Abstract
Amyotrophic lateral sclerosis (ALS) is a fatal neurodegenerative disorder and the most common form of motor neurone disease (MND) which is characterized by the damage and death of motor neurons in the brain and spinal cord of affected individuals. Due to the heterogeneity of the disease, a better understanding of the interaction between genetics and biochemistry with the identification of biomarkers is crucial for therapy development. In this study, we used cerebrospinal fluid (CSF) RNA-sequencing data from the New York Genome Center (NYGC) ALS Consortium and analyzed differential gene expression between 47 MND individuals and 29 healthy controls. Pathway analysis showed that the affected genes are enriched in many pathways associated with ALS, including nucleocytoplasmic transport, autophagy, and apoptosis. Moreover, we assessed differential expression on both gene- and transcript-based levels and demonstrate that the expression of previously identified potential biomarkers, including CAPG, CCL3, and MAP2, was significantly higher in MND individuals. Ultimately, this study highlights the transcriptomic composition of CSF which enables insights into changes in the brain in ALS and therefore increases the confidence in the use of CSF for biomarker development.
Keywords: Cerebrospinal fluid, amyotrophic lateral sclerosis, motor neurone disease, transcriptome, RNA-seq, biomarker
Impact Statement
In this study, we describe significant expression profile changes and enrichment of key ALS pathways in MND individuals compared to healthy controls using RNA-sequencing data. We demonstrate that protein biomarkers in CSF are reflected by their corresponding increased expression in CSF RNA-sequencing profiles highlighting CSF RNA data as a valuable resource for biomarker development. We also suggest another potential biomarker in form of the COPA gene which plays important roles in protein transport between endoplasmic reticulum and Golgi apparatus, a key pathway in ALS pathology. Overall, this study highlights the importance of studying RNA-sequencing data for both biomarker development and complementation of CSF proteomic analysis.
Introduction
Motor neurone disease (MND), the most common form being amyotrophic lateral sclerosis (ALS), is a rare progressive neurodegenerative disease which results in the damage and death of motor neurons in the brain and spinal cord of affected individuals. This degeneration of neurons leads to the weakening and stiffening of muscles, which eventually results in individuals losing the ability to walk and breathe with sufferers dying between three and five years following symptom onset.1,2 Starting with the discovery of pathogenic variants in SOD1, FUS, TARDBP, and C9ORF72, there are now over 100 genes associated with the disease which reflects the heterogeneity of ALS.3–6 Several pathological mechanisms, such as oxidative stress, inflammation, protein aggregation, impairment of autophagy, or RNA processing and aberrant retrotransposon function, have been hypothesized to be involved in disease pathogenesis.7–9 Therefore, a better understanding of the interaction between genetics and biochemistry including the identification of biomarkers is crucial to progress therapy development and effective clinical trial design by allowing stratification of cohorts into more homogeneous groups. The focus on biomarker development has shifted to the use of cerebrospinal fluid (CSF) due to its contact with the borders/extracellular space of the brain and neuroglia cell enriched composition which facilitates diagnosis and monitoring of diseases, as previously shown by the increased application in diagnosis of Alzheimer’s disease (AD).10,11 ALS-specific biomarkers are urgently needed for the early confirmation of ALS; to date several proteomic approaches and immunoassays having been developed and several molecules have been identified with the potential to act as progression and prognosis biomarkers. 12 These include, for example, neurofilaments, proteins implicated in neuroinflammation, cytokines, chemokines, and cytoplasmic protein indicators, including TDP-43 which is a hallmark of ALS pathology.3,12 However, large-scale studies are still needed to validate the use of these potential biomarkers and to correlate corresponding biomarker concentrations with medical status and progression of ALS. Furthermore, as yet, CSF RNA-sequencing analysis has not been explored as a source for identification of novel biomarkers in ALS/MND, we hypothesize it will provide both transcriptomic information that overlaps with the previous proteomic approaches employed but also provide further unique insights and potential biomarkers.
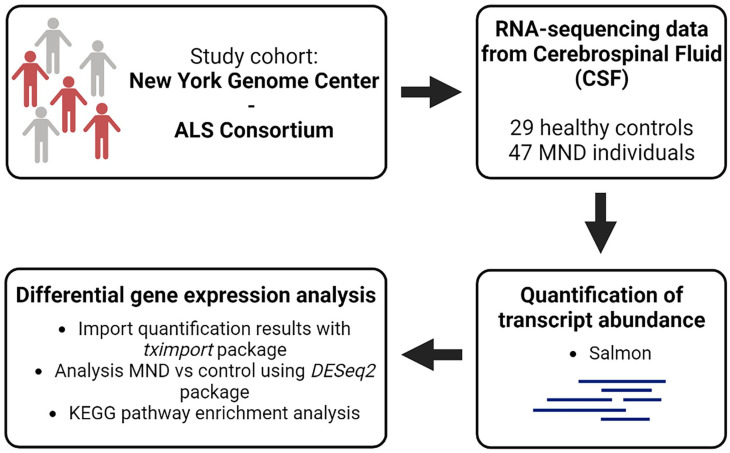
In this study, we made use of transcriptomic data from CSF of 47 MND and 29 healthy control individuals from the New York Genome Center (NYGC) ALS Consortium (Figure 1). The aim was to assess differential expression of genes encoding previously determined ALS biomarkers (extensively reviewed in Dreger et al. 12 ) between individuals with MND and controls and to integrate that data into pathway analysis which has not been commonly studied within neurological conditions such as MND (Figure 1). We demonstrated that the differentially expressed genes were enriched in many pathways associated with ALS, including nucleocytoplasmic transport, autophagy, and apoptosis. In addition, the expression of previously identified potential CSF biomarkers, including CAPG, CCL3, and MAP2 among others, 12 was significantly higher in MND individuals. The results of this study demonstrate that protein or immunological biomarkers in CSF are often correlated with a corresponding increased expression in CSF RNA-sequencing data highlighting CSF RNA data as a valuable resource for biomarker development and potentially expanding or complementing those identified from proteomic studies.
Figure 1.
Overview of study. In this study, transcriptomic data set from the New York Genome Center ALS Consortium was used. RNA-seq data from cerebrospinal fluid of 47 MND and 29 healthy control individuals were available. The aim of the study was to perform differential gene expression analysis between MND and control individuals with a focus on biomarker expression and explore enriched pathways.
Materials and methods
Overview of study
In this study, we used transcriptomic data from the NYGC ALS Consortium cohort (https://www.nygenome.org/als-consortium/). We analyzed CSF data from 47 MND and 29 age-matched healthy control individuals. MND individuals include 45 classic/typical ALS subjects complemented by one case of primary lateral sclerosis and one case of progressive muscular atrophy. CSF from MND and neurologically normal controls was collected at Stony Brook University Hospital. Healthy control individuals were recruited to participate in the study as part of a hip or knee arthroplasty, whereby patients gave consent to collect CSF.
Differential transcriptome and pathway analysis
To evaluate the changes in the transcriptome and transcriptional involvement in disease pathways, we performed differential gene expression analysis by comparing the MND data to controls. Quantification of transcriptomic data from the NYGC ALS consortium cohort on a gene- and transcript-based level was performed using the Salmon tool (https://salmon.readthedocs.io). The tximport function from the tximport package 13 was used to import salmon generated quantification files into R. Raw counts were extracted with the DESeqDataSetFromTximport function and normalized using the median-of-ratios method, specifically by dividing the raw counts by sample-specific size factors which represent the median ratio of gene counts to the geometric mean per gene. Differential gene expression analysis was performed between MND and healthy control individuals using the DESeq2 package in R. 14 Resulting P values for differentially expressed genes and transcripts were adjusted by Benjamini and Hochberg False Discovery Rate (FDR) and only FDR-adjusted values < 0.05 were considered significant. After formal statistical comparison, KEGG (Kyoto Encyclopedia of Genes and Genomes) pathway enrichment analysis was performed using the clusterProfiler package in R 15 and only P-adjusted (P-adj.) values ⩽ 0.05 were considered as significant.
Results
Pathway analysis shows enrichment for ALS disease pathway
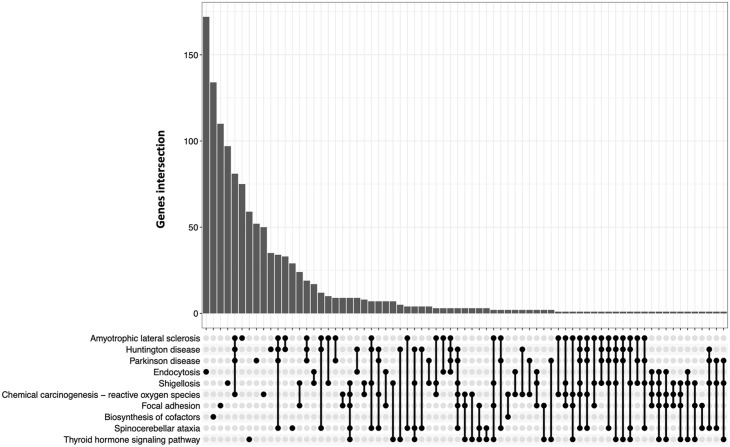
We used transcriptomic data from CSF and compared expression changes between MND and healthy control individuals (Figure 1). Following this analysis, we performed KEGG (Kyoto Encyclopedia of Genes and Genomes) pathway analysis which utilizes a database with integrated genomic, chemical, and systemic functional information 16 to identify pathways associated with gene expression changes in the individuals with MND. This analysis revealed the enrichment of 37 pathways including multiple nervous system-associated pathways (Table 1). Interestingly, one of these pathways represents ALS with a gene ratio of 310/6292 (P-adj. = 9.35E−05). We expanded the analysis by generating an UpSet plot visualizing the association between affected genes and overlapping gene sets associated with certain pathways/diseases (Figure 2). Increased gene overlapping was detected between the neurodegenerative diseases: ALS, Parkinson’s disease and Huntington’s disease indicating common disease–related pathways were affected (Figure 2). However, 75 genes were solely associated with ALS pathway demonstrating the over-representation of affected genes in the pathway of interest (Figure 2). To further this analysis, we specifically looked at the effect of this differential gene expression data on the ALS pathway (Supplementary Data 4). This identified several genes involved in key disease pathways including nucleocytoplasmic transport, autophagy, apoptosis, regulation of actin cytoskeleton, or protein processing in endoplasmic reticulum which were up- or down-regulated in MND individuals compared to healthy controls (Supplementary Data 4). Interestingly, increased TARDBP expression encoding TDP-43, a hallmark of ALS pathology, was detected in MND individuals, as previously described (Supplementary Data 4).12,17
Table 1.
Enriched KEGG pathways from CSF in MND individuals compared to healthy controls. The gene ratio indicates the number of differentially expressed genes associated with the corresponding pathway divided by the total number of differentially expressed genes (6292).
| ID | Description | Gene ratio | P-adjusted |
|---|---|---|---|
| hsa04510 | Focal adhesion | 184/6292 | 4.03E−06 |
| hsa05016 | Huntington’s disease | 266/6292 | 2.90E−05 |
| hsa04919 | Thyroid hormone signaling pathway | 112/6292 | 6.56E−05 |
| hsa04144 | Endocytosis | 219/6292 | 9.35E−05 |
| hsa05014 | Amyotrophic lateral sclerosis | 310/6292 | 9.35E−05 |
| hsa05131 | Shigellosis | 215/6292 | 1.21E−04 |
| hsa05017 | Spinocerebellar ataxia | 129/6292 | 1.81E−04 |
| hsa05012 | Parkinson’s disease | 229/6292 | 2.73E−04 |
| hsa05208 | Chemical carcinogenesis – reactive oxygen species | 194/6292 | 2.78E−04 |
| hsa01240 | Biosynthesis of co-factors | 136/6292 | 4.94E−04 |
| hsa05132 | Salmonella infection | 214/6292 | 5.15E−04 |
| hsa05020 | Prion disease | 233/6292 | 5.91E−04 |
| hsa04666 | Fc gamma R-mediated phagocytosis | 89/6292 | 5.96E−04 |
| hsa04974 | Protein digestion and absorption | 94/6292 | 5.96E−04 |
| hsa04142 | Lysosome | 118/6292 | 6.50E−04 |
| hsa03013 | Nucleocytoplasmic transport | 98/6292 | 6.52E−04 |
| hsa05022 | Pathways of neurodegeneration – multiple diseases | 393/6292 | 8.40E−04 |
| hsa04814 | Motor proteins | 165/6292 | 8.40E−04 |
| hsa05010 | Alzheimer’s disease | 319/6292 | 1.56E−03 |
| hsa05100 | Bacterial invasion of epithelial cells | 71/6292 | 1.81E−03 |
| hsa04110 | Cell cycle | 137/6292 | 1.91E−03 |
| hsa04218 | Cellular senescence | 136/6292 | 2.09E−03 |
| hsa05415 | Diabetic cardiomyopathy | 174/6292 | 2.09E−03 |
| hsa04918 | Thyroid hormone synthesis | 69/6292 | 2.29E−03 |
| hsa05166 | Human T-cell leukemia virus 1 infection | 189/6292 | 2.31E−03 |
| hsa01521 | EGFR tyrosine kinase inhibitor resistance | 72/6292 | 3.34E−03 |
| hsa04211 | Longevity regulating pathway | 80/6292 | 4.69E−03 |
| hsa04072 | Phospholipase D signaling pathway | 128/6292 | 5.72E−03 |
| hsa04152 | AMPK signaling pathway | 106/6292 | 5.72E−03 |
| hsa05135 | Yersinia infection | 119/6292 | 5.72E−03 |
| hsa05171 | Coronavirus disease – COVID-19 | 195/6292 | 6.43E−03 |
| hsa05222 | Small cell lung cancer | 82/6292 | 6.43E−03 |
| hsa04360 | Axon guidance | 155/6292 | 6.43E−03 |
| hsa04724 | Glutamatergic synapse | 100/6292 | 6.43E−03 |
| hsa00830 | Retinol metabolism | 62/6292 | 6.55E−03 |
| hsa05215 | Prostate cancer | 86/6292 | 6.55E−03 |
| hsa03018 | RNA degradation | 71/6292 | 7.47E−03 |
Figure 2.
UpSet plot to visualize intersections of differentially expressed genes. The bar chart represents the size of the gene set. The number of differentially expressed genes for single (filled-in cells) and intersecting (filled-in cells with connecting lines) pathways/diseases is shown. A full list of genes affected in the corresponding disease/pathway is listed in Supplementary Data 3.
Expression of ALS biomarkers is elevated in MND individuals from CSF transcriptomic data
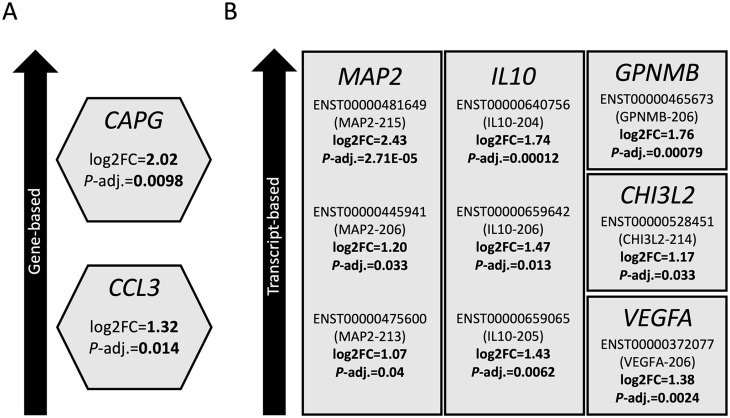
Having demonstrated that differential gene expression in MND individuals is significantly associated with the enrichment of the ALS pathway (Table 1 and Figure 3), we next aimed to assess specific expression changes on a gene- and transcript-based level (Tables 2 and 3, Supplemental Data 1 and 2). The 20 most significant transcripts and genes are shown in Tables 2 and 3. We focused on changes in genes encoding previously characterized biomarkers, specifically capping actin protein, gelsolin like (CAPG) and C-C motif chemokine ligand 3 (CCL3) which showed significantly higher expression in MND individuals, with 2.02 (P-adj. = 0.0098) and 1.32 (P-adj. = 0.014) log2 fold changes (log2FC) obtained (Figure 3(A)). In addition, we demonstrated transcript-based changes in gene expression of potential ALS biomarkers. One of these represented microtubule-associated protein 2 (MAP2); we found that three transcripts (MAP2-215, MAP2-206, and MAP2-213) were significantly more highly expressed in MND individuals (Figure 3(B)); log2FC of 2.43 (P-adj. = 2.71E−05), 1.20 (P-adj. = 0.033), and 1.07 (P-adj. = 0.04), respectively, were obtained. Other transcripts significantly elevated in MND individuals encoded for the cytokine interleukin-10 (IL-10), including IL10-204 (log2FC = 1.74, P-adj. = 0.00012), IL10-206 (log2FC = 1.47, P-adj. = 0.013), IL10-205 (log2FC = 1.43, P-adj. = 0.0062), the vascular endothelial growth factor (VEGF), including VEGF-206 (log2FC = 1.38, P-adj. = 0.0024), and the proteins chitinase-3 like 2/CHI3L2 (CHI3L2-214, log2FC = 1.17, P-adj. = 0.033) and glycoprotein NMB/GPNMB (GPNMB-206, log2FC = 1.76, P-adj. = 0.00079), involved in inflammatory processes (Figure 3(B)). It should be noted that some of these transcripts (MAP2-215, MAP2-213, IL10-204, and GPNMB-206) are alternatively spliced transcripts of a protein-coding gene for which the coding sequence has not been defined yet. However, these data confirm the validity of using CSF for biomarker discovery in ALS and illustrate the plethora of expression changes on a gene- and isoform-based level obtained from RNA-sequencing data of CSF. In addition, the top hits obtained in this analysis, including the transcript ENST00000648280 (COPA-212, P-adj. = 6.06E−13), have not been investigated as biomarkers to date.
Figure 3.
Several genes encoding potential biomarker proteins are significantly upregulated in CSF from MND individuals. Differential gene expression analysis was performed using transcriptomic data sets from 47 MND and 29 healthy control individuals. Expression changes were analyzed on a gene (A)- and transcript (B)-based level. Several genes which have been previously identified to encode potential biomarker proteins were significantly more highly expressed in MND individuals. Log2 fold changes (log2FC) and P-adjusted values are indicated. For transcript-based analysis, corresponding transcript IDs are shown.
Table 2.
Differentially expressed transcripts from MND CSF. Top 20 hits are shown.
| Transcript ID | Transcript name | P-adjusted |
|---|---|---|
| ENST00000648280 | COPA-212 | 6.06E−13 |
| ENST00000513295 | CEP63-210 | 2.07E−11 |
| ENST00000659698 | LINC00467-214 | 2.67E−11 |
| ENST00000409039 | DNAH10-201 | 9.14E−11 |
| ENST00000328843 | DNAH11-201 | 1.11E−10 |
| ENST00000493526 | RHBDD1-214 | 1.09E−09 |
| ENST00000628444 | LINC02203-204 | 1.39E−09 |
| ENST00000422452 | TENM1-202 | 1.65E−09 |
| ENST00000444250 | ECT2-212 | 2.27E−09 |
| ENST00000590416 | LSM14A-208 | 4.79E−09 |
| ENST00000476513 | APOBEC3F-203 | 6.37E−09 |
| ENST00000646052 | RP11-114N19.3-002 | 6.46E−09 |
| ENST00000432504 | TMEM50B-202 | 8.99E−09 |
| ENST00000492563 | DCUN1D1-206 | 9.26E−09 |
| ENST00000410031 | TBC1D14-202 | 9.94E−09 |
| ENST00000566391 | ARL6IP1-206 | 1.11E−08 |
| ENST00000531607 | CHEK1-210 | 1.11E−08 |
| ENST00000511794 | LINC02057-202 | 1.11E−08 |
| ENST00000406359 | TEK-202 | 1.12E−08 |
| ENST00000532638 | POU2F3-203 | 1.43E−08 |
Table 3.
Differentially expressed genes from MND CSF. Top 20 hits are shown.
| Gene ID | Gene name | P-adjusted |
|---|---|---|
| ENSG00000140688 | RUSF1 | 3.31E−11 |
| ENSG00000128283 | CDC42EP1 | 2.70E−09 |
| ENSG00000185220 | PGBD2 | 2.70E−09 |
| ENSG00000231183 | AC007003.1 | 3.03E−09 |
| ENSG00000277741 | GOLGA6L17P | 6.21E−09 |
| ENSG00000114349 | GNAT1 | 1.06E−08 |
| ENSG00000216360 | RP1-182O16.2 | 1.09E−08 |
| ENSG00000231784 | DBIL5P | 1.27E−08 |
| ENSG00000266777 | SH3GL1P1 | 1.40E−08 |
| ENSG00000230701 | FBXW4P1 | 1.73E−08 |
| ENSG00000256420 | OSBPL9P4 | 1.77E−08 |
| ENSG00000009724 | MASP2 | 1.82E−08 |
| ENSG00000162191 | UBXN1 | 1.82E−08 |
| ENSG00000260123 | CARMAL | 1.82E−08 |
| ENSG00000110665 | C11orf21 | 6.08E−08 |
| ENSG00000287398 | RP11-642N14.4 | 6.08E−08 |
| ENSG00000280432 | AP000962.1 | 8.54E−08 |
| ENSG00000287379 | RP1-296G17.4 | 1.06E−07 |
| ENSG00000204934 | ATP6V0E2-AS1 | 1.82E−07 |
| ENSG00000256500 | RP11-73M18.2 | 1.99E−07 |
Discussion
To date, there is no cure for ALS, highlighting the urgent need to understand the interaction between genetics and biochemistry including the identification of biomarkers. Advances in the development of biomarkers would help to deepen the knowledge both of the preclinical disease phase and to progress therapy development and design of effective clinical trials by stratification of patients into more homogeneous groups.
Several CSF protein biomarkers have already been established for ALS such as neurofilaments, synucleins, or tau.12,18,19 Recent studies including proteomic analyses have identified further novel biomarkers for ALS which include MAP2, CAPG, and GPNMB plus others involved in neuroinflammation (CHI3L2) or with neuroprotective roles (VEGF).12,20
CHI3L2 is part of the chitinase-like proteins and secreted by astrocytes/microglia. This protein may lead to neuronal death in ALS as a direct correlation between its CSF concentration in ALS individuals, and disease progression rate was found.21,22 CCL3, also termed macrophage inflammatory protein 1 alpha, is involved in the accumulation of microglia and has functions in inflammatory responses and therefore indicates neuroinflammation in ALS. 12 CCL3 has been shown to inversely correlate with disease progression rate. 23 Other proteins, including CAPG and GPNBM, have also been associated with inflammatory processes.20,24,25 More specifically, GPNMB expression has been linked to neurodegeneration by the observation that ALS patients were characterized by a shorter survival time with high GPNMB CSF levels and the correlation with the disease severity Amyotrophic Lateral Sclerosis Functional Rating Scale (ALSFRS) score. 20 Tanaka et al. 26 confirmed the increased GPNMB levels in the CSF. MAP2 is part of the family of microtubule-associated proteins which have crucial roles in modulating the microtubule network. Oeckl and colleagues showed a significant increase of MAP2 in the CSF of ALS individuals and the potential to act as a marker of motor neuron degeneration. 20 It has been demonstrated that MAP2 can induce neurites, 27 and the corresponding increased MAP2 expression in the CSF may be an adjustment following axonal loss. Therefore, MAP2 could act as a marker of motor neuron loss. The cytokine VEGF has been associated with faster disease progression (and shorter survival) in patients with lower VEGF levels which may therefore represent a positive prognostic measure. 23 Interestingly, VEGF CSF levels positively correlated with levels of PaO2 in ALS individuals, which suggests a hypoxia response dysfunction in ALS patients. 28
Our data are consistent with previous CSF level studies, confirming the findings by transcriptomic data analysis focused on the expression of the corresponding gene targets. By doing so, it also conferred greater confidence in the other novel targets; we identified thus highlighting the utility/validity of transcriptomic analysis in such studies.
Transcriptomic analysis of CSF is not commonly performed in neurological conditions, and here, we provide new data analyzing RNA-sequencing data from 47 MND and 29 healthy control individuals and assessing expression changes between the two groups. Utilizing data derived from this biofluid, we demonstrated on the gene-based level that CAPG expression was significantly higher in MND individuals (log2FC = 2.02, P-adj. = 0.0098) (Figure 3(A)). Furthermore, MND individuals showed a 1.32 log2FC increase (P-adj. = 0.014) in CCL3 which is line with previously reported elevated levels of the protein in ALS CSF.12,23,29 Moreover, our study highlighted transcript-specific changes in isoform expression of ALS biomarkers, for example, for MAP2, three transcripts (MAP2-215, MAP2-206, and MAP2-213) were significantly more highly expressed in MND individuals (Figure 3(B)), and log2FC of 2.43 (P-adj. = 2.71E−05), 1.20 (P-adj. = 0.033), and 1.07 (P-adj. = 0.04) were obtained. These results highlight the plethora and specificity of transcriptomic changes obtained from CSF transcriptomic data sets which can be a tool to confirm advances in biomarker development and their involvement in specific signaling pathways. In this study, the gene demonstrating the greatest difference in cases versus controls was a specific protein-coding transcript of the gene COPA (ENST00000648280) which was significantly more highly expressed in MND individuals (log2FC = 3.98, P-adj. = 6.06E−13) compared to controls. This gene encodes the COP1 protein which has been shown to be involved in protein transport between endoplasmic reticulum and Golgi apparatus, with gene mutations being associated with an inflammatory syndrome showing lung, renal, and joint involvement.30,31
High sensitivity of measurement is crucial to detect small changes in a low concentration range; thus, this sensitive RNA-seq analysis may complement proteomic analysis to support biomarker development. This view is reinforced by the KEGG pathway enrichment analysis which demonstrated that the differentially expressed genes were enriched in the ALS pathway (hsa05014, gene ratio 310/6292, P-adj. = 9.35E−05), with numerous pathways involved in ALS pathology affected, including nucleocytoplasmic transport, autophagy, apoptosis, regulation of actin cytoskeleton, or protein processing in endoplasmic reticulum (Supplementary Data 4). We were able to detect significant changes in RNA expression for known CSF biomarker proteins which confirms CSF RNA-seq is a suitable method for this type of analysis. CSF analysis presents several challenges: CSF is challenging to collect, and it is inconvenient and has risks as a procedure; however, CSF contains cell-free RNA, and this can be analyzed by modern sensitive analytical tools. Future additional larger and longitudinal studies utilizing CSF are needed to validate and reproduce these findings and to increase precision of biomarker development. Another limitation of the study is that the cut-off for the transcript-based level has not been identified yet and this limits the use of these as biomarkers.
Supplemental Material
Supplemental material, sj-pdf-1-ebm-10.1177_15353702231209427 for Transcriptomic profiling of cerebrospinal fluid identifies ALS pathway enrichment and RNA biomarkers in MND individuals by Alexander Fröhlich, Abigail L Pfaff, Vivien J Bubb, John P Quinn and Sulev Koks in Experimental Biology and Medicine
Acknowledgments
The authors acknowledge the ALS Consortium/Target ALS Human Postmortem Tissue Core, New York Genome Center for Genomics of Neurodegenerative Disease, Amyotrophic Lateral Sclerosis Association, and TOW Foundation for the RNA-sequencing sequencing data used in this publication. This work was supported by resources provided by the Pawsey Supercomputing Centre with funding from the Australian Government and the Government of Western Australia.
Footnotes
Authors’ contributions: AF and SK contributed to the conceptualization, methodology, and formal analysis. AF, ALP, VJB, JPQ, and SK contributed to the data interpretation. AF contributed to the writing – original draft preparation. AF, ALP, VJB, JPQ, and SK contributed to the writing – review and editing. All authors have read and agreed to the published version of the manuscript.
The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding: The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: AF and JPQ are funded by the Andrzej Wlodarski Memorial Research Fund. AF is the recipient of Andrzej Wlodarski Memorial Research PhD scholarship. ALP and SK are funded by Multiple Sclerosis Western Australia and Perron Institute for Neurological and Translational Science. VJB and JPQ are funded by the Darby Rimmer Foundation. VJB, JPQ, ALP, and SK are funded by the MNDA (ref Quinn/Apr20/875-791).
ORCID iDs: Alexander Fröhlich  https://orcid.org/0000-0001-9083-7485
https://orcid.org/0000-0001-9083-7485
Vivien J Bubb  https://orcid.org/0000-0003-2763-7004
https://orcid.org/0000-0003-2763-7004
Sulev Koks  https://orcid.org/0000-0001-6087-6643
https://orcid.org/0000-0001-6087-6643
Supplemental material: Supplemental material for this article is available online.
References
- 1. Ajroud-Driss S, Siddique T. Sporadic and hereditary amyotrophic lateral sclerosis (ALS). Biochim Biophys Acta 2015;1852:679–84 [DOI] [PubMed] [Google Scholar]
- 2. Grad LI, Rouleau GA, Ravits J, Cashman NR. Clinical spectrum of amyotrophic lateral sclerosis (ALS). Cold Spring Harb Perspect Med 2017;7:a024117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Shatunov A, Al-Chalabi A. The genetic architecture of ALS. Neurobiol Dis 2021;147:105156. [DOI] [PubMed] [Google Scholar]
- 4. Theunissen F, Flynn LL, Anderton RS, Mastaglia F, Pytte J, Jiang L, Hodgetts S, Burns DK, Saunders A, Fletcher S, Wilton SD, Akkari PA. Structural variants may be a source of missing heritability in sALS. Front Neurosci 2020;14:47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Veldink JH. ALS genetic epidemiology “how simplex is the genetic epidemiology of ALS?” J Neurol Neurosurg Psychiatry 2017;88:537. [DOI] [PubMed] [Google Scholar]
- 6. Wroe R, Wai-Ling Butler A, Andersen PM, Powell JF, Al-Chalabi A. ALSOD: the amyotrophic lateral sclerosis online database. Amyotroph Lateral Scler 2008;9:249–50 [DOI] [PubMed] [Google Scholar]
- 7. Ghasemi M, Brown RH. Genetics of amyotrophic lateral sclerosis. Cold Spring Harb Perspect Med 2018;8:a024125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Ranganathan R, Haque S, Coley K, Shepheard S, Cooper-Knock J, Kirby J. Multifaceted genes in amyotrophic lateral sclerosis-frontotemporal dementia. Front Neurosci 2020;14:684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Savage AL, Schumann GG, Breen G, Bubb VJ, Al-Chalabi A, Quinn JP. Retrotransposons in the development and progression of amyotrophic lateral sclerosis. J Neurol Neurosurg Psychiatry 2019;90:284–93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Niemantsverdriet E, Valckx S, Bjerke M, Engelborghs S. Alzheimer’s disease CSF biomarkers: clinical indications and rational use. Acta Neurol Belg 2017;117:591–602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Paterson RW, Slattery CF, Poole T, Nicholas JM, Magdalinou NK, Toombs J, Chapman MD, Lunn MP, Heslegrave AJ, Foiani MS, Weston PSJ, Keshavan A, Rohrer JD, Rossor MN, Warren JD, Mummery CJ, Blennow K, Fox NC, Zetterberg H, Schott JM. Cerebrospinal fluid in the differential diagnosis of Alzheimer’s disease: clinical utility of an extended panel of biomarkers in a specialist cognitive clinic. Alzheimers Res Ther 2018;10:32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Dreger M, Steinbach R, Otto M, Turner MR, Grosskreutz J. Cerebrospinal fluid biomarkers of disease activity and progression in amyotrophic lateral sclerosis. J Neurol Neurosurg Psychiatry 2022;93:422–35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Soneson C, Love MI, Robinson MD. Differential analyses for RNA-seq: transcript-level estimates improve gene-level inferences. F1000Research 2016;4:1521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 2014;15:550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Yu G, Wang LG, Han Y, He QY. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS 2012;16:284–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Kanehisa M. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res 2000;28:27–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Majumder V, Gregory JM, Barria MA, Green A, Pal S. TDP-43 as a potential biomarker for amyotrophic lateral sclerosis: a systematic review and meta-analysis. BMC Neurol 2018;18:90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Agah E, Saleh F, Sanjari Moghaddam H, Saghazadeh A, Tafakhori A, Rezaei N. CSF and blood biomarkers in amyotrophic lateral sclerosis: protocol for a systematic review and meta-analysis. Syst Rev 2018;7:237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Verber NS, Shepheard SR, Sassani M, McDonough HE, Moore SA, Alix JJP, Wilkinson ID, Jenkins TM, Shaw PJ. Biomarkers in motor neuron disease: a state of the art review. Front Neurol 2019;10:291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Oeckl P, Weydt P, Thal DR, Weishaupt JH, Ludolph AC, Otto M. Proteomics in cerebrospinal fluid and spinal cord suggests UCHL1, MAP2 and GPNMB as biomarkers and underpins importance of transcriptional pathways in amyotrophic lateral sclerosis. Acta Neuropathol 2020;139:119–34 [DOI] [PubMed] [Google Scholar]
- 21. Liu L, Yang Y, Duan H, He J, Sun L, Hu W, Zeng J. CHI3L2 is a novel prognostic biomarker and correlated with immune infiltrates in gliomas. Front Oncol 2021;11:611038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Thompson AG, Gray E, Bampton A, Raciborska D, Talbot K, Turner MR. CSF chitinase proteins in amyotrophic lateral sclerosis. J Neurol Neurosurg Psychiatry 2019;90:1215–20 [DOI] [PubMed] [Google Scholar]
- 23. Guo J, Yang X, Gao L, Zang D. Evaluating the levels of CSF and serum factors in ALS. Brain Behav 2017;7:e00637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Van Der Lienden M, Gaspar P, Boot R, Aerts J, Van Eijk M. Glycoprotein non-metastatic protein B: an emerging biomarker for lysosomal dysfunction in macrophages. Int J Mol Sci 2018;20:66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Witke W, Li W, Kwiatkowski DJ, Southwick FS. Comparisons of CapG and gelsolin-null macrophages. J Cell Biol 2001;154:775–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Tanaka H, Shimazawa M, Kimura M, Takata M, Tsuruma K, Yamada M, Takahashi H, Hozumi I, Niwa J, Iguchi Y, Nikawa T, Sobue G, Inuzuka T, Hara H. The potential of GPNMB as novel neuroprotective factor in amyotrophic lateral sclerosis. Sci Rep 2012;2:573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Dehmelt L, Halpain S. The MAP2/Tau family of microtubule-associated proteins. Genome Biol 2004;6:204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Moreau C. Paradoxical response of VEGF expression to hypoxia in CSF of patients with ALS. J Neurol Neurosurg Psychiatry 2006;77:255–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Huang F, Zhu Y, Hsiao-Nakamoto J, Tang X, Dugas JC, Moscovitch-Lopatin M, Glass JD, Brown RH, Ladha SS, Lacomis D, Harris JM, Scearce-Levie K, Ho C, Bowser R, Berry JD. Longitudinal biomarkers in amyotrophic lateral sclerosis. Ann Clin Transl Neurol 2020;7:1103–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Kumrah R, Mathew B, Vignesh P, Singh S, Rawat A. Genetics of COPA syndrome. Appl Clin Genet 2019;12:11–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Lepelley A, Martin-Niclós MJ, Le Bihan M, Marsh JA, Uggenti C, Rice GI, Bondet V, Duffy D, Hertzog J, Rehwinkel J, Amselem S, Boulisfane-El Khalifi S, Brennan M, Carter E, Chatenoud L, Chhun S, Coulomb l’Hermine A, Depp M, Legendre M, Mackenzie KJ, Marey J, McDougall C, McKenzie KJ, Molina TJ, Neven B, Seabra L, Thumerelle C, Wislez M, Nathan N, Manel N, Crow YJ, Frémond M-L. Mutations in COPA lead to abnormal trafficking of STING to the Golgi and interferon signaling. J Exp Med 2020;217:e20200600 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental material, sj-pdf-1-ebm-10.1177_15353702231209427 for Transcriptomic profiling of cerebrospinal fluid identifies ALS pathway enrichment and RNA biomarkers in MND individuals by Alexander Fröhlich, Abigail L Pfaff, Vivien J Bubb, John P Quinn and Sulev Koks in Experimental Biology and Medicine