Abstract
Single-molecule force spectroscopy provides access to the mechanics of biomolecules. Recently, magnetic and laser optical tweezers were applied in the studies of chaperones and their interaction with protein clients. Various aspects of the chaperone–client interactions can be revealed based on the mechanical probing strategies. First, when a chaperone is probed under load, one can examine the inner workings of the chaperone while it interacts with and works on the client protein. Second, when protein clients are probed under load, the action of chaperones on folding clients can be studied in great detail. Such client folding studies have given direct access to observing actions of chaperones in real-time, like foldase, unfoldase, and holdase activity. In this review, we introduce the various single molecule mechanical techniques and summarize recent single molecule mechanical studies on heat shock proteins, chaperone-mediated folding on the ribosome, SNARE folding, and studies of chaperones involved in the folding of membrane proteins. An outlook on significant future developments is given.
I. INTRODUCTION
Single-molecule force spectroscopy is a novel method for monitoring biological processes, which can directly measure the distances and forces involved in conformational changes of proteins at high spatio-temporal resolution. As force and distances and their mathematical product, energy, are fundamental characteristics of biological processes, force spectroscopy provides direct insight into the energy landscape of conformational transitions in biomolecules. The main focus of this review is on mechanical single-molecule studies of chaperones and clients. In this context, we understand the term “chaperone” in its conservative definition as a protein factor transiently interacting with proteinous clients but not being a part of the final functional form of the client. We did not include nucleic acid chaperones (e.g., retroviral nucleocapsid proteins,1 Orf1p,2 and others) and small-molecule pharmacological chaperones3 here. The review is organized as follows: In Sec. II, we shall start with a description of chaperone systems and their large abundance. Section III focuses on single-molecule mechanical studies of chaperone–client interactions and includes studies of heat shock proteins, the effect of chaperones on the folding of ribosome-bound proteins, as well as membrane protein chaperones. In Sec. IV, we summarize recent advances in single-molecule force spectroscopy of proteins, which we assume will drive future development of force-spectroscopy assays and thus provide even more insight into multifaceted chaperone–client interactions.
II. COMPLEXITY OF CHAPERONES AND THEIR CLIENTS
The diverse classes of chaperones are collectively summarized under the concept of the chaperome, which is related to the ensemble of chaperones and co-chaperones interacting in a complex network of molecular folding machines to regulate proteome function.4 The chaperone is central to the proteostasis network in the cell by providing supportive activity, preventing misfolding, helping non-native intermediates, and getting the native state and other roles.5 As a vital part of protein quality control mechanisms, the chaperome protects proteome functionality and prevents a toxic accumulation of mutant, misfolded, and damaged proteins.6
The complexity of the chaperome is overwhelming. This complexity arises mainly from two factors: first, the number of clients per chaperone ranges from one to hundreds depending on the selectivity of the specific chaperone-type. For example, NarW—a NarJ homolog, is a chaperone exclusive to the nitrate reductase subunits,7 whereas, for the DnaK system, more than seven hundred clients have been identified.8 A second factor contributing to complexity is the large number of chaperones possessing enormous networking capacity and buffering ability due to overlapping pools of clients. While in Escherichia coli, more than 70 proteins have been identified with chaperone activities (https://ecocyc.org after manual correction), even higher complexity is seen in eukaryotes. In humans, for example, 332 genes were identified and divided into nine chaperone gene families.9 In the human chaperome, 88 genes are functionally classified as genes encoding chaperones and 244 as co-chaperones. The larger number of chaperones in eukaryotes is likely owed to larger and more complex multi-domain proteins. In contrast, in E. coli,10 the average protein size is 310 aa in bacterium and 560 aa in humans.
Historically, many chaperones were named according to their function as heat shock proteins (Hsps). Based on their observed molecular weights, they were divided into five major classes: Hsp60, Hsp70, Hsp90, Hsp104, and the small Hsps.11 However, this definition needs extension as more and more chaperones and chaperone functions are being discovered. Initially, chaperones were assumed to be folding helpers that helped nascent client chains to achieve effective folding by rescuing aggregation-prone partially folded states. Nowadays, the function of chaperones is much richer, including foldase, unfoldase, and holdase activities. In a broader sense, chaperones are involved when a protein client conformation needs to be controlled. Chaperones can be categorized using a vocabulary of gene ontology terms,12 which, however, are of limited use for practical experimental studies. A meaningful classification of chaperones is based on the need for energy input provided mainly by ATP, which is used for conformational cycling between high and low-affinity states. Of the 88 chaperones identified in humans, 50 are ATP-dependent and 38 ATP-independent.9 ATP-dependent chaperones and their functional cycling between states are often further regulated by additional proteins called co-chaperones.
One particular issue related to the diversity of chaperones and co-chaperones is the confusing and often inconsistent nomenclature, which arises from the mixing of genetic and biochemical names. For example, Hsp70 homologs can have several names, e.g., DnaK, Ssc1, Bip, and mortalin. Similarly, Hsp40 co-chaperones have many alternate names: DnaJ, DnaJB1, HspF1, Hdj1, and Sis1, to name a few.
Given the diversity of chaperome and their clients, our knowledge of chaperone principles is just emerging. While for understanding the ever-increasing complexity, structural biology has been and will remain essential; however, novel biophysical methods are needed to report on the dynamics of the protein function. The single-molecule force studies covered in this review provide just an initial glimpse of what we can hope for in the future from such methods to understand the chaperome.
III. SINGLE-MOLECULE MECHANICAL STUDIES OF CHAPERONES
While single-molecule studies of some chaperones have also been covered in several previous reviews,18–28 this section provides a short description of mechanical interrogation strategies, instruments for force experiments, and our perspective on selected contributions in chaperone–client interactions that have been published since 2015. The subsections are divided into heat shock proteins, the study of the impact of chaperones on folding ribosome-bound proteins, DsbA–client interactions, SNARE proteins with Mun/Munc chaperones, and chaperone-assisted folding of the membrane proteins.
A. Instruments for protein mechanical studies
In general, to study chaperone–client interactions using force spectroscopy, two general mechanical interrogation strategies are at hand:
-
(1)
a tethered client is probed under load in the presence of a free chaperone, or
-
(2)
a tethered chaperone is probed under load in the presence of a free client.
In most cases, strategy (1) is applied, and the client protein is held under load while chaperones are added free in solution, and their effect on folding is observed. Information can be obtained about the folding process of the client as well as the state of the folding protein, which is recognized by the chaperone. For example, the chaperone may bind to the fully unfolded or to partially folded, misfolded, or aggregated states. Moreover, the binding stoichiometry, binding/unbinding rates, and relative affinities between chaperone and client can be determined by studying the concentration dependence. Since force spectroscopy offers a simple structural readout through the measured length of given protein conformations, all the binding parameters can be directly attributed to those structural states, thus providing a unique combination of structural as well as dynamic insight. In strategy (2), the chaperone itself is probed under load, and the effect of adding free substrate is monitored. Here, information can be obtained about the inner workings of the chaperone while it interacts with and works on the client protein. Also, using client concentration titrations, kinetics and thermodynamics of chaperone/substrate binding can be determined.
Several technical implementations of single-molecule force experiments have been developed over the years, including atomic force microscopy (AFM), laser optical tweezers, magnetic tweezers, and acoustic and centrifugal force spectroscopy. Each of the technical realizations has its advantages and disadvantages.29 In AFM force spectroscopy, sensitive movable piezo stages enable mechanical stretching of proteins while measuring force by the deflection of the cantilever needle. AFM was established as an effective technique for studying protein folding mechanics in 1997.30 Applying forces to concatenated multiple copies of proteins or domains leads to a characteristic saw-tooth pattern that can be used to identify single-molecule events from a typically large background of nonspecific and multiple molecule events. A significant advantage of AFM is that high forces up to several nanonewtons (nN) can be applied to allow even studying ultra-stable proteins exhibiting unfolding forces in the range of breaking forces of covalent bonds.31,32
Optical tweezers use highly focused laser beams for optical trapping of two dielectric microparticles tethered by a single DNA–protein–DNA construct. In one possible technical realization, laser beams are used to project the back focal plane of the condenser onto a position-sensitive photodetector.33 Upon calibration, displacement of the beads from the trap center gives a direct readout for the force assuming a harmonic regime. Optical tweezers are ideally suited for the low-to-intermediate force regime (0.5 – ∼100 pN) and have been applied in numerous protein folding studies since 1997.34,35 Force spectroscopy by optical tweezers allows for a detailed analysis of protein folding pathways, transition path times at the microsecond time range,36,37 and subnanometer precision.38
Magnetic tweezers force spectroscopy39 uses magnetic field gradients to apply pulling forces to biomolecules tethered to superparamagnetic beads40 and gives access to long timescales of several hours or even days41 on tens of thousands of molecules in parallel.42,43 In acoustic force microscopy,44,45 a piezo-element is driven by an oscillating voltage to resonantly excite a planar acoustic standing wave over a flow cell. A microsphere subjected to this standing wave experiences a force along the vertical direction toward an acoustic pressure node. In the Centrifuge Force Microscope,46–49 microspheres in an orbiting sample are subjected to a calibration-free, macroscopically uniform force field while their motion is observed. Magnetic, acoustic, and centrifugal force microscopy have a unique advantage in the possibility of multiplexing and massive parallel detection of several single-molecule tethers during one pulling cycle, which introduces a high-throughput potential for mechanical force experiments.
In standard mechanical single molecule studies, different protocols are employed: constant velocity, constant distance/force and force jump/quench experiments. In a constant velocity experiment, the protein is stretched at constant pulling velocity, typically 20–2000 nm/s. At a certain point, the protein or a part unfolds, leading to a sudden drop in force and an increase in protein extension. After applying a polymer elasticity model to account for non-Hookean polymer elasticity, contour lengths can be calculated to quantify the measured unfolding patterns50 by calculating the number of residues involved in the conformational transition. This protocol is particularly useful for assessing non-equilibrium properties of protein folding and unfolding. The constant distance protocol (passive mode) applies a constant pre-tension on a molecule at some narrow force range to observe the hopping of the molecule between individual states displaying different lengths. As the molecule hops, it spends a varying amount of time in different states. The analysis of hopping traces by a hidden Markov model yields transition probabilities and, hence, the microscopic rate constants. The further distinction among different states possessing the same length can be obtained by analyzing deviations from single-exponential dwell-time distributions. Corrections for events missed due to a limited time resolution can be applied.51 The constant distance/force protocol is ideally suited for assessing kinetic networks52 in and near thermodynamic equilibrium and can be used to deconvolve the protein's energy landscape under load.53–56 Signal-pair and autocorrelation analysis can be applied as well.36,57 In the force quench protocol, the load on the protein is changed abruptly.58 Force jumps can probe, for example, the folding status of a refolding protein59 or transient populations of kinetically rare species such as unfolded states with cis-proline isomer.60
B. Studies of heat shock proteins and clients
Heat shock proteins (Hsps) are proteins that upregulate their levels at elevated temperatures. The heat shock response is essential for the survival of bacteria, and the expression of the associated Hsps is controlled by a specific σ factor, σ32, encoded by the rpoH gene.61 Many single-molecule studies covered in this review were conducted on bacterial Hsps, reflecting their overall importance as paradigms in the field of chaperone research.
1. Mechanics of the Hsp70 chaperone and multifaceted interaction with clients
The Hsp70 (heat shock protein of 70 kDa) family of chaperones is ubiquitous, displaying ATP-regulated chaperone function.8 The Hsp70 chaperones are conserved across all domains of life—from bacteria to humans. Hsp70 from E. coli is called DnaK and is the most prominent member of the Hsp70 family.62 It consists of two domains with different functions: a nucleotide-binding domain (NBD) and a substrate-binding domain (SBD). The domains are connected by a short, flexible linker (for structure, see Refs. 13, 14, and 63 and Fig. 1). The NBD binds MgADP and MgATP with nanomolar affinity;64 the SBD binds an extensive number of protein clients and confers chaperone function.8,65 The affinity and kinetics of client binding are strongly coupled to the nucleotide state of the NBD, which is triggered by allosteric communication between the domains. Disruption of the ATPase activity or interdomain communication impairs biological function in vivo.66–68 Thus, the binding of nucleotides and regulation of ATPase activity plays a central role in the biological function of DnaK. The chaperone activity of DnaK is further enhanced by the co-chaperones DnaJ and GrpE;5,69,70 both proteins regulate the nucleotide turnover of the NBD at different checkpoints. DnaJ can recruit clients and speeds up ATP hydrolysis rate after binding to DnaK. GrpE plays a role as a nucleotide exchange factor and accelerates exchange of ADP by ATP by >5000-fold.71
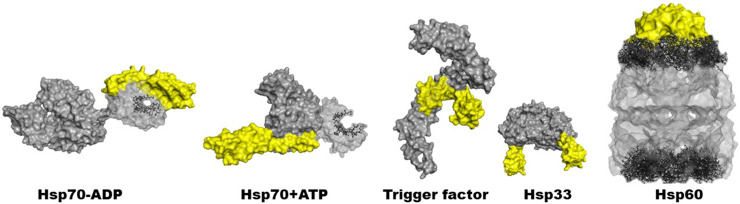
FIG. 1.
3D structures of a few selected chaperones, from left to right: E. coli Hsp70 in the ADP/ATP form, helical lid shown in yellow, residues involved in peptide binding are shown as ball-and-sticks (PDB codes: 2KHO, NMR-RDC/x-ray structure hybrid,13 4B9Q14) E. coli trigger factor and its touching arms in yellow (PDB code: 1W2615), the dimeric form of Bacillus subtilis Hsp33 with yellow C-terminal redox sensing domain (PDB code: 1VZY16), and E. coli Hsp60 and yellow Hsp10 aka GroEL/GroES structure with highlighted apical domain, residues 191–376, as ball-and-sticks.17
a. Internal mechanics of Hsp70 and Hsp40
The Hsp70 chaperone is a model for the functional coupling between ATP hydrolysis and binding affinity and kinetics of two functionally distinct domains. The heart of the coupling is in the nucleotide-binding domain of Hsp70, which performs regulatory functions by conformational switching during the ATP/ADP cycles. Such switching relies on the coupling of the mechanics of the internal structures and the nucleotide status. In a study by Bauer et al.,72 the NBD structural elements were probed under load to investigate the internal mechanics and the role of allosteric coupling. Mechanical pulling at the termini of the NBD revealed that the overall mechanical stability along this direction does not depend on the nucleotide state; instead, the experiments revealed that nucleotide binding differentially stabilizes internal substructures of the NBD. In the presence of ATP/ADP, lobe II gained significant stability, possibly due to the strong stabilization of the bound nucleotide. Coarse-grained simulations were used to enhance the structural interpretation of these experiments and confirmed that the unfolding pathways differ in the apo vs the nucleotide-bound state due to lobe II binding the nucleotide. The authors found the key event triggering NBD unfolding is the unfolding of a highly buried C-terminal helix forming the lobe I/lobe II interface. The apparent insensitivity of the unfolding forces on nucleotide-binding was surprising but highlighted the importance of the mechanical pulling direction. Apparently, when pulled at the termini, the reaction coordinate is insensitive to the presence of the bound nucleotide. Indeed, in a follow-up investigation, Meinhold et al.73 found that using different pulling directions where force is applied across the lobe I/lobe II interface, unfolding forces are highly sensitive to the nucleotide type and even the presence of the bound GrpE co-chaperones. These studies highlight that regarding nucleotide binding, care has to be taken when choosing pulling directions because only some may be informative projections. Those NBD mechanical studies highlighted the importance of lobe II for the interactions with the nucleotide. In another study by Bauer et al., optical tweezers were used to monitor refolding of the NBD, and refolding intermediates were identified that were nucleotide-binding competent.74 Using loop insertions, a coarse structural model of a minimal ATP binding domain was suggested from single molecule experiments. The 3D structure, as well as its ATP binding properties, could then be successfully determined. The authors could show that the formation of this minimal ATP binding domain is a key step for the folding of the NBD. In fact, an incompetent folding homolog of the Hsp70 NBD from yeast mitochondria lacks this important folding intermediate, leading to misfolding.
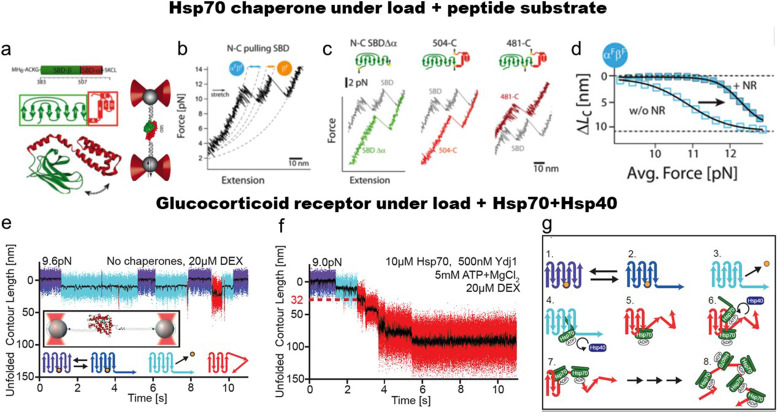
Single-molecule studies of the substrate-binding domain of Hsp70 (SBD) revealed significant fluctuations at the α/β interface dividing the SBD into the substrate-binding site [Fig. 2(a)] and the α-helical subdomain, including the lid [Fig. 2(b)].75 The SBD was interrogated by tethering the N and C terminus. Opening/closing fluctuations within the SBD were found depending on the folding state of the α-helical subdomain. The αβ fluctuations represent opening/closing fluctuations of helix A as well as β7-β8 when both subdomains are still folded [Fig. 2(b)]. When the α-helical subdomain unfolds, only fluctuation corresponding to the opening of β7-β8 was observed [orange arrow in Fig. 2(b)]. These experiments helped to identify a flexible hinge structure within the β domain that appears monolithic in the crystal structure. The authors used different force application points to pinpoint the hinge to strands β7-β8 [Fig. 2(c)]. Moreover, the binding of the substrate peptide mediates significant stabilization of the αβ fluctuations [Fig. 2(d)], while β7/8 fluctuations alone remained nearly unaffected. The remaining core β-subdomain showed significant mechanical stabilization with bound peptide as demonstrated by higher unfolding forces. Thus, substrate-binding increases the energy needed for the opening of the interface while hinge sheets are not affected, highlighting their lack of structural cooperativity with the core. This study demonstrated how the binding of the peptide substrate is propagated and distributed within the internal structures of the chaperones.
FIG. 2.
Internal mechanics of the Hsp70 chaperone domain and mechanics of the glucocorticoid receptor ligand-binding domain, GR-LBD, as a client for Hsp70-Hsp40. (a) Scheme of optical tweezers and pulling off the DNA tethered SBD. (a) Force-extension trace of the SBD of Hsp70. (c) Mechanical pulling of the SBD along with different directions (shown in color, SBD N-to-C pulling in gray). (d) Force-dependence fluctuations between folded α- and β-subdomain after the contour-length transformation. (e) Fluctuations of holo GR-LBD show fast opening and closing of the N-terminal lid (fast transitions between purple and dark-blue state), ligand dissociation (transition to light-blue state), and ligand rebinding (return to purple/dark-blue flipping), including rare partial unfoldings. Inset: scheme for the single-molecule optical tweezers experiment. (f) Sample trace of Hsp70/40 unfolding apo GR-LBD completely via five intermediates within a few seconds. Unfolding sets in within 1 s after DEX dissociation. The red dashed line marks the 32 nm of unfolded contour length, at which the first chaperone-induced unfolding intermediate is located. (g) A scheme of the GR-LBD unfolding in the presence of co/chaperones. Reproduced with permission from Mandal et al., Proc. Natl. Acad. Sci. U. S. A. 114, 6040 (2017) and Moessmer et al., Proc. Natl. Acad. Sci. U. S. A. 119, e2119076119 (2022). Copyright 2022 National Academy of Sciences. Copyright 2017 National Academy of Sciences.75,80
In addition to studies focused on internal mechanics of the Hsp70 domains, truncated variants of multi-domain Hsp40 were also mechanically investigated.76 The full-length E. coli DnaJ (1–376) consists of four domains; domain II is a so-called zinc-binding domain. Atomic force microscopy (AFM) study of a truncated DnaJΔ107 was sandwiched between two protein L copies and stretched. Single-molecule experiments showed that Zinc fingers in this domain display unexpected mechanical lability (∼90 pN). The mechanics of the zinc finger is finely regulated by the interplay between zinc binding and disulfide bond formation. Furthermore, the study finds that the peptide substrate binding to DnaJ significantly increases the mechanical stability of domain I.
b. How the Hsp70 system modulates folding of the clients under load
Mashaghi et al.77 investigated the effect of Hsp70 binding to a tethered model substrate. Four copies of maltose-binding protein (4×MBP) were tethered in series in an optical trap and probed in the absence and presence of a chaperone. The unfolding of 4×MBP first produced a gradual unfolding of the C-terminal segments of all four MBP proteins, followed by distinct unfolding events of the four remaining core structures. Subsequently, the protein was relaxed to low forces and probed for refolding. In the absence of chaperones, stretching cycles revealed that 4×MBP misfolds into structures of different mechanical stabilities. Native-like core structures were found when the complete chaperone system DnaK/J/E + MgATP was added. However, misfolding still occurred, albeit with a substantial difference in the misfolding species and preferably mechanically weak misfolding substructures were found. Single-molecule force experiments were conducted on a single MBP under load to get insights into details of the chaperone–client interactions. Surprisingly, the authors found that, in the absence of the co-chaperones, DnaK + MgADP or +MgATP can bind to partially folded structures and stabilize them. The lid plays a crucial role in observing the stabilizing action of DnaK on the folded structures and suppressing aggregation. From these experiments on DnaK, a picture emerges of an Hsp70 functional repertoire that is broad and suggests that Hsp70 can also guide and organize late stages of folding by, for example, limiting inter-domain contacts. Since MBP is rather a model protein but not a natural substrate of Hsp70, it remains to be seen whether those interesting results will also hold for natural substrates.
The interaction between the eukaryotic Hsp70 chaperone, the so-called Bip chaperone and a model client—an archaeal protein MJ0366 was examined.78 Bip is an immunoglobulin binding protein involved in protein folding in the endoplasmic reticulum. MJ0366 is a hypothetical cell-expressed knotted protein from Methanocaldococcus jannaschii, containing one putative binding site for BiP, simplifying the analysis and interpretation. Another favorable property of the chosen client protein is a robust reversible unfolding/refolding behavior during constant velocity cycles. In the presence of 1 μM Bip+ATP, refolding of the client MJ0336 drops to ca. 70%. When 1 μM Bip + 2 mM ADP + 0.33 mM ATP is added, the refolding yield drops to 16.8%. Mechanical data indicate that chaperone BiP binds to the unfolded client and controls its folding. It is possible that the presence of the complete Hsp70 system would rescue the interaction between the chaperone and MJ0366, as observed in other studies.
An AFM study employing the complete DnaK/J/E chaperone system was conducted by Perales-Calvo et al.79 As a model client, ubiquitin was used. As in the previous study, the choice of the chaperone–client pair does not match the occurrence of their physiological interactions. Nevertheless, this study can be viewed as a model case study of chaperone–client interactions. For the experiments, a polyprotein was prepared composed of nine copies of ubiquitin. Mechanical folding of ubiquitin was probed using the force-quench assay, whereby an initial high-force 120 pN pulse unfolds the protein manifested by 20-nm steps, and after quenching the force for variable time intervals, the protein is stretched again at high force to probe successful refolding during the quench time. Successful refolding was measured in the absence and presence of DnaK/J/E chaperones. In the presence of 5 μM DnaJ, ubiquitin refolding efficiency drops 2.5-fold, from 75% (no DnaJ) to 30%, indicating the binding to unfolded ubiquitin chains. No DnaJ binding to the native client was observed, as expected.
Further experiments showed that DnaJ binds with high affinity to the mechanically extended ubiquitin and force-dependent binding. A plausible explanation is given with the help of molecular dynamics (MD) simulations of the DnaJ-bound fragment of ubiquitin corresponding to the putative binding site. For effective DnaJ binding, remodeling of the dihedral angles of the bound ubiquitin fragment is needed, which may account for the non-trivial force-dependence of the DnaJ binding to stretched ubiquitin chain. DnaJ alone decreases the client's refolding; however, other components of the DnaK chaperone system are also present in the cell and may help release this unproductive complex. A drastic drop in the refolding yield was observed when experiments were performed with 5 μM DnaK + MgADP, closed state with high affinity for unfolded clients, a drastic drop in refolding yield was observed (∼30%). Hence, DnaK-ADP and DnaJ were corrupting the refolding of the client. Only the complete DnaKJE + MgATP chaperone system was able to increase refolding efficiency of ubiquitin.
A study of Hsp70 interacting and unfolding one of its natural substrates, the glucocorticoid receptor (GR), was performed by Moessmer et al.80 GR is a steroid hormone receptor that, when activated, acts as a transcription factor regulating important signal cascades involved in inflammation.81,82 It is one of the most important drug targets. The activation of GR is tightly regulated by the Hsp70/40 and 90 chaperone systems, also involving numerous co-chaperones. In single-molecule mechanics experiments, the authors could show that hormone binding to the ligand-binding domain of GR [GR-LBD, Fig. 2(e)] is tightly linked to the opening and closing of a helix involving the first 33 residues of GR.59,80 In Fig. 2(e), passive mode experiments are shown where rapid opening and closing of this N-terminal helix can be observed (transition between purple and dark blue states). Those purple/dark blue phases are interrupted by long events colored in light blue where this helix remains open because the hormone has left its binding pocket. When the hormone rebinds, the fast opening/closing dynamics continue. A schematic of various states can be seen at the bottom of Fig. 2(e).
When Hsp70, Hsp40, and ATP are added to the solution, Moessmer et al.80 could show that as soon as the hormone has left the binding pocket for the first time [Fig. 2(f)], the chaperone system attacks the hormone structure and actively unfolds it in up to five consecutive steps. Each step is associated with a new hsp70 molecule binding to GR and inducing further unfolding upon Hsp40 stimulated ATP hydrolysis. This result provides direct evidence for the Hsp70/40 system acting as an unfoldase.
The mechanism through which Hsp70 can unfold the protein has been extensively debated. Some studies have provided evidence for Hsp70 binding and holding to the already unfolded portion of the substrate,83–85 thus decreasing the accessible conformational states for the folded protein leading to an entropic effect termed “entropic pulling,” which eventually unfolds the protein. Moessmer et al.,80 however, provided evidence that Hsp70 may also be capable of directly interacting with the folded core of GR and thus inducing unfolding [see the model in Fig. 2(g)].
2. GroEL system accelerates client folding by modulating its chain collapse
Chaperonin protein GroEL and its co-chaperonin GroES use ATP to fold proteins.86–90 GroEL forms an 800 kDa double toroid consisting of two heptameric rings of 57 kDa subunits91 (for structure, see Fig. 1). GroES forms a lid on the GroEL chamber, which can bind to either end of the GroEL complex. The lid is formed by assembling a heptamer of 10 kDa subunits.17 The binding of GroES capped a large cavity. A highly polar inner surface of the cavity provides a suitable environment, which supports the folding of the fully unfolded or partially folded substrate. Accelerated folding is attributed to the sterical confinement of the unfolded chain and a reduction in polypeptide chain entropy in the net negatively charged chaperonin cavity.92,93 GroEL-ES chaperone function can be approximated by an iterative annealing model whereby GroEL unfolds and refolds misfolded polypeptides in multiple cycles.94
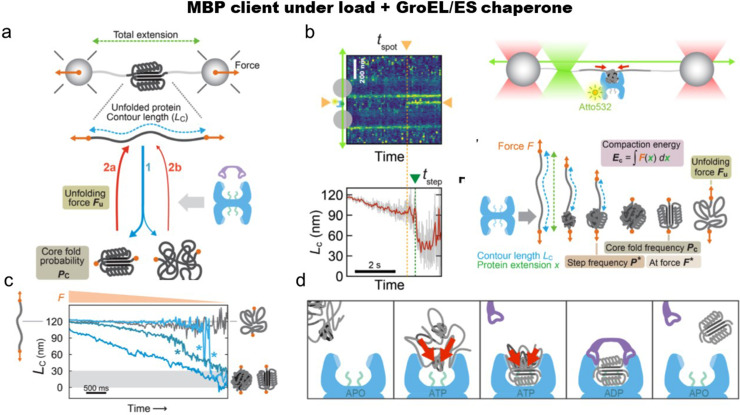
Naqvi et al.95 employed laser traps and single-molecule fluorescence to examine the effect of the GroEL-ES chaperone on MBP during its refolding reaction [Figs. 3(a)–3(d)]. First, they examined whether the folding of isolated MBPs is affected by GroEL-ES. MBP has been studied previously as a GroEL-ES client.21,30 To quantify the observed effects, they counted the cycles with complete folding of the MBP core [Fig. 3(a)] and determined the fraction. In the presence of GroEL/GroES and ATP, they found only a weak improvement in the complete folding. The authors then re-designed the assay using a less stable construct of MBP that folds more slowly than the wild-type. Such protein may resemble destabilized proteins. In addition, the authors put MBP under load at 2 pN, destabilizing protein further. In the apo-state of GroEL, the chaperone interacts with the unfolded chain and stabilizes it. To directly observe GroEL–substrate interactions, the authors used Atto532-labeled chaperone and lateral laser fluorescence scanning [Fig. 3(b)].
FIG. 3.
MBP as a model substrate for GroEL/GroES chaperone. (a) A scheme of optical tweezers experiments. Assay to find out the refolding efficiency of the MBP. (b) Experimental technically orthogonal single-molecule assay for watching MBP-GroEL complex in real time: laser traps are shown in red, scanning fluorescence excitation is shown in green. For fluorescence experiments, 15 nM Atto532-labeled GroEL + ADP was used in the assay. Time-dependent fluorescence scan during the force relaxation. A spot appearing at tspot corresponds to a single GroEL binding and, as shown below, time-dependent Lc. (c) Lc of MBP as a function of time and decreased force in the presence of GroEL and ADP. Here, one can see compaction of MBP is visible (blue traces). Stars * point out folding steps. No detectable compaction can be seen in the absence of GroEL (shown in gray). (d) Suggested effect of GroEL on protein substrate—driving polypeptide chain collapse and folding. Reproduced with permission from Naqvi et al., Sci. Adv. 8, eabl6293 (2022) Copyright 2022 Author(s) licensed under a Creative Commons Attribution (CC BY) License.95
Additionally, MBP in the presence of GroEL and ADP deviates from the expected worm-like chain model. This may indicate that the binding of GroEL collapses MBP [Figs. 3(c) and 3(d)]. Hence, in summary, the interaction of GroEL and MBP is twofold: first, the unfolded substrate is bound and immobilized, and second, the client is compacted by attractive forces. Interestingly, suggested GroEL-ES effects, such as steric confinement and misfolds unfolding do not assume collapse modulation of the unfolded substrate and hence this modulation presents a truly new mechanism of the chaperone action.
3. Single-molecule force studies of processive client translocation by ClpB disaggregase
Avellaneda et al.96,97 studied the effect of disaggregase ClpB, a member of the Hsp100 chaperone family, on the folding of single-molecule MBP. After mechanical MBP unfolding, the force was set to 5 and 10 pN preventing a spontaneous MBP refolding. For ATPase-activated Y503D ClpB variant+MgATP, the contraction was observed, which was interpreted as the result of processive translocation of the MBP chain by ClpB until the loss of the grip. After release, the applied force stretches the unfolded MBP, and a new ClpB translocation can be initiated. Alternative translocation models were tested by combining optical tweezers experiments with ClpB tracking at sub-wavelength resolution using single-molecule fluorescence imaging.
Interestingly, optical tweezers with fluorescence reveal ClpB translocation of both loop arms; hence, polypeptide loop extrusion is one possible mode of action. It might shed light on the disaggregation activity of Hsp100 since internal segments of aggregated proteins are targeted more readily and translocated as loops. The authors also explain how successfully folded client structures presented in cis and trans sides of ClpB can affect translocation dynamics in a looped topology.
4. Monitoring of the anti-aggregation activity of the Hsp33 chaperone
Hsp33 is the zinc-dependent, redox-regulated chaperone, which binds tightly to unfolding proteins under stress conditions with subsequent release to chaperone “foldases” when non-stress conditions resume (for structure, see Fig. 1). Hsp33 can toggle between reduced and oxidized forms; chaperone activity is activated under oxidizing conditions.98 Moayed et al.99 studied aggregation behavior at the molecular level of individual protein constructs composed of 4 MBP and analyzed the effects of Hsp33 in the folding and unfolding of MBPs. Upon unfolding of 4 MBP construct in the absence of Hsp33, refolding at zero force was inefficient, and often, only one of the four MBP cores was refolded. In most traces, the authors observed distinct length changes larger than for one MBP core and unfolding forces higher than for native monomeric MBP. These findings indicate the need to disrupt non-native aggregated structures consisting of multiple MBPs. For single-molecule experiments with a chaperone, a constitutively active Hsp33 mutant Y12E was used because the conditions when the wild-type chaperone is active are not compatible with the assay. In the presence of a chaperone, the occurrence of partially folded or aggregated structures decreased ca. fivefold. In the next experiments, the effects of Hsp33 on a single MBP monomer were evaluated as well; they found that Hsp33 suppresses folding in single isolated substrates. A statistical mechanical model was developed to describe the behavior of 4×MBP in the presence of a chaperone.
5. Folding of Hsp90 chaperone
While the mechanism of the substrate interaction and chaperoning is already very well understood for the Hsp60 and Hsp70 chaperone systems, it is much less clear how the large dimeric chaperone Hsp90 and its co-chaperones achieve their function. In brief, Hsp90 consists of three domains: the N-terminal domain containing the ATPase site, a middle domain (M domain) involved in client-binding, and a C-terminal domain leading to dimerization. Jahn et al.100 studied the structural mechanics and folding of this large protein machine. They found that the N and M domain dock dynamically to each other through a so-called “charged-linker” element.101 This charged linker element can have different mechanical stabilities in different Hsp90 homologs.102 The application of higher forces leads to the consecutive unfolding of C, N, and M domains. While the individual domains can readily unfold, the authors found that refolding of the full Hsp90 is substantially hampered by non-native aggregates forming from unfolded stretches across different domains. The degree of misfolding was shown to vary in Hsp90 isoforms.103 Applying a small mechanical force can keep the aggregation-prone sequences apart, thus speeding up successful folding. Tych et al.104 found that the stability of the C-terminal association of the Hsp90 dimer is ATP-dependent, despite the C-terminal dimerization interface being far from the ATP binding site.
C. Studies of other chaperone-client pairs
1. Chaperones and their roles in disulfide bond formation of the clients
Using magnetic tweezers-based force spectroscopy, chaperone activity of PDI and DsbA on protein clients was examined.105,106 For the mechanical unfolding of proteins in the presence of oxidoreductases, a model titin immunoglobulin domain, I27C32–C75, was used that contains a single disulfide bond. The unfolding of the disulfide bond I27C32–C75 domain has a characteristic extension of 11 nm, while upon reduction of the disulfide bond, an additional 14 nm can be detected. Eckels et al. observed that a single disulfide bond shifts titin folding to higher forces. The formation of disulfide bonds was followed by a refolding assay of polyproteins containing eight I27C32–C75 domains. After the complete unfolding of all domains, the force was reduced to 5.2 pN for 150 s (to enable folding), followed by a subsequent force jump to 77 pN. This assay allows for the counting of re-oxidized I27C32–C75 domains. In the presence of a TCEP reducer, only one domain was refolded. In contrast, seven domains were refolded in the presence of DsbA, and six contained disulfide bonds. Next, the redox-dependent interaction of the DsbA chaperones and cysteine-free substrate was examined. The authors found that oxidized DsbA is a much more effective chaperone for the model substrate and the binding of peptide inhibitor blocks the chaperone activities of oxidized DsbA. A concept was suggested that the DsbA enhanced folding of domains on the periplasmic site of the Sec pore generates a force that transfers its strain to the polypeptide in the translocon tunnel to any portion still in the cytosol. Chaperone-assisted folding on the periplasmic side of the membrane would ease the protein translocation. Using a different oxidoreductase called protein disulfide isomerase, PDI, Eckels et al.106 showed that this enzyme can reversibly induce disulfide formation at forces as high as 5 pN and possesses additional chaperone activity to assist the folding.
Bacteria use the pili type to attach to cells; hence, pilus integrity is essential. The pilus consists of four different subunit types, FimA-FimF-FimG-FimH. To assemble a pilus, the subunits oxidatively fold, which can be catalyzed by the oxidoreductase DsbA. This enzyme encounters the subunits in the periplasm as they are secreted in an extended state by the SecYEG pathway. Alonso-Caballero et al.107 monitored the oxidative folding of a single Fim domain assisted by periplasmic FimC and the oxidoreductase DsbA. They found that pilus domains bear high mechanical stability following a hierarchy by which domains close to the tip are weaker than those close to or at the pilus rod. During folding, this remarkable stability is achieved by the intervention of DsbA, which forms strategic disulfide bonds and serves as a chaperone assisting the folding of the domains.
2. Client folding on ribosomes and the chaperone mechanism of trigger factor (TF)
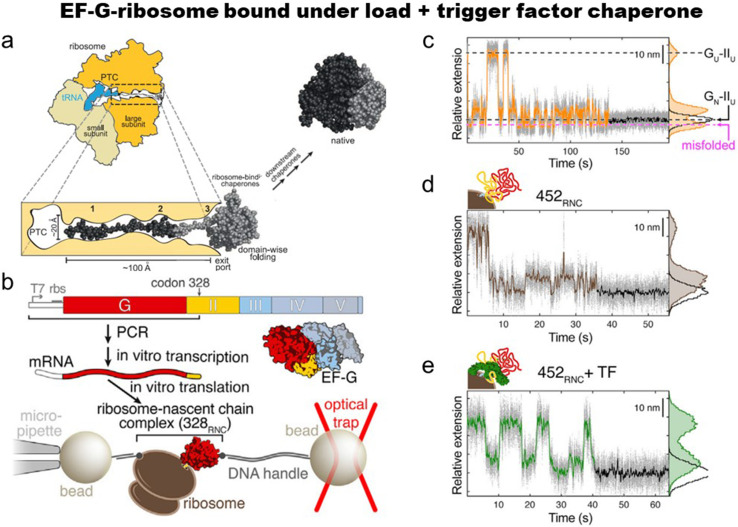
Liu et al.108 studied how the ribosome [Fig. 4(a)] and trigger factor (for structure, Fig. 1) affect the folding of elongation factor G, EF-G [Figs. 4(b)–4(e)]. In their carefully designed experiments, the authors produce stalled ribosome-nascent chain complexes (RNCs) of EF-G. Such a molecular system enables the examination of co-translational events at the stalled ribosome. The experiment is designed so that the translation stops at the positions of 328 of the EF-G coding sequence (328RNC). The entire N-terminal G-domain (amino acids 1–293 of EF-G) is present, whereas the following 35 residues (amino acids 294–328) of domain II are within the exit tunnel in the large ribosomal subunit [Fig. 4(b)]. Surprisingly, when longer nascent chains were produced, the folding was slower. The adverse effect of a longer protein chain was interpreted as the result of domain-domain interactions. Further analysis of G domain refolding, the authors found that such adverse intramolecular domain-domain reactions can be relieved by the ribosome and TF [Fig. 4(e)]. In summary, the study shows that the TF chaperone (1) helps to reduce unproductive domain-domain interactions and (2) protects the folded G-domain by the unfolded domain II.
FIG. 4.
EF-G folding on ribosomes. (a) The polypeptide exit tunnel of the large ribosomal subunit is magnified. (b) Experimental scheme for EF-G folding on the ribosome. For optical tweezer experiments, ribosome-protein chain complexes containing mRNAs lacking stop-codon are connected by two polystyrene beads. (c) G-domain folding at 3.5 pN: 1 kHz data (gray dots) and 10 Hz averaged data (line). Shown are states before and after the folding (black dashed lines) as well as misfolded state (magenta dashed line). (d) Refolding transitions 452-RNC without and (e) with trigger factor. The population of the compact misfolded species is reduced, as apparent from the extension-time trace and the extension histogram. (a) Reproduced with permission from Balchin et al., Science 353, aac4354 (2016). Copyright 2016 The American Association for the Advancement of Science.10 (b)–(e) Reproduced with permission from Liu et al., Mol. Cell 74, 310 (2019). Copyright 2019 Elsevier.108
The importance of the ribosome–client interactions was also highlighted in another study of folding the small, 28-residue long zinc finger called ADR1a domain.109 By combining optical tweezers with single-molecule FRET and molecular dynamics simulations, ADR1a folding was investigated at different locations of the ribosomal tunnel. The tunnel accelerates folding and stabilizes the folded state.
A single-molecule magnetic tweezers study by Haldar et al. examined the force-dependent folding dynamics of protein L in the presence of a trigger factor.110 Here, the trigger factor prominently increases the probability of folding against force and accelerates the refolding kinetics. Trigger factor as a chaperone becomes less efficient as forces increase. The authors proposed that the trigger factor can work as foldase under force, a mechanism that could be physiologically relevant.
In a theoretical study, all-atom MD simulations were conducted to provide insights into the chaperone function of the trigger factor, TF.111 The authors suggest that the tips of the fingerlike tentacles of TF play a vital role in the early interactions with unfolded chains and/or partially folded structures. When bound to TF, unfolded clients are kinetically trapped and reduce transient, non-native intramolecular contacts. Mechanical flexibility allows TF to hold partially folded structures with two tips and to stabilize them by wrapping around its appendages.
3. Studies of SNARE chaperones
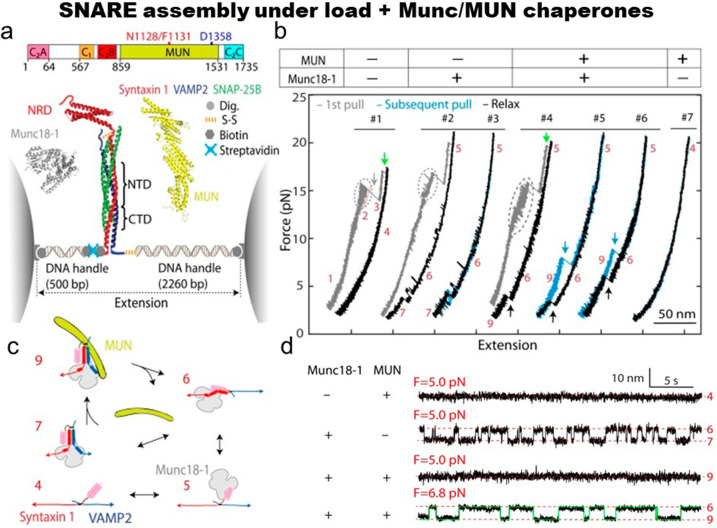
Synaptic vesicle fusion plays an essential role in neurotransmission.114 The fusion involves several proteins such as membrane-anchored SNARE proteins, syntaxin 1, and SNAP-25 on the plasma membrane and VAMP2 (or synaptobrevin 2) on the vesicle membrane and at least five regulatory proteins, Munc13–1, Munc18–1, synaptotagmin, complexin, and N-ethylmaleimide sensitive factor (NSF). SNARE proteins consist of 60 aa long SNARE motifs, which are intrinsically disordered in solution and, hence, coupled folding and assembly of the four SNARE motifs in the three SNAREs into a four-helix bundle pull their associated membranes into proximity and induce a membrane fusion.115 The Zhang group112,113 used single-molecule force spectroscopy and found that the SM protein Munc18–1 catalyzes step-by-step zippering of three synaptic SNAREs (syntaxin, VAMP2, and SNAP-25) into a four-helix bundle [Figs. 5(a)–5(d)]. The formation of an intermediate template complex in which Munc18–1 binds to the load-free N-terminal regions of the SNARE motifs of syntaxin and VAMP2, while keeping their C-terminal regions separated. SNAP-25 binds efficiently only when Munc18–1 is presented in the ternary complex of Munc18–1 • syntaxin 1•VAMP-2 and it induces a full SNARE zippering [Fig. 5(b)]. In the absence of SNAP-25B, the full SNARE assemblies were rare. Munc18–1 inhibits spontaneous, non-templated SNARE complex formation by suppressing the formation of the complex intermediate. In addition, they found that the NRD of syntaxin is stabilizing the template complex. In another study, the same group discovered that the MUN domain of Munc13–1 stabilizes the template complex [Fig. 5(d)]. The MUN-bound template complex enhances SNAP-25 binding to the templated SNAREs and subsequent full SNARE assembly.113
FIG. 5.
Mechanics of the SNARE assembly in the presence of chaperones. (a) Schematic diagram of Munc1-13 and the optical tweezers setup. A single SNARE complex was pulled from the C termini of syntaxin 1 A (red) and VAMP2 (blue), while Munc18-1 and the MUN domain of Munc13-1 were added to the solution. SNARE proteins were cross-linked via a disulfide bond. The syntaxin 1 A molecule contains the N-terminal regulatory domain (NRD). (b) Force-extension curves in the presence (+) or absence (−) of chaperones (color codes: gray for pulling the initial purified SNARE complex, cyan for subsequent pulls, and black for relaxations). The state numbers indicate states at different stages. (c) Schematic diagrams of different SNARE folding and protein binding states: 4, fully unfolded SNARE motifs; 5, unfolded SNARE motifs with Munc18-1. (d) Time trajectories of SNARE extensions at indicated constant mean forces in the absence or presence of 1 μM Munc18-1 or 1 μM MUN domain. Reproduced with permission from Shu et al., Proc. Natl. Acad. Sci. U. S. A. 117, 1036 (2020). Copyright 2019 National Academy of Sciences.113
D. Chaperones for membrane protein folding
The picture of how chaperones assist in the folding of membrane proteins has emerged in the past years, investigated mainly using atomic force microscopy experiments in the group of Daniel Mueller.116–119
The chaperone-assisted folding of single ferric hydroxamate uptake receptors (FhuAs) in E. coli lipid membranes was examined using AFM and NMR spectroscopy.119 They observed that, after partial unfolding, unfolded β-barrels remained stably in the membrane; however, in the absence of chaperones, refolding to the native state did not occur; instead, non-native, misfolded structures were detected. In fact, FhuA misfolded with a high probability (60%), remained unfolded in 33% of the events, and only 7% showed native β-hairpins recorded after a refolding time of 1 s. In the presence of the natural periplasmic holdase chaperone SurA, refolding to the native FhuA occurred due to the successful reinsertion of single β-hairpins into the lipid membrane. Skp decreased the probability of misfolding events to 12%, 73% of the FhuA receptors remained unfolded, and 15% folded β-hairpins. SurA decreased the FhuA misfolding to 14%, 46% of the FhuA receptors remained unfolded, and the folding of native β-hairpins increased to 40%. Adding both SurA and Skp to the refolding assay resulted in 11% of the FhuA showing correctly folded β-hairpins, 8% misfolded form, and 81% unfolded substrates. In this assay, the effect of Skp thus dominated that of SurA. In summary, the authors concluded that chaperones SurA and Skp prevent FhuA from misfolding and that SurA facilitates the insertion of β-hairpins into the lipid membrane.
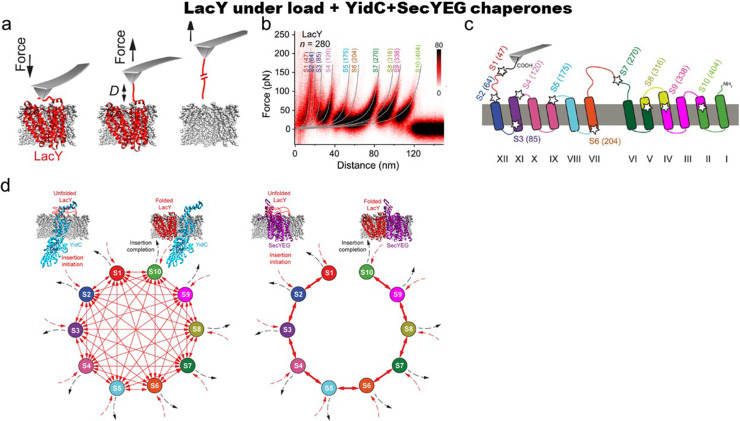
In another study, single-molecule mechanical experiments were conducted with reconstituted LacY into phospholipid membranes that compositionally mimics E. coli membrane.116 Under these conditions, LacY assumes a native conformation that is functionally active. In this conformation, both termini are placed at the cytoplasmic membrane.120 Pulling experiments revealed a “fingerprint” for native LacY as the unfolding of secondary structures was demonstrated by characteristic force peaks [Figs. 6(a)–6(c)]. After partial, transient unfolding, LacY can refold efficiently as indicated by the native unfolding pattern.
FIG. 6.
Folding of membrane protein in the presence of chaperones. (a) Mechanical unfolding of native LacY. Schematics of the unfolding of a single LacY from the phospholipid membrane. LacY unfolds stepwise until wholly extracted from the membrane.(b) Density plot of 280 superimposed LacY force-distance curves. Mean contour lengths are given at the top of each WLC curve to define the ending of the previously unfolded structural segment and the beginning of the next segment to be unfolded. (c) Structural segments S1 to S10 mapped to the secondary structure of LacY as unfolded beginning from the C terminus. (d) SecYEG and YidC inset and fold the membrane protein LacY along different pathways. (a)–(c) Reproduced with permission from Serdiuk et al., Nano Lett. 17, 4478 (2017). Copyright 2017 American Chemical Society. (d) Reproduced with permission from Serdiuk et al., Sci. Adv. 5, eaau6824 (2019). Copyright 2019 Author(s), licensed under a Creative Commons Attribution (CC BY) License.
Folding of LacY and its insertion into the membrane was characterized by the pull-and-paste single-molecule method:117 first, they picked up the elongated C terminus, then unfolded and extracted from the membrane a large portion of LacY consisting of the C terminus, ten transmembrane α-helices, and the intervening loops, leaving the first two N-terminal transmembrane α-helices in the membrane.
By placing unfolded protein close to membrane, they allowed LacY to insert and fold for few seconds, and, in the final step, they probed LacY by pulling it out from the membrane. After two seconds of refolding, the authors find that 6% LacY stayed unfolded, and roughly the half of refolded LacY exhibited unfolding forces, which are different compared to the native pattern. The other half showed force peaks corresponding to the native fingerprint and were classified as having folded some of the native structural segments. Although LacY refolded individual structures into the membrane, the full folding was not reached. To fold correctly in the membrane, LacY may need the help of other proteins such as YidC.121,122 Using AFM assays, the authors found that YidC prevents LacY from misfolding by stabilizing the unfolded state. From there, LacY inserts substructures into the membrane in a stepwise manner until folding is completed [Fig. 6(d)]. During stepwise insertion, YidC and the membrane together stabilize the transient folds. The sequence of the insertion events seems random, indicating heterogenous pathways toward the native structure. The folding of LacY was examined further in the presence of YidC insertase and SecYEG translocon. They found that both YidC and SecYEG initiate folding of the completely unfolded polypeptide by inserting a single structural segment. YidC then inserts the remaining substructures in random order, whereas SecYEG inserts them sequentially [Fig. 6(e)]. Each insertion process proceeds until LacY folding is complete. When YidC and SecYEG cooperate, the folding pathway of the membrane protein is dominated by the translocase.
IV. ADVANCES IN SINGLE-MOLECULE FORCE SPECTROSCOPY OF PROTEINS
In the past years, significant advances in single-molecule mechanical studies will shape the development of chaperone-client studies. For single-molecule force studies, several bottlenecks exist (1) chemical coupling of proteins, (2) DNA handles, (3) better time resolution, (4) modeling of experiments, and (5) automation and high-throughput experiments and analysis.
Briefly, in laser optical tweezers experiments using differential detection, the readout is based on monitoring the position of the functionalized beads. These beads are interconnected by a single DNA–protein–DNA tether. The combination of DNA–protein is effective for several reasons. First, proteins alone often stick and attach nonspecifically to surfaces, which affects their physico-chemical properties. Second, using long DNA handles it is possible to probe protein far from the beads' surface and laser foci, which may produce damaging oxygen radicals. Several different strategies have been developed for protein-DNA covalent linking. In the first approach, a single cysteine residue was introduced in a protein coupled with thiol- or maleimide- containing single-stranded oligonucleotides.123,124 Oligonucleotides were then hybridized with an overhang presented in longer DNA handles.125 Other chemical couplings have been developed (reviewed, for example, Ref. 126), including click chemistry, unnatural amino acids, and others.127–130 Using different coupling strategies enables the attachment of linkers of different mechanical elastic properties, which may affect the quality of the signal. For example, the mechanical stiffness of DNA handles is critical for the signal-to-noise ratio of the single-molecule measurement, and stiffer handles can improve the measured signal.131 In addition to the signal-to-noise ratio, a high temporal resolution can yield insights into microscopic details of ultrafast processes and deconvolute a complex free energy folding landscape.132–136
In addition, temperature dependences in so-called calorimetric force experiments can determine the heat capacity of the conformational changes, which complete the thermodynamic description.137 Along with the analysis of single-molecule processes, conceptual frameworks are important to understanding underlying physical processes, as highlighted by the application of Ising-like models for folding consensus-designed superhelical arrays of short helix-turn-helix motifs.138 Force-jump experiments can yield hidden information about different cis/trans proline isomeric states of proteins.60 While single-molecule mechanical experiments report a 1D projection of pulling coordinate, new information can be gained in parallel by using orthogonal fluorescence detection.95,97,109,139 The folding of membrane proteins can be further extended by using nanodisc as a membrane surrogate, making access to optical and magnetic tweezer studies.
Single-molecule force spectroscopy can implement microfluidics with several laminar flow channels, enabling watching a single molecule under several buffer conditions and programmed order of events, e.g., the presence of different chaperones in various sequences of additions. Another potential weak point of single-molecule approaches is the low throughput of the experiment, and often only a few tens-hundreds of proteins can be investigated in a reasonable time frame. Using multiplexing and parallel experiments, e.g., magnetic tweezers or centrifugal force microscopy, a significant number of single-molecule tethers can be examined simultaneously. Multiplexing and high-throughput single-molecule experiments demand the development of full automation of the detection and analysis of experimental data. Recently, machine learning models have started to be used for categorization and approach the fully automated data analysis.140–142
V. OUTLOOK
Based on current achievements, we foresee several goals (i) to increase the investigated chaperone repertoire, (ii) to scrutinize complex dynamics of multiple chaperone–substrate interactions during different stages of cycles, (iii) to develop assay and seamless molecular tethering strategies for complex multimeric and cysteine-rich chaperones, and (iv) to combine several detection techniques with microfluidics to examine substrate passage from chaperone-co-chaperone and/or chaperone–chaperone hand-over mechanisms of supramolecular protein assemblies.
Current single-molecule force studies are conducted using well-known canonical chaperones. Further extension toward different isoforms and less-studied chaperone systems will greatly benefit our understanding of internal chaperone mechanics and how they function and move during their functional cycles. We anticipate that such studies can also help identify minimal functional chaperone systems. Owing to the intrinsic complexity of chaperone–client interactions, more insights into chaperone–substrate dynamics are expected from mechanical studies, including the question of whether chaperones can randomly diffuse through the unfolded chain and bind transiently to several binding motifs. The realm of complex multimeric and cysteine-rich chaperones has remained largely unexplored, primarily due to the complexity of molecular constructs and the high reactivity of cysteine residues. We expect that assays using genetically concatenated proteins with embedded suitable flexible linkers can provide a reasonable strategy for examining multimeric chaperones.
Further progress in the development of orthogonal labeling strategies may further expand the toolkit to achieve stable tethering between molecular systems and microscopic beads. In the cell, client proteins are often handed over between different chaperones and co-chaperones. It is challenging to understand how these dynamical supramolecular complexes communicate and how these complexes are regulated. Such complex many-body interactions require approaches utilizing a combination of detection techniques such as fluorescence and force and the necessity to control the external conditions, which can be, in principle, achieved by measurements in multiple laminar flow stream channels inside of the microfluidic device. We envision that understanding the chaperome will greatly benefit from the proposed enhancements cutting-edge single molecule methods.
ACKNOWLEDGMENTS
This work was supported by the German Research Foundation, Sonderforschungsbereich 1035, Projektnummer 201302640, Project A5 (to M.R.). G.Z. was supported by the Research Grant from the Grant Provided by Slovak Research and Development Agency (Grant No. APVV-18–0285), the Slovak Grant Agency VEGA No. 1/0024/22, KEGA No. 005UPJŠ-4/2021, and the Project BioPickmol, ITMS2014+: 313011AUW6 supported by the Operational Programme Integrated Infrastructure, funded by the ERDF. The collaboration between M.R./G.Z. was supported by European Union's Horizon 2020 Research and Innovation Programme under Grant Agreement No. 952333, Project CasProt (Fostering high scientific quality in protein science in Eastern Slovakia).
AUTHOR CONTRIBUTIONS
Conflict of Interest
The authors have no conflicts to disclose.
Author Contributions
Matthias Rief: Conceptualization (equal); Funding acquisition (equal); Writing – original draft (equal); Writing – review & editing (equal). Gabriel Žoldák: Conceptualization (equal); Funding acquisition (equal); Writing – original draft (equal); Writing – review & editing (equal).
DATA AVAILABILITY
Data sharing is not applicable to this article as no new data were created or analyzed in this study.
References
- 1. McCauley M. J. et al. , “ Targeted binding of nucleocapsid protein transforms the folding landscape of HIV-1 TAR RNA,” Proc. Natl. Acad. Sci. U. S. A. 112(44), 13555–13560 (2015). 10.1073/pnas.1510100112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Naufer M. N. and Williams M. C., “ Characterizing complex nucleic acid interactions of LINE1 ORF1p by single molecule force spectroscopy,” Methods Mol. Biol. 2106, 283–297 (2020). 10.1007/978-1-0716-0231-7 [DOI] [PubMed] [Google Scholar]
- 3. Gupta A. N. et al. , “ Pharmacological chaperone reshapes the energy landscape for folding and aggregation of the prion protein,” Nat. Commun. 7, 12058 (2016). 10.1038/ncomms12058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Albanese V. et al. , “ Systems analyses reveal two chaperone networks with distinct functions in eukaryotic cells,” Cell 124(1), 75–88 (2006). 10.1016/j.cell.2005.11.039 [DOI] [PubMed] [Google Scholar]
- 5. Hartl F. U., Bracher A., and Hayer-Hartl M., “ Molecular chaperones in protein folding and proteostasis,” Nature 475(7356), 324–332 (2011). 10.1038/nature10317 [DOI] [PubMed] [Google Scholar]
- 6. Balch W. E. et al. , “ Adapting proteostasis for disease intervention,” Science 319(5865), 916–919 (2008). 10.1126/science.1141448 [DOI] [PubMed] [Google Scholar]
- 7. Pinchbeck B. J. et al. , “ A dual functional redox enzyme maturation protein for respiratory and assimilatory nitrate reductases in bacteria,” Mol. Microbiol. 111(6), 1592–1603 (2019). 10.1111/mmi.14239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Calloni G. et al. , “ DnaK functions as a central hub in the E. coli chaperone network,” Cell Rep. 1(3), 251–264 (2012). 10.1016/j.celrep.2011.12.007 [DOI] [PubMed] [Google Scholar]
- 9. Brehme M. et al. , “ A chaperome subnetwork safeguards proteostasis in aging and neurodegenerative disease,” Cell Rep. 9(3), 1135–1150 (2014). 10.1016/j.celrep.2014.09.042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Balchin D., Hayer-Hartl M., and Hartl F. U., “ In vivo aspects of protein folding and quality control,” Science 353(6294), aac4354 (2016). 10.1126/science.aac4354 [DOI] [PubMed] [Google Scholar]
- 11. Bascos N. A. D. and Landry S. J., “ A history of molecular chaperone structures in the protein data bank,” Int. J. Mol. Sci. 20(24), 6195 (2019). 10.3390/ijms20246195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Ashburner M. et al. , “ Gene ontology: Tool for the unification of biology. The gene ontology consortium,” Nat. Genet. 25(1), 25–29 (2000). 10.1038/75556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Bertelsen E. B. et al. , “ Solution conformation of wild-type E. coli Hsp70 (DnaK) chaperone complexed with ADP and substrate,” Proc. Natl. Acad. Sci. U. S. A. 106(21), 8471–8476 (2009). 10.1073/pnas.0903503106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kityk R. et al. , “ Structure and dynamics of the ATP-bound open conformation of Hsp70 chaperones,” Mol. Cell 48(6), 863–874 (2012). 10.1016/j.molcel.2012.09.023 [DOI] [PubMed] [Google Scholar]
- 15. Ferbitz L. et al. , “ Trigger factor in complex with the ribosome forms a molecular cradle for nascent proteins,” Nature 431(7008), 590–596 (2004). 10.1038/nature02899 [DOI] [PubMed] [Google Scholar]
- 16. Janda I. et al. , “ The crystal structure of the reduced, Zn2+-bound form of the B. subtilis Hsp33 chaperone and its implications for the activation mechanism,” Structure 12(10), 1901–1907 (2004). 10.1016/j.str.2004.08.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Xu Z., Horwich A. L., and Sigler P. B., “ The crystal structure of the asymmetric GroEL-GroES-(ADP)7 chaperonin complex,” Nature 388(6644), 741–750 (1997). 10.1038/41944 [DOI] [PubMed] [Google Scholar]
- 18. Wruck F. et al. , “ Protein folding mediated by trigger factor and Hsp70: New insights from single-molecule approaches,” J. Mol. Biol. 430(4), 438–449 (2018). 10.1016/j.jmb.2017.09.004 [DOI] [PubMed] [Google Scholar]
- 19. Choudhary D. et al. , “ Studying heat shock proteins through single-molecule mechanical manipulation,” Cell Stress Chaperones 25(4), 615–628 (2020). 10.1007/s12192-020-01096-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Thoma J., Sapra K. T., and Muller D. J., “ Single-molecule force spectroscopy of transmembrane beta-barrel proteins,” Annu. Rev. Anal. Chem. 11(1), 375–395 (2018). 10.1146/annurev-anchem-061417-010055 [DOI] [PubMed] [Google Scholar]
- 21. Avellaneda M. J. et al. , “ The chaperone toolbox at the single-molecule level: From clamping to confining,” Protein Sci. 26(7), 1291–1302 (2017). 10.1002/pro.3161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Banerjee S. et al. , “ Cutting-edge single-molecule technologies unveil new mechanics in cellular biochemistry,” Annu. Rev. Biophys. 50, 419–445 (2021). 10.1146/annurev-biophys-090420-083836 [DOI] [PubMed] [Google Scholar]
- 23. Bustamante C. et al. , “ Single-molecule studies of protein folding with optical tweezers,” Annu. Rev. Biochem. 89, 443–470 (2020). 10.1146/annurev-biochem-013118-111442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Schonfelder J. et al. , “ The life of proteins under mechanical force,” Chem. Soc. Rev. 47(10), 3558–3573 (2018). 10.1039/C7CS00820A [DOI] [PubMed] [Google Scholar]
- 25. Petrosyan R., Narayan A., and Woodside M. T., “ Single-molecule force spectroscopy of protein folding,” J. Mol. Biol. 433(20), 167207 (2021). 10.1016/j.jmb.2021.167207 [DOI] [PubMed] [Google Scholar]
- 26. Ritchie D. B. and Woodside M. T., “ Probing the structural dynamics of proteins and nucleic acids with optical tweezers,” Curr. Opin. Struct. Biol. 34, 43–51 (2015). 10.1016/j.sbi.2015.06.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Heidarsson P. O. and Cecconi C., “ From folding to function: Complex macromolecular reactions unraveled one-by-one with optical tweezers,” Essays Biochem. 65(1), 129–142 (2021). 10.1042/EBC20200024 [DOI] [PubMed] [Google Scholar]
- 28. Bustamante C. J. et al. , “ Optical tweezers in single-molecule biophysics,” Nat. Rev. Methods Primers 1, 25 (2021). 10.1038/s43586-021-00021-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Zoldak G. and Rief M., “ Force as a single molecule probe of multidimensional protein energy landscapes,” Curr. Opin. Struct. Biol. 23(1), 48–57 (2013). 10.1016/j.sbi.2012.11.007 [DOI] [PubMed] [Google Scholar]
- 30. Rief M. et al. , “ Reversible unfolding of individual titin immunoglobulin domains by AFM,” Science 276(5315), 1109–1112 (1997). 10.1126/science.276.5315.1109 [DOI] [PubMed] [Google Scholar]
- 31. Milles L. F. et al. , “ Molecular mechanism of extreme mechanostability in a pathogen adhesin,” Science 359(6383), 1527–1533 (2018). 10.1126/science.aar2094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Grandbois M. et al. , “ How strong is a covalent bond?,” Science 283(5408), 1727–1730 (1999). 10.1126/science.283.5408.1727 [DOI] [PubMed] [Google Scholar]
- 33. Svoboda K. et al. , “ Direct observation of kinesin stepping by optical trapping interferometry,” Nature 365(6448), 721–727 (1993). 10.1038/365721a0 [DOI] [PubMed] [Google Scholar]
- 34. Kellermayer M. S. et al. , “ Folding-unfolding transitions in single titin molecules characterized with laser tweezers,” Science 276(5315), 1112–1116 (1997). 10.1126/science.276.5315.1112 [DOI] [PubMed] [Google Scholar]
- 35. Tskhovrebova L. et al. , “ Elasticity and unfolding of single molecules of the giant muscle protein titin,” Nature 387(6630), 308–312 (1997). 10.1038/387308a0 [DOI] [PubMed] [Google Scholar]
- 36. Zoldak G. et al. , “ Ultrafast folding kinetics and cooperativity of villin headpiece in single-molecule force spectroscopy,” Proc. Natl. Acad. Sci. U. S. A. 110(45), 18156–18161 (2013). 10.1073/pnas.1311495110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Hoffer N. Q. and Woodside M. T., “ Probing microscopic conformational dynamics in folding reactions by measuring transition paths,” Curr. Opin. Chem. Biol. 53, 68–74 (2019). 10.1016/j.cbpa.2019.07.006 [DOI] [PubMed] [Google Scholar]
- 38. Pelz B. et al. , “ Subnanometre enzyme mechanics probed by single-molecule force spectroscopy,” Nat. Commun. 7, 10848 (2016). 10.1038/ncomms10848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Kriegel F., Ermann N., and Lipfert J., “ Probing the mechanical properties, conformational changes, and interactions of nucleic acids with magnetic tweezers,” J. Struct. Biol. 197(1), 26–36 (2017). 10.1016/j.jsb.2016.06.022 [DOI] [PubMed] [Google Scholar]
- 40. Tapia-Rojo R., Eckels E. C., and Fernandez J. M., “ Ephemeral states in protein folding under force captured with a magnetic tweezers design,” Proc. Natl. Acad. Sci. U. S. A. 116(16), 7873–7878 (2019). 10.1073/pnas.1821284116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Popa I. et al. , “ A HaloTag anchored ruler for week-long studies of protein dynamics,” J. Am. Chem. Soc. 138(33), 10546–10553 (2016). 10.1021/jacs.6b05429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Agarwal R. and Duderstadt K. E., “ Multiplex flow magnetic tweezers reveal rare enzymatic events with single molecule precision,” Nat. Commun. 11(1), 4714 (2020). 10.1038/s41467-020-18456-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Lof A. et al. , “ Multiplexed protein force spectroscopy reveals equilibrium protein folding dynamics and the low-force response of von Willebrand factor,” Proc. Natl. Acad. Sci. U. S. A. 116(38), 18798–18807 (2019). 10.1073/pnas.1901794116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Sitters G. et al. , “ Acoustic force spectroscopy,” Nat. Methods 12(1), 47–50 (2015). 10.1038/nmeth.3183 [DOI] [PubMed] [Google Scholar]
- 45. Kamsma D. et al. , “ Tuning the music: Acoustic force spectroscopy (AFS) 2.0,” Methods 105, 26–33 (2016). 10.1016/j.ymeth.2016.05.002 [DOI] [PubMed] [Google Scholar]
- 46. Yang D. and Wong W. P., “ Repurposing a benchtop centrifuge for high-throughput single-molecule force spectroscopy,” Methods Mol. Biol. 1665, 353–366 (2018). 10.1007/978-1-4939-7271-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Abraham Punnoose J. et al. , “ Wi-Fi live-streaming centrifuge force microscope for benchtop single-molecule experiments,” Biophys. J. 119(11), 2231–2239 (2020). 10.1016/j.bpj.2020.10.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. LeFevre T. B. et al. , “ Measuring colloid-surface interaction forces in parallel using fluorescence centrifuge force microscopy,” Soft Matter 17(26), 6326–6336 (2021). 10.1039/D1SM00461A [DOI] [PubMed] [Google Scholar]
- 49. Hoang T., Patel D. S., and Halvorsen K., “ A wireless centrifuge force microscope (CFM) enables multiplexed single-molecule experiments in a commercial centrifuge,” Rev. Sci. Instrum. 87(8), 083705 (2016). 10.1063/1.4961477 [DOI] [PubMed] [Google Scholar]
- 50. Puchner E. M. et al. , “ Comparing proteins by their unfolding pattern,” Biophys. J. 95(1), 426–434 (2008). 10.1529/biophysj.108.129999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Stigler J. and Rief M., “ Hidden Markov analysis of trajectories in single-molecule experiments and the effects of missed events,” Chemphyschem 13(4), 1079–1086 (2012). 10.1002/cphc.201100814 [DOI] [PubMed] [Google Scholar]
- 52. Stigler J. et al. , “ The complex folding network of single calmodulin molecules,” Science 334(6055), 512–516 (2011). 10.1126/science.1207598 [DOI] [PubMed] [Google Scholar]
- 53. Gebhardt J. C., Bornschlogl T., and Rief M., “ Full distance-resolved folding energy landscape of one single protein molecule,” Proc. Natl. Acad. Sci. U. S. A. 107(5), 2013–2018 (2010). 10.1073/pnas.0909854107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Ramm B. et al. , “ Sequence-resolved free energy profiles of stress-bearing vimentin intermediate filaments,” Proc. Natl. Acad. Sci. U. S. A. 111(31), 11359–11364 (2014). 10.1073/pnas.1403122111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Woodside M. T. et al. , “ Direct measurement of the full, sequence-dependent folding landscape of a nucleic acid,” Science 314(5801), 1001–1004 (2006). 10.1126/science.1133601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Hinczewski M. et al. , “ From mechanical folding trajectories to intrinsic energy landscapes of biopolymers,” Proc. Natl. Acad. Sci. U. S. A. 110(12), 4500–4505 (2013). 10.1073/pnas.1214051110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Hoffmann A. and Woodside M. T., “ Signal-pair correlation analysis of single-molecule trajectories,” Angew. Chem. Int. Ed. Engl. 50(52), 12643–12646 (2011). 10.1002/anie.201104033 [DOI] [PubMed] [Google Scholar]
- 58. Fernandez J. M. and Li H., “ Force-clamp spectroscopy monitors the folding trajectory of a single protein,” Science 303(5664), 1674–1678 (2004). 10.1126/science.1092497 [DOI] [PubMed] [Google Scholar]
- 59. Suren T. et al. , “ Single-molecule force spectroscopy reveals folding steps associated with hormone binding and activation of the glucocorticoid receptor,” Proc. Natl. Acad. Sci. U. S. A. 115(46), 11688–11693 (2018). 10.1073/pnas.1807618115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Sengupta A. et al. , “ SlyD Accelerates trans-to-cis prolyl isomerization in a mechanosignaling protein under load,” J. Phys. Chem. B 125(31), 8712–8721 (2021). 10.1021/acs.jpcb.1c03648 [DOI] [PubMed] [Google Scholar]
- 61. Yura T., “ Regulation of the heat shock response in Escherichia coli: History and perspectives,” Genes Genet. Syst. 94(3), 103–108 (2019). 10.1266/ggs.19-00005 [DOI] [PubMed] [Google Scholar]
- 62. Genevaux P., Georgopoulos C., and Kelley W. L., “ The Hsp70 chaperone machines of Escherichia coli: A paradigm for the repartition of chaperone functions,” Mol. Microbiol. 66(4), 840–857 (2007). 10.1111/j.1365-2958.2007.05961.x [DOI] [PubMed] [Google Scholar]
- 63. Qi R. et al. , “ Allosteric opening of the polypeptide-binding site when an Hsp70 binds ATP,” Nat. Struct. Mol. Biol. 20(7), 900–907 (2013). 10.1038/nsmb.2583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Taneva S. G. et al. , “ Energetics of nucleotide-induced DnaK conformational states,” Biochemistry 49(6), 1338–1345 (2010). 10.1021/bi901847q [DOI] [PubMed] [Google Scholar]
- 65. Zhuravleva A. and Gierasch L. M., “ Allosteric signal transmission in the nucleotide-binding domain of 70-kDa heat shock protein (Hsp70) molecular chaperones,” Proc. Natl. Acad. Sci. U. S. A. 108(17), 6987–6992 (2011). 10.1073/pnas.1014448108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Tomoyasu T. et al. , “ Genetic dissection of the roles of chaperones and proteases in protein folding and degradation in the Escherichia coli cytosol,” Mol. Microbiol. 40(2), 397–413 (2001). 10.1046/j.1365-2958.2001.02383.x [DOI] [PubMed] [Google Scholar]
- 67. Barthel T. K., Zhang J., and Walker G. C., “ ATPase-defective derivatives of Escherichia coli DnaK that behave differently with respect to ATP-induced conformational change and peptide release,” J. Bacteriol. 183(19), 5482–5490 (2001). 10.1128/JB.183.19.5482-5490.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Vogel M., Mayer M. P., and Bukau B., “ Allosteric regulation of Hsp70 chaperones involves a conserved interdomain linker,” J. Biol. Chem. 281(50), 38705–38711 (2006). 10.1074/jbc.M609020200 [DOI] [PubMed] [Google Scholar]
- 69. Mayer M. P., “ Hsp70 chaperone dynamics and molecular mechanism,” Trends Biochem. Sci. 38(10), 507–514 (2013). 10.1016/j.tibs.2013.08.001 [DOI] [PubMed] [Google Scholar]
- 70. Mayer M. P. and Bukau B., “ Hsp70 chaperones: Cellular functions and molecular mechanism,” Cell. Mol. Life Sci. 62(6), 670–684 (2005). 10.1007/s00018-004-4464-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Packschies L. et al. , “ GrpE accelerates nucleotide exchange of the molecular chaperone DnaK with an associative displacement mechanism,” Biochemistry 36(12), 3417–3422 (1997). 10.1021/bi962835l [DOI] [PubMed] [Google Scholar]
- 72. Bauer D. et al. , “ Nucleotides regulate the mechanical hierarchy between subdomains of the nucleotide binding domain of the Hsp70 chaperone DnaK,” Proc. Nat. Acad. Sci. U. S. A. 112(33), 10389–10394 (2015). 10.1073/pnas.1504625112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Meinhold S. et al. , “ An active, ligand-responsive pulling geometry reports on internal signaling between subdomains of the DnaK nucleotide-binding domain in single-molecule mechanical experiments,” Biochemistry 58(47), 4744–4750 (2019). 10.1021/acs.biochem.9b00155 [DOI] [PubMed] [Google Scholar]
- 74. Bauer D. et al. , “ A folding nucleus and minimal ATP binding domain of Hsp70 identified by single-molecule force spectroscopy,” Proc. Natl. Acad. Sci. U. S. A. 115(18), 4666–4671 (2018). 10.1073/pnas.1716899115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Mandal S. S. et al. , “ Nanomechanics of the substrate binding domain of Hsp70 determine its allosteric ATP-induced conformational change,” Proc. Natl. Acad. Sci. U. S. A. 114(23), 6040–6045 (2017). 10.1073/pnas.1619843114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Perales-Calvo J., Lezamiz A., and Garcia-Manyes S., “ The Mechanochemistry of a structural zinc finger,” J. Phys. Chem. Lett. 6(17), 3335–3340 (2015). 10.1021/acs.jpclett.5b01371 [DOI] [PubMed] [Google Scholar]
- 77. Mashaghi A. et al. , “ Alternative modes of client binding enable functional plasticity of Hsp70,” Nature 539(7629), 448–451 (2016). 10.1038/nature20137 [DOI] [PubMed] [Google Scholar]
- 78. Ramirez M. P. et al. , “ Single molecule force spectroscopy reveals the effect of BiP chaperone on protein folding,” Protein Sci. 26(7), 1404–1412 (2017). 10.1002/pro.3137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Perales-Calvo J. et al. , “ The force-dependent mechanism of DnaK-mediated mechanical folding,” Sci. Adv. 4(2), eaaq0243 (2018). 10.1126/sciadv.aaq0243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Moessmer P. et al. , “ Active unfolding of the glucocorticoid receptor by the Hsp70/Hsp40 chaperone system in single-molecule mechanical experiments,” Proc. Natl. Acad. Sci. U. S. A. 119(15), e2119076119 (2022). 10.1073/pnas.2119076119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Weikum E. R. et al. , “ Glucocorticoid receptor control of transcription: Precision and plasticity via allostery,” Nat. Rev. Mol. Cell Biol. 18(3), 159–174 (2017). 10.1038/nrm.2016.152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Chrousos G. P. and Kino T., “ Glucocorticoid signaling in the cell. Expanding clinical implications to complex human behavioral and somatic disorders,” Ann. N. Y. Acad. Sci. 1179, 153–166 (2009). 10.1111/j.1749-6632.2009.04988.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. De Los Rios P. and Goloubinoff P., “ Hsp70 chaperones use ATP to remodel native protein oligomers and stable aggregates by entropic pulling,” Nat. Struct. Mol. Biol. 23(9), 766–769 (2016). 10.1038/nsmb.3283 [DOI] [PubMed] [Google Scholar]
- 84. Goloubinoff P. and De Los Rios P., “ The mechanism of Hsp70 chaperones: (entropic) Pulling the models together,” Trends Biochem. Sci. 32(8), 372–380 (2007). 10.1016/j.tibs.2007.06.008 [DOI] [PubMed] [Google Scholar]
- 85. Goloubinoff P. et al. , “ Chaperones convert the energy from ATP into the nonequilibrium stabilization of native proteins,” Nat. Chem. Biol. 14(4), 388–395 (2018). 10.1038/s41589-018-0013-8 [DOI] [PubMed] [Google Scholar]
- 86. Hemmingsen S. M. et al. , “ Homologous plant and bacterial proteins chaperone oligomeric protein assembly,” Nature 333(6171), 330–334 (1988). 10.1038/333330a0 [DOI] [PubMed] [Google Scholar]
- 87. Kusukawa N. et al. , “ Effects of mutations in heat-shock genes groES and groEL on protein export in Escherichia coli,” EMBO J. 8(11), 3517–3521 (1989). 10.1002/j.1460-2075.1989.tb08517.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Goloubinoff P. et al. , “ Reconstitution of active dimeric ribulose bisphosphate carboxylase from an unfoleded state depends on two chaperonin proteins and Mg-ATP,” Nature 342(6252), 884–889 (1989). 10.1038/342884a0 [DOI] [PubMed] [Google Scholar]
- 89. Martin J. et al. , “ Chaperonin-mediated protein folding at the surface of groEL through a 'molten globule'-like intermediate,” Nature 352(6330), 36–42 (1991). 10.1038/352036a0 [DOI] [PubMed] [Google Scholar]
- 90. Horwich A. L. et al. , “ Folding in vivo of bacterial cytoplasmic proteins: Role of GroEL,” Cell 74(5), 909–917 (1993). 10.1016/0092-8674(93)90470-B [DOI] [PubMed] [Google Scholar]
- 91. Braig K. et al. , “ The crystal structure of the bacterial chaperonin GroEL at 2.8 A,” Nature 371(6498), 578–586 (1994). 10.1038/371578a0 [DOI] [PubMed] [Google Scholar]
- 92. Tang Y. C. et al. , “ Structural features of the GroEL-GroES nano-cage required for rapid folding of encapsulated protein,” Cell 125(5), 903–914 (2006). 10.1016/j.cell.2006.04.027 [DOI] [PubMed] [Google Scholar]
- 93. Gupta A. J. et al. , “ Active cage mechanism of chaperonin-assisted protein folding demonstrated at single-molecule level,” J. Mol. Biol. 426(15), 2739–2754 (2014). 10.1016/j.jmb.2014.04.018 [DOI] [PubMed] [Google Scholar]
- 94. Thirumalai D. and Lorimer G. H., “ Chaperonin-mediated protein folding,” Annu. Rev. Biophys. Biomol. Struct. 30, 245–269 (2001). 10.1146/annurev.biophys.30.1.245 [DOI] [PubMed] [Google Scholar]
- 95. Naqvi M. M. et al. , “ Protein chain collapse modulation and folding stimulation by GroEL-ES,” Sci. Adv. 8(9), eabl6293 (2022). 10.1126/sciadv.abl6293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Avellaneda M. J. et al. , “ Publisher correction: Processive extrusion of polypeptide loops by a Hsp100 disaggregase,” Nature 578(7796), E23 (2020). 10.1038/s41586-020-2017-2 [DOI] [PubMed] [Google Scholar]
- 97. Avellaneda M. J. et al. , “ Processive extrusion of polypeptide loops by a Hsp100 disaggregase,” Nature 578(7794), 317–320 (2020). 10.1038/s41586-020-1964-y [DOI] [PubMed] [Google Scholar]
- 98. Jakob U. et al. , “ Chaperone activity with a redox switch,” Cell 96(3), 341–352 (1999). 10.1016/S0092-8674(00)80547-4 [DOI] [PubMed] [Google Scholar]
- 99. Moayed F. et al. , “ The anti-aggregation holdase Hsp33 Promotes the formation of folded protein structures,” Biophys. J. 118(1), 85–95 (2020). 10.1016/j.bpj.2019.10.040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Jahn M. et al. , “ Folding and assembly of the large molecular machine Hsp90 studied in single-molecule experiments,” Proc. Natl. Acad. Sci. U. S. A. 113(5), 1232–1237 (2016). 10.1073/pnas.1518827113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Jahn M. et al. , “ The charged linker of the molecular chaperone Hsp90 modulates domain contacts and biological function,” Proc. Natl. Acad. Sci. U. S. A. 111(50), 17881–17886 (2014). 10.1073/pnas.1414073111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Jahn M. et al. , “ Folding and domain interactions of three orthologs of Hsp90 studied by single-molecule force spectroscopy,” Structure 26(1), 96–105 (2018). 10.1016/j.str.2017.11.023 [DOI] [PubMed] [Google Scholar]
- 103. Girstmair H. et al. , “ The Hsp90 isoforms from S. cerevisiae differ in structure, function and client range,” Nat. Commun. 10(1), 3626 (2019). 10.1038/s41467-019-11518-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Tych K. M. et al. , “ Nucleotide-dependent dimer association and dissociation of the chaperone Hsp90,” J. Phys. Chem. B 122(49), 11373–11380 (2018). 10.1021/acs.jpcb.8b07301 [DOI] [PubMed] [Google Scholar]
- 105. Eckels E. C. et al. , “ DsbA is a redox-switchable mechanical chaperone,” Chem. Sci. 12(33), 11109–11120 (2021). 10.1039/D1SC03048E [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Eckels E. C. et al. , “ The mechanical power of titin folding,” Cell Rep. 27(6), 1836–1847 (2019). 10.1016/j.celrep.2019.04.046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Alonso-Caballero A. et al. , “ Mechanical architecture and folding of E. coli type 1 pilus domains,” Nat. Commun. 9(1), 2758 (2018). 10.1038/s41467-018-05107-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Liu K., Maciuba K., and Kaiser C. M., “ The ribosome cooperates with a chaperone to guide multi-domain protein folding,” Mol. Cell 74(2), 310–319 (2019). 10.1016/j.molcel.2019.01.043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Wruck F. et al. , “ The ribosome modulates folding inside the ribosomal exit tunnel,” Commun. Biol. 4(1), 523 (2021). 10.1038/s42003-021-02055-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Haldar S. et al. , “ Trigger factor chaperone acts as a mechanical foldase,” Nat. Commun. 8(1), 668 (2017). 10.1038/s41467-017-00771-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Singhal K. et al. , “ The trigger factor chaperone encapsulates and stabilizes partial folds of substrate proteins,” PLoS Comput. Biol. 11(10), e1004444 (2015). 10.1371/journal.pcbi.1004444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Jiao J. et al. , “ Munc18-1 catalyzes neuronal SNARE assembly by templating SNARE association,” Elife 7, e41771 (2018). 10.7554/eLife.41771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Shu T. et al. , “ Munc13-1 MUN domain and Munc18-1 cooperatively chaperone SNARE assembly through a tetrameric complex,” Proc. Natl. Acad. Sci. U. S. A. 117(2), 1036–1041 (2020). 10.1073/pnas.1914361117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Sudhof T. C. and Rothman J. E., “ Membrane fusion: Grappling with SNARE and SM proteins,” Science 323(5913), 474–477 (2009). 10.1126/science.1161748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Gao Y. et al. , “ Single reconstituted neuronal SNARE complexes zipper in three distinct stages,” Science 337(6100), 1340–1343 (2012). 10.1126/science.1224492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Serdiuk T. et al. , Nat. Chem. Biol. 12(11), 911–917 (2016). 10.1038/nchembio.2169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Serdiuk T., Mari S. A., and Muller D. J., “ Pull-and-paste of single transmembrane proteins,” Nano Lett. 17(7), 4478–4488 (2017). 10.1021/acs.nanolett.7b01844 [DOI] [PubMed] [Google Scholar]
- 118. Serdiuk T. et al. , “ Insertion and folding pathways of single membrane proteins guided by translocases and insertases,” Sci. Adv. 5(1), eaau6824 (2019). 10.1126/sciadv.aau6824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119. Thoma J. et al. , “ Impact of holdase chaperones Skp and SurA on the folding of beta-barrel outer-membrane proteins,” Nat. Struct. Mol. Biol. 22(10), 795–802 (2015). 10.1038/nsmb.3087 [DOI] [PubMed] [Google Scholar]
- 120. Abramson J. et al. , “ Structure and mechanism of the lactose permease of Escherichia coli,” Science 301(5633), 610–615 (2003). 10.1126/science.1088196 [DOI] [PubMed] [Google Scholar]
- 121. Nagamori S., Smirnova IN., and Kaback H. R., “ Role of YidC in folding of polytopic membrane proteins,” J. Cell Biol. 165(1), 53–62 (2004). 10.1083/jcb.200402067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122. Zhu L., Kaback H. R., and Dalbey R. E., “ YidC protein, a molecular chaperone for LacY protein folding via the SecYEG protein machinery,” J. Biol. Chem. 288(39), 28180–28194 (2013). 10.1074/jbc.M113.491613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123. Cecconi C. et al. , “ Protein-DNA chimeras for single molecule mechanical folding studies with the optical tweezers,” Eur. Biophys. J. 37(6), 729–738 (2008). 10.1007/s00249-007-0247-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124. Cecconi C. et al. , “ DNA molecular handles for single-molecule protein-folding studies by optical tweezers,” Methods Mol. Biol. 749, 255–271 (2011). 10.1007/978-1-61779-142-0 [DOI] [PubMed] [Google Scholar]
- 125. Tych K. and Zoldak G., “ Stable substructures in proteins and how to find them using single-molecule force spectroscopy,” Methods Mol. Biol. 1958, 263–282 (2019). 10.1007/978-1-4939-9161-7 [DOI] [PubMed] [Google Scholar]
- 126. van der Sleen L. M. and Tych K. M., “ Bioconjugation strategies for connecting proteins to DNA-linkers for single-molecule force-based experiments,” Nanomaterials 11(9), 2424 (2021). 10.3390/nano11092424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127. Synakewicz M. et al. , “ Bioorthogonal protein-DNA conjugation methods for force spectroscopy,” Sci. Rep. 9(1), 13820 (2019). 10.1038/s41598-019-49843-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128. Maciuba K., Zhang F., and Kaiser C. M., “ Facile tethering of stable and unstable proteins for optical tweezers experiments,” Biophys. J. 120(13), 2691–2700 (2021). 10.1016/j.bpj.2021.05.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129. Moayed F. et al. , “ Protein tethering for folding studies,” Methods Mol. Biol. 1665, 43–51 (2018). 10.1007/978-1-4939-7271-5_3 [DOI] [PubMed] [Google Scholar]
- 130. Jiao J. et al. , “ Single-molecule protein folding experiments using high-precision optical tweezers,” Methods Mol. Biol. 1486, 357–390 (2017). 10.1007/978-1-4939-6421-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131. Pfitzner E. et al. , “ Rigid DNA beams for high-resolution single-molecule mechanics,” Angew. Chem. Int. Ed. Engl. 52(30), 7766–7771 (2013). 10.1002/anie.201302727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132. Mehlich A. et al. , “ Slow transition path times reveal a complex folding barrier in a designed protein,” Front. Chem. 8, 587824 (2020). 10.3389/fchem.2020.587824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133. Satija R., Berezhkovskii A. M., and Makarov D. E., “ Broad distributions of transition-path times are fingerprints of multidimensionality of the underlying free energy landscapes,” Proc Natl Acad Sci U S A 117(44), 27116–27123 (2020). 10.1073/pnas.2008307117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134. Hoffer N. Q. et al. , “ Measuring the average shape of transition paths during the folding of a single biological molecule,” Proc. Natl. Acad. Sci. U S. A. 116(17), 8125–8130 (2019). 10.1073/pnas.1816602116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135. Alemany A. et al. , “ Mechanical folding and unfolding of protein barnase at the single-molecule level,” Biophys. J. 110(1), 63–74 (2016). 10.1016/j.bpj.2015.11.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136. Edwards D. T., LeBlanc M. A., and Perkins T. T., “ Modulation of a protein-folding landscape revealed by AFM-based force spectroscopy notwithstanding instrumental limitations,” Proc. Natl. Acad. Sci. U. S. A. 118(12), e2015728118 (2021). 10.1073/pnas.2015728118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137. Rico-Pasto M. et al. , “ Molten globule-like transition state of protein barnase measured with calorimetric force spectroscopy,” Proc. Natl. Acad. Sci. U. S. A. 119(11), e2112382119 (2022). 10.1073/pnas.2112382119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138. Synakewicz M. et al. , “ Unraveling the mechanics of a repeat-protein nanospring: From folding of individual repeats to fluctuations of the superhelix,” ACS Nano 16(3), 3895–3905 (2022). 10.1021/acsnano.1c09162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139. Ganim Z. and Rief M., “ Mechanically switching single-molecule fluorescence of GFP by unfolding and refolding,” Proc. Natl. Acad. Sci. U. S. A. 114(42), 11052–11056 (2017). 10.1073/pnas.1704937114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140. Maksudov F., Jones L. K., and Barsegov V., “ Statistical learning from single-molecule experimentsSupport vector machines and expectation-maximization approaches to understanding protein unfolding data,” J. Phys. Chem. B 125(22), 5794–5808 (2021). 10.1021/acs.jpcb.1c02334 [DOI] [PubMed] [Google Scholar]
- 141. Lin Z. et al. , “ Learning-based event locating for single-molecule force spectroscopy,” Biochem. Biophys. Res. Commun. 556, 59–64 (2021). 10.1016/j.bbrc.2021.03.159 [DOI] [PubMed] [Google Scholar]
- 142. Horvath D. and Zoldak G., “ Entropy-based strategies for rapid pre-processing and classification of time series data from single-molecule force experiments.,” Entropy 22(6), 701 (2020). 10.3390/e22060701 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data sharing is not applicable to this article as no new data were created or analyzed in this study.