Abstract
UV irradiation leads to severe damage, such as cutaneous inflammation, immunosuppression, and cancer, but it also results in a gene induction protective response termed the UV response. The signal triggering the UV response was thought to originate from DNA damage; recent findings, however, have shown that it is initiated at or near the cell membrane and transmitted via cytoplasmic kinase cascades to induce gene transcription. Urokinase-type plasminogen activator (uPA) was the first protein shown to be UV inducible in xeroderma pigmentosum DNA repair-deficient human cells. However, the underlying molecular mechanisms responsible for the induction were not elucidated. We have found that the endogenous murine uPA gene product is transcriptionally upregulated by UV in NIH 3T3 fibroblast and F9 teratocarcinoma cells. This induction required an activator protein 1 (AP1) enhancer element located at −2.4 kb, since deletion of this site abrogated the induction. We analyzed the contribution of the three different types of UV-inducible mitogen-activated protein (MAP) kinases (ERK, JNK/SAPK, and p38) to the activation of the murine uPA promoter by UV. MEKK1, a specific JNK activator, induced transcription from the uPA promoter in the absence of UV treatment, whereas coexpression of catalytically inactive MEKK1(K432M) and of cytoplasmic JNK inhibitor JIP-1 inhibited UV-induced uPA transcriptional activity. In contrast, neither dominant negative MKK6 (or SB203580) nor PD98059, which specifically inhibit p38 and ERK MAP kinase pathways, respectively, could abrogate the UV-induced effect. Moreover, our results indicated that wild-type N-terminal c-Jun, but not mutated c-Jun (Ala-63/73), was able to mediate UV-induced uPA transcriptional activity. Taken together, we show for the first time that kinases of the JNK family can activate the uPA promoter. This activation links external UV stimulation and AP1-dependent uPA transcription, providing a transcription-coupled signal transduction pathway for the induction of the murine uPA gene by UV.
Stress responses allow cells and organisms to rapidly respond to extreme conditions such as hyperosmolarity, oxidants, and heat shock; infection and invasion by parasites, viruses, and bacteria; and exposure to X rays and UV light (4, 45). Solar UV light represents one of the major environmental impacts for humans. It stimulates melanogenesis, causes skin reddening and inflammation, induces proinflammatory cytokines, and promotes premature skin aging and skin cancer. However, UV light not only leads to cell destruction but also induces the transcription of many genes, including genes coding for transcription factors, growth factors, viral proteins, and proteases. This induction response is known as the “mammalian UV response” (25, 81). Recent evidence suggests that the UV response can play a protective function other than DNA repair (26). The main target of UV light was regarded to be chromosomal DNA damage, which in turn would provide the primary signal triggering the response (76, 83). However, the rapid UV activation of Ras, Src, and other molecules located at or near the plasma membrane argues against DNA damage as the primary signal, suggesting that a nuclear signal is not always required for the UV response (26, 27). In fact, the first cellular reaction detectable in UV-irradiated cells is the phosphorylation of different cell membrane growth factor receptors at tyrosine residues (77). The UV response could be inhibited by prior down modulation of growth factor receptor signaling by growth factor prestimulation, by suramin, a fairly specific inhibitor of the action of growth factor receptor, or by expression of a dominant negative epidermal growth factor receptor mutant (77). In addition, it has recently been shown that UV irradiation inhibits tyrosine phosphatases, and this UV-induced inhibition causes elevated levels of growth factor receptor phosphorylation, leading to signaling and to transcription of UV-responsive genes (44). Altogether, the signaling cascades induced by UV appear to be complex. Wherever generated, the UV signal seems to reach the cell membrane and to activate three groups of mitogen-activated protein kinases (MAPKs) involved in three distinct signaling pathways, which mediate UV-induced transcription factor activation. Activation of the extracellular regulated kinases 1 and 2 (ERK1/2) or p42-44 MAPK signaling pathway by UV was detected first (68). It involves activation of plasma membrane tyrosine kinases following activation of Ras and Raf. Raf activates MEK (MAP kinase-ERK kinase), which in turn activates ERK. ERKs phosphorylate various transcription factors including TCF/Elk-1, ATF2, and c-Myc (reviewed in reference 19). This pathway is a critical regulatory element of cell growth and differentiation, mediating the response to agonists of tyrosine kinase receptors. Stresses, however, more strongly induce the MAP kinases JNK/SAPK (c-Jun N-terminal kinase–stress-activated protein kinase) and p38, which were both implicated in the induction of apoptosis triggered by growth factor removal (89). JNK was first described as a UV light-activated kinase that activated c-Jun by phosphorylation at Ser-63 and Ser-73 (26, 38). Like all MAPKs, the JNKs are activated by a MAPKK named JNKK1 (55) (SEK [80] or MKK4 [24]), but not by MEK; JNKK1, in turn, is phosphorylated and activated by a MAPKKK named MEKK1 (55, 80). Very recently, a cytoplasmic inhibitor of JNK, JNK-interacting protein 1 (JIP-1), has been identified, establishing protein targeting as an alternative mechanism that regulates signaling by stress-activated MAP kinases (28). The third MAPK subfamily, p38 (also known as CSBP, RK, Mxi2, and SAPK2a), is a homolog of the yeast MAPK Hog-1 (high-osmolarity glycerol response 1) kinase (35, 51, 75, 90) and is phosphorylated and activated by MEK family member MKK3 (24). In addition to the original isoform of p38 (now referred to as p38α), three other p38 kinase family members have been identified and are designated p38β (SAPK2b) (41), p38γ (SAPK3/ERK6) (49, 54, 59), and p38δ (SAPK4) (31, 42, 86); all of them can be phosphorylated and activated by a novel MAP kinase kinase named MKK6 (36, 70). Besides phosphorylating c-Jun protein, JNK can also phosphorylate ATF2 and TCF/Elk-1 (11, 33, 56), whereas p38 can activate substrates such as ATF2 (41), MAPKAP-K2/K3 (13, 58, 75), Mnk1/2 (32, 88), CHOP (87), and Elk-1 (67). It has been suggested that UV-induced JNK triggers a protective response through the activation of genes coding for protective proteins (23, 26, 48). p38 appears to be also involved in the production of cytokines by stimulated monocytes and to mediate the aggregation of platelets in response to collagen (51, 78).
Urokinase-type plasminogen activator (uPA) was the first protein shown to be inducible in xeroderma pigmentosum cells at much lower UV doses than in parental heterozygotic cells (64), suggesting that DNA damage might be contributing to this induction. Moreover, a UV-induced secreted factor was proposed to mediate uPA induction in low-repair-capacity human fetal fibroblasts (72, 74). These reports, however, contrast with previously mentioned findings showing that UV activates cytoplasmic kinases through pathways used by growth factors, independently of UV-induced DNA damage (26, 27, 77). Therefore, it remained to be resolved how UV irradiation and uPA induction were linked at the molecular level.
uPA is a secreted serine protease that converts the zymogen plasminogen to plasmin, a trypsin-like serine protease capable of degrading extracellular matrix components and of activating other proteinases (79). Although gene inactivation studies have demonstrated that uPA is not essential for embryonic development, increasing evidence supports a role for uPA in physiological processes such as fibrinolysis and angiogenesis, as well as in pathological events such as muscle regeneration, wound healing, inflammation, and tumor invasiveness (8–10, 18, 34, 47, 65, 82). In addition to these proteolytic functions, recent studies have demonstrated mitogenic and chemotactic properties of uPA through interaction with its cell surface high-affinity receptor (7, 71). Reflecting its wide spectrum of functions, uPA expression is regulated by numerous extracellular stimuli depending on the cell type. uPA gene transcription can be induced by growth factors, phorbol esters, cytokines, cytoskeletal reorganization, and several oncoproteins (5, 6, 21, 40, 52, 53, 57, 63, 66, 73). Most of these inductions were found to occur via the ERK pathway of MAP kinases. In contrast, in the present study we have found that the uPA gene can be also induced by activation of the JNK signaling pathway. Specifically, we have investigated the mechanism(s) of UV-induced uPA transcription. This induction required two activator protein 1 (AP1) enhancer sequences, which are located at −2.4 kb from the transcription start site, and was mediated by the JNK pathway.
MATERIALS AND METHODS
Cell culture.
The murine fibroblastic NIH 3T3 cell line was obtained from the American Type Culture Collection and grown in Dulbecco modified Eagle medium containing 10% fetal bovine serum (FBS). The murine teratocarcinoma F9 cell line was grown in Dulbecco modified Eagle medium-Ham’s F-12 (1:1), 10% FBS, and 100 μM β-mercaptoethanol. For UV stimulation, cells were kept in 0.5% FBS medium for 16 h. The next day, the medium was removed and retained, and the cells were washed with phosphate-buffered saline (PBS) and UVC (254 nm) (irradiated 30 J/m2). The retained medium was added back to the cells, and analyses were performed at different lengths of time postirradiation as indicated in the figure legends. Alternatively, cells were treated with the phorbol ester TPA (12-O-tetradecanoylphorbol-13-acetate) at a final concentration of 100 ng/ml.
Northern blot analysis.
Total RNA was extracted from NIH 3T3 cells by using the commercial Ultraspec RNA isolation system (Biotecx) based on the Chomczynski method (12). Poly(A)+ mRNA was obtained from F9 cells with the QuickPrep Micro mRNA purification kit (Pharmacia). RNAs were size fractionated by electrophoresis on 1% agarose gels, transferred to nylon membranes, and UV cross-linked. Membrane prehybridization, probe hybridization, washes, and autoradiography were performed as described previously (65). The cDNA probes used for RNA hybridizations were labeled with [α-32P]dCTP by a standard random oligonucleotide-primed reaction by using the commercial Megaprime DNA labeling system (Amersham). The uPA and GAPDH (glyceraldehyde-3-phosphate dehydrogenase) (65) and the c-jun and c-fos (25) probes were described previously.
Reverse transcriptase (RT) PCR and Southern blotting.
For the analysis of uPA induction at early time points post-UV radiation, cells were plated at 6 × 105 cells/100-mm-diameter dish, grown for 12 h in 10% FBS, and transferred to 0.5% FBS for 16 h, after which the cells were irradiated with UVC. Total RNA was extracted at different time points postradiation, and 2-μg samples were used to synthesize cDNA with the First Strand cDNA synthesis kit (Pharmacia) in a final volume of 15 μl. Then, 5 μl was used in a multiplex PCR with the oligonucleotides 5′-TAGAGCCTTCTGGCCACACTG-3′ and 5′-GGCAGTGTACTTGGAGCTCCT-3′ mapping to murine uPA exons 3 and 7 (22), respectively, and murine G3PDH primers (Clontech) used as an internal control in a final volume of 50 μl (20 mM Tris-HCl, pH 8.4; 50 mM KCl; 1.5 mM MgCl2; 0.2 mM concentrations of each deoxynuclcoside triphosphate [dNTP]; 0.5 μM concentrations of each primer; and 2 U of Taq polymerase). Amplification parameters were 94°C for 1 min, 55°C for 1 min, and 72°C for 3 min. Two 10-μl aliquots were removed from the assay during the linear amplification range (15 cycles) and run in two independent 1% agarose gels. Gels were soaked in denaturing solution (0.4 N NaOH, 1 M NaCl) for 1 h, further neutralized (1 M Tris-HCl, pH 7.4; 1.5 M NaCl) for 1 h, blotted, and hybridized with α-32P-labeled uPA and G3PDH probes. The remaining PCR mixture was allowed to proceed for another 20 cycles, run on a 1% agarose, and ethidium bromide stained to verify correct amplification products.
Plasmids.
The p-6.6Luc murine uPA-promoter luciferase reporter plasmid contains 6.6 kb of murine uPA promoter region and has been previously described (6). p-2.0Luc (−2 kb of murine uPA promoter) and p-4.2Luc (−4.2 kb of murine uPA promoter) were derived from p-6.6Luc, which was cut with SmaI and either NheI or HindIII, respectively, and the backbone was filled in with T4 DNA polymerase and religated. p-4.2(ΔPEA3/AP1A)Luc was derived by combining DNA fragments from p-6.6(ΔPEA3/AP1A)Luc (6) and p-4.2Luc. A thymidine kinase-driven luciferase reporter vector, ptk-Luc, was constructed by cloning an EcoRI-BglII fragment corresponding to map positions −80 to 52 bp of the thymidine kinase gene into pSP73 and subcloning a SacI-BglII fragment into pGL2-basic (Promega). The polylinker SmaI site of ptk-Luc was further used to clone single or multimerized copies of double-stranded oligonucleotides. p3xAP1B-tk-Luc contains three copies of murine AP1B oligonucleotide (5′-GGCCATGTGAATCACGACAGCCTG-3′, map positions: −2369 and −2345), and p6xPEA3/AP1A-tk-Luc contains six copies of oligonucleotide (5′-GAGGAAATGAGGTCATCTTGCTCTG-3′, map positions: −2454 and −2429). Both of these oligonucleotides were used to PCR amplify the murine uPA AP1-enhancer containing the whole PEA3/AP1A, COM, and AP1B elements to generate pPEA3/AP1A-COM-AP1B-tk-Luc. p3xAP1(col)-tk-Luc contains three copies of the human collagenase AP1-binding site inserted upstream of the minimal promoter of the thymidine kinase gene. Expression vectors pcDNAIII-HA-JNK1, pcDNAIII-HA-ERK-2, pSRα-MEKK1(K432M), pCEVZ9-MEKK1, pcDNAIII-MKK6b(A), pcDNAIII-JIP-1, pRSV-cJun, and pRSV-cJun(Ala-63/73) have all been previously described (14, 15, 23, 28, 36, 61).
Western blotting.
Cells were cultured in 0.5% FBS and, at the indicated time points postirradiation, whole-cell extracts (WCE) were prepared by lysing the cells in 20 mM HEPES (pH 7.5), 10 mM EGTA, 40 mM β-glycerophosphate, 1% Nonidet P-40, 2.5 mM MgCl2, 2 mM orthovanadate, 1 mM dithiothreitol (DTT), 1 mM phenylmethylsulfonyl fluoride (PMSF), and aprotinin and leupeptin (1 μg of each per ml). JNK1 protein was detected in WCE by immunoblotting with the specific antibody sc-474 from Santa Cruz Biotechnologies. Immunoblots were performed and developed with the ECL detection system (Amersham).
Protein kinase assays.
JNK was immunoprecipitated from WCE with an anti-JNK1 antibody, sc-474, and immunocomplexes were recovered with protein A-Sepharose. Beads were sequentially washed three times with 1% Nonidet P-40 and 2 mM sodium orthovanadate in PBS, once with 100 mM Tris-HCl (pH 7.5)–0.5 M LiCl, and once with kinase buffer (20 mM HEPES-NaOH, pH 7.6; 2 mM DTT, 20 mM β-glycerophosphate, 20 mM MgCl2; 100 μM sodium orthovanadate). Phosphorylation reactions were performed in a 30-μl volume containing kinase buffer supplemented with 20 μM ATP, 0.5 μCi of [γ-32P]ATP, and 1 μg of glutathione-S-transferase (GST)–c-Jun substrate at 30°C for 30 min, stopped by the addition of 4× Laemmli sample buffer, and resolved by sodium dodecyl sulfate–10% polyacrylamide gel electrophoresis.
Transfection assays.
Reporter plasmids were transfected with the liposome-mediated transfection reagent DOTAP (Boehringer Mannheim). First, 2.5 × 104 cells were seeded overnight on 24-well dishes. The next day, cells were cotransfected with 300 ng of uPA-luciferase plasmid and 5 ng of the renilla luciferase plasmid pRL-SV40 as an internal control. After transfection, cells were cultured in 0.5% FBS for 16 h before UVC stimulation (254 nm, 30 J/m2) and reporter activities were analyzed after 16 or 24 h. When indicated, cells were cotransfected with 150 ng of reporter plasmid and 150 ng of expression plasmids or with empty vector alone, together with 5 ng of internal control. Inhibition of MEK and p38 kinase was performed by pretreating transfected cells with 50 μM PD98059 and 10 μM SB203580 (Calbiochem), respectively, for 30 min prior to UVC irradiation. Firefly luciferase activities were standardized for renilla luciferase activity, which was used as the internal control. All transfections were repeated at least three times and showed less than 25% variability. A Student’s t test was used to validate the results.
Electrophoretic mobility shift assay (EMSA).
Nuclear extracts were obtained from NIH 3T3 cells after UV irradiation. The extraction of nuclear proteins was performed as described by De Cesare et al. (20). Briefly, cells were washed twice in cold PBS and then scraped, and the cellular pellet was resuspended in 10 mM HEPES (pH 7.9)–10 mM KCl–1.5 mM MgCl2–0.1 mM EGTA–0.5 mM DTT on ice. Cells were passed five times through a 26-gauge needle and centrifuged to collect the nuclei, which were subsequently resuspended in an equal volume of 10 mM HEPES (pH 7.9)–0.4 M NaCl–1.5 mM MgCl2–0.1 mM EGTA–0.5 mM DTT–5% glycerol to allow elution of nuclear proteins by gentle shaking at 4°C for 30 min. Nuclei were pelleted for 5 min at 14,000 rpm and 4°C, and the supernatant was then aliquoted, snap frozen in liquid nitrogen, and stored at −80°C until use. All solutions contained the protease inhibitors leupeptin and aprotinin at 1 μg/ml, PMSF (0.5 mM), and benzamidine (1 mM). A Bio-Rad protein assay was used to determine protein concentration. For band-shift assays, 5-μg portions of nuclear extracts were incubated in 50 mM Tris-HCl (pH 7.9)–12.5 mM MgCl2–1 mM EDTA–1 mM DTT–20% glycerol–0.5 mM PMSF–2 μg of poly(dI-dC) for 10 min at room temperature to titrate out nonspecific binding before the addition of 15,000 to 20,000 cpm of labeled oligonucleotide; the reaction mixture was then further incubated for 20 min. When unlabeled competing oligonucleotides or antibodies were added, nuclear extracts were preincubated for 30 min at room temperature or for 16 h on ice, respectively, before the addition of the labeled probe. Samples were loaded on a pre-run 5% polyacrylamide gel (29:1 in 0.25× TBE) and electrophoresed at 200 V. Gels were dried and autoradiographed at −80°C. Antibodies against c-Jun, c-Fos, ATF2, and myogenin were obtained from Santa Cruz Biotechnologies.
The sequences of the sense strands of the oligonucleotides used in the EMSAs were as follows: PEA3/AP1A, 5′-GAGGAAATGAGGTCATCTTGCTCTG-3′; mut AG, 5′-GAGGAAATGAGGagATCTTGCTCTG-3′; ΔG, 5′-GAGGAAATGAGTCATCTTGCTCTG-3′; mut PEA3, 5′-GAccAAATGAGGTCATCTTGCTCTG-3′; AP1B, 5′-GGCCATGTGAATCACGACAGCCTG-3′; col TRE, 5′-CGCTTGATGAGTCAGCCGGAA-3′; cjun2 TRE, 5′-AGCATTACCTCATCCC-3′; and IgkB, 5′-CAGAGGGGACTTTCCGAG-3′.
RESULTS
UV induces murine uPA mRNA expression.
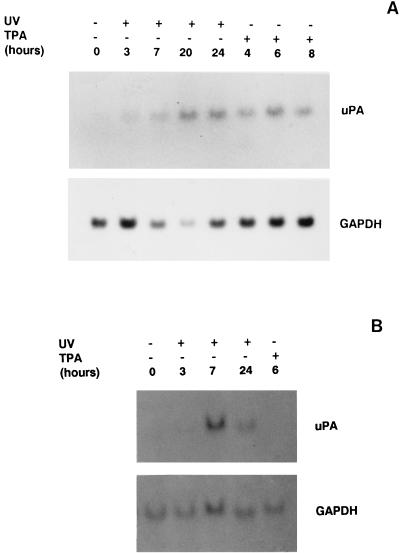
As shown in Fig. 1A, NIH 3T3 fibroblast cells expressed low detectable levels of a 2.7-kb transcript corresponding to murine uPA mRNA, which were increased after UVC irradiation. The increase in uPA gene induction was time dependent since it was first observed at 3 h postirradiation, further increasing at 7 h and reaching its maximum at about 20 to 24 h after UV treatment. In contrast, uPA mRNA was maximally induced by UV at about 7 h postirradiation in F9 murine teratocarcinoma cells (Fig. 1B), suggesting that murine uPA gene induction by UV followed distinct induction kinetics depending on the cell type. This induction was not due to an unspecific upregulation of RNA synthesis, since UV irradiation did not significantly modify the levels of GAPDH mRNA in either cell line. The phorbol ester TPA, known to induce the uPA gene in distinct cell types (21, 52, 66), was included in these experiments for comparison. TPA treatment led to a maximal induction of uPA mRNA at 6 h in NIH 3T3 cells, a finding in agreement with previous results (6), whereas it was unable to induce uPA expression in F9 cells (Fig. 1). These results suggested the existence of different mechanisms of action between that of UV and other extracellular stimuli, such as TPA, to induce the endogenous uPA gene.
FIG. 1.
uPA induction by UV irradiation. (A) Analysis of uPA mRNA expression in UV-stimulated NIH 3T3 fibroblast cells. NIH 3T3 cells were cultured in 0.5% FBS for 16 h and then irradiated with UVC or treated with TPA. Total RNA was extracted at the indicated time points after UVC and TPA stimulation and analyzed by Northern blotting with the mouse uPA and GAPDH cDNA probes as indicated. (B) Analysis of uPA mRNA expression in UV-stimulated F9 teratocarcinoma cells. F9 cells were cultured in 0.5% FBS for 16 h and then exposed to UVC or TPA. Poly(A)+ mRNA was extracted at the indicated time points and analyzed by Northern blotting as for panel A.
Transcriptional analysis of murine uPA gene induction by UV in NIH 3T3 cells.
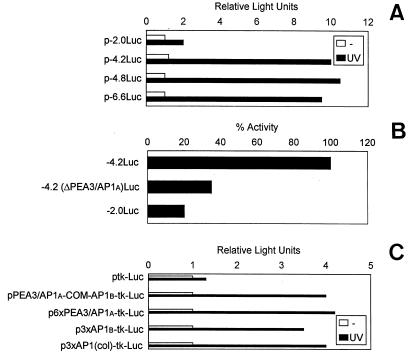
We next examined the effect of UV irradiation on the activity of the murine uPA gene promoter. To do this, a murine uPA genomic fragment (−6.6 kb to −398 bp), ligated upstream of the firefly luciferase reporter gene, p-6.6Luc (6), was assessed for luciferase activity in NIH 3T3 cells. A comparison of luciferase activities generated by p-6.6Luc between unstimulated and UV-stimulated cells showed that the uPA promoter activity was augmented an average 9.5-fold following UV treatment (Fig. 2), indicating that the murine uPA promoter contains UV-responsive sequences that might account, at least in part, for the UV-mediated induction of uPA in these cells. To begin identifying the main regions involved in the uPA transcriptional response to UV, different murine uPA promoter-deletion luciferase constructs were generated from p-6.6Luc, and their UV-inducible luciferase activities were analyzed upon transient transfection in NIH 3T3 cells. As shown in Fig. 2A, deletion of 1.8 and 2.4 kb from the −6.6-kb WPA promoter (generating plasmids p-4.8Luc and p-4.2Luc, respectively) did not alter the luciferase induction of the full-length −6.6-kb promoter plasmid (P > 0.05), suggesting that these upstream promoter regions were irrelevant for uPA transcriptional induction by UV in these cells. However, whereas p-4.2Luc retained full UV-induced luciferase activity, p-2.0Luc, a plasmid containing 2 kb of the murine uPA promoter, showed 80% reduced luciferase activity (P < 0.01) (Fig. 2B), clearly indicating the location within this region of cis-acting element(s) relevant for UV-induced uPA transcription.
FIG. 2.
Transcriptional activity of murine uPA promoter deletion constructs in response to UV. Requirement of an AP1 enhancer element. (A) Transcriptional induction of uPA promoter deletion constructs by UV irradiation. NIH 3T3 cells were transiently transfected with uPA promoter-luciferase constructs of different lengths and then grown in 0.5% FBS for about 16 h before UVC stimulation. Luciferase activity was analyzed at 24 h postirradiation, expressed relative to the activity found with the −2-kb uPA promoter construct (p-2.0Luc) in uninduced cells, and standardized for renilla luciferase activity. All values represented the average of five experiments. The length of the uPA 5′-deletion constructs is indicated below each bar. (B) Requirement of PEA3/AP1A and AP1B sites for uPA promoter induction by UV. NIH 3T3 cells were transiently transfected with different uPA promoter-luciferase constructs lacking either the PEA3/AP1A site only [p-4.2(ΔPEA3/AP1A)Luc] or both the PEA3/AP1A and the AP1B sites (p-2.0Luc) and irradiated as described for panel A. Luciferase activities are represented as a percentage of the activity of the uPA promoter plasmid (p-4.2Luc) containing both AP1 sites in UVC-irradiated cells, which has been given a value of 100%. (C) UV inducibility of multimerized uPA-AP1 sites on heterologous promoters. NIH 3T3 cells were transiently transfected by different reporter constructs containing the complete uPA-AP1 enhancer (pPEA3/AP1A-COM-AP1B), six tandem repeats of the uPA 5′-TRE (p6xPEA3/AP1A), or three tandem copies of the uPA 3′-TRE (p3xAP1B), as well as three copies of the collagenase AP1 element [p3xAP1(col)-tk-Luc], ligated to thymidine kinase-driven luciferase reporter plasmids. Luciferase values were standardized for renilla activities and expressed relative to the activity found with the unresponsive construct ptk-Luc, to which a value of 1.0 has been assigned. All normalized activities represent a minimum of five experiments, showing less than 25% variability.
An AP1 enhancer element is involved in the induction of murine uPA promoter by UV.
The presence in the murine uPA promoter area of an AP1 enhancer element, located at −2.4 kb and known to respond to several extracellular stimuli, suggested that this element might also be responsive to UV stimulation. The uPA AP1 enhancer is composed of two phorbol ester responsive elements (TRE): the combined PEA3/AP1A site (5′-TRE) and the downstream AP1B site (3′-TRE), separated by an intervining element (COM) (see Fig. 3). Of these elements, the PEA3/AP1A element has been shown to be essential for the response to most inductions (reviewed in reference 7). Figure 2B shows that deletion of the PEA3/AP1A enhancer element in the 4.2(ΔPEA3/AP1A)Luc construct reduced UV induction by 65%, whereas deletion of both the PEA3/AP1A and the AP1B sites in the 2.0Luc construct further reduced UV-induced luciferase activity, indicating the contribution of both sites to uPA induction by UV. Thus, we assessed the ability of each AP1 site of the uPA enhancer to confer UV inducibility on a heterologous promoter. A 109-bp fragment (from −2345 to −2454 bp) comprising the complete PEA3/AP1A-COM-AP1B murine enhancer was ligated upstream of a thymidine kinase-driven luciferase gene (ptk-Luc) to generate plasmid pPEA3/AP1A-COM-AP1B-tk-Luc and tested for UV inducibility. Similarly, we assessed the UV-induced transcriptional activity of plasmids p6xPEA3/AP1A-tk-Luc and p3xAP1B-tk-Luc, containing six and three copies of the PEA3/AP1A and AP1B elements, respectively. The full-enhancer-containing plasmid yielded an approximately fourfold greater luciferase activity than the control plasmid ptk-Luc (Fig. 2B), whereas tk-Luc activity was enhanced 4.2- and 3.5-fold by the multimerized PEA3/AP1A and AP1B sites, respectively (Fig. 2C). This indicated that both TREs of the AP1-enhancer were contributing to the UV-induced uPA promoter activity. In agreement with these results, a tk-Luc reporter plasmid containing three copies of the AP1 binding site of the collagenase gene, i.e., plasmid p3xAP1(col)-tk-Luc, was also induced by UV in NIH 3T3 cells.
FIG. 3.
Schematic representation of the murine uPA AP1 enhancer. The PEA3/AP1A and AP1B transactivating sites and the intervening COM (cooperative mediator) sequence, located at position −2.4 kb (relative to the mRNA start site), are shown.
PEA3/AP1A- and AP1B-binding activities in UV-treated NIH 3T3 cells.
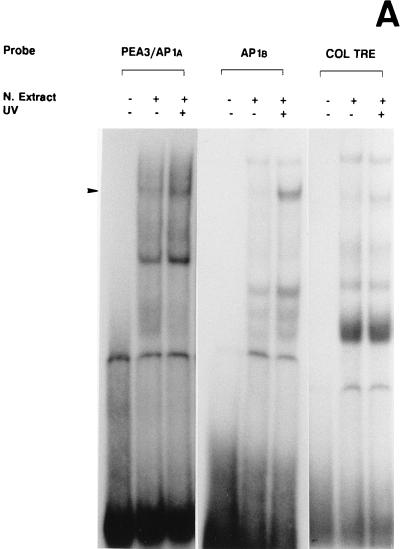
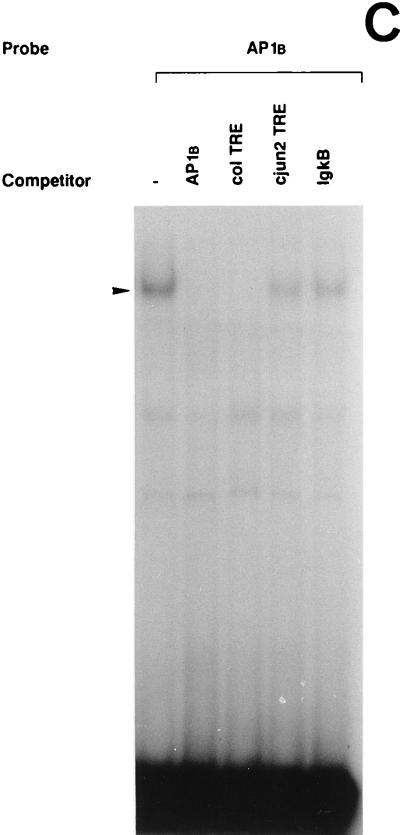
The 5′-TRE and 3′-TRE sequences of the uPA AP1 enhancer bound a major nuclear protein complex, respectively, whose intensity increased following UV treatment of NIH 3T3 cells, as assessed by EMSA (Fig. 4A). Both complexes comigrated with the complex formed by the canonical heptameric AP1 site of the collagenase gene (col TRE), which was used as a reference. As shown in Fig. 4B and C, when each oligonucleotide was used as an unlabeled competitor, its corresponding protein binding complex was totally competed. In contrast, an excess of an unrelated competitor (the κB site of the Igκ enhancer, IgκB) sequence did not affect the formation of either complex.
FIG. 4.
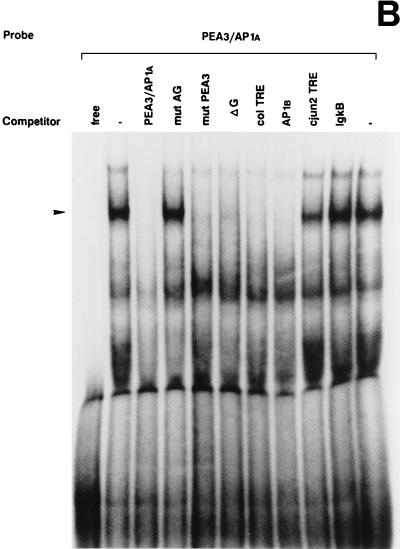
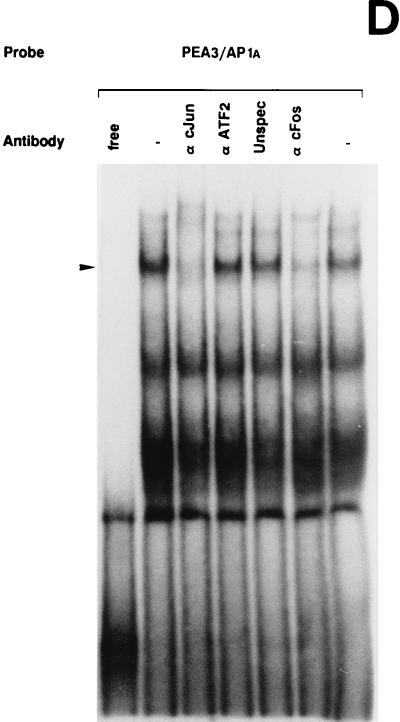
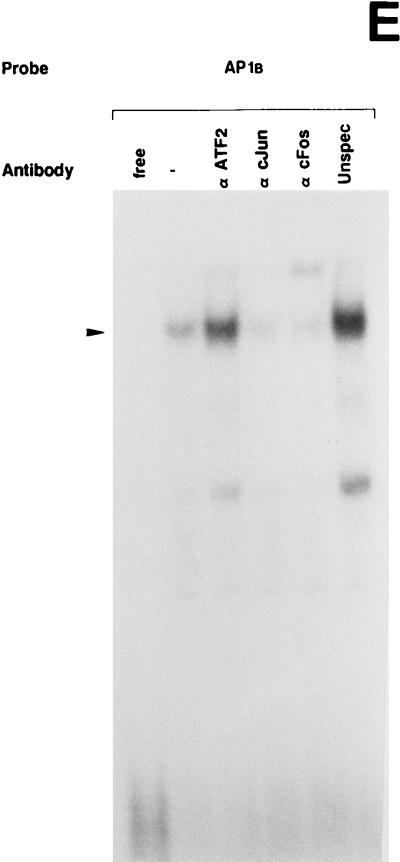
UV irradiation induces AP1 binding activity to the uPA enhancer elements. (A) Induction of uPA 5′-TRE- and 3′-TRE-binding activities by UV irradiation: comparison with the collagenase TRE. NIH 3T3 cells were grown in 0.5% FBS for 16 h and either exposed (+) or not (−) to UVC. Nuclear extracts were prepared at 4 h postirradiation and incubated with 20,000 cpm of the indicated 32P-labeled probes, which correspond to the 5′-TRE (PEA3/AP1A) and 3′-TRE (AP1B) elements of the uPA promoter and to the canonical TRE of the collagenase promoter (COL TRE). EMSAs were performed as described previously (20). The arrowhead indicates specific TRE-binding complexes. (B) Specificity of the induced uPA PEA3/AP1A binding activity. NIH 3T3 nuclear extract induced for 4 h was incubated with the labeled PEA3/AP1A oligonucleotide in the absence or presence of a 150-fold molar excess of unlabeled competitors (as described in Materials and Methods): PEA3/AP1A, mut AG, mut PEA3, and ΔG (corresponding to variants of the uPA PEA3/AP1A site), col TRE (AP1 binding site of the human collagenase promoter), AP1B (uPA AP1B site), cjun2 TRE (distal AP1 binding site of the human c-jun promoter) and IgκB (κB site of the Igκ enhancer). (C) Specificity of the induced uPA AP1B binding activity. NIH 3T3 nuclear extract induced for 4 h was incubated with the labeled AP1B oligonucleotide site in the presence of a 150-fold molar excess of the indicated competitors as described for panel B. (D) The uPA PEA3/AP1A element binds members of the AP1 family of transcription factors upon UV stimulation. EMSAs with a labeled oligonucleotide containing the uPA PEA3/AP1A site and 4-h-induced NIH 3T3 nuclear extracts were performed in the absence or presence of different antibodies as indicated. Extracts were preincubated with specific antibodies directed against the murine c-Jun (α cJun), the murine ATF2 protein (α ATF2), the rat myogenin protein (unspec), the murine c-Fos protein (α cFos), or no antibody (−). (E) UV-induced binding of AP1 transcription factors to the uPA AP1B site. EMSAs with a labeled oligonucleotide containing the uPA AP1B element were performed as described for panel D.
The uPA 5′-TRE is formed by two motifs, the PEA3 site and the octameric AP1A site. In order to assess the relevance of each motif in the 5′-TRE-binding activity, we synthesized an oligonucleotide in which the PEA3 (GGAA) sequence was mutated in two positions. As shown in Fig. 4B, the oligonucleotide containing the AP1A site, flanked by the mutated PEA3 (mut PEA3), fully competed for the binding to the radiolabeled wild-type PEA3/AP1A site. Furthermore, the electrophoretic mobilities of the UV-induced complexes formed by the radiolabeled wild-type PEA3/AP1A oligonucleotide and the one containing the mutated PEA3, respectively, were identical (data not shown). In contrast, an oligonucleotide containing 2-bp mutations inside the AP1A element which destroyed the consensus TRE (mut AG) was unable to compete for the binding of UV-induced nuclear factors, indicating the specificity of the association. Altogether, we concluded that the ability to form a UV-induced complex resided within the central octanucleotide of the uPA 5′-TRE and that the PEA3 flanking element did not affect the binding. Moreover, complex formation could be inhibited by heptameric oligonucleotides corresponding to the uPA 3′-TRE (AP1B) and the collagenase TRE (col TRE), respectively, but not by the octameric distal TRE of the c-jun promoter (cjun2 TRE). In agreement with these results, we observed that a 1-bp deletion inside the AP1A site (ΔG), a mutation transforming the octameric (TGAGGTCA) sequence into the heptameric (TGAGTCA) sequence, which coincided with the collagenase TRE, totally competed with the wild-type 5′-TRE for binding.
The uPA 3′-TRE is formed by a heptameric AP1B site. As shown in Fig. 4A, this site bound a UV-inducible nuclear complex that comigrated with those formed by the uPA PEA3/AP1A and the col TRE, respectively. When the AP1B oligonucleotide was used as unlabeled competitor sequence, the UV-inducible complex was totally competed, whereas an excess of unrelated competitor sequence (IgκB) did not affect AP1B-complex formation (Fig. 4C). Moreover, this complex disappeared in the presence of the col TRE competing sequence but not in the presence of the cjun2 TRE competing sequence. These results suggested that both AP1 sites of the uPA enhancer bound UV-induced complexes of similar properties.
We next investigated the composition of the uPA 5′- and 3′-TRE nuclear complexes by using antibodies to the AP1 components c-Jun and c-Fos, respectively, as well as to the related ATF2 protein. As shown in Fig. 4D and E, both 5′- and 3′-TRE-binding activities were inhibited by the anti-c-Jun (αcJun) and anti-c-Fos (α cFos) antibodies, while the anti-ATF2 (α ATF2) antibody had no effect. As controls, the addition of an irrelevant antibody (anti-myogenin) did not inhibit the binding (unspec). These data indicated that Jun and Fos proteins, but not ATF2, participated in the formation of the UV-inducible AP1 complex binding to uPA 5′-TRE and 3′-TRE, respectively.
Involvement of the JNK pathway in uPA gene induction by UV.
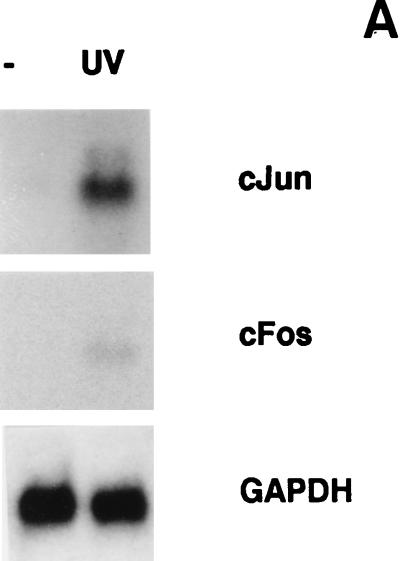
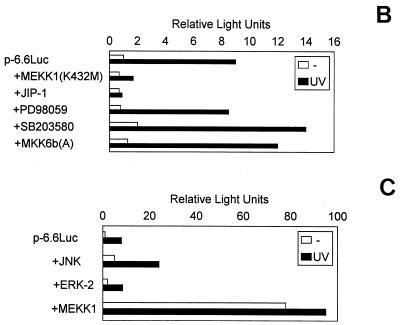
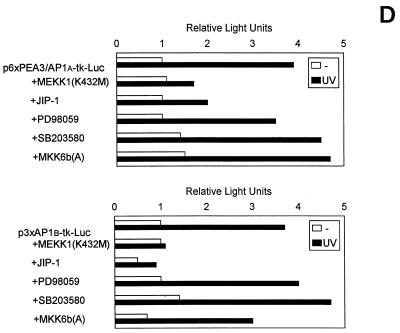
UV irradiation induces the synthesis (Fig. 5A) and the activation of transcription factors such as c-Jun and c-Fos (25, 68, 81). As demonstrated above, an AP1 enhancer element is required for uPA promoter induction by UV (Fig. 2 and 4). Therefore, we hypothesized that the signaling pathway(s) leading to uPA induction might involve activation of either of the three types of UV-inducible MAPKs (ERK, JNK, and p38), two of which can activate the AP1 transcription factor. Accordingly, we examined the ability of specific inhibitors of these pathways to block the induction of uPA promoter activity by UV. MEKK1 has been shown to specifically activate JNK (61). MEKK1(K432M) (61), a dominant negative form of MEKK1, was overexpressed in cotransfection experiments with p-6.6Luc in NIH 3T3 cells, with or without UV irradiation. As shown in Fig. 5B, the uPA promoter activity induced by UV was strongly suppressed by the catalytically inactive mutant form of MEKK1. Furthermore, overexpression of JIP-1, a cytoplasmic protein that causes retention of JNK in the cytoplasm and subsequent inhibition of the JNK pathway (28), also downregulated UV-induced uPA promoter activity in these cells (Fig. 5B). In contrast, PD98059, a specific inhibitor of the MAPK kinase-ERK kinase (MEK) pathway (29), had no significant effect on uPA promoter induction by UV (Fig. 5B). Moreover, overexpression of wild-type JNK but not ERK-2 enhanced uPA promoter induction by UV in NIH 3T3 cells, and overexpression of a constitutively active form of MEKK1 strongly induced uPA promoter activity in the absence of UV treatment (Fig. 5C), indicating that the induction of the uPA gene by UV was mediated, at least in part, by the MEKK1-JNK pathway. Because p38 MAP kinase has also been shown to mediate stress responses, including UV (69), we assessed the potential involvement of this pathway in mediating the induction of the uPA gene promoter by UV. SB203580, a specific inhibitor of two p38 isoforms (p38α and p38β) (17, 31), was unable to block the UV-induced activation of the uPA promoter (Fig. 5A) in NIH 3T3 cells (interestingly, SB203580 further enhanced uPA promoter induction by UV). However, since two newly identified p38 isoforms (p38γ and p38δ) (42, 49, 54, 59, 86) have been shown to be activated by UV but are insensitive to SB203580 (16, 31), the potential involvement of these latter kinases on uPA induction by UV had to be assessed. Accordingly, MKK6b(A), a dominant negative form of MKK6b, which has been shown to diminish the activation of all p38 isoforms (36, 70), was cotransfected together with p-6.6Luc in NIH 3T3 cells. As shown in Fig. 5B, the uPA promoter activity induced by UV was not suppressed by the catalytically inactive mutant form of MKK6b. In agreement with this, only JNK inhibitors MEKK1(K432M) and JIP-1, but not MEK inhibitor PD98059 nor p38 inhibitors SB203580 and MKK6b(A), downregulated the UV inducibility of the multimerized PEA3/AP1A and AP1B elements, respectively, in the tk-Luc plasmids (Fig. 5D). Taken together, these results indicated that the JNK pathway was directly involved in the transcriptional activation of the murine uPA gene by UV in NIH 3T3 cells via its AP1 enhancer element. Consistent with this, the collagenase TRE reporter plasmid, p3xAP1(col)-tk-Luc, was not induced by UV in the presence of MEKK1(K432M) or JIP-1, whereas it was highly inducible in the presence of an irrelevant expression plasmid (data not shown). The effectiveness of PD98059 and SB203580 in downregulating ERK and p38 kinase-dependent reporter activities, respectively, in NIH 3T3 cells was demonstrated according to standard assays (data not shown).
FIG. 5.
Requirement of the MEKK1-JNK pathway for murine uPA induction by UV. (A) UV induces c-Fos and c-Jun expression. NIH 3T3 cells were cultured in 0.5% FBS for 16 h and then irradiated with UVC. Total RNA was extracted at 45 min postirradiation, and sequential blot hybridizations were performed with the c-Jun, c-Fos, and GAPDH cDNA probes as indicated. (B) Effect of specific inhibitors of the JNK, ERK, and p38 MAPK pathways on the activation of the murine uPA promoter by UV. NIH 3T3 cells were transiently transfected with the p-6.6Luc reporter plasmid, together with expression vectors for dominant negative MEKK1 [MEKK1(K432M)], JIP-1, dominant negative MKK6b [MKK6b(A)], or vector alone. Alternatively, the MEK and p38 inhibitors PD98059 (50 μM) and SB203580 (10 μM), respectively, were added to the culture medium 30 min before UVC irradiation. Luciferase activities are expressed relative to the activity of p-6.6Luc in unstimulated NIH 3T3 cells; this activity was given an arbitrary value of 1. Results obtained represent the average of at least three independent experiments. (C) Effect of MAPK signaling components on UV-induced promoter activity. NIH 3T3 cells were transiently transfected with p-6.6Luc reporter plasmid, together with expression vectors for wild-type JNK and ERK2, constitutively active MEKK1, or vector alone. Luciferase activities were determined following UVC irradiation as for panel B. (D) Inhibition of the UV-induced transcriptional activity of the uPA AP1 sites by MEKK1(K432M) and JIP-1. NIH 3T3 cells were transiently transfected with the multimerized PEA3/AP1A or AP1B sites of the uPA enhancer elements ligated to the tk-Luc reporter as described in Materials and Methods, together with expression vectors for dominant negative MEKK1 [MEKK1(K432M)], JIP-1, dominant negative MKK6b [MKK6b(A)], or vector alone. PD98059 and SB203580 were added to the culture medium 30 min before UVC irradiation at final concentrations of 50 and 10 μM, respectively. Luciferase activities are expressed relative to the activity of ptk-Luc in unstimulated NIH 3T3 cells; this activity was given an arbitrary value of 1. All normalized activities represent a minimum of three experiments and show less than 25% variability.
The confirmation of JNK as the MAP kinase mediating uPA gene induction by UV was obtained by using a different approach. It is known that JNK is activated by stress conditions, including UV, but not by other types of stimuli, such as TPA (62). Activated JNK then phosphorylates c-Jun N-terminal Ser-63 and Ser-73, inducing c-Jun transactivation (23, 38). Thus, if c-Jun phosphorylation by JNK has any role in uPA gene induction by UV, mutations that decrease the phosphorylation of c-Jun should attenuate uPA promoter induction. Accordingly, cotransfection experiments with the uPA full promoter plasmid p-6.6Luc, along with plasmid pRSV-cJun, carrying the c-Jun activation domain, were performed in F9 cells, which are known to transduce JNK- but not ERK-dependent inductions (3, 84). As shown in Fig. 6, UV treatment induced uPA transcriptional activity in F9 cells. Moreover, overexpression of c-Jun further increased the UV-mediated induction of the uPA promoter. In contrast, overexpression of a mutated version of the c-Jun protein, carrying double mutation Ala-63 and Ala-73, almost abrogated uPA promoter induction, strongly indicating that the observed activation of the uPA gene by UV was dependent on c-Jun N-terminal phosphorylation by JNK. On the other hand, TPA was not able to induce uPA promoter activity in F9 cells, a finding that correlates with its inability to induce uPA mRNA in these cells (Fig. 1B) and is in agreement with the fact that phorbol esters do not induce AP1 activity in F9 cells (3). Moreover, TPA could not enhance the uPA promoter activity induced by overexpression of c-Jun in F9 cells (Fig. 6), which is in agreement with previous reports demonstrating that TPA induction is not mediated by the JNK pathway but rather preferentially through the ERK pathway (62). Similar results were obtained when the multimerized AP1 sites of the uPA enhancer linked to thymidine kinase-driven reporters were used in cotransfection assays with c-Jun expression plasmids (data not shown). As an experimental control, the ability of Gal4–c-Jun chimeric expression vectors to activate the Gal4-dependent reporter 5xGal4-Luc was examined in nonirradiated and UV-irradiated F9 cells. Whereas wild-type c-Jun resulted in an increase in basal and UV responsiveness of the Gal4 reporter, mutations of amino acids 63 and 73 resulted in the abolition of these effects (data not shown).
FIG. 6.
Involvement of the c-Jun transactivation domain in uPA promoter induction by UV. p-6.6Luc was transiently cotransfected with wild-type c-Jun or mutated c-Jun (Ala-63/73) expression plasmids or with empty vector in F9 murine teratocarcinoma cells. Cells were left untreated or exposed to UVC or TPA, and reporter activity was then measured at 16 h poststimulation. Luciferase activity was expressed relative to the activity found in the uninduced empty-vector-cotransfected cells and standardized for renilla luciferase activity; results represent the averages of three experiments.
Kinetics of JNK, AP1, and uPA induction by UV.
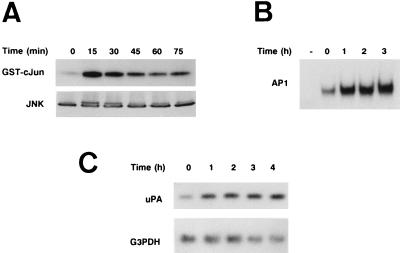
Overall, our results suggest that activation of the JNK pathway and AP1 transcriptional activity may link external UV irradiation and uPA expression. The initial Northern analysis showed that uPA RNA levels were induced by 3 h after UV irradiation (see Fig. 1A). Therefore, a more detailed time course analysis of this induction was performed during the first few hours postirradiation. Figure 7A shows that JNK activity was strongly induced between 15 and 30 min after UV irradiation of NIH 3T3 cells and decreased thereafter, as demonstrated by JNK activation and Western blot analyses. This result correlated with the increased AP1 binding to the 3′-TRE of the uPA enhancer, detected 1 h postirradiation of parallel cell cultures (Fig. 7B). Next, the kinetics of UV-induced uPA mRNA expression during this early period postirradiation were analyzed by RT PCR followed by Southern blotting, a more sensitive transcript detection technique than the Northern blotting method used in Fig. 1. As shown in Fig. 7C, UV-induced uPA expression was detected as early as 1 h after UV irradiation, whereas G3PDH RNA levels, which were used as an RT PCR control, remained constant. Altogether, the timing of these three events (UV-induced JNK activation, AP1 binding, and uPA expression) reinforces the idea that the uPA gene is a direct target for the JNK pathway mediating UV induction.
FIG. 7.
Induction kinetics of c-Jun phosphorylation, AP1 binding activities, and uPA mRNA expression in NIH 3T3 cells upon UV radiation. NIH 3T3 cells were cultured in 0.5% FBS for 16 h and then irradiated with UVC. WCE, nuclear extracts, and total RNA were obtained from parallel cultures at the indicated time points postirradiation. (A) Time course of UV light-stimulated JNK activation in NIH 3T3 cells. WCE were prepared as detailed in Materials and Methods at the indicated time points after UV stimulation. (Top panel) JNK activity was determined by immunoprecipitation of JNK from WCE and kinase assay with the substrate GST–c-Jun (1–223). (Bottom panel) Western blot showing total JNK amount present in each cell extract (50 μg/lane). (B) Time course of UV-induced AP1 binding to the uPA 3′-TRE. EMSA was performed with nuclear extracts (5 μg/lane) prepared from NIH 3T3 cells at the indicated time points postirradiation. (C) Time course of uPA RNA levels in response to UV light. Total RNA was purified at the indicated time points postirradiation of NIH 3T3 cells and analyzed by RT PCR followed by Southern blotting with specific uPA and G3PDH murine primers.
DISCUSSION
Irradiation of cells with short-wavelength UV induces the transcription of many genes. In this study we demonstrate that the endogenous murine uPA gene product can be induced by UV at the level of gene transcription via the JNK pathway of MAP kinases. In particular, we found that an AP1 enhancer element, which is conserved in murine, porcine, and human uPA promoters, is required for the induction by UV, since deletion of this element abrogated the induction.
We have shown that the endogenous murine uPA gene product can be induced in the first hours following UV irradiation in F9 and NIH 3T3 cells, although the induction was maintained for a longer period of time in the latter cell type. Moreover, uPA promoter induction by UV could be completely blocked by inhibitors of cytoplasmic signaling cascades, which are initiated at or near the plasma membrane (26) (Fig. 5 and 6), suggesting that the uPA gene could be activated, at least in part, by a cytoplasm-transduced signal. These results have to be reconciled with studies showing that UV caused a late (occurring a few days postirradiation) induction of human uPA expression in normal and xeroderma pigmentosum cells after inducing a signal originating from DNA damage (64, 74). At present, we can only speculate on these apparently different origins of UV induction. Possibly, uPA gene activation by UV is under the control of pathways from both the plasma membrane and the nucleus.
The availability of a well-characterized murine uPA promoter plasmid and of inhibitors of three MAPK pathways known to transduce extracellular stimuli such as UV into intracellular responses facilitated the analysis of the mechanism(s) underlying uPA gene induction by UV. Promoter deletion analysis revealed that the murine AP1 enhancer element located at −2.4 kb was required for uPA promoter induction by UV, since its deletion abrogated the induction. The uPA enhancer contains two TREs: the combined PEA3/AP1A site (5′-TRE) and the downstream AP1B site (3′-TRE), both of which were able to confer UV inducibility on a heterologous promoter, a finding correlating with their induced AP1-binding activity following UV irradiation of NIH 3T3 cells. Moreover, the UV-inducible complexes formed by the two uPA TREs were formed by c-Jun and c-Fos and not by ATF2.
Transient-transfection assays with coexpression of specific inhibitors for proteins implicated in distinct MAPK signaling pathways suggested that JNK, but not ERK or p38, is involved in UV-dependent uPA gene induction. Expression of MEKK1 results in efficient JNK activation without a significant increase in ERK1/2 or p38 activities (55, 61). Our present work shows that overexpression of MEKK1(K432M), a catalytically inactive form of MEKK1, was capable of abrogating the UV-induced upregulation of the murine uPA promoter in NIH 3T3 cells. The implication of JNK in UV-induced uPA transcription was further evidenced by the inhibitory effect caused by overexpression of JIP-1, a cytoplasmic inhibitor of JNK, which has been shown to inhibit JNK-regulated gene expression (28). Moreover, constitutively active MEKK1 was able to activate uPA gene transcription in the absence of UV stimulation, suggesting that the uPA gene is a target for this MAPK pathway. We do not know, however, whether MEKK1 simultaneously activates other signaling routes which cooperate with the MEKK1-JNK pathway in uPA gene induction. This possibility is worth considering because MEKK1 has recently been shown to activate NFκB by activating the IκBα kinase complex in vivo and in vitro (50, 60). Interestingly, an NFκB-like element has been identified in the human uPA promoter mediating induction by TPA in HepG2 and HT1080 cells (37), although this site is not conserved in the murine uPA promoter.
We next demonstrated that c-Jun was the subsequent substrate in the JNK pathway mediating UV induction of uPA. JNK phosphorylation of c-Jun occurs at the N-terminal Ser-63 and Ser-73 sites (23, 38). Whereas cotransfection of intact c-Jun augmented uPA promoter induction by UV in F9 cells, a mutated version of the c-Jun protein carrying a double Ala-63–Ala-73 mutation could not mediate this effect. In contrast, TPA could not upregulate the basal or the c-Jun-activated uPA promoter in F9 cells, a finding in agreement both with previous results showing that the TPA-mediated induction of the uPA gene occurs through the ERK pathway (6) and with the inability of TPA to induce uPA mRNA in this cell line (Fig. 1B). Altogether, these results indicated that uPA transcriptional induction by UV was dependent on c-Jun phosphorylation by JNK. Moreover, the kinetics of JNK activation in response to UV were shown to correlate with the early induction of AP1 binding to the uPA 3′-TRE and with the early upregulation of uPA transcript levels, reinforcing the idea that the uPA gene is a direct target of the JNK pathway mediating UV stimulation. Therefore, one pathway proposed for UV-dependent uPA activation was likely to proceed as follows: MEKK1 → JNKK1 → JNK → c-Jun → AP1 → uPA (TREs). Interestingly, it has been shown that in addition to c-Jun, JNK also phosphorylates and activates ATF2 and TCF/Elk-1, one of the transcription factors regulating the activity of the c-Fos promoter (11). Whereas ATF2 is not found in the UV-induced complex binding to the uPA AP1 enhancer, c-fos is, together with c-Jun, one of the proteins participating in this complex (Fig. 4). Therefore, it could be speculated that a potential UV-induced JNK pathway activating c-Fos via Elk-1 might be indirectly contributing to the induced c-Fos binding to the uPA enhancer following UV stimulation, although this question has not been addressed in the present study. Recent evidence also suggested that UV-induced DNA damage can lead to JNK activation (1), in contrast to the previously accepted idea of cell membrane-initiated JNK activation by UV, as has been demonstrated in enucleated cells (27). Whether this novel mechanism of DNA-induced JNK activation might be somehow involved in the uPA induction by UV in human DNA-repair-deficient cells should be determined in future studies.
Whether DNA damage provides the primary signal for the mammalian UV response, it seems likely that this response might enhance the DNA repair capacity of the irradiated cell in a way similar to that of the bacterial SOS response (85). However, none of the UV-inducible genes identified in mammalian cells appears to be involved in DNA repair (39, 43). Recently, several reports have suggested that the mammalian UV response was involved in a protective function other than DNA repair (26, 30). In fact, the inhibition of the UV response by tyrosine kinase inhibitors has confirmed its protective role (26). Additionally, c-fos−/− cells and c-jun−/− cells are hypersensitive to UV irradiation (81), suggesting that the UV-inducible c-Jun and c-Fos are essential components of the cellular defense mechanisms against the cytotoxic effects of UV irradiation. What is the physiological significance of the uPA induction by UV? UV light can cause severe damage, such as the induction and promotion of cancer, cutaneous inflammation, and immunosuppression (46). Exposure of cells to UV and most other DNA-damaging agents results in the damage of biomembranes, proteins, and nucleic acids either directly or by generating oxidative stress (2). A simple protective mechanism against damage to such components would consist of replacing them with newly synthesized ones. Accordingly, the uPA gene and other AP1-regulated genes, such as collagenase I and stromelysin I and II, code for proteolytic enzymes which are induced following UV irradiation (81). These proteases have complementary functions in the degradation of the extracellular matrix, a mechanism playing a role in conditions such as wound healing, cancer, and inflammation, situations in which cell migration is required (reviewed in references 7 and 18). In particular, uPA was hypothesized to provide proteolytic activity that enabled inflammatory cells to traverse tissues during recruitment and was implicated as a cytokine modulator. Recent work with uPA−/− mice has in fact demonstrated that uPA is required for the pulmonary inflammatory response to Cryptococcus neoformans, since a lack of uPA resulted in inadequate cellular recruitment, uncontrolled infection, and death (34). In addition, endogenously produced uPA could amplify tumor necrosis factor alpha neosynthesis by mononuclear phagocytes, representing a novel mechanism by which a phagocyte-derived protease contributes to generating proinflammatory signals (82). It is therefore tempting to speculate that uPA might play a similar role in the inflammation caused by UV light. At present, uPA-deficient mice are available. The sensitivity of the uPA−/− cells to UV irradiation will greatly clarify the role of this protease in the mammalian UV response.
ACKNOWLEDGMENTS
F. Miralles and M. Parra contributed equally to this work.
We are grateful to M. Karin, J. Han, R. Davis, and S. Gutkind for generously providing us with various plasmids. We also thank A. Martin-Corbacho for excellent secretarial assistance.
This work was supported in part by DGES PM97:0088.
REFERENCES
- 1.Adler V, Fuchs S Y, Kim J, Kraft A, King M P, Pelling J, Ronai Z. jun-NH2-terminal kinase activation mediated by UV-induced DNA lesions in melanoma and fibroblast cells. Cell Growth Differ. 1995;6:1437–1446. [PubMed] [Google Scholar]
- 2.Angel P. The role and regulation of Jun proteins in response to phorbol ester and UV light. In: Baeuerle P, editor. Inducible gene expression and cytoplasmic/nuclear signal transduction. Boston, Mass: Birkhauser Verlag; 1995. pp. 62–92. [Google Scholar]
- 3.Auer H-P, Konig H, Litfin M, Stein B, Rahmsdorf H J. Ultraviolet irradiation, although it activates the transcription factor AP-1 in F9 teratocarcinoma stem cells, does not induce the full complement of differentiation-associated genes. Exp Cell Res. 1994;214:131–138. doi: 10.1006/excr.1994.1241. [DOI] [PubMed] [Google Scholar]
- 4.Bender K, Blattner C, Knebel A, Iordanov M, Herrlich P, Rahmsdorf H J. UV-induced signal transduction. J Photochem Photobiol B Biol. 1997;37:1–17. doi: 10.1016/s1011-1344(96)07459-3. [DOI] [PubMed] [Google Scholar]
- 5.Besser D, Bardelli A, Didichenko S, Thelen M, Comoglio P M, Ponzetto C, Nagamine Y. Regulation of the urokinase-type plasminogen activator gene by the oncogene Tpr-Met involves GRB2. Oncogene. 1997;14:705–711. doi: 10.1038/sj.onc.1200879. [DOI] [PubMed] [Google Scholar]
- 6.Besser D, Presta M, Nagamine Y. Elucidation of a signaling pathway induced by FGF-2 leading to uPA gene expression in NIH3T3 fibroblasts. Cell Growth Differ. 1995;6:1009–1017. [PubMed] [Google Scholar]
- 7.Besser D, Verde P, Nagamine Y, Blasi F. Signal transduction and the uPA/uPAR system. Fibrinolysis. 1996;10:215–237. [Google Scholar]
- 8.Blasi F, Verde P. Urokinase-dependent cell surface proteolysis and cancer. Cancer Biol. 1990;1:117–126. [PubMed] [Google Scholar]
- 9.Carmeliet P, Collen D. Targeted gene manipulation and transfer of the plasminogen and coagulation systems in mice. Fibrinolysis. 1996;10:195–213. doi: 10.1055/s-2007-999055. [DOI] [PubMed] [Google Scholar]
- 10.Carmeliet P, Schoonjans L, Kieckens L, Ream B, Degen J, Bronson R, De Vos R, Van den Oord J J, Collen D, Mulligan R C. Physiological consequences of loss of plasminogen activator gene function in mice. Nature. 1994;368:419–424. doi: 10.1038/368419a0. [DOI] [PubMed] [Google Scholar]
- 11.Cavigelli M, Dolfi F, Claret F-X, Karin M. Induction of c-fos expression through JNK-mediated TCF/Elk-1 phosphorylation. EMBO J. 1995;14:5957–5964. doi: 10.1002/j.1460-2075.1995.tb00284.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chomczynski P, Sacchi N. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chlorophorm extraction. Anal Biochem. 1987;162:156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- 13.Clifton A D, Young P R, Cohen P. A comparison of the substrate specificity of MAPKAP kinase-2 and MAPKAP kinase-3 and their activation by cytokines and cellular stress. FEBS Lett. 1996;392:209–214. doi: 10.1016/0014-5793(96)00816-2. [DOI] [PubMed] [Google Scholar]
- 14.Coso O A, Chiariello M, Yu J-C, Teramoto H, Crespo P, Xu N, Miki T, Gutkind J S. The small GTP-binding proteins Rac1 and Cdc42 regulate the activity of the JNK/SAPK signaling pathway. Cell. 1995;81:1137–1146. doi: 10.1016/s0092-8674(05)80018-2. [DOI] [PubMed] [Google Scholar]
- 15.Crespo P, Xu N, Simonds W F, Gutkind J S. Ras-dependent activation of MAP kinase pathway mediated by G-protein βγ subunits. Nature. 1994;369:418–420. doi: 10.1038/369418a0. [DOI] [PubMed] [Google Scholar]
- 16.Cuenda A, Cohen P, Buée-Scherrer V, Goedert M. Activation of stress-activated protein kinase-3 (SAPK3) by cytokines and cellular stresses is mediated via SAPKK3 (MKK6): comparison of the specificities of SAPK3 and SAPK2 (RK/p38) EMBO J. 1997;16:295–305. doi: 10.1093/emboj/16.2.295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cuenda A, Rouse J, Doza Y N, Meier R, Cohen P, Gallagher T F, Young P R, Lee J C. SB203580 is a specific inhibitor of a MAP kinase homologue which is stimulated by cellular stresses and interleukin-1. FEBS Lett. 1995;364:229–233. doi: 10.1016/0014-5793(95)00357-f. [DOI] [PubMed] [Google Scholar]
- 18.Dano K, Andreasen P A, Grondahl-Hansen J, Kristensen P, Nielsen L S, Skriver L. Plasminogen activators, tissue degradation, and cancer. Adv Cancer Res. 1985;44:139–265. doi: 10.1016/s0065-230x(08)60028-7. [DOI] [PubMed] [Google Scholar]
- 19.Davis R J. The mitogen-activated protein kinase signal transduction pathway. J Biol Chem. 1993;268:14553–14556. [PubMed] [Google Scholar]
- 20.De Cesare D, Vallone D, Caracciolo A, Sassone-Corsi P, Nerlov C, Verde P. Heterodimerization of c-Jun with ATF-2 and c-Fos is required for positive and negative regulation of the human urokinase enhancer. Oncogene. 1995;11:365–376. [PubMed] [Google Scholar]
- 21.Degen J L, Estensen R D, Nagamine Y, Reich E. Induction and desensitization of plasminogen activator gene expression by tumor promoters. J Biol Chem. 1985;260:12426–12433. [PubMed] [Google Scholar]
- 22.Degen S J F, Heckel J L, Reich E, Degen J L. The murine urokinase-type plasminogen activator gene. Biochemistry. 1987;26:8270–8279. doi: 10.1021/bi00399a038. [DOI] [PubMed] [Google Scholar]
- 23.Derijard B, Hibi M, Wu I-H, Barrett T, Su B, Deng T, Karin M, Davis R J. JNK1: a protein kinase stimulated by UV light and Ha-Ras that binds and phosphorylates the c-Jun activation domain. Cell. 1994;76:1025–1037. doi: 10.1016/0092-8674(94)90380-8. [DOI] [PubMed] [Google Scholar]
- 24.Derijard B, Raingeaud J, Barrett T, Wu I H, Han J, Ulevitch R J, Davis R J. Independent human MAP-kinase signal transduction pathways defined by MEK and MKK isoforms. Science. 1995;267:682–685. doi: 10.1126/science.7839144. [DOI] [PubMed] [Google Scholar]
- 25.Devary Y, Gottlieb R A, Lau L F, Karin M. Rapid and preferential activation of the c-jun gene during the mammalian UV response. Mol Cell Biol. 1991;11:2804–2811. doi: 10.1128/mcb.11.5.2804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Devary Y, Gottlieb R A, Smeal T, Karin M. The mammalian ultraviolet response is triggered by activation of Src tyrosine kinases. Cell. 1992;71:1081–1091. doi: 10.1016/s0092-8674(05)80058-3. [DOI] [PubMed] [Google Scholar]
- 27.Devary Y, Rosette C, DiDonato J A, Karin M. NF-κB activation by ultraviolet light not dependent on a nuclear signal. Science. 1993;261:1442–1445. doi: 10.1126/science.8367725. [DOI] [PubMed] [Google Scholar]
- 28.Dickens M, Rogers J S, Cavanagh J, Raitano A, Xia Z, Halpern J R, Greenberg M E, Sawyers C L, Davis R J. A cytoplasmic inhibitor of the JNK signal transduction pathway. Science. 1997;277:693–696. doi: 10.1126/science.277.5326.693. [DOI] [PubMed] [Google Scholar]
- 29.Dudley D T, Pang L, Decker S J, Bridges A J, Saltiel A R. A synthetic inhibitor of the mitogen-activated protein kinase cascade. Proc Natl Acad Sci USA. 1995;92:7686–7689. doi: 10.1073/pnas.92.17.7686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Engelberg D, Klein C, Martinetto H, Struhl K, Karin M. The UV response involving the Ras signaling pathway and AP-1 transcription factors is conserved between yeast and mammals. Cell. 1994;77:381–390. doi: 10.1016/0092-8674(94)90153-8. [DOI] [PubMed] [Google Scholar]
- 31.Goedert M, Cuenda A, Craxton M, Jakes R, Cohen P. Activation of the novel stress-activated protein kinase SAPK4 by cytokines and cellular stresses is mediated by SKK3 (MKK6): comparison of its substrate specificity with that of other SAP kinases. EMBO J. 1997;16:3563–3571. doi: 10.1093/emboj/16.12.3563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Groot R P, van Dijk T B, Caldenhoven E, Coffer P J, Raaijmakers J A M, Lammers J-W J, Koenderman L. Activation of 12-O-tetradecanoylphorbol-13-acetate response element- and dyad symmetry element-dependent transcription by interleukin-5 is mediated by Jun N-terminal kinase/stress-activated protein kinase kinases. J Biol Chem. 1997;272:2319–2325. doi: 10.1074/jbc.272.4.2319. [DOI] [PubMed] [Google Scholar]
- 33.Gupta S, Campbell D, Derijard B, Davis J. Transcription factor ATF2 regulation by the JNK signal transduction pathway. Science. 1995;267:389–393. doi: 10.1126/science.7824938. [DOI] [PubMed] [Google Scholar]
- 34.Gyetko M R, Chen G H, McDonald R A, Goodman R, Huffnagle G B, Wilkinson C C, Fuller J A, Toews G B. Urokinase is required for the pulmonary inflammatory response to Cryptococcus neoformans. A murine transgenic model. J Clin Invest. 1996;97:1818–1826. doi: 10.1172/JCI118611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Han J, Bibbs L, Ulevitch R J. A MAP kinase targeted by endotoxin and hyperosmolarity in mammalian cells. Science. 1994;265:808–811. doi: 10.1126/science.7914033. [DOI] [PubMed] [Google Scholar]
- 36.Han J, Lee J-D, Jiang Y, Li Z, Feng L, Ulevitch R J. Characterization of the structure and function of a novel MAP kinase kinase (MKK6) J Biol Chem. 1996;271:2886–2891. doi: 10.1074/jbc.271.6.2886. [DOI] [PubMed] [Google Scholar]
- 37.Hansen S K, Nerlov C, Zabel U, Verde P, Johnsen M, Baeuerle P A, Blasi F. A novel complex between the p65 subunit of NF-kappaB and c-Rel binds to a DNA element involved in the phorbol ester induction of the human urokinase gene. EMBO J. 1992;11:205–213. doi: 10.1002/j.1460-2075.1992.tb05043.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hibi M, Lin A, Smeal T, Minden A, Karin M. Identification of an oncoprotein- and UV-responsive protein kinase that binds and potentiates the c-jun activation domain. Genes Dev. 1993;7:2135–2148. doi: 10.1101/gad.7.11.2135. [DOI] [PubMed] [Google Scholar]
- 39.Holbrook N J, Fornace A J J. Response to adversity: molecular control of gene activation following genotoxic stress. New Biol. 1991;3:825–833. [PubMed] [Google Scholar]
- 40.Irigoyen J, Besser D, Nagamine Y. Cytoskeleton reorganization induces the urokinase-type plasminogen activator gene via the Ras/Extracellular signal-regulated kinase (ERK) signaling pathway. J Biol Chem. 1997;272:1904–1909. doi: 10.1074/jbc.272.3.1904. [DOI] [PubMed] [Google Scholar]
- 41.Jiang Y, Chen C, Li Z, Guo W, Gegner J A, Lin S, Han J. Characterization of the structure and function of a new mitogen-activated protein kinase (p38β) J Biol Chem. 1996;271:17920–17926. doi: 10.1074/jbc.271.30.17920. [DOI] [PubMed] [Google Scholar]
- 42.Jiang Y, Gram H, Zhao M, New L, Gu J, Feng L, Di Padova F, Ulevitch R J, Han J. Characterization of the structure and function of the fourth member of p38 group mitogen-activated protein kinases, p. 38δ. J Biol Chem. 1997;272:30122–30128. doi: 10.1074/jbc.272.48.30122. [DOI] [PubMed] [Google Scholar]
- 43.Karin M, Herrlich P. cis- and trans-acting genetic elements responsible for induction of specific genes by tumor promoters, serum factors and stress. In: Colburn N H, editor. Genes and signal transduction in multistage carcinogenesis. New York, N.Y: Marcel Dekker, Inc.; 1989. pp. 415–440. [Google Scholar]
- 44.Knebel A, Rahmsdorf H J, Ullrich A, Herrlich P. Dephosphorylation of receptor tyrosine kinases as target of regulation by radiation, oxidants or alkylating agents. EMBO J. 1996;15:5314–5325. [PMC free article] [PubMed] [Google Scholar]
- 45.Koj A. Initiation of acute phase response and synthesis of cytokines. Biochim Biophys Acta. 1996;1317:84–94. doi: 10.1016/s0925-4439(96)00048-8. [DOI] [PubMed] [Google Scholar]
- 46.Kripke M L. Immunology and photocarcinogenesis. J Am Acad Dermatol. 1986;14:149–155. doi: 10.1016/s0190-9622(86)70017-0. [DOI] [PubMed] [Google Scholar]
- 47.Kwaan H C. The plasminogen-plasmin system in malignancy. Cancer Metastasis Rev. 1992;11:291–311. doi: 10.1007/BF01307184. [DOI] [PubMed] [Google Scholar]
- 48.Kyriakis J M, Banerjee P, Nikolakaki E, Dai T, Rubie E A, Ahmad M F, Avruch J, Woodgett J R. The stress-activated protein kinase subfamily of c-Jun kinases. Nature. 1994;369:156–160. doi: 10.1038/369156a0. [DOI] [PubMed] [Google Scholar]
- 49.Lechner C, Zahalka M A, Giot J-F, Moller M P, Ullrich A. ERK6, a mitogen-activated protein kinase involved in C2C12 myoblast differentiation. Proc Natl Acad Sci USA. 1996;93:4355–4359. doi: 10.1073/pnas.93.9.4355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lee F S, Hagler J, Chen Z J, Maniatis T. Activation of the IκBα kinase complex by MEKK1, a kinase of the JNK pathway. Cell. 1997;88:213–222. doi: 10.1016/s0092-8674(00)81842-5. [DOI] [PubMed] [Google Scholar]
- 51.Lee J C, Laydon J T, McDonnell P C, Gallagher T F, Kumar S, Green D, McNulty D, Blumenthal M J, Heys J R, Landvatter S W, Strickler J E, McLaughlin M M, Siemens I R, Fisher S M, Livi G P, White J R, Adams J L, Young P R. A protein kinase involved in the regulation of inflammatory cytokine biosynthesis. Nature. 1994;372:739–746. doi: 10.1038/372739a0. [DOI] [PubMed] [Google Scholar]
- 52.Lee J S, Von der Ahe D, Kiefer B, Nagamine Y. Cytoskeletal reorganization and TPA differently modify AP-1 to induce the urokinase-type plasminogen activator gene in LLC-PK1 cells. Nucleic Acids Res. 1993;21:3365–3372. doi: 10.1093/nar/21.15.3365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lengyel E, Klostergaard J, Boyd D. Stimulation of urokinase expression by TNF-α requires the activation of binding sites for the AP-1 and PEA3 transcription factors. Biochim Biophys Acta. 1995;1268:65–72. doi: 10.1016/0167-4889(95)00050-3. [DOI] [PubMed] [Google Scholar]
- 54.Li Z, Jiang Y, Ulevitch R J, Han J. The primary structure of p38γ: a new member of p38 group of MAP kinases. Biochem Biophys Res Commun. 1996;228:334–340. doi: 10.1006/bbrc.1996.1662. [DOI] [PubMed] [Google Scholar]
- 55.Lin A, Minden A, Martinetto H, Claret F-X, Lange-Carter C, Mercurio F, Johnson G L, Karin M. Identification of a dual specificity kinase that activates the Jun kinases and p38-Mpk2. Science. 1995;268:286–290. doi: 10.1126/science.7716521. [DOI] [PubMed] [Google Scholar]
- 56.Livingstone C, Patel G, Jones N. ATF-2 contains a phosphorylation-dependent transcriptional activation domain. EMBO J. 1995;14:1785–1797. doi: 10.1002/j.1460-2075.1995.tb07167.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Marshall B C, Xu Q P, Rao N V, Hoidal J R. Pulmonary epithelial cell urokinase-type plasminogen activator: induction by interleukin-1 beta and tumor necrosis factor α. J Biol Chem. 1992;267:11462–11469. [PubMed] [Google Scholar]
- 58.McLaughlin M M, Kumar S, McDonnell P C, Van Horn S, Lee J C, Livi G P, Young P R. Identification of mitogen-activated protein (MAP) kinase-activated protein kinase-3, a novel substrate of CSBP p38 MAP kinase. J Biol Chem. 1996;271:8488–8492. doi: 10.1074/jbc.271.14.8488. [DOI] [PubMed] [Google Scholar]
- 59.Mertens S, Craxton M, Goedert M. SAP kinase-3, a new member of the family mammalian stress-activated protein kinases. FEBS Lett. 1996;383:273–276. doi: 10.1016/0014-5793(96)00255-4. [DOI] [PubMed] [Google Scholar]
- 60.Meyer C F, Wang X, Chang C, Templeton D, Tan T-H. Interaction between c-Rel and the mitogen-activated protein kinase kinase kinase 1 signaling cascade in mediating κB enhancer activation. J Biol Chem. 1996;271:8971–8976. doi: 10.1074/jbc.271.15.8971. [DOI] [PubMed] [Google Scholar]
- 61.Minden A, Lin A, McMahon M, Lange-Carter C, Derijard B, Davis R J, Johnson G L, Karin M. Differential activation of ERK and JNK mitogen-activated protein kinases by Raf-1 and MEKK. Science. 1994;266:1719–1723. doi: 10.1126/science.7992057. [DOI] [PubMed] [Google Scholar]
- 62.Minden A, Lin A, Smeal T, Derijard B, Cobb M, Davis R, Karin M. c-Jun N-terminal phosphorylation correlates with activation of the JNK subgroup but not the ERK subgroup of mitogen-activated protein kinases. Mol Cell Biol. 1994;14:6683–6688. doi: 10.1128/mcb.14.10.6683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Miralles F, Ron D, Baiget M, Felez J, Muñoz-Canoves P. Differential regulation of urokinase-type plasminogen activator expression by basic fibroblast growth factor and serum in myogenesis. J Biol Chem. 1998;273:2052–2058. doi: 10.1074/jbc.273.4.2052. [DOI] [PubMed] [Google Scholar]
- 64.Miskin R, Ben-Ishai R. Induction of plasminogen activator by UV light in normal and xeroderma pigmentosum fibroblasts. Proc Natl Acad Sci USA. 1981;78:6236–6240. doi: 10.1073/pnas.78.10.6236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Muñoz-Canoves P, Miralles F, Baiget M, Felez J. Inhibition of urokinase-type plasminogen activator (uPA) abrogates myogenesis in vitro. Thromb Haemostasis. 1997;77:526–534. [PubMed] [Google Scholar]
- 66.Nerlov C, De Cesare D, Pergola F, Caracciolo A, Blasi F, Johnsen M, Verde P. A regulatory element that mediates co-operation between a PEA3-AP-1 element and an AP-1 site is required for phorbol ester induction of urokinase enhancer activity in HepG2 hepatoma cells. EMBO J. 1992;11:4573–4582. doi: 10.1002/j.1460-2075.1992.tb05559.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Price M A, Cruzalegui F H, Treisman R. The p38 and ERK MAP kinase pathways cooperate to activate ternary complex factors and c-fos transcription in response to UV light. EMBO J. 1996;15:6552–6563. [PMC free article] [PubMed] [Google Scholar]
- 68.Radler-Pohl A, Sachsenmaier C, Gebel S, Auer H-P, Bruder J T, Rapp U, Angel P, Rahmsdorf H J, Herrlich P. UV-induced activation of AP-1 involves obligatory extranuclear steps including Raf-1 kinase. EMBO J. 1993;12:1005–1012. doi: 10.1002/j.1460-2075.1993.tb05741.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Raingeaud J, Gupta S, Rogers J S, Dickens M, Han J, Ulevitch R J, Davis R J. Pro-inflammatory cytokines and environmental stress cause p38 mitogen-activated protein kinase activation by dual phosphorylation on tyrosine and threonine. J Biol Chem. 1995;270:7420–7426. doi: 10.1074/jbc.270.13.7420. [DOI] [PubMed] [Google Scholar]
- 70.Raingeaud J, Whitmarsh A J, Barrett T, Derijard B, Davis R J. MKK3 and MKK6-regulated gene expression is mediated by the p38 mitogen-activated protein kinase signal transduction pathway. Mol Cell Biol. 1996;16:1247–1255. doi: 10.1128/mcb.16.3.1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Resnati M, Guttinger M, Valcamonica S, Sidenius N, Blasi F, Fazioli F. Proteolytic cleavage of the urokinase receptor substitutes for the agonist-induced chemotactic effect. EMBO J. 1996;15:1572–1582. [PMC free article] [PubMed] [Google Scholar]
- 72.Ron-Ishai R, Sharon R, Rothman M, Miskin R. DNA repair and induction of plasminogen activator in human fetal cells treated with ultraviolet light. Carcinogenesis. 1984;5:357–362. doi: 10.1093/carcin/5.3.357. [DOI] [PubMed] [Google Scholar]
- 73.Rorth P, Nerlov C, Blasi F, Johnsen M. Transcription factor PEA3 participates in the induction of urokinase plasminogen activator transcription in murine keratinocytes stimulated with epidermal growth factor or phorbol-ester. Nucleic Acids Res. 1990;18:5009–5017. doi: 10.1093/nar/18.17.5009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Rotem N, Axelrod J H, Miskin R. Induction of urokinase-type plasminogen activator by UV light in human fetal fibroblasts is mediated through a UV-induced secreted protein. Mol Cell Biol. 1987;7:622–631. doi: 10.1128/mcb.7.2.622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Rouse J, Cohen P, Trigon S, Morange M, Alonso-Llamazares A, Zamanillo D, Hunt T, Nebreda A R. A novel kinase cascade triggered by stress and heat shock that stimulates MAPKAP kinase-2 and phosphorylation of the small and heat shock proteins. Cell. 1995;78:1027–1037. doi: 10.1016/0092-8674(94)90277-1. [DOI] [PubMed] [Google Scholar]
- 76.Sachsenmaier C, Radler-Pohl A, Muller A, Herrlich P, Rahmsdorf H J. Damage to DNA by UV light and activation of transcription factors. Biochem Pharmacol. 1994;47:129–136. doi: 10.1016/0006-2952(94)90446-4. [DOI] [PubMed] [Google Scholar]
- 77.Sachsenmaier C, Radler-Pohl A, Zinck R, Nordheim A, Herrlich P, Rahmsdorf H J. Involvement of growth factor receptors in the mammalian UVC response. Cell. 1994;78:963–972. doi: 10.1016/0092-8674(94)90272-0. [DOI] [PubMed] [Google Scholar]
- 78.Saklatvala J, Rawlinson L, Waller R J, Sarsfield S, Lee J C, Morton L F, Barnes M J, Farndale R W. Role for p38 mitogen-activated protein kinase in platelet aggregation caused by collagen or a thromboxane analogue. J Biol Chem. 1996;271:6586–6589. doi: 10.1074/jbc.271.12.6586. [DOI] [PubMed] [Google Scholar]
- 79.Saksela O, Rifkin D B. Cell-associated plasminogen activation: regulation and physiological functions. Annu Rev Cell Biol. 1988;4:93–126. doi: 10.1146/annurev.cb.04.110188.000521. [DOI] [PubMed] [Google Scholar]
- 80.Sanchez I, Hughes R T, Mayer B J, Yee K, Woodgett J R, Avruch J, Kyriakis J M, Zon L I. Role of SAPK/ERK kinase-1 in the stress-activated pathway regulating transcription factor c-Jun. Nature. 1994;372:794–798. doi: 10.1038/372794a0. [DOI] [PubMed] [Google Scholar]
- 81.Schreiber M, Baumann B, Cotten M, Angel P, Wagner E F. Fos is an essential component of the mammalian UV response. EMBO J. 1995;14:5338–5349. doi: 10.1002/j.1460-2075.1995.tb00218.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Sitrin R G, Shollenberger S B, Strieter R M, Gyetko M R. Endogenously produced urokinase amplifies tumor necrosis factor-alpha secretion by THP-1 mononuclear phagocytes. J Leukocyte Biol. 1996;59:302–311. doi: 10.1002/jlb.59.2.302. [DOI] [PubMed] [Google Scholar]
- 83.Stein B, Rahmsdorf H J, Steffen A, Litfin M, Herrlich P. UV-induced DNA damage is an intermediate Step in UV-induced expression of human immunodeficiency virus type 1, collagenase, c-fos, and metallothionein. Mol Cell Biol. 1989;9:5169–5181. doi: 10.1128/mcb.9.11.5169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.van Dam H, Wilhelm D, Herr I, Steffen A, Herrlich P, Angel P. ATF-2 is preferentially activated by stress-activated protein kinases to mediate c-jun induction in response to genotoxic agents. EMBO J. 1995;14:1798–1811. doi: 10.1002/j.1460-2075.1995.tb07168.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Walker G C. Inducible DNA repair systems. Annu Rev Biochem. 1985;54:425–457. doi: 10.1146/annurev.bi.54.070185.002233. [DOI] [PubMed] [Google Scholar]
- 86.Wang X S, Diener K, Manthey C L, Wang S-W, Rosenzweig B, Bray J, Delaney J, Cole C N, Chan-Hui P-Y, Mantlo N, Lichenstein H S, Zukowski M, Yao Z. Molecular cloning and characterization of a novel p38 mitogen-activated protein kinase. J Biol Chem. 1997;272:23668–23674. doi: 10.1074/jbc.272.38.23668. [DOI] [PubMed] [Google Scholar]
- 87.Wang X Z, Ron D. Stress-induced phosphorylation and activation of the transcription factor CHOP (GADD153) by p38 MAP kinase. Science. 1996;272:1347–1348. doi: 10.1126/science.272.5266.1347. [DOI] [PubMed] [Google Scholar]
- 88.Waskiewicz A J, Flynn A, Proud C G, Cooper J A. Mitogen-activated protein kinases activate the serine/threonine kinases Mnk1 and Mnk2. EMBO J. 1997;16:1909–1920. doi: 10.1093/emboj/16.8.1909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Xia Z, Dickens M, Raingeaud J, Davis R J, Greenberg M E. Opposing effects of ERK and JNK-p38 MAP kinases on apoptosis. Science. 1995;270:1326–1331. doi: 10.1126/science.270.5240.1326. [DOI] [PubMed] [Google Scholar]
- 90.Zervos A S, Faccio L, Gatto J P, Kyriakis J M, Brent R. Mxi2, a mitogen-activated protein kinase that recognizes and phosphorylates Max protein. Proc Natl Acad Sci USA. 1995;92:10531–10534. doi: 10.1073/pnas.92.23.10531. [DOI] [PMC free article] [PubMed] [Google Scholar]