Abstract
The demand for novel antimicrobial compounds is rapidly growing due to the phenomenon of antibiotic resistance in bacteria. In response, numerous alternative approaches are being taken including use of polymers, metals, combinatorial approaches, and antimicrobial peptides (AMPs). AMPs are a naturally occurring part of the immune system of all higher organisms and display remarkable broad-spectrum activity and high selectivity for bacterial cells over host cells. However, despite good activity and safety profiles, AMPs have struggled to find success in the clinic. In this review, we outline the fundamental properties of AMPs that make them effective antimicrobials and extend this into three main approaches being used to help AMPs become viable clinical options. These three approaches are the incorporation of non-natural amino acids into the AMP sequence to impart better pharmacological properties, the incorporation of AMPs in hydrogels, and the chemical modification of surfaces with AMPs for device applications. These approaches are being developed to enhance the biocompatibility, stability, and/or bioavailability of AMPs as clinical options.
I. INTRODUCTION
Peptides are “short” polymeric chains of amino acids linked via peptide bonds between the carboxyl group of one amino acid and the amino group of another amino acid. Peptides have a variety of functions in biological systems, frequently serving as hormones, signaling molecules, components of the immune system, and G protein-coupled receptor (GPCR) modulators. These naturally occurring peptides typically range from 10 to 50 amino acids, although there is no definitive length designation that delineates peptides from being categorized as proteins. The physiochemical properties of peptides are inherently determined by the amino acids that comprise the sequence, which varies based on the source and/or the intended function of the peptide. Similarly, the secondary and tertiary structures of peptides vary and are inherently linked to the primary amino acid sequence of the peptide.
Studies of peptides dramatically increased with the advent of solid-phase peptide synthesis methods pioneered by Merrifield.1 This method centers on the covalent linkage between an inert resin to the COOH terminus of an amino acid, which is sequentially modified to form the desired peptide sequence. Incoming amino acids are NH2-terminal protected with a labile group to prevent unwanted polymerization in solution, thus driving the reaction of the incoming amino acid to the free-amide of the peptide chain on the resin. After the reaction is complete, the solid-phase resin is easily separated from unreacted materials and reaction byproducts by simple filtration. The process is then repeated to add the next amino acid in sequence. The iterative process is easily automated, allowing commercial peptide synthesizers to effectively operate continuously, increasing the ease of obtaining peptides for further investigation.2,3
Due to the relative ease of synthesis, peptides have emerged as a widely used model system for investigating basic biological, biochemical, and biophysical interactions. In particular, biophysicists have widely adopted synthetic peptides as a model system due to the uniformity, purity, and compatibility with numerous physical and spectroscopic approaches. Peptide model systems have been extensively used in the investigation of protein secondary structure, specifically looking at amino acid propensities for secondary structure formation.4–8 Hydrophobic peptides have served as an attractive model system to study protein-lipid and protein-protein interactions in a lipid bilayer environment as an alternative to large membrane proteins that are traditionally difficult to work with in the laboratory.9–15 Peptides have also been an informative system for the de novo design of peptides with specific functions due to the flexibility of the synthetic model.11,16–20
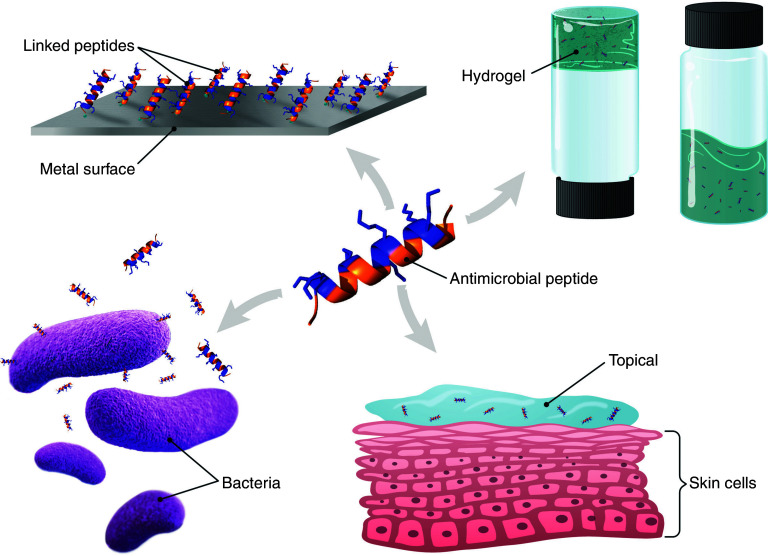
In this article, a background on antimicrobial peptides will be presented, with a focus on the application of these molecules. Specifically, a discussion of strategies to improve and expand the utilization of these molecules, both in a traditional clinical/therapeutic arena and as components of materials such as hydrogels and surface coatings, will be presented (Fig. 1).
FIG. 1.
Schematic example of different applications of AMPs such as topical applications, tethering to surfaces, components of hydrogels, or direct action on bacteria. Cationic residues of the model peptide are shown as blue and hydrophobic residues are orange, to highlight the facile amphiphilicity of helical AMPs. The schematic portrays helical peptides as they are the majority of AMPs; however, AMPs that adopt β-sheet structures are more frequently employed in hydrogel applications.
A. Peptide roles in biology
In biological systems, one of the primary roles of peptides is in signaling between cells and tissues. Specifically, peptide hormones are a common messenger between tissues. These peptides are secreted from cells in one tissue and are transported to distal tissues via the bloodstream. Once at the target tissue, these peptide hormones interact with a cell surface receptor on the target cells, beginning an intracellular signaling cascade. The most widely characterized peptide hormone, insulin, has been a focus of intensive study due to its direct role in the manifestation of diabetes. Other well-known peptide hormones include vasoactive intestinal peptide (VIP), follicle-stimulating hormone (FSH), glucagon, and calcitonin.21–24 All these peptide hormones have different structural conformations and binding targets based on amino acid composition.
Beyond hormones, peptides play an integral role in the innate immune and adaptive immune system of higher organisms. As they are involved in the immune response, these peptides are often referred to as host-defense peptides (HDPs). These HDPs are across both the innate and adaptive systems and serve direct cytotoxic roles against infectious agents as well as immunomodulatory roles to promote the host response to the infection. The innate immune system is the first line of defense against bacterial or viral infections, and a major component of this system is antimicrobial peptides. These peptides act directly on invading bacteria and/or viruses, leading to cell death or facilitating attack from macrophages (see below). The adaptive immune system also relies on peptides for numerous functions. T-cells, which function in a variety of roles in the immune system, present peptide fragments of antigens via major histocompatibility complexes (MHCs) on the cell surface to help immune cells to distinguish “self” and “non-self” cells during the identification of infection. HDPs also function in an immunomodulatory role, playing a role in affecting chemotaxis, chemokine and cytokine release, reactive oxygen species (ROS) production, and activation of leukocytes.25,26
B. Peptides as materials
Despite the significant improvements in peptide synthesis, purification, and characterization over the past 50–75 years, there has been a relatively small amount of translation beyond selected therapeutic applications. One of the major challenges to the application of peptides as therapeutics or materials is the limitations around scale-up to typical industrial quantities.27,28 The solid-phase synthetic routes used in the laboratory are difficult to scale into batch volumes due to mixing issues and solubility issues of protected amino acids. Additionally, the solid-phase approach is generally atom-inefficient, not in the target reaction, but in that there is significant waste generated at each step as a result of the numerous protecting groups, activator byproducts, as well as the solid-phase resin itself that is discarded after cleavage from the final, complete peptide. Peptides also suffer from the expected drawbacks of any protein material including temperature sensitivity, pH sensitivity, and sensitivity to proteases.
However, the applications landscape is not barren regarding peptidic materials. The flexibility of the solid-phase synthetic approach to incorporate non-proteinogenic (natural amino acids not normally incorporated into proteins/peptides) and non-natural amino acids greatly expands the chemical space available for functional group interactions. Peptide synthesis also allows for site-specific modification of peptides with abiotic functionalities such as fluorophores for binding/recognition, detection, or installing specific reactive groups.
Numerous groups have been working on peptide-based materials at bench-scale, which have proven to be successful in multiple applications. Many of these applications have focused on incorporating biocompatibility, biodegradability, and greener materials as an alternative to conventionally used materials. Due to the biological parallels, there is a significant number of peptides that have been designed and developed that can bind metal ions and/or porphyrin molecules.29–38 These systems display significant diversity in the peptide sequences, ligands, ligand binding approach, and peptide secondary structure which, taken together, highlight the flexibility of using peptides as a scaffold for binding functional ligands. Peptide amphiphiles, peptides which can self-assemble into micelles, have been developed to bind metals and porphyrin molecules for electron transfer applications.39,40 Peptides as functional materials in theranostics have been widely applied by exploiting native biological interactions.41,42 These theranostic materials predominantly involve peptide-modified liposomes, polymers, or nanoparticles. Other peptide-modified materials including carbon nanotubes have been used to create patterning of proteins on surfaces.43 Peptides have also been widely used to create supramolecular aggregates and structures, with tremendous flexibility on shape, which can be exploited for a variety of functions.42,44–46 The literature also contains numerous examples of peptides used as scaffolds or nucleation sites for crystal formation of non-biological materials.47,48
II. ANTIMICROBIAL PEPTIDES
Antimicrobial peptides are short, typically cationic and amphiphilic molecules that have been isolated from organisms throughout the evolutionary timeline from bacteria through humans. These peptides exhibit remarkable selectivity for bacteria over host cells, have low cytotoxicity, and have shown little potential for resistance development. Only recently have these molecules begun to emerge as components of materials to impart antimicrobial characteristics, rather than being viewed as a direct alternative/replacement to existing or traditional small-molecule antibiotic therapeutics.
A. Background and importance of antimicrobial peptides
Antibiotics have been described as a major contributing factor in the advancement of human health in the 20th century. After the discovery, purification, and widespread application of penicillin beginning in the 1940s, a “golden age” of antibiotic discovery occurred between the 1950s and 1970s. Antibiotics were developed and applied to treat a wide variety of infectious diseases and became a fundamental tool in the modern medical toolkit. Some of these antibiotics were broad spectrum, treating numerous infections caused by a variety of bacteria, while others were more narrow spectrum, with more specific targets or types of bacteria that were susceptible (Gram-positive vs. Gram-negative, for example).
Despite the leap forward in modern medicine that antibiotics represented, there was a flipside to the coin. Shortly after widespread clinical use, strains of bacteria that were resistant to antibiotics began to be identified. For example, penicillin was approved for use in 1941, and the first resistant strain of Staphylococcus aureus (S. aureus) was isolated in 1942, with resistant strains of Streptococcus pneumoniae isolated in 1967.49 Many resistant strains showed reduced sensitivity to the antibiotic treatment, while others were completely resistant to the antibiotics.50–53 These resistant bacteria present a significant hurdle to the treatment of once-routine infections and highlight the need for novel antimicrobial molecules, compounds, and approaches.
According the United States Center for Disease Control's (US CDC) 2019 Antibiotic Resistance threat report, ∼2.8 million antibiotic-resistant infections occur annually in the United States.54 Both the US CDC and the World Health Organization (WHO) identify antibiotic resistance as a major and significant threat to public health.54,55 These organizations have identified a number of high-priority bacterial strains that are already exhibiting some level of resistance and must be addressed due to the significant impact on human health. These strains include carbapenem-resistant Acetinobacter, Clostridioides difficile (C. difficile), Neisseria gonorrhoeae (N. gonorrhoeae), vancomycin-resistant Enterococci (VRE), multidrug-resistant Pseudomonas aeruginosa (P. aeruginosa), and methicillin-resistant S. aureus (MRSA), among others.54,55 The significant challenge posed by these strains, and emerging ones, is that antibiotic resistance often arises much faster than the development of novel antibiotics with similar or improved efficacy.56,57 Notably, there are often cross-resistance phenotypes observed between different molecules within a class of antibiotics, which means resistance development often differentially impacts multiple therapeutics.58,59 Thus, development of novel molecules with novel targets or mechanisms of action is the solution to the antibiotic-resistance phenomenon.
Antimicrobial peptides (AMPs) have emerged as an attractive option for development in the fight against resistance.60 The first antimicrobial peptides that were isolated and characterized were the gramicidins, isolated from the bacterium Bacillus brevis.61–63 Later, the first true AMP isolated from higher organisms was magainin, a component of skin secretions from Xenopus laevis.64 Since that time, thousands of AMPs have been identified and characterized throughout the evolutionary ladder from insects to fish to humans.65–68 The conservation of these molecules throughout evolution indicates that bacteria cannot effectively evolve a fully resistant phenotype to these molecules. This is not surprising as many AMPs are known to broadly target integral cellular components such as the cell membrane or cellular nucleic acids (see below). While reports of increased resistance are present, there is little to no evidence of complete insensitivity to AMPs, thus making them an attractive target for further development.69,70
B. Antimicrobial peptides mechanism of action
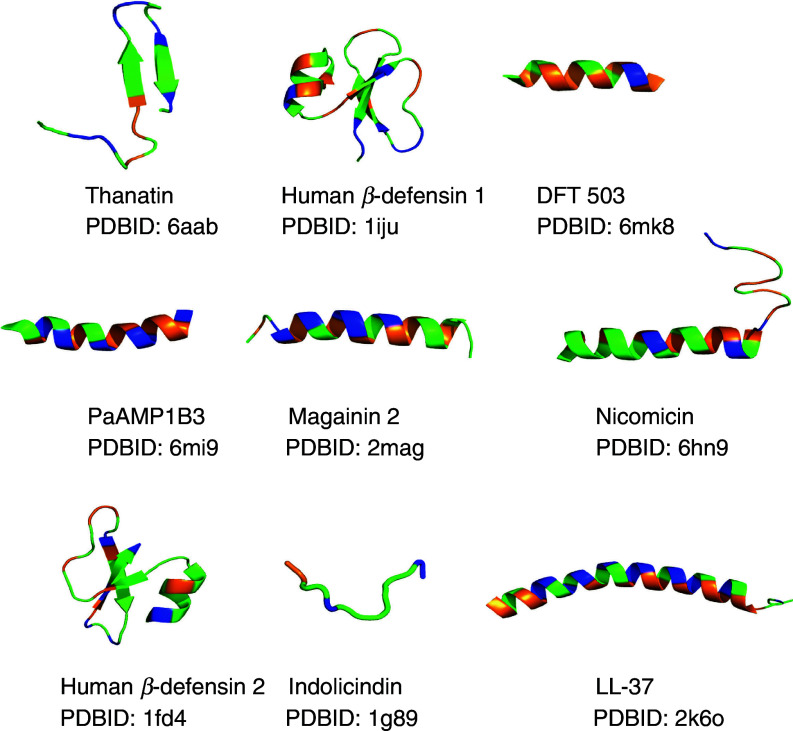
The small size of most AMPs makes them an easily tractable system to investigate, and thus significant amount of experimentation was quickly devoted to determining the mechanism of action. It became clear that there was no traditional consensus sequence that imparted antimicrobial characteristics to these peptides, but rather a more comprehensive set of physiochemical properties. A majority of AMPs were found to be cationic-amphiphilic sequences that contained significant fractions of both cationic and hydrophobic amino acids. The cationic net charge imparts the selectivity to bacteria over host cells, as bacterial cell surfaces present a net-negative charge while mammalian and other higher organisms generally have a net-neutral cell surface. This selection step driven by favorable electrostatic attraction between the cationic peptide and anionic bacterial cell surface is also the first stage of the mechanism.71–74 Upon binding to the cell surface, AMPs often undergo a conformational rearrangement facilitating the burial of the hydrophobic residues in the bacterial cell membrane. This structural rearrangement has been most often observed as a coil-to-helix transition. Indeed, many of the naturally identified AMPs adopt a facially amphiphilic structure, that is, one in which the cationic residues segregate to one face of the α-helix while the hydrophobic residues segregate to the opposite face of the helix.72,73,75–78 This structure allows the cationic groups to maintain interactions with the anionic bilayer surface while the hydrophobes become buried within the nonpolar core of the bilayer. However, not all naturally occurring AMPs are helical, with more and more examples containing β-hairpin and other β-sheet structures. Figure 2 shows several high-resolution structures of AMPs that have been experimentally determined by nuclear magnetic resonance (NMR) or x-ray methods.
FIG. 2.
Representative 3D structures of AMPs from the RCSB (rcsb.org). High-resolution structures were solved either by NMR or x-ray crystallography methods. The peptide name and PDBID accession numbers are listed under each peptide. Colors are related to amino acid properties: blue, cationic (Arg, Lys); orange, hydrophobic (Leu, Val, Ile, Phe, Ala, Phe); green, all other amino acids. Structures were visualized using PyMol.
At this stage, many AMPs have been shown to induce membrane disruption or destabilization.72,73,75,79,80 This destabilization can cause breakdown of the proton-motive force (PMF) required for ATP production, leakage of cellular material, disruption of ion balance, and changes in osmotic pressure in the bacterial cell, which can all contribute to cell death. Not surprisingly, as with AMP primary sequences, there is also no consensus on the molecular mechanism that causes membrane disruption by these peptides. Generally, there are three well-established models for the membrane active mechanism of AMPs: the barrel-stave pore model, the toroidal pore model, and the carpet model.81–84 The barrel-stave model involves the AMP inserting into the bacterial membrane in a traditional transmembrane orientation and subsequently self-associating into an oligomer with an aqueous pore/channel at the center of the proteinaceous oligomer. The toroidal pore model is similar to the barrel-stave pore model except that this structure also involves the local deformation of the lipid bilayer structure, causing a “bending” of the bilayer and resulting in a pore lined with both peptides and lipid head groups. In both the barrel-stave and toroidal pore mechanisms, the facially amphiphilic structures of the AMPs allow the hydrophobic groups to maintain contact with the nonpolar core of the bilayer, while the cationic groups are involved in lining the aqueous interior of the newly formed pore. The third model, often referred to as the “carpet” model, involves the coating of the bacterial cell surface with AMPs. This results in transient pores driven by a mass imbalance between the outer and inner leaflets of the bilayer or through a detergent-like removal of lipids from the membrane (although there is growing evidence that the carpet model and the detergent-like model may be two distinct mechanistic pathways). All of these mechanisms are affected by peptide primary and secondary structure, membrane lipid composition, and bound peptide concentration. The authors suggest recent reviews by Raheem and Straus85 as well as Bechinger and co-workers86 for a thorough review of the current literature on AMPs mechanism of action on membranes.
There are also numerous AMPs that have been shown to act through other non-membrane-disruptive mechanisms. Defensins have been demonstrated to bind to lipid II, a precursor in the synthesis of the bacterial cell wall.87 Other AMPs are known to translocate through the lipid membrane without significant bilayer disruption and subsequently interact with cellular components such as RNA and DNA.87–89 Notably, there are numerous reports in the literature regarding peptides from the Trp-rich class of AMPs that do not induce membrane permeabilization to significant degrees but are active antimicrobials.88,90 The current hypothesis is that the aromatic Trp residues promote interaction with the nucleotide bases of RNA or DNA, disrupting transcription or translation. It should be noted that other members of this Trp-rich class do exhibit membrane permeabilization, so it does not appear to be a universal mechanism within this class of peptides.91,92 The AMPs lactoferricin B and C18G have been indicated in the disruption of intracellular signaling through two-component signaling systems.93,94 Many AMPs are also known to act in an immunomodulatory role, promoting the activation and mobilization of factors in both the adaptive and innate immune systems.25
C. Applications and limitations of AMPs
Despite the high selectivity, low cytotoxicity, and low propensity for resistance development, AMPs have not been particularly successful in translating into clinical applications. Despite the thousands of identified AMPs and synthetically designed AMP mimetics, only about 10 molecules have made it through the entire FDA approval process in the USA, and two of that class are antiviral compounds (telaprevir and enfuvirtide).95–97 Notably, none of these are traditional linear AMPs, but instead have cyclic structures and often contain nonstandard amino acids in their structures, such as polymyxin, vancomycin, and daptomycin. Equally important are the formulations and route of administration for these approved peptide antimicrobials which, aside from vancomycin, all require injection or topical application. Intravenous and intramuscular injections reduce the widespread utility of AMPs as routine antibiotic therapies for outpatient use.
An additional two to three dozen clinical trials of AMPs have recently emerged, but of those trials, approximately 10 ended in withdrawn or discontinued trials due to lack of improved efficacy or adverse events.95–97 Interestingly, some of the ongoing or recent trials have focused on traditional, linear AMPs and target a number of different types of bacterial and fungal infections.95 The majority of these ongoing trials involve topical or intravenous applications, although two trials on orally delivered peptides, delmitide acetate and NVB-302, are still ongoing.98,99 Also within this group of ongoing trials are molecules that act as traditional membrane-disruptive AMPs,100,101 immunomodulators,102 and synthetic AMP mimetics.103
With a review of the successes and failures in the clinic, several important aspects of AMP translation can be gleaned. One of the major hurdles to AMP success is lack of bioavailability, specifically via oral administration. Traditional linear AMPs are readily digested by proteases in the gut, making it impossible for them to reach the target of infection.104–106 There is also an apparent reduction in efficacy observed for many AMPs between in vitro and in vivo studies, likely caused by a combination of pH, ionic strength/salt concentration, and nonspecific binding to serum proteins, which is not faithfully replicated in the in vitro screens.105,107,108
Taken together, one common characteristic of many of the AMPs successful in the clinic is the presence of non-natural or non-proteinogenic amino acids in the sequences. Published reports show that both overcoming ionic strength/salt challenges77,109,110 and increasing protease resistance92,111–113 can be achieved through the incorporation of nonstandard amino acids. This approach is gaining popularity due to the accessible chemical routes and expanded chemical space available using atypical amino acids.
D. Expanding the chemical space of AMPs
Based on the limited success thus far in the clinic, what can be done to improve the successful translation of these molecules into therapeutics? The AMP platform provides a number of advantages for the development of novel therapeutics and materials. One such advantage is the low propensity for resistance development as mentioned above.69,70 Another significant advantage for wider development of AMPs is the facile synthesis of these molecules. Specifically, the ability to synthesize these molecules in vitro allows for incorporation of non-biological functional groups, amino acids, and spectroscopic labels. This dramatically expands the chemical space available to these molecules, well beyond the limitations of the 20 naturally occurring amino acids. The commercial availability of numerous non-natural amino acids compatible with solid-phase peptide synthesis (SPPS) chemistry further aids in the synthesis and characterization of these AMPs.
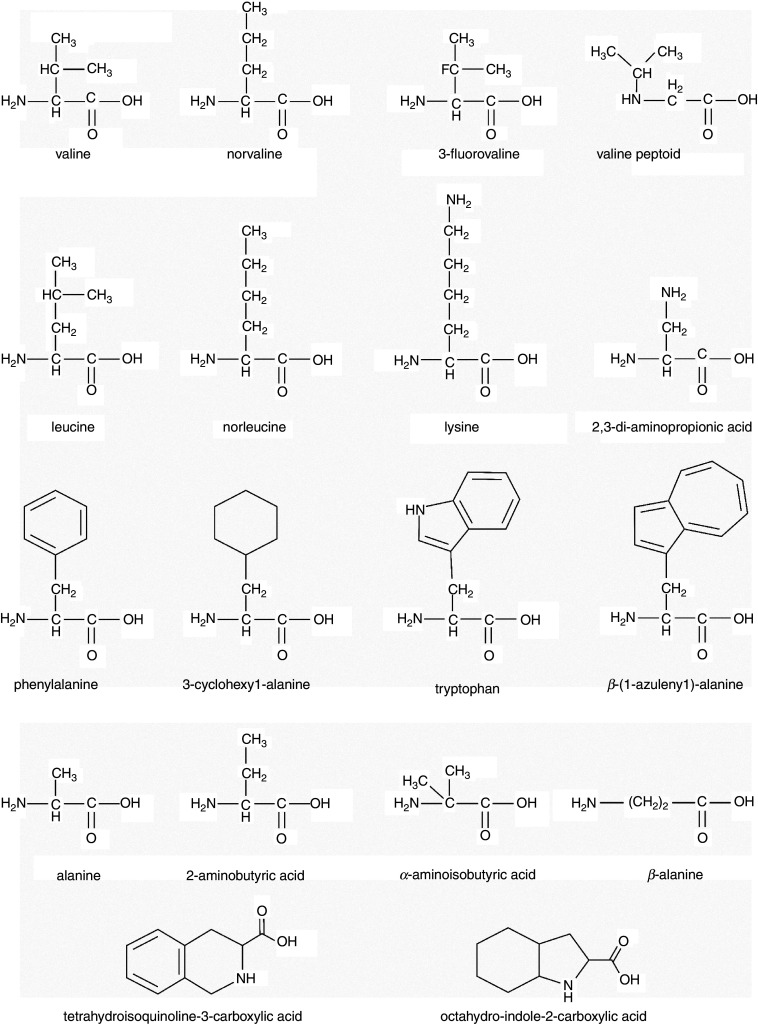
One strategy is to incorporate non-natural amino acids into the AMP sequence. This approach, while taking advantage of the SPPS method, has parallels in nature, as numerous naturally occurring AMPs have non-canonical amino acids in their sequences.114–116 Incorporation of these non-natural amino acids range from isosteres and isomers of naturally occurring amino acids to structures that are significantly different compared to natural amino acids. Conservative replacements in AMP sequences have allowed a systematic approach to the investigation of the structure-activity relationship (SAR) in AMPs,117–119 while other SAR approaches employ more significant modifications to peptide structures.120–122 Not surprisingly, the majority of research has focused on non-natural replacements of either cationic or hydrophobic amino acids in AMPs. A number of examples of these amino acids can be found in Fig. 3.
FIG. 3.
Chemical structures of natural and non-natural amino acids used in AMP sequences, highlighting related structures side by side. Images were created in ChemDraw.
Replacement of cationic amino acids with non-natural ones can generally fall into two categories: changing the overall shape/structure of the amino acid side chain or changing the cationic moiety that imparts the charge. These modifications can be as conservative as the lengthening or shortening of an amino acid side chain by a single methyl group.123–126 Our own work along with that of other groups have shown the shortening of the Lys side chain (4 methylenes) by replacing it with ornithine (Orn, 3 methylenes), di-aminobutyric acid (Dab, 2 methylenes), or di-aminopropionic acid (Dap, 1 methylene) reduces the antimicrobial activity of several AMPs,123,124 while it has little impact on others although improving cytotoxicity.92,125 Mechanistically, the shortening of the side chain appears to force the peptide into a shallower location in the bilayer, potentially reducing the ability to destabilize the membrane.123,126,127 This mechanism is consistent with what has been known regarding cationic amino acids “snorkeling” out of the nonpolar core of the bilayer to allow hydrophobic amino acids to remain buried while the charged group is allowed to interact with the polar membrane surface and aqueous milieu.14,127–130
The replacement of hydrophobic amino acids in AMPs with non-natural versions has been even more widely studied than the cationic replacements. This is likely due to a greater variety of nonpolar amino acids that are commercially available in SPPS-compatible forms. One of the most common approaches is to investigate the effect of non-natural amino acids that are isomers or isosteres on a naturally occurring residue, such as leucine and norleucine (Nle), valine and norvaline (Nva), or phenylalanine and cyclohexanoic acid (Cha). These variants are generally conservative to the original residue, but allow a fine-tuning of structural parameters for function or SAR studies.72,131–136 Cyclic and aromatic residues contribute to the hydrophobic character of AMPs, but also, the bulky structure of these residues likely participates in different membrane interactions than standard alkyl chains.137–140 Non-natural aromatic and cyclic residues have only recently begun to be explored in SAR of Trp-rich peptides. Our groups have undertaken investigations of the Trp isostere β-azulenyl-alanine (AzAla), which exhibits unique and beneficial fluorescence properties compared to Trp.141,142 AzAla was shown to be a conservative replacement in the helical membrane active peptide melittin.140 The residues Tic (tetrahydroisoquinoline-3-carboxylic acid) and Oic (octahydro-indole-2-carboxylic acid) as well as Tic-Oic dipeptides have been extensively studied in a number of AMP sequences for SAR studies, as these residues add hydrophobicity and structural restrictions due to the conformationally restricted backbone structures of the amino acids.120,143–145 A number of peptides containing these residues were demonstrated to have good activity against the high-threat “ESKAPE” pathogens (Enterococcus faecium, S. aureus, Klebsiella pnemoniae, Acinetobacter baumannii, P. aeruginosa, and Enterobacter species).144
The expansive chemical space associated with non-natural or non-proteinogenic amino acids is only a small fraction of the novel structures based on AMPs. An early approach involved the incorporation of one or more d-enantiomers of amino acids into the AMP sequence.111,146–150 In many cases, d-enantiomers of AMPs exhibit similar antimicrobial activity to the natural l-enantiomer, but have exhibited increased resistance to protease degradation.146–148 Beyond chirality, β-amino acids, which contain an extra -CH2 group in the peptide backbone, have been investigated both as α/β hybrid AMPs and as pure β-peptides.151–156 β-Amino acids are inherently more hydrophobic than α-peptides, due to the additional backbone methyl, which may play a role in the membrane binding of these AMPs. Other more non-natural modifications to AMPs such as incorporation of amino acids containing nonstandard atoms such as fluorine, bromine, or silicon are also commonly studied. These atoms change the properties of molecules and can add steric bulk, hydrophobicity, polarity, or electronegativity depending on the atom(s) incorporated.157–160
Further beyond the traditional peptide backbone, numerous groups have developed peptidomimetic structures and backbones designed to mimic the facile amphiphilicity of AMPs but offer some alternative advantages to the traditional α-amino acid structure, such as structural differences, protease resistance, or otherwise. Peptoids, which have the side chain attached to the amino nitrogen instead of the α-carbon, also have a wide range of R-groups available and a different set of constraints driving structure formation due to the loss of the amide hydrogen involved in helix stabilization. These peptoids exhibit the same selective binding of bacteria based on electrostatics and hydrophobic membrane interaction as the peptide counterparts.161–165 Small, polymeric peptidomimetics based on arylamide, methacrylate, ethyleneimine, thiophenes, polynorbornenes, and others have been investigated.166–176 These polymers are typically synthesized in solution resulting in greater polydispersity of the product molecules and lack of a uniform 3D structure but are effectively immune to proteolytic degradation. The arylamide peptidomimetics are generally smaller than typical AMPs or the other polymers and have yielded one molecule in clinical trials (brilacidin).103,170,171 The polymethacrylates have been extensively examined for SAR regarding antimicrobial activity due to the availability and flexibility of functional groups that can be incorporated into this polymeric backbone structure. These polymers exhibit a clear link between overall hydrophobicity and cytotoxicity, as well as similar SAR patterns in cationic and hydrophobic content to AMPs.172–176
Overall, there are a number of approaches researchers are taking to improve the activity and overcome drawbacks of antimicrobial peptides as therapeutic alternatives. These molecules have strong potential for future application. However, combining AMPs with other materials and platforms is another emerging approach to gain applied functionality from this class of molecules.
III. ANTIMICROBIAL HYDROGELS
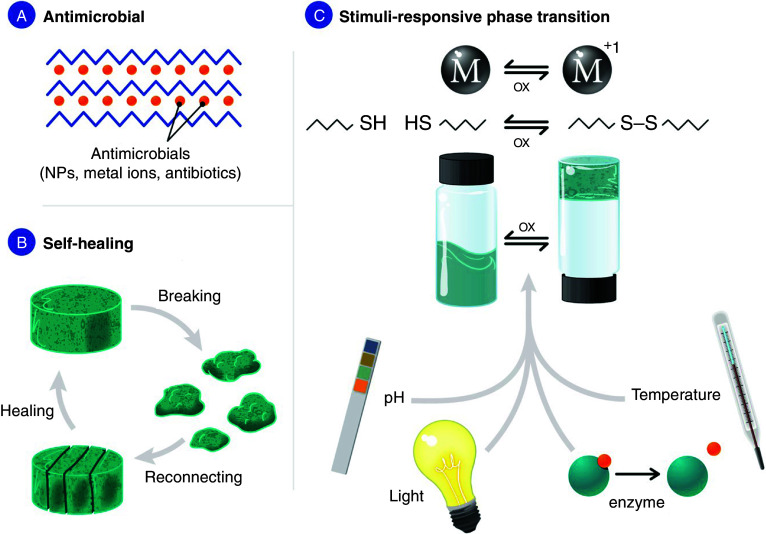
Hydrogels are highly hydrated biomaterials that are often produced from peptides or natural/non-natural polymers. Hydrogels have a great potential for tissue engineering applications and drug delivery.177–179 An especially attractive area of application is wound dressings because hydrogels provide a moist environment, facilitating a wound-healing process. However, high water content in hydrogels could also lead to microbial infection, and therefore, antimicrobial properties in hydrogels are desirable. Hydrogels with antimicrobial properties can be obtained by (1) the inclusion of known antimicrobial agent, or (2) the peptide itself can be designed to be antimicrobial [Fig. 4(a)].
FIG. 4.
Hydrogels and the properties that make them attractive for biomedical application (including wound healing). (a) Peptide-based hydrogels could be made antimicrobial through use of AMPs for gelation or incorporation of antimicrobial agents (NPs, metal ions, or antibiotics). (b) Self-healing hydrogels could be delivered by syringe. (c) Stimuli-responsive hydrogels could change their state between liquid and gel in response to pH, light, temperature, redox state, or biological stimuli (enzyme).
Peptide hydrogels can serve as a platform to deliver antimicrobial agents (such as antibiotics). Many examples of hydrogels loaded with antibiotics have been developed.180,181 Another widely used strategy is incorporation of metal nanoparticles (NPs; such as silver nanoparticles, AgNPs) in the hydrogel; however, AMPs are never used as a scaffold for such hydrogels, and antimicrobial properties come from NPs.180,182–185 As an antimicrobial agent, silver nitrate is commonly used186–188 and has recently gained more attention as antibiotic-resistant bacteria are on the rise.189–194 Silver-containing antimicrobials are attractive for topical applications, including wound healing and burns.195,196 However, high concentration of silver has been shown to be toxic to mammalian cells,197,198 limiting the use of silver-containing products.199 For example, some recent studies have shown toxicity of silver sulfadiazine (commonly used to treat burn wounds) for various host cells and a delayed wound-healing process.200–202 To minimize silver toxicity while harnessing its desirable antimicrobial properties, our group developed an antimicrobial hydrogel material that binds to Ag(I) and releases only small quantities of it, providing sufficient concentration of the metal ion for bactericidal activity but at a safe concentration for the host organism.203 In our approach, we incorporated non-natural amino acid pyridyl alanine into a peptide and used its power to coordinate Ag(I) ions in order to release only small amounts of this metal into the solution. Using pyridyl alanine for silver coordination provides a major benefit over histidine and cysteine previously utilized in similar applications204,205 due to stronger association of Ag(I) with pyridyl ligands. Additionally, the low pKa value of the non-natural amino acid allows for operation at reduced pH values often found in healing wounds.206
Some hydrogels have inherently antimicrobial properties due to gelation of AMPs. Schneider and Pochan labs developed peptides that contain positively-charged residues (Lys, Arg) that self-assemble into β-sheet rich hydrogels, which are active against both Gram-negative and Gram-positive bacteria.207–209 We, and others, have demonstrated that Arg-rich peptide-based hydrogels have antimicrobial and fracture-healing properties.203,210
A. Self-healing properties
Self-healing hydrogels have reversible cross-links and could be injected.211,212 Injectable hydrogels represent a novel class of biomaterials with potential application in tissue engineering [Fig. 4(b)]. A major advantage of peptide-based materials is biocompatibility, easy incorporation of sequence modifications (such as cell adhesion motif RGD); these materials can often be made biodegradable and antimicrobial. One of the strategies to prepare self-healing hydrogels is the use of non-covalent cross-links, such as formation of metal complexes, inspired by the material that mussels use underwater to adhere to surfaces.187,213–217 In addition to serving as a cross-link, metal ions can confer antimicrobial properties onto peptide-based hydrogels.203–205,218 The observed self-healing behavior is likely due to the plasticity of the coordination bonds between the metal and the peptides and the absence of covalent cross-links. This opens the path for delivery of the preset hydrogel into any wound shape using a syringe.
B. Stimuli-responsive peptide-based hydrogels
Biomaterials that change their properties in response to environmental stimuli [Fig. 4(c)] are being increasingly studied for application in regenerative medicine (also in wound healing). In addition to the practical application of such materials, the fundamental knowledge generated by studying switchable hydrogels will contribute to development of the wider fields of dynamic nanomaterials, drug delivery, and biosensing.219–222 While many hydrogels consist of polymers221,223–227 (natural or unnatural), here we will focus on hydrogels made from peptides. Hydrogels have been designed to respond to various environmental stimuli such as light, temperature, pH, enzymes, and redox state.181,219,220,228–231 Peptide-based hydrogels often change their supramolecular structure in response to pH. The peptide designed in the Schneider and Pochan labs called MAX1 consists of alternating valine and lysine residues; the latter are protonated at low pH (Table I). Gelation of this peptide can be triggered by neutralization of lysine residues through increasing the pH to 9.0.232 Peptide amphiphiles consisting of histidine and serine amino acids and palmitoyl tail were reported to form a hydrogel above pH 6.5 (233). The resulting hydrogel has been demonstrated to be compatible with fibroblast cells (fibroblasts are a major cell type involved in skin reparatory processes234). A 21-residue helical peptide has been reported to form a hydrogel in pH range 5.8–6.0235 that later has been modified to gel at physiological conditions (pH = 7.4), giving a hydrogel nontoxic to fibroblast cells.236 Antimicrobial peptide has been developed to undergo a transition from a random coil to a hydrogel when exposed to stimuli such as pH.237
TABLE I.
Responsive system based on peptide hydrogels.
| Entry | Name | Peptide sequence | Stimulus | References |
|---|---|---|---|---|
| 1 | MAX1 | VKVKVKVK-VDPPT-KVKVKVKV | pH | 232 |
| 2 | C16GSH | SSSGGK*GGHHH, where K* is conjugated to a fatty acid | pH | 233 |
| 3 | AFD36 | LKELAKV LHELAKL VKEALHA | pH | 235, 236 |
| 4 | ASCP1 | (KIGAKI)3-TDPPG-(KIGAKI)3 | pH | 237 |
| 5 | — | C(FKFE)2CG | Redox | 239 |
| 6 | Pep5 | E(CLSL)3E | Redox | 240 |
| 7 | 2 | Nap-GFFYE-CS-EERGD, where Nap is naphthalene and CS is cystamine succinate | Redox | 241 |
| 8 | — | Fc-F | Redox | 243 |
| 9 | 1 | Fc-(CO-WW-OMe)2 | Redox, t° | 244 |
| 10 | 1 | Fc-(CO-VFF-OMe) | Redox | 245 |
| 11 | 3 | (Nap-FFK)2-dicarboxyl-bipyridine-Ru | Redox | 246 |
| 12 | 1 | Fmoc-DA-DA | t° | 247 |
| 13 | hSAFQQQ | K IQQLKQK IQQLKQE IQQLEQE NQQLEQ | t° | 248 |
| 14 | MAX7CNB | VKVKVKVK-VDPPT-KVKXKVKV, where X is Cys(α-carboxy-2-nitrobenzyl) | Light | 249 |
| 15 | — | FmocFFGGGY + Ru(bpy)32+ | Light | 250 |
| 16 | FEK16 | (FEFEFKFK)2 + Ca2+-encapsulated vesicles | Light, t° | 251 |
| 17 | 2 | Fmoc-Y(P*), where P* is phosphate | AP | 254 |
| 18 | 1 | Nap-FF(KA*)Y(P*), where A* is azobenzene | AP, light | 255 |
| 19 | — | FF + Fmoc-F | Thermolysin | 256 |
| 20 | — | FEFK | Thermolysin | 257 |
| 21 | — | GCRD-GPQGIWGQ-DRCG + PEG | MMP | 258 |
| 22 | PA1 | (Palmitic acid)-GTAGLIGQES | MMP | 259 |
| 23 | D1 | Acceptor-AAPV-donor | Elastase | 260 |
| 24 | 4 | PEG4-DR-DR-DSP-LTPR-gelator | Thrombin | 261 |
| 25 | MDP4 | KSLSLSLRGSLSLSLK | Collagenase | 262 |
The transition to hydrogel can be also caused by reduction-oxidation. The most common example is formation of disulfide bonds between peptides, which could be reduced by a number of reductants. This strategy has been demonstrated using a cysteine derivative, which formed a gel due to disulfide bond formation, but the gel decomposed in the presence of reductant [tris(2-carboxyethyl)phosphine (TCEP)].238 The Nilsson lab developed a peptide Ac-C(FKFE)2CG-NH2 that cyclizes through disulfide bonding; however, when this bond is reduced by TCEP, the peptide adopts a linear form and assembles into hydrogel.239 Disulfide bond formation was also used to modulate strength of another peptide-based hydrogel, formed by Ac-E(CLSL)3E-NH2.240 Disulfide linker has been developed to induce gelation upon reduction by dithiothreitol (DTT) or glutathione (GSH), and these gels could be used to support 3D culture of fibroblasts.241,242 Ferrocene (Fc) has also been used to create redox-responsive hydrogels.219 The first hydrogel with Fc and redox response has been developed by the Zhang group.243 The redox properties of Fc were used to create a dipeptide-Fc that forms a gel upon reduction of iron.244 Another example of a redox-triggered transition from hydrogel to solution is reported for Fc-VFF.245 Reduction of metal to modulate the gelation of peptide derivatives has been demonstrated by the use of Ru(III)-tripeptide conjugate.246
The ferrocene complex with Trp dipeptide also has been shown to be temperature-dependent, as cooling leads to gel formation but heating dissolves the gel.244 Thermoresponsive materials have been of great interest for medical use because physiological temperature (37 °C) is common in many organisms and presents a convenient point for phase transition, which could be used to release biomolecules or cells. Another example of temperature-responsive gel came from the Bing Xu lab, where it was reported that Fmoc-DA-DA peptide formed a hydrogel at lower temperature, but higher temperature induced dissolution of the gel.247 The Woolfson lab reported a design of α-helical peptide dimer that displays transition to liquid with an increase in temperature.248
Pochan and Schneider labs modified MAX1 peptide, replacing one of the valines with cysteine that was photocaged by α-carboxy-2-nitrobenzyl. Exposure of the resulting MAX7 peptide to light of ∼300 nm for 30 min resulted in release of photocage and assembly of peptide into a hydrogel. The surface of the hydrogel was shown to be nontoxic to fibroblast cells,249 which is the first step for application in wound healing. Another peptide-based hydrogel has been developed to undergo cross-linking of the tyrosine in the presence of Ru complex. After illumination with white light, the resulting hydrogel increases its mechanical stability by 10,000-fold.250 Most of the light-sensitive hydrogels need UV light to trigger the solution to gel transition, and thus, application is limited due to low penetration of this light through tissues. In comparison, near-infrared (NIR) light is less damaging and can penetrate through tissues. The Messersmith lab developed a 16-residue peptide (FEK16) that self-assembles into a hydrogel in response to a photochemical trigger.251 When a mixture of peptide and light-sensitive liposomes with Ca(II) was exposed to NIR light, the vesicles became leaky and released Ca(II), which led to peptide gelation.
Enzymes trigger hydrogelation by converting precursor into active hydrogelator that undergoes self-assembly or enzymes can degrade the existing hydrogel.252,253 One of the classic examples of enzyme-triggered hydrogelation is the use of alkaline phosphatase (AP), which hydrolyzes the phosphate group,254 a strategy that was later expanded to create a peptide that forms a gel in response to both AP and light.255 The enzyme thermolysin and reverse hydrolysis have been used to form a bond between diphenylalanine peptide and Fmoc-Phe to make a tripeptide hydrogel,256 which was also used to drive self-assembly of tetrapeptide into an octapeptide that formed a hydrogel.257 Hydrogels that can report protease activity have been reported. An early example by the Hubbell group describes a matrix metalloproteinase (MMP)-responsive hydrogel that consists of polyethylene glycol (PEG) gel cross-linked with peptides.258 Hydrogels that can indicate protease activity have also been reported in the literature.259 Later hydrogels have been reported for detection of MMP and elastase,260 enzymes that are overexpressed in chronic wounds. Enzyme-triggered gelation has been shown to detect protease activity, and thrombin-activated gelation could be used to trigger artificial blood clots.261 Another example of enzyme-sensitive hydrogel was provided by the Hartgerink lab.262 Incorporation of enzyme-sensitive motif into peptide sequence allowed to obtain hydrogel sensitive to degradation with collagenase IV and MMP-2.
C. Wound-healing applications
Antimicrobial hydrogels not only treat infection but also provide a moist hydrated environment to facilitate wound healing;263,264 thus, hydrogels that inherently retain moisture have been successfully used in wound dressings.227,265–268 In addition to being antimicrobial, hydrogels should also possess self-healing properties, which would allow them to be delivered using a syringe and re-anneal in the wound bed. One of the advantages of using peptides over polymeric materials is ease of RGD motif incorporation. RGD269–271 is an integrin binding sequence present in extracellular matrix (ECM) proteins, which has been widely utilized to enhance cell adhesion toward mammalian cells.272–278 For example, peptide RADA16 has been functionalized with RGD, and the resulting hydrogel helps proliferation and migration of the skin cells279 (fibroblasts and keratinocytes). Short peptides functionalized with RGD also have been shown to form a hydrogel that promotes adhesion of fibroblasts and subsequent cell spreading and proliferation.275 The use of self-healing peptide-based hydrogels is a relatively new area of research, and their application in wound healing is limited.280 Several peptide-based hydrogels for cell encapsulation have been developed by the Schneider and Pochan labs and demonstrated to be nontoxic to mesenchymal stem cells.281 Short peptide-based hydrogels with self-healing properties have been reported for primary cortical neuron transplantation. Most cells were live after the injection of cells in hydrogel.282 Although the latter hydrogel has been designed for neurological injury repair, it shows that self-healing peptide-based hydrogels present a good scaffold for cell delivery. Peptide-based hydrogel formed from MAX1 peptide has been shown to be cytocompatible with fibroblasts, even supporting cells without serum in the media.283
The integration of protease-sensitive peptides into the hydrogel was shown by the Hubbell lab. Incorporation of peptides had resulted in faster decomposition of the hydrogels upon exposure to MMP and resulted in increased fibroblast spreading and invasion, important properties for wound-healing application.284
IV. PEPTIDE-MODIFIED SURFACES
There is a significant body of literature regarding the functionalization and modification of surfaces for specific applications, which has been a major focus of materials scientists for years. These surface modifications range from metals to small organics to large proteins. Similarly, the applications of these modified surfaces include photovoltaics, biosensors, antimicrobials, and electron conductors, among others.285–288 These surface coatings are another emerging area for the application of AMPs.
In general, modifications of surfaces to impart antimicrobial properties are of great interest to the medical device community. Providing an antimicrobial surface on hospital devices and surfaces is known to reduce and prevent the spread of nosocomial infections in the healthcare setting.289,290 Metal and metal-alloy coatings on medical devices have been widely studied using metals such as gold, silver, copper, and zinc.289–295 These coatings are usually very thin films but, depending on the substrate material and coating methodology, may result in changes to material properties of the device or ineffective adhesion to the device. This avenue of investigation was further expanded to include silver nanoparticles, which have also shown some success.296 Notably, some metallic coatings present an issue of cytotoxicity, which has limited wider application of these coatings.297 Polymer coatings are another alternative to impart antimicrobial character to a surface. These can be inherently antimicrobial or can be a suitable matrix to store and deliver another molecule that exerts antimicrobial activity. Polymers such as PEG and heparin have been demonstrated to resist bacterial adhesion and biofilm formation.298,299 Inert porous polymer materials have been loaded with antimicrobials such as chitosan, biosurfactants, or traditional small-molecule antimicrobials300–302 as well as have been covalently modified to attach antimicrobial functionalities.303
Peptide and protein interactions with surfaces have long been an interest, both in creating protein-functionalized surfaces and in the prevention of nonspecific protein binding to a surface. Peptides have been used as a functional material adhered to surfaces to promote tissue adhesion, act as biosensors, promote biocompatibility of implanted medical devices, and serve as components of semiconductors.288,304–307 In the context of antimicrobial surfaces, the attachment of AMPs to a material surface has been of great interest to many research groups, primarily because it avoids one of the major roadblocks facing AMP translation into clinical application: delivery. These functionalized surfaces could provide a long-lived, biocompatible, antimicrobial surface with little concern of local resistance development.
When discussing any peptide-functionalized surfaces, the first aspect is surface chemistry and the method by which the peptide is attached to the surface. Naturally, this depends on the surface material and chemistry, but is also often limited by the functional groups available in the peptide. As most helical AMPs require facile amphiphilicity for antibacterial activity, disrupting this 3D orientation would impede function. Gold has been widely used as a surface material in both electronic and medical devices and is thus a common material for functionalization by proteins or peptides. Covalent attachment of peptides and proteins to gold surfaces is often achieved using gold functionalized with maleimide reactive groups, which react with the sulfhydryl of Cys residues forming a stable thioether linkage;308 however, an oxidized gold surface can also react directly with Cys, forming an Au–S bond.309 Single-walled carbon nanotubes (SWCNTs) have also been functionalized with proteins and peptides, both covalently and non-covalently.310 Chemically harsh treatments are required to introduce carboxyl groups to the SWCNT structure, allowing reaction with peptide amine groups. Generally, non-covalent methods are preferred for SWCNTs, as the introduction of the reactive carboxyls often interferes with the materials properties of the SWCNT.311–313 Instead, patterned, designed, non-covalent interactions between peptides and SWCNTs can be used as a scaffold for further protein binding.43,314 Polymer-based materials have inherently more chemical flexibility, which is determined by the polymer matrix and any additives. The chemical attachment of peptides to a polymer material is inherently dependent on the functional groups present in the polymer but is also open to introduction of small amounts of reactive monomers to allow functionalization. Some groups have taken an approach similar to the non-covalent coating of SWCNTs by adsorbing a layer of bulk protein onto the polymer surface, which is then used as the platform for attachment of peptides of interest,315,316 while other groups rely on porous polymers to imbed peptides within the matrix.317–319
The fabrication of AMP-modified surfaces is taking advantage of all of the approaches mentioned above. Cys and DOPA residues, respectively, can directly bind metal surfaces such as gold and titanium. Cecropin P1 was shown to retain 3D structure upon binding to gold surfaces, while the peptide Tet-124 efficiently bound to Ti surfaces but lost activity upon binding.320,321 The peptide minTBP-1 has been shown to promote binding to titanium surfaces, and AMPs conjugated to the minTBP-1 motif retained antimicrobial efficacy.322 More recently, the one-pot synthesis of P-13 AMP-modified silver nanoparticles was reported and demonstrated both enhanced antimicrobial activity and reduced cytotoxicity,323 while similar results were shown for a synthetic AMP attached to a silver nanoparticle via click chemistry.324 AMPs have also been modified to attach to silver nanoparticles using a peptide-polymer linker (see below). This is especially promising as there have been several reports in the literature regarding synergistic interactions between silver and AMPs.325,326
Polymeric materials are also very widely studied, primarily because of the sheer volume of applications that polymers and plastics have in manufacturing. Embedding of AMPs within electrospun fibers of poly(ethylene oxide) (PEO) mixed with polycaprolactone (PCL) or poly(d,l‐lactide) (PDLLA) have shown to be effective in the encapsulation, release, and delivery of active AMPs using a number of different AMP sequences.317,327–329 Other approaches use a polymer layer-by-layer assembly in which the AMP is physically deposited between layers of polymers such as alginic acid, poly(-amino ester), chondroitin sulfate, or poly(methylmethacrylate).330,331 The insect-derived AMP Ponericin G1 showed significant efficacy and good release profiles using this approach.330 Similar to the protein layers described above, poly(DOPA-r-PEGMEMA-r-FuMaMA) [dopamine methacrylamide (DOPA), furan-protected maleimide methacrylate (FuMaMA), and poly(ethylene glycol) methyl ether methacrylate (PEGMEMA)] polymers have been applied as the coating agent in which dopamine bound directly to a titanium substrate, and the maleimide was used to bind a Cys in the AMPs.332 However, the most widely studied approach is direct covalent attachment of AMPs to polymeric materials. One common approach, providing the appropriate polymeric reactive groups, is to directly modify the polymer surface with the peptide. Four different peptides (melimine, lactoferricin, LL-37, and Mel-4) were directly linked to poly-hydroxyethylmethacrylate (pHEMA) surfaces and retained activity and did not result in cytotoxicity.333 The peptide GL13K was attached to a Ti microgrooved surface that was subsequently silanized. This coating retained antimicrobial activity but also promoted adhesion and patterned growth of human gingival fibroblasts.334 Another example demonstrated that the AMP E6 was conjugated to polyurethane catheter surfaces that were first modified with the brush copolymer PDMA-co-APMA [N,N-dimethylacrylamide and N-(3-aminopropyl)methacrylamide hydrochloride, respectively]. This copolymer-brush coupled to the E6 peptide significantly reduced infections in a murine urinary tract infection model and showed no indications of cytotoxicity.335 An alternative approach to functionalizing poly(urethane) surfaces was achieved by adding mixed poly(3-[dimethyl-[2-(2-methylprop-2-enoyloxy)ethyl]azaniumyl]propane-1-sulfonate) (PDMAPS) and poly(methacrylic acid) (PMAA) brushes to which the AMP HHC36 was covalently attached. This mixed-polymer brush coating retained in vitro and in vivo anti-infective activity while exhibiting good biocompatibility.336
PEG is a very commonly used polymeric linker between the primary surface because of the favorable biocompatibility, FDA approval for use in humans, and the compatibility with peptide synthetic chemistry methods. Soluble compounds of AMPs modified with PEG have shown enhanced activity for the peptide Maximin H5.337 The AMP KR12 was attached to a PEG linker that was bound to a carbohydrate sponge and was shown to have broad-spectrum antimicrobial activity over a multi-day period.338 Similarly, the AMP 6K8L was attached to a polystyrene surface using a PEG linker and exhibited both broad-spectrum activity and a robust performance over a wide range of pH and temperature conditions.339 Recent work has also demonstrated the synthesis and efficacy of multifunctional peptides attached to silver nanoparticles. This work demonstrated the targeted use of the PEG-modified AMP tachyplesin-1 coupled to a protease cleavage sequence that linked the PEG peptide to the nanoparticle.340
Taken together, AMPs appear to have multiple available routes for surface modification toward development of antibacterial surfaces. These hybrid approaches using non-natural amino acids, polymers, and other linkers are generating renewed interest in AMPs as functional material components.
V. FUTURE PERSPECTIVE
One of the most challenging aspects of translating AMPs into clinical application has been the fact that AMPs are, inherently, too biocompatible. These molecules can be broken down and degraded before ever reaching the target site of infection. In response, researchers have developed approaches using non-natural amino acids and peptidomimetics, hydrogels, and surface modification approaches to facilitate the application of these effective and selective AMPs for infection control. In many cases, these approaches overlap, combining effective methods to enhance antimicrobial activity and biocompatibility. The most successful applications leverage the inherent properties that drive antimicrobial activity in the peptides, namely, cationic and amphiphilic properties, with physical or chemical modifications that enhance delivery or reduce protease sensitivity.
There are still a number of poorly understood aspects of AMPs that need significant further research to develop into a mature field. First, greater clarity on the mechanism of action and how amino acid sequence length, composition, and properties determine the mechanism of action is necessary. This will allow for greater control over activity in designed peptides and peptidomimetics, allowing for tailored applications. Next, the role of structural and dynamic plasticity in these molecules is also of interest, specifically relating to the applications where the AMPs may be physically tethered or restricted by another material. Does this limit the ability of the AMP to adopt the “active” 3D structure, interact with the bacterial membrane, or access internal cytoplasmic targets and thereby limit the efficacy?
Overall, while AMPs have struggled to find widespread efficacy in the clinic as stand-alone antimicrobial therapeutics, the expansion of these molecules as components of materials appears to be a potential avenue for success. The ongoing work in this field will undoubtedly bring AMPs into new clinical trials in the near future.
ACKNOWLEDGMENTS
G.A.C. would like to thank Kenichi Kuroda and Jeffrey Hettinger for helpful discussions. O.V.M. would like to thank the Innovative and Interdisciplinary Research Grant from Syracuse University for support. Both authors would like to thank Karlee Rogers for assistance in preparation of figures.
Contributor Information
Olga V. Makhlynets, Email: mailto:ovmakhly@syr.edu.
Gregory A. Caputo, Email: mailto:caputo@rowan.edu.
DATA AVAILABILITY
Data sharing is not applicable to this article as no new data were created or analyzed in this article.
References
- 1. Merrifield B., Science 232, 341–347 (1986). 10.1126/science.3961484 [DOI] [PubMed] [Google Scholar]
- 2. Varnava K. G. and Sarojini V., Chem. Asian J. 14, 1088–1097 (2019). 10.1002/asia.201801807 [DOI] [PubMed] [Google Scholar]
- 3. Jaradat D. M. M., Amino Acids 50, 39–68 (2018). 10.1007/s00726-017-2516-0 [DOI] [PubMed] [Google Scholar]
- 4. Pace C. N. and Scholtz J. M., Biophys. J. 75, 422–427 (1998). 10.1016/S0006-3495(98)77529-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Li S.-C. and Deber C. M., Nat. Struct. Biol. 1, 368–373 (1994). 10.1038/nsb0694-368 [DOI] [PubMed] [Google Scholar]
- 6. Blaber M., Zhang X. J., and Matthews B. W., Science 260, 1637 (1993). 10.1126/science.8503008 [DOI] [PubMed] [Google Scholar]
- 7. Fisher B. F., Hong S. H., and Gellman S. H., J. Am. Chem. Soc. 139, 13292–13295 (2017). 10.1021/jacs.7b07930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Neil K. T. and DeGrado W. F., Science 250, 646 (1990). 10.1126/science.2237415 [DOI] [PubMed] [Google Scholar]
- 9. Choma C., Gratkowski H., Lear J. D., and DeGrado W. F., Nat. Struct. Biol. 7, 161–166 (2000). 10.1038/72440 [DOI] [PubMed] [Google Scholar]
- 10. Gratkowski H., Lear J. D., and DeGrado W. F., Proc. Natl. Acad. Sci. U S A 98, 880–885 (2001). 10.1073/pnas.98.3.880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Yin H., Slusky J. S., Berger B. W., Walters R. S., Vilaire G. et al. , Science 315, 1817–1822 (2007). 10.1126/science.1136782 [DOI] [PubMed] [Google Scholar]
- 12. Shahidullah K., Krishnakumar S. S., and London E., J. Mol. Biol. 396, 209–220 (2010). 10.1016/j.jmb.2009.11.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Shahidullah K. and London E., J. Mol. Biol. 379, 704–718 (2008). 10.1016/j.jmb.2008.04.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Caputo G. A. and London E., Biochemistry 42, 3275–3285 (2003). 10.1021/bi026697d [DOI] [PubMed] [Google Scholar]
- 15. Melnyk R. A., Kim S., Curran A. R., Engelman D. M., Bowie J. U., and Deber C. M., J. Biol. Chem. 279, 16591–16597 (2004). 10.1074/jbc.M313936200 [DOI] [PubMed] [Google Scholar]
- 16. Beesley J. L. and Woolfson D. N., Curr. Opin. Biotechnol. 58, 175–182 (2019). 10.1016/j.copbio.2019.03.017 [DOI] [PubMed] [Google Scholar]
- 17. Lombardi A., Pirro F., Maglio O., Chino M., and DeGrado W. F., Accounts Chem. Res. 52, 1148–1159 (2019). 10.1021/acs.accounts.8b00674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Marshall L. R., Zozulia O., Lengyel-Zhand Z., and Korendovych I. V., ACS Catal. 9, 9265–9275 (2019). 10.1021/acscatal.9b02509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Korendovych I. V. and DeGrado W. F., Q. Rev. Biophys. 53, e3 (2020). 10.1017/S0033583519000131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Ng A. H., Nguyen T. H., Gómez-Schiavon M., Dods G., Langan R. A. et al. , Nature 572, 265–269 (2019). 10.1038/s41586-019-1425-7 [DOI] [PubMed] [Google Scholar]
- 21. Nieves J. W., Komar L., Cosman F., and Lindsay R., Am. J. Clin. Nutr. 67, 18–24 (1998). 10.1093/ajcn/67.1.18 [DOI] [PubMed] [Google Scholar]
- 22. Tfelt-Hansen P. and Le H., J. Headache Pain 10, 137–143 (2009). 10.1007/s10194-009-0112-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Bousfield G. R. and Harvey D. J., Endocrinology 160, 1515–1535 (2019). 10.1210/en.2019-00001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Said S. I., J. Endocrinol. Invest. 9, 191–200 (1986). 10.1007/BF03348097 [DOI] [PubMed] [Google Scholar]
- 25. Hilchie A. L., Wuerth K., and Hancock R. E., Nat. Chem. Biol. 9, 761–768 (2013). 10.1038/nchembio.1393 [DOI] [PubMed] [Google Scholar]
- 26. Krantic S., Peptides 21, 1941–1964 (2000). 10.1016/S0196-9781(00)00347-8 [DOI] [PubMed] [Google Scholar]
- 27. Lee A. C., Harris J. L., Khanna K. K., and Hong J. H., Int. J. Mol. Sci. 20, 2383 (2019). 10.3390/ijms20102383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Bray B. L., Nat. Rev. Drug Discov. 2, 587–593 (2003). 10.1038/nrd1133 [DOI] [PubMed] [Google Scholar]
- 29. Koo J., Park J., Tronin A., Zhang R., Krishnan V. et al. , Langmuir 28, 3227–3238 (2012). 10.1021/la205002f [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Fry H. C., Lehmann A., Sinks L. E., Asselberghs I., Tronin A. et al. , J. Am. Chem. Soc. 135, 13914–13926 (2013). 10.1021/ja4067404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Fry H. C., Solomon L. A., Diroll B. T., Liu Y., Gosztola D. J., and Cohn H. M., J. Am. Chem. Soc. 142, 233–241 (2020). 10.1021/jacs.9b09935 [DOI] [PubMed] [Google Scholar]
- 32. Snyder R. A., Betzu J., Butch S. E., Reig A. J., DeGrado W. F., and Solomon E. I., Biochemistry 54, 4637–4651 (2015). 10.1021/acs.biochem.5b00324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Snyder R. A., Butch S. E., Reig A. J., DeGrado W. F., and Solomon E. I., J. Am. Chem. Soc. 137, 9302–9314 (2015). 10.1021/jacs.5b03524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Lee M., Wang T., Makhlynets O. V., Wu Y., Polizzi N. F. et al. , Proc. Natl. Acad. Sci. U S A 114, 6191–6196 (2017). 10.1073/pnas.1706179114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Korendovych I. V., Senes A., Kim Y. H., Lear J. D., Fry H. C. et al. , J. Am. Chem. Soc. 132, 15516–15518 (2010). 10.1021/ja107487b [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Farid T. A., Kodali G., Solomon L. A., Lichtenstein B. R., Sheehan M. M. et al. , Nat Chem Biol 9, 826–833 (2013). 10.1038/nchembio.1362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Kuciauskas D., Kiskis J., Caputo G. A., and Gulbinas V., J. Phys. Chem. B 114, 16029–16035 (2010). 10.1021/jp108685n [DOI] [PubMed] [Google Scholar]
- 38. Kuciauskas D. and Caputo G. A., J. Phys. Chem. B 113, 14439–14447 (2009). 10.1021/jp905468y [DOI] [PubMed] [Google Scholar]
- 39. Fry H. C., Garcia J. M., Medina M. J., Ricoy U. M., Gosztola D. J. et al. , J. Am. Chem. Soc. 134, 14646–14649 (2012). 10.1021/ja304674d [DOI] [PubMed] [Google Scholar]
- 40. Solomon L. A., Kronenberg J. B., and Fry H. C., J. Am. Chem. Soc. 139, 8497–8507 (2017). 10.1021/jacs.7b01588 [DOI] [PubMed] [Google Scholar]
- 41. Accardo A., Tesauro D., and Morelli G., Polym. J. 45, 481–493 (2013). 10.1038/pj.2012.215 [DOI] [Google Scholar]
- 42. Li L.-L., Qiao Z.-Y., Wang L., and Wang H., Adv. Mater. 31, 1804971 (2019). 10.1002/adma.201804971 [DOI] [PubMed] [Google Scholar]
- 43. Grigoryan G., Kim Y. H., Acharya R., Axelrod K., Jain R. M. et al. , Science 332, 1071–1076 (2011). 10.1126/science.1198841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Matson J. B., Zha R. H., and Stupp S. I., Curr. Opin. Solid State Mater. Sci. 15, 225–235 (2011). 10.1016/j.cossms.2011.08.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Ueda M., Makino A., Imai T., Sugiyama J., and Kimura S., Polym. J. 45, 509–515 (2013). 10.1038/pj.2013.4 [DOI] [Google Scholar]
- 46. Koss K. M. and Unsworth L. D., Materials 11, 1539 (2018). 10.3390/ma11091539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Kim K.-H., Ko D.-K., Kim Y.-T., Kim N. H., Paul J. et al. , Nat. Commun. 7, 11429 (2016). 10.1038/ncomms11429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Bong D. T. and Ghadiri M. R., Angew. Chem. Int. Ed. 40, 2163–2166 (2001). [DOI] [PubMed] [Google Scholar]
- 49. Plough H. H., Am. J. Clin. Pathol. 15, 446–451 (1945). 10.1093/ajcp/15.10.446 [DOI] [PubMed] [Google Scholar]
- 50. Bengtsson-Palme J., Kristiansson E., and Larsson D. G. J., FEMS Microbiol. Rev. 42, fux053 (2018). 10.1093/femsre/fux053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Dodds D. R., Biochem. Pharmacol. 134, 139–146 (2017). 10.1016/j.bcp.2016.12.005 [DOI] [PubMed] [Google Scholar]
- 52. Haaber J., Penadés J. R., and Ingmer H., Trends Microbiol. 25, 893–905 (2017). 10.1016/j.tim.2017.05.011 [DOI] [PubMed] [Google Scholar]
- 53. Klopper M., Warren R. M., Hayes C., Gey van Pittius N. C., Streicher E. M. et al. , Emerg. Infect. Dis. 19, 449–455 (2013). 10.3201/eid1903.120246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.CDC, U.S. Department of Health and Human Services. 2019.
- 55.WHO. 2014.
- 56.WHO. 2017.
- 57. Lepore C., Silver L., Theuretzbacher U., Thomas J., and Visi D., Nat. Rev. Drug Discov. 18, 739 (2019). 10.1038/d41573-019-00130-8 [DOI] [PubMed] [Google Scholar]
- 58. Dopcea G. N., Dopcea I., Nanu A. E., Diguţă C. F., and Matei F., J. Global Antimicrobial Resistance 21, 399–404 (2020). 10.1016/j.jgar.2019.10.021 [DOI] [PubMed] [Google Scholar]
- 59. de Souza Cândido E. S., de Barros E., Cardoso M. H., and Franco O. L., Curr. Opin. Pharmacol. 48, 76–81 (2019). 10.1016/j.coph.2019.05.004 [DOI] [PubMed] [Google Scholar]
- 60. Fosgerau K. and Hoffmann T., Drug Discov. Today 20, 122–128 (2015). 10.1016/j.drudis.2014.10.003 [DOI] [PubMed] [Google Scholar]
- 61. Dubos R. J. and Cattaneo C., J. Exp. Med. 70, 249–256 (1939). 10.1084/jem.70.3.249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Dubos R. J., J. Exp. Med. 70, 11–17 (1939). 10.1084/jem.70.1.11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Dubos R. J., J. Exp. Med. 70, 1–10 (1939). 10.1084/jem.70.1.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Zasloff M., Proc. Natl. Acad. Sci. U S A 84, 5449–5453 (1987). 10.1073/pnas.84.15.5449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Lazzaro B. P., Zasloff M., and Rolff J., Science 368, eaau5480 (2020). 10.1126/science.aau5480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Shabir U., Ali S., Magray A. R., Ganai B. A., Firdous P. et al. , Microbial Pathogenesis 114, 50–56 (2018). 10.1016/j.micpath.2017.11.039 [DOI] [PubMed] [Google Scholar]
- 67. Boparai J. K. and Sharma P. K., Protein Peptide Lett. 27, 4–16 (2019). 10.2174/0929866526666190822165812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Selsted M. E. and Ouellette A. J., Nat. Immunol. 6, 551–557 (2005). 10.1038/ni1206 [DOI] [PubMed] [Google Scholar]
- 69. Joo H.-S., Fu C.-I., and Otto M., Philos. Trans. R. Soc. B Biol. Sci. 371, 20150292 (2016). 10.1098/rstb.2015.0292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Andersson D. I., Hughes D., and Kubicek-Sutherland J. Z., Drug Resistance Updates 26, 43–57 (2016). 10.1016/j.drup.2016.04.002 [DOI] [PubMed] [Google Scholar]
- 71. Bobone S. and Stella L., in Antimicrobial Peptides: Basics for Clinical Application, edited by Matsuzaki K. ( Singapore: Springer Singapore, 2019), pp. 175–214. [Google Scholar]
- 72. Hitchner M. A., Necelis M. R., Shirley D., and Caputo G. A., Probiotics Antimicrob Proteins (published online 2020). 10.1007/s12602-020-09701-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Saint Jean K. D., Henderson K. D., Chrom C. L., Abiuso L. E., Renn L. M., and Caputo G. A., Probiotics Antimicrob Proteins 10, 408–419 (2018). 10.1007/s12602-017-9345-z [DOI] [PubMed] [Google Scholar]
- 74. Nourbakhsh S., Taheri-Araghi S., and Ha B.-Y., Soft Matter 15, 7509–7526 (2019). 10.1039/C9SM00930B [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Krauson A. J., He J., and Wimley W. C., Biochim. Biophys. Acta 1818, 1625–1632 (2012). 10.1016/j.bbamem.2012.02.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Yount N. Y., Weaver D. C., Lee E. Y., Lee M. W., Wang H. et al. , Proc. Natl. Acad. Sci. U S A 116, 6944–6953 (2019). 10.1073/pnas.1819250116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Yu H. Y., Tu C. H., Yip B. S., Chen H. L., Cheng H. T. et al. , Antimicrob. Agents Chemother. 55, 4918–4921 (2011). 10.1128/AAC.00202-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Zhang S.-K., Song J.-w., Gong F., Li S.-B., Chang H.-Y. et al. , Sci. Rep. 6, 27394 (2016). 10.1038/srep27394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Rashid M. M. O., Moghal M. M. R., Billah M. M., Hasan M., and Yamazaki M., Biochim. Biophys. Acta Biomembr. 1862, 183381 (2020). 10.1016/j.bbamem.2020.183381 [DOI] [PubMed] [Google Scholar]
- 80. Hasan M., Karal M. A. S., Levadnyy V., and Yamazaki M., Langmuir 34, 3349–3362 (2018). 10.1021/acs.langmuir.7b04219 [DOI] [PubMed] [Google Scholar]
- 81. Kumar P., Kizhakkedathu J. N., and Straus S. K., Biomolecules 8, 4 (2018). 10.3390/biom8010004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Brogden K. A., Nat. Rev. Microbiology 3, 238–250 (2005). 10.1038/nrmicro1098 [DOI] [PubMed] [Google Scholar]
- 83. Shai Y., Peptide Sci. 66, 236–248 (2002). 10.1002/bip.10260 [DOI] [PubMed] [Google Scholar]
- 84. Nguyen L. T., Haney E. F., and Vogel H. J., Trends Biotechnol. 29, 464–472 (2011). 10.1016/j.tibtech.2011.05.001 [DOI] [PubMed] [Google Scholar]
- 85. Raheem N. and Straus S. K., Front. Microbiol. 10, 2866 (2019). 10.3389/fmicb.2019.02866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Aisenbrey C., Marquette A., and Bechinger B., in Antimicrobial Peptides: Basics for Clinical Application, edited by Matsuzaki K. ( Singapore: Springer Singapore, 2019), pp. 33–64. [Google Scholar]
- 87. Malanovic N. and Lohner K., Pharmaceuticals 9, 59 (2016). 10.3390/ph9030059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Haney E. F., Petersen A. P., Lau C. K., Jing W., Storey D. G., and Vogel H. J., Biochim. Biophys. Acta (BBA) - Biomembranes 1828, 1802–1813 (2013). 10.1016/j.bbamem.2013.03.023 [DOI] [PubMed] [Google Scholar]
- 89. Park C. B., Kim H. S., and Kim S. C., Biochem. Biophys. Res. Commun. 244, 253–257 (1998). 10.1006/bbrc.1998.8159 [DOI] [PubMed] [Google Scholar]
- 90. Necelis M. R., Santiago-Ortiz L. E., and Caputo G. A., Protein Pept. Lett. 27, 1–15 (2020). 10.2174/0929866527666200813202918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Bozelli J. C., Yune J., Dang X., Narayana J. L., Wang G., and Epand R. M., Biochim. Biophys. Acta (BBA) - Biomembranes 1862, 183280 (2020). 10.1016/j.bbamem.2020.183280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Arias M., Piga K. B., Hyndman M. E., and Vogel H. J., Biomolecules 8, 19 (2018). 10.3390/biom8020019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Ho Y.-H., Sung T.-C., and Chen C.-S., Mol. Cell. Proteomics 11, M111.014720 (2012). 10.1074/mcp.M111.014720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Bader M. W., Sanowar S., Daley M. E., Schneider A. R., Cho U. et al. , Cell 122, 461–472 (2005). 10.1016/j.cell.2005.05.030 [DOI] [PubMed] [Google Scholar]
- 95. Mookherjee N., Anderson M. A., Haagsman H. P., and Davidson D. J., Nat. Rev. Drug Discovery 19, 311–332 (2020). 10.1038/s41573-019-0058-8 [DOI] [PubMed] [Google Scholar]
- 96. Pinheiro da Silva F. and Machado M. C. C., Peptides 36, 308–314 (2012). 10.1016/j.peptides.2012.05.014 [DOI] [PubMed] [Google Scholar]
- 97. Lei J., Sun L., Huang S., Zhu C., Li P. et al. , Am. J. Transl. Res. 11, 3919–3931 (2019). [PMC free article] [PubMed] [Google Scholar]
- 98. Bayés M., Rabasseda X., and Prous J. R., Methods Find. Exp. Clin. Pharmacol. 28, 233–277 (2006). [PubMed] [Google Scholar]
- 99. Crowther G. S., Baines S. D., Todhunter S. L., Freeman J., Chilton C. H., and Wilcox M. H., J. Antimicrob. Chemother. 68, 168–176 (2013). 10.1093/jac/dks359 [DOI] [PubMed] [Google Scholar]
- 100. Kaplan C. W., Sim J. H., Shah K. R., Kolesnikova-Kaplan A., Shi W., and Eckert R., Antimicrob. Agents Chemother. 55, 3446–3452 (2011). 10.1128/AAC.00342-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Toney J. H., Curr. Opin. Investig. Drugs 3, 225–228 (2002). [PubMed] [Google Scholar]
- 102. Kudrimoti M., Curtis A., Azawi S., Worden F., Katz S. et al. , J. Biotechnol. 239, 115–125 (2016). 10.1016/j.jbiotec.2016.10.010 [DOI] [PubMed] [Google Scholar]
- 103. Mensa B., Howell G. L., Scott R., and DeGrado W. F., Antimicrob. Agents Chemother. 58, 5136–5145 (2014). 10.1128/AAC.02955-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Arenas I., Villegas E., Walls O., Barrios H., Rodríguez R., and Corzo G., Molecules 21, 225 (2016). 10.3390/molecules21020225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Mahlapuu M., Håkansson J., Ringstad L., and Björn C., Front. Cell Infect. Microbiol. 6, 194 (2016). 10.3389/fcimb.2016.00194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Mohamed M. F., Abdelkhalek A., and Seleem M. N., Sci. Rep. 6, 29707 (2016). 10.1038/srep29707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Falanga A., Lombardi L., Franci G., Vitiello M., Iovene M. R. et al. , Int. J. Mol. Sci. 17, 785 (2016). 10.3390/ijms17050785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Koo H. B. and Seo J., Peptide Sci. 111, e24122 (2019). 10.1002/pep2.24122 [DOI] [Google Scholar]
- 109. Friedrich C., Scott M. G., Karunaratne N., Yan H., and Hancock R. E. W., Antimicrob. Agents Chemother. 43, 1542–1548 (1999). 10.1128/AAC.43.7.1542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Chu H. L., Yu H. Y., Yip B. S., Chih Y. H., Liang C. W. et al. , Antimicrob. Agents Chemother. 57, 4050–4052 (2013). 10.1128/AAC.00252-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Zhao Y., Zhang M., Qiu S., Wang J., Peng J. et al. , AMB Express 6, 122 (2016). 10.1186/s13568-016-0295-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Mardirossian M., Pompilio A., Degasperi M., Runti G., Pacor S. et al. , Front. Chem. 5, 40 (2017). 10.3389/fchem.2017.00040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Vasilchenko A. S., Vasilchenko A. V., Pashkova T. M., Smirnova M. P., Kolodkin N. I. et al. , J. Peptide Sci. 23, 855–863 (2017). 10.1002/psc.3049 [DOI] [PubMed] [Google Scholar]
- 114. Steenbergen J. N., Alder J., Thorne G. M., and Tally F. P., J. Antimicrob. Chemother. 55, 283–288 (2005). 10.1093/jac/dkh546 [DOI] [PubMed] [Google Scholar]
- 115. Velkov T., Thompson P. E., Nation R. L., and Li J., J. Med. Chem. 53, 1898–1916 (2010). 10.1021/jm900999h [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Cooper L. E., Li B., and van der Donk W. A., in Comprehensive Natural Products II, edited by Liu H.-W. and Mander L. ( Elsevier, Oxford, 2010), pp. 217–256. [Google Scholar]
- 117. Blondelle S. E., Takahashi E., Weber P. A., and Houghten R. A., Antimicrob. Agents Chemother. 38, 2280–2286 (1994). 10.1128/AAC.38.10.2280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Baumann T., Nickling J. H., Bartholomae M., Buivydas A., Kuipers O. P., and Budisa N., Front. Microbiol. 8, 124 (2017). 10.3389/fmicb.2017.00124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119. Oliva R., Chino M., Pane K., Pistorio V., De Santis A. et al. , Sci. Rep. 8, 8888 (2018). 10.1038/s41598-018-27231-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120. Russell A. L., Kennedy A. M., Spuches A. M., Gibson W. S., Venugopal D. et al. , Chem. Phys. Lipids 164, 740–758 (2011). 10.1016/j.chemphyslip.2011.09.003 [DOI] [PubMed] [Google Scholar]
- 121. Fields F. R., Manzo G., Hind C. K., Janardhanan J., Foik I. P. et al. , ACS Pharmacol. Transl. Sci. 3, 418–424 (2020). 10.1021/acsptsci.0c00001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122. Abdel Monaim S. A. H., Jad Y. E., El-Faham A., de la Torre B. G., and Albericio F., Bioorganic Medicinal Chem. 26, 2788–2796 (2018). 10.1016/j.bmc.2017.09.040 [DOI] [PubMed] [Google Scholar]
- 123. Kohn E. M., Shirley D. J., Arotsky L., Picciano A. M., Ridgway Z. et al. , Molecules 23, 329 (2018). 10.3390/molecules23020329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124. Abdel Monaim S. A. H., Ramchuran E. J., El-Faham A., Albericio F., and de la Torre B. G., J. Med. Chem. 60, 7476–7482 (2017). 10.1021/acs.jmedchem.7b00834 [DOI] [PubMed] [Google Scholar]
- 125. Mant C. T., Jiang Z., Gera L., Davis T., Nelson K. L. et al. , J. Med. Chem. 62, 3354–3366 (2019). 10.1021/acs.jmedchem.8b01785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126. Senetra A. S., Necelis M. R., and Caputo G. A., Peptide Sci. 112, e24162 (2020). 10.1002/pep2.24162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127. Jafari M., Mehrnejad F., and Doustdar F., PLOS One 12, e0187216 (2017). 10.1371/journal.pone.0187216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128. Strandberg E. and Killian J. A., FEBS Lett. 544, 69–73 (2003). 10.1016/S0014-5793(03)00475-7 [DOI] [PubMed] [Google Scholar]
- 129. Öjemalm K., Higuchi T., Lara P., Lindahl E., Suga H., and von Heijne G., Proc. Nat. Acad. Sci. U. S. A. 113, 10559–10564 (2016). 10.1073/pnas.1606776113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130. Keskin A., Akdoğan E., and Dunn C. D., Genetics 205, 691–705 (2017). 10.1534/genetics.116.196428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131. Radchenko D. S., Kattge S., Kara S., Ulrich A. S., and Afonin S., Biochim. Biophys. Acta (BBA) - Biomembranes 1858, 2019–2027 (2016). 10.1016/j.bbamem.2016.06.002 [DOI] [PubMed] [Google Scholar]
- 132. Nešuta O., Hexnerová R., Buděšínský M., Slaninová J., Bednárová L. et al. , J. Natural Prod. 79, 1073–1083 (2016). 10.1021/acs.jnatprod.5b01129 [DOI] [PubMed] [Google Scholar]
- 133. Postma T. M. and Liskamp R. M. J., RSC Adv. 6, 94840–94844 (2016). 10.1039/C6RA17944D [DOI] [Google Scholar]
- 134. Mourtada R., Herce H. D., Yin D. J., Moroco J. A., Wales T. E. et al. , Nat. Biotechnol. 37, 1186–1197 (2019). 10.1038/s41587-019-0222-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135. Ng S. M. S., Yap J. M., Lau Q. Y., Ng F. M., Ong E. H. Q. et al. , Eur. J. Med. Chem. 150, 479–490 (2018). 10.1016/j.ejmech.2018.03.027 [DOI] [PubMed] [Google Scholar]
- 136. Henriksen J. R., Etzerodt T., Gjetting T., and Andresen T. L., PLOS One 9, e91007 (2014). 10.1371/journal.pone.0091007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137. Haug B. E., Stensen W., Stiberg T., and Svendsen J. S., J. Med. Chem. 47, 4159–4162 (2004). 10.1021/jm049582b [DOI] [PubMed] [Google Scholar]
- 138. Lau Q. Y., Ng F. M., Cheong J. W. D., Yap Y. Y. A., Tan Y. Y. F. et al. , Eur. J. Med. Chem. 105, 138–144 (2015). 10.1016/j.ejmech.2015.10.015 [DOI] [PubMed] [Google Scholar]
- 139. Haug B. E., Skar M. L., and Svendsen J. S., J. Peptide Sci. 7, 425–432 (2001). 10.1002/psc.338 [DOI] [PubMed] [Google Scholar]
- 140. Ridgway Z., Picciano A. L., Gosavi P. M., Moroz Y. S., Angevine C. E. et al. , Biopolymers 104, 384–394 (2015). 10.1002/bip.22624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141. Moroz Y. S., Binder W., Nygren P., Caputo G. A., and Korendovych I. V., Chem. Commun. 49, 490–492 (2013). 10.1039/C2CC37550H [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142. Gosavi P. M., Moroz Y. S., and Korendovych I. V., Chem Commun 51, 5347–5350 (2015). 10.1039/C4CC08720H [DOI] [PubMed] [Google Scholar]
- 143. Hicks R. P. and Russell A. L., in Unnatural Amino Acids: Methods and Protocols, edited by Pollegioni L. and Servi S. ( Humana, Totowa, NJ, 2012), pp. 135–167. [Google Scholar]
- 144. Hicks R. P., Abercrombie J. J., Wong R. K., and Leung K. P., Bioorganic Med. Chem. 21, 205–214 (2013). 10.1016/j.bmc.2012.10.039 [DOI] [PubMed] [Google Scholar]
- 145. Hicks R. P., Bhonsle J. B., Venugopal D., Koser B. W., and Magill A. J., J. Med. Chem. 50, 3026–3036 (2007). 10.1021/jm061489v [DOI] [PubMed] [Google Scholar]
- 146. Hamamoto K., Kida Y., Zhang Y., Shimizu T., and Kuwano K., Microbiol. Immunol. 46, 741–749 (2002). 10.1111/j.1348-0421.2002.tb02759.x [DOI] [PubMed] [Google Scholar]
- 147. Adaligil E., Patil K., Rodenstein M., and Kumar K., ACS Chem. Biol. 14, 1498–1506 (2019). 10.1021/acschembio.9b00234 [DOI] [PubMed] [Google Scholar]
- 148. Cardoso M. H., Cândido E. S., Oshiro K. G. N., Rezende S. B., and Franco O. L., in Peptide Applications in Biomedicine, Biotechnology and Bioengineering, edited by Koutsopoulos S ( Woodhead, 2018), pp. 131–155. [Google Scholar]
- 149. Wade D., Boman A., Wåhlin B., Drain C. M., Andreu D. et al. , Proc. Natl. Acad. Sci. U S A 87, 4761–4765 (1990). 10.1073/pnas.87.12.4761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150. Lee D. L., Powers J. P. S., Pflegerl K., Vasil M. L., Hancock R. E. W., and Hodges R. S., J. Peptide Res. 63, 69–84 (2004). 10.1046/j.1399-3011.2003.00106.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151. Godballe T., Nilsson L. L., Petersen P. D., and Jenssen H., Chem. Biol. Drug Des. 77, 107–116 (2011). 10.1111/j.1747-0285.2010.01067.x [DOI] [PubMed] [Google Scholar]
- 152. Wani N. A., Singh G., Shankar S., Sharma A., Katoch M., and Rai R., Peptides 97, 46–53 (2017). 10.1016/j.peptides.2017.09.016 [DOI] [PubMed] [Google Scholar]
- 153. Porter E. A., Wang X., Lee H.-S., Weisblum B., and Gellman S. H., Nature 404, 565 (2000). 10.1038/35007145 [DOI] [PubMed] [Google Scholar]
- 154. Steer D. L., Lew R. A., Perlmutter P., Smith A., and Aguilar M. I., Curr. Med. Chem. 9, 811–822 (2002). 10.2174/0929867024606759 [DOI] [PubMed] [Google Scholar]
- 155. Raguse T. L., Porter E. A., Weisblum B., and Gellman S. H., J. Am. Chem. Soc. 124, 12774–12785 (2002). 10.1021/ja0270423 [DOI] [PubMed] [Google Scholar]
- 156. DeGrado W. F., Schneider J. P., and Hamuro Y., J. Peptide Res. 54, 206–217 (2008). 10.1034/j.1399-3011.1999.00131.x [DOI] [PubMed] [Google Scholar]
- 157. Jia F., Zhang Y., Wang J., Peng J., Zhao P. et al. , Peptides 112, 56–66 (2019). 10.1016/j.peptides.2018.11.006 [DOI] [PubMed] [Google Scholar]
- 158. Arias M., Aramini J. M., Riopel N. D., and Vogel H. J., Biochim. Biophys. Acta (BBA) - Biomembranes 1862, 183260 (2020). 10.1016/j.bbamem.2020.183260 [DOI] [PubMed] [Google Scholar]
- 159. Huhmann S. and Koksch B., Eur. J. Organic Chem. 2018, 3667–3679. 10.1002/ejoc.201800803 [DOI] [Google Scholar]
- 160. Madsen J. L. H., Hjørringgaard C. U., Vad B. S., Otzen D., and Skrydstrup T., Chem. Eur. J. 22, 8358–8367 (2016). 10.1002/chem.201600445 [DOI] [PubMed] [Google Scholar]
- 161. Goodson B., Ehrhardt A., Ng S., Nuss J., Johnson K. et al. , Antimicrob. Agents Chemother. 43, 1429–1434 (1999). 10.1128/AAC.43.6.1429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162. Chongsiriwatana N. P., Patch J. A., Czyzewski A. M., Dohm M. T., Ivankin A. et al. , Proc. Nat. Acad. Sci.U S A 105, 2794–2799 (2008). 10.1073/pnas.0708254105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163. Molchanova N., Wang H., Hansen P. R., Høiby N., Nielsen H. M., and Franzyk H., Front. Microbiol. 10, 275 (2019). 10.3389/fmicb.2019.00275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164. Yoo B. and Kirshenbaum K., Curr. Opin. Chem. Biol. 12, 714–721 (2008). 10.1016/j.cbpa.2008.08.015 [DOI] [PubMed] [Google Scholar]
- 165. Andreev K., Martynowycz M. W., Huang M. L., Kuzmenko I., Bu W. et al. , Biochim. Biophys. Acta (BBA) - Biomembranes 1860, 1414–1423 (2018). 10.1016/j.bbamem.2018.03.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166. Zhou Z., Ergene C., Lee J. Y., Shirley D. J., Carone B. R. et al. , ACS Appl. Mater. Interfaces 11, 1896–1906 (2019). 10.1021/acsami.8b19098 [DOI] [PubMed] [Google Scholar]
- 167. Ergene C. and Palermo E. F., J. Mater. Chem. B 6, 7217–7229 (2018). 10.1039/C8TB01632A [DOI] [PubMed] [Google Scholar]
- 168. deRonde B. M., Birke A., and Tew G. N., Chem. Eur. J. 21, 3013–3019 (2015). 10.1002/chem.201405381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169. Colak S., Nelson C. F., Nüsslein K., and Tew G. N., Biomacromolecules 10, 353–359 (2009). 10.1021/bm801129y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170. Mensa B., Kim Y. H., Choi S., Scott R., Caputo G. A., and DeGrado W. F., Antimicrob. Agents Chemother. 55, 5043–5053 (2011). 10.1128/AAC.05009-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171. Choi S., Isaacs A., Clements D., Liu D., Kim H. et al. , Proc. Natl. Acad. Sci. U S A 106, 6968–6973 (2009). 10.1073/pnas.0811818106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172. Kuroda K., Caputo G. A., and DeGrado W. F., Chem. Eur. J. 15, 1123–1133 (2009). 10.1002/chem.200801523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173. Thoma L. M., Boles B. R., and Kuroda K., Biomacromolecules 15, 2933–2943 (2014). 10.1021/bm500557d [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174. Palermo E. F., Vemparala S., and Kuroda K., Biomacromolecules 13, 1632–1641 (2012). 10.1021/bm300342u [DOI] [PubMed] [Google Scholar]
- 175. Michl T. D., Hibbs B., Hyde L., Postma A., Tran D. T. T. et al. , Acta Biomater. 108, 168–177 (2020). 10.1016/j.actbio.2020.03.011 [DOI] [PubMed] [Google Scholar]
- 176. Locock K. E. S., Michl T. D., Stevens N., Hayball J. D., Vasilev K. et al. , ACS Macro. Lett. 3, 319–323 (2014). 10.1021/mz5001527 [DOI] [PubMed] [Google Scholar]
- 177. Liu C., Zhang Q., Zhu S., Liu H., and Chen J., RSC Adv. 9, 28299–28311 (2019). 10.1039/C9RA05934B [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178. Koutsopoulos S., J. Biomed. Mater. Res. A 104, 1002–1016 (2016). 10.1002/jbm.a.35638 [DOI] [PubMed] [Google Scholar]
- 179. Qureshi D., Nayak S. K., Maji S., Anis A., Kim D., and Pal K., Eur. Pol. J. 120, 109220 (2019). 10.1016/j.eurpolymj.2019.109220 [DOI] [Google Scholar]
- 180. Yang K., Han Q., Chen B., Zheng Y., Zhang K. et al. , Int. J. Nanomedicine 13, 2217–2263 (2018). 10.2147/IJN.S154748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181. Gonzalez-Henriquez C. M., Sarabia-Vallejos M. A., and Rodriguez-Hernandez J., Materials 10, 232 (2017). 10.3390/ma10030232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182. Ng V. W., Chan J. M., Sardon H., Ono R. J., Garcia J. M. et al. , Adv. Drug Deliv. Rev. 78, 46–62 (2014). 10.1016/j.addr.2014.10.028 [DOI] [PubMed] [Google Scholar]
- 183. Veiga A. S. and Schneider J. P., Biopolymers 100, 637–644 (2013). 10.1002/bip.22412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184. Simonson A. W., Aronson M. R., and Medina S. H., Molecules 25, 2751 (2020). 10.3390/molecules25122751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185. Wahid F., Zhong C., Wang H. S., Hu X. H., and Chu L. Q., Polymers 9, 636 (2017). 10.3390/polym9120636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186. Eckhardt S., Brunetto P. S., Gagnon J., Priebe M., Giese B., and Fromm K. M., Chem. Rev. 113, 4708–4754 (2013). 10.1021/cr300288v [DOI] [PubMed] [Google Scholar]
- 187. Shi L., Ding P., Wang Y., Zhang Y., Ossipov D., and Hilborn J., Macromol. Rapid Commun. 40, e1800837 (2019). 10.1002/marc.201800837 [DOI] [PubMed] [Google Scholar]
- 188. Chernousova S. and Epple M., Angew. Chem. Int. Ed. Engl. 52, 1636–1653 (2013). 10.1002/anie.201205923 [DOI] [PubMed] [Google Scholar]
- 189. Chastre J., Clin. Microbiol. Infect. 14(Suppl 3), 3–14 (2008). 10.1111/j.1469-0691.2008.01958.x [DOI] [PubMed] [Google Scholar]
- 190. Slama T. G., Critical Care 12, S4 (2008). 10.1186/cc6820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191. Marston H. D., Dixon D. M., Knisely J. M., Palmore T. N., and Fauci A. S., JAMA 316, 1193–1204 (2016). 10.1001/jama.2016.11764 [DOI] [PubMed] [Google Scholar]
- 192. Wright J. B., Lam K., and Burrell R. E., Am. J. Infect. Control 26, 572–577 (1998). 10.1053/ic.1998.v26.a93527 [DOI] [PubMed] [Google Scholar]
- 193. Regiel-Futyra A., Dąbrowski J. M., Mazuryk O., Śpiewak K., Kyziol A. et al. , Coordination Chem. Rev. 351, 76–117 (2017). 10.1016/j.ccr.2017.05.005 [DOI] [Google Scholar]
- 194. Mohler J. S., Sim W., Blaskovich M. A. T., Cooper M. A., and Ziora Z. M., Biotechnol. Adv. 36, 1391–1411 (2018). 10.1016/j.biotechadv.2018.05.004 [DOI] [PubMed] [Google Scholar]
- 195. C. L. Fox, Jr. , Arch. Surg. 96, 184–188 (1968). 10.1001/archsurg.1968.01330200022004 [DOI] [PubMed] [Google Scholar]
- 196. Monafo W. W. and Freedman B., Surg. Clin. North Am. 67, 133–145 (1987). 10.1016/S0039-6109(16)44137-X [DOI] [PubMed] [Google Scholar]
- 197. Hidalgo E., Bartolomé R., Barroso C., Moreno A., and Domínguez C., Skin Pharmacol. Appl. Skin Physiol. 11, 140–151 (1998). 10.1159/000029820 [DOI] [PubMed] [Google Scholar]
- 198. Miura N. and Shinohara Y., Biochem. Biophys. Res. Commun. 390, 733–737 (2009). 10.1016/j.bbrc.2009.10.039 [DOI] [PubMed] [Google Scholar]
- 199. Poon V. K. and Burd A., Burns 30, 140–147 (2004). 10.1016/j.burns.2003.09.030 [DOI] [PubMed] [Google Scholar]
- 200. Cho Lee A. R., Leem H., Lee J., and Park K. C., Biomaterials 26, 4670–4676 (2005). 10.1016/j.biomaterials.2004.11.041 [DOI] [PubMed] [Google Scholar]
- 201. Fraser J. F., Cuttle L., Kempf M., and Kimble R. M., ANZ J. Surg. 74, 139–142 (2004). 10.1046/j.1445-2197.2004.02916.x [DOI] [PubMed] [Google Scholar]
- 202. Atiyeh B. S., Costagliola M., Hayek S. N., and Dibo S. A., Burns 33, 139–148 (2007). 10.1016/j.burns.2006.06.010 [DOI] [PubMed] [Google Scholar]
- 203. D'Souza A., Yoon J. H., Beaman H., Gosavi P., Lengyel-Zhand Z. et al. , ACS Appl. Mater. Interfaces 12, 17091–17099 (2020). 10.1021/acsami.0c01154 [DOI] [PubMed] [Google Scholar]
- 204. Guo Y., Wang S., Du H., Chen X., and Fei H., Biomacromolecules 20, 558–565 (2019). 10.1021/acs.biomac.8b01480 [DOI] [PubMed] [Google Scholar]
- 205. Hu Y., Xu W., Li G., Xu L., Song A., and Hao J., Langmuir 31, 8599–8605 (2015). 10.1021/acs.langmuir.5b02036 [DOI] [PubMed] [Google Scholar]
- 206. Schneider L. A., Korber A., Grabbe S., and Dissemond J., Arch. Dermatol. Res. 298, 413–420 (2007). 10.1007/s00403-006-0713-x [DOI] [PubMed] [Google Scholar]
- 207. Salick D. A., Kretsinger J. K., Pochan D. J., and Schneider J. P., J. Am. Chem. Soc. 129, 14793–14799 (2007). 10.1021/ja076300z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208. Salick D. A., Pochan D. J., and Schneider J. P., Adv. Mater. 21, 4120–4123 (2009). 10.1002/adma.200900189 [DOI] [Google Scholar]
- 209. Veiga A. S., Sinthuvanich C., Gaspar D., Franquelim H. G., Castanho M. A. R. B., and Schneider J. P., Biomaterials 33, 8907–8916 (2012). 10.1016/j.biomaterials.2012.08.046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210. Tripathi J. K., Pal S., Awasthi B., Kumar A., Tandon A. et al. , Biomaterials 56, 92–103 (2015). 10.1016/j.biomaterials.2015.03.046 [DOI] [PubMed] [Google Scholar]
- 211. Guvendiren M., Lu H. D., and Burdick J. A., Soft Matter 8, 260–272 (2012). 10.1039/C1SM06513K [DOI] [Google Scholar]
- 212. Yan C. and Pochan D. J., Chem. Soc. Rev. 39, 3528–3540 (2010). 10.1039/b919449p [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213. Krogsgaard M., Nue V., and Birkedal H., Chem. Eur. J. 22, 844–857 (2016). 10.1002/chem.201503380 [DOI] [PubMed] [Google Scholar]
- 214. Li C. H. and Zuo J. L., Adv. Mater. 32, e1903762 (2020). 10.1002/adma.201903762 [DOI] [PubMed] [Google Scholar]
- 215. Holten-Andersen N., Jaishankar A., Harrington M., Fullenkamp D. E., DiMarco G. et al. , J. Mater. Chem. B 2, 2467–2472 (2014). 10.1039/C3TB21374A [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216. Shao T., Falcone N., and Kraatz H. B., ACS Omega 5, 1312–1317 (2020). 10.1021/acsomega.9b03939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217. Fages F., Angew. Chem. Int. Ed. Engl. 45, 1680–1682 (2006). 10.1002/anie.200503704 [DOI] [PubMed] [Google Scholar]
- 218. Xu C., Cai Y., Ren C., Gao J., and Hao J., Sci. Rep. 5, 7753 (2015). 10.1038/srep07753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219. Falcone N. and Kraatz H. B., Chem. Eur. J. 24, 14316–14328 (2018). 10.1002/chem.201801247 [DOI] [PubMed] [Google Scholar]
- 220. Mart R. J., Osborne R. D., Stevens M. M., and Ulijn R. V., Soft Matter 2, 822–835 (2006). 10.1039/b607706d [DOI] [PubMed] [Google Scholar]
- 221. Koetting M. C., Peters J. T., Steichen S. D., and Peppas N. A., Mater. Sci. Eng. R. Rep. 93, 1–49 (2015). 10.1016/j.mser.2015.04.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222. Li J. and Mooney D. J., Nat. Rev. Mater. 1, 16071 (2016). 10.1038/natrevmats.2016.71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223. Yan X., Wang F., Zheng B., and Huang F., Chem. Soc. Rev. 41, 6042–6065 (2012). 10.1039/c2cs35091b [DOI] [PubMed] [Google Scholar]
- 224. Saunders L. and Ma P. X., Macromol. Biosci. 19, e1800313 (2019). 10.1002/mabi.201800313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225. Dong R., Zhou Y., Huang X., Zhu X., Lu Y., and Shen J., Adv. Mater. 27, 498–526 (2015). 10.1002/adma.201402975 [DOI] [PubMed] [Google Scholar]
- 226. Chen G. and Jiang M., Chem. Soc. Rev. 40, 2254–2266 (2011). 10.1039/c0cs00153h [DOI] [PubMed] [Google Scholar]
- 227. Rodríguez-Rodríguez R., Espinosa-Andrews H., Velasquillo-Martínez C., and García-Carvajal Z. Y., Int. J. Polym. Mater. Polym. Biomater. 69, 1–20 (2020). 10.1080/00914037.2019.1581780 [DOI] [Google Scholar]
- 228. Hoque J., Sangaj N., and Varghese S., Macromol. Biosci. 19, e1800259 (2019). 10.1002/mabi.201800259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229. Du X., Zhou J., Shi J., and Xu B., Chem. Rev. 115, 13165–13307 (2015). 10.1021/acs.chemrev.5b00299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230. Lu Y., Aimetti A. A., Langer R., and Gu Z., Nat. Rev. Mater. 2, 16075 (2017). 10.1038/natrevmats.2016.75 [DOI] [Google Scholar]
- 231. Wong S., Shim M. S., and Kwon Y. J., J. Mater. Chem. B 2, 595–615 (2014). 10.1039/C3TB21344G [DOI] [PubMed] [Google Scholar]
- 232. Lamm M. S., Rajagopal K., Schneider J. P., and Pochan D. J., J. Am. Chem. Soc. 127, 16692–16700 (2005). 10.1021/ja054721f [DOI] [PubMed] [Google Scholar]
- 233. Lin B. F., Megley K. A., Viswanathan N., Krogstad D. V., Drews L. B. et al. , J. Mater. Chem. 22, 19447–19454 (2012). 10.1039/c2jm31745a [DOI] [Google Scholar]
- 234. Bainbridge P., J. Wound Care 22 410–412 (2013). 10.12968/jowc.2013.22.8.407 [DOI] [PubMed] [Google Scholar]
- 235. Fletcher N. L., Lockett C. V., and Dexter A. F., Soft Matter 7, 10210–10218 (2011). 10.1039/c1sm06261a [DOI] [Google Scholar]
- 236. Dexter A. F., Fletcher N. L., Creasey R. G., Filardo F., Boehm M. W., and Jack K. S., RSC Adv. 7, 27260–27271 (2017). 10.1039/C7RA02811C [DOI] [Google Scholar]
- 237. Liu Y., Yang Y., Wang C., and Zhao X., Nanoscale 5, 6413–6421 (2013). 10.1039/c3nr00225j [DOI] [PubMed] [Google Scholar]
- 238. Wojciechowski J. P., Martin A. D., and Thordarson P., J. Am. Chem. Soc. 140, 2869–2874 (2018). 10.1021/jacs.7b12198 [DOI] [PubMed] [Google Scholar]
- 239. Bowerman C. J. and Nilsson B. L., J. Am. Chem. Soc. 132, 9526–9527 (2010). 10.1021/ja1025535 [DOI] [PubMed] [Google Scholar]
- 240. Aulisa L., Dong H., and Hartgerink J. D., Biomacromolecules 10, 2694–2698 (2009). 10.1021/bm900634x [DOI] [PubMed] [Google Scholar]
- 241. Ren C., Song Z., Zheng W., Chen X., Wang L. et al. , Chem. Commun. 47, 1619–1621 (2011). 10.1039/C0CC04135A [DOI] [PubMed] [Google Scholar]
- 242. Lv L., Liu H., Chen X., and Yang Z., Colloids Surfaces B Biointerfaces 108, 352–357 (2013). 10.1016/j.colsurfb.2013.03.013 [DOI] [PubMed] [Google Scholar]
- 243. Sun Z., Li Z., He Y., Shen R., Deng L. et al. , J. Am. Chem. Soc. 135, 13379–13386 (2013). 10.1021/ja403345p [DOI] [PubMed] [Google Scholar]
- 244. Falcone N., Basak S., Dong B., Syed J., Ferranco A. et al. , Chempluschem 82, 1282–1289 (2017). 10.1002/cplu.201700407 [DOI] [PubMed] [Google Scholar]
- 245. Adhikari B. and Kraatz H. B., Chem. Commun. 50, 5551–5553 (2014). 10.1039/C3CC49268K [DOI] [PubMed] [Google Scholar]
- 246. Zhang Y., Zhang B., Kuang Y., Gao Y., Shi J. et al. , J. Am. Chem. Soc. 135, 5008–5011 (2013). 10.1021/ja402490j [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247. Zhang Y., Gu H., Yang Z., and Xu B., J. Am. Chem. Soc. 125, 13680–13681 (2003). 10.1021/ja036817k [DOI] [PubMed] [Google Scholar]
- 248. Banwell E. F., Abelardo E. S., Adams D. J., Birchall M. A., Corrigan A. et al. , Nat. Mater. 8, 596–600 (2009). 10.1038/nmat2479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 249. Haines L. A., Rajagopal K., Ozbas B., Salick D. A., Pochan D. J., and Schneider J. P., J. Am. Chem. Soc. 127, 17025–17029 (2005). 10.1021/ja054719o [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250. Ding Y., Li Y., Qin M., Cao Y., and Wang W., Langmuir 29, 13299–13306 (2013). 10.1021/la4029639 [DOI] [PubMed] [Google Scholar]
- 251. Collier J. H., Hu B. H., Ruberti J. W., Zhang J., Shum P. et al. , J. Am. Chem. Soc 123, 9463–9464 (2001). 10.1021/ja011535a [DOI] [PubMed] [Google Scholar]
- 252. Yang Z., Liang G., and Xu B., Acc. Chem. Res. 41, 315–326 (2008). 10.1021/ar7001914 [DOI] [PubMed] [Google Scholar]
- 253. Hu J., Zhang G., and Liu S., Chem. Soc. Rev. 41, 5933–5949 (2012). 10.1039/c2cs35103j [DOI] [PubMed] [Google Scholar]
- 254. Yang Z., Gu H., Fu D., Gao P., Lam J. K., and Xu B., Adv. Mater. 16, 1440–1444 (2004). 10.1002/adma.200400340 [DOI] [Google Scholar]
- 255. Li X., Gao Y., Kuang Y., and Xu B., Chem. Commun. 46, 5364–5366 (2010). 10.1039/c0cc00163e [DOI] [PubMed] [Google Scholar]
- 256. Toledano S., Williams R. J., Jayawarna V., and Ulijn R. V., J. Am. Chem. Soc. 128, 1070–1071 (2006). 10.1021/ja056549l [DOI] [PubMed] [Google Scholar]
- 257. Guilbaud J. B., Vey E., Boothroyd S., Smith A. M., Ulijn R. V. et al. , Langmuir 26, 11297–11303 (2010). 10.1021/la100623y [DOI] [PubMed] [Google Scholar]
- 258. Lutolf M. P., Raeber G. P., Zisch A. H., Tirelli N., and Hubbell J. A., Adv. Mater. 15, 888–892 (2003). 10.1002/adma.200304621 [DOI] [Google Scholar]
- 259. Jun H. W., Paramonov S. E., Dong H., Forraz N., McGuckin C., and Hartgerink J. D., J. Biomater. Sci. Polym. Ed. 19, 665–676 (2008). 10.1163/156856208784089625 [DOI] [PubMed] [Google Scholar]
- 260. Patrick A. G. and Ulijn R. V., Macromol. Biosci. 10, 1184–1193 (2010). 10.1002/mabi.200900457 [DOI] [PubMed] [Google Scholar]
- 261. Bremmer S. C., Chen J., McNeil A. J., and Soellner M. B., Chem. Commun. 48, 5482–5484 (2012). 10.1039/c2cc31537h [DOI] [PMC free article] [PubMed] [Google Scholar]
- 262. Galler K. M., Aulisa L., Regan K. R., D'Souza R. N., and Hartgerink J. D., J. Am. Chem. Soc. 132, 3217–3223 (2010). 10.1021/ja910481t [DOI] [PMC free article] [PubMed] [Google Scholar]
- 263. Junker J. P., Kamel R. A., Caterson E. J., and Eriksson E., Adv. Wound Care 2, 348–356 (2013). 10.1089/wound.2012.0412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 264. Field F. K. and Kerstein M. D., Am. J. Surg 167, 2S–6S (1994). 10.1016/0002-9610(94)90002-7 [DOI] [PubMed] [Google Scholar]
- 265. Hamedi H., Moradi S., Hudson S. M., and Tonelli A. E., Carbohydr. Polym. 199, 445–460 (2018). 10.1016/j.carbpol.2018.06.114 [DOI] [PubMed] [Google Scholar]
- 266. Li J., Xing R., Bai S., and Yan X., Soft Matter 15, 1704–1715 (2019). 10.1039/C8SM02573H [DOI] [PubMed] [Google Scholar]
- 267. Francesko A., Petkova P., and Tzanov T., Curr. Med. Chem. 25, 5782–5797 (2019). 10.2174/0929867324666170920161246 [DOI] [PubMed] [Google Scholar]
- 268. Op 't Veld R. C., Walboomers X. F., Jansen J. A., and Wagener F. A. D. T., Tissue Eng. Part B: Reviews 20, 230–248 (2020). 10.1089/ten.teb.2019.0281 [DOI] [PubMed] [Google Scholar]
- 269. Bellis S. L., Biomaterials 32, 4205–4210 (2011). 10.1016/j.biomaterials.2011.02.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 270. Pierschbacher M. D. and Ruoslahti E., Nature 309, 30–33 (1984). 10.1038/309030a0 [DOI] [PubMed] [Google Scholar]
- 271. Ruoslahti E., Annu. Rev. Cell Dev. Biol. 12, 697–715 (1996). 10.1146/annurev.cellbio.12.1.697 [DOI] [PubMed] [Google Scholar]
- 272. Hern D. L. and Hubbell J. A., J. Biomed. Mater. Res. 39, 266–276 (1998). [DOI] [PubMed] [Google Scholar]
- 273. Zhou M., Ulijn R. V., and Gough J. E., J. Tissue Eng. 5, 204173141453159 (2014). 10.1177/2041731414531593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 274. Hersel U., Dahmen C., and Kessler H., Biomaterials 24, 4385–4415 (2003). 10.1016/S0142-9612(03)00343-0 [DOI] [PubMed] [Google Scholar]
- 275. Zhou M., Smith A. M., Das A. K., Hodson N. W., Collins R. F. et al. , Biomaterials 30, 2523–2530 (2009). 10.1016/j.biomaterials.2009.01.010 [DOI] [PubMed] [Google Scholar]
- 276. Storrie H., Guler M. O., Abu-Amara S. N., Volberg T., Rao M. et al. , Biomaterials 28, 4608–4618 (2007). 10.1016/j.biomaterials.2007.06.026 [DOI] [PubMed] [Google Scholar]
- 277. Guler M. O., Hsu L., Soukasene S., Harrington D. A., Hulvat J. F., and Stupp S. I., Biomacromolecules 7, 1855–1863 (2006). 10.1021/bm060161g [DOI] [PMC free article] [PubMed] [Google Scholar]
- 278. Li Y., Rodrigues J., and Tomás H., Chem. Soc. Rev. 41, 2193–2221 (2012). 10.1039/C1CS15203C [DOI] [PubMed] [Google Scholar]
- 279. Bradshaw M., Ho D., Fear M. W., Gelain F., Wood F. M., and Iyer K. S., Sci. Rep. 4, 6903 (2015). 10.1038/srep06903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 280. Dimatteo R., Darling N. J., and Segura T., Adv. Drug Deliv. Rev. 127, 167–184 (2018). 10.1016/j.addr.2018.03.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 281. Geisler I. M. and Schneider J. P., Adv. Funct. Mater. 22, 529–537 (2012). 10.1002/adfm.201102330 [DOI] [Google Scholar]
- 282. Wang Y., He X., Bruggeman K. F., Gayen B., Tricoli A. et al. , Adv. Funct. Mater. 30, 1900390 (2020). 10.1002/adfm.201900390 [DOI] [Google Scholar]
- 283. Kretsinger J. K., Haines L. A., Ozbas B., Pochan D. J., and Schneider J. P., Biomaterials 26, 5177–5186 (2005). 10.1016/j.biomaterials.2005.01.029 [DOI] [PubMed] [Google Scholar]
- 284. Patterson J. and Hubbell J. A., Biomaterials 31, 7836–7845 (2010). 10.1016/j.biomaterials.2010.06.061 [DOI] [PubMed] [Google Scholar]
- 285. Brinkmann J., Wasserberg D., and Jonkheijm P., Eur. Polym. J. 83, 380–389 (2016). 10.1016/j.eurpolymj.2016.03.029 [DOI] [Google Scholar]
- 286. Enciso P., Woerner M., and Cerdá M. F., MRS Adv. 3, 3557–3562 (2018). 10.1557/adv.2018.533 [DOI] [Google Scholar]
- 287. Janna Olmos J. D., Becquet P., Gront D., Sar J., Dąbrowski A. et al. , RSC Adv. 7, 47854–47866 (2017). 10.1039/C7RA10895H [DOI] [Google Scholar]
- 288. Zhang Q., Lu Y., Li S., Wu J., and Liu Q., in Peptide Applications in Biomedicine, Biotechnology and Bioengineering, edited by Koutsopoulos S. ( Woodhead, 2018), pp. 565–601. [Google Scholar]
- 289. Geyao L., Yang D., Wanglin C., and Chengyong W., Procedia CIRP 89, 250–262 (2020). 10.1016/j.procir.2020.05.149 [DOI] [Google Scholar]
- 290. Greenhalgh R., Dempsey-Hibbert N. C., and Whitehead K. A., Int. Biodeterioration Biodegradation 136, 1–14 (2019). 10.1016/j.ibiod.2018.10.005 [DOI] [Google Scholar]
- 291. Goderecci S. S., Kaiser E., Yanakas M., Norris Z., Scaturro J. et al. , Molecules 22, 1487 (2017). 10.3390/molecules22091487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 292. Kannan M. B., Moore C., Saptarshi S., Somasundaram S., Rahuma M., and Lopata A. L., Sci. Rep. 7, 15605 (2017). 10.1038/s41598-017-15873-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 293. Fang Y., Xing C., Zhan S., Zhao M., Li M., and Liu H., J. Mater. Chem. B 7, 1933–1944 (2019). 10.1039/C8TB03331E [DOI] [PubMed] [Google Scholar]
- 294. Wang L., Erişen D. E., Yang K., Zhang B., Guan H., and Chen S., J. Biomed. Mater. Res. B Appl. Biomater. 108, 2868–2877 (2020). 10.1002/jbm.b.34618 [DOI] [PubMed] [Google Scholar]
- 295. Huang Q., Ouyang Z., Tan Y., Wu H., and Liu Y., Acta Biomater. 100, 415–426 (2019). 10.1016/j.actbio.2019.09.030 [DOI] [PubMed] [Google Scholar]
- 296. Stevens K. N. J., Croes S., Boersma R. S., Stobberingh E. E., van der Marel C. et al. , Biomaterials 32, 1264–1269 (2011). 10.1016/j.biomaterials.2010.10.042 [DOI] [PubMed] [Google Scholar]
- 297. Hodgkinson V. and Petris M. J., J. Biol. Chem. 287, 13549–13555 (2012). 10.1074/jbc.R111.316406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 298. Ruggieri M. R., Hanno P. M., and Levin R. M., J. Urol. 138, 423–426 (1987). 10.1016/S0022-5347(17)43177-6 [DOI] [PubMed] [Google Scholar]
- 299. Ostuni E., Chapman R. G., Liang M. N., Meluleni G., Pier G. et al. , Langmuir 17, 6336–6343 (2001). 10.1021/la010552a [DOI] [Google Scholar]
- 300. Carlson R. P., Taffs R., Davison W. M., and Stewart P. S., J. Biomaterials Sci., Polym. Ed. 19, 1035–1046 (2008). 10.1163/156856208784909372 [DOI] [PubMed] [Google Scholar]
- 301. Velraeds M. M. C., Van Der Mei H. C., Reid G., and Busscher H. J., Urology 49, 790–794 (1997). 10.1016/S0090-4295(97)00065-4 [DOI] [PubMed] [Google Scholar]
- 302. Luo J., Chen Z., and Sun Y., J. Biomed. Mater. Res. Part A 77, 823–831 (2006). 10.1002/jbm.a.30689 [DOI] [PubMed] [Google Scholar]
- 303. Xu J., Zhao H., Xie Z., Ruppel S., Zhou X. et al. , Adv. Healthcare Mater. 8, 1900232 (2019). 10.1002/adhm.201900232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 304. Zamuner A., Brun P., Scorzeto M., Sica G., Castagliuolo I., and Dettin M., Bioactive Mater. 2, 121–130 (2017). 10.1016/j.bioactmat.2017.05.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 305. Politi J., Zappavigna S., Rea I., Grieco P., Caliò A. et al. , J. Sens. 2017, 1. 10.1155/2017/6792396 [DOI] [Google Scholar]
- 306. Matmor M., Lengyel G. A., Horne W. S., and Ashkenasy N., Phys. Chem. Chem. Phys. 19, 5709–5714 (2017). 10.1039/C6CP07198H [DOI] [PubMed] [Google Scholar]
- 307. Place E. S., Evans N. D., and Stevens M. M., Nat. Mater. 8, 457–470 (2009). 10.1038/nmat2441 [DOI] [PubMed] [Google Scholar]
- 308. Marchesan S. and Prato M., Chem. Commun. 51, 4347–4359 (2015). 10.1039/C4CC09173F [DOI] [PubMed] [Google Scholar]
- 309. Xue Y., Li X., Li H., and Zhang W., Nat. Commun. 5, 4348 (2014). 10.1038/ncomms5348 [DOI] [PubMed] [Google Scholar]
- 310. Budhathoki-Uprety J., Harvey J. D., Isaac E., Williams R. M., Galassi T. V. et al. , J. Mater. Chem. B 5, 6637–6644 (2017). 10.1039/C7TB00695K [DOI] [PubMed] [Google Scholar]
- 311. Jung Y. C., Muramatsu H., Fujisawa K., Kim J. H., Hayashi T. et al. , Small 7, 3292–3297 (2011). 10.1002/smll.201100668 [DOI] [PubMed] [Google Scholar]
- 312. Yaron P. N., Holt B. D., Short P. A., Lösche M., Islam M. F., and Dahl K. N., J. Nanobiotechnol. 9, 45 (2011). 10.1186/1477-3155-9-45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 313. Ebrahim-Habibi M.-B., Ghobeh M., Mahyari F. A., Rafii-Tabar H., and Sasanpour P., Sci. Rep. 9, 1273 (2019). 10.1038/s41598-018-37311-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 314. Kurppa K., Jiang H., Szilvay G. R., Nasibulin A. G., Kauppinen E. I., and Linder M. B., Angew. Chem. Int. Ed. Engl. 46, 6446–6449 (2007). 10.1002/anie.200702298 [DOI] [PubMed] [Google Scholar]
- 315. Corrales-Ureña Y. R., Souza-Schiaber Z., Lisboa-Filho P. N., Marquenet F., Michael Noeske P.-L. et al. , RSC Adv. 10, 376–386 (2020). 10.1039/C9RA07380A [DOI] [PMC free article] [PubMed] [Google Scholar]
- 316. Nilebäck L., Hedin J., Widhe M., Floderus L. S., Krona A. et al. , Biomacromolecules 18, 846–854 (2017). 10.1021/acs.biomac.6b01721 [DOI] [PubMed] [Google Scholar]
- 317. Dart A., Bhave M., and Kingshott P., Macromol. Biosci. 19, 1800488 (2019). 10.1002/mabi.201800488 [DOI] [PubMed] [Google Scholar]
- 318. Chen L., Shao L., Wang F., Huang Y., and Gao F., RSC Adv. 9, 10494–10507 (2019). 10.1039/C8RA08788A [DOI] [PMC free article] [PubMed] [Google Scholar]
- 319. Li X., Contreras-Garcia A., LoVetri K., Yakandawala N., Wertheimer M. R. et al. , J. Biomed. Mater. Res. Part A 103, 3736–3746 (2015). 10.1002/jbm.a.35511 [DOI] [PubMed] [Google Scholar]
- 320. Corrales Ureña Y. R., Wittig L., Vieira Nascimento M., Faccioni J. L., Lisboa Filho P. N., and Rischka K., Appl. Adhesion Sci. 3, 7 (2015). 10.1186/s40563-015-0032-6 [DOI] [Google Scholar]
- 321. Uzarski J. R., Tannous A., Morris J. R., and Mello C. M., Colloids Surf. B: Biointerfaces 67, 157–165 (2008). 10.1016/j.colsurfb.2008.07.011 [DOI] [PubMed] [Google Scholar]
- 322. Yoshinari M., Kato T., Matsuzaka K., Hayakawa T., and Shiba K., Biofouling 26, 103–110 (2010). 10.1080/08927010903216572 [DOI] [PubMed] [Google Scholar]
- 323. Gao J., Na H., Zhong R., Yuan M., Guo J. et al. , Colloids Surf. B: Biointerfaces 186, 110704 (2020). 10.1016/j.colsurfb.2019.110704 [DOI] [PubMed] [Google Scholar]
- 324. Gakiya-Teruya M., Palomino-Marcelo L., Pierce S., Angeles-Boza A. M., Krishna V., and Rodriguez-Reyes J. C. F., J. Nanopart. Res. 22, 90 (2020). 10.1007/s11051-020-04799-6 [DOI] [Google Scholar]
- 325. Mohanty S., Jena P., Mehta R., Pati R., Banerjee B. et al. , Antimicrob. Agents Chemother. 57, 3688–3698 (2013). 10.1128/AAC.02475-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 326. Ruden S., Hilpert K., Berditsch M., Wadhwani P., and Ulrich A. S., Antimicrob. Agents Chemother. 53, 3538–3540 (2009). 10.1128/AAC.01106-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 327. Heunis T. D., Botes M., and Dicks L. M., Probiotics Antimicrob. Prot. 2, 46–51 (2010). 10.1007/s12602-009-9024-9 [DOI] [PubMed] [Google Scholar]
- 328. Ahire J. J. and Dicks L. M. T., Probiotics Antimicrob. Prot. 7, 52–59 (2015). 10.1007/s12602-014-9171-5 [DOI] [PubMed] [Google Scholar]
- 329. Han D., Sherman S., Filocamo S., and Steckl A. J., Acta Biomater. 53, 242–249 (2017). 10.1016/j.actbio.2017.02.029 [DOI] [PubMed] [Google Scholar]
- 330. Shukla A., Fleming K. E., Chuang H. F., Chau T. M., Loose C. R. et al. , Biomaterials 31, 2348–2357 (2010). 10.1016/j.biomaterials.2009.11.082 [DOI] [PubMed] [Google Scholar]
- 331. Etienne O., Gasnier C., Taddei C., Voegel J. C., Aunis D. et al. , Biomaterials 26, 6704–6712 (2005). 10.1016/j.biomaterials.2005.04.068 [DOI] [PubMed] [Google Scholar]
- 332. Gevrek T. N., Yu K., Kizhakkedathu J. N., and Sanyal A., ACS Appl. Polym. Mater. 1, 1308–1316 (2019). 10.1021/acsapm.9b00117 [DOI] [Google Scholar]
- 333. Dutta D., Kumar N., and Willcox M. D. P., Biofouling 32, 429–438 (2016). 10.1080/08927014.2015.1129533 [DOI] [PubMed] [Google Scholar]
- 334. Zhou L., Lai Y., Huang W., Huang S., Xu Z. et al. , Colloids Surf. B: Biointerfaces 128, 552–560 (2015). 10.1016/j.colsurfb.2015.03.008 [DOI] [PubMed] [Google Scholar]
- 335. Yu K., Lo J. C. Y., Yan M., Yang X., Brooks D. E. et al. , Biomaterials 116, 69–81 (2017). 10.1016/j.biomaterials.2016.11.047 [DOI] [PubMed] [Google Scholar]
- 336. Zhang X.-Y., Zhao Y.-Q., Zhang Y., Wang A., Ding X. et al. , Biomacromolecules 20, 4171–4179 (2019). 10.1021/acs.biomac.9b01060 [DOI] [PubMed] [Google Scholar]
- 337. Ortiz-Gómez V., Rodríguez-Ramos V. D., Maldonado-Hernández R., González-Feliciano J. A., and Nicolau E., ACS Appl. Mater. Interfaces 12, 46991–47001 (2020). 10.1021/acsami.0c13492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 338. Yang X., Liu W., Xi G., Wang M., Liang B. et al. , Carbohydrate Polymers 222, 115012 (2019). 10.1016/j.carbpol.2019.115012 [DOI] [PubMed] [Google Scholar]
- 339. Appendini P. and Hotchkiss J. H., J. Appl. Polym. Sci. 81, 609–616 (2001). 10.1002/app.1476 [DOI] [Google Scholar]
- 340. Li W., Li Y., Sun P., Zhang N., Zhao Y. et al. , RSC Adv. 10, 38746–38754 (2020). 10.1039/D0RA05640E [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data sharing is not applicable to this article as no new data were created or analyzed in this article.