Abstract
Motivation
Genomic intervals are one of the most prevalent data structures in computational genome biology, and used to represent features ranging from genes, to DNA binding sites, to disease variants. Operations on genomic intervals provide a language for asking questions about relationships between features. While there are excellent interval arithmetic tools for the command line, they are not smoothly integrated into Python, one of the most popular general-purpose computational and visualization environments.
Results
Bioframe is a library to enable flexible and performant operations on genomic interval dataframes in Python. Bioframe extends the Python data science stack to use cases for computational genome biology by building directly on top of two of the most commonly-used Python libraries, NumPy and Pandas. The bioframe API enables flexible name and column orders, and decouples operations from data formats to avoid unnecessary conversions, a common scourge for bioinformaticians. Bioframe achieves these goals while maintaining high performance and a rich set of features.
Availability and implementation
Bioframe is open-source under MIT license, cross-platform, and can be installed from the Python Package Index. The source code is maintained by Open2C on GitHub at https://github.com/open2c/bioframe.
1 Introduction
Operations on genomic intervals, also known as genomic ranges, are fundamental to bioinformatic analyses. These operations can be used to answer questions that include: Where is the closest enhancer to a gene of interest? How do chromatin states change across cell types? Which repeat elements contain binding sites for transcription factor motifs? Which annotations are enriched for SNVs associated with various diseases? Which promoters contain eQTLs? Given the ubiquity of these sorts of queries in genomic analysis, specialized interval arithmetic tools have been developed for the command line (Quinlan and Hall 2010, Neph et al. 2012), which operate on genomic interval text files such as the Browser Extensible Data (BED) format. To facilitate interactive and programmatic use cases, there are also implementations for popular programming environments, including Python (Dale et al. 2011, Stovner and Sætrom 2020, Russell and Fiddes 2021), and R (Lawrence et al. 2013, Akalin et al. 2015, Lee et al. 2019).
The rich and robust set of data science and machine learning libraries makes Python a popular choice for computational biology and data science more broadly. Core libraries in the Python data science stack, including Pandas (The pandas development team 2023), NumPy (Harris et al. 2020), Matplotlib (Hunter 2007), and Jupyter notebooks (Kluyver et al. 2016), offer nearly seamless integration. Other disciplines have developed tools to leverage the Python data science infrastructure, e.g. GeoPandas (den Bossche et al. 2024), SpatialPandas (Pothina et al. 2020), and BioPandas (Raschka 2017) (for molecular structures). However, Python libraries for genomic intervals do not yet meet this high standard of integration.
Current Python packages providing support for genomic interval operations have limitations that impede smooth integration into Python data science workflows. For example, pybedtools (Dale et al. 2011), the wrapper for bedtools (Quinlan and Hall 2010), relies on interconversion between in-memory objects and text files stored on disk because data processing is delegated to a command line program. Furthermore, it inherits an API designed for the command line, and is restricted by rigid genomic interval schemas designed for storage (e.g. BED files). This leads to lower expressivity and less flexibility than what can be accomplished using Python-native (e.g. NumPy and Pandas) data structures and operations, as well as terse arguments with unintuitive names (e.g. -wao). The consequences include decreased performance, more boilerplate code, loss of metadata (such as column names), and code that is more difficult to read and debug. More recently, PyRanges (Stovner and Sætrom 2020) addresses many of these shortcomings, in particular by providing a 10–50 speed increase, but still has an API that is somewhat insulated from the data science stack. Conversions are required to switch between the custom PyRanges object used to perform genomic interval operations and standard Pandas DataFrames, and PyRanges columns have relatively strict naming conventions.
The growth of the Python data science ecosystem presents an opportunity to re-imagine the implementation of genomic interval operations to smoothly interface with the full data science stack. Integration with this ecosystem can enable new avenues for data visualization, modeling, and insight into genomic data. Here we present bioframe, a Python library for operating on genomic interval sets built directly on top of the Pandas data analysis library. As a result, bioframe is fast and Pythonic, providing immediate access to a rich set of DataFrame operations. This in turn enables complex workflows as well as rapid iteration, inspection and visualization of genomic analyses.
2 Methods
2.1 Design principles
The goal of bioframe is to enable in-memory, programmatic workflows on sets of genomic intervals using Pandas DataFrames, integrating smoothly with the Python data science stack. With this in mind, we aimed to:
Reuse existing data structures: We encode interval sets using DataFrames and avoid introducing new custom objects, e.g. ones based on interval trees.
Reuse existing methods: We delegate generic DataFrame operations to Pandas, whenever possible, and aim at the principle of least surprise for experienced Pandas and NumPy users.
Permit flexible schemas: We avoid hard-coded column names, numbers and orderings.
2.2 Definitions
To implement genomic interval operations in Python, we required formal definitions, which we did not readily find in one place in the literature. We thus put together definitions, starting from an interval (https://bioframe.readthedocs.io/en/latest/guide-definitions.html). We implemented these definitions as specifications (https://bioframe.readthedocs.io/en/latest/api-validation.html) for properties of genomic interval dataframes.
While aligning reads to the full set of scaffolds in an assembly is typically advisable, using a subset of scaffolds and/or breaking scaffolds into semantic subintervals (e.g. chromosome arms) is often crucial for downstream genomic analyses. In bioframe we thus introduce the concept of a genomic view to specify a unique genomic coordinate sub-system. A genomic view is an ordered set of uniquely-named non-overlapping genomic intervals known as regions. There can be more than one region from the same scaffold and multiple scaffolds represented in a view. Defining a view allows a user to focus analysis on a well-characterized portion of an assembly and specify the order of scaffolds and regions for effective visualization. Indeed, defining a view for downstream analysis can be more important for non-model organisms, where assembly quality is often lower and thus requires judicious choice of order and subset of scaffolds to analyze.
3 Implementation
Bioframe is implemented using the machinery of NumPy and Pandas, making for a lightweight set of dependencies. For example, to determine overlaps between intervals (bioframe.overlap), as well as find pairs of nearby intervals (bioframe.closest), bioframe uses sorting-based algorithms. First, intervals are split into subsets by chromosome and optional columns like “strand” (DataFrame.groupby). These interval subsets are then sorted (numpy.lexsort), and overlaps (or neighbors) are detected via bisection search operations (numpy.searchsorted). All user-facing operations are imported into the base bioframe namespace.
Building directly on Pandas allows bioframe to readily generalize the genomic interval model used for BED files. Bioframe requires only genomic coordinate columns—the equivalent of chrom, chromStart, chromEnd in the BED specification—with flexible names, for a valid BED-like DataFrame, or “BedFrame”. Almost any number of additional annotations can be added to a set of intervals.
4 Functionality
The core genomic interval operations in bioframe are: overlap, cluster, closest, and complement. Bioframe additionally provides frequently-used operations that can be expressed as combinations of these core operations and Pandas DataFrame operations, including: coverage, count_overlaps, expand, merge, select, subtract, setdiff, and trim. Building from the definition of genomic views, bioframe provides functions to: assign intervals in a bedframe to regions in a genomic view, assign_view, and sort a BedFrame based on the order of regions specified in a view, sort_bedframe.
Building on Pandas enables flexible control over column usage and selection. Bioframe includes a context manager for setting default column-names for genomic coordinate columns. This flexibility shines when dealing with BED-like files that can have variable headers or conventions for the genomic coordinate columns (e.g. chrom or chromStart or chr or CHR#), and variable orders of other interval metadata columns (e.g. score, color, or strand). Since operations like overlap are performed following a DataFrame groupby operation, bioframe also flexibly generalizes genomic operations that consider strand to any list of common columns present in a pair of DataFrames.
In addition to these features, bioframe provides functions for genomic interval DataFrame construction, checks, string operations, and I/O. For example, there are wrappers and schemas for reading and writing common binary and text genomic file formats to and from DataFrames.
5 Performance
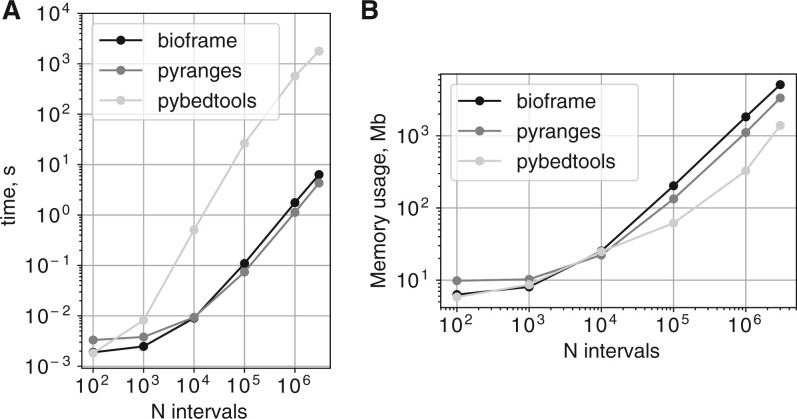
We profiled speed and memory usage for typical use cases using bioframe in an interactive Jupyter notebook, and compared performance with that of pybedtools and PyRanges. We intersected sets of random genomic intervals stored as pandas DataFrames, and included the format conversions needed in pybedtools and PyRanges. For overlaps of up to intervals, bioframe and PyRanges have comparable speeds (Figure 1A), while pybedtools can be more than 100 slower. The memory consumption of bioframe is similar to PyRanges and both are higher than pybedtools (Figure 1B). We note that for chained operations, PyRanges offers further speedups by caching per-chromosome interval tables, with the tradeoff of storing intervals as a custom object with its own API layer. We also note that while the other libraries use signed 64-bit integers to represent genomic coordinates, bioframe has the flexibility to use any numerical data type, including both NumPy types and Pandas integer extension types that support NA missing values. To conclude, both bioframe and PyRanges offer reasonable performance ( for intervals) on genomic interval data frames. For much larger sets of genomic intervals (), users may also want to consider other high-performance options (Neph et al. 2012, Li and Rong 2021).
Figure 1.
Performance comparison of bioframe v0.6.1, PyRanges v0.0.129, and pybedtools v0.9.1 (bedtools v2.30.0) for detecting overlapping intervals between pairs of DataFrames of randomly generated genomic intervals. (A) Run time and (B) Peak memory consumption of bioframe overlap vs. PyRanges join show comparable performance up to millions of intervals and comparable memory usage. Pybedtools intersect shows slower performance. Code for this performance comparison is available at https://bioframe.readthedocs.io/en/latest/guide-performance.html.
6 Conclusion
In summary, bioframe provides a pure Python library for genomic interval operations. Bioframe presents a Python-centered API for these operations, as opposed to inheriting syntax from the command line. Working in Python with Pandas DataFrames enables flexible generalization of the BED format, including flexible naming for genomic interval columns. Bioframe has already proven useful for Pandas-heavy genomic workflows, like cooltools (Venev et al. 2023). In the future, providing tools specific to binned genomic intervals, paired genomic intervals, and out-of-core dataframe operations, e.g. with Dask (Rocklin 2015) or Modin (Petersohn et al. 2020), would be valuable extensions to bioframe.
Acknowledgments
The authors thank Endre Bakken Stovner, Sameer Abraham, Luis Chumpitaz, George Spracklin, Aafke van den Berg, and Vedat Yilmaz for helpful comments and suggestions.
Contributor Information
Open2C, https://open2c.github.io.
Nezar Abdennur, Department of Genomics and Computational Biology, UMass Chan Medical School, Worcester, MA 01605, United States; Department of Systems Biology, UMass Chan Medical School, Worcester, MA 01605, United States.
Geoffrey Fudenberg, Department of Quantitative and Computational Biology, University of Southern California, Los Angeles, CA 90089, United States.
Ilya M Flyamer, Friedrich Miescher Institute for Biomedical Research, 4058 Basel, Switzerland.
Aleksandra A Galitsyna, Institute for Medical Engineering and Science, Massachusetts Institute of Technology, Cambridge, MA 02139, United States.
Anton Goloborodko, Institute of Molecular Biotechnology of the Austrian Academy of Sciences (IMBA), Vienna BioCenter (VBC), 1030 Vienna, Austria.
Maxim Imakaev, Institute for Medical Engineering and Science, Massachusetts Institute of Technology, Cambridge, MA 02139, United States.
Sergey Venev, Department of Systems Biology, UMass Chan Medical School, Worcester, MA 01605, United States.
Author contributions
NA, GF, IMF, AAG, AG, MI, and SV made contributions as detailed in the Open2C authorship policy guide. All authors are listed alphabetically, and read, and approved the manuscript.
Conflict of interest
No competing interest is declared.
Funding
A.G. is supported by IMBA and the Austrian Academy of Sciences (OeAW). G.F. is supported by the National Institute of General Medical Sciences R35 GM143116-01. N.A. acknowledges support from the National Institutes of Health Common Fund 4D Nucleome Program (DK107980). I.F. acknowledges funding support from the Medical Research Council University Unit grant MC_UU_00007/2.
Data availability
Source code is available at https://github.com/open2c/bioframe.
References
- Akalin A, Franke V, Vlahoviček K. et al. Genomation: a toolkit to summarize, annotate and visualize genomic intervals. Bioinformatics 2015;31:1127–9. [DOI] [PubMed] [Google Scholar]
- Dale RK, Pedersen BS, Quinlan AR.. Pybedtools: a flexible python library for manipulating genomic datasets and annotations. Bioinformatics 2011;27:3423–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- den Bossche JV, Jordahl K, Fleischmann M. et al. geopandas/geopandas: v0.14.3. 2024. 10.5281/zenodo.2585848 [DOI]
- Harris CR, Millman KJ, van der Walt SJ. et al. Array programming with NumPy. Nature 2020;585:357–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter JD. Matplotlib: a 2D graphics environment. Comput Sci Eng 2007;9:90–5. [Google Scholar]
- Kluyver T, Ragan-Kelley B, Pérez F. et al. Jupyter notebooks-a publishing format for reproducible computational workflows. Elpub 2016;2016:87–90. [Google Scholar]
- Lawrence M, Huber W, Pagès H. et al. Software for computing and annotating genomic ranges. PLoS Comput Biol 2013;9:e1003118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S, Cook D, Lawrence M.. Plyranges: a grammar of genomic data transformation. Genome Biol 2019;20:4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Rong J.. Bedtk: finding interval overlap with implicit interval tree. Bioinformatics June 2021;37:1315–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neph S, Kuehn MS, Reynolds AP. et al. BEDOPS: high-performance genomic feature operations. Bioinformatics July 2012;28:1919–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersohn D, Macke S, Xin D. et al. Towards scalable dataframe systems. Proc VLDB Endow 2020;13:2033–46. [Google Scholar]
- Pothina D, Pevey K, Lewis A. Spatial algorithms at scale with spatialpandas. In: Proceedings of the Python in Science Conference, Austin, TX, USA, 2020. 10.25080/Majora-342d178e-026 [DOI]
- Quinlan AR, Hall IM.. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 2010;26:841–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raschka S. BioPandas: working with molecular structures in pandas DataFrames. JOSS 2017;2:279. [Google Scholar]
- Rocklin M. Dask: Parallel computation with blocked algorithms and task scheduling. In: Proceedings of the 14th python in science conference, Austin, TX, USA, vol. 130, p. 136, 2015. [Google Scholar]
- Russell PH, Fiddes IT. Biocantor: a python library for genomic feature arithmetic in arbitrarily related coordinate systems. bioRxiv, 10.1101/2021.07.09.451743, 2021, preprint: not peer reviewed. [DOI]
- Stovner EB, Sætrom P.. PyRanges: efficient comparison of genomic intervals in python. Bioinformatics 2020;36:918–9. [DOI] [PubMed] [Google Scholar]
- The pandas development team. pandas-dev/pandas: Pandas. 2023. 10.5281/zenodo.3509134 [DOI]
- Venev S, Abdennur N, Goloborodko A. et al. open2c/cooltools: v0.6.1. 2023. 10.5281/zenodo.3553139 [DOI]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Source code is available at https://github.com/open2c/bioframe.