In this study, Binet et al. show that the transcription factor MITF plays a nontranscriptional role in suppressing the DNA damage response (DDR) in melanoma cells. S325-phosphorylated MITF, similar to the melanoma predisposing MITF-E318K SUMOylation mutant, interacts with and destabilizes the MRN complex essential for DDR, impairing its recruitment to sites of DNA damage and increasing genomic instability during cancer initiation.
Keywords: MITF, SUMOylation, E318K, DNA damage repair, melanoma, NBS1, DNA replication, homologous recombination, replication stress
Abstract
Since genome instability can drive cancer initiation and progression, cells have evolved highly effective and ubiquitous DNA damage response (DDR) programs. However, some cells (for example, in skin) are normally exposed to high levels of DNA-damaging agents. Whether such high-risk cells possess lineage-specific mechanisms that tailor DNA repair to the tissue remains largely unknown. Using melanoma as a model, we show here that the microphthalmia-associated transcription factor MITF, a lineage addition oncogene that coordinates many aspects of melanocyte and melanoma biology, plays a nontranscriptional role in shaping the DDR. On exposure to DNA-damaging agents, MITF is phosphorylated at S325, and its interactome is dramatically remodeled; most transcription cofactors dissociate, and instead MITF interacts with the MRE11–RAD50–NBS1 (MRN) complex. Consequently, cells with high MITF levels accumulate stalled replication forks and display defects in homologous recombination-mediated repair associated with impaired MRN recruitment to DNA damage. In agreement with this, high MITF levels are associated with increased single-nucleotide and copy number variant burdens in melanoma. Significantly, the SUMOylation-defective MITF-E318K melanoma predisposition mutation recapitulates the effects of DNA-PKcs-phosphorylated MITF. Our data suggest that a nontranscriptional function of a lineage-restricted transcription factor contributes to a tissue-specialized modulation of the DDR that can impact cancer initiation.
Preserving the integrity of the genome is critical for survival and the faithful inheritance of genetic information following DNA replication. Since the genome is subjected to a wide range of insults, including DNA replication stress, irradiation, and exposure to DNA-damaging agents including chemotherapeutic drugs, cells have evolved an arsenal of sophisticated mechanisms to repair DNA damage and maintain genome integrity. Failure of the repair pathways to accurately resolve DNA damage can lead to cell death or disease, including cancer. One of the most detrimental types of DNA damage is double-strand breaks (DSBs), which are repaired by two principal mechanisms: nonhomologous end joining (NHEJ) and homologous recombination-mediated repair (HRR) (Ceccaldi et al. 2016). In NHEJ, the Ku70/80/DNA-PK complex protects exposed DNA ends before the ligase IV/XRCC4/XLF complex mediates end rejoining. This process can occur in all phases of the cell cycle but is error-prone and may consequently cause detrimental mutations or deletions. In contrast, HRR relies on an initial resection step controlled by the MRE11/RAD50/NBS1 (MRN) complex and the nuclease CtIP (Buis et al. 2012; Paull 2018). Newly generated 3′ overhangs are then protected by RPA, which serves as a platform to recruit the RAD51 recombinase, which in turn searches for a template DNA to invade and copy. As a result, HRR is restricted to S and G2 phases of the cell cycle but mediates more faithful repair than NHEJ. Although DNA damage repair mechanisms appear to be largely conserved in different tissues, accumulating evidence suggests that their function might be modulated by tissue-restricted factors that would tailor repair pathways to reflect the demands of specific cell types (Chao and Lipkin 2006; D'Errico et al. 2007; Swope et al. 2014; Herbert et al. 2019). However, how cells might shape a tissue-restricted DNA damage response (DDR) is poorly understood.
Melanocytes, specialized pigment-producing cells in the skin, play a key role in photoprotection but can also give rise to melanoma, the most lethal form of skin cancer, because of mutations that promote proliferation and suppress senescence (Shain and Bastian 2016). Much of melanocyte and melanoma biology is coordinated by the microphthalmia-associated transcription factor (MITF) (Goding and Arnheiter 2019). MITF, a lineage survival oncogene (Garraway and Sellers 2006), controls cell survival (McGill et al. 2002) and autophagy (Ploper et al. 2015; Möller et al. 2019) and promotes proliferation and differentiation (Widlund et al. 2002; Carreira et al. 2005; Loercher et al. 2005) but also suppresses invasion/migration (Carreira et al. 2006) and senescence (Giuliano et al. 2010; Ohanna et al. 2011). MITF also has a key role in regulating cell metabolism, including mitochondrial biogenesis (Haq et al. 2013; Vazquez et al. 2013), the TCA cycle (Louphrasitthiphol et al. 2019), and fatty acid and lipid metabolism (Leclerc et al. 2019; Vivas-Garcia et al. 2020). Significantly, MITF can promote genome integrity by transcriptionally activating DNA damage repair genes, and depletion of MITF can trigger DNA damage (Beuret et al. 2011; Strub et al. 2011).
MITF is not a common target for melanoma-associated mutations. The notable exception, however, is a germline E318K mutation that is associated with increased melanoma (Bertolotto et al. 2011; Yokoyama et al. 2011) and renal cell carcinoma (Bertolotto et al. 2011) predisposition in humans and disease progression in a mouse melanoma model (Bonet et al. 2017). The E318K mutation prevents efficient SUMOylation of MITF-K316 (Bertolotto et al. 2011; Yokoyama et al. 2011), but although it has been suggested that SUMOylation might affect the ability of MITF to regulate transcription (Miller et al. 2005; Murakami and Arnheiter 2005; Bertolotto et al. 2011) and suppress senescence (Bonet et al. 2017), how SUMOylation mechanistically affects MITF function is poorly understood. The range of biological processes controlled by SUMO-MITF is also not known.
Here we reveal that after exposure to DNA-damaging agents, MITF is phosphorylated by ATM/DNA-PK and dissociates from its transcription cofactors. Phosphorylated MITF is stabilized at sites of DNA damage, interacts with the MRN complex, limits HR-mediated repair, and triggers replication stress, therefore increasing genome instability. Furthermore, we describe how the E318K mutation recapitulates the effects of MITF phosphorylation and propose a new mechanism for the observed increase in melanoma predisposition.
Results
MITF expression correlates with replication stress and genome instability
Previous analysis of a potential link between MITF levels or activity and single-nucleotide variation (SNV) load failed to detect any significant correlation (Herbert et al. 2019). Here we used a different approach based on the rheostat model for MITF function (Carreira et al. 2006) that describes how (1) low MITF is associated with invasiveness and a stem cell-like phenotype, (2) moderate activity is found in proliferative cells, and (3) high levels or altered activity of MITF corresponds to a more differentiated phenotype (Carreira et al. 2006). Examination of the TCGA melanoma cohort stratified into five bins corresponding to different MITF levels provided an intriguing insight into the accumulation of SNVs in relation to MITF expression. These data show that both high (MITFHigh) and low (MITFLow) levels of MITF correlate with an increased mutational landscape (Fig. 1A, top panel). Previous work has indicated that MITF can control expression of genes implicated in DNA damage repair, and consequently depletion of MITF can induce DNA damage (Beuret et al. 2011; Strub et al. 2011). Using the set of replication, recombination, and DDR-associated MITF target genes described by Strub et al. (2011) we confirmed their down-regulation upon MITF targeting siRNA (siMITF) transfection into 501mel cells (Supplemental Fig. S1A,B). However, the increase in SNV load in MITFHigh tumors was unexpected. Using the same data set, we further investigated the link between MITF levels and copy number variations (CNVs). Remarkably, in contrast to SNVs, only higher levels of MITF correlated with an increase in CNVs (Fig. 1A, bottom panel). We next asked whether an association between a change in MITF activity and accumulation of genomic alterations could also be observed in a nonpathological model comparing human pluripotent stem cell (hPSC)-derived melanoblasts, which do not express a range of MITF target genes associated with differentiation, and melanocytes, which do. In this model, melanoblasts and mature melanocytes originate from the differentiation of the same population of hPSCs (Chambers et al. 2009; Lee et al. 2010; Mica et al. 2013; Baggiolini et al. 2021). Targeted sequencing of each population showed that only melanocytes displayed a significant amount of copy number alterations, while melanoblasts were similar to the control (Fig. 1B).
Figure 1.
MITF associates with mutational burden and replication stress. (A) Box plots showing the distribution of single-nucleotide variants (SNVs) per bin (top) and the distribution of copy number variations (CNVs) per bin (bottom) from 428 melanoma samples from ICGC, plotted after log transformation computed as log10(0.1+SNV) in five bins by their MITF expression value. The P-value was computed using the paired Wilcoxon rank sum test. Medians are indicated in red. (B, top) Description of the differentiation process from hPSCs to melanoblasts and melanocytes (created with BioRender.com). (Middle and bottom) Total copy number log ratio (logR) of melanoblasts (middle) and melanocytes (bottom). The orange horizontal line indicates the inferred diploid state, and red lines show copy number segments along each chromosome. Melanoblasts display a flat profile, indicating no copy number alterations, whereas melanocytes show copy number alterations on chromosomes 1, 7, and 10. (C, top) Timeline of the DNA fiber experiment. Cells were transfected for 48 h before being treated sequentially for 30 min with BrdU and for 30 min with EdU, before DNA extraction. (Bottom) Examples of patterns used to quantify the percentage of stalled forks, ongoing forks, and new origins in the DNA fiber assay. BrdU-containing fibers are stained red, and EdU-containing fibers are stained green. (D) Graph expressing the percentage of ongoing forks (OF) plus new origins (NO) and stalled replication forks (SF) in 501mel cells transfected with siMITF (black) or the nontargeting siRNA (gray). The P-value was determined using Fisher's exact test. For siMITF, P = 9.453417 × 10−6. (E) Immunofluorescence analysis of the accumulation of ssDNA in 501mel cells transfected with siMITF (gray) of the nontargeting siRNA (black). Cells incorporated BrdU for 24 h and were treated with 10 µM camptothecin (CPT) for 1 h. ssDNA foci were detected using an anti-BrdU antibody. (Left) Bee swarm plots representing the quantification of the number of BrdU foci. The P-value was computed using the Wilcoxon rank sum test. For siMITF, P = 1.442 × 10−6. The medians are indicated in red. (Right) Representative images of ssDNA staining. Scale bar, 5 μm. (F) Immunofluorescence analysis of the accumulation of ssDNA after CPT treatment in HEK293 and 501mel cells transfected with HA-MITF or the corresponding empty vector. All cells had incorporated BrdU for 24 h and were treated with 10 µM camptothecin (CPT) for 1 h. ssDNA foci were detected using an anti-BrdU antibody. (Left) Bee swarm plots representing the quantification of the number of BrdU foci. The P-values were computed using the Wilcoxon rank sum test. For HEK293, P = 0.002372; for 501mel, P = 6.014 × 10−6. The medians are indicated in red. (Right) Representative images of ssDNA staining in HEK293 and 501mel cells as indicated. Scale bars, 5 μm. (G) Graph expressing the percentage of ongoing forks (OF) plus new origins (NO) and stalled replication forks (SF) in HEK293 cells overexpressing MITF WT (black) or the corresponding empty vector (gray). P-values were determined using Fisher's exact test. For MITF WT, P = 0.0109008. (H) Graph expressing the percentage of ongoing forks (OF) plus new origins (NO) and stalled replication forks (SF) in 501mel cells overexpressing MITF WT (black), ΔR217 (blue), or the corresponding empty vector (gray). P-values were determined using Fisher's exact test. For MITF WT, P = 2.461506 × 10−6; for del217, P = 1.960337 × 10−19. (I) Diagram depicting the position of the basic domain of MITF and the ΔR217 deletion.
Since MITF can promote both proliferation and differentiation (Widlund et al. 2002; Carreira et al. 2005, 2006; Loercher et al. 2005), we asked whether higher levels of MITF protein could trigger replication stress that might explain the genome instability that we observed in melanocytes and melanoma. We performed a series of DNA fiber experiments in MITF-expressing 501mel melanoma cells. In this assay, cells were exposed first to BrdU for 30 min and then to EdU for the same duration to allow the two thymidine analogs to integrate into replicating DNA (Fig. 1C). After the second pulse, DNA was spread onto microscope slides and fixed, and each analog was revealed using Click-It chemistry for EdU (green) and a specific antibody for BrdU (red). As a result, replicating DNA tracks could be visualized and discriminated into ongoing forks containing both BrdU and EdU, stalled forks that did not incorporate EdU (red only), and new origins that incorporated only EdU (green only). We first performed the DNA fiber assay in 501mel cells transfected with an MITF targeting or control siRNA (siCTL) (Fig. 1D; Supplemental Fig. S1B). Qualitative analysis showed a marked reduction of the percentage of stalled forks, from 15% with the control nontargeting siRNA to 5% with siMITF. This difference did not arise as a consequence of altered replication fork speed, since the distribution of EdU track lengths in each sample did not reveal any difference (Supplemental Fig. S1C). In an orthogonal approach, we measured the accumulation of single-strand DNA (ssDNA) as a marker of replication stress after exposure to the topoisomerase poison camptothecin (CPT). 501mel cells were transfected with siMITF or siCTL, allowed to incorporate BrdU for 24 h, and then exposed to CPT, and any exposed BrdU, reflecting ssDNA, was detected using a specific antibody under nondenaturing conditions. After quantification, the number of BrdU foci was lower in MITF-depleted cells, indicating a lower level of replication stress (Fig. 1E). In a reciprocal experiment, we next transfected a HA-tagged MITF or a corresponding empty expression vector into the MITF-negative nonmelanoma HEK293 cell line or MITF-positive 501mel melanoma cells and measured the amount of CPT-induced ssDNA and the DNA replication tracks. In this context, MITF expression correlated with an increased amount of ssDNA in both HEK293 and 501mel cells (Fig. 1F) and a higher percentage of stalled replication forks (Fig. 1G,H). To further support the association between MITF and replicative stress, we measured the levels of phosphorylation of CHK1 on Thr68 as a readout of ATR activity in melanoma cell lines with high (501mel and IGR37) or undetectable (IGR39 and WM793) levels of MITF. Cells were exposed to UV, and the increase in pCHK1 was measured over time (Supplemental Fig. S1D). The results indicated that the two MITFHigh cell lines have moderately more pCHK1 at steady state than the MITFLow cell lines, in accordance with a higher replicative stress, and that accumulation of pCHK1 following UV exposure is higher in both 501mel and IGR37 compared with the MITFLow IGR39 and WM793 cell lines. In the MITFHigh cell lines, the level of pCHK1 remained high 3 h after UV exposure, by which time pCHK1 levels started to decrease in the MITFLow lines. Collectively, these data, together with the DNA fiber assay and the ssDNA detection experiments, indicate that MITF expression correlates with the level of replication stress in melanoma cells.
MITF-induced genome instability is independent of its transcriptional activity
Because it is unclear whether the difference in the percentage of stalled forks arose owing to the transcriptional activity of MITF, we generated a non-DNA-binding MITF mutant by deletion of arginine 217 (ΔR217) in the basic domain as described (Fig. 1I; Hemesath et al. 1994). We then transfected 501mel cells with HA-MITF-ΔR217 and performed the DNA fiber assay (Fig. 1H, blue bars). The results revealed 9% of stalled replication forks in the control cells, 16% in the MITF transfected cells, and 25% with the ΔR217 mutant, indicating that replication fork stalling caused by MITF expression is independent of its ability to bind DNA and, consequently, its capacity to regulate transcription. Moreover, MITF expression was associated with globally decreased fork speed, as shown by a shift of the peak of EdU track lengths to the left (Supplemental Fig. S1E), with an even greater shift being observed with MITF-ΔR217 (Supplemental Fig. S1F). These results support the hypothesis that the effect of MITF on the replication machinery is nontranscriptional.
MITF sensitizes cells to DNA damage
Knowing that MITF expression is associated with replication stress, we hypothesized that MITF-expressing cells would be more sensitive to DNA damage. We exposed MITF-negative HEK293 cells transfected or not with HA-MITF to UV, CPT, or the DNA-alkylating agent cisplatin (CisPt). All three stresses have a deleterious effect on DNA replication that can lead to formation of DSBs. Quantification of γH2AX, a marker of DNA damage, showed that expression of MITF increased γH2AX in response to UV and after exposure to CPT and, to a lesser extent, CisPt (Fig. 2A). We next performed a time course of γH2AX activation after UV in HEK293 cells transfected with HA-tagged MITF or a HA-only vector. More MITF-expressing cells displayed an elevated γH2AX signal 1 h after UV exposure, and although by 24 h after UV in the control cells the γH2AX signal was close to that in unirradiated cells, in the MITF-expressing population, the γH2AX signal remained high (Fig. 2B). A comparable shift of the γH2AX activation dynamic was observed using CPT in HEK293 cells (Supplemental Fig. S2A) and in the MITFLow melanoma cell line IGR39 transfected with MITF (Supplemental Fig. S2B). We also performed the reciprocal experiment and silenced MITF in the MITFHigh melanoma cell line IGR37. As expected, after UV damage, γH2AX levels are reduced in the siMITF transfected cells (Supplemental Fig. S2C). The experiment was repeated with the transcriptionally inactive MITF-ΔR217 mutant, and γH2AX was detected over 4 h following UV irradiation (Fig. 2C). The ΔR217 mutant was similarly able to stimulate the activation of γH2AX in comparison with empty vector, even though its expression was slightly reduced compared with MITF WT by Western blot (Fig. 2D). We conclude that direct DNA binding was not necessary for the enhanced induction of γH2AX observed using MITF WT following UV.
Figure 2.
MITF-positive cells are sensitive to different types of DNA damage. (A) Immunofluorescence analysis of γH2AX activation in HEK293 cells transfected with HA-MITF (black) or the corresponding empty vector (gray). Cells were exposed to 24 J/m2 UV or treated for 2 h with 10 µM camptothecin (CPT) or 300 ng/mL cisplatin (CisPt). Bee swarm plots represent the distribution of γH2AX intensities per cells. The P-values were computed using the Wilcoxon rank sum test. (****) P < 0.0001. The medians are indicated in red. (B) Immunofluorescence analysis of γH2AX activation over time after 24 J/m2 UV irradiation. HEK293 cells were transfected with HA-MITF (black) or the corresponding empty vector (gray). (Left) Representative images. Scale bar, 40 μm. (Right) Box plots representing the distribution of γH2AX intensities per cells. The P-values were computed using the Wilcoxon rank sum test. (*) P < 0.05, (***) P < 0.001, (****) P < 0.0001. The medians are indicated in red. (C) Immunofluorescence analysis of γH2AX activation over time after 24 J/m2 UV irradiation. HEK293 cells were transfected with HA-MITF (black), the ΔR217 mutant (blue), or the corresponding empty vector (gray). (Left) Representative images. Scale bar, 100 μm. (Right) Box plots representing the distribution of γH2AX intensities per cells. The P-values were computed using the Wilcoxon rank sum test. (**) P < 0.01, (****) P < 0.0001. The medians are indicated in red. (D) Western blot of HEK293 cells transfected with HA-tagged MITF WT, the ΔR217 mutant, or the control vector and exposed to 100 J/m2 UV. GAPDH was used as a loading control. (E) Western blot of melanoma cell lines exposed to 100 J/m2 UV and harvested at the indicated times. Phospho-ATM was used as a DDR activation control, and GAPDH was used as a loading control. (F) Immunofluorescence analysis of the formation of RAD51 foci after 2 Gy of IR (X-rays) in U2-OS cells transfected with HA-MITF or the corresponding empty vector. Cells were fixed and processed at the indicated times after IR. (Left) Representative images. Scale bar, 20 μm. (Right) Box plots representing the distribution of the number of RAD51 foci per cell. The P-values were computed using the Wilcoxon rank sum test. (****) P < 0.0001. The medians are indicated in red. (G) Immunofluorescence analysis of the formation of 53BP1 foci after 2 Gy of IR (X-rays) in U2-OS cells transfected with HA-MITF or the corresponding empty vector. Cells were fixed and processed at the indicated times after IR. (Left) Representative images. Scale bar, 20 μm. (Right) Box plots representing the distribution of the number of 53BP1 foci per cell. The P-values were computed using the Wilcoxon rank sum test. (***) P < 0.001, (****) P < 0.0001. The medians are indicated in red. (H) Cartoon depicting the principle of the U2-OS-DR-GFP reporter system. (I, left) Graph showing the relative efficiency of homologous recombination using the U2-OS-DR-GFP reporter system. Data represent the mean (±SEM) from three independent experiments and are normalized against the I-SceI-only samples. The P-values were computed using the Wilcoxon rank sum test. (ns) Nonsignificant, (**) P < 0.01. (Right): Western blot of U2-OS-DR-GFP reporter cells transfected with MITF WT, the ΔR217 mutant, or the control vector. GAPDH was used as a loading control. (J) Western blot of FLAG-MITF induction with doxycycline. GAPDH was used as a loading control. (K) Timeline of the experiment. Cells were induced with doxycycline for 4 h before being exposed to 24 J/m2 UV. Immunofluorescence analysis was performed after 24 h. (L) Immunofluorescence analysis of the persistence of 53BP1 foci (red) in FLAG-MITF-expressing cells (green). Scale bar, 30 μm. Violin plot representing the distribution of the number of 53BP1 foci per cells in inducible HEK293 cells expressing FLAG-MITF (gray) or the corresponding empty FLAG (white) 24 h after being exposed to UV. The P-values were computed using the Wilcoxon rank sum test. (ns) Nonsignificant, (****) P < 0.0001. The medians are indicated in red.
These data suggest that MITF levels affect the kinetics of the DDR. We therefore selected four melanoma cell lines—two MITFHigh (501mel and IGR37) and two MITFLow (WM278 and IGR39)—and evaluated by Western blot the dynamics of UV-induced ATM Ser1981 phosphorylation (pATM), a marker of active ATM (Fig. 2E). All cell lines displayed an activation of pATM within 2 h after UV exposure, except for IGR37, where the kinase was already activated in steady-state conditions. After the initial activation, pATM levels returned close to baseline 6 h after irradiation only in the MITFLow cell lines WM278 and IGR39 while remaining high in 501mel and IGR37. We performed a similar experiment using HEK293 cells transfected with MITF WT (Supplemental Fig. S2D) or MITFLow IGR39 melanoma cells carrying an inducible MITF construct (Supplemental Fig. S2E). The results revealed that in HEK293 cells, MITF expression suppressed CHK2 phosphorylation by ATM, while in IGR39 cells, the effect of MITF induction was less apparent; however, KAP1 phosphorylation, another ATM target, was reduced, confirming reduced ATM activity in MITF-expressing cells. Although only a correlation, these results are consistent with MITF expression perturbing the normal course of UV-induced damage repair.
As ATM and γH2AX are well-characterized markers of DSB repair, we investigated the involvement of MITF in the two major DSB repair pathways: HRR and NHEJ. First, we examined the formation of RAD51 foci to explore the impact of MITF expression on HRR activation in U2-OS cells, a well-defined model for the study of the formation of DDR foci, and a cell line negative for endogenous MITF. U2-OS cells transfected with HA-MITF or a control vector were exposed to X-rays (IR), and the number of RAD51 foci was determined over time (Fig. 2F). With the control vector, the number of RAD51 foci reached a peak 4 h after IR and then started to decrease. In the presence of HA-MITF, the peak of RAD51 foci was delayed and was observed only after 6 h. Note that the expression of MITF itself also moderately increased the basal number of RAD51 foci, possibly reflecting the replicative stress induced by MITF expression (Fig. 1D–H). By 8 h after IR, both samples displayed a similar recovery. A similar delay in resolution of RAD51 foci was observed in MITF-expressing HEK293 cells (Supplemental Fig. S2F) or MITF-inducible IGR39 cells exposed to CPT (Supplemental Fig. S2G). We then asked whether the delay in RAD51 foci formation would be compensated for by an increase in another DSB repair mechanism and assessed the dynamics of formation of 53BP1 foci, a well-known marker of NHEJ-mediated repair. After IR, the number of 53BP1 foci increased between 4 and 6 h in the control cells and started to decrease at 8 h (Fig. 2G). In the presence of MITF, the timing of activation was similar; however, the number of foci per cell was higher than with the empty vector at both 4 and 6 h. These results suggest that MITF expression triggers a shift from HRR to NHEJ in response to IR, possibly reflecting a deficiency in HRR. To confirm this, we used the U2-OS-DR-GFP cell reporter system (Pierce et al. 1999). In this assay, engineered U2-OS cells contained a GFP cDNA containing a blunt-ended I-SceI digestion site and another fragment of GFP spanning the cleavage site. After enzymatic digestion with I-SceI, cells used the second GFP fragment as a template to restore a functional fluorescent GFP (Fig. 2H). We cotransfected the reporter cells with expression vectors for I-SceI and MITF or an empty vector and measured GFP expression (Fig. 2I). In the control cells, GFP expression was markedly increased upon expression of I-SceI. Coexpression of MITF WT moderately but consistently reduced the efficiency of HRR by ∼20% compared with I-SceI alone. As a control, depletion of BRCA2 decreased HRR efficiency to ∼20%. Notably, the non-DNA-binding MITF-ΔR217 mutant also reduced the efficiency of HRR, confirming that the effects of MITF on HRR are likely nontranscriptional.
As MITF-expressing cells are more sensitive to DNA damage and appear to be HRR-deficient, we anticipated that MITFHigh cells would accumulate damage over time, explaining the increase in SNVs/CNVs observed in melanomas expressing high levels of MITF (Fig. 1A). We therefore used a HEK293 cell line stably expressing a FLAG-tagged MITF (or its empty FLAG counterpart) under a doxycycline-inducible promoter. Using a range of doxycycline concentrations for 24 h, we determined that FLAG-MITF was induced at 10 ng/mL, and its expression increased with increased doxycycline (Fig. 2J). We chose to work with 100 ng/mL doxycycline to induce an intermediate level of MITF consistent with that observed in MITFHigh melanoma cell lines. For the persistent damage assay, we induced FLAG-MITF for 4 h prior to exposing cells to UV and then allowed cells to rest for 24 h while maintaining MITF induction with doxycycline. Cells were then analyzed by immunofluorescence for persistent 53BP1 foci indicative of unrepaired DSBs (Fig. 2K). Twenty-four hours after UV, no significant accumulation of 53BP1 foci was observed in the FLAG-only cells, while FLAG-MITF-expressing cells displayed a significant increase in persistent 53BP1 foci (Fig. 2L). This result confirmed that MITF expression decreases the efficiency of repair of UV-induced DNA damage, leading to the accumulation of unrepaired damage.
DNA damage remodels the MITF interactome
Our results so far have established that MITF delays the resolution of UV-induced damage without the need to directly bind DNA. We therefore hypothesized that MITF may have a nontranscriptional function in the regulation of DNA repair by interacting with known DNA repair factors. To test this, a stable HEK293 cell line expressing a doxycycline-inducible BirA*-FLAG-MITF (Chauhan et al. 2022) was used in an unbiased search for MITF interactors using biotin–streptavidin-based purification coupled to mass spectrometry (BioID) (Lambert et al. 2015). HEK293 cells were used because we have robust control data from this cell line (Herbert et al. 2019; Lu et al. 2021), and while MITF-interacting factors might exhibit some variation between cell lines, the many different MITF isoforms can be expressed widely in different tissues and cell types. We therefore expect that the cofactors identified here will be broadly conserved between tissues/cell types. BioID was performed 20 h after induction of MITF using doxycycline in the presence or absence of CPT. This identified 70 MITF-interacting proteins in the absence of CPT, including the other members of the MiT family (TFEB and TFE3) that can heterodimerize with MITF (Fig. 3A,B; Chauhan et al. 2022).
Figure 3.

The MITF interactome is remodeled by DNA damage. (A) Dot plot of BioID data showing the significant proximity partners of MITF in nontreated versus CPT-treated HEK293 cells. The color of the dots represents the average spectral count. The size of the dots represents the relative abundance between conditions. The gray intensity of the line encircling the dots represents the Bayesian false discovery rate (BFDR) cutoff. Association with any of the six categories (DNA replication and recombination, RNA processing, ubiquitin system, chromatin, transcription, and membrane trafficking) is depicted by the colored lines at the right of each box. Components of the MRN complex are indicated in red. (B) Diagram representing the BioID results according to the effect of CPT on MITF interaction. Where possible, proteins from A were classified using the KEGG BRITE database (https://www.kegg.jp) and information from UniProt (https://www.uniprot.org) into six categories (transcription regulation, chromatin regulators, RNA processing, ubiquitin system, membrane trafficking, and DNA replication and recombination) and segregated into columns highlighting (from left to right) a decreased, unchanged, or increased interaction with MITF after CPT treatment. Components of the MRN complex are indicated in red.
The majority of MITF interaction partners are transcription factors and presumably represent DNA binding partners that might facilitate high-affinity MITF binding to sequence elements recognized by both proteins. This set includes TFAP2A, a transcription factor that genetically interacts with MITF to control melanocyte development and differentiation and that genome-wide co-occupies a subset of sites bound by MITF (Seberg et al. 2017; Kenny et al. 2022). The second most represented proteins are chromatin regulators, including the histone acetyltransferases CREBBP and p300, known coactivators for MITF (Sato et al. 1997; Price et al. 1998) that also mediate MITF acetylation (Louphrasitthiphol et al. 2020, 2023). Also identified was SMARCA2, a member of the SWI/SNF complex that functions as a MITF cofactor (de la Serna et al. 2006; Saladi et al. 2013; Laurette et al. 2015), as well as several other potential cofactors not previously identified as MITF interactors. MITF also interacts significantly with RNA binding and processing factors, suggesting a new role in RNA biology beyond transcription. We confirmed the previously known interaction with the deubiquitinase USP11 and the E3 ubiquitin ligase TRIM24 (Laurette et al. 2015), as well as the E3 SUMO protein ligase PIAS1 (Murakami and Arnheiter 2005). The BioID analysis also discovered lysosome-associated factors (VPS33B/C14orf133) in line with the association of some MITF family members with lysosomes (Martina and Puertollano 2013) and the role of MITF in autophagy and lysosome biogenesis (Möller et al. 2019). Also found, albeit less represented, were factors involved in cytokinesis (ARF6, LZTS2, and RAE1), consistent with a role for MITF in spindle formation and chromosome segregation (Strub et al. 2011). More interestingly, we revealed interactions between MITF and the DNA repair protein PAXIP1, which is also involved in transcription regulation, as well as RAD50 and NBS1. Together with MRE11, RAD50 and NBS1 are both members of the MRN complex, which plays a major role in replication fork restart as well as in the early steps of DSB repair and is required for the activation of ATM (Uziel et al. 2003).
After treating cells with CPT, only 10 of the 70 interacting proteins detected were found equally in both control and CPT conditions, including TFE3, CREBBP, EP300, NCOA2, and NCOA6. Remarkably, apart from TFE3 (which can dimerize with MITF), all transcription factors and most chromatin regulators exhibited decreased interaction with MITF, suggesting that MITF transcriptional activity is attenuated. An exception was BRPF1, which regulates the acetylation of histone H3K23 (Yan et al. 2017) and showed a significant association with MITF only after CPT treatment but has not been previously linked to the DDR. RNA processing factors KRI1 and MRPS17, associated with the ribosome biogenesis, are also overrepresented after CPT treatment (Sasaki et al. 2000; Kenmochi et al. 2001). Notably, we saw decreased interaction with TET1, a methyl cytosine dioxygenase that is implicated in DNA demethylation but is also associated with spliceosomes (Ito et al. 2010). We noted that SUMO1 is detected only after CPT treatment. Since SUMOylated peptides are difficult to detect using mass spectrometry, our interpretation is that we may be detecting SUMO1 that is interacting with, rather than modifying, MITF.
MITF interacts with components of the MRN complex
Of the MITF-interacting partners that were more abundant after damage, those implicated in DNA replication and the DDR—TOP2B, RAD50, and NBS1—were especially interesting. TOP2B is a type II topoisomerase required to relax the topology of the DNA double helix prior to transcription and replication and is not inhibited by CPT. Given the potential for MITF to function in DNA replication and damage repair, we focused on its interaction with RAD50 and NBS1. Intriguingly, while NBS1, RAD50, and MRE11 function in a ternary complex, we never observed MRE11 as a significant MITF proximity partner (SAINTexpress FDR ≤ 1%).
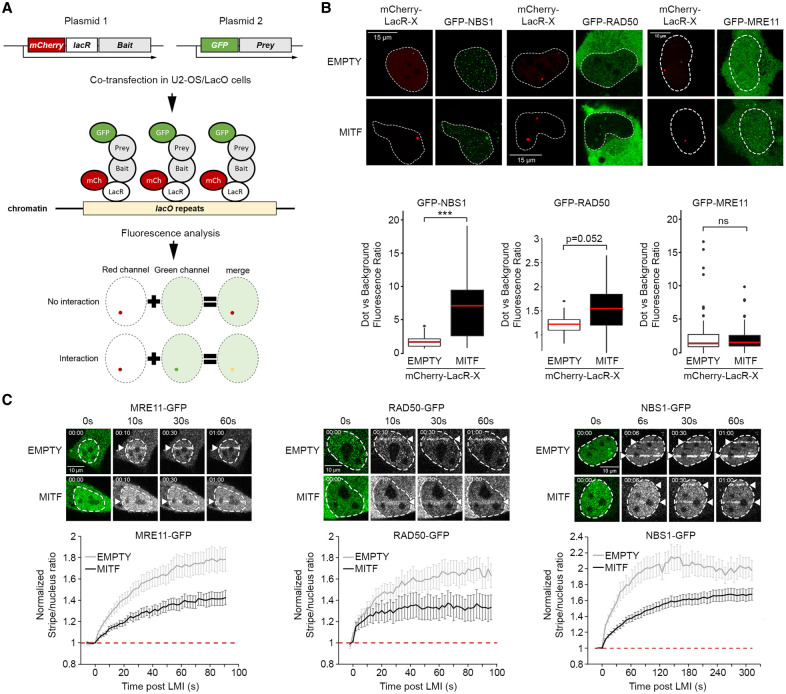
To confirm the BioID results using an orthogonal approach in an in vivo setting, we examined the interaction between MITF and components of the MRN complex using the LacR/LacO nuclear tethering assay (NTA) (Luijsterburg et al. 2012; Lu et al. 2021). This live-cell-based assay uses a U2-OS cell line containing an array of 256 repeats of the Lac operator (LacO) DNA sequence integrated into the genome to tether a bait fusion protein containing the Lac repressor (LacR) tagged with mCherry to produce a single red fluorescent nuclear focus in G1 cells (Lambert et al. 2019). Cells are then transfected with the putative interactor protein tagged with GFP, and colocalization of the GFP fusion with the mCherry nuclear focus indicates interaction (Fig. 4A). Since MITF can form homodimers, by using mCherry-LacR-MITF as bait, we first verified that we could detect an interaction using MITF-GFP as a positive control. The results showed a clear colocalization of tethered mCherry MITF with MITF-GFP, consistent with MITF dimerization (Supplemental Fig. S3A). No significant interaction was observed if we used LacR-mCherry-MITF with GFP only or with both empty vectors. Using again LacR-mCherry-MITF as bait, we next monitored the interaction with GFP-tagged NBS1, RAD50, or MRE11. Only NBS1-GFP was primarily nuclear and accumulated significantly at the MITF-rich nuclear mCherry dot with a median enrichment of sevenfold (Fig. 4B, left panel). No colocalization of NBS1-GFP was detected using an mCherry bait lacking MITF. RAD50-GFP, despite its predominantly cytoplasmic expression, also displayed interaction (Fig. 4B, middle panel). On the other hand, MRE11-GFP did not accumulate at the LacO array irrespective of whether we used the control vector or mCherry-LacR-MITF (Fig. 4B, right panel). This last observation supports the results from the BioID experiment in which MRE11 was not detected as a significant MITF proximity partner. We also controlled the interaction between each member of the MRN complex (Supplemental Fig. S3B). As expected, MRE11-GFP recruitment was enriched in the presence of mCherry-LacR-RAD50 or NBS1, but the interaction between NBS1-GFP and mCherry-LacR-RAD50 was not significant, reflecting the dynamics of interactions inside the MRN complex as described previously (Lafrance-Vanasse et al. 2015).
Figure 4.
(A) Cartoon depicting the nuclear tethering assay. (B) Representative images and quantification of the nuclear tethering assay showing interaction between MITF and NBS1, RAD50, or MRE11. (Top) The left panels show the localization of mCherry-LacR-NLS or mCherry-LacR-MITF dots in the nuclei of U2OS-LacO#13 cells, and the right panels show GFP-NBS1, GFP-RAD50, and GFP-MRE11. Scale bar, 15 μm. (Bottom) The quantification is expressed as the ratio between the GFP fluorescence measured inside the area delimited by the mCherry dot and in the rest of the nucleus. The P-values were computed using the Wilcoxon rank sum test. (ns) Nonsignificant, (***) P < 0.001. The medians are indicated in red. (C, top) Still images of MRE11-GFP, RAD50-GFP, and NBS1-GFP LMI. Nuclei are delimited using the preirradiation images. The positions of the irradiated lines are indicated with an arrowhead. Scale bar, 10 μm. (Bottom) Quantification of MRE11-GFP, RAD50-GFP, and NBS1-GFP recruitment in U2-OS cells after LMI when cotransfected with an empty vector or HA-MITF. The graphs represent the mean ± SEM of the stripe/nucleus ratio over time. Values were normalized against the pre-LMI measurements. The baselines are indicated with red dotted lines.
Under steady-state conditions, MRE11 and RAD50 form a cytoplasmic complex with NBS1 that is required to bring the complex into the nucleus (Tauchi et al. 2002), while the possibility of a complex between RAD50 and NBS1 lacking MRE11 has only been described in vitro (van der Linden et al. 2009). The physical interaction between MITF and RAD50/NBS1, but not MRE11, prompted us to ask whether it was of functional significance. Knowing that MITF limits HRR activation and that the MRN complex is implicated in DSB repair pathway choice by promoting HRR and inhibiting NHEJ, we asked whether MITF, by interacting with MRN, could affect recruitment of the MRN complex to DSBs and thereby impair HRR. We used laser microirradiation (LMI) with a near-infrared (NIR) laser emitting at 750 nm to induce damage in a strictly defined area of the nucleus and performed a series of NIR-LMI experiments coupled with video microscopy in U2-OS cells that lack endogenous MITF to analyze the dynamics of recruitment to DNA damage of the different components of the complex (Supplemental Fig. S3C). We then compared the relative recruitment of each factor. All members of the MRN complex are recruited rapidly in a few seconds, but their recruitment was substantially impaired by coexpression of MITF WT (Fig. 4C; Supplemental Movies S1–S3).
Phosphorylation-dependent MITF recruitment to DNA damage
Given the DNA-binding-independent effect of MITF on γH2AX activation, and MITF's interaction with the MRN complex, we next asked whether MITF could be directly involved in the DDR pathway using 501mel cells stably expressing HA-MITF at endogenous levels (Louphrasitthiphol et al. 2020). We performed LMI using a UV-emitting laser and detected the localization of MITF by immunofluorescence. Twenty minutes after UV-LMI, MITF colocalized with γH2AX (Fig. 5A). To measure more precisely the dynamics of MITF recruitment, we repeated this experiment and irradiated adjacent regions every 10 sec for 15 min before staining. We observed that HA-MITF accumulated at the damage sites by 100 sec after UV (Supplemental Fig. S4A). We repeated the experiments with NIR-LMI and confirmed that, like with UV, HA-MITF accumulated at the sites of damage (Fig. 5B). We also excluded a cell line-specific effect, as we equally observed at DNA damage the accumulation of GFP-MITF transfected in IGR37 cells (Supplemental Fig. S4B). We then assessed the role of key DDR factors in MITF recruitment. We generated a stable 501mel cell line expressing GFP-MITF and performed live confocal microscopy after NIR-LMI. This revealed that recruitment of MITF to DNA damage was fully prevented by the DNA-PK inhibitor NU7441 (Fig. 5C; Supplemental Movie S4) but only marginally attenuated by the ATM inhibitor KU55933 or ATR inhibitors VE821 and VE822 (Supplemental Fig. S4C; Supplemental Movie S5). Inhibition of poly-ADP ribose polymerase (PARP) using olaparib also prevented MITF recruitment to DNA damage (Fig. 5C; Supplemental Movie S6).
Figure 5.
MITF recruitment to DNA damage sites. (A) Immunofluorescence of stable 501mel⋅HA-MITF cells after UV-LMI. The irradiated area was identified using an anti-γH2AX antibody (red) and MITF using anti-HA (green). Scale bar, 40 μm. (B) Immunofluorescence of stable 501mel⋅HA-MITF cells after NIR-LMI. The irradiated area was identified using an anti-γH2AX antibody (red). MITF was detected with an anti-HA antibody (green). DAPI was used to stain the nuclei. Scale bar, 20 μm. (C) Still images and quantification of live video microscopy showing recruitment of MITF in 501mel cells stably expressing GFP-MITF after NIR-LMI. Cells were treated with DNA-PK (1 µM NU7441) or PARP (10 µM olaparib) inhibitors for 24 h before irradiation. Scale bar, 10 μm. The graphs represent the mean ± SEM of the stripe/nucleus ratio over time. Values were normalized against the pre-LMI measurements. For clarity, each individual graph represents the same control curve (black) against only one type of inhibitor (DNAPKi [turquoise] or PARPi [pink]). The baselines are indicated with red dotted lines. (D, top) Diagram depicting the position of the bHLH-LZ domain of MITF and the position of the SUMOylation sites K182 and K316 (green), the phosphorylation site S325 (red), and the familial mutation E318K (blue). (Bottom) Sequence of the 314–333 peptides showing that residues K316, E318, and S325 are conserved from humans to zebrafish. (E) Western blot of HEK293 cells transiently transfected with the indicated mutants of HA-MITF or the corresponding empty vector. Phosphorylation of MITF on serine 325 was detected by a phospho-specific antibody. GAPDH was used as a loading control. (F) In vitro kinase assay performed on a peptide array. Peptide-bound anti-pS325 antibodies were detected using chemiluminescence. The size and intensity of the dots are proportional to the amount of bound antibodies. The kinases used are indicated above. (G) Western blot of HEK293 cells transiently transfected with HA-MITF. Cells were UV-irradiated and harvested at different time points to measure phosphorylation of S325. The ATM inhibitor KU55933 was used to confirm the role of the kinase. GAPDH was used as a loading control. (H, left) Still images of GFP-MITF recruitment in 501mel cells stably expressing GFP-MITF WT and S325 and E318 mutants after LMI. Nuclei were delimited using the preirradiation images. The positions of the irradiated lines are indicated with arrowheads. Scale bar, 10 μm. (Right) Quantification. The graph represents the mean ± SEM of the stripe/nucleus ratio over time. Values were normalized against the pre-LMI measurements. The baseline is indicated with a red dotted line. (I) Still images of GFP-MITF behavior in 501mel cells stably expressing GFP-MITF wild type and S325 and E318 mutants after LMI as quantified in J and K. Nuclei were delimited using the preirradiation images. Scale bar, 10 μm. (J) Details of GFP-MITF behavior in 501mel cells stably expressing GFP-MITF WT and S325 and E318 mutants after LMI. Quantification of GFP intensities at (stripes) and away from (nuclei) the LMI sites. The graph represents the mean ± SEM of GFP fluorescence over time. Values were normalized against the pre-LMI measurements. The dotted lines represent the key values as described in K. The baselines are indicated with red dotted lines. (K) Key values extracted from the quantification in J. “Stripe—time ≥100% intensity” represents the time GFP fluorescence remained ≥1 after LMI, “stripe—time to decrease to 80% intensity” represents the time required to see the GFP intensity at the stripe drop to 80% of the pre-LMI value from the time of irradiation, “stripe—from 100% to 80%” represents the time required to see the GFP intensity at the stripe drop to 80% of the pre-LMI value after it started to decrease, and “nucleus—time to decrease to 70% intensity” represents the time required to see the GFP intensity away from the stripe drop to 70% of the pre-LMI value.
Considering the effect of the DNA-PK inhibitor on MITF recruitment, we asked whether MITF might be a direct target of the kinases implicated in the DDR. By examining evolutionarily conserved sequences within MITF, we identified, C-terminal to the MITF leucine zipper, a highly conserved SQ motif (Fig. 5D) characteristic of targets of the ATM and DNA-PK kinases (Kim et al. 1999). Initial attempts to identify phosphorylation at this residue using a mass spectrometry approach were unsuccessful because although post-translational modifications at many other sites were identified, we were unable to generate peptide coverage of the region containing S325. As an alternative strategy, we generated a phosphorylation-specific polyclonal antibody against phospho-S325 and validated its specificity using a peptide array spanning mouse or human MITF amino acids 314–333 that contained either the WT sequence, S325 variants, or, as an additional control, acetyl-K316 (Supplemental Fig. S4D). The results confirmed that the antibody was highly specific, recognizing pS325 in both the mouse and human sequences but not the nonphosphorylated version or S325A or S325T mutants (Supplemental Fig. S4E). A pT325 variant was recognized less well than pS325, and acetylation at K316 did not affect antibody recognition. To confirm the specificity of the anti-pS325 antibody in a cellular context, we transfected HEK293 cells with plasmids expressing HA-tagged MITF WT or S325A (null) or S325E (phosphomimetic) phosphorylation site mutants. Following transfection, cells were analyzed by Western blot using the pS325 antibody. The results (Fig. 5E) revealed that MITF WT, but not the S325 mutants, was recognized by the anti-pS325 antibody, confirming both the specificity of the antibody for pS325 and that S325 is phosphorylated in cells.
We next investigated the ability of DDR kinases to phosphorylate MITF-S325 using two different approaches. First, we used commercially available purified CHK2 and DNA-PK in a nonradioactive in vitro kinase assay on a peptide array. In this assay, only the DNA-PK substantially increased the signal on the nonphosphorylated S325 residue and weakly on T325 (Fig. 5F). Because commercially available purified ATM was unavailable, we next used a specific ATM inhibitor, KU-55933, to treat HEK293 cells transfected with HA-tagged MITF prior to UV irradiation and examined the extent of MITF-S325 phosphorylation over time by Western blot (Fig. 5G). The results confirmed that S325 phosphorylation can be detected in cells and increases after UV irradiation. The Western blot also revealed that the ATM inhibitor was able to reduce the levels of S325 phosphorylation 2 h after UV but did not reduce the basal level of phosphorylation. These results suggest that DNA-PK can target MITF-S325 for phosphorylation. However, as we could not confirm the direct involvement of ATM in a direct in vitro kinase assay, whether ATM phosphorylates MITF in addition to DNA-PK remains incompletely resolved.
We next asked whether phosphorylation of S325 could affect MITF recruitment to DNA damage. We generated a series of stable 501mel cell lines expressing GFP-tagged MITF WT and mutants and performed live-cell NIR-LMI. We then quantified the dynamics of recruitment of GFP-MITF WT and S325A and S325E mutants (Fig. 5H; Supplemental Movie S7). GFP-MITF WT accumulated slowly at the damage sites and became detectable after 30 sec, reaching a maximum intensity between 120 and 240 sec. By comparison, the recruitment of the MRN complex, an early factor in DSB repair, happened in as little as 5 sec (Fig. 4C). Compared with MITF WT, recruitment of the S325A mutant was significantly reduced. In contrast, the S325E mutant exhibited substantially enhanced recruitment, reaching a maximum at 360 sec, although already at 240 sec its presence at the damage stripe was well above that of the WT protein. These data suggest that phosphorylation at S325 represents a key regulator of MITF association with DNA damage sites. Since MITF colocalization with DNA damage is dependent on DNA-PK activity that may be one of the kinases responsible for MITF-S325 phosphorylation, we postulated that the NU7441 inhibitor would have a reduced effect on the S325E mutant compared with MITF WT. We repeated the NIR-LMI experiment on 501mel GFP-MITF-S325E cells pretreated with the DNA-PK inhibitor or olaparib as a positive control. The DNA-PK inhibitor NU7441 still reduced MITF-S325E colocalization with DNA damage but was much less potent compared with its effect on MITF WT (Supplemental Fig. S4F; Supplemental Movie S8), supporting the role of the DNA-PK in MITF phosphorylation. As expected, the PARP inhibitor olaparib still abolished recruitment.
Intriguingly, residue S325 is close to the germline mutation site E318K, which is known to increase melanoma risk (Bertolotto et al. 2011; Yokoyama et al. 2011) and prevent SUMOylation on K316 to affect the transcriptional program of MITF. We therefore generated a 501mel cell line expressing GFP-MITF-E318K and measured the recruitment of MITF-E318K to the NIR-LMI-generated damage (Fig. 5H). Strikingly, MITF-E318K was rapidly recruited, and the quantification showed a dynamic similar to that observed with MITF-S325E, with both mutations exhibiting a significantly increased recruitment compared with MITF WT, suggesting that SUMOylation on K316 is also a regulator of MITF function in the DDR.
In closely examining the dynamics of MITF recruitment, we observed that rather than MITF accumulating at sites of DNA damage, the increased stripe/nucleus ratio was mainly due to a rapid decrease in the amount of GFP-MITF away from the stripe immediately after LMI (Fig. 5I–5K). To confirm this, we measured separately the intensity of GFP-MITF at the damage sites (Fig. 5J, blue curves) and the intensity away from damage (Fig. 5J, yellow curves). The analysis showed that MITF WT stably stayed at the stripe for ∼150 sec before diminishing (Fig. 5J, green dotted line), while its nuclear intensity decreased immediately after irradiation (Fig. 5J, left panel). To compare the behaviors of all the mutants, we marked the time required to reach a 20% decrease in fluorescence at the stripe (Fig. 5J, dotted blue lines) and a 30% decrease in the nucleus (Fig. 5J, dotted yellow lines). MITF WT stripe intensity decreased to 80% in 325 sec, and its nuclear intensity decreased to 70% in 237.5 sec (Fig. 5K). By comparison, the MITF-S325E mutant is more stable at the stripe (162.5 sec to 100% and 387.5 sec to 80%), and MITF-S325A is more stable in the nucleus (fluorescence value remained >70% during the course of the experiment), showing that S325 phosphorylation helps stabilize MITF at the stripe while promoting a decrease in the nucleus. These observations explained the previously noted differences in the “recruitment” dynamics (Fig. 5H). On the other hand, the SUMOylation-defective E318K is the only mutant showing increased recruitment and stability at the stripe up to 225 sec after LMI. At the same time, the decrease of E318K fluorescence in the nucleus is slower than the WT (30% decrease in 387.5 sec). We also analyzed the LMI data of MITF in the presence of the DNA-PK inhibitor NU7441. Similar to the S325A mutant, MITF was more stable in the nucleus in DNA-PK inhibitor-treated cells (20% decrease in 100 sec in NU7441-treated vs. 50 sec in control) but was rapidly lost at the stripe, with the two curves overlapping over the course of the experiment (Supplemental Fig. S4G). These results suggest that the main contributor to the apparent accumulation of MITF at sites of DNA damage is the stability of the protein at the damage site. Using the protein synthesis inhibitor cycloheximide (CHX) to arrest de novo MITF synthesis, we measured the half-life of endogenous MITF after treatment with inhibitors of DNA-PK and ATM, the two kinases suggested to phosphorylate MITF-S325, and observed that both inhibitors reduced MITF protein stability (Supplemental Fig. S4H). As MITF stability is regulated by ubiquitination and the proteasome pathway (Zhao et al. 2011), we evaluated the impact of a proteasome inhibitor, MG132, on MITF's apparent accumulation at sites of DNA damage and confirmed that MG132 abolished the formation of MITF-GFP stripes after LMI (Supplemental Fig. S4I). Taken together, these results suggest that the DNA damage-dependent phosphorylation of MITF-S325 stabilizes the protein at the site of DNA damage but promotes its degradation in the rest of the nucleus.
MITF phosphorylation and the E318K mutation regulate its role in homologous recombination-mediated repair
Given the roles of S325 and K316/E318 in controlling MITF recruitment to DNA damage, we next investigated the effect of their respective mutations on HRR. Examination by immunofluorescence of RAD51 foci after γ-irradiation in U2-OS cells transfected with HA-MITF, HA-MITF-E318K, or the corresponding control plasmid at 4 and 6 h after irradiation (Fig. 6A) revealed that in the control cells, the accumulation of RAD51 foci peaked at 4 h before decreasing at 6 h. In contrast, expression of either HA-MITF or MITF-E318K provoked a delay, with RAD51 foci reaching a maximum at 6 h. In the same experiment, the phospho-mimetic S325E also delayed the accumulation of RAD51 foci, with no increase observed at 4 h, while the phospho-null S325A mutant behaved as though no MITF were present. These data indicate that S325 phosphorylation and the absence of K316 SUMOylation or presence of the E318K mutation delay HRR. We also transfected HA-MITF or the HA-MITF-S325A and HA-MITF-S325E mutants into HEK293 cells and measured the accumulation of γH2AX after up to 6 h after UV exposure (Supplemental Fig. S5A,B). As before, expression of HA-MITF triggered accumulation of γH2AX 2 h after UV irradiation. Expression of the phospho-mimetic S325E further enhanced the accumulation of γH2AX, while the MITF-S325A mutant was largely able to revert the effect of MITF. We performed a similar experiment using MITF-E318K and determined that this mutant was able to activate γH2AX fourfold more than MITF WT (Supplemental Fig. S5C,D). To eliminate any transcriptional role of MITF in the accumulation of γH2AX after UV irradiation, we also generated the double mutant E318K/ΔR217 and observed the same γH2AX increase after UV as seen using MITF WT (Supplemental Fig. S5E).
Figure 6.
The effects of S325 and E318 mutations on MITF-mediated genome instability. (A) Immunofluorescence images (top) and quantification (bottom) of the formation of RAD51 foci after 2 Gy of IR (X-rays) in U2-OS cells transfected with HA-MITF WT (gray/black), S325A (purple), S235E (gold), or E318K (red) or the corresponding empty vector (white). Cells were fixed and processed at the indicated times after IR. Scale bar, 30 μm. The P-values were computed using the paired Wilcoxon rank sum test. (ns) Nonsignificant, (*) P < 0.05, (**) P < 0.01, (****) P < 0.0001. The medians are indicated in red. (B, left) Still images of MRE11-GFP LMI in the presence of MITF WT and E318K and ΔBasic mutants or the corresponding empty vector. Nuclei are delimited using the preirradiation images. The positions of the irradiated lines are indicated with arrowheads. Scale bar, 10 μm. (Right) Quantification of MRE11-GFP recruitment. The graph represents the mean ± SEM of the stripe/nucleus ratio over time. Values were normalized against the pre-LMI measurements. The baseline is indicated with a red dotted line. (C, left) Still images of MRE11-GFP LMI in the presence of MITF WT or S325A or S325E mutants. Nuclei are delimited using the preirradiation images. The positions of the irradiated lines are indicated with arrowheads. Scale bar, 10 μm. (Right) Quantification MRE11-GFP recruitment. The graph represents the mean ± SEM of the stripe/nucleus ratio over time. Values were normalized against the pre-LMI measurements. The baseline is indicated with a red dotted line. (D,E) Nuclear tethering assay showing the effects of MITF (D) and MITF-E318K (E) on the NBS1/MRE11 or RAD50/MRE11 interactions. The quantification is expressed as the ratio between the GFP fluorescence measured inside the area delimited by the mCherry dot and in the rest of the nucleus. Scale bars: D, 5 μm; E, 10 μm. The P-values were computed using the Wilcoxon rank sum test. (ns) Nonsignificant, (**) P < 0.01. The medians are indicated in red. (F) Graph expressing the percentage of ongoing forks (OF)/new origins (NO) and stalled replication forks (SF) in 501mel cells overexpressing MITF WT (black) or E318K (red) or the corresponding empty vector (gray). P-values were determined using Fisher's exact test. For MITF WT, P = 2.461506 × 10−6; for E318K, P = 1.605343 × 10−20. Empty HA and MITF WT are the same as in Figure 1H. (G) Graph expressing the percentage of ongoing forks (OF)/new origins (NO) and stalled replication forks (SF) in 501mel cells overexpressing MITF WT (black), S325A (purple), or S325E (gold) or the corresponding empty vector (gray). P-values were determined using Fisher's exact test. For MITF WT, P = 6.942773 × 10−7; for S325A, P = 6.332160 × 10−2; for S325E, P = 2.172679 × 10−7. (H) GFP-TRAP experiment in HEK293 cells transfected with GFP-MITF WT or MITF K316R, K316R/S325A, or E318K mutants and exposed to 100 J/m2 UV. An anti-phospho-SQ antibody was used to measure MITF phosphorylation after immunoprecipitation. Ten percent of the protein extract was kept as input.
Having determined that MITF WT could block MRE11 recruitment to DNA damage, which would explain the deficiency in HRR and the delayed accumulation of RAD51 foci, we examined the effects of the MITF mutants. MRE11-GFP together with MITF WT, the E318K mutant, or the control HA-only vector, was expressed in U2-OS cells that were subjected to LMI (Fig. 6B). We again observed a moderate reduction in MRE11 accumulation at DNA damage when MITF WT was expressed. Strikingly, this was more pronounced using the SUMOylation-defective E318K mutant. To rule out that the effect of MITF on MRE11 recruitment was transcriptional, we also used a ΔBasic mutant lacking the basic region that enables MITF DNA binding (Louphrasitthiphol et al. 2020). The result revealed that this mutant suppressed MRE11-GFP recruitment to DNA damage even more efficiently than the E318K mutant. We then repeated this assay using the S325A and S325E mutants. In the presence of the phospho-null MITF-S325A mutant, MRE11 showed a slight increase in the accumulation of damage compared with MITF WT. In contrast, the phospho-mimetic S325E largely blocked MRE11 recruitment (Fig. 6C). Taken together, our LMI data show that the negative effect of MITF on MRE11 recruitment is dependent on S325 phosphorylation and potentiated by the SUMOylation-inhibiting E318K mutation.
To elucidate further the dynamics between the MRN complex and MITF, we used the NTA to measure the impact of MITF post-translational modifications on its interaction with NBS1. We observed that the MITF–NBS1 interaction is dependent on S325 status, with low levels of interaction between NBS1 and MITF-S325A (Supplemental Fig. S5F). At the same time, the S325E and E318K mutants retained the ability to bind NBS1. These results correlated with the results from the LMI experiments and the ability of MITF WT and mutants to suppress MRE11 recruitment to sites of damage, indicating that the MITF–NBS1 interaction may be involved in the inhibition of MRE11 recruitment. We therefore hypothesized that MITF might disrupt the MRN complex. In the NTA system, we analyzed the interactions between NBS1 and MRE11 or RAD50 and MRE11 while coexpressing MITF. In the control experiment undertaken in the absence of any DNA-damaging agent, the presence of MITF did not affect either interaction (Fig. 6D). In contrast, using the MITF-E318K mutant, although the interaction between NBS1 and MRE11 was only minimally affected, we observed a significant decrease in the RAD50/MRE11 interaction (Fig. 6E). Taken together, these results indicate that the E318K mutant may destabilize the MRN complex and inhibit MRE11 recruitment to DNA damage.
Given these results, we would predict that the phosphorylation and SUMOylation mutants would affect HRR and MITF-induced replication stress. Using the U2-OS DR-GFP HRR reporter assay, cells were transfected with the I-SceI-expressing vector and cotransfected with either the empty HA vector, WT HA-MITF, or the SUMOylation-defective MITF-E318K mutant (Supplemental Fig. S5G). We confirmed the 20% reduction in HRR efficiency in the presence of HA-MITF compared with I-SceI alone and observed an equivalent reduction in HRR efficiency when expressing MITF-E318K. A BRCA2-specific siRNA was used as a control. We then performed the DNA fiber assays in 501mel cells expressing MITF WT or either mutant. We confirmed that expression of MITF WT increased the percentage of stalled replication forks (Fig. 6F,G). Strikingly, MITF-E318K-expressing cells displayed an even higher proportion of stalled forks (Fig. 6F). On the other hand, cells expressing the S325E mutant displayed a proportion of stalled forks similar to that of MITF WT, while S325A-expressing cells showed no increase (Fig. 6G). Notably, MITF-E318K-expressing cells have a slower replication speed overall, which likely reflects the increase in stalled forks (Supplemental Fig. S5H). Overall, we conclude that the functions of MITF in HRR and DNA replication are governed by post-translational modifications on K316 and S325.
The MITF-S325E mutant phenocopies the MITF-E318K mutant
Since both S325 phosphorylation and K316 SUMOylation regulate the role of MITF in HRR and replication stress, it was possible that one modification may regulate the other. Examination of the amino acid sequence of human MITF in the vicinity of S325 revealed that the sequence I-K-Q-E-P-V-L-E-N-C-S-Q-D-D resembles a negatively charged amino acid-dependent SUMO conjugation motif (NDSM), ψ-K-x-E-x-x-x-E-x-x-S-x-D-D, raising the possibility that ATM-induced S325 phosphorylation could regulate MITF K316 SUMOylation (Hietakangas et al. 2006; Yang et al. 2006). This is important given the role of the SUMOylation-defective E318K melanoma predisposition mutant identified previously (Bertolotto et al. 2011; Yokoyama et al. 2011). Using SUMO1-RFP or SUMO2-RFP fusion proteins coexpressed with HA-MITF, we confirmed that in HEK293, SUMO1 is more able to modify MITF, as seen with a slow-migrating MITF band corresponding to SUMOylated MITF only when SUMO1-RFP was present (Supplemental Fig. S6A). Because MITF can be modified by either SUMO1 or SUMO2 (Miller et al. 2005; Murakami and Arnheiter 2005; Bertolotto et al. 2011; Yokoyama et al. 2011), we also used the deconjugation-defective mutant SUMO2-Q90P (Békés et al. 2011; Garvin et al. 2013) to confirm that SUMO2 is able to target MITF but with lower efficiency. We generated a HA-MITF K182R expression vector that had no effect on the SUMO-MITF band, confirming that SUMOylation must happen most favorably on K316 with WT HA-tagged MITF in HEK293 cells. We then generated double mutants of MITF to study the relationship between S325 phosphorylation and K316 SUMOylation. Preliminary results indicated that the introduction of either S325 mutation did not impact the level of MITF SUMOylation. An alternative possibility is that MITF SUMOylation affects S325 phosphorylation. In this case, the MITF-K316R and MITF-E318K mutants would show higher levels of phospho-S325 in response to UV irradiation. After transfecting GFP-MITF WT and mutants into HEK293 cells and exposing them to UV, we purified the GFP-containing complexes and measured phospho-S325 using an anti-phospho-SQ antibody. The results show that both K316R and E318K displayed higher levels of S325 phosphorylation, consistent with the comparable behavior of E318K and S325E mutants in our previous experiments (Fig. 6H).
Previous work reported that K316 SUMOylation can affect MITF transcriptional activity on specific promoters and that E318K mutant cells are more proliferative than their wild-type counterparts (Bertolotto et al. 2011). We therefore asked whether the MITF-S325E mutant, which behaves like E318K in the context of DNA and replication fork progression, would also recapitulate the effect of E318K on transcription and proliferation. To evaluate this, we cotransfected MITF WT or mutants with a MET promoter luciferase reporter, a well-characterized proliferation-related target for MITF (McGill et al. 2006). After adjusting for the expression levels of MITF WT and mutants, we confirmed that the MET promoter is activated by MITF WT (Supplemental Fig. S6B), as well as by the MITF-S325A mutant that permits MITF SUMOylation at K316. In contrast, the SUMOylation-defective E318K mutant reduced reporter activation, as did the phospho-mimetic S325E mutant. Note that the E318K-mediated decrease in MET promoter activity observed in MITF-negative HEK293 differs from previous observations made in MITFHigh 501mel cells (Bertolotto et al. 2011). It is possible that the difference may be attributable to the presence of endogenous WT MITF in 501mel cells that could dimerize with transfected MITF and mitigate the effect of the E318K mutation. Using a colony formation assay in which cells were plated at low density, expression of the SUMOylation-defective E318K mutant gave a growth advantage to HEK293 cells compared with cells expressing MITF WT, as previously described (Bertolotto et al. 2011). The S325E mutant also displayed a significant increase in colonies compared with MITF WT or the S325A mutant (Supplemental Fig. S6C). We performed the same experiment in the 501mel melanoma cells and obtained a similar result in which the S325E and E318K mutants exhibited an increased growth rate compared with MITF WT and the S325A mutant (Supplemental Fig. S6D). Overall, our data revealed that the MITF-S325E phospho-mimetic mutant recapitulates much of the effect of the germline E318K SUMOylation-defective mutant on transcription and proliferation.
Discussion
Maintenance of the integrity of the genome is regarded as one of the most important cellular functions because failure of DNA repair mechanisms may lead to an increased mutation burden with major deleterious consequences, including senescence or malignant transformation. On the other hand, under stress conditions, generation of genetic diversity arising from inefficient DNA damage repair may present an advantage by ensuring that some cells within a population may be better adapted to survive. However, although many of the mechanisms underpinning DNA damage repair are common to all cells, some cell types exposed to distinct types of DNA damage or microenvironmental stresses or located in specific anatomical positions may superimpose lineage-restricted modulation of their DNA repair functions. In melanocytes and melanoma, MITF has received considerable attention as a critical coordinator of many cell functions (Goding and Arnheiter 2019). Previous work established a transcriptional role for MITF in regulating genes implicated in DNA damage repair (Strub et al. 2011), recapitulated here, as well as in promoting survival and pigmentation following UV exposure of the skin. MITF expression after UV irradiation exhibits a damped oscillation pattern (Malcov-Brog et al. 2018) in which it seems likely that a MITF negative feedback loop may participate (Louphrasitthiphol et al. 2019). However, although examination of SNVs in melanoma previously failed to identify any correlation between MITF expression and accumulated DNA damage (Herbert et al. 2019), here we revealed that both low and high levels of MITF correlated with an increased tumor mutational burden (TMB). While the detection of increased SNVs in MITFLow cells might be explained by reduced expression of MITF target genes involved in the DDR such as BRCA1, LIG1, RPA1, FANCA, or FANCC (Strub et al. 2011), why MITFHigh cells might also have a high SNV and CNV burden was not clear. We considered two possibilities. First, MITFHigh cells may have a survival advantage that allows them to resist cell death following DNA damage, leading to an increased TMB in the surviving cells. Consistent with this, MITF can activate expression of the antiapoptotic effector BCL2 within a few hours following UV irradiation (McGill et al. 2002; Malcov-Brog et al. 2018). A second, but not mutually exclusive, possibility is that in melanoma cells driven to proliferate owing to activation of BRAF or NRAS, expression of higher levels of MITF would trigger replication stress that consequently leads to accumulation of DNA damage. The ability of MITF to cause replication stress may be related to its capacity to induce expression of proproliferative genes such as CDK2 (Du et al. 2004), MET (McGill et al. 2006), E2F1 (Chauhan et al. 2022), and SCD (Vivas-Garcia et al. 2020) and to suppress antiproliferative factors such as p27 (CDKN1B) and p16 (CDKN2A) (Loercher et al. 2005; Carreira et al. 2006).
Our results also indicate that following DNA damage, the MITF interactome is dramatically remodeled. Consistent with recent observations (Elkoshi et al. 2023) indicating that UVB exposure reshapes the MITF interactome, we found that MITF interaction with the majority of transcription cofactors was greatly reduced on exposure to camptothecin, with the notable exception of EP300 and CREBBP, which can bind and acetylate MITF on multiple lysines (Louphrasitthiphol et al. 2020, 2023). We view it as likely that the loss of interaction with transcription cofactors arises following DNA damage as a consequence of the rapid degradation of the majority of MITF that is not retained at the DNA damage sites. These observations suggest that the ability of MITF to regulate transcription may be impaired, at least at a subset of genes where specific cofactors may be required. At first sight, this may seem at odds with the known ability of MITF to activate transcription of pigmentation gene programs after UV irradiation as part of the tanning response designed to prevent UV-mediated DNA damage to skin cells (Malcov-Brog et al. 2018). However, in this respect, the timing of the activation of pigmentation genes by MITF may be relevant. Following UV exposure, keratinocytes mediate the activation of MC1R (Guida et al. 2022), which is expressed in melanocytes. Signaling downstream from MC1R can promote DNA damage repair (Swope et al. 2014; Robles-Espinoza et al. 2016) but can also increase MITF levels (Khaled et al. 2003), which may subsequently lead later to increased pigmentation (Malcov-Brog et al. 2018). Consistent with this, the transcriptional response of the pigmentation-related MITF targets appears to occur 7–10 h after UV (Malcov-Brog et al. 2018). It therefore seems likely that at early times (within minutes), DNA damage decreases MITF association with transcription cofactors, but at later times (within hours), the ability of MITF to activate transcription of its downstream target genes may be restored.
As the mechanisms uncovered here would not have evolved to facilitate melanoma progression, why would DNA damage promote loss of MITF's transcriptional competence? One possibility, highlighted in the model outlined in Figure 7, is that following UV irradiation of the skin, degradation of MITF associated with transcription cofactors would lead to transient melanocyte dedifferentiation and proliferation, as has been seen both in human skin (Rosen et al. 1987; Bacharach-Buhles et al. 1999; van Schanke et al. 2005) and in zebrafish models, where loss of MITF function can lead to differentiated melanocytes entering the cell cycle (Taylor et al. 2011). Since MITF binding sites found in differentiation-associated promoters are lower affinity than those present in genes regulating cell proliferation (Louphrasitthiphol et al. 2023), even moderate levels of MITF degradation in nonproliferating melanocytes may trigger dedifferentiation and promote proliferation. Furthermore, by binding the MRN complex and blunting its recruitment to DNA damage, MITF would prevent MRN-mediated cell cycle arrest. In this scenario, the penalty imposed by the potential genome instability caused by reduced MRN complex function when bound by MITF would be offset by increased melanocyte proliferation and their ultimate differentiation, which would protect against subsequent DNA damage. In contrast, in melanoma cells that are already driven to proliferate owing to BRAF/NRAS mutation, the ability of elevated MITF levels to promote differentiation will be counterbalanced by MAPK-driven MITF acetylation (Louphrasitthiphol et al. 2020, 2023) and consequently lead to slow recruitment of MRN to stalled replication forks and genomic instability, as observed in melanomas with high MITF expression. In addition, degradation of MITF in response to DNA damage (for example, in melanoma cells with a hypermutation phenotype) may decrease MITF expression and transcriptional activity and consequently increase the probability of a phenotype switch toward an invasive state.
Figure 7.
The impact of MITF expression on DNA damage repair. (I) In cells that do not express MITF, the MRN complex senses DNA damage, binds to DSBs, and then activates ATM to amplify the DDR and trigger a CHK2-dependent cell cycle arrest and DNA repair by HRR. (II) In melanocytes and melanoma cells, MITF is rapidly phosphorylated by DNA-PK and interacts with NBS1–RAD50 but not MRE11, destabilizing the MRN complex. As a result, the rate of MRN recruitment is decreased, slowing down HR-mediated repair and promoting the formation of SNVs/CNVs. The delay in MRN recruitment is emphasized in cells expressing the mutated MITF-E318K. (III) Subsequently, MITF is rapidly degraded, releasing its transcription cofactors, which results in cell cycle entry and proliferation of SNV-harboring melanocytes/melanoma cells. Later, UV-activated MC1R signaling restores MITF expression, promoting a differentiation program characterized by the expression of pigmentation and survival genes. Elevated melanocyte numbers combined with increased pigment production will provide robust protection against future UV-mediated DNA damage.
Although MITF loses its interaction with the majority of transcription cofactors following DNA damage, MITF maintains its association with two components of the MRN complex, RAD50 and NBS1, whose role is to facilitate the localization of the MRN complex to DNA damage sites, which contributes to the optimal activation of ATM (Uziel et al. 2003). We could detect the MITF–NBS1/RAD50 interaction in steady-state conditions, although more interaction was detected after camptothecin treatment. This suggests that MITF may titrate NBS1/RAD50 away from the damage sites and diminish the availability of the complex after DNA damage. Indeed, we observed that expression of MITF impacts the intensity of recruitment of the MRN complex at laser-induced double-strand breaks. Two recent studies proposed an absolute quantitation of most proteins in human cells (Beck et al. 2011) or tissues (Jiang et al. 2020). In the report from Jiang et al. (2020), it was shown that in the skin, NBS1 is the limiting factor among the MRN complex, with an average of 784 copies of NBS1, 1345 copies of MRE11, and 5000 copies of RAD50. Beck et al. (2011) measured the proteome of U2-OS cells and quantified between 5480 and 8030 copies of MRN proteins. In contrast, in 501mel cells, a high-MITF-expressing cell line, the number of MITF dimers is estimated at up to 250,000 copies (Louphrasitthiphol et al. 2020). Thus, titration of NBS1 by an excess of MITF is feasible and would have an effect on the recruitment of all three MRN components. As a consequence, MITF may limit the recruitment of the MRN complex to sites of damage, leading to reduced HRR, increased replication stress, and higher TMB. By limiting MRN activity, MITF may create a tolerance to DNA damage that may be important to allow cells to survive and proliferate, especially in the case of prolonged sun exposure, when MITF-dependent melanocyte proliferation and melanin production are essential (Goding 2007).
Unexpectedly, MITF is retained at damage sites as a consequence of a mechanism controlled by DNA damage-associated phosphorylation, while in the rest of the nucleus, MITF levels decrease. This is fundamentally different from active recruitment as observed with bona fide repair proteins like NBS1 or MRE11 and may indicate that MITF is a modulator of the DDR rather than an essential player. Interestingly, the E318K melanoma predisposition mutation seems to affect the behavior of MITF both at and away from the damage sites. Only the MITF-E318K mutant showed an increase at the laser-induced DNA damage sites, suggesting its active recruitment in addition to retention. Moreover, following UV irradiation, the overall nuclear decrease in MITF-E318K mutant levels was much slower than the WT protein or the phosphorylation mutants. Because MITF-E318K is also able to interact with NBS1/RAD50 and can efficiently disrupt the MRN complex to prevent MRE11 recruitment and function, we observed an even higher accumulation of γH2AX due to a delayed HRR in MITF-E318K-expressing cells that also have a higher percentage of stalled replication forks and slower fork speed and resist senescence-induced terminal growth arrest (Bonet et al. 2017; Leclerc et al. 2017). This combination of delayed HRR and tolerance to replication stress may therefore explain the increased melanoma risk associated with the MITF-E318K mutation. Importantly, we also revealed that the S325E phospho-mimetic phenocopies the SUMOylation-inhibiting E318K mutation, with both accumulating more at sites of DNA damage and both able to bind NBS1 and reduce replication fork speed. Most likely, the similarity in the behaviors of the S325E and E318K mutants in all assays reflects the ability of the E318K mutant to increase S325 phosphorylation following UV irradiation.
MITF is a tissue-restricted transcription factor, and we reveal that it can play a nontranscriptional role in the DDR. It is likely that MITF is not unique in this respect and that more transcription factors regulate the DDR in a context-specific manner. Here, the MITF-dependent regulation of HRR is mediated by the interaction with RAD50/NBS1 and regulated by DNA damage-activated kinases. Although several transcription factors can be phosphorylated by DDR-associated kinases or interact with DDR factors, only a few have been implicated in DNA repair (Bhoumik et al. 2005; Rubin et al. 2007; Beishline et al. 2012; Park et al. 2015; Wysokinski et al. 2015), and a study by Izhar et al. (2015) suggested that the recruitment of transcription factors to DNA damage was an artifact of their affinity for PARP1. Here instead we showed not only that MITF retention at sites of damage is regulated, but that its effects on replication fork progression and DNA damage repair are mediated though a specific interaction with RAD50/NBS1. The ability of MITF to influence DDR resembles ATF2, which is also found at damage sites. However, in contrast to MITF, ATF2 enhances the recruitment of the MRN complex (Bhoumik et al. 2005). Interestingly, ATF2 is a coactivator of Jun whose transcriptional program is associated with melanoma dedifferentiation and low-MITF cells (Riesenberg et al. 2015; Verfaillie et al. 2015; Comandante-Lou et al. 2022), raising the possibility that DDR is affected differently in distinct phenotypic states. Thus, we propose that the transcription factor-dependent regulation of DDR may be a general mechanism that allows fine-tuning of the repair processes, depending on the tissue or the context.
In summary, our results provide a key insight into how a tissue-restricted transcription regulator and lineage survival oncogene, MITF, can shape the response to DNA damage and may provide a potential mechanistic explanation for how the E318K mutation impacts melanoma predisposition.
Materials and methods
SNV count
Gene expression, simple somatic mutation, copy number variation, and donor data were downloaded from the International Cancer Genome Consortium (ICGC) data portal for the Skin Cutaneous Melanoma–ICGC project (SKCMUS; https://dcc.icgc.org/releases/current/Projects/SKCM-US). The 428 samples were sorted by their MITF expression value and split into five bins of approximately equal size. The distribution of single-nucleotide variants (SNVs) per sample was plotted after log transformation, computed as log10(0.1 + SNV). Total number of copy number variations (CNVs) per sample was computed as the sum of absolute value of the CNV segment mean. The distributions of total SNVs and CNVs were plotted as a box plot in each of the five MITF expression bins. Paired Wilcoxon rank sum test was used to compare the values between the bins.
Melanoblast/melanocyte differentiation protocol
The procedure to differentiate melanoblasts and melanocytes from hPSCs has been described previously (Baggiolini et al. 2021).
DNA extraction
DNA from frozen cells was isolated with the DNeasy blood and tissue kit (Qiagen 69504) according to the manufacturer's protocol and modified by replacing AW2 buffer with 80% ethanol. DNA was eluted in 50 µL of 0.5× buffer AE heated to 55°C.
Integrated mutation profiling of actionable cancer targets (IMPACT)
After PicoGreen quantification and quality control by Agilent BioAnalyzer, 100 ng of DNA was used to prepare libraries using the KAPA Hyper preparation kit (Kapa Biosystems KK8504) with eight cycles of PCR. Two-hundred nanograms to 400 ng of each barcoded library was captured by hybridization in a pool of five samples using the IMPACT (integrated mutation profiling of actionable cancer targets) assay (Cheng et al. 2015) (Nimblegen SeqCap), designed to capture all protein-coding exons and select introns of 468 commonly implicated oncogenes, tumor suppressor genes, and members of pathways deemed actionable by targeted therapies. Captured pools were sequenced on a HiSeq 4000 in a PE100 run using the HiSeq 3000/4000 SBS kit (Illumina), producing an average of 866× coverage per tumor and 538× per normal. Sequence reads were aligned to human genome hg19 using BWA MEM (Li and Durbin 2010). ABRA was used to realign reads around indels to reduce alignment artifacts, and the Genome Analysis Toolkit was used to recalibrate base quality scores (McKenna et al. 2010; Mose et al. 2014). Duplicate reads were marked for removal, and the resulting BAM files were used for subsequent analysis. The sequencing data analysis pipeline is available at https://github.com/soccin/BIC-variants_pipeline. Copy number analysis was performed using FACETS (version 0.3.9) (Shen and Seshan 2016) using a set of pooled normals for each cell line.
Cell culture and drugs
Melanoma cell lines were grown in RPMI containing 10% FBS at 37°C and 5% CO2. All other cell lines were grown in DMEM containing 10% FBS at 37°C and 5% CO2. U2-OS-DR-GFP, 501-HA-MITF wild type and mutants, and U2-OS-LacO#13 were maintained under puromycin selection. MITF-GFP cells (wild type and mutants) expressing 501mel were selected and maintained in geneticin. HEK293 cells were transfected with Fugene 6 (Promega). All other cell lines were transfected using Lipofectamine 2000 (Thermo Fisher Scientific). All transfections were performed following the manufacturer's instructions. Doxycycline was used for all inducible cell lines. The drugs and chemicals used in this study are listed in Table 1.
Table 1.
Drugs/chemicals used in this study
| Drug/Chemical | Supplier | Reference |
|---|---|---|
| Doxycycline | Sigma-Aldrich | D9891 |
| Puromycin | Sigma-Aldrich | P8833 |
| Geneticin | Thermo Fisher Scientific | 10131027 |
| KU55933 | Tocris | 3544 |
| VE821 | Selleckchem | S8007 |
| VE822 | Selleckchem | S7102 |
| NU7441 | Selleckchem | S2638 |
| Olaparib | Selleckchem | S1060 |
| Camptothecin | Cambridge Bioscience | 1039-1 |
| CHK2 purified kinase | Life Technologies | PV3367 |
| DNA-PKcs purified kinase | Life Technologies | PV5866 |
| Cycloheximide | Merck | C4859 |
Plasmids and oligonucleotides
All plasmids have been described elsewhere (Table 2). Point mutations were performed using the QuikChange Lightning site-directed mutagenesis kit (Agilent Technologies) according to the manufacturer's instructions. Mutagenesis primers (Table 3) were designed using the Agilent online tool (https://www.agilent.com/store/primerDesignProgram.jsp). For MITF silencing, we used a previously validated sequence (Carreira et al. 2006) and transfected cells using Lipofectamine RNAiMax (Thermo Fisher Scientific) as per the manufacturer's instructions.
Table 2.
Plasmids used in this study
| Plasmid | Source |
|---|---|
| pCMV-3XHA | Tony Kouzarides, Cambridge |
| pEGFP-C1-MITF | Ngeow et al. 2018 |
| RFP-SUMO1/2 WT and mutants | Jo Morris, Birmingham |
| GFP-MRE11 | Tim Humphrey, Oxford |
| mCherry-LacR-NLS | Roger Greenberg, Philadelphia |
| I-SceI | Tim Humphrey, Oxford |
| pcDNA3.1(+)-C-eGFP-NBS1 | GenScript |
| pcDNA3.1(+)-C-eGFP-RAD50 | GenScript |
| pLUC-Met | Generated in our laboratory |
Table 3.
Mutagenesis primers used in this study
| Mutation | Primers |
|---|---|
| S325A | 5′-GCTGAAGGAGGTCTTGGGCGCAGTTCTCAAGAACGG-3′ 5′-CCGTTCTTGAGAACTGCGCCCAAGACCTCCTTCAGC-3′ |
| S325E | 5′-GATGCTGAAGGAGGTCTTGCTCGCAGTTCTCAAGAACGGGT-3′ 5′-ACCCGTTCTTGAGAACTGCGAGCAAGACCTCCTTCAGCATC-3′ |
| K316R | 5′-ACGGGTTCTTGCCTGATGATCCGATTCACCAAATC-3′ 5′-GATTTGGTGAATCGGATCATCAGGCAAGAACCCGT-3′ |
| E318K | 5′-CAAGAACGGGTTTTTGCTTGATGATCCGATTCACC-3′ 5′-GGTGAATCGGATCATCAAGCAAAAACCCGTTCTTG-3′ |
| K182R | 5′-GTGAGCTCCCTTCTTATGTTGGGAAGGTTGGCTGG-3′ 5′-CCAGCCAACCTTCCCAACATAAGAAGGGAGCTCAC-3′ |
| ΔR217 | 5′-ACAACCTGATTGAACGAAGAAGATTTAACATAAATGACCGCATTAA-3′ 5′-TTAATGCGGTCATTTATGTTAAATCTTCTTCGTTCAATCAGGTTGT-3′ |
| siMITF | Carreira et al. 2006 |
| siBRCA2 | Tim Humphrey |
Peptide arrays for kinase assays
Cellulose-bound peptide arrays were prepared using standard Fmoc solid-phase peptide synthesis on a MultiPep-RSi-Spotter (Intavis) according to the SPOT synthesis method provided by the manufacturer, as previously described (Picaud and Filippakopoulos 2015). In brief, 21-residue-long peptides based on MITF residues 314–333 were synthesized on amino-functionalized cellulose membranes (Whatman chromatography paper grade 1CHR; GE Healthcare Life Sciences 3001-878), and the presence of SPOTed peptides was confirmed by ultraviolet light (UV; λ = 280 nm). The wild-type sequence was mutated or modified where indicated. The membrane was quickly washed in 100% EtOH and then equilibrated overnight in kinase buffer (50 mM HEPES at pH 7.5, 200 mM NaCl, 10 mM MgCl2, 10 mM KCl). For in vitro kinase assays, the membrane was then incubated in kinase buffer containing 500 ng of kinase for 1 h at 30°C. After washing, the membranes were analyzed by Western blot. The wild-type sequence was mutated or modified where indicated. The membrane was briefly incubated in 100% EtOH followed by an overnight incubation in kinase buffer (50 mM HEPES at pH 7.5, 200 mM NaCl, 10 mM MgCl2, 10 mM KCl). For the in vitro kinase assays, the membrane was then incubated in kinase buffer containing 500 ng of kinase for 1 h at 30°C. After washing, the membrane was analyzed by Western blot.
GFP-TRAP
Cells were suspended in lysis buffer (50 mM HEPES-NaOH at pH 8.0, 100 mM KCl, 2 mM EDTA, 0.1% NP40, 10% glycerol, 1 mM PMSF, 1 mM DTT), flash-frozen, sonicated in a Diagenode Bioruptor for 5 min (cycling 30 sec on/15 sec off) at 4°C, incubated with 1 U/μL benzonase for 1 h at 4°C, and centrifuged at 10,000g for 30 min. From the supernatant, 10% of the volume was kept aside as input. The remaining was incubated overnight at 4°C on a rotating wheel with washed ChromoTek GFP-Trap magnetic agarose beads at 1:10 (vol:vol) ratio. The unbound fraction was kept aside, and the beads were washed twice with lysis buffer and prepared for SDS-PAGE.
Western blot and antibodies
Cells were lysed with 1× Laemmli sample buffer (50 mM Tris-Cl at pH 6.8, 100 mM DTT, 2% SDS, 12.5% glycerol, 0.1% Bromophenol blue), sonicated in a Diagenode Bioruptor for 5 min (cycling 30 sec on/15 sec off) at 4°C, and boiled for 15 min at 95°C. Protein separation was performed by SDS–polyacrylamide gel electrophoresis (SDS-PAGE) using 12.5% 200:1 bis-acrylamide gels. Proteins were transferred onto nitrocellulose membranes and saturated in 3% PBS-BSA for 1 h. The membranes were incubated overnight with the primary antibody at 1 µg/mL in 1% PBS-BSA, followed by 1 h of incubation with the HRP-conjugated secondary antibody diluted 1/10,000 (mouse) or 1/20,000 (rabbit) in 0.05% PBS-Tween. Detection of the chemo-luminescent signal was performed using Amersham ECL Western blot detection (GE Healthcare) according to the manufacturer's instructions. Images were acquired on a Bio-Rad ChemiDoc XRS+ system and analyzed/quantified with Image Lab software. The antibodies used are shown in Table 4.
Table 4.
Antibodies used in this study
| Target | Supplier | Reference |
|---|---|---|
| 53BP1 | Cell Signaling Technology | 4937 |
| FLAG | Merck (Sigma-Aldrich) | F1804 |
| GAPDH | Merck (Sigma-Aldrich) | G9545 |
| GFP | Abcam | ab290 |
| HA tag | Merck (Sigma-Aldrich) | H3663 |
| HA tag | Cell Signaling Technology | 3724 |
| MITF | Cambridge Bioscience | HPA003259 |
| MITF D5 | Abcam | ab3201 |
| MITF phospho-S325 | Custom-made | |
| Phospho-ATM (Ser1981) | Abcam | ab81292 |
| Phospho-ATM/ATR substrate (S*Q) | Cell Signaling Technology | 9607 |
| Phospho-Chk1 (Ser345) | Cell Signaling Technology | 2348 |
| Phospho-Chk2 (Thr68) | Cell Signaling Technology | 2197 |
| Phospho-H2AX (Ser139) | Merck (Millipore) | 05-636 |
| Phospho-H2AX (Ser139) | Cell Signaling Technology | 9718 |
| RAD51 | Cell Signaling Technology | 8875 |
| RFP | Abcam | ab152123 |
| Antimouse HRP | Bio-Rad | 1706516 |
| Antirabbit HRP | Bio-Rad | 1706515 |
| Antimouse Alexa fluor 488 | Thermo Fisher Scientific | A11001 |
| Antimouse Alexa fluor 546 | Thermo Fisher Scientific | A10036 |
| Antirabbit Alexa fluor 488 | Thermo Fisher Scientific | A11008 |
| Antirabbit Alexa fluor 546 | Thermo Fisher Scientific | A10040 |
Immunofluorescence
For immunofluorescence-based experiments, cells were seeded on coverslips or in eight-chamber glass-bottom slides (Ibidi). Cells were fixed in 4% formaldehyde and permeabilized in 0.1% Triton X-100/1× PBS. For LMI or focus experiments, cells were first incubated in CSK-T buffer (10 mM PIPES at pH 7, 100 mM NaCl, 300 mM sucrose, 3 mM MgCl2, 0.5% Triton X-100) and then fixed in formaldehyde. Samples were saturated in 3% PBS-BSA for 1 h and then incubated with the primary antibody in 1% PBS-BSA from 1 h to overnight, followed by 1 h of incubation with the fluorochrome-conjugated secondary antibody. When required, the nuclear content was counterstained with DAPI or Hoechst. Coverslips were mounted on glass slides using VectaShield Antifade mounting medium (Vector Laboratories). When eight-chamber slides were used, samples were covered with 9-mm coverslips. Analysis of the fluorescent patterns was performed on a Zeiss LSM710 confocal microscope. Quantification of the DDR foci was automatized using ImageJ2 software and self-created macros.
Colony formation assay
Cells were transfected in six-well plates. After 24 h, cells were counted and plated at low density (500, 1000, or 2000 cells per well) in a separate six-well plate and left to grow for 10 d. Newly formed colonies were fixed in PFA, stained with Crystal Violet, and dried. Plates were scanned using a FLA-5100 fluorescent image analyzer (Fujifilm), and colonies were manually counted.
Luciferase reporter
HEK293 cells were seeded in 12-well plates and cotransfected with the MET-Luc plasmid and HA-MITF WT, mutant, or empty-HA vector. After 48 h, measurement of luciferase activity was performed using the Dual-Glo luciferase assay system (Promega) according to the manufacturer's instructions in a GloMax multidetection system (Promega).
Cycloheximide treatment
501mel cells were seeded in six-well plates and treated with the indicated inhibitor for 6 h. Cycloheximide was added at a final concentration of 100 μg/mL, and cells were collected at the indicated time points for SDS-PAGE.
Laser microirradiation
For live-cell LMI, cells were cultured in 35-mm µ-Dishes (Ibidi 81158). Twenty-four hours after transfections, cells were presensitized with 1 µg/mL Hoechst 33342 for 30 min. The medium was immediately replaced with FluoroBrite DMEM (Thermo Fisher Scientific) supplemented with FBS and glutamine. For UV-LMI, UV irradiation was performed on a Nikon TE-2000 with a Nikon Plan Fluor 20×/0.45 objective (Nikon) using a Team Photonic SNV-04P-100 laser (Cairn Research), a plan-apochromat 20×/0.8 M27 objective, and a Prime camera (Photometrics). For NIR-LMI, irradiation was performed on a Zeiss LSM710 confocal microscope using a MaiTai multiphoton laser (Spectra Physics) and a plan-apochromat 63×/1.40 oil DICM27 objective. We used a 750-nm laser, which at 100% provides 0.5 W at the objective. Cells were exposed to 5%–12% power depending on the application. Analysis and quantification of the fluorescent stripes were automatized using ImageJ2 software and self-created macros. When the samples were subjected to IF, cells were plated on large coverslips, presensitized, irradiated through a plan-apochromat 20×/0.8 M27 objective, further incubated for the required time, and processed as indicated.
Nuclear tethering
U2OS-LacO#13 cells (Lu et al. 2021) were grown on coverslips and transfected with the indicated combinations of plasmids for 24 h. Samples were fixed with formaldehyde and mounted as previously indicated. Single-cell images were acquired using a Zeiss LSM980 confocal microscope, and the intensities of colocalizations were determined manually.
DNA fiber assay
Cells were incubated with 10 μM BrdU for 30 min followed by incubation with 10 μM EdU for 30 min and collected. The DNA fiber spreads were prepared as recommended (Nieminuszczy et al. 2016). After DNA denaturation and blocking, EdU was detected using Click-iT EdU imaging kit 488 (Thermo Fisher Scientific) using 250 μL of Click-It reaction mix per slide for 2 h. After another blocking step, BrdU was detected with a mouse anti-BrdU antibody clone 3D4 (BD Bioscience 555627) and antimouse Alexa fluor 546. Images were acquired using a Zeiss LSM710 confocal microscope. Double-labeled replication forks were analyzed manually using ImageJ2. For each sample, 300–400 forks were recorded. To determine the forks’ speed, the length of the EdU part of the ongoing forks was measured using the formula V(kb/min) = [(x × 0.132 µm) × 2.59 kb/µm]/t(min), where x is the length of EdU.
γ-Irradiation and UV irradiation
For γ-irradiation, cells were exposed to X-rays at a final dose of 1–10 Gy using a cesium-137 irradiator at a dose rate of 1.87 Gy/min. For UV irradiation, the medium was replaced with PBS, and plates were placed in a CL-1000 UV cross-linker containing 254-nm tubes. After exposure to a final dose of 24–100 J/m2, the culture medium was added back, and the cells were cultivated for the time required.
DSB repair reporter assays
The procedure for the HR-DR-GFP reporter assay has been described elsewhere (Ahrabi et al. 2016). U2-OS-DR-GFP cells were cotransfected with an I-SceI plasmid plus the indicated expression vector or siRNA using Lipofectamine 2000 (Invitrogen) for 72 h. The cells were trypsinized, washed, and resuspended in cold PBS before flow cytometry analysis using a BD FACScanto (BD Bioscience). Results were generated using FlowJo software (BD Bioscience).
Proximity-dependent biotinylation mass spectrometry
The methods for the generation of MITF-tagged cell lines and the proximity-dependent biotinylation mass spectrometry have been described in detail elsewhere (Chauhan et al. 2022). Briefly, HEK293 cells stably expressing BirA*-FLAG-MITF or BirA*-FLAG-NLS were pelleted from two 150-mm plates. To induce DNA damage, cells were treated with 20 nM CPT for 4 h. After extraction, biotinylated proteins were captured using streptavidin-Sepharose beads for 3 h at 4°C, washed, and recovered by incubation in a trypsin solution overnight at 37°C. The tryptic peptides were then stored at −80°C. For mass spectrometry analysis, eluted samples were analyzed on a TripleTOF 5600 instrument (AB SCIEX) set to data-dependent acquisition (DDA) mode. MS data storage, handling, and analysis have been described by Chauhan et al. (2022). For statistical scoring, we used significance analysis of interactome (SAINTexpress version 3.6.1; Teo et al. 2014) to model the background proteins observed in our BioID analysis and enforced a 1% false discovery rate threshold to identify significant proximity partners of MITF.
The MS files used in this report were deposited to MassIVE (http://massive.ucsd.edu) under accession number MSV000089109 and to ProteomeXchange (http://www.proteomexchange.org) under accession number PXD032772. They are available at ftp://MSV000089109@massive.ucsd.edu.
RNA-seq
RNA was extracted using the RNeasy kit (Qiagen 74106), and quality control was performed on a Bioanalyzer. Samples with RIN ≥9.5 were used for library preparation and sequencing using Wellcome Trust Genomic Service, Oxford, as previously described (Louphrasitthiphol et al. 2019). Briefly, siMITF libraries were prepared using the QuantSeq Forward kit (Lexogen 015.96) using 500 ng of starting material to minimize the PCR amplification step with ERCC ExFold RNA spike-in mixes (Ambion) and sequenced on a HiSeq 4000. The output raw Fastq files were trimmed of poly-A using CutAdapt (Martin 2011) and mapped using STAR 2.5.1b (Dobin et al. 2013) against hg38 (Genome Research Consortium human build 38, 2015). Counts per gene from STAR were used as input for differential gene expression analysis using EdgeR (Robinson et al. 2010). Reads for each sample set were first filtered for genes whose expression was less than one count per million. GSVAs were performed using the Bioconductor package GSVA (Hänzelmann et al. 2013). The GSVA matrices and gene-specific expression table were clustered and displayed as a heat map using Pheatmap (https://cran.r-project.org/web/packages/pheatmap/index.html).
Data availability
RNA-seq data have been previously published (Louphrasitthiphol et al. 2019). MS data are available at MassIVE (http://massive.ucsd.edu; accession no. MSV000089109; ftp://massive.ucsd.edu/MSV000089109) and the ProteomeXchange Consortium (http://proteomecentral.proteomexchange.org; accession no. PXD032772).
Supplementary Material
Acknowledgments
We thank Dr. Jo Morris and Dr. Alexander Garvin (Institute of Cancer and Genomics Sciences, University of Birmingham) for the SUMO expression vectors. This work was funded by the Ludwig Institute for Cancer Research (to C.R.G., R.B., P.L., and M.T.), Graduate Program in Areas of Basic and Applied Biology (GABBA; to D.D.), Marie Curie Cancer Care (to C.R.G. and S.C.), J.-P.L. was supported by a Junior 2 salary award from the Fonds de Recherche du Québec-Santé and operating grants from the Cancer Research Society and Genome Quebec (25123 and 935296). P.F. and S.P. were supported by the Medical Research Council (MRC; MR/N010051/1). S.S. and T.C.H. are funded by a Medical Research Council Programme grant (MR/X006778/1 to T.C.H.). A.B. was supported by the Swiss National Science Foundation Postdoctorate Mobility Fellowship (P2ZHP3_171967 and P400PB_180672). R.W. was supported by the National Institutes of Health Director's New Innovator Award (DP2CA186572).
Author contributions: R.B. and C.R.G. conceived the experiments. J.-P.L. performed the BioID analysis. S.P. and P.F. prepared the peptide arrays. D.D. performed the FACS experiments. M.T. performed the SNV analysis. R.W., A.B., and S.T. performed the hPSC-based experiments. P.L. provided the RNA-seq data and analysis. S.S. performed the γ-irradiation-based experiments. S.C. performed preliminary experiments. T.C.H., P.F., R.W., and C.R.G. provided supervision and funding. R.B. and C.R.G. wrote the manuscript. All authors discussed the experiments and reviewed the manuscript.
Footnotes
Supplemental material is available for this article.
Article published online ahead of print. Article and publication date are online at http://www.genesdev.org/cgi/doi/10.1101/gad.350740.123.
Freely available online through the Genes & Development Open Access option.
Competing interest statement
The authors declare no competing interests.
References
- Ahrabi S, Sarkar S, Pfister SX, Pirovano G, Higgins GS, Porter AC, Humphrey TC. 2016. A role for human homologous recombination factors in suppressing microhomology-mediated end joining. Nucleic Acids Res 44: 5743–5757. 10.1093/nar/gkw326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bacharach-Buhles M, Lubowietzki M, Altmeyer P. 1999. Dose-dependent shift of apoptotic and unaltered melanocytes into the dermis after irradiation with UVA 1. Dermatology 198: 5–10. 10.1159/000018056 [DOI] [PubMed] [Google Scholar]
- Baggiolini A, Callahan SJ, Montal E, Weiss JM, Trieu T, Tagore MM, Tischfield SE, Walsh RM, Suresh S, Fan Y, et al. 2021. Developmental chromatin programs determine oncogenic competence in melanoma. Science 373: eabc1048. 10.1126/science.abc1048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beck M, Schmidt A, Malmstroem J, Claassen M, Ori A, Szymborska A, Herzog F, Rinner O, Ellenberg J, Aebersold R. 2011. The quantitative proteome of a human cell line. Mol Syst Biol 7: 549. 10.1038/msb.2011.82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beishline K, Kelly CM, Olofsson BA, Koduri S, Emrich J, Greenberg RA, Azizkhan-Clifford J. 2012. Sp1 facilitates DNA double-strand break repair through a nontranscriptional mechanism. Mol Cell Biol 32: 3790–3799. 10.1128/MCB.00049-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Békés M, Prudden J, Srikumar T, Raught B, Boddy MN, Salvesen GS. 2011. The dynamics and mechanism of SUMO chain deconjugation by SUMO-specific proteases. J Biol Chem 286: 10238–10247. 10.1074/jbc.M110.205153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertolotto C, Lesueur F, Giuliano S, Strub T, de Lichy M, Bille K, Dessen P, d'Hayer B, Mohamdi H, Remenieras A, et al. 2011. A SUMOylation-defective MITF germline mutation predisposes to melanoma and renal carcinoma. Nature 480: 94–98. 10.1038/nature10539 [DOI] [PubMed] [Google Scholar]
- Beuret L, Ohanna M, Strub T, Allegra M, Davidson I, Bertolotto C, Ballotti R. 2011. BRCA1 is a new MITF target gene. Pigment Cell Res 24: 725–727. 10.1111/j.1755-148X.2011.00862.x [DOI] [PubMed] [Google Scholar]
- Bhoumik A, Takahashi S, Breitweiser W, Shiloh Y, Jones N, Ronai Z. 2005. ATM-dependent phosphorylation of ATF2 is required for the DNA damage response. Mol Cell 18: 577–587. 10.1016/j.molcel.2005.04.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonet C, Luciani F, Ottavi JF, Leclerc J, Jouenne FM, Boncompagni M, Bille K, Hofman V, Bossis G, Marco de Donatis G, et al. 2017. Deciphering the role of oncogenic MITFE318K in senescence delay and melanoma progression. J Natl Cancer Inst 109: djw340. 10.1093/jnci/djw340 [DOI] [PubMed] [Google Scholar]
- Buis J, Stoneham T, Spehalski E, Ferguson DO. 2012. Mre11 regulates CtIP-dependent double-strand break repair by interaction with CDK2. Nat Struct Mol Biol 19: 246–252. 10.1038/nsmb.2212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carreira S, Goodall J, Aksan I, La Rocca SA, Galibert MD, Denat L, Larue L, Goding CR. 2005. Mitf cooperates with Rb1 and activates p21Cip1 expression to regulate cell cycle progression. Nature 433: 764–769. 10.1038/nature03269 [DOI] [PubMed] [Google Scholar]
- Carreira S, Goodall J, Denat L, Rodriguez M, Nuciforo P, Hoek KS, Testori A, Larue L, Goding CR. 2006. Mitf regulation of Dia1 controls melanoma proliferation and invasiveness. Genes Dev 20: 3426–3439. 10.1101/gad.406406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ceccaldi R, Rondinelli B, D'Andrea AD. 2016. Repair pathway choices and consequences at the double-strand break. Trends Cell Biol 26: 52–64. 10.1016/j.tcb.2015.07.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chambers SM, Fasano CA, Papapetrou EP, Tomishima M, Sadelain M, Studer L. 2009. Highly efficient neural conversion of human ES and iPS cells by dual inhibition of SMAD signaling. Nat Biotechnol 27: 275–280. 10.1038/nbt.1529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chao EC, Lipkin SM. 2006. Molecular models for the tissue specificity of DNA mismatch repair-deficient carcinogenesis. Nucleic Acids Res 34: 840–852. 10.1093/nar/gkj489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chauhan JS, Hölzel M, Lambert JP, Buffa FM, Goding CR. 2022. The MITF regulatory network in melanoma. Pigment Cell Res 35: 517–533. 10.1111/pcmr.13053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng DT, Mitchell TN, Zehir A, Shah RH, Benayed R, Syed A, Chandramohan R, Liu ZY, Won HH, Scott SN, et al. 2015. Memorial Sloan Kettering-integrated mutation profiling of actionable cancer targets (MSK-IMPACT): a hybridization capture-based next-generation sequencing clinical assay for solid tumor molecular oncology. J Mol Diagn 17: 251–264. 10.1016/j.jmoldx.2014.12.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comandante-Lou N, Baumann DG, Fallahi-Sichani M. 2022. AP-1 transcription factor network explains diverse patterns of cellular plasticity in melanoma cells. Cell Rep 40: 111147. 10.1016/j.celrep.2022.111147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de la Serna IL, Ohkawa Y, Higashi C, Dutta C, Osias J, Kommajosyula N, Tachibana T, Imbalzano AN. 2006. The microphthalmia-associated transcription factor requires SWI/SNF enzymes to activate melanocyte-specific genes. J Biol Chem 281: 20233–20241. 10.1074/jbc.M512052200 [DOI] [PubMed] [Google Scholar]
- D'Errico M, Lemma T, Calcagnile A, De Santis LP, Dogliotti E. 2007. Cell type and DNA damage specific response of human skin cells to environmental agents. Mutat Res 614: 37–47. 10.1016/j.mrfmmm.2006.06.009 [DOI] [PubMed] [Google Scholar]
- Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR. 2013. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29: 15–21. 10.1093/bioinformatics/bts635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du J, Widlund HR, Horstmann MA, Ramaswamy S, Ross K, Huber WE, Nishimura EK, Golub TR, Fisher DE. 2004. Critical role of CDK2 for melanoma growth linked to its melanocyte-specific transcriptional regulation by MITF. Cancer Cell 6: 565–576. 10.1016/j.ccr.2004.10.014 [DOI] [PubMed] [Google Scholar]
- Elkoshi N, Parikh S, Malcov-Brog H, Parikh R, Manich P, Netti F, Maliah A, Elkoshi H, Haj M, Rippin I, et al. 2023. Ataxia telangiectasia mutated signaling delays skin pigmentation upon UV exposure by mediating MITF function toward DNA repair mode. J Invest Dermatol 143: 2494–2506.e4. 10.1016/j.jid.2023.03.1686 [DOI] [PubMed] [Google Scholar]
- Garraway LA, Sellers WR. 2006. Lineage dependency and lineage-survival oncogenes in human cancer. Nat Rev Cancer 6: 593–602. 10.1038/nrc1947 [DOI] [PubMed] [Google Scholar]
- Garvin AJ, Densham RM, Blair-Reid SA, Pratt KM, Stone HR, Weekes D, Lawrence KJ, Morris JR. 2013. The deSUMOylase SENP7 promotes chromatin relaxation for homologous recombination DNA repair. EMBO Rep 14: 975–983. 10.1038/embor.2013.141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giuliano S, Cheli Y, Ohanna M, Bonet C, Beuret L, Bille K, Loubat A, Hofman V, Hofman P, Ponzio G, et al. 2010. Microphthalmia-associated transcription factor controls the DNA damage response and a lineage-specific senescence program in melanomas. Cancer Res 70: 3813–3822. 10.1158/0008-5472.CAN-09-2913 [DOI] [PubMed] [Google Scholar]
- Goding CR. 2007. Melanocytes: the new black. Int J Biochem Cell Biol 39: 275–279. 10.1016/j.biocel.2006.10.003 [DOI] [PubMed] [Google Scholar]
- Goding CR, Arnheiter H. 2019. MITF—the first 25 years. Genes Dev 33: 983–1007. 10.1101/gad.324657.119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guida S, Guida G, Goding CR. 2022. MC1R functions, expression, and implications for targeted therapy. J Invest Dermatol 142: 293–302.e1. 10.1016/j.jid.2021.06.018 [DOI] [PubMed] [Google Scholar]
- Hänzelmann S, Castelo R, Guinney J. 2013. GSVA: gene set variation analysis for microarray and RNA-seq data. BMC Bioinformatics 14: 7. 10.1186/1471-2105-14-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haq R, Shoag J, Andreu-Perez P, Yokoyama S, Edelman H, Rowe GC, Frederick DT, Hurley AD, Nellore A, Kung AL, et al. 2013. Oncogenic BRAF regulates oxidative metabolism via PGC1α and MITF. Cancer Cell 23: 302–315. 10.1016/j.ccr.2013.02.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemesath TJ, Steingrimsson E, McGill G, Hansen MJ, Vaught J, Hodgkinson CA, Arnheiter H, Copeland NG, Jenkins NA, Fisher DE. 1994. Microphthalmia, a critical factor in melanocyte development, defines a discrete transcription factor family. Genes Dev 8: 2770–2780. 10.1101/gad.8.22.2770 [DOI] [PubMed] [Google Scholar]
- Herbert K, Binet R, Lambert JP, Louphrasitthiphol P, Kalkavan H, Sesma-Sanz L, Robles-Espinoza CD, Sarkar S, Suer E, Andrews S, et al. 2019. BRN2 suppresses apoptosis, reprograms DNA damage repair, and is associated with a high somatic mutation burden in melanoma. Genes Dev 33: 310–332. 10.1101/gad.314633.118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hietakangas V, Anckar J, Blomster HA, Fujimoto M, Palvimo JJ, Nakai A, Sistonen L. 2006. PDSM, a motif for phosphorylation-dependent SUMO modification. Proc Natl Acad Sci 103: 45–50. 10.1073/pnas.0503698102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito S, D'Alessio AC, Taranova OV, Hong K, Sowers LC, Zhang Y. 2010. Role of Tet proteins in 5mC to 5hmC conversion, ES-cell self-renewal and inner cell mass specification. Nature 466: 1129–1133. 10.1038/nature09303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izhar L, Adamson B, Ciccia A, Lewis J, Pontano-Vaites L, Leng Y, Liang AC, Westbrook TF, Harper JW, Elledge SJ. 2015. A systematic analysis of factors localized to damaged chromatin reveals PARP-dependent recruitment of transcription factors. Cell Rep 11: 1486–1500. 10.1016/j.celrep.2015.04.053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang L, Wang M, Lin S, Jian R, Li X, Chan J, Dong G, Fang H, Robinson AE, Consortium GT, et al. 2020. A quantitative proteome Map of the human body. Cell 183: 269–283.e19. 10.1016/j.cell.2020.08.036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenmochi N, Suzuki T, Uechi T, Magoori M, Kuniba M, Higa S, Watanabe K, Tanaka T. 2001. The human mitochondrial ribosomal protein genes: mapping of 54 genes to the chromosomes and implications for human disorders. Genomics 77: 65–70. 10.1006/geno.2001.6622 [DOI] [PubMed] [Google Scholar]
- Kenny C, Dilshat R, Seberg HE, Van Otterloo E, Bonde G, Helverson A, Franke CM, Steingrímsson E, Cornell RA. 2022. TFAP2 paralogs facilitate chromatin access for MITF at pigmentation and cell proliferation genes. PLoS Genet 18: e1010207. 10.1371/journal.pgen.1010207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khaled M, Larribere L, Bille K, Ortonne JP, Ballotti R, Bertolotto C. 2003. Microphthalmia associated transcription factor is a target of the phosphatidylinositol-3–kinase pathway. J Invest Dermatol 121: 831–836. 10.1046/j.1523-1747.2003.12420.x [DOI] [PubMed] [Google Scholar]
- Kim ST, Lim DS, Canman CE, Kastan MB. 1999. Substrate specificities and identification of putative substrates of ATM kinase family members. J Biol Chem 274: 37538–37543. 10.1074/jbc.274.53.37538 [DOI] [PubMed] [Google Scholar]
- Lafrance-Vanasse J, Williams GJ, Tainer JA. 2015. Envisioning the dynamics and flexibility of Mre11-Rad50-Nbs1 complex to decipher its roles in DNA replication and repair. Prog Biophys Mol Biol 117: 182–193. 10.1016/j.pbiomolbio.2014.12.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambert JP, Tucholska M, Go C, Knight JD, Gingras AC. 2015. Proximity biotinylation and affinity purification are complementary approaches for the interactome mapping of chromatin-associated protein complexes. J Proteomics 118: 81–94. 10.1016/j.jprot.2014.09.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambert JP, Picaud S, Fujisawa T, Hou H, Savitsky P, Uusküla-Reimand L, Gupta GD, Abdouni H, Lin ZY, Tucholska M, et al. 2019. Interactome rewiring following pharmacological targeting of BET bromodomains. Mol Cell 73: 621–638 e617. 10.1016/j.molcel.2018.11.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laurette P, Strub T, Koludrovic D, Keime C, Le Gras S, Seberg H, Van Otterloo E, Imrichova H, Siddaway R, Aerts S, et al. 2015. Transcription factor MITF and remodeller BRG1 define chromatin organisation at regulatory elements in melanoma cells. Elife 4: e68057. 10.7554/eLife.06857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leclerc J, Ballotti R, Bertolotto C. 2017. Pathways from senescence to melanoma: focus on MITF sumoylation. Oncogene 36: 6659–6667. 10.1038/onc.2017.292 [DOI] [PubMed] [Google Scholar]
- Leclerc J, Garandeau D, Pandiani C, Gaudel C, Bille K, Nottet N, Garcia V, Colosetti P, Pagnotta S, Bahadoran P, et al. 2019. Lysosomal acid ceramidase ASAH1 controls the transition between invasive and proliferative phenotype in melanoma cells. Oncogene 38: 1282–1295. 10.1038/s41388-018-0500-0 [DOI] [PubMed] [Google Scholar]
- Lee G, Chambers SM, Tomishima MJ, Studer L. 2010. Derivation of neural crest cells from human pluripotent stem cells. Nat Protoc 5: 688–701. 10.1038/nprot.2010.35 [DOI] [PubMed] [Google Scholar]
- Li H, Durbin R. 2010. Fast and accurate long-read alignment with Burrows–Wheeler transform. Bioinformatics 26: 589–595. 10.1093/bioinformatics/btp698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loercher AE, Tank EM, Delston RB, Harbour JW. 2005. MITF links differentiation with cell cycle arrest in melanocytes by transcriptional activation of INK4A. J Cell Biol 168: 35–40. 10.1083/jcb.200410115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Louphrasitthiphol P, Ledaki I, Chauhan J, Falletta P, Siddaway R, Buffa FM, Mole DR, Soga T, Goding CR. 2019. MITF controls the TCA cycle to modulate the melanoma hypoxia response. Pigment Cell Res 32: 792–808. 10.1111/pcmr.12802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Louphrasitthiphol P, Siddaway R, Loffreda A, Pogenberg V, Friedrichsen H, Schepsky A, Zeng Z, Lu M, Strub T, Freter R, et al. 2020. Tuning transcription factor availability through acetylation-mediated genomic redistribution. Mol Cell 79: 472–487.e10. 10.1016/j.molcel.2020.05.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Louphrasitthiphol P, Loffreda A, Pogenberg V, Picaud S, Schepsky A, Friedrichsen H, Zeng Z, Lashgari A, Thomas B, Patton EE, et al. 2023. Acetylation reprograms MITF target selectivity and residence time. Nat Commun 14: 6051. 10.1038/s41467-023-41793-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu S, Louphrasitthiphol P, Goradia N, Lambert JP, Schmidt J, Chauhan J, Rughani MG, Larue L, Wilmanns M, Goding CR. 2021. TBX2 controls a proproliferative gene expression program in melanoma. Genes Dev 35: 1657–1677. 10.1101/gad.348746.121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luijsterburg MS, Acs K, Ackermann L, Wiegant WW, Bekker-Jensen S, Larsen DH, Khanna KK, van Attikum H, Mailand N, Dantuma NP. 2012. A new non-catalytic role for ubiquitin ligase RNF8 in unfolding higher-order chromatin structure. EMBO J 31: 2511–2527. 10.1038/emboj.2012.104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malcov-Brog H, Alpert A, Golan T, Parikh S, Nordlinger A, Netti F, Sheinboim D, Dror I, Thomas L, Cosson C, et al. 2018. UV-protection timer controls linkage between stress and pigmentation skin protection systems. Mol Cell 72: 444–456.e7. 10.1016/j.molcel.2018.09.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin M. 2011. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnetjournal 17: 3. 10.14806/ej.17.1.200 [DOI] [Google Scholar]
- Martina JA, Puertollano R. 2013. Rag GTPases mediate amino acid-dependent recruitment of TFEB and MITF to lysosomes. J Cell Biol 200: 475–491. 10.1083/jcb.201209135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGill GG, Horstmann M, Widlund HR, Du J, Motyckova G, Nishimura EK, Lin YL, Ramaswamy S, Avery W, Ding HF, et al. 2002. Bcl2 regulation by the melanocyte master regulator Mitf modulates lineage survival and melanoma cell viability. Cell 109: 707–718. 10.1016/S0092-8674(02)00762-6 [DOI] [PubMed] [Google Scholar]
- McGill GG, Haq R, Nishimura EK, Fisher DE. 2006. c-Met expression is regulated by Mitf in the melanocyte lineage. J Biol Chem 281: 10365–10373. 10.1074/jbc.M513094200 [DOI] [PubMed] [Google Scholar]
- McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, Garimella K, Altshuler D, Gabriel S, Daly M, et al. 2010. The genome analysis toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res 20: 1297–1303. 10.1101/gr.107524.110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mica Y, Lee G, Chambers SM, Tomishima MJ, Studer L. 2013. Modeling neural crest induction, melanocyte specification, and disease-related pigmentation defects in hESCs and patient-specific iPSCs. Cell Rep 3: 1140–1152. 10.1016/j.celrep.2013.03.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller AJ, Levy C, Davis IJ, Razin E, Fisher DE. 2005. Sumoylation of MITF and its related family members TFE3 and TFEB. J Biol Chem 280: 146–155. 10.1074/jbc.M411757200 [DOI] [PubMed] [Google Scholar]
- Möller K, Sigurbjornsdottir S, Arnthorsson AO, Pogenberg V, Dilshat R, Fock V, Brynjolfsdottir SH, Bindesboll C, Bessadottir M, Ogmundsdottir HM, et al. 2019. MITF has a central role in regulating starvation-induced autophagy in melanoma. Sci Rep 9: 1055. 10.1038/s41598-018-37522-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mose LE, Wilkerson MD, Hayes DN, Perou CM, Parker JS. 2014. ABRA: improved coding indel detection via assembly-based realignment. Bioinformatics 30: 2813–2815. 10.1093/bioinformatics/btu376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murakami H, Arnheiter H. 2005. Sumoylation modulates transcriptional activity of MITF in a promoter-specific manner. Pigment Cell Res 18: 265–277. 10.1111/j.1600-0749.2005.00234.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ngeow KC, Friedrichsen HJ, Li L, Zeng Z, Andrews S, Volpon L, Brunsdon H, Berridge G, Picaud S, Fischer R, et al. 2018. BRAF/MAPK and GSK3 signaling converges to control MITF nuclear export. Proc Natl Acad Sci USA 115: E8668–E8677. 10.1073/pnas.1810498115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nieminuszczy J, Schwab RA, Niedzwiedz W. 2016. The DNA fibre technique—tracking helicases at work. Methods 108: 92–98. 10.1016/j.ymeth.2016.04.019 [DOI] [PubMed] [Google Scholar]
- Ohanna M, Giuliano S, Bonet C, Imbert V, Hofman V, Zangari J, Bille K, Robert C, Bressac-de Paillerets B, Hofman P, et al. 2011. Senescent cells develop a PARP-1 and nuclear factor-κB-associated secretome (PNAS). Genes Dev 25: 1245–1261. 10.1101/gad.625811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park S, Kang JM, Kim SJ, Kim H, Hong S, Lee YJ, Kim SJ. 2015. Smad7 enhances ATM activity by facilitating the interaction between ATM and Mre11–Rad50–Nbs1 complex in DNA double-strand break repair. Cell Mol Life Sci 72: 583–596. 10.1007/s00018-014-1687-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paull TT. 2018. 20 years of Mre11 biology: no end in sight. Mol Cell 71: 419–427. 10.1016/j.molcel.2018.06.033 [DOI] [PubMed] [Google Scholar]
- Picaud S, Filippakopoulos P. 2015. SPOTing acetyl-lysine dependent interactions. Microarrays (Basel) 4: 370–388. 10.3390/microarrays4030370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierce AJ, Johnson RD, Thompson LH, Jasin M. 1999. XRCC3 promotes homology-directed repair of DNA damage in mammalian cells. Genes Dev 13: 2633–2638. 10.1101/gad.13.20.2633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ploper D, Taelman VF, Robert L, Perez BS, Titz B, Chen HW, Graeber TG, von Euw E, Ribas A, De Robertis EM. 2015. MITF drives endolysosomal biogenesis and potentiates Wnt signaling in melanoma cells. Proc Natl Acad Sci 112: E420–E429. 10.1073/pnas.1424576112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price ER, Ding HF, Badalian T, Bhattacharya S, Takemoto C, Yao TP, Hemesath TJ, Fisher DE. 1998. Lineage-specific signaling in melanocytes. C-kit stimulation recruits p300/CBP to microphthalmia. J Biol Chem 273: 17983–17986. 10.1074/jbc.273.29.17983 [DOI] [PubMed] [Google Scholar]
- Riesenberg S, Groetchen A, Siddaway R, Bald T, Reinhardt J, Smorra D, Kohlmeyer J, Renn M, Phung B, Aymans P, et al. 2015. MITF and c-Jun antagonism interconnects melanoma dedifferentiation with pro-inflammatory cytokine responsiveness and myeloid cell recruitment. Nat Commun 6: 8755. 10.1038/ncomms9755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson MD, McCarthy DJ, Smyth GK. 2010. Edger: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26: 139–140. 10.1093/bioinformatics/btp616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robles-Espinoza CD, Roberts ND, Chen S, Leacy FP, Alexandrov LB, Pornputtapong N, Halaban R, Krauthammer M, Cui R, Timothy Bishop D, et al. 2016. Germline MC1R status influences somatic mutation burden in melanoma. Nat Commun 7: 12064. 10.1038/ncomms12064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosen CF, Seki Y, Farinelli W, Stern RS, Fitzpatrick TB, Pathak MA, Gange RW. 1987. A comparison of the melanocyte response to narrow band UVA and UVB exposure in vivo. J. Invest. Dermatol 88: 774–779. 10.1111/1523-1747.ep12470474 [DOI] [PubMed] [Google Scholar]
- Rubin E, Wu X, Zhu T, Cheung JC, Chen H, Lorincz A, Pandita RK, Sharma GG, Ha HC, Gasson J, et al. 2007. A role for the HOXB7 homeodomain protein in DNA repair. Cancer Res 67: 1527–1535. 10.1158/0008-5472.CAN-06-4283 [DOI] [PubMed] [Google Scholar]
- Saladi SV, Wong PG, Trivedi AR, Marathe HG, Keenen B, Aras S, Liew ZQ, Setaluri V, de la Serna IL. 2013. BRG1 promotes survival of UV-irradiated melanoma cells by cooperating with MITF to activate the melanoma inhibitor of apoptosis gene. Pigment Cell Melanoma Res 26: 377–391. 10.1111/pcmr.12088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki T, Toh-e A, Kikuchi Y. 2000. Yeast Krr1p physically and functionally interacts with a novel essential Kri1p, and both proteins are required for 40S ribosome biogenesis in the nucleolus. Mol. Cell. Biol 20: 7971–7979. 10.1128/MCB.20.21.7971-7979.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato S, Roberts K, Gambino G, Cook A, Kouzarides T, Goding CR. 1997. CBP/p300 as a co-factor for the Microphthalmia transcription factor. Oncogene 14: 3083–3092. 10.1038/sj.onc.1201298 [DOI] [PubMed] [Google Scholar]
- Seberg HE, Van Otterloo E, Cornell RA. 2017. Beyond MITF: multiple transcription factors directly regulate the cellular phenotype in melanocytes and melanoma. Pigment Cell Res 30: 454–466. 10.1111/pcmr.12611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shain AH, Bastian BC. 2016. From melanocytes to melanomas. Nat Rev Cancer 16: 345–358. 10.1038/nrc.2016.37 [DOI] [PubMed] [Google Scholar]
- Shen R, Seshan VE. 2016. FACETS: allele-specific copy number and clonal heterogeneity analysis tool for high-throughput DNA sequencing. Nucleic Acids Res 44: e131. 10.1093/nar/gkw520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strub T, Giuliano S, Ye T, Bonet C, Keime C, Kobi D, Le Gras S, Cormont M, Ballotti R, Bertolotto C, et al. 2011. Essential role of microphthalmia transcription factor for DNA replication, mitosis and genomic stability in melanoma. Oncogene 30: 2319–2332. 10.1038/onc.2010.612 [DOI] [PubMed] [Google Scholar]
- Swope V, Alexander C, Starner R, Schwemberger S, Babcock G, Abdel-Malek ZA. 2014. Significance of the melanocortin 1 receptor in the DNA damage response of human melanocytes to ultraviolet radiation. Pigment Cell Res 27: 601–610. 10.1111/pcmr.12252 [DOI] [PubMed] [Google Scholar]
- Tauchi H, Matsuura S, Kobayashi J, Sakamoto S, Komatsu K. 2002. Nijmegen breakage syndrome gene, NBS1, and molecular links to factors for genome stability. Oncogene 21: 8967–8980. 10.1038/sj.onc.1206136 [DOI] [PubMed] [Google Scholar]
- Taylor KL, Lister JA, Zeng Z, Ishizaki H, Anderson C, Kelsh RN, Jackson IJ, Patton EE. 2011. Differentiated melanocyte cell division occurs in vivo and is promoted by mutations in Mitf. Development 138: 3579–3589. 10.1242/dev.064014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teo G, Liu G, Zhang J, Nesvizhskii AI, Gingras AC, Choi H. 2014. SAINTexpress: improvements and additional features in significance analysis of interactome software. J Proteomics 100: 37–43. 10.1016/j.jprot.2013.10.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uziel T, Lerenthal Y, Moyal L, Andegeko Y, Mittelman L, Shiloh Y. 2003. Requirement of the MRN complex for ATM activation by DNA damage. EMBO J 22: 5612–5621. 10.1093/emboj/cdg541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Linden E, Sanchez H, Kinoshita E, Kanaar R, Wyman C. 2009. RAD50 and NBS1 form a stable complex functional in DNA binding and tethering. Nucleic Acids Res 37: 1580–1588. 10.1093/nar/gkn1072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Schanke A, Jongsma MJ, Bisschop R, van Venrooij GMCAL, Rebel H, de Gruijl FR. 2005. Single UVB overexposure stimulates melanocyte proliferation in murine skin, in contrast to fractionated or UVA-1 exposure. J. Invest. Dermatol 124: 241–247. 10.1111/j.0022-202X.2004.23551.x [DOI] [PubMed] [Google Scholar]
- Vazquez F, Lim JH, Chim H, Bhalla K, Girnun G, Pierce K, Clish CB, Granter SR, Widlund HR, Spiegelman BM, et al. 2013. PGC1α expression defines a subset of human melanoma tumors with increased mitochondrial capacity and resistance to oxidative stress. Cancer Cell 23: 287–301. 10.1016/j.ccr.2012.11.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verfaillie A, Imrichova H, Atak ZK, Dewaele M, Rambow F, Hulselmans G, Christiaens V, Svetlichnyy D, Luciani F, Van den Mooter L, et al. 2015. Decoding the regulatory landscape of melanoma reveals TEADS as regulators of the invasive cell state. Nat Commun 6: 6683. 10.1038/ncomms7683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vivas-Garcia Y, Falletta P, Liebing J, Louphrasitthiphol P, Feng Y, Chauhan J, Scott DA, Glodde N, Chocarro-Calvo A, Bonham S, et al. 2020. Lineage-restricted regulation of SCD and fatty acid saturation by MITF controls melanoma phenotypic plasticity. Mol Cell 77: 120–137.e9. 10.1016/j.molcel.2019.10.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Widlund HR, Horstmann MA, Price ER, Cui J, Lessnick SL, Wu M, He X, Fisher DE. 2002. β-Catenin-induced melanoma growth requires the downstream target Microphthalmia-associated transcription factor. J Cell Biol 158: 1079–1087. 10.1083/jcb.200202049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wysokinski D, Pawlowska E, Blasiak J. 2015. RUNX2: a master bone growth regulator that may be involved in the DNA damage response. DNA Cell Biol 34: 305–315. 10.1089/dna.2014.2688 [DOI] [PubMed] [Google Scholar]
- Yan K, Rousseau J, Littlejohn RO, Kiss C, Lehman A, Rosenfeld JA, Stumpel CTR, Stegmann APA, Robak L, Scaglia F, et al. 2017. Mutations in the chromatin regulator gene BRPF1 cause syndromic intellectual disability and deficient histone acetylation. Am J Human Genet 100: 91–104. 10.1016/j.ajhg.2016.11.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang SH, Galanis A, Witty J, Sharrocks AD. 2006. An extended consensus motif enhances the specificity of substrate modification by SUMO. EMBO J 25: 5083–5093. 10.1038/sj.emboj.7601383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokoyama S, Woods SL, Boyle GM, Aoude LG, MacGregor S, Zismann V, Gartside M, Cust AE, Haq R, Harland M, et al. 2011. A novel recurrent mutation in MITF predisposes to familial and sporadic melanoma. Nature 480: 99–103. 10.1038/nature10630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X, Fiske B, Kawakami A, Li J, Fisher DE. 2011. Regulation of MITF stability by the USP13 deubiquitinase. Nat Commun 2: 414. 10.1038/ncomms1421 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
RNA-seq data have been previously published (Louphrasitthiphol et al. 2019). MS data are available at MassIVE (http://massive.ucsd.edu; accession no. MSV000089109; ftp://massive.ucsd.edu/MSV000089109) and the ProteomeXchange Consortium (http://proteomecentral.proteomexchange.org; accession no. PXD032772).