Abstract
Current advances in DNA nanotechnology pinpoint exciting perspectives for the design of customized, patient-specific treatments. This advance is made possible by the exceptionally high precision and specificity that are typical for DNA base pairing on the one hand and our growing ability to harness those features in synthetic, DNA-based constructs on the other hand. Modern medicine may soon benefit from recent developments in this field, especially regarding the targeted delivery of drugs and the rational interference of synthetic DNA strands with cellular oligonucleotides. In this Review, we summarize selected examples from the area of DNA nanotechnology, where the development of precisely controlled, advanced functional mechanisms was achieved. To demonstrate the high versatility of these rationally designed structures, we categorize the dynamic DNA-based materials suggested for precision medicine according to four fundamental tasks: “hold & release,” “heal,” “detect & measure,” as well as “guide & direct.” In all the biomedical applications we highlight, DNA strands not only constitute structural building blocks but allow for creating stimuli-responsive objects, serve as an active cargo, or act as molecular control/guidance tools. Moreover, we discuss several issues that need to be considered when DNA-based structures are designed for applications in the field of precision medicine. Even though the majority of DNA-based objects have not been used in clinical settings yet, recent progress regarding the stability, specificity, and control over the dynamic behavior of synthetic DNA structures has advanced greatly. Thus, medical applications of those nanoscopic objects should be feasible in the near future.
I. INTRODUCTION
Leonardo da Vinci, both an artist and an engineer, had a strong opinion on what mankind will be able to achieve in terms of designing functional objects: “Human subtlety will never devise an invention more beautiful, more simple, or more direct than does nature because in her inventions nothing is lacking, and nothing is superfluous.” Up to now, he might still be right; yet, our understanding of how nature uses molecules as high-precision tools with specific functions is increasing day by day. Thanks to one of the most important biomacromolecules found in all life forms, i.e., DNA, the field of structural nanotechnology was made possible. Since Seeman1 introduced the DNA nanotechnology in 1982, and Rothemund2 showed that it is possible to fold DNA to form nanoscopic patterns, numerous objects have been generated by employing artificial DNA strands as building blocks; examples range from funny-looking objects (smiley-faces) over nanorockets and robots to smart containers and bipedal systems. Up to now, very informative reviews have been published about structural DNA nanotechnology, and these works cover the different aspects of this subject.3,4 However, the complementarity of Watson–Crick base pairing not only allows for the controlled self-assembly of DNA strands into complex shapes: by the rational integration of non-ideal, imperfect hybridizations in combination with offering alternative binding options with a higher degree of complementarity, strand displacement reactions were introduced that enabled a precise control over the dynamic behavior of DNA-based objects.
In the last decade, significant progress was made in the field of DNA-based nanotechnology regarding the development of functional objects. For instance, DNA-PAINT, a technique that is based on transient binding of DNA constructs to molecular target, reduces the need for complex detection hardware, and enables imaging of subcellular elements at molecular-scale resolution.5–7 Other examples are DNA-based nanomotors mimicking the function of motor proteins8 and DNA microchips encoding digital information.9 In combination with advances in the field of materials science, DNA nanotechnology has assumed an indispensable role for the creation of artificial, dynamic systems–especially for those designed for biomedical applications. Recent examples of dynamic DNA-based devices can be programmed to interact with each other, respond to specific targets, and perform logical operations even inside living organisms. In this Review, we highlight such dynamic DNA-based devices, which establish control mechanisms for applications in precision medicine; examples we discuss here include targeted delivery, real-time monitoring of therapies, and medical diagnostics. Conventional medicine may greatly benefit from the unique opportunities made possible by DNA nanotechnology: a new generation of DNA-based nanodevices is able to interfere with specific subsets of the cellular machinery, can control the delivery and release of drugs, can detect early markers of diseases, and can perform therapeutic actions on their own. Further advances in this particular area of DNA nanotechnology may soon revolutionize the way we diagnose and treat diseases.
II. DYNAMIC DNA NANOMATERIALS PERFORM FOUR MAJOR TASKS IN NANOMEDICINE
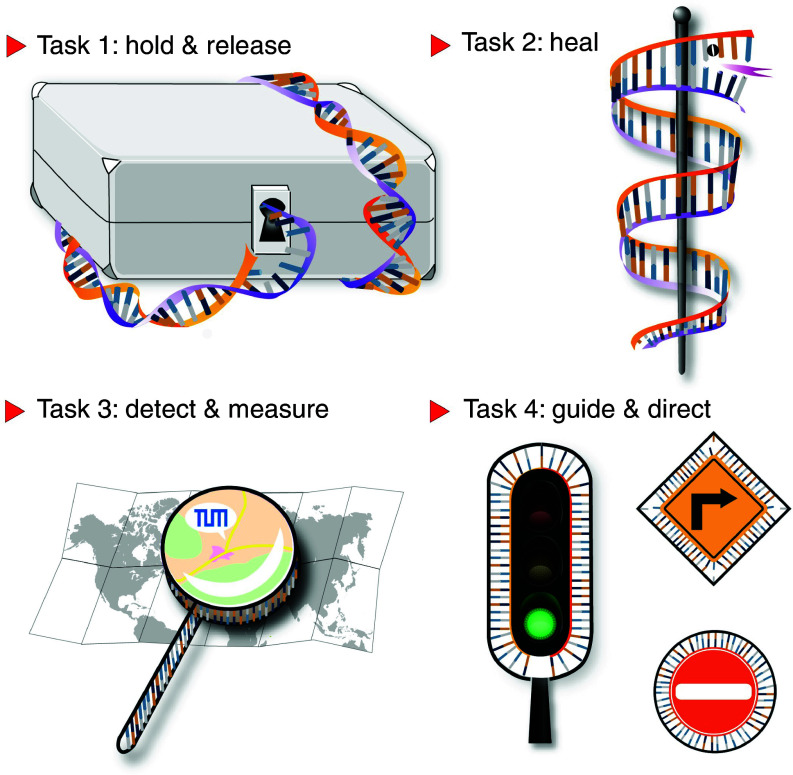
In the following, we will discuss four main functions of dynamic DNA constructs that are important in the context of nanomedicine (Fig. 1): holding and releasing drug molecules in a controlled fashion, serving as therapeutic agents that directly interact with molecules from the cellular metabolism, detecting and visualizing cellular dysfunction based on the presence of molecular indicators, and acting as control tools that guide the spatio-temporal sequence of microscopic processes. For each of those functions, we discuss selected examples that highlight how those functions are implemented in the DNA-based constructs and how certain areas from the fields of nanomedicine, diagnostics, or drug delivery may benefit from them.
FIG. 1.
Schematic representation of the four main functions dynamic DNA-based objects can fulfill in the context of healthcare-related applications: They can hold and release pharmaceuticals, perform healing actions by direct binding, detect and measure the status of the pathological conditions, and guide biological or synthetic objects to selected targets or direct the temporal sequence of pre-programmed events.
In the last decade, the implementation of synthetic DNA has experienced strong growth through structural nanotechnology, which allows for the design of various shapes using DNA oligonucleotides as building blocks.10 With the use of the robustness of the Watson–Crick nucleic acid hybridization principle, oligonucleotide bases can be used with the simplicity of Lego blocks, and self-assembled templates with various shapes were generated with extremely high precision. One of the impressive advantages is that numerous modifications (e.g., attachment of a fluorescent molecule, ligands, or magnetically controllable nanoparticles) can be applied to these building blocks to achieve the desired function. The DNA origami approach can be used in the design of functional materials such as hydrogels, nanoparticles, or nanotubes. This unprecedented control over the size and shape of nanoarchitecture has led to successful transport of those DNA-based constructs even across the blood-brain barrier11,12 or into lymph nodes.13 Yet, the developments in this area are not limited to the creation of static materials, but also allow for the generation of active, dynamic objects that can undergo configurational changes, i.e., they can switch from one defined state to another in a pre-programmed fashion. In the following section, we highlight a few examples of such dynamic DNA structures that comprise a smart response mechanism useful for drug delivery.
A. Task 1: Hold and release on demand
After the development of the first functional DNA tetrahedral cage loaded with one molecule of cytochrome c,14 the use of DNA nanomaterials in on-demand delivery applications has gained a lot of interest. Whereas the first DNA-based cage could serve as a container, it did not possess a mechanism that would allow for a controlled release of its cargo. Such control was achieved first in a DNA-based nanobox, which contained a programmable lid made from DNA strands.15 Here, a strand displacement mechanism was employed for the first time as a trigger to open a container [Fig. 2(a)]. To date, a variety of dynamic DNA structures have been developed that were inspired by this DNA-based opening mechanism and serve as carrier objects for molecules.16
FIG. 2.
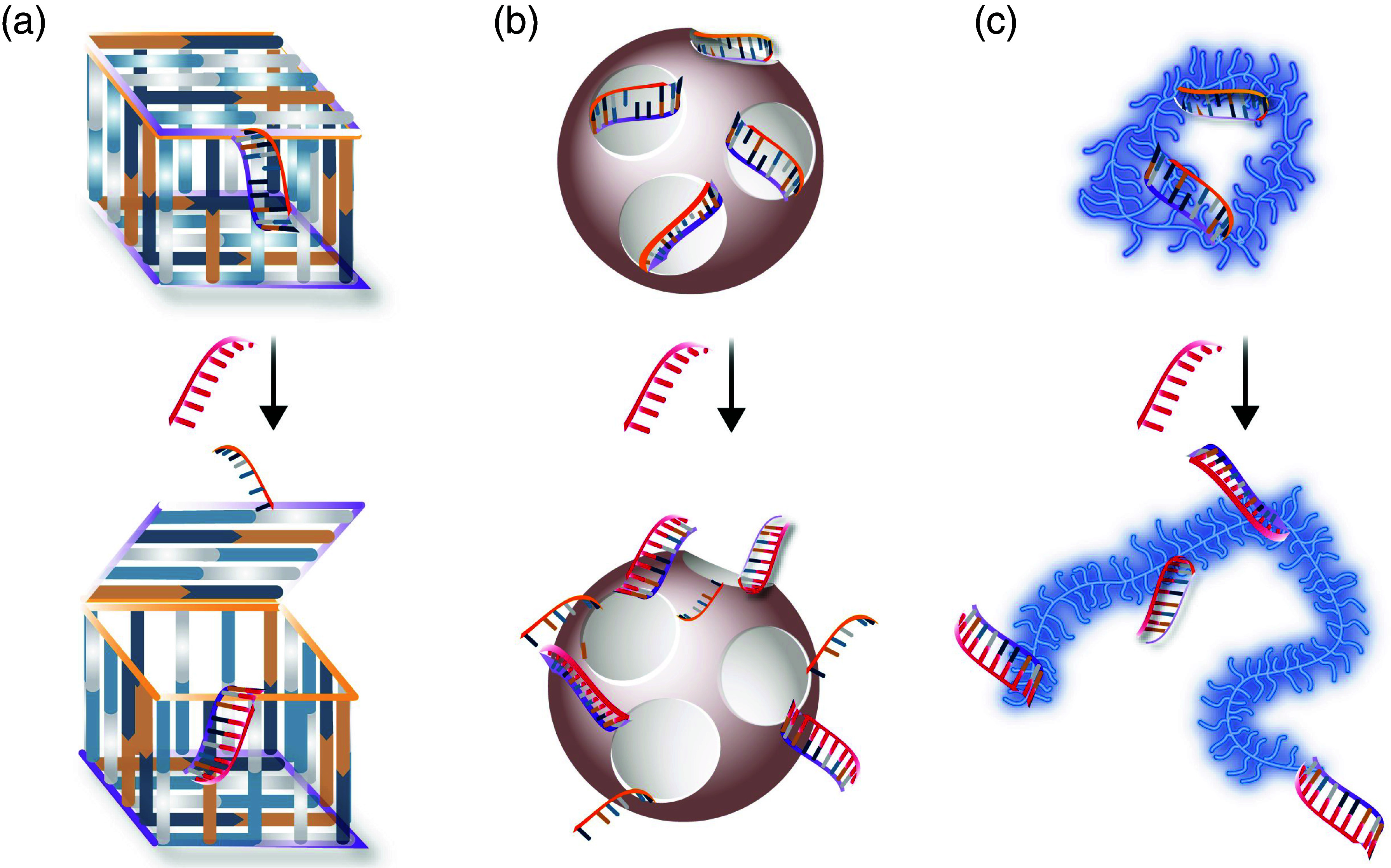
Examples of nanocarriers where synthetic DNA strands keep the carrier in a closed state until a correct trigger, i.e., a single-stranded DNA sequence, is introduced to the system. (a) DNA-based nanobox. (b) Mesoporous silica nanoparticles with DNA strands acting as control gates. (c) Condensed mucin glycoproteins that are transiently stabilized by DNA strands.
A key advantage of DNA structures is their ability to undergo a conformational change in response to a pre-defined trigger. In early designs, control over this structural change was obtained by supplying an external, synthetic trigger molecule that binds to the DNA scaffold. Recent examples from the field of DNA-based drug encapsulation and release, however, are now able to manipulate the shape of structures by making use of the certain site- or disease-specific biological conditions, thus abolishing the need for externally adding a trigger substance. A global destabilization of DNA structures can be achieved, e.g., in response to strong pH alterations17 or elevated glutathione (GSH) concentrations.18 In a way, those attempts open a piggy bank by cracking it open with a hammer–efficient, but not very subtle. More recent examples make use of biological triggers to induce drug release events from DNA-stabilized nanostructures by locally unlinking stabilizing elements in the DNA construct. For instance, the Linko lab has recently developed a DNA-based dynamic nanocarrier that can be loaded with various types of loads.19 There, even small pH changes (i.e., on the order of ∼0.5 pH units) were sufficient to induce a configurational change of the “latch” strands, which, in turn, allows the nanocapsule to release its cargo.
Examples of these biological molecules that induce such a controlled alteration in the drug carrier structure make use of pathological marker molecules, e.g., overexpressed microRNAs or aptamers. For instance, mesoporous silica-coated quantum dots, loaded with drug molecules, were successfully sealed with a DNA hybrid that can serve as a dynamic gate closing the nanoparticle pores [Fig. 2(b)]. When exposed to the correct trigger strands, these DNA-based gates opened, and the nanoparticles released their load.20 In this study, cellular DNA sequences could act as keys to initiate a drug release process. However, to maintain the therapeutic efficiency, cellular key strands had to be amplified to achieve efficient nanoparticle opening. A recent study made use of mucin-based drug carrier nanoparticles that were partially stabilized by self-complementary DNA strands.21 There, the DNA sequences were designed such that the drug-loaded nanoparticles maintained their stability, thus, holding their cargo, under normal physiological conditions [Fig. 2(c)]. However, in the presence of sufficiently high concentrations of cellular miR-21 (and the nucleotide levels in the target cells were high enough), drug release inside the target cells was enabled–but not in control cells where the expression level of miR-21 was significantly lower. In another example, human telomerase expression was successfully employed as a “key” to elongate the primers of the drug-loaded DNA icosahedron, and–by design–this unlocking event can release platinum nanodrugs to cisplatin-resistant tumor cells.22
The examples discussed so far required one type of key molecule only to trigger the release event from the carrier. However, by making use of multivalent interactions, DNA nanostructures can be turned into “vault-like” structures that only grant access if several keys are presented at the same time. For example, Douglas et al.23 engineered a hexagonal DNA barrel such that it responds to the correct combination of protein cues; only if the correct combination of keys, i.e., cell-surface antigens, is present, the configurational change–subsequently–a selective interface with target cells to deliver cargo materials, is initiated. Furthermore, these robotic DNA mechanisms allow for combining external and internal cues: for instance, an external DNA sequence (that prevents opening of the structure by further locking it) can be mounted onto another DNA-based construct. Then, opening the vault structure is only possible in the absence of the locking agent.24 With this implementation, logical gates were implemented that combine different pieces of molecular information to decide if and where cargo release should occur. Amazingly, this complex combination of molecular interaction events could even be realized in living animals (in cockroaches), which demonstrates that such precise control mechanisms to control the therapeutics and bioprocesses are actually possible at physiological conditions.
B. Task 2: Heal by binding
In addition to holding, transporting, and releasing drugs, rationally designed DNA nanostructures can also directly perform therapeutic actions within cells. This ability to act as a “healing” agent is mostly based on the DNA strands' ability to interfere or replace specific disease-related genes. Especially in modern research, since several cancer-related issues have been identified to be associated with irregularities in RNA expression levels, synthetic oligonucleotides are used as therapeutic molecules. Compared to conventional drugs that cannot easily be guided to their required molecular target only, synthetic oligonucleotides can be programmed such that they perform therapeutic actions on the envisioned target gene, e.g., by silencing, amplifying, or even degrading a selected cellular nucleotide sequence.
In 1998, Fire and Mello coined the phrase “RNA interference” (RNAi) and demonstrated that non-coding RNA can be a central gene expression regulator in multicellular organisms:25 Small interfering RNA (siRNA) regulate gene expression by inducing messenger RNA (mRNA) cleavage, and this prevents the production of the encoded proteins. Three years after this discovery, successful use of siRNA in the treatment of hepatitis C was achieved in vivo.26 To date, the therapeutic efficacy of synthetic siRNA has been verified for the treatment of various diseases; examples include hypercholesterolemia, liver cirrhosis, viruses-induced diseases, and certain cancer types.27,28
Another strategy to achieve synthetic oligonucleotide-based genetic intervention targets microRNAs (miRNAs), i.e., short, non-coding RNA molecules. Since each miRNA strand can interact with multiple mRNAs through complementary base pairing, interfering with the cellular machinery on the level of miRNA strands is a very efficient strategy. About 50% of the miRNA sequences discovered so far are predominantly located in cancer-associated genomic regions or at such sites where an unphysiological up- or downregulation negatively impacts the immune response.29 Nowadays, it is possible to target specific miRNA strands by binding synthetic, well-matching nucleotides to them or by mimicking them with engineered constructs. As a consequence of such intracellular mediations, the biological fate of a cell can be altered: for instance, it is possible to suppress tumor growth, to block cell invasion into neighboring tissues, to prevent metastasis, and to promote apoptosis.30 Some examples of signature miRNA strands, the pathological scenario they are associated with, and a typical therapeutic approach in which they are manipulated are listed in Table I.
TABLE I.
Specific modulation of disease-related expression of miRNAs.
| Target | Expression alteration | Physiological role | Related diseases | Delivery techniques for the modulating agent |
|---|---|---|---|---|
| miR-1 | ↑/↓ | Calcium signaling controller in heart muscles41 | Coronary artery disease, gastric cancer | Intramuscular injection42 |
| miR-10b | ↑ | Metastasis activator43 | Cancer (e.g., breast, glioblastoma, melanoma) | Polymer nanoparticles (NPs), systemic injection, locked nucleic acid oligonucleotides (LNAs)44,45 |
| miR-15/16 cluster | ↓ | Tumor suppressor, impairs migration, decreases proliferation46 | Cancer (e.g., ovarian, prostate) and multiple myeloma | LNAs, viral vectors47,48 |
| miR-21, miR 221/222 cluster | ↑ | Cell growth, migration, proliferation stimulator, suppression of apoptosis49,50 | Cancer (e.g., cervical, breast, esophagus, liver, pancreas), cardiac hypertrophy, lupus, kidney fibrosis | Polymer/metal/silica NPs, stent coatings, LNAs, hydrogels, packaging RNA51–55 |
| miR-29 | ↓ | Cell survival regulator, collagen expressor, modulator for the reactivation of silenced tumor suppressor genes56 | Scleroderma, cancer (e.g., lung, breast, cholangiocarcinoma), acute myeloid leukemia | Lipopolyplex nanoparticles, systemic delivery57 |
| miR-33 | ↑ | Regulator of the genes involved in cellular cholesterol export, fatty acid oxidation, insulin signaling, glucose production58 | Atherosclerosis | LNAs, polyplexes58 |
| miR-34 family | ↓ | Tumor suppressor, immune evasion, and cell cycle modulator59 | Cancer (e.g., bladder, colon, brain), B cell lymphoma, myeloma | Polymer/lipid/silica NPs, viral vectors, dendrimers, micelles59,60 |
| miR-103, miR-107 | ↑ | Regulator of multiple genes involved in insulin signaling61 | Type II diabetes, obesity, colorectal cancer | Lipid NPs62 |
| miR-122 | ↑/↓ | Upregulates the replication of the hepatitis C virus RNA genome, tumor suppressor63 | Cancer, hepatitis C infection and related liver diseases | Locked nucleic acid antisense oligonucleotides, liposomes64–66 |
| miR-132, miR-134 | ↓ | Synapse formation and maturation67 | Alzheimer's disease, schizophrenia, bipolar disorder | Polymer NPs68,69 |
| miR-141 | ↑ | Epithelial-mesenchymal transition (EMT), chemotherapy resistance stimulator50 | Cancer (e.g., prostate, lung, colorectal) | Hydrogels, carbon nanotubes70,71 |
| miR-143 | ↓ | Cell proliferation and invasion inhibitor72 | Cancer (e.g., head, neck tumors), lymphoid leukemia | Liposomes, polymer NPs73,74 |
| miR-155 | ↑ | Formation of blood cells, immune system, malignant growth, homeostasis regulator75 | Cancer (e.g., liver, thyroid, kidney), viral infections, cardiovascular diseases, Burkitt lymphoma, inflammation | Peptide nucleic acids, polymer NPs76–78 |
| miR-192 | ↓ | Targets e-cadherin (regulator of epithelial cell morphology), angiogenesis regulator79 | Diabetes-related kidney complications, cancer (e.g., ovarian, renal) | Liposomes80 |
| miR-200 family | ↓ | Modulator of tumor metastasis, invasion inhibitor, plays role in EMT, reactive oxygen species signaling regulator81 | Cancer (e.g., breast, ovarian, lung) | Xenografts, liposomes36,82 |
| miR-506 | ↓ | Modulator the expression of proteins involved in tumor metastasis, cellular senescence, DNA damage response83 | Cancer (e.g., ovarian, lung) | Liposomes, polymeric NPs83,84 |
| miR-520 | ↓ | Tumor suppressor, modulator for the expression of proteins involved in metastasis, tumor growth85 | Cancer (e.g., breast, ovarian) | Liposomes86 |
One of the therapeutic interventions applied to silence the upregulated miRNAs is the delivery of anti-microRNA (antimiR) strands. Those can form selective and stable base pairings with the target, thus suppressing its activity. For example, Dahl and co-workers31 used FDA-approved poly(lactic-co-glycolic acid) (PLGA) nanoparticles as antimiR-21 and antimiR-10b carriers to reprogram tumor cells and increase their sensitivity to chemotherapy. By facilitating the cellular uptake of these DNA-functionalized nanoparticles via ultrasound and microbubbles, enhanced antimiR delivery to deeper areas of liver and kidney tissues was achieved in vivo without affecting the neighboring, healthy tissue. Alternatively, DNA nanotechnology can also produce miR-mimics that increase the cellular concentrations of selected, downregulated miRNAs sequences. Combinatory approaches, where a specific miR-mimic and an antimiR are delivered simultaneously, can provide an efficient expression modulation to combat aggressive tumors both in vitro and in vivo.32 Such an approach is highly relevant as insufficient levels of tumor suppressor miRNAs are a hallmark of cancer development. For example, the simultaneous delivery of different potent, endogenous tumor-suppressive miRNAs (e.g., from the miR-24a and miR-34 family) is a promising approach to obtain synergistic effects in the fight of tumor growth, and this strategy has already successfully been implemented both in vitro and in vivo.33,34 Another example from the area of cancer therapy tackles the resistance to medical treatment;35 here, miR-based approaches have also been developed. For instance, the nucleotides from the miR-200 family are downregulated in certain tumors, which is responsible for a high aggressiveness and metastasis propensity of the tumor. Yet, preclinical models could demonstrate that the delivery of miR-200c mimics (e.g., via lipid-based carriers) enhances the radiosensitivity of lung cancer cells, thus facilitating their eradication.36 A similar approach can also increase the treatment efficiency of other targets: examples include diabetes-related kidney complications, cardiac regeneration, and myocardial infarction.37–40
C. Task 3: Detect and measure
Both the correct selection of a therapeutic strategy and the site-specific treatment of a disease rely on the accurate detection of unphysiological alterations. Identifying such alterations is based on our ability to distinguish the “target” cells/tissues from others by a suitable parameter (or a combination of several parameters). Indeed, DNA-based devices can also detect biological signals and convert them into a measurable output. Such strategies can be based on various mechanisms, such as an electrical, fluorescent, or topological response. Examples include the use of AFM, electrochemical aptasensors, magnetic microparticles, fiber-optic surface plasmon resonance (FO-SPR), and fluorimetry, e.g., via FRET-pairs.87 Incorporating a sensitive and specific detection mechanism into dynamic DNA assemblies can contribute to decoding the molecular basis of diseases, allow for monitoring their progress, and provide information on the treatment success.
Aptamers (originating from a combination of the Latin word aptus–fit and the Greek word meros–part) are oligonucleotide or peptide molecules that can serve as molecular investigators. They selectively bind to intracellular, extracellular, or cell-surface markers (e.g., proteins, small molecules, metal ions). Even though their function is not based on selective hybridization processes, aptamers are able to recognize their ligand through non-covalent interactions with high specificity.
Indeed, the integration of aptamers into DNA-based nanomaterials has been shown to enhance the accuracy of the detection–and this property allows them to be used for detection, bioimaging, and targeted delivery.88 For instance, Jin et al.89 developed a DNA-based nanosensor based on Förster resonance energy transfer (FRET) and employed it to detect pathogenic bacteria in tap/pond water and milk samples. There, two metallic nanoparticles bound to each other via DNA hybridization, and one hybridization partner is an aptamer sequence. In the “off” state of this DNA-based sensor, the two nanoparticles are connected via imperfect hybridization [Fig. 3(a)]. In the presence of target bacteria (here, E. coli), however, this connection is broken since the aptamer strands preferably bind to the bacteria. As a result, a shift in the color of the fluorescent signal occurs, which can be quantified. A similar strategy was also used for cocaine90 or mycotoxin detection,91 and much shorter processing times and higher sensitivities were achieved compared to conventional methods.
FIG. 3.
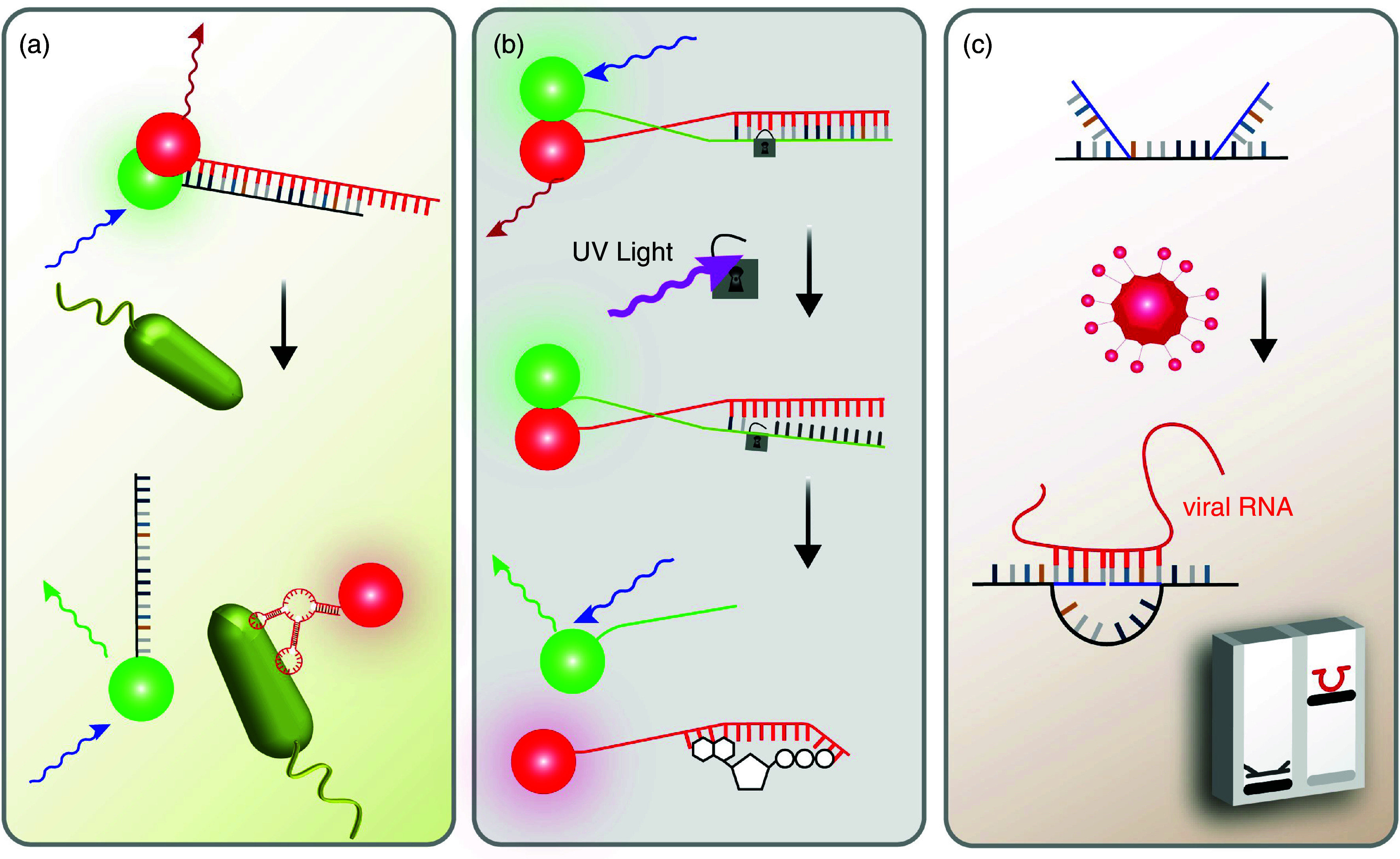
DNA-based tools enable the detection of a variety of nanoscopic/microscopic targets: (a) pathogenic microorganisms, (b) ATP, and (c) viral DNA. Those strategies make use of fluorescence signals modulated by the presence or absence of Förster transfer [(a) and (b)] or alter the electrophoretic mobility of their targets (c).
Another FRET-based sensor was introduced by Zhao et al.:92 they developed a DNA nanodevice that can measure the cytoplasmic ATP levels in living cells [Fig. 3(b)]. To prevent off-target (i.e., extracellular) signal detection, a photocleavable DNA sequence was integrated into the structure to stabilize the aptamer in its “off” state (i.e., it cannot bind to ATP). Then, after the target cells successfully internalized this device, near-infrared light (NIR) was used to remove the locking strand via photolysis to degrade the stabilizing strand into short DNA fragments that do not interfere with the function of the aptamer sequence. As a consequence, the aptamer molecule was made accessible so it can bind to ATP–and such binding was quantified as a change in fluorescence based on FRET.
Of course, metabolic imbalances cannot only occur in the cytoplasm but also in cellular compartments; yet, subcellular dysfunctions are more difficult to detect. However, there is an increasing effort to make use of DNA-based detection methods in different subcellular microenvironments. Indeed, a DNA-based device designed by Leung et al.93 allows for such a microscopic analysis by conducting a spatio-temporal mapping of lysosomal ion concentrations and pH levels in living cells. This makes it possible to discriminate between healthy and diseased cells from biopsies conducted on patients with certain genetic diseases. Such quantitative information obtained on subcellular conditions may enable monitoring the current status of a disease. As another design approach, cellular membrane-bound DNA constructs were also applied to detect both intra- and extracellular ion levels, which have important roles in cellular metabolism.94,95
There are also examples where the diagnostic abilities of DNA-based sensor templates rely on the detection of a specific target molecule (e.g., an oligonucleotide sequence, antibody, or polypeptide). Here also, successful binding of the DNA sensor to its target is typically detected optically, e.g., via an amplified fluorescent signal. However, when a specific microRNA sequence is selected as a target, this is not always trivial: specific miRNA overexpression in the context of two (or more) different pathological conditions can lead to an ambiguous result. For example, the overexpression of miR-21 can be detected in both cancer and some cardiovascular diseases. Therefore, a logic gate would be required to allow for distinguishing the two diseases. Zhu et al.96 developed a programmable nanodevice containing a “cancer assessment operator” in which only a combined overexpression of miR-21 and downregulation of miR-145 in the same cell returned a positive signal. Of course, such a strategy requires a more complex design of the DNA-based sensor, and current applications of this approach have not yet been attempted in vivo.
When considering the growing impact of global pandemics, high-cost and time-consuming laboratory detection methods are certainly among the biggest obstacles that make it difficult to control spreading of the diseases. Still, there is a growing need for low-cost detection tools to conduct analysis in a quick and reliable manner. To overcome the limitations of current detection techniques, DNA nanoswitches can be considered as powerful, yet simple platforms. For example, by making use of circular dichroism, Funck et al.97 detected viral RNA (from hepatitis C viruses) even at sub-nanomolar levels. This was made possible by following a chiral orientation change triggered by strand displacement reactions between the viral RNA and surface-bound, synthetic DNA strands. Alternatively, by attaching a specific molecule to the DNA sensor, a structural transformation from linear to looped state can be triggered.98 The resulting difference in electrophoretic mobility enables the detection of a variety of molecules; examples include antibodies, small molecules, cell receptors, and enzymes.99 Zhou et al.100 recently reported a DNA nanoswitch that can detect viral RNA [Fig. 3(c)]. By using Zika and Dengue viruses as model targets, they succeeded in detecting viral oligonucleotides at concentrations in the attomolar level. Furthermore, the detection of SARS-CoV-2 RNA in human saliva was made possible at clinically relevant concentrations. This high sensitivity–paired with unmatched precision and accuracy–is certainly a hallmark of DNA nanotechnology that is hard to match with other existing strategies.
D. Task 4: Guide and direct
An application area where DNA nanomaterials have only rarely been used yet is to guide the trafficking of molecules/particles or to initiate a series of events in a biological environment. However, as we highlight below, in such scenarios, specific base-pairing and strand-displacement can also be powerful tools to achieve control over the spatio-temporal positioning of nano/microscopic objects. Again, aptamers can be employed here; through conjugation of specific aptamers to drug carriers/molecules, the aptamers can guide them to a particular target location. For example, to combat wet macular degeneration in the ocular compartment, vascular endothelial growth factor (VEGF)-binding aptamers have successfully been employed to increase the bioavailability of the drugs that they are conjugated to.101
Artificial DNA sequences can also play a pivotal role in guiding sequential release events, thus enabling control over the order in which drug carriers are liberated from a hydrogel environment [Fig. 4(a)]. By adjusting the intrinsic properties of these semi-stable structures (e.g., using hybridizations with different melting temperatures),102 or by designing DNA-stabilized clusters where the liberation/addition of an initiator sequence initiates a cascade of strand displacement reactions, a step-by-step release of different molecules/particles can be programmed.103–105 Further adaption of such DNA-based control strategies may provide an important stepping stone toward automated medication management [Fig. 4(b)].
FIG. 4.
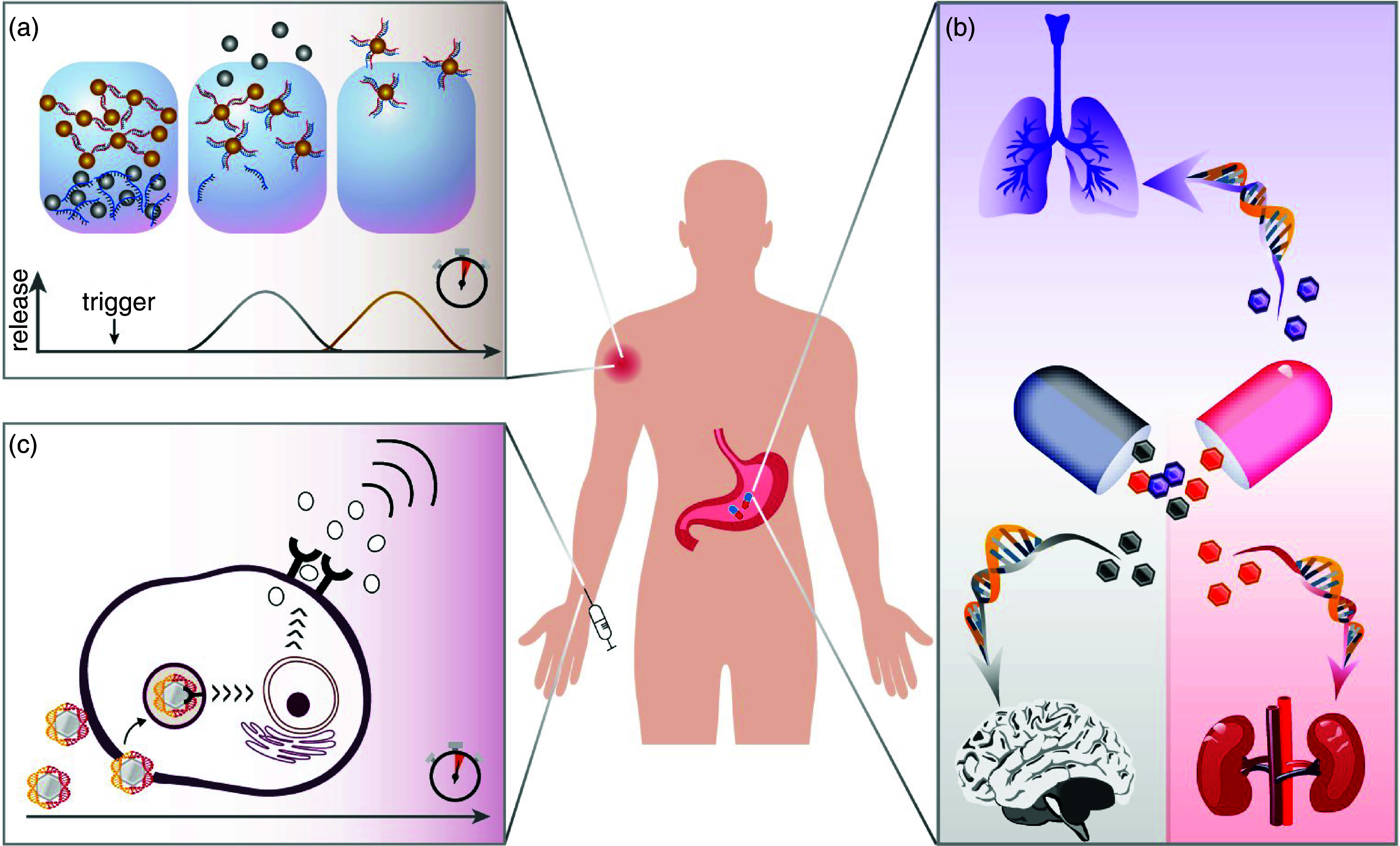
Synthetic DNA strands can achieve spatio-temporal control over the distribution of molecules and particles across complex barriers: They can orchestrate the sequential release of nanoparticles from hydrogels (a), stimulate the immune response (b), and guide pharmaceuticals to pre-defined target tissues (c).
The guided transportation of molecules by DNA-based strategies does, of course, not have to be limited to active pharmaceutical ingredients, or their carriers. Also, direct immunomodulation strategies can benefit from structural DNA nanotechnology. In mammals, pathogen-associated molecular patterns (PAMPs) are recognized by endosomal Toll-like receptors (TLR). TLRs detect unmethylated cytosine-phosphate-guanosine oligodeoxynucleotides (CpGs, those typically appear in prokaryotic DNA but are rare in eukaryotic DNA106) and initiate a cellular signaling cascade that leads to the expression of specific membrane molecules and the release of proinflammatory cytokines−both of which stimulate further steps in the immune response.107 And, indeed, there are examples of how synthetic DNA-based objects can modulate this immune response [Fig. 4(c)]. An important strategy was introduced by Liedl and co-workers,108 who generated DNA nanotubes carrying synthetic CpGs, thus mimicking the presence of a prokaryotic PAMP. In this study, the successful uptake of such CpG-functionalized DNA structures by mouse splenocytes induced the expression of early markers of immune activation in vitro. Importantly, these DNA constructs caused lower cytotoxic effects than commonly used transfection agents, which underscores their good biocompatibility. A similar strategy was implemented with other DNA-based immune modulators, e.g., peptide-modified DNA dendrimers,109 nucleic acid nanoparticles,110 and DNA wires,111 and made it possible to stimulate cells such that cytokine secretion was boosted.
The final example we highlight here makes use of synthetic DNA strands to guide a CRISPR/Cas9 system to edit/repair a specific location on the cellular genome, which can result in a permanent change. Guided by a single-guide RNA (sgRNA), the Cas9 protein is directed to a specific location in the genome where it can create a single- or double-stranded break by acting as molecular scissors.112 This technique, derived from the prokaryotic adaptive immune system, was developed into a facile genome-editing method. CRISPR/Cas9-mediated genome editing–whose inventors were was just awarded the Nobel Prize–has stimulated completely novel approaches to treat a wide range of medical conditions that originate from genetic disorders; examples include Alzheimer's disease, hepatitis B, and cancer.
Consequently, recent studies implementing CRISPR/Cas-based strategies showed very promising results in vivo.113–116 However, it remains an open question how the target-specific delivery of such synthetic CRISPR/Cas genome-editing machineries can be ensured at high accuracy and efficiency. From a therapeutic point of view, this is important to prevent undesired modifications of random genomic loci, which can lead to off-target mutations. Current approaches mostly rely on liposomal formulations and viral vectors to facilitate the transport of synthetic CRISPR/Cas systems across the cellular membrane. With the examples we discussed in Secs. IIA and IIC, it seems very likely that DNA-based transport strategies may also be a promising tool here: DNA-based nanomaterials offer good stability (e.g., within endosomes or during circulation in the bloodstream), specific cell targeting, and cell-specific release from carriers–all of those aspects can help improve the gene-editing efficiency of CRISPR-based strategies. For example, a yarn-like DNA structure designed by Sun et al.117 was shown to form a partially complementary complex with sgRNA to guide the CRISPR/Cas complex into a cell. In another very recent example, an aptamer-anchored tetrahedral DNA structure was employed to guide the gene-editing tools to specific cells.118 Certainly, with the rapid progress made in the field of DNA nanotechnology, many more examples are to come, which will render the feasibility of clinical CRISPR-based therapies much more likely.
III. CAVEATS WHEN DESIGNING FUNCTIONAL DNA CONSTRUCTS FOR APPLICATIONS IN THE HUMAN BODY
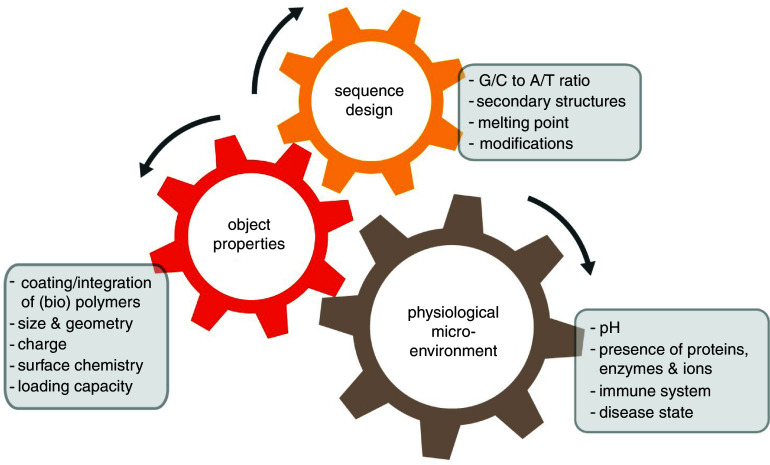
To date, the possibilities to use specific base pairing for generating objects with a precisely defined geometry have been explored extensively. However, in order to design functional DNA-based nanomaterials, several additional parameters must be considered that go beyond controlling the shape of the generated nanodevice. For instance, even today, most of the rationally designed drug delivery systems suffer from low efficiency and off-target accumulation: e.g., only 0.7% of intravenously administered gold nanoparticles can be delivered into the tumor environment; even worse, although they were functionalized with targeting ligands, less than 14 out of 1 million could reach the cytosol of cancer cells in vivo.119 Such a low targeting efficiency could have severe consequences in nucleic acid therapy, genome editing, and immunotherapy. As the examples we discussed in Secs. II A–II D indicate, there are however strategies to address this issue, and–at least in the lab–they returned very promising results. Of course, to ensure both high functionality and delivery efficiency/precision at the same time, a DNA-based nanomaterial has to be designed very carefully. Sequence design, object properties, and the biological microenvironment influence each other, and the correct combination of those aspects needs to be identified to ensure that the engineered object can correctly fulfill its envisioned function (Fig. 5). Thus, to allow for a rational planning procedure, the boundary conditions of the target environment as well as the administration route must be known in detail.
FIG. 5.
The sequence design of a functional DNA nanomaterial not only dictates the physico-chemical properties of an artificial DNA-based object but also defines how this object will behave in a certain physiological microenvironment.
Among all the parameters that need to be considered, the stability of a DNA-based nanomaterial constitutes one of the biggest challenges. To ensure sufficient stability, the melting temperature (Tm, i.e., the temperature at which 50% of the base pairs are open) is a key parameter. Tm depends on the length of the oligonucleotide sequence, its content of C/G and A/T pairs, and the presence of cations in the buffer. In particular, divalent ions efficiently screen the negative charges on the DNA backbone and allow single-stranded DNA strands to hybridize. Of course, other ions can also induce such charge screening effects, and the efficiency of this process depends on the ion valency and concentration.120 In addition, neighboring bases also play a role in the stability of a base pair, and such nearest neighbor effects can be included into mathematical models to estimate Tm. With the help of such software tools and considering the aspects discussed above, the Tm of simple oligonucleotide sequences can be accurately predicted; however, complex designs involving chemically modified sequences require more advanced models.121 In addition, it is possible to estimate the probability that the designed sequence forms secondary structures–which might be an undesired side effect interfering with the envisioned function of the designed sequence.
Some of the examples we discussed make use of DNA strands linked to a specific cargo (e.g., drug molecules, CpG motifs, genome-editing agents) or to a carrier object–the latter strategy is typically applied to therapeutic DNA strands (e.g., siRNA or antisense oligonucleotides). Such a linkage between oligonucleotides and other objects can be established via several methods such as direct covalent/non-covalent binding interactions, molecular ligands, or intercalating agents. The particular choice of a linking strategy depends on whether tunable stability of the generated functional object is required, e.g., if it is supposed to be sensitive to a specific trigger. At this point, several physicochemical properties of the created object such as size, geometry, stiffness, chemical composition, surface chemistry, and surface charge are pivotal for controlling the behavior of those nanodevices in biological settings.122 For instance, low Mg2+ concentrations and nuclease-mediated degradation adversely affect the stability of DNA-based materials–but those conditions are present in a physiological environment. This issue is particularly important for DNA-based objects that are designed to perform targeted delivery tasks; here, insufficient stability of the DNA structure would entail premature cargo release and thus increase the risk of side effects. To tackle this problem, strategies to increase the mechanical stiffness of DNA-based structures have been proposed. Chemical and enzymatic modifications of DNA, i.e., covalent conjugation and enzymatic ligation, increase the stability of a DNA object;123–126 however, this comes at a price as highly stable DNA structures tend to be less dynamic. Yet, it is possible to obtain sufficiently high but tunable stability by employing oligopeptide/protein coatings,127–130 or by using bioinspired, lipid-based envelopes131,132 to protect DNA constructs from low salt denaturation and nuclease-triggered degradation.
A major challenge a DNA-based device faces in vivo is that it will encounter very different conditions at the organ, sub-organ, and subcellular level:133 depending on the particular microenvironment, a DNA-based object will not only be affected by the biological world it is exposed to, it will also influence biological processes. For example, whereas cationic objects are efficiently internalized by cells, they not only tend to show increased cytotoxicity but have also been found to be more sensitive toward serum proteins that tend to create a protein corona around the object. Moreover, the propensity of such a corona formation process also depends on the shape and size of the object. Thus, the overall fate of an artificial, DNA-based device regarding a putative immune response, its biodistribution, and cellular uptake depends on several parameters. Insufficient cellular entry and endosomal entrapment are key issues that need to be addressed if the DNA-based object is supposed to take an action in the cytosol. Importantly, overcoming the natural barriers of the human body can be even more difficult when pathological alterations are present; examples include an increased viscosity and altered composition of the extracellular matrix in the tumor environment.134
Several suggestions have already been made on how to overcome the most common problems in this area. However, when designing functional DNA-based nanomaterials, there is always a trade-off between addressing the particular issue and not creating additional risks. Table II lists some examples of those problems along with possible solutions as indicated in the literature.122,135 Due to the complexity of this multi-faceted issue, it was suggested to utilize decision matrices to identify key design parameters specific to the particular object and its application.136,137
TABLE II.
Important considerations for the design of DNA-based nanodevices that are envisioned to perform a function in a complex biological setting.
| Problem | Solution | Associated risk |
|---|---|---|
| Off-target biodistribution (accumulation in organs, kidney filtration) | Increasing the molecular size by complex formation with other molecules; altering the object size, surface charge, or shape | Cellular uptake will be more difficult |
| Off-target action | Conjugation with target molecules (aptamer, antibody, peptide, ligand, protein), choosing highly specific gene targets | Local overdose, lower efficiency of drugs due to conjugation |
| Toxicity | Using biodegradable vectors | Premature release from the DNA construct before it has reached its target |
| Immunogenic reactions | Pretreatment using corticosteroids and anti-allergy medications, 2′-O-methyl base modifications | Overstimulation of the immune system |
| Quick degradation in a biological environment (e.g., by serum RNase), endosomal degradation | Altering the oligonucleotide chemistry, (e.g., methylation, peptide conjugation, phosphorothioate modification), protective coatings (oligolysine, PEG, polypeptides), using endosomolytic agents (e.g., melittin), carriers (polymer or lipid based) | Cytotoxic effects, reduced activity (e.g., for siRNA) |
| Low intracellular uptake | Using cationic carrier particles, PEGylation, transfection vectors, electroporation, lipid conjugation | Aggregation with serum proteins, accumulation of transport material in the cell |
In conclusion, understanding different biological barriers at all levels is equally as important for guaranteeing the functionality of a DNA-based device as the meticulous design of its physicochemical properties. The field of structural nanotechnology can create beautiful objects such as happy smiley-faces, even a nanoscopic Mona Lisa portrait made from DNA staples with extremely high precision. However, although medical nanotechnology also relies on the high level of precision and control made possible by base pairing, guaranteeing the functionality of the engineered devices in a biological setting is certainly the key challenge the field has to address in future research.
ACKNOWLEDGMENTS
The authors thank Iris König-Decker for providing graphics for this article and Matthias Marczynski for valuable comments on the manuscript draft.
The authors declare no conflict of interest.
DATA AVAILABILITY
Data sharing is not applicable to this article as no new data were created or analyzed in this study.
References
- 1. Seeman N. C., “ Nucleic acid junctions and lattices,” J. Theor. Biol. 99, 237−247 (1982). 10.1016/0022-5193(82)90002-9 [DOI] [PubMed] [Google Scholar]
- 2. Rothemund P. W., “ Folding DNA to create nanoscale shapes and patterns,” Nature 440, 297−302 (2006). 10.1038/nature04586 [DOI] [PubMed] [Google Scholar]
- 3. Jiang Q., Liu S., Liu J., Wang Z. G., and Ding B., “ Rationally designed DNA‐origami nanomaterials for drug delivery in vivo,” Adv. Mater. 31, 1804785 (2019). 10.1002/adma.201804785 [DOI] [PubMed] [Google Scholar]
- 4. Saccà B. and Niemeyer C. M., “ DNA origami: The art of folding DNA,” Angew. Chem. Int. Ed. Engl. 51, 58−66 (2012). 10.1002/anie.201105846 [DOI] [PubMed] [Google Scholar]
- 5. Jungmann R. et al. , “ Single-molecule kinetics and super-resolution microscopy by fluorescence imaging of transient binding on DNA origami,” Nano Lett. 10, 4756−4761 (2010). 10.1021/nl103427w [DOI] [PubMed] [Google Scholar]
- 6. Schnitzbauer J., Strauss M. T., Schlichthaerle T., Schueder F., and Jungmann R., “ Super-resolution microscopy with DNA-PAINT,” Nature Protocols 12, 1198 (2017). 10.1038/nprot.2017.024 [DOI] [PubMed] [Google Scholar]
- 7. Schueder F. et al. , “ Multiplexed 3D super-resolution imaging of whole cells using spinning disk confocal microscopy and DNA-PAINT,” Nature Commun. 8, 2090 (2017). 10.1038/s41467-017-02028-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Yehl K. et al. , “ High-speed DNA-based rolling motors powered by RNase H,” Nature Nanotechnol. 11, 184−190 (2016). 10.1038/nnano.2015.259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Church G. M., Gao Y., and Kosuri S., “ Next-generation digital information storage in DNA,” Science 337, 1628−1628 (2012). 10.1126/science.1226355 [DOI] [PubMed] [Google Scholar]
- 10. Castro C. E. et al. , “ A primer to scaffolded DNA origami,” Nature Methods 8, 221 (2011). 10.1038/nmeth.1570 [DOI] [PubMed] [Google Scholar]
- 11. García-Romero N. et al. , “ DNA sequences within glioma-derived extracellular vesicles can cross the intact blood-brain barrier and be detected in peripheral blood of patients,” Oncotarget 8, 1416 (2017). 10.18632/oncotarget.13635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Mead B. P. et al. , “ Targeted gene transfer to the brain via the delivery of brain-penetrating DNA nanoparticles with focused ultrasound,” J. Controlled Release 223, 109−117 (2016). 10.1016/j.jconrel.2015.12.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Kim K.-R. et al. , “ Sentinel lymph node imaging by a fluorescently labeled DNA tetrahedron,” Biomaterials 34, 5226−5235 (2013). 10.1016/j.biomaterials.2013.03.074 [DOI] [PubMed] [Google Scholar]
- 14. Erben C. M., Goodman R. P., and Turberfield A. J., “ Single‐molecule protein encapsulation in a rigid DNA cage,” Angew. Chem. Int. Ed. Engl. 45, 7414−7417 (2006). 10.1002/anie.200603392 [DOI] [PubMed] [Google Scholar]
- 15. Andersen E. S. et al. , “ Self-assembly of a nanoscale DNA box with a controllable lid,” Nature 459, 73−76 (2009). 10.1038/nature07971 [DOI] [PubMed] [Google Scholar]
- 16. Shi B. et al. , “ Challenges in DNA delivery and recent advances in multifunctional polymeric DNA delivery systems,” Biomacromolecules 18, 2231−2246 (2017). 10.1021/acs.biomac.7b00803 [DOI] [PubMed] [Google Scholar]
- 17. Zhang Q. et al. , “ DNA origami as an in vivo drug delivery vehicle for cancer therapy,” ACS Nano 8, 6633−6643 (2014). 10.1021/nn502058j [DOI] [PubMed] [Google Scholar]
- 18. Paiphansiri U. et al. , “ Glutathione‐responsive DNA‐based nanocontainers through an ‘interfacial click’ reaction in inverse miniemulsion,” Macromol. Chem. Phys. 215, 2457−2462 (2014). 10.1002/macp.201400374 [DOI] [Google Scholar]
- 19. Ijäs H., Hakaste I., Shen B., Kostiainen M. A., and Linko V., “ Reconfigurable DNA origami nanocapsule for pH-controlled encapsulation and display of cargo,” ACS Nano 13, 5959−5967 (2019). 10.1021/acsnano.9b01857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Zhang P. et al. , “ DNA‐hybrid‐gated multifunctional mesoporous silica nanocarriers for dual‐targeted and microRNA‐responsive controlled drug delivery,” Angew. Chem. Int. Ed. Engl. 126, 2403−2407 (2014). 10.1002/ange.201308920 [DOI] [PubMed] [Google Scholar]
- 21. Kimna C. et al. , “ DNA strands trigger the intracellular release of drugs from mucin-based nanocarriers,” ACS Nano (published online). [DOI] [PubMed] [Google Scholar]
- 22. Ma Y. et al. , “ A telomerase‐responsive DNA icosahedron for precise delivery of platinum nanodrugs to cisplatin‐resistant cancer,” Angew. Chem. Int. Ed. Engl. 57, 5389−5393 (2018). 10.1002/anie.201801195 [DOI] [PubMed] [Google Scholar]
- 23. Douglas S. M., Bachelet I., and Church G. M., “ A logic-gated nanorobot for targeted transport of molecular payloads,” Science 335, 831−834 (2012). 10.1126/science.1214081 [DOI] [PubMed] [Google Scholar]
- 24. Amir Y. et al. , “ Universal computing by DNA origami robots in a living animal,” Nature Nanotechnol. 9, 353−357 (2014). 10.1038/nnano.2014.58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Fire A. et al. , “ Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans,” Nature 391, 806−811 (1998). 10.1038/35888 [DOI] [PubMed] [Google Scholar]
- 26. McCaffrey A. P. et al. , “ RNA interference in adult mice,” Nature 418, 38−39 (2002). 10.1038/418038a [DOI] [PubMed] [Google Scholar]
- 27. Whitehead K. A., Langer R., and Anderson D. G., “ Knocking down barriers: Advances in siRNA delivery,” Nature Rev. Drug Discovery 8, 129−138 (2009). 10.1038/nrd2742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kandil R. et al. , “ Coming in and finding out: Blending receptor‐targeted delivery and efficient endosomal escape in a novel bio‐responsive siRNA delivery system for gene knockdown in pulmonary T cells,” Adv. Therap. 2, 1900047 (2019). 10.1002/adtp.201900047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Calin G. A. et al. , “ Human microRNA genes are frequently located at fragile sites and genomic regions involved in cancers,” Proc. Natl. Acad. Sci. USA 101, 2999−3004 (2004). 10.1073/pnas.0307323101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Conde J., Edelman E. R., and Artzi N., “ Target-responsive DNA/RNA nanomaterials for microRNA sensing and inhibition: The jack-of-all-trades in cancer nanotheranostics?” Adv. Drug Delivery Rev. 81, 169−183 (2015). 10.1016/j.addr.2014.09.003 [DOI] [PubMed] [Google Scholar]
- 31. Di Ianni T. et al. , “ Ultrasound/microbubble-mediated targeted delivery of anticancer microRNA-loaded nanoparticles to deep tissues in pigs,” J. Controlled Release 309, 1−10 (2019). 10.1016/j.jconrel.2019.07.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Conde J., Oliva N., Atilano M., Song H. S., and Artzi N., “ Self-assembled RNA-triple-helix hydrogel scaffold for microRNA modulation in the tumour microenvironment,” Nature Mater. 15, 353−363 (2016). 10.1038/nmat4497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Deng X. et al. , “ Hyaluronic acid-chitosan nanoparticles for co-delivery of MiR-34a and doxorubicin in therapy against triple negative breast cancer,” Biomaterials 35, 4333−4344 (2014). 10.1016/j.biomaterials.2014.02.006 [DOI] [PubMed] [Google Scholar]
- 34. Yao C. et al. , “ Reducible self-assembling cationic polypeptide-based micelles mediate co-delivery of doxorubicin and microRNA-34a for androgen-independent prostate cancer therapy,” J. Controlled Release 232, 203−214 (2016). 10.1016/j.jconrel.2016.04.034 [DOI] [PubMed] [Google Scholar]
- 35. Li R., Hehlman R., Sachs R., and Duesberg P., “ Chromosomal alterations cause the high rates and wide ranges of drug resistance in cancer cells,” Cancer Genet. Cytogenet. 163, 44−56 (2005). 10.1016/j.cancergencyto.2005.05.003 [DOI] [PubMed] [Google Scholar]
- 36. Cortez M. A. et al. , “ Therapeutic delivery of miR-200c enhances radiosensitivity in lung cancer,” Mol. Ther. 22, 1494−1503 (2014). 10.1038/mt.2014.79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Montgomery R. L. et al. , “ Therapeutic inhibition of miR-208a improves cardiac function and survival during heart failure,” Circulation 124, 1537−1547 (2011). 10.1161/CIRCULATIONAHA.111.030932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Hullinger T. G. et al. , “ Inhibition of miR-15 protects against cardiac ischemic injury,” Circ. Res. 110, 71−81 (2012). 10.1161/CIRCRESAHA.111.244442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Long J., Wang Y., Wang W., Chang B. H., and Danesh F. R., “ MicroRNA-29c is a signature microRNA under high glucose conditions that targets Sprouty homolog 1, and its in vivo knockdown prevents progression of diabetic nephropathy,” J. Biol. Chem. 286, 11837−11848 (2011). 10.1074/jbc.M110.194969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Pramanik D. et al. , “ Restitution of tumor suppressor microRNAs using a systemic nanovector inhibits pancreatic cancer growth in mice,” Mol. Cancer Therap. 10, 1470−1480 (2011). 10.1158/1535-7163.MCT-11-0152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Han C. et al. , “ MicroRNA-1 (miR-1) inhibits gastric cancer cell proliferation and migration by targeting MET,” Tumor Biol. 36, 6715−6723 (2015). 10.1007/s13277-015-3358-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Yang B. et al. , “ The muscle-specific microRNA miR-1 regulates cardiac arrhythmogenic potential by targeting GJA1 and KCNJ2,” Nature Med. 13, 486−491 (2007). 10.1038/nm1569 [DOI] [PubMed] [Google Scholar]
- 43. Ma L., Teruya-Feldstein J., and Weinberg R. A., “ Tumour invasion and metastasis initiated by microRNA-10b in breast cancer,” Nature 449, 682−688 (2007). 10.1038/nature06174 [DOI] [PubMed] [Google Scholar]
- 44. Ma L. et al. , “ Therapeutic silencing of miR-10b inhibits metastasis in a mouse mammary tumor model,” Nature Biotechnol. 28, 341−347 (2010). 10.1038/nbt.1618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Devulapally R. et al. , “ Polymer nanoparticles mediated codelivery of antimiR-10b and antimiR-21 for achieving triple negative breast cancer therapy,” ACS Nano 9, 2290−2302 (2015). 10.1021/nn507465d [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Pekarsky Y. and Croce C. M., “ Role of miR-15/16 in CLL,” Cell Death Differentiation 22, 6−11 (2015). 10.1038/cdd.2014.87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Calin G. A. et al. , “ MiR-15a and miR-16-1 cluster functions in human leukemia,” Proc. Natl. Acad. Sci. USA 105, 5166−5171 (2008). 10.1073/pnas.0800121105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Reid G. et al. , “ Targeted delivery of a synthetic microRNA-based mimic as an approach to cancer therapy,” Cancer Res. 75, 3976–3976 (2015). [Google Scholar]
- 49. Yan L.-X. et al. , “ MicroRNA miR-21 overexpression in human breast cancer is associated with advanced clinical stage, lymph node metastasis and patient poor prognosis,” RNA 14, 2348−2360 (2008). 10.1261/rna.1034808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Barger J. F. and Nana-Sinkam S. P., “ MicroRNA as tools and therapeutics in lung cancer,” Respir. Med. 109, 803−812 (2015). 10.1016/j.rmed.2015.02.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Seo Y.-E. et al. , “ Nanoparticle-mediated intratumoral inhibition of miR-21 for improved survival in glioblastoma,” Biomaterials 201, 87−98 (2019). 10.1016/j.biomaterials.2019.02.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Wang D. et al. , “ Local microRNA modulation using a novel anti-miR-21–eluting stent effectively prevents experimental in-stent restenosis,” Arterioscler. Thromb. Vasc. Biol. 35, 1945−1953 (2015). 10.1161/ATVBAHA.115.305597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Li F., Tang J., Geng J., Luo D., and Yang D, “ Polymeric DNA hydrogel: Design, synthesis and applications,” Prog. Polymer Science 98, 101163 (2019). 10.1016/j.progpolymsci.2019.101163 [DOI] [Google Scholar]
- 54. Shu D. et al. , “ Systemic delivery of anti-miRNA for suppression of triple negative breast cancer utilizing RNA nanotechnology,” ACS Nano 9, 9731−9740 (2015). 10.1021/acsnano.5b02471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Bertucci A. et al. , “ Tumor-targeting, microRNA-silencing porous silicon nanoparticles for ovarian cancer therapy,” ACS Appl. Materials Interfaces 11, 23926−23937 (2019). 10.1021/acsami.9b07980 [DOI] [PubMed] [Google Scholar]
- 56. Maurer B. et al. , “ MicroRNA‐29, a key regulator of collagen expression in systemic sclerosis,” Arthritis Rheumatism 62, 1733−1743 (2010). 10.1002/art.27443 [DOI] [PubMed] [Google Scholar]
- 57. Huang X. et al. , “ Targeted delivery of microRNA-29b by transferrin-conjugated anionic lipopolyplex nanoparticles: A novel therapeutic strategy in acute myeloid leukemia,” Clin. Cancer Res. 19, 2355−2367 (2013). 10.1158/1078-0432.CCR-12-3191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Ibrahim A. F. et al. , “ MicroRNA replacement therapy for miR-145 and miR-33a is efficacious in a model of colon carcinoma,” Cancer Res. 71, 5214−5224 (2011). 10.1158/0008-5472.CAN-10-4645 [DOI] [PubMed] [Google Scholar]
- 59. Misso G. et al. , “ Mir-34: a new weapon against cancer?” Mol. Ther. Nucleic Acids 3, e195 (2014). 10.1038/mtna.2014.47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Li H. et al. , “ Rational design of polymeric hybrid micelles with highly tunable properties to co‐deliver microRNA‐34a and vismodegib for melanoma therapy,” Adv. Funct. Mater. 25, 7457−7469 (2015). 10.1002/adfm.201503115 [DOI] [Google Scholar]
- 61. Herrera B. et al. , “ Global microRNA expression profiles in insulin target tissues in a spontaneous rat model of type 2 diabetes,” Diabetologia 53, 1099−1109 (2010). 10.1007/s00125-010-1667-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Trajkovski M. et al. , “ MicroRNAs 103 and 107 regulate insulin sensitivity,” Nature 474, 649−653 (2011). 10.1038/nature10112 [DOI] [PubMed] [Google Scholar]
- 63. Jopling C. L., Yi M., Lancaster A. M., Lemon S. M., and Sarnow P., “ Modulation of hepatitis C virus RNA abundance by a liver-specific microRNA,” Science 309, 1577−1581 (2005). 10.1126/science.1113329 [DOI] [PubMed] [Google Scholar]
- 64. Esau C. et al. , “ miR-122 regulation of lipid metabolism revealed by in vivo antisense targeting,” Cell Metab. 3, 87−98 (2006). 10.1016/j.cmet.2006.01.005 [DOI] [PubMed] [Google Scholar]
- 65. Krützfeldt J. et al. , “ Silencing of microRNAs in vivo with ‘antagomirs’,” Nature 438, 685−689 (2005). 10.1038/nature04303 [DOI] [PubMed] [Google Scholar]
- 66. Elmén J. et al. , “ LNA-mediated microRNA silencing in non-human primates,” Nature 452, 896−899 (2008). 10.1038/nature06783 [DOI] [PubMed] [Google Scholar]
- 67. Yu H.-c. et al. , “ Alterations of miR-132 are novel diagnostic biomarkers in peripheral blood of schizophrenia patients,” Prog. Neuro-Psychopharmacol. Biol. Psychiatry 63, 23−29 (2015). 10.1016/j.pnpbp.2015.05.007 [DOI] [PubMed] [Google Scholar]
- 68. Su Y. et al. , “ Intranasal delivery of targeted nanoparticles loaded with mir-132 to brain for the treatment of neurodegenerative diseases,” Front. Pharmacol. 11, 1165 (2020). 10.3389/fphar.2020.01165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Devalliere J. et al. , “ Sustained delivery of proangiogenic microRNA‐132 by nanoparticle transfection improves endothelial cell transplantation,” FASEB J. 28, 908−922 (2014). 10.1096/fj.13-238527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Kim M.-K. et al. , “ Tumor-suppressing miR-141 gene complex-loaded tissue-adhesive glue for the locoregional treatment of hepatocellular carcinoma,” Theranostics 8, 3891 (2018). 10.7150/thno.24056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Tran H. et al. , “ Label-free and reagentless electrochemical detection of microRNAs using a conducting polymer nanostructured by carbon nanotubes: Application to prostate cancer biomarker miR-141,” Biosensors Bioelectronics 49, 164−169 (2013). 10.1016/j.bios.2013.05.007 [DOI] [PubMed] [Google Scholar]
- 72. Zhang Q., Feng Y., Liu P., and Yang J., “ MiR-143 inhibits cell proliferation and invasion by targeting DNMT3A in gastric cancer,” Tumor Biol. 39, 1010428317711312 (2017). 10.1177/1010428317711312 [DOI] [PubMed] [Google Scholar]
- 73. Jiang Q. et al. , “ Therapeutic delivery of microRNA-143 by cationic lipoplexes for non-small cell lung cancer treatment in vivo,” J. Cancer Res. Clin. Oncol. 145, 2951−2967 (2019). 10.1007/s00432-019-03051-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Shimbo K. et al. , “ Exosome-formed synthetic microRNA-143 is transferred to osteosarcoma cells and inhibits their migration,” Biochem. Biophys. Res. Commun. 445, 381−387 (2014). 10.1016/j.bbrc.2014.02.007 [DOI] [PubMed] [Google Scholar]
- 75. O'Connell R. M., Chaudhuri A. A., Rao D. S., and Baltimore D., “ Inositol phosphatase SHIP1 is a primary target of miR-155,” Proc. Natl. Acad. Sci. USA 106, 7113−7118 (2009). 10.1073/pnas.0902636106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Cheng C. J. et al. , “ MicroRNA silencing for cancer therapy targeted to the tumour microenvironment,” Nature 518, 107−110 (2015). 10.1038/nature13905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Reshetnyak Y. K., Andreev O. A., Lehnert U., and Engelman D. M., “ Translocation of molecules into cells by pH-dependent insertion of a transmembrane helix,” Proc. Natl. Acad. Sci. USA 103, 6460−6465 (2006). 10.1073/pnas.0601463103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Babar I. A. et al. , “ Nanoparticle-based therapy in an in vivo microRNA-155 (miR-155)-dependent mouse model of lymphoma,” Proc. Natl. Acad. Sci. USA 109, E1695−E1704 (2012). 10.1073/pnas.1201516109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Gu Y. et al. , “ miR-192-5p silencing by genetic aberrations is a key event in hepatocellular carcinomas with cancer stem cell features,” Cancer Res. 79, 941−953 (2019). 10.1158/0008-5472.CAN-18-1675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Wu S. Y. et al. , “ A miR-192-EGR1-HOXB9 regulatory network controls the angiogenic switch in cancer,” Nature Commun. 7, 11169 (2016). 10.1038/ncomms11169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. De Craene B. and Berx B., “ Regulatory networks defining EMT during cancer initiation and progression,” Nature Rev. Cancer 13, 97−110 (2013). 10.1038/nrc3447 [DOI] [PubMed] [Google Scholar]
- 82. Pecot C. V. et al. , “ Tumour angiogenesis regulation by the miR-200 family,” Nature Commun. 4, 2427 (2013). 10.1038/ncomms3427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Yang D. et al. , “ Integrated analyses identify a master microRNA regulatory network for the mesenchymal subtype in serous ovarian cancer,” Cancer Cell 23, 186−199 (2013). 10.1016/j.ccr.2012.12.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Hossian A. N. et al. , “ Delivery of miR-143 and miR-506 with novel nano carrier to arrest cell cycle in lung cancer,” Cancer Res. 79, 4429–4429 (2019). [Google Scholar]
- 85. Keklikoglou I. et al. , “ MicroRNA-520/373 family functions as a tumor suppressor in estrogen receptor negative breast cancer by targeting NF-κB and TGF-β signaling pathways,” Oncogene 31, 4150−4163 (2012). 10.1038/onc.2011.571 [DOI] [PubMed] [Google Scholar]
- 86. Nishimura M. et al. , “ Therapeutic synergy between microRNA and siRNA in ovarian cancer treatment,” Cancer Discovery 3, 1302−1315 (2013). 10.1158/2159-8290.CD-13-0159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Mathur D. and Medintz I. L., “ The growing development of DNA nanostructures for potential healthcare‐related applications,” Advanced Healthcare Materials 8, 1801546 (2019). 10.1002/adhm.201801546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Meng H.-M. et al. , “ Aptamer-integrated DNA nanostructures for biosensing, bioimaging and cancer therapy,” Chem. Soc. Rev. 45, 2583−2602 (2016). 10.1039/C5CS00645G [DOI] [PubMed] [Google Scholar]
- 89. Jin B. et al. , “ Upconversion nanoparticles based FRET aptasensor for rapid and ultrasenstive bacteria detection,” Biosensors Bioelectronics 90, 525−533 (2017). 10.1016/j.bios.2016.10.029 [DOI] [PubMed] [Google Scholar]
- 90. He M., Li Z., Ge Y., and Liu Z., “ Portable upconversion nanoparticles-based paper device for field testing of drug abuse,” Anal. Chem. 88, 1530−1534 (2016). 10.1021/acs.analchem.5b04863 [DOI] [PubMed] [Google Scholar]
- 91. Dai S., Wu S., Duan N., and Wang Z., “ A luminescence resonance energy transfer based aptasensor for the mycotoxin Ochratoxin A using upconversion nanoparticles and gold nanorods,”Microchim. Acta 183, 1909−1916 (2016). 10.1007/s00604-016-1820-9 [DOI] [Google Scholar]
- 92. Zhao J. et al. , “ Upconversion luminescence-activated DNA nanodevice for ATP sensing in living cells,” J. Am. Chem. Soc. 140, 578−581 (2018). 10.1021/jacs.7b11161 [DOI] [PubMed] [Google Scholar]
- 93. Leung K., Chakraborty K., Saminathan A., and Krishnan Y., “ A DNA nanomachine chemically resolves lysosomes in live cells,” Nature Nanotechnol 14, 176−183 (2019). 10.1038/s41565-018-0318-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Xiong M. et al. , “ A membrane-anchored fluorescent probe for detecting K+ in the cell microenvironment,” Chem. Commun. 52, 4679−4682 (2016). 10.1039/C6CC00192K [DOI] [PubMed] [Google Scholar]
- 95. Qiu L. et al. , “ Cell membrane-anchored biosensors for real-time monitoring of the cellular microenvironment,” J. Am. Chem. Soc. 136, 13090−13093 (2014). 10.1021/ja5047389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Zhang P. et al. , “ In situ amplification of intracellular microRNA with MNAzyme nanodevices for multiplexed imaging, logic operation, and controlled drug release,” ACS Nano 9, 789−798 (2015). 10.1021/nn506309d [DOI] [PubMed] [Google Scholar]
- 97. Funck T., Nicoli F., Kuzyk A., and Liedl T., “ Sensing picomolar concentrations of RNA using switchable plasmonic chirality,” Angew. Chem. Int. Ed. Engl. 130, 13683−13686 (2018). 10.1002/ange.201807029 [DOI] [PubMed] [Google Scholar]
- 98. Koussa M. A., Halvorsen K., Ward A., and Wong W. P., “ DNA nanoswitches: A quantitative platform for gel-based biomolecular interaction analysis,” Nature Methods 12, 123−126 (2015). 10.1038/nmeth.3209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Ranallo S., Porchetta A., and Ricci F., “ DNA-based scaffolds for sensing applications,” Anal. Chem. 91, 44−59 (2019). 10.1021/acs.analchem.8b05009 [DOI] [PubMed] [Google Scholar]
- 100. Zhou L. et al. , “ Programmable low-cost DNA-based platform for viral RNA detection,” bioRxiv (2020). [DOI] [PMC free article] [PubMed]
- 101. Ng E. W. et al. , “ Pegaptanib, a targeted anti-VEGF aptamer for ocular vascular disease,” Nature Rev. Drug Discovery 5, 123−132 (2006). 10.1038/nrd1955 [DOI] [PubMed] [Google Scholar]
- 102. Díaz J. A. and Gibbs‐Davis J. M., “ Sharpening the thermal release of DNA from nanoparticles: Towards a sequential release strategy,” Small 9, 2862−2871 (2013). 10.1002/smll.201202278 [DOI] [PubMed] [Google Scholar]
- 103. Kimna C. and Lieleg O., “ Engineering an orchestrated release avalanche from hydrogels using DNA-nanotechnology,” J. Controlled Release 304, 19−28 (2019). 10.1016/j.jconrel.2019.04.028 [DOI] [PubMed] [Google Scholar]
- 104. Nowald C., Käsdorf B., and Lieleg O., “ Controlled nanoparticle release from a hydrogel by DNA-mediated particle disaggregation,” J. Controlled Release 246, 71−78 (2017). 10.1016/j.jconrel.2016.12.015 [DOI] [PubMed] [Google Scholar]
- 105. Scalise D. et al. , “ Programming the sequential release of DNA,” ACS Synthetic Biol. 9, 749−755 (2020). 10.1021/acssynbio.9b00398 [DOI] [PubMed] [Google Scholar]
- 106. Razin A. and Friedman J., “ DNA methylation and its possible biological roles,” Prog. Nucleic Acid Res. Mol. Biol. 25, 33−52 (1981). 10.1016/S0079-6603(08)60482-1 [DOI] [PubMed] [Google Scholar]
- 107. Medzhitov R., “ Recognition of microorganisms and activation of the immune response,” Nature 449, 819−826 (2007). 10.1038/nature06246 [DOI] [PubMed] [Google Scholar]
- 108. Schuller V. J. et al., “ Cellular immunostimulation by CpG-sequence-coated DNA origami structures,” ACS Nano 5, 9696−9702 (2011). 10.1021/nn203161y [DOI] [PubMed] [Google Scholar]
- 109. Qu Y. et al. , “ Self-assembled DNA dendrimer nanoparticle for efficient delivery of immunostimulatory CpG motifs,” ACS Appl. Materials Interfaces 9, 20324−20329 (2017). 10.1021/acsami.7b05890 [DOI] [PubMed] [Google Scholar]
- 110. Hong E. et al. , “ Structure and composition define immunorecognition of nucleic acid nanoparticles,” Nano Lett. 18, 4309−4321 (2018). 10.1021/acs.nanolett.8b01283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Yu W. et al. , “ Enhanced immunostimulatory activity of a cytosine-phosphate-guanosine immunomodulator by the assembly of polymer DNA wires and spheres,” ACS Appl. Materials Interfaces 12, 17167−17176 (2020). 10.1021/acsami.9b21075 [DOI] [PubMed] [Google Scholar]
- 112. Javed M. R. et al. , “ CRISPR-Cas system: History and prospects as a genome editing tool in microorganisms,” Curr. Microbiol. 75, 1675−1683 (2018). 10.1007/s00284-018-1547-4 [DOI] [PubMed] [Google Scholar]
- 113. Wu Y. et al. , “ Correction of a genetic disease in mouse via use of CRISPR-Cas9,” Cell Stem Cell 13, 659−662 (2013). 10.1016/j.stem.2013.10.016 [DOI] [PubMed] [Google Scholar]
- 114. Long C. et al. , “ Prevention of muscular dystrophy in mice by CRISPR/Cas9-mediated editing of germline DNA,” Science 345, 1184−1188 (2014). 10.1126/science.1254445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Lee B. et al. , “ Nanoparticle delivery of CRISPR into the brain rescues a mouse model of fragile X syndrome from exaggerated repetitive behaviours,” Nature Biomed. Eng. 2, 497−507 (2018). 10.1038/s41551-018-0252-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Xue W. et al. , “ CRISPR-mediated direct mutation of cancer genes in the mouse liver,” Nature 514, 380−384 (2014). 10.1038/nature13589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Sun W. et al. , “ Self‐assembled DNA nanoclews for the efficient delivery of CRISPR–Cas9 for genome editing,” Angew. Chem. Int. Ed. Engl. 127, 12197−12201 (2015). 10.1002/ange.201506030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Zhuang J. et al. , “ Extracellular vesicles engineered with valency-controlled DNA nanostructures deliver CRISPR/Cas9 system for gene therapy,” Nucleic Acids Res. 48, 8870−8882 (2020). 10.1093/nar/gkaa683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119. Dai Q. et al. , “ Quantifying the ligand-coated nanoparticle delivery to cancer cells in solid tumors,” ACS Nano 12, 8423−8435 (2018). 10.1021/acsnano.8b03900 [DOI] [PubMed] [Google Scholar]
- 120. Marras A. E. et al. , “ Cation-activated avidity for rapid reconfiguration of DNA nanodevices,” ACS Nano 12, 9484−9494 (2018). 10.1021/acsnano.8b04817 [DOI] [PubMed] [Google Scholar]
- 121. McTigue P. M., Peterson R J., and Kahn J. D., “ Sequence-dependent thermodynamic parameters for locked nucleic acid (LNA)-DNA duplex formation,” Biochemistry 43, 5388−5405 (2004). 10.1021/bi035976d [DOI] [PubMed] [Google Scholar]
- 122. Blanco E., Shen H., and Ferrari M., “ Principles of nanoparticle design for overcoming biological barriers to drug delivery,” Nature Biotechnol. 33, 941 (2015). 10.1038/nbt.3330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123. Madsen M. and Gothelf K. V., “ Chemistries for DNA nanotechnology,” Chem. Rev. 119, 6384−6458 (2019). 10.1021/acs.chemrev.8b00570 [DOI] [PubMed] [Google Scholar]
- 124. Cassinelli V. et al. , “ One‐step formation of ‘chain‐armor’‐stabilized DNA nanostructures,” Angew. Chem. Int. Ed. Engl. 54, 7795−7798 (2015). 10.1002/anie.201500561 [DOI] [PubMed] [Google Scholar]
- 125. Gerling T., Kube M., Kick B., and Dietz H., “ Sequence-programmable covalent bonding of designed DNA assemblies,” Science Adv. 4, eaau1157 (2018). 10.1126/sciadv.aau1157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126. Raniolo S. et al. , “ Cellular uptake of covalent and non-covalent DNA nanostructures with different sizes and geometries,” Nanoscale 11, 10808−10818 (2019). 10.1039/C9NR02006C [DOI] [PubMed] [Google Scholar]
- 127. Ponnuswamy N. et al. , “ Oligolysine-based coating protects DNA nanostructures from low-salt denaturation and nuclease degradation,” Nature Commun. 8, 15654 (2017). 10.1038/ncomms15654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128. Anastassacos F. M., Zhao Z., Zeng Y., and Shih W. M., “ Glutaraldehyde cross-linking of oligolysines coating DNA origami greatly reduces susceptibility to nuclease degradation,” J. Am. Chem. Soc. 142, 3311−3315 (2020). 10.1021/jacs.9b11698 [DOI] [PubMed] [Google Scholar]
- 129. Auvinen H. et al. , “ Protein coating of DNA nanostructures for enhanced stability and immunocompatibility,” Advanced Healthcare Materials 6, 1700692 (2017). 10.1002/adhm.201700692 [DOI] [PubMed] [Google Scholar]
- 130. Lacroix A. l., Edwardson T G., Hancock M. A., Dore M. D., and Sleiman H. F., “ Development of DNA nanostructures for high-affinity binding to human serum albumin,” J. Am. Chem. Soc. 139, 7355−7362 (2017). 10.1021/jacs.7b02917 [DOI] [PubMed] [Google Scholar]
- 131. Mikkila J. et al. , “ Virus-encapsulated DNA origami nanostructures for cellular delivery,” Nano Lett. 14, 2196−2200 (2014). 10.1021/nl500677j [DOI] [PubMed] [Google Scholar]
- 132. Perrault S. D. and Shih W. M., “ Virus-inspired membrane encapsulation of DNA nanostructures to achieve in vivo stability,” ACS Nano 8, 5132−5140 (2014). 10.1021/nn5011914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133. Poon W., Kingston B. R., Ouyang B., Ngo W., and Chan W. C., “ A framework for designing delivery systems,” Nature Nanotechnol. 15, 1−11 (2020). 10.1038/s41565-020-0759-5 [DOI] [PubMed] [Google Scholar]
- 134. Netti P. A., Berk D. A., Swartz M. A., Grodzinsky A. J., and Jain R. K., “ Role of extracellular matrix assembly in interstitial transport in solid tumors,” Cancer Res. 60, 2497−2503 (2000). [PubMed] [Google Scholar]
- 135. Madhanagopal B. R., Zhang S., Demirel E., Wady H., and Chandrasekaran A. R., “ DNA nanocarriers: Programmed to deliver,” Trends Biochem. Sci. 43, 997−1013 (2018). 10.1016/j.tibs.2018.09.010 [DOI] [PubMed] [Google Scholar]
- 136. Sykes E. A. et al. , “ Tailoring nanoparticle designs to target cancer based on tumor pathophysiology,” Proc. Natl. Acad. Sci USA 113, E1142−E1151 (2016). 10.1073/pnas.1521265113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137. Poon W. et al. , “ Elimination pathways of nanoparticles,” ACS Nano 13, 5785−5798 (2019). 10.1021/acsnano.9b01383 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data sharing is not applicable to this article as no new data were created or analyzed in this study.