Abstract
Glucose inhibits meiosis in Saccharomyces cerevisiae at three different steps (IME1 transcription, IME2 transcription, and entry into late stages of meiosis). Because many of the regulatory effects of glucose in yeast are mediated through the inhibition of Snf1 kinase, a component of the glucose repression pathway, we determined the role of SNF1 in regulating meiosis. Deleting SNF1 repressed meiosis at the same three steps that were inhibited by glucose, suggesting that glucose blocks meiosis by inhibiting Snf1. For example, the snf1Δ mutant completely failed to induce IME1 transcripts in sporulation medium. Furthermore, even when this block was bypassed by expression of IME1 from a multicopy plasmid, IME2 transcription and meiotic initiation occurred at only 10 to 20% of the levels seen in wild-type cells. The addition of glucose did not further inhibit IME2 transcription, suggesting that Snf1 is the primary mediator of glucose controls on IME2 expression. Finally, in snf1Δ cells in which both blocks on meiotic initiation were bypassed, early stages of meiosis (DNA replication and commitment to recombination) occurred, but later stages (chromosome segregation and spore formation) did not, suggesting that Snf1 controls later stages of meiosis independently from the two controls on meiotic initiation. Because Snf1 is known to activate the expression of genes required for acetate metabolism, it may also serve to connect glucose and acetate controls on meiotic differentiation.
Diploid Saccharomyces cerevisiae may undergo either vegetative growth or meiosis and spore formation; the choice depends on the nutritional environment. Three criteria determine which path is taken (reviewed in reference 14). First, meiosis requires that cells be starved for at least one essential growth nutrient (16, 17). Second, meiosis depends on the presence of a nonfermentable carbon source, such as acetate (12, 20). Third, meiosis is blocked by the presence of a fermentable carbon source, such as glucose (36). The choice between meiosis and growth depends on each of these criteria. For example, acetate does not promote meiosis unless cells are also starved for an essential growth nutrient and glucose is absent.
One way in which these nutritional conditions regulate meiosis is by controlling the expression of the IME1 gene. The product of this gene is a transcription factor that is expressed very early in the meiotic program and that is required for initiation of this program; the Ime1 protein binds to and activates the expression of the IME2 gene (reviewed in reference 37). Nutritional controls on IME1 expression are mediated in part through the Ras-cyclic AMP pathway (34, 51); IME1 is also regulated by cell type such that only diploid cells may enter meiosis (8). A number of other genes involved in meiotic initiation have been identified—e.g., IME2, IME4, MCK1, RIM1, and UME6 (15, 31, 38, 40, 45, 53)—but it is not yet known which signal transduction pathways connect different nutrient signals to the choice between growth and meiosis or how these different signals are integrated.
Meiosis is characterized by two sequential phases of global genomic change (reviewed in reference 2). The first phase involves recombination between homologous chromosomes; the second involves two rounds of chromosome segregation to yield haploid products. The necessity for coordinating these two phases is revealed by yeast mutants that are defective in meiotic recombination (Rec− mutants) (reviewed in references 29 and 41). Many mutants with early defects in synapsis and/or recombination can still undergo chromosome segregation (e.g., spo11Δ, hop1Δ, mei4Δ, and rec104Δ), and this process invariably leads to high levels of chromosome nondisjunction, indicating that synapsis and/or recombination are required for proper meiotic segregation (1, 7, 22, 35). A second class of mutants (e.g., dmc1Δ, rad51Δ, and zip1Δ) begins but does not complete recombination (3, 47, 54). As a result, these cells arrest (or delay) meiosis before chromosome segregation. This arrest is thought to result from checkpoint functions that recognize specific recombination intermediates and delay the onset of meiotic segregation until replication and recombination are complete (18, 33, 58, 59). By this argument, the first class of Rec− mutants does not arrest in meiosis because these intermediates are not yet generated. In further support of the idea of meiotic checkpoints, meiotic arrest in dmc1Δ and zip1Δ mutants was found to be dependent on several genes (RAD17, RAD24, and MEC1) that are also required for a mitotic checkpoint function (33).
Despite the coordination between recombination and segregation, recent evidence suggests that nutrients control the two corresponding phases of meiosis separately (30). For example, cells expressing IME1 from a multicopy plasmid can initiate meiosis even when acetate is absent, but these cells still fail to complete the meiotic program. Specifically, these cells undergo chromosome replication, commitment to recombination, and the formation and dissolution of synaptonemal complexes; however, they arrest in meiosis before chromosome segregation and spore formation. Transfer of these arrested cells to sporulation medium (which contains acetate) releases the arrest, allowing the completion of meiosis. These results suggest that nutrients control the late phase of meiosis through a pathway distinct from their controls on IME1 expression and the early phase.
This report focuses on the repression of meiosis by glucose. Many of the responses to glucose in yeast are mediated through a signal transduction network referred to as the glucose repression pathway (reviewed in references 27 and 42); a central component of this pathway is Snf1 kinase. Glucose represses the transcription of a variety of genes (e.g., GAL1 and SUC2) by inactivating Snf1 kinase (25, 26). Previously, it was shown that snf1Δ mutants fail to form spores (6); here we extend this result to show that SNF1 is required for the induction of high levels of IME1 and IME2 transcripts under sporulation conditions. In addition, there is a separate requirement for Snf1 kinase in controlling the late stages of meiosis, suggesting that SNF1 may coordinately activate early and late phases of the meiotic program. Because SNF1 is required for the expression of genes involved in acetate metabolism, our results also suggest a link between glucose and acetate controls on meiosis.
MATERIALS AND METHODS
Yeast strains and plasmids.
All strains used in this study were isogenic relative to SH777, a W303 derivative with the following genotype: MATa/MATα ade2-1/ade2-1 can1R::ADE2::CAN1S/can1R::ADE2::CAN1S his3-11,13/his3-11,13 lys2(3′Δ)::HIS3::/lys2(5′Δ)/LYS2 trp1-1/trp1-3′Δ ura3-1/ura3-1 (30). The snf1Δ::URA3 strains were constructed by a one-step disruption with a KpnI-HindIII fragment of pST70 (provided by K. Tatchell, Louisiana State University Medical Center [55]); the disruption was verified by Southern blotting. YEp351-IME1 was constructed by inserting the BglII-BamHI fragment of the IME1 gene into the BamHI site of YEp351. The IME1 fragment used in this plasmid contains the complete open reading frame (ORF) but lacks negative regulatory regions present in the genomic copy (21). pS405 was constructed by inserting a 1.7-kb BglII-EcoRI fragment containing the IME2 ORF into the BamHI and EcoRI sites of pRS304 (48).
Media.
Synthetic complete (SC) medium, minimal (MIN) medium, and presporulation medium (YPA, a rich growth medium containing acetate as a carbon source) were described elsewhere (24, 43). Sporulation medium contained 2% potassium acetate and 0.17% yeast nitrogen base without amino acids and ammonium sulfate (YNB). Yeast cells do not require YNB for sporulation, but these components do not inhibit meiosis, and they are necessary to maintain viability in snf1Δ strains. Media lacking glucose, nitrogen, or both nutrients retained all other components of MIN medium. All growth and sporulation media were supplemented with leucine (100 mg/ml), tryptophan (50 mg/ml), and uracil (20 mg/ml) as necessary to complement auxotrophies. For strains bearing the YEp351-IME1 plasmid or the control plasmid (YEp351), the media used to assay recombination and commitment lacked leucine; thus, these measurements included only cells that retained the plasmid.
Growth and sporulation conditions.
Except as noted, growth and sporulation conditions were as follows. Cells were inoculated at a concentration of 2 × 105 cells/ml in 10 to 50 ml of growth medium. When the strains contained a plasmid bearing the LEU2 marker (YEp351 or YEp351-IME1), the growth medium was SC medium lacking leucine; when no plasmid was present, growth was in SC medium. Cells were grown for 36 h at 30°C with constant aeration, harvested, washed, transferred to an equal volume of YPA medium, and incubated for 4 h. Growth in YPA medium increases the efficiency and synchrony of the subsequent sporulation. Cells were harvested from YPA medium, washed, and transferred to sporulation medium or other media.
Assays for meiotic landmarks.
DNA replication was monitored by flow cytometry after cells had been sonicated, fixed in ethanol, and stained with propidium iodide (44). Flow cytometry was done with a Becton-Dickinson FACSCAN 4 apparatus, and the data were analyzed with CellFIT 2.0 software.
The frequency of intragenic recombination was measured as Trp+ prototrophs/CFU. These prototrophs result from recombination between trp1-1 and trp1-3′Δ heteroalleles. In addition, since LYS2 is disrupted with HIS3 on one copy of chromosome II, diploid recombinants can be specifically selected by plating on His− Lys− Trp− medium.
Intergenic recombination in the intervals from CEN3 to MATa/MATα and CEN2 to HIS3/LYS2 was determined by detecting loss of heterozygosity as described previously (23). In brief, recombination followed by mitotic segregation leads to cosegregation of recombinant and nonrecombinant chromatids (and loss of heterozygosity) 50% of the time. Thus, the expected frequency of loss of heterozygosity when cells undergo meiotic recombination and then return to the growth cycle is estimated as (0.5 · map distance)/100 cM.
Commitment to meiotic chromosome segregation was measured as described previously (30). In brief, the parent strain (SH777) contains a tandem duplication of a CAN1S allele and a can1R allele on each copy of chromosome V. Because the CAN1S allele confers sensitivity to the drug canavanine, efficient generation of CanR isolates requires two events: (i) recombination between the duplicated alleles on one copy of chromosome V, leading to a loss of the CAN1S allele on one chromatid, and (ii) meiotic chromosome segregation, leading to four haploid products, one of which will be CanR. Thus, the frequency of cells committed to meiotic segregation is directly proportional to the fraction that is resistant to canavanine. Spore formation was assayed by light microscopy. The meiotic divisions were monitored by staining nuclei with 4′,6-diamidino-2-phenylindole (DAPI) and visualizing mononucleate, binucleate, and tetranucleate cells with fluorescence microscopy (23). For both light microscopy and fluorescence microscopy, at least 300 cells were counted for each determination. All values for commitment to recombination, commitment to meiotic chromosome segregation, meiotic divisions, and spore formation given in this study are the averages of three experiments and are expressed as mean ± standard error of the mean.
Transcript measurement.
RNA was isolated by vortexing 2 × 108 yeast cells with glass beads and phenol as described previously (10). S1 nuclease protection was used to measure levels of the IME1, IME2, and DED1 transcripts. The DED1 transcript is found at constant levels throughout meiosis and growth and serves as a loading control. Both 32P-labeled probes were present in a 5 to 10-fold excess over the maximum level of the protected transcript. The probe for IME1 expression protected a 0.23-kb PstI-SacI region of the IME1 ORF, the IME2 probe protected a 0.3-kb EcoRI-BamHI region of the IME2 ORF, and the control probe protected a 0.25-kb BamHI-AflII region of the DED1 ORF. The probes were prepared by SP6 in vitro transcription of EcoRI-linearized pPL136 or AflII-linearized pDED1 (30) or by T3 in vitro transcription of BamHI-linearized pS405.
RESULTS
Both spore formation and meiotic recombination are inhibited by glucose.
Glucose is known to repress meiosis and sporulation in Saccharomyces cerevisiae. To separate this regulation from other nutritional controls on the meiotic program, we examined the effect of adding glucose to otherwise optimal sporulation medium (sporulation medium contains acetate and lacks nitrogen). Our results showed that even a relatively low concentration of glucose (0.4%) could dramatically inhibit the meiosis and sporulation pathway (Table 1, compare rows 2 and 3). Furthermore, when glucose was absent, spore formation was complete by 24 h; however, when glucose was present, spore formation was minimal even after 4 days (6% ± 3% of total cells were asci).
TABLE 1.
Effect of glucose or snf1Δ on meiosis
| Genotypea | Mediumb | Frequency of intragenic recombinants (10−5)c | % of cells forming sporesd |
|---|---|---|---|
| Wild type | Growth | 0.6 ± 0.2 | 0 |
| OAc | 60 ± 11 | 47 ± 2 | |
| OAc + Glu | 0.4 ± 0.1 | 0 | |
| snf1Δ | OAc | 0.4 ± 0.1 | 0 |
| OAc + Glu | 0.6 ± 0.2 | 0 | |
| Wild type(YEp-IME1) | OAc | 67 ± 2 | 48 ± 3 |
| Glu | 17 ± 3 | 6 ± 3 | |
| snf1Δ(YEp-IME1) | OAc | 4.2 ± 0.9 | 0 |
| Glu | 7.8 ± 0.8 | 0.2 ± 0.1 |
The wild type is SH777, snf1Δ is snf1Δ::URA3/snf1Δ::URA3, and wild type (YEp-IME1) and snf1Δ(YEp-IME1) contain the YEp351-IME1 plasmid. Strains are otherwise isogenic.
Growth, YPA medium; OAc, sporulation medium; OAc + Glu, sporulation medium with 0.4% glucose added; Glu, 2% glucose.
Commitment to intragenic recombination between trp1 heteroalleles (Trp+ prototrophs/CFU) after 36 h in growth medium or after an additional 24 h in OAc, OAc + Glu, or Glu. Percentages of cell viability for each of the nine cultures were as follows (from top to bottom): 38 ± 2, 37 ± 1, 49 ± 1, 33 ± 1, 38 ± 3, 34 ± 5, 30 ± 6, 33 ± 1, and 19 ± 1.
Percentages of the total population of cells that formed asci, measured at the same times as recombination.
Glucose might prevent spore formation, the final step in the meiotic pathway in yeast, by blocking any previous stage in the program. To define where this block was occurring, a relatively early event in meiosis, commitment to DNA recombination, was monitored; this event can be detected even if later stages of meiosis are blocked (see Materials and Methods). As expected, high levels of commitment to intragenic recombination at the trp1 locus occurred in sporulation medium. In contrast, only background levels of recombination were evident when 0.4% glucose was added to sporulation medium (Table 1, compare rows 2 and 3). These results suggest that glucose blocks meiosis at an early stage, before commitment to recombination.
Full induction of the IME1 transcript is prevented by glucose.
Since IME1 is required for the initiation of meiosis and induction of the IME1 transcript is the first detectable event in the meiotic program, the effect of glucose and other nutrients on IME1 transcript levels was examined (Fig. 1A). Diploid cells from a mid-log-phase culture were transferred to sporulation medium or to various other media and incubated with constant shaking. Samples were removed at various times, and IME1 transcript levels were determined. As reported previously (28), the IME1 transcript was undetectable in cells from mid-log-phase cultures (Fig. 1A, lane 2) but were induced to high levels several hours after transfer to sporulation medium (lanes 3 to 5). As a negative control, samples transferred instead to growth medium for the same amounts of time did not show a detectable level of the IME1 transcript (Fig. 1A, lanes 6 to 8).
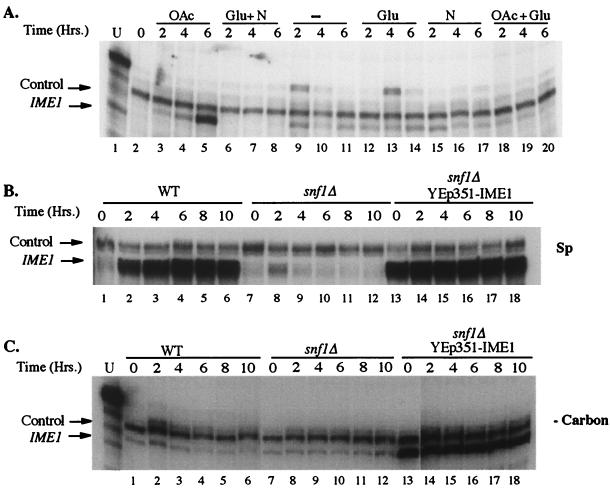
FIG. 1.
Effect of nutritional conditions and the snf1Δ mutation on IME1 transcript levels. Cells were assayed for the presence of IME1 and control (DED1) RNAs by S1 nuclease protection. (A) Log-phase wild-type yeast cells were transferred to different media for the indicated times. Lane 1, undigested probe at 10% of the amount used in the protection assays; lane 2, log-phase cells; lanes 3 to 5, sporulation medium; lanes 6 to 8, SC medium; lanes 9 to 11, YNB; lanes 12 to 14, 2% glucose; lanes 15 to 17, 0.5% ammonium sulfate; lanes 18 to 20, sporulation medium with 1% glucose. (B) Cells were transferred to sporulation medium (Sp) for the indicated times. Lanes 1 to 6, wild type; lanes 7 to 12, snf1Δ; lanes 13 to 18, snf1Δ plus YEp351-IME1. (C) Same as panel B, except that cells were transferred to medium containing only YNB (i.e, lacking a carbon source [−Carbon]) and the first lane contained undigested probe at 10% of the amount used in protection assays.
Although the IME1 transcript accumulated to high levels when cells were exposed to acetate alone (Fig. 1A, lanes 3 to 5), IME1 was only expressed to moderate levels when glucose and acetate were both present (lanes 18 to 20). This result suggests that glucose blocks the ability of acetate to induce the IME1 transcript. Interestingly, moderate levels of the IME1 transcript were also observed in cells exposed to any medium that promoted neither growth nor sporulation. Specifically, these media lacked a nonfermentable carbon source (e.g., acetate), which is essential for meiosis, and a second nutrient (e.g., nitrogen), which is essential for growth. For example, cells placed in medium containing only glucose (Fig. 1A, lanes 12 to 14) or medium containing only nitrogen (lanes 15 to 17) displayed moderate levels of the IME1 transcript, and the same was true for cells placed in medium containing neither a carbon nor a nitrogen source (lanes 9 to 11). As expected, none of these conditions promoted either growth or sporulation. A comparison of different autoradiograph exposures indicated that the moderate level of the IME1 transcript was approximately 5 to 10% of the level seen under sporulation conditions. These results are consistent with earlier studies suggesting that IME1 transcription is promoted by both respiration, which is induced by nonfermentable carbon sources, and cell cycle arrest, which is induced by starvation for nutrients (46, 52, 56). Our experiments do not yet distinguish whether the moderate levels of the IME1 transcript that we observed reflected equal expression in all cells or higher expression in a subpopulation.
In summary, there may be two different controls of IME1 transcript levels: in growing cells, the IME1 transcript is completely repressed, whereas in nongrowing cells, glucose and/or acetate determine whether moderate or high levels of the transcript accumulate.
Snf1 kinase is required for the initiation of meiosis.
Snf1 kinase is repressed by glucose (25, 26) and is required for spore formation (6), suggesting the possibility that glucose prevents the accumulation of high levels of the IME1 transcript by repressing Snf1 activity. We confirmed that a snf1Δ::URA3/snflΔ::URA3 mutant (referred to in this paper as snf1Δ) is defective in spore formation (Table 1, row 4). In addition, we determined that the snf1Δ mutant does not undergo either meiotic DNA replication (Fig. 2A) or recombination in sporulation medium (Table 1, row 4). Thus, the effect of deleting the SNF1 gene is similar to the effect of adding glucose to wild-type cells; this correlation suggests that glucose may repress meiosis by inhibiting Snf1. Consistent with this idea, the snf1Δ mutant does not initiate meiosis any better when glucose is absent than when it is present (Table 1, compare rows 4 and 5).
FIG. 2.
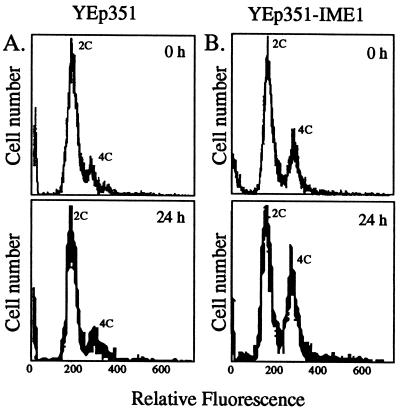
Effect of IME1 overexpression on DNA content in a snf1Δ mutant. To ensure that most cells started in G1, cultures were grown in SC medium lacking leucine for 36 h, sonicated, and incubated in the spent medium for a further 12 h. Cells were transferred to sporulation medium, and DNA content was analyzed by flow cytometry after 0 h (upper panels) or 24 h (lower panels). Peaks represent unreplicated (2C) or fully replicated (4C) DNA. (A) snf1Δ cells containing YEp351 (control plasmid); (B) snf1Δ cells containing YEp351-IME1.
Since early and late meiotic events do not occur in the snf1Δ strain, we examined whether the IME1 transcript can be induced in this mutant. In contrast to the induction of the IME1 transcript seen in wild-type cells (Fig. 1B, lanes 1 to 6), the IME1 transcript did not accumulate to high levels in the snf1Δ mutant (lanes 7 to 12). A moderate level of IME1 transcript was detected reproducibly in the mutant after 2 h in sporulation medium (Fig. 1B, lane 8) and then disappeared at later times (lanes 9 to 12). The moderate level of IME1 transcript expressed under these conditions was similar to the levels seen for wild-type cells that were neither growing nor sporulating (Fig. 1A, lanes 9 to 20).
Because moderate levels of the IME1 transcript were also observed in wild-type cells in the absence of any carbon source (Fig. 1A, lanes 9 to 11), we examined IME1 expression in the snf1Δ mutant under the same conditions (Fig. 1C, lanes 7 to 12). We found that IME1 transcripts were present at moderate but stable levels in the snf1Δ mutant. These levels of IME1 transcript were similar to the levels observed in wild-type cells under the same conditions (Fig. 1C, lanes 1 to 6). Thus, neither the deletion of SNF1 (Fig. 1C) nor the addition of glucose (Fig. 1A, lanes 12 to 14) affected the moderate IME1 expression that occurred in the absence of carbon. Interestingly, IME1 transcript levels in the snf1Δ mutant were actually more stable in the absence of any carbon source than in sporulation medium (compare Fig. 1B and C). Thus, when SNF1 is deleted such that IME1 cannot be induced, sporulation conditions may actually destabilize IME1 transcript levels.
Snf1 kinase has a second and nonessential role in meiotic initiation.
Is the regulation of IME1 transcript levels the only target for Snf1 kinase in meiosis? To examine the effect of snf1Δ on later aspects of meiosis, a multicopy plasmid bearing the IME1 gene, YEp351-IME1, was placed in the snf1Δ mutant (see Materials and Methods). The plasmid caused high levels of IME1 transcript to be expressed in both growth and sporulation cultures of this mutant (Fig. 1B, lanes 13 to 18). These high levels of IME1 transcript allowed 15 to 20% of snf1Δ cells to undergo DNA replication in sporulation medium (Fig. 2B). In addition, recombination frequency was increased 10-fold relative to that of the snf1Δ mutant not containing the plasmid (Table 1, compare rows 4 and 8) or containing only the vector (data not shown). The frequency of recombination observed in snf1Δ(YEp351-IME1) cells was approximately 10 to 20% that observed in wild-type cells. Thus, when the requirement for Snf1 to induce the IME1 transcript was bypassed, the initiation of meiosis occurred in 10 to 20% of snf1Δ cells.
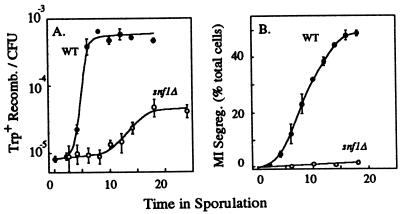
Because the levels of replication and recombination achieved in snf1Δ(YEp351-IME1) cells were only 10 to 20% of those obtained in wild-type cells, we compared the timing of recombination in these two types of cells (Fig. 3A). Recombination was delayed in the snf1Δ strain by 10 to 15 h relative to the SNF1+ control, and the maximum level of recombination achieved was again approximately 10-fold lower in the snf1Δ mutant than in the wild type. The delayed timing and diminished frequency of recombination in the snf1Δ mutant, which occurred even when high levels of IME1 transcript were present, suggested that in addition to being required for the induction of the IME1 transcript, Snf1 is required for some subsequent step in meiotic initiation. As described later, this subsequent step is required for induction of the IME2 transcript. Furthermore, this second role for Snf1 in meiotic initiation is not absolutely required, since some cells can initiate meiosis even though SNF1 is deleted.
FIG. 3.
Effect of the YEp351-IME1 plasmid on recombination (Trp+ Recomb.) and meiosis I chromosome segregation (MI Segreg.) in the snf1Δ strain. At various times in sporulation medium, the wild-type (WT) (•) or the snf1Δ (○) strain was assayed for recombination at the trp1 locus as the frequency of the Trp+ prototrophs in the culture (A) or for meiosis I chromosome segregation as the percentage of cells in binucleate or later stages of meiosis (B). The data shown are the means of three determinations, and the error bars represent the standard errors of the means.
Crossover recombination is rare during vegetative growth and very common during meiosis. To verify that the increase in the number of Trp+ recombinants in the snf1Δ(YEp351-IME1) strain indeed resulted from commitment to meiotic recombination, we examined these Trp+ colonies for evidence of crossover recombination at two different intervals (CEN2 to HIS3/LYS2 and CEN3 to MATa/MATα); both of these intervals are unlinked to TRP1. Recombination in the interval from CEN2 to HIS3/LYS2 was detected in 10% of the Trp+ colonies tested (26 of 259), and recombination in the interval from CEN3 to MATa/MATα was detected in 6.3% of the colonies (17 to 270). As a control, Trp+ colonies from a stationary-phase wild-type culture were also examined. As expected, since crossover recombination occurs relatively infrequently in vegetative cells, no crossover recombination was detected at either interval among 341 Trp+ colonies tested. Furthermore, crossover recombination depended on IME1 expression; a snf1Δ mutant containing only the YEp351 vector yielded no evidence of recombination in either interval among 120 Trp+ colonies tested. Since the frequency of crossover recombination was much higher in the snf1Δ(YEp351-IME1) strain than in either control culture, this recombination likely derived from the meiotic pathway. Unexpectedly, crossover recombination in the snf1Δ(YEp351-IME1) strain occurred two to three times less often than predicted (25% recombination for CEN2 to HIS3/LYS2 and 14% recombination for CEN3 to MATa/MATα) based on known map distances (see Materials and Methods). It is possible that even in the snf1Δ(YEp351-IME1) cells that initiate meiosis (i.e., that give rise to Trp+ colonies), crossover recombination occurs two to three times less efficiently than it does in wild-type cells.
Effects of glucose and Snf1 on the expression of the IME2 gene.
The above results suggest the possibility that glucose can repress meiotic initiation even after the IME1 transcript has accumulated. One potential candidate for this later control is transcriptional activation of the IME2 gene. As described above, Ime1 is a transcription factor that directly activates IME2 transcription (5, 50). To test whether Snf1 controls IME2 transcription separately from its effect on IME1 expression, we first measured IME2 transcript accumulation in wild-type and snf1Δ cells under different conditions. As expected from previous studies (51, 60), the IME2 transcript was not expressed at detectable levels during growth (Fig. 4, lane 2) and was strongly induced after transfer to sporulation conditions (lanes 3 and 4). Furthermore, like IME1, IME2 was expressed at moderate levels when no carbon source was present (Fig. 4, lanes 5 and 6, bottom). However, the addition of glucose repressed IME2 transcript levels (unlike IME1 transcript levels) below the levels observed in the absence of a carbon source (compare Fig. 1A, lanes 9 to 14, to Fig. 4, lanes 5 to 8). Consistent with this experiment, deletion of SNF1 did not affect IME1 transcript levels when acetate was absent (Fig. 1C, lanes 1 to 12), whereas this deletion did affect IME2 transcript levels, whether acetate was present or not (Fig. 4, lanes 13 to 14, and data not shown). These results suggest that Snf1 is involved in IME2 transcription separately from its role in activating accumulation of the IME1 transcript.
FIG. 4.
Effect of nutritional conditions, IME1 overexpression, and the snf1Δ mutation on IME2 transcript levels. RNA was extracted from various cultures, and the amounts of IME2 and control (DED1) transcripts were measured by S1 nuclease protection. (Top) 1-day exposure. (Bottom) 7-day exposure. Lane 1, control reaction in which RNA was omitted; lanes 2 to 8, wild-type cells containing the YEp351 vector in the log phase (lane 2) and 3 or 6 h after transfer to sporulation medium (lanes 3 and 4), to medium lacking carbon and nitrogen (lanes 5 and 6), or to sporulation medium containing 0.4% glucose (lanes 7 and 8); lanes 9 to 12, wild-type cells containing the YEp351-IME1 plasmid 3 or 6 h after transfer to medium lacking both carbon and nitrogen (lanes 9 and 10) or to medium containing 2% glucose (lanes 11 and 12); lanes 13 and 14, snf1Δ mutant cells containing the YEp351 vector 3 or 6 h after transfer to sporulation medium; lanes 15 to 18, snf1Δ mutant cells containing the YEp351-IME1 plasmid 3 or 6 h after transfer to sporulation medium (lanes 15 and 16) or to medium containing 2% glucose (lanes 17 and 18); lane 19, undigested probe at 5% of the concentration used in the protection assays.
We also measured IME2 expression in the snf1Δ mutant when the normal controls on IME1 transcript levels were bypassed. As mentioned above, expression of IME1 from a plasmid activated maximum levels of the IME1 transcript in the snf1Δ mutant, even in the absence of a carbon source (Fig. 1C, lanes 13 to 18). In the snf1Δ mutant under sporulation conditions, the IME2 transcript was also induced by the YEp351-IME1 plasmid (Fig. 4, compare lanes 13 and 14 to lanes 15 and 16); nevertheless, IME2 transcript levels in these cells were approximately 10-fold lower than those observed in SNF1+ cells (lanes 3 and 4). These results support the idea of glucose repression operating not only at the level of IME1 transcript accumulation but also between induction of the IME1 transcript and induction of the IME2 transcript.
When IME1 was expressed from the multicopy plasmid, glucose inhibited IME2 transcript levels to approximately the same degree as the deletion of SNF1 inhibited this transcript (Fig. 4, lanes 11, 12, 15, and 16) and, importantly, these effects were not additive (lanes 17 and 18). This latter result strongly suggests that the primary effect of glucose on IME2 transcript accumulation is through the repression of Snf1 activity.
As described above, when glucose was added to sporulation medium or when the SNF1 gene was deleted, IME1 and IME2 transcripts were expressed at moderate levels. Interestingly, these moderate levels of expression were equal to the levels expressed in cells deprived of acetate. Thus, the inactivation of Snf1 by glucose may block meiotic initiation primarily by preventing acetate from inducing IME1 and IME2 transcription.
Snf1 kinase is required independently for the initiation and the completion of meiosis.
Although the YEp351-IME1 plasmid can partially suppress the requirement for Snf1 in meiotic replication and recombination, we found that this plasmid was not sufficient to allow snf1Δ cells to form spores. In the control strain, approximately half of the cells formed asci by 24 h in sporulation medium; however, in the snf1Δ(YEp351-IME1) strain, no asci were observed by 24 h (Table 1, compare rows 6 and 8) or 5 days (data not shown). In addition, the IME1 plasmid was not sufficient to allow detectable levels of chromosome segregation in the snf1Δ mutant, as measured by observation of the number of nuclear masses per cell (Fig. 3B).
Two additional genetic tests confirmed that the snf1Δ mutant did not induce late stages of meiosis. First, we used an assay for commitment to the completion of meiosis (see Materials and Methods). This test revealed that, after 24 h in sporulation medium, the frequency of committed cells in the snf1Δ mutant was only 0.01% of the frequency in the SNF1+ control [(4.4 ± 0.1) × 10−6 and (4.8 ± 0.3) × 10−2 CanR cells/CFU for the snf1Δ mutant and the wild type, respectively)]. Second, we measured the decline in the number of diploid recombinants which occurs as cells undergo meiotic chromosome segregation to form haploids. In the SNF1+ control, by 24 h the frequency of diploid Trp+ recombinants/CFU [(5.4 ± 0.7) × 10−6] had declined to only approximately 1% of the total Trp+ recombination frequency (Table 1, row 6). Conversely, in the snf1Δ mutant, the frequency of diploid Trp+ recombinants/CFU [(3.4 ± 0.8) × 10−5] at this time was approximately the same as the total Trp+ recombinants/CFU (Table 1, row 8), indicating that little or no formation of haploids had occurred.
Roles of glucose and Snf1 in late controls on meiosis.
Because glucose acts through Snf1 in repressing IME1 transcript accumulation, we examined whether it also acts through Snf1 in later controls on meiosis. Glucose controls on IME1 transcript accumulation were bypassed when IME1 was expressed from the multicopy plasmid (Fig. 1B, lane 13) (30). Nevertheless, the addition of glucose to wild-type cells bearing this plasmid decreased the levels of both recombination and spore formation (Table 1, compare rows 6 and 7). That is, the addition of glucose to a strain bearing the YEp351-IME1 plasmid inhibited meiosis in the same way as the deletion of SNF1 (row 8).
Because glucose and the snf1Δ mutation were equally effective in repressing the IME1 transcript, it was surprising that later controls on meiosis were inhibited much more strongly by snf1Δ than by glucose (Table 1, compare rows 7 and 8). An explanation is suggested by comparing the effects of glucose and sporulation medium on commitment to recombination in the snf1Δ(YEp351-IME1) mutant; strikingly, this strain initiated meiosis twice as efficiently in the presence of glucose as it did in sporulation medium (Table 1, compare rows 8 and 9). These results can be explained if glucose has two opposing roles in later controls on meiosis. As discussed above, its major role is to repress Snf1 activity. Deleting the SNF1 gene obviates this role, revealing a second effect of glucose on meiosis, which is stimulatory. For example, the snf1Δ mutant metabolizes glucose more efficiently than acetate (11), so it is possible that the energy derived from glucose metabolism stimulates meiosis (at least weakly) in this mutant. In wild-type cells, the strong inhibitory effect of glucose on meiosis would outweigh its relatively modest stimulatory effect.
DISCUSSION
Glucose inhibits the initiation of meiosis by repressing Snf1.
Glucose has two independent roles in blocking the initiation of meiosis: first, it blocks induction of the IME1 transcript, and second, it inhibits induction of the IME2 transcript. Strikingly, both roles are paralleled by the phenotype of the snf1Δ mutation. These results, together with the recent finding that glucose blocks the activity of Snf1 kinase (26), strongly suggest that glucose prevents the initiation of meiosis primarily through inactivating Snf1. A direct test of this idea was possible for glucose control on IME2 transcription. This control was separated from the earlier control by overexpression of IME1 from a plasmid, thus bypassing the first control. Under these conditions, either the addition of glucose or the deletion of SNF1 partially inhibited both induction of the IME2 transcript and initiation of meiosis. Significantly, a snf1Δ mutant exposed to glucose was no more repressed than the same mutant under optimal sporulation conditions. This result strongly suggests that glucose acts primarily through the repression of Snf1 activity, at least with respect to the control of IME2 expression. Earlier studies demonstrated that IME1 is regulated not only transcriptionally but also through posttranslational modification (4, 46), and recent evidence suggests that glucose blocks the interaction of Ime1 and another transcription factor, Ume6 (57). Thus, it is possible that Snf1 kinase regulates IME2 transcription by directly or indirectly controlling the posttranslational activation of Ime1.
Snf1 kinase links glucose control and acetate control on meiosis.
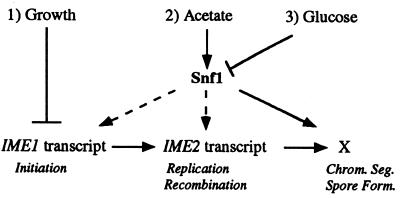
How are different controls on the same differentiation program coordinated? In budding yeast, three separate criteria determine the choice between growth and meiosis (Fig. 5): growth conditions repress meiosis, the presence of glucose represses meiosis, and nonfermentable carbon sources (such as acetate) stimulate meiosis. These criteria converge on at least three different targets—IME1 transcription, IME2 transcription, and an unknown regulator that controls entry into the late phase of meiosis.
FIG. 5.
Model for the role of Snf1 kinase in coordinating glucose and acetate controls on early and late phases of meiotic differentiation. Negative regulation is represented by perpendicular lines, and positive regulation is indicated by arrows; broken arrows indicate that activation is not fully dependent on the upstream signal. X, unidentified regulatory gene(s). Meiosis is controlled by at least three different signaling pathways: (i) growth—in growing cells, IME1 transcript levels are fully repressed; (ii) acetate—acetate or other nonfermentable carbon sources increase IME1 and IME2 transcript levels, leading to early meiotic events, such as DNA replication and recombination; acetate separately activates late meiotic events (chromosome segregation and spore formation); and both early regulation and late regulation by acetate requires Snf1 kinase; (iii) glucose—glucose represses meiosis at both early and late stages by repressing Snf1 kinase activity. Other nutritional controls on meiosis, in addition to the ones shown on the diagram, are also possible. Chrom. Seg., chromosome segregation; Form., formation.
Snf1 is required for the transcription of a large number of genes, in particular, the genes needed for gluconeogenesis and respiratory growth (reviewed in references 27 and 42). These metabolic pathways are essential for the utilization of nonfermentable carbon sources such as acetate, and it has also been shown that the genes in these pathways are required for sporulation. Indeed, respiration is required for IME1 expression (56). Thus, a simple model for the interaction of glucose control and acetate control on meiosis is that glucose represses meiosis by inactivating Snf1 kinase, hence blocking acetate metabolism (Fig. 5). That is, the glucose repression pathway serves as a gate for the signaling pathway by which acetate induces the meiotic program. Our results do not address whether other carbon sources (e.g., galactose or glycerol) may regulate meiosis through additional signaling pathways.
Snf1 kinase may connect the regulation of early stages of meiosis to the regulation of the late stages.
In theory, differentiation programs could be regulated by extracellular signals entirely at the initiation of the program. That is, initiation could trigger an obligatory progression of different cellular events which follow one another until differentiation is complete. However, the control of meiosis in S. cerevisiae clearly does not follow this simple paradigm: if cells are placed in sporulation medium long enough to progress through the early stages of meiosis (DNA replication and meiotic recombination) and then transferred back into growth medium, they reenter the growth cycle without undergoing the later stages (meiotic chromosome segregation and spore formation) (19). The completion of meiosis only becomes obligatory (termed commitment to meiosis) at approximately the same time as the initiation of chromosome segregation (13, 30, 49). The reversibility of meiotic differentiation throughout the early stages results, in part, from separate nutritional controls on early and late stages (30). Here we suggest that both of these phases are controlled independently by the same signal transduction component—Snf1 kinase (Fig. 5).
Once meiotic replication and recombination have initiated, checkpoint functions ensure that segregation and spore formation are not induced until these earlier events are completed. However, as described above, entry into the late stages of meiosis also requires the continued presence of appropriate nutritional signals. As a result, early and late meiotic events are coordinately regulated but are not interdependent. The dual requirement for Snf1 at both early and late phases of meiosis may allow nutritional signals to coordinately control both phases.
Snf1 mediates many different cellular responses to glucose, and the different targets of these responses are controlled by different effectors. For example, Mig1 is a transcriptional repressor which is inactivated by Snf1 and which acts on some but not all genes repressed by glucose. As a result, a snf1Δ mig1Δ double mutant is able to grow almost normally on galactose but is still unable to grow on gluconeogenic sources such as raffinose (42). It is possible that different effectors of Snf1 kinase act at each of the different stages of meiosis. In this regard, it is interesting to note that the regulatory region of IME1 contains a putative binding site for Mig1, whereas the upstream region of IME2 does not. The Mig1 site in the IME1 gene is 1.7 kb upstream of the start codon and has the sequence ATTTACGCGGGG, which matches the consensus sequence [(A/T)5N(G/C)PyG4] defined by homology, DNA footprints, and mutational analysis (32). This site is within a 2.2-kb region that has been identified as being involved in the nutritional regulation of IME1 (21).
During development in complex organisms, cross talk between different signal transduction pathways allows specialized cell fates to be chosen from among multiple possibilities (e.g., 9, 39). In S. cerevisiae, a related situation exists; a single binary choice between meiosis and growth is precisely regulated under a wide range of extracellular conditions. The work presented in this paper suggests that this precise regulation may be accomplished by combinatorial interactions among relatively few signaling pathways.
ACKNOWLEDGMENTS
We are grateful to K. Tatchell for plasmids, N. Gonchoroff for flow cytometry, E. Maine and N. Kleckner for comments on the manuscript and helpful suggestions, T. Schedl for stimulating this study, and C. Raymond and P. Dominguez for technical support.
REFERENCES
- 1.Atcheson C L, DiDomenico B, Frackman S, Esposito R E, Elder R T. Isolation, DNA sequence, and regulation of a meiosis-specific eukaryotic recombination gene. Proc Natl Acad Sci USA. 1987;84:8035–8039. doi: 10.1073/pnas.84.22.8035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baker B S, Carpenter A T, Esposito M S, Esposito R E, Sandler L. The genetic control of meiosis. Annu Rev Genet. 1976;10:53–134. doi: 10.1146/annurev.ge.10.120176.000413. [DOI] [PubMed] [Google Scholar]
- 3.Bishop D K, Park D, Xu L, Kleckner N. DMC1: a meiosis-specific yeast homolog of E. coli recA required for recombination, synaptonemal complex formation, and cell cycle progression. Cell. 1992;69:439–456. doi: 10.1016/0092-8674(92)90446-j. [DOI] [PubMed] [Google Scholar]
- 4.Bowdish K S, Yuan H E, Mitchell A P. Analysis of RIM11, a yeast protein kinase that phosphorylates the meiotic activator IME1. Mol Cell Biol. 1994;14:7909–7919. doi: 10.1128/mcb.14.12.7909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bowdish K S, Yuan H E, Mitchell A P. Positive control of yeast meiotic genes by the negative regulator UME6. Mol Cell Biol. 1995;15:2955–2961. doi: 10.1128/mcb.15.6.2955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Carlson M, Osmond B C, Botstein D. Mutants of yeast defective in sucrose utilization. Genetics. 1981;98:25–40. doi: 10.1093/genetics/98.1.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cool M, Malone R E. Molecular and genetic analysis of the yeast early meiotic recombination genes REC102 and REC107/MER2. Mol Cell Biol. 1992;12:1248–1256. doi: 10.1128/mcb.12.3.1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Covitz P A, Mitchell A P. Repression by the yeast meiotic inhibitor RME1. Genes Dev. 1993;7:1598–1608. doi: 10.1101/gad.7.8.1598. [DOI] [PubMed] [Google Scholar]
- 9.Duffy J B, Perrimon N. Recent advances in understanding signal transduction pathways in worms and flies. Curr Opin Cell Biol. 1996;8:231–238. doi: 10.1016/s0955-0674(96)80070-6. [DOI] [PubMed] [Google Scholar]
- 10.Elder R T, Loh E Y, Davis R W. RNA from yeast transposable element Ty1 has both ends in the direct repeats, a structure similar to retrovirus RNA. Proc Natl Acad Sci USA. 1983;80:2432–2436. doi: 10.1073/pnas.80.9.2432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Entian K D, Zimmermann F K. New genes involved in carbon catabolite repression and derepression in the yeast Saccharomyces cerevisiae. J Bacteriol. 1982;151:1123–1128. doi: 10.1128/jb.151.3.1123-1128.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Esposito M S, Esposito R E, Arnaud M, Halvorson H O. Acetate utilization and macromolecular synthesis during sporulation of yeast. J Bacteriol. 1969;100:180–186. doi: 10.1128/jb.100.1.180-186.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Esposito R E, Esposito M S. Genetic recombination and commitment to meiosis in Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 1974;71:3172–3176. doi: 10.1073/pnas.71.8.3172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Esposito R E, Klapholz S. Meiosis and ascospore development. In: Strathern J N, Jones E W, Broach J R, editors. Molecular biology of the yeast Saccharomyces: life cycle and inheritance. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1981. pp. 211–287. [Google Scholar]
- 15.Foiani M, Nadjar-Boger E, Capone R, Sagee S, Hashimshoni T, Kassir Y. A meiosis-specific protein kinase, Ime2, is required for the correct timing of DNA replication and for spore formation in yeast meiosis. Mol Gen Genet. 1996;253:278–288. doi: 10.1007/s004380050323. [DOI] [PubMed] [Google Scholar]
- 16.Freese E B, Chu M I, Freese E. Initiation of yeast sporulation by partial carbon, nitrogen, or phosphate deprivation. J Bacteriol. 1982;149:840–851. doi: 10.1128/jb.149.3.840-851.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Freese E B, Olempska B Z, Hartig A, Freese E. Initiation of meiosis and sporulation of Saccharomyces cerevisiae by sulfur or guanine deprivation. Dev Biol. 1984;102:438–451. doi: 10.1016/0012-1606(84)90209-4. [DOI] [PubMed] [Google Scholar]
- 18.Galbraith A M, Bullard S A, Jiao K, Nau J J, Malone R E. Recombination and the progression of meiosis in Saccharomyces cerevisiae. Genetics. 1997;146:481–489. doi: 10.1093/genetics/146.2.481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ganesan A T, Holter H, Roberts C. Some observations on sporulation in Saccharomyces. C R Trav Lab Carlsberg. 1958;13:1–6. [PubMed] [Google Scholar]
- 20.Gorts C P. Role of acetate metabolism in sporulation of Saccharomyces carlsbergensis. Antonie Leeuwenhoek. 1975;41:265–271. doi: 10.1007/BF02565062. [DOI] [PubMed] [Google Scholar]
- 21.Granot D, Margolskee J P, Simchen G. A long region upstream of the IME1 gene regulates meiosis in yeast. Mol Gen Genet. 1989;218:308–314. doi: 10.1007/BF00331283. [DOI] [PubMed] [Google Scholar]
- 22.Hollingsworth N M, Byers B. HOP1: a yeast meiotic pairing gene. Genetics. 1989;121:445–462. doi: 10.1093/genetics/121.3.445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Honigberg S, Esposito R E. Reversal of cell determination in yeast meiosis: post-commitment arrest allows return to mitotic growth. Proc Natl Acad Sci USA. 1994;91:6559–6563. doi: 10.1073/pnas.91.14.6559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Honigberg S M, Conicella C, Esposito R E. Commitment to meiosis in Saccharomyces cerevisiae: involvement of the SPO14 gene. Genetics. 1992;130:703–716. doi: 10.1093/genetics/130.4.703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jiang R, Carlson M. Glucose regulates protein interactions within the yeast Snf1 protein kinase complex. Genes Dev. 1996;10:3105–3115. doi: 10.1101/gad.10.24.3105. [DOI] [PubMed] [Google Scholar]
- 26.Jiang R, Carlson M. The Snf1 protein kinase and its activating subunit, Snf4, interact with distinct domains of the Sip1/Sip2/Gal83 component in the kinase complex. Mol Cell Biol. 1997;17:2099–2106. doi: 10.1128/mcb.17.4.2099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Johnston H M, Carlson M. Regulation of carbon and phosphate utilization. In: Jones E W, Pringle J R, Broach J R, editors. The molecular and cellular biology of the yeast Saccharomyces: gene expression. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1992. pp. 193–282. [Google Scholar]
- 28.Kassir Y, Granot D, Simchen G. IME1, a positive regulator of meiosis in S. cerevisiae. Cell. 1988;52:853–862. doi: 10.1016/0092-8674(88)90427-8. [DOI] [PubMed] [Google Scholar]
- 29.Kupiec M, Byers B, Esposito R E, Mitchell A P. Meiosis and sporulation in Saccharomyces cerevisiae. In: Pringle J R, Broach J R, Jones E W, editors. The molecular and cellular biology of the yeast Saccharomyces: cell cycle and cell biology. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1997. pp. 889–1036. [Google Scholar]
- 30.Lee R H, Honigberg S M. Nutritional regulation of late meiotic events in Saccharomyces cerevisiae through a pathway distinct from initiation. Mol Cell Biol. 1996;16:3222–3232. doi: 10.1128/mcb.16.6.3222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li W, Mitchell A P. Proteolytic activation of Rim1p, a positive regulator of yeast sporulation and invasive growth. Genetics. 1997;145:63–73. doi: 10.1093/genetics/145.1.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lundin M, Nehlin J O, Ronne H. Importance of a flanking AT-rich region in target site recognition by the GC box-binding zinc finger protein Mig1. Mol Cell Biol. 1994;14:1979–1985. doi: 10.1128/mcb.14.3.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lydall D, Nikolsky Y, Bishop D K, Weinert T. A meiotic recombination checkpoint controlled by mitotic checkpoint genes. Nature. 1996;383:840–843. doi: 10.1038/383840a0. [DOI] [PubMed] [Google Scholar]
- 34.Matsuura A, Treinin M, Mitsuzawa H, Kassir Y, Uno I, Simchen G. The adenylate cyclase/protein kinase cascade regulates entry into meiosis in Saccharomyces cerevisiae through the gene IME1. EMBO J. 1990;9:3225–3232. doi: 10.1002/j.1460-2075.1990.tb07521.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Menees T M, Roeder G S. MEI4, a yeast gene required for meiotic recombination. Genetics. 1989;123:675–682. doi: 10.1093/genetics/123.4.675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Miller J J. Sporulation in Saccharomyces cerevisiae. In: Rose A H, Harrison J S, editors. The yeasts. San Diego, Calif: Academic Press Ltd.; 1989. pp. 489–550. [Google Scholar]
- 37.Mitchell A P. Control of meiotic gene expression in Saccharomyces cerevisiae. Microbiol Rev. 1994;58:56–70. doi: 10.1128/mr.58.1.56-70.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mitchell A P, Driscoll S E, Smith H E. Positive control of sporulation-specific genes by the IME1 and IME2 products in Saccharomyces cerevisiae. Mol Cell Biol. 1990;10:2104–2110. doi: 10.1128/mcb.10.5.2104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Moon R T, Brown J D, Torres M. WNTs modulate cell fate and behavior during vertebrate development. Trends Genet. 1997;13:157–162. doi: 10.1016/s0168-9525(97)01093-7. [DOI] [PubMed] [Google Scholar]
- 40.Neigeborn L, Mitchell A P. The yeast MCK1 gene encodes a protein kinase homolog that activates early meiotic gene expression. Genes Dev. 1991;5:533–548. doi: 10.1101/gad.5.4.533. [DOI] [PubMed] [Google Scholar]
- 41.Roeder G S. Meiotic chromosomes: it takes two to tango. Genes Dev. 1997;11:2600–2621. doi: 10.1101/gad.11.20.2600. [DOI] [PubMed] [Google Scholar]
- 42.Ronne H. Glucose repression in fungi. Trends Genet. 1995;11:12–17. doi: 10.1016/s0168-9525(00)88980-5. [DOI] [PubMed] [Google Scholar]
- 43.Rose M D, Winston F, Hieter P. Methods in yeast genetics: a laboratory course manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1990. [Google Scholar]
- 44.Sazer S, Sherwood S W. Mitochondrial growth and DNA synthesis occur in the absence of nuclear DNA replication in fission yeast. J Cell Sci. 1990;97:509–516. doi: 10.1242/jcs.97.3.509. [DOI] [PubMed] [Google Scholar]
- 45.Shah J C, Clancy M J. IME4, a gene that mediates MAT and nutritional control of meiosis in Saccharomyces cerevisiae. Mol Cell Biol. 1992;12:1078–1086. doi: 10.1128/mcb.12.3.1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sherman A, Shefer M, Sagee S, Kassir Y. Post-transcriptional regulation of IME1 determines initiation of meiosis in Saccharomyces cerevisiae. Mol Gen Genet. 1993;237:375–384. doi: 10.1007/BF00279441. [DOI] [PubMed] [Google Scholar]
- 47.Shinohara A, Ogawa H, Ogawa T. Rad51 protein involved in repair and recombination in S. cerevisiae is a RecA-like protein. Cell. 1992;69:457–470. doi: 10.1016/0092-8674(92)90447-k. [DOI] [PubMed] [Google Scholar]
- 48.Sikorski R S, Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Simchen G, Pinon R, Salts Y. Sporulation in Saccharomyces cerevisiae: premeiotic DNA synthesis, readiness and commitment. Exp Cell Res. 1972;75:207–218. doi: 10.1016/0014-4827(72)90538-1. [DOI] [PubMed] [Google Scholar]
- 50.Smith H E, Driscoll S E, Sia R A, Yuan H E, Mitchell A P. Genetic evidence for transcriptional activation by the yeast IME1 gene product. Genetics. 1993;133:775–784. doi: 10.1093/genetics/133.4.775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Smith H E, Mitchell A P. A transcriptional cascade governs entry into meiosis in Saccharomyces cerevisiae. Mol Cell Biol. 1989;9:2142–2152. doi: 10.1128/mcb.9.5.2142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Smith H E, Su S S, Neigeborn L, Driscoll S E, Mitchell A P. Role of IME1 expression in regulation of meiosis in Saccharomyces cerevisiae. Mol Cell Biol. 1990;10:6103–6113. doi: 10.1128/mcb.10.12.6103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Strich R, Surosky R T, Steber C, Dubois E, Messenguy F, Esposito R E. UME6 is a key regulator of nitrogen repression and meiotic development. Genes Dev. 1994;8:796–810. doi: 10.1101/gad.8.7.796. [DOI] [PubMed] [Google Scholar]
- 54.Sym M, Engebrecht J, Roeder G S. ZIP1 is a synaptonemal complex protein required for meiotic chromosome synapsis. Cell. 1993;72:365–378. doi: 10.1016/0092-8674(93)90114-6. [DOI] [PubMed] [Google Scholar]
- 55.Thompson-Jaeger S, Francois J, Gaughran J P, Tatchell K. Deletion of SNF1 affects the nutrient response of yeast and resembles mutations which activate the adenylate cyclase pathway. Genetics. 1991;129:697–706. doi: 10.1093/genetics/129.3.697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Treinin M, Simchen G. Mitochondrial activity is required for the expression of IME1, a regulator of meiosis in yeast. Curr Genet. 1993;23:223–227. doi: 10.1007/BF00351500. [DOI] [PubMed] [Google Scholar]
- 57.Vidan S, Mitchell A P. Stimulation of yeast meiotic gene expression by the glucose-repressible protein kinase Rim15p. Mol Cell Biol. 1997;17:2688–2697. doi: 10.1128/mcb.17.5.2688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Weber L, Byers B. A RAD9-dependent checkpoint blocks meiosis of cdc13 yeast cells. Genetics. 1992;131:55–63. doi: 10.1093/genetics/131.1.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Xu L, Weiner B M, Kleckner N. Meiotic cells monitor the status of the interhomolog recombination complex. Genes Dev. 1997;11:106–118. doi: 10.1101/gad.11.1.106. [DOI] [PubMed] [Google Scholar]
- 60.Yoshida M, Kawaguchi H, Sakata Y, Kominami K, Hirano M, Shima H, Akada R, Yamashita I. Initiation of meiosis and sporulation in Saccharomyces cerevisiae requires a novel protein kinase homologue. Mol Gen Genet. 1990;221:176–186. doi: 10.1007/BF00261718. [DOI] [PubMed] [Google Scholar]