Abstract
Intron-encoded U17a and U17b RNAs are members of the H/ACA-box class of small nucleolar RNAs (snoRNAs) participating in rRNA processing and modification. We have investigated the organization and expression of the U17 locus in human cells and found that intronic U17a and U17b sequences are transcribed as part of the three-exon transcription unit, named U17HG, positioned approximately 9 kb upstream of the RCC1 locus. Comparison of the human and mouse U17HG genes has revealed that snoRNA-encoding intron sequences but not exon sequences are conserved between the two species and that neither human nor mouse spliced U17HG poly(A)+ RNAs have the potential to code for proteins. Analyses of polysome profiles and effects of translation inhibitors on the abundance of U17HG RNA in HeLa cells indicated that despite its cytoplasmic localization, little if any U17HG RNA is associated with polysomes. This distinguishes U17HG RNA from another non-protein-coding snoRNA host gene product, UHG RNA, described previously (K. T. Tycowski, M. D. Shu, and J. A. Steitz, Nature 379:464–466, 1996). Determination of the 5′ terminus of the U17HG RNA revealed that transcription of the U17HG gene starts with a C residue followed by a polypyrimidine tract, making this gene a member of the 5′-terminal oligopyrimidine (5′TOP) family, which includes genes encoding ribosomal proteins and some translation factors. Interestingly, other known snoRNA host genes, including the UHG gene (Tycowski et al., op. cit.), have features of the 5′TOP genes. Similar characteristics of the transcription start site regions in snoRNA host and ribosomal protein genes raise the possibility that expression of components of ribosome biogenesis and translational machineries is coregulated.
Nucleoli of eukaryotic cells contain a large number of distinct small nucleolar RNAs (snoRNAs) which are involved in various aspects of rRNA processing and modification (reviewed in references 45, 61, 63, and 66). These RNAs can be subdivided into two major classes. Members of one class contain short conserved sequence elements referred to as boxes C and D/D′ and are associated with the phylogenetically conserved protein fibrillarin (45, 63). Most of them function as guide RNAs specifying sites of 2′-O-methylation in rRNA (13, 38, 69). Another class of snoRNAs contains conserved sequence elements known as boxes H and ACA (5, 25). Members of this group function as guide RNAs in site-specific pseudouridylation of rRNA (24, 52). For the yeast Saccharomyces cerevisiae, it has been shown that the H/ACA snoRNAs are associated with the nucleolar protein GAR1 (5, 7, 25).
Biogenesis of snoRNAs can follow many distinct pathways. Some of the snoRNAs, e.g., U3, U8, U13, and 7-2/MRP RNAs, are transcribed from independent transcription units and contain either a 5′-terminal trimethylguanosine cap (polymerase [pol] II transcripts such as U3, U8, or U13) or a 5′-triphosphate (the pol III-transcribed 7-2/MRP RNA) (reviewed in reference 45). Most of the snoRNAs studied to date in vertebrates and some snoRNAs in yeast are, however, encoded within introns of the mosaic pol II-transcribed genes (43, 45). Generally, these snoRNAs are processed exonucleolytically from excised introns (12, 15, 36), although maturation of some intronic snoRNAs, such as U16 and U18, involves endonucleases (10, 11). Being posttranscriptionally processed from larger transcripts, all intron-encoded snoRNAs are not capped but contain a monophosphate group at the 5′ end (37, 68). The biogenesis of most snoRNAs in plants and some snoRNAs in yeast follows yet another pathway. In these organisms, snoRNAs are clustered and transcribed as polycistronic pre-snoRNA transcripts that are subsequently cleaved into single snoRNAs in a process involving both endo- and exonucleases (19, 42, 45, 72).
Characterization of genes that act as hosts for intronic snoRNAs has produced interesting findings. Most snoRNA host genes analyzed to date encode proteins essential for ribosome biogenesis or function such as ribosomal and nucleolar proteins or translational factors (reviewed in reference 45). This observation has led to the speculation that cotranscription of snoRNAs with mRNAs for nucleolar proteins or translational components may provide a regulatory mechanism to coordinate the accumulation of different molecules required for the assembly and function of ribosomes (45, 62). More recently, Tycowski et al. (67) discovered that the UHG gene, which harbors intronic snoRNAs U22 and U25 to U31, is distinct from all other snoRNA hosts characterized to date. The spliced poly(A)+ RNAs produced from UHG genes in humans, mice, and frogs are not conserved in sequence and have no apparent protein-coding potential. The finding that in UHG genes snoRNA-encoding introns and not exons are evolutionarily conserved and express functional RNAs requires a modification of the current description of exons as the main information-carrying regions of a gene (67, 69).
We are interested in the function and biogenesis of the H/ACA class snoRNA U17 (also referred to as E1 [16, 37, 57]). This evolutionarily conserved RNA has been implicated in the early processing event in the 5′ external transcribed spacer upstream of the 18S rRNA region and also has been shown to psoralen cross-link to 18S rRNA and the spacer in vivo (19, 47, 56). The U17 RNA has all the features of guide RNAs which specify sites of pseudouridylation, but its potential target sequence in rRNA is not readily apparent (16, 24, 54a). It is possible that U17 RNA catalyzes modification of some other, as yet unidentified RNA or that its function in rRNA-processing reactions does not involve pseudouridylation. The observation that in human cells the U17 RNA is more abundant than other RNAs of the H/ACA-box family (reference 35a and this work) is consistent with these possibilities. U17 RNA was previously characterized in many vertebrate species (14, 16, 37, 48, 57, 60). In Xenopus laevis (16) and fugu fish (14), six U17 sequence variants reside in introns of the ribosomal protein S7 (formerly referred to as S8) gene. In humans, two U17 RNAs, U17a and U17b (23, 37, 48), were postulated to originate from the 5′-proximal introns of the multiexon 5′ untranslated region (5′UTR) of the gene encoding the guanine nucleotide exchange factor RCC1, which participates in control of nucleocytoplasmic transport (reviewed in reference 27).
In this study we demonstrate that introns containing U17a and U17b sequences in humans do not reside in the RCC1 gene but are part of an independent transcription unit positioned approximately 9 kb upstream of the RCC1 locus. Comparisons of the human U17 host gene, named U17HG (U17 host gene), with its mouse counterpart (mU17HG) have revealed that these genes have very similar exon/intron organizations but are unlikely to encode proteins. Hence, U17HG is another example, in addition to UHG (67), of a snoRNA host gene whose only apparent function is to act as a vehicle for the expression of intron-located snoRNAs. Characterizations of human and mouse U17HG genes have also revealed that their transcription starts with a C residue followed by an oligopyrimidine tract. This feature makes these genes members of the 5′-terminal oligopyrimidine (5′TOP) family, which includes genes encoding ribosomal proteins and some other essential housekeeping proteins (reviewed in references 1 and 46). Interestingly, other known snoRNA host genes, including the non-protein-coding UHG gene characterized previously by Tycowski et al. (67), also have features of the 5′TOP family. 5′TOP sequences in mRNAs have been shown to play a role in coordinating their translation in response to cell growth conditions (1, 46). Identification of non-protein-coding snoRNA host genes as new members of the 5′TOP family is consistent with the +1 oligopyrimidine tracts having an additional, most likely transcriptional function, in regulating gene expression.
MATERIALS AND METHODS
Unless stated otherwise, all procedures for manipulating DNA and RNA were carried out according to Sambrook et al. (59) and Ausubel et al. (3).
Cloning of the human U17HG cDNAs.
A 282-bp fragment corresponding to the first four noncoding exons of the U17/RCC1 transcription unit (Fig. 1) was obtained by PCR using cDNA prepared with a HeLa cell total RNA as a template and GATTCGCAGTGGTCGCTTCTTCTC and CTCTCCAAGTTTACCTCTGCCTCC oligonucleotides as upstream and downstream primers, respectively. This PCR fragment was used as a probe to screen a human Namalwa (Burkitt lymphoma) cell cDNA library in plasmid pRS314/UNVP16 (65) (kind gift of P. Matthias of this institute). The probe was labeled with [α-32P]dATP (3,000 Ci/mmol; Amersham) by the random priming method (21). Hybridization was carried out at 55°C in 6× SSC (1× SSC is 0.15 M NaCl plus 15 mM sodium citrate) containing 0.5% nonfat dry milk. The washes were at 55°C twice for 10 min in 2× SSC–0.1% sodium dodecyl sulfate (SDS) and twice for 10 min in 0.2× SSC–0.1% SDS. The isolated positive clones were analyzed by restriction mapping and sequenced on both strands by the dideoxy method using appropriate primers.
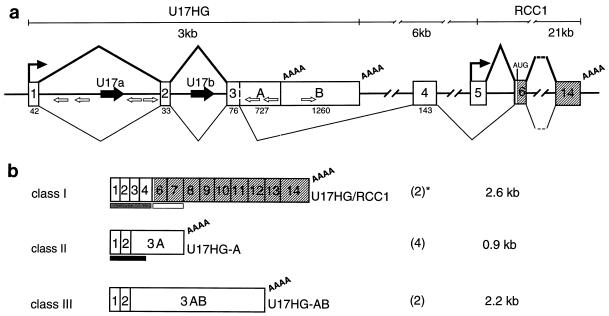
FIG. 1.
Expression of the human U17/RCC1 locus. (a) Schematic structure of the upstream portion of the human locus. Numbering of exons, shown as open and gray boxes, is according to Furuno et al. (23). Extensions of exon 3 identified in class II and class III cDNAs are denoted by A and B. Only terminal RCC1 protein-coding exons are shown. U17a and U17b sequences (black arrows), Alu sequences (open arrows), transcription start sites, and polyadenylation sites (AAAA) are indicated. Major splicing patterns supported by results presented in this work are shown in thick lines above the gene structure; the minor splicing pathway is indicated by thin lines. (b) Structures of clones isolated from the human Namalwa B-cell cDNA library. Numbers in parentheses indicate the number of clones corresponding to each class. Clone sizes are indicated. Bars below the cDNA schemes show the fragments used as probes for the cDNA library screening (gray) and for Northern analysis (black and open boxes represent U17HG- and RCC1-specific probes, respectively, used for experiments shown in Fig. 2). ∗, cDNA library screen might have been biased in favor of isolating class I clones. These clones hybridize to the probe over its entire length (296 bp, exons 1 through 4), while class II and III clones hybridize only to 153 bp (exons 1 to 3) of the probe.
Cloning of the mU17HG cDNA and genomic sequences.
Two oligonucleotides, GGGGAGACAAACCATGCAGG and CCAACGTTGTGGAAAGGGAC, specific for conserved regions of the human and Xenopus U17 snoRNAs and corresponding to positions 180 to 199 and 90 to 109 in the human U17 RNA, respectively (37), were used as primers for PCR performed with mouse genomic DNA. The amplified 413-bp fragment was cloned into the HincII site of pBluescript KS+ and sequenced. A candidate 36-nucleotide (nt) exon sequence separating the two U17 RNA-containing introns was identified by using the HEXON exon-finding program from the Baylor College of Medicine Gene Finder package (http://dot.imgen.bcm.tmc.edu:9331/gene-finder/gf.html). Three partially overlapping oligonucleotides, TCCATCAACAAAGTACCTGAAATC, TCAACAAAGTACCTGAAATCATTG, and AAGTACCTGAAATCATTGACCGGG, complementary to the exon sequence, were used as gene-specific primers for nested PCR in the 5′RACE (rapid amplification of cDNA ends) protocol, and two partially overlapping oligonucleotides, CCCGGTCAATGATTTCAGGTACTTTG and GTCAATGATTTCAGGTACTTTGTTG, were used as primers in the 3′RACE reactions. Both the 3′RACE and the 5′RACE experiments were performed with mouse NIH 3T3 total RNA, by using a 5′/3′RACE kit (Boehringer Mannheim) as instructed by the manufacturer. The obtained 5′RACE and 3′RACE PCR fragments were cloned into the HincII site of the pBluescript KS+, giving clones pmU17HG/5′RACE and pmU17HG/3′RACE, respectively. The genomic sequence of the mU17HG locus was obtained by PCR using mouse genomic DNA as a template and oligonucleotides TGACATTCAAATGCTTTAATTCAG and TCTCTCTAGGCGTCGCTCTCTTGG, specific for terminal sequences of the mU17HG cDNA, as primers. The obtained 1.6-kb PCR fragment was cloned into the HincII site of pBluescript KS+, resulting in pmU17HG/gene, and sequenced.
Cell culture.
Human HeLa and mouse NIH 3T3 cells were grown in monolayers at 5% CO2 in Dulbecco modified Eagle medium (Gibco) containing 10% fetal calf serum (Gibco) and 2 mM glutamine. Namalwa cells were grown at 5% CO2 in RPMI 1640 medium (Gibco) containing 10% fetal calf serum. Cycloheximide (20 μg/ml), pactamycin (560 ng/ml), or puromycin (50 μg/ml) was added to cells 4 h prior to harvesting where indicated. HeLa suspension culture was grown in spinner flasks in Joklik’s minimal essential medium (Gibco) containing 5% newborn calf serum (Amimed) as described elsewhere (22).
Cell fractionation and RNA isolation.
HeLa cells grown in suspension were fractionated as described previously (17). Total RNA from human and mouse cultured cells and from cytoplasmic and nuclear fractions was isolated by the guanidinium thiocyanate-phenol-chloroform method (26). Poly(A)+ RNA was isolated directly from cell lysates by using an Oligotex oligo(dT) affinity matrix (Qiagen) according to the manufacturer’s protocol. Mouse (BALB/c) genomic DNA was a kind gift of R. Neve of this institute.
Analysis of HeLa cell extracts on sucrose gradients.
HeLa cell suspension culture (3 × 107 cells, 5 × 105 cells/ml) was treated for 5 min with cycloheximide (100 μg/ml), and then washed once with 20 ml of phosphate-buffered saline containing cycloheximide (100 μg/ml) and twice with 20 ml of buffer A (5 mM Tris-HCl [pH 7.4], 1.5 mM KCl, 2.5 mM MgCl2, 100 μg of cycloheximide per ml). The cells were lysed in 400 μl of buffer A containing 3 mM dithiothreitol, 30 U of RNasin (Promega), 0.5% Triton X-100, and 0.5% sodium deoxycholate. The lysates were centrifuged for 8 min at 4°C at 3,000 × g. The supernatant was collected and loaded onto 17 to 51% linear sucrose density gradients prepared in 20 mM Tris-HCl (pH 7.4)–80 mM NaCl–5 mM MgCl2. The lysates were centrifuged at 4°C for 2 h at 36,000 rpm in an SW41 rotor (Beckman). RNA was isolated from individual fractions by the proteinase K method.
Northern analysis.
Total RNA and poly(A)+ RNA were separated on a 1.4% agarose-formaldehyde gel, blotted onto a Hybond-N nylon membrane (Amersham) by using 20× SSPE (1× SSPE is 0.18 M NaCl, 10 mM NaH2PO4, and 1 mM Na2EDTA [pH 7.7]), and UV cross-linked to the membrane. A KpnI-HindIII fragment of phU17HG/123a, corresponding to nt 20 to 237 of the U17HG-A cDNA, was used as a U17HG-specific probe. phU17HG/123a was constructed by cloning a PCR fragment corresponding to nt 20 to 237 of the human U17HG cDNA into the HincII site of pBluescriptII KS+. A KpnI-HindIII fragment of plasmid pRCC1, containing exons 6 and 7 of the human RCC1 cDNA, was used as the RCC1-specific probe. pRCC1 was constructed by cloning a PCR fragment, extending from position −9 (relative to the initiator AUG) in exon 6 to position +261 in exon 7 of the RCC1 protein-coding sequence, into the HincII site of pBluescriptII KS+. Blots were hybridized overnight at 42°C in 5× SSPE containing 50% formamide, 10% dextran sulfate, 1% SDS, and 50 g of salmon sperm DNA per ml. They were washed in 2× SSC–0.1% SDS for 30 min at 42°C and subsequently in 0.2× SSC–0.1% SDS for 30 min at 55°C.
RNase A/T1 mapping.
RNase A/T1 mapping was performed as described by Goodall et al. (26). The following plasmids were used to generate probes. The human U17HG-specific probe was transcribed with T3 polymerase from plasmid phU17HG/123a linearized with EcoRI. The UHG-specific probe was a T7 RNA polymerase transcript from a plasmid containing the human UHG cDNA insert (gift of K. Tycowski, Yale University). This probe covers positions 932 to 1118 of the UHG cDNA (67). The human U17a-specific probe was transcribed from a plasmid containing the ClaI-XhoI fragment of plasmid pA (36), cloned in the ClaI and XhoI sites of pBluescriptII KS+. The plasmid, referred to as pU17a(C-X), was cut with BamHI and transcribed with T3 RNA polymerase. The human β-actin gene-specific probe was transcribed by T3 polymerase, by using plasmid ATCC65129 (obtained from the American Type Culture Collection) cut with DraIII. The human U22 snoRNA-specific probe was transcribed by SP6 RNA polymerase from plasmid pU22 (obtained from K. Tycowski) linearized with MunI. This probe is complementary to the entire 125-nt-long U22 snoRNA. The 18S rRNA-specific probe, complementary to nt 715 to 793 of the human 18S rRNA, was transcribed from plasmid pT7/RNA/18S (Ambion). The plasmid was cut with HindIII and transcribed with T7 RNA polymerase. The 18S rRNA-specific probe was labeled with [α-32P]UTP with a specific activity of 30 mCi/mmol; all other probes were labeled with [α-32P]UTP with a specific activity of 30 Ci/mmol. An amount of probe corresponding to 105 cpm was used in each mapping experiment. Protected fragments were separated on 6% polyacrylamide–8 M urea gels. Where indicated, radioactivity of protected fragments was quantitated with a Storm 860 PhosphorImager (Molecular Dynamics).
Primer extension and RACE analysis.
For primer extension analysis, three independent partially overlapping primers complementary to exon 1 of the human U17HG RNA (primer A [AGCGACCACTGCGAATCTGTCTCC], primer B [GAAGAAGCGACCACTGCGAATCTG], and primer C [CAAGGAGAAGAAGCGACCACTGCG] [see Fig. 3b]) were end labeled with [γ-32P]ATP (3,000 Ci/mmol; Amersham) and T4 polynucleotide kinase and gel purified. Annealing reaction mixtures contained 20 pmol of each primer and 15 μg of RNA in 160 mM HEPES-KOH (pH 7.7)–1 M NaCl–1 mM Na2EDTA. Following annealing overnight at 48°C, the mixture was ethanol precipitated and dissolved in 20 μl of the first-strand synthesis buffer for Superscript II reverse transcriptase (Gibco-BRL) containing 10 mM dithiothreitol, a 1 mM concentration of each deoxynucleoside triphosphate, and 400 U of Superscript II reverse transcriptase. Samples were incubated at 50°C for 50 min and processed for the analysis on a 20% acrylamide–8 M urea gel.
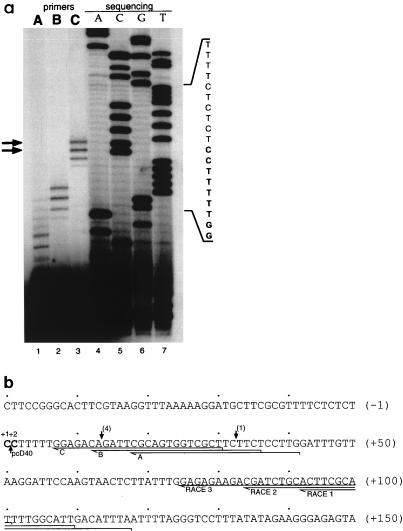
FIG. 3.
Characterization of the transcription start site of the U17HG RNA. (A) Determination of the 5′ end of U17HG RNA by primer extension. Lanes 1 to 3, primer extension reactions with primers A to C, respectively (for positions of oligonucleotide primers, see panel b). Lanes 4 to 7, sequencing reactions performed with the primer C and pU17HG/PE1 as a template. The relevant portion of the sequence is shown on the right. Reverse transcription products obtained with primer C, corresponding to cDNAs extended to the C residues at positions +1 and +2, are indicated by arrows. (b) Sequence of the −50 to +150 region of the U17HG locus. Two adjacent C residues, +1 and +2, identified as transcription start sites are in bold. Regions corresponding to oligonucleotides A, B, and C used as primers in primer extension experiments, as well as oligonucleotides RACE 1, RACE 2, and RACE 3 used in successive steps of the RACE experiment (see Materials and Methods), are underlined. 5′ ends of cDNA clones isolated in this work are indicated by arrows; numbers of clones ending at a particular site are shown in parentheses. The 5′ end of the cDNA clone pcD40, described by Ohtsubo et al. (54), is also indicated.
For the 5′RACE analysis, three partially overlapping primers (RACE 1 [TAAATGTCAATGCCAAAATGCGAAG], RACE 2 [ATGCCAAAATGCGAAGTGCAGATCG], and RACE 3 [TGCGAAGTGCAGATCGTCTTCTCTC] [see Fig. 3b]), complementary to exon 3 of the human U17HG RNA, were used as gene-specific nested backward primers in successive 5′RACE steps, using HeLa total RNA and a 5′/3′RACE kit. The PCR/RACE product was sequenced directly with an automated DNA sequencer or cloned into the HincII site of pBluescript KS+ in order to sequence individual clones.
Nucleotide sequence accession numbers.
The nucleotide sequences of U17HG-A and U17HG-AB cDNAs have been submitted to the EMBL/GenBank/DDBJ nucleotide sequence libraries under accession no. AJ006834 and AJ006835, respectively. The nucleotide sequences of the mU17HG cDNA and mU17HG gene have been submitted under accession no. AJ006837 and AJ006836, respectively.
RESULTS
Expression of the U17/RCC1 host transcription unit in human cells.
Previous studies on expression of the RCC1 gene in human cells have indicated that transcription of this gene is initiated at two different promoters positioned approximately 9 kb apart (23, 54). Initiation at the downstream promoter produced a pre-mRNA in which a 5′-terminal single noncoding exon (exon 5 in Fig. 1) is spliced to downstream exons encoding the RCC1 protein. Initiation at the upstream promoter would yield a transcript containing four short noncoding exons (exons 1 to 4; in Fig. 1) spliced to the coding part of mRNA; U17a and U17b snoRNA sequences are located in introns 1 and 2 of this transcript (23, 37, 48) (Fig. 1). As pre-mRNAs containing so many noncoding exons are unusual and the splicing pattern of the RCC1 transcript initiating at the upstream promoter was deduced from a single cDNA clone (54), we have investigated the expression of the U17/RCC1 locus in human cells in more detail.
A human Namalwa B-cell cDNA library was screened with a probe corresponding to the four 5′-proximal noncoding exons of the transcript initiated at the upstream RCC1 promoter. Nine identified positive clones can be grouped into three classes (Fig. 1B). Only class I, represented by two clones, is consistent with the previously proposed expression pattern of the U17/RCC1 locus. Class II is represented by four clones called U17HG-A. In these clones, exons 1, 2, and 3 are spliced together as expected but exon 3 extends for an additional 600 nt and terminates with poly(A). Class III, comprising two clones called U17HG-AB, is similar to class II except that the clones represent RNAs that are polyadenylated another 1.26 kb further downstream in the extended exon 3 (Fig. 1). One additional isolated clone contained an insert corresponding to a fragment of unspliced U17HG transcript (data not shown). A search of sequence databases, with the 3′-end portions of U17HG-A and U17HG-AB RNAs, identified three independent expressed sequence tag (EST) entries representing RNA polyadenylated at the U17HG-A site (GenEMBL accession no. H54400, N25905, and AA522843) and three corresponding to RNA polyadenylated at the U17HG-AB site (GenEMBL accession no. H19509, H46276, and AA563638), further confirming the use of both upstream polyadenylation sites. These findings show that transcription from the upstream U17/RCC1 gene promoter gives rise to three different classes of transcripts, all hosting the U17a and U17b RNAs.
To determine the relative abundances of different transcripts initiated at the upstream U17/RCC1 promoter, Northern analysis was performed. Total and poly(A)+ RNAs from HeLa cells and Namalwa B cells were probed with two different cDNA probes, one specific for the upstream exons 1, 2, and 3 and another specific for the RCC1 protein-coding exons 6 and 7 (Fig. 1B). Hybridization with the exon 1 to 3 probe revealed the presence of the 0.9-kb transcript corresponding in length to the U17HG-A RNA (Fig. 2A). This probe detected neither the 2.2-kb human U17HG-AB RNA nor the 2.6-kb RCC1 mRNA initiated at the upstream promoter (Fig. 2B), indicating that U17 host transcripts polyadenylated at the U17HG-AB site or spliced into the RCC1 coding region are much less abundant than the U17HG-A RNA. Additional RNase A/T1 mapping experiments carried out with a probe which distinguishes U17HG/RCC1 spliced mRNA from the sum of U17HG-A and U17HG-AB RNAs have indicated that the former RNA constitutes less than 10% (10% corresponds to the detection limit) of the total spliced RNA initiated at the upstream promoter (data not shown). Hybridization of the Northern blot shown in Fig. 2A with the probe specific for the RCC1 coding exons 6 and 7 revealed a 2.6-kb band, the size expected for the RCC1 mRNA initiated at the downstream promoter (Fig. 2B). Comparison of Northern blots shown in Fig. 2A and B (72 and 15 h of autoradiography, respectively) indicates that the 2.6-kb U17HG/RCC1 mRNA constitutes at most 5 to 10% of the total RCC1 mRNA present in HeLa or Namalwa B cells.
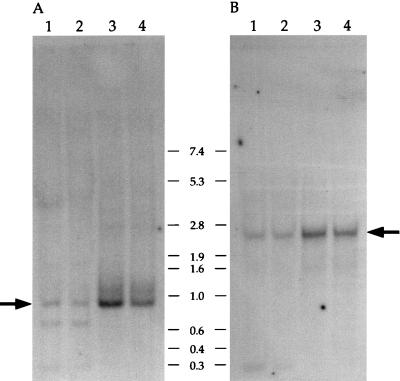
FIG. 2.
Expression of the U17HG (A) and RCC1 (B) genes in human cell lines determined by Northern analysis. An RNA blot containing 15 μg of total (lanes 1 and 2) and 2 μg of poly(A)+ (lanes 3 and 4) RNA from Namalwa B cells (lanes 2 and 4) and HeLa cells (lanes 1 and 3) was probed first with the cDNA fragments corresponding to upstream exons 1 to 3 (A) and then with the probe specific for the RCC1 protein-coding exons 6 and 7 (B). The probes, schematically shown in Fig. 1b, were of comparable length and specific activity. Major transcripts identified by each probe are indicated by arrows. Positions of RNA size markers (in kilobases) are also shown. Autoradiography was for 72 h (A) and 15 h (B).
Characterization of the 5′ end of transcripts initiated at the upstream promoter.
Primer extension and RACE analysis were performed to characterize the 5′ terminus of the human U17HG RNA. Three different oligonucleotides complementary to the 5′-proximal exon of the U17HG RNA were used as primers for reverse transcription. Each of the oligonucleotides yielded two major extended cDNA products, consistent with transcription of the U17HG RNA being initiated at C residues positioned 14 or 15 nt upstream of the 5′ end of the longest cDNA clone characterized in this work (Fig. 3). To further verify the sequence of the 5′ end of the U17HG RNA, RACE analysis was performed. Direct sequencing of the RACE product placed the start site at the upstream C residue (C+1 [Fig. 3b]) identified by primer extension. Sequencing of individual clones resulting from cloning of the RACE product in pBluescriptII KS+ vector has identified two clones terminating at the C+1 and two clones terminating at the C+2 (data not shown).
Based on the results presented in this study and the fact that the single previously characterized cDNA clone (54) (Fig. 3b) also starts at the second of the two C residues, the start site of transcription from this promoter is assigned to C residues +1 and +2. These C residues are preceded and followed by tracts of 10 and 5 pyrimidines, respectively (Fig. 3b), which makes the human U17HG gene similar to the 5′TOP genes (see Discussion).
Human and murine U17 host poly(A)+ RNAs have no apparent coding potential.
With the exception of several Alu repeats (Fig. 1A), U17HG-A and U17HG-AB RNAs show no significant homology to other sequences present in public databases. Moreover, both RNAs show very little protein-coding potential (Fig. 4B). The 5′-proximal ATG, starting at position 230, is in a very poor translation initiation context [CATTTCATGC; a single position conforming with the consensus GCC(A/G)CCATGG (39) is underlined]. The longest open reading frame of the U17HG-A RNA starts at the second ATG (positions 436 to 438, context CTAGGAATGC) and has the potential to encode a 29-amino-acid polypeptide. Within the consensus sequence, two highly conserved nucleotides have the strongest effect on translation initiation: a purine, preferably A, at position −3 and a G following the ATG codon (40). Hence, the G at position −3 makes the second ATG of the U17HG RNA potentially adequate for initiation of translation (40). We note, however, that all other nucleotides surrounding this ATG do not fit the extended consensus (39).
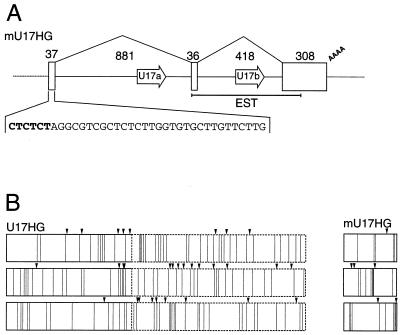
FIG. 4.
Schematic structure of the mouse U17HG gene (A) and analysis of the protein-coding potential of human and mouse U17HG RNAs (B). (A) Positions of exons and snoRNA regions are denoted by open boxes and open arrows, respectively. Lengths of exons and introns and the splicing pattern are indicated. The sequence of exon 1 is shown with the 5′ polypyrimidine tract in bold. The region corresponding to the EST clone representing the unspliced mouse U17HG RNA (GenEMBL accession no. AA198537) is indicated with a line. (B) Coding potentials of human (left) and mouse (right) U17HG RNAs. Positions of AUG codons (arrowheads) and stop codons (vertical lines) are indicated for all reading frames of the spliced mouse and human U17HG RNAs. The region corresponding to region B of exon 3 in human RNA (Fig. 1) is represented by dashed lines.
To verify that U17HG RNAs are indeed unlikely to code for proteins, we characterized the U17HG RNA and its gene from mice. On the assumption that two U17 RNAs are also encoded in adjacent introns in the mouse genome, the region separating U17 sequences was PCR amplified by using mouse genomic DNA and oligonucleotide primers specific for the sequences conserved in vertebrate U17 RNAs. The 36-bp exon sequence in the amplified DNA was predicted with the help of the exon-finding computer software and used for designing appropriate primers for characterization of the complete cDNA by 5′RACE and 3′RACE. Finally, oligonucleotides complementary to the 5′ and 3′ ends of the cDNA were used as primers for PCR with genomic DNA as a template (see Materials and Methods). The structure of the mouse U17HG (mU17HG) gene is schematically shown in Fig. 4A. Like the human U17HG gene, the mouse gene comprises two short 5′-proximal exons and a longer 3′ exon. The mU17HG cDNA starts with a C residue followed by five pyrimidines. The spliced mU17HG RNA is 382 nt long, excluding the poly(A) tail. Each of the two mU17HG introns encodes one U17 RNA. We investigated the expression of mU17a and mU17b RNAs by RNase A/T1 mapping and found that U17b but not U17a accumulates in mouse NIH 3T3 cells (data not shown). The failure to detect the U17a variant may be due to a 17-nt deletion in the 5′ region of the RNA (corresponding to positions 15 to 31 in human U17a RNA [37]) that may interfere with proper RNA folding and/or packaging into RNP.
Despite being similar in gene organization, the mU17HG RNA shows very little sequence similarity to its human counterpart. While intron-encoded U17a and U17b RNA sequences show 83 and 89% identity, respectively, between humans and mice, pairwise comparisons of individual exons indicated only 41 to 51% identity. This level of sequence conservation is similar to that of corresponding segments of intron sequences positioned either upstream or downstream of the snoRNA-coding regions (35 to 48% identity). Like human U17HG RNA, the mouse RNA shows no apparent protein-coding potential (Fig. 4B). The 5′-proximal ATG, starting at position 47, is in a very weak translational context (CGGTCAATGA). The longest open reading frame is 38 amino acids long, and due to the G present at position −3, its ATG is in the potentially adequate context, AGGTTATGT (39, 40).
There is no significant similarity between peptides potentially encoded by the human and mouse U17HG RNAs. Moreover, no sequences similar to these peptides were identified in public sequence databases. Taken together, these findings suggest that the only conserved function of both mouse and human U17HG genes is the generation of U17 snoRNAs.
Cellular localization of U17HG RNA and effects of translation inhibitors.
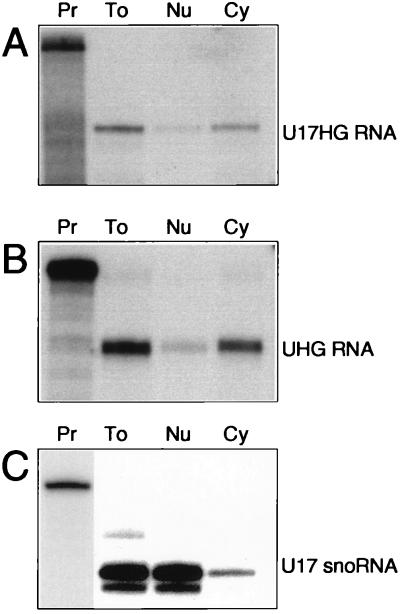
We investigated the cellular localization of the U17HG RNA. Cell fractionation experiments indicated that U17HG RNA is enriched in the cytoplasm (Fig. 5A), similar to UHG RNA, another noncoding snoRNA host having no protein-coding potential (67) (Fig. 5B). As expected, U17 snoRNA, used as a control, was found mainly in the nuclear fraction (Fig. 5C). Treatment of HeLa cells with a translation inhibitor such as cycloheximide, pactamycin, or puromycin did not cause any detectable increase in the level of U17HG RNA (Fig. 6A), in marked contrast to the UHG RNA, the level of which increased in response to inhibitors (67) (Fig. 6B and legend). Kinetic experiments in which HeLa cells were grown in the presence of cycloheximide, pactamycin, or puromycin for up to 6 h also failed to show any changes in accumulation of U17HG RNA, although they demonstrated a time-dependent increase in the level of the control UHG RNA (data not shown). To better assess the relative abundance of U17HG and UHG host RNAs and snoRNAs encoded in their introns, we performed RNase A/T1 mapping experiments using total HeLa cell RNA and probes specific for U17HG and UHG RNAs (Fig. 6C) and U17a and U22 snoRNAs (Fig. 6D). The abundances of U17HG, UHG, U17a, and U22 RNAs were calculated to be 7, 65, 15,000, and 11,000 molecules per cell, respectively, taking 18S rRNA (4 × 106 ribosomes/cell [34]) as a reference. Hence, despite the fact that the levels of U17a and U22 snoRNAs are comparable, U17HG RNA is significantly less abundant than UHG RNA.
FIG. 5.
Cellular localization of the human U17HG RNA. RNA isolated from the HeLa total cell extract (lane To) and from nuclear (lane Nu) and cytoplasmic (lane Cy) fractions was analyzed by RNase A/T1 mapping using probes specific for human U17HG RNA (A), UHG RNA (B), and U17 snoRNA (C). RNA amounts (10, 1, and 9 μg of total, nuclear, and cytoplasmic RNAs, respectively) corresponding to approximately equal numbers of cells were used in each mapping experiment. Lane Pr, aliquots of undigested probes. Exposure times were 24 h (A and B) and 2 h (C).
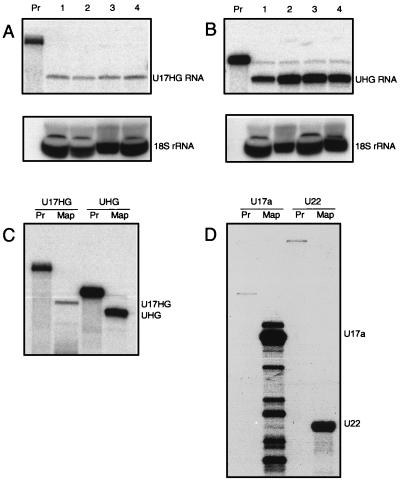
FIG. 6.
Influence of translation inhibitors on the levels of U17HG (A) and UHG (B) RNAs in HeLa cells and comparison of steady-state levels of U17HG and UHG RNAs (C) with levels of U17 and U22 snoRNAs (D). (A and B) RNA (5 μg) isolated from control HeLa cells (lane 1) or cells treated for 4 h with cycloheximide (20 μg/ml; lane 2), pactamycin (560 ng/ml; lane 3), or puromycin (50 μg/ml; lane 4) was analyzed by RNase A/T1 mapping using a mixture of the host gene-specific and 18S rRNA-specific probes. Lane Pr, aliquots of undigested U17HG- or UHG-specific probe (amounts of probe loaded in lanes Pr are not proportional to the amounts used in mapping experiments; see Materials and Methods). Host gene-specific and 18S rRNA-specific protected fragments are shown in upper and lower panels, respectively. The 18S rRNA-specific probe used in the RNase A/T1 mapping experiment was labeled with [α-32P]UTP of 1,000-times-lower specific activity than that used for other probes (see Materials and Methods). Exposure times were 24 and 1 h for upper and lower panels, respectively. In panel A, the relative levels of U17HG RNA in lanes 1, 2, 3, and 4 are 1, 0.9, 0.8, and 0.9, respectively; in panel B, the relative levels of UHG RNA in lanes 1, 2, 3, and 4 are 1, 4.9, 3.2, and 2.2, respectively. All values were normalized by using the 18S rRNA mapping as a loading control. (C and D) HeLa cell total RNA (4 μg) was analyzed by RNase A/T1 mapping using indicated antisense probes specific for U17HG, UHG, U17a, and U22 RNAs. All probes were labeled with [α-32P]UTP of the same specific activity. Lanes Pr in each pair contained an aliquot of the probe used for mapping (shown in lane Map). Exposure times were 24 h and 30 min for panels C and D, respectively. The relative abundances of the RNAs were calculated as 1, 9, 2,100, and 1,550 for U17HG, UHG, U17a, and U22 RNAs, respectively. Quantitation values from PhosphorImager analysis were corrected for the number of U residues present in protected fragments.
Distribution of U17HG RNA on sucrose gradients.
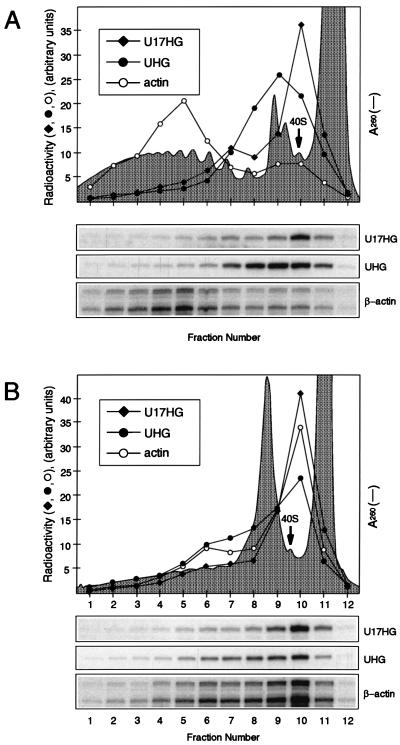
UHG RNA, although it has no apparent protein-coding potential and contains numerous stop codons, is found in association with translating ribosomes (67). The most plausible explanation for why levels of UHG RNA increase upon treatment of cells with protein synthesis inhibitors is that these compounds, by blocking translation, also suppress nonsense codon-mediated decay of this RNA (44, 67). For a possible explanation as to why translation inhibitors have no effect on U17HG RNA accumulation, we have investigated whether this RNA is associated with translating ribosomes. Cytoplasmic extracts were prepared from HeLa cells grown in suspension and analyzed on sucrose gradients. The U17HG RNA was detected mainly in a fraction sedimenting close to the top of the gradient in the 40S region, while most of the UHG RNA was found in association with 80S ribosomes (Fig. 7A). The cosedimentation of a small amount of U17HG RNA with light polysomes was not reproducibly observed. Treatment of HeLa cells with the translation initiation inhibitor NaF (32) resulted in runoff of ribosomes from UHG RNA and actin mRNA but had little effect on the distribution of U17HG RNA (Fig. 7B). This result further supports the conclusion that most of this RNA is not associated with translating ribosomes. Analysis of the U17HG RNA distribution in extracts prepared from HeLa cells grown in monolayers yielded similar results. Furthermore, experiments performed with sucrose gradients, which allow better resolution of ribosomal subunits, showed that U17HG is not associated with 40S subunits but sediments significantly slower than 40S particles (data not shown).
FIG. 7.
U17HG RNA is not associated with translating ribosomes in HeLa cells. Cytoplasmic extracts prepared from control HeLa cells grown in suspension (A) or cells treated with 30 mM NaF for 25 min (B) were fractionated on linear 17 to 51% sucrose gradients. RNA was isolated from individual fractions and analyzed by RNase A/T1 mapping using U17HG-, UHG-, and β-actin gene-specific probes. Only relevant parts of autoradiograms, containing gene-specific protected bands, are shown below the sucrose gradient profiles. Radioactivity in protected fragments was quantified with a PhosphorImager and plotted in the panels which also show A260 profiles of the gradients. Positions of 40S ribosomal subunits are indicated.
DISCUSSION
Investigation of the transcription and splicing patterns of the human U17/RCC1 locus carried out in this work has shown that, contrary to previous conclusions (23, 37, 48), the introns hosting U17a and U17b RNA sequences comprise part of the independent U17HG transcription unit positioned ∼9 kb upstream of the RCC1 gene. Nevertheless, perhaps as much as 10% of transcripts initiated at the U17HG promoter undergo alternative splicing resulting in a product containing U17HG exons 1 and 2 and a part of exon 3, joined to the downstream exons of the RCC1 gene. This splicing event produces a variant RCC1 mRNA with the 5′UTR different from the 5′UTR present in mRNA initiated at the downstream RCC1 promoter (54) (Fig. 1). Northern analysis suggests that this hybrid U17HG/RCC1 mRNA comprises no more than 5 to 10% of the total RCC1 mRNA present in either HeLa or Namalwa B cells. Whether its formation is of any biological significance remains to be established. In light of the important role played by the RCC1 protein in controlling transport of macromolecules through the nuclear pore (reviewed in reference 27), further investigation of its alternative mRNA forms would be interesting. In this context, it is important to note that RCC1 mRNAs initiated at either of the two promoters start with a C residue followed by a stretch of pyrimidines (23, 54) (this work), making them similar to the 5′TOP family of mRNAs (reviewed in references 1 and 46). It is not known whether translation of RCC1 mRNA responds to conditions known to affect translation of the previously characterized 5′TOP mRNAs.
Comparison of human and mouse U17HG genes has revealed their very similar organizations. The genes in both species consist of two short 5′-proximal exons and one long 3′-proximal exon. Each of the two introns contains a single U17 RNA coding sequence. While both U17 RNA variants accumulate in human cells (37), only the intron 2-encoded U17 RNA was detected in mouse fibroblasts (this work). Since the mouse intron 1-encoded variant lacks 17 nt in a region that is relatively highly conserved among vertebrate U17 RNAs (14, 16, 37, 57, 60), it is likely that this RNA is not productively processed and/or packaged into the RNP.
Despite the similarities in gene organization, corresponding exons in human and mouse U17HG RNAs show no significant sequence similarity. Likewise, neither of the two RNAs has the capacity to code for peptides longer than 38 amino acids, and none of the peptides potentially encoded by the human RNA has a counterpart with a significant similarity in the mouse. Hence, it is very likely that the function of U17HG, like that of the snoRNA host gene UHG (67) and two other non-protein-coding snoRNA hosts, gas5 (60a) and U19HG (6), is to act exclusively as a vehicle for expression of intron-located snoRNAs. The conclusion that U17HG RNA does not code for a protein is further supported by the observation that little, if any, of this RNA is found in association with 80S ribosomes or polysomes in HeLa cells. It is not known why human U17HG RNA, in contrast to UHG and gas5 non-protein-coding host RNAs (60a, 67), does not associate with translating ribosomes. Since both the U17HG 5′-proximal ATG and the second ATG, which is in a better translational context, are positioned very far from the 5′ end of the RNA, it is possible that RNA secondary structure or proteins interacting with the 5′-terminal region interfere with the 40S ribosome scanning and initiation of translation of U17HG RNA.
The levels of UHG and gas5 non-protein-coding host RNAs were found to increase markedly following treatment of cells with translation inhibitors, most likely as a result of suppression of the translation-dependent nonsense codon-mediated RNA decay (44, 60a, 67). In contrast, we have found that treatment of HeLa cells with cycloheximide, pactamycin, or puromycin has no effect on the level of U17HG RNA. This finding is consistent with the observation that little of this RNA is found in association with translating ribosomes. Although U17HG RNA does not appear to be subject to nonsense codon-mediated degradation, its very low concentration (7 molecules per HeLa cell) compared to U17a RNA (15,000 molecules/cell) or UHG RNA (65 molecules/cell) suggests that it decays very rapidly. We have inspected the human U17HG RNA sequence for the presence of AUUUA-type destabilizing elements. U17HG-A RNA contains four AUUUA stretches, but none of them form part of a longer sequence known to act as a functional AUUUA-type destabilizing element (20, 41). The reasons for inefficient accumulation of U17HG RNA remain to be established.
We have investigated whether human and mouse U17HG RNAs share secondary structure elements which might suggest their possible function as regulatory or structural RNAs. No obvious conserved structures were found by using the RNA folding program Mfold version 2.3 (73). Likewise, Northern hybridizations performed with poly(A)+ RNAs originating from different human organs did not offer any clue as to a possible function of the U17HG-A RNA; the RNA was present in all tissues investigated (54a).
Both human and mouse U17HG RNAs at the 5′ terminus contain a C residue which is followed by an oligopyrimidine tract of five and six residues, respectively. This feature makes these RNAs members of the 5′TOP family. Most interestingly, another snoRNA host lacking protein-coding potential, the UHG RNA, also has features of 5′TOP RNAs (67). Inspection of the transcription start site regions of other genes known to act as hosts for intronic snoRNAs indicated that most of them belong to the 5′TOP category (Table 1). All characterized ribosomal protein genes known to host snoRNAs contain 5′-terminal oligopyrimidine tracts (sequences not shown; see Table 1, footnote a). Thirteen characterized non-ribosomal protein hosts, including non-protein-coding genes UHG (67), gas5 (60a), U19HG (6), and U17HG (this work), also start with a C residue followed by stretches of pyrimidines (Table 1). The human hsc70 gene starts with C but its start site is not distinctly pyrimidine rich, while mouse eIF4AI and eIF4AII genes and the human ATP synthase-β gene start with G or A located within oligopyrimidine sequences. Two snoRNA host genes, mouse hsc70 and human eIF4AII, have no apparent properties of the 5′TOP genes.
TABLE 1.
Transcription start sites in mammalian U-snoRNA host genesa
| Host gene | Organism | snoRNA | Transcription start site regionb | Reference (GenEMBL accession no.) |
|---|---|---|---|---|
| Nucleolin | Human | U20 | CTCTCGCTGGCTTCGGGTGT | 64 (M60858) |
| EF-2 | Hamster | U37 | CGTCAGCGGTCTCTTCCGCC | 50 (Not available) |
| EF-1-bc | Human | U51 | CTTTTTCCTC | 33 (D28350, AA371392) |
| Q1Z 7F5/QM protein | Human | U70 | CTCTTTCCCT | 70 (M81806, D28410) |
| eIF4AIc | Human | U67 | CTAGTTTCTA | 35 (D13748, AA113744) |
| Laminin binding proteind | Human | E2 | TTTCCTGCTGCCTGTCTTTT | 31 (U43901) |
| UHG | Human | U22, U25–U31 | GAGATTCGTTCTCATTTTTC | 67 (U40580) |
| mUHG | Mouse | U22, U25–U31 | TCTTGTCGGCCTCATTTTTC | 67 (U40654) |
| U17HGe | Human | U17 | TTTTCTCTCTCCTTTTTGGA | This work |
| mU17HG | Mouse | U17 | CTCTCTAGGC | This work |
| gas5 | Human | U44, 47, U73-80 | CTTTTCG | 60a |
| gas5e | Mouse | U44, 47, U73-80 | CTCGGCCTTTCGGAG | 60a |
| U19HG | Human | U19 | CTGCGCCCTG | 6 |
| eIF4AII | Human | E3 | GTGGTTTTTC | 49 (D30655) |
| eIF4AII | Mouse | E3 | GTCTTTTCAG | 53 (X14422) |
| ATP synthase-β | Human | U59 | TGCAGCCTTCAGTCTCCACC | 51 (M2713) |
| eIF4AI | Mouse | U67 | GCGGCACTCCGCCCTAGATT | 55 (M22873) |
| hsc70 | Human | U14 | GGGCGGAAACCGGTGCTCAG | 18 (Y00371) |
| hsc70 | Mouse | U14 | ATTGAACGCGGAGGCAGCTG | 30 |
Mammalian snoRNA host genes encoding ribosomal proteins S3 (GenEMBL accession no. D28344), L1a (X06552), S8 (X67247), L5 (D10737), L7a (X61923), and L13a (X51528) are not included. All of these genes belong to the 5′TOP family.
5′ ends of transcripts are shown in bold, with pyrimidine residues underlined. The 5′ terminus in each gene listed is supported by a method(s) other than just cDNA cloning.
Intronic localization based on intron-containing ESTs corresponding to genes with characterized transcription start sites.
Possible additional transcription start site was mapped at T at position −2. According to Kato et al. (33), transcription most likely initiates at C+6, resulting in the 5′-terminus CTTTTCCGTG.
Transcription starts at two adjacent C residues.
The 5′-terminal C residue and a tract of 4 to 13 pyrimidines immediately following it are the key structural features distinguishing mRNAs for ribosomal proteins and some translational factors, collectively referred to as 5′TOP mRNAs, from most other cellular messengers. In addition, 5′TOP mRNAs usually contain relatively short and unstructured 5′UTRs, terminated with an AUG positioned in a favorable initiation context. The polypyrimidine tracts, in combination with sequences positioned downstream in the 5′UTR, are responsible for upregulation of translation of the 5′TOP mRNAs in response to growth factors or other conditions which require increased, and coordinated, synthesis of proteins making up the translational apparatus (reviewed in references 1 and 46). Besides their role in protein synthesis, the oligopyrimidine tracts, usually present not only downstream but also upstream of the start site C residue (46), are likely to play a role in transcription of the 5′TOP genes. Perry and coworkers (28, 58) have reported that integrity of the +1 oligopyrimidine tracts in mouse ribosomal protein genes S16 and L30 is important for efficient and precise initiation of their transcription.
The observation that all characterized non-protein-coding and several other snoRNA host genes belong to the 5′TOP family raises the possibility that expression of these genes, and their resident snoRNAs participating in ribosome biogenesis, is coregulated with expression of genes that encode components of the translational machinery. It is more plausible to propose that coordination of expression of snoRNA hosts and the genes coding for translational components occurs at the transcriptional rather than translational level. Processing of snoRNAs from introns is a nuclear event (reviewed in reference 45). In addition, it is rather unlikely that 5′-terminal oligopyrimidine tracts have been conserved in U17HG- and UHG-like RNAs in order to regulate translation of these apparently non-protein-coding RNAs. Although it could be argued that 5′TOP sequences in UHG and gas5 RNAs are conserved in order to regulate binding of these RNAs to ribosomes and subsequent RNA degradation, such an explanation would not apply to U17HG RNA, which does not appear to associate with ribosomes. A transcriptional rather than translational role of the +1 oligopyrimidine tracts in snoRNA host gene expression might also explain why oligopyrimidine sequences in some of the genes tolerate insertions of several purines (Table 1). It has been previously shown that replacement by purines of three or more pyrimidines in the cap site polypyrimidine stretch in mouse ribosomal protein genes is required to reduce their transcription (28, 58). In contrast, regulation of translation of the 5′TOP mRNA by cell growth conditions is severely affected by even a single C-to-A mutation at the 5′ end of the tract (1, 4, 46). Although we consider the transcriptional role for the +1 oligopyrimidine tract as the most probable one, it is possible that these sequences have a different function, more directly related to the processing of snoRNAs from primary transcripts.
In view of the rather widespread occurrence of non-protein-coding snoRNA host genes, it is interesting to speculate on their possible origins. Several scenarios can be envisaged. (i) Exons of these genes originally encoded a protein, but this property was lost during evolution. (ii) The host exons may have encoded a structural or regulatory RNA whose function became obsolete with time or which we have not yet identified. In this context, it is important to note that several intron-containing genes, the spliced products of which code for a known or putative regulatory RNA rather than an mRNA, have been characterized (2, 8, 9, 29, 71). There is no evidence that introns in these genes encode any functional RNAs. (iii) The UHG- and U17HG-like genes originated from polycistronic snoRNA genes, similar to genes expressed in plants and yeast (42, 54b, 72), by conversion of the inter-snoRNA spacers into spliceable exons. Loss of an endonuclease activity required for processing poly-snoRNAs could be a factor triggering such an evolutionary event. The last scenario would imply that transcriptional units encoding poly-snoRNAs are evolutionarily old prototype genes. Such genes could have originally arisen by duplication of a single-unit DNA (or RNA) segment at a time when the complexity of rRNA modification was increasing. Characterization of snoRNA transcription units in additional distantly related organisms might shed more light on the possible origins of the non-protein-coding snoRNA host genes.
ACKNOWLEDGMENTS
We thank P. Matthias for the cDNA library, K. Tycowski for the human UHG cDNA clone, C. Smith and J. A. Steitz for sharing unpublished results, F. Dragon, B. Hohn, Z. Lorkovic, and G. Thomas for critical reading of the manuscript, and H. Angliker, F. Fisher, and P. Müller for oligonucleotide synthesis and DNA sequencing.
REFERENCES
- 1.Amaldi F, Pierandrei-Amaldi P. TOP genes: a translationally controlled class of genes including those coding for ribosomal proteins. Prog Mol Subcell Biol. 1997;18:1–17. doi: 10.1007/978-3-642-60471-3_1. [DOI] [PubMed] [Google Scholar]
- 2.Amrein H, Axel R. Genes expressed in neurons of adult male Drosophila. Cell. 1997;88:459–469. doi: 10.1016/s0092-8674(00)81886-3. [DOI] [PubMed] [Google Scholar]
- 3.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current protocols in molecular biology. New York, N.Y: John Wiley & Sons; 1994. [Google Scholar]
- 4.Avni D, Shama S, Loreni F, Meyuhas O. Vertebrate mRNAs with a 5′-terminal pyrimidine tract are candidates for translational repression in quiescent cells: characterization of the translational cis-regulatory element. Mol Cell Biol. 1994;14:3822–3833. doi: 10.1128/mcb.14.6.3822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Balakin A G, Smith L, Fournier M J. The RNA world of the nucleolus: two major families of small RNAs defined by different box elements with related functions. Cell. 1996;86:823–834. doi: 10.1016/s0092-8674(00)80156-7. [DOI] [PubMed] [Google Scholar]
- 6.Bortolin M, Kiss T. Human U19 intron-encoded snRNA is processed from a long primary transcript that possesses little potential for protein coding. RNA. 1998;4:445–454. [PMC free article] [PubMed] [Google Scholar]
- 7.Bousquet-Antonelli C, Henry Y, G’elugne J P, Caizergues-Ferrer M, Kiss T. A small nucleolar RNP protein is required for pseudouridylation of eukaryotic ribosomal RNAs. EMBO J. 1997;16:4770–4776. doi: 10.1093/emboj/16.15.4770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brannan C I, Dees E C, Ingram R S, Tilghman S M. The product of the H19 gene may function as an RNA. Mol Cell Biol. 1990;10:28–36. doi: 10.1128/mcb.10.1.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brockdorff N, Ashworth A, Kay G F, McCabe V M, Norris D P, Cooper P J, Swift S, Rastan S. The product of the mouse Xist gene is a 15 kb inactive X- specific transcript containing no conserved ORF and located in the nucleus. Cell. 1992;71:515–526. doi: 10.1016/0092-8674(92)90519-i. [DOI] [PubMed] [Google Scholar]
- 10.Caffarelli E, Arese M, Santoro B, Fragapane P, Bozzoni I. In vitro study of processing of the intron-encoded U16 small nucleolar RNA in Xenopus laevis. Mol Cell Biol. 1994;14:2966–2974. doi: 10.1128/mcb.14.5.2966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Caffarelli E, Fatica A, Prislei S, De Gregorio E, Fragapane P, Bozzoni I. Processing of the intron-encoded U16 and U18 snoRNAs: the conserved C and D boxes control both the processing reaction and the stability of the mature snoRNA. EMBO J. 1996;15:1121–1131. [PMC free article] [PubMed] [Google Scholar]
- 12.Cavaille J, Bachellerie J P. Processing of fibrillarin-associated snoRNAs from pre-mRNA introns: an exonucleolytic process exclusively directed by the common stem-box terminal structure. Biochimie. 1996;78:443–456. doi: 10.1016/0300-9084(96)84751-1. [DOI] [PubMed] [Google Scholar]
- 13.Cavaille J, Nicoloso M, Bachellerie J P. Targeted ribose methylation of RNA in vivo directed by tailored antisense RNA guides. Nature. 1996;383:732–735. doi: 10.1038/383732a0. [DOI] [PubMed] [Google Scholar]
- 14.Cecconi F, Crosio C, Mariottini P, Cesareni G, Giorgi M, Brenner S, Amaldi F. A functional role for some Fugu introns larger than the typical short ones: the example of the gene coding for ribosomal protein S7 and snoRNA U17. Nucleic Acids Res. 1996;24:3167–3172. doi: 10.1093/nar/24.16.3167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cecconi F, Mariottini P, Amaldi F. The Xenopus intron-encoded U17 snoRNA is produced by exonucleolytic processing of its precursor in oocytes. Nucleic Acids Res. 1995;23:4670–4676. doi: 10.1093/nar/23.22.4670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cecconi F, Mariottini P, Loreni F, Pierandrei-Amaldi P, Campioni N, Amaldi F. U17XS8, a small nucleolar RNA with a 12 nt complementarity to 18S rRNA and coded by a sequence repeated in the six introns of Xenopus laevis ribosomal protein S8 gene. Nucleic Acids Res. 1994;22:732–741. doi: 10.1093/nar/22.5.732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dignam J D, Lebovitz R M, Roeder R G. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 1983;11:1475–1489. doi: 10.1093/nar/11.5.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dworniczak B, Mirault M E. Structure and expression of a human gene coding for a 71 kd heat shock ‘cognate’ protein. Nucleic Acids Res. 1987;15:5181–5197. doi: 10.1093/nar/15.13.5181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Enright C A, Maxwell E S, Eliceiri G L, Sollner-Webb B. 5′ETS rRNA processing facilitated by four small RNAs: U14, E3, U17, and U3. RNA. 1996;2:1094–1099. [PMC free article] [PubMed] [Google Scholar]
- 20.Fan X C, Myer V E, Steitz J A. AU-rich elements target small nuclear RNAs as well as mRNAs for rapid degradation. Genes Dev. 1997;11:2557–2568. doi: 10.1101/gad.11.19.2557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Feinberg A P, Vogelstein B. A technique for radiolabeling DNA restriction endonuclease fragments to high specific activity. Anal Biochem. 1984;137:266–267. doi: 10.1016/0003-2697(84)90381-6. [DOI] [PubMed] [Google Scholar]
- 22.Filipowicz W, Vicente O. RNA 3′-terminal phosphate cyclase from HeLa cells. Methods Enzymol. 1990;181:499–510. doi: 10.1016/0076-6879(90)81147-m. [DOI] [PubMed] [Google Scholar]
- 23.Furuno N, Nakagawa K, Eguchi U, Ohtsubo M, Nishimoto T, Soeda E, Ohtubo M. Complete nucleotide sequence of the human RCC1 gene involved in coupling between DNA replication and mitosis. Genomics. 1991;11:459–461. doi: 10.1016/0888-7543(91)90156-9. [DOI] [PubMed] [Google Scholar]
- 24.Ganot P, Bortolin M L, Kiss T. Site-specific pseudouridine formation in preribosomal RNA is guided by small nucleolar RNAs. Cell. 1997;89:799–809. doi: 10.1016/s0092-8674(00)80263-9. [DOI] [PubMed] [Google Scholar]
- 25.Ganot P, Caizergues-Ferrer M, Kiss T. The family of box ACA small nucleolar RNAs is defined by an evolutionarily conserved secondary structure and ubiquitous sequence elements essential for RNA accumulation. Genes Dev. 1997;11:941–956. doi: 10.1101/gad.11.7.941. [DOI] [PubMed] [Google Scholar]
- 26.Goodall G J, Wiebauer K, Filipowicz W. Analysis of pre-mRNA processing in transfected plant protoplasts. Methods Enzymol. 1990;181:148–161. doi: 10.1016/0076-6879(90)81117-d. [DOI] [PubMed] [Google Scholar]
- 27.Gorlich D, Mattaj I W. Nucleocytoplasmic transport. Science. 1996;271:1513–1518. doi: 10.1126/science.271.5255.1513. [DOI] [PubMed] [Google Scholar]
- 28.Hariharan N, Perry R P. Functional dissection of a mouse ribosomal protein promoter: significance of the polypyrimidine initiator and an element in the TATA-box region. Proc Natl Acad Sci USA. 1990;87:1526–1530. doi: 10.1073/pnas.87.4.1526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hogan N C, Traverse K L, Sullivan D E, Pardue M L. The nucleus-limited Hsr-omega-n transcript is a polyadenylated RNA with a regulated intranuclear turnover. J Cell Biol. 1994;125:21–30. doi: 10.1083/jcb.125.1.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hunt, C. R. 1996. GenBank accession no. U73744 (direct submission).
- 31.Jackers P, Minoletti F, Belotti D, Clausse N, Sozzi G, Sobel M E, Castronovo V. Isolation from a multigene family of the active human gene of the metastasis-associated multifunctional protein 37LRP/p40 at chromosome 3p21.3. Oncogene. 1996;13:495–503. [PubMed] [Google Scholar]
- 32.Jefferies H B, Thomas G. Elongation factor-1 alpha mRNA is selectively translated following mitogenic stimulation. J Biol Chem. 1994;269:4367–4372. [PubMed] [Google Scholar]
- 33.Kato S, Sekine S, Oh S W, Kim N S, Umezawa Y, Abe N, Yokoyama-Kobayashi M, Aoki T. Construction of a human full-length cDNA bank. Gene. 1994;150:243–250. doi: 10.1016/0378-1119(94)90433-2. [DOI] [PubMed] [Google Scholar]
- 34.Kiledian M, Burd C G, Gorlach M, Portman D S, Dreyfuss G. Structure and function of hnRNP proteins. In: Nagai K, Mattaj I W, editors. RNA-protein interactions. Oxford, England: Oxford University Press; 1994. pp. 125–149. [Google Scholar]
- 35.Kim N S, Kato T, Abe N, Kato S. Nucleotide sequence of human cDNA encoding eukaryotic initiation factor 4AI. Nucleic Acids Res. 1993;21:2012. doi: 10.1093/nar/21.8.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35a.Kiss, T. Personal communication.
- 36.Kiss T, Filipowicz W. Exonucleolytic processing of small nucleolar RNAs from pre-mRNA introns. Genes Dev. 1995;9:1411–1424. doi: 10.1101/gad.9.11.1411. [DOI] [PubMed] [Google Scholar]
- 37.Kiss T, Filipowicz W. Small nucleolar RNAs encoded by introns of the human cell cycle regulatory gene RCC1. EMBO J. 1993;12:2913–2920. doi: 10.1002/j.1460-2075.1993.tb05953.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kiss-Laszlo Z, Henry Y, Bachellerie J P, Caizergues-Ferrer M, Kiss T. Site-specific ribose methylation of preribosomal RNA: a novel function for small nucleolar RNAs. Cell. 1996;85:1077–1088. doi: 10.1016/s0092-8674(00)81308-2. [DOI] [PubMed] [Google Scholar]
- 39.Kozak M. An analysis of 5′-noncoding sequences from 699 vertebrate messenger RNAs. Nucleic Acids Res. 1987;15:8125–8148. doi: 10.1093/nar/15.20.8125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kozak M. Interpreting cDNA sequences: some insights from studies on translation. Mamm Genome. 1996;7:563–574. doi: 10.1007/s003359900171. [DOI] [PubMed] [Google Scholar]
- 41.Lagnado C A, Brown C Y, Goodall G J. AUUUA is not sufficient to promote poly(A) shortening and degradation of an mRNA: the functional sequence within AU-rich elements may be UUAUUUA(U/A)(U/A) Mol Cell Biol. 1994;14:7984–7995. doi: 10.1128/mcb.14.12.7984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Leader D J, Clark G P, Watters J, Beven A F, Shaw P J, Brown J W S. Clusters of multiple different small nucleolar RNA genes in plants are expressed as and processed from polycistronic pre-snoRNAs. EMBO J. 1997;16:5742–5751. doi: 10.1093/emboj/16.18.5742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Leverette R D, Andrews M T, Maxwell E S. Mouse U14 snRNA is a processed intron of the cognate hsc70 heat shock pre-messenger RNA. Cell. 1992;71:1215–1221. doi: 10.1016/s0092-8674(05)80069-8. [DOI] [PubMed] [Google Scholar]
- 44.Maquat L E. When cells stop making sense: effects of nonsense codons on RNA metabolism in vertebrate cells. RNA. 1995;1:453–465. [PMC free article] [PubMed] [Google Scholar]
- 45.Maxwell E S, Fournier M J. The small nucleolar RNAs. Annu Rev Biochem. 1995;64:897–934. doi: 10.1146/annurev.bi.64.070195.004341. [DOI] [PubMed] [Google Scholar]
- 46.Meyuhas O, Avni D, Shama S. Translational control of ribosomal protein mRNA in eukaryotes, translational control. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1996. [Google Scholar]
- 47.Mishra R K, Eliceiri G L. Three small nucleolar RNAs that are involved in ribosomal RNA precursor processing. Proc Natl Acad Sci USA. 1997;94:4972–4977. doi: 10.1073/pnas.94.10.4972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nag M K, Thai T T, Ruff E A, Selvamurugan N, Kunnimalaiyaan M, Eliceiri G L. Genes for E1, E2, and E3 small nucleolar RNAs. Proc Natl Acad Sci USA. 1993;90:9001–9005. doi: 10.1073/pnas.90.19.9001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nakamura A, Amikura R, Mukai M, Kobayashi S, Lasko P F. Requirement for a noncoding RNA in Drosophila polar granules for germ cell establishment. Science. 1996;274:2075–2079. doi: 10.1126/science.274.5295.2075. [DOI] [PubMed] [Google Scholar]
- 50.Nakanishi T, Kohno K, Ishiura M, Ohashi H, Uchida T. Complete nucleotide sequence and characterization of the 5′- flanking region of mammalian elongation factor 2 gene. J Biol Chem. 1988;263:6384–6391. [PubMed] [Google Scholar]
- 51.Neckelmann N, Warner C K, Chung A, Kudoh J, Minoshima S, Fukuyama R, Maekawa M, Shimizu Y, Shimizu N, Liu J D, et al. The human ATP synthase beta subunit gene: sequence analysis, chromosome assignment, and differential expression. Genomics. 1989;5:829–843. doi: 10.1016/0888-7543(89)90125-0. [DOI] [PubMed] [Google Scholar]
- 52.Ni J, Tien A L, Fournier M J. Small nucleolar RNAs direct site-specific synthesis of pseudouridine in ribosomal RNA. Cell. 1997;89:565–573. doi: 10.1016/s0092-8674(00)80238-x. [DOI] [PubMed] [Google Scholar]
- 53.Nielsen P J, Trachsel H. The mouse protein synthesis initiation factor 4A gene family includes two related functional genes which are differentially expressed. EMBO J. 1988;7:2097–2105. doi: 10.1002/j.1460-2075.1988.tb03049.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ohtsubo M, Kai R, Furuno N, Sekiguchi T, Sekiguchi M, Hayashida H, Kuma K, Miyata T, Fukushige S, Murotsu T, et al. Isolation and characterization of the active cDNA of the human cell cycle gene (RCC1) involved in the regulation of onset of chromosome condensation. Genes Dev. 1987;1:585–593. doi: 10.1101/gad.1.6.585. [DOI] [PubMed] [Google Scholar]
- 54a.Pelczar, P., and W. Filipowicz. Unpublished data.
- 54b.Petfalski E, Dandekar T, Henry Y, Tollervey D. Processing of the precursors to small nucleolar RNAs and rRNAs requires common components. Mol Cell Biol. 1998;18:1181–1189. doi: 10.1128/mcb.18.3.1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Reddy N S, Roth W W, Bragg P W, Wahba A J. Isolation and mapping of a gene for protein synthesis initiation factor 4A and its expression during differentiation of murine erythroleukemia cells. Gene. 1988;70:231–243. doi: 10.1016/0378-1119(88)90195-3. [DOI] [PubMed] [Google Scholar]
- 56.Rimoldi O J, Raghu B, Nag M K, Eliceiri G L. Three new small nucleolar RNAs that are psoralen cross-linked in vivo to unique regions of pre-rRNA. Mol Cell Biol. 1993;13:4382–4390. doi: 10.1128/mcb.13.7.4382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ruff E A, Rimoldi O J, Raghu B, Eliceiri G L. Three small nucleolar RNAs of unique nucleotide sequences. Proc Natl Acad Sci USA. 1993;90:635–638. doi: 10.1073/pnas.90.2.635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Safrany G, Perry R P. The relative contributions of various transcription factors to the overall promoter strength of the mouse ribosomal protein L30 gene. Eur J Biochem. 1995;230:1066–1072. doi: 10.1111/j.1432-1033.1995.tb20657.x. [DOI] [PubMed] [Google Scholar]
- 59.Sambrook J, Fritsch F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 60.Selvamurugan N, Joost O H, Haas E S, Brown J W, Galvin N J, Eliceiri G L. Intracellular localization and unique conserved sequences of three small nucleolar RNAs. Nucleic Acids Res. 1997;25:1591–1596. doi: 10.1093/nar/25.8.1591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60a.Smith, C., and J. A. Steitz. Submitted for publication.
- 61.Smith C M, Steitz J A. Sno storm in the nucleolus: new roles for myriad small RNPs. Cell. 1997;89:669–672. doi: 10.1016/s0092-8674(00)80247-0. [DOI] [PubMed] [Google Scholar]
- 62.Sollner-Webb B. Novel intron-encoded small nucleolar RNAs. Cell. 1993;75:403–405. doi: 10.1016/0092-8674(93)90374-y. [DOI] [PubMed] [Google Scholar]
- 63.Sollner-Webb B, Tycowski K T, Steitz J A. Ribosomal RNA. Structure, evolution, processing and function in protein biosynthesis. In: Zimmerman R A, Dahlberg A E, editors. Ribosomal processing in eukaryotes. Boca Raton, Fla: CRC Press; 1996. pp. 469–490. [Google Scholar]
- 64.Srivastava M, McBride O W, Fleming P J, Pollard H B, Burns A L. Genomic organization and chromosomal localization of the human nucleolin gene. J Biol Chem. 1990;265:14922–14931. [PubMed] [Google Scholar]
- 65.Strubin M, Newell J W, Matthias P. OBF-1, a novel B cell-specific coactivator that stimulates immunoglobulin promoter activity through association with octamer-binding proteins. Cell. 1995;80:497–506. doi: 10.1016/0092-8674(95)90500-6. [DOI] [PubMed] [Google Scholar]
- 66.Tollervey D, Kiss T. Function and synthesis of small nucleolar RNAs. Curr Opin Cell Biol. 1997;9:337–342. doi: 10.1016/s0955-0674(97)80005-1. [DOI] [PubMed] [Google Scholar]
- 67.Tycowski K T, Shu M D, Steitz J A. A mammalian gene with introns instead of exons generating stable RNA products. Nature. 1996;379:464–466. doi: 10.1038/379464a0. [DOI] [PubMed] [Google Scholar]
- 68.Tycowski K T, Shu M D, Steitz J A. A small nucleolar RNA is processed from an intron of the human gene encoding ribosomal protein S3. Genes Dev. 1993;7:1176–1190. doi: 10.1101/gad.7.7a.1176. [DOI] [PubMed] [Google Scholar]
- 69.Tycowski K T, Smith C M, Shu M D, Steitz J A. A small nucleolar RNA requirement for site-specific ribose methylation of rRNA in Xenopus. Proc Natl Acad Sci USA. 1996;93:14480–14485. doi: 10.1073/pnas.93.25.14480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.van den Ouweland A M, Verdijk M, Mannens M M, van Oost B A. The QM gene is X-linked and therefore not involved in suppression of tumorigenesis in Wilms’ tumor. Hum Genet. 1992;90:144–146. doi: 10.1007/BF00210759. [DOI] [PubMed] [Google Scholar]
- 71.Wevrick R, Francke U. An imprinted mouse transcript homologous to the human imprinted in Prader-Willi syndrome (IPW) gene. Hum Mol Genet. 1997;6:325–332. doi: 10.1093/hmg/6.2.325. [DOI] [PubMed] [Google Scholar]
- 72.Zagorski J, Tollervey D, Fournier M J. Characterization of an SNR gene locus in Saccharomyces cerevisiae that specifies both dispensable and essential small nuclear RNAs. Mol Cell Biol. 1988;8:3282–3290. doi: 10.1128/mcb.8.8.3282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Zucker, M., and D. H. Turner.http://www.ibc.wustl.edu.