Abstract
Carotenoids are health-promoting plastidial isoprenoids with essential functions in plants as photoprotectants and photosynthetic pigments in chloroplasts. They also accumulate in specialized plastids named chromoplasts, providing color to non-photosynthetic tissues such as flower petals and ripe fruit. Carotenoid accumulation in chromoplasts requires specialized structures and proteins such as fibrillins (FBNs). The FBN family includes structural components of carotenoid sequestering structures in chromoplasts and members with metabolic roles in chloroplasts and other plastid types. However, the association of FBNs with carotenoids in plastids other than chromoplasts has remained unexplored. Here, we show that Arabidopsis (Arabidopsis thaliana) FBN6 interacts with phytoene synthase (PSY), the first enzyme of the carotenoid pathway. FBN6, but not FBN4 (a FBN that does not interact with PSY), enhances the activity of plant PSY (but not of the bacterial PSY crtB) in Escherichia coli cells. Overexpression of FBN6 in Nicotiana benthamiana leaves results in a higher production of phytoene, the product of PSY activity, whereas loss of FBN6 activity in Arabidopsis mutants dramatically reduces the production of carotenoids during seedling de-etiolation and after exposure to high light. Our work hence demonstrates that FBNs promote not only the accumulation of carotenoids in chromoplasts but also their biosynthesis in chloroplasts.
Binding of FIBRILLIN6 to the carotenoid biosynthetic enzyme phytoene synthase promotes carotenogenesis in bacteria and chloroplasts.
Introduction
Carotenoids are a family of plant isoprenoids with roles in photosynthesis and photoprotection in green tissues. They also function as pigments in non-photosynthetic tissues such as flower petals and ripe fruits to attract animals for pollination and seed dispersal. Carotenoids are also precursors of bioactive molecules in plants (including hormones such as abscisic acid and strigolactones) and animals (including retinoids such as vitamin A) (Rodriguez-Concepcion et al. 2018). Carotenoids accumulate at high levels in chloroplasts, where they are mostly associated with the photosynthetic apparatus. However, they are most abundant in chromoplasts, which are plastids specialized in the sequestration of these highly lipophilic compounds found in carotenoid-accumulating non-photosynthetic tissues (Rodriguez-Concepcion et al. 2018). Chromoplast ultrastructure changes depending on the species, organ, and carotenoid type they accumulate. For example, chromoplasts from carrot (Daucus carota) roots accumulate carotenoids as large crystals whereas those of red pepper (Capsicum annuum) contain a large number of plastoglobules (PG) with fibrillar extensions surrounded by an outer layer of proteins referred to as plastoglobulins or fibrillins (FBNs) (Egea et al. 2010).
FBNs are encoded by gene families in plants and algae that can be grouped into 12 clades (Singh and McNellis 2011; Kim and Kim 2022). They include members with diverse molecular masses, pI values, hydrophobicity profiles, and homology with lipocalins (small proteins involved in the binding and transport of small lipophilic compounds), suggesting that each FBN family member has specific biological function(s) (Singh and McNellis 2011; Lundquist et al. 2012; Kim and Kim 2022). In agreement, plastid types vary in their FBN composition, suggesting that specific FBNs might have specialized functions in different classes of plastids (Singh and McNellis 2011; Kim and Kim 2022). However, their mechanisms of action remain poorly understood in most cases. In tomato (Solanum lycopersicum), the PG-associated FBN1, FBN2, and FBN4 isoforms accumulate at high levels during fruit ripening, when carotenoids are actively produced and stored in chromoplasts (Barsan et al. 2012; Suzuki et al. 2015). FBN1 homologs from rapeseed (Brassica juncea) and melon (Cucumis melo) contribute to carotenoid accumulation in chromoplasts (Li et al. 2023; Zhou et al. 2023). By contrast, the Arabidopsis (Arabidopsis thaliana) homologs FBN1a, FBN1b, and FBN2 are involved in photoprotection and stress responses (Torres-Romero et al. 2022) whereas FBN4 is likely involved in the partitioning of metabolites between the PG and thylakoid membranes of chloroplasts (Singh et al. 2012). Arabidopsis FBN1a, FBN1b, and FBN2 interact with each other around the PG surface, but FBN2 also plays FBN1-independent roles through interaction with other proteins (Torres-Romero et al. 2022). Arabidopsis FBN5 is a stromal protein involved not only in the acclimation to photooxidative stress but also in plastoquinone-9 biosynthesis by binding to solanesyl diphosphate synthases (Kim et al. 2015). FBN6 is needed for plants to acclimate to light stress, and it contributes to reactive oxygen species (ROS) scavenging (Lee et al. 2020). The role of other FBNs remains poorly studied (Singh and McNellis 2011; Kim and Kim 2022).
Arabidopsis FBN6 was found to be among the proteins co-immunoprecipitated with a GFP-tagged version of phytoene synthase (PSY), the first and main rate-determining enzyme of the carotenoid pathway (Welsch et al. 2018). Furthermore, FBN6 is the only FBN enriched in purified envelope fractions where PSY is also found (Ferro et al. 2010; Bouchnak et al. 2019). Therefore, we questioned whether this particular isoform modulates PSY activity, thereby potentially impacting the metabolic flux of the carotenoid pathway in chloroplasts. Besides confirming binding and co-localization of Arabidopsis FBN6 and PSY, we demonstrate the functional relevance of such an interaction.
Results and discussion
Arabidopsis FBN6 and PSY physically interact in particular subplastidial locations
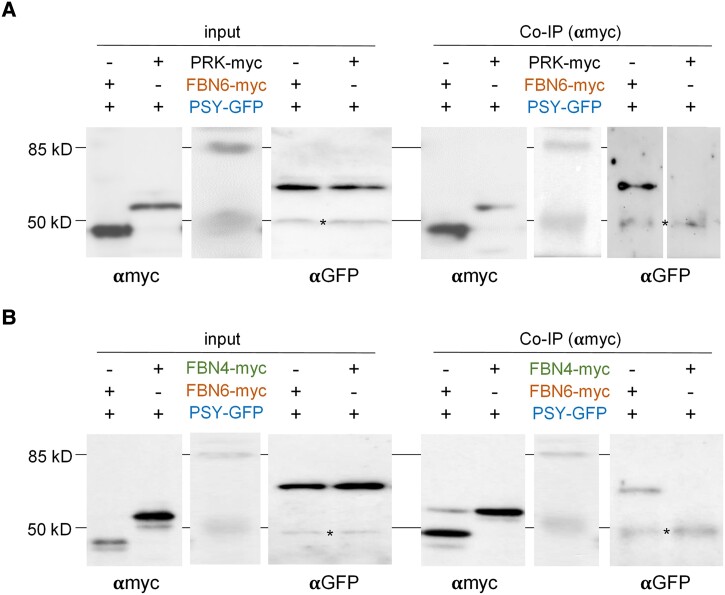
FBN6 was one of the proteins pulled down with a GFP antibody in Arabidopsis lines overexpressing the fusion protein PSY-GFP (Welsch et al. 2018), suggesting that FBN6 and PSY physically interact in vivo. To confirm this previously observed interaction and test its specificity, we performed co-immunoprecipitation experiments in Nicotiana benthamiana leaves. Constructs encoding C-terminal myc-tagged FBN6 and GFP-tagged PSY were co-agroinfiltrated in leaves, and 4 d later, protein extracts were used for immunoprecipitation using anti-myc antibodies (Fig. 1). As a negative control, we used a myc-tagged version of Arabidopsis phosphoribulokinase (PRK), a stromal enzyme of the Calvin cycle (Barja et al. 2021). Immunoblot analysis of immunoprecipitated proteins with anti-GFP antibodies detected PSY-GFP in FBN6-myc but not in PRK-myc samples (Fig. 1A). The specificity of the FBN6-PSY interaction was then tested by using a myc-tagged version of the FBN4 protein as a negative control (Torres-Romero et al. 2022). Results (Fig. 1B) showed that Arabidopsis FBN4 does not interact with PSY, and this confirmed the specificity of FBN6 and PSY interaction.
Figure 1.
FBN6 and PSY can be immunoprecipitated together. N. benthamiana leaves were co-agroinfiltrated with the indicated combinations of proteins with C-terminal myc or GFP tags. Four days later, agroinfiltrated leaves were collected and used for protein extraction and analysis. Part of the protein extracts was used to test protein production (input) by immunoblot analyses with antibodies against myc (αmyc) or GFP (αGFP; asterisk marks the position of unspecific bands). The remaining protein extracts were used for co-immunoprecipitation (Co-IP) experiments using αmyc followed by immunoblot analyses with immunoblot analyses with αmyc (to confirm successful IP) and αGFP (to detect the presence of co-immunoprecipitated PSY-GFP protein). A) Experiment using the Calvin cycle enzyme PRK fused to the myc tag as a negative control. B) Experiment using the PG-associated FBN4 protein fused to the myc tag to test for the specificity of the FBN6-PSY interaction.
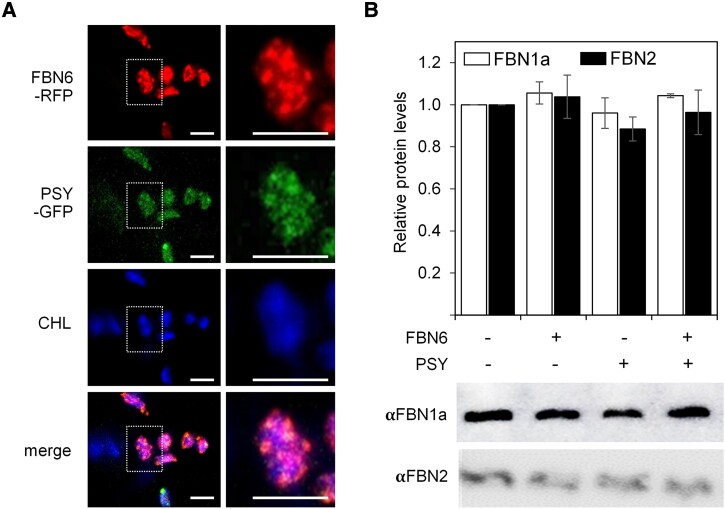
When analyzing the co-immunoprecipitation assays, we noticed that the PSY-GFP protein concentrated in fluorescent dots within chloroplasts (Fig. 2A). These dots are similar to those observed for PG-associated proteins (Lundquist et al. 2012; Gámez-Arjona et al. 2014; Morelli et al. 2023) and for PSY enzymes (Shumskaya et al. 2012) when fused to fluorescent protein tags. Interestingly, GFP-tagged Arabidopsis FBN6 shows a spotted distribution within chloroplasts despite the fact that the protein is not found in PG but localized in thylakoid and envelope membranes (Ferro et al. 2010; Lundquist et al. 2012; Bouchnak et al. 2019; Lee et al. 2020). To test whether the fluorescent dots observed in chloroplasts harboring FBN6-GFP and PSY-GFP proteins corresponded to the same or different subplastidial locations, we fused the red fluorescent protein (RFP) to FBN6 (FBN6-RFP construct) and co-agroinfiltrated it with the PSY-GFP construct. Confocal microscopy of the leaves 4 d after agroinfiltration showed a clear overlapping of RFP and GFP fluorescence signals (Fig. 2A), indicating that FBN6 and PSY co-localize within chloroplasts, as expected for interacting proteins.
Figure 2.
FBN6 and PSY co-localize in chloroplasts. N. benthamiana leaves were co-agroinfiltrated with constructs encoding FBN6-RFP and PSY-GFP, and 4 d later, samples were taken for confocal and immunoblot analyses. A) Representative confocal microscopy images of a leaf section transiently expressing the indicated fluorescent fusion proteins. All panels in the same column show the same field. RFP fluorescence is shown in the top panels, GFP fluorescence in the second row, and chlorophyll autofluorescence in the third row. Bottom panels show a merge of the 3 fluorescence channels. Right panels are a magnification of the area boxed in white in the left panels. Bars, 5 µm. B) Immunoblot analysis of the accumulation of FBNs identified using antibodies against Arabidopsis PG-associated FBN1a and FBN2 proteins in leaves expressing the indicated constructs or a GFP control (−/−). Quantification was made by relativizing each band intensity to the one obtained in the control. Representative results are shown in the lower panels. Barplot shows the mean ± Sd values of quantitative data from n = 3 leaves.
Despite the fact that FBN6-GFP has been previously found to co-localize with PG proteins fused to fluorescent tags in co-expression experiments similar to the results reported here, subsequent analysis of chloroplast membrane fractions argued against a localization of FBN6 in PG (Lee et al. 2020), a conclusion that is supported by proteomic data (Ferro et al. 2010; Lundquist et al. 2012; Bouchnak et al. 2019). While overexpression of FBNs and other PG-associated proteins typically results in PG proliferation (Shanmugabalaji et al. 2013; Van Wijk and Kessler 2017), overproduction of FBN6 or/and PSY proteins in agroinfiltrated leaf chloroplasts did not cause changes in PG core-associated proteins such as FBN1 or FBN2 homologs (Fig. 2B). These results provide further indirect evidence that FBN6 and PSY are not PG-associated proteins. Together, we conclude that Arabidopsis FBN6 and PSY interact and preferentially co-localize in particular locations of the chloroplast such as envelope or thylakoid membrane domains different from PG.
FBN6 promotes PSY activity
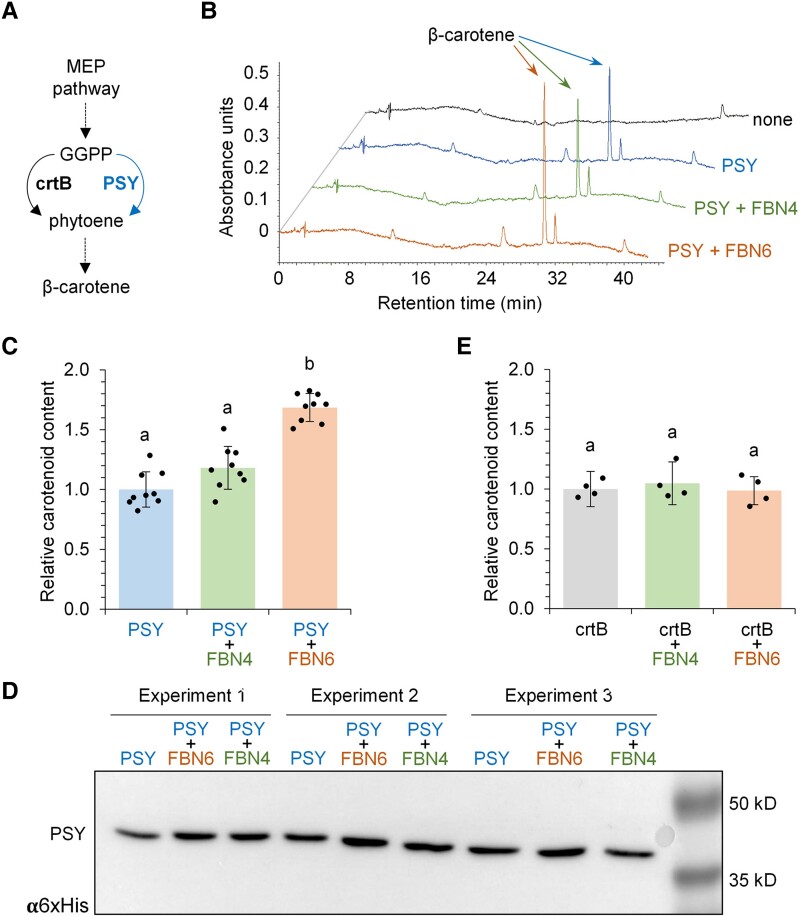
To test the possible relevance of FBN6 binding for PSY activity, we used a bacterial system free of other plant proteins that might influence the outcome of the interaction. Specifically, we used an Escherichia coli strain carrying a synthetic operon that only lacks a gene encoding PSY to produce the orange carotenoid β-carotene from endogenous bacterial precursors (Ahrazem et al. 2019; Fig. 3A). Constructs to produce versions of plant proteins lacking their N-terminal plastid-targeting sequences were generated using appropriate gene-specific primers (Supplemental Table S1). Strains co-transformed with such constructs harboring Arabidopsis PSY and FBN6 sequences were found to produce significantly more β-carotene than those expressing only PSY or co-expressing PSY and FBN4 (Fig. 3, B and C), despite that PSY protein levels remained very similar in all the cases (Fig. 3D). By contrast, FBN6 had no effect when crtB, a PSY from the bacterium Pantoea ananatis, was used instead of PSY to produce carotenoids in the bacterial cells (Fig. 3E). These results strongly suggest that the presence of FBN6 specifically promotes the activity of the plant PSY enzyme in this bacterial system (i.e., in the absence of other plastid proteins that might indirectly improve PSY activity).
Figure 3.
FBN6 enhances PSY activity. A)E. coli cells were transformed with constructs to produce β-carotene from precursors supplied by the endogenous methylerythritol 4-phosphate pathway when plant or bacterial (PSYs or crtB, respectively) were exogenously provided. GGPP, geranylgeranyl diphosphate. B) Representative HPLC chromatograms showing the carotenoids produced in cells co-transformed with constructs encoding PSY, FBN4 or/and FBN6. A control not expressing PSY or FBNs is also shown (marked as “none”). The main carotenoid product (β-carotene) is indicated. C) Total carotenoid levels in cells co-transformed with constructs encoding PSY, FBN4, or/and FBN6. D) Immunoblot analysis of the His-tagged PSY protein used in the assays. The blot shows the levels of the recombinant protein in 3 different experiments of the indicated combinations. E) Total carotenoid levels in cells expressing crtB instead of PSY together with FBN4 or FBN6. In C) and E), mean ± Sd are represented relative to carotenoid levels in samples with PSY (C, n = 9) or crtB (E, n = 4) alone; dots represent individual data points and letters represent statistically significant differences (P < 0.05) among means according to one-way ANOVA followed by post hoc Tukey's tests.
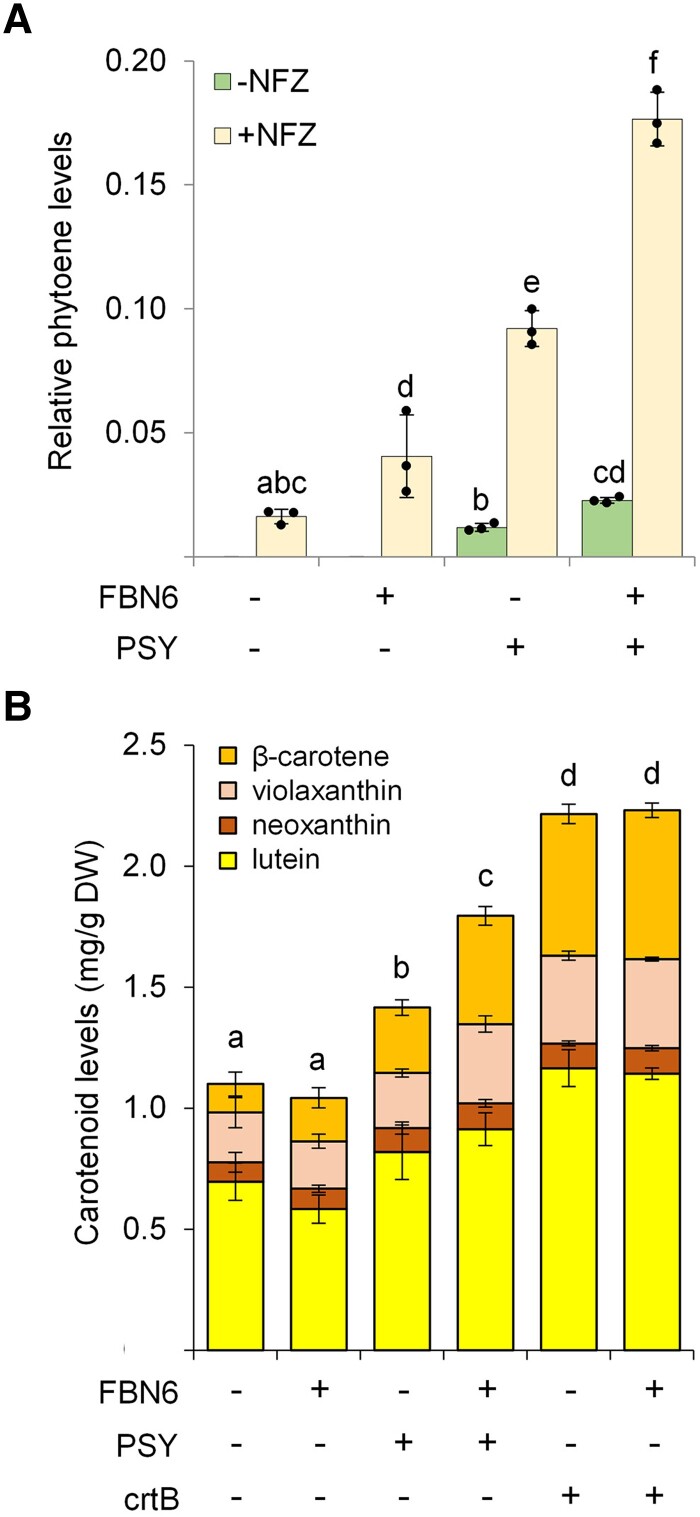
To verify whether FBN6 could also promote PSY activity in the chloroplasts of plant cells, we analyzed the production of phytoene (the direct product of PSY activity) and downstream carotenoids in N. benthamiana leaves agroinfiltrated with constructs to transiently express the Arabidopsis genes encoding FBN6, PSY, or both (Fig. 4 and Supplemental Tables S2 and S3). Control leaves were agroinfiltrated with a GFP construct. While phytoene was not detected in control or FBN6 samples, it was present in PSY samples, and it increased in samples agroinfiltrated with both FBN6 and PSY (Fig. 4A). To investigate whether overexpression of FBN6 could also impact the endogenous PSY enzymes in N. benthamiana leaves, we used norflurazon (NFZ). This inhibitor prevents phytoene conversion into downstream carotenoids, thereby allowing indirect estimation of PSY activity through quantifying the amount of phytoene accumulation in NFZ-treated tissues. As expected, phytoene was detected in all the samples treated with NFZ. FBN6 samples showed a slight but statistically significant increase in phytoene compared with GFP controls, in agreement with the conclusion that FBN6 is a promoter of endogenous PSY activity. In PSY-overexpressing samples, the presence of FBN6 led to a much more obvious increase in phytoene content (Fig. 4A). In the case of total carotenoids, FBN6 alone did not lead to increased levels but a combination of FBN6 and PSY resulted in more carotenoids than PSY alone (Fig. 4B). Consistent with the results in bacterial systems, the effect of FBN6 in enhancing PSY-promoted carotenoid accumulation was not observed when crtB was used instead of the plant PSY enzyme (Fig. 4B).
Figure 4.
FBN6 promotes phytoene and downstream carotenoid production. N. benthamiana leaves were agroinfiltrated with the indicated constructs, and 4 d later, samples were taken for HPLC analysis of carotenoids. A GFP construct was used as a control. A) Phytoene levels in leaf samples either treated (+) or not (−) with NFZ 1 day after agroinfiltration with GFP, PSY, or/and FBN6 constructs. Values are represented relative to total carotenoid levels in untreated control leaves and they correspond to the mean ± Sd of n = 3 leaf samples. B) Levels of downstream carotenoids in untreated samples. DW, dry weight. Values correspond to the mean ± Sd of n = 3 leaf samples. In both plots, different letters represent statistically significant differences (P < 0.05) among means of total carotenoids according to one-way ANOVA followed by post hoc Tukey's tests. Supplemental Tables S2 and S3 show absolute values of carotenoid and chlorophyll levels.
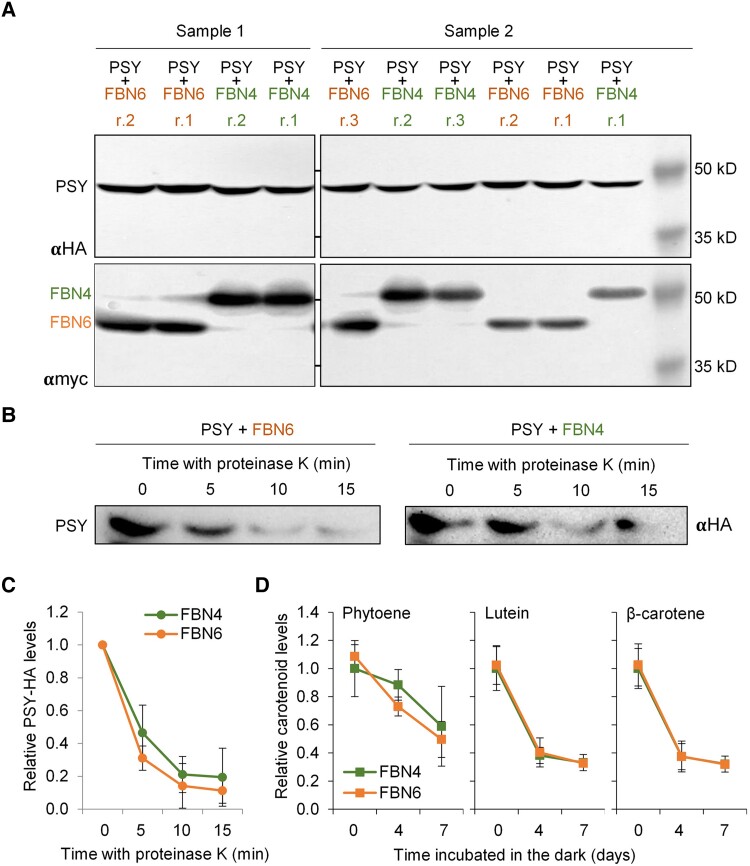
While these results support a positive and specific effect of FBN6 on PSY activity, it is also possible that FBN6 might promote the accumulation of PSY-derived phytoene and other carotenoids in chloroplasts by playing other (additional) roles, e.g., by modulating PSY folding or carotenoid stability. To test whether PSY folding or/and degradation rate could be altered by FBN6 in planta, we overexpressed the 2 Arabidopsis proteins in N. benthamiana leaves (or PSY and FBN4 as a control) and used total protein extracts in proteolytic assays (Fig. 5). Levels of PSY protein were similar in extracts from leaves co-expressing FBN6 or FBN4 constructs (Fig. 5A), arguing against a possible role for FBN6 in promoting PSY accumulation in chloroplasts. Next, PSY folding and aggregation status was estimated from accessibility to cleavage by proteinase K. Consistent with the conclusion that FBN6 does not change PSY protein structure, PSY degradation by proteinase K occurred similarly in extracts from leaves co-agroinfiltrated with FBN6 or FBN4 (Fig. 5, B and C). These results suggest that PSY folding or degradation rate is not altered by interaction with FBN6. We further investigated the possible influence of FBN6 on the stability of the carotenoid products by analyzing the rate of carotenoid removal during leaf senescence (Fig. 5D). In this case, N. benthamiana leaves were agroinfiltrated with constructs harboring either FBN6 or FBN4 as a control together with crtB to boost phytoene and downstream carotenoid production (Morelli et al. 2023). Four days later, agroinfiltrated leaves were cut from the plant and incubated in the dark for up to 1 wk. Degradation of phytoene and downstream carotenoids was virtually identical in samples carrying FBN6 or FBN4 (Fig. 5D), indicating that these FBNs do not have a differential role in carotenoid storage or stability. Together, the results in bacterial and plant systems strongly support the conclusion that FBN6 interaction with PSY leads to a higher enzymatic activity.
Figure 5.
FBN6 does not influence PSY or carotenoid accumulation. N. benthamiana leaves were co-agroinfiltrated with constructs to express combinations of myc-tagged FBN6 (or FBN4 as a control) with HA-tagged PSY (or crtB as a control), and 4 days later, samples were collected to prepare protein or pigment extracts. A) Immunoblot analysis of PSY-HA, FBN4-myc and FBN6-myc protein levels in several replicates (i.e. plants) from 2 different samples (i.e., experiments). B) Immunoblot analysis of PSY-HA protein in extracts additionally containing either FBN6-myc or FBN4-myc after incubation with proteinase K for the indicated times (minutes). C) Quantification of PSY-HA protein abundance in extracts treated with proteinase K as described in B). Mean ± Sd values of n = 3 independent experiments are represented relative to the starting levels, i.e. before incubation with the protease (Time 0). D) Carotenoid levels in N. benthamiana leaves co-expressing crtB together with either FBN6 or FBN4, cut from the plant and incubated in the dark for the indicated times (days). Mean ± Sd values of n = 3 independent experiments are represented relative to the starting levels, i.e. before placing the cut leaves in the dark (Day 0). The y-axis is the same for the 3 plots. Statistical analyses (2-way ANOVA) detected no significant differences (2-way ANOVA, P > 0.05) between FBN6 and FBN4 samples in the experiments shown in C) and D).
Genetic evidence confirms the functional relevance of FBN6 for PSY activity
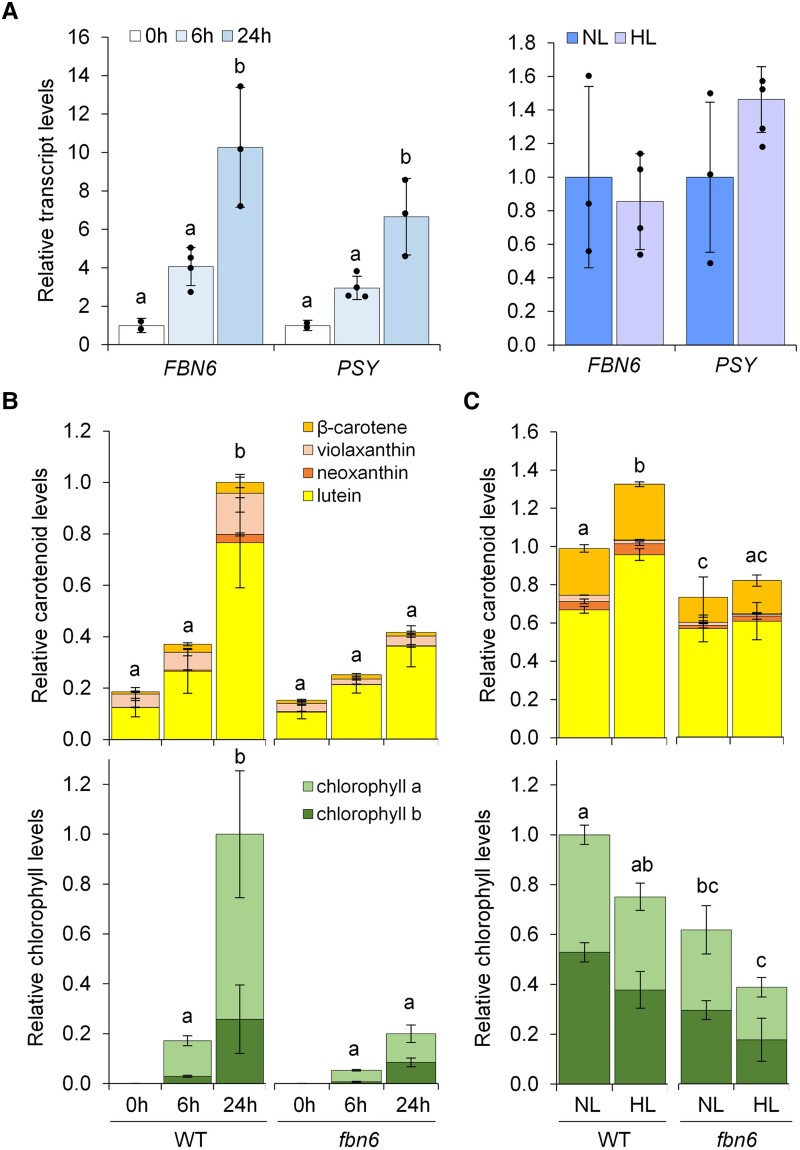
If FBN6 is a positive regulator of PSY activity, loss of FBN6 in mutants would be expected to prevent normal carotenoid synthesis. FBN6-defective Arabidopsis fbn6-1 mutants were found to have reduced chlorophyll levels compared with wild-type (WT) controls, but no data on carotenoid levels were reported (Lee et al. 2020). In Arabidopsis, the most active production of carotenoids takes place during de-etiolation to protect the emerging photosynthetic apparatus against photooxidative damage caused by excess incoming light (Rodríguez-Villalón et al. 2009). The boost in carotenoid synthesis during de-etiolation is mediated in part by increased expression of the PSY-encoding gene in Arabidopsis (Rodríguez-Villalón et al. 2009; Toledo-Ortiz et al. 2010). Interestingly, the gene encoding FBN6 was also highly induced during this process (Fig. 6A). Because co-expression is only associative evidence of a possible common role, we next used genetics to test whether FBN6 activity was required for normal carotenoid production in etiolated and de-etiolating seedlings. Compared with the WT, the fbn6-1 mutant contains similar levels of carotenoids in etiolated seedlings, but it shows a strongly attenuated production during de-etiolation (up to 24 h after illumination). As a result, greening (chlorophyll accumulation) was delayed (Fig. 6B and Supplemental Table S4). After a few days under normal light (NL), the differences in carotenoid (and chlorophyll) levels between WT and fbn6-1 plants were attenuated but still detectable (Fig. 6C and Supplemental Table S5). To test the putative influence of increased FBN6 on carotenoid biosynthesis in etiolated and de-etiolating seedlings, we transformed Arabidopsis plants with a construct to overexpress an RFP-tagged FBN6 protein. Most transformants showed no RFP fluorescence, and the ones producing detectable levels of FBN6-RFP protein showed levels of total FBN6 transcripts (endogenous and transgenic) that were similar or lower than those in untransformed controls (Supplemental Fig. S1). These results suggest that accumulation of FBN6-encoding transcripts is tightly regulated, perhaps to maintain a balanced ratio with PSY transcripts (Fig. 6A).
Figure 6.
FBN6 is required for carotenoid production in Arabidopsis. De-etiolation experiments were carried out with Arabidopsis WT and FBN6-defective mutant seeds germinated in the dark for 3 d; the resulting etiolated seedlings were then exposed to white light for 6 and 24 h. For HL experiments, WT and fbn6 seedlings germinated and grown under NL conditions for 9 d were exposed for 24 h to either NL (as a control) or HL. A) Expression profiles of genes encoding FBN6 and PSY during de-etiolation (left) or HL exposure (right) of WT seedlings. Transcript levels are normalized using the UBC21 gene and shown relative to etiolated (0 h) or NL-exposed samples, respectively. B) Carotenoid and chlorophyll levels in de-etiolating WT and fbn6 seedlings. Values are represented relative to total pigment levels in WT 24 h samples. C) Carotenoid and chlorophyll levels in HL-exposed WT and fbn6 seedlings. Values are represented relative to total pigment levels in WT NL samples. In all plots, mean ± Sd of n ≥ 3 replicates are represented. Letters represent statistically significant differences (P < 0.05) among means of total carotenoids and chlorophylls according to one-way ANOVA followed by post hoc Tukey's tests. Supplemental Tables S4 and S5 show absolute values of carotenoid and chlorophyll levels.
Carotenoid levels, but not PSY expression, increase when Arabidopsis plants are exposed to high light (HL), suggesting an up-regulation of PSY enzyme activity to increase the metabolic flux toward carotenoid synthesis (Kreslavski et al. 2021). Similar to PSY, transcripts encoding FBN6 also showed unchanged levels when 9-d-old Arabidopsis seedlings were exposed for 24 h to HL compared with controls left at NL for the same time (Fig. 6A). Most interestingly, the HL-triggered increase in carotenoid levels was highly attenuated in FBN6-defective plants compared with the WT controls (Fig. 6C). By contrast, the reduction in chlorophylls associated with HL exposure was similar in WT and fbn6-1 seedlings in absolute terms but seemingly stronger in the mutant in relative terms (Fig. 6C), likely because disrupted up-regulation of carotenoid contents in fbn6-1 seedlings results in defective photoprotection and a higher chlorophyll degradation rate. We conclude that FBN6 is required to support the increase in carotenoid levels that takes place in response to HL, a role that is consistent with its proposed PSY-promoting activity.
An alternative connection between FBNs and carotenoids
FBN gene expression often parallels the carotenoid content of non-photosynthetic plant tissues, e.g. during fruit ripening, where their role is assumed to be mainly structural (Singh and McNellis 2011). Indeed, FBNs were initially identified as proteins specifically associated with carotenoid accumulation in fruit and petal chromoplasts, as components of “fibrils” and PG structures (Deruère et al. 1994; Vainstein et al. 1994). Similarly, green algae FBNs were also proposed to be involved in the formation and stabilization of the chloroplast eyespot, a carotenoid-rich light sensing system (Schmidt et al. 2006; Davidi et al. 2015). More recently, rapeseed and melon FBN1 homologs were shown to contribute to carotenoid accumulation by interacting with other proteins and promoting PG proliferation in flower and fruit chromoplasts (Li et al. 2023; Zhou et al. 2023). However, in plant chloroplasts, carotenoids are not normally concentrated in PG or any other lipid body but are distributed in thylakoids and, to a lower extent, envelope membranes (Morelli et al. 2023). Despite the fact that chloroplast FBNs are presumed to maintain photosynthetic function and are present in all the compartments where carotenoids are synthesized and accumulated, a direct functional association between FBNs and carotenoid biosynthesis has remained unexplored.
Studies on FBN-interacting proteins have substantially improved our understanding of the function of different FBN family members in chloroplasts. Arabidopsis FBN1a, FBN1b, and FBN2 interact with each other, presumably forming a network around the PG surface that might be important to recruit other FBN-interacting proteins to PG (Gámez-Arjona et al. 2014; Torres-Romero et al. 2022). For example, these FBNs interact with allene oxide synthase, which catalyzes one of the first steps of jasmonic acid (JA) biosynthesis. FBN2 also interacts with other proteins involved in different metabolic processes, including other enzymes of the JA pathway and the carotenoid cleavage dioxygenase CCD4, a PG-associated enzyme involved in carotenoid degradation (Torres-Romero et al. 2022). It was proposed that FBN2 might mediate the recruitment of JA biosynthetic enzymes to the PG under stress conditions, but the functional relevance of the interaction with CCD4 was not explored (Torres-Romero et al. 2022). FBN1a and FBN1b also interact with starch synthase isoform 4 (SS4), but the elimination of the FBNs does not alter the initiation of starch granule formation (Gámez-Arjona et al. 2014). FBN4 interacts with the major ferredoxin protein in Arabidopsis (Fd2) and with harpin protein HrpN, an elicitor secreted by pathogenic bacteria that activates plant defense and resistance (Kim and Kim 2022). FBN5 participates in the production of the plastidial isoprenoid plastoquinone through interaction with solanesyl diphosphate synthase isoforms 1 and 2 (SPS1 and SPS2), which catalyze the first committed step in the production of the prenyl chain of the molecule (Kim et al. 2015). Based on the presence of a lipocalin domain involved in binding to and transporting small hydrophobic metabolites, it was proposed that FBN5 might bind the solanesyl diphosphate product of SPS1 and SPS2 and remove it from the active site of the enzymes, thereby contributing to increasing their enzymatic activity (Kim et al. 2015).
The lipocalin motif of FBN4 likely contributes to the role of this isoform in the trafficking of plastoquinone and other small lipids between the thylakoids and the PG (Singh et al. 2012). Interestingly, FBN6 also contains a lipocalin motif (Singh and McNellis 2011). Based on that proposed for FBN5, a non-PG FBN protein, we speculate that direct interaction of FBN6 with PSY might result in improved activity of the enzyme by facilitating removal of the phytoene product from the active site. Regardless of the specific mechanism involved, our data support the conclusion that FBN6 plays a physiologically relevant role in carotenoid biosynthesis in Arabidopsis chloroplasts by physically binding to PSY to promote its enzymatic activity. Therefore, we demonstrate that FBNs promote not only the accumulation of carotenoids but also their biosynthesis. The unveiled role of FBN6 in carotenogenesis reported here can explain the phenotypes previously reported for the fbn6-1 mutant, which were similar to those of mutants disrupted in ROS homeostasis (Lee et al. 2020). Because carotenoids are essential photoprotectants that dissipate excess light energy as heat and detoxify excess ROS in chloroplasts (Rodriguez-Concepcion et al. 2018), we conclude that the reduced acclimation to light stress and impaired ROS scavenging phenotypes observed in the fbn6-1 mutant are likely a primary result of defective carotenoid biosynthesis. Lower growth, increased glutathione levels, or resistance to cadmium stress would be secondary effects of impaired ROS homeostasis.
Similar to the up-regulation of PSY and FBN6 genes during Arabidopsis de-etiolation (Fig. 6A), the burst in carotenoid synthesis that takes place during tomato fruit ripening is also associated with increased expression of genes encoding the corresponding PSY and FBN6 homologs (Barja et al. 2021; Sun et al. 2022). This observation suggests that FBN6-dependent promotion of PSY activity and carotenoid biosynthesis might not be restricted to chloroplasts but can also occur in chromoplasts. Further experiments with FBN6-defective tomato plants should confirm this possibility and provide additional insights into the possible biotechnological use of this FBN isoform.
Materials and methods
Plant material and growth conditions
N. benthamiana and Arabidopsis (A. thaliana) plants were grown as described (Morelli et al. 2023). The fbn6-1 mutant (GABI_159E10) (Lee et al. 2020) was obtained from the Nottingham Arabidopsis Stock Centre (NASC). Homozygous lines were identified by genomic PCR with T-DNA and gene-specific primers (Supplemental Table S1). Transgenic lines overexpressing an RFP-tagged version of Arabidopsis FBN6 were generated in the Columbia-0 background using construct pGWB454-FBN6-RFP. Lines with a single T-DNA insertion were identified based on the Mendelian segregation of the kanamycin resistance marker gene, and transformants showing different levels of FBN6-RFP protein (as deduced from RFP fluorescence intensity) were selected for analysis. For the de-etiolation experiments, seeds were surface-sterilized and sown on Petri dishes containing solid 0.5× MS medium without sucrose. After stratification for 3 d at 4°C in the dark, seeds were exposed to white light (50 μmol photons·m−2 s−1, referred to as normal light or NL) for 3 h at 22°C to stimulate synchronized germination. Then, plates were covered in aluminum foil and incubated at 22°C in the dark for 3 d for seeds to germinate and produce etiolated seedlings. Such etiolated seedlings were either collected in the dark or exposed to NL to allow de-etiolation. For the intense light experiments, seedlings grown on plates for 9 d under NL were exposed for 24 h to 600 μmol photons·m−2 s−1 (referred to as high light or HL) or left under NL for the same time. N. benthamiana leaves were agroinfiltrated as described (Morelli et al. 2023). Some leaves were later treated with the commercial herbicide Zorial, containing 80% NFZ. The herbicide was diluted in a 0.05% (v/v) solution of Tween 20 in water to a final concentration of 2 μm NFZ. The solution was then infiltrated with a syringe on the leaf areas that had been agroinfiltrated with constructs of interest 24 h earlier (so the agroinfiltration halo was properly absorbed). For the carotenoid degradation experiment, agroinfiltrated leaves were left in the plant for 4 d and then cut and placed on top of solid 0.5× MS medium without sucrose in Petri dishes. Samples were taken before and after incubating the plates in the dark at 22°C for 4 or 7 d.
Gene constructs
The full coding region of the Arabidopsis FBN6 (AT5G19940) cDNA was PCR-amplified using primers AtFBN6-attB1-F and AtFBN6-attB2-R (Supplemental Table S1) and cloned into Gateway pDONR207 and subsequently pGWB420 (to create the FBN6-myc fusion) and pGWB454 (to create the FBN6-RFP fusion). Similarly, the FBN4-myc fusion was generated by PCR-mediated amplification of a cDNA sequence encoding full-length FBN4 (AT3G23400) with primers AtFBN4-attB1-F and AtFBN4-attB2-R (Supplemental Table S1) and cloned into Gateway pGWB420 via pDONR207. Similarly, the PSY-HA fusion was generated in pGWB414 following PCR amplification with primers AtPSY-attB1-F and AtPSY-attB2-R (Supplemental Table S1). Constructs encoding PRK-myc (Barja et al. 2021) and PSY-GFP (Welsch et al. 2018) were previously available in the lab. For the experiments in bacteria, plant cDNA sequences were PCR-amplified using gene-specific primers to remove their plastid transcript peptide (Supplemental Table S1). The Arabidopsis PSY sequence was cloned into a Gateway-compatible version of pET28 fused to an N-terminal 6× His tag, whereas the FBN-encoding cDNAs were cloned in pET23 (Novogen).
Activity assays in bacteria
Competent TOP10 cells were transformed with either plasmid pAC-BETA (with the P. ananatis genes crtE, crtB, crtI, and crtY required for β-carotene production in E. coli) or with the derived plasmid pAC-85b, which lacks the crtB gene encoding PSY (Cunningham and Gantt 2007). As indicated, cells were also co-transformed with plasmids to produce His-tagged plant PSY either alone or in combination with FBN6 or FBN4 proteins lacking their N-terminal plastid-targeting domains. Transformants were selected on Luria broth (LB) solid medium supplemented with antibiotics to final concentrations of 30 μg/ml chloramphenicol (to select for the pAC plasmids), 25 μg/ml kanamycin (to select for the pET28-His-PSY construct), or/and 100 μg/ml carbenicillin (to select for the pET23 plasmids encoding FBNs). To assess carotenoid production, overnight-grown 5 ml precultures of positive transformants were used to inoculate 50 ml of fresh LB media supplemented with the indicated antibiotics. After growth at 20°C and 200 rpm for 3 d, cells were harvested by centrifugation (6,000 × g, 10 min) and resuspended in 4 ml of acetone. The mixture was shaken at 55°C in darkness for 15 min in a ThermoMixer (Eppendorf) and then spin at 4°C (13,000 × g, 10 min). The supernatant was transferred to a new tube and evaporated using a SpeedVac. The remaining residue was resuspended in 1 ml DMSO for spectrophotometric quantification of carotenoids by measuring absorbance at 472 nm. Some samples diluted in acetone were used for HPLC-DAD analysis of the carotenoid composition (Barja et al. 2021). Levels were normalized to optical density at 600 nm (OD600).
Protein degradation
For protease accessibility assays, N. benthamiana leaves agroinfiltrated with constructs to produce PSY-HA together with FBN6-myc or FBN4-myc were collected after 4 d. The tissue was flash-frozen and ground in liquid nitrogen to a fine powder. About 75 mg of the resulting powder was mixed with 0.3 ml of lysis buffer containing 50 mm Tris–HCl pH = 7.5, 150 mm NaCl, 1 mm DTT, 1 mm MgCl2, 5% (v/v) glycerol, 1% (v/v) NP-40, and 2% (w/v) PVPP. The mixture was vortexed and centrifugated at 14,000 × g, 2 min. The supernatant was collected, and the amount of total protein content was quantified using a commercial Bradford assay (Pierce). Immediately after preparation, protein extracts were diluted with 50 mm Tris–HCl pH = 7.5 to a final concentration of 2 µg/µl of protein and incubated at 37°C with 0.1 μg/ml proteinase K (Invitrogen). After stopping the reaction with SDS-PAGE loading buffer, extracts were used for immunoblot analysis.
Microscopy
Fluorescent proteins transiently expressed in agroinfiltrated N. benthamiana leaves or stably produced in transgenic Arabidopsis lines were analyzed with a Leica TCS SP5 Confocal Laser Scanning Microscope using a 63× immersion objective. GFP fluorescence was detected using a PMT detector (gain 980) from 500 to 560 nm after argon laser excitation at 488 nm (29%), while RFP was excited by a Diode laser 561 (20%) and signal was detected using a PMT detector (gain 76) from 570 to 630 nm. Chlorophyll autofluorescence was detected using a PMT detector (gain 41) from 650 to 750 nm after argon laser exitation at 488 nm (29%).
Co-immunoprecipitation and immunoblot assays
N. benthamiana leaves were agroinfiltrated with the appropriate constructs and samples were collected after 4 d for co-immunoprecipitation experiments performed as described (Barja et al. 2021). Immunoblot analysis of plant protein extracts was carried out as described (Paulišić et al. 2021). In the case of E. coli, extracts for immunoblot analysis were made by directly boiling pelleted cells in 12.5 mm Tris–HCl pH = 6.8, 4% (w/v) SDS. Blots were incubated with available antibodies against FBN1a and FBN2 (Morelli et al. 2023) diluted 1:5,000 or commercial antibodies against GFP (Invitrogen, 1:2,000), Myc (Calbiochem, 1:5,000), HA (Sigma, 1:1,000) and 6× His (Proteintech, 1:5,000) epitopes. Quantification of the signals was carried out as described (Morelli et al. 2023).
Gene expression and metabolite analysis
RNA isolation, cDNA synthesis, and reverse transcription quantitative PCR experiments were carried out as described (Barja et al. 2021) using the primers listed in Supplemental Table S1. Normalized transcript levels were calculated using Arabidopsis UBC21 (At5g25760) as the reference gene. Carotenoids and chlorophylls were extracted from plant tissues, separated, identified, and quantified by HPLC-DAD (Barja et al. 2021).
Statistical analyses
One-way ANOVA followed by Tukey's multiple comparisons test and Student’s t-test analyses were used to determine statistically significant differences.
Accession numbers
Sequence data from this article can be found under the following accession numbers: At5g17230 (PSY), At5g19940 (FBN6), At3g23400 (FBN4), Solyc09g090330 (SlFBN4), At1g32060 (PRK), At5g25760 (UBC21), and ADD79329 (crtB).
Supplementary Material
Acknowledgments
We thank Venkatasalam Shanmugabalaji and Felix Kessler (University of Neuchatel, Switzerland) for the kind gift of the fibrillin antibodies, Tsuyoshi Nakagawa (Shimane University, Japan) for the Gateway vectors, Lourdes Gómez Gómez for the pAC-85b plasmid, and the Nottingham Arabidopsis Stock Centre (NASC) for providing fbn6-1 seeds. We also thank members of our laboratory for helpful discussions.
Contributor Information
Ariadna Iglesias-Sanchez, Institute for Plant Molecular and Cell Biology (IBMCP), CSIC-Universitat Politècnica de València, Valencia 46022, Spain; Centre for Research in Agricultural Genomics (CRAG) CSIC-IRTA-UAB-UB, Campus UAB Bellaterra, Barcelona 08193, Spain.
Juan Navarro-Carcelen, Institute for Plant Molecular and Cell Biology (IBMCP), CSIC-Universitat Politècnica de València, Valencia 46022, Spain.
Luca Morelli, Institute for Plant Molecular and Cell Biology (IBMCP), CSIC-Universitat Politècnica de València, Valencia 46022, Spain; Centre for Research in Agricultural Genomics (CRAG) CSIC-IRTA-UAB-UB, Campus UAB Bellaterra, Barcelona 08193, Spain.
Manuel Rodriguez-Concepcion, Institute for Plant Molecular and Cell Biology (IBMCP), CSIC-Universitat Politècnica de València, Valencia 46022, Spain.
Author contributions
A.I.-S., J.N.-C., L.M., and M.R.-C. designed the research and analyzed data; A.I.-S., J.N.-C., and L.M. conducted the experiments; M.R.-C. wrote the paper.
Supplemental data
The following materials are available in the online version of this article.
Supplemental Figure S1 . Characterization of 35S:FBN6-RFP lines.
Supplemental Table S1 . Primers used in this work.
Supplemental Table S2 . Phytoene levels in control and NFZ-treated N. benthamiana leaves agroinfiltrated with FBN6 and/or PSY constructs.
Supplemental Table S3 . Photosynthetic pigment levels in N. benthamiana leaves agroinfiltrated with FBN6, PSY, and/or crtB constructs.
Supplemental Table S4 . Photosynthetic pigment levels in etiolated and de-etiolating WT and fbn6 Arabidopsis seedlings.
Supplemental Table S5 . Photosynthetic pigment levels in WT and fbn6 Arabidopsis seedlings exposed to HL.
Funding
This work was funded by grants from Spanish MCIN/AEI/10.13039/501100011033 and European NextGeneration EU/PRTR and PRIMA programs to M.R.-C. (PID2020-115810GB-I00 and UToPIQ-PCI2021-121941). M.R.-C. is also supported by Generalitat Valenciana (PROMETEU/2021/056 and AGROALNEXT/2022/067). Our group is a member of CaRed (Spanish Carotenoid Network) funded by MCIN/AEI (RED2022-134577-T). A.I.-S. and J.N.-C. received predoctoral fellowships from MCIN/AEI (PRE2018-083610 and PRE2021-098681, respectively).
Dive Curated Terms
The following phenotypic, genotypic, and functional terms are of significance to the work described in this paper:
References
- Ahrazem O, Diretto G, Argandoña Picazo J, Fiore A, Rubio-Moraga Á, Rial C, Varela RM, Macías FA, Castillo R, Romano E, et al. The specialized roles in carotenogenesis and apocarotenogenesis of the phytoene synthase gene family in saffron. Front Plant Sci. 2019:10:249. 10.3389/fpls.2019.00249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barja MV, Ezquerro M, Beretta S, Diretto G, Florez-Sarasa I, Feixes E, Fiore A, Karlova R, Fernie AR, Beekwilder J, et al. Several geranylgeranyl diphosphate synthase isoforms supply metabolic substrates for carotenoid biosynthesis in tomato. New Phytol. 2021:231(1):255–272. 10.1111/nph.17283 [DOI] [PubMed] [Google Scholar]
- Barsan C, Zouine M, Maza E, Bian W, Egea I, Rossignol M, Bouyssie D, Pichereaux C, Purgatto E, Bouzayen M, et al. Proteomic analysis of chloroplast-to-chromoplast transition in tomato reveals metabolic shifts coupled with disrupted thylakoid biogenesis machinery and elevated energy-production components. Plant Physiol. 2012:160(2):708–725. 10.1104/pp.112.203679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouchnak I, Brugière S, Moyet L, Le Gall S, Salvi D, Kuntz M, Tardif M, Rolland N. Unraveling hidden components of the chloroplast envelope proteome: opportunities and limits of better MS sensitivity. Mol Cell Proteomics. 2019:18(7):1285–1306. 10.1074/mcp.RA118.000988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cunningham FX Jr, Gantt E. A portfolio of plasmids for identification and analysis of carotenoid pathway enzymes: adonis aestivalis as a case study. Photosynth Res. 2007:92(2):245–259. 10.1007/s11120-007-9210-0 [DOI] [PubMed] [Google Scholar]
- Davidi L, Levin Y, Ben-Dor S, Pick U. Proteome analysis of cytoplasmatic and plastidic β-carotene lipid droplets in Dunaliella bardawil. Plant Physiol. 2015:167(1):60–79. 10.1104/pp.114.248450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deruère J, Römer S, D’Harlingue A, Backhaus RA, Kuntz M, Camara B. Fibril assembly and carotenoid overaccumulation in chromoplasts: a model for supramolecular lipoprotein structures. Plant Cell. 1994:6(1):119–133. 10.1105/tpc.6.1.119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egea I, Barsan C, Bian W, Purgatto E, Latché A, Chervin C, Bouzayen M, Pech JC. Chromoplast differentiation: current status and perspectives. Plant Cell Physiol. 2010:51(10):1601–1611. 10.1093/pcp/pcq136 [DOI] [PubMed] [Google Scholar]
- Ferro M, Brugière S, Salvi D, Seigneurin-Berny D, Court M, Moyet L, Ramus C, Miras S, Mellal M, Le Gall S, et al. AT-CHLORO, a comprehensive chloroplast proteome database with subplastidial localization and curated information on envelope proteins. Mol Cell Proteomics. 2010:9(6):1063–1084. 10.1074/mcp.M900325-MCP200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gámez-Arjona FM, Raynaud S, Ragel P, Mérida Á. Starch synthase 4 is located in the thylakoid membrane and interacts with plastoglobule-associated proteins in Arabidopsis. Plant J. 2014:80(2):305–316. 10.1111/tpj.12633 [DOI] [PubMed] [Google Scholar]
- Kim I, Kim HU. The mysterious role of fibrillin in plastid metabolism: current advances in understanding. J Exp Bot. 2022:73(9):2751–2764. 10.1093/jxb/erac087 [DOI] [PubMed] [Google Scholar]
- Kim EH, Lee Y, Kim HU. Fibrillin 5 is essential for plastoquinone-9 biosynthesis by binding to solanesyl diphosphate synthases in arabidopsis. Plant Cell. 2015:27(10):2956–2971. 10.1105/tpc.15.00707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreslavski VD, Khudyakova AY, Strokina VV, Shirshikova GN, Pashkovskiy PP, Balakhnina TI, Kosobryukhov AA, Kuznetsov VV, Allakhverdiev SI. Impact of high irradiance and UV-B on the photosynthetic activity, pro-/antioxidant balance and expression of light-activated genes in Arabidopsis thaliana hy4 mutants grown under blue light. Plant Physiol Biochem. 2021:167:153–162. 10.1016/j.plaphy.2021.07.030 [DOI] [PubMed] [Google Scholar]
- Lee K, Lehmann M, Paul MV, Wang L, Luckner M, Wanner G, Geigenberger P, Leister D, Kleine T. Lack of FIBRILLIN6 in Arabidopsis thaliana affects light acclimation and sulfate metabolism. New Phytol. 2020:225(4):1715–1731. 10.1111/nph.16246 [DOI] [PubMed] [Google Scholar]
- Li R, Zeng Q, Zhang X, Jing J, Ge X, Zhao L, Yi B, Tu J, Fu T, Wen J, et al. Xanthophyll esterases in association with fibrillins control the stable storage of carotenoids in yellow flowers of rapeseed (Brassica juncea). New Phytol. 2023:240(1):285–301. 10.1111/nph.18970 [DOI] [PubMed] [Google Scholar]
- Lundquist PK, Poliakov A, Bhuiyan NH, Zybailov B, Sun Q, van Wijk KJ. The functional network of the arabidopsis plastoglobule proteome based on quantitative proteomics and genome-wide coexpression analysis. Plant Physiol. 2012:158(3):1172–1192. 10.1104/pp.111.193144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morelli L, Torres-Montilla S, Glauser G, Shanmugabalaji V, Kessler F, Rodriguez-Concepcion M. Novel insights on the contribution of plastoglobules and reactive oxygen species to chromoplast differentiation. New Phytol. 2023:237(5):1696–1710. 10.1111/nph.18585 [DOI] [PubMed] [Google Scholar]
- Paulišić S, Qin W, Arora Verasztó H, Then C, Alary B, Nogue F, Tsiantis M, Hothorn M, Martínez-García JF. Adjustment of the PIF7-HFR1 transcriptional module activity controls plant shade adaptation. EMBO J. 2021:40(1):e104273. 10.15252/embj.2019104273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez-Concepcion M, Avalos J, Bonet ML, Boronat A, Gomez-Gomez L, Hornero-Mendez D, Limon MC, Meléndez-Martínez AJ, Olmedilla-Alonso B, Palou A, et al. A global perspective on carotenoids: metabolism, biotechnology, and benefits for nutrition and health. Prog Lipid Res. 2018:70:62–93. 10.1016/j.plipres.2018.04.004 [DOI] [PubMed] [Google Scholar]
- Rodríguez-Villalón A, Gas E, Rodríguez-Concepción M. Phytoene synthase activity controls the biosynthesis of carotenoids and the supply of their metabolic precursors in dark-grown Arabidopsis seedlings. Plant J. 2009:60(3):424–435. 10.1111/j.1365-313X.2009.03966.x [DOI] [PubMed] [Google Scholar]
- Schmidt M, Gessner G, Luff M, Heiland I, Wagner V, Kaminski M, Geimer S, Eitzinger N, Reissenweber T, Voytsekh O, et al. Proteomic analysis of the eyespot of Chlamydomonas reinhardtii provides novel insights into its components and tactic movements. Plant Cell. 2006:18(8):1908–1930. 10.1105/tpc.106.041749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shanmugabalaji V, Besagni C, Piller LE, Douet V, Ruf S, Bock R, Kessler F. Dual targeting of a mature plastoglobulin/fibrillin fusion protein to chloroplast plastoglobules and thylakoids in transplastomic tobacco plants. Plant Mol Biol. 2013:81(1–2):13–25. 10.1007/s11103-012-9977-z [DOI] [PubMed] [Google Scholar]
- Shumskaya M, Bradbury LMT, Monaco RR, Wurtzel ET. Plastid localization of the key carotenoid enzyme phytoene synthase is altered by isozyme, allelic variation, and activity. Plant Cell. 2012:24(9):3725–3741. 10.1105/tpc.112.104174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh DK, Laremore TN, Smith PB, Maximova SN, McNellis TW. Knockdown of FIBRILLIN4 gene expression in apple decreases plastoglobule plastoquinone content. PLoS One. 2012:7(10):1–10. 10.1371/journal.pone.0047547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh DK, McNellis TW. Fibrillin protein function: the tip of the iceberg? Trends Plant Sci. 2011:16(8):432–441. 10.1016/j.tplants.2011.03.014 [DOI] [PubMed] [Google Scholar]
- Sun H, Ren M, Zhang J. Genome-wide identification and expression analysis of fibrillin (FBN) gene family in tomato (Solanum lycopersicum L.). PeerJ. 2022:10:e13414. 10.7717/peerj.13414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki M, Takahashi S, Kondo T, Dohra H, Ito Y, Kiriiwa Y, Hayashi M, Kamiya S, Kato M, Fujiwara M, et al. Plastid proteomic analysis in tomato fruit development. PLoS One. 2015:10(9):e0137266. 10.1371/journal.pone.0137266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toledo-Ortiz G, Huq E, Rodríguez-Concepción M. Direct regulation of phytoene synthase gene expression and carotenoid biosynthesis by phytochrome-interacting factors. Proc Natl Acad Sci U S A. 2010:107(25):11626–11631. 10.1073/pnas.0914428107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres-Romero D, Gómez-Zambrano Á, Serrato AJ, Sahrawy M, Mérida Á. Arabidopsis fibrillin 1–2 subfamily members exert their functions via specific protein–protein interactions. J Exp Bot. 2022:73(3):903–914. 10.1093/jxb/erab452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vainstein A, Halevy AH, Vishnevetsky M. Chromoplast biogenesis in Cucumis sativus corollas. Plant Physiol. 1994:104(2):321–326. 10.1104/pp.104.2.321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Wijk KJ, Kessler F. Plastoglobuli: plastid microcompartments with integrated functions in metabolism, plastid developmental transitions, and environmental adaptation. Annu Rev Plant Biol. 2017:68(1):253–289. 10.1146/annurev-arplant-043015-111737 [DOI] [PubMed] [Google Scholar]
- Welsch R, Zhou X, Yuan H, Álvarez D, Sun T, Schlossarek D, Yang Y, Shen G, Zhang H, Rodriguez-Concepcion M, et al. Clp protease and OR directly control the proteostasis of phytoene synthase, the crucial enzyme for carotenoid biosynthesis in Arabidopsis. Mol Plant. 2018:11(1):149–162. 10.1016/j.molp.2017.11.003 [DOI] [PubMed] [Google Scholar]
- Zhou X, Sun T, Owens L, Yang Y, Fish T, Wrightstone E, Lui A, Yuan H, Chayut N, Burger J, et al. Carotenoid sequestration protein FIBRILLIN participates in CmOR-regulated β-carotene accumulation in melon. Plant Physiol. 2023:193(1):643–660. 10.1093/plphys/kiad312 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.