Abstract
Emerging resistance to currently used antibiotics is a global public health crisis. As most of the biosynthetic capacity within the bacterial kingdom has remained silent in previous antibiotic discovery efforts, uncharacterized biosynthetic gene clusters found in bacterial genome sequencing studies remain an appealing source of antibiotics with distinctive modes of action. Here we report the discovery of a naturally inspired lipopeptide antibiotic termed cilagicin, which we chemically synthesized based on a detailed bioinformatic analysis of the cil biosynthetic gene cluster. Cilagicin’s ability to sequester two distinct, indispensable undecaprenyl phosphates used in cell wall biosynthesis, together with the absence of detectable resistance in laboratory tests and among multi-drug resistant clinical isolates, makes it an appealing candidate for combating antibiotic resistant pathogens.
The discovery and therapeutic development of natural product antibiotics, especially those produced by microbes, has dramatically reduced mortality caused by bacterial infections.(1) Unfortunately, this situation is challenged by antibiotic resistance, which is rising at a faster rate than the introduction of molecules with modes of action (MOAs) capable of circumventing existing resistance mechanisms.(2, 3) From a clinical development standpoint, non-ribosomal peptide synthetase (NRPS) encoded lipopeptides are an appealing potential source of future antibiotics as they have a history of inhibiting bacterial growth by diverse MOAs.(4, 5) Bacterial genome sequencing efforts have uncovered a large number of biosynthetic gene clusters (BGCs) that do not appear to encode for known natural products, including many BGCs that are predicted to encode undescribed lipopeptides. These BGCs likely contain genetic instructions for the biosynthesis of antibiotics with diverse MOAs that could help to replenish antibiotic discovery pipelines. Unfortunately, the vast majority of sequenced BGCs remain silent in the laboratory, and the molecules they encode remain a mystery. Here we used a phylogenetic analysis of condensation starter (Cs) domain sequences, which introduce the acyl substituent into lipopeptides, to identify the cryptic cil BGC as a potential source of an uncharacterized lipopeptide antibiotic.
To identify BGCs that might encode lipopeptide antibiotics with distinctive MOAs, we collected NRPS BGCs from ~10,000 sequenced bacterial genomes. Clinically relevant lipopeptide antibiotics (e.g., polymyxin, daptomycin, etc.) have historically tended to be larger macrocyclic structures, and therefore BGCs predicted to encode peptides with fewer than 5 amino acids (i.e., containing less than 5 adenylation domains) were removed from this collection. A recent screen of this collection for BGCs that were predicted to encode congeners of known antibiotics guided our synthesis of macolacin, a colistin analog that is effective against pathogens that express the mcr-1 resistance gene.(6) In the current analysis we searched for uncharacterized BGC families. The key conserved feature across lipopeptides is the presence of an N-terminal lipid that is installed by a Cs domain.(7–9) Among the sequenced large NRPS BGCs we collected, we identified 3,426 that contained a Cs domain. Cs domain sequences from these BGCs were used to construct a phylogenetic tree that guided our discovery efforts. As we have seen with other biosynthetic genes, sequences arising from BGCs sharing close common ancestors and hence the same MOA are likely to group together into the same clade.(10–12) By extension, clades that do not contain any sequences from characterized BGCs would represent candidates for identifying structurally and mechanistically new antibiotics. The Cs domain phylogenetic tree contained a number of clades that were not associated with any characterized lipopeptides; however, one was particularly intriguing as it fell into a larger group of sequences where most other clades were associated with antibiotic biosynthesis. These included BGCs for a number of clinically used, as well as clinically appealing, antibiotics (e.g., polymyxins, tridecaptins and brevicidines). This “orphan” Cs clade we identified contained three closely related sequences that arose from the same BGC found in two different sequenced Paenibacillus mucilaginosus strains (KNP414 and K02) (Fig. 1A). Based on gene content and gene organization, this BGC, which we have called the cil BGC, did not resemble any characterized BGCs. The vast majority of sequenced BGCs remain silent in the laboratory, even when interrogated with advanced synthetic biology tools.(13). With the power of modern synthetic organic chemistry and the increasing accuracy of natural product structure prediction algorithms, it is now possible to generate a bioactive molecule from the genetic instructions found in the primary sequence of a BGC by first bioinformatically predicting the encoded structure and then chemically synthesizing the predicted structure (i.e., produce a synthetic-bioinformatic natural product, syn-BNP).(14–16) In this study, we used a syn-BNP approach to generate lipopeptide structures based on the cil BGC and then tested these small molecules for antibacterial activity.
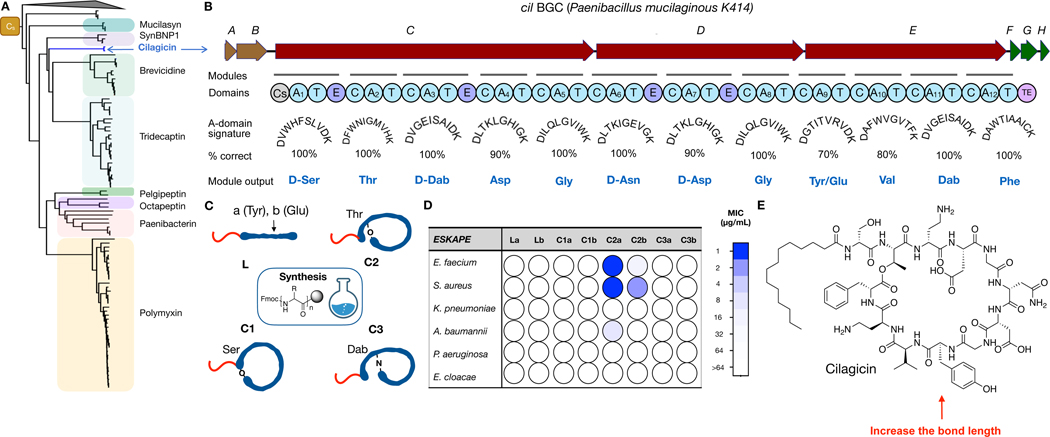
Fig. 1.
Discovery of cilagicin. (A) Condensation starter (Cs) domains from sequenced NRPS BGCs were used to construct a phylogenetic tree. Clades associated with characterized antibiotic BGCs are labeled. The “orphan” cilagicin clade is labeled in blue. (B) The cil BGC contains three NRPS open reading frames (cilC-E). Biosynthesis of cilagicin is predicted to start from a Cs domain in CilC. It is then expected to proceed using one A (adenylation) and T (thiolation) domain containing initiation module followed by 11 C (condensation), A and T domain containing extender modules. The substrate specificity of the cil BGC A-domains was predicted based on a comparison of each A-domain’s substrate binding pocket to the 10 amino acid A-domain signature sequences found in functional characterized BGCs. E (epimerization) domains in modules 1, 3, 6 and 7 are predicted to result in the incorporation of D amino acids. The TE (thioesterase) domain at the end of CilE releases the mature structure from the final T domain as either a linear or cyclic product. (C) Cartoons of the four different peptide topologies that were synthesized from the linear peptide predicted to arise from the cil BGC. Position nine was either Tyr (a) or Glu (b). L is a linear peptide. C1, C2 and C3 are cyclized through the C-terminal carboxyl group and Ser-1, Thr-2 or Dab-3, respectively. Dab, 2,4-diaminobutyric acids. (D) MIC data against the ESKAPE pathogens for the eight predicted synthetic structures depicted in (C). Concentrations tested ranged from 1 µg/mL (blue) to 64 µg/mL (white). Data are representative of 3 independent experiments. (E) Structure of the antibiotic cilagicin, which corresponds to C2a in (C).
The cil BGC contains three NRPS open reading frames (cil C-E) that encode 12 distinct modules (Fig. 1B, Table S1). The biosynthesis of a 12-mer lipopeptide is predicted to begin with the Cs domain at the N-terminus of CilC and end with the thioesterase at the C-terminus of CilE. The composition of each module’s A-domain substrate binding pocket (i.e., substrate signature based on positions 235, 236, 239, 278, 299, 301, 322, 330, 331, 517 of the A-domain) was used to predict the 12 monomers used by this BGC.(17) Eleven A-domains had perfect or near perfect (80%) matches to characterized A-domains, and therefore we could make high confidence predictions for the amino acid incorporated by these modules. The A-domain substrate signature from module 9 had equally good matches (70%) to two amino acids (Tyr and Glu). This analysis gave us two potential predicted linear lipopeptide products for the cil BGC (La and Lb) (Fig. 1c). Epimerization domains found in modules 1, 3, 6 and 7, indicated that these amino acids appear in the D-configuration in the cil BGC product. The absence of any genes predicted to encode tailoring enzymes (i.e., methyltransferase, hydroxylation, amino transferase, glycosyl transferase, etc.) within 10 kB of the cil NRPS genes suggested that the product of the cil BGC was not modified beyond the NRPS produced lipopeptide.(18)
Naturally occurring lipopeptides appear as either linear or cyclic structures. The predicted cil linear peptide contains three amino acids that could serve as nucleophiles (D-Ser-1, Thr-2, D-Dab-3) for cyclization through the C-terminal carboxyl. Bringing together our linear peptide prediction and three potential cyclization sites yielded eight structures (2 linear, 6 cyclic) that we predicted could arise from the cil BGC (Fig. 1C). Each of the eight potential BGC products was generated by solid-phase peptide synthesis (Table S2). The cil BGC does not contain any lipid biosynthetic genes and therefore we predicted that the lipid found on the product of the cil BGC would arise directly from native fatty acid biosynthesis. Among characterized bacterial lipopeptides, myristic acid is one of the most frequently seen simple lipids and therefore all synthetic peptides were N-terminal acylated with myristic acid.
All eight synthetic structures were assayed for activity against the ESKAPE pathogens (Enterococcus faecium, Staphylococcus aureus, Klebsiella pneumoniae, Acinetobacter baumannii, Pseudomonas aeruginosa, and Enterobacter species) (Fig. 1D, Table S3). The 11 amino acid macrocycle that was cyclized through the hydroxyl of Thr-2 and contained tyrosine at position 9 (compound-C2a) showed potent antibacterial activity against the two Gram-positive ESKAPE pathogens (MIC 1 µg/mL). None of the remaining close analogs we synthesized showed any significant activity against these strains. We have called the active structure cilagicin (Fig. 1E).
Cilagicin was active against all Gram-positive pathogens that we tested (Table 1). Cilagicin was active against a number of difficult-to-treat vancomycin-resistant Enterococci pathogens as well as Clostridioides difficile, both of which are considered urgent and serious threat pathogens by the CDC.(19) It was also active against all antibiotic resistant Gram-positive pathogens that we tested. It maintained potent activity against a panel of 19 S. aureus strains that show different patterns of resistance to clinically relevant families of antibiotics (Table S4), and in stark contrast to FDA approved antibiotics, it maintained potent activity against all strains found in a panel of 30 vancomycin-resistant Enterococci clinical isolates (Table S5). This collection is highly enriched in multidrug resistant (MDR) isolates with more than half exhibiting resistance to between five and eight different clinically used antibiotics. Cilagicin was largely inactive against Gram-negative bacteria with the exception of A. baumannii (Table S6) and outer membrane permeabilized E. coli BAS849, suggesting that the outer membrane of Gram-negative bacteria blocks cilagicin’s access to its target. Even at the highest concentration we tested cilagicin did not show human cell line cytotoxicity (Table 1).
Table 1.
Activity of cilagicin against microorganisms and human cells.
| Pathogens/Human cells | Cilagicin MIC (μg/mL)a |
|---|---|
|
| |
| Gram-positive | |
| Staphylococcus aureus USA300 (MRSA) | 1 |
| Staphylococcus aureus BAA1717(BRSA) | 0.5 |
| Enterococcus faecium EF18 (VRE) | 0.5 |
| Enterococcus faecalis AR785 (VRE) | 0.5 |
| Enterococcus gallinarum AR784 (VRE) | 0.5 |
| Enterococcus casseliflavus AR798 (VRE) | 0.125 |
| Streptococcus pneumoniae Rb | 0.5 |
| Streptococcus pneumoniae Tigr4b | 0.25 |
| Clostridium difficile HM89c | 2 |
| Clostridium difficile HM746c | 2 |
| Streptococcus pyrogens ATCC19615 | 0.125 |
| Streptococcus agalactiae BAA2675 | 1 |
| Streptococcus agalactiae BAA1176 | 1 |
| Bacillus subtilis 168A1 | 2 |
| Gram-negative | |
| Acinetobacter baumannii ATCC17978 | 8 |
| Escherichia coli BAS849 | 4 |
| Escherichia coli ATCC25922 | >64 |
| Klebsiella pneumoniae ATCC13833 | >64 |
| Pseudomonas aeruginosa PAO1 | >64 |
| Enterobacter cloacae ATCC13047 | >64 |
| Human cell line | |
| HEK293 | >64c |
The MIC was tested by broth microdilution.
Bacteria were cultured under 5% CO2;
Bacteria were cultured under anaerobic condition; MRSA, methicillin-resistant S. aureus; BRSA, bacitracin-resistant S. aureus; VRE, vancomycin-resistant Enterococci.
IC50
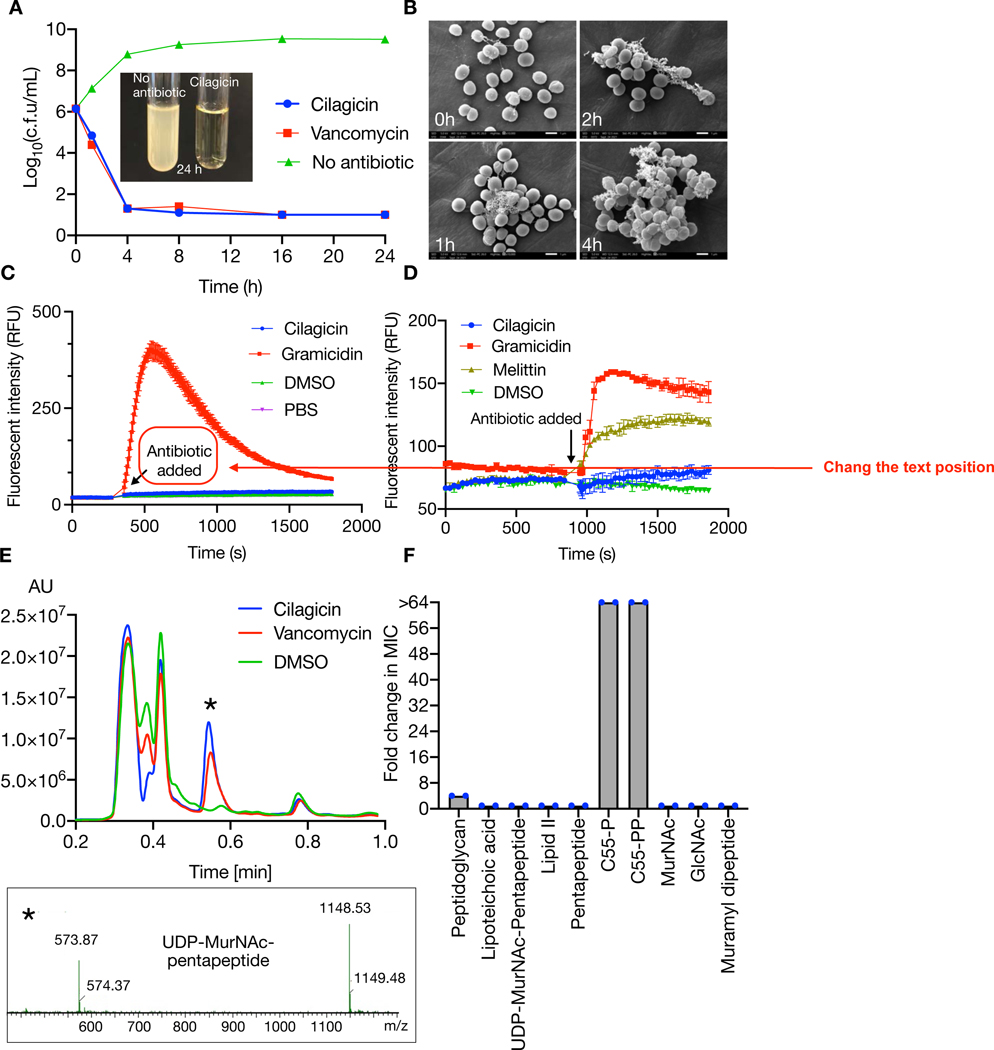
In a time-dependent killing curve analysis, cilagicin was found to be bactericidal and to reduce the number of viable bacteria by more than four orders of magnitude after 4 hours (Fig. 2A). Electron microscopy images of cilagicin treated cells showed cell collapse over time (Fig. 2B). In an effort to elucidate cilagicin’s MOA we tried to raise mutants by direct plating of S. aureus on cilagicin. In these direct plating experiments we never observed any colonies that showed more than a 1-fold increase in MIC. To explore the possibility of cilagicin having a detergent-like activity, we tested cilagicin for membrane depolarization and cell lytic activities using 3,3’-dipropylthiadicarboncyanine iodide [DiSC3(5)] and SYTOX based fluorescence assays, respectively (Fig. 2C, D).(20–22) No response was detected in either assay when S. aureus was exposed to even 8-fold cilagicin’s MIC, ruling out membrane disruption as its MOA.
Fig. 2. Cilagicin mode of action.
(A) Survival of S. aureus USA300 after timed exposure to 10x the MIC of cilagicin. DMSO and Vancomycin (10x MIC) were included as controls. CFU were counted three independent times and plotted as mean ± SD. (B) Scanning electron microscopy image of S. aureus USA300 cultures treated with cilagicin. Conditions were the same as in (A). (C) Cell lysis or (D) membrane depolarization in cilagicin treated S. aureus cultures was monitored using SYTOX and DiSC3(5) dyes, respectively. Data are presented as the mean of three independent experiments ± SD. (E) Accumulation of UDP-MurNAc-pentapeptide after treating S. aureus cultures with cilagicin (1x MIC) was monitored by LCMS. DMSO and vancomycin (10x MIC) treated cultures were examined as controls. UDP-MurNAc-pentapeptide corresponds to [M-H]- = 1148.53 and [M-2H]2- =573.87. (F) Fold change in cilagicin MIC upon treatment of S. aureus with 5-fold molar excess of different lipid II intermediates. The peptidoglycan mixture was added at 100 µg/mL. Data are representative of two independent experiments.
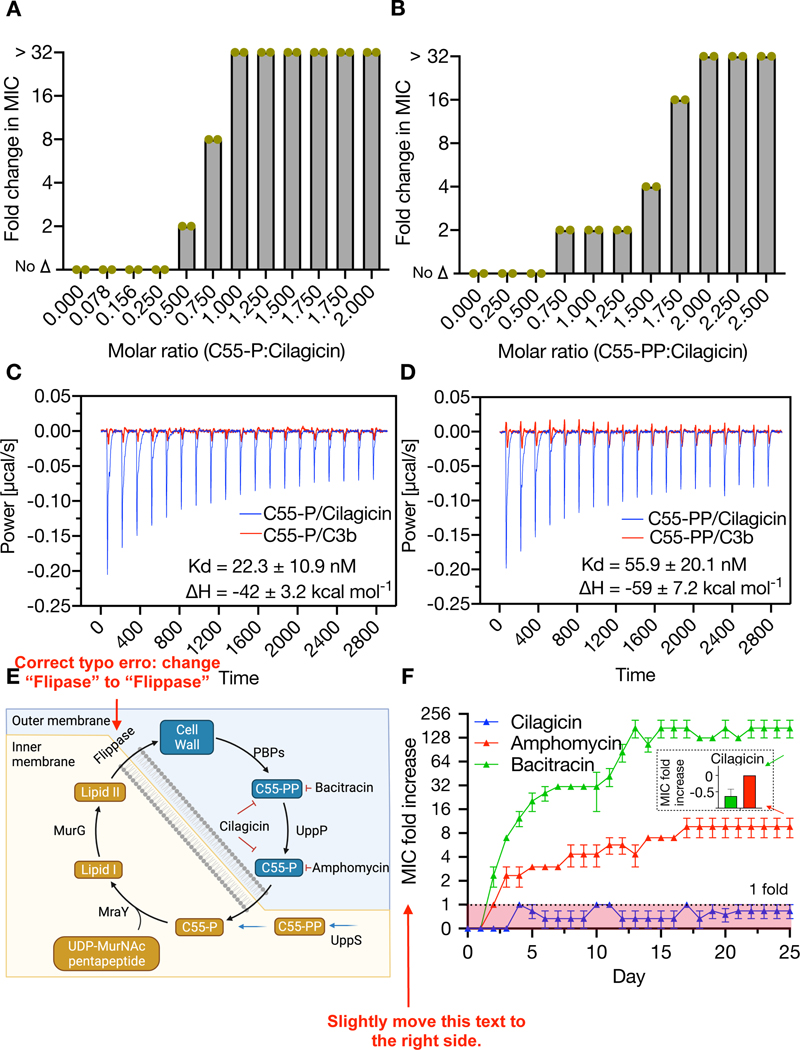
Cilagicin is a zwitterion with two positively charged residues (3-D-Dab, 11-D-Dab) and two negatively charged residues (4-Asp, 7-D-Asp). Charged lipopeptide antibiotics often do not enter the cell and instead function by disrupting synthesis of the cell wall outside the cell membrane.(11, 23, 24) Antibiotics that block peptidoglycan biosynthesis lead to the accumulation of the lipid II precursor UDP-MurNAc-pentapeptide, which is easily detected by LCMS in antibiotic exposed cultures.(11, 25–27) LCMS analysis of S. aureus cultures exposed to cilagicin (1x MIC) showed an obvious accumulation of UDP-MurNAc-pentapeptide (Fig. 2E). As it is often much more difficult to alter a small molecule target than a protein target through genomic mutations, our inability to identify cilagicin resistant mutants hinted at the binding of a small molecule instead of a protein as the mode of inhibiting cell wall biosynthesis. To identify metabolite(s) that interact with cilagicin we screened a series of lipid II intermediates for the ability to suppress cilagicin’s antibacterial activity (Fig. 2F). In these studies, the MIC of cilagicin against S. aureus USA300 was determined in the presence of a 5-fold molar excess of each metabolite. Two of the compounds we tested, undecaprenyl phosphate (C55-P) and undecaprenyl pyrophosphate (C55-PP), showed dose dependent inhibition of cilagicin’s antibacterial activity (Fig. 3A, 3B).
Fig. 3. Interaction of cilagicin with C55-P and C55-PP.
Fold change in MIC of cilagicin treated cultures of S. aureus USA300 in the presence of different concentrations of C55-P (A) or C55-PP (B). The highest concentration tested was 32x the MIC. Data from two independent experiments are presented. Isothermal titration calorimetry data for cilagicin or its inactive analog C3b interacting with either C55-P (C) or C55-PP (D). Two independent experiments were performed with similar results. (E) Diagram of the role of C55-P and C55-PP in Gram-positive cell wall biosynthesis. (F) Resistance acquisition during serial passaging of S. aureus USA300 in the presence of sub-MIC levels of cilagicin, bacitracin or amphomycin. Data shown represent the mean of three independent experiments ± SEM. Inset: The MIC fold increase of cilagicin against bacitracin (green) and amphomycin (red) resistant strains on 25 days.
C55-P is a monophosphorylated 55 carbon-long isoprene that is essential for transporting intermediates in the biosynthesis of cell wall carbohydrate polymers (e.g., peptidoglycan, O antigen, teichoic acids and others) across the bacteria cell membrane.(28, 29) C55-PP is the di-phosphorylated version of the same 55 carbon isoprene. It is produced both de novo and recycled from C55-P during the biosynthesis of the cell wall. Its dephosphorylation by membrane-embedded pyrophosphatases generate the cellular pool of C55-P that is required for cell wall synthesis.(30). Using isothermal titration calorimetry (ITC), we observed that cilagicin binds C55P and C55PP (Fig. 3C, 3D), but a representative inactive analog from our initial synthesis studies (cilagicin-3b) did not bind either compound. Collectively, our MOA studies are consistent with cilagicin being able to sequester both C55-P and C55-PP and thus acting as a bifunctional antimicrobial.
Bacteria only have a small pool of free undecaprenyl carrier lipids (~105 molecules per cell) to use in the transfer of critical biosynthetic intermediates across the cell membrane.(31) Although disruption of this process is an appealing antibacterial MOA it remains underexploited clinically as only a few antibiotics have been identified that bind even one undecaprenyl phosphate. These include the antibiotics bacitracin and tripropeptin, which specifically bind C55-PP in zinc and calcium dependent manners, respectively.(32, 33) The only known antibiotics that bind C55-P are the calcium dependent lipopeptide friulimicin and its close congeners (e.g., amphomycin, laspartomycin).(26, 27, 34) Binding C55-P directly reduces the amount of available C55-P, whereas sequestering C55-PP indirectly reduces C55-P by preventing C55-PP dephosphorylation. In either case, this disrupts the flow of peptidoglycan precursors into the cell-wall, ultimately leading to cell death (Fig. 3E).
Bacitracin is used clinically as a topical antibiotic and friulimicin is in development for use in animal health. Unfortunately, bacteria exposed to antibiotics that bind a single undecaprenyl phosphate are reported to readily develop resistance. Antibiotics with multiple molecular targets tend to have reduced rates of resistance because of the difficulty associated with altering multiple targets simultaneously. We predicted therefore that in the case of cilagicin, its ability to bind both undecaprenyl phosphates (i.e., two distinct small molecules) would lead to a reduced resistance rate compared to antibiotics that bind a single phosphorylated undecaprenyl moiety. As we had failed to identify mutants resistant to cilagicin by direct plating on antibiotic containing media, we attempted to raise S. aureus resistant mutants by daily serial passage in the presence of sub-MIC levels of antibiotic. We did this using cilagicin, bacitracin or the friulimicin congener amphomycin to allow a direct comparison of resistance rates for antibiotics that bind either one or two phosphorylated undecaprenyl moieties. S. aureus rapidly developed resistance to both bacitracin and amphomycin. MICs for these antibiotics increased by 8- to 256-fold, respectively during the course of the serial passage experiment. In contrast, after 25 days of constant exposure to cilagicin we observed no higher than a doubling of its MIC (Fig 3F). In addition, neither the highly bacitracin resistant mutants nor the highly amphomycin resistant mutants that we generated showed cross resistance to cilagicin. Interestingly, the cil BGC is found in the genome of P. mucilaginosus. The genus Paenibacillus is Gram-variable. P. mucilaginosus’s reported negative Gram staining suggests it contains an outer membrane that could protect it from cilagicin’s toxicity thus potentially eliminating the need for self-resistance elements to have evolved in nature.(35, 36)
Bacitracin resistance arises from either the release of C55-PP from bacitracin by the ABC transporter BceAB, increased production of C55-PP by the undecaprenyl-pyrophosphate phosphatase BcrC or by a still ill-defined mechanism associated with phage shock protein-like protein expression (LiaI and LiaH).(37) In the case of C55-P binding, antibiotic resistance is associated with the expression of the cell envelope stress response regulator (38), which provides only low level resistance compared to what is seen for bacitracin resistance mechanisms (<8 vs >254 fold MIC increases, respectively). As the binding of C55-P alone appears to be difficult to overcome, it was not surprising that sequestration of the entire pool of undecaprenyl phosphates by cilagicin would further reduce the propensity for resistance to develop. In general, the sequestration of an essential extracellular metabolite is an appealing MOA as the development of resistance has often proved difficult in laboratory experiments using individual pathogens.(39) Historically, once these antibiotics are exposed to the global pool of resistance determinants present in the clinical setting resistance might appear, albeit often at a much slower rate than is seen for other MOAs. Although we have not yet observed cilagicin resistance in MDR clinical isolates this does not rule out the eventual appearance of resistance with broader environmental exposure.
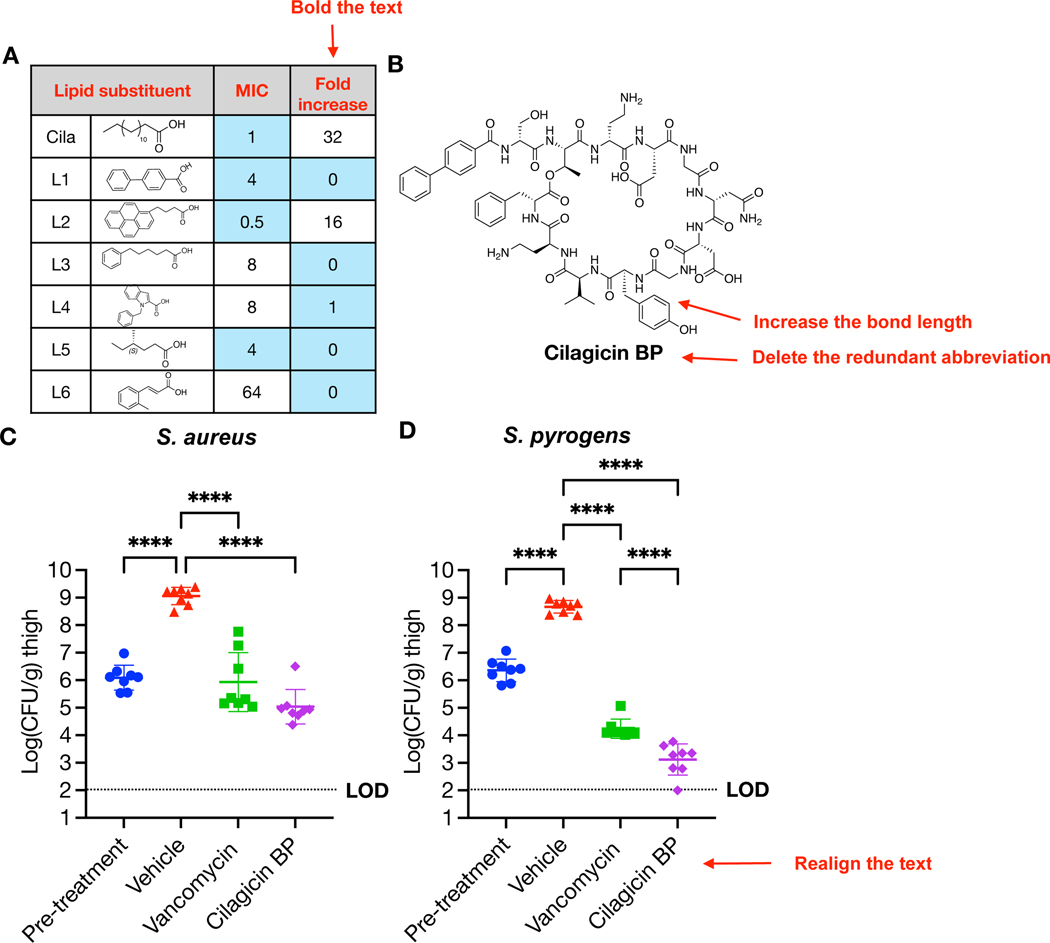
Our initial pharmacological assessment in mice showed cilagicin had high plasma bioavailability when delivered by intraperitoneal (IP) injection (Fig. S2). However, it did not reduce bacterial burden in an animal infection model. We subsequently observed that cilagicin’s antibacterial activity was significantly suppressed in the presence of serum suggesting that high serum binding might have also limited its in vivo activity (Fig. 4A). We therefore attempted to generate a cilagicin analog with reduced serum binding. To do this we created a collection of cilagicin analogs with different N-terminal lipids and compared their MICs in the presence and absence of serum. An analog containing a biphenyl N-terminal substituent (cilagicin BP) maintained good antimicrobial activity and showed no increase in MIC in the presence of serum (Fig. 4B, Table S7). Similar biphenyl substituents are found in a number of synthetically optimized natural product antibiotics including glycopeptides (oritavancin) and other lipopeptides (macolacin).(6, 40) This change of lipid substituent did not alter the antibiotics MOA (Fig. S3A), and cilagicin BP continued to show no hemolytic activity and no human cell cytotoxicity (Fig. S3B, 3C). We assessed the in vivo efficacy of cilagicin BP using a neutropenic mouse thigh model. At 24 hours post infection, cilagicin BP showed significant antibacterial activity against S. aureus USA300 at 40 mg/kg (thrice daily, TID), resulting in an almost 4 log10 reduction in colony forming units (CFU) compared to the vehicle control (Fig. 4C). Next, we evaluated cilagicin BP against S. pyrogens ATCC19615 in the same neutropenic thigh model. In this case, cilagicin BP showed an even more impressive reduction (> 5 log10) in bacterial burden compared to the vehicle control, which was consistent with the lower MIC values for this pathogen in vitro (Fig. 4D). Against S. pyrogens, cilagicin BP resulted in more than a log greater reduction of bacterial burden than vancomycin.
Fig. 4. Cilagicin BP activity in a murine neutropenic thigh infection model.
(A) Anti S. aureus activity of cilagicin analogs with different lipid substituents in the presence of 10% serum. Blue: MIC < 4 µg/mL or no change in MIC in the presence of serum. (B) Structure of cilagicin BP (L1). (C) Neutropenic thigh infection model using S. aureus USA300. (D) Neutropenic thigh infection model using S. pyrogens ATCC19615. Two hours post infection with a fresh bacterial suspension (1×106 CFU), vehicle (10% DMSO, TID), vancomycin (40 mg/kg, TID) or cilagicin BP (40 mg/kg, TID) were delivered by IP injection. After 24 hours post infection, CFUs were determined from homogenized thigh tissue samples. Significant differences between groups were analyzed by one-way analysis of variance (ANOVA) (***P=0.0001, ****P<0.0001) (n=4 mice, n=8 thighs). Mean CFU counts and SD are shown.
Cilagicin BP’s mode of action, absence of detectable resistance, and in vivo activity make it an appealing lead structure for the development of a next generation antibiotic that is capable of helping to address the growing antibiotic resistance crisis. As seen with the characterization of biologically produced natural products, the study of syn-BNPs that are inspired by unexplored BGC families should prove to be a productive strategy for identifying distinctive scaffolds that can serve as lead structures for developing antibiotics with diverse MOAs.
Supplementary Material
Acknowledgments:
We thank John C. Vederas for providing synthetic lipid II(lys), the CDC for providing vancomycin resistant strains, the Rockefeller University High Throughput Screening and Spectroscopy Resource Center for assistance with ITC experiments, the Rockefeller University Comparative Bioscience Center for their help with our animal studies, and The Rockefeller University Electron Microscopy Resource Center for collecting SEM data.
Funding:
This work was supported by National Institutes of Health grant 1U19AI142731 and 5R35GM122559.
Footnotes
Competing interests: A patent covering the structure and activity of cilagicin has been filed by Rockefeller University. SFB consulted for Lodo Therapeutics and Zymergen and has Zymergen stock options.
Data and materials availability:
The genome sequences of Paenibacillus mucilaginosus K02 and KNP414 are available in GenBank under the accession numbers NC_017672.3 and NC_015690.1, respectively. BGCs were collected from antiSMASH-db (version 2.0). The Perl script used to identify lipopeptide BGCs can be accessed through Zenodo (41). NMR spectra for cilagicin and cilagicin BP are presented as Supplementary Information. All data are available in the main text or the supplementary materials.
References and Notes
- 1.Rossiter SE, Fletcher MH, Wuest WM, Natural products as platforms to overcome antibiotic resistance. Chemical reviews 117, 12415–12474 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Davies J, Davies D, Origins and evolution of antibiotic resistance. Microbiol Mol Biol Rev 74, 417–433 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aslam B et al. , Antibiotic resistance: a rundown of a global crisis. Infect Drug Resist 11, 1645–1658 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hamley IW, Lipopeptides: from self-assembly to bioactivity. Chem Commun (Camb) 51, 8574–8583 (2015). [DOI] [PubMed] [Google Scholar]
- 5.Raaijmakers JM, De Bruijn I, Nybroe O, Ongena M, Natural functions of lipopeptides from Bacillus and Pseudomonas: more than surfactants and antibiotics. FEMS Microbiol Rev 34, 1037–1062 (2010). [DOI] [PubMed] [Google Scholar]
- 6.Wang Z et al. , A naturally inspired antibiotic to target multidrug-resistant pathogens. Nature 601, 606–611 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bloudoff K, Schmeing TM, Structural and functional aspects of the nonribosomal peptide synthetase condensation domain superfamily: discovery, dissection and diversity. Biochim Biophys Acta Proteins Proteom 1865, 1587–1604 (2017). [DOI] [PubMed] [Google Scholar]
- 8.Rausch C, Hoof I, Weber T, Wohlleben W, Huson DH, Phylogenetic analysis of condensation domains in NRPS sheds light on their functional evolution. BMC Evol Biol 7, 78 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Roongsawang N et al. , Phylogenetic analysis of condensation domains in the nonribosomal peptide synthetases. FEMS Microbiol Lett 252, 143–151 (2005). [DOI] [PubMed] [Google Scholar]
- 10.Kang HS, Brady SF, Arixanthomycins A-C: Phylogeny-guided discovery of biologically active eDNA-derived pentangular polyphenols. ACS Chem Biol 9, 1267–1272 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hover BM et al. , Culture-independent discovery of the malacidins as calcium-dependent antibiotics with activity against multidrug-resistant Gram-positive pathogens. Nat Microbiol 3, 415–422 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang Z et al. , Metagenome-Guided Analogue Synthesis Yields Improved Gram-Negative-Active Albicidin-and Cystobactamid-Type Antibiotics. Angewandte Chemie 133, 22346–22351 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tomm HA, Ucciferri L, Ross AC, Advances in microbial culturing conditions to activate silent biosynthetic gene clusters for novel metabolite production. J Ind Microbiol Biotechnol 46, 1381–1400 (2019). [DOI] [PubMed] [Google Scholar]
- 14.Chu J et al. , Discovery of MRSA active antibiotics using primary sequence from the human microbiome. Nat Chem Biol 12, 1004–1006 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chu J et al. , Synthetic-Bioinformatic Natural Product Antibiotics with Diverse Modes of Action. J Am Chem Soc 142, 14158–14168 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chu J, Vila-Farres X, Brady SF, Bioactive Synthetic-Bioinformatic Natural Product Cyclic Peptides Inspired by Nonribosomal Peptide Synthetase Gene Clusters from the Human Microbiome. J Am Chem Soc 141, 15737–15741 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Stachelhaus T, Mootz HD, Marahiel MA, The specificity-conferring code of adenylation domains in nonribosomal peptide synthetases. Chem Biol 6, 493–505 (1999). [DOI] [PubMed] [Google Scholar]
- 18.Walsh CT et al. , Tailoring enzymes that modify nonribosomal peptides during and after chain elongation on NRPS assembly lines. Current opinion in chemical biology 5, 525–534 (2001). [DOI] [PubMed] [Google Scholar]
- 19.CDC. Antibiotic Resistance Threats in the United States, 2019. Altlanta, GA: U.S. Department of Health and Human Services, CDC; (2019). [Google Scholar]
- 20.Roth BL, Poot M, Yue ST, Millard PJ, Bacterial viability and antibiotic susceptibility testing with SYTOX green nucleic acid stain. Appl Environ Microbiol 63, 2421–2431 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cabrini G, Verkman AS, Potential-sensitive response mechanism of diS-C3-(5) in biological membranes. J Membr Biol 92, 171–182 (1986). [DOI] [PubMed] [Google Scholar]
- 22.Wu M, Maier E, Benz R, Hancock RE, Mechanism of interaction of different classes of cationic antimicrobial peptides with planar bilayers and with the cytoplasmic membrane of Escherichia coli. Biochemistry 38, 7235–7242 (1999). [DOI] [PubMed] [Google Scholar]
- 23.Wu C, Shang Z, Lemetre C, Ternei MA, Brady SF, Cadasides, Calcium-Dependent Acidic Lipopeptides from the Soil Metagenome That Are Active against Multidrug-Resistant Bacteria. J Am Chem Soc 141, 3910–3919 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cochrane SA et al. , Antimicrobial lipopeptide tridecaptin A1 selectively binds to Gram-negative lipid II. Proc Natl Acad Sci U S A 113, 11561–11566 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ling LL et al. , A new antibiotic kills pathogens without detectable resistance. Nature 517, 455–459 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schneider T et al. , The lipopeptide antibiotic Friulimicin B inhibits cell wall biosynthesis through complex formation with bactoprenol phosphate. Antimicrob Agents Chemother 53, 1610–1618 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kleijn LH et al. , Total Synthesis of Laspartomycin C and Characterization of Its Antibacterial Mechanism of Action. J Med Chem 59, 3569–3574 (2016). [DOI] [PubMed] [Google Scholar]
- 28.Touz ET, Mengin-Lecreulx D, Undecaprenyl Phosphate Synthesis. EcoSal Plus 3, (2008). [DOI] [PubMed] [Google Scholar]
- 29.van Heijenoort J, Recent advances in the formation of the bacterial peptidoglycan monomer unit. Nat Prod Rep 18, 503–519 (2001). [DOI] [PubMed] [Google Scholar]
- 30.El Ghachi M et al. , Crystal structure of undecaprenyl-pyrophosphate phosphatase and its role in peptidoglycan biosynthesis. Nat Commun 9, 1078 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hernandez-Rocamora VM et al. , Coupling of polymerase and carrier lipid phosphatase prevents product inhibition in peptidoglycan synthesis. Cell Surf 2, 1–13 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hashizume H et al. , Tripropeptin C blocks the lipid cycle of cell wall biosynthesis by complex formation with undecaprenyl pyrophosphate. Antimicrob Agents Chemother 55, 3821–3828 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stone KJ, Strominger JL, Mechanism of action of bacitracin: complexation with metal ion and C 55 -isoprenyl pyrophosphate. Proc Natl Acad Sci U S A 68, 3223–3227 (1971). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Singh M, Chang J, Coffman L, Kim SJ, Solid-state NMR characterization of amphomycin effects on peptidoglycan and wall teichoic acid biosyntheses in Staphylococcus aureus. Sci Rep 6, 31757 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hu XF et al. , Transfer of Bacillus mucilaginosus and Bacillus edaphicus to the genus Paenibacillus as Paenibacillus mucilaginosus comb. nov. and Paenibacillus edaphicus comb. nov. Int J Syst Evol Microbiol 60, 8–14 (2010). [DOI] [PubMed] [Google Scholar]
- 36.Goswami D, Parmar S, Vaghela H, Dhandhukia P, Thakker JN, Describing Paenibacillus mucilaginosus strain N3 as an efficient plant growth promoting rhizobacteria (PGPR). Cogent Food & Agriculture 1, 1000714 (2015). [Google Scholar]
- 37.Radeck J et al. , Anatomy of the bacitracin resistance network in Bacillus subtilis. Mol Microbiol 100, 607–620 (2016). [DOI] [PubMed] [Google Scholar]
- 38.Wecke T et al. , Daptomycin versus Friulimicin B: in-depth profiling of Bacillus subtilis cell envelope stress responses. Antimicrob Agents Chemother 53, 1619–1623 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Larsson DGJ, Flach CF, Antibiotic resistance in the environment. Nat Rev Microbiol, (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Brade KD, Rybak JM, Rybak MJ, Oritavancin: A New Lipoglycopeptide Antibiotic in the Treatment of Gram-Positive Infections. Infect Dis Ther 5, 1–15 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang et al. , Code for the paper: “Bioinformatic prospecting and synthesis of a bifunctional lipopeptide antibiotic that evades resistance ”. Zenodo; (2022); 10.5281/zenodo.5793911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Blin K et al. , The antiSMASH database version 2: a comprehensive resource on secondary metabolite biosynthetic gene clusters. Nucleic Acids Res 47, D625–D630 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Medema MH et al. , antiSMASH: rapid identification, annotation and analysis of secondary metabolite biosynthesis gene clusters in bacterial and fungal genome sequences. Nucleic Acids Res 39, W339–346 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Carmichael J, DeGraff WG, Gazdar AF, Minna JD, Mitchell JB, Evaluation of a tetrazolium-based semiautomated colorimetric assay: assessment of chemosensitivity testing. Cancer Res 47, 936–942 (1987). [PubMed] [Google Scholar]
- 45.Colosimo DA et al. , Mapping Interactions of Microbial Metabolites with Human G-Protein-Coupled Receptors. Cell Host Microbe 26, 273–282 e277 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The genome sequences of Paenibacillus mucilaginosus K02 and KNP414 are available in GenBank under the accession numbers NC_017672.3 and NC_015690.1, respectively. BGCs were collected from antiSMASH-db (version 2.0). The Perl script used to identify lipopeptide BGCs can be accessed through Zenodo (41). NMR spectra for cilagicin and cilagicin BP are presented as Supplementary Information. All data are available in the main text or the supplementary materials.