Abstract
Background:
An increased risk of neurocognitive deficits, anxiety, and depression has been reported in childhood cancer survivors.
Methods:
We analyzed associations of neurocognitive deficits, as well as anxiety and depression, with common and rare genetic variants derived from whole-exome sequencing data of acute lymphoblastic leukemia (ALL) survivors from the PETALE cohort. In addition, significant associations were assessed using stratified and multivariable analyses. Next, top-ranking common associations were analyzed in an independent SJLIFE replication cohort of ALL survivors.
Results:
Significant associations were identified in the entire discovery cohort (N=229) between the AK8 gene and changes in neurocognitive function, whereas PTPRZ1, MUC16, TNRC6C-AS1 were associated with anxiety. Following stratification according to sex, the ZNF382 gene was linked to a neurocognitive deficit in males, whereas APOL2 and C6orf165 were associated with anxiety and EXO5 with depression. Following stratification according to prognostic risk groups, the modulatory effect of rare variants on depression was additionally found in the CYP2W1 and PCMTD1 genes.
In the replication SJLIFE cohort (N=688), the male-specific association in the ZNF382 gene was not significant, however, a p-value<0.05 was observed when the entire SJLIFE cohort was analyzed. ZNF382 was significant in males in the combined cohorts as shown by meta-analyses as well as the depression-associated gene EXO5.
Conclusions:
Further research is needed to confirm whether the current findings, along with other known risk factors, may be valuable in identifying patients at increased risk of these long-term complications.
Impact:
Our results suggest that specific genes may be related to increased neuropsychological consequences.
INTRODUCTION
The survival rates in children diagnosed with acute lymphoblastic leukemia (ALL)(1, 2), the most frequent childhood cancer(3), have dramatically increased over the past decades due to the introduction of multi-agent risk-adapted treatment regimens and outstanding improvements in care delivery. However, exposure to cytotoxic therapy during a vulnerable period of child development can have long-term consequences, including impaired neurocognitive functions(4, 5), and mood disorders(6, 7). Furthermore, childhood and adolescence are periods characterized by intensive development of the central nervous system(8, 9), which is pertinent in the context of the impact of cancer treatment on the integrity of the white matter(10–14). Indeed, numerous studies conducted in childhood ALL survivors(4, 15, 16) have reported an increased risk of neurocognitive deficits(6) in attention(15–20), working memory(21), processing speed(16, 22, 23), and executive functions, such as verbal fluency and cognitive flexibility(24); as well as depression, anxiety, behavioral difficulties, distress, and post-traumatic symptoms compared with siblings(25–30).
Varying degrees of neurocognitive dysfunction and levels of emotional distress associated with cancer treatment have been observed that differ by patient characteristics, such as age and sex, and possibly reflecting different underlying mechanisms(5, 31, 32). Moreover, while some survivors may not experience any of these complications, others may have more than one. Factors contributing to this variability, include the type of treatment, the characteristics of the malignancy, the lifestyle, and the genetic makeup of the patient(33).
We examined whether common and rare genetic polymorphisms contribute to this variability by altering the risk of treatment-related neurocognitive deficits, as well as anxiety and depression in combination with non-genetic factors.
We previously analyzed these complications in a well-described cohort of ALL survivors (PETALE)(34) using a candidate gene approach(33); two associations between the MTR and CACNB2 genes and neurocognitive deficit were validated in an independent St. Jude Lifetime Cohort (SJLIFE) replication cohort (St. Jude Children’s Research Hospital, Memphis, USA)(33).
Here, the association analyses are extended to a hypothesis-free approach – an exome-wide association study, which could identify additional genes as potential risk modulators of these complications.
MATERIALS AND METHODS
Discovery cohort
The discovery cohort included 229 patients diagnosed and treated for childhood ALL according to Dana Farber Cancer Institute (DFCI) ALL 87-01 to 05-01 protocols at Sainte-Justine University Health Center (SJUHC), Montreal, (Quebec), Canada. The participants were recruited during 2013–2015 in the context of the PETALE study(34). Written informed consent was obtained from every patient or parent/legal guardian. The study was conducted in accordance with the Declaration of Helsinki and the protocol was approved by the Ethics Committee of SJUHC.
Eligible participants were of European descent, younger than 19 years old at diagnosis and older than 12 years at evaluation, at least 5 years after diagnosis of ALL, without a history of relapse or refractory ALL or Down syndrome and had not received a hematopoietic stem cell transplant. The median age of patients at the time of diagnosis was 4 years, the time from the end of treatment to evaluation ranged from 3–24 years with a median of 13 years (for 76.0% of participants, it was ≥10 years), both sexes were equally represented (51.1% of females). The patients were classified to standard (SR) and high relapse risk (HR) groups based on prognostic factors, including age, white blood cell count, immunophenotype, and central nervous system (CNS) status at diagnosis(35, 36). The frequency of patients assigned to SR and HR groups during the treatment was 45.9% and 54.1%, respectively.
Neuropsychological evaluation
A neurocognitive evaluation was performed using standardized testing procedures and scores from the Delis-Kaplan Executive Function System (D-KEFS)(37) and the Wechsler Adult Intelligence Scale-Fourth Edition (WAIS-IV)(38), and included the Trail Making Test (D-KEFS) score, the Verbal Fluency (D-KEFS) score, and the Digit span (WAIS-IV) total score, as described previously(39). The conversion of raw scores to age-adjusted scaled scores was based on population means(40); subsequently, neurocognitive outcomes were transformed into dichotomous variables and studied accordingly. For each of these variables, scores lower than one and a half standard deviations below the mean of the normative dataset were indicative of impairment(41), all other scores were considered non-impaired.
Anxiety and Depression
Categorization of participants into anxiety or depression groups was based on their symptoms exceeding age-specific norms as described previously(39). For participants under the age of 19, we employed the anxiety and depression modules of the Beck Youth Inventories - Second Edition (BYI) (42). For older participants (≥19 years) the Brief Symptom Inventory-18 (BSI-18 anxiety and depression score) was applied (43). The Cronbach’s alpha coefficients for internal consistency demonstrated satisfactory levels, all exceeding 0.80(44). Scores adjusted for age that were one standard deviation above the population mean were regarded as indicative of impairment.
Sequencing and quality control
Whole-exome sequencing (WES) was performed on germline DNA, extracted from peripheral blood samples from participants in the PETALE cohort, using standard protocols as described previously(33). Whole exomes were captured in solution with Agilent’s SureSelect Human All Exon 50Mb kits and sequenced on either Life Technologies SOLiD System 4.0 (mean coverage = 40X) or Illumina HiSeq 2500 platform (mean coverage = 113.1X) at SJUHC integrated clinical genomic center in pediatrics as described previously(39, 45). Only missense, nonsense, and splicing common and rare variants with predicted functional impact (Sift (<0.1) and/or PolyPhen2 (≥0.85)) were considered(46, 47). Variants were defined as rare (minor allele frequency, MAF<5%) and common (MAF≥5%) according to the reported frequency for European populations in public datasets(48). Variants exceeding a missing rate of 20%, not in Hardy-Weinberg Equilibrium (p=0.05/number of tests)(49), and common variants with pairwise linkage disequilibrium (LD, r2≥0.8) were excluded. Therefore, 5312 common genetic variants corresponding to 3793 genes, and 58924 rare genetic variants corresponding to 11441 genes that satisfied the above-mentioned filtering criteria entered the association analyses.
Association analyses
The P-value threshold of 5 × 10−8, commonly used to identify an association between a common genetic variant and an outcome of interest in a typical GWAS(50–52) is not applicable to our study since variants for analysis were selected from WES data to focus on those predicted by various instruments to affect coding protein function(46, 47). As a result, 5312 common variants were analyzed, which is less than the typical number of GWAS variants; and the Benjamini-Hochberg procedure for false discovery rate (FDR)(53, 54) was used to adjust for the number of variants tested with an adjusted cut-off value of <5% considered to be statistically significant(52). The analyses between common genetic variants and neurocognitive outcomes, and anxiety/depression were performed by the allelic chi-square or Fisher’s exact test implemented in PLINK v.1.07(55, 56). Analyses were performed in 229 sequenced patients and stratified by sex and risk groups with different treatment intensities because these factors have an established role in modulating neurocognitive outcomes(57, 58). For top-ranking associations, the best genotyping model was subsequently used in the univariate model as well as in multivariable logistic regression analysis to assess the effect of each genotype when controlling for non-genetic covariates. In cases when several genetic variants were associated with the same outcome, additional multivariable analyses were performed in which all genetic variants associated with that outcome were analyzed with non-genetic covariates in a single model. Non-genetic co-variables included: age at the time of diagnosis (continuous variable); time since the end of treatment (continuous variable); sex: males/females (categorical variable); DFCI Protocol: 87-01=1, 91-01=2, 95-01=3, 00-01=4, 05-01=5 (categorical variable); risk: SR/HR (categorical variable); treatment variable with patients who received only chemotherapy and patients who received chemotherapy and cranial radiation (categorical variable). The associations that remained statistically significant through multivariable regression models were retained for further analyses. The effect of genotype was quantified by odds ratio (OR) with 95% CI according to the best-fitting genetic model, which was either dominant or recessive in all cases. In addition, the potential additive effect of combining risk loci by recoding genotypes as having none, one, or two and more risk alleles has also been explored.
The effects of rare variants were evaluated using the SKAT-O test (Optimal Sequence Kernel Association Test)(59–61) implemented in SKAT package v.2.0.1(62). Only genes with at least two variants that satisfied filtering criteria were retained for the SKAT-O test. To evaluate individual variant contributions to association signals, a collapsing approach(63, 64), with the iterative exclusion of every single variant, was additionally executed in SPSS v.25.0. Similar to the common variant analyses, genetic associations were assessed for each of the neurocognitive outcomes in the entire cohort, in different sex groups, as well as in SR and HR groups, and through multivariable models adjusted for the covariates described above. The multiple test adjustment (FDR) for the number of genes tested was included in all analyses.
Replication cohort
The replication cohort consisted of 688 childhood ALL survivors of European ancestry enrolled in the SJLIFE study with whole-genome sequencing data. The maximum number of participants in the replication cohort with available outcome data was 675; the total number varied depending on the outcomes and subgroup studied. Participants were selected to resemble the discovery cohort based on demographics and treatment characteristics. They were younger than 19 years at diagnosis, older than 12 years at evaluation, with no history of relapse within 5 years of the primary ALL diagnosis date, Down syndrome, or hematopoietic stem cell transplantation. The median age at diagnosis was 4.9 years, the median time since 5-year survival from ALL diagnosis was 25.8 years for neurocognitive evaluation, and 25.6 years for anxiety and depression evaluation; 50.4% of participants were males. All outcome measures were the same as in the discovery cohort with the exception of the Patient-Reported Outcomes Measurement Information System (PROMIS) Anxiety and Depression Scales(65, 66) that were used to assess anxiety and depression in participants under 18 years of age. Associations that remained significant in the discovery cohort using multivariable regression models were analyzed in the replication cohort by Fisher’s exact test for allelic contingency tables and logistic regression adjusting for continuous age at diagnosis, sex, the continuous time between the date of becoming a 5-year ALL survivor and date of test measurement, whether the survivor was treated with chemotherapy only versus chemotherapy plus radiation, and the top 20 principal components adjusting for genetic ancestry. Variant rs750295511 (MUC16) did not pass quality control and was excluded from the analysis. Stratification by the risk group designation was not available for the SJLIFE cohort. Rare variant replication analyses were not performed since rare variant associations in the discovery cohort were only detected in the risk group stratified analyses. The combined effect in the discovery and replication cohort was addressed through meta-analysis performed using the Mantel-Haenszel method implemented in MedCalc software and assuming a fixed-effects model.
Data Availability
The datasets used and/or analyzed during the current study are available from the corresponding author upon request and revision of the projects for which the data might be used.
RESULTS
Discovery cohort characteristics
Demographics and clinical characteristics of PETALE participants are presented in Table 1. The analyses were performed in either all patients or subgroups. These included patients assigned to standard (SR, 45.9%) and high risk (HR, 54.1%) groups, males (48.9%) and females (51.1%). The median age of ALL survivors at the time of evaluation was 21 years. The most prevalent deficit in neurocognitive test performance was noted for digit span (19.7%) followed by verbal fluency (19.2%) and trail making test (8.7%). Moderate-severe anxiety was noted in 9.2% survivors, whereas 10.5% of survivors were affected by moderate-severe depression.
Table 1.
Patient demographics and clinical characteristics, Discovery PETALE cohort, N=229.
| N | % | ||
|---|---|---|---|
| Sex | |||
| Males | 112 | 48.9 | |
| Females | 117 | 51.1 | |
| Prognostic risk group | |||
| Standard risk | 105 | 45.9 | |
| High risk | 124 | 54.1 | |
| Criteria for High-risk stratification * | |||
| Age | 41 | 33.1 | |
| WBC | 33 | 26.6 | |
| T-cell markers | 5 | 4.1 | |
| Combination Age and/or WBC and/or T-cell markers | 21 | 16.9 | |
| CNS involvement | 16 | 12.9 | |
| MRD+ | 2 | 1.6 | |
| Other** | 6 | 4.8 | |
| DFCI protocol | |||
| (87–1) | 18 | 7.9 | |
| (91–1) | 47 | 20.5 | |
| (95–01) | 68 | 29.7 | |
| (00–1) | 72 | 31.4 | |
| (5–1) | 24 | 10.5 | |
| Cranial radiation therapy (median 18 Gy) | |||
| Yes | 134 | 58.5 | |
| No | 95 | 41.5 | |
| Age at diagnosis, median (range) | |||
| 4 (0–18) | |||
| Time since the end of treatment | |||
| More than 10 years | 174 | 76 | |
| Less than 10 years | 55 | 24 | |
| Median (range) | 13 (3–24) | ||
| Tested outcomes | |||
| Affected | Unaffected | Missing | |
| Neurocognitive outcomes | |||
| Trail making test | 20 (8.7%) | 209 (91.3%) | 0 (0.0%) |
| Verbal fluency | 44 (19.2 %) | 184 (80.4%) | 1 (0.4%) |
| Digit span | 45 (19.7%) | 187 (80.3%) | 0 (0.0%) |
| Emotional distress outcomes | |||
| Moderate-severe anxiety | 21 (9.2%) | 180 (78.6%) | 28 (12.2%) |
| Moderate-severe depression | 24 (10.5%) | 177 (77.3%) | 28 (12.2%) |
DFCI, Dana-Farber Cancer Institute. WBC: White Blood Cell; CNS: Central Nervous System; MRD: Minimal Residual Disease.
Criteria for High-risk stratification was mainly attributed based on age, white blood cell count, immunophenotype (presence of T-cell markers) and combination of these factors; as well as central nervous system (CNS) status and Minimal residual disease at diagnosis.
This category represents patients with chromosomal abnormalities and/or combination of factors.
Common variants
Among the common genetic variants, the top-ranking associations obtained using PLINK (Supplementary Tables S1–S3) were assessed through multivariate regression models that also included non-genetic co-variables. Only the associations that remained significant in these models were retained for further analyses (Supplementary Tables S4–S7).
Accordingly, significant associations were detected between Trail making test and rs17407084 variant in the AK8 gene (OR=7.3, 95% CI, 2.7–19.7; p=4.52E-04), as well as between Moderate-severe anxiety and the following variants: rs740965 in PTPRZ1 (OR=5.1, 95% CI, 1.98–12.9; p=1.00E-03), rs750295511 in MUC16 (OR=8.3, 95% CI, 3.1–22.5; p=3.10E-05), and rs2748431in TNRC6C-AS1 (OR=6.1, 95% CI, 2.0–18.1; p=1.00E-03). These association are presented in their best genetic models (Tables 2 and 3) as well as by Manhattan plots (Supplementary Figures S1 and S2). Additionally, the combined effect of the rs740965 (PTPRZ1), rs750295511 (MUC16), and rs2748431 (TNRC6C-AS1) variants was identified by recoding genotypes as having none, one or two and more alleles at risk (Supplementary Figure S3). Following stratification according to sex we identified variant rs61732180 in the ZNF382 gene that was associated with the increased risk of deficit scores in the Trail making test in male participants (OR=20.2, 95% CI, 4.3–95.4; p=2.62E-04, Table 2). Male carriers of the variant alleles in the rs7285167 (APOL2) and rs61731441 (C6orf165) genes were more prone to Moderate-severe anxiety (OR=9.6, 95% CI, 2.3–40.5; p=3.00E-03 and OR=10.9, 95% CI, 2.5–47.0; p=2.00E-03, respectively, Table 3), whereas male carriers of the variant allele in the rs35672330 (EXO5) gene were more prone to Moderate-severe depression (OR=20.3, 95% CI, 4.1–99.8; p=4.90E-04, Table 4). Additionally, the combined effect of the rs7285167 (APOL2) and rs61731441 (C6orf165) variants was seen when they were tested by recoding genotypes as having none, one or two and more alleles at risk (Supplementary Figure S4).
Table 2.
Top-ranking associations of the common variants analysis regarding the performance of the Trail making test.
| Genotype | Case*, N (%) | Control*, N (%) | Model | Case*, N (%) | Control*, N (%) | P value** | OR [95%-CI] | P value adj*** |
|---|---|---|---|---|---|---|---|---|
|
AK8 rs17407084
| ||||||||
|
All cohort, N=229
| ||||||||
| TT | 11 (55.0) | 188 (90.0) | TT | 11 (55.0) | 188 (90.0) | 2.16E-04d | 7.3 [2.7–19.7] | 4.52E-04 |
| TC | 7 (35.0) | 21 (10.0) | TC+CC | 9 (45.0) | 21 (10.0) | |||
| CC | 2 (10.0) | 0 (0.0) | ||||||
|
ZNF382 rs61732180 | ||||||||
|
Males, N=112
| ||||||||
| CC | 3 (27.3) | 65 (64.4) | CC+CT | 6 (54.5) | 97 (96.0) | 3.58E-04r | 20.2 [4.3–95.4] | 2.62E-04 |
| CT | 3 (27.3) | 32 (31.6) | TT | 5 (45.5) | 4 (4.0) | |||
| TT | 5 (45.4) | 4 (4.0) | ||||||
|
AK8 rs17407084 | ||||||||
| Standard risk, N=105 | ||||||||
| TT | 2 (28.6) | 90 (91.8) | TT | 2 (28.6) | 90 (91.8) | 2.44E-04d | 28.1 [4.7–168.8] | 4.40E-04 |
| TC | 4 (57.1) | 8 (8.2) | TC+CC | 5 (71.4) | 8 (8.2) | |||
| CC | 1 (14.3) | 0 (0.0) | ||||||
AK8: Adenylate Kinase 8; ZNF: Zinc Finger Protein 382; FDR: false discovery rate; OR: odds ratio. TT indicates homozygosity for the T allele; TC represents heterozygosity, with one copy of the T allele and one copy of the C allele; CC indicates homozygosity for the C allele.
Participants with and without indicated complications are defined as cases and controls, respectively.
P values are calculated by chi-square or Fisher exact test, as appropriate. The most representative genetic model used is indicated (a: Additive; d: Dominant, r: Recessive).
P value adj: p value from logistic regression adjusted for age at diagnosis, sex, time since the end of treatment, protocol, and treatment variable (chemotherapy only or chemotherapy and radiotherapy).
Table 3.
Top-ranking associations between common variants and Moderate-severe anxiety.
| Genotype | Case*, N (%) | Control*, N (%) | Model | Case*, N (%) | Control*, N (%) | P value** | OR [95%-CI] | P value adj*** |
|---|---|---|---|---|---|---|---|---|
|
PTPRZ1 rs740965
| ||||||||
|
All cohort, N=200
| ||||||||
| TT | 10 (47.6) | 147 (82.1) | TT | 10 (47.6) | 147 (82.1) | 1.00E-03 | 5.1 [1.98–12.9] | 1.00E-03 |
| TG | 8 (38.1) | 31 (17.3) | TG+GG | 11 (52.4) | 32 (17.9) | |||
| GG | 3 (14.3) | 1 (0.6) | ||||||
|
MUC16 rs750295511 | ||||||||
|
All cohort, N=197
| ||||||||
| AA | 10 (50.0) | 158 (89.3) | AA | 10 (50.0) | 158 (89.3) | 7.20E-05 | 8.3 [3.1–22.5] | 3.10E-05 |
| TA | 10 (50.0) | 19 (10.7) | AT+TT | 10 (50.0) | 19 (10.7) | |||
| TT | 0 (0.0) | 0 (0.0) | ||||||
|
TNRC6C-AS1 rs2748431 | ||||||||
|
All cohort, N=167
| ||||||||
| GG | 12 (63.2) | 135 (91.2) | GG | 12 (63.2) | 135 (91.2) | 2.00E-03 | 6.1 [2.0–18.1] | 1.00E-03 |
| GA | 5 (26.3) | 11 (7.4) | GA+AA | 7 (36.8) | 13 (8.8) | |||
| AA | 2 (10.5) | 2 (1.4) | ||||||
|
APOL2 rs7285167 | ||||||||
|
Males only, N=95
| ||||||||
| GG | 5 (50.0) | 77 (90.6) | GG | 5 (50.0) | 77 (90.6) | 4.00E-03 | 9.6 [2.3–40.5] | 3.00E-03 |
| GA | 4 (40.0) | 8 (9.4) | GA+AA | 5 (50.0) | 8 (9.4) | |||
| AA | 1 (10.0) | 0 (0.0) | ||||||
|
C6orf165 rs61731441 | ||||||||
|
Males only, N=95
| ||||||||
| GG | 3 (30.0) | 70 (82.4) | GG | 3 (30.0) | 70 (82.4) | 1.00E-03 | 10.9 [2.5–47.0] | 2.00E-03 |
| GA | 6 (60.0) | 15 (17.6) | GA+AA | 7 (70.0) | 15 (17.6) | |||
| AA | 1 (10.0) | 0 (0.0) | ||||||
PTPRZ1: Protein Tyrosine Phosphatase Receptor Type Z1; MUC16: Mucin 16, Cell Surface Associated; TNRC6C-AS1: TNRC6C antisense RNA 1; APOL2: Apolipoprotein L2; C6orf165: Cilia And Flagella Associated Protein 206; FDR: false discovery rate; OR: odds ratio. TT indicates homozygosity for the T allele; TG represents heterozygosity, with one copy of the T allele and one copy of the G allele; GG indicates homozygosity for the G allele; AA indicates homozygosity for the A allele; TA represents heterozygosity, with one copy of the T allele and one copy of the A allele; GA represents heterozygosity, with one copy of the A allele and one copy of the G allele.
Participants with and without indicated complications are defined as cases and controls, respectively.
P values are calculated by chi-square or Fisher exact test, as appropriate. Given the low frequency of homozygotes for minor alleles, genetic model in all cases was dominant.
P value adj: p value from logistic regression adjusted for age at diagnosis, sex, time since the end of treatment, protocol, and treatment variable (chemotherapy only or chemotherapy and radiotherapy).
Table 4.
Top-ranking associations between common variants and Moderate-severe depression.
| Genotype | Case*, N (%) | Control*, N (%) | Model | Case*, N (%) | Control*, N (%) | P value** | OR [95%-CI] | P value adj*** |
|---|---|---|---|---|---|---|---|---|
|
EXO5 rs35672330
| ||||||||
|
Males only, N=94
| ||||||||
| TT | 4 (44.4) | 81(94.2) | TT | 4 (44.4) | 81(94.2) | 4.52E-04d | 20.3 [4.1–99.8] | 4.90E-04 |
| TC | 4 (44.4) | 5 (5.8) | TC+CC | 5 (55.6) | 5 (5.8) | |||
| CC | 1 (11.2) | 0 (0.0) | ||||||
EXO5: Exonuclease 5; FDR: false discovery rate; OR: odds ratio. TT indicates homozygosity for the T allele; TC represents heterozygosity, with one copy of the T allele and one copy of the C allele; CC indicates homozygosity for the C allele.
Participants with and without indicated complications are defined as cases and controls, respectively.
P values are calculated by chi-square or Fisher exact test, as appropriate. Given the low frequency of homozygotes for minor alleles, genetic model in all cases was dominant.
P value adj: p value from logistic regression adjusted for age at diagnosis, sex, time since the end of treatment, protocol, and treatment variable (chemotherapy only or chemotherapy and radiotherapy).
We did not find any significant common variant association that would satisfy multiple testing adjustments when performing an exome-wide association with the remaining neurocognitive phenotypes either in entire group or following stratification.
Replication results.
Demographics and clinical characteristics of participants from the replication SJLIFE cohort are presented in Supplementary Table S8. Of the eight common variant associations that remained statistically significant in the discovery cohort using multivariable regression models, seven variants passed SJLIFE quality control (except for a variant rs750295511 in MUC16) and were further analyzed for an association using the respective phenotype measures in the independent cohort of ALL survivors in the SJLIFE cohort (Tables 5). An association between deficit in the trail making test performance and the minor allele of ZNF382 rs61732180 was not observed in males, however, it was seen in all survivors (OR=1.4; 95% CI, 1.04–1.90; p=0.025) with statistical significance in both allelic Fisher’s exact test and adjusted logistic regression analyses (Supplementary Table S9). Additionally, variant rs61732180 in the ZNF382 gene remained statistically significant in male participants of the combined discovery and replication set when assessed through meta-analysis (Figure 1a).
Table 5.
Replication results, SJLIFE cohort, N=688*.
| Gene | rs number | EAF | cases | controls | Allelic Fisher exact test | Logistic regression** | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| n.2 | n.1 | n.0 | n.2 | n.1 | n.0 | OR [95%CI] | P value | OR [95%CI] | LRT P value | |||
|
Trail making test
| ||||||||||||
|
All cohort, N= 675
*
| ||||||||||||
| AK8 | rs17407084 | 0.05 | 0 | 12 | 140 | 1 | 52 | 470 | 0.76 [0.36–1.45] | 0.45 | 0.75 [0.36–1.44] | 0.39 |
|
Males only, N= 337 | ||||||||||||
| ZNF382 | rs61732180 | 0.26 | 7 | 36 | 40 | 14 | 94 | 146 | 1.36 [0.90–2.04] | 0.13 | 1.40 [0.92– 2.15] | 0.12 |
|
Moderate-severe anxiety | ||||||||||||
|
All cohort, N= 675
*
| ||||||||||||
| PTPRZ1 | rs740965 | 0.15 | 2 | 20 | 45 | 13 | 150 | 445 | 1.29 [0.77–2.09] | 0.31 | 1.52 [0.90–2.50] | 0.11 |
| TNRC6C-AS1 | rs2748431 | 0.07 | 0 | 9 | 58 | 5 | 77 | 526 | 0.93 [0.40–1.92] | 1.00 | 0.86 [0.38–1.70] | 0.67 |
|
Males only, N= 340 | ||||||||||||
| APOL2 | rs7285167 | 0.08 | 0 | 2 | 32 | 0 | 52 | 254 | 0.33 [0.04–1.29] | 0.15 | 0.24 [0.04–0.94] | 0.040 |
| C6orf165 | rs61731441 | 0.1 | 1 | 3 | 30 | 1 | 61 | 244 | 0.69 [0.21–1.80] | 0.53 | 0.71 [0.23–1.83] | 0.50 |
|
Moderate-severe depression | ||||||||||||
|
Males only, N= 339
| ||||||||||||
| EXO5 | rs35672330 | 0.06 | 0 | 10 | 45 | 0 | 33 | 251 | 1.62 [0.69–3.50] | 0.20 | 2.18 [0.91–4.97] | 0.08 |
AK8: Adenylate Kinase 8; ZNF: Zinc Finger Protein 382; PTPRZ1: Protein Tyrosine Phosphatase Receptor Type Z1; TNRC6C-AS1: TNRC6C antisense RNA 1; APOL2: Apolipoprotein L2; C6orf165: Cilia And Flagella Associated Protein 206; EXO5: Exonuclease 5.
EAF: Effect allele frequency; n.2: 2 copies of the effect allele; n.1: 1 copy of the effect allele; n.0: 0 copies of the effect allele; OR: odds ratio for each additional copy of the effect allele; CI: confidence interval; LRT P value: Likelihood ratio test P value.
While the total replication SJLIFE cohort consisted of 688 participants, it’s noteworthy that when participants with non-missing data were selected for the Trail making test and Moderate-severe anxiety regressions, the number included happened to be identical, with 675 survivors in each analysis. However, these participants were not the exact same survivors in both analyses.
Logistic regression analyses adjusted for continuous age at diagnosis, sex, continuous time between date of becoming a 5-year ALL survivor and date of test measurement, whether the survivor was treated with chemotherapy only versus chemotherapy plus radiation, and the top 20 principal components adjusting for genetic ancestry.
Figure 1.
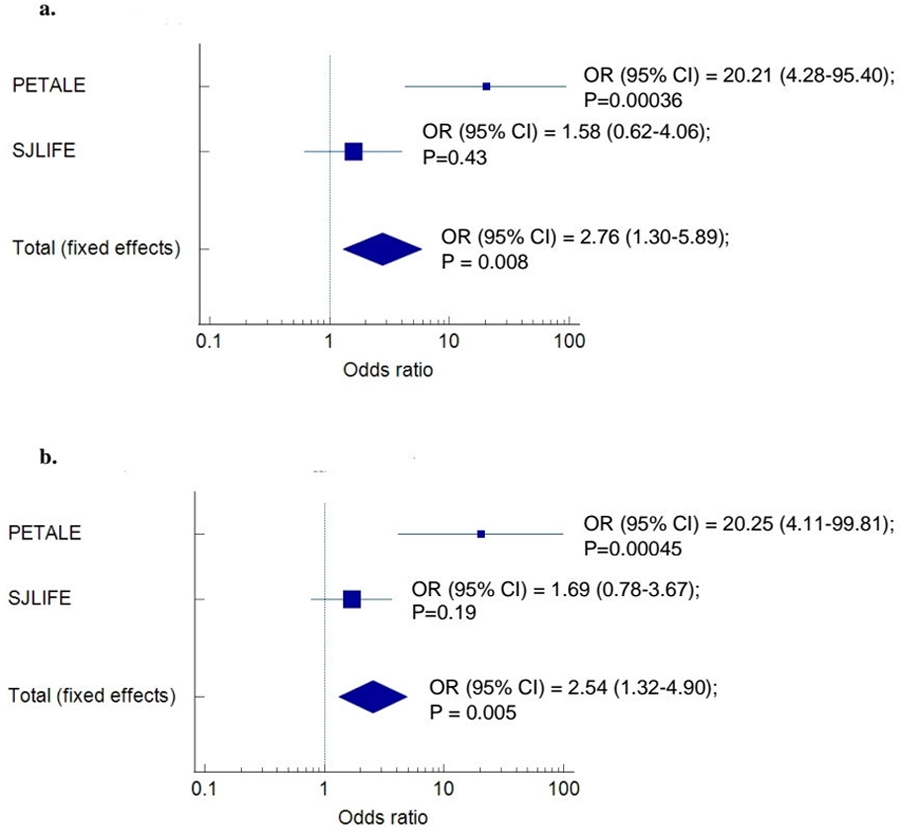
Meta-analysis of the effect of the variant rs61732180 ZNF382 on the neurocognitive deficit (a) and rs35672330 EXO5 on moderate-severe depression (b) in male participants of combined discovery and replication set.
Plot represents the associations in the discovery cohort (PETALE), the replication cohort (SJLIFE) and the combined cohort (Total). Odd ratios (OR) comparing carriers to non-carriers, along with the 95% confidence intervals (95% CI) and the p-values of the associations are provided next to the combined patient set.
An allelic association between moderate-severe anxiety and APOL2 rs7285167 was seen with p-value<0.05 in the male-restricted adjusted logistic regression analysis and the full SJLIFE cohort allelic and adjusted logistic regression analyses (Table 5 and Supplementary Table S9); however, it had the opposite effect and therefore cannot be considered as replicated. In addition, although an association between moderate-severe depression and the minor allele of EXO5 rs35672330 was not observed in males, the effect of this allele was detected in the pooled discovery and replication cohort through the meta-analysis (Figure 1b).
Rare variants
The analysis of functionally predicted rare variants in PETALE cohort led to the detection of the associations between the moderate-severe anxiety and rare variants in the PCMTD1 and CYP2W1 genes in the HR patients (p=9.4E-6 and p=1.3E-5, respectively, Table 6). Using the collapsing approach, we explored variant combinations that contributed to the observed association signal, consequently identifying two variants in PCMTD1 (rs201786115 and rs200377849), and one in CYP2W1 (rs3735684). No significant association was obtained for moderate-severe depression and neurocognitive outcomes through the rare variants’ analysis.
Table 6.
Top-ranking associations of the rare variants identified through the SKAT-O test regarding the Moderate-severe anxiety in HR patients, N=124.
| Gene | SNVs tested |
MAF | P value SKAT-O | FDR-BH | 11/12+22 | 11 | 12+22 | P value Fisher test | OR [95% CI] | ||
|---|---|---|---|---|---|---|---|---|---|---|---|
| position (hg19) | rs number | ||||||||||
| PCMTD1 | chr8:52732981 * | rs201786115 | 0.013 | 9.4E-06 | 0.03 | 21.5 (4.4–105.9) * | |||||
| chr8:52733110 | 0.008 | ||||||||||
| chr8:52733164 | rs149898988 | 0.021 | Significant combination of collapsed variants ** | ||||||||
| chr8:52733209 |
rs202074278 |
0.022 |
91/9 | Affected |
5 (50.0%) |
5 (50.0%) |
4E-04 | ||||
| chr8:52733214 * | rs200377849 | 0.025 | Unaffected | 86 (95.6%) | 4 (4.4%) | ||||||
| chr8:52733227 * | 0.016 | ||||||||||
|
| |||||||||||
| CYP2W1 |
chr7:1024855
*
|
rs3735684 |
0.038 | 1.3E-05 | 0.03 |
Individual contribution
*
|
21.9 (4.2–114.8)
*
|
||||
| chr7:1024874 | 0.005 | 85/8 | Affected |
6 (54.5%) |
5 (45.5%) |
4E-04 | |||||
| chr7:1024921 | 0.005 | Unaffected | 79 (96.3%) | 3 (3.7%) | |||||||
PCMTD1: Protein-L-Isoaspartate (D-Aspartate) O-Methyltransferase Domain Containing 1; CYP2W1: Cytochrome P450 Family 2 Subfamily W Member 1; SNV: single nucleotide variation; MAF: minor allele frequency; FDR-BH: Benjamini–Hochberg false discovery rate; OR: odds ratio; CI: confidence interval.
SNVs that are identified as the most important contributors to the association signal are highlighted.
Collapsed variants (carriers of at least one of rare variants were included into the model, variants with missing values were excluded.
Genotypes were recoded as follows:11-homozygote wild type; 12-heterozygote variant; 22-homozygote variant.
The detailed description of common and rare significant variants, including rs identifiers and their functional implication, is provided in Supplementary Table S10.
DISCUSSION
Neurocognitive function
In our study, functionally predicted germline common variants in the AK8 and ZNF382 genes were found to be significantly associated with deficits in the performance of the trail making test in PETALE participants. The association with the AK8 gene was detected when the entire cohort of survivors was analyzed and also in the standard-risk group, whereas that of ZNF382 was context-dependent, and was detected upon sex stratification. The detailed description of the AK8 gene function is provided in Supplementary Material (Supplementary Material S1).
Although the male specific ZNF382 association was not observed in males alone, it was detected within the entire replication cohort. Zinc finger protein 382, encoded by the ZNF382 gene, is a member of the largest family of transcriptional regulators - Krüppel-associated box domain (KRAB) zinc finger proteins(67). It plays critical roles as a transcription inhibitor and has been suggested to be a tumor suppressor in various types of human cancer, including pediatric acute myeloid leukemia(68, 69). Interestingly, ZNF382 inhibits the activating protein 1 (AP-1) and nuclear factor kappa-B (NF-kB) signaling. NF-κB involvement was detected in the different categories of neurons including both excitatory (glutamatergic) and inhibitory (GABAergic), as well as in the neural sub-compartment of the synapse; thus suggesting that neuronal NF-κB signaling pathway functions under normal physiological conditions to promote synaptic growth and to improve synaptic activity and long-lasting forms of plasticity(70). Moreover, its activation by excitatory neurotransmission and participation in multiple forms of structural and synaptic plasticity is probably at the basis of the function of this transcription factor in cognitive behaviors(70). On the other hand, NF-κB, a key regulator of innate immunity, is over-activated in a number of neurodegenerative diseases, including Alzheimer’s disease(71). Although the male-specific association between the deficit score in neurocognitive test performance and the minor rs61732180 ZNF382 allele identified in our study was not observed in males from the SJLIFE replication cohort, a p-value<0.05 was obtained for the full SJLIFE population and in males of the pooled discovery and replication cohorts. Therefore, the involvement of the ZNF382 gene in neurocognitive function may warrant further investigation since the genetic variant might potentially play a role in the outcome regardless of gender.
Сhildhood ALL patients who have received CNS-directed chemotherapy demonstrate persistent and significant neurocognitive impairment that manifests after treatment(5, 72). Specifically, treatment of ALL may result in smaller gray and white matter volumes as well as alterations in white matter microstructure, often associated with decreased neurocognitive performance(73). Our previous work identified, using a candidate gene approach, a panel of several genes that showed an effect on neurocognitive decline in the PETALE cohort. Two variants, rs1805087 in the MTR gene and rs58225473 in the CACNB2 gene, deserve mention as these associations were confirmed in the independent SJLIFE replication cohort.
It is worth noting that standard-risk childhood ALL patients typically receive less intensive treatment than high-risk patients(4, 74, 75) and the modulation of the genetic association by the treatment might be more obvious in the high-risk group. On the other hand, some of the drug doses did not differ between the two groups(74, 76). Moreover, epidemiological and neuroimaging results indicate that ALL survivors who received contemporary therapeutic protocols (which consist of intensified intravenous and intrathecal administration of chemotherapeutic drugs for standard risk patients(74, 77)) were still at risk of neurocognitive problems(4).
Interestingly, genes identified in the present study through a hypothesis-free approach regarding the impaired neurocognitive function are associated to varying degrees with oxidative stress (AK8 gene) or with immune regulation (ZNF382 gene); and all of them, in one way or another, are also related to the function of the CNS. There is a lot of evidence to suggest that the brain is very susceptible to oxidative damage due to its high metabolic demand(78, 79). The oxidative stress, which is related to elevated intracellular levels of reactive oxygen species (ROS) is a key mediator of neuroinflammation, metabolic changes, bioenergetic deficiency, and neuronal apoptosis (80). ROS generated during chemotherapy may be associated with various harmful events, including neurotoxicity(81). For example, methotrexate (MTX) promotes oxidative stress in several organs, including the brain(81). In addition, MTX inhibits the activation of NF-κB, the already mentioned protein complex, which, among other functions, plays a central role in DNA transcription and the regulation of inflammation(82, 83).
Anxiety and Depression
We found that functionally predicted germline common variants in the PTPRZ1, MUC16, TNRC6C-AS1, APOL2, and C6orf165 genes were significantly associated with moderate-severe anxiety in ALL survivors; whereas the EXO5 gene was associated with the increased risk of moderate-severe depression. Noteworthy, most of the associations were sex-specific and were detected in males, which may be partly explained by the fact that self-reported anxiety/depression includes more social variance in females than in males(84). Additionally, an association was found between the moderate-severe anxiety in HR patients and rare variants enrichment in the PCMTD1 and CYP2W1 genes. The EXO5 association deserves special mention since its effect was also seen in the pooled discovery and replication cohort.
EXO5 (Exonuclease 5) is a single-stranded DNA-specific bidirectional exonuclease that functions in the repair of nuclear DNA(85). In a recent study, the EXO5 gene was identified as a risk gene involved in prostate tumorigenesis(86). Although there is no data available on the potential involvement of EXO5 in mood disorders or CNS function, it is interesting to note that the effect of the EXO5 gene detected in our cohort was also gender-specific and has been identified in male individuals.
The detailed description of the gene functions of the remaining associations is provided in Supplementary Material (Supplementary Material S1).
Emotional challenges that arise from diagnosis and treatment of cancer in childhood can be seen as a highly traumatic event and can have long-term consequences. There is growing evidence that exposure to psychological distress at an early age can seriously affect brain maturation and development(87); in addition, childhood stress can increase vulnerability to later development of mental disorders, such as depression and anxiety(88). Particularly, during peri-adolescence, brain areas critical for emotional regulation, such as the prefrontal cortex, hippocampus, and amygdala are still developing and are highly sensitive to stress(87). In a similar stressful environment, individuals will respond to stress differently as only part of them will demonstrate vulnerability, while others will remain resilient(30, 89).
The loci reported in the current study have not previously been identified as potential risk predictors of anxiety and/or depression susceptibility, however, given their important biological role, may warrant further study. In addition, the significant combined effect demonstrated in our study indicates that a single marker and/or single genotype may not be sufficient to explain the etiology of psychological distress phenotypes, given their complexity and environmental influences.
We acknowledge that our study has certain limitations. For example, limited sample size may affect the accuracy of the results, since the power of the study and magnitude of the effect might be affected in the context of a stratified analysis. Some phenotypes, such as trail making test, appear to occur with the similar frequency as in the general population, but the cause of their development, including genetic predisposition, may differ between patients exposed to the treatment and untreated individuals(90). At the same time, it is also important to note that survivors may tend to systematically report lower or normal distress rates as a result of a tendency to over normalize their situation. If this is the case in the present group, then it is likely that those with moderate-severe levels experience feelings to an even more significant degree. Thus, despite the small number of affected individuals in the study group it is nevertheless legitimate to explore why some individuals are more vulnerable. The association results obtained for rare variants in the discovery cohort (not evaluated in the SJLIFE cohort) should be taken with caution given their low number. Most of the common variant associations found in the PETALE cohort were not replicated in the SJLIFE cohort. This can be explained by several reasons. Mainly, despite the use of similar inclusion/exclusion criteria and the use of similar outcomes between the two cohorts, it is possible that the small sample sizes in both cohorts, differences in treatment protocols, lack of evaluation according to risk groups and/or timing between ALL diagnosis and evaluation contributed to the observed discrepancies. Finally, we can not disregard the possibility that some of the associations observed in the discovery cohort could have been obtained by chance.
In conclusion, using WES data and a hypothesis-free approach, we identified several genes as potential modulators of the risk of developing treatment-related neurocognitive complications, as well as anxiety and depression. The association between deficit in the trail making test and variant rs61732180 in the ZNF382 and rs35672330 in EXO5 genes are of particular interest since associations were also found in the replication cohort or meta-analysis of discovery and replication cohorts.
Multiple evidence has been collected nowadays for potential genetic and epigenetic risk markers of the long-term treatment-related neurocognitive and emotional complications in survivors of childhood cancer. In addition, accumulating data suggest that genetic factors contribute significantly to resilient responses to trauma and stress(91). Large genome-wide association studies on the genetic architecture of mental disorders indicate its polygenic nature(92–94). Therefore, future studies will be required not only to verify current results, but also for multilevel integration of several approaches including polygenic score models along with other non-genetic factors, in order to identify markers of neurocognitive and emotional disorders and implement them into clinical practice.
Supplementary Material
ACKNOWLEDGEMENTS
Funding
Institute of Cancer Research (ICR) of the Canadian Institutes of Health Research (CIHR) grant number: 118694 awarded to Dr Daniel Sinnett, Dr Maja Krajinovic, Dr Caroline Laverdière, Dr Philippe Robaey, Dr Sarah Lippé, Dr Serge Sultan (http://www.cihr-irsc.gc.ca); C17 Council, Cancer Research Society (CRS), Canadian Cancer Society Research Institute (CCSRI), Ontario Institute for Cancer Research (OICR), Pediatric Oncology Group of Ontario (POGO), Garron Family Cancer Centre at SickKids Hospital (Ontario), The Terry Fox foundation, grant number: TFF - 105266 awarded to Dr Daniel Sinnett and Dr Maja Krajinovic (https://www.terryfox.org/), FRQS Applied Medical Genetics Network and Sainte-Justine Hospital Foundation supported the PETALE study. Kateryna Petrykey is a scholar of the Cole Foundation and the Université de Montréal. This work was supported by the National Cancer Institute, the Cancer Center Support grant [5P30CA021765-33], the St. Jude Lifetime Cohort Study Grant [U01CA195547], and the American Lebanese Syrian Associated Charities. Registered at Clinicaltrials.gov (#NCT00760656).
The authors would like to thank all childhood ALL survivors and their parents for the participation in the PETALE study, as well as all study collaborators for their valuable contribution.
We would like to thank Dr. Kevin R. Krull from the St. Jude Children’s Research Hospital for a critical review of the manuscript. We also thank Aziz Rezgui for his contributions to patient data processing for the PETALE cohort.
LIST OF ABBREVIATIONS
- ALL
Acute lymphoblastic leukemia
- PETALE
Prévenir les Effets tardifs des Traitements de la leucémie Aiguë Lymphoblastique chez l’Enfant
- SJUHC
Sainte-Justine University Health Center
- SJLIFE
St. Jude Lifetime Cohort
- WES
Whole exome sequencing
- DFCI
Dana-Faber Cancer Institute
- SJLIFE
St-Jude Lifetime cohort
- D-KEFS
Delis-Kaplan Executive Function System
- WAIS-IV
Wechsler Adult Intelligence Scale-Fourth Edition
- BYI
Beck Youth Inventory
- BSI-18
Brief Symptom Inventory-18
- PROMIS
Patient-Reported Outcomes Measurement Information System
- MAF
Minor allele frequency
- EAF
Effect allele frequency
- SKAT-O test
Optimal Sequence Kernel Association Test
- FDR
False discovery rate
- SR
standard risk
- HR
high risk
- OR
odds ratio
- CI
confidence interval
- CNS
central nervous system
- NF-kB
nuclear factor kappa-B
- ROS
reactive oxygen species
- ZNF382
Zinc finger protein 382
- EXO5
Exonuclease 5
- AK8
Adenylate Kinase 8 ATP-AMP Transphosphorylase 8
- KIR3DL1
Killer Cell Immunoglobulin Like Receptor
- PPARG
Peroxisome Proliferator Activated Receptor Gamma
- PLAUR
Urokinase Plasminogen Activator Surface Receptor
- DUOXA1
Dual Oxidase Maturation Factor 1
- DUOX2
Dual Oxidase 2
- PTPRZ1
Protein Tyrosine Phosphatase Receptor Type Z1
- MUC16
Mucin 16, Cell Surface Associated
- TNRC6C-AS1
TNRC6C antisense RNA 1
- MICB
MHC Class I Polypeptide-Related Sequence B
- ARL16
ADP Ribosylation Factor Like GTPase 16
- APOL2
Apoliprotein L2
- C6orf165
Cilia And Flagella Associated Protein 206
Footnotes
The authors declare no potential conflicts of interest.
REFERENCES
- 1.Gaynon PS, Angiolillo AL, Carroll WL, Nachman JB, Trigg ME, Sather HN, et al. Long-term results of the children’s cancer group studies for childhood acute lymphoblastic leukemia 1983–2002: a Children’s Oncology Group Report. Leukemia 2010;24(2):285–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Salzer WL, Devidas M, Carroll WL, Winick N, Pullen J, Hunger SP, et al. Long-term results of the pediatric oncology group studies for childhood acute lymphoblastic leukemia 1984–2001: a report from the children’s oncology group. Leukemia 2010;24(2):355–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bhojwani D, Yang JJ, Pui CH. Biology of childhood acute lymphoblastic leukemia. Pediatr Clin North Am 2015;62(1):47–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cheung YT, Krull KR. Neurocognitive outcomes in long-term survivors of childhood acute lymphoblastic leukemia treated on contemporary treatment protocols: A systematic review. Neurosci Biobehav Rev 2015;53:108–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Krull KR, Brinkman TM, Li C, Armstrong GT, Ness KK, Srivastava DK, et al. Neurocognitive outcomes decades after treatment for childhood acute lymphoblastic leukemia: a report from the St Jude lifetime cohort study. J Clin Oncol 2013;31(35):4407–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jacola LM, Edelstein K, Liu W, Pui CH, Hayashi R, Kadan-Lottick NS, et al. Cognitive, behaviour, and academic functioning in adolescent and young adult survivors of childhood acute lymphoblastic leukaemia: a report from the Childhood Cancer Survivor Study. Lancet Psychiatry 2016;3(10):965–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zeltzer LK, Chen E, Weiss R, Guo MD, Robison LL, Meadows AT, et al. Comparison of psychologic outcome in adult survivors of childhood acute lymphoblastic leukemia versus sibling controls: a cooperative Children’s Cancer Group and National Institutes of Health study. J Clin Oncol 1997;15(2):547–56. [DOI] [PubMed] [Google Scholar]
- 8.Giedd JN, Blumenthal J, Jeffries NO, Castellanos FX, Liu H, Zijdenbos A, et al. Brain development during childhood and adolescence: a longitudinal MRI study. Nature Neuroscience 1999;2(10):861–3. [DOI] [PubMed] [Google Scholar]
- 9.Lenroot RK, Giedd JN. Brain development in children and adolescents: insights from anatomical magnetic resonance imaging. Neurosci Biobehav Rev 2006;30(6):718–29. [DOI] [PubMed] [Google Scholar]
- 10.Konrad K, Firk C, Uhlhaas PJ. Brain development during adolescence: neuroscientific insights into this developmental period. Dtsch Arztebl Int 2013;110(25):425–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pääkkö E, Vainionpää L, Lanning M, Laitinen J, Pyhtinen J. White matter changes in children treated for acute lymphoblastic leukemia. Cancer 1992;70(11):2728–33. [DOI] [PubMed] [Google Scholar]
- 12.Vázquez E, Lucaya J, Castellote A, Piqueras J, Sainz P, Olivé T, et al. Neuroimaging in Pediatric Leukemia and Lymphoma: Differential Diagnosis. RadioGraphics 2002;22(6):1411–28. [DOI] [PubMed] [Google Scholar]
- 13.Campbell LK, Scaduto M, Sharp W, Dufton L, Van Slyke D, Whitlock JA, et al. A meta-analysis of the neurocognitive sequelae of treatment for childhood acute lymphocytic leukemia. Pediatric Blood & Cancer 2007;49(1):65–73. [DOI] [PubMed] [Google Scholar]
- 14.Reddick WE, Shan ZY, Glass JO, Helton S, Xiong X, Wu S, et al. Smaller white-matter volumes are associated with larger deficits in attention and learning among long-term survivors of acute lymphoblastic leukemia. Cancer 2006;106(4):941–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Campbell LK, Scaduto M, Sharp W, Dufton L, Van Slyke D, Whitlock JA, et al. A meta-analysis of the neurocognitive sequelae of treatment for childhood acute lymphocytic leukemia. Pediatr Blood Cancer 2007;49(1):65–73. [DOI] [PubMed] [Google Scholar]
- 16.Iyer NS, Balsamo LM, Bracken MB, Kadan-Lottick NS. Chemotherapy-only treatment effects on long-term neurocognitive functioning in childhood ALL survivors: a review and meta-analysis. Blood 2015;126(3):346–53. [DOI] [PubMed] [Google Scholar]
- 17.Ashford J, Schoffstall C, Reddick WE, Leone C, Laningham FH, Glass JO, et al. Attention and working memory abilities in children treated for acute lymphoblastic leukemia. Cancer 2010;116(19):4638–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kingma A, van Dommelen RI, Mooyaart EL, Wilmink JT, Deelman BG, Kamps WA. Slight cognitive impairment and magnetic resonance imaging abnormalities but normal school levels in children treated for acute lymphoblastic leukemia with chemotherapy only. J Pediatr 2001;139(3):413–20. [DOI] [PubMed] [Google Scholar]
- 19.Lofstad GE, Reinfjell T, Hestad K, Diseth TH. Cognitive outcome in children and adolescents treated for acute lymphoblastic leukaemia with chemotherapy only. Acta Paediatr 2009;98(1):180–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rodgers J, Marckus R, Kearns P, Windebank K. Attentional ability among survivors of leukaemia treated without cranial irradiation. Arch Dis Child 2003;88(2):147–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Daams M, Schuitema I, van Dijk BW, van Dulmen-den Broeder E, Veerman AJ, van den Bos C, et al. Long-term effects of cranial irradiation and intrathecal chemotherapy in treatment of childhood leukemia: a MEG study of power spectrum and correlated cognitive dysfunction. BMC Neurol 2012;12:84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Edelmann MN, Krull KR, Liu W, Glass JO, Ji Q, Ogg RJ, et al. Diffusion tensor imaging and neurocognition in survivors of childhood acute lymphoblastic leukaemia. Brain 2014;137(Pt 11):2973–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mennes M, Stiers P, Vandenbussche E, Vercruysse G, Uyttebroeck A, De Meyer G, et al. Attention and information processing in survivors of childhood acute lymphoblastic leukemia treated with chemotherapy only. Pediatr Blood Cancer 2005;44(5):478–86. [DOI] [PubMed] [Google Scholar]
- 24.Carey ME, Haut MW, Reminger SL, Hutter JJ, Theilmann R, Kaemingk KL. Reduced frontal white matter volume in long-term childhood leukemia survivors: a voxel-based morphometry study. AJNR Am J Neuroradiol 2008;29(4):792–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schultz KA, Ness KK, Whitton J, Recklitis C, Zebrack B, Robison LL, et al. Behavioral and social outcomes in adolescent survivors of childhood cancer: a report from the childhood cancer survivor study. J Clin Oncol 2007;25(24):3649–56. [DOI] [PubMed] [Google Scholar]
- 26.Zeltzer LK, Recklitis C, Buchbinder D, Zebrack B, Casillas J, Tsao JC, et al. Psychological status in childhood cancer survivors: a report from the Childhood Cancer Survivor Study. J Clin Oncol 2009;27(14):2396–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Michel G, Rebholz CE, von der Weid NX, Bergstraesser E, Kuehni CE. Psychological distress in adult survivors of childhood cancer: the Swiss Childhood Cancer Survivor study. J Clin Oncol 2010;28(10):1740–8. [DOI] [PubMed] [Google Scholar]
- 28.Stuber ML, Meeske KA, Krull KR, Leisenring W, Stratton K, Kazak AE, et al. Prevalence and predictors of posttraumatic stress disorder in adult survivors of childhood cancer. Pediatrics 2010;125(5):e1124–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Baytan B, Asut C, Cirpan Kantarcioglu A, Sezgin Evim M, Gunes AM. Health-Related Quality of Life, Depression, Anxiety, and Self-Image in Acute Lymphocytic Leukemia Survivors. Turk J Haematol 2016;33(4):326–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pepin AJ, Lippe S, Krajinovic M, Laverdiere C, Michon B, Sinnett D, et al. How to interpret high levels of distress when using the Distress Thermometer in the long-term follow-up clinic? A study with Acute Lymphoblastic Leukemia survivors. Pediatr Hematol Oncol 2017;34(3):133–7. [DOI] [PubMed] [Google Scholar]
- 31.Oancea SC, Brinkman TM, Ness KK, Krull KR, Smith WA, Srivastava DK, et al. Emotional distress among adult survivors of childhood cancer. J Cancer Surviv 2014;8(2):293–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jain N, Brouwers P, Okcu MF, Cirino PT, Krull KR. Sex-specific attention problems in long-term survivors of pediatric acute lymphoblastic leukemia. Cancer 2009;115(18):4238–45. [DOI] [PubMed] [Google Scholar]
- 33.Petrykey K, Lippé S, Robaey P, Sultan S, Laniel J, Drouin S, et al. Influence of genetic factors on long-term treatment related neurocognitive complications, and on anxiety and depression in survivors of childhood acute lymphoblastic leukemia: The Petale study. PLoS One 2019;14(6):e0217314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Marcoux S, Drouin S, Laverdiere C, Alos N, Andelfinger GU, Bertout L, et al. The PETALE study: Late adverse effects and biomarkers in childhood acute lymphoblastic leukemia survivors. Pediatr Blood Cancer 2017;64(6). [DOI] [PubMed] [Google Scholar]
- 35.Moghrabi A, Levy DE, Asselin B, Barr R, Clavell L, Hurwitz C, et al. Results of the Dana-Farber Cancer Institute ALL Consortium Protocol 95–01 for children with acute lymphoblastic leukemia. Blood 2007;109(3):896–904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Silverman LB, Stevenson KE, O’Brien JE, Asselin BL, Barr RD, Clavell L, et al. Long-term results of Dana-Farber Cancer Institute ALL Consortium protocols for children with newly diagnosed acute lymphoblastic leukemia (1985–2000). Leukemia 2010;24(2):320–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Delis DC, Kaplan E, & Kramer JH Delis-Kaplan Executive Function System (D-KEFS). APA PsycTests; 2001. [Google Scholar]
- 38.Wechsler D Wechsler Adult Intelligence Scale--Fourth Edition (WAIS-IV). APA PsycTests; 2008. [Google Scholar]
- 39.Petrykey K, Lippe S, Robaey P, Sultan S, Laniel J, Drouin S, et al. Influence of genetic factors on long-term treatment related neurocognitive complications, and on anxiety and depression in survivors of childhood acute lymphoblastic leukemia: the Petale study. PloS one 2019;14(6):e0217314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Harrison AG, Armstrong IT, Harrison LE, Lange RT, Iverson GL. Comparing Canadian and American normative scores on the Wechsler Adult Intelligence Scale-Fourth Edition. Arch Clin Neuropsychol 2014;29(8):737–46. [DOI] [PubMed] [Google Scholar]
- 41.Boulet-Craig A, Robaey P, Laniel J, Bertout L, Drouin S, Krajinovic M, et al. DIVERGT screening procedure predicts general cognitive functioning in adult long-term survivors of pediatric acute lymphoblastic leukemia: A PETALE study. Pediatr Blood Cancer 2018;65(9):e27259. [DOI] [PubMed] [Google Scholar]
- 42.Beck JSBA, Jolly JB, Steer RA. Beck Youth Inventories-Second Edition for children and adolescent’s manual San Antonio, TX: PsychCorp; 2005. [Google Scholar]
- 43.Derogatis LR. Brief Symptom Inventory (BSI) 18: Administration, scoring, and procedures manual Minneapolis, MN: NCS Pearson; 2000. [Google Scholar]
- 44.Streiner DL. Starting at the beginning: an introduction to coefficient alpha and internal consistency. J Pers Assess 2003;80(1):99–103. [DOI] [PubMed] [Google Scholar]
- 45.Petrykey K, Rezgui AM, Le Guern M, Beaulieu P, St-Onge P, Drouin S, et al. Genetic factors in treatment-related cardiovascular complications in survivors of childhood acute lymphoblastic leukemia. Pharmacogenomics 2021;22(14):885–901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kumar P, Henikoff S, Ng PC. Predicting the effects of coding non-synonymous variants on protein function using the SIFT algorithm. Nat Protoc 2009;4(7):1073–81. [DOI] [PubMed] [Google Scholar]
- 47.Ng PC, Henikoff S. Predicting the effects of amino acid substitutions on protein function. Annu Rev Genomics Hum Genet 2006;7:61–80. [DOI] [PubMed] [Google Scholar]
- 48.Auton A, Abecasis GR, Altshuler DM, Durbin RM, Abecasis GR, Bentley DR, et al. A global reference for human genetic variation. Nature 2015;526(7571):68–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rohlfs RV, Weir BS. Distributions of Hardy-Weinberg equilibrium test statistics. Genetics 2008;180(3):1609–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Fadista J, Manning AK, Florez JC, Groop L. The (in)famous GWAS P-value threshold revisited and updated for low-frequency variants. European Journal of Human Genetics 2016;24(8):1202–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sham PC, Purcell SM. Statistical power and significance testing in large-scale genetic studies. Nature Reviews Genetics 2014;15(5):335–46. [DOI] [PubMed] [Google Scholar]
- 52.Storey JD, Tibshirani R. Statistical significance for genomewide studies. Proc Natl Acad Sci U S A 2003;100(16):9440–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Benjamini Y, Drai D, Elmer G, Kafkafi N, Golani I. Controlling the false discovery rate in behavior genetics research. Behav Brain Res 2001;125(1–2):279–84. [DOI] [PubMed] [Google Scholar]
- 54.Benjamini Y, Yekutieli D. Quantitative trait Loci analysis using the false discovery rate. Genetics 2005;171(2):783–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Whole genome association analysis toolset [Available from: http://zzz.bwh.harvard.edu/plink/index.shtml.
- 56.Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet 2007;81(3):559–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Buizer AI, de Sonneville LM, van den Heuvel-Eibrink MM, Veerman AJ. Chemotherapy and attentional dysfunction in survivors of childhood acute lymphoblastic leukemia: effect of treatment intensity. Pediatr Blood Cancer 2005;45(3):281–90. [DOI] [PubMed] [Google Scholar]
- 58.Waber DP, Tarbell NJ, Kahn CM, Gelber RD, Sallan SE. The relationship of sex and treatment modality to neuropsychologic outcome in childhood acute lymphoblastic leukemia. J Clin Oncol 1992;10(5):810–7. [DOI] [PubMed] [Google Scholar]
- 59.Lee S, Abecasis Gonçalo R, Boehnke M, Lin X. Rare-Variant Association Analysis: Study Designs and Statistical Tests. The American Journal of Human Genetics 2014;95(1):5–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lee S, Emond MJ, Bamshad MJ, Barnes KC, Rieder MJ, Nickerson DA, et al. Optimal unified approach for rare-variant association testing with application to small-sample case-control whole-exome sequencing studies. Am J Hum Genet 2012;91(2):224–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lee S, Wu MC, Lin X. Optimal tests for rare variant effects in sequencing association studies. Biostatistics 2012;13(4):762–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.CRAN. Package SKAT: SNP-Set (Sequence) Kernel Association Test [Available from: https://cran.r-project.org/web/packages/SKAT/index.html.
- 63.Dering C, Hemmelmann C, Pugh E, Ziegler A. Statistical analysis of rare sequence variants: an overview of collapsing methods. Genet Epidemiol 2011;35 Suppl 1(Suppl 1):S12–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sung YJ, Korthauer KD, Swartz MD, Engelman CD. Methods for collapsing multiple rare variants in whole-genome sequence data. Genet Epidemiol 2014;38 Suppl 1(0 1):S13–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Cella D, Riley W, Stone A, Rothrock N, Reeve B, Yount S, et al. The Patient-Reported Outcomes Measurement Information System (PROMIS) developed and tested its first wave of adult self-reported health outcome item banks: 2005–2008. J Clin Epidemiol 2010;63(11):1179–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Schalet BD, Pilkonis PA, Yu L, Dodds N, Johnston KL, Yount S, et al. Clinical validity of PROMIS Depression, Anxiety, and Anger across diverse clinical samples. J Clin Epidemiol 2016;73:119–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ecco G, Imbeault M, Trono D. KRAB zinc finger proteins. Development 2017;144(15):2719–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Tao YF, Hu SY, Lu J, Cao L, Zhao WL, Xiao PF, et al. Zinc finger protein 382 is downregulated by promoter hypermethylation in pediatric acute myeloid leukemia patients. Int J Mol Med 2014;34(6):1505–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Chen S, Xiao Z, Zhou J, Yang M, Feng S, Huang Q, et al. ZNF382: A transcription inhibitor down-regulated in multiple tumors due to promoter methylation. Clin Chim Acta 2020;500:220–5. [DOI] [PubMed] [Google Scholar]
- 70.Dresselhaus EC, Meffert MK. Cellular Specificity of NF-κB Function in the Nervous System. Frontiers in Immunology 2019;10(1043). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Jones SV, Kounatidis I. Nuclear Factor-Kappa B and Alzheimer Disease, Unifying Genetic and Environmental Risk Factors from Cell to Humans. Frontiers in Immunology 2017;8(1805). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Jacola LM, Krull KR, Pui CH, Pei D, Cheng C, Reddick WE, et al. Longitudinal Assessment of Neurocognitive Outcomes in Survivors of Childhood Acute Lymphoblastic Leukemia Treated on a Contemporary Chemotherapy Protocol. J Clin Oncol 2016;34(11):1239–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Boulet-Craig A, Robaey P, Barlaam F, Laniel J, Oswald V, Jerbi K, et al. Visual short-term memory activation patterns in adult survivors of childhood acute lymphoblastic leukemia. Cancer 2019;125(20):3639–48. [DOI] [PubMed] [Google Scholar]
- 74.Waber DP, Queally JT, Catania L, Robaey P, Romero I, Adams H, et al. Neuropsychological outcomes of standard risk and high risk patients treated for acute lymphoblastic leukemia on Dana-Farber ALL consortium protocol 95–01 at 5 years post-diagnosis. Pediatr Blood Cancer 2012;58(5):758–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Burns MA, Place AE, Stevenson KE, Gutiérrez A, Forrest S, Pikman Y, et al. Identification of prognostic factors in childhood T-cell acute lymphoblastic leukemia: Results from DFCI ALL Consortium Protocols 05–001 and 11–001. Pediatr Blood Cancer 2021;68(1):e28719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Vrooman LM, Blonquist TM, Harris MH, Stevenson KE, Place AE, Hunt SK, et al. Refining risk classification in childhood B acute lymphoblastic leukemia: results of DFCI ALL Consortium Protocol 05–001. Blood Adv 2018;2(12):1449–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Pui CH, Campana D, Pei D, Bowman WP, Sandlund JT, Kaste SC, et al. Treating childhood acute lymphoblastic leukemia without cranial irradiation. N Engl J Med 2009;360(26):2730–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Ma MW, Wang J, Zhang Q, Wang R, Dhandapani KM, Vadlamudi RK, et al. NADPH oxidase in brain injury and neurodegenerative disorders. Molecular Neurodegeneration 2017;12(1):7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Salim S Oxidative Stress and the Central Nervous System. J Pharmacol Exp Ther 2017;360(1):201–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Areti A, Yerra VG, Naidu V, Kumar A. Oxidative stress and nerve damage: role in chemotherapy induced peripheral neuropathy. Redox Biol 2014;2:289–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Taylor OA, Hockenberry MJ, McCarthy K, Gundy P, Montgomery D, Ross A, et al. Evaluation of Biomarkers of Oxidative Stress and Apoptosis in Patients With Severe Methotrexate Neurotoxicity: A Case Series. J Pediatr Oncol Nurs 2015;32(5):320–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Mattson MP, Meffert MK. Roles for NF-kappaB in nerve cell survival, plasticity, and disease. Cell Death Differ 2006;13(5):852–60. [DOI] [PubMed] [Google Scholar]
- 83.Liu T, Zhang L, Joo D, Sun S-C. NF-κB signaling in inflammation. Signal Transduction and Targeted Therapy 2017;2(1):17023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Shi P, Yang A, Zhao Q, Chen Z, Ren X, Dai Q. A Hypothesis of Gender Differences in Self-Reporting Symptom of Depression: Implications to Solve Under-Diagnosis and Under-Treatment of Depression in Males. Frontiers in Psychiatry 2021;12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Sparks JL, Kumar R, Singh M, Wold MS, Pandita TK, Burgers PM. Human exonuclease 5 is a novel sliding exonuclease required for genome stability. J Biol Chem 2012;287(51):42773–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Ali S, Zhang Y, Zhou M, Li H, Jin W, Zheng L, et al. Functional deficiency of DNA repair gene EXO5 results in androgen-induced genomic instability and prostate tumorigenesis. Oncogene 2020;39(6):1246–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Breton JM, Barraza M, Hu KY, Frias SJ, Long KLP, Kaufer D. Juvenile exposure to acute traumatic stress leads to long-lasting alterations in grey matter myelination in adult female but not male rats. Neurobiology of Stress 2021;14:100319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Heim C, Nemeroff CB. The role of childhood trauma in the neurobiology of mood and anxiety disorders: preclinical and clinical studies. Biol Psychiatry 2001;49(12):1023–39. [DOI] [PubMed] [Google Scholar]
- 89.Anestin AS, Lippé S, Robaey P, Bertout L, Drouin S, Krajinovic M, et al. Psychological risk in long-term survivors of childhood acute lymphoblastic leukemia and its association with functional health status: A PETALE cohort study. Pediatr Blood Cancer 2018;65(11):e27356. [DOI] [PubMed] [Google Scholar]
- 90.Chantziara S, Musoro J, Rowsell AC, Sleurs C, Coens C, Pe M, et al. Quality of life of long-term childhood acute lymphoblastic leukemia survivors: Comparison with healthy controls. Psychooncology 2022;31(12):2159–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Wu G, Feder A, Cohen H, Kim J, Calderon S, Charney D, et al. Understanding resilience. Frontiers in Behavioral Neuroscience 2013;7(10). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Musliner KL, Mortensen PB, McGrath JJ, Suppli NP, Hougaard DM, Bybjerg-Grauholm J, et al. Association of Polygenic Liabilities for Major Depression, Bipolar Disorder, and Schizophrenia With Risk for Depression in the Danish Population. JAMA Psychiatry 2019;76(5):516–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Power RA, Tansey KE, Buttenschøn HN, Cohen-Woods S, Bigdeli T, Hall LS, et al. Genome-wide Association for Major Depression Through Age at Onset Stratification: Major Depressive Disorder Working Group of the Psychiatric Genomics Consortium. Biol Psychiatry 2017;81(4):325–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Schulze TG, Akula N, Breuer R, Steele J, Nalls MA, Singleton AB, et al. Molecular genetic overlap in bipolar disorder, schizophrenia, and major depressive disorder. World J Biol Psychiatry 2014;15(3):200–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author upon request and revision of the projects for which the data might be used.


