Summary
Infectious severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) research, conducted in high-containment laboratories, requires transferring samples to lower containment labs for downstream applications, mandating sample inactivation. Here, we present a stepwise protocol for chemical inactivation of SARS-CoV-2 virus in culture supernatants or within infected cells and organoids, using eight chemical reagents validated via plaque assays. Additionally, we describe steps for troubleshooting virus inactivation, titer calculation, and log reduction. This protocol offers valuable resources for the COVID-19 research community, providing essential tools to advance research on this virus.
Subject areas: Cell Biology, Cell culture, Cell-based Assays, Microbiology
Graphical abstract
Highlights
-
•
Protocol for chemical inactivation of SARS-CoV-2 virus
-
•
Steps for virus inactivation in culture supernatants or within cells and organoids
-
•
Details on optimization of the protocol, limitations, and possible solutions
-
•
Details steps to calculate and interpret the efficacy of inactivation reagents
Publisher’s note: Undertaking any experimental protocol requires adherence to local institutional guidelines for laboratory safety and ethics.
Infectious severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) research, conducted in high-containment laboratories, requires transferring samples to lower containment labs for downstream applications, mandating sample inactivation. Here, we present a stepwise protocol for chemical inactivation of SARS-CoV-2 virus in culture supernatants or within infected cells and organoids, using eight chemical reagents validated via plaque assays. Additionally, we describe steps for troubleshooting virus inactivation, titer calculation, and log reduction. This protocol offers valuable resources for the COVID-19 research community, providing essential tools to advance research on this virus.
Before you begin
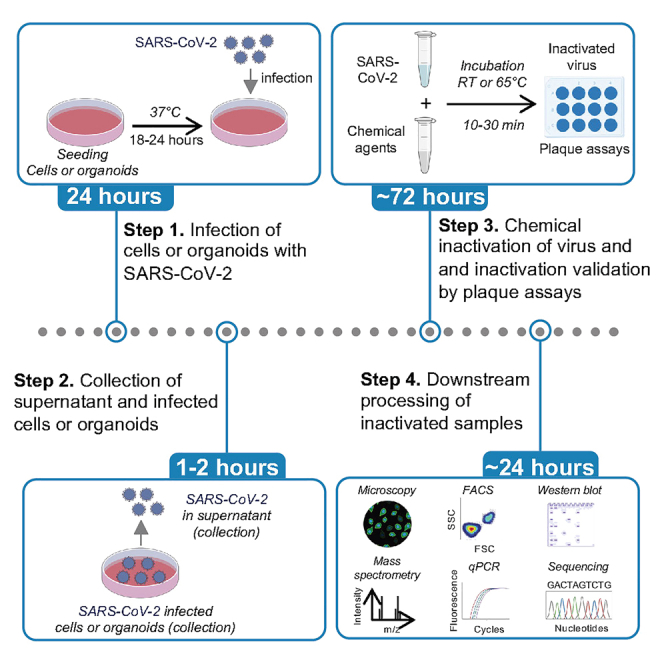
Current available methods for SARS-CoV-2 inactivation primarily rely on RT-qPCR-based detection and quantification of viral RNA transcripts; however, it falls short in detecting infectious viral particles and assessing chemical-induced toxicity in cells. Moreover, a comprehensive spectrum of inactivation methods is lacking in the field. Here, we provide a complete end-to-end working protocol, covering eight SARS-CoV-2 inactivation agents for different sample types, which is contingent upon the specific experimental goals, whereas the type of inactivating method/agent used depends on the desired downstream application. In this study, first, we describe detailed steps for virus inactivation of four types of infectious samples (SARS-CoV-2 virus, cell and organoid supernatant, and infected cells) using eight different agents, including heat, RIPA Buffer, Triton X-100, Paraformaldehyde (PFA), Molecular Transport Medium (MTM), Isopropanol, methanol, and Parse reagents (refer to graphical abstract). Then we outline a detailed cell-based plaque assay protocol, which provides a sensitive and reliable readout for viral load. Finally, we show the validation of all listed inactivation methods by enumerating log reduction of viral titer.
A suitable cell model susceptible to SARS-CoV-2 virus is essential for conducting plaque assays. Here, we have used various cell lines for SARS-CoV-2 infection, including Calu-3 (human epithelial cells), VeroE6, and VeroE6-TMPRSS2 (monkey kidney cells), along with adult stem cell-derived lung organoids (ALOs),1 which have previously demonstrated susceptibility to SARS-CoV-2.2 It is crucial to note that these procedures necessitate both Biosafety Level 2 (BSL2) and Biosafety Level 3 (BSL3) facilities.
Institutional permissions
All SARS-CoV-2-related work was conducted in BSL3 at Stanford University with institutional permission for this research. Users must obtain appropriate training and approvals before working with live SARS-CoV-2 virus.
Preparation of VeroE6, VeroE6-TMPRSS2, and Calu-3 cell line or adult stem cell-derived lung organoid culture
Timing: 2 days
-
1.Prepare VeroE6, VeroE6-TMPRSS2, or Calu-3 cell lines for infection (BSL2).
-
a.Maintain VeroE6 cells (ATCC, #Cat CRL-1586) and VeroE6-TMPRSS2 (JCRB cell bank, #Cat JCRB1819) in Dulbecco’s Modified Eagle Medium (DMEM) containing 10% Fetal bovine serum (FBS), designated as D10 and D10 with 1 mg/mL G418 respectively. Maintain Calu-3 cells (ATCC, #Cat HTB-55) DMEM supplemented with 10% FBS, 1% Penicillin-Streptomycin (Pen-Strep), and 1% Non-Essential Amino Acids (NEAA).
-
b.Cells were maintained in a humidified incubator with 5% CO2 at 37°C and tested negative for mycoplasma by MycoAlert (Lonza, Morristown, NJ).
-
c.Trypsinize cells using Trypsin-EDTA (0.05% v/v) or TrypLE for 5 min (min) at 37°C.
-
d.Neutralize trypsin with 10 mL of D10.
-
e.Transfer cells to a falcon and centrifuge cells at 500 g.
-
f.Resuspend cells in 10 mL of D10.
-
g.Seed 1 × 104, 1 × 105, 6 × 105, or 1 × 106 cells per well of a 96-, 24-, 12- or 6-well plate respectively at a volume of 0.1, 0.5, 1 and 2 mL per well respectively.
-
h.Place the plate at 37°C for 18–24 h (h).
-
a.
-
2.Prepare adult stem cell-derived lung organoids (ALOs) for infection (BSL2).
-
a.Seed ALOs in a 96-well plate, with 1 × 105 cells per well and maintain in complete PneumaCult-Ex Plus media (STEMCELL Technologies, #cat05040).
-
b.Place the plate at 37°C for 48 h.
-
a.
CRITICAL: It is crucial to use early cell passages for infection assays. The passage number of cell lines might influence the results of the plaque assay due to its impact on cell health and subsequently, viral replication. Moreover, upon reaching their Hayflick limit, the adherent cells tend to detach, therefore, cells that have been passaged multiple times might give false positives. Therefore, initiating experiments with low passage cell cultures and limiting their usage to 20–25 passages will be beneficial, especially when cells display altered characteristics like a failure to adhere or changes in cell morphology.
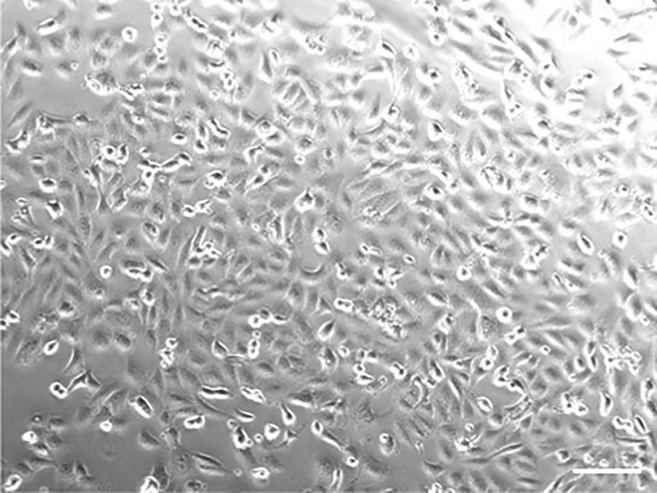
CRITICAL: It is critical that cells are grown in monolayer and 90%–100% confluent the next day before infection. A representative light microscopy image of 90%–100% confluent monolayer of VeroE6 cells is illustrated in Figure 1.
CRITICAL: Cell culture and seeding are performed in the BSL2 lab.
Figure 1.
Light microscopy image of 90%–100% confluent monolayers of VeroE6 cells
Seed VeroE6 cells in a 24-well plate with a cell number of 1 × 105 cells per well in D10 medium and incubate the plate at 37°C. 18–24 h later, check cells for confluency and take images of cells using light microscopy. Scale bar: 10 μm.
Viral infection
Timing: 24 h, all steps are performed inside the BSC of the BSL3 laboratory
-
3.
Thaw the SARS-CoV-2 virus ideally at 20°C–25°C inside the biosafety cabinet (BSC) in the BSL3 facility.
-
4.
Pipet off complete DMEM into liquid waste containing 100% bleach.
-
5.
Gently wash cells from step 1h or ALOs from step 2b, twice with PBS.
-
6.
Infect cells or ALOs from step 5 with SARS-CoV-2 at a multiplicity of infection (MOI) of 1 and/or 0.5 or 0.1 in modified (2%) DMEM or complete PneumaCult-Ex Plus media respectively at a volume of 50, 100, 250 and 500 μL in a 96-, 24-, 12- or 6-well plate respectively.
-
7.
Incubate for 1 h for cells or 4 h for ALOs at 37°C respectively.
-
8.
Following infection, remove the virus, wash cells twice with PBS, and replenish with fresh medium according to the plate size as mentioned in step 1g.
-
9.
Incubate for 24 h at 37°C.
-
10.
Collect supernatant for inactivation of virus in supernatant or collect infected cells and ALOs to perform inactivation of the virus within these cells.
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Bacterial and virus strains | ||
| SARS-CoV-2 USA-WA1/2020 | BEI Resources | NR-52281 |
| Chemicals, peptides, and recombinant proteins | ||
| Trypsin-EDTA | Fisher Scientific | #Cat25052CI |
| TrypLE | Gibco | #Cat12604-021 |
| Geneticin selective antibiotic (G418 sulfate) | Thermo Scientific | #Cat10131035 |
| PFA | Fisher Scientific | #Cat15714-S |
| RIPA lysis and extraction buffer | Thermo Fisher Scientific | #Cat89900 |
| Triton X-100 | Sigma | #CatT8787 |
| MTM | PrimeStore | #CatLH102 |
| Isopropanol | Fisher Scientific | #CatA416500 |
| Methanol certified ACS | Fisher Scientific/RIMS | #Cat A4124 BPA412-4 |
| Parse reagent | Parse Biosciences | #CatECF2001 |
| CMC | Millipore | #Cat17851 |
| Avicel PH-101 | Sigma-Aldrich | #Cat11365 |
| Crystal violet 1% solution | MilliporeSigma | #CatV5265 |
| Experimental models: Cell lines | ||
| VeroE6 | ATCC | #CatCRL-1586 |
| VeroE6-TMPRSS2 | JCRB Cell Bank | #CatJCRB1819 |
| Calu-3 | ATCC | #CatHTB-55 |
| ALO | HUMANOID Center of Research Excellence (CoRE), UCSD, USA | N/A |
Materials and equipment
DMEM 10% (D10) medium
| Reagent | Final concentration | Amount |
|---|---|---|
| DMEM | N/A | 500 mL |
| FBS | 10% | 50 mL |
| L-glutamate | 1% each | 5 mL each |
| Pen-Strep | ||
| NEAA | ||
| HEPES | ||
| Na Pyruvate | ||
| Total | N/A | 575 mL |
Note: D2 composition is the same as D10 except the FBS percentage is 2%. Store at 4°C, and warm at 37°C before use. Can be stored at 4°C for three months. n/a = Not applicable
2X Minimum Essential Medium (MEM) medium
| Reagent | Final concentration | Amount |
|---|---|---|
| MEM | N/A | 500 mL |
| Bovine Serum Albumin (BSA) | 3% | 6 mL |
| L-glutamate | 2% each | 10 mL each |
| Pen-Strep | ||
| HEPES | ||
| Sodium Bicarbonate (NaHCO3) | 5% | 16 mL |
| Total | N/A | 552 mL |
Note: Store at 4°C, and warm at 37°C before use. Can be stored at 4°C for three months. n/a = Not applicable.
Overlay
| Reagent | Amount |
|---|---|
| 2X MEM medium | 37 mL |
| 2% carboxymethylcellulose (CMC) | 13 mL |
| Total | 50 mL |
2% CMC Can be stored at 4°C for months six months.
Note: The overlay must be prepared immediately before use. Do not store overlay.
Step-by-step method details
Here we describe chemical inactivation of virus from two different sources. For the inactivation of viruses within cells or organoids, follow steps 1–2 (a-c) and for inactivation of virus stock or virus collected from infected cell supernatant, follow steps 3–4 (a-e) as described below.
Inactivation of virus-infected cells and organoids
Timing: 1 h, all steps require a BSL3 facility
This section describes a stepwise process to optimize the inactivation of SARS-CoV-2 virus within cells or organoids using different chemical reagents (Figure 2). This multi-step process will help to collect inactivated cells or organoids for various downstream processing applications highlighted in the graphical abstract.
-
1.
From step 9, wash virus-infected cells twice with PBS.
-
2.Follow the steps as mentioned below depending on the inactivation chemical used, as summarized in Table 1.
-
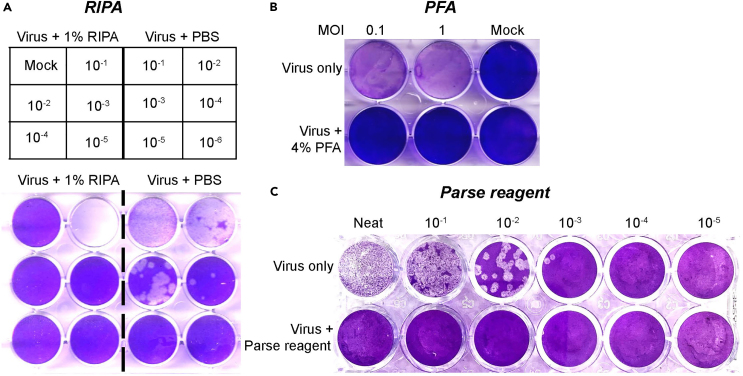
a.RIPA buffer (NP-40) inactivation of infected cells, related to Figure 2A.
-
i.Condition 1- experimental tube with RIPA: Infected cells from step 1, collect cells in 150 μL medium and lyse in 150 μL of RIPA buffer.
-
ii.Condition 2- control tube with PBS: Infected cells from step 1, collect in 150 μL of medium and add 150 μL of PBS (instead of RIPA) as control/no treatment.
-
iii.Incubate both steps (i) and (ii) samples for 30 min inside BSC at 20°C–25°C.
-
i.
-
b.PFA-based inactivation of infected cells, related to Figure 2B.
-
i.Infect cells from step 1h of “preparation of VeroE6, VeroE6-TMPRSS2, and Calu-3 cell line or adult stem cell-derived lung organoid culture”, with SARS-CoV-2 with MOI of 0.1 and 1.0 and take uninfected mock control and follow steps 3–10 of “viral infection”.
-
ii.24 h later, wash cells twice with PBS.
-
iii.Condition 1- fix infected cells with 500 μL of 4% PFA.
-
iv.Condition 2- incubate uninfected mock control and non-inactivated control with PBS.
-
v.Incubate for 30 min at 20°C–25°C inside BSC.
-
vi.Wash cells twice with PBS and replenish with fresh medium.
-
vii.72 h later, harvest culture supernatants to validate the PFA inactivation by plaque assay.
-
i.
-
c.Parse reagent inactivation of infected ALOs, related to Figure 2C.
-
i.In ALOs, from step 10, condition 1- fix infected cells with Cell fixation solution (Parse, Evercode Fixation, #catECF2001, Part number WF303) for 10 min at 20°C–25°C.
-
ii.Condition 2- Incubate unfixed cells (a.k.a non-inactivated/ control) with 1X PneumaCult-Ex Plus media.
-
iii.Following fixation, incubate fixed cells with Cell permeabilization Solution (Parse, Evercode Fixation, #catECF2001, Part number WF305) for 3 min at 20°C–25°C.
-
iv.Following permeabilization, wash cells twice with PBS from each condition and add PneumaCult-Ex Plus media (150 μL) into cells for an additional 96 h of incubation at 37°C in a humidified, 5% CO2 incubator.
-
v.Collect culture supernatants to validate the inactivation method by plaque assay.
-
i.
-
a.
Figure 2.
Plaque reduction assay for evaluating inactivation of SARS-CoV-2 virus in virus-infected cells using different chemical agents
The SARS-CoV-2 WA1/2020 strain from virus-infected cells was inactivated using (A) RIPA buffer (NP-40), (B) PFA, and (C) virus-infected ALO’s were inactivated using Parse reagent, and untreated controls followed by titration on VeroE6 or VeroE6-TMPRSS2 cell lines at different dilution. Cells were fixed and stained at 72 h post-infection with crystal violet. The experiment for each inactivation was repeated twice, and representative images are shown.
Table 1.
Effective concentration of different chemical reagents in inactivating SARS-CoV-2 virus, their incubation time and temperature
| Chemical Reagents/Conditions | Final concentration | Incubation time | Temperature | Reference |
|---|---|---|---|---|
| Heat | N/A | 30 min | 65°C | Batéjat et al.3 |
| Triton X-100 | 1% | 20 min | 20°C–25°C | Case et al.4 |
| Isopropanol | 30% | 30 min | 20°C–25°C | Kratzel et al.,5 Meyers et al.6 |
| Methanol | 50% | 10 min | 20°C–25°C | Kariwa et al.,7 Welch et al.,8 Patterson et al.9 |
| MTM | 1:3 v/v | 10 min | 20°C–25°C | Welch et al.8 |
| RIPA (NP-40) | 0.5% | 30 min | 20°C–25°C | Welch et al.8 |
| PFA | 4% | 20 min | 20°C–25°C | Möller et al.,10 Eddins et al.11 |
| Parse reagent | 100% | 10 min | 20°C–25°C | Flamier et al.12 |
n/a = Not applicable.
Inactivation of virus stock or infected-cell supernatant
Timing: 1–2 h, requires BSL3 facility
This section describes a stepwise process to optimize the inactivation of SARS-CoV-2 virus in the supernatant using different chemical reagents (Figure 3). This multi-step process will help to collect inactivated cells or organoids for various downstream processing applications highlighted in the graphical abstract.
-
3.
From step 10, collect the virus supernatant to inactivate the virus present in the supernatant, or inactivate viruses from step 3 of “viral infection”.
-
4.Follow the steps as mentioned below depending on the inactivation chemical used, as summarized in Table 1.
-
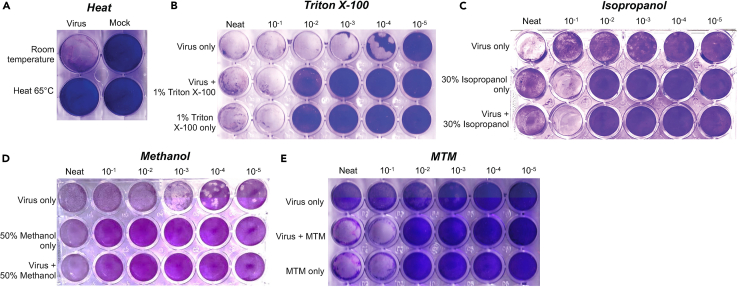
a.Heat inactivation of virus, related to Figure 3A.
-
i.Condition 1- experimental tube: Incubate virus stock from step 3.
-
ii.Condition 2- control tube: Incubate virus stock from step 3.
-
iii.Incubate the mixtures for 30 min at 65°C (i) or at 20°C–25°C (ii) inside BSC.
-
i.
-
b.Triton X-100 inactivation of virus, related to Figure 3B.
-
i.Condition 1- experimental tube: Incubate 450 μL virus stock from step 3 and 50 μL 10% Triton X-100 (1% final conc.).
-
ii.Condition 2- control tube I: Incubate 450 μL of virus stock from step 3 and 50 μL of PBS.
-
iii.Condition 3- control tube II: Incubate 450 μL of D2 and 50 μL 10% Triton X-100 (1% final conc.).
-
iv.Incubate the mixtures for 20 min at 20°C–25°C inside BSC.
-
i.
-
c.Isopropanol-based inactivation of virus, related to Figure 3C.
-
i.Condition 1- experimental tube: Incubate 100 μL of virus stock from step 3 and 400 μL of D2.
-
ii.Condition 2- control tube I: Incubate 350 μL D2 and 150 μL of 100% isopropanol (30% final conc.).
-
iii.Condition 3- control tube II: Incubate 100 μL of virus stock from step 3, 250 μL D2 and 150 μL of 100% isopropanol (30% final conc.).
-
iv.Incubate the mixtures for 20 min at 20°C–25°C inside BSC.
-
i.
-
d.Methanol-based inactivation of virus stock, related to Figure 3D.
-
i.Condition 1- experimental tube: Incubate 250 μL of virus stock from step 3 and 250 μL of D2.
-
ii.Condition 2- control tube I: Incubate 250 μL D2 and 250 μL of 100% methanol (50% final conc.).
-
iii.Condition 3- control tube II: Incubate 250 μL of virus stock from step 3 and 250 μL of 100% methanol (50% final conc.).
-
iv.Incubate the mixtures for 20 min at 20°C–25°C inside BSC.
-
i.
-
e.MTM-based inactivation of virus stock, related to Figure 3E.
-
i.Condition 1- experimental tube: Incubate 150 μL of virus stock from step 3 and 50 μL of MTM.
-
ii.Condition 2- control tube I: Incubate 150 μL virus stock from step 3 and 50 μL of PBS.
-
iii.Condition 3- control tube II: Incubate 150 μL of D2 and 50 μL of MTM.
-
iv.Incubate the mixtures for 10 min at 20°C–25°C inside BSC.
-
i.
-
a.
Figure 3.
Plaque reduction assay for evaluating inactivation of SARS-CoV-2 virus in stock or virus-infected culture supernatants using different chemical agents
The SARS-CoV-2 WA1/2020 strain from viral stock or culture supernatant was inactivated using (A) Heat at 65°C, (B) Triton X-100, (C) Isopropanol, (D) Methanol, and (E) MTM or respective conditions only and untreated controls followed by titration on VeroE6 or VeroE6-TMPRSS2 cell lines at different dilution. Cells were fixed and stained at 72 h-post infection with crystal violet. The experiment for each inactivation was repeated twice, and representative images are shown.
Plaque assay
Timing: 1–2 h followed by 72 h
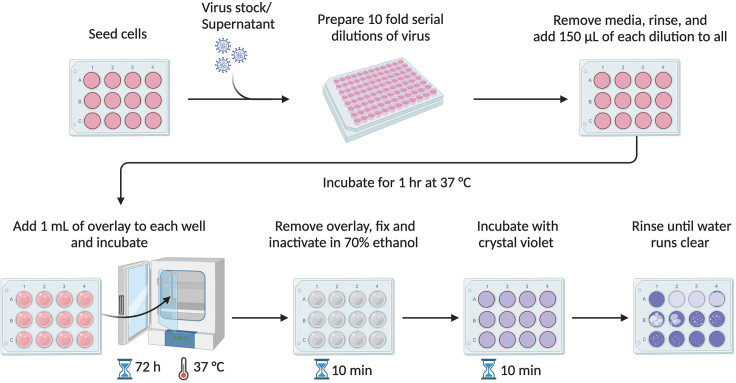
A schematic of the plaque assay procedure is illustrated in Figure 4.
-
5.
Thaw the samples (inactivated and controls) from step 2 of “inactivation of virus-infected cells” and step 4 of “inactivation of virus stock or infected-cell supernatant” ideally at 20°C–25°C inside the BSC.
-
6.
Perform 10-fold serial dilutions of viral stocks in modified (2%) DMEM, for instance, add 20 μL of virus stock in 180 μL of D2 medium in a 96-well plate. See Figure 4.
CRITICAL: Use new tips for each dilution to avoid cross-contamination between wells.
-
7.
Pipet off complete DMEM into liquid waste containing 100% bleach.
-
8.
Gently wash cells once with PBS with calcium and magnesium.
CRITICAL: Remove medium from a few wells at a time to avoid cells getting dry. If dried, the cells will come off the plate and will not provide a monolayer for infection and plaque assays.
-
9.
Inoculate VeroE6 or VeroE6-TMPRSS2 cells from step 1h of “preparation of VeroE6, VeroE6-TMPRSS2, and Calu-3 cell line or adult stem cell-derived lung organoid culture” with 150 μL of the diluted virus per well from step 6.
CRITICAL: Rock the plate gently after adding virus dilutions to each plate to avoid drying.
-
10.Incubate at 37°C on a shaker for 1 h in appropriate containment.
-
a.During the incubation, warm Minimum Essential Medium (MEM) and carboxymethylcellulose (CMC) or Avicel in a water bath.
-
b.Prepare "overlay mixture" (1.2% Avicel in 1X MEM) or 2% CMC in MEM (13 mL CMC in 37 mL MEM).
-
a.
-
11.
Add 1 mL of the overlay into each well.
-
12.
Gently shake your plate for mixing of virus and overlay before incubating at 37°C for 72 h.
CRITICAL: Do not allow the plates to be moved or disturbed at the incubator, as it might create a smear and will affect the final result.
CRITICAL: Infection and plaque assays are performed in the BSL3 lab. Cells that were seeded a day before are transported from BSL2 into BSL3 lab using a cell transporter.
Figure 4.
Schematic of plaque assays and crystal violet staining
The procedure includes seeding of cells in a 6-, 12-, or 24-well plate for the experiment. The inactivated virus and their respective controls are serially diluted (wherever mentioned) and inoculated to cells and incubate to facilitate viral attachment (virus adsorption), add an avicel or CMC overlay. Incubate the plate in the incubator for 72 h at 37°C, to allow plaque formation; Fix infected cells with ethanol and stain the cells with crystal violet.
Crystal violet staining for plaque assay
Timing: 1–2 h
Crystal violet staining is a common method used to make the plaques visible, as it stains the cells in the monolayer. It helps to visualize the plaques formed by infectious viral particles, making it easier to count them and quantify the viral titer/infectivity in the original sample. A schematic of the crystal violet staining is illustrated in Figure 4.
-
13.
72 h later, slowly take out the plates from the incubator, and carefully transfer them to BSC.
-
14.
Gently pipet the overlay out of the wells and into the waste bucket containing 100% bleach.
-
15.
Remove residual overlay by washing the cells once with 1 mL warm PBS in each well.
-
16.
Fix cells and decontaminate plate: Submerge the entire plate and lid in 70% ethanol for 10 min.
-
17.
Remove the plate from BSC.
-
18.
Dry the plate on the benchtop.
-
19.
Add 0.5 mL or 1 mL of 0.3% crystal violet to each well and incubate for 10 min at 20°C–25°C.
CRITICAL: The crystal violet must be prepared immediately before use. Do not store, as it will precipitate.
-
20.
Gently wash with tap water to remove crystal violet dye.
-
21.
Leave the plate on the bench top to allow it to air dry.
-
22.
Scan the plate to visualize and count plaques to determine the titer.
Expected outcomes
We have described detailed methods for virus inactivation using several chemical reagents and their validation through plaque assays. Following successful inactivation, we expect to see exponential drops in infectious viral titer. Plaque assay, used for quantification of the infectious viral titer in a sample, is expressed in terms of plaque-forming units per unit volume (PFU/mL), as explained in “quantification and statistical analysis” section.
The outcome of chemical inactivation of the SARS-CoV-2 virus-infected cells and the virus in cells or organoids culture supernatants are shown in Figures 2 and 3 respectively. Our results demonstrate that incubation of the virus with RIPA buffer for 30 min and Parse reagent for 10 min at 20°C–25°C leads to a successful reduction of viral titer by > 5 logs and >4 logs, respectively (Figures 2A and 2C; Table 2). Additionally, the viral population is completely inactivated when incubated at 65°C for 30 min, where the entire sample was plaqued (as recommended), leading to complete elimination of the virus demonstrating complete inactivation of virus (Figure 3A). Similarly, the virus treated with Triton X-100 and isopropanol inactivation led to >5 logs reduction (Figures 3B and 3C; Table 2), and methanol and MTM show reductions of >6 logs and >4 logs respectively (Figures 3D and 3E; Table 2).
Table 2.
Calculation of viral titer and the effectiveness of different chemical reagents in inactivating SARS-CoV-2 virus
| Chemical Reagents/Conditions | Virus only/+PBS condition |
Number of plaques in virus + inactivating agents | Log10 reduction | ||
|---|---|---|---|---|---|
| Number of plaques | Dilutions (10-fold) | Titers (PFU/mL) | |||
| Heat | N/A | N/A | N/A | n/d | N/A |
| Triton X-100 | 1 | 5 | 6.66 × 105 | n/d | >5 logs |
| Isopropanol | 4 | 4 | 2.66 × 105 | n/d | >5 logs |
| Methanol | 19 | 4 | 1.27 × 106 | n/d | >6 logs |
| MTM | 6 | 3 | 4.00 × 104 | n/d | >4 logs |
| RIPA (NP-40) | 3 | 4 | 2.00 × 105 | n/d | >5 logs |
| PFA | N/A | N/A | N/A | n/d | N/A |
| Parse reagent | 2 | 3 | 1.33 × 104 | n/d | >4 logs |
n/a = Not applicable: entire volume of sample was used for plaque assay that led to the complete elimination of viral population and no dilution series was carried out.
n/d = Not detected, due to complete inactivation of viruses.
Certain chemicals such as Triton X-100, isopropanol, methanol and MTM, induce toxicity in cells when used at high concentrations and cells survive at lower concentrations (Figures 3B–3E). Hence, we included an additional control group (chemicals-only group), to differentiate between chemical-induced cell death/toxicity versus virus infection or a combination of virus and chemical. In addition, the observed differences in plaques between the chemical + virus and virus only conditions, validates the efficacy of the chemical reagent in inactivating the virus. This dual approach not only confirms the successful virus inactivation but also provides insights into potential cytotoxic effects induced by the inactivating chemical agents. Chemical agents such as RIPA buffer and PFA, used in this inactivation protocol, were previously used for downstream processing of inactivated samples including microscopy, western blot, cytokine measurement, flow cytometry etc.2,13 Moreover, our report provides sufficient data on time and concentration for successful inactivation of infectious virus particles (Table 1), and our present protocol will serve as a reference for further modification in validation methods.
Quantification and statistical analysis
The formula for titer calculation
The viral titer is calculated based on the number of plaques counted at different virus dilutions. The formula for calculating titer is expressed as Plaque-Forming Units per unit volume (PFU/mL).
Titer (PFU/mL) = Number of Plaques X Dilution Factor X Dilution at which plaques are counted.
Dilution Factor = 1000/ Volume of diluted virus added.
Plaques are counted visually, either manually or using automated methods. A plaque is defined as a clear zone in the cell monolayer, and each plaque represents one infectious viral particle.
For example, in Figure 3D, we count 19 plaques at a dilution of 10-4 in the virus-only condition, using 150 μL volume of virus inoculum per well (Table 2).
So, virus titer = 19 × 6.66 × 104 = 1.27 × 106 PFU/ml.
Log reduction calculation
Virus log reduction is a measure of the decrease in virus concentration or load achieved, commonly expressed in logarithmic units. The formula for calculating virus log reduction is:
Log reduction = log10 (Viral titer in virus only condition/Virus titer in Methanol treated condition).
For example: in Figure 3D (Methanol inactivation plaque assay).
Given that we found no plaques in Methanol treated condition while virus-only treatment has 1.27 × 106 PFU/ml, therefore, there are 6 log reductions in viral titer using methanol-based inactivation (Table 2).
Limitations
One potential limitation is the possible interference of inactivation agents with plaque formation and enumeration. Certain chemicals are toxic to cells and may affect the ability of viruses to form visible plaques on cell monolayers, resulting in inaccurate quantification of viral infectivity. These mechanical limitations underscore the importance of careful method development and validation when using plaque assays for virus inactivation assessment. However, the advantages outweigh these limitations, as the assay is unique in allowing quantification of the absolute number of viral particles in a sample, rendering an accurate measure of the effectiveness of the inactivation method.
Troubleshooting
Here are some of the problems that might arise during chemical inactivation of SARS-CoV-2 using plaque assays. Although plaque assay troubleshooting has been discussed for conducting experiments in regular BSL2 labs,14 we will highlight a few main challenges and precise solutions from a BSL3 lab perspective to minimize repetitions.
Problem 1
Visibility challenges of viral plaques, variations in plaque size, and the potential merging of larger plaques with nearby ones.
Potential solution
-
•
Adjust the incubation period for plaque assays and optimize the overlay concentration to achieve optimal plaque size and visibility. This is critical, especially when viruses form larger plaques and fuse with others, reducing incubation time can prevent this fusion. On the contrary, if the plaque sizes are too small, increasing incubation time will promote the formation of larger plaques.
Problem 2
Failure of complete virus inactivation.
Potential solution
-
•
Evaluate and adjust inactivation conditions by experimenting with different concentrations of inactivation agents and incubation time to ensure complete inactivation of the virus is achieved. Thereafter, follow with plaque development with each time point and concentration. Calculate proper log reduction using the formula provided (related to Table 2).
Problem 3
Failure to establish a successful infection assay in setting up the inactivation assay.
Potential solution
-
•
Explore diverse cell culture conditions and identify suitable host cells to enhance assay performance. VeroE6 and VeroE6-TMPRSS2 are widely accepted cell types for conducting SARS-CoV-2 plaque assays (related to step 9 of “plaque assay”).
Problem 4
Inconsistent results between different dilutions.
Potential solution
-
•
Standardize procedures, validate methods rigorously, and monitor experimental conditions for consistency.
-
•
Perform inactivation and plaque assays in replicates to ensure accuracy (related to Table 1, showing a variety of initial conditions).
Problem 5
Toxicity of chemical agents interferes with the plaque assay outcome.
Potential solution
-
•
Dilute chemicals with DMEM and perform 10-fold serial dilution, followed by plaque assays using the diluted chemical+virus. Start with a high virus titer and include a control to determine the point at which the diluted chemical shows no toxicity on monolayers and cell survives. This is critical particularly for chemical agents that cause cellular cytotoxicity, as doing so clearly distinguishes complete inactivation, which is detected beyond toxicity, as in non-toxic lower dilution of chemical, no plaques were detected (related to Figures 2 and 3, showing visual representation).
Problem 6
Unclear plaque borders.
Potential solution
-
•
Gently add overlay with less shaky movements, avoid any movement or shaking during incubation for plaque assays (related to step 12 of “plaque assay”).
-
•
In BSL3 lab, dedicate an incubator solely for plaque assays which are not open multiple times by other users.
Problem 7
Peeling of cell monolayer.
Potential solution
-
•
Use corning cell culture plates for better adherence of monolayers, some cells are better adherent than others, which can be selectively used. Ensure 18–24 h incubation post-seeding.
-
•
Fix cells longer before removing the overlay will help.
-
•
Rinse the plate gently as in step 20 of “crystal violet staining for plaque assay”.
Problem 8
Drying of monolayer cells during the addition/incubation of the virus inoculum.
Potential solution
-
•
Gently swirl the plate after the addition of diluted virus every few wells as in step 12 of “plaque assay” or incubate cells with submerging volume for longer on a rocker within a sealed box (following BSL3 lab standard operating procedure) instead of a regular incubator.
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Jaishree Garhyan (jgarhyan@stanford.edu).
Technical contact
All technical questions related to SARS-CoV-2 inactivition should be directed to and will be fulfilled by the technical contact, Marwah Karim (mkarim@stanford.edu).
Materials availability
All reagents used in this study are available from the lead contact with a completed materials transfer agreement.
Data and code availability
This protocol does not report any original code. Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
Acknowledgments
This work was supported by awards W81XWH2210283 and W81XWH-16-1-0691 from the Department of Defense/Congressionally Directed Medical Research Programs, award HDTRA11810039 from the Defense Threat Reduction Agency/Fundamental Research to Counter Weapons of Mass Destruction, and grant 1R01AI158569-01 from the National Institutes of Health to S.E. S.E. is a Chan Zuckerberg Biohub investigator. M.K. was supported by a PhRMA Foundation Postdoctoral Fellowship in Translational Medicine. We thank the Stanford In vitro BSL3 Service Center and its Director Jaishree Garhyan for assistance with this research. Figure 4 was created using BioRender.
Author contributions
Conceptualization, M.K. and J.G.; methodology, M.K., A.A.P., C.W.L., and J.G.; investigation, M.K., A.A.P., and C.W.L.; data interpretation, M.K. and J.G.; writing and editing, M.K., S.E., and J.G.; funding and resources, S.E. and J.G.; supervision, M.K. and J.G.
Declaration of interests
The authors declare no competing interests.
Contributor Information
Marwah Karim, Email: mkarim@stanford.edu.
Jaishree Garhyan, Email: jgarhyan@stanford.edu.
References
- 1.Tindle C., Fuller M., Fonseca A., Taheri S., Ibeawuchi S.-R., Beutler N., Katkar G.D., Claire A., Castillo V., Hernandez M., et al. Adult stem cell-derived complete lung organoid models emulate lung disease in COVID-19. Elife. 2021;10 doi: 10.7554/eLife.66417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Saul S., Karim M., Ghita L., Huang P.-T., Chiu W., Durán V., Lo C.-W., Kumar S., Bhalla N., Leyssen P., et al. Anticancer pan-ErbB inhibitors reduce inflammation and tissue injury and exert broad-spectrum antiviral effects. J. Clin. Invest. 2023;133 doi: 10.1172/JCI169510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Batéjat C., Grassin Q., Manuguerra J.-C., Leclercq I. Heat inactivation of the severe acute respiratory syndrome coronavirus 2. J. Biosaf. Biosecur. 2021;3:1–3. doi: 10.1016/j.jobb.2020.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Case J.B., Bailey A.L., Kim A.S., Chen R.E., Diamond M.S. Growth, detection, quantification, and inactivation of SARS-CoV-2. Virology. 2020;548:39–48. doi: 10.1016/j.virol.2020.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kratzel A., Todt D., V’kovski P., Steiner S., Gultom M., Thao T.T.N., Ebert N., Holwerda M., Steinmann J., Niemeyer D., et al. Inactivation of severe acute respiratory syndrome coronavirus 2 by WHO-recommended hand rub formulations and alcohols. Emerg. Infect. Dis. 2020;26:1592–1595. doi: 10.3201/eid2607.200915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Meyers C., Kass R., Goldenberg D., Milici J., Alam S., Robison R. Ethanol and isopropanol inactivation of human coronavirus on hard surfaces. J. Hosp. Infect. 2021;107:45–49. doi: 10.1016/j.jhin.2020.09.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kariwa H., Fujii N., Takashima I. Inactivation of SARS coronavirus by means of povidone-iodine, physical conditions and chemical reagents. Dermatology. 2006;212:119–123. doi: 10.1159/000089211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Welch S.R., Davies K.A., Buczkowski H., Hettiarachchi N., Green N., Arnold U., Jones M., Hannah M.J., Evans R., Burton C., et al. Analysis of inactivation of SARS-CoV-2 by specimen transport media, nucleic acid extraction reagents, detergents, and fixatives. J. Clin. Microbiol. 2020;58 doi: 10.1128/JCM.01713-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Patterson E.I., Prince T., Anderson E.R., Casas-Sanchez A., Smith S.L., Cansado-Utrilla C., Solomon T., Griffiths M.J., Acosta-Serrano Á., Turtle L., Hughes G.L. Methods of inactivation of SARS-CoV-2 for downstream biological assays. J. Infect. Dis. 2020;222:1462–1467. doi: 10.1093/infdis/jiaa507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Möller L., Schünadel L., Nitsche A., Schwebke I., Hanisch M., Laue M. Evaluation of virus inactivation by formaldehyde to enhance biosafety of diagnostic electron microscopy. Viruses. 2015;7:666–679. doi: 10.3390/v7020666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Eddins D.J., Bassit L.C., Chandler J.D., Haddad N.S., Musall K.L., Yang J., Kosters A., Dobosh B.S., Hernández M.R., Ramonell R.P., et al. Inactivation of SARS-CoV-2 and COVID-19 patient samples for contemporary immunology and metabolomics studies. Immunohorizons. 2022;6:144–155. doi: 10.4049/immunohorizons.2200005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Flamier A., Bisht P., Richards A., Tomasello D.L., Jaenisch R. Human iPS cell-derived sensory neurons can be infected by SARS-CoV-2. iScience. 2023;26 doi: 10.1016/j.isci.2023.107690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Karim M., Saul S., Ghita L., Sahoo M.K., Ye C., Bhalla N., Lo C.-W., Jin J., Park J.-G., Martinez-Gualda B., et al. Numb-associated kinases are required for SARS-CoV-2 infection and are cellular targets for antiviral strategies. Antiviral Res. 2022;204 doi: 10.1016/j.antiviral.2022.105367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Baer A., Kehn-Hall K. Viral concentration determination through plaque assays: using traditional and novel overlay systems. J. Vis. Exp. 2014;93 doi: 10.3791/52065. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
This protocol does not report any original code. Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.