Abstract
The chicken cardiac troponin T (cTNT) gene contains a single 30-nucleotide alternative exon that is included in embryonic striated muscle and skipped in the adult. Transient-transfection analysis of cTNT minigenes in muscle and fibroblast cell cultures previously identified four muscle-specific splicing enhancers (MSEs) that promote exon inclusion specifically in embryonic striated muscle cultures. Three MSEs located in the intron downstream from the alternative exon were sufficient for muscle-specific exon inclusion. In the present study, the boundaries of these MSEs were defined by scanning mutagenesis, allowing analysis of individual elements in gain-of-function experiments. Concatamers of MSE2 were necessary and sufficient to promote muscle-specific inclusion of a heterologous exon, indicating that it is a target for muscle-specific regulation. Sequences present in MSE2 are also found in MSE4, suggesting that these two MSEs act in a similar manner. MSE3 appears to be different from MSE2 and MSE4 yet is able to functionally replace both of these elements, demonstrating functional redundancy of elements that are likely to bind different factors. MSE2 and MSE4 each contain a novel sequence motif that is found adjacent to a number of alternative exons that undergo regulated splicing in striated muscle, suggesting a common role for this element in muscle-specific regulation.
A large number of genes express multiple protein isoforms as a result of alternative pre-mRNA splicing (2, 23). For many genes, the choice of splicing pathway is regulated according to cell-specific patterns (e.g., according to differentiated cell type, developmental stage, or gender or in response to an external stimulus). Investigations into the mechanism of alternative splicing have addressed two major questions: (i) what is the basis for nonconstitutive usage of some splice sites and (ii) what is the basis for cell-specific modulation of splice site usage? An answer to the first question has emerged: alternative splice sites are generally recognized less efficiently than constitutive splice sites due to several features of the pre-mRNA, such as splice site sequences that diverge from the consensus, small exon size, relative strength of competing splice sites, or secondary structure. For some exons, exonic or intronic splicing enhancers are required to prevent the exon from being ignored, while for other exons, repressor elements can contribute to nonconstitutive use of splice sites. The question of cell-specific regulation has been more difficult to answer. Paradigms for regulated splicing have been well defined in Drosophila melanogaster; however, progress has been slower in vertebrate experimental systems. A large number of pre-mRNA cis elements have been shown to affect splicing efficiency; however, only a few of these appear to be targets for cell-specific regulation (4).
Our laboratory is using the chicken cardiac troponin T (cTNT) gene to investigate the mechanisms of regulated splicing in striated muscle. cTNT is transcribed in embryonic skeletal muscle and in embryonic and adult cardiac muscle (8, 9). One exon (exon 5) within the pre-mRNA undergoes developmentally regulated splicing such that it is included in embryonic skeletal and cardiac muscle and is excluded in the adult (9). Transient-transfection analysis using cTNT minigenes has demonstrated that the default splicing pattern in nonmuscle cells is exon skipping. Exon inclusion in embryonic striated muscle cultures is mediated by a positive mechanism that requires muscle-specific splicing enhancers (MSEs) located in the adjacent introns and trans-acting factors that are induced as part of the myogenic program (27).
The last 99 nucleotides of intron 4 and the first 142 nucleotides of intron 5 were sufficient to mediate enhanced inclusion of a heterologous exon in embryonic skeletal muscle cultures (27). Regulation with this cTNT genomic fragment was comparable to that observed with a larger genomic fragment containing exons 1 to 6, indicating that the proximal regions were sufficient for the maximal level of regulation observed by transfection. Substitution and deletion analysis of this small genomic fragment defined four MSEs within the flanking introns, at least three of which are required for regulation. MSE1 is located between the branch site and the 3′ splice site in intron 4. MSE2, 3, and 4 are located downstream from the alternative exon in intron 5. Each MSE was defined by one mutation (substitution or deletion of a block of 13 to 32 nucleotides) that reduced the level of exon inclusion in muscle to or to less than the default level of exon inclusion observed in fibroblast cultures. Importantly, these mutations did not affect the default level of inclusion in fibroblasts demonstrating muscle-specific recognition. MSE3 was initially identified by its conservation in both sequence and position in three genes that undergo similarly regulated alternative splicing in striated muscle. The functional boundaries for the three MSEs within intron 5 were unknown. In addition, since at least three MSEs were required for muscle-specific exon inclusion, it was unknown whether each element was able to direct muscle-specific regulation or whether only some MSEs were targets for muscle-specific regulation while the others served an essential function through recognition by the constitutive splicing machinery.
In the present study, boundaries for the MSEs in intron 5 were defined by scanning mutagenesis, providing the basis for gain-of-function analysis of individual elements. Sequence and functional comparisons of MSE2 and MSE3 demonstrated that these are distinct elements with redundant functions. MSE2 and MSE4 appear to be repeats of a single element and contain a motif that is located adjacent to several alternative exons that undergo regulated splicing in muscle. Gain-of-function experiments using concatamerized elements indicated that MSE2 alone can promote strong muscle-specific inclusion of a heterologous exon. The ability to regulate splicing by using a single element will greatly simplify analysis of the factors that promote muscle-specific exon inclusion.
MATERIALS AND METHODS
Minigene constructs.
All clones were derived from RTB33.51 in which a cassette containing cTNT genomic fragments flanking a heterologous alternative exon was inserted into the second intron of a minigene derived from the constitutively spliced skeletal troponin I gene (reference 27 and Fig. 1). The cTNT cassette contains (from 5′ to 3′) the last 99 nucleotides of cTNT intron 4, a 46-nucleotide heterologous exon (see below), and the first 142 nucleotides of cTNT intron 5. Cloning was facilitated by three restriction sites that were unique to the RTB33.51 plasmid: a SalI site at the 5′ end of the cTNT fragment, a BstBI site within the heterologous alternative exon, and an SpeI site at the 3′ end of the cTNT fragment (Fig. 1). The heterologous exon (GGTTCACAACCATCTACGCATTCGAACCAAGCAAGATGTCTGACAG) contains the 30-nucleotide sTNI exon 2 (nonunderlined text), a 12-nucleotide random insert (underlined once) containing a BstBI site, and the consensus first and last exon nucleotides (underlined twice). The cTNT splice sites in RTB33.51 were converted to globin sites [R35(46)] (Fig. 1) by using PCR primers that contained the nucleotide substitutions. To generate a series of exon sizes in the R35(46) plasmid, PCR was used to amplify the cTNT insert as two halves that could be ligated together at different sites within the heterologous exon (6). The EI4 oligonucleotide (GCCTTCATCGATTCGAACCGGTCGATGGTTGTGAACCCT) was used to prime the upstream half of the cTNT genomic fragment (Fig. 1), and the EI5 oligonucleotide (GCCGGCATCGATGGCGCCTCGAGATCTCTGACAGGT) was used to prime the downstream half. The underlined regions of these oligonucleotides anneal within the heterologous exon. The regions not underlined contain multiple compatible restriction sites with CG 5′ overhangs. The upstream and downstream PCR products were digested with these enzymes in separate reactions, and different combinations of upstream and downstream fragments were ligated together to reassemble the cTNT genomic fragment, producing heterologous exons of different sizes. The scanning mutations (Fig. 2) were introduced by the megaprimer approach (28). MSE2 and MSE3 fragments used for concatamerization were generated by PCR. The priming oligonucleotides introduced compatible restriction sites (XbaI or NheI) at the ends of the PCR product so that the DNA could be digested, gel isolated, and concatamerized by ligation. Ligation reaction products were then digested with both restriction enzymes to obtain ligations in only the head-to-tail orientation (in the head-to-tail orientation, both restriction sites are destroyed; junctions of non-head-to-tail ligations regenerate one of the restriction sites). Concatamers were then blunt-ended with T4 DNA polymerase and were gel isolated. To construct MSE2(×3) (a construct with three copies of MSE2), MSE3(×3), MSE3(×6), and MSE3mut(×6) (see Fig. 4), concatamers were inserted into R5.15-21 between the AatII (introduced by the mutation) (Fig. 2) and SpeI sites (after blunt-ending with T4 DNA polymerase), placing the concatemers 16 nucleotides downstream from the exon. The globin 3′ splice site in R5.15-21 was replaced with a SalI/BstBI fragment containing the cTNT intron 4 segment. To construct M2/M2, the blunt-ended MSE2(×3) concatamer was blunt-end ligated onto a PCR product containing the first nucleotide of the exon to the SpeI site, and this ligated fragment was cloned between the SalI (filled-in) and SpeI sites in the RTB33.51 plasmid (Fig. 1). To construct M2/M2TB, a 12-nucleotide synthetic double-stranded fragment (CGCTCGAGCAAT) was ligated into the unique BstBI site within the exon. All constructs were confirmed by sequencing.
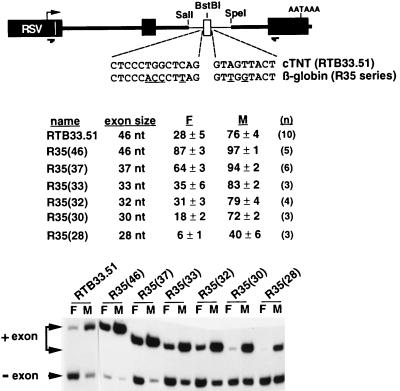
FIG. 1.
cTNT MSEs regulate splicing of a heterologous exon flanked by heterologous splice sites. The diagram illustrates that a 46-nucleotide exon (open box) flanked by the last 99 nucleotides of cTNT intron 4 and the first 142 nucleotides of cTNT intron 5 (thin lines) was inserted into a constitutively spliced minigene derived from skeletal troponin T (thick lines and filled boxes). The relative positions of the oligonucleotides used for RT-PCR are indicated by arrows below the diagram. Nucleotide substitutions within the cTNT splice sites (underlined) were used to introduce the 3′ and 5′ splice sites of human β-globin intron 1. RTB33.51 contains the natural cTNT splice sites. The R35 series constructs contain the globin splice sites flanking an exon of the size indicated in parentheses. Minigenes were transiently transfected into QT35 fibroblasts (F) and primary chicken embryo skeletal muscle (M) cultures and assayed and quantitated as described in Materials and Methods. n, number of independent transfections.
FIG. 2.
Scanning mutagenesis of the MSEs in cTNT intron 5. Shown for each clone are the name, the percent of spliced mRNA that includes the exon in fibroblast (F) and muscle (M) cultures (with standard deviation), the number of transfections (n), and whether splicing is regulated in muscle (REG). Constructs marked with a minus sign do not express statistically significant higher levels of exon inclusion in muscle cultures than in nonmuscle cultures. For example, the difference between the average levels of exon inclusion for construct RTB76-81 is not significant due to the high variability of the results indicated by the large standard deviations. Several mutations alter basal splicing efficiency (see text). The focus of this analysis is the difference in level of exon inclusion between fibroblasts and muscle for each construct. The significance of construct-to-construct variation in the basal level of exon inclusion is less clear since different mutations of the same nucleotides have very different effects on basal splicing efficiency (see Discussion). Nucleotide substitutions are indicated in boldface type. All constructs contain two nucleotide substitutions (positions +71 and +74), which created an Asp718 cloning site. Previously defined MSEs are also indicated in boldface type in the genomic sequence. MSEs are redefined based on the results of the scanning mutation analysis presented in this figure. The revised boundaries are indicated by overlining above the genomic sequence. A boundary is defined as the nucleotide adjacent to a mutation that did not affect enhanced splicing in muscle. For MSE2, the 5′ splice site is excluded from the 5′ boundary. The primary goal for defining these boundaries was to identify elements for gain-of-function experiments. MSE2 and MSE3 segments used for gain-of-function studies are underlined in the genomic sequence. na, not applicable.
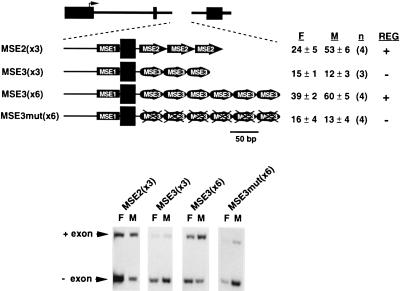
FIG. 4.
Gain-of-function assays for MSE2 and MSE3. MSE2(×3) contains three copies of MSE2 (positions +3 to +46 of intron 5) (Fig. 2) placed 16 nucleotides downstream from the 46-nucleotide heterologous exon. Upstream of the exon are the last 99 nucleotides of cTNT intron 4, which contains MSE1. MSE3(×3) and MSE3(×6) contain three or six copies of MSE3 (positions +57 to +90 of intron 5) (Fig. 2) placed 16 nucleotides downstream of the alternative exon. The MSE3 concatamers in MSE3mut(×6) contain the RTB66-81 mutation (Fig. 2). The percent of spliced mRNA that includes the alternative exon in fibroblasts (F) and muscle (M) is indicated with standard deviation and number of transfections (n). Whether splicing is (+) or is not (−) regulated in muscle (REG) is also indicated.
Transient transfection and RT-PCR.
Preparation of primary skeletal muscle cultures from chicken embryos and transient transfection were performed as described previously (32). Total RNA was extracted 40 to 48 h following addition of DNA to the cells by using guanidinium thiocyanate (31). RNA was DNase (Worthington) treated prior to reverse transcription (RT)-PCR. RT was performed on total RNA (approximately 10 μg) from one-quarter of a 60-mm-diameter plate by using 10 ng of the reverse primer, annealed at 65°C for 10 min in annealing buffer (300 mM NaCl, 40 mM Tricine [pH 8.0], and 0.1 mM EDTA). An equal volume of extension cocktail was added to give final concentrations of 100 mM Tris (pH 8.0), 12 mM MgCl2, 10 mM dithiothreitol, 1 mM dNTPs, and 4 U of avian myeloblastosis virus reverse transcriptase. Extension was performed at 42°C for 1 h. RNA was hydrolyzed in 50 mM NaOH–6.3 mM EDTA at 100°C for 5 min and then neutralized by 50 mM HCl and 0.375 M sodium acetate (pH 7.0), and the cDNA was ethanol precipitated. One-quarter to one-half of the cDNA was added to 40-μl PCR mixtures with Taq polymerase buffers (Promega), 2 ng of the forward primer 32P labeled by polynucleotide kinase, 120 ng each of unlabeled forward and reverse primers, and 2.5 U of Taq DNA polymerase. Eighteen cycles were performed with annealing and extension temperatures of 70°C and 72°C, respectively. Forward and reverse oligonucleotides were CATTCACCACATTGGTGTGC and AGGTGCTGCCGCCGGGCGGTGGCTG, respectively. PCR products were resolved on 5% nondenaturing polyacrylamide gels. Bands were quantitated directly from the gel by using a phosphoimager. The percent exon inclusion is the percent of spliced RNA that contains the exon and is calculated as follows: counts per minute of the inclusion band ÷ (counts per minute of the inclusion band + counts per minute of the exclusion band) × 100. Each result is presented with its standard deviation and the number of transfections. Representative RT-PCR results are presented in the figures. The quantitative nature of these RT-PCR conditions was established by using in vitro-transcribed RNAs synthesized from the cloned RT-PCR products of construct RTB5.1.
RESULTS
Intronic segments to be tested for MSE activity were cloned adjacent to an artificial heterologous exon (see Materials and Methods), and the resulting intron-exon cassettes were inserted into the second intron of a minigene derived from the constitutively spliced skeletal troponin I gene (Fig. 1 and reference 27). All constructs described in this work use heterologous exons that do not contain exonic splicing enhancers. Minigenes were transiently transfected into primary chicken embryo skeletal muscle cultures and a QT35 quail fibroblast cell line. Primary skeletal muscle cultures were prepared from embryonic day-11 skeletal muscle in which >85% of the endogenous cTNT mRNAs included the alternative exon. Both the endogenous and transfected genes express mRNAs that predominantly include the exon in these cultures, indicating that regulated splicing is maintained in differentiated muscle cells in culture and minigene pre-mRNAs respond appropriately to regulatory factors expressed in these cells. In contrast to the muscle cultures, fibroblast cultures express a default preference for exon skipping (10, 32). The analysis here focused on mutations that reduced the level of exon inclusion in muscle to the default level observed in fibroblasts in order to distinguish elements that are recognized specifically in muscle from those that affect splicing in both cell types.
The cTNT MSEs function independently of the cTNT splice sites.
Previous results from this laboratory demonstrated that regulated splicing of cTNT exon 5 requires a weak 5′ splice site (10, 32); however, it was unclear whether splicing could be regulated in the absence of both of the natural cTNT 3′ and 5′ splice sites. For example, the 5′ splice site of NCAM exon 18 is required for neuron-specific exon inclusion and can promote neuron-specific splicing to a heterologous substrate (29). A requirement for a specific splice site sequence would suggest that the initial regulatory event occurs concomitantly with splice site recognition. Alternatively, the ability of the MSEs to regulate utilization of two heterologous splice sites would suggest that the MSEs are the sole targets for the factors that mediate cell-specific regulation and that the regulatory event is distinct from (and may precede) splice site recognition. To determine whether the natural splice sites flanking exon 5 were required for regulated splicing, they were replaced with those of human β-globin intron 1 in construct RTB33.51 (Fig. 1). In anticipation that the stronger globin splice sites would result in constitutive inclusion of the 46-nucleotide heterologous exon, the splice site substitutions were tested on a series of artificial heterologous exons of decreasing size (see Materials and Methods). The expectation was that smaller exons would decrease the basal splicing efficiency (3, 11, 32), without disrupting regulatory elements, and reveal whether exon inclusion was regulated in muscle cultures. Indeed, the globin splice sites resulted in nearly constitutive inclusion of the 46-nucleotide exon in both cell types [R35(46)] (Fig. 1); however, muscle-specific regulation was revealed by decreasing the size of the exon. For example, when exon size was reduced to 30, 32, or 33 nucleotides [R35(30), R35(32), and R35(33)], muscle-specific inclusion was as strong as that of the 46-nucleotide exon flanked by cTNT splice sites (RTB33.51). Therefore, the specific sequences of the splice sites flanking the alternative exon are not required for regulated splicing, indicating that the MSEs are true auxiliary elements able to regulate a heterologous exon flanked by heterologous splice sites.
Scanning mutagenesis of intron 5.
The first 142 nucleotides of intron 5 contain MSE2, MSE3, and MSE4 and are sufficient to provide a low but consistent level of regulated splicing to a heterologous exon (RTB5.1) (Fig. 2 and reference 27). Scanning mutagenesis was performed through this region to identify the nucleotides that were critical for MSE activity and to define the boundaries of the MSEs within intron 5. Because MSE1 provides redundant MSE activity (27), it was necessary to do this analysis in RTB5.1, in which the upstream intron containing MSE1 was replaced with a comparable segment of human β-globin intron 1. While this substitution eliminated the functional redundancy, it also reduced the level of enhanced exon inclusion observed in muscle. In addition, the stronger β-globin 3′ splice site increases the level of inclusion in fibroblasts so that overall, the level of regulation is weaker than that in constructs containing all four MSEs. Still, consistent results were obtained in multiple transfections of each mutant. Levels of exon inclusion with standard deviations and numbers of transfections for each mutation are presented in Fig. 2.
Only 5 of the 16 mutations tested did not disrupt muscle-specific exon inclusion. The results in Fig. 2 indicate that most of the first 130 nucleotides of intron 5 are required for enhanced inclusion in embryonic skeletal muscle. Mutations that did not affect regulation were assigned as boundaries between MSEs. An exception was one mutation (RTB66-69) within MSE3 that did not eliminate regulation. This mutation was unlikely to define a boundary between MSEs because it is within a motif that appears to be a functional unit, since it is conserved in sequence and relative position downstream from six similarly regulated alternative exons (reference 27 and data not shown). The MSEs thus defined provided the basis for the analysis of individual units of activity described below.
Some of the mutations had a striking effect on the level of exon inclusion in both cell types. For example, R5.15-21 resulted in high levels of inclusion in fibroblasts and muscle cells, suggesting that the mutation either disrupted a general repressor of exon inclusion or fortuitously introduced a sequence that enhanced splicing efficiency in both cell types. Consistent with the first possibility, the mutated region contains the sequence UCUU in a pyrimidine-rich region which is a preferred binding site for PTB/hnRNP I (24), a protein known to repress splicing in other experimental systems (1, 5, 22, 24). However, the presence of a repressor element would be inconsistent with our previous results obtained with a 12-nucleotide substitution including the same nucleotides that resulted in decreased exon inclusion in muscle cells and had no effect on the level of exon inclusion in fibroblasts (RBT5.2) (reference 27 and Fig. 2). To determine whether the mutation resulted in a loss of repressor function or a gain of enhancer function, the same nucleotides were mutated by four additional nucleotide substitutions (R5.15AG, R5.15AT, R5.15GG, and R5.15GT) (Fig. 2). Consistent with the designation of this region as part of an MSE, all five mutations expressed a level of exon inclusion in muscle cells lower than that observed in fibroblasts. However, different mutations resulted in significantly different basal levels of exon inclusion in fibroblasts, demonstrating that the sequence of a mutation introduced into this region can have strong effects on the basal level of exon recognition. Taken together, the results from the five mutations in this region are not completely consistent with the presence of a general repressor in this region since not all mutations promote exon inclusion. However, the fact that the mutations upstream (RTB10-14) and downstream (R5.26-31) from this region also increased the level of exon inclusion in both cell types indicates that this possibility cannot be completely ruled out (see Discussion).
MSE2 and MSE3 are functionally redundant.
Once functional MSEs were defined, they were compared for sequence and functional similarities. The sequences of MSE2 and MSE4 could be aligned, strongly suggesting that they are repeats of the same element (Fig. 3). Both MSE2 and MSE4 contain an intronic motif that is found adjacent to alternative exons that undergo regulated splicing in muscle (see below). In contrast, MSE3 does not contain sequence motifs in common with MSE2 or MSE4, suggesting that MSE3 may be a target for different regulatory factors. To compare muscle-specific splicing activities of MSE2 and MSE3, gain-of-function experiments were performed. MSE2 and MSE3 were individually concatamerized and placed 16 nucleotides downstream from the heterologous exon. In all of these constructs, the intron upstream of the alternative exon contains the last 99 nucleotides of cTNT intron 4, which contains MSE1 (Fig. 4). A reasonable level of regulation was observed when the intron 5 segment was replaced by three copies of MSE2 [MSE2(×3)] (Fig. 4). This result was not surprising, since MSE1, MSE2, and MSE4 were sufficient to regulate splicing (27) and MSE2 and MSE4 are likely to be repeats of a single element. Three copies of MSE3 in the same construct was not sufficient for regulated splicing, but six copies did provide regulated splicing [MSE3(×3) and MSE3(×6), respectively] (Fig. 4). Six copies of the same MSE3 segment containing the 66-81 mutation (Fig. 2) did not show enhanced exon inclusion in muscle cultures [MSE3mut(×6) (Fig. 4)], demonstrating sequence specificity of the MSE activities. Therefore, multiple copies of MSE2 or MSE3 can functionally replace MSEs 2, 3, and 4. These results demonstrate that while MSEs 2 and 3 appear to be different elements, they are redundant in function.
FIG. 3.
Sequence alignment of MSE2 and MSE4. Matching nucleotides are underlined. A motif in positions 25 to 32 is repeated in positions 33 to 40 and 113 to 121. The sequence in boldface type is found within introns that flank alternative exons that are regulated in muscle (see Fig. 6).
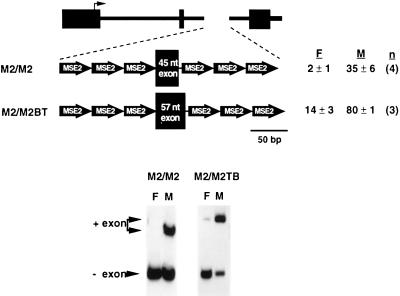
MSE2 is sufficient for regulated splicing of a heterologous exon.
To determine whether a single element could mediate muscle-specific inclusion of a heterologous exon, three copies of MSE2 were placed upstream and downstream of the 46-nucleotide heterologous exon (M2/M2) (Fig. 5). MSE2 is pyrimidine rich and contains several adenosine residues, so it was anticipated that this element could serve as a pyrimidine tract and provide a branch site. The 3′ splice site was created from a filled-in XbaI restriction site at the 3′ end of the MSE2 concatamer (TCTAG) that was blunt-end ligated to the first nucleotide of the exon (the underlined nucleotides are the last two nucleotides of the intron). As shown in Fig. 5, six copies of MSE2 were sufficient for robust regulated splicing of the 46-nucleotide exon. Since inclusion of the 46-nucleotide alternative exon was barely detectable in fibroblasts, a 57-nucleotide exon was also tested to evaluate the full level of regulated splicing (M2/M2TB). The level of regulation is actually greater for M2/M2TB than for constructs regulated by the natural flanking introns (e.g., RTB33.51) (Fig. 1). These results demonstrate that multiple copies of MSE2 alone are sufficient to mediate enhanced inclusion of heterologous exons in embryonic skeletal muscle.
FIG. 5.
MSE2 is sufficient for muscle-specific exon inclusion. Three copies of MSE2 were placed upstream and downstream of a 45- or 57-nucleotide heterologous exon. The percent of spliced mRNA that includes the alternative exon in fibroblasts (F) and muscle (M) is indicated with standard deviation and number of transfections (n).
MSE2 and MSE4 contain a novel sequence motif found adjacent to alternative exons that are regulated in striated muscle.
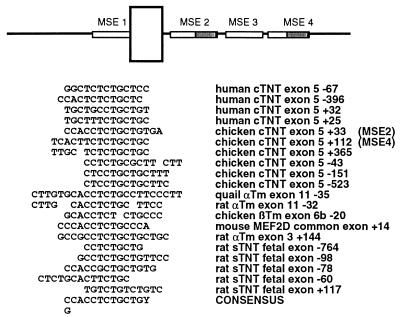
A different approach to identifying sequences that are critical for regulated splicing was to identify intronic sequences that are common to similarly regulated alternative exons. Intron sequences upstream and downstream of alternative exons that are regulated in striated or smooth muscle were searched for: (i) sequence motifs that were repeated within each gene and (ii) sequence motifs that were common to different genes. From this analysis a number of repeated and/or potentially conserved sequence motifs were identified. The significance of most of these motifs remains unknown due to their presence in only one or two genes and a lack of functional analysis. However, one motif found within MSE2 and MSE4 was found in introns adjacent to many exons that are regulated in striated or smooth muscle (Fig. 6). Of particular interest is that in three genes, human cTNT exon 5, chicken cTNT exon 5, and rat skeletal TNT exon 8a, variations of this motif are present in multiple copies located upstream and downstream of the alternative exon. Mutation analysis has demonstrated that this sequence is important for regulation in both chicken (this work) and human (25) cTNT genes. In the chicken cTNT minigene, mutations in either copy of this motif disrupted regulated splicing (R5.35-41 and R5.119-124) (Fig. 2). We have recently demonstrated that a human cTNT genomic fragment containing the homologous 30-nucleotide alternative exon 5 and 300 and 372 nucleotides of the upstream and downstream introns, respectively, is appropriately regulated in chicken primary skeletal muscle cultures. Nucleotide substitutions within the common motif (positions 20, 32, 35, 39, and 42) (Fig. 6) disrupts regulation (25). Therefore, this variably conserved sequence motif has been demonstrated to contribute to regulated splicing in two genes.
FIG. 6.
Sequence comparisons of introns flanking alternative exons that undergo regulated splicing in striated or smooth muscle has identified a novel motif. The position of the motif in MSE2 and MSE4 is indicated by shading in the diagram. The number indicates the position of the first nucleotide of the sequence from the regulated alternative exon. Negative numbers are upstream and positive numbers are downstream from the alternative exon. Abbreviations: αTm, α-tropomyosin; βTm, β-tropomyosin; and sTNT, skeletal troponin T.
DISCUSSION
This laboratory previously defined four MSEs required for enhanced inclusion of cTNT exon 5 in embryonic skeletal muscle cultures. Each element was defined by one mutation that decreased the level of exon inclusion in muscle compared to the default level observed in fibroblasts (27). In the present study, scanning mutagenesis was used to define the boundaries of the MSEs in intron 5. Most mutations in intron 5 disrupted regulation; those that did not were designated as boundaries between MSEs (an exception was an MSE3 mutation which split a conserved motif). Defining MSEs in this way provided units of activity for analysis in gain-of-function experiments. Sequence comparisons suggested that MSE2 and MSE4 are repeats of a single element and that MSE3 is distinct from MSE2 and MSE4. In gain-of-function experiments, MSE2 and MSE3 were shown to be functionally redundant since concatamers of either element could substitute for MSEs 2, 3, and 4 and promote muscle-specific exon inclusion. These results suggest the possibility that distinct elements that assemble different complexes are able to perform the same muscle-specific enhancer function when placed downstream of the exon in context with MSE1. Analyses of the complexes that assemble on MSE2 and MSE3 in muscle nuclear extracts are necessary to determine whether they contain components in common. However, functional differences between MSE2 and MSE3 were also apparent since three copies of MSE2 downstream of the exon were sufficient to regulate splicing while more than three copies of MSE3 were required to perform the same function (Fig. 4). In addition, one, two, three, or six copies of MSE3 were unable to substitute for MSE1, even when a consensus branch site was introduced upstream of the MSE3 insertion (data not shown) while three copies of MSE2 could functionally replace MSE1 (Fig. 5).
At least three MSEs are required for muscle-specific regulation; however, it is unclear whether all MSEs are muscle-specific splicing elements. It is possible that only one element is the target for muscle-specific recognition and the others are required for recognition by the constitutive splicing machinery. Mutations that disrupt either a muscle-specific or a required ubiquitous complex would disrupt muscle-specific regulation. The results presented in this work demonstrate that MSE2 is a direct target for muscle-specific regulation since this element alone is sufficient for muscle-specific regulation of exon inclusion.
Given the sizes of MSEs 2, 3, and 4 (40, 35, and 37 nucleotides, respectively), it is likely that each of these units contains multiple binding sites and assembles a multicomponent complex. For example, the 13-nucleotide Drosophila dsx element assembles a complex containing at least three components (18). A 33-nucleotide intronic element that regulates neuron-specific inclusion of the c-src N1 exon binds a complex containing multiple components (19, 20). Even purine-rich exon splicing enhancers, which initially appeared to be low in sequence complexity, have recently been shown to contain multiple components. Chimeric swaps between the caldesmon (32 nucleotides) and cTNT (30 nucleotides) purine-rich exon splicing enhancers have demonstrated that the 3′ one-quarter of each of these two elements contains distinct components that are responsible for distinct functions (12). The 3′ one-third of MSE2 contains a motif found near many alternative exons that are regulated in muscle (see below), which also supports the contention that the MSEs are made up of smaller components.
It is unclear whether MSE2 and MSE3 could be further subdivided into smaller functional units or whether the components of these MSEs must remain associated. Ideally, a bona fide element can be functionally defined as the minimal sequence that will promote regulation (when concatamerized if necessary). One example of this is a hexanucleotide originally identified in the fibronectin gene (16, 17) which has recently been shown to promote the neuron-specific inclusion of c-src when concatamerized (21). Presumably, a minimal element promotes assembly of a core complex (composed of a single or multiple components) sufficient for regulation. The results presented in this work and a previous work (27) demonstrate that three copies but not one copy of MSE2 in place of MSEs 2, 3, and 4 regulate splicing, indicating that a single type of complex is sufficient for regulation as long as it is present in multiple copies. Since heterologous pairs of weakly recognized splice sites can be regulated by the MSEs (Fig. 1), the MSEs are likely to function by recruiting and or stabilizing binding of constitutive splicing factors to inefficiently recognized splice sites. The fact that multiple elements are required to perform this function suggests that cooperativity between units is required and/or that the number of assembled complexes must reach a threshold level to successfully capture components of the constitutive splicing machinery.
A novel sequence motif has been identified within MSE2 by sequence comparisons of introns flanking alternative exons that are regulated in striated muscle (Fig. 6). It remains to be determined how many of the sequences shown in Fig. 6 actually function in regulated splicing; however, point mutations in three of these elements affect regulated splicing in muscle (chicken cTNT exon 5 positions +33 and +112 and human cTNT position +32) (Fig. 2, Fig. 6, and reference 25). In the human cTNT gene, this motif has recently been shown to have MSE activity and is a target for positive regulation of exon inclusion by a novel hnRNP protein called CUG-binding protein (CUG-BP). CUG-BP is thought to play a role in the pathogenesis of myotonic dystrophy, a disease caused by a CTG expansion in the 3′ untranslated region of a protein kinase gene, by a trans-dominant effect on posttranscriptional processing of several genes (26, 30). Consistent with this proposal, cTNT is aberrantly spliced in striated muscle from myotonic dystrophy patients (25). Therefore, other genes listed in Fig. 6 may be regulated posttranscriptionally by CUG-BP and their expression may also be affected in myotonic dystrophy.
An unanticipated result was the observation that different substitutions between positions +15 and +21 showed significant differences in the level of exon inclusion in both regulating and nonregulating cell types. This was not an isolated effect, since multiple substitutions within a 5-nucleotide region in MSE1 showed dramatic effects in both cell types (data not shown), making it impossible to draw reliable conclusions regarding nucleotides required for regulation. These observations may partially explain the inconsistent results that have made it so difficult to define cell-specific splicing elements in vertebrates. For example, if each of the five mutations between positions +15 and +21 of cTNT intron 5 was analyzed in the absence of the others, four different conclusions could be argued: (i) enhanced inclusion in both muscle and fibroblasts (R5.15-21) suggests the presence of a general repressor; (ii) enhanced inclusion only in fibroblasts (R5.15AG and R5.15GG) suggests the presence of a fibroblast-specific repressor; (iii) decreased inclusion in fibroblasts and muscle (R5.15GT) suggests the presence of a general enhancer; and (iv) decreased inclusion specifically in muscle (R5.15AT) suggests the presence of a muscle-specific enhancer. Since all five mutations in this region prevent enhanced exon inclusion in muscle compared to fibroblasts, the only clear conclusion is that the region between positions +15 and +21 is required for muscle-specific exon inclusion.
A role for fibroblast-specific repressors (suggested by R5.15AG and R5.15GG [Fig. 2]), needs to be considered since a role for negative splicing elements in cell-specific splicing has been demonstrated in several experimental systems (5, 13, 14). One important difference between cTNT and these systems is that the cTNT pre-mRNA is not expressed in the cell type that would repress exon inclusion. It is unlikely that this gene would contain an element specifically recognized in a cell type in which it is not expressed. It is possible, however, that a repressor in this region is utilized during the switch to exon skipping in adult heart and that similarities in repressive activities of constitutive splicing factors such as PTB in fibroblasts and adult heart result in repressor activity in fibroblasts. Still, it is difficult to explain why only some mutations of the same nucleotides showed an increased level of exon inclusion if this region contains a repressor. Overall, this lack of consistency suggests that some nucleotide substitutions fortuitously introduced sequences that enhanced splicing. Our laboratory has previously shown that the general splicing efficiency is stronger in the fibroblast cultures. A stronger splicing machinery in fibroblasts may be more responsive to fortuitously introduced enhancers and lead to the higher levels of exon inclusion observed in these cells.
Some common themes are emerging regarding the cis-acting elements that regulate cell-specific splicing (6): (i) these elements are intronic (with one exception [33]; all other exonic splicing elements identified so far are general splicing enhancers [7]); (ii) a genomic fragment containing 100 to 300 nucleotides upstream and downstream of the regulated exon is likely to contain sufficient information in cis for appropriate cell-specific regulation (however, additional distal elements with redundant function are also likely); (iii) regulation is often mediated by multiple elements with different sequence motifs; (iv) multiple repeats of each element may be located upstream and downstream of the regulated exon; (v) elements are often common to different genes that undergo a similar cell-specific regulatory pattern and are conserved in the homologous gene from different species; and (vi) both positive- and negative-acting elements are likely to be involved. These observations suggest a straightforward approach to streamlining the identification of elements that regulate cell-specific splicing: (i) perform sequence comparisons of introns that flank similarly regulated alternative exons for repeated and conserved sequence elements to identify the most obvious regulatory elements; (ii) functionally define the minimal genomic segment that is necessary and sufficient to mediate cell-specific splicing to remove redundant elements that can obscure the effects in loss-of-function experiments; (iii) perform mutation analysis on this genomic fragment to identify elements by loss-of-function mutations; and (iv) use the critical elements to perform gain-of-function experiments on heterologous alternative exons and splice sites.
Overall, the picture that is emerging from results from Drosophila and vertebrate systems demonstrates strong parallels between the regulation of splicing and the regulation of transcription (15). Both are mediated by multiple distinct auxiliary elements present in multiple copies, and regulation involves assembly of a complex which ultimately serves to recruit the basal machinery. These complexes contain components that are ubiquitously expressed and are required for constitutive functions (18–20). A question that is key to understanding the mechanism of regulated splicing is whether cell-specific (or at least cell-restricted) factors are also a component of the complex. Thus far, the factors that have been shown to bind to cell-specific splicing elements in vertebrates are ubiquitously expressed, indicating either that cell-specific factors remain to be isolated or that splicing is regulated by a committee of general splicing factors. Given the complexity of the mechanism of cell-specific splicing, the ability to use multiple copies of a single element rather than multiple distinct elements to regulate muscle-specific splicing will allow simplified analysis of the factors that mediate muscle-specific exon inclusion.
ACKNOWLEDGMENTS
I thank Claire Lo for outstanding technical assistance, Kathy Ryan for thoughtful discussions, and Gil Cote and Miles Wilkinson for critical comments on the manuscript.
This work was supported by grants from the National Institutes of Health and the Muscular Dystrophy Association.
REFERENCES
- 1.Ashiya M, Grabowski P J. A neuron-specific splicing switch mediated by an array of pre-mRNA repressor sites: evidence of a regulatory role for the polypyrimidine tract binding protein and a brain-specific PTB counterpart. RNA. 1997;3:996–1015. [PMC free article] [PubMed] [Google Scholar]
- 2.Black D. Finding splice sites within a wilderness of RNA. RNA. 1995;1:763–771. [PMC free article] [PubMed] [Google Scholar]
- 3.Black D L. Does steric interference between splice sites block the splicing of a short c-src neuron-specific exon in non-neuronal cells? Genes Dev. 1991;5:389–402. doi: 10.1101/gad.5.3.389. [DOI] [PubMed] [Google Scholar]
- 4.Chabot B. Directing alternative splicing: cast and scenarios. Trends Genet. 1996;12:472–478. doi: 10.1016/0168-9525(96)10037-8. [DOI] [PubMed] [Google Scholar]
- 5.Chan R C, Black D L. The polypyrimidine tract binding protein binds upstream of neural cell-specific c-src exon N1 to repress the splicing of the intron downstream. Mol Cell Biol. 1997;17:4667–4676. doi: 10.1128/mcb.17.8.4667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cooper, T. A. Strategies for defining pre-mRNA cis elements that regulate cell specific splicing. Methods Mol. Biol., in press. [DOI] [PubMed]
- 7.Cooper T A, Mattox W. The regulation of splice site selection and its role in human disease. Am J Hum Genet. 1997;61:259–266. doi: 10.1086/514856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cooper T A, Ordahl C P. A single cardiac troponin T gene regulated by different programs in cardiac and skeletal muscle development. Science. 1984;226:979–982. doi: 10.1126/science.6095446. [DOI] [PubMed] [Google Scholar]
- 9.Cooper T A, Ordahl C P. A single cardiac troponin T gene generates embryonic and adult isoforms via developmentally regulated alternate splicing. J Biol Chem. 1985;260:11140–11148. [PubMed] [Google Scholar]
- 10.Cooper T A, Ordahl C P. Nucleotide substitutions within the cardiac troponin T alternative exon disrupt pre-mRNA alternative splicing. Nucleic Acids Res. 1989;17:7905–7921. doi: 10.1093/nar/17.19.7905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dominski Z, Kole R. Selection of splice sites in pre-mRNAs with short internal exons. Mol Cell Biol. 1991;11:6075–6083. doi: 10.1128/mcb.11.12.6075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Elrick L L, Humphrey M B, Cooper T A, Berget S M. A short sequence within two purine-rich enhancers determines 5′ splice site specificity. Mol Cell Biol. 1998;18:343–352. doi: 10.1128/mcb.18.1.343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gooding C, Roberts G C, Moreau G, Nadal-Ginard B, Smith C W J. Smooth muscle-specific switching of alpha-tropomyosin mutually exclusive exon selection by specific inhibition of the strong default exon. EMBO J. 1994;13:3861–3872. doi: 10.1002/j.1460-2075.1994.tb06697.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Graham I R, Hamshere M, Eperon I C. Alternative splicing of a human α-tropomyosin muscle-specific exon: identification of determining sequences. Mol Cell Biol. 1992;12:3872–3882. doi: 10.1128/mcb.12.9.3872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hertel K J, Lynch K W, Maniatis T. Common themes in the function of transcription and splicing enhancers. Curr Opin Cell Biol. 1997;9:350–357. doi: 10.1016/s0955-0674(97)80007-5. [DOI] [PubMed] [Google Scholar]
- 16.Huh G S, Hynes R O. Elements regulating an alternatively spliced exon of the rat fibronectin gene. Mol Cell Biol. 1993;13:5301–5314. doi: 10.1128/mcb.13.9.5301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Huh G S, Hynes R O. Regulation of alternative pre-mRNA splicing by a novel repeated hexanucleotide element. Genes Dev. 1994;8:1561–1574. doi: 10.1101/gad.8.13.1561. [DOI] [PubMed] [Google Scholar]
- 18.Lynch K W, Maniatis T. Assembly of specific SR protein complexes on distinct regulatory elements of the Drosophila doublesex splicing enhancer. Genes Dev. 1996;10:2089–2101. doi: 10.1101/gad.10.16.2089. [DOI] [PubMed] [Google Scholar]
- 19.Min H, Turck C W, Nikolic J M, Black D L. A new regulatory protein, KSRP, mediates exon inclusion through an intronic splicing enhancer. Genes Dev. 1997;11:1023–1036. doi: 10.1101/gad.11.8.1023. [DOI] [PubMed] [Google Scholar]
- 20.Min H S, Chan R C, Black D L. The generally expressed hnRNP F is involved in a neural-specific pre-mRNA splicing event. Genes Dev. 1995;9:2659–2671. doi: 10.1101/gad.9.21.2659. [DOI] [PubMed] [Google Scholar]
- 21.Modafferi E F, Black D L. A complex intronic splicing enhancer from the c-src pre-mRNA activates inclusion of a heterologous exon. Mol Cell Biol. 1997;17:6537–6545. doi: 10.1128/mcb.17.11.6537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mulligan G J, Guo W, Wormsley S, Helfman D M. Polypyrimidine tract binding protein interacts with sequences involved in alternative splicing of beta-tropomyosin pre-mRNA. J Biol Chem. 1992;267:25480–25487. [PubMed] [Google Scholar]
- 23.Norton P A. Alternative pre-mRNA splicing—factors involved in splice site selection. J Cell Sci. 1994;107:1–7. doi: 10.1242/jcs.107.1.1. [DOI] [PubMed] [Google Scholar]
- 24.Perez I, Lin C H, McAfee J G, Patton J G. Mutation of PTB binding sites causes misregulation of alternative 3′ splice site selection in vivo. RNA. 1997;3:764–778. [PMC free article] [PubMed] [Google Scholar]
- 25.Philips A V, Timchenko L T, Cooper T A. Disruption of splicing regulated by a CUG-binding protein in myotonic dystrophy. Science. 1998;280:737–741. doi: 10.1126/science.280.5364.737. [DOI] [PubMed] [Google Scholar]
- 26.Roberts R, Timchenko N A, Miller J W, Reddy S, Caskey C T, Swanson M S, Timchenko L T. Altered phosphorylation and intracellular distribution of a (CUG)n triplet repeat RNA-binding protein in patients with myotonic dystrophy and in myotonin protein kinase knockout mice. Proc Natl Acad Sci USA. 1997;94:13221–13226. doi: 10.1073/pnas.94.24.13221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ryan K R, Cooper T A. Muscle-specific splicing enhancers regulate inclusion of the cardiac troponin T alternative exon in embryonic skeletal muscle. Mol Cell Biol. 1996;16:4014–4023. doi: 10.1128/mcb.16.8.4014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sarkar G, Sommer S S. The “megaprimer” method of site-directed mutagenesis. BioTechniques. 1990;8:404–407. [PubMed] [Google Scholar]
- 29.Tacke R, Goridis C. Alternative splicing in the neural cell adhesion molecule pre-mRNA: regulation of exon 18 skipping depends on the 5′-splice site. Genes Dev. 1991;5:1416–1429. doi: 10.1101/gad.5.8.1416. [DOI] [PubMed] [Google Scholar]
- 30.Timchenko L T, Miller J W, Timchenko N A, Devore D R, Datar K V, Lin L J, Roberts R, Caskey C T, Swanson M S. Identification of a (CUG)n triplet repeat RNA-binding protein and its expression in myotonic dystrophy. Nucleic Acids Res. 1996;24:4407–4414. doi: 10.1093/nar/24.22.4407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Xie W, Rothblum L I. Rapid, small-scale RNA isolation from tissue culture cells. BioTechniques. 1991;11:325–327. [PubMed] [Google Scholar]
- 32.Xu R, Teng J, Cooper T A. The cardiac troponin T alternative exon contains a novel purine-rich positive splicing element. Mol Cell Biol. 1993;13:3660–3674. doi: 10.1128/mcb.13.6.3660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang L, Ashiya M, Sherman T G, Grabowski P J. Essential nucleotides direct neuron-specific splicing of gamma(2) pre-mRNA. RNA. 1996;2:682–698. [PMC free article] [PubMed] [Google Scholar]