Abstract
Nanomedicine has a great potential to revolutionize the therapeutic landscape. However, up-to-date results obtained from in vitro experiments predict the in vivo performance of nanoparticles weakly or not at all. There is a need for in vitro experiments that better resemble the in vivo reality. As a result, animal experiments can be reduced, and potent in vivo candidates will not be missed. It is important to gain a deeper knowledge about nanoparticle characteristics in physiological environment. In this context, the protein corona plays a crucial role. Its formation process including driving forces, kinetics, and influencing factors has to be explored in more detail. There exist different methods for the investigation of the protein corona and its impact on physico-chemical and biological properties of nanoparticles, which are compiled and critically reflected in this review article. The obtained information about the protein corona can be exploited to optimize nanoparticles for in vivo application. Still the translation from in vitro to in vivo remains challenging. Functional in vitro screening under physiological conditions such as in full serum, in 3D multicellular spheroids/organoids, or under flow conditions is recommended. Innovative in vivo screening using barcoded nanoparticles can simultaneously test more than hundred samples regarding biodistribution and functional delivery within a single mouse.
I. INTRODUCTION
Nanomedicine is an interesting, emerging field of modern medical care.1–4 Many nanotherapeutics are in clinical trials, and the number of approved nanoparticulate drug products is continuously growing. Liposomal doxorubicin (DOXIL®) was the first nanodrug to be approved by the United States Food and Drug Administration (FDA) in 1995.5 Currently, messenger RNA (mRNA) vaccines are the big hope in the SARS-CoV-2 pandemic,6 small-interfering RNA (siRNA) lipid nanoparticles (LNPs) reached the medical market,7 and Cas9 mRNA LNPs have been applied for the first successful in vivo genetic correction by CRISPR Cas9/single-guide RNA in patients.8
Despite the enormous potential, the progress in the development and application of nanoparticles as delivery vehicles for (bio)pharmaceutics (e.g., chemotherapeutics, therapeutic nucleic acids, or proteins) is rather slow and moderate. The production of such nanotherapeutics in pharmaceutical grade and scale is challenging.4 Moreover, different cargos place different demands on their carriers.4,9,10 Important is especially that the delivery system comprise extracellular stability and intracellular release of the cargo in its active form at the site of action. Bio-/stimuli responsiveness can be a helpful tool for creating delivery systems, which change their properties in a dynamic mode upon specific endogenous or exogenous stimuli (e.g., changes in pH, redox potential, or temperature).11 The probably most relevant reason for the slow progress, however, is the often weak to absent translatability from obtained in vitro data to the in vivo situation,12–18 which makes it hard to draw conclusions about the in vivo performance of nanoparticles from in vitro experiments.
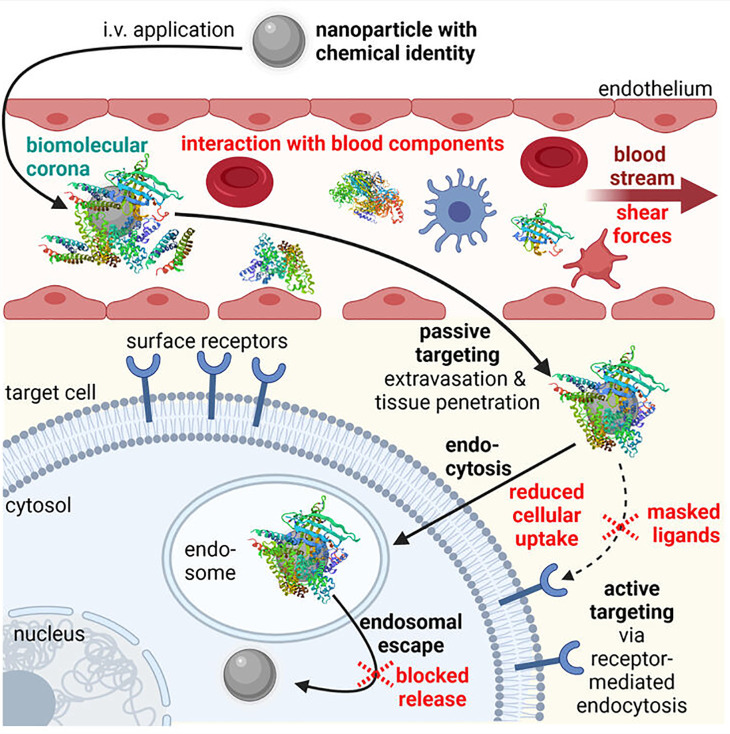
After intravenous administration, the nanoparticles have to face several obstacles that differ from in vitro (Fig. 1). First, they get in contact with blood components. Usually, the nanoparticles are then covered by a biomolecular multi-layer (so-called protein corona or biomolecular corona),19,20 which creates a biological identity,20,21 thereby altering the physico-chemical properties, the pharmacokinetics, and toxicity profile of the nanoparticles.22 Interaction with electrolytes, plasma proteins, and blood cells (e.g., erythrocytes and thrombocytes) can cause nanoparticle dissociation, self-aggregation, or aggregation with, for example, erythrocytes.10,23 Cationic nanoparticles bind and activate complement blood proteins, thereby inducing innate immune responses with serious side effects.24,25 In addition, destabilizing shear forces within the bloodstream act on the nanoparticles.26,27 Functionalization of the nanoparticle surface with shielding agents [e.g., polyethylene glycol (PEG), poly(2-ethyl-2-oxazoline) (PEtOx), or polysarcosine (pSar)] can reduce, but not completely prevent the protein corona formation and create a “stealth” character, by this hindering dissociation or aggregation.21,28–32 Second, the nanoparticles have to extravasate, penetrate, and accumulate in the right tissue, followed by uptake in the target cells. Endothelial targeting as well as passive [via the so-called enhanced permeation and retention (EPR) effect] and/or active targeting can be helpful for efficient delivery in vivo.10,11,13,33 However, it has to be considered that the biomolecular corona can mask targeting ligands, thereby reducing the targeting capability.22,31,34 Third, endosomal escape and cargo release are necessary. Both processes are more or less comparable between in vitro and in vivo (single-cell level). However, also here masked nanoparticles may be blocked, for example, in their lytic activity, resulting in reduced release from the endosomes and thus reduced transfection efficiency.12
FIG. 1.
Obstacles (in red) in the in vivo delivery process of intravenously (i.v.) applied nanoparticles. Created with BioRender.com.
Also, for other application routes than intravenous (e.g., inhalative, intravitreal, or transdermal) several biological barriers have to be overcome and the delivery system has to fulfill certain requirements.35–37 Moreover, there can be difficulties to reach the target organ. The most prominent example is the systemic delivery to the brain, where the blood–brain barrier (BBB) is a major hurdle.38
Due to the huge discrepancy between the in vitro and the in vivo situation, there is the need for in vitro assays that resemble the in vivo situation more realistically. In this context, it is also necessary to gain a deeper knowledge about the interactions at the nano-bio interface, the protein corona formation, and the impact of physiological milieu on the nanoparticles. With better predictions of the in vivo efficacy, potent in vivo candidates that show only minor activity in standard in vitro assays will not be missed.14,15 In addition, such assays are also advantageous in regard to animal welfare and protection as ineffective carriers can be excluded from in vivo studies with greater certainty at an earlier point in time.
In recent years, several in vitro and ex vivo models have been developed, which mimic the in vivo situation with relevant biological barriers. However, these are not subject of the current review article as detailed reviews about such models can be found elsewhere—for example, about microfluidic organ-on-chip technology,39–41 about lung and inhalation models,35,42 about skin models for transdermal application,43,44 and about BBB models.38 To briefly mention a practical example, Onyema et al. established a BBB model based on human induced pluripotent stem cells, which is suitable to study interactions with nanoparticles in correlation with their material, size, and protein corona composition.45
In the following, different methods for the characterization of the protein corona and its impact on nanoparticles' properties will be discussed and critically reflected. Moreover, a more advanced in vivo screening method using barcoded nanoparticles will be illustrated. With this method, more than hundred samples can be simultaneously screened within a single mouse.46 By this, the number of animals in the in vivo studies can be reduced, which is in line with a more reasonable and ethical use of animals.
II. CHARACTERIZATION OF THE PROTEIN CORONA AND ITS IMPACT ON NANOPARTICLE PROPERTIES
In the physiological environment, the nanoparticle surface gets coated inter alia with proteins,47–50 forming a so-called protein corona.19,51 The protein adsorption phenomenon was first described by Vroman et al. in 1962.52 In 2012, the extended term “biomolecular corona” was introduced by Dawson and co-workers to underline the complexity of the nanoparticle corona in biological fluids, consisting not only of proteins but also of other biomolecules.20 The chemical identity of the nanoparticles is changed toward a biological identity.20,21 Physico-chemical properties of the nanoparticles as well as their pharmacokinetics (e.g., blood circulation time, clearance, biodistribution, targeting capability, and drug release profiles) and biosafety (hemocompatibility, toxicity, immune response) are altered in biofluids.21,22,31,53–56 The corona formation is a dynamic process comprising physico-chemical interactions and thermodynamic exchanges,47 which evolves over time.56–65 The multi-layered structure of the protein corona can be subdivided into the inner tight hard corona (protein–nanoparticle interactions) and the outer looser soft corona (protein–protein interactions).47 The properties of the formed protein corona depend on the nanoparticle composition,31,66–68 but also on additional nanoparticle properties such as size, surface charge, shape, nanoparticle surface, and functionalization (e.g., PEGylation).47–50,67,69–71 Moreover, experimental conditions (e.g., biofluid, temperature, time, static vs dynamic incubation) can influence the protein adsorption.49
A. General considerations for the experimental setup of protein corona investigations
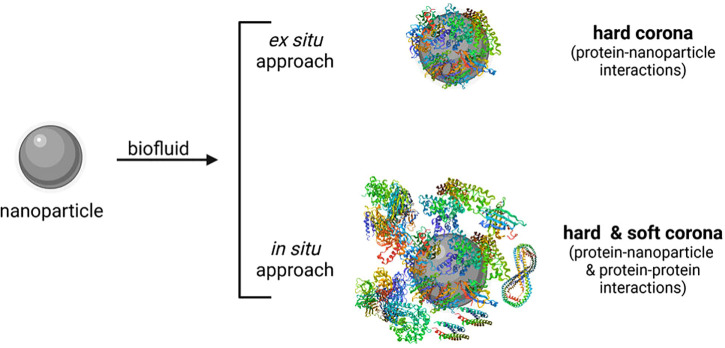
In general, the investigation of the hard corona is much easier and more accessible compared to the soft corona as the latter is of an unstable nature and difficult to isolate.47 Thus, there exist only a few attempts to evaluate the soft corona along with the hard corona using in situ techniques, where the protein corona is investigated immediately in the biofluid without prior separation of unbound proteins47,65,72–84 (Fig. 2). The classical ex situ approach consists of three steps: sample incubation in physiological fluids, followed by isolation and purification of nanoparticle–protein complexes from free proteins, and subsequent proteomic analysis and/or evaluation of physico-chemical and biological nanoparticle properties before and after protein interaction.47,85 With the ex situ approach, mainly the hard corona can be analyzed (Fig. 2). Sample incubation conditions have to be carefully chosen,47 that is, (i) sample concentration; (ii) biofluid—type (blood, plasma, serum, protein solutions, simulated body fluid, etc.), origin (e.g., murine, bovine, human), and amount; (iii) temperature; (iv) time; and (v) shaking speed. Dynamic incubation conditions (e.g., peristaltic pumps for adjusting flow rates) may simulate more realistically the in vivo situation.26,27,86,87 Some research was also conducted investigating the in vivo protein corona of nanoparticles after blood circulation in mice.59,88–93 The different possible techniques for the isolation and purification of the nanoparticle–protein complexes with all their advantages and limitations are reviewed in detail by Weber et al.72 In short, the mostly used method is centrifugation,85 often performed in the presence of a sucrose cushion.72 However, high centrifugal forces may alter the protein corona. Alternatively, chromatography-based methods such as asymmetrical flow field-flow fractionation (AF4), hydrodynamic chromatography (HDC), and size exclusion chromatography (SEC) can be utilized to purify the nanoparticle–protein complexes.72 The very mild separation conditions of AF4 may allow to also isolate weakly bound proteins of the soft corona.94 In the case of SEC, relatively high shear stress occurs that may influence the protein corona composition.95 However, with this method dissociation rates can be investigated.72 Moreover, special nanocarrier properties can be used for isolating the nanoparticle–protein complexes.47,72 Magnetic separation, for instance, can be an option for magnetic nanocarriers.96–101
FIG. 2.
Protein corona investigation using ex situ or in situ approaches. While with the ex situ approach mainly the hard corona can be evaluated, in situ technologies allow for the characterization of the hard and soft corona. Created with BioRender.com.
After successful separation from free, unbound proteins, the protein corona can be analyzed with various techniques. Figure 3 illustrates a typical experimental setup for a protein corona investigation. Overall, in order to obtain as complete a picture as possible, a combination of in situ and ex situ technologies for the protein corona characterization is recommended, since each preparation method can have an influence on the tested system and findings.72,102
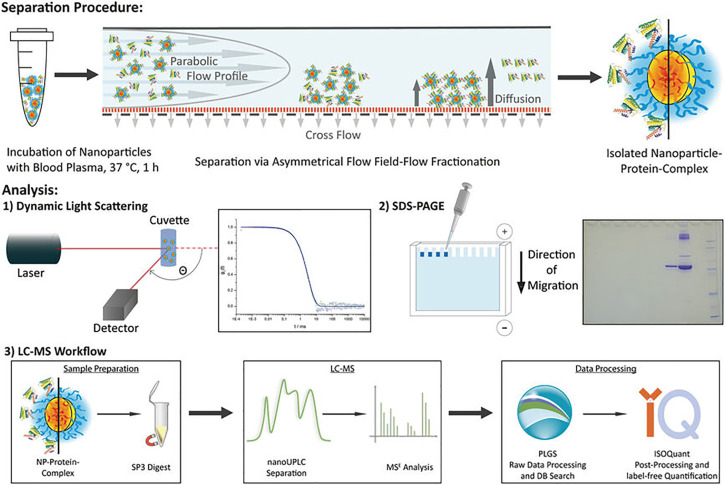
FIG. 3.
A typical experimental setup for protein corona investigations. After incubation of nanoparticles in human blood plasma, nanoparticle–protein complexes are isolated by asymmetrical flow field-flow fractionation (AF4). The sizes can be assessed by dynamic light scattering (DLS). The protein corona composition and amount are analyzed by SDS-PAGE (sodium dodecyl sulfate polyacrylamide gel electrophoresis) and label-free quantitative proteomic analysis. LC-MS, liquid chromatography-mass spectrometry; UPLC, ultra-performance liquid chromatography. Reproduced with permission from Small 16, 1907574 (2020).28 Copyright 2021 John Wiley and Sons.
In the following, such characterization methods will be discussed in detail with a focus on their advantages and limitations regarding protein corona analysis. Moreover, technologies for studying protein–nanoparticle interactions will be illustrated.
B. Investigation of protein–nanoparticle interactions
To gain a deeper knowledge about the protein corona formation process, it is important to comprehend the driving forces and kinetics of protein binding on nanoparticle surfaces. There exist several techniques to investigate single protein–nanoparticle interactions. Thermodynamic parameters such as affinity and stoichiometry of protein binding can be studied via isothermal titration calorimetry (ITC).51,103–105 Surface plasmon resonance (SPR) can help to assess association and dissociation rates.51,61,106,107 Multi-parametric SPR was utilized by Kari et al. for the in situ investigation of protein binding on biosensor-immobilized liposomes in undiluted protein solutions or human plasma.74,108 Oh et al. developed a new technique based on atomic force microscopy (AFM)-derived recognition imaging to determine binding affinities by visualizing molecular bindings at the nanoscale, as demonstrated by means of DNA hybridization.109 They state that this method is applicable to any receptor/ligand combination (e.g., interaction between nanoparticles and plasma proteins), thus representing a potent alternative for next-generation affinity sensors. Furthermore, different fluorescence spectroscopy-based methods can be used to characterize protein–nanoparticle interactions.110–113 Boulos et al. studied the bovine serum albumin (BSA) adsorption on gold nanoparticles (kinetics, binding constants) via steady-state fluorescence spectroscopy, taking advantage of the fluorescence quenching capability of the tested gold nanoparticles.111 They found that the nanoparticle shape and surface as well as PEGylation had no impact on the BSA binding affinity. This was confirmed by affinity capillary electrophoresis (ACE). Comparison of the binding constants derived from the two different methods, however, revealed orders of magnitude difference. Moreover, both techniques have their limitations: While fluorescence spectroscopy suffers from inner filter effects and gold nanoparticle optical interference, inner capillary wall effects are an issue in ACE. Therefore, combination of several methods is required to determine binding affinities as accurately as possible. A fluorescence polarization assay was developed by Gaus et al. to study protein binding toward antisense oligonucleotides (ASO).112 In a follow-up study, ASO–fatty acid conjugates showed in the fluorescence polarization assay improved protein binding affinities with increasing carbon chain length (optimum C16-C22).113 The activity of ASO-fatty acid conjugates correlated with their affinity to albumin. The tighter the BSA binding, the greater the improvement in muscle activity.
When a protein binds on a nanoparticle, it can alter its structure/conformation in response to the nanoparticle surface, resulting in altered functionality.49,114,115 Such changes can be examined by various spectroscopic methods. Circular dichroism (CD) spectroscopy can detect changes in the secondary structure of proteins in real time and in situ.49,114,116 By using synchrotron-radiation (SR) as light source for CD spectroscopy, measurements access much more of the vacuum ultraviolet (UV) wavelength range down to the extreme UV and even x-ray range.117,118 The SR-CD spectra are therefore richer in information than conventional CD spectra, including even electronic transitions of the polypeptide backbone.118 By this, more precise determinations of changes within the secondary structure of proteins can be made. Sanchez-Guzman et al. used SR-CD to investigate changes in structure and stability of weakly bound proteins on nanoparticles in situ.119 In combination with computer simulation (molecular dynamics) and thermodynamic analysis, they concluded that nanoparticles altered weakly bound proteins by shifting the equilibrium toward the unfolded states at physiological temperature. In addition to CD spectroscopy, several other techniques are used in the literature to examine structural changes in proteins, for example, Fourier transform infrared (FT-IR) spectroscopy,120,121 Raman spectroscopy,122 surface-enhanced Raman scattering (SERS),123 differential scanning calorimetry (DSC),48 nuclear magnetic resonance (NMR) spectroscopy,124 or UV-vis spectroscopy.125,126
C. Computational simulations of protein–nanoparticle interactions
In addition to the aforementioned experimental techniques for the characterization of nanoparticle–protein interactions, in silico predictions via mathematical, theoretical modeling, and computational simulations can be helpful to gain a better understanding of the complex processes happening at the nano-bio interface upon nanoparticle interaction with the physiological environment. There exist different simulation methods to comprehend the mechanistic properties (binding sites, functional units), (thermo-)dynamics, and kinetics of protein–nanoparticle interactions. These are discussed in detail elsewhere.48,50,127–129 However, since only single protein–nanoparticle interaction can be simulated, these computational studies fall short to completely depict the reality in a complex biological environment.129 Great effort has been put into the development of simulation methods better resembling reality. To outline a few of those methods, (i) atomistic and coarse-grained molecular dynamics or Monte Carlo simulations help to predict details of nanoparticle–protein interactions at the molecular-to-particle level;107,129–136 (ii) the adopted Hill model can be utilized to assess equilibrium dissociation and kinetic coefficients for one or two protein species binding with one nanoparticle type;137 (iii) with dynamic modeling, Dell'Orco et al. predicted the corona formation process (evolution and equilibrium composition) based on affinities, stoichiometries, and rate constants;61 and (iv) statistical modeling can be another option for in silico predictions. Examples for this are statistical modeling of quantitative structure–activity relationships (QSAR) via linear and non-linear regression models,138,139 and statistical modeling of a so-called biological surface adsorption index (BSAI) based on multiple linear regression analysis and experimentally obtained binding coefficients.140 With the rapid progress in electronic development and the improvement of computing power, in silico predictions will gain more importance for protein corona investigations in the future.50
D. Identification and quantification of protein corona components
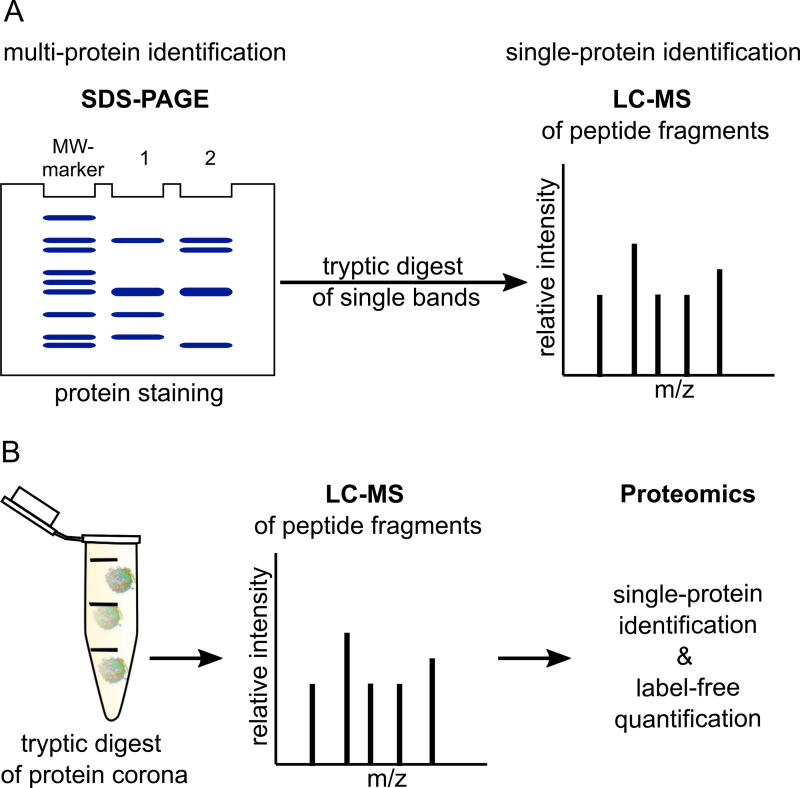
The commonly used techniques for the identification of protein corona components are SDS-PAGE (sodium dodecyl sulfate polyacrylamide gel electrophoresis) and MS (mass spectrometry)49,141 (Fig. 4). The corona proteins have to be first eluted from the nanoparticles' surface under denaturing conditions and heating.85 Then, in the case of SDS-PAGE, the detergent SDS (often together with a reducing agent like dithiothreitol) completely unfolds the proteins.142 The negatively charged SDS–protein complexes are then separated by molecular weight. PAGE can be performed one-dimensionally (1D-PAGE, standard SDS-PAGE)143 or two-dimensionally (2D-PAGE).144 In the case of the latter, usually separation using isoelectric focusing (IEF) is followed by separation based on protein size (SDS-PAGE). This two-step separation process allows for the separation of complex protein mixtures into discrete spots but is more time consuming than 1D-PAGE.144 Protein detection in the polyacrylamide gels is normally done by Coomassie Blue staining, staining with SYPRO Ruby (a preformulated, noncovalent fluorescent stain), or Silver staining.145 By this, multi-protein identification is achieved; for the specific detection of single proteins, immunoblotting can be performed after SDS-PAGE.141 Moreover, SDS-PAGE can be combined with liquid chromatography (LC)-MS or LC-tandem MS (LC-MS/MS).49,63,71 For this, a trypsin-digest of the proteins has to be done beforehand.146–148 Often used MS methods are such with soft ionization sources, for example, matrix-assisted laser desorption/ionization time-of-flight MS (MALDI-TOF MS),149 quadrupole TOF MS,63,150 or electrospray ionization MS (ESI-MS).71,151 LC-MS of digested proteins152 can be also performed without prior separation by SDS-PAGE.28,87,141,151 Advantage is the small sample volumes that are required.47,151 For the subsequent proteomics analysis, bioinformatic tools for data processing and database search (e.g., Sequest,153 Mascot154 or ProteinLynx Global Server28,70) and specialized analyzing software (e.g., Scaffold155 or Progenesis156) are inevitable.47,157 In the case of LC-MS, absolute in-sample amounts of proteins can be obtained by label-free quantification, for example, as described in Ref. 158, whereas in the case of SDS-PAGE, only the quantity of proteins with similar molecular weight can be assessed.49 Alberg et al. quantified the human serum albumin (HSA) amount attached on nanoparticles by performing a comparative SDS-PAGE-based analysis with free, pure HSA at different concentrations.28
FIG. 4.
Classical approaches for the identification and quantification of protein corona components. (a) SDS-PAGE (sodium dodecyl sulfate polyacrylamide gel electrophoresis) followed by protein staining allows for multi-protein identification. Tryptic digest of single bands and subsequent LC-MS (liquid chromatography-mass spectrometry) analysis enables the identification of single proteins. (b) Trypsinized corona proteins can also be directly analyzed by LC-MS. With subsequent data processing and proteomic analysis, besides single-protein identification also label-free quantification is possible. MW-marker, molecular weight-marker. Nanoparticle–protein complexes (b) are created with BioRender.com.
Bicinchoninic acid (BCA) assay is a standard protein quantification assay.159,160 With this assay, the total amount of corona proteins can be determined.26,93 Cysteines, tryptophans, tyrosines, and the peptide bonds reduce CuII to CuI in an alkaline milieu, which then forms complexes with BCA.161 However, drawbacks are the relatively low sensitivity and possible interference of the nanoparticles with this assay.49,161,162
Quartz crystal microbalance (QCM) can be used to predict the surface coverage on nanoparticles with proteins.163,164 This method is ultra-sensitive to total mass changes (down to the femto- to attogram range73) as measured by changes in the resonance frequency on a piezo-electric crystal.165,166 QCM can be measured in vacuo or in fluids.166 With the further developed method termed QCM with dissipation monitoring (QCM-D), additional information about the viscoelastic properties of the adlayer can be derived in real time and in situ.165 By the way, QCM-D can be also used to study interactions between nanoparticles and membranes/lipid bilayers and how the protein corona influences these interactions.167–169 Sebastiani et al. utilized QCM-D to screen various LNPs for their binding affinities to serum proteins in order to find the most promising candidates for subsequent in vitro and in vivo experiments.170 For this purpose, they used gold sensors functionalized with antibodies against human ApoE or PEG. By this, binding affinities of LNPs toward ApoE as well as of PEGylated LNPs to serum proteins could be investigated.
Inductively coupled plasma mass spectrometry (ICP-MS) can be used to determine the stoichiometric composition of the protein corona of inorganic nanoparticles.73 With ICP-MS, the metal (e.g., gold) of the inorganic nanoparticles and sulfur of cysteine residues of proteins are detected, allowing for the calculation of the total concentrations of both elements.171,172 Together with results derived from classical protein quantification like BCA assay, the number of proteins per molecule can be determined.172,173
E. Impact of the protein corona on the physico-chemical properties of nanoparticles
The protein corona determines inter alia physico-chemical properties (e.g., size, surface charge, and stability) of the nanoparticles in physiological environment.21,49,174–176 The influence on the physico-chemical properties of the nanoparticles can be investigated using different characterization methods (Table I). These can be divided into three general categories:174 (i) scattering and correlation methods, (ii) microscopy-based methods, and (iii) fractionating methods based on hydrodynamic separation.
TABLE I.
Physico-chemical characterization methods for nanoparticles—practical examples with regard to protein corona investigations. AF4, asymmetrical flow field-flow fractionation; AFM, atomic force microscopy; AUC, analytical ultra-centrifugation; DCS, differential centrifugal sedimentation; DDLS, depolarized dynamic light scattering; DLS, dynamic light scattering; ELS, electrophoretic light scattering; FCS, fluorescence correlation spectroscopy; PTV, particle tracking velocimetry; SANS, small-angle neutron scattering; SAXS, small-angle x-ray scattering; SEC, size-exclusion chromatography; SEM, scanning electron microscopy; SLS, static light scattering; TEM, transmission electron microscopy; TRPS, tunable resistive pulse sensing.
| Characterization method | Information about | Practical examples—references | |
|---|---|---|---|
| Scattering- and correlation-based | DLS, DDLS, SLS | Size | 28, 30, 64, 65, 80, and 176–182 |
| ELS | Zeta-potential | 57, 65, 71, 86, 97, 176, and 181 | |
| SAXS, SANS | Size | 181 and 183–186 | |
| FCS | Size | 75, 78, and 187–189 | |
| PTV | Size | 179 and 190 | |
| Microscopy-based | (Cryo-)TEM | Size, shape | 29, 65, 93, 119, 178, 180, 181, and 191–194 |
| SEM | 195 | ||
| AFM | 107, 196, and 197 | ||
| Fractionating | AF4 | Size distribution | 28, 30, 178, 180, and 198 |
| SEC | 51, 112, and 199 | ||
| AUC | 196 | ||
| DCS | 65, 132, 176, and 200 | ||
| Others | TRPS | Size, zeta-potential | 201–204 |
In the following, a closer look at the different characterization methods is taken. Practical examples for the methods with regard to protein corona investigations are listed in Table I. A selection of these examples is also described in more detail in the text. Table II summarizes the advantages and limitations of the different methods.
TABLE II.
Advantages and limitations of the physico-chemical characterization methods. AF4, asymmetrical flow field-flow fractionation; AFM, atomic force microscopy; AUC, analytical ultra-centrifugation; DCS, differential centrifugal sedimentation; DLS, dynamic light scattering; ELS, electrophoretic light scattering; FCS, fluorescence correlation spectroscopy; NP, nanoparticle; PC, protein corona; PTV, particle tracking velocimetry; SANS, small-angle neutron scattering; SAXS, small-angle x-ray scattering; SEC, size-exclusion chromatography; SEM, scanning electron microscopy; SLS, static light scattering; TEM, transmission electron microscopy; TRPS, tunable resistive pulse sensing.
| Characterization method | Advantages | Limitations | References |
|---|---|---|---|
| DLS | Nonperturbative, fast, and accurate method to determine hydrodynamic radii (size range 1 nm–10 μm); no calibration required | Hydrodynamic radii influenced by the formation of hydration shells, particle shapes, and counterion binding; high sensitivity toward the presence of larger particles; inability to characterize highly polydisperse systems | 129 |
| ELS | Straight forward method to measure surface charge and changes in surface charge; indicator of stability of NP dispersions | Minimum ionic strength required; only for monodisperse NPs (calculation of charge/size ratio) | 129 |
| SLS | Absolute method, no calibration required | Average values for Mw and Rg influenced by the sample's polydispersity | 205 |
| SAXS, SANS | High-resolved, in-depth structural characterization at the nanoscale | X rays: possible radiation damage to the samples (not the case for nondestructive neutrons) | 206 |
| FCS | Sensitive method to measure minute changes in NP diffusivity; ability to quantify PC formation with high accuracy in the presence of free protein | Fluorescent label required | 129 and 188 |
| PTV | Tracking of single NPs; potentially less sensitive to bigger particles (compared to, e.g., DLS) | Limited to analytes with low particle concentrations; loss of sensitivity in the case of small NP distances; sensitive to background scattering | 207–209 |
| TEM | Visualization of protein adsorption onto the NP surface | Negative staining of protein needed; shrinkage of vesicles due to drying process; cost- and time-intensive | 49 and 207 |
| Cryo-TEM | Investigation of NPs in their natural surroundings | Cost- and time-intensive; contrast reduction caused by the water film | 191, 207, 210, and 211 |
| SEM | Detailed 3D images; less prone to overestimate NP size (compared to, e.g., DLS) | Difficulties in the detection of proteins on the NP surface; staining with heavy metals required; shrinkage of vesicles during the drying process; cost- and time-intensive | 49 and 207 |
| AFM | Visualization of the surface topography and properties (e.g., hardness, texture, protein adsorption) with atomic resolution | Impossibility to distinguish hard and/or soft PC formation; difficult sample preparation; time-consuming; matching of probe and operating mode to the specific sample required; various sources of artifacts (e.g., tip and scanner) | 49, 207, 212, and 213 |
| AF4 | Characterization of soluble and insoluble sample specimen, and also of complex mixtures of colloids, particles or even cells (size range 1 nm–100 μm); higher selectivity and greater resolution over a wider size range than SEC; low shear forces (soft corona investigation) | Strong dilution of the sample in the carrier liquid and incomplete particle recollection depending on the crossflow | 94, 207, and 214 |
| SEC | Simplicity of the method; high speed and precision in separation; very small amounts of sample needed | Limited dynamic range; nonspecific interactions with the chromatographic material and column hardware; inaccuracy due to alteration of size distribution (e.g., for reversible aggregates) | 95 |
| AUC | Hydrodynamic and thermodynamic characterization of macromolecules or colloids in situ; high resolution (Ångström range for NP size); no calibration necessary | Calculation of NP size distribution heavily depended on the knowledge of the sample's density; no fraction collection possible | 215 |
| DCS | Applicable to complex biological systems, without the need for fluorescent labels; high-resolution separation and detection of a small percentage of particle populations (size range 3 nm–60 μm) within polydisperse colloidal samples | Identification of the “true” NP size relies on a simple core-shell model considering the new NP density; no fraction collection possible | 65 and 200 |
| TRPS | High resolution and accuracy in measurement of multimodal samples; measurement in complex biological media | Limited speed and detectable size range; no fraction collection possible | 201, 216, and 217 |
1. Scattering and correlation methods
In the case of static light scattering (SLS), a laser beam passes through the sample and the mean value of the intensity of the scattered light is measured at different angles. For particles larger than λ/20, the scattered light intensity depends on the detection angle θ. Using the Zimm equation, one can derive the z-average of the squared radius of gyration ⟨Rg2⟩z.218 In dynamic light scattering (DLS) or quasi-elastic light scattering (QELS), the fluctuation of the scattered light intensity is measured and autocorrelated. The autocorrelation function yields the z-average of the diffusion coefficient ⟨D⟩z. Using the Stokes–Einstein equation, the z-average of the reciprocal hydrodynamic radius ⟨1/Rh⟩z can be calculated.218 As a practical example, Alberg et al. could show by multiangle light scattering that incubation with 50% (v/v) human plasma did not affect the hydrodynamic radii and polydispersity of the tested polymeric micelles, which were surface modified with different hydrophilic shielding agents [PEG, poly(N-2-hydroxypropylmethacrylamide) (pHPMA), pSar].28 Consecutive proteomic analysis confirmed that only a neglectable amount of plasma proteins was attached to the nanoparticles. This is especially preferable for in vivo applications. A relatively new improvement of DLS is depolarized DLS (DDLS).219 Hereby, the scattered light is divided into two beams (horizontally and vertically polarized), allowing the simultaneous determination of the translational and rotational diffusion. This enables the observation of nanoparticles with an optical anisotropy in a complex biological matrix, as the contribution to the scattered light intensity for particles without an optical anisotropy is negligible.80
The zeta-potential or effective charge of the nanoparticle can be measured with electrophoretic light scattering (ELS).220–222 Sakulkhu and co-workers investigated the effect of serum on the zeta-potential of different polyvinyl alcohol (PVA)-coated superparamagnetic iron oxide nanoparticles (SPIONs).97 The surface charge dropped in all cases due to the net-negative-charged layer of adsorbed serum proteins. At the same time, the hydrodynamic size of the SPIONs increased as determined by DLS measurements.
Small-angle x-ray scattering (SAXS) and small-angle neutron scattering (SANS) are also static light scattering experiments like SLS; the advantage of using neutrons or x rays is that the de Broglie wavelengths are on the order of 0.1–10 nm as opposed to around 400–700 nm for the wavelength of visible light. This allows for a more detailed observation of structures.223,224 SAXS measurements were utilized, for instance, by Orts-Gil et al. to evaluate colloidal stability of silica nanoparticles in the presence of BSA.181 The derived SAXS structure factor indicated a short-range attractive potential in the binary silica-BSA system, which is in line with observed agglomeration in DLS measurements. The authors hypothesized that protein bridging might be an explanation for the observed agglomeration. Sebastiani and co-workers examined the influence of ApoE binding to mRNA-LNPs via SANS.183 They found that ApoE binding led to the re-arrangement of components both at the surface as well as in the LNP core.
Fluorescence correlation spectroscopy (FCS) autocorrelates the fluctuations of the intensity of fluorescent light, analog to DLS, caused by the diffusion of fluorescent nanoparticles through the observation volume.188,225 This method is limited to fluorescent particles but offers the advantage that non fluorescent components of the sample do not influence the autocorrelation function.47 Negwer et al. developed a new method utilizing FCS for the direct characterization of nanoparticles in flowing blood.75 For this purpose, they labeled the nanoparticles with near-infrared (NIR) fluorescent dyes and fitted the autocorrelation functions with an analytical model accounting for the presence of blood cells.
Particle tracking velocimetry (PTV) can be used to track the Brownian motion of a single nanoparticle and calculate its diffusion coefficient from the obtained data.208,209 In this setup, the particle suspension is illuminated, and the motion of individual scattering centers is tracked with multiple charge-coupled device (CCD) cameras.209 Due to the measurement principle, only analytes with a low particle concentration can be investigated by PTV. Di Silvio and co-workers used particle tracking analysis to investigate different nanoparticle–protein complexes isolated from complex biofluids by sucrose gradient ultracentrifugation.179 The sizes of these complexes were comparable to that determined in situ. In contrast, isolation via conventional centrifugation had a bigger impact on the nanoparticle–protein complexes. The results were confirmed by DLS measurements. The authors concluded that ultracentrifugation could isolate and recover nanoparticle–protein complexes in stable form with high size resolution.
2. Microscopy-based methods
Nanoparticles cannot be observed using light microscopy because they are a lot smaller than the abbe limit for visible light. Transmission electron microscopy (TEM) overcomes this limitation by using electrons instead of visible light, which have a very short de Broglie wavelength (around 3 pm depending on the acceleration voltage of the electron source) and therefore lower the abbe limit. This makes it possible to visualize structures with a resolution of a few nanometers.226 One problem that arises with the typical dry preparation of samples for TEM imaging is the formation of artifacts formed during the drying process (e.g., compaction of sample constituents in spots). The dry preparation is especially a problem for samples containing biological materials as such samples significantly change their appearance during the drying process. Cryo-TEM offers a solution to this problem.191,210,211 Here, the sample is vitrified in a water film, which makes it possible to investigate the sample in its natural surroundings. A disadvantage of this preparation method is that the water film reduces the contrast of the sample. Hadjidemetriou et al., for example, compared the structure and morphology of liposomes before and after protein corona formation in vitro and in vivo by TEM and cryo-TEM.93 Structural integrity of the liposomes remained after isolation from both blood (in vivo) and plasma (in vitro), but the morphology of the protein coronas differed. The in vitro protein corona consisted of a network of linear fibrillary structures, whereas the in vivo corona appeared more compact but not covering the whole liposome surface. The authors assumed that these morphological differences were due to the different protein corona compositions in vitro and in vivo (higher content of fibrinogen molecules in the case of in vitro protein coronas). Additional DLS measurements revealed a shrinkage of liposomes upon protein corona formation, which was more pronounced for the in vivo protein corona. This effect was most likely osmotically driven.227
A further microscopy-based method is scanning electron microscopy (SEM).195 Here, the sample is scanned with an electron beam, and the intensity of the backscattered electrons is analyzed to create a topographic view of the sample.228 This method again has certain limitations since the sample itself has to be conductive or it has to be coated with a thin layer of gold or carbon. Mirshafiee et al. used SEM to evaluate the influence of human plasma on the size of silica nanoparticles, which were either uncoated or pre-coated with γ-globulin (GG) or human serum albumin (HSA).195 They found that the size changed only slightly (9 and 3 nm in the case of uncoated and HSA-coated nanoparticles) to not at all (in the case of GG-coated nanoparticles), but that there were less clustered nanoparticles, indicating that a protein corona was formed. Comparative analysis via DLS showed no increase but a decrease in size for HSA- and GG-coated nanoparticles, confirming the hypothesis of protein corona formation. In a next step, protein corona profiles were examined revealing an opsonin-rich corona for GG-coated but not for the other two nanoparticles. The expected enhanced uptake of GG-coated nanoparticles in macrophages, however, could not be observed, most probably due to unspecific absorption of other blocking plasma proteins.
Atomic force microscopy (AFM) is another method to obtain a topographic image of a sample.213 Hereby, a small tip with a radius of around 10 nm is attached to a cantilever and rastered over the sample surface using piezo-drives. The deflection of the cantilever is measured with a laser beam and a photo diode to calculate a topographic image of the sample. AFM can be used in a variety of different modes. The three most common modes are contact mode, non-contact mode, and tapping mode. Dobrovolskaia et al. applied different size characterization methods (DLS, TEM, AFM) to investigate the influence of human plasma on the size of colloidal gold nanoparticles.197 They found an almost twofold increase in the hydrodynamic size as measured by DLS but no effect on the nanoparticle size as determined by TEM and AFM. The authors supposed that this is most probably due to the different sample preparation techniques.
3. Fractionating methods based on hydrodynamic separation
The family of field-flow fractionation (FFF) methods contains a wide variety of methods.175,229,230 In general, the separation is achieved by the application of a physical field perpendicular to the direction the sample travels through a thin channel. This field leads to particle clouds of different heights for individual sample components. This in turn leads to different retention times because thicker particle clouds reach into regions of higher flow velocity in the parabolic flow profile, which is formed in the separation channel due to its small height.
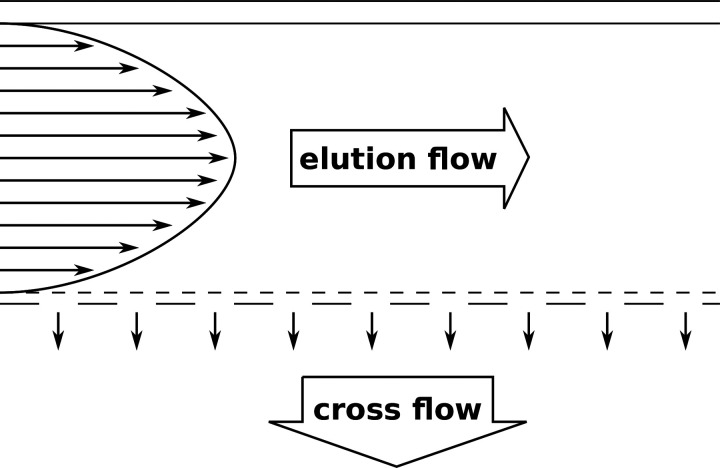
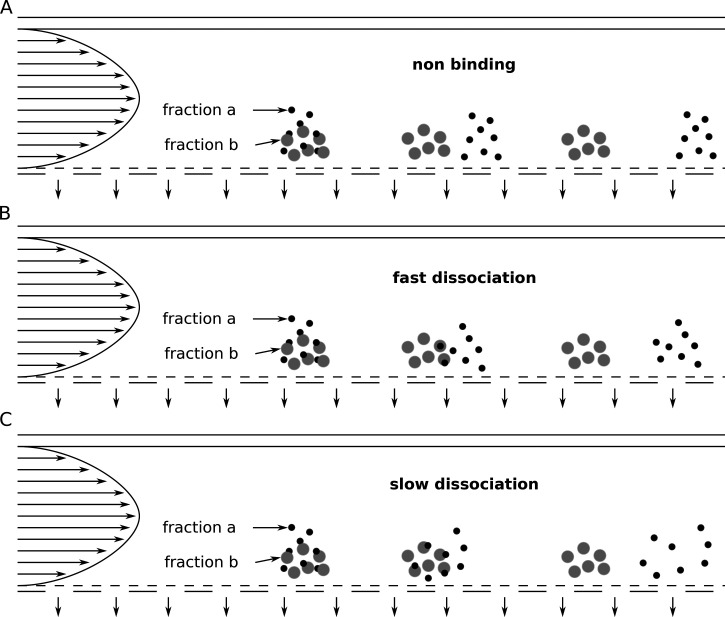
Examples for FFF methods are thermal FFF (Th-FFF),205,231–233 sedimentation FFF (Sd-FFF),234 electric FFF (E-FFF),235 and flow FFF (Fl-FFF).236 One particularly interesting FFF method is asymmetrical flow field-flow fractionation (AF4), which is a variant of Fl-FFF.180,198,232,237–242 Here, the fractionation of the sample components is achieved by a flow field. The AF4 setup consists of a separation channel with one permeable wall (the accumulation wall) through which a part of the eluent flows through and so creates the perpendicular flow field (Fig. 5). Depending on the particles' diffusion coefficients (typically for particles smaller than 1 μm), the sample components accumulate in particle clouds of different thicknesses. The retention times of individual sample components depend on the height of the particle cloud (components with higher diffusion coefficients form higher particle clouds). A higher particle cloud reaches into regions of higher flow velocity in the parabolic flow profile within the separation channel and thus elutes earlier. Figure 6 illustrates the impact of the dissociation rates of nanoparticle–protein complexes on the elution profile. When there is no interaction between the proteins (fraction a) and the nanoparticles (fraction b), the two fractions can be separated completely [Fig. 6(a)]. For rapidly dissociating complexes, the particle cloud of fraction a (= proteins) is broadened a bit [Fig. 6(b)]. In the case of slow dissociation, this effect is even more pronounced [Fig. 6(c)]. With AF4, also weakly bound proteins and thus the soft corona can be investigated.94 To mention a practical example, Bantz et al. used AF4, DLS, and TEM to investigate the stability of various silicon oxide-based nanoparticles with different surface properties (different surface charges and polarities, PEGylation) under physiological conditions.180 They showed that negatively charged silica nanoparticles were stable at physiological salt concentrations (150 mM sodium chloride) but aggregated in the presence of serum proteins. In contrast, positively charged poly(organosiloxane) nanoparticles macroscopically precipitated under physiological salt concentrations. In the presence of serum proteins, this was inhibited, but still large particles were formed. PEGylation hindered aggregation to a great extent. Alkylation of secondary amines led to increased sizes at physiological salinity, which were not observable in the presence of serum. All in all, this study showed that different surface properties of the nanoparticles have a huge impact on the stability under physiological conditions, which has in the last consequence also an influence on the biological performance.
FIG. 5.
The principle of asymmetrical flow field-flow fractionation (AF4).
FIG. 6.
Impact of different dissociation rates of nanoparticle–protein complexes on the elution profile in asymmetrical flow field-flow fractionation (AF4). Fraction a in black represents proteins, and fraction b in gray represents nanoparticles.
Size exclusion chromatography (SEC) is another fractionating method.95 Here, the fractionation takes place in a column packed with porous beads made from polystyrene crosslinked with divinylbenzene. Smaller sample components can diffuse deeper into these pores and therefore remain in the column longer than larger particles. Particles of a certain size depending on the packing material of the column cannot diffuse into these pores, and therefore, no fractionation takes place.95 This is called the upper exclusion limit. This problem does principally not occur in FFF methods due to the nature of its separation mechanism.214,230 A further advantage of FFF methods is the lower shear stress induced on the sample,72,214 which is especially relevant for sensitive samples such as biological cells or nanoparticle–protein complexes. Nevertheless, Gaus et al. successfully utilized SEC to investigate protein association with phosphorothioate-modified antisense oligonucleotides (ASOs) in plasma of different origin.112 To identify binding of ASOs to plasma fractions, ASOs were spiked with 125I-labeled ASOs; detection was done by UV-vis and β-RAM. The obtained binding profiles revealed species-specific differences. In the case of murine and human plasma, ASOs were mostly associated with albumin and histidine-rich glycopeptide (HRG). In contrast, ASOs complexed predominantly with HRG in monkey plasma. The authors found that HRG bound to ASOs with a very high affinity. They claimed that this could be of relevance for in vivo efficacy especially in monkeys, which showed highest HRG plasma levels among the tested species.
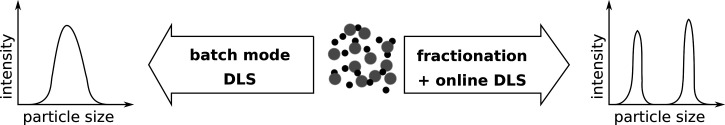
For both methods (FFF, SEC), the sample detection is done with a detector in line with the channel respectively column. Depending on the sample and the physical property of interest, a variety of different detection methods can be used, for example, UV-absorbance, fluorescence detection, refractive index (RI), multi-angle laser light scattering (MALLS), SLS, DLS, or ICP-MS.174 Comparing classical DLS (batch mode) with AF4 online DLS, the latter allows for a better assessment of the batch-to-batch variability and changes in the nanoparticle size induced by the interaction with serum proteins.243 In general, fractionation coupled with light scattering methods has the advantage over batch mode measurements that the size distribution can be determined more realistically (Fig. 7).
FIG. 7.
Comparison of the size determination of heterogeneous particle mixtures by dynamic light scattering (DLS) via batch mode measurement (left) vs prior fractionation (right).
Analytical ultra-centrifugation (AUC) is an absolute analytical method, which does not require a calibration with standards.215,244 In the experimental setup, the sample is spun at high rotational speeds and the sample concentration along the radius is measured using, for example, a UV-absorbance detector. The sample components start to move away from the rotational axis due to centrifugal forces, which causes a change of the sample concentration along the radius. In general, AUC can be used in two modes. The first mode is called sedimentation-diffusion equilibrium (SE) ultra-centrifugation. Here, the sample is spun until a concentration equilibrium between sedimentation and back diffusion is reached. The sample's molecular weight can be derived from this equilibrium concentration profile. The second mode is called sedimentation velocity (SV) ultra-centrifugation. In this mode, sedimentation coefficients can be determined by observing the change of the concentration profile in the sample cell over time. Schaefer et al. characterized the agglomeration state of four batches (A-C synthesized via flame pyrolysis; D synthesized via precipitation) of cerium oxide (CeO2) nanoparticles in fetal calf serum by AUC with interference detection.196 Two of these batches (A, B) showed low agglomeration tendency, indicating an effectively formed protein corona as adsorbed proteins are thought to promote stabilization. In contrast, the other two batches (C, D) showed a higher tendency to agglomerate, suggesting less effective protein corona formation. These results were in line with findings of AFM measurements with a BSA-modified tip. The first two batches (A, B) had a higher affinity toward BSA compared to the other two batches (C, D). In addition, densitometry revealed smaller Hill slopes for batches C and D compared to A and B, indicating once more a lower adsorption behavior. Altogether, the study demonstrated that there were big variations in the interaction with proteins between the different CeO2 nanoparticle batches. This could not be explained by their intrinsic physico-chemical properties (hydrodynamic diameter, zeta potential, pH value) as these were only minimally different. The authors concluded that it is of great importance to investigate the in situ properties of nanoparticles with a combination of various proper characterization methods.
Differential centrifugal sedimentation (DCS) is a further fractionating analytical method, which can be used to determine the size distribution of a given sample.245,246 Here, the sample is injected in the center of a spinning disk containing a concentration gradient of an aqueous sucrose solution. A UV-absorbance detector located close to the outer circumference of the disk is used to measure the sedimentation time of the sample and its concentration. DCS can be used as a detection method not only for the hard corona but also for the soft corona.246 Walczyk et al. applied various techniques (DCS, DLS, TEM) to characterize different polystyrene and silica nanoparticles in human plasma without (in situ) and with prior separation from excess plasma proteins (ex situ).65 The results of all three methods were consistent, demonstrating a robust protein coating on the nanoparticle surfaces upon plasma incubation with no significant difference between the in situ and the ex situ approach. This indicated that the nanoparticle–protein complexes could be physically isolated without changing the structure. Time-resolved studies, however, revealed a size increase for the in situ approach but no changes in nanoparticle size for the ex vivo approach.
Notably, in contrast to the other fractionating methods, AUC and DCS typically do not allow the collection of the fractionated sample.
4. Other methods for the characterization of physico-chemical nanoparticle properties influenced by biofluids
Tunable resistive pulse sensing (TRPS) is a high-resolution technique for size and zeta-potential measurements of nanoparticle dispersions in complex media (such as blood serum or plasma)201,202,204 and has proved to be an alternative to other characterization methods as described in Secs. II E 1–II E 3.202–204,216 It works according to the Coulter Counter principle.217,247,248 Hereby, changes in the ionic current, caused by (nano)particles passing through a single size-tunable elastomeric pore (“blockade” event), are detected particle-by-particle.204 By monitoring the changes in blockade width, magnitude and frequency, zeta-potential, size, and concentration of colloidal dispersions can be determined in situ.201 The sensitivity can be improved by adjusting the pore size.249 Limitations of light scattering techniques (e.g., DLS or PTV) like the high sensitivity toward the presence of larger particles/agglomerates as well as the inability to characterize highly polydisperse systems do not play a role in TRPS measurements.202,204,216
Agarose gel electrophoresis can be used to assess serum stability of nucleic acid delivery systems.12,13,250–252 Free nucleic acid can be monitored by staining with, for example, ethidium bromide or GelRed. Berger et al., for example, tested the stability of plasmid DNA (pDNA) complexes in 90% serum.12 The serum stability was strongly dependent on the backbone of the peptide-like carriers as well as the length of the lipidic unit within the carriers. Cysteine-containing carriers led through crosslinking to more stable complexes; and longer fatty acids provided higher stability by hydrophobic interactions. Karimov et al. formed complexes from tyrosine-modified linear polyethylenimine (LPEI) 10 kDa and siRNA, which displayed good stability in 50% serum as determined via agarose gel shift assay.251 By incubation of the complexes in tumor tissue and cell lysates, they demonstrated that the complexes at least partially disassembled at these conditions, confirming the possibility of siRNA release from the complexes upon cellular internalization.
A serum stability turbidity assay253 was utilized by Kaczmarek et al. to detect serum-induced nanoparticle precipitation.254 For this purpose, absorbance measurements at 660 nm (no interference with serum components at this wavelength) were done after indicated serum incubation times. A decrease in optical transmittance corresponded to nanoparticle precipitation, as confirmed by the quantification of the cargo mRNA in the supernatant. With this assay, they optimized co-formulations of poly(β-amino esters) (PBAEs) with PEG-lipid for mRNA delivery. Serum stability was achieved by higher amounts of PEG-lipid. The optimized nanoparticles exhibited increased in vitro mRNA transfection efficiency and functional mRNA delivery to the lungs upon systemic application in mice.
X-ray based techniques (imaging, spectroscopy, scattering) to investigate the nano-bio interface are reviewed in detail by Sanchez-Cano et al.255 The authors expect that with improved compact x-ray sources such methods can be applied to study the fate of nanoparticles in situ in animals and even humans.
III. IMPACT OF THE PROTEIN CORONA ON THE BIOLOGICAL ACTIVITY OF NANOPARTICLES
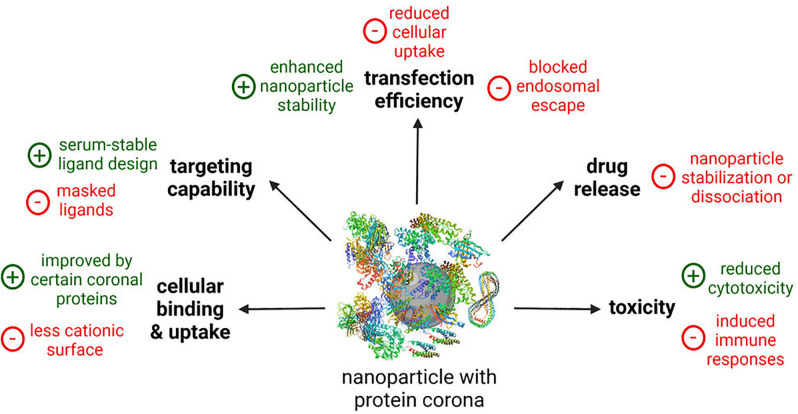
In addition to the aforementioned impact of the protein corona on physico-chemical properties of the nanoparticles in physiological environment (Sec. II), this protein corona also dictates the biological activity of the nanoparticles.22 The in vivo performance of the nanoparticles can completely differ from their in vitro behavior. Pharmacokinetics and biosafety can be completely changed compared to the in vitro experimental results. Therefore, more efforts have been made in recent years to develop in vitro assays that better describe the in vivo situation. In this context, the focus is on the effects of the protein corona on cellular binding and internalization, targeting capability, drug release, transfection efficiency, and toxicity of the nanoparticles (Fig. 8). Table III gives an overview over selected examples, which are also described in more detail in the text.
FIG. 8.
Effects of the protein corona on the biological activity of nanoparticles. Created with BioRender.com.
TABLE III.
Examples investigating the biological activity of nanoparticles in the presence of biofluids. ApoA4/C3/H, apolipoprotein A4/C3/H; BPEI, branched polyethylenimine; pDNA, plasmid DNA; FBS, fetal bovine serum; FCS, fetal calf serum; hb-PG, hyper-branched polyglycerol; HP, human plasma; HS, human serum; i.v., intravenous; LNPs, lipid nanoparticles; LPEI, linear polyethylenimine; ND, not determined; NPs, nanoparticles; mAB, monoclonal antibody; PC, protein corona; PEG, polyethylene glycol; PEI, polyethylenimine; siRNA, small interfering RNA; SPIONs, superparamagnetic iron oxide NPs; Tf, transferrin; hTf, human Tf. Cell lines: 1321N1, human brain astrocytoma cell line; ECV 30, spontaneously transformed, human umbilical vein endothelial cell line; H441, human lung adenocarcinoma cell line; HEK293, human embryonic kidney cells; HeLa, human cervix carcinoma cells; hMSCs, human mesenchymal stem cells; Huh7, human hepatocellular carcinoma cell line; J774, murine macrophage cell line; MCF-7, human breast adenocarcinoma; N2a, murine neuroblastoma cell line Neuro2a; PC3, human prostate carcinoma cell line; RAW264.7, murine monocyte/macrophage-like cell line.
| Nanoparticles | Biofluid | Protein corona | Biological impact | Reference |
|---|---|---|---|---|
| Cellular uptake and internalization | ||||
| Polystyrene NPs | 10% FBS, static vs dynamic incubation | Dynamic incubation: protein-enriched corona (esp. plasminogen) | Dynamic incubation: reduced binding to HeLa cells | 26 |
| Unmodified LNPs | 50% FBS, static vs dynamic incubation | Dynamic incubation: increased levels of α1-antitrypsin in the PC | Dynamic incubation: increased uptake in HeLa cells, whereas decreased uptake in MCF-7 cells | 27 |
| Differently functionalized polystyrene NPs | HS, ratio of total particle-surface area to serum concentration = 5 ml/m2 | ApoH-enriched PC | Increased uptake in HeLa cells and hMSCs | 193 |
| ApoA4- and ApoC3-enriched PC | Decreased uptake in HeLa cells and hMSCs | |||
| Liposomes modified with PEG or hb-PG | 5% and 100% HP | neglectable PC formation | PEG: decreased uptake in RAW264.7 cells; hb-PG: increased uptake in RAW264.7 cells | 29 |
| Targeting capability | ||||
| hTf-functionalized silica NPs | 10% FBS | ND | Abolished targeting efficiency | 34 |
| Tf-modified virus-like nanoparticles | 55% FBS, murine, or chicken serum | Minor PC formation | No effect on targeting efficiency | 256 |
| 55% HS | Reduced targeting efficiency (competing hTf) | |||
| mAB-conjugated liposomes | CD-1 mouse plasma in vitro vs in vivo | Amount of adsorbed proteins comparable for in vitro and in vivo PC, but in vivo PC composition more complex | No full inhibition of the targeting efficiency for the in vivo PC, but for the in vitro PC | 93 |
| Drug release | ||||
| 4-nitroanisole loaded polymeric nanocapsules | 10% or 100% FBS | ND | Minimal change in release profile | 53 |
| Tamoxifen-loaded SPIONs | 10% or 100% FBS | ND | Reduced burst effect | |
| Albumin-bound paclitaxel drug (Abraxane®) | 10% or 100% FBS or HP | ND | ||
| Transfection efficiency | ||||
| Carboxymethyl poly(L-histidine)/poly(β-amino ester) (PbAE)/pDNA ternary complexes | 5 – 50% FBS | ND | HEK293 cells: improved serum resistance and gene transfer | 257 |
| PEG-coated polyplex micelles loaded with bundled mRNA | 50% FBS | ND | Improved serum stability and transfection efficiency (Huh7 cells) | 250 |
| T-shape oligoaminoamides/pDNA complexes | 45%, 90% FBS | ND | N2a and Huh7 cells: decreased transfection efficiency in high serum due to inhibited lytic activity | 12 |
| Gene transfectants histone H1 and cationic lipid DOSPER | > 10% FCS | ND | ECV 304 cells: inhibited transfection efficiency, remedy by addition of calcium ions or chloroquine | 258 |
| Tyrosine-modified LPEI 10 kDa/siRNA complexes | 50% FCS | ND | H441 cells: no decrease in transfection efficiency | 251 |
| Tyrosine-modified disulfide-crosslinked BPEI 2 kDa/pDNA complexes | 50% FCS | ND | PC3 cells: no decrease in transfection efficiency | 252 |
| Toxicity | ||||
| Cationic polystyrene NPs | 10% fluorescent labeled FBS | ND | 1321N1 cells: reduced cytotoxicity due to masked cationic charges | 259 |
| Silica and polystyrene NPs | 90% HP | Rapidly formed PC containing >300 different proteins | Reduced hemolysis, thrombocyte activation, and endothelial cell death | 56 |
| Magnetic NPs | 2.5%, 10%, 40% HS | ND | No hemolytic effect | 260 |
| Silver NPs | 1 or 10% FBS | Strongly attached PC | J774 cells: decreased cytotoxicity due to sulfidation | 55 |
| PEI 5 kDa; PEI 25 kDa PEG-free and PEGylated (2 kDa, 20 kDa; different grafting degrees (1; 10)) | In vitro: HS; in vivo: pig model, i.v. injection | In vitro: formation of the complement terminal complex (SC5b-9); in vivo: cardiopulmonary changes in pigs | In vitro: complement activation only for PEG-free PEI 25 kDa; in vivo: PEG of ≥ 20 kDa may be favorable in terms of less complement activation | 25 |
A. Cellular binding and uptake
The impact of the protein corona on the cellular uptake of nanoparticles can be studied by flow cytometry and confocal microscopy.26,29,193,261,262 Silica nanoparticles for instance showed a stronger cell adhesion and enhanced cellular internalization under serum-free conditions as determined via flow cytometry, confocal, and electron microscopy.261 Another work evaluated the impact of static vs dynamic incubation with 10% fetal bovine serum (FBS) on protein corona formation and cellular binding efficiency of polystyrene nanoparticles.26 The protein corona composition was highly dependent on the initial mixing. Incubation under flow resulted in nanoparticle–protein complexes with protein-enriched (especially plasminogen) corona. The binding of these complexes to human cervical cancer cells (HeLa) was reduced compared to static incubation, as determined via flow cytometry. Palchetti et al. also found that changes in the protein corona induced by dynamic incubation with 50% FBS affected the uptake of unmodified LNPs in HeLa cells.27 However, in this study dynamic flow led to an increased internalization in HeLa cells most likely due to increased levels of α1-antitrypsin in the protein corona under flow conditions, an important promoter of nanoparticle–cell association.138 These results were cell line-dependent. In a second tested cell line, human breast adenocarcinoma (MCF-7) cells, circulating FBS (especially at longer exposure time), had less effect on the cellular uptake, leading only to a small decrease.27 Ritz et al. identified that cellular internalization of differently functionalized polystyrene nanoparticles is regulated by certain corona proteins.193 First, they determined the relative protein corona composition via quantitative LC-MS and correlated these findings with the cellular uptake into human cancer and bone marrow stem cells. In a next step, they validated key candidate proteins by artificially coating nanoparticles with the individual proteins. Apolipoprotein ApoH was found to increase cellular uptake, while apolipoproteins ApoA4 and ApoC3 acted rather as masking proteins and significantly decreased internalization. The effect of surface functionalization with two different hydrophilic polymers [PEG and hyper-branched polyglycerol (hb-PG)] on protein corona formation and internalization of liposomes in macrophages was investigated by Weber et al.29 The low protein adsorption was comparable for unmodified as well as both functionalized liposomal formulations, whereas the cellular uptake completely differed. PEGylation led to the expected decreased internalization. Functionalization with hb-PG, however, surprisingly enhanced the uptake independent of the protein corona. Thus, it is assumed that this is a liposome-specific rather than a protein corona effect.
Protein corona may also be positively required for functional delivery, as highlighted for the case of LNPs with diffusible PEG-lipids.8,263,264 In vivo accumulation and transfection potency of LNPs require apolipoprotein E (ApoE) adsorption to the nanoparticle surface in the bloodstream, resulting in receptor-mediated uptake by ApoE-dependent low-density lipoprotein (LDL) receptors on the sinusoidal surface of hepatocytes. In an ApoE knockout mouse model, the transfection activity was abolished.
B. Targeting capability
Ligands (especially peptide– and protein–ligands) are often not sufficiently stable in physiological environment and rapidly degraded (e.g., by serum proteases). Cyclization, amino acid modifications, or a so-called “retro-enantio” approach can be advantageous in terms of increased protease resistance and serum stability.265,266 In other cases, targeting ligands can be masked/blocked by components of the protein corona, which results in a reduced ligand recognition by receptors on the cell surface.31,267 By this, the targeting capability of nanoparticles can be drastically diminished in biological environment as proven in a model targeting reaction.268 Both the presence of 10% (mimicking in vitro conditions) and 100% (mimicking in vivo conditions) serum inhibited the copper-free click reaction between fluorescent cycloalkyne-functionalized nanoparticles and azide-bearing silicon substrate monolayer as determined via fluorescent and scanning electron microscopy. Salvati et al. demonstrated via flow cytometry that the targeting specificity of silica nanoparticles functionalized with human transferrin disappeared in the presence of already 10% fetal bovine serum (FBS).34 In contrast, Zackova Suchanova et al. showed that the protein corona formed in 55% FBS, mouse, or chicken serum did not influence transferrin-receptor (TfR) targeting of transferrin-modified virus-like nanoparticles.256 Serum proteins adsorbed only to a small extent as determined via SDS-PAGE, DLS, and TEM. However, in human serum a decreased targeting capability was observed due to the high content of competing human transferrin. TfR targeting in the presence of serum (different types and amounts) with and without human transferrin competition was evaluated via an enzyme-linked immunosorbent assay (ELISA) as well as by uptake studies in TfR-expressing cells via flow cytometry and confocal microscopy. A comparison between the in vitro and in vivo formed protein coronas and their impact on the targeting capability of monoclonal antibody-conjugated liposomes was conducted by Hadjidemetriou and colleagues.93 Both protein coronas significantly reduced cellular internalization as visualized with confocal microscopy. Interestingly, the in vivo protein corona, unlike the in vitro corona, did not lead to a full inhibition of the targeting efficiency. Finally, standard 2D cell culture systems do not at all represent the physiological real situation, with continuous blood flow and cells growing in all three dimensions. The ability of ligands to find and bind with their cellular targets can be more realistically evaluated in cellular adhesion models under flow conditions.269 Accessibility of target cells is better simulated in 3D multicellular spheroids and organoids.270–273 Such 3D culture systems display heterogeneous cell populations, cell-to-cell, and cell-to-extracellular matrix interactions.272 Thus, they recapitulate the in vivo situation to a greater extent compared to 2D cell monolayers.272 Spheroids are mostly used in cancer research, whereas, the more advanced organoids, derived from pluripotent stem cells, progenitor cells of specific tissues or patients,272,274 can be used for different disease models.275–278 Assembloids are generated by spatial organization of multiple cell types and are considered to even better mimic in vivo tissues.274,279 A combination of 3D cell culture and microfluidics can be realized with the “organ-on-a-chip” technology.270,272,280 Bioengineering can help to construct more physiologically relevant spheroid/organoid models, for example, by incorporating vasculature,280,281 microenvironment,272 or even the immune system.282,283 The microfluidic “organ-on-a-chip” systems enable high-throughput screening and are seen as potential alternative to animal models.272,278 This will speed up efficient preclinical research in the areas of drug discovery as well as precision, regenerative, and personalized medicine.274
C. Drug release
Controlled drug release is important for successful delivery, and an instant release has to be avoided.53 The protein corona can affect the drug release profile of nanocarriers in two contrary directions by (i) destabilizing the delivery system leading to disassembly or aggregation,84 or (ii) by additional shielding and stabilization.53 Instability in physiological environment can result in immediate drug release. In the case of therapeutic nucleic acids, this would lead to rapid clearance and ineffectiveness,10 whereas severe toxic effects would be the consequence in the case of chemotherapeutics (i.e., burst effect).53 Buyens et al. developed a fluorescence fluctuation spectroscopy-based in situ method to quantitatively investigate the integrity of siRNA–nanocarrier complexes in complex biological fluids like full human serum.84 Amin et al. evaluated the stability of liposomal doxorubicin nanoformulations in 30% serum by measuring the amount of free, released drug using high-performance liquid chromatography (HPLC).284 The detection of mRNA intactness after serum incubation via quantitative reverse transcriptase polymerase chain reaction (qRT-PCR) can be a method to assess serum stability (more precisely, serum-RNase resistance) of mRNA–nanocarrier systems.250,285–291 A detailed investigation of the influence of the protein corona on drug release profiles was performed by Behzadi and co-workers.53 They evaluated (i) tamoxifen-loaded SPIONs in 10% or 100% FBS; (ii) 4-nitroanisole loaded polymeric nanocapsules in 10% or 100% FBS; and (iii) albumin-bound paclitaxel drug (Abraxane®) in 10% or 100% FBS or human plasma. Drug release was determined by UV (in the case of tamoxifen) or HPLC (in the case of 4-nitroanisole and paclitaxel) after centrifugation or in situ. This study demonstrated that the drug release profiles are affected by the protein corona (i.e., types and amounts of corona proteins), but to a different extent for the different nanocarrier types. In the case of SPIONs and Abraxane®, the protein corona could strongly reduce the burst effect. For polymeric nanocapsules, the protein corona only slightly changed the release profile.
D. Transfection efficiency
Transfection experiments in high serum can help to better predict in vivo efficacies of nucleic acid nanocarriers.12,258,292 Read-out is done by reporter assays such as luciferase12,250–252,258,292 or eGFP (enhanced green fluorescent protein) expression assays.257,293 In vitro protocols often recommend transfection under serum-free conditions for optimum gene transfer, as transfection efficiency of lipidic nanocarriers was found to be inhibited even at the standard moderate amounts of ∼10% serum in the transfection medium.262,292,294 As prescreen for subsequent in vivo application, the use of 10% serum-supplemented medium has been frequently applied.293,295–297 However, optimization of nanocarriers for efficient delivery at high serum content (50% and higher), as done for instance by Chan et al. for their cationic liposome-DNA complexes,292 is advisable for a more reliable prediction of the in vivo transfection performance. Gu et al. improved the serum-resistance and gene-transfer efficiency in 50% serum of DNA-poly(β-amino ester) (PBAE) complexes through electrostatic coating with carboxymethyl poly(L-histidine) (CM-PLH), as demonstrated via flow cytometry and fluorescence microscopy.257 Koji et al. were able to improve serum stability and transfection efficiency in 50% serum by loading PEG-coated polyplex micelles with bundled mRNA (i.e., sterically stabilized, tight mRNA structure).250 Olden et al. needed higher polymer content in their mRNA and pDNA nanoformulations to achieve transfection efficiency in medium supplemented with 10% FBS.293 This can be explained by the fact that free polymers, known to facilitate gene transfer,298 are partially blocked by serum components. Berger et al. made a similar observation that serum-treated (45% and 90% serum) carriers exhibited reduced transfection efficiency.12 Cell culture screening in standard 10% serum-supplemented medium had identified lipo-peptide nanocarriers with high gene transfer potency; subsequent evaluation in full serum blocked the lytic potential of such lipo-peptide carriers. As endosomolytic potential is one of the most important promoters for endosomal escape and thus gene delivery, a decreased in vitro gene-transfer performance was observed in full serum. This also explained the moderate in vivo results, which fell short of the expectations of standard in vitro transfections (in the presence of 10% serum). Notably, in vitro gene transfer of gold standard LPEI 22 kDa was less to not affected by high serum content,12 which is in line with the good in vivo performance as demonstrated in many publications.298–303 The endosomal release of LPEI complexes apparently works according to mechanisms different from membranolytic interactions.304,305 Haberland et al. also found for other gene transfectants (histone H1 and cationic lipid DOSPER) that the endosomal escape was responsible for the reduced transfection efficiency in serum of 10% and higher.258 This serum inhibition could be overcome by calcium ions (in the form of nascent calcium phosphate micro-precipitates) and chloroquine in the cell culture medium, which both promote endosomal/lysosomal release through their fusogenic and membranolytic activity.306 Consistent with the above-mentioned good performance of LPEI in serum, Karimov et al. showed that the gene silencing efficiency of siRNA complexed with tyrosine-modified LPEI 10 kDa was not decreased in the presence of 50% serum.251 On the contrary, serum may even be advantageous regarding preservation of bioactivity during prolonged storage of the complexes, as shown for storage for three days at 4 °C, room temperature, and 37 °C. Similar findings were also made for pDNA complexed with disulfide-crosslinked, tyrosine-modified branched PEI 2 kDa.252
E. Toxicity
The protein corona impacts the biosafety and toxicity profile of nanoparticles.54,56 In particular, cationic nanoparticles are prone to interfere with the (predominately) negatively charged bio membranes, resulting in membrane disruption at multiple stages.10,304 The formed protein corona can more or less shield the nanoparticles and by this reduce interactions with cell membranes (e.g., of thrombocytes, erythrocytes, or endothelial cells). This protective effect of the protein corona was demonstrated for instance by Dawson and his team.259 The adsorbed serum proteins on cationic polystyrene nanoparticles prevented cell damage induced by the bare nanoparticle surface until the protein corona was enzymatically degraded in the lysosomes. In the context of pathophysiology, the protein corona can lower the risk for nanoparticle-induced thrombocyte activation/aggregation, erythrocyte aggregation or hemolysis, and cell death in general. Tenzer et al. demonstrated this and showed that the rapidly formed protein corona strongly improved the toxicity profile of the tested nanoparticles.56 Cytotoxicity can be assessed inter alia by cell viability assays (e.g., quantification of ATP)12,56 or by microscopic observation of cell morphology.307 Thrombocyte aggregation can be evaluated via aggregometry measurements.56 An assay to visualize nanoparticle-induced erythrocyte aggregation was developed by Ogris et al.23 Yallapu et al. investigated the interaction of magnetic nanoparticles with erythrocytes via a hemolysis assay (spectrophotometric quantification of hemoglobin release) and SEM.260 Both the nanoparticles without and with protein coronas showed no hemolytic activity. A detailed study on membrane interactions of gold nanoparticles was conducted by Wang et al.136 They found a protective effect of the serum protein corona against cell membrane damage. Eventual cell membrane damage was evaluated by environmental SEM as well as TEM, and quantification of LDH (lactate dehydrogenase) release. Cytotoxicity was measured by an apoptosis/necrosis ratio analysis using flow cytometry, a CCK-8 assay to determine the activity of the mitochondrial dehydrogenase, and a live/dead assay.
The protein corona can also alter the biotransformation of the nanoparticles as found for instance for silver nanoparticles.55 In this study, the hard corona mediated sulfidation, resulting in decreased cytotoxicity.
In contrast, immune responses (innate as well as adaptive) may be triggered by protein corona components (e.g., by stimulation of immune cells or by complement activation), which may lead to immunotoxic effects.54 Cationic nanoparticles, for example, can directly bind complement proteins and activate the alternative pathway of complement, or subsequent to protein binding the classical pathway, often resulting in serious toxicity.24,25
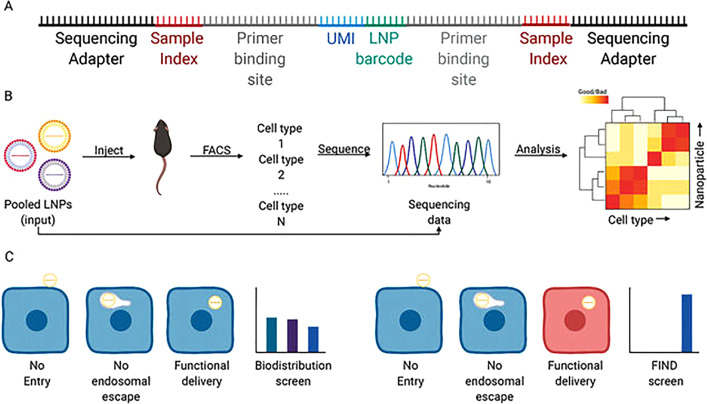
IV. IN VIVO SCREENING USING BARCODED NANOPARTICLES
Despite of all the additional information about nanoparticle properties in biofluids that can be obtained using the in vitro methods described in Secs. II and III, there are still uncertainties about the in vivo performance. Biodistribution and corresponding off-target effects, for example, can be hardly predicted with in vitro experiments alone, making in vivo studies inevitable. Dahlman and co-workers developed a high-throughput in vivo screening method, where simultaneously hundreds of nanoparticles can be tested within a single mouse.46,308 This not only accelerates the discovery of potent in vivo delivery systems and reduces the costs of in vivo studies but also is beneficial in view of the so-called “3R principle” (i.e., replace, reduce, refine) for a rational use of animals. This technology utilizes DNA barcodes, which are individually formulated into chemically distinct nanoparticles. DNA barcodes are single-stranded DNA oligonucleotides (∼60 nucleotides) with terminal stabilizing phosphorothioate modifications, 8–10 central nucleotides serving as individual barcode, and the 3′- and 5′-ends as priming sites for next-generation Illumina deep sequencing46 [Fig. 9(a)]. The different barcoded nanoparticles are then co-administrated in mice and later on quantified simultaneously [Fig. 9(b)]. Initially, Dahlman et al. demonstrated the predictability of the in vivo biodistribution of siRNA LNPs by this DNA barcoding system.46 In a follow-up study, this technique, which is named Joint Rapid DNA Analysis of Nanoparticles (JORDAN), proved to enable analysis of hundreds of nanoparticles at the same time.14,309 Subsequently, improvements were made regarding DNA barcode stability310 and optimization of DNA-amplification (e.g., QUANT barcodes for a more sensitive multiplexed analysis by Droplet Digital PCR).311 In early works, only biodistribution was investigated,14,46,308,311 whereas later on functional testing was possible by using a high-throughput method to quantify functional mRNA312–314 or siRNA delivery,315,316 which is termed Fast Identification of Nanoparticle Delivery (FIND). Individual DNA barcodes and the functional nucleic acid (e.g., specific mRNA or siRNA) were co-formulated in single nanoparticles and applied in appropriate reporter mouse models [Fig. 9(c)]. After isolating successfully transfected cells by fluorescence activated cell sorting (FACS), they were deep sequenced to identify the bioactive nanoparticles. To sum up, with this new high-throughput barcoding system, screening of several hundreds of nanoparticles at once in vivo is possible. It allows the investigation of functional delivery alongside biodistribution.317 By this, knowledge about the on-target to off-target ratio of nanoparticulate delivery systems can be gained, which is important for developing and improving therapeutics such as RNA therapeutics for COVID or cystic fibrosis.317,318
FIG. 9.
Principle of the high-throughput barcoding system developed by Dahlman and co-workers. (a) Structure of the DNA barcodes with 8–10 central nucleotides (green) as barcode region. (b) Several hundred different barcoded nanoparticles (e.g., lipid nanoparticles, LNPs) are co-administrated in mice. With next-generation sequencing, the in vivo biodistribution of the distinct barcoded nanoparticles can be analyzed simultaneously. This technology is termed Joint Rapid DNA Analysis of Nanoparticles (JORDAN). (c) JORDAN does not allow for functional delivery screening, as this method does not discriminate between nanoparticles outside or inside the cells (left). Fast Identification of Nanoparticle Delivery (FIND) provides a remedy (right). Here, successfully transfected reporter cells are identified. Reproduced with permission from Adv. Healthcare Mater. 10, e2002022 (2021).317 Copyright 2021 John Wiley and Sons.
Another barcoding system was developed by Yaari et al. to detect the therapeutic potential of different anticancer drugs (gemcitabine, doxorubicin, cisplatin) even at the single-cell level.319 They loaded liposomes with various chemotherapeutics and corresponding double-stranded DNA barcodes, which differed in length, sequence, and primers. Detection of the distinct DNA barcodes was done—in contrast to the next-generation sequencing used by Dahlman and co-workers—by real-time PCR, gel electrophoresis, and conventional sequencing. A correlation was found between the barcode distribution in cells and the therapeutic potency.
All in all, these barcoding techniques have the potential to make the preclinical pipeline more efficient320 and, as a diagnostic tool, to lead to personalized therapy.319
V. CONCLUSION
When predicting in vivo performance from in vitro data, it has to be considered that each of the above-mentioned characterization methods has advantages and limitations. None of these methods is able to completely illustrate the nanoparticle properties in physiological environment.174 The experimental setup and the characterization technique chosen can have a huge impact on the outcome.72,102 Combination of several analytical and biological as well as ex situ and in situ methods is advisable to get a better and more detailed insight into the in vivo characteristics and behavior of the nanoparticles. In addition, the choice of the biofluid (serum, plasma, or full blood; animal origin) is of great importance as different biological fluids can have a huge impact on the resulting protein corona composition and thus also on the physico-chemical and biological properties of the nanoparticle. This has been demonstrated in several publications.321–328 However, up until now, this aspect has been rather neglected and biofluids have been used inconsistently and interchangeably. For the future, it is recommended that the biofluid source for in vitro investigation is selected matching to the in vivo studies.321,323,324 Furthermore, testing in human plasma is considered to be one step closer to the translatability of the nanoparticles' performance in humans.321,323
Physiologically relevant in vitro settings include, for example, screening in (i) full serum,12 (ii) 3D multicellular spheroid and organoid cell culture,272,273,280 (iii) static and dynamic air–liquid interfaces,35,329–332 (iv) BBB models38,45 and other disease models,35,40–42 or (v) cellular adhesion models under flow conditions.269 Moreover, the gained knowledge about protein corona formation can be exploited to optimize carriers for nanomedical application.22
However, some information like in vivo biodistribution and off-target effects cannot be obtained from in vitro experiments. Consequently, in vivo studies are still necessary. With new high-throughput in vivo screening methods like the barcoding system of Dahlman and co-workers,46 such in vivo investigations can be more effective, economical, and ethical.
The main goal is to generate large datasets about nanoparticle characteristic in physiological environment and analyze them appropriately.320 In this respect, the establishment of standardized protocols is of great importance for more consistent, robust, and comprehensive pre-clinical studies (in vitro characterization, animal models) with reproducible and reliable results.212 By this, structure–activity relationships and in vitro–in vivo correlations can be derived.17,320 This knowledge can then be transferred to the rational design of nanoparticles for specific cargos and specific target cell types.
However, there is still an uncertainty about translatability from small to large animals and humans.320,333–335 So far, there are no systematic studies available, which address this subject. Species- and strain-dependent biological factors can influence the nanoparticle delivery. In the future, the question how well preclinical animal models predict nanoparticle performance in humans has to be investigated in more detail. Bioinformatics could help to identify best fitting animal models for certain diseases as recently shown for SARS-CoV-2.336 Alternatives to animal models such as in ovo testing, microfluidic “human-organ-on-a-chip” technology as well as in silico predictions can be promising strategies for replacing animal studies in the future.334,337,338
Another aspect, which has to be considered, is that the protein corona differs between individuals and is disease-specific.339–341 In this context, pharmacogenomics and personalized, patient-specific nanomedicine will gain importance.31
ACKNOWLEDGMENTS
The authors are greatly thankful for the support of their work by the German Research Foundation (DFG) via the Collaborative Research Centre SFB 1066. Moreover, acknowledgements to the DFG SFB1032 sub-project B4 for the financial support of Professor Dr. E. Wagner and his team.
AUTHOR DECLARATIONS
Conflict of Interest
The authors have no conflicts of interest to disclose.
DATA AVAILABILITY
Data sharing is not applicable as no new data were generated.
References
- 1. Cheng Z., Li M., Dey R., and Chen Y., J. Hematol. Oncol. 14, 85 (2021). 10.1186/s13045-021-01096-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Shi J., Kantoff P. W., Wooster R., and Farokhzad O. C., Nat. Rev. Cancer 17, 20 (2017). 10.1038/nrc.2016.108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Hager S., Fittler F. J., Wagner E., and Bros M., Cells 9, 2061 (2020). 10.3390/cells9092061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Freitag F. and Wagner E., Adv. Drug Delivery Rev. 168, 30 (2021). 10.1016/j.addr.2020.03.005 [DOI] [PubMed] [Google Scholar]
- 5. Barenholz Y., J. Controlled Release 160, 117 (2012). 10.1016/j.jconrel.2012.03.020 [DOI] [PubMed] [Google Scholar]
- 6. Verbeke R., Lentacker I., De Smedt S. C., and Dewitte H., J. Controlled Release 333, 511 (2021). 10.1016/j.jconrel.2021.03.043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Adams D., Gonzalez-Duarte A., O'Riordan W. D., Yang C. C., Ueda M., Kristen A. V., Tournev I., Schmidt H. H., Coelho T., Berk J. L., Lin K. P., Vita G., Attarian S., Planté-Bordeneuve V., Mezei M. M., Campistol J. M., Buades J., Brannagan T. H. III, Kim B. J., Oh J., Parman Y., Sekijima Y., Hawkins P. N., Solomon S. D., Polydefkis M., Dyck P. J., Gandhi P. J., Goyal S., Chen J., Strahs A. L., Nochur S. V., Sweetser M. T., Garg P. P., Vaishnaw A. K., Gollob J. A., and Suhr O. B., N. Engl. J. Med. 379, 11 (2018). 10.1056/NEJMoa1716153 [DOI] [PubMed] [Google Scholar]
- 8. Gillmore J. D., Gane E., Taubel J., Kao J., Fontana M., Maitland M. L., Seitzer J., O'Connell D., Walsh K. R., Wood K., Phillips J., Xu Y., Amaral A., Boyd A. P., Cehelsky J. E., McKee M. D., Schiermeier A., Harari O., Murphy A., Kyratsous C. A., Zambrowicz B., Soltys R., Gutstein D. E., Leonard J., Sepp-Lorenzino L., and Lebwohl D., N. Engl. J. Med. 385, 493 (2021). 10.1056/NEJMoa2107454 [DOI] [PubMed] [Google Scholar]
- 9. Peng L. and Wagner E., Biomacromolecules 20, 3613 (2019). 10.1021/acs.biomac.9b00999 [DOI] [PubMed] [Google Scholar]
- 10. Lächelt U. and Wagner E., Chem. Rev. 115, 11043 (2015). 10.1021/cr5006793 [DOI] [PubMed] [Google Scholar]
- 11. Hager S. and Wagner E., Expert Opin. Drug Delivery 15, 1067 (2018). 10.1080/17425247.2018.1526922 [DOI] [PubMed] [Google Scholar]
- 12. Berger S., Krhač Levačić A., Hörterer E., Wilk U., Benli-Hoppe T., Wang Y., Öztürk Ö., Luo J., and Wagner E., Biomacromolecules 22, 1282 (2021). 10.1021/acs.biomac.0c01779 [DOI] [PubMed] [Google Scholar]
- 13. Luo J., Schmaus J., Cui M., Hörterer E., Wilk U., Höhn M., Däther M., Berger S., Benli-Hoppe T., Peng L., and Wagner E., J. Controlled Release 329, 919 (2021). 10.1016/j.jconrel.2020.10.022 [DOI] [PubMed] [Google Scholar]
- 14. Paunovska K., Sago C. D., Monaco C. M., Hudson W. H., Castro M. G., Rudoltz T. G., Kalathoor S., Vanover D. A., Santangelo P. J., Ahmed R., Bryksin A. V., and Dahlman J. E., Nano Lett. 18, 2148 (2018). 10.1021/acs.nanolett.8b00432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Leng Q., Chou S. T., Scaria P. V., Woodle M. C., and Mixson A. J., J. Gene Med. 16, 317 (2014). 10.1002/jgm.2807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Zugates G. T., Peng W., Zumbuehl A., Jhunjhunwala S., Huang Y. H., Langer R., Sawicki J. A., and Anderson D. G., Mol. Ther. 15, 1306 (2007). 10.1038/sj.mt.6300132 [DOI] [PubMed] [Google Scholar]
- 17. Whitehead K. A., Matthews J., Chang P. H., Niroui F., Dorkin J. R., Severgnini M., and Anderson D. G., ACS Nano 6, 6922 (2012). 10.1021/nn301922x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Negron K., Khalasawi N., Lu B., Ho C. Y., Lee J., Shenoy S., Mao H. Q., Wang T. H., Hanes J., and Suk J. S., J. Controlled Release 303, 1 (2019). 10.1016/j.jconrel.2019.04.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Lynch I., Cedervall T., Lundqvist M., Cabaleiro-Lago C., Linse S., and Dawson K. A., Adv. Colloid Interface Sci. 134–135, 167 (2007). 10.1016/j.cis.2007.04.021 [DOI] [PubMed] [Google Scholar]
- 20. Monopoli M. P., Aberg C., Salvati A., and Dawson K. A., Nat. Nanotechnol. 7, 779 (2012). 10.1038/nnano.2012.207 [DOI] [PubMed] [Google Scholar]
- 21. Schöttler S., Becker G., Winzen S., Steinbach T., Mohr K., Landfester K., Mailänder V., and Wurm F. R., Nat. Nanotechnol. 11, 372 (2016). 10.1038/nnano.2015.330 [DOI] [PubMed] [Google Scholar]
- 22. Cai R. and Chen C., Adv. Mater. 31, e1805740 (2019). 10.1002/adma.201805740 [DOI] [PubMed] [Google Scholar]
- 23. Ogris M., Brunner S., Schüller S., Kircheis R., and Wagner E., Gene Ther. 6, 595 (1999). 10.1038/sj.gt.3300900 [DOI] [PubMed] [Google Scholar]
- 24. Plank C., Mechtler K., F. C. Szoka, Jr. , and Wagner E., Hum. Gene Ther. 7, 1437 (1996). 10.1089/hum.1996.7.12-1437 [DOI] [PubMed] [Google Scholar]
- 25. Merkel O. M., Urbanics R., Bedocs P., Rozsnyay Z., Rosivall L., Toth M., Kissel T., and Szebeni J., Biomaterials 32, 4936 (2011). 10.1016/j.biomaterials.2011.03.035 [DOI] [PubMed] [Google Scholar]
- 26. Jayaram D. T., Pustulka S. M., Mannino R. G., Lam W. A., and Payne C. K., Biophys. J. 115, 209 (2018). 10.1016/j.bpj.2018.02.036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Palchetti S., Pozzi D., Capriotti A. L., Barbera G., Chiozzi R. Z., Digiacomo L., Peruzzi G., Caracciolo G., and Laganà A., Colloids Surf. B 153, 263 (2017). 10.1016/j.colsurfb.2017.02.037 [DOI] [PubMed] [Google Scholar]
- 28. Alberg I., Kramer S., Schinnerer M., Hu Q., Seidl C., Leps C., Drude N., Möckel D., Rijcken C., Lammers T., Diken M., Maskos M., Morsbach S., Landfester K., Tenzer S., Barz M., and Zentel R., Small 16, 1907574 (2020). 10.1002/smll.201907574 [DOI] [PubMed] [Google Scholar]
- 29. Weber C., Voigt M., Simon J., Danner A. K., Frey H., Mailänder V., Helm M., Morsbach S., and Landfester K., Biomacromolecules 20, 2989 (2019). 10.1021/acs.biomac.9b00539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Koshkina O., Westmeier D., Lang T., Bantz C., Hahlbrock A., Würth C., Resch-Genger U., Braun U., Thiermann R., Weise C., Eravci M., Mohr B., Schlaad H., Stauber R. H., Docter D., Bertin A., and Maskos M., Macromol. Biosci. 16, 1287 (2016). 10.1002/mabi.201600074 [DOI] [PubMed] [Google Scholar]
- 31. Caracciolo G., Nanoscale 10, 4167 (2018). 10.1039/C7NR07450F [DOI] [PubMed] [Google Scholar]
- 32. Pozzi D., Colapicchioni V., Caracciolo G., Piovesana S., Capriotti A. L., Palchetti S., De Grossi S., Riccioli A., Amenitsch H., and Laganà A., Nanoscale 6, 2782 (2014). 10.1039/c3nr05559k [DOI] [PubMed] [Google Scholar]
- 33. Maeda H., Adv. Drug. Delivery Rev. 91, 3 (2015). 10.1016/j.addr.2015.01.002 [DOI] [PubMed] [Google Scholar]
- 34. Salvati A., Pitek A. S., Monopoli M. P., Prapainop K., Bombelli F. B., Hristov D. R., Kelly P. M., Åberg C., Mahon E., and Dawson K. A., Nat. Nanotechnol. 8, 137 (2013). 10.1038/nnano.2012.237 [DOI] [PubMed] [Google Scholar]
- 35. Selo M. A., Sake J. A., Kim K. J., and Ehrhardt C., Adv. Drug Delivery Rev. 177, 113862 (2021). 10.1016/j.addr.2021.113862 [DOI] [PubMed] [Google Scholar]
- 36. Martens T. F., Vercauteren D., Forier K., Deschout H., Remaut K., Paesen R., Ameloot M., Engbersen J. F., Demeester J., De Smedt S. C., and Braeckmans K., Nanomedicine (London, U. K.) 8, 1955 (2013). 10.2217/nnm.12.202 [DOI] [PubMed] [Google Scholar]
- 37. Neupane R., Boddu S. H. S., Abou-Dahech M. S., Bachu R. D., Terrero D., Babu R. J., and Tiwari A. K., Pharmaceutics 13, 960 (2021). 10.3390/pharmaceutics13070960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Hanafy A. S., Dietrich D., Fricker G., and Lamprecht A., Adv. Drug Delivery Rev. 176, 113859 (2021). 10.1016/j.addr.2021.113859 [DOI] [PubMed] [Google Scholar]
- 39. Bhatia S. N. and Ingber D. E., Nat. Biotechnol. 32, 760 (2014). 10.1038/nbt.2989 [DOI] [PubMed] [Google Scholar]
- 40. Zheng F., Fu F., Cheng Y., Wang C., Zhao Y., and Gu Z., Small 12, 2253 (2016). 10.1002/smll.201503208 [DOI] [PubMed] [Google Scholar]
- 41. Wang D., Cong Y., Deng Q., Han X., Zhang S., Zhao L., Luo Y., and Zhang X., Micromachines 12, 1106 (2021). 10.3390/mi12091106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Primavessy D., Metz J., Schnur S., Schneider M., Lehr C.-M., and Hittinger M., Eur. J. Pharm. Biopharm. 168, 62 (2021). 10.1016/j.ejpb.2021.08.005 [DOI] [PubMed] [Google Scholar]
- 43. Abd E., Yousef S. A., Pastore M. N., Telaprolu K., Mohammed Y. H., Namjoshi S., Grice J. E., and Roberts M. S., Clin. Pharmacol. 8, 163 (2016). 10.2147/CPAA.S64788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Garcia M. T. J., de Vasconcellos F. L. L., Raffier C. P., Roberts M. S., Grice J. E., Benson H. A. E., and Leite-Silva V. R., Curr. Top. Med. Chem. 18, 287 (2018). 10.2174/1568026618666180412153214 [DOI] [PubMed] [Google Scholar]
- 45. Onyema H. N., Berger M., Musyanovych A., Bantz C., Maskos M., and Freese C., Biointerphases 16, 021004 (2021). 10.1116/6.0000889 [DOI] [PubMed] [Google Scholar]
- 46. Dahlman J. E., Kauffman K. J., Xing Y., Shaw T. E., Mir F. F., Dlott C. C., Langer R., Anderson D. G., and Wang E. T., Proc. Natl. Acad. Sci. U. S. A. 114, 2060 (2017). 10.1073/pnas.1620874114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. García-Álvarez R. and Vallet-Regí M., Nanomaterials 11, 888 (2021). 10.3390/nano11040888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Ghosh G. and Panicker L., Soft Matter 17, 3855 (2021). 10.1039/D0SM02050H [DOI] [PubMed] [Google Scholar]
- 49. Baimanov D., Cai R., and Chen C., Bioconjugate Chem. 30, 1923 (2019). 10.1021/acs.bioconjchem.9b00348 [DOI] [PubMed] [Google Scholar]
- 50. Ke P. C., Lin S., Parak W. J., Davis T. P., and Caruso F., ACS Nano 11, 11773 (2017). 10.1021/acsnano.7b08008 [DOI] [PubMed] [Google Scholar]
- 51. Cedervall T., Lynch I., Lindman S., Berggård T., Thulin E., Nilsson H., Dawson K. A., and Linse S., Proc. Natl. Acad. Sci. U. S. A. 104, 2050 (2007). 10.1073/pnas.0608582104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Vroman L., Nature 196, 476 (1962). 10.1038/196476a0 [DOI] [PubMed] [Google Scholar]
- 53. Behzadi S., Serpooshan V., Sakhtianchi R., Müller B., Landfester K., Crespy D., and Mahmoudi M., Colloids Surf. B 123, 143 (2014). 10.1016/j.colsurfb.2014.09.009 [DOI] [PubMed] [Google Scholar]
- 54. Corbo C., Molinaro R., Parodi A., Toledano Furman N. E., Salvatore F., and Tasciotti E., Nanomedicine (London, U. K.) 11, 81 (2016). 10.2217/nnm.15.188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Miclăuş T., Beer C., Chevallier J., Scavenius C., Bochenkov V. E., Enghild J. J., and Sutherland D. S., Nat. Commun. 7, 11770 (2016). 10.1038/ncomms11770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Tenzer S., Docter D., Kuharev J., Musyanovych A., Fetz V., Hecht R., Schlenk F., Fischer D., Kiouptsi K., Reinhardt C., Landfester K., Schild H., Maskos M., Knauer S. K., and Stauber R. H., Nat. Nanotechnol. 8, 772 (2013). 10.1038/nnano.2013.181 [DOI] [PubMed] [Google Scholar]
- 57. Casals E., Pfaller T., Duschl A., Oostingh G. J., and Puntes V., ACS Nano 4, 3623 (2010). 10.1021/nn901372t [DOI] [PubMed] [Google Scholar]
- 58. Pisani C., Gaillard J. C., Odorico M., Nyalosaso J. L., Charnay C., Guari Y., Chopineau J., Devoisselle J. M., Armengaud J., and Prat O., Nanoscale 9, 1840 (2017). 10.1039/C6NR04765C [DOI] [PubMed] [Google Scholar]
- 59. Hadjidemetriou M., Al-Ahmady Z., and Kostarelos K., Nanoscale 8, 6948 (2016). 10.1039/C5NR09158F [DOI] [PubMed] [Google Scholar]
- 60. Weiss A. C. G., Krüger K., Besford Q. A., Schlenk M., Kempe K., Förster S., and Caruso F., ACS Appl. Mater. Interfaces 11, 2459 (2019). 10.1021/acsami.8b14307 [DOI] [PubMed] [Google Scholar]
- 61. Dell'Orco D., Lundqvist M., Oslakovic C., Cedervall T., and Linse S., PLoS One 5, e10949 (2010). 10.1371/journal.pone.0010949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Lundqvist M., Nat. Nanotechnol. 8, 701 (2013). 10.1038/nnano.2013.196 [DOI] [PubMed] [Google Scholar]
- 63. Lundqvist M., Stigler J., Cedervall T., Berggård T., Flanagan M. B., Lynch I., Elia G., and Dawson K., ACS Nano 5, 7503 (2011). 10.1021/nn202458g [DOI] [PubMed] [Google Scholar]
- 64. Barrán-Berdón A. L., Pozzi D., Caracciolo G., Capriotti A. L., Caruso G., Cavaliere C., Riccioli A., Palchetti S., and Laganà A., Langmuir 29, 6485 (2013). 10.1021/la401192x [DOI] [PubMed] [Google Scholar]
- 65. Walczyk D., Bombelli F. B., Monopoli M. P., Lynch I., and Dawson K. A., J. Am. Chem. Soc. 132, 5761 (2010). 10.1021/ja910675v [DOI] [PubMed] [Google Scholar]
- 66. Chen D., Parayath N., Ganesh S., Wang W., and Amiji M., Nanoscale 11, 18806 (2019). 10.1039/C9NR05788A [DOI] [PubMed] [Google Scholar]
- 67. Chen D., Ganesh S., Wang W., and Amiji M., Nanoscale 11, 8760 (2019). 10.1039/C8NR09855G [DOI] [PubMed] [Google Scholar]
- 68. Pozzi D., Caracciolo G., Capriotti A. L., Cavaliere C., La Barbera G., Anchordoquy T. J., and Laganà A., J. Proteomics 119, 209 (2015). 10.1016/j.jprot.2015.02.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Treuel L., Docter D., Maskos M., and Stauber R. H., Beilstein J. Nanotechnol. 6, 857 (2015). 10.3762/bjnano.6.88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Tenzer S., Docter D., Rosfa S., Wlodarski A., Kuharev J., Rekik A., Knauer S. K., Bantz C., Nawroth T., Bier C., Sirirattanapan J., Mann W., Treuel L., Zellner R., Maskos M., Schild H., and Stauber R. H., ACS Nano 5, 7155 (2011). 10.1021/nn201950e [DOI] [PubMed] [Google Scholar]
- 71. Lundqvist M., Stigler J., Elia G., Lynch I., Cedervall T., and Dawson K. A., Proc. Natl. Acad. Sci. U. S. A. 105, 14265 (2008). 10.1073/pnas.0805135105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Weber C., Morsbach S., and Landfester K., Angew. Chem., Int. Ed. 58, 12787 (2019). 10.1002/anie.201902323 [DOI] [PubMed] [Google Scholar]
- 73. Carrillo-Carrion C., Carril M., and Parak W. J., Curr. Opin. Biotechnol. 46, 106 (2017). 10.1016/j.copbio.2017.02.009 [DOI] [PubMed] [Google Scholar]
- 74. Kari O. K., Ndika J., Parkkila P., Louna A., Lajunen T., Puustinen A., Viitala T., Alenius H., and Urtti A., Nanoscale 12, 1728 (2020). 10.1039/C9NR08186K [DOI] [PubMed] [Google Scholar]
- 75. Negwer I., Best A., Schinnerer M., Schäfer O., Capeloa L., Wagner M., Schmidt M., Mailänder V., Helm M., Barz M., Butt H. J., and Koynov K., Nat. Commun. 9, 5306 (2018). 10.1038/s41467-018-07755-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Carril M., Padro D., Del Pino P., Carrillo-Carrion C., Gallego M., and Parak W. J., Nat. Commun. 8, 1542 (2017). 10.1038/s41467-017-01826-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Frost R., Langhammer C., and Cedervall T., Nanoscale 9, 3620 (2017). 10.1039/C6NR06399C [DOI] [PubMed] [Google Scholar]
- 78. Shang L. and Nienhaus G. U., Acc. Chem. Res. 50, 387 (2017). 10.1021/acs.accounts.6b00579 [DOI] [PubMed] [Google Scholar]
- 79. Lo Giudice M. C., Herda L. M., Polo E., and Dawson K. A., Nat. Commun. 7, 13475 (2016). 10.1038/ncomms13475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Balog S., Rodriguez-Lorenzo L., Monnier C. A., Obiols-Rabasa M., Rothen-Rutishauser B., Schurtenberger P., and Petri-Fink A., Nanoscale 7, 5991 (2015). 10.1039/C4NR06538G [DOI] [PubMed] [Google Scholar]
- 81. Nuhn L., Gietzen S., Mohr K., Fischer K., Toh K., Miyata K., Matsumoto Y., Kataoka K., Schmidt M., and Zentel R., Biomacromolecules 15, 1526 (2014). 10.1021/bm500199h [DOI] [PubMed] [Google Scholar]
- 82. Pitek A. S., O'Connell D., Mahon E., Monopoli M. P., Baldelli Bombelli F., and Dawson K. A., PLoS One 7, e40685 (2012). 10.1371/journal.pone.0040685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Rausch K., Reuter A., Fischer K., and Schmidt M., Biomacromolecules 11, 2836 (2010). 10.1021/bm100971q [DOI] [PubMed] [Google Scholar]
- 84. Buyens K., Lucas B., Raemdonck K., Braeckmans K., Vercammen J., Hendrix J., Engelborghs Y., De Smedt S. C., and Sanders N. N., J. Controlled Release 126, 67 (2008). 10.1016/j.jconrel.2007.10.024 [DOI] [PubMed] [Google Scholar]
- 85. Monopoli M. P., Pitek A. S., Lynch I., and Dawson K. A., in Nanomaterial Interfaces in Biology: Methods and Protocols, edited by Bergese P. and Hamad-Schifferli K. ( Humana Press, Totowa, NJ, 2013), p. 137. [Google Scholar]
- 86. Pozzi D., Caracciolo G., Digiacomo L., Colapicchioni V., Palchetti S., Capriotti A. L., Cavaliere C., Zenezini Chiozzi R., Puglisi A., and Laganà A., Nanoscale 7, 13958 (2015). 10.1039/C5NR03701H [DOI] [PubMed] [Google Scholar]
- 87. Palchetti S., Colapicchioni V., Digiacomo L., Caracciolo G., Pozzi D., Capriotti A. L., Barbera G. L., and Laganà A., Biochim. Biophys. Acta, Biomembr. 1858, 189 (2016). 10.1016/j.bbamem.2015.11.012 [DOI] [PubMed] [Google Scholar]
- 88. Simon J., Kuhn G., Fichter M., Gehring S., Landfester K., and Mailänder V., Cells 10, 132 (2021). 10.3390/cells10010132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Bai X., Wang J., Mu Q., and Su G., Front. Bioeng. Biotechnol. 9, 646708 (2021). 10.3389/fbioe.2021.646708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. García-Álvarez R., Hadjidemetriou M., Sánchez-Iglesias A., Liz-Marzán L. M., and Kostarelos K., Nanoscale 10, 1256 (2018). 10.1039/C7NR08322J [DOI] [PubMed] [Google Scholar]
- 91. Bertrand N., Grenier P., Mahmoudi M., Lima E. M., Appel E. A., Dormont F., Lim J. M., Karnik R., Langer R., and Farokhzad O. C., Nat. Commun. 8, 777 (2017). 10.1038/s41467-017-00600-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Corbo C., Molinaro R., Taraballi F., Toledano Furman N. E., Hartman K. A., Sherman M. B., De Rosa E., Kirui D. K., Salvatore F., and Tasciotti E., ACS Nano 11, 3262 (2017). 10.1021/acsnano.7b00376 [DOI] [PubMed] [Google Scholar]
- 93. Hadjidemetriou M., Al-Ahmady Z., Mazza M., Collins R. F., Dawson K., and Kostarelos K., ACS Nano 9, 8142 (2015). 10.1021/acsnano.5b03300 [DOI] [PubMed] [Google Scholar]
- 94. Weber C., Simon J., Mailänder V., Morsbach S., and Landfester K., Acta Biomater. 76, 217 (2018). 10.1016/j.actbio.2018.05.057 [DOI] [PubMed] [Google Scholar]
- 95. Brusotti G., Calleri E., Colombo R., Massolini G., Rinaldi F., and Temporini C., Chromatographia 81, 3 (2018). 10.1007/s10337-017-3380-5 [DOI] [Google Scholar]
- 96. Sakulkhu U., Maurizi L., Mahmoudi M., Motazacker M., Vries M., Gramoun A., Ollivier Beuzelin M.-G., Vallée J.-P., Rezaee F., and Hofmann H., Nanoscale 6, 11439 (2014). 10.1039/C4NR02793K [DOI] [PubMed] [Google Scholar]
- 97. Sakulkhu U., Mahmoudi M., Maurizi L., Salaklang J., and Hofmann H., Sci. Rep. 4, 5020 (2014). 10.1038/srep05020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Hu Z., Zhang H., Zhang Y., Wu R., and Zou H., Colloids Surf. B 121, 354 (2014). 10.1016/j.colsurfb.2014.06.016 [DOI] [PubMed] [Google Scholar]
- 99. Raesch S. S., Tenzer S., Storck W., Rurainski A., Selzer D., Ruge C. A., Perez-Gil J., Schaefer U. F., and Lehr C. M., ACS Nano 9, 11872 (2015). 10.1021/acsnano.5b04215 [DOI] [PubMed] [Google Scholar]
- 100. Bonvin D., Chiappe D., Moniatte M., Hofmann H., and Mionić Ebersold M., Analyst 142, 3805 (2017). 10.1039/C7AN00646B [DOI] [PubMed] [Google Scholar]
- 101. Cursi L., Vercellino S., McCafferty M. M., Sheridan E., Petseva V., Adumeau L., and Dawson K. A., Nanoscale 13, 16324 (2021). 10.1039/D1NR04582B [DOI] [PubMed] [Google Scholar]
- 102. Winzen S., Schoettler S., Baier G., Rosenauer C., Mailaender V., Landfester K., and Mohr K., Nanoscale 7, 2992 (2015). 10.1039/C4NR05982D [DOI] [PubMed] [Google Scholar]
- 103. Ding Z., Ma H., and Chen Y., RSC Adv. 4, 55290 (2014). 10.1039/C4RA09613D [DOI] [Google Scholar]
- 104. Fleischer C. C. and Payne C. K., J. Phys. Chem. B 118, 14017 (2014). 10.1021/jp502624n [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Lindman S., Lynch I., Thulin E., Nilsson H., Dawson K. A., and Linse S., Nano Lett. 7, 914 (2007). 10.1021/nl062743+ [DOI] [PubMed] [Google Scholar]
- 106. de Puig H., Federici S., Baxamusa S. H., Bergese P., and Hamad-Schifferli K., Small 7, 2477 (2011). 10.1002/smll.201100530 [DOI] [PubMed] [Google Scholar]
- 107. Chong Y., Ge C., Yang Z., Garate J. A., Gu Z., Weber J. K., Liu J., and Zhou R., ACS Nano 9, 5713 (2015). 10.1021/nn5066606 [DOI] [PubMed] [Google Scholar]
- 108. Kari O. K., Rojalin T., Salmaso S., Barattin M., Jarva H., Meri S., Yliperttula M., Viitala T., and Urtti A., Drug Delivery Transl. Res. 7, 228 (2017). 10.1007/s13346-016-0320-0 [DOI] [PubMed] [Google Scholar]
- 109. Oh Y. J., Koehler M., Lee Y., Mishra S., Park J. W., and Hinterdorfer P., Nano Lett. 19, 612 (2019). 10.1021/acs.nanolett.8b04883 [DOI] [PubMed] [Google Scholar]
- 110. Röcker C., Pötzl M., Zhang F., Parak W. J., and Nienhaus G. U., Nat. Nanotechnol. 4, 577 (2009). 10.1038/nnano.2009.195 [DOI] [PubMed] [Google Scholar]
- 111. Boulos S. P., Davis T. A., Yang J. A., Lohse S. E., Alkilany A. M., Holland L. A., and Murphy C. J., Langmuir 29, 14984 (2013). 10.1021/la402920f [DOI] [PubMed] [Google Scholar]
- 112. Gaus H. J., Gupta R., Chappell A. E., Østergaard M. E., Swayze E. E., and Seth P. P., Nucl. Acids Res. 47, 1110 (2019). 10.1093/nar/gky1260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Prakash T. P., Mullick A. E., Lee R. G., Yu J., Yeh S. T., Low A., Chappell A. E., Østergaard M. E., Murray S., Gaus H. J., Swayze E. E., and Seth P. P., Nucl. Acids Res. 47, 6029 (2019). 10.1093/nar/gkz354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Park S. J., Int. J. Nanomed. 15, 5783 (2020). 10.2147/IJN.S254808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Wang J., Jensen U. B., Jensen G. V., Shipovskov S., Balakrishnan V. S., Otzen D., Pedersen J. S., Besenbacher F., and Sutherland D. S., Nano Lett. 11, 4985 (2011). 10.1021/nl202940k [DOI] [PubMed] [Google Scholar]
- 116. Greenfield N. J., Nat. Protoc. 1, 2876 (2006). 10.1038/nprot.2006.202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Sutherland J. C., in Circular Dichroism and the Conformational Analysis of Biomolecules, edited by Fasman G. D. ( Springer US, Boston, MA, 1996), p. 599. [Google Scholar]
- 118. Wallace B. A., Nat. Struct. Biol. 7, 708 (2000). 10.1038/78915 [DOI] [PubMed] [Google Scholar]
- 119. Sanchez-Guzman D., Giraudon-Colas G., Marichal L., Boulard Y., Wien F., Degrouard J., Baeza-Squiban A., Pin S., Renault J. P., and Devineau S., ACS Nano 14, 9073 (2020). 10.1021/acsnano.0c04165 [DOI] [PubMed] [Google Scholar]
- 120. Yang H., Yang S., Kong J., Dong A., and Yu S., Nat. Protoc. 10, 382 (2015). 10.1038/nprot.2015.024 [DOI] [PubMed] [Google Scholar]
- 121. Wang M., Fu C., Liu X., Lin Z., Yang N., and Yu S., Nanoscale 7, 15191 (2015). 10.1039/C5NR04498G [DOI] [PubMed] [Google Scholar]
- 122. Shashilov V. A., Sikirzhytski V., Popova L. A., and Lednev I. K., Methods 52, 23 (2010). 10.1016/j.ymeth.2010.05.004 [DOI] [PubMed] [Google Scholar]
- 123. Zhang D., Neumann O., Wang H., Yuwono V. M., Barhoumi A., Perham M., Hartgerink J. D., Wittung-Stafshede P., and Halas N. J., Nano Lett. 9, 666 (2009). 10.1021/nl803054h [DOI] [PubMed] [Google Scholar]
- 124. Assfalg M., Ragona L., Pagano K., D'Onofrio M., Zanzoni S., Tomaselli S., and Molinari H., Biochim. Biophys. Acta Proteins Proteomics 1864, 102 (2016). 10.1016/j.bbapap.2015.04.024 [DOI] [PubMed] [Google Scholar]
- 125. Deka J., Paul A., and Chattopadhyay A., Nanoscale 2, 1405 (2010). 10.1039/c0nr00154f [DOI] [PubMed] [Google Scholar]
- 126. Xu L., Dong S., Hao J., Cui J., and Hoffmann H., Langmuir 33, 3047 (2017). 10.1021/acs.langmuir.6b04591 [DOI] [PubMed] [Google Scholar]
- 127. Casalini T., Limongelli V., Schmutz M., Som C., Jordan O., Wick P., Borchard G., and Perale G., Front. Bioeng. Biotechnol. 7, 268 (2019). 10.3389/fbioe.2019.00268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128. Kharazian B., Hadipour N. L., and Ejtehadi M. R., Int. J. Biochem. Cell Biol. 75, 162 (2016). 10.1016/j.biocel.2016.02.008 [DOI] [PubMed] [Google Scholar]
- 129. Mahmoudi M., Lynch I., Ejtehadi M. R., Monopoli M. P., Bombelli F. B., and Laurent S., Chem. Rev. 111, 5610 (2011). 10.1021/cr100440g [DOI] [PubMed] [Google Scholar]
- 130. Tavanti F., Pedone A., and Menziani M. C., Int. J. Mol. Sci. 20, 3539 (2019). 10.3390/ijms20143539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131. Brancolini G. and Tozzini V., Curr. Opin. Colloid Interface Sci. 41, 66 (2019). 10.1016/j.cocis.2018.12.001 [DOI] [Google Scholar]
- 132. Vilanova O., Mittag J. J., Kelly P. M., Milani S., Dawson K. A., Rädler J. O., and Franzese G., ACS Nano 10, 10842 (2016). 10.1021/acsnano.6b04858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133. Settanni G., Zhou J., Suo T., Schöttler S., Landfester K., Schmid F., and Mailänder V., Nanoscale 9, 2138 (2017). 10.1039/C6NR07022A [DOI] [PubMed] [Google Scholar]
- 134. Settanni G., Zhou J., and Schmid F., J. Phys.: Conf. Ser. 921, 012002 (2017). 10.1088/1742-6596/921/1/012002 [DOI] [Google Scholar]
- 135. Settanni G., Schäfer T., Muhl C., Barz M., and Schmid F., Comput. Struct. Biotechnol. J. 16, 543 (2018). 10.1016/j.csbj.2018.10.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136. Wang L., Li J., Pan J., Jiang X., Ji Y., Li Y., Qu Y., Zhao Y., Wu X., and Chen C., J. Am. Chem. Soc. 135, 17359 (2013). 10.1021/ja406924v [DOI] [PubMed] [Google Scholar]
- 137. Pino P. d, Pelaz B., Zhang Q., Maffre P., Nienhaus G. U., and Parak W. J., Mater. Horiz. 1, 301 (2014). 10.1039/C3MH00106G [DOI] [Google Scholar]
- 138. Liu R., Jiang W., Walkey C. D., Chan W. C., and Cohen Y., Nanoscale 7, 9664 (2015). 10.1039/C5NR01537E [DOI] [PubMed] [Google Scholar]
- 139. Bigdeli A., Palchetti S., Pozzi D., Hormozi-Nezhad M. R., Baldelli Bombelli F., Caracciolo G., and Mahmoudi M., ACS Nano 10, 3723 (2016). 10.1021/acsnano.6b00261 [DOI] [PubMed] [Google Scholar]
- 140. Xia X. R., Monteiro-Riviere N. A., and Riviere J. E., Nat. Nanotechnol. 5, 671 (2010). 10.1038/nnano.2010.164 [DOI] [PubMed] [Google Scholar]
- 141. Docter D., Distler U., Storck W., Kuharev J., Wünsch D., Hahlbrock A., Knauer S. K., Tenzer S., and Stauber R. H., Nat. Protoc. 9, 2030 (2014). 10.1038/nprot.2014.139 [DOI] [PubMed] [Google Scholar]
- 142. Svasti J. and Panijpan B., J. Chem. Educ. 54, 560 (1977). 10.1021/ed054p560 [DOI] [PubMed] [Google Scholar]
- 143. Gallagher S. R., Curr. Protoc. Protein Sci. 68, 10.1.1 (2012). 10.1002/0471140864.ps1001s68 [DOI] [PubMed] [Google Scholar]
- 144. Harper S. and Speicher D. W., Curr. Protoc. Protein Sci. 96, e87 (2019). 10.1002/cpps.87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145. Beer L. A. and Speicher D. W., Curr. Protoc. Protein Sci. 91, 10.5.1 (2018). 10.1002/cpps.48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146. Rosenfeld J., Capdevielle J., Guillemot J. C., and Ferrara P., Anal. Biochem. 203, 173 (1992). 10.1016/0003-2697(92)90061-B [DOI] [PubMed] [Google Scholar]
- 147. Jeno P., Mini T., Moes S., Hintermann E., and Horst M., Anal. Biochem. 224, 75 (1995). 10.1006/abio.1995.1010 [DOI] [PubMed] [Google Scholar]
- 148. Shevchenko A., Wilm M., Vorm O., and Mann M., Anal. Chem. 68, 850 (1996). 10.1021/ac950914h [DOI] [PubMed] [Google Scholar]
- 149. Wang H., Shang L., Maffre P., Hohmann S., Kirschhöfer F., Brenner-Weiß G., and Nienhaus G. U., Small 12, 5836 (2016). 10.1002/smll.201602283 [DOI] [PubMed] [Google Scholar]
- 150. Cedervall T., Lynch I., Foy M., Berggård T., Donnelly S. C., Cagney G., Linse S., and Dawson K. A., Angew. Chem., Int. Ed. 46, 5754 (2007). 10.1002/anie.200700465 [DOI] [PubMed] [Google Scholar]
- 151. Gaspari M. and Cuda G., Methods Mol. Biol. 790, 115 (2011). 10.1007/978-1-61779-319-6_9 [DOI] [PubMed] [Google Scholar]
- 152. Sielaff M., Kuharev J., Bohn T., Hahlbrock J., Bopp T., Tenzer S., and Distler U., J. Proteome Res. 16, 4060 (2017). 10.1021/acs.jproteome.7b00433 [DOI] [PubMed] [Google Scholar]
- 153. Eng J. K., McCormack A. L., and Yates J. R., J. Am. Soc. Mass Spectrom. 5, 976 (1994). 10.1016/1044-0305(94)80016-2 [DOI] [PubMed] [Google Scholar]
- 154. Perkins D. N., Pappin D. J., Creasy D. M., and Cottrell J. S., Electrophoresis 20, 3551 (1999). [DOI] [PubMed] [Google Scholar]
- 155. Searle B. C., Proteomics 10, 1265 (2010). 10.1002/pmic.200900437 [DOI] [PubMed] [Google Scholar]
- 156. Zhang R., Barton A., Brittenden J., Huang J. T.-J., and Crowther D., J. Proteomics Bioinf. 3, 260 (2010). 10.4172/jpb.1000149 [DOI] [Google Scholar]
- 157. Pozzi D., Caracciolo G., Capriotti A. L., Cavaliere C., Piovesana S., Colapicchioni V., Palchetti S., Riccioli A., and Laganà A., Mol. BioSyst. 10, 2815 (2014). 10.1039/C4MB00292J [DOI] [PubMed] [Google Scholar]
- 158. Silva J. C., Gorenstein M. V., Li G. Z., Vissers J. P., and Geromanos S. J., Mol. Cell Proteomics 5, 144 (2006). 10.1074/mcp.M500230-MCP200 [DOI] [PubMed] [Google Scholar]
- 159. Brown R. E., Jarvis K. L., and Hyland K. J., Anal. Biochem. 180, 136 (1989). 10.1016/0003-2697(89)90101-2 [DOI] [PubMed] [Google Scholar]
- 160. Smith P. K., Krohn R. I., Hermanson G. T., Mallia A. K., Gartner F. H., Provenzano M. D., Fujimoto E. K., Goeke N. M., Olson B. J., and Klenk D. C., Anal. Biochem. 150, 76 (1985). 10.1016/0003-2697(85)90442-7 [DOI] [PubMed] [Google Scholar]
- 161. Wiechelman K. J., Braun R. D., and Fitzpatrick J. D., Anal. Biochem. 175, 231 (1988). 10.1016/0003-2697(88)90383-1 [DOI] [PubMed] [Google Scholar]
- 162. Kessler R. J. and Fanestil D. D., Anal. Biochem. 159, 138 (1986). 10.1016/0003-2697(86)90318-0 [DOI] [PubMed] [Google Scholar]
- 163. Kaufman E. D., Belyea J., Johnson M. C., Nicholson Z. M., Ricks J. L., Shah P. K., Bayless M., Pettersson T., Feldotö Z., Blomberg E., Claesson P., and Franzen S., Langmuir 23, 6053 (2007). 10.1021/la063725a [DOI] [PubMed] [Google Scholar]
- 164. Brewer S. H., Glomm W. R., Johnson M. C., Knag M. K., and Franzen S., Langmuir 21, 9303 (2005). 10.1021/la050588t [DOI] [PubMed] [Google Scholar]
- 165. Dixon M. C., J. Biomol. Tech. 19, 151 (2008). [PMC free article] [PubMed] [Google Scholar]
- 166. Chen Q., Xu S., Liu Q., Masliyah J., and Xu Z., Adv. Colloid Interface Sci. 233, 94 (2016). 10.1016/j.cis.2015.10.004 [DOI] [PubMed] [Google Scholar]
- 167. Di Silvio D., Maccarini M., Parker R., Mackie A., Fragneto G., and Baldelli Bombelli F., J. Colloid Interface Sci. 504, 741 (2017). 10.1016/j.jcis.2017.05.086 [DOI] [PubMed] [Google Scholar]
- 168. Wang X., Wang M., Lei R., Zhu S. F., Zhao Y., and Chen C., ACS Nano 11, 4606 (2017). 10.1021/acsnano.7b00200 [DOI] [PubMed] [Google Scholar]
- 169. Wang Q., Lim M., Liu X., Wang Z., and Chen K. L., Environ. Sci. Technol. 50, 2301 (2016). 10.1021/acs.est.5b04694 [DOI] [PubMed] [Google Scholar]
- 170. Sebastiani F., Yanez Arteta M., Lindfors L., and Cárdenas M., J. Colloid Interface Sci. 610, 766–774 (2021). 10.1016/j.jcis.2021.11.117 [DOI] [PubMed] [Google Scholar]
- 171. Matczuk M., Legat J., Shtykov S. N., Jarosz M., and Timerbaev A. R., Electrophoresis 37, 2257 (2016). 10.1002/elps.201600152 [DOI] [PubMed] [Google Scholar]
- 172. Fernández-Iglesias N. and Bettmer J., Nanoscale 7, 14324 (2015). 10.1039/C5NR02625C [DOI] [PubMed] [Google Scholar]
- 173. Costa-Fernández J. M., Menéndez-Miranda M., Bouzas-Ramos D., Encinar J. R., and Sanz-Medel A., TrAC Trends Anal. Chem. 84, 139 (2016). 10.1016/j.trac.2016.06.001 [DOI] [Google Scholar]
- 174. Eslahian K. A., Lang T., Bantz C., Keller R., Sperling R., Docter D., Stauber R., and Maskos M., in Measuring Biological Impacts of Nanomaterials, edited by Wegener J. ( Springer International Publishing, Cham, 2016), p. 1. [Google Scholar]
- 175. Maskos M. and Stauber R. H., in Comprehensive Biomaterials, edited by Ducheyne P., Healy K. E., Hutmacher D. W.. et al. ( Elsevier, 2011), Vol. 3, p. 329. [Google Scholar]
- 176. Monopoli M. P., Walczyk D., Campbell A., Elia G., Lynch I., Bombelli F. B., and Dawson K. A., J. Am. Chem. Soc. 133, 2525 (2011). 10.1021/ja107583h [DOI] [PubMed] [Google Scholar]
- 177. Grun M. K., Suberi A., Shin K., Lee T., Gomerdinger V., Moscato Z. M., Piotrowski-Daspit A. S., and Saltzman W. M., Biomaterials 272, 120780 (2021). 10.1016/j.biomaterials.2021.120780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178. Koshkina O., Lang T., Thiermann R., Docter D., Stauber R. H., Secker C., Schlaad H., Weidner S., Mohr B., Maskos M., and Bertin A., Langmuir 31, 8873 (2015). 10.1021/acs.langmuir.5b00537 [DOI] [PubMed] [Google Scholar]
- 179. Di Silvio D., Rigby N., Bajka B., Mayes A., Mackie A., and Baldelli Bombelli F., Nanoscale 7, 11980 (2015). 10.1039/C5NR02618K [DOI] [PubMed] [Google Scholar]
- 180. Bantz C., Koshkina O., Lang T., Galla H.-J., Kirkpatrick C. J., Stauber R. H., and Maskos M., Beilstein J. Nanotechnol. 5, 1774 (2014). 10.3762/bjnano.5.188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181. Orts-Gil G., Natte K., Thiermann R., Girod M., Rades S., Kalbe H., Thünemann A. F., Maskos M., and Österle W., Colloids Surf. B 108, 110 (2013). 10.1016/j.colsurfb.2013.02.027 [DOI] [PubMed] [Google Scholar]
- 182. Bello Roufaï M. and Midoux P., Bioconjugate Chem. 12, 92 (2001). 10.1021/bc0000738 [DOI] [PubMed] [Google Scholar]
- 183. Sebastiani F., Yanez Arteta M., Lerche M., Porcar L., Lang C., Bragg R. A., Elmore C. S., Krishnamurthy V. R., Russell R. A., Darwish T., Pichler H., Waldie S., Moulin M., Haertlein M., Forsyth V. T., Lindfors L., and Cárdenas M., ACS Nano 15, 6709 (2021). 10.1021/acsnano.0c10064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184. Waldie S., Sebastiani F., Moulin M., Del Giudice R., Paracini N., Roosen-Runge F., Gerelli Y., Prevost S., Voss J. C., Darwish T. A., Yepuri N., Pichler H., Maric S., Forsyth V. T., Haertlein M., and Cárdenas M., Front. Chem. 9, 630152 (2021). 10.3389/fchem.2021.630152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185. Marichal L., Giraudon-Colas G., Cousin F., Thill A., Labarre J., Boulard Y., Aude J. C., Pin S., and Renault J. P., Langmuir 35, 10831 (2019). 10.1021/acs.langmuir.9b01373 [DOI] [PubMed] [Google Scholar]
- 186. Meissner J., Wu Y., Jestin J., Shelton W. A., Findenegg G. H., and Bharti B., Soft Matter 15, 350 (2019). 10.1039/C8SM01909F [DOI] [PubMed] [Google Scholar]
- 187. Klapper Y., Maffre P., Shang L., Ekdahl K. N., Nilsson B., Hettler S., Dries M., Gerthsen D., and Nienhaus G. U., Nanoscale 7, 9980 (2015). 10.1039/C5NR01694K [DOI] [PubMed] [Google Scholar]
- 188. Maffre P., Brandholt S., Nienhaus K., Shang L., Parak W. J., and Nienhaus G. U., Beilstein J. Nanotechnol. 5, 2036 (2014). 10.3762/bjnano.5.212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189. Milani S., Bombelli F. B., Pitek A. S., Dawson K. A., and Rädler J., ACS Nano 6, 2532 (2012). 10.1021/nn204951s [DOI] [PubMed] [Google Scholar]
- 190. Kittler S., Greulich C., Gebauer J., Diendorf J., Treuel L., Ruiz L., Gonzalez-Calbet J., Vallet-Regi M., Zellner R., and Köller M., J. Mater. Chem. 20, 512 (2010). 10.1039/B914875B [DOI] [Google Scholar]
- 191. Sheibani S., Basu K., Farnudi A., Ashkarran A., Ichikawa M., Presley J. F., Bui K. H., Ejtehadi M. R., Vali H., and Mahmoudi M., Nat. Commun. 12, 573 (2021). 10.1038/s41467-020-20884-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192. Klepac D., Kostková H., Petrova S., Chytil P., Etrych T., Kereïche S., Raška I., Weitz D. A., and Filippov S. K., Nanoscale 10, 6194 (2018). 10.1039/C7NR09355A [DOI] [PubMed] [Google Scholar]
- 193. Ritz S., Schöttler S., Kotman N., Baier G., Musyanovych A., Kuharev J., Landfester K., Schild H., Jahn O., Tenzer S., and Mailänder V., Biomacromolecules 16, 1311 (2015). 10.1021/acs.biomac.5b00108 [DOI] [PubMed] [Google Scholar]
- 194. Schäffler M., Semmler-Behnke M., Sarioglu H., Takenaka S., Wenk A., Schleh C., Hauck S. M., Johnston B. D., and Kreyling W. G., Nanotechnology 24, 265103 (2013). 10.1088/0957-4484/24/26/265103 [DOI] [PubMed] [Google Scholar]
- 195. Mirshafiee V., Kim R., Park S., Mahmoudi M., and Kraft M. L., Biomaterials 75, 295 (2016). 10.1016/j.biomaterials.2015.10.019 [DOI] [PubMed] [Google Scholar]
- 196. Schaefer J., Schulze C., Marxer E. E., Schaefer U. F., Wohlleben W., Bakowsky U., and Lehr C. M., ACS Nano 6, 4603 (2012). 10.1021/nn202657q [DOI] [PubMed] [Google Scholar]
- 197. Dobrovolskaia M. A., Patri A. K., Zheng J., Clogston J. D., Ayub N., Aggarwal P., Neun B. W., Hall J. B., and McNeil S. E., Nanomedicine (N. Y., NY, U. S.) 5, 106 (2009). 10.1016/j.nano.2008.08.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198. Ashby J., Schachermeyer S., Pan S., and Zhong W., Anal. Chem. 85, 7494 (2013). 10.1021/ac401485j [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199. Osborn M. F., Coles A. H., Biscans A., Haraszti R. A., Roux L., Davis S., Ly S., Echeverria D., Hassler M. R., Godinho B., Nikan M., and Khvorova A., Nucl. Acids Res. 47, 1070 (2019). 10.1093/nar/gky1232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200. Perez-Potti A., Lopez H., Pelaz B., Abdelmonem A., Soliman M. G., Schoen I., Kelly P. M., Dawson K. A., Parak W. J., Krpetic Z., and Monopoli M. P., Sci. Rep. 11, 6443 (2021). 10.1038/s41598-021-84029-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201. Blundell E., Healey M. J., Holton E., Sivakumaran M., Manstana S., and Platt M., Anal. Bioanal. Chem. 408, 5757 (2016). 10.1007/s00216-016-9678-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202. Sikora A., Shard A. G., and Minelli C., Langmuir 32, 2216 (2016). 10.1021/acs.langmuir.5b04160 [DOI] [PubMed] [Google Scholar]
- 203. Sikora A., Bartczak D., Geißler D., Kestens V., Roebben G., Ramaye Y., Varga Z., Palmai M., Shard A. G., Goenaga-Infante H., and Minelli C., Anal. Methods 7, 9835 (2015). 10.1039/C5AY02014J [DOI] [Google Scholar]
- 204. Pal A. K., Aalaei I., Gadde S., Gaines P., Schmidt D., Demokritou P., and Bello D., ACS Nano 8, 9003 (2014). 10.1021/nn502219q [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205. Eslahian K. A., Majee A., Maskos M., and Würger A., Soft Matter 10, 1931 (2014). 10.1039/c3sm52779d [DOI] [PubMed] [Google Scholar]
- 206. Di Cola E., Grillo I., and Ristori S., Pharmaceutics 8, 10 (2016). 10.3390/pharmaceutics8020010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207. Mast M.-P., Modh H., Champanhac C., Wang J.-W., Storm G., Krämer J., Mailänder V., Pastorin G., and Wacker M. G., Adv. Drug Delivery Rev. 179, 113829 (2021). 10.1016/j.addr.2021.113829 [DOI] [PubMed] [Google Scholar]
- 208. Matsuura Y., Nakamura A., and Kato H., Sens. Actuators, B 256, 1078 (2018). 10.1016/j.snb.2017.10.054 [DOI] [Google Scholar]
- 209. Maas H., Gruen A., and Papantoniou D., Exp. Fluids 15, 133 (1993). 10.1007/BF00190953 [DOI] [Google Scholar]
- 210. Subramaniam S., Earl L. A., Falconieri V., Milne J. L., and Egelman E. H., Curr. Opin. Struct. Biol. 41, 194 (2016). 10.1016/j.sbi.2016.07.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211. Milne J. L., Borgnia M. J., Bartesaghi A., Tran E. E., Earl L. A., Schauder D. M., Lengyel J., Pierson J., Patwardhan A., and Subramaniam S., FEBS J. 280, 28 (2013). 10.1111/febs.12078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212. Mahmoudi M., Nat. Commun. 12, 5246 (2021). 10.1038/s41467-021-25584-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213. Giessibl F. J., Rev. Mod. Phys. 75, 949 (2003). 10.1103/RevModPhys.75.949 [DOI] [Google Scholar]
- 214. Fraunhofer W. and Winter G., Eur. J. Pharm. Biopharm. 58, 369 (2004). 10.1016/j.ejpb.2004.03.034 [DOI] [PubMed] [Google Scholar]
- 215. Planken K. L. and Cölfen H., Nanoscale 2, 1849 (2010). 10.1039/c0nr00215a [DOI] [PubMed] [Google Scholar]
- 216. Anderson W., Kozak D., Coleman V. A., Jämting Å. K., and Trau M., J. Colloid Interface Sci. 405, 322 (2013). 10.1016/j.jcis.2013.02.030 [DOI] [PubMed] [Google Scholar]
- 217. Kozak D., Anderson W., Vogel R., and Trau M., Nano Today 6, 531 (2011). 10.1016/j.nantod.2011.08.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218. Schärtl W., Light Scattering from Polymer Solutions and Nanoparticle Dispersions, 1st ed. ( Springer-Verlag, Berlin/Heidelberg, 2007). [Google Scholar]
- 219. Meyer A., Dierks K., and Betzel C., Acta Crystallogr., Sect. A 70, C1749 (2014). 10.1107/S2053273314082503 [DOI] [Google Scholar]
- 220. Delgado Á. V., González-Caballero F., Hunter R., Koopal L., and Lyklema J., J. Colloid Interface Sci. 309, 194 (2007). 10.1016/j.jcis.2006.12.075 [DOI] [PubMed] [Google Scholar]
- 221. Hunter R. J., Introduction to Modern Colloid Science ( Oxford University Press, 1993). [Google Scholar]
- 222. Varenne F., Botton J., Merlet C., Vachon J.-J., Geiger S., Infante I. C., Chehimi M. M., and Vauthier C., Colloids Surf. A 486, 218 (2015). 10.1016/j.colsurfa.2015.08.044 [DOI] [Google Scholar]
- 223. Guinier A., Fournet G., and Yudowitch K. L., Small-Angle Scattering of X-Rays ( John Wiley & Sons Inc., 1955). [Google Scholar]
- 224. Glatter O., J. Appl. Crystallogr. 10, 415 (1977). 10.1107/S0021889877013879 [DOI] [Google Scholar]
- 225. Wang H., Lin Y., Nienhaus K., and Nienhaus G. U., Wiley Interdiscip. Rev.: Nanomed. Nanobiotechnol. 10, e1500 (2018). 10.1002/wnan.1500 [DOI] [PubMed] [Google Scholar]
- 226. Pennycook S. J., David B., and Williams C. B., Microsc. Microanal. 16, 111 (2010). 10.1017/S1431927609991140 [DOI] [Google Scholar]
- 227. Wolfram J., Suri K., Yang Y., Shen J., Celia C., Fresta M., Zhao Y., Shen H., and Ferrari M., Colloids Surf. B Biointerfaces 114, 294 (2014). 10.1016/j.colsurfb.2013.10.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228. Smith K. and Oatley C., Br. J. Appl. Phys. 6, 391 (1955). 10.1088/0508-3443/6/11/304 [DOI] [Google Scholar]
- 229. Schimpf M. E., Caldwell K., and Giddings J. C., Field-Flow Fractionation Handbook ( John Wiley & Sons, 2000). [Google Scholar]
- 230. Giddings J. C., Science 260, 1456 (1993). 10.1126/science.8502990 [DOI] [PubMed] [Google Scholar]
- 231. Eslahian K. A. and Maskos M., in Encyclopedia of Analytical Chemistry, edited by Meyers R. A. ( John Wiley & Sons, Ltd, 2015), p. 1. [Google Scholar]
- 232. Lang T., Eslahian K. A., and Maskos M., Macromol. Chem. Phys. 213, 2353 (2012). 10.1002/macp.201200132 [DOI] [Google Scholar]
- 233. Liu G. and Giddings J., Chromatographia 34, 483 (1992). 10.1007/BF02290241 [DOI] [Google Scholar]
- 234. Giddings J. C., Karaiskakis G., Caldwell K. D., and Myers M. N., J. Colloid Interface Sci. 92, 66 (1983). 10.1016/0021-9797(83)90117-0 [DOI] [Google Scholar]
- 235. Tri N., Caldwell K., and Beckett R., Anal. Chem. 72, 1823 (2000). 10.1021/ac990822i [DOI] [PubMed] [Google Scholar]
- 236. Giddings J. C., Yang F. J., and Myers M. N., Anal. Biochem. 81, 395 (1977). 10.1016/0003-2697(77)90710-2 [DOI] [PubMed] [Google Scholar]
- 237. Berger M., Scherer C., Noskov S., Bantz C., Nickel C., Schupp W., and Maskos M., J. Chromatogr. A 1640, 461941 (2021). 10.1016/j.chroma.2021.461941 [DOI] [PubMed] [Google Scholar]
- 238. Nickel C., Scherer C., Noskov S., Bantz C., Berger M., Schupp W., and Maskos M., J. Chromatogr. A 1637, 461840 (2021). 10.1016/j.chroma.2020.461840 [DOI] [PubMed] [Google Scholar]
- 239. Noskov S., Scherer C., and Maskos M., J. Chromatogr. A 1274, 151 (2013). 10.1016/j.chroma.2012.12.001 [DOI] [PubMed] [Google Scholar]
- 240. Scherer C., Noskov S., Utech S., Bantz C., Mueller W., Krohne K., and Maskos M., J. Nanosci. Nanotechnol. 10, 6834 (2010). 10.1166/jnn.2010.2973 [DOI] [PubMed] [Google Scholar]
- 241. Jungmann N., Schmidt M., and Maskos M., Macromolecules 34, 8347 (2001). 10.1021/ma0106752 [DOI] [Google Scholar]
- 242. Wahlund K. G. and Giddings J. C., Anal. Chem. 59, 1332 (1987). 10.1021/ac00136a016 [DOI] [PubMed] [Google Scholar]
- 243. Caputo F., Arnould A., Bacia M., Ling W. L., Rustique E., Texier I., Mello A. P., and Couffin A.-C., Mol. Pharmaceutics 16, 756 (2019). 10.1021/acs.molpharmaceut.8b01033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244. Scott D. J. and Schuck P., A Brief Introduction to the Analytical Ultracentrifugation of Proteins for Beginners ( RSC Publishing, 2005), p. 1. [Google Scholar]
- 245. Krpetić Z., Davidson A. M., Volk M., Lévy R., Brust M., and Cooper D. L., ACS Nano 7, 8881 (2013). 10.1021/nn403350v [DOI] [PubMed] [Google Scholar]
- 246. Davidson A. M., Brust M., Cooper D. L., and Volk M., Anal. Chem. 89, 6807 (2017). 10.1021/acs.analchem.7b01229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247. Bayley H. and Martin C. R., Chem. Rev. 100, 2575 (2000). 10.1021/cr980099g [DOI] [PubMed] [Google Scholar]
- 248. Henriquez R. R., Ito T., Sun L., and Crooks R. M., Analyst 129, 478 (2004). 10.1039/b404251b [DOI] [PubMed] [Google Scholar]
- 249. Kozak D., Anderson W., Vogel R., Chen S., Antaw F., and Trau M., ACS Nano 6, 6990 (2012). 10.1021/nn3020322 [DOI] [PubMed] [Google Scholar]
- 250. Koji K., Yoshinaga N., Mochida Y., Hong T., Miyazaki T., Kataoka K., Osada K., Cabral H., and Uchida S., Biomaterials 261, 120332 (2020). 10.1016/j.biomaterials.2020.120332 [DOI] [PubMed] [Google Scholar]
- 251. Karimov M., Schulz M., Kahl T., Noske S., Kubczak M., Gockel I., Thieme R., Büch T., Reinert A., Ionov M., Bryszewska M., Franke H., Krügel U., Ewe A., and Aigner A., Nanomedicine (N. Y., NY, U. S.) 36, 102403 (2021). 10.1016/j.nano.2021.102403 [DOI] [PubMed] [Google Scholar]
- 252. Karimov M., Appelhans D., Ewe A., and Aigner A., Eur. J. Pharm. Biopharm. 161, 56 (2021). 10.1016/j.ejpb.2021.02.005 [DOI] [PubMed] [Google Scholar]
- 253. Lauraeus S., Holopainen J. M., Taskinen M.-R., and Kinnunen P. K. J., Biochim. Biophys. Acta Biomembr. 1373, 147 (1998). 10.1016/S0005-2736(98)00102-3 [DOI] [PubMed] [Google Scholar]
- 254. Kaczmarek J. C., Patel A. K., Kauffman K. J., Fenton O. S., Webber M. J., Heartlein M. W., DeRosa F., and Anderson D. G., Angew. Chem., Int. Ed. 55, 13808 (2016). 10.1002/anie.201608450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 255. Sanchez-Cano C., Alvarez-Puebla R. A., Abendroth J. M., Beck T., Blick R., Cao Y., Caruso F., Chakraborty I., Chapman H. N., Chen C., Cohen B. E., Conceição A. L. C., Cormode D. P., Cui D., Dawson K. A., Falkenberg G., Fan C., Feliu N., Gao M., Gargioni E., Glüer C. C., Grüner F., Hassan M., Hu Y., Huang Y., Huber S., Huse N., Kang Y., Khademhosseini A., Keller T. F., Körnig C., Kotov N. A., Koziej D., Liang X. J., Liu B., Liu S., Liu Y., Liu Z., Liz-Marzán L. M., Ma X., Machicote A., Maison W., Mancuso A. P., Megahed S., Nickel B., Otto F., Palencia C., Pascarelli S., Pearson A., Peñate-Medina O., Qi B., Rädler J., Richardson J. J., Rosenhahn A., Rothkamm K., Rübhausen M., Sanyal M. K., Schaak R. E., Schlemmer H. P., Schmidt M., Schmutzler O., Schotten T., Schulz F., Sood A. K., Spiers K. M., Staufer T., Stemer D. M., Stierle A., Sun X., Tsakanova G., Weiss P. S., Weller H., Westermeier F., Xu M., Yan H., Zeng Y., Zhao Y., Zhao Y., Zhu D., Zhu Y., and Parak W. J., ACS Nano 15, 3754 (2021). 10.1021/acsnano.0c09563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 256. Zackova Suchanova J., Hejtmankova A., Neburkova J., Cigler P., Forstova J., and Spanielova H., Bioconjugate Chem. 31, 1575 (2020). 10.1021/acs.bioconjchem.0c00240 [DOI] [PubMed] [Google Scholar]
- 257. Gu J., Chen X., Xin H., Fang X., and Sha X., Int. J. Pharm. 461, 559 (2014). 10.1016/j.ijpharm.2013.12.029 [DOI] [PubMed] [Google Scholar]
- 258. Haberland A., Knaus T., Zaitsev S. V., Buchberger B., Lun A., Haller H., and Böttger M., Pharm. Res. 17, 229 (2000). 10.1023/A:1007581700996 [DOI] [PubMed] [Google Scholar]
- 259. Wang F., Yu L., Monopoli M. P., Sandin P., Mahon E., Salvati A., and Dawson K. A., Nanomedicine (N. Y., NY, U. S.) 9, 1159 (2013). 10.1016/j.nano.2013.04.010 [DOI] [PubMed] [Google Scholar]
- 260. Yallapu M. M., Chauhan N., Othman S. F., Khalilzad-Sharghi V., Ebeling M. C., Khan S., Jaggi M., and Chauhan S. C., Biomaterials 46, 1 (2015). 10.1016/j.biomaterials.2014.12.045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 261. Lesniak A., Fenaroli F., Monopoli M. P., Åberg C., Dawson K. A., and Salvati A., ACS Nano 6, 5845 (2012). 10.1021/nn300223w [DOI] [PubMed] [Google Scholar]
- 262. Zelphati O., Uyechi L. S., Barron L. G., and F. C. Szoka, Jr. , Biochim. Biophys. Acta 1390, 119 (1998). 10.1016/S0005-2760(97)00169-0 [DOI] [PubMed] [Google Scholar]
- 263. Kulkarni J. A., Witzigmann D., Chen S., Cullis P. R., and van der Meel R., Acc. Chem. Res. 52, 2435 (2019). 10.1021/acs.accounts.9b00368 [DOI] [PubMed] [Google Scholar]
- 264. Akinc A., Maier M. A., Manoharan M., Fitzgerald K., Jayaraman M., Barros S., Ansell S., Du X., Hope M. J., Madden T. D., Mui B. L., Semple S. C., Tam Y. K., Ciufolini M., Witzigmann D., Kulkarni J. A., van der Meel R., and Cullis P. R., Nat. Nanotechnol. 14, 1084 (2019). 10.1038/s41565-019-0591-y [DOI] [PubMed] [Google Scholar]
- 265. Prades R., Oller-Salvia B., Schwarzmaier S. M., Selva J., Moros M., Balbi M., Grazú V., de La Fuente J. M., Egea G., Plesnila N., Teixidó M., and Giralt E., Angew. Chem., Int. Ed. 54, 3967 (2015). 10.1002/anie.201411408 [DOI] [PubMed] [Google Scholar]
- 266. Cardle I. I., Jensen M. C., Pun S. H., and Sellers D. L., J. Biol. Chem. 296, 100657 (2021). 10.1016/j.jbc.2021.100657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 267. Mahon E., Salvati A., Baldelli Bombelli F., Lynch I., and Dawson K. A., J. Controlled Release 161, 164 (2012). 10.1016/j.jconrel.2012.04.009 [DOI] [PubMed] [Google Scholar]
- 268. Mirshafiee V., Mahmoudi M., Lou K., Cheng J., and Kraft M. L., Chem. Commun. (Cambridge, U. K.) 49, 2557 (2013). 10.1039/c3cc37307j [DOI] [PMC free article] [PubMed] [Google Scholar]
- 269. Broda E., Mickler F. M., Lächelt U., Morys S., Wagner E., and Bräuchle C., J. Controlled Release 213, 79 (2015). 10.1016/j.jconrel.2015.06.030 [DOI] [PubMed] [Google Scholar]
- 270. Jensen C., Shay C., and Teng Y., in Physical Exercise and Natural and Synthetic Products in Health and Disease, edited by Guest P. C. ( Springer US, New York, NY, 2022), p. 3. [Google Scholar]
- 271. Elter J. K., Quader S., Eichhorn J., Gottschaldt M., Kataoka K., and Schacher F. H., Biomacromolecules 22, 1458 (2021). 10.1021/acs.biomac.0c01661 [DOI] [PubMed] [Google Scholar]
- 272. Wu Y., Zhou Y., Qin X., and Liu Y., Biomicrofluidics 15, 061503 (2021). 10.1063/5.0062697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 273. Han M., Bae Y., Nishiyama N., Miyata K., Oba M., and Kataoka K., J. Controlled Release 121, 38 (2007). 10.1016/j.jconrel.2007.05.012 [DOI] [PubMed] [Google Scholar]
- 274. Wu X., Su J., Wei J., Jiang N., and Ge X., Stem Cells Int. 2021, 9477332. 10.1155/2021/9477332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 275. Chew L., Añonuevo A., and Knock E., Methods Mol. Biol. 2389, 177 (2022). 10.1007/978-1-0716-1783-0_15 [DOI] [PubMed] [Google Scholar]
- 276. Salem T., Frankman Z., and Churko J. M., Tissue Eng., Part B (published online 2021). 10.1089/ten.TEB.2021.0088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 277. Brighi C., Cordella F., Chiriatti L., Soloperto A., and Di Angelantonio S., Front. Neurosci. 14, 655 (2020). 10.3389/fnins.2020.00655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 278. Rossi G., Manfrin A., and Lutolf M. P., Nat. Rev. Genet. 19, 671 (2018). 10.1038/s41576-018-0051-9 [DOI] [PubMed] [Google Scholar]
- 279. Vogt N., Nat. Methods 18, 27 (2021). 10.1038/s41592-020-01026-x [DOI] [PubMed] [Google Scholar]
- 280. Mansouri M. and Leipzig N. D., Biophys. Rev. 2, 021305 (2021). 10.1063/5.0048837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 281. Bhat S. M., Badiger V. A., Vasishta S., Chakraborty J., Prasad S., Ghosh S., and Joshi M. B., J. Cancer Res. Clin. Oncol. 147, 3477 (2021). 10.1007/s00432-021-03814-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 282. Yuki K., Cheng N., Nakano M., and Kuo C. J., Trends Immunol. 41, 652 (2020). 10.1016/j.it.2020.06.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 283. Sherman H., Gitschier H. J., and Rossi A. E., Front. Immunol. 9, 857 (2018). 10.3389/fimmu.2018.00857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 284. Amin M., Mansourian M., Koning G. A., Badiee A., Jaafari M. R., and Ten Hagen T. L. M., J. Controlled Release 220, 308 (2015). 10.1016/j.jconrel.2015.10.039 [DOI] [PubMed] [Google Scholar]
- 285. Uchida H., Itaka K., Nomoto T., Ishii T., Suma T., Ikegami M., Miyata K., Oba M., Nishiyama N., and Kataoka K., J. Am. Chem. Soc. 136, 12396 (2014). 10.1021/ja506194z [DOI] [PubMed] [Google Scholar]
- 286. Yoshinaga N., Cho E., Koji K., Mochida Y., Naito M., Osada K., Kataoka K., Cabral H., and Uchida S., Angew. Chem., Int. Ed. 58, 11360 (2019). 10.1002/anie.201905203 [DOI] [PubMed] [Google Scholar]
- 287. Yoshinaga N., Uchida S., Naito M., Osada K., Cabral H., and Kataoka K., Biomaterials 197, 255 (2019). 10.1016/j.biomaterials.2019.01.023 [DOI] [PubMed] [Google Scholar]
- 288. Miyazaki T., Uchida S., Nagatoishi S., Koji K., Hong T., Fukushima S., Tsumoto K., Ishihara K., Kataoka K., and Cabral H., Adv. Healthcare Mater. 9, e2000538 (2020). 10.1002/adhm.202000538 [DOI] [PubMed] [Google Scholar]
- 289. Miyazaki T., Uchida S., Miyahara Y., Matsumoto A., and Cabral H., Mater. Proc. 4, 5 (2021). 10.3390/IOCN2020-07857 [DOI] [Google Scholar]
- 290. Uchida S., Koji K., Yoshinaga N., Mochida Y., Hong T., and Cabral H., Mater. Proc. 4, 82 (2021). 10.3390/IOCN2020-07789 [DOI] [Google Scholar]
- 291. Yoshinaga N., Naito M., Tachihara Y., Boonstra E., Osada K., Cabral H., and Uchida S., Pharmaceutics 13, 800 (2021). 10.3390/pharmaceutics13060800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 292. Chan C. L., Ewert K. K., Majzoub R. N., Hwu Y. K., Liang K. S., Leal C., and Safinya C. R., J. Gene Med. 16, 84 (2014). 10.1002/jgm.2762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 293. Olden B. R., Cheng Y., Yu J. L., and Pun S. H., J. Controlled Release 282, 140 (2018). 10.1016/j.jconrel.2018.02.043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 294. Turek J., Dubertret C., Jaslin G., Antonakis K., Scherman D., and Pitard B., J. Gene Med. 2, 32 (2000). [DOI] [PubMed] [Google Scholar]
- 295. Kursa M., Walker G. F., Roessler V., Ogris M., Roedl W., Kircheis R., and Wagner E., Bioconjugate Chem. 14, 222 (2003). 10.1021/bc0256087 [DOI] [PubMed] [Google Scholar]
- 296. Warriner L. W., Duke J. R., Pack D. W., and DeRouchey J. E., Biomacromolecules 19, 4348 (2018). 10.1021/acs.biomac.8b01248 [DOI] [PubMed] [Google Scholar]
- 297. Uddin N., Warriner L. W., Pack D. W., and DeRouchey J. E., Mol. Pharmaceutics 18, 3452 (2021). 10.1021/acs.molpharmaceut.1c00368 [DOI] [PubMed] [Google Scholar]
- 298. Boeckle S., von Gersdorff K., van der Piepen S., Culmsee C., Wagner E., and Ogris M., J. Gene Med. 6, 1102 (2004). 10.1002/jgm.598 [DOI] [PubMed] [Google Scholar]
- 299. Boussif O., Lezoualc F. h, Zanta M. A., Mergny M. D., Scherman D., Demeneix B., and Behr J. P., Proc. Natl. Acad. Sci. U. S. A. 92, 7297 (1995). 10.1073/pnas.92.16.7297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 300. Wightman L., Kircheis R., Rössler V., Carotta S., Ruzicka R., Kursa M., and Wagner E., J. Gene Med. 3, 362 (2001). 10.1002/jgm.187 [DOI] [PubMed] [Google Scholar]
- 301. Kircheis R., Wightman L., Schreiber A., Robitza B., Rössler V., Kursa M., and Wagner E., Gene Ther. 8, 28 (2001). 10.1038/sj.gt.3301351 [DOI] [PubMed] [Google Scholar]
- 302. Coll J. L., Chollet P., Brambilla E., Desplanques D., Behr J. P., and Favrot M., Hum. Gene Ther. 10, 1659 (1999). 10.1089/10430349950017662 [DOI] [PubMed] [Google Scholar]
- 303. Zou S. M., Erbacher P., Remy J. S., and Behr J. P., J. Gene. Med. 2, 128 (2000). [DOI] [PubMed] [Google Scholar]
- 304. Hall A., Lächelt U., Bartek J., Wagner E., and Moghimi S. M., Mol. Ther. 25, 1476 (2017). 10.1016/j.ymthe.2017.01.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 305. Sonawane N. D., F. C. Szoka, Jr. , and Verkman A. S., J. Biol. Chem. 278, 44826 (2003). 10.1074/jbc.M308643200 [DOI] [PubMed] [Google Scholar]
- 306. Haberland A., Knaus T., Zaitsev S. V., Stahn R., Mistry A. R., Coutelle C., Haller H., and Böttger M., Biochim. Biophys. Acta 1445, 21 (1999). 10.1016/S0167-4781(99)00017-2 [DOI] [PubMed] [Google Scholar]
- 307. Docter D., Bantz C., Westmeier D., Galla H. J., Wang Q., Kirkpatrick J. C., Nielsen P., Maskos M., and Stauber R. H., Beilstein J. Nanotechnol. 5, 1380 (2014). 10.3762/bjnano.5.151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 308. Lokugamage M. P., Sago C. D., and Dahlman J. E., Curr. Opin. Biomed. Eng. 7, 1 (2018). 10.1016/j.cobme.2018.08.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 309. Paunovska K., Gil C. J., Lokugamage M. P., Sago C. D., Sato M., Lando G. N., Gamboa Castro M., Bryksin A. V., and Dahlman J. E., ACS Nano 12, 8341 (2018). 10.1021/acsnano.8b03640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 310. Sago C. D., Kalathoor S., Fitzgerald J. P., Lando G. N., Djeddar N., Bryksin A. V., and Dahlman J. E., J. Mater. Chem. B 6, 7197 (2018). 10.1039/C8TB01642A [DOI] [PMC free article] [PubMed] [Google Scholar]
- 311. Sago C. D., Lokugamage M. P., Lando G. N., Djeddar N., Shah N. N., Syed C., Bryksin A. V., and Dahlman J. E., Nano Lett. 18, 7590 (2018). 10.1021/acs.nanolett.8b03149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 312. Sago C. D., Lokugamage M. P., Paunovska K., Vanover D. A., Monaco C. M., Shah N. N., Gamboa Castro M., Anderson S. E., Rudoltz T. G., Lando G. N., Munnilal Tiwari P., Kirschman J. L., Willett N., Jang Y. C., Santangelo P. J., Bryksin A. V., and Dahlman J. E., Proc. Natl. Acad. Sci. U. S. A. 115, E9944 (2018). 10.1073/pnas.1811276115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 313. Paunovska K., Da Silva Sanchez A. J., Sago C. D., Gan Z., Lokugamage M. P., Islam F. Z., Kalathoor S., Krupczak B. R., and Dahlman J. E., Adv. Mater. 31, e1807748 (2019). 10.1002/adma.201807748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 314. Gan Z., Lokugamage M. P., Hatit M. Z. C., Loughrey D., Paunovska K., Sato M., Cristian A., and Dahlman J. E., Bioeng. Transl. Med. 5, e10161 (2020). 10.1002/btm2.10161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 315. Sago C. D., Lokugamage M. P., Islam F. Z., Krupczak B. R., Sato M., and Dahlman J. E., J. Am. Chem. Soc. 140, 17095 (2018). 10.1021/jacs.8b08976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 316. Lokugamage M. P., Sago C. D., Gan Z., Krupczak B. R., and Dahlman J. E., Adv. Mater. 31, e1902251 (2019). 10.1002/adma.201902251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 317. Dobrowolski C., Paunovska K., Hatit M. Z. C., Lokugamage M. P., and Dahlman J. E., Adv. Healthcare Mater. 10, e2002022 (2021). 10.1002/adhm.202002022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 318. Sanchez A. D. S., Paunovska K., Cristian A., and Dahlman J. E., Hum. Gene Ther. 31, 940 (2020). 10.1089/hum.2020.137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 319. Yaari Z., da Silva D., Zinger A., Goldman E., Kajal A., Tshuva R., Barak E., Dahan N., Hershkovitz D., Goldfeder M., Roitman J. S., and Schroeder A., Nat. Commun. 7, 13325 (2016). 10.1038/ncomms13325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 320. Paunovska K., Loughrey D., Sago C. D., Langer R., and Dahlman J. E., Adv. Mater. 31, e1902798 (2019). 10.1002/adma.201902798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 321. Yang K., Reker-Smit C., Stuart M. C. A., and Salvati A., Adv. Healthcare Mater. 10, e2100370 (2021). 10.1002/adhm.202100370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 322. Lee S. Y., Son J. G., Moon J. H., Joh S., and Lee T. G., Biointerphases 15, 061002 (2020). 10.1116/6.0000396 [DOI] [PubMed] [Google Scholar]
- 323. Müller L. K., Simon J., Rosenauer C., Mailänder V., Morsbach S., and Landfester K., Biomacromolecules 19, 374 (2018). 10.1021/acs.biomac.7b01472 [DOI] [PubMed] [Google Scholar]
- 324. Lundqvist M., Augustsson C., Lilja M., Lundkvist K., Dahlbäck B., Linse S., and Cedervall T., PLoS One 12, e0175871 (2017). 10.1371/journal.pone.0175871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 325. Solorio-Rodríguez A., Escamilla-Rivera V., Uribe-Ramírez M., Chagolla A., Winkler R., García-Cuellar C. M., and De Vizcaya-Ruiz A., Nanoscale 9, 13651 (2017). 10.1039/C7NR04685E [DOI] [PubMed] [Google Scholar]
- 326. Schöttler S., Klein K., Landfester K., and Mailänder V., Nanoscale 8, 5526 (2016). 10.1039/C5NR08196C [DOI] [PubMed] [Google Scholar]
- 327. Mirshafiee V., Kim R., Mahmoudi M., and Kraft M. L., Int. J. Biochem. Cell Biol. 75, 188 (2016). 10.1016/j.biocel.2015.11.019 [DOI] [PubMed] [Google Scholar]
- 328. Laurent S., Burtea C., Thirifays C., Rezaee F., and Mahmoudi M., J. Colloid Interface Sci. 392, 431 (2013). 10.1016/j.jcis.2012.10.005 [DOI] [PubMed] [Google Scholar]
- 329. Dvorak A., Tilley A. E., Shaykhiev R., Wang R., and Crystal R. G., Am. J. Respir. Cell Mol. Biol. 44, 465 (2011). 10.1165/rcmb.2009-0453OC [DOI] [PMC free article] [PubMed] [Google Scholar]
- 330. Baldassi D., Gabold B., and Merkel O. M., Adv. Nanobiomed. Res. 1, 2000111 (2021). 10.1002/anbr.202000111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 331. Bennet T. J., Randhawa A., Hua J., and Cheung K. C., Cells 10, 1602 (2021). 10.3390/cells10071602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 332. Carius P., Dubois A., Ajdarirad M., Artzy-Schnirman A., Sznitman J., Schneider-Daum N., and Lehr C.-M., Front. Bioeng. Biotechnol. 9, 743236 (2021). 10.3389/fbioe.2021.743236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 333. McGonigle P. and Ruggeri B., Biochem. Pharmacol. 87, 162 (2014). 10.1016/j.bcp.2013.08.006 [DOI] [PubMed] [Google Scholar]
- 334. Mak I. W., Evaniew N., and Ghert M., Am. J. Transl. Res. 6, 114 (2014). [PMC free article] [PubMed] [Google Scholar]
- 335. Perel P., Roberts I., Sena E., Wheble P., Briscoe C., Sandercock P., Macleod M., Mignini L. E., Jayaram P., and Khan K. S., BMJ 334, 197 (2007). 10.1136/bmj.39048.407928.BE [DOI] [PMC free article] [PubMed] [Google Scholar]
- 336. Liu B., Liu S., Zhang S., Bai L., and Liu E., Heliyon 6, e05725 (2020). 10.1016/j.heliyon.2020.e05725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 337. Preis E., Schulze J., Gutberlet B., Pinnapireddy S. R., Jedelská J., and Bakowsky U., Adv. Drug Delivery Rev. 174, 317 (2021). 10.1016/j.addr.2021.04.022 [DOI] [PubMed] [Google Scholar]
- 338. Schulze J., Lehmann J., Agel S., Amin M. U., Schaefer J., and Bakowsky U., ACS Appl. Bio. Mater. 4, 7764 (2021). 10.1021/acsabm.1c01016 [DOI] [PubMed] [Google Scholar]
- 339. Hajipour M. J., Laurent S., Aghaie A., Rezaee F., and Mahmoudi M., Biomater. Sci. 2, 1210 (2014). 10.1039/C4BM00131A [DOI] [PubMed] [Google Scholar]
- 340. Hajipour M. J., Raheb J., Akhavan O., Arjmand S., Mashinchian O., Rahman M., Abdolahad M., Serpooshan V., Laurent S., and Mahmoudi M., Nanoscale 7, 8978 (2015). 10.1039/C5NR00520E [DOI] [PubMed] [Google Scholar]
- 341. Corbo C., Molinaro R., Tabatabaei M., Farokhzad O. C., and Mahmoudi M., Biomater. Sci. 5, 378 (2017). 10.1039/C6BM00921B [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data sharing is not applicable as no new data were generated.