Abstract
A microbiome consists of viruses, bacteria, archaea, fungi, and other microeukaryotes. It influences host immune systems and contributes to the development of various diseases, such as obesity, diabetes, asthma, and skin diseases, including atopic dermatitis and seborrheic dermatitis. The skin is the largest organ in the human body and has various microorganisms on its surface. Several studies on skin microbiomes have illustrated the effects of their composition, metabolites, and interactions with host cells on diseases. However, most studies have focused on the bacterial microbiome rather than the fungal microbiome, namely, mycobiome, although emerging evidence indicates that fungi also play a critical role in skin microbiomes through interactions with the host cells. I briefly summarize the current progress in the analysis of mycobiomes on human skin. I focused on alteration of the skin mycobiome caused by atopic and seborrheic dermatitis, with an emphasis on the Malassezia genus, which are the most dominant fungi residing here.
INTRODUCTION
Most multicellular eukaryotes are closely associated with microorganisms that they host and are together considered metaorganisms (Berg et al., 2020). A microbial community in the human body is termed microbiome and is composed of viruses, bacteria, archaea, fungi, and other microeukaryotes. Microbiomes in living organisms are considered an inseparable part of the hosts and play functional roles that affect their health (Berg et al., 2020). Disruption of the diversity of a microbiome in the human body due to intrinsic or extrinsic factors is called dysbiosis. It influences host immune systems and causes various diseases, such as obesity, diabetes, and asthma (Hooks and O'Malley, 2017; Bello et al., 2018, Upadhyay et al., 2022; and Zubeldia-Varela et al., 2022). Therefore, the microbiome compositions in different organs, such as the respiratory tract, uterus, vagina, and digestive tract, have recently been analyzed extensively.
The skin is the largest organ in the human body, and various microorganisms are present on its surface. However, the composition of microbiomes differs in different skin regions because of variations in anatomical and chemical factors, such as temperature, humidity, and sebum content (Oh et al., 2014; Cundell, 2018). Several studies regarding skin microbiomes have highlighted the effect of their composition, metabolites, and interactions with host cells on human skin diseases, such as atopic dermatitis (AD) and seborrheic dermatitis (SD). However, most studies were concentrated on bacterial communities on the skin although other microorganisms, including fungi, play a critical role in the interactions between the skin microbiome and host cells. A metagenome analysis revealed that fungi, mainly Malassezia restricta and M. globosa, constituted 3.9%–5.0% of the microbiomes in most skin sites, while the external auditory canal, retroauricular crease, and glabella, had 16.86%–5.1%, 7.56%–4.2%, and 7.16%–4.0% fungi in the skin microbial communities, respectively (Oh et al., 2014). Moreover, dysbiosis of the fungal microbiome, termed mycobiome hereafter, can cause many skin diseases, including AD and SD (Paulino, 2017; Schoch et al., 2019; and Szczepanska et al., 2022).
The importance of skin mycobiome in skin homeostasis and disease development is just emerging; therefore, only a limited number of studies regarding the structure of fungal communities on the human skin and their influence on diseases or the impact diseases have on them is available. We briefly summarize the current progress in the analysis of mycobiome of the human skin, with a focus on Malassezia fungi and their role in the alteration of skin mycobiome due to AD and SD when compared with that of healthy controls.
MYCOBIOME IN ATOPIC DERMATITIS
AD is a chronic and often relapsing inflammatory skin disease characterized by eczematous lesions, pruritus, and skin dryness. AD is common worldwide with a prevalence of 10%–20% and 3%–5% in children and the general population, respectively (Williams et al., 2008; Ring et al., 2019; and Fasseeh et al., 2022). The pathophysiology of the disease is complicating and caused by multifactorial factors, including an abnormality in the epidermal barrier function that could be genetic or acquired and an immunologic disorder due to abnormal immune responses to antigens that often occur with increased T helper 2 (Th2) cells, which express interleukin-4 (IL-4)/IL-13, Th22, and Th17 cytokine activations and increased serum immunoglobulin E (IgE) levels (Leung and Guttman-Yassky, 2014). AD is often characterized by disruption of the epidermal barrier function, which causes decreased hydration, altered lipid composition, abnormal lamellar organization, increased skin pH, and decreased diversity of skin microbiota accompanied by an increased abundance of Staphylococcus aureus (Weidinger and Novak, 2016). The null mutation in the filaggrin (FLG) gene encoding filaggrin (FLG) is reported to be significantly associated with epidermal barrier impairment in patients with AD. However, how filaggrin deficiency contributes to AD is still not fully understood (Moosbrugger-Martinz et al., 2022). FLG deficiency is associated with altered diversity of skin microbiota, i.e., reduced abundance of proteolytic gram-positive anaerobic cocci (GPAC), such as Finegoldia, Anaerococcus, and Peptoniphilus genera, which utilize histidine-rich FLG as a nutrient (Zeeuwen et al., 2017). Another study revealed reduced abundance of Proteobacteria, such as Acinetobacter, Enhydrobacter, and Microvirgula, along with an increased abundance of Cutibacterium and Firmicutes, Staphylococcus in particular. Moreover, skin microbiomes in FLG-deficient individuals demonstrated greater similarity to those of patients with AD than those of individuals with normal FLG did (Baurecht et al., 2018). A previous study suggested that AD is strongly associated with dysbiosis of the skin microbiome, which is evident in the reduced richness and diversity of bacterial communities and increased abundance of S. aureus (Alam et al., 2022). S. aureus produces virulence factors, such as phenol-soluble modulins and proteases, which disrupt the host's skin barrier (Hirasawa et al., 2010; Nakagawa et al., 2017). Moreover, increased abundance of S. aureus is often related to reduction in skin commensal Staphylococcus species, such as S. epidermidis and S. hominis, which selectively produce an antimicrobial peptide against S. aureus (Nakatsuji et al., 2017).
Most research on skin microbiomes to study impaired skin barrier functions due to microbes is concentrated on bacteria. However, recent studies have demonstrated the critical role played by fungi in skin microbiomes. A shotgun metagenome analysis showed that fungal presence in healthy human skin was lower (0.36%–16.86%) than the bacterial one, with a predominance of Malassezia species, especially M. restricta and M. globosa (Oh et al., 2014). Malassezia is a lipophilic yeast; hence, its chief residing site is skin affected by SD. A total of 18 Malassezia species have been reported, and the genomes of most of them have been sequenced. Their analysis revealed that the lipid dependency is caused by the absence of the gene encoding fatty acid synthase (Xu et al., 2007; Park et al., 2017; and Cho et al., 2022).
Several studies have suggested a strong association between Malassezia and AD. For example, sensitization rates against Malassezia were higher in patients with AD; moreover, they exhibited Malassezia-specific serum IgE antibodies (Kieffer et al., 1990; Ring et al., 1992). Furthermore, various immunogenic proteins secreted by Malassezia induced specific IgE responses (Glatz et al., 2015).
Although accumulated evidence suggests a critical role of fungi in AD, studies to analyze mycobiomes using a culture-independent method in the lesional and nonlesional skin sites of patients with AD having different geological and ethical backgrounds are limited. An early study using polymerase chain reaction (PCR) with specific primers for each Malassezia species in a Japanese cohort with AD showed that M. globosa and M. restricta were detected more frequently than other Malassezia species, such as M. furfur and M. sympodialis, in the lesional skin sites. Moreover, AD skin lesions demonstrated a higher diversity of Malassezia than healthy skin did (Sugita et al., 2001). Similarly, while patients with mild and moderate head and neck AD exhibited a predominance of M. restricta over M. globosa, those with severe head and neck AD demonstrated an equal abundance of these species (Kaga et al., 2011).
A whole metagenome analysis of the antecubital fossae of individuals in Singapore having a history of AD, designated as AD-susceptible individuals, was performed between flares, and the results were compared with those obtained from the analysis of healthy controls. Overall, fungal abundance was higher in skin affected by AD than in healthy controls, and M. globosa and M. restricta were predominant in all the investigated skin sites. Moreover, M. dermatis and M. sympodialis were predominant in skin affected by AD, while M. globosa was relatively more abundant in healthy controls. Thus, fungal diversity in skin mycobiome varies between AD-susceptible individuals and healthy controls (Chng et al., 2016).
Amplicon sequencing analysis of multiple skin lesions in a cohort with AD in Denmark was performed using the 18S rRNA gene. M. restricta and M. globosa were most prevalent in all the tested skin sites, although a decreased abundance of M. restricta in skin affected by AD was observed. Moreover, an increased abundance of the microscopic mite Demodex folliculorum and the fungus Geotrichum candidum was found in skin affected by AD than in healthy skin (Edslev et al., 2021). Similarly, another study in which amplicon sequencing analysis was performed using internal transcribed spacer region (ITS) primers in a Korean cohort with AD showed that the Malassezia species were the most detected fungi among fungal communities in both AD and healthy skin. M. restricta and M. globosa were most dominant; moreover, M. sympodialis were found in both groups (Han et al., 2018). Furthermore, fungal community diversity was higher in skin affected by AD than in healthy skin. AD-specific Malassezia species, including M. slooffiae, M. obtusa, and M. yamatoensis, were identified (Han et al., 2018). Additionally, a recent study involving a cohort with AD in Switzerland showed that M. restricta and M. globosa were the two most dominant fungi in the human skin, regardless of AD severity. However, the abundance of Malassezia was decreased in severe AD skin lesions than in mild and moderate AD skin lesions and healthy skin. In contrast, diversity of other fungal genera, including Candida, Debaryomyces, Aureobasidium, and Penicillium were high in severe AD skin lesions (Schmid et al., 2022).
Scalp mycobiome in Korean patients with AD having scalp dermatitis was determined by amplicon sequencing analysis using the ITS region. Malassezia was the most predominant fungus, and fungal diversity was higher in AD skin lesions than in healthy controls. M. restricta was most abundant followed by M. globosa in both AD-affected and healthy skin samples. Furthermore, a significantly higher abundance of Saccharomyces and Cladosporium was observed in AD samples than in healthy controls (Woo et al., 2022).
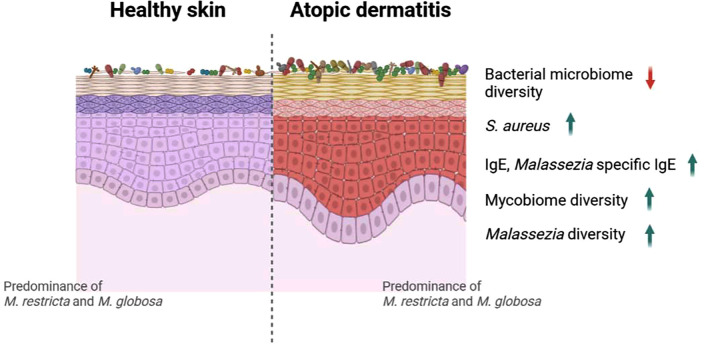
Although the number of the AD-associated mycobiome studies are limited, and cohorts from different geological and ethnical backgrounds were involved, several common features were noticed. First, regardless of the severity of AD symptoms, Malassezia is the dominant fungus found on the human skin, with M. restricta and M. globosa demonstrating the highest detection rates in both diseased and healthy skin. Second, in the AD skin lesions, fungal diversity is significantly increased while the abundance of Malassezia is decreased (Fig. 1). However, more systematic investigations are required since the reviewed studies had relatively small sample sizes and used different sampling and sequencing methods.
FIG. 1.
Graphical depiction of altered microbiome in skin affected by atopic dermatitis (AD). Several studies on bacterial microbiomes demonstrated reduced diversity of bacterial communities and increased abundance of Staphylococcus aureus in AD-affected skin than in healthy skin. In contrast, studies suggested increased diversity of mycobiomes, specifically that of Malassezia species. Malassezia-specific immunoglobulin (Ig) E increased, and high IgE levels were observed in patients with AD. Furthermore, most mycobiome studies commonly showed that M. restricta and M. globosa were predominant in both healthy and AD-affected skin. The image was prepared using Biorender.com.
SKIN MYCOBIOME IN SEBORRHEIC DERMATITIS
SD is a common chronic inflammatory skin disorder presenting in areas rich in sebaceous glands (scalp, back, face, and chest). Approximately 3% of the adult population experience SD, and a higher prevalence is reported in patients with immune deficiency and neurologic diseases, such as Parkinson's disease (Kim et al., 2016), with more than 50% of patients with Parkinson's disease developing SD (Rietcheck et al., 2021). Several underlying factors, including alteration of production and composition of sebum and disruption of epidermal barrier function, are considered to contribute to SD. Moreover, Malassezia is regarded as one of the main predisposing factors for SD because increased fungal counts are often observed while the disease progresses, and antifungal treatment alleviates the symptoms (Wikramanayake et al., 2019). Although the pathological role of Malassezia in SD is not yet fully understood, several of its characteristics likely contribute to the progress of SD. Malassezia fungi secrete multiple lipases that hydrolyze triglycerides in human sebum and produce unsaturated fatty acids, such as oleic acid, which may cause dandruff development resulting in flaking of skin lesions affected by SD (DeAngelis et al., 2007; Park et al., 2021). Moreover, the scalp and facial lesions in patients with SD exhibited hyphae formation by Malassezia cells, whose morphological characteristics may contribute to disease development (Li et al., 2022).
Innate immune response against Malassezia may also be involved in SD. When the skin barrier function is impaired, Malassezia may be able to penetrate the epithelial tissue to interact with antigen-presenting cells, such as neutrophils, macrophages, and dendritic cells, and induce activation of the NLRP3 inflammasome leading to IL-1β secretion, which also contribute to the development and progress of SD (Kistowska et al., 2014). A recent study suggested that Malassezia induces IL-17 cytokine production because Mpzl3-knockout mice displayed SD-like phenotype, including increased IL-17 cytokine production (Wikramanayake et al., 2017; Sparber et al., 2019).
Dandruff is a type of SD that mainly affects the scalp, making it itchy and scaly but often without inflammation (Borda and Wikramanayake, 2015). The effect of mycobiome on SD was first investigated by comparing fungal and bacterial communities in dandruff-affected and healthy scalps. The analysis of dandruff samples from a cohort in France using fungal-specific PCR amplification, cloning, and sequencing methods revealed that the most abundant fungal species on scalps with and without dandruff was M. restricta (84% and 97%, respectively), and M. globosa and M. sympodialis accounted for less than 1% in both groups. Furthermore, M. restricta was more abundant in scalps affected by dandruff than in the healthy controls (Clavaud, et al., 2013). Considering bacterial species, the population of S. epidermidis rose, while that of Cutibacterium acnes reduced in scalps affected by dandruff than in the healthy controls. Moreover, the M. restricta/C. acnes ratio was 0.37, which was significantly higher in scalps affected by dandruff than in healthy scalps, in which the ratio was 0.012 (Clavaud et al., 2013). ITS amplicon sequencing analysis to compare microbiome structures between individuals with dandruff and healthy controls in China showed that the most predominant fungi were M. restricta (88.5%) and M. globosa (5.1%). Moreover, compared with the fungal mycobiome, bacterial microbiome was more strongly associated with dandruff severity. While the abundance of Cutibacterium was significantly reduced, that of Staphylococcus was high in the same scalp samples with dandruff (Xu et al., 2016). The recent mycobiome analysis conducted via ITS amplicon sequencing in Korea showed that M. restricta and M. globosa were predominant on the scalp. The population of M. restricta was high and that of M. globosa was low in scalps affected by dandruff, suggesting a strong association between M. restricta and higher disease incidence. Moreover, dysbiosis of fungal communities may contribute to scalp SD development (Park et al., 2017). Similarly, another study in China indicated that Malassezia is one of the most predominant fungi on the scalp. Furthermore, the fungal diversity was higher in healthy scalps than in those affected by dandruff, with an increased abundance of Malassezia in the scalp affected by dandruff than in the healthy scalp. This observation was common to that in other studies although the tested samples differed geologically and ethnically (Lin et al., 2021).
A novel Malassezia species, M. arunalokei has been recently reported (Honnavar et al., 2016). A study conducted to compare the distribution of M. arunalokei between three skin sites (scalp, forehead, and cheeks) of patients with SD and healthy individuals showed that the abundance of M. arunalokei was similar in the patient and healthy individual groups. However, the concentration of M. arunalokei was higher on the forehead and cheeks than on the scalp in both the groups, suggesting that the fungus is not associated with SD but rather with the skin site (Cho et al., 2022).
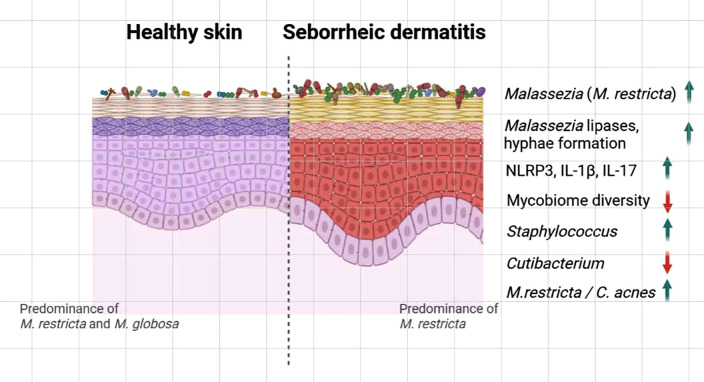
Apart from research to study abundance, a study to investigate the alteration of facial mycobiome in patients with SD after antifungal drug treatment was conducted (Tao et al., 2022). Topical ketoconazole, which is widely considered the most effective treatment for fungus-associated skin conditions, such as SD, had a clinical efficacy of 63%–90% (Choi et al., 2019). Malassezia is the most predominant fungal genus on both SD-affected and healthy facial skin. The basal structures of facial mycobiome in patients with facial SD were analyzed, and the abundance of Malassezia was significantly higher in the lesional skin sites than in the healthy controls. Moreover, fungal diversity was significantly lower with increased populations of Candida and Aspergillus in both lesional and nonlesional skin sites of patients with facial SD than in the healthy controls. Fungal diversity significantly increased after ketoconazole treatment when compared with the baseline (Tao et al., 2022). Monitoring of scalp mycobiome before and after ketoconazole treatment revealed increased fungal diversity and reduction of Malassezia on the scalp of patients with SD patients than in the healthy controls (Massiot et al., 2022). Overall, most culture-independent mycobiome analyses of scalp dandruff and facial SD suggest that the abundance of Malassezia, M. restricta in particular, increases and mycobiome diversity reduces with disease progression (Fig. 2).
FIG. 2.
Graphical depiction of altered microbiome in skin affected by seborrheic dermatitis (SD). Studies demonstrated reduced diversity of fungal communities and increased abundance of Malassezia restricta in SD-affected skin than in healthy skin. Studies also suggested increased expression of lipases and hyphae formation in Malassezia. Activation of the NLRP3 inflammasome leading to IL-1β secretion and increased IL-17 cytokine production was also observed in the SD skin lesion. Furthermore, bacterial microbiome analysis demonstrated increased abundance of Staphylococcus, while Cutibacterium was reduced. The higher M. restricta/C. acnes ratio in scalps affected by SD compared to that in healthy scalps was also reported. The image was prepared using Biorender.com.
CONCLUSION
Evidence from previous research showed that AD and SD, which are the most common skin diseases, are closely associated with skin microbiomes, and the fungal constituents of microbial communities play a critical role in homeostasis, host interactions, and disease development. However, most culture-independent microbiome analyses of the human skin are focused on bacterial rather than fungal communities. Therefore, the effects of mycobiomes on human skin health and diseases are largely unknown.
The number of studies conducted to understand the role of mycobiome in the skin environment is limited; nevertheless, several remarkable findings have been revealed. While AD increased fungal diversity, SD decreased it, when compared with that on the healthy skin. Whether this is related to the different influences of the diseases or the differences in the fungal communities is not clear. Although the currently available data on skin mycobiomes are from persons of different geological and ethnical backgrounds, almost all the studies suggested that Malassezia is the most predominant fungal genus in the skin mycobiota. Moreover, amplicon sequencing and metagenome analysis revealed that abundance of Malassezia is highly and positively correlated with development of AD and SD. However, our knowledge regarding Malassezia in the skin environment needs to be expanded. Uncertainties regarding the roles fungi play as essential members in skin microbiomes and the consequences of host–fungus interactions remains, and further research is required to clarify them.
ACKNOWLEDGMENTS
This study was supported by the Basic Science Research Program through the National Research Foundation of Korea (NRF), funded by the Ministry of Science and ICT (Grant Nos. 2021M3A9I4021431 and 2022R1F1A1065306).
AUTHOR DECLARATIONS
Conflict of Interest
The authors have no conflicts to disclose.
Author Contributions
Won Hee Jung: Conceptualization (lead); Data curation (lead); Formal analysis (lead); Funding acquisition (lead); Investigation (lead); Resources (lead); Visualization (lead); Writing – original draft (lead); Writing – review & editing (lead).
DATA AVAILABILITY
All data are publicly available.
References
- 1. Alam, M. J. , Xie, L. , Yap, Y. A. , Marques, F. Z. , and Robert, R. , “ Manipulating microbiota to treat atopic dermatitis: Functions and therapies,” Pathogens 11, 642 (2022). 10.3390/pathogens11060642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Baurecht, H. , Ruhlemann, M. C. , Rodriguez, E. , Thielking, F. , Harder, I. , Erkens, A.-S. , Stölzl, D. , Ellinghaus, E. , Hotze, M. , Lieb, W. , Wang, S. , Heinsen-Groth, F.-A. , Franke, A. , and Weidinger, S. , “ Epidermal lipid composition, barrier integrity, and eczematous inflammation are associated with skin microbiome configuration,” J. Allergy Clin. Immunol. 141, 1668–1676 (2018). 10.1016/j.jaci.2018.01.019 [DOI] [PubMed] [Google Scholar]
- 3. Bello, M. G. D. , Knight, R. , Gilbert, J. A. , and Blaser, M. J. , “ Preserving microbial diversity,” Science 362, 33–34 (2018). 10.1126/science.aau8816 [DOI] [PubMed] [Google Scholar]
- 4. Berg, G. , Rybakova, D. , Fischer, D. , Cernava, T. , Vergès, M.-C. C. , Charles, T. , Chen, X. , Cocolin, L. , Eversole, K. , Corral, G. H. , Kazou, M. , Kinkel, L. , Lange, L. , Lima, N. , Loy, A. , Macklin, J. A. , Maguin, E. , Mauchline, T. , McClure, R. , Mitter, B. , Ryan, M. , Sarand, I. , Smidt, H. , Schelkle, B. , Roume, H. , Kiran, G. S. , Selvin, J. , de Souza, R. S. C. , van Overbeek, L. , Singh, B. K. , Wagner, M. , Walsh, A. , Sessitsch, A. , and Schloter, M. , “ Microbiome definition re-visited: Old concepts and new challenges,” Microbiome 8, 103 (2020). 10.1186/s40168-020-00875-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Borda, L. J. and Wikramanayake, T. C. , “ Seborrheic dermatitis and dandruff: A comprehensive review,” J. Clin. Invest. Dermatol. 3(2), 10 (2015). 10.13188/2373-1044.1000019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Chng, K. R. , Tay, A. S. , Li, C. , Ng, A. H. Q. , Wang, J. , Suri, B. K. , Matta, S. A. , McGovern, N. , Janela, B. , Wong, X. F. C. C. , Sio, Y. Y. , Au, B. V. , Wilm, A. , De Sessions, P. F. , Lim, T. C. , Tang, M. B. Y. , Ginhoux, F. , Connolly, J. E. , Lane, E. B. , Chew, F. T. , Common, J. E. A. , and Nagarajan, N. , “ Whole metagenome profiling reveals skin microbiome-dependent susceptibility to atopic dermatitis flare,” Nat. Microbiol. 1, 16106 (2016). 10.1038/nmicrobiol.2016.106 [DOI] [PubMed] [Google Scholar]
- 7. Cho, Y. J. , Kim, T. , Croll, D. , Park, M. , Kim, D. , Keum, H. L. , Sul, W. J. , and Jung, W. H. , “ Genome of malassezia arunalokei and its distribution on facial skin,” Microbiol. Spectrum 10, e0050622 (2022). 10.1128/spectrum.00506-22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Choi, F. D. , Juhasz, M. L. W. , and Mesinkovska, N. A. , “ Topical ketoconazole: A systematic review of current dermatological applications and future developments,” J. Dermatol. Treat. 30, 760–771 (2019). 10.1080/09546634.2019.1573309 [DOI] [PubMed] [Google Scholar]
- 9. Clavaud, C. , Jourdain, R. , Bar-Hen, A. , Tichit, M. , Bouchier, C. , Pouradier, F. , Rawadi, C. E. , Guillot, J. , Ménard-Szczebara, F. , Breton, L. , Latge, J.-P. , and Mouyna, I. , “ Dandruff is associated with disequilibrium in the proportion of the major bacterial and fungal populations colonizing the scalp,” PLoS One 8, e58203 (2013). 10.1371/journal.pone.0058203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Cundell, A. M. , “ Microbial ecology of the human skin,” Microb. Ecol. 76, 113–120 (2018). 10.1007/s00248-016-0789-6 [DOI] [PubMed] [Google Scholar]
- 11. DeAngelis, Y. M. , Saunders, C. W. , Johnstone, K. R. , Reeder, N. L. , Coleman, C. G. , Kaczvinsky, J. R., Jr. , Gale, C. , Walter, R. , Mekel, M. , Lacey, M. P. , Keough, T. W. , Fieno, A. , Grant, R. A. , Begley, B. , Sun, Y. , Fuentes, G. , Youngquist, R. S. , Xu, J. , and Dawson, T. L., Jr. , “ Isolation and expression of a malassezia globosa lipase gene, LIP1,” J. Invest. Dermatol. 127, 2138–2146 (2007). 10.1038/sj.jid.5700844 [DOI] [PubMed] [Google Scholar]
- 12. Edslev, S. M. , Andersen, P. S. , Agner, T. , Saunte, D. M. L. , Ingham, A. C. , Johannesen, T. B. , and Clausen, M.-L. , “ Identification of cutaneous fungi and mites in adult atopic dermatitis: Analysis by targeted 18S rRNA amplicon sequencing,” BMC Microbiol. 21, 72 (2021). 10.1186/s12866-021-02139-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Fasseeh, A. N. , Elezbawy, B. , Korra, N. , Tannira, M. , Dalle, H. , Aderian, S. , Abaza, S. , and Kaló, Z. , “ Burden of atopic dermatitis in adults and adolescents: A systematic literature review,” Dermatol. Ther. 12, 2653–2668 (2022). 10.1007/s13555-022-00819-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Glatz, M. , Bosshard, P. P. , Hoetzenecker, W. , and Schmid-Grendelmeier, P. , “ The role of malassezia spp. in atopic dermatitis,” J. Clin. Med. 4, 1217–1228 (2015). 10.3390/jcm4061217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Han, S. H. , Cheon, H. I. , Hur, M. S. , Kim, M. J. , Jung, W. H. , Lee, Y. W. , Choe, Y. B. , and Ahn, K. J. , “ Analysis of the skin mycobiome in adult patients with atopic dermatitis,” Exp. Dermatol. 27, 366–373 (2018). 10.1111/exd.13500 [DOI] [PubMed] [Google Scholar]
- 16. Hirasawa, Y. , Takai, T. , Nakamura, T. , Mitsuishi, K. , Gunawan, H. , Suto, H. , Ogawa, T. , Wang, X.-L. , Ikeda, S. , Okumura, K. , and Ogawa, H. , “ Staphylococcus aureus extracellular protease causes epidermal barrier dysfunction,” J. Invest. Dermatol. 130, 614–617 (2010). 10.1038/jid.2009.257 [DOI] [PubMed] [Google Scholar]
- 17. Honnavar, P. , Prasad, G. S. , Ghosh, A. , Dogra, S. , Handa, S. , and Rudramurthy, S. M. , “ Malassezia arunalokei sp. nov., a novel yeast species isolated from seborrheic dermatitis patients and healthy individuals from India,” J. Clin. Microbiol. 54, 1826–1834 (2016). 10.1128/JCM.00683-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Hooks, K. B. and O'Malley, M. A. , “ Dysbiosis and its discontents,” mBio 8, e01492 (2017). 10.1128/mBio.01492-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kaga, M. , Sugita, T. , Nishikawa, A. , Wada, Y. , Hiruma, M. , and Ikeda, S. , “ Molecular analysis of the cutaneous malassezia microbiota from the skin of patients with atopic dermatitis of different severities,” Mycoses 54, e24–e28 (2011). 10.1111/j.1439-0507.2009.01821.x [DOI] [PubMed] [Google Scholar]
- 20. Kieffer, M. , Bergbrant, I. M. , Faergemann, J. , Jemec, G. B. , Ottevanger, V. , Skov, P. S. , and Svejgaard, E. , “ Immune reactions to Pityrosporum ovale in adult patients with atopic and seborrheic dermatitis,” J. Am. Acad. Dermatol. 22, 739–742 (1990). 10.1016/0190-9622(90)70100-V [DOI] [PubMed] [Google Scholar]
- 21. Kim, K. M. , Kim, H. S. , Yu, J. , Kim, J. T. , and Cho, S. H. , “ Analysis of dermatologic diseases in neurosurgical in-patients: A retrospective study of 463 cases,” Ann. Dermatol. 28, 314–320 (2016). 10.5021/ad.2016.28.3.314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kistowska, M. , Fenini, G. , Jankovic, D. , Feldmeyer, L. , Kerl, K. , Bosshard, P. , Contassot, E. , and French, L. E. , “ Malassezia yeasts activate the NLRP3 inflammasome in antigen-presenting cells via Syk-kinase signalling,” Exp. Dermatol. 23, 884–889 (2014). 10.1111/exd.12552 [DOI] [PubMed] [Google Scholar]
- 23. Leung, D. Y. and Guttman-Yassky, E. , “ Deciphering the complexities of atopic dermatitis: Shifting paradigms in treatment approaches,” J. Allergy Clin. Immunol. 134, 769–779 (2014). 10.1016/j.jaci.2014.08.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Li, J. , Feng, Y. , Liu, C. , Yang, Z. , de Hoog, S. , Qu, Y. , Chen, B. , Li, D. , Xiong, H. , and Shi, D. , “ Presence of malassezia hyphae is correlated with pathogenesis of seborrheic dermatitis,” Microbiol. Spectrum 10, e0116921 (2022). 10.1128/spectrum.01169-21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Lin, Q. , Panchamukhi, A. , Li, P. , Shan, W. , Zhou, H. , Hou, L. , and Chen, W. , “ Malassezia and Staphylococcus dominate scalp microbiome for seborrheic dermatitis,” Bioprocess. Biosyst. Eng. 44, 965–975 (2021). 10.1007/s00449-020-02333-5 [DOI] [PubMed] [Google Scholar]
- 26. Massiot, P. , Clavaud, C. , Thomas, M. , Ott, A. , Gueniche, A. , Panhard, S. , Muller, B. , Michelin, C. , Kerob, D. , Bouloc, A. , and Reygagne, P. , “ Continuous clinical improvement of mild-to-moderate seborrheic dermatitis and rebalancing of the scalp microbiome using a selenium disulfide-based shampoo after an initial treatment with ketoconazole,” J. Cosmet. Dermatol. 21, 2215–2225 (2022). 10.1111/jocd.14362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Moosbrugger-Martinz, V. , Leprince, C. , Méchin, M.-C. , Simon, M. , Blunder, S. , Gruber, R. , and Dubrac, S. , “ Revisiting the roles of filaggrin in atopic dermatitis,” Int. J. Mol. Sci. 23, 5318 (2022). 10.3390/ijms23105318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Nakagawa, S. , Matsumoto, M. , Katayama, Y. , Oguma, R. , Wakabayashi, S. , Nygaard, T. , Saijo, S. , Inohara, N. , Otto, M. , Matsue, H. , Nunez, G. , and Nakamura, Y. , “ Staphylococcus aureus virulent PSMalpha peptides induce keratinocyte alarmin release to orchestrate IL-17-dependent skin inflammation,” Cell Host Microbe 22, 667–677 (2017). 10.1016/j.chom.2017.10.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Nakatsuji, T. , Chen, T. H. , Narala, S. , Chun, K. A. , Two, A. M. , Yun, T. , Shafiq, F. , Kotol, P. F. , Bouslimani, A. , Melnik, A. V. , Latif, H. , Kim, J.-N. , Lockhart, A. , Artis, K. , David, G. , Taylor, P. , Streib, J. , Dorrestein, P. C. , Grier, A. S. , Gill, R. , Zengler, K. , Hata, T. R. , Leung, D. Y. M. , and Gallo, R. L. , “ Antimicrobials from human skin commensal bacteria protect against Staphylococcus aureus and are deficient in atopic dermatitis,” Sci. Transl. Med. 9, eaah4680 (2017). 10.1126/scitranslmed.aah4680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Oh, J. , Byrd, A. L. , Deming, C. , Conlan, S. , NISC Comparative Sequencing Program, Kong, H. H. , and Segre, J. , “ Biogeography and individuality shape function in the human skin metagenome,” Nature 514, 59–64 (2014). 10.1038/nature13786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Park, M. , Cho, Y.-J. , Lee, Y. W. , and Jung, W. H. , “ Whole genome sequencing analysis of the cutaneous pathogenic yeast malassezia restricta and identification of the major lipase expressed on the scalp of patients with dandruff,” Mycoses 60, 188–197 (2017). 10.1111/myc.12586 [DOI] [PubMed] [Google Scholar]
- 32. Park, M. , Park, S. , and Jung, W. H. , “ Skin commensal fungus malassezia and its lipases,” J. Microbiol. Biotechnol. 31, 637–644 (2021). 10.4014/jmb.2012.12048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Park, T. , Kim, H. J. , Myeong, N. R. , Lee, H. G. , Kwack, I. , Lee, J. , Kim, B. J. , Sul, W. J. , and An, S. , “ Collapse of human scalp microbiome network in dandruff and seborrhoeic dermatitis,” Exp. Dermatol. 26, 835–838 (2017). 10.1111/exd.13293 [DOI] [PubMed] [Google Scholar]
- 34. Paulino, L. C. , “ New perspectives on dandruff and seborrheic dermatitis: Lessons we learned from bacterial and fungal skin microbiota,” Eur. J. Dermatol. 27, 4–7 (2017). 10.1684/ejd.2017.3038 [DOI] [PubMed] [Google Scholar]
- 35. Rietcheck, H. R. J. , Maghfour, C. W. , Rundle, S. S. , Husayn, C. L. , Presley, S. H. , Sillau, Y. , Liu, M. A. , Rietcheck, H. R. , Maghfour, J. , Rundle, C. W. , Husayn, S. S. , Presley, C. L. , Sillau, S. H. , Liu, Y. , Leehey, M. A. , Dunnick, C. A. , and Dellavalle, R. P. , “ A review of the current evidence connecting seborrheic dermatitis and Parkinson's disease and the potential role of oral cannabinoids,” Dermatology 237, 872–877 (2021). 10.1159/000512189 [DOI] [PubMed] [Google Scholar]
- 36. Ring, J. , Abeck, D. , and Neuber, K. , “ Atopic eczema: Role of microorganisms on the skin surface,” Allergy 47, 265–269 (1992). 10.1111/j.1398-9995.1992.tb02051.x [DOI] [PubMed] [Google Scholar]
- 37. Ring, J. , Zink, A. , Arents, B. W. M. , Seitz, I. A. , Mensing, U. , Schielein, M. C. , Wettemann, N. , de Carlo, G. , and Fink-Wagner, A. , “ Atopic eczema: Burden of disease and individual suffering-results from a large EU study in adults,” J. Eur. Acad. Dermatol. Venereol. 33, 1331–1340 (2019). 10.1111/jdv.15634 [DOI] [PubMed] [Google Scholar]
- 38. Schmid, B. , Kunstner, A. , Fahnrich, A. , Bersuch, E. , Schmid-Grendelmeier, P. , Busch, H. , Glatz, M. , and Bosshard, P. P. , “ Dysbiosis of skin microbiota with increased fungal diversity is associated with severity of disease in atopic dermatitis,” J. Eur. Acad. Dermatol. Venereol. 36, 1811–1819 (2022). 10.1111/jdv.18347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Schoch, J. J. , Monir, R. L. , Satcher, K. G. , Harris, J. , Triplett, E. , and Neu, J. , “ The infantile cutaneous microbiome: A review,” Pediatr. Dermatol. 36, 574–580 (2019). 10.1111/pde.13870 [DOI] [PubMed] [Google Scholar]
- 40. Sparber, F. , Gregorio, C. D. , Steckholzer, S. , Ferreira, F. M. , Dolowschiak, T. , Ruchti, F. , Kirchner, F. R. , Mertens, S. , Prinz, I. , Joller, N. , Buch, T. , Glatz, M. , Sallusto, F. , and LeibundGut-Landmann, S. , “ The skin commensal yeast malassezia triggers a type 17 response that coordinates anti-fungal immunity and exacerbates skin inflammation,” Cell Host Microbe 25, 389–403 (2019). 10.1016/j.chom.2019.02.002 [DOI] [PubMed] [Google Scholar]
- 41. Sugita, T. , Suto, H. , Unno, T. , Tsuboi, R. , Ogawa, H. , Shinoda, T. , and Nishikawa, A. , “ Molecular analysis of malassezia microflora on the skin of atopic dermatitis patients and healthy subjects,” J. Clin. Microbiol. 39, 3486–3490 (2001). 10.1128/JCM.39.10.3486-3490.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Szczepanska, M. , Blicharz, L. , Nowaczyk, J. , Makowska, K. , Goldust, M. , Waskiel-Burnat, A. , Czuwara, J. , Samochocki, Z. , and Rudnicka, L. , “ The role of the cutaneous mycobiome in atopic dermatitis,” J. Fungi 8, 1153 (2022). 10.3390/jof8111153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Tao, R. , Wang, R. , Wan, Z. , Song, Y. , Wu, Y. , and Li, R. , “ Ketoconazole 2% cream alters the skin fungal microbiome in seborrhoeic dermatitis: A cohort study,” Clin. Exp. Dermatol. 47, 1088–1096 (2022). 10.1111/ced.15115 [DOI] [PubMed] [Google Scholar]
- 44. Upadhyay, K. G. , Desai, D. C. , Ashavaid, T. F. , and Dherai, A. J. , “ Microbiome and metabolome in inflammatory bowel disease,” J. Gastroenterol. Hepatol. 38, 34–43 (2022). 10.1111/jgh.16043 [DOI] [PubMed] [Google Scholar]
- 45. Weidinger, S. and Novak, N. , “ Atopic dermatitis,” Lancet 387, 1109–1122 (2016). 10.1016/S0140-6736(15)00149-X [DOI] [PubMed] [Google Scholar]
- 46. Wikramanayake, T. C. , Borda, L. J. , Kirsner, R. S. , Wang, Y. , Duffort, S. , Reyes-Capo, A. , Barsam, A. , Urbieta, M. , and Perez, V. L. , “ Loss of MPZL3 function causes seborrhoeic dermatitis-like phenotype in mice,” Exp. Dermatol. 26, 736–738 (2017). 10.1111/exd.13150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Wikramanayake, T. C. , Borda, L. J. , Miteva, M. , and Paus, R. , “ Seborrheic dermatitis-looking beyond malassezia,” Exp. Dermatol. 28, 991–1001 (2019). 10.1111/exd.14006 [DOI] [PubMed] [Google Scholar]
- 48. Williams, H. , Stewart, A. , von Mutius, E. , Cookson, W. , Anderson, H. R. and International Study of Asthma and Allergies in Childhood (ISAAC) Phase One and Three Study Groups, “ Is eczema really on the increase worldwide?,” J. Allergy Clin. Immunol. 121, 947–954 (2008). 10.1016/j.jaci.2007.11.004 [DOI] [PubMed] [Google Scholar]
- 49. Woo, Y. R. , Cho, M. , Han, Y. , Lee, S. H. , Cho, S. H. , Lee, J. D. , and Kim, H. S. , “ Characterization of distinct microbiota associated with scalp dermatitis in patients with atopic dermatitis,” J. Clin. Med. 11, 1735 (2022). 10.3390/jcm11061735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Xu, J. , Saunders, C. W. , Hu, P. , Grant, R. A. , Boekhout, T. , Kuramae, E. E. , Kronstad, J. W. , Deangelis, Y. M. , Reeder, N. L. , Johnstone, K. R. , Leland, M. , Fieno, A. M. , Begley, W. M. , Sun, Y. , Lacey, M. P. , Chaudhary, T. , Keough, T. , Chu, L. , Sears, R. , Yuan, B. , and Dawson, T. L., Jr. , “ Dandruff-associated malassezia genomes reveal convergent and divergent virulence traits shared with plant and human fungal pathogens,” Proc. Natl. Acad. Sci. U.S.A. 104, 18730–18735 (2007). 10.1073/pnas.0706756104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Xu, Z. , Wang, Z. , Yuan, C. , Liu, X. , Yang, F. , Wang, T. , Wang, J. , Manabe, K. , Qin, O. , Wang, X. , Zhang, Y. , and Zhang, M. , “ Dandruff is associated with the conjoined interactions between host and microorganisms,” Sci. Rep. 6, 1–9 (2016). 10.1038/srep24877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Zeeuwen, P. L. , Ederveen, T. H. , van der Krieken, D. A. , Niehues, H. , Boekhorst, J. , Kezic, S. , Hanssen, D. A. , Otero, M. E. , van Vlijmen-Willems, I. M. , Rodijk-Olthuis, D. , Falcone, D. , van den Bogaard, E. H. , Kamsteeg, M. , de Koning, H. D. , Zeeuwen-Franssen, M. E. , van Steensel, M. A. , Kleerebezem, M. , Timmerman, H. M. , van Hijum, S. A. , and Schalkwijk, J. , “ Gram-positive anaerobe cocci are underrepresented in the microbiome of filaggrin-deficient human skin,” J. Allergy Clin. Immunol. 139, 1368–1371 (2017). 10.1016/j.jaci.2016.09.017 [DOI] [PubMed] [Google Scholar]
- 53. Zubeldia-Varela, E. , Barker-Tejeda, T. C. , Obeso, D. , Villasenor, A. , Barber, D. , and Perez-Gordo, M. , “ Microbiome and allergy: New insights and perspectives,” J. Invest. Allergol. Clin. Immunol. 32, 327–344 (2022). 10.18176/jiaci.0852 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data are publicly available.