Abstract
N6-methyladenosine (m6A) RNA methylation plays a pivotal role in immune responses and the onset and advancement of cancer. Nonetheless, the precise impact of m6A modification in lung adenocarcinoma (LUAD) and its associated tumor microenvironment (TME) remains to be fully elucidated. Here, we distinguished distinct m6A modification patterns within two separate LUAD cohorts using a set of 21 m6A regulators. The TME characteristics associated with these two patterns align with the immune-inflamed and immune-excluded phenotypes, respectively. We identified 2064 m6A-related genes, which were used as a basis to divide all LUAD samples into three distinct m6A gene clusters. We applied a scoring system to evaluate the m6A gene signature of the m6A modification pattern in individual patients. To authenticate the categorization significance of m6A modification patterns, we established a correlation between m6A score and TME infiltration profiling, tumor somatic mutations, and responses to immunotherapy. A high level of m6A modification may be associated with the aggressiveness and poor prognosis of LUAD. Further studies should investigate the mechanism of action of m6A regulators and m6A-related genes to improve the diagnosis and treatment of patients with LUAD.
Keywords: m6A, tumor microenvironment, stroma, lung adenocarcinoma, tumor somatic mutation, immunotherapy
Impact Statement
Here, we distinguished two distinct m6A modification patterns within two lung adenocarcinoma (LUAD) cohorts using 21 m6A regulators. The attributes of the tumor microenvironment (TME) corresponding to these patterns aligned with the immune-excluded and immune-inflamed phenotypes, respectively. We applied a scoring system, called m6A score, to evaluate the m6A gene signature of the m6A modification pattern in individual patients. The m6A score was correlated with TME infiltration characterization, tumor somatic mutation, and patients’ response to immunotherapy. Further studies should be conducted on the mechanism of action of m6A regulators and m6A-related genes for improved diagnosis and treatment of LUAD patients.
Introduction
Cumulative evidence suggests the presence of diverse post-transcriptional RNA modifications across nearly all organisms. 1 RNA modifications, including N6-methyladenosine (m6A) modification, hold a crucial role within the field of RNA epigenetics.2,3 Already in the 1970s, m6A was recognized as the predominant modification found in mRNA and other non-coding RNAs as the most common modifications in mRNA and other non-coding within eukaryotic organisms.1,4–7 Important biological functions regulated by m6A modification are reversible and rely on a series of enzymes, encompassing methyltransferase (writers), demethylase (erasers), and binding proteins (readers). 8 Writers include RBM15, ZC3H13, MeTTTL3, MeTTTL14, WTAP, and KIAA1429, which add functional sites of m6A. Erasers include ALKBH5 and FTO, which can remove functional sites of m6A. Readers encompassing YTHDF1/2/3, FMR1, LRPPRC, YTHDC1/2, HNRNPA2B1, and LRPPRC preferentially bind to the functional site of m6A and play regulatory roles. Thus far, a large amount of evidence has confirmed that m6A modification regulates a series of tumor cells’ malignant biological behavior, encompassing proliferation, invasion, and metastasis, alongside its involvement in immune regulation.9–14 Studies have established a connection between m6A modification and the pathogenesis of lung adenocarcinoma (LUAD).15,16
LUAD has high morbidity and mortality rates worldwide and ranks among the top in malignant tumors. Its pathogenesis is complicated, and early diagnosis is difficult. 17 Despite the development of approaches, such as molecular targeted therapy, chemotherapy, surgical resection, and radiotherapy, for LUAD in recent decades, it has not significantly improved patients’ outcome with the condition.18,19 Therefore, attaining a holistic comprehension of the molecular mechanisms driving LUAD initiation and progression is imperative to unearth innovative therapeutic strategies and advance early diagnosis, treatment efficacy, and prognostic outcomes. A number of RNA methylation modifications, including pseudoguanosine, 5-methylcytosine (m5C), and m6A, 20 are extensively present in LUAD RNA and exert significant influences on the initiation, advancement, metastasis, therapy, and prognosis of LUAD. 21 Continuous research has found the engagement of m6A modification in tumor regulation through its impact on the tumor microenvironment (TME).12,22 This environment is vital for tumor survival and contains an array of stromal cells, immune cells, tumor cells, and various cytokines, which are important in tumor growth, migration, and invasion.23,24 The interaction between various cells and cytokines is an important factor for TME to affect the malignant biological behavior of tumors. 25
A rapid advancement in high throughput detection technology has led to increasing insights into m6A modification’s biochemical and biological functions. 26 Reversibility of the m6A modification is becoming increasingly evident. 26 m6A regulators target different genes, the levels of m6A regulatory factors vary, and the methylated target mRNA changes differently; as such, biological functions regulated by m6A modification can be affected, enabling it to play distinct protumor or antitumor roles. 27 In summary, unraveling the regulatory influence of m6A modification on LUAD tumors and their TME is essential for identifying novel serological or histological predictive indicators for therapeutic interventions, prognosis, and diagnosis. However, the comprehensive scope of m6A modification’s role in LUAD and its TME remains unclear.
Our study aims to investigate the roles played by m6A regulators and m6A-related genes within LUAD and its TME, using publicly available databases. The gene expression data and clinical details of LUAD patients were acquired from The Cancer Genome Atlas (TCGA) and Gene Expression Omnibus (GEO) repositories. 28 Two distinct patterns of m6A modification were identified. The TME characteristics of the two patterns correspond to the immune-inflamed and immune-excluded phenotypes. We detected three tumor subtypes of m6A-related genes. Given the inherent tumor heterogeneity, we devised a scoring system capable of quantifying individual m6A modification patterns. Findings validated the potential of the m6A score in influencing the staging, treatment approaches, and prognosis of LUAD.
Materials and methods
Dataset source and preprocessing
Supplemental Figure 1 presents the research process. GSE68465 gene expression profiles were retrieved from the GEO database and used as training cohort. 29 Microarray data were generated using the Affymetrix Human Genome U133A Array. GSE50081, GSE37745, GSE31210, GSE30219, and GSE72094 consist of 181, 196, 226, 293, and 442 patients with lung cancer, respectively, with survival data available in a total of 1294 cases. All data were gathered as validation cohort. The matrix files were normalized for further analysis. The RNA sequencing data (expressed as Fragments Per Kilobase of transcript per Million mapped reads (FPKM) values) for TCGA-LUAD were acquired from the TCGA data. 30 The FPKM values underwent conversion to transcripts per kilobase million (TPM) values. Somatic mutation information was acquired from TCGA database. The correction of batch effects was performed using the ComBat algorithm from the R package “sva.” 31 The copy number variation (CNV) dataset was sourced from the TCGA database. Genes within CNV regions and corresponding CNV values were extracted using hg19 annotation data from University of Colomboschool of Computing (UCSC) for CNV analysis. 32 To depict the CNV landscape of the 21 m6A regulators across 23 chromosome pairs, we used the “RCircos” R package for visualization.33,34
Unsupervised clustering for m6A clusters
The expression matrix of the 21 m6A regulators was retrieved from the GEO datasets to discern distinct m6A modification patterns influenced by these regulators. These m6A regulators comprised 11 readers (YTHDF1/2/3, HNRNPA2B1, IGF2BP1, HNRNPC, YTHDC1/2, LRPPRC, ELAVL1, FMR1), 8 writers (RBM15/15B, METTL3/14, KIAA1429, ZC3H13, WTAP, CBLL1), and 2 erasers (ALKBH5, FTO). 35 Employing unsupervised cluster analysis, we identified diverse m6A modification patterns through the analysis of expression profiles associated with these m6A regulators. We applied the consensus clustering algorithm to ensure stable and reliable outcomes, using the “ConsensuClusterPlus” R package for unsupervised clustering, with 1000 repetitions. 36
Gene set variation analysis
We employed the “Gene set variation analysis (GSVA)” R package (version 1.34.0) to conduct GSVA enrichment analysis, aiming to explore distinctions in biological processes among the various m6A modification patterns. 37 GSVA, a widely used approach, allows the evaluation of changes in biological process and pathway activity at the level of individual samples. This gene set is based on the “c2.cp.kegg.v7.1.symbols.gmt” collection in the molecular signature database (MSigDB). 38 Statistically significant differences were indicated by adjusted P values less than 0.05. Functional annotation of m6A-related genes was performed using the “clusterProfiler” package. 39 Significance was established at a false discovery rate (FDR) < 0.05. Subsequently, heatmaps were generated through the “pheatmap” R package (version 1.0.12) to visually represent distinct attributes within the m6A modification patterns. 40
Single-sample gene set enrichment analysis for TME cell infiltration
The “GSVA” R package was employed for single-sample gene set enrichment analysis (ssGSEA) analysis to assess the abundance of immune cell infiltration types within the LUAD TME. Charoentong et al. 41 provided gene markers for various immune cell types within the TME, including dendritic cells (DCs), comprising activated DCs and immature DCs, and plasmacytoid DCs. In addition, markers were available for mast cells, eosinophils, macrophages, natural killer cells (NKs), NK CD56bright cells, NK CD56dim cells, neutrophils, and adaptive immune cells, such as T helper cells, T helper 1 cells, Th2 cells, T helper 17 cells, T gamma delta cells, CD8 + T cells, T central memory cells, T effector memory cells, T follicular helper cells, regulatory T (Treg) cells, T cells, B cells, and cytotoxic cells. 42
DEGs associated with m6A clusters
Based on the results of unsupervised clustering involving 21 m6A regulators, patients were categorized into two distinct clusters, reflecting diverse m6A modification patterns. The “limma” R package, employing an empirical Bayesian approach, was used for identifying differentially expressed genes (DEGs). Designated as either m6A phenotype-associated genes or m6A gene signatures, these DEGs were used to quantify the m6A modification patterns within individual samples. 43
Generation of m6A score
To assess the m6A signature in individual patients and quantify their m6A modification patterns, we implemented a scoring system known as the m6A score. To establish m6A gene signatures, we identified DEGs between two m6A clusters in all LUAD cohort sample, postextraction of normalized and overlapping genes. The DEGs underwent unsupervised clustering to segment patients into distinct gene clusters, facilitating subsequent analyses. For evaluating prognosis, the Univariate Cox regression model was applied to each m6A gene signature, identifying genes with noteworthy prognostic implications. Principal component analysis (PCA) was then employed to construct the m6A-related gene signature, 34 using i as the m6A phenotype-associated genes’ expression
Correlation between m6A score and somatic alteration data
To assess the tumor mutation burden (TMB) in LUAD, we tallied the total count of non-synonymous mutations within patients from the TCGA-LUAD cohort, using corresponding mutation data obtained from the TCGA database. 44 The somatic changes in driver genes for LUAD were assessed within high and low m6A score subgroups. To pinpoint LUAD driver genes, we employed the “waterfall” function from the “maftools” R package. 45 We employed an OncoPrint to visualize the top 25 driver genes with the highest alteration frequencies. 44
Immunotherapy response and m6A score
We obtained patient immunophenoscores (IPSs) through the Cancer Immunome Atlas framework (TCIA). The IPS serves as a marker for immune checkpoint inhibitor (ICI) response and is derived in an unbiased manner, considering four categories: major histocompatibility complex (MHC)–related molecules, suppressive cells (Tregs and Myeloid-derived suppressor cells [MDSCs]), effector cells (effector memory/activated CD8 + T cells and effector memory/activated CD4 + T cells), and checkpoints or immunomodulators, color-coded along the outer rim (red for positive Z scores, blue for negative Z scores). 41 This weighted averaged Z-score is used to calculate the IPS, which ranges from 0 to 10. An IPS of 10 corresponds to a Z-score ⩾ 3, while an IPS of 0 corresponds to a z-score ⩽ 0. 46 To assess the predictive potential of m6A scores, we generated a violin plot to compare IPSs between high- and low-score m6A groups.
Statistical analysis
R 4.0.5 was used to process the data. We used Spearman and distance correlation analyses to calculate the correlation coefficients. 47 Differences between the two groups were assessed using Wilcoxon test and one-way analysis of variance (ANOVA). Further comparison between the groups was conducted using the Kruskal–Wallis test. Based on the correlation between m6A score and overall survival (OS), we used the “survMiner” R package to establish subgroup cutoff points. The maximum rank statistics was found by the “Surv-Cutpoint” function through repeated tests at all potential point cuts. According to the maximum log-rank selection, the m6A score was dichotomized and patients were categorized into high- and low-score m6A groups, which helped mitigate the influence of batch processing on calculations. Prognostic analysis was performed using Kaplan–Meier survival curves and log-rank test. The hazard ratios (HRs) of m6A regulators and phenotype-related genes were computed using the univariate Cox regression model. Patients were then subjected to final multivariate prognostic analysis. Statistical significance was determined at a two-tailed alpha level of P < 0.05. 48
Results
Genetic variations in m6A regulators within LUAD
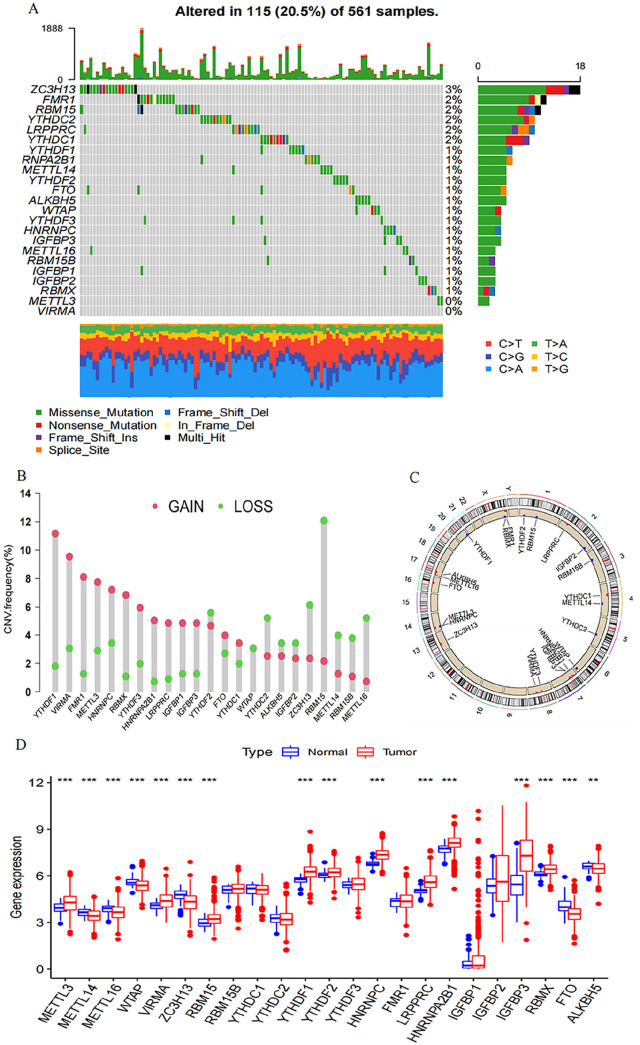
We finally identified 21 m6A regulators. Specific mRNAs are regulated by m6A regulators, indicating the relation between those regulators and the mechanism of cell proliferation, differentiation, and cancer progression. A compilation of LUAD’s 21 m6A regulators was undertaken to determine somatic mutations and CNVs. A comprehensive count of 115 mutations was detected within the dataset of 561 samples. The most frequent mutation was found in ZC3H13 (3%), followed by YTHDC1/2, LRPPRC, RBM15, and FMR1 (2%); meanwhile, METTL3 and VIRMA did not show any mutation (Figure 1(A)). Further analysis showed significant co-occurrences, including ZC3H13 with YTHDC2, YTHDC1 with YTHDF1, YTHDC1 with LRPPRC, YTHDC1 with IGFBP1, and YTHDC2 with LRPPRC (Supplemental Figure S1(B)). We assessed the frequency of CNV changes, observing prevalent CNV alterations among the 21 m6A regulators, with a predominant focus on copy number amplification. The deletion frequencies of ALKBH5, IGFBP2, METTL14/16, RBM15/15B, ZC3H13, YTHDF2, YTHDC2, and WTAP CNV had a widespread frequency (Figure 1(B)). Figure 1(C) illustrates the chromosome locations with CNV alterations among the m6A regulators. To examine the link between the genetic variations mentioned above and m6A regulatory expression among LUAD patients, we investigated those mRNA expression levels in normal and LUAD samples. m6A regulatory expression may be mainly interfered with CNV changes. LUAD tissues express m6A regulators with CNV amplification at a significantly higher level than normal lung tissues (e.g. YTHFF1 and VIRMA), while the opposite trend was observed for others (e.g. WTAP and ZC3H13) (Figure 1(D)).
Figure 1.
The genetic variation landscape of m6A regulators in LUAD. (A) Mutation frequencies of 21 m6A regulators in 561 LUAD patients from GSE68465 and TCGA-LUAD meta-cohorts. Each column represents an individual patient. The right-side numbers depict the mutation frequency for each regulator, while the proportion of each variant type is shown in the right barplot. Upper barplot indicates TMB, and the stacked barplot below displays the fraction of conversions for individual sample. (B) The CNV variation frequency of each m6A regulator (TCGA-LUAD cohort). Each column’s height corresponds to the alteration frequency, with green dots indicating deletion frequency and red dots signifying amplification frequency. (C) A circular diagram was employed to depict the location of CNV alterations of m6A regulators across the 23 chromosomes (TCGA-LUAD cohort). (D) The comparison of expression levels for the 21 m6A regulators (between tumor and normal tissues) was illustrated using red and blue boxes for tumor and normal samples, respectively. The boxes represented the interquartile range, with median values depicted as lines within the boxes, and outliers shown as black dots. The statistical P value was calculated (Kruskal–Wallis test), denoted by *P < 0.05, **P < 0.01, and ***P < 0.001.
Patterns of modification facilitated by the 21 m6A regulators
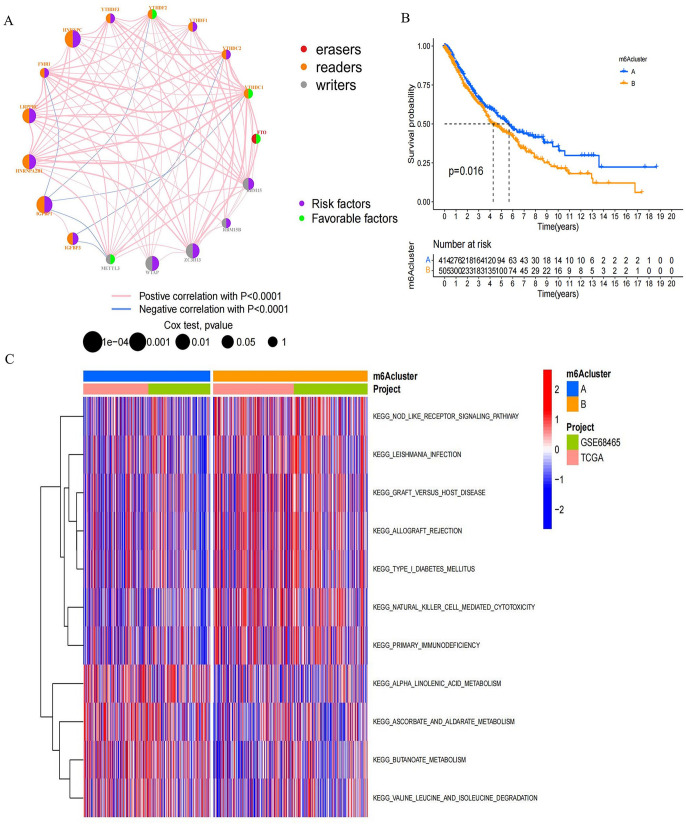
A GEO dataset with survival data and clinical characteristics (GSE68465) and the TCGA-LUAD dataset were included in a meta-cohort. The significance of the 21 m6A regulators in predicting the prognosis of LUAD patients was demonstrated through the univariate Cox regression model (Supplemental Figure S1(C) and Supplemental Table S1). The m6A regulatory networks in Figure 2(A) depict the global mode of m6A regulator connections, interactions, and prognostic value for patients with LUAD (Supplemental Table S2). Readers, writers, and erasers showed significant expression correlations not only among m6A regulators within a functional class but also among those in different functional classes. The simultaneous presence of high expression in writer genes and low expression in eraser genes in tumors was not universal, as it depended on specific genes (Supplemental Figure S2(A) to (I)). Tumors exhibiting elevated expression of RBM15B and ZC3H13 showed concurrent high expression of the alpha-ketoglutarate dependent dioxygenase (FTO), contrasting with tumors marked by high expression of the reader gene heterogeneous nuclear ribonucleoprotein C (HNRNPC). The reader genes (FMR1, IGFBP2, YTHDC1, YTHDC2, YTHDF2) and FTO exhibited high expression in the tumor (Supplemental Figure S2). Given the comparatively elevated mutation frequency of the ZC3H13 (a writer gene), we assessed the differences in FTO expression between the ZC3H13 mutant and the wild type. The results showed no significant difference. In this regard, we speculated that the crosstalk among 21 reader, writer, and eraser m6A regulators formed different m6A modification modes in LUAD. These patterns exhibited significant correlations with the TME’s cell infiltration characteristics. As a result of the expression of 21 molecules that regulate the m6A, patients with LUAD were stratified into distinct patterns of m6A modifications. Employing an unsupervised clustering approach, we identified two distinct modification patterns. (415 cases in Pattern A and 505 in B). Survival analysis of the two major m6A modification subtypes showed the significant survival advantage of m6A cluster A, also called m6A modification Pattern A (Figure 2(B)).
Figure 2.
Distinctive m6A modification patterns orchestrated by the 21 m6A regulators. (A) Interplay among m6A regulators in LUAD. The circle sizes denoted the impact of regulators on prognosis, evaluated using Cox-rank test values: P < 0.0001, P < 0.0001, P < 0.01, P < 0.05, and P < 1. Purple dots signified risk factors, while green dots indicated favorable prognosis factors. Interactions were visualized by linking lines, with their thickness reflecting correlation strength. Positive correlations were highlighted in red and negative correlations in blue. Writers, readers, and erasers were denoted by gray, orange, and red, respectively. (B) OS of all LUAD patients in distinct m6A clusters. Log-rank test showed an overall P = 0.016. All LUAD patients included 919 cases from one GEO cohort (GSE68465) and TCGA-LUAD cohort. (C) GSVA analysis reveals activation status of biological pathways in each m6A modification patterns. Heatmap representation of biological processes displays activated pathways in red and inhibited pathways in blue. Annotations from LUAD cohorts were applied for sample categorization.
Association of m6A modification patterns and TME
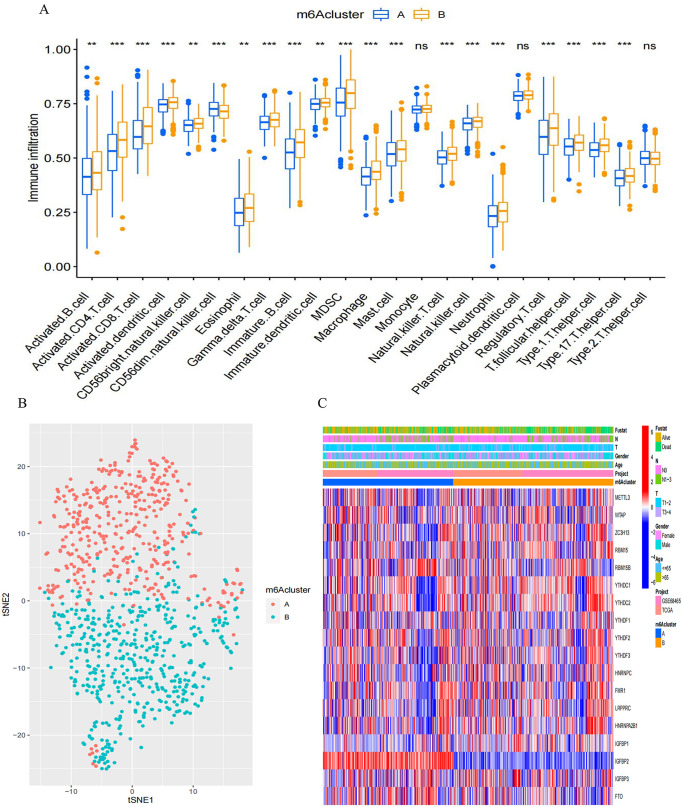
We employed GSVA analysis to explore the biological characteristics associated with distinct modification patterns. Illustrated in Figure 2(C) and Supplemental Table S3, cluster B displayed prominent enrichments in pathways, such as nod-like receptor signaling and primary immunodeficiency. Cluster A demonstrated enriched pathways linked to α-linolenic acid metabolism, ascorbic acid and uronic acid metabolism, butanoate metabolism, and isoleucine, valine, and leucine degradation. We then performed TME cell infiltration analysis and found the remarkably enriched infiltration of innate immune cells in m6A cluster B; these cells include plasmacytoid DCs, eosinophils, MDSCs, NKs, and macrophages (Figure 3(A)). However, patients in this modification pattern showed poor outcomes in survival analysis, without a corresponding survival advantage (Figure 2(B)). Research indicates the presence of numerous immune cells in the immune rejection phenotype. However, these cells remain within the stromal area surrounding the tumor cell nest and are impeded from infiltrating the parenchyma due to TME-induced stromal activation, leading to immunosuppression. We hypothesized that this phenomenon could contribute to the unfavorable prognosis observed in m6A cluster A. The two modification patterns had significantly different characteristics of TME cell infiltration. CIBERSORT was used to compare the immune cell composition of the two modification patterns. No significant difference in TME cell type composition was found between the two modification patterns, indicating that m6A methylation may not change the infiltrating cell types in TME (Supplemental Figure S2(J)). The t-distributed stochastic neighbor embedding (t-SNE) analysis indicated significant distinction in m6A transcription profiles between the two patterns (Figure 3(B)). Within m6A cluster A, FTO and IGFBP3 exhibited heightened expression, whereas the remaining m6A regulators displayed diminished variability. m6A cluster B highly expressed RBM15, YTHDC1, YTHDC2, YTHDF2, and FMR1 (Figure 3(C)). Therefore, tumors in m6A cluster B were associated with interstitial activation, elevated malignancy rates, and unfavorable prognosis.
Figure 3.
Distinct m6A modification patterns and their impact on TME infiltration and transcriptome traits. (A) Distribution of tumor-infiltrating cells in each m6A modification pattern. Interquartile ranges were depicted by the upper and lower box ends. Median values were indicated by lines within the boxes, while outliers were represented by black dots. Statistical significance was assessed (Kruskal–Wallis test). *P < 0.05; **P < 0.01; ***P < 0.001. (B) A significant contrast in transcriptome profiles between the two m6A modification patterns, visualized using t-SNE. (C) The unsupervised clustering of all LUAD patients based on the 21 m6A regulators. Patient annotations included survival status, N staging, T staging, gender, age, project, and m6A cluster. High expression of m6A regulators was denoted by red, while low expression was denoted by blue.
Discovery of m6A gene signatures
Next, we identified 2062 DEGs associated with m6A phenotypes, aiming to unveil the potential biological features of each modification pattern. These DEGs underwent gene ontology (GO) enrichment analysis through the “clusterProfiler” package (Supplemental Table S4 and Supplemental Figure S3(A)). As expected, the enrichment analysis of biological processes highlighted a substantial association between m6A and immunity, confirming the pivotal role of their regulation within the TME.
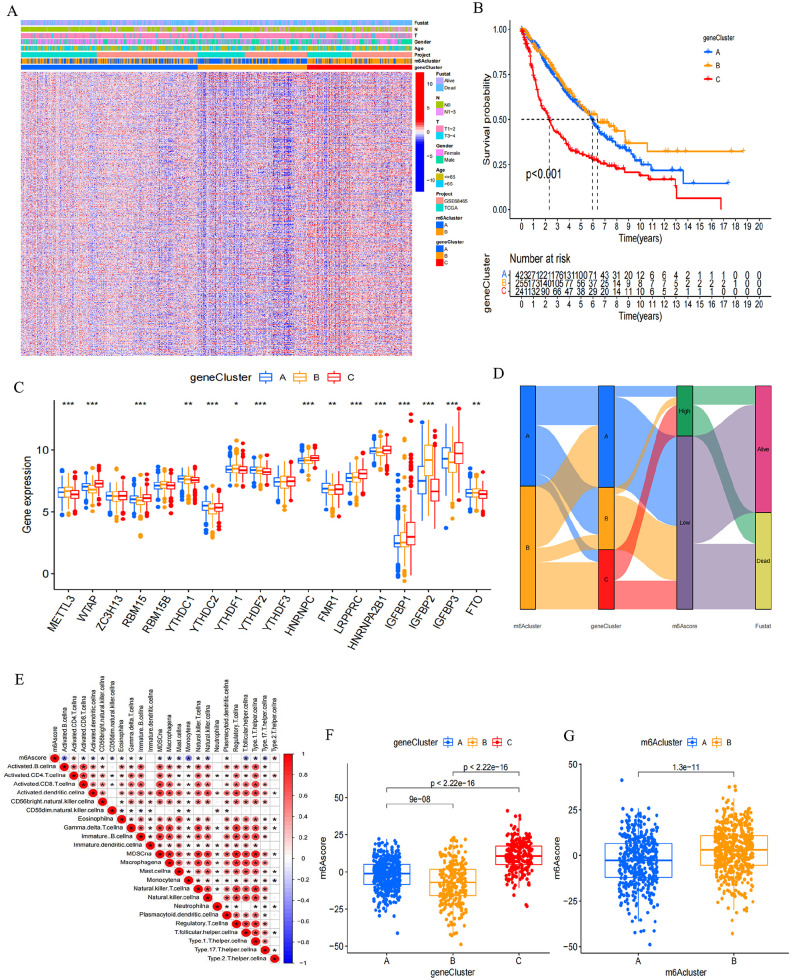
Subsequently, based on the 2062 m6A phenotype–associated genes, an unsupervised cluster analysis was executed to categorize patients into distinct subtypes. Using these two patterns, three distinct genomic phenotypes of m6A modification were identified, designated as m6A gene clusters A–C (Supplemental Figure S3(B) to (D) and Figure 4(A)). Among the 920 LUAD patients, 242 in m6A gene cluster C exhibited a poorer prognosis, while the most favorable outcomes were observed in the 256 patients belonging to gene cluster B, followed by those in gene cluster A (Figure 4(B)). Gene clusters displayed distinct characteristics (Figure 4(A)), and m6A regulator expression was significantly different among them (Figure 4(C)).
Figure 4.
Identification of m6A gene signatures. (A) Using unsupervised clustering, overlapping m6A phenotype-related genes were clustered in all LUAD cohorts, leading to the categorization of patients into three distinct genomic subtypes known as m6A gene clusters A–C. Survival status, N staging, T staging, gender, age, project, and m6A cluster for patient annotations. High expression of m6A regulators was visualized in red, while low expression was depicted in blue. (B) For all LUAD patients within different m6A gene clusters, Kaplan–Meier curves were generated to depict OS outcomes. The log-rank test indicated a significant overall P value of < 0.001. The collective cohort consisted of 919 cases from both the GEO cohort (GSE68465) and the TCGA-LUAD cohort. (C) The proportion of tumor-infiltrating cells was assessed across the three m6A gene clusters (Kruskal–Wallis test). In the box plots, the interquartile range was depicted by the upper and lower edges, the median value was represented by lines within the boxes, and outliers were indicated by black dots. The statistical significance was denoted as *P < 0.05, **P < 0.01, and ***P < 0.001. (D) An alluvial diagram illustrating the distribution of m6A clusters among various groups based on distinct m6A gene clusters, survival status, and m6A scores. (E) Analyzing the correlation between m6A score and each type of TME infiltration cell through Spearman analysis. Positive correlations were denoted by red, while negative correlations were indicated by blue (*P < 0.05; **P < 0.01). (F) Using the Kruskal–Wallis test, we examined the variations in m6A scores across the three gene clusters among all LUAD patients (P < 0.001). (G) Employing the Wilcoxon test, we assessed the disparities in m6A scores within the two m6A clusters across all LUAD patients, highlighting a substantial statistical distinction (P < 0.001).
Difference among m6A-related phenotypes
A chemokine and cytokine expression analysis within all gene clusters examined how m6A-related phenotypes influence TME immune regulation. Cytokines and chemokines were extracted from previous studies; of which PD-1, PD-L1/L2, CTLA-4, CD80/86, IDO1, TIGIT, LAG3, TNFRSF9, and HAVCR2 were confirmed to be correlated with immune checkpoints. CD8A, CXCL9/10, TNF, PRF1, TBX2, IFNG, GZMA, and GZMB are associated with immune activation. In addition to being involved in the epithelial–mesenchymal transition (EMT) and transforming growth factor-β pathways, ZEB1 also plays a role in proliferation. COL4A1, SMAD9, TGRB1, TWIST1, TGFBR2, CLDN3, ACTA2, VIM, and ZEB1 are related to the pathway of epithelial–mesenchymal transition (EMT) and TGF-β. 49 A significant increase in TGF-β/EMT pathway mRNAs was observed in gene cluster C, indicating its activation. Furthermore, gene cluster C displayed heightened mRNAs expression linked to immune activation. Thus, gene cluster C could be categorized as part of the immune-activated subgroup (Supplemental Figure S4(A)). However, individuals within gene cluster C exhibited the poorest prognosis, confirming that immune activation could not effectively predict the prognosis of patients; as such, more accurate indicators are needed.
The alluvial diagram portrayed the alterations in individual patient characteristics (Figure 4(D)). Furthermore, the m6A signature was employed to establish its correlation with immune cell type to further illustrate its utility (Figure 4(E)). The m6A score exhibited notable differences among m6A gene clusters as indicated by the Kruskal–Wallis test. Scores ranged from lowest in gene cluster B to highest in cluster C, with m6A cluster B having a higher score than m6A cluster A (Figure 4(F) and (G)). This suggests that higher m6A scores in tumors may indicate increased stromal activation, allowing for effective evaluation of both individual tumor m6A modification patterns and TME cell infiltration.
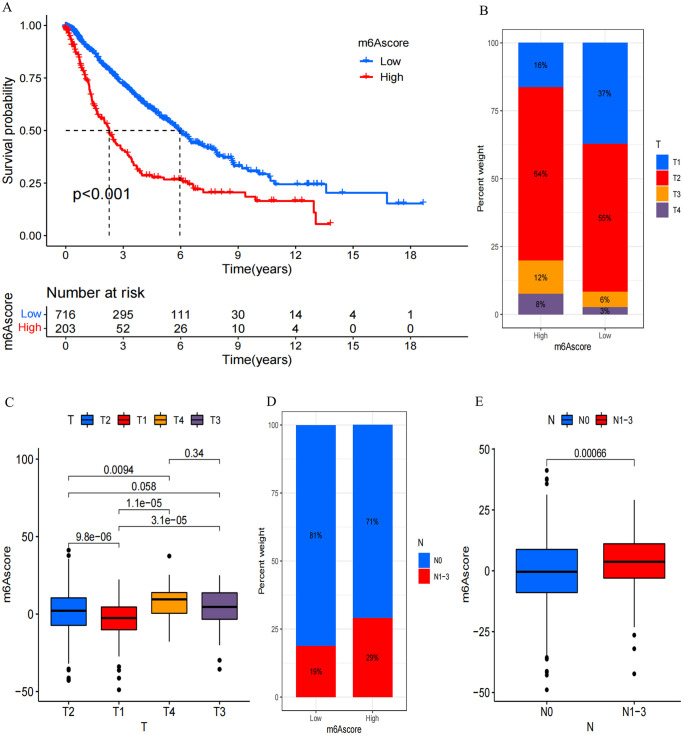
We then determined the clinical significance of m6A score in predicting patient outcomes. Using the Surv_cutpoint function, we determined the optimal cutoff value as 10.28667, leading to the division of patients into low- and high-score m6A groups. Subsequent prognostic analysis revealed that low m6A score correlated with extended survival, whereas high m6A score indicated poorer survival (P < 0.001, Figure 5(A)). Notably, patients exhibiting a low m6A score displayed a median survival time (MST) of 5.956, while for those with high m6A score, it was markedly reduced to 2.265. Univariate and multivariate Cox regression model analyses involving age, T staging, N staging, gender, and m6A score reaffirmed the m6A score’s potential as a robust and independent prognostic biomarker for evaluating patient outcomes (Supplemental Figure S4(B) and (C)). Furthermore, the m6A score effectively captured clinical attributes, such as T and N staging (Figure 5(B) to (E)), and predicted prognosis in various subgroups based on age (⩾ 65 years or < 65 years), N staging (N0 or N1–3), T staging (T1–2 or T3–4), and gender (male or female). It was observed that patients with elevated m6A scores consistently faced unfavorable overall prognoses (Supplemental Figure S5(A) to (G)).
Figure 5.
Distinctive traits of m6A modification. (A) Kaplan–Meier survival analysis for OS of all LUAD patients between patients with a high m6A score and those with a low m6A score. The log-rank test indicated a significant overall P value of < 0.001. All LUAD patients included 919 cases from one GEO cohort (GSE68465) and TCGA-LUAD cohort. (B) The proportion of T staging 1–4 in high and low m6A score group. T1, blue; T2, red; T3, yellow; T4, purple. (C) Variations in m6A scores among patients with different T staging. The Kruskal–Wallis test was employed to assess the statistical significance across four T-staging groups, yielding a significant P value of <0.0001. (D) The proportion of N staging 0 and N staging 1–3 in high and low m6A score group. N0, blue; N1–3, red. (E) Variations in m6A scores were investigated between patients categorized as N staging 0 and those with N staging 1–3, revealing a highly significant P value of < 0.0001.(Kruskal–Wallis test).
GSE50081, GSE37745, GSE31210, GSE30219, and GSE72094, which included 1294 lung cancer samples, were used as validation data sets to characterize the performance of the m6A score model. Individuals exhibiting low m6A scores experienced extended survival periods, in contrast to those with elevated m6A scores who encountered poorer survival rates (P < 0.001, Supplemental Figure S6(A)), aligning with findings from the training set (GSE68465). The high-score m6A group had more patients aged above 65 years, more deaths recorded, and more patients with T3–4 stage and lymph node metastasis (Supplemental Figure S6(B) to (E)), In addition, deceased patients exhibited markedly higher m6A scores compared to those who survived (P = 0.035, Supplemental Figure S6(F)). In addition, patients in T stage T3–4 displayed notably elevated scores in contrast to those in T stage T1–2 (P < 0.001, Supplemental Figure S6(G)). Furthermore, patients afflicted with lymph node metastases demonstrated notably elevated scores relative to those without (P < 0.001, Supplemental Figure S6(H)).
Somatic mutation and m6A modification in TCGA tumors
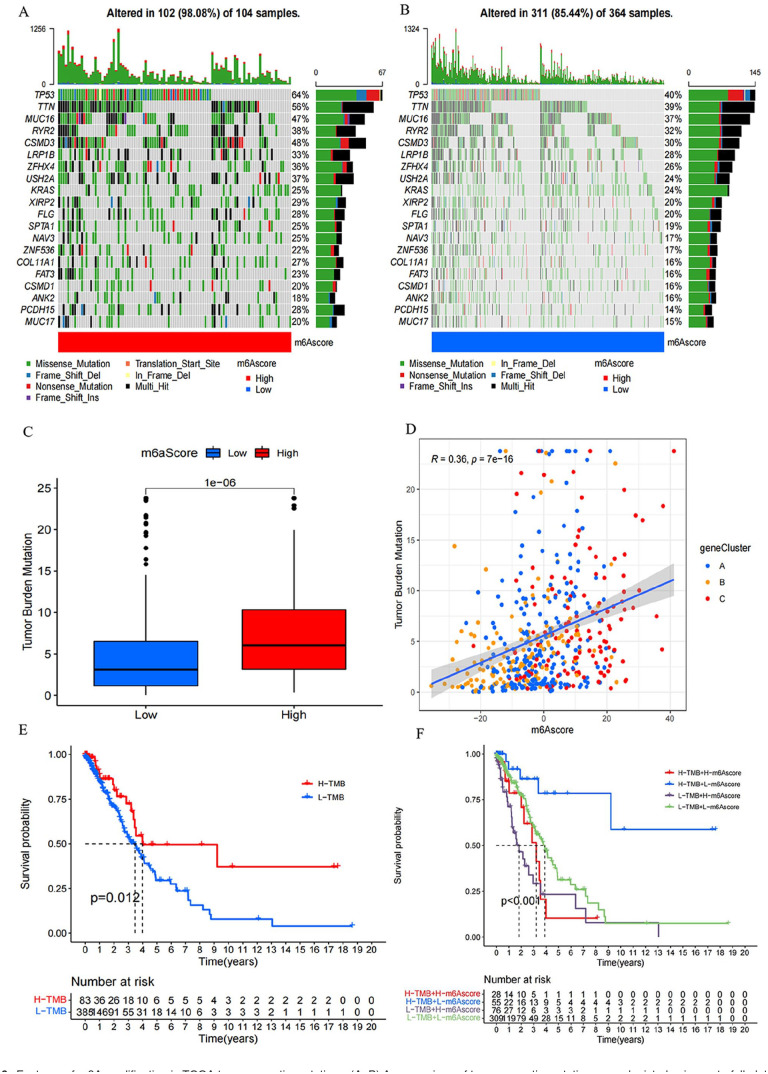
The “maftools” package was employed to assess variations in somatic mutation distribution between the two distinct m6A groups within the TCGA-LUAD cohort. The high-score m6A group exhibited a higher extent of TMB compared to the low-score m6A group. Notably, the highest expression gene displayed mutation rates of 64% and 40% in the high and low groups, and the fifth gene’s mutation rates were 48% and 30%, respectively (Figure 6(A) and (B)). Quantitative analyses confirmed a clear association between high m6A scores and elevated TMB, while the opposite was observed (Figure 6(C)). Furthermore, an affirmative correlation was identified between the m6A score and TMB (Figure 6(D)). Despite this, patients with high TMB experienced better prognoses (Figure 6(E)). Further exploration revealed the independence of m6A score as a distinct prognostic index from TMB (Figure 6(F)).
Figure 6.
Features of m6A modification in TCGA tumor somatic mutations. (A, B) A comparison of tumor somatic mutations was depicted using waterfall plots for individuals with (A) high m6A score and (B) low m6A score. Each column represented an individual patient. The upper barplot displayed the TMB, while the right barplot illustrated the proportion of each variant type. The mutation frequency for each gene was indicated by the number on the right. (C) Statistical analysis using the Wilcoxon test revealed a significant difference in TMB between the subgroups with high and low m6A scores (P < 0.0001). (D) In the TCGA-LUAD cohort, scatterplots illustrate a positive correlation between m6A scores and mutation load, with a Spearman correlation coefficient of 0.36 and a P value of < 0.0001. (E) OS of the TCGA-LUAD cohort stratified by high and low TMB. Log-rank test showed a P = 0.012. (F) Kaplan–Meier survival curves in the TCGA-LUAD cohort were plotted to stratify patients based on both TMB and m6A scores. The log-rank test revealed a significant P value of < 0.001 for OS in this stratification.
Response to immunotherapy is correlated with m6A score
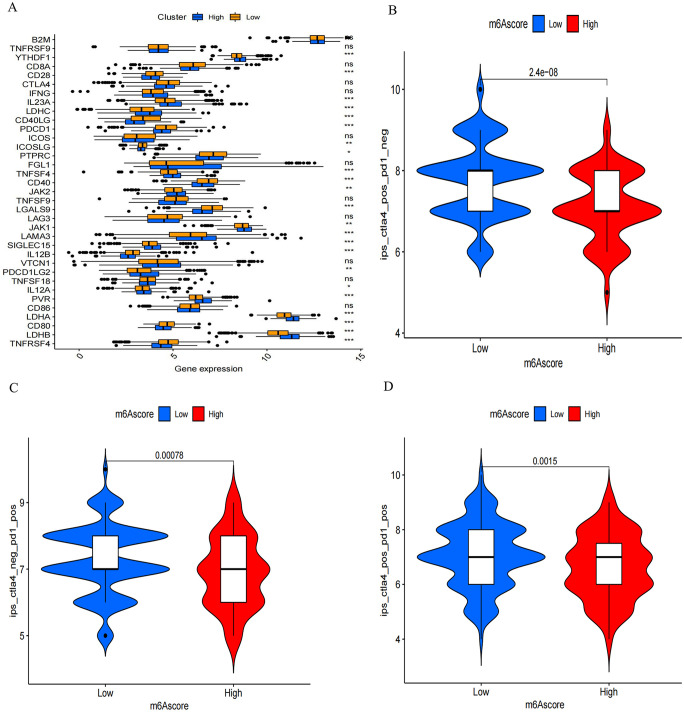
We conducted a comparison of checkpoint gene expression in patients with varying m6A scores, including high and low scores. Overall, 38 immune checkpoint genes were selected from previous reports. Distinct variations in the expression of the majority of checkpoint genes were evident between patients exhibiting high and low m6A scores (Figure 7(A)). To assess the connection between m6A modification patterns and immunotherapy response, we analyzed data obtained from TCIA. Based on the variations in immune scores between the two m6A subgroups, patients with lower m6A scores exhibited greater benefits from immunotherapy against PD-1/L1 or CTLA4 or a combination of both (Figure 7(B) to (D)).
Figure 7.
M6A score and the response to immunotherapy. (A) Comparison of checkpoint gene expression in high and low m6A score subgroups was conducted. The boxes’ left and right ends denoted the interquartile range, median values were indicated by lines within, and outliers were depicted as black dots. (*P < 0.05; **P < 0.01; ***P < 0.001; Kruskal–Wallis test). In the figure, the high m6A score subgroup is denoted by blue, while the low m6A score subgroup is represented by yellow. (B–D) Response disparities to different treatments were observed between high and low m6A score subgroups. Specifically, there were differences in response to (B) anti-CTLA4, (C) anti-CTLA4, and (D) the combination of anti-CTLA4 with anti-CTLA4, P < 0.0001, P = 0.00078, P = 0.0015, and P = 0.0001, respectively.(Kruskal–Wallis test).
Discussion
According to the most recent data from the Global Burden of Disease Research report, almost 2 million lung cancer deaths were reported in 2017 alone, with an estimated 2.3 million new cases. 50 Lung cancer’s emergence and progression stem from a myriad of factors and genes, following an intricate and multistage process. Research supports the connection between DNA-methylation patterns and the development, invasion, and metastasis of lung cancer. 51 Abnormal methylation patterns in the genome of lung cells can actively or passively accelerate the occurrence and development of LUAD.52–55 Further research found that a higher cell malignancy rate is related to a more significant degree of hypomethylation in the whole genome of the cell.56,57 However, no broad consensus has been established on the exact relationship between DNA aberrant methylation and lung cancer.
The importance of m6A modification in RNA epigenetics in vivo is being revealed in a growing number of studies.6,58 In this era of rapid development of bioinformatics technology, the biological significance and potential mechanism of m6A modification in tumors are gradually emerging. Studies have confirmed that RNA stability, RNA splicing, translation efficiency, and RNA–protein interaction are interfered by m6A modification. This disruption influences the expression of targeted genes, consequently altering the malignant behavior of tumor cells.9,11 m6A RNA methylation also holds a significant role in inflammation, immunity, tumor development, TME, and antitumor. 59 Hence, m6A modification has the potential to function as a predictive biomarker for treatment response and a viable therapeutic target. The clinical diagnosis of lung cancer is mainly performed using the combination of CT and serum tumor markers. However, the two methods, either alone or in combination, cannot achieve the purpose of early detection and accurate diagnosis. Shi et al. 60 collected patients’ bronchoalveolar lavage fluid and detected the methylation status of RASSF1 and ASHOX2; they found that this index had high sensitivity and specificity in diagnosing lung cancer. Another study used plasma and sputum DNA promoter hypermethylation to detect lung cancer early, and the method had a specificity of 98%. 61 In the experimental stage, an extremely sensitive non-invasive diagnostic assay was created to assess the methylation patterns of circulating tumor DNA. Its purpose was to distinguish between lung cancer and benign nodules, enabling early detection of cancer. 62 In addition, increasing lines of evidence indicate a dual role for m6A, that is, m6A regulators may promote and inhibit tumorigenesis in different cancers or different types or levels of m6A regulators may promote and inhibit tumorigenesis in cancers. Niu et al. 63 revealed that the γ-aminobutyric acid B2 receptor could be a novel epigenetic target for induction therapy of epidermal growth factor receptor 19-deficient LUAD. In vitro and in vivo investigations demonstrated that TMU-35435 and DNA demethylation reagent 5-aza-dC can achieve synergistic antitumor effects by reactivating tumor suppressor genes and genes inhibiting the Wnt pathway without causing obvious adverse reactions. 64 The m6A methylation of specific genes can also provide guidance for assessing lung cancer treatment drugs, treatment options, and patient prognosis. The methylation status of the T-BOX transcription factor 2 subfamily exhibits a dual function in predicting the responsiveness of non–small cell lung cancer to the antitumor drug 5-azacytidine. 65 LRP12 methylation status has predictive value for carboplatin resistance in lung cancer, 66 and the axon arrestin gene and miR-9 promoter methylation status are crucial in forecasting tumor response to radiation therapy. 67 Moreover, distinct gene methylation statuses reveal the prognosis of patients with the same histological type of lung cancer. Specifically, the methylation statuses of the RASSF1A promoter, TMEM196, the homeodomain protein homeobox gene, and the TGF-β-inducible gene promoter are associated with lung cancer and independent prognostic indicator in patients with adenocarcinoma.68–71 Therefore, the modification mode of m6A in different tumors should be further investigated. Yet, many prior investigations have primarily concentrated on individual infiltrating cell types within the TME and specific m6A regulators.72–74 The complete regulatory interplay between the immune cell infiltration landscape in LUAD and m6A regulators remains incompletely understood. Investigating distinct m6A modification patterns and m6A-related genes within the TME can enhance early tumor diagnosis, deepen comprehension of antitumor immune responses, unveil new therapeutic targets, inform treatment approaches, and forecast treatment outcomes for patients.
This study delineated two distinct m6A modification patterns in LUAD using 21 m6A regulators found in previous studies. Differences in TME cell infiltration were pronounced between the distinct patterns. m6A cluster A exhibited adaptive immune activation, aligning with the immune inflammatory phenotype, while m6A cluster B displayed innate immunity and interstitial activation, indicative of the immune rejection phenotype. Patients in m6A cluster A experienced improved survival outcomes. Although the immunorejection phenotype also showed massive immune cell infiltrations, even more than that in the TME of the immunoinflammatory phenotype, the immune cells remained in the stroma and cannot penetrate the tumor parenchyma due to excessive stromal activation. Stroma does not exist only around the tumor, and it may even infiltrate the core of the tumor, leading to a false understanding that immune cells are inside the tumor. 75 This phenomenon might clarify the association between activated innate immunity and poor prognosis among patients in m6A cluster B; moreover, the protumor or antitumor effects of the TME cannot be explained by a single cell. In addition, the two different patterns had significantly different immune-related biological pathways, thereby confirming the rationality and reliability of immunophenotypic classification of different m6A methylation modification patterns.
We further identified 2062 DEGs, which were called m6A-related genes, in patients with two m6A methylation modification patterns. Using these DEGs, we divided patients with LUAD into three different m6A gene clusters (A–C). Significant distinctions in immune and matrix activation were identified within the three gene clusters, highlighting the association between m6A methylation patterns and the infiltration of TME cells. Survival analysis affirmed the highest survival rate in m6A gene cluster B, then A, with C indicating the least favorable prognosis. Considering that such TME landscapes are population-based and the heterogeneity of individual tumors, we constructed the m6A score model to quantify TME cell infiltration patterns in each patient and confirmed the reliability of the model in the validation set. The m6A score exhibits a negative correlation with immune activation, aligning with the immune rejection phenotype m6A modification pattern and correlating with poorer survival prognosis. A lower m6A score is consistent with the immunoinflammatory phenotype and indicates better survival prognosis. We also established that the m6A score stood independently of TMB in forecasting prognosis. Favorable outcomes were observed in patients with high TMB and low m6A score, whereas the poorest outcomes were linked to low TMB and high m6A score. Moreover, individuals with low m6A score derived greater benefit from anti-PD-1/L1 treatment, anti-CTLA4 therapy, or a combination thereof.
Through a series of analyses, we found that m6A modification pattern be bound up with the formation of different TME landscapes. The established m6A score model holds significance in predicting cancer stage, prognosis, TMB, and efficacy of anti-immunotherapy. According to accumulated research, regulation of m6A modulators (activators/ inhibitors) can reverse tumor immune responses, which may enhance the efficacy of traditional anticancer drugs and provide new therapeutic prospects.76–78 The abnormal methylation of some specific genes serve as biomarkers for the early detection of tumors. Further experimental research should be conducted to develop a mature non-invasive early diagnosis technology of lung cancer for early detection and treatment. In addition, the methylation of specific genes is expected to become an ideal biological target for lung cancer treatment because it can evaluate treatment drugs and plans for lung cancer and improve the prognosis of patients; it holds the potential to serve as a dependable prognostic marker for forecasting the likelihood of lung cancer recurrence and metastasis. It has important clinical value to guide the clinical treatment of patients, thereby prolonging the relapse-free survival (RFS) and OS of patients with lung cancer.
Although drugs targeting histone modifications and DNA have entered the clinical application stage, therapeutic strategies targeting RNA epigenetics are just beginning to be studied. Before applying models to clinical practice, they should have undergone thorough validation and testing using an independent dataset to assess their accuracy and performance. Cross-validation should also be conducted to verify the robustness of the models. Integrating the models into clinical decision support systems provides physicians with easy access and utilization of the model’s outcomes. The process can also assist doctors in swiftly obtaining predictive information during the clinical decision-making process, leading to better personalized treatment strategies. Moreover, we should ensure that clinical staff are well informed on the proper utilization and interpretation of the model’s results. During practical implementation, the performance and application outcomes of the model should be continuously monitored. Collection of feedback information and iterative improvement and optimization of the model should be conducted based on feedback to ensure its sustained effectiveness in clinical practice. Further studies should be performed on the mechanism of m6A regulators and m6A-related genes in LUAD. To establish a solid theoretical foundation for drug discovery, it is essential to gain a deeper and more comprehensive insight into the underlying mechanisms of m6A’s mode of action. Overall, m6A regulators and m6A-related genes should be used for early diagnosis and treatment of patients with LUAD.
Supplemental Material
Supplemental material, sj-pdf-1-ebm-10.1177_15353702231214266 for Link between m6A modification and infiltration characterization of tumor microenvironment in lung adenocarcinoma by Sha Yang, Ke Li, Jiqin Zhang, Jian Liu, Lin Liu, Ying Tan and Chuan Xu in Experimental Biology and Medicine
Footnotes
Authors’ contributions: SY participated in methodology, validation, software, conceptualization, visualization, and writing – original draft and review. KL, JL, and YT participated in methodology, visualization, validation, software, and writing – review and editing. JQZ, LL, CX, and YT participated in conceptualization, funding acquisition, and supervision.
The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding: The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: Funding for this study was provided by several sources, including the National Natural Science Foundation of China (Grant Nos. 81960454 to YT, 81960344 to JQZ, and 82260533 to JL), the Guizhou Provincial People’s Hospital National Science Foundation (Grants GPPH-NSFC-2019-18 and GPPH-NSFC-D-2019-17 to YT and GPPH-NSFC-2019-09 to JQZ), the Guizhou Provincial People’s Hospital Doctor Foundation (Grant Nos. (2018)06 to YT and (2018)03 to JQZ), and the Guizhou Provincial Science and Technology Projects (Grant No. (2020)1Z066 to YT).
Data availability statement: The data sets supporting the findings of this study are accessible from the TCGA-GDC (https://portal.gdc.cancer.gov/) repository and the gene expression omnibus database (GEO: https://www.ncbi.nlm.nih.gov/geo/), which were used as the training cohort.
ORCID iD: Ying Tan  https://orcid.org/0000-0001-8792-5396
https://orcid.org/0000-0001-8792-5396
Supplemental material: Supplemental material for this article is available online.
References
- 1. Lavi U, Fernandez-Muñoz R, Darnell JE., Jr. Content of N-6 methyl adenylic acid in heterogeneous nuclear and messenger RNA of HeLa cells. Nucleic Acids Res 1977;4:63–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Sun T, Wu R, Ming L. The role of m6A RNA methylation in cancer. Biomed Pharmacother 2019;112:108613. [DOI] [PubMed] [Google Scholar]
- 3. Zhuang Z, Chen L, Mao Y, Zheng Q, Li H, Huang Y, Hu Z, Jin Y. Diagnostic, progressive and prognostic performance of m6A methylation RNA regulators in lung adenocarcinoma. Int J Biol Sci 2020;16:1785–97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Ma Y, Liu X, Bi Y, Wang T, Chen C, Wang Y, Han D, Cao F. Alteration of N6-methyladenosine mRNA methylation in a human stem cell-derived cardiomyocyte model of tyrosine kinase inhibitor-induced cardiotoxicity. Front Cardiovasc Med 2022;9:849175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Ma S, Chen C, Ji X, Liu J, Zhou Q, Wang G, Yuan W, Kan Q, Sun Z. The interplay between m6A RNA methylation and noncoding RNA in cancer. J Hematol Oncol 2019;12:121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Huang H, Weng H, Chen J. m6A modification in coding and non-coding RNAs: roles and therapeutic implications in cancer. Cancer Cell 2020;37:270–88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Coker H, Wei G, Brockdorff N. m6A modification of non-coding RNA and the control of mammalian gene expression. Biochim Biophys Acta Gene Regul Mech 2019;1862:310–8 [DOI] [PubMed] [Google Scholar]
- 8. Lv W, Wang Y, Zhao C, Tan Y, Xiong M, Yi Y, He X, Ren Y, Wu Y, Zhang Q. Identification and validation of m6A-related lncRNA signature as potential predictive biomarkers in breast cancer. Front Oncol 2021;11:745719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Ni XF, Xie QQ, Zhao JM, Xu YJ, Ji M, Hu WW, Wu J, Wu CP. The hepatic microenvironment promotes lung adenocarcinoma cell proliferation, metastasis, and epithelial-mesenchymal transition via METTL3-mediated N6-methyladenosine modification of YAP1. Aging 2021;13:4357–69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Chen X, Xu M, Xu X, Zeng K, Liu X, Pan B, Li C, Sun L, Qin J, Xu T, He B, Pan Y, Sun H, Wang S. METTL14-mediated N6-methyladenosine modification of SOX4 mRNA inhibits tumor metastasis in colorectal cancer. Mol Cancer 2020;19:106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Chen Y, Pan C, Wang X, Xu D, Ma Y, Hu J, Chen P, Xiang Z, Rao Q, Han X. Silencing of METTL3 effectively hinders invasion and metastasis of prostate cancer cells. Theranostics 2021;11:7640–57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Tang R, Zhang Y, Liang C, Xu J, Meng Q, Hua J, Liu J, Zhang B, Yu X, Shi S. The role of m6A-related genes in the prognosis and immune microenvironment of pancreatic adenocarcinoma. PeerJ 2020;8:e9602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Xu S, Tang L, Dai G, Luo C, Liu Z. Expression of m6A regulators correlated with immune microenvironment predicts therapeutic efficacy and prognosis in gliomas. Front Cell Dev Biol 2020;8:594112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Zhang Y, Liu X, Liu L, Li J, Hu Q, Sun R. Expression and prognostic significance of m6A-related genes in lung adenocarcinoma. Med Sci Monit 2020;26:e919644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Li Y, Gu J, Xu F, Zhu Q, Chen Y, Ge D, Lu C. Molecular characterization, biological function, tumor microenvironment association and clinical significance of m6A regulators in lung adenocarcinoma. Brief Bioinform 2021;22:bbaa225 [DOI] [PubMed] [Google Scholar]
- 16. Li N, Zhan X. Identification of pathology-specific regulators of m6A RNA modification to optimize lung cancer management in the context of predictive, preventive, and personalized medicine. EPMA J 2020;11:485–504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. National Lung Screening Trial Research Team. Lung cancer incidence and mortality with extended follow-up in the National Lung Screening Trial. J Thorac Oncol 2019;14:1732–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Blake-Cerda M, Lozano-Ruíz F, Maldonado-Magos F, De la Mata-Moya D, Díaz-García D, Lara-Mejía L, Zatarain-Barrón ZL, Cuevas-Góngora MF, Barron-Barron F, Corona-Cruz JF, Cabrera-Miranda L, Arroyo-Hernández M, Gerson R, Arrieta O. Consolidative stereotactic ablative radiotherapy (SABR) to intrapulmonary lesions is associated with prolonged progression-free survival and overall survival in oligometastatic NSCLC patients: a prospective phase 2 study. Lung Cancer 2021;152:119–26 [DOI] [PubMed] [Google Scholar]
- 19. Ruiz-Cordero R, Devine WP. Targeted therapy and checkpoint immunotherapy in lung cancer. Surg Pathol Clin 2020;13:17–33 [DOI] [PubMed] [Google Scholar]
- 20. Zheng J, Zhao Z, Wan J, Guo M, Wang Y, Yang Z, Li Z, Ming L, Qin Z. N-6 methylation-related lncRNA is potential signature in lung adenocarcinoma and influences tumor microenvironment. J Clin Lab Anal 2021;35:e23951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Sun L, Liu WK, Du XW, Liu XL, Li G, Yao Y, Han T, Li WY, Gu J. Large-scale transcriptome analysis identified RNA methylation regulators as novel prognostic signatures for lung adenocarcinoma. Ann Transl Med 2020;8:751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Lin S, Xu H, Zhang A, Ni Y, Xu Y, Meng T, Wang M, Lou M. Prognosis analysis and validation of m6A signature and tumor immune microenvironment in glioma. Front Oncol 2020;10:541401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Arora L, Pal D. Remodeling of stromal cells and immune landscape in microenvironment during tumor progression. Front Oncol 2021;11:596798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Pan X, Zheng L. Epigenetics in modulating immune functions of stromal and immune cells in the tumor microenvironment. Cell Mol Immunol 2020;17:940–53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Berraondo P, Sanmamed MF, Ochoa MC, Etxeberria I, Aznar MA, Pérez-Gracia JL, Rodríguez-Ruiz ME, Ponz-Sarvise M, Castañón E, Melero I. Cytokines in clinical cancer immunotherapy. Br J Cancer 2019;120:6–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Gao R, Ye M, Liu B, Wei M, Ma D, Dong K. m6A modification: a double-edged sword in tumor development. Front Oncol 2021;11:679367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Song H, Song J, Cheng M, Zheng M, Wang T, Tian S, Flavell RA, Zhu S, Li HB, Ding C, Wei H, Sun R, Peng H, Tian Z. METTL3-mediated m6A RNA methylation promotes the anti-tumour immunity of natural killer cells. Nat Commun 2021;12:5522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Miao TW, Yang DQ, Chen FY, Zhu Q, Chen X. A ferroptosis-related gene signature for overall survival prediction and immune infiltration in lung squamous cell carcinoma. Biosci Rep 2022;42:BSR20212835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Li Z, Wei J, Zheng H, Gan X, Song M, Zhang Y, Kong L, Zhang C, Yang J, Jin Y. m6A regulator-mediated methylation modification patterns and tumor immune microenvironment in sarcoma. Aging 2022;14:330–53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Cao R, Ma B, Wang G, Xiong Y, Tian Y, Yuan L. Characterization of hypoxia response patterns identified prognosis and immunotherapy response in bladder cancer. Mol Ther Oncolytics 2021;22:277–93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Leek JT, Johnson WE, Parker HS, Jaffe AE, Storey JD. The sva package for removing batch effects and other unwanted variation in high-throughput experiments. Bioinformatics 2012;28:882–3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Lu X, Ye K, Zou K, Chen J. Identification of copy number variation-driven genes for liver cancer via bioinformatics analysis. Oncol Rep 2014;32:1845–52 [DOI] [PubMed] [Google Scholar]
- 33. Zhang H, Meltzer P, Davis S. RCircos: an R package for Circos 2D track plots. BMC Bioinformatics 2013;14:244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Luo Y, Sun X, Xiong J. Characterization of m6A regulator-mediated methylation modification patterns and tumor microenvironment infiltration in ovarian cancer. Front Cell Dev Biol 2022;9:794801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Wang L, Cao H, Zhong Y, Ji P, Chen F. The role of m6A regulator-mediated methylation modification and tumor microenvironment infiltration in glioblastoma multiforme. Front Cell Dev Biol 2022;10:842835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Wilkerson MD, Hayes DN. ConsensusClusterPlus: a class discovery tool with confidence assessments and item tracking. Bioinformatics 2010;26:1572–3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Hänzelmann S, Castelo R, Guinney J. GSVA: gene set variation analysis for microarray and RNA-seq data. BMC Bioinformatics 2013;14:7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Luo H, Ye M, Hu Y, Wu M, Cheng M, Zhu X, Huang K. DNA methylation regulator-mediated modification patterns and tumor microenvironment characterization in glioma. Aging 2022;14:7824–50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Yu G, Wang LG, Han Y, He QY. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS 2012;16:284–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Ma C, Kang W, Yu L, Yang Z, Ding T. AUNIP expression is correlated with immune infiltration and is a candidate diagnostic and prognostic biomarker for hepatocellular carcinoma and lung adenocarcinoma. Front Oncol 2020;10:590006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Charoentong P, Finotello F, Angelova M, Mayer C, Efremova M, Rieder D, Hackl H, Trajanoski Z. Pan-cancer immunogenomic analyses reveal genotype-immunophenotype relationships and predictors of response to checkpoint blockade. Cell Rep 2017;18:248–62 [DOI] [PubMed] [Google Scholar]
- 42. Cao R, Yuan L, Ma B, Wang G, Tian Y. Immune-related long non-coding RNA signature identified prognosis and immunotherapeutic efficiency in bladder cancer (BLCA). Cancer Cell Int 2020;20:276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, Smyth GK. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res 2015;43:e47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Zhang X, Shi M, Chen T, Zhang B. Characterization of the immune cell infiltration landscape in head and neck squamous cell carcinoma to aid immunotherapy. Mol Ther Nucleic Acids 2020;22:298–309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Mayakonda A, Lin DC, Assenov Y, Plass C, Koeffler HP. Maftools: efficient and comprehensive analysis of somatic variants in cancer. Genome Res 2018;28:1747–56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Givechian KB, Wnuk K, Garner C, Benz S, Garban H, Rabizadeh S, Niazi K, Soon-Shiong P. Identification of an immune gene expression signature associated with favorable clinical features in Treg-enriched patient tumor samples. NPJ Genom Med 2018;3:14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Yang Q, Xu F, Jian A, Yu H, Ye T, Hu W. m6A regulator-mediated methylation modification patterns and tumor microenvironment cell-infiltration characterization in head and neck cancer. Front Cell Dev Biol 2022;9:803141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Qiu Y, Liu S, Chen HT, Yu CH, Teng XD, Yao HT, Xu GQ. Upregulation of caveolin-1 and SR-B1 in mice with non-alcoholic fatty liver disease. Hepatobiliary Pancreat Dis Int 2013;12:630–6 [DOI] [PubMed] [Google Scholar]
- 49. Zhang Y, Liang X, Zhang L, Wang D. Metabolic characterization and metabolism-score of tumor to predict the prognosis in prostate cancer. Sci Rep 2021;11:22486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Global Burden of Disease 2019 Cancer Collaboration. Cancer incidence, mortality, years of life lost, years lived with disability, and disability-adjusted life years for 29 cancer groups from 2010 to 2019: a systematic analysis for the global burden of disease study 2019. JAMA Oncol 2022;8:420–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Chao YL, Pecot CV. Targeting epigenetics in lung cancer. Cold Spring Harb Perspect Med 2021;11:a038000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Zhang L, Lu Q, Chang C. Epigenetics in health and disease. Adv Exp Med Biol 2020;1253:3–55 [DOI] [PubMed] [Google Scholar]
- 53. Asano T. Drug resistance in cancer therapy and the role of epigenetics. J Nippon Med Sch 2020;87:244–51 [DOI] [PubMed] [Google Scholar]
- 54. Villanueva L, Álvarez-Errico D, Esteller M. The contribution of epigenetics to cancer immunotherapy. Trends Immunol 2020;41:676–91 [DOI] [PubMed] [Google Scholar]
- 55. Ilango S, Paital B, Jayachandran P, Padma PR, Nirmaladevi R. Epigenetic alterations in cancer. Front Biosci 2020;25:1058–109 [DOI] [PubMed] [Google Scholar]
- 56. Sheaffer KL, Elliott EN, Kaestner KH. DNA hypomethylation contributes to genomic instability and intestinal cancer initiation. Cancer Prev Res 2016;9:534–46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Mari-Alexandre J, Diaz-Lagares A, Villalba M, Juan O, Crujeiras AB, Calvo A, Sandoval J. Translating cancer epigenomics into the clinic: focus on lung cancer. Transl Res 2017;189:76–92 [DOI] [PubMed] [Google Scholar]
- 58. Cao G, Li HB, Yin Z, Flavell RA. Recent advances in dynamic m6A RNA modification. Open Biol 2016;6:160003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Xu Z, Peng B, Cai Y, Wu G, Huang J, Gao M, Guo G, Zeng S, Gong Z, Yan Y. N6-methyladenosine RNA modification in cancer therapeutic resistance: current status and perspectives. Biochem Pharmacol 2020;182:114258. [DOI] [PubMed] [Google Scholar]
- 60. Shi J, Chen X, Zhang L, Fang X, Liu Y, Zhu X, Zhang H, Fan L, Gu J, Zhang S, She B, Han H, Yi X. Performance evaluation of SHOX2 and RASSF1A methylation for the aid in diagnosis of lung cancer based on the analysis of FFPE specimen. Front Oncol 2020;10:565780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Hulbert A, Jusue-Torres I, Stark A, Chen C, Rodgers K, Lee B, Griffin C, Yang A, Huang P, Wrangle J, Belinsky SA, Wang TH, Yang SC, Baylin SB, Brock MV, Herman JG. Early detection of lung cancer using DNA promoter hypermethylation in plasma and sputum. Clin Cancer Res 2017;23:1998–2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Liang W, Zhao Y, Huang W, Gao Y, Xu W, Tao J, Yang M, Li L, Ping W, Shen H, Fu X, Chen Z, Laird PW, Cai X, Fan JB, He J. Non-invasive diagnosis of early-stage lung cancer using high-throughput targeted DNA methylation sequencing of circulating tumor DNA (ctDNA). Theranostics 2019;9:2056–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Niu X, Liu F, Zhou Y, Zhou Z, Zhou D, Wang T, Li Z, Ye X, Yu Y, Weng X, Zhang H, Ye J, Liao M, Liu Y, Chen Z, Lu S. Genome-wide DNA methylation analysis reveals GABBR2 as a novel epigenetic target for EGFR 19 deletion lung adenocarcinoma with induction erlotinib treatment. Clin Cancer Res 2017;23:5003–14 [DOI] [PubMed] [Google Scholar]
- 64. Shieh JM, Tang YA, Hu FH, Huang WJ, Wang YJ, Jen J, Liao SY, Lu YH, Yeh YL, Wang TW, Lin P, Wang YC. A histone deacetylase inhibitor enhances expression of genes inhibiting Wnt pathway and augments activity of DNA demethylation reagent against nonsmall-cell lung cancer. Int J Cancer 2017;140:2375–86 [DOI] [PubMed] [Google Scholar]
- 65. Nehme E, Rahal Z, Sinjab A, Khalil A, Chami H, Nemer G, Kadara H. Epigenetic suppression of the T-box subfamily 2 (TBX2) in human non-small cell lung cancer. Int J Mol Sci 2019;20:1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Grasse S, Lienhard M, Frese S, Kerick M, Steinbach A, Grimm C, Hussong M, Rolff J, Becker M, Dreher F, Schirmer U, Boerno S, Ramisch A, Leschber G, Timmermann B, Grohé C, Lüders H, Vingron M, Fichtner I, Klein S, Odenthal M, Büttner R, Lehrach H, Sültmann H, Herwig R, Schweiger MR. Epigenomic profiling of non-small cell lung cancer xenografts uncover LRP12 DNA methylation as predictive biomarker for carboplatin resistance. Genome Med 2018;10:55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Yang LH, Han Y, Li G, Xu HT, Jiang GY, Miao Y, Zhang XP, Zhao HY, Xu ZF, Stoecker M, Wang E, Xu K, Wang EH. Axin gene methylation status correlates with radiosensitivity of lung cancer cells. BMC Cancer 2013;13:368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Pankova D, Jiang Y, Chatzifrangkeskou M, Vendrell I, Buzzelli J, Ryan A, Brown C, O’Neill E. RASSF1A controls tissue stiffness and cancer stem-like cells in lung adenocarcinoma. EMBO J 2019;38:e100532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Seok Y, Lee WK, Park JY, Kim DS. TGFBI promoter methylation is associated with poor prognosis in lung adenocarcinoma patients. Mol Cells 2019;42:161–5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Chen Y, Yang L, Cui T, Pacyna-Gengelbach M, Petersen I. HOPX is methylated and exerts tumour-suppressive function through Ras-induced senescence in human lung cancer. J Pathol 2015;235:397–407 [DOI] [PubMed] [Google Scholar]
- 71. Liu WB, Han F, Huang YS, Chen HQ, Chen JP, Wang DD, Jiang X, Yin L, Cao J, Liu JY. TMEM196 hypermethylation as a novel diagnostic and prognostic biomarker for lung cancer. Mol Carcinog 2019;58:474–87 [DOI] [PubMed] [Google Scholar]
- 72. Zhang C, Huang S, Zhuang H, Ruan S, Zhou Z, Huang K, Ji F, Ma Z, Hou B, He X. YTHDF2 promotes the liver cancer stem cell phenotype and cancer metastasis by regulating OCT4 expression via m6A RNA methylation. Oncogene 2020;39:4507–18 [DOI] [PubMed] [Google Scholar]
- 73. Song P, Feng L, Li J, Dai D, Zhu L, Wang C, Li J, Li L, Zhou Q, Shi R, Wang X, Jin H. β-catenin represses miR455-3p to stimulate m6A modification of HSF1 mRNA and promote its translation in colorectal cancer. Mol Cancer 2020;19:129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Du C, Lv C, Feng Y, Yu S. Activation of the KDM5A/miRNA-495/YTHDF2/m6A-MOB3B axis facilitates prostate cancer progression. J Exp Clin Cancer Res 2020;39:223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Bremnes RM, Dønnem T, Al-Saad S, Al-Shibli K, Andersen S, Sirera R, Camps C, Marinez I, Busund LT. The role of tumor stroma in cancer progression and prognosis: emphasis on carcinoma-associated fibroblasts and non-small cell lung cancer. J Thorac Oncol 2011;6:209–17 [DOI] [PubMed] [Google Scholar]
- 76. Zeng C, Huang W, Li Y, Weng H. Roles of METTL3 in cancer: mechanisms and therapeutic targeting. J Hematol Oncol 2020;13:117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Han D, Liu J, Chen C, Dong L, Liu Y, Chang R, Huang X, Liu Y, Wang J, Dougherty U, Bissonnette MB, Shen B, Weichselbaum RR, Xu MM, He C. Anti-tumour immunity controlled through mRNA m6A methylation and YTHDF1 in dendritic cells. Nature 2019;566:270–4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Ma Z, Ji J. N6-methyladenosine (m6A) RNA modification in cancer stem cells. Stem Cells. Epub ahead of print 27 September 2020. DOI: 10.1002/stem.3279. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental material, sj-pdf-1-ebm-10.1177_15353702231214266 for Link between m6A modification and infiltration characterization of tumor microenvironment in lung adenocarcinoma by Sha Yang, Ke Li, Jiqin Zhang, Jian Liu, Lin Liu, Ying Tan and Chuan Xu in Experimental Biology and Medicine