that characterizes entry into G0 (stationary phase), including the induction of polymerase (Pol) II genes typically expressed only in G0. Normally, progression into G0 is characterized by transcriptional repression of the Pol I and III genes. Here, we show that rapamycin treatment also causes the transcriptional repression of Pol I and III genes. The down-regulation of Pol III transcription is TOR dependent. While it coincides with translational repression by rapamycin, transcriptional repression is due in part to a translation-independent effect that is evident in extracts from a conditional tor2 mutant. Biochemical experiments reveal that RNA Pol III and probably transcription initiation factor TFIIIB are targets of repression by rapamycin. In view of previous evidence that TFIIIB and Pol III are inhibited when protein phosphatase 2A (PP2A) function is impaired, and that PP2A is a component of the TOR pathway, our results suggest that TOR signaling regulates Pol I and Pol III transcription in response to nutrient growth signals.
Gradual nutrient depletion in Saccharomyces cerevisiae provokes a broad spectrum of morphological and biochemical changes that result in a terminal cell cycle arrest phenotype called G0 or stationary phase (reviewed in references 44 and 45). Stationary-phase cells have a 1n DNA content, are uniformly large and unbudded, and display a prominent vacuole. The G0 state is further characterized by reduced protein synthesis, and the pattern of RNA polymerase (Pol) II transcripts is distinct in cycling and G0 cells. Thus, more than 95% of Pol II genes are repressed in G0, and a subset of Pol II genes whose products promote survival under conditions of nutrient limitation are massively induced at the transcriptional level (8). Induction of the G0 pattern of Pol II transcription in yeast accompanies the repression of transcription of the large rRNA genes by Pol I and the tRNA and 5S rRNA genes by Pol III (9, 26, 33, 36). Since tRNA and rRNA synthesis accounts for about 70% of nuclear transcription, this regulatory mechanism may enhance survival in G0 by limiting the energetically costly production of relatively stable RNA products not immediately required for viability.
While there is striking repression of translation in G0, some critical aspects of the stationary-phase response are not simply downstream consequences of a decreased rate of protein synthesis. For example, treatment of cultures with cycloheximide does not cause the accumulation of large unbudded cells or cells with a 1n DNA content (6), and in some strains there is no inhibition of Pol I or Pol III transcription in extracts from cells treated with cycloheximide (9). Key physiological steps in the differentiation of a stationary-phase cell are therefore likely to involve signaling mechanisms that act independently of, but in parallel with, effects on translation.
The signaling pathway involved in setting the G0 pattern of Pol II transcription is the TOR (for target of rapamycin) pathway, which is comprised of the highly conserved TOR kinases (Tor1p and Tor2p), protein phosphatase 2A (PP2A), and the type 2A-related phosphatase Sit4p (4, 10). As its name indicates, the TOR pathway was originally defined by the sensitivity of the TOR kinases to the macrolide antibiotic rapamycin, and indeed, rapamycin treatment of yeast induces many aspects of the normal response to nutrient limitation, including the induction of G0-specific Pol II genes (4, 10). The observation that TOR signaling is involved in setting the G0 pattern of Pol II transcription raises the possibility that the repression of Pol I and Pol III transcription in stationary phase is also under TOR control. This possibility was tested by the experiments described in the present study. We find that rapamycin represses transcription initiation by RNA Pol I and Pol III. The down-regulation of Pol III transcription is TOR-dependent, and, although coincident with inhibition of protein synthesis by rapamycin, it includes an effect that is independent of translational repression.
We have explored the biochemical basis of Pol III repression by rapamycin. In all species, the core Pol III transcription machinery consists of RNA Pol III, TFIIIC, a sequence-specific DNA binding factor, and TFIIIB, which is recruited to the promoter by TFIIIC and in turn recruits Pol III (13, 47). Biochemical experiments reveal that RNA Pol III and possibly TFIIIB are repressed in extracts from rapamycin-treated cells. The same components of the Pol III transcriptional machinery are known to be inhibited when the regulation of PP2A, a component of the TOR pathway, is perturbed by inactivation of its noncatalytic A subunit (10, 41). Our results are consistent with the proposal that Pol III transcription is regulated according to nutrient availability by a signal transduction pathway that includes the TOR kinases.
MATERIALS AND METHODS
Strains and extract preparation.
Extracts were prepared from S. cerevisiae W303-1A (a ade2-1 trp1-1 can1-100 leu2-3,112 his3-11,15 ura3-1 [38]), JK9-3Dα (α ura3-52 leu2-3,112 his4 trp1-1 rme1 HMLa [7]), JHY3-3B (JK9-3Dα fpr1::URA3 [19]), MH346-1a TOR2 (Δtor1 Δtor2/TOR2 [7]), MH346-1a tor2-ts4 (Δtor1 Δtor2/tor2-ts4 [7]), and cdc28-1 (α cdc28-1 ade1 gal1 lys2 met14 his7 tyr1 [21] [Yeast Genetic Stock Center]) after breaking of frozen cells with a motorized mortar grinder (large-scale extracts) or a hand-held coffee mill (34).
Yeast culture.
For rapamycin experiments, extracts were prepared from strains W303-1A, JK9-3Dα, and JHY3-3B. Rapamycin treatment induced G0 arrest and transcriptional repression in all FPR1 strain backgrounds, although the repression was more pronounced in extracts from rapamycin-treated JK9-3Dα cells than in extracts from treated W303-1A cells (not shown). Cells were cultured in YPD (2% yeast extract–1% Bacto Peptone–2% dextrose) at 30°C with vigorous shaking. The medium was supplemented at indicated points in the growth cycle with the drug vehicle (in control cultures) or rapamycin (added from a 2 mM stock solution in 90% ethanol–10% Tween 20). One hour of rapamycin treatment was performed by adding the drug to a final concentration of 10 μg/ml when the cells reached an optical density at 600 nm (OD600) of 1.0. For 24-h treatments, rapamycin was added to a final concentration of 1 μg/ml when the cells reached an OD600 of 0.1. Extracts from 24-h-treated cells were about half the protein concentration of control extracts, indicating that cells are harder to disrupt after rapamycin treatment. This phenotype is similar to the increased zymolyase resistance of G0 cells (reviewed in reference 44). Strain cdc28-1 was grown in YPD at 30°C to an OD600 of 1.0 and then incubated at 37°C for 4 h and harvested. Strains MH346-1a TOR2 and MH346-1a tor2-ts4 were grown in YPD at 25°C to an OD600 of 0.9 and harvested as usual.
Cell viability assay following rapamycin treatment.
Strain W303-1A was grown to an OD600 of 0.1 and treated with either rapamycin to 1 μg/ml or the drug vehicle for 24 h. The OD600 was taken and aliquots (nominally 100 and 200 cells) of each culture were plated on YPD. The plates were incubated at 30°C for 3 days, and the number of colonies on each plate was counted. This assay was performed in triplicate.
Measurement of bulk protein synthesis in vivo.
In vivo labeling of total yeast proteins was performed according to the method of Barbet et al. (4) with slight modifications. One-milliliter cultures of JK9-3Dα were grown to an OD600 of 1.0 and treated with either 10 μg of rapamycin per ml or the drug vehicle for 1 h at 30°C. The cells were then washed with ice-cold water and resuspended in 1 ml of synthetic glucose-minus-methionine medium plus fresh rapamycin or vehicle. Then, 500 μCi of [35S]methionine (NEN) was added, and the cells were labeled with shaking for 20 min at 30°C. After incubation, 120 μl of each culture was removed and transferred into separate screw-capped tubes containing 250 μl of 1 mM NaN3. Proteins were isolated as described by Werner-Washburne et al. (43), modified as follows. The cells were pelleted and washed with 250 μl of ice-cold NaN3 (1 mM) and with 250 μl of ice-cold water. They were then resuspended in 300 μl of breaking buffer (10 mM Tris-HCl [pH 7.9]–5 mM MgCl2–50 μg of RNase A per ml) with ∼0.3 g of glass beads (0.3 to 0.5 mm in diameter) and lysed by vortexing vigorously six times for 30 s each, with a 30-s incubation on ice after each vortexing. The mixture was centrifuged and the supernatant was acetone precipitated. The protein precipitate was resuspended in 50 μl of lysis buffer (9.5 M urea–2% [wt/vol] Nonidet P-40–5% β-mercaptoethanol) and quantitated by scintillation counting. To control for the amount of cell breakage, nonradioactive samples were prepared identically and measured for protein concentration by the method of Bradford (5). The protein concentrations of samples obtained from rapamycin and control cultures were found to be similar.
In vitro transcription reactions.
Multiple-round transcription reactions were performed as described previously (22, 33, 35). The templates for 5S rRNA and tRNA runoff transcription were pY5S (400 ng/20-μl reaction mixture) and pGE2, which contains the tRNALeu gene (25ng/20-μl reaction mixture). RNA Pol III reactions were performed for 30 min at 30°C with [α-32P]UTP (3,000 Ci/mmol; NEN). RNA Pol I transcription was assayed with pBYr11AL, which contains the yeast 35S rDNA promoter (400 ng/20-μl reaction mixture). pBYr11AL is identical to pBYr11A (33) except for the insertion of a linker oligonucleotide (XbaI, SalI, BglII, BamHI) between the XbaI and BamHI sites. The specific template was added after preincubation of the extracts with 400 ng of HpaII-digested pBluescript II KS+ for 10 min at room temperature. Reactions were stopped after 45 min at room temperature. Extract amounts are noted in the figures. The products from pBYr11AL were detected by S1 nuclease protection analysis using an end-labeled oligonucleotide probe (33). Pol III reactions with MH346-1a TOR2 and MH346-1a tor2-ts4 extracts were performed by incubating the extract with all components except the template and nucleotides at the indicated temperature (22 or 37°C) for 5 min. A template-nucleotide mix was then added, and transcription was performed at the indicated temperature for 30 min. In one pair of extracts, the temperature-dependent repression of Pol III transcription in tor2-ts4 extracts was accentuated by performing the reactions with fivefold less unlabeled UTP than usual (10 μM final concentration). Quantitation of specific transcription was performed by phosphorimager analysis (Fujix Bas 1000 bioimaging analyzer; MacBAS software). Background was subtracted from all signals.
Purification and assay of Pol III transcription factors.
Transcription factors were prepared from large-scale whole-cell extracts of a YPH250 derivative, YDH6 (18), grown to an OD600 of 2, and from the protease-deficient strain BJ5626 (25), grown to an OD600 of 3. The methods of purification were adopted from Kassavetis et al. (27), as described by Ghavidel and Schultz (15). TFIIIB was purified to the hydroxyapatite step, and TFIIIC was obtained by oligonucleotide affinity chromatography and monitored for activity by a standard gel mobility shift assay. After collection of the 300 mM Pol III–TFIIIC fraction from DEAE-Sepharose, as described previously (15), the resin was eluted with 600 mM KCl to obtain crude RNA Pol III. The specific activities of Pol III in the Pol III–TFIIIC and 600 mM fractions are similar (nonspecific activity assay, as described below); however, the 600 mM fraction is devoid of TFIIIB and TFIIIC (determined by Western blotting and gel mobility shift assay, respectively). The 600 mM fraction has no specific initiation activity on its own. The effect of wild-type transcription factors added to extracts from rapamycin-treated cells was tested by incubating whole-cell extracts with the indicated amount of factor for 5 min at room temperature prior to adding the template and nucleotides. Nonspecific (bulk) RNA Pol III reactions were performed according to published methods (17, 32) for 20 min at 22°C in 25-μl mixtures containing 50 mM Tris-HCl (pH 8), 200 mM (NH4)2SO4, 0.6 mM (each) ATP, CTP, and GTP, 0.06 mM UTP, 16 μg of sheared calf thymus DNA per ml, 2 mM MnCl2, 50 μg of α-amanatin per ml, and 1 μl of [5,6-3H]UTP (36.1 Ci/mmol; NEN). Twenty microliters of each reaction was stopped by spotting onto DE-81 paper and was processed according to the method of Roeder (32).
CKII assay.
The casein kinase II (CKII) assay was performed essentially as described previously (reference 22, with modifications in reference 15). Reactions were performed in 25-μl volumes with 6.25 μg of extract protein under the conditions for Pol I and III transcription, except that α-amanatin was omitted. Each data point is the mean result of four reactions.
RESULTS
Repression of translation and transcription resulting from treatment of cells with rapamycin.
The effects of rapamycin on transcription were tested with extracts from cells treated for 1 or 24 h with the drug. Control extracts (cells treated only with the rapamycin solvent) were prepared in parallel from cells harvested at the density recorded for cultures treated with rapamycin. Extracts were made by a method that preserves transcription by RNA Pol I, II, and III (33, 35), chromatin assembly (34), and various physiological responses including down-regulation of the Pol I and Pol III transcription that occurs normally when yeast cells enter G0 (33, 49). The extracts were assayed in parallel for the capacity to support transcription with exogenous tRNA and 5S rRNA genes and a 35S rDNA promoter construct. Pol III transcription was measured by a runoff assay, and Pol I transcripts were detected by S1 nuclease protection analysis. In order to establish that rapamycin was eliciting the expected cellular responses, we variously monitored gross morphology, plating efficiency, and translation for comparison with effects that have been extensively documented elsewhere (4, 19, 20).
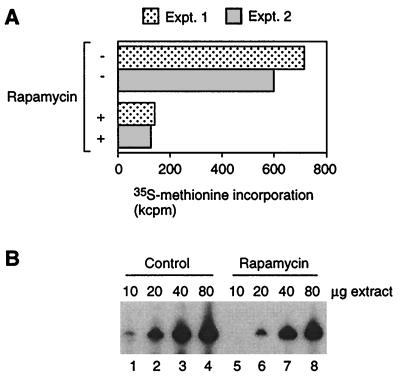
As reported in the literature (4), rapamycin treatment for 1 h inhibited translation in vivo by approximately 80% (Fig. 1A) without changing the budding index of the cell population as a whole (not shown). Extracts from 1-h-treated cells were two- to fourfold less active for Pol III transcription than were control extracts (Fig. 1B), suggesting that the transcriptional machinery responds quickly to rapamycin treatment, although in magnitude the effect is not comparable to repression of translation. Rapamycin treatment for 24 h resulted in a uniform morphological phenotype (>90% large unbudded cells, consistent with arrest in G0) but no cell death; plating efficiencies determined at the conclusion of the treatment period were identical between rapamycin-treated and control cells (not shown). Compared to extracts from cells treated for 1 h, a substantially more severe repression effect on Pol III transcription was observed in extracts from cells treated with rapamycin for 24 h (Fig. 2A and B). We conclude that rapamycin treatment in vivo leads to repression of Pol III transcription in vitro and that repression occurs progressively rather than instantaneously.
FIG. 1.
Translation and Pol III transcription are repressed by short-term treatment (1 h) of cells with rapamycin. (A) Repression of translation. Translation was measured as [35S]methionine incorporation during a 20-min labeling period at 30°C. Results are shown for two experiments. (B) Repression of Pol III transcription. 5S rRNA transcription was measured in increasing amounts of extracts from control and rapamycin-treated cells.
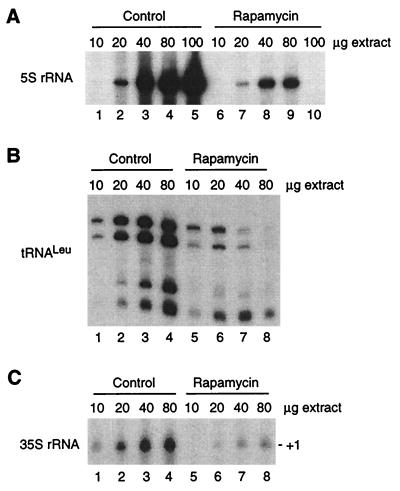
FIG. 2.
Transcription by RNA Pol I and III is strongly repressed in extracts from cells treated for 24 h with rapamycin. 5S rRNA (A), tRNALeu (B), and 35S rRNA (Pol I) (C) transcription was assayed in increasing amounts of extracts from control and treated cells. Note that the extracts from rapamycin-treated cells support Pol III transcription over only a narrow range of protein concentrations and that there is an unusually sharp decline in 5S rRNA transcription from 80 to 100 μg of extracts; these effects were reproducible but remain unexplained.
Since Pol I and Pol III transcription are coordinately regulated under many circumstances (see reference 9), we tested whether Pol I transcription is also affected in extracts from rapamycin-treated cells, compared to control cells. As shown, Pol I transcription is repressed in extracts from cells treated for 24 h with rapamycin (Fig. 2C). Furthermore, the magnitude of this effect is similar to that observed for Pol III (Fig. 2B and C). We conclude that long-term rapamycin treatment globally represses transcription of the nuclear genes encoding the most abundant nontranslated RNAs.
Rapamycin treatment does not result in the appearance of a Pol III transcription inhibitor in extracts.
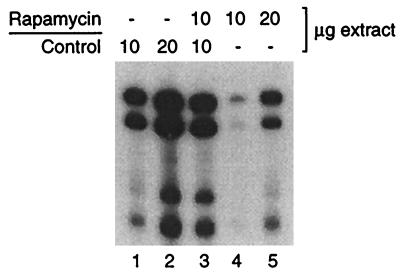
The results in Fig. 1 and 2 could be explained if an excess of nonspecific inhibitor of the transcriptional machinery is generated in the extract by rapamycin treatment of cells (see, for example, reference 41). In order to test this possibility, we performed a mixing experiment using extracts from 24-h-treated and control cells (Fig. 3). This is a severe test because indirect inhibitory effects are expected to be most pronounced in cells subjected to prolonged exposure to rapamycin. When 10 μg of extracts from rapamycin-treated cells is mixed with 10 μg of control extracts, the level of transcription (Fig. 3, lane 3) is intermediate between the levels obtained with 10 and 20 μg of control extracts (Fig. 3, lanes 1 and 2) and is greater than the sum of the signal from 10 μg of rapamycin and control extracts individually (Fig. 3, lanes 1 and 4). A similar result was obtained when a mixture of 20 μg of each extract was compared to 40 μg of rapamycin and control extracts alone (not shown). Therefore, the extracts from rapamycin-treated cells do not contain an excess of a transcription inhibitor, suggesting that a component of the transcription machinery is directly repressed by rapamycin. We further note that rapamycin extracts support normal levels of a variety of biochemical activities, including chromatin assembly (not shown). Transcriptional repression in extracts, therefore, is not due to a general inhibitory effect such as would result from the activation of DNases or vacuolar proteases.
FIG. 3.
Repression of Pol III transcription is not due to an excess of inhibitor in extracts from rapamycin-treated cells. tRNALeu transcription was compared in the indicated amounts of extracts from control cells and cells cultured in the presence of rapamycin for 24 h.
Rapamycin treatment does not mimic a G1 effect on transcription.
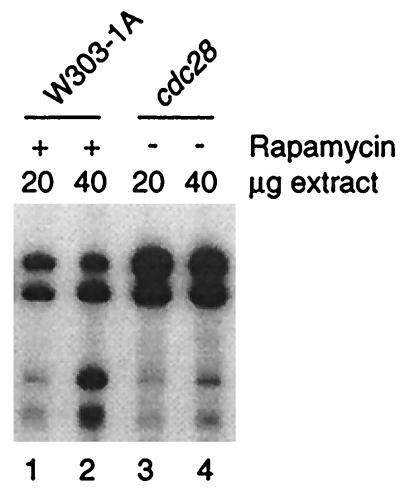
Since rapamycin induces a transient G1 arrest in yeast (4), our results may reflect inhibition of Pol III transcription in G1 of the cell cycle. Indeed, White et al. (46) have demonstrated in a mammalian system that Pol III activity is very low in G1 extracts compared to extracts from asynchronously growing cells. To test if rapamycin treatment mimics a G1 Pol III transcription phenotype in yeast, we compared transcription in extracts from rapamycin-treated cells and cdc28ts cells cultured at the restrictive temperature for 4 h, at which time ∼90% of cdc28 cells are arrested in G1. As shown in Fig. 4, extracts from cdc28ts cells support a significantly higher level of transcription than extracts from rapamycin treated-cells. We conclude that in yeast, rapamycin treatment does not mimic the phenotype of Pol III transcription in G1.
FIG. 4.
Repression of pol III transcription in extracts from rapamycin-treated cells does not reproduce the transcription phenotype of G1 extracts. tRNALeu transcription was compared in extracts from rapamycin-treated (24 h) wild-type cells and from G1-arrested cdc28 cells.
The rapamycin effect is TOR dependent.
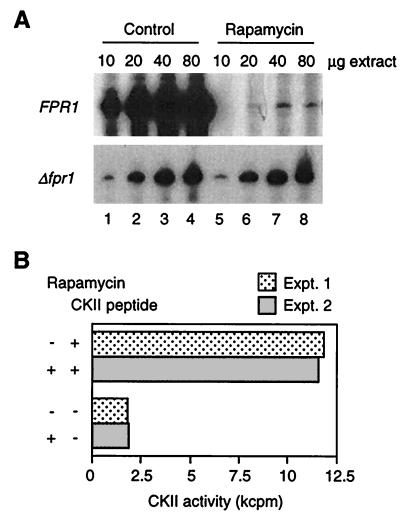
The inhibition of TOR-dependent cellular functions by rapamycin requires the presence of the intracellular ligand for rapamycin, FKBP12 (encoded by FPR1 in yeast [7, 19, 30, 37, 50; reviewed in reference 28]). Therefore, rapamycin phenotypes that normally result from interference with TOR signaling become insensitive to rapamycin when FKBP12 is absent. Based upon this fact, it was possible to examine the TOR dependency of the rapamycin effect on Pol III transcription by comparing transcription in extracts from rapamycin-treated wild-type (FPR1) and fpr1 null (Δfpr1) strains. As expected, rapamycin treatment of wild-type cells for 24 h strongly inhibits Pol III transcription in vitro (Fig. 5A, top panel). In contrast, extracts from Δfpr1 cells show no decline in transcription in response to rapamycin treatment for 24 h (Fig. 5A, bottom panel). We conclude that the effect of rapamycin on Pol III transcription results from interference with the TOR signaling pathway.
FIG. 5.
(A) Repression of Pol III transcription by rapamycin treatment in vivo results from interference with TOR signaling. 5S rRNA transcription is compared in parallel extracts from FPR1 wild-type and fpr1 null cells that were or were not treated with rapamycin (24 h). An overexposure is shown for the FPR1 experiment to demonstrate the low level of transcription in extracts from treated cells (control extracts from FPR1 and Δfpr1 cells supported the same level of transcription). (B) Inhibition of CKII does not account for the repression of Pol III transcription by rapamycin. CKII activity is expressed as kilocounts per minute incorporated during a 5-min reaction at 22°C in the presence (+) or absence (−) of the specific peptide substrate for CKII. Results are shown for two experiments using the FPR1 extract analyzed in panel A.
In mammalian cells (24), the rapamycin binding protein FKBP25 is found in a complex with the highly conserved protein kinase CKII (reviewed in references 2, 29, 31, and 48). CKII in yeast phosphorylates one member of the FKBP family, Fpr3p (48), and is required for efficient Pol III transcription in vivo and in vitro (15, 22). This evidence raises the possibility that rapamycin interferes with Pol III transcription by inhibiting CKII. We therefore tested whether the repression of Pol III transcription in extracts from rapamycin-treated cells is associated with reduced CKII activity. Figure 5B shows that bulk CKII activity, as measured with a specific peptide substrate of CKII (15, 22), is identical in extracts from control and treated cells. This result argues that rapamycin does not exert its effect on Pol III transcription by inhibiting CKII.
Conditional repression of Pol III transcription in extracts from a temperature-sensitive tor2 mutant.
Repression of Pol III transcription in extracts from rapamycin-treated cells may result from the inhibitory effect of rapamycin on translation, resulting, for example, in inadequate synthesis of a labile transcription factor in treated cells (11, 39, 40). We refer to this as a “passive” mechanism of repression. However, our results do not rule out the possibility that interference with TOR signaling represses transcription by a “direct” mechanism that is independent of interference with translation. We therefore tested whether interference with TOR function is involved in translation-independent repression by assaying transcription in extracts from a temperature-sensitive mutant of tor2 and an isogenic wild-type strain. Extracts were prepared in parallel from TOR2 and tor2ts cells harvested at the permissive temperature; at this temperature, Tor2p activity and abundance are identical in these strains (7). Since the transcription reactions do not support translation and are insensitive to 100 μg of cycloheximide per ml (16), any difference in transcription between wild-type and mutant extracts is independent of protein synthesis.
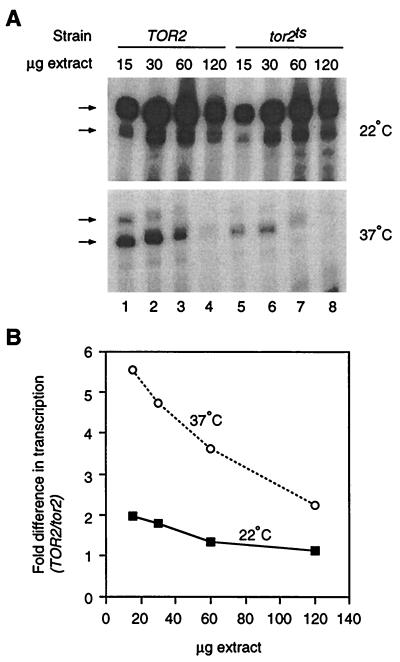
Figure 6 shows transcription in wild-type TOR2 and mutant tor2ts extracts prepared and assayed in parallel. The extracts support a high level of transcription when reactions are performed at 22°C, and the wild-type extract is slightly more active than the mutant at 15 and 30 μg of input protein. At 37°C, the difference in activity between the extracts is substantially enhanced. This effect is most clearly revealed by plotting the ratio of transcription in wild-type/mutant extract against the amount of extract used (Fig. 6B). At 37°C, there is up to a 5.5-fold inhibition of transcription in tor2ts extract, compared to 2.0-fold inhibition at 22°C in reactions with the same amount of protein. Similar results were obtained with two additional pairs of extracts prepared independently from those used in the experiments depicted in Fig. 6. We conclude that interference with TOR function in vitro represses Pol III transcription by a mechanism that is independent of TOR-mediated effects on translation.
FIG. 6.
Temperature-dependent repression of Pol III transcription in extracts from a tor2ts mutant. (A) Pol III transcription is inhibited at the restrictive temperature in tor2 extracts. tRNALeu transcription at 22 and 37°C was compared in extracts from TOR2 and tor2ts cells harvested at the permissive temperature. Although transcription is inhibited at 37°C in wild-type and mutant extracts, the effect is more pronounced in the mutant. (B) Quantitation of the experiment represented in panel A (bands indicated by the arrows were analyzed; see references 3 and 12). Data are plotted as the ratio of transcription in wild-type/mutant extracts for the indicated amounts of protein. A quantifiable signal was detected in lane 8 for the 37°C reaction upon long exposure of the film and phosphorimager plate.
In the absence of the data in Fig. 6, it remained formally possible that rapamycin represses transcription by an FPR1-dependent but TOR-independent mechanism: for example, direct binding of the drug to a component of the Pol III transcription machinery. This scenario seems highly unlikely, given that all documented effects of rapamycin act at the level of the TOR kinases (see the introduction) and that we observed repression in vitro using extracts from tor2ts cells without rapamycin treatment.
Biochemical analysis of the transcription defect in extracts from cells treated with rapamycin.
Interference with TOR function in vivo is likely to affect Pol III transcription by a passive mechanism involving repression of translation and by a direct mechanism that is independent of the inhibition of translation. In order to gain an appreciation of the full spectrum of effects on the Pol III transcriptional machinery (i.e., passive and direct) resulting from short-term interference with TOR function, we investigated the biochemical basis of the modest transcription defect (two- to threefold in this experiment) in extracts from cells treated for 1 h with rapamycin.
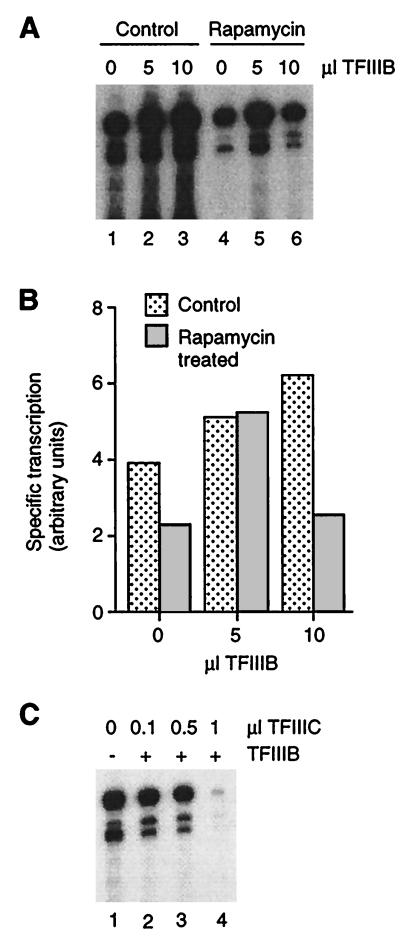
Since the pattern of labeled transcription products in extracts from treated and untreated cells is virtually identical in standard 8% polyacrylamide sequencing gels, we conclude that interference with TOR function affects transcription at the level of initiation rather than of pausing, termination, or start site selection. The effect of rapamycin is therefore likely to impinge on the core Pol III transcription machinery, namely RNA Pol III, TFIIIB, and TFIIIC. TFIIIB was considered a likely target, since it is significantly down-regulated when cells enter G0 (36) and, at least under some conditions, when cells are treated with cycloheximide (11; compare with reference 9). We therefore tested whether TFIIIB purified from untreated cells could overcome the transcription defect of extracts from rapamycin-treated cells. As shown in Fig. 7A and B, TFIIIB slightly stimulates transcription in control extract, consistent with reports that TFIIIB is normally limiting in vivo and in vitro (see reference 36). An amount of TFIIIB that stimulates wild-type extract by 31% rescues transcription in extract from rapamycin-treated cells to the level observed in control extract supplemented with the same amount of TFIIIB (Fig. 7B). This result is consistent with repression of TFIIIB as a result of short-term treatment with rapamycin.
FIG. 7.
Identification of TFIIIB as a possible target of repression by rapamycin. (A) Extracts (40 μg) from control and rapamycin-treated (1 h) cells were supplemented with TFIIIB (0, 5, and 10 μl) or buffer. (B) Quantitation of the gel in panel A. (C) TFIIIC does not restore transcription in extracts from rapamycin-treated (1 h) cells. TFIIIC was added in increasing amounts to extracts (25 μg) in the presence (+) or absence (−) of a saturating amount of TFIIIB (as defined in the experiments represented in panel A).
Interestingly, a larger amount of TFIIIB further stimulates control extracts but not rapamycin-treated extracts (Fig. 7A; compare lanes 2 and 3 to lanes 5 and 6). Indeed, large amounts of TFIIIB added to extracts from treated cells inhibit transcription, perhaps by unproductive interaction of soluble TFIIIB with another factor that has become limiting (see Fig. 4A in reference 36 for a similar example). In other words, inhibition of extracts from rapamycin-treated cells by large amounts of TFIIIB suggests that a component of the transcription machinery that is in excess of TFIIIB in control extracts is repressed in treated extracts. We tested whether this component is TFIIIC by adding a saturating amount of TFIIIB and increasing amounts of affinity-purified TFIIIC to extracts from treated cells. As shown in Fig. 7C, TFIIIC does not stimulate transcription in treated extracts supplemented with a saturating amount of TFIIIB. Therefore, rapamycin probably does not repress TFIIIC.
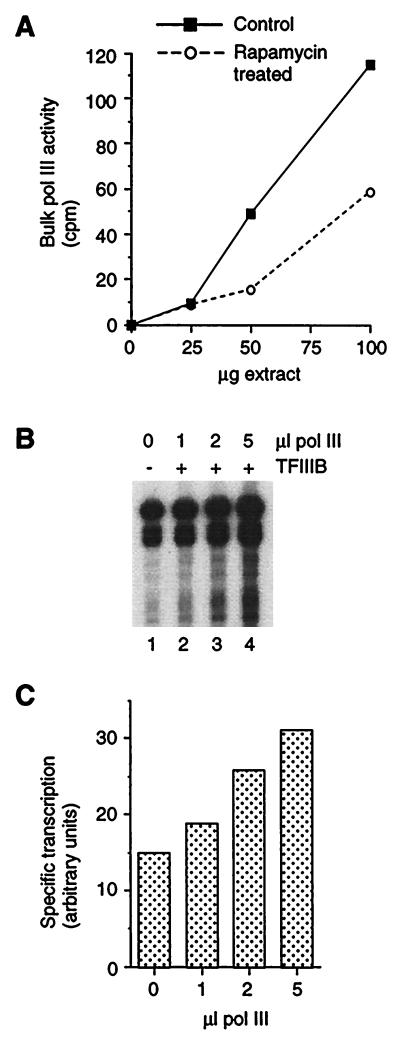
Our next step was to test whether RNA Pol III is repressed by rapamycin. Measurements of bulk Pol III activity in extracts from treated and untreated cells revealed that Pol III is significantly inhibited in extracts from cells treated for 1 h with rapamycin (by 49% at the 100-μg point in the titration in Fig. 8A). Bulk Pol III activity in 100 μg of extract from cells exposed to rapamycin for 24 h is also inhibited (by 59% [not shown]). To confirm that Pol III is repressed by rapamycin, we titrated a crude Pol III preparation from untreated cells into rapamycin-treated extracts (1 h) supplemented with a saturating amount of TFIIIB (Fig. 8B). In marked contrast to the result obtained with TFIIIC, Pol III does stimulate transcription in the presence of excess TFIIIB. Quantitation reveals a 2.1-fold effect (Fig. 8C). Therefore, rapamycin down-regulates Pol III transcription in part by an effect on RNA Pol III.
FIG. 8.
Identification of RNA Pol III as a target of repression by rapamycin. (A) Measurement of bulk Pol III activity in extracts from control and rapamycin-treated (1 h) cells. (B) Stimulation of specific transcription by addition of Pol III to extracts (25 μg) from rapamycin-treated (1 h) cells. (C) Quantitation of the gel in panel B.
DISCUSSION
A TOR-dependent mechanism induces the G0 program of transcription in yeast.
Previous studies have revealed that interference with TOR signaling induces all aspects of the stationary-phase response so far examined, namely, cell cycle arrest with 1n DNA content, repression of translation, cell wall thickening, glycogen deposition, transcriptional induction of G0-specific Pol II genes, vacuolar enlargement, and increased thermotolerance (4, 7, 10). Our experiments with rapamycin and a conditional tor2 mutant further demonstrate repression of Pol I and Pol III transcription when TOR function is perturbed. Taken together, the available data support the notion that TOR signaling in yeast occupies a central position in the global regulation of nuclear transcription in response to nutrient availability. Since a universal component of growth control in eukaryotes is the down-regulation of Pol I and Pol III transcription in G0, and since the kinase domain of a mammalian TOR homolog can provide rapamycin-sensitive TOR function in yeast (1), it is attractive to consider that in all eukaryotes TOR signaling may be involved in establishing the pattern of nuclear transcription in G0.
Transcriptional repression by a Tor2p-dependent mechanism that does not involve translational repression.
Interference with Tor2p function in extracts from a temperature-sensitive tor2 mutant represses transcription by a mechanism that is independent of TOR effects on translation. Considering that TOR kinases regulate a broad spectrum of cellular responses associated with entry into stationary phase and our observation that rapamycin treatment of cells represses Pol III transcription in vitro, we take the tor2ts result to indicate that Pol III down-regulation during entry into G0 is not exclusively a passive consequence of the inhibition of translation. Repression in stationary phase indeed may result exclusively from direct regulation of the transcriptional machinery by a signaling network involving the TOR kinases.
Effects of PP2A and TOR on transcription: how are they related?.
The defect in Pol III transcription in yeast mutants of the regulatory A subunit of PP2A stems from the inhibition of Pol III and TFIIIB under restrictive conditions (41). In this instance, misregulated PP2A is proposed to dephosphorylate and activate an inhibitor of transcription. Considering that PP2A is a component of the TOR pathway (10) and that Pol III and probably TFIIIB are under TOR control in yeast, repression of Pol III transcription by rapamycin may reflect interference with a TOR-PP2A signal transduction pathway that regulates Pol III transcription. An appealing extension of this model is that a TOR-responsive kinase and PP2A act on the same transcriptional inhibitor, the TOR-dependent kinase as a repressor of the inhibitor and PP2A as an activator. To date, however, we have no evidence for such an inhibitor in extracts from cells exposed to rapamycin, even in the extreme case of a 24-h treatment. The precise relationship between the dominant inhibitory effect involving PP2A and TOR-dependent transcriptional repression, therefore, remains to be fully elucidated.
Since the TOR-PP2A pathway regulates the pattern of Pol II transcription in G0 (4), alterations in PP2A function that perturb Pol III transcription might also be expected to affect Pol II transcription. That the transcription of some Pol II genes indeed is impaired in PP2A mutants (41) lends support to the hypothesis that TOR-PP2A signaling is involved in setting the overall pattern of nuclear transcription in G0.
What other protein kinases might be involved in TOR effects on pol III transcription?
The only protein kinase previously shown to influence pol III transcription in yeast is CKII (14, 15, 22). Our results, however, argue against an involvement of CKII in TOR signaling to the Pol III transcriptional machinery. We were unable to detect any change in bulk CKII activity in extracts from cells treated with rapamycin compared to control extracts, and the elongation capacity of Pol III is not affected when CKII is inactivated (15).
Recent evidence suggests that TOR functions related to the control of progression into stationary phase involve genes implicated in protein kinase C signaling (20). tor2 mutations affecting translation and cell cycle arrest in yeast are suppressed by overexpression of (i) MSS4, a phosphatidylinositol 4-phosphate kinase homolog that may be upstream of the yeast protein kinase C (Pkc1p), and (ii) PLC1, a phosphoinositide-specific phospholipase C homolog that is likely to function in the same pathway as Pkc1p. These observations, in view of reports that Pol III transcription is sensitive to protein kinase C in metazoans (23, 42), suggest the intriguing possibility that a TOR-protein kinase C signaling pathway impinges on the Pol III transcriptional machinery in yeast.
Mechanism of repression at the level of transcriptional machinery.
Our working model is that TOR signaling regulates Pol III transcription by modulating the activity of TFIIIB and Pol III. At present, however, it remains to be shown that TFIIIB is defective in extracts in which TOR function is impaired, and the molecular basis of the repression of Pol III is not known. Theoretically, the decreased Pol III activity in extracts from rapamycin-treated cells could be a secondary consequence of the inhibition of TFIIIB. For example, when TFIIIB is inhibited, Pol III could be released into the nucleoplasm, where it would be susceptible to inactivation by soluble factors. We do not favor this possibility because in other cases where TFIIIB is inhibited in vivo, there is no decline in Pol III activity in vitro (11, 15, 36). Rather, based upon the observation that some subunits of Pol III are phosphoproteins in vivo (see reference 41), we favor a model in which TOR-dependent signaling regulates the phosphorylation state and therefore the activity of Pol III. It remains possible, however, that TOR signaling impinges on the Pol III transcriptional machinery not by affecting the phosphorylation status of components of the transcriptional machinery but by activating other regulatory mechanisms such as, for example, targeted degradation pathways (see reference 11).
Previous studies of Pol III transcriptional repression in yeast have not provided any evidence for regulation of the polymerase (11, 36) except in a case in which perturbation of PP2A function apparently induces a transcriptional inhibitor (41). In these studies, transcription was analyzed in nuclear extracts or in whole-cell extracts made by cell disruption nominally at 4°C. On the other hand, using whole-cell extracts prepared from cells broken open while they are frozen, we find that interference with TOR function represses Pol III by a mechanism that is distinct from the dominant-negative effect in PP2A mutants (41). Our demonstration that Pol III activity is sensitive to TOR kinases, which control all aspects of the stationary-phase response so far examined, justifies reexamination of the physiological regulation of Pol III in yeast.
ACKNOWLEDGMENTS
Bernard Leduc and Suren Sehgal of Wyeth-Ayerst Research are thanked for supplying rapamycin. Valuable technical assistance was afforded by Darren Hockman, and Brent Altheim kindly provided extracts from cdc28 cells.
This work was supported by an establishment grant from the Alberta Heritage Foundation for Medical Research and by operating grants from the Canadian NCI and MRC. M.C.S. is a scholar of the MRC. D.Z. was supported in part by a 75th Anniversary Scholarship from the University of Alberta.
REFERENCES
- 1.Alarcon C M, Cardenas M E, Heitman J. Mammalian RAFT1 kinase domain provides rapamycin-sensitive TOR function in yeast. Genes Dev. 1996;10:279–288. doi: 10.1101/gad.10.3.279. [DOI] [PubMed] [Google Scholar]
- 2.Allende J E, Allende C C. Protein kinase CK2: an enzyme with multiple substrates and a puzzling regulation. FASEB J. 1995;9:313–323. doi: 10.1096/fasebj.9.5.7896000. [DOI] [PubMed] [Google Scholar]
- 3.Baker R E, Hall B D. Structural features of yeast tRNA genes which affect transcription factor binding. EMBO J. 1984;3:2793–2800. doi: 10.1002/j.1460-2075.1984.tb02211.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barbet N C, Schneider U, Helliwell S B, Stansfield I, Tuite M F, Hall M N. TOR controls translation initiation and early G1 progression in yeast. Mol Biol Cell. 1996;7:25–42. doi: 10.1091/mbc.7.1.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bradford M M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle or protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 6.Burke D J, Church D. Protein synthesis requirements for nuclear division, cytokinesis, and cell separation in Saccharomyces cerevisiae. Mol Cell Biol. 1991;11:3691–3698. doi: 10.1128/mcb.11.7.3691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cardenas M E, Heitman J. FKBP12-rapamycin target TOR2 is a vacuolar protein with an associated phosphatidylinositol 4-kinase activity. EMBO J. 1995;14:5892–5907. doi: 10.1002/j.1460-2075.1995.tb00277.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Choder M. A general topoisomerase I-dependent transcriptional repression in the stationary phase in yeast. Genes Dev. 1991;5:2315–2326. doi: 10.1101/gad.5.12a.2315. [DOI] [PubMed] [Google Scholar]
- 9.Clarke E M, Peterson C L, Brainard A V, Riggs D L. Regulation of the RNA polymerase I and III transcription systems in response to growth conditions. J Biol Chem. 1996;271:22189–22195. doi: 10.1074/jbc.271.36.22189. [DOI] [PubMed] [Google Scholar]
- 10.Di Como C J, Arndt K T. Nutrients, via the Tor proteins, stimulate the association of Tap42 with type 2A phosphatases. Genes Dev. 1996;10:1904–1916. doi: 10.1101/gad.10.15.1904. [DOI] [PubMed] [Google Scholar]
- 11.Dieci G, Duimio L, Peracchia G, Ottonello S. Selective inactivation of two components of the multiprotein transcription factor TFIIIB in cycloheximide growth-arrested yeast cells. J Biol Chem. 1995;270:13476–13482. doi: 10.1074/jbc.270.22.13476. [DOI] [PubMed] [Google Scholar]
- 12.Fabrizio P, Coppo A, Fruscoloni P, Benedetti P, Di Segni G, Tocchini-Valentini G P. Comparative mutational analysis of wild-type and stretched tRNA3Leu gene promoters. Proc Natl Acad Sci USA. 1987;84:8763–8767. doi: 10.1073/pnas.84.24.8763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Geiduschek E P, Kassavetis G A. RNA polymerase III transcription complexes. In: McKnight S L, Yamamoto K, editors. Transcriptional regulation. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1992. pp. 247–280. [Google Scholar]
- 14.Ghadivel, A., D. J. Hockman, and M. C. Schultz. A review of progress towards elucidating the role of protein kinase CK2 in polymerase III transcription: regulation of the TATA binding protein. Mol. Cell. Biochem., in press. [PubMed]
- 15.Ghavidel A, Schultz M C. Casein kinase II regulation of yeast TFIIIB is mediated by the TATA binding protein. Genes Dev. 1997;11:2780–2789. doi: 10.1101/gad.11.21.2780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ghavidel, A., and M. C. Schultz. Unpublished data.
- 17.Hammond C I, Holland M J. Purification of yeast RNA polymerases using heparin agarose affinity chromatography. Transcriptional properties of the purified enzymes on defined templates. J Biol Chem. 1983;258:3230–3241. [PubMed] [Google Scholar]
- 18.Hanna D E, Rethinaswamy A, Glover C V C., III Casein kinase II is required for cell cycle progression during G1 and G2/M in Saccharomyces cerevisiae. J Biol Chem. 1995;270:25905–25914. doi: 10.1074/jbc.270.43.25905. [DOI] [PubMed] [Google Scholar]
- 19.Heitman J, Movva N R, Hall M N. Targets for cell cycle arrest by the immunosuppressant rapamycin in yeast. Science. 1991;253:905–909. doi: 10.1126/science.1715094. [DOI] [PubMed] [Google Scholar]
- 20.Helliwell S B, Howald I, Barbet N, Hall M N. TOR2 is part of two related signaling pathways coordinating cell growth in Saccharomyces cerevisiae. Genetics. 1998;148:99–112. doi: 10.1093/genetics/148.1.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hereford L M, Hartwell L H. Sequential gene function in the initiation of Saccharomyces cerevisiae DNA synthesis. J Mol Biol. 1974;84:445–461. doi: 10.1016/0022-2836(74)90451-3. [DOI] [PubMed] [Google Scholar]
- 22.Hockman D J, Schultz M C. Casein kinase II is required for efficient transcription by RNA polymerase III. Mol Cell Biol. 1996;16:892–898. doi: 10.1128/mcb.16.3.892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.James C B L, Carter T H. Activation of protein kinase C inhibits adenovirus VA gene transcription in vitro. J Gen Virol. 1992;73:3133–3139. doi: 10.1099/0022-1317-73-12-3133. [DOI] [PubMed] [Google Scholar]
- 24.Jin Y J, Burakoff S J. The 25-kDa FK506-binding protein is localized in the nucleus and associates with casein kinase II and nucleolin. Proc Natl Acad Sci USA. 1993;90:7769–7773. doi: 10.1073/pnas.90.16.7769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jones E W. Tackling the protease problem in Saccharomyces cerevisiae. Methods Enzymol. 1991;194:428–453. doi: 10.1016/0076-6879(91)94034-a. [DOI] [PubMed] [Google Scholar]
- 26.Ju Q, Warner J R. Ribosome synthesis during the growth cycle of Saccharomyces cerevisiae. Yeast. 1994;10:151–157. doi: 10.1002/yea.320100203. [DOI] [PubMed] [Google Scholar]
- 27.Kassavetis G A, Riggs D L, Negri R, Nguyen L H, Geiduschek E P. Transcription factor IIIB generates extended DNA interactions in RNA polymerase III transcription complexes on tRNA genes. Mol Cell Biol. 1989;9:2551–2566. doi: 10.1128/mcb.9.6.2551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kay J E. Structure-function relationships in the FK506-binding protein (FKBP) family of peptidylprolyl cis-trans isomerases. Biochem J. 1996;314:361–385. [PMC free article] [PubMed] [Google Scholar]
- 29.Litchfield D W, Lüscher B. Casein kinase II in signal transduction and cell cycle regulation. Mol Cell Biochem. 1993;127/128:187–199. doi: 10.1007/BF01076770. [DOI] [PubMed] [Google Scholar]
- 30.Lorenz M C, Heitman J. TOR mutations confer rapamycin resistance by preventing interaction with FKBP12 rapamycin. J Biol Chem. 1995;270:27531–27537. doi: 10.1074/jbc.270.46.27531. [DOI] [PubMed] [Google Scholar]
- 31.Pinna L A. Casein kinase 2: an ‘eminence grise’ in cellular regulation? Biochim Biophys Acta. 1990;1054:267–284. doi: 10.1016/0167-4889(90)90098-x. [DOI] [PubMed] [Google Scholar]
- 32.Roeder R G. Multiple forms of deoxyribonucleic acid-dependent ribonucleic acid polymerase in Xenopus laevis. Isolation and partial characterization. J Biol Chem. 1974;249:241–248. [PubMed] [Google Scholar]
- 33.Schultz M C, Choe S Y, Reeder R H. Specific initiation by RNA polymerase I in a whole-cell extract from yeast. Proc Natl Acad Sci USA. 1991;88:1004–1008. doi: 10.1073/pnas.88.3.1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schultz M C, Hockman D J, Harkness T A A, Garinther W I, Altheim B A. Chromatin assembly in a yeast whole-cell extract. Proc Natl Acad Sci USA. 1997;94:9034–9039. doi: 10.1073/pnas.94.17.9034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schultz M C, Reeder R H, Hahn S. Variants of the TATA-binding protein can distinguish subsets of RNA polymerase I, II, and III promoters. Cell. 1992;69:697–702. doi: 10.1016/0092-8674(92)90233-3. [DOI] [PubMed] [Google Scholar]
- 36.Sethy I, Moir R D, Librizi M, Willis I M. In vitro evidence for growth regulation of tRNA transcription in yeast. A role for transcription factor (TF) IIIB70 and TFIIIC. J Biol Chem. 1995;270:28463–28470. doi: 10.1074/jbc.270.47.28463. [DOI] [PubMed] [Google Scholar]
- 37.Stan R, McLaughlin M M, Cafferkey R, Johnson R K, Rosenberg M, Livi G P. Interaction between FKBP12-rapamycin and TOR involves a conserved serine residue. J Biol Chem. 1994;269:32027–32030. [PubMed] [Google Scholar]
- 38.Thomas B J, Rothstein R. Elevated recombination rates in transcriptionally active DNA. Cell. 1989;56:619–630. doi: 10.1016/0092-8674(89)90584-9. [DOI] [PubMed] [Google Scholar]
- 39.Tower J, Sollner-Webb B. Transcription of mouse rDNA is regulated by an activated subform of RNA polymerase I. Cell. 1987;50:873–883. doi: 10.1016/0092-8674(87)90514-9. [DOI] [PubMed] [Google Scholar]
- 40.Tower J, Sollner-Webb B. Polymerase III transcription factor B activity is reduced in extracts of growth-restricted cells. Mol Cell Biol. 1988;8:1001–1005. doi: 10.1128/mcb.8.2.1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.van Zyl W, Huang W, Sneddon A A, Stark M, Camier S, Werner M, Marck C, Sentenac A, Broach J R. Inactivation of the protein phosphatase 2A regulatory subunit A results in morphological and transcriptional defects in Saccharomyces cerevisiae. Mol Cell Biol. 1992;12:4946–4959. doi: 10.1128/mcb.12.11.4946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang H-D, Yuh C-H, Dang C V, Johnson D L. The hepatitis B virus X protein increases the cellular level of TATA-binding protein, which mediates transactivation of RNA polymerase III genes. Mol Cell Biol. 1995;15:6720–6728. doi: 10.1128/mcb.15.12.6720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Werner-Washburne M, Brown D, Braun E. Bcy1, the regulatory subunit of cAMP-dependent protein kinase A in yeast, is differentially modified in response to the physiological status of the cell. J Biol Chem. 1991;266:19704–19709. [PubMed] [Google Scholar]
- 44.Werner-Washburne M, Braun E, Johnston G C, Singer R A. Stationary phase in the yeast Saccharomyces cerevisiae. Microbiol Rev. 1993;57:383–401. doi: 10.1128/mr.57.2.383-401.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Werner-Washburne M, Braun E L, Crawford M E, Peck V M. Stationary phase in Saccharomyces cerevisiae. Mol Microbiol. 1996;19:1159–1166. doi: 10.1111/j.1365-2958.1996.tb02461.x. [DOI] [PubMed] [Google Scholar]
- 46.White R J, Gottlieb T M, Downes C S, Jackson S P. Cell cycle regulation of RNA polymerase III transcription. Mol Cell Biol. 1995;15:6653–6662. doi: 10.1128/mcb.15.12.6653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Willis I M. RNA polymerase III. Genes, factors and transcriptional specificity. Eur J Biochem. 1993;212:1–11. doi: 10.1111/j.1432-1033.1993.tb17626.x. [DOI] [PubMed] [Google Scholar]
- 48.Wilson L K, Dhillon N, Thorner J, Martin G S. Casein kinase II catalyzes tyrosine phosphorylation of the yeast nucleolar immunophilin Fpr3. J Biol Chem. 1997;272:12961–12967. doi: 10.1074/jbc.272.20.12961. [DOI] [PubMed] [Google Scholar]
- 49.Zaragoza, D., and M. C. Schultz. Unpublished data.
- 50.Zheng X-F, Fiorentino D, Chen J, Crabtree G R, Schreiber S L. TOR kinase domains are required for two distinct functions, only one of which is inhibited by rapamycin. Cell. 1995;82:121–130. doi: 10.1016/0092-8674(95)90058-6. [DOI] [PubMed] [Google Scholar]