Abstract
We identified a number of upregulated genes by differential screening of interleukin-9-stimulated T-helper lymphocytes. Interestingly, two of these messengers encode proteins that are similar to proteins of the gelsolin family. The first displays a typical structure of six homologous domains and shows a high level of identity (90%) with bovine adseverin (or scinderin) and may therefore be considered the murine adseverin homolog. The second encodes a protein with only five segments. Sequence comparison shows that most of the fifth segment and a short amino-terminal part of the sixth segment (amino acids 528 to 628 of adseverin) are missing, and thus, this form may represent an alternatively spliced product derived from the same gene. The corresponding protein is called mouse adseverin (D5). We expressed both proteins in Escherichia coli and show that mouse adseverin displays the typical characteristics of all members of the gelsolin family with respect to actin binding (capping, severing, and nucleation) and its regulation by Ca2+. In contrast, mouse adseverin (D5) fails to nucleate actin polymerization, although like mouse adseverin and gelsolin, it severs and caps actin filaments in a Ca2+-dependent manner. Adseverin is present in all of the tissues and most of the cell lines tested, although at low concentrations. Mouse adseverin (D5) was found only in blood cells and in cell lines derived from T-helper lymphocytes and mast cells, where it is weakly expressed. In a gel filtration experiment, we demonstrated that mouse adseverin forms a 1:2 complex with G actin which is stable only in the presence of Ca2+, while no stable complex was observed for mouse adseverin (D5).
Gelsolins form a highly conserved family of multifunctional actin binding proteins. Most of the properties of these proteins were derived from in vitro and in vivo studies on plasma and macrophage gelsolin. Gelsolin is characterized by a typical organization of six homologous domains, each containing approximately 125 amino acids residues (19, 40). Two G-actin binding domains and a single F-actin binding site have been assigned to segments S1, S4 to S6, and S2 and S3, respectively (20, 41, 45). The amino-terminal half of gelsolin (S1 to S3) is important for severing and capping, while the carboxy-terminal part (S4 to S6) is necessary for efficient nucleation (41). The interactions with actin are regulated by Ca2+ and phosphatidylinositol 4,5-bisphosphate (15, 16). This led to the concept that the cytoplasmic form of gelsolin is an important regulator of subcortical actin cytoskeleton organization, connecting the phosphoinositide status with actin polymerization, and that it is a key player in signal transduction (9, 12, 18).
Gelsolin has been implicated in a number of pathologies. For instance, familiar Finnish-type amyloidosis results from a single point mutation at position 654 of gelsolin, where Asp replaces Asn (29). Human cell carcinomas of the bladder have been correlated with deletion of the gelsolin gene. Consequently, tumor suppression was obtained by gelsolin transfection (36). In similar experiments, the tumorigenicity of ras-transformed cells was suppressed by genetic transfection with gelsolin-His321, a mutant with a Pro-to-His substitution at position 321 (27). Transgenic gelsolin null mice show normal embryonic development and longevity. However, the observed phenotypes emphasize the importance of gelsolin for rapid motile processes in cell types involved in stress response such as homostasis, inflammation, and wound healing (42). Members of the gelsolin family are also found in invertebrates, e.g., Physarum polycephalum (2), Dictyostelium sp. (43), Homerus americanus (23), and Lumbricus terrestris (11). Here, the predominant forms consist of only three segments, but they appear to share all of the actin binding properties of vertebrate gelsolin, including severing, capping, and nucleation (2, 3, 10, 38).
Interleukin-9 (IL-9) is a pleiotropic cytokine produced by activated T-helper type 2 lymphocytes and was originally identified by its ability to stimulate the proliferation of murine T-cell clones and mast cell lines (14, 39). More recently, in an attempt to better characterize the activity of IL-9 on mouse T-helper lymphocytes, we identified four genes whose expression is induced by IL-9, but not by IL-2 or IL-3, in cytokine-dependent T-cell clones and mast cells (22). Three of these genes correspond to granzymes A and B and the α chain of the high-affinity receptor for immunoglobulin E (IgE) (FcɛRlα). The fourth gene encodes a previously unknown murine protein.
In this report, we present the sequence and functional characterization of this IL-9-induced protein, comparison of whose sequence suggests that it is the murine homolog of adseverin or scinderin. We found that mouse adseverin displays most of the typical gelsolin properties. It is able to form a stable 1:2 complex with G actin, but unlike that formed by gelsolin, the complex completely dissociates upon Ca2+ chelation. In addition, we isolated another IL-9-upregulated protein which we call mouse adseverin (D5). It is a novel gelsolin family member with only five segments, lacking most of the fifth domain and part of the sixth segment. It has lost its ability to nucleate actin polymerization and to form a stable complex with G actin; however, it still displays Ca2+-dependent capping and severing activities.
MATERIALS AND METHODS
Cell culture.
TS2 and TS3 are factor-dependent T-helper cell clones derived from clones TUC5.37 and TUC7.33, respectively, by culturing cells in the absence of antigen in medium supplemented with IL-9 and IL-3 (22, 39). Cultures were maintained in Dulbecco’s modified Eagle medium supplemented with 10% fetal calf serum, 50 μM 2-mercaptoethanol, 0.55 mM l-arginine, 0.24 mM l-asparagine, and 1.25 mM l-glutamine.
Cell lines were kindly provided as follows: T-helper clone ST2K9 (33) by E. Schmitt (Johannes Gutenberg-Universität, Mainz, Federal Republic of Germany); IL-9-dependent mast cell lines L138 and MC-9 by L. Hültner (Forschungszentrum für Umwelt und Gesundheit GmbH, Munich, Federal Republic of Germany) and C. Petit-Frère (Institut Henri Beaufour, Les Ullys, France), respectively; macrophage cell line PU5.8 by L. Franssen (Innogenetics, Gent, Belgium); and the EL4 lymphoma cell line by H. R. MacDonald (Ludwig Institute for Cancer Research, Lausanne, Switzerland).
Mouse bone marrow-derived mast cells were obtained by culturing bone marrow from BALB/c mice for 2 to 4 weeks in enriched medium (RPMI 1640 medium containing 0.1 mM nonessential amino acids, 2 mM l-glutamine, 100 μg/ml penicillin, 100-μg/ml streptomycin, 10-μg/ml gentamicin, 50 μM 2-mercaptoethanol, and 20% fetal calf serum supplemented with either 1-ng/ml IL-3 [Biogen, Geneva, Switzerland] alone or in combination with IL-9 [5 ng/ml]). Fluorescence-activated cell sorter analysis of these cells showed homogeneous staining by biotinylated IgE and no staining with anti-Mac1, anti-Mac2, anti-Mac3, and Thy1 antibodies.
Construction and screening of a cDNA library.
The differential hybridization approach used for isolation of IL-9-induced genes has been described previously (22). Briefly, TS2 cells are washed and deprived of growth factor for 14 h before stimulation with IL-9 for 24 h. A cDNA library was constructed in a BstXI- and NotI-digested plasmid derived from pSVK3 (Pharmacia, Uppsala, Sweden). Aliquots from the cDNA library containing about 1,000 individual colonies were plated onto nitrocellulose membranes (Schleicher & Schuell, Inc.). Triplicates of each membrane were prepared, and two of them were treated as previously described (4) to allow hybridization with either a negative or a positive probe consisting of a single-stranded, 32P-labelled cDNA probe prepared from poly(A) RNA isolated from TS2 cells stimulated with IL-2 (negative probe) or IL-9 (positive probe). Colonies exhibiting differential hybridization to the positive and negative probes were selected for further analysis by Northern blot hybridization.
Preparation of mRNA and Northern analysis.
Total cellular mRNA prepared by the guanidinium-CsCl method (4) was fractionated by electrophoresis in a 1.3% agarose gel containing 2.2 M formaldehyde and transferred to Hybond nylon. cDNA inserts were labelled with the Multiprime DNA labelling kit from Amersham. The filters were hybridized at 60°C in a solution of 10% (wt/vol) dextran sulfate; 1 M NaCl, 1% sodium dodecyl sulfate (SDS), and 200-μg/ml denatured salmon sperm. After autoradiography, blots were reprobed with a β-actin-specific probe to check the amount of RNA.
DNA sequencing and analysis.
Sequencing was performed by the dideoxy sequencing technique, using the T7 sequencing kit (Pharmacia) on CsCl-purified plasmid DNA with primers flanking the cloning sites. Further sequence analysis was performed on clones resulting from exonuclease III digestion using the Erase-a-Base kit (Promega). Characterization of the 5′ end of mouse adseverin cDNA sequences was done by using the 5′ Amplifinder Race kit (Clontech). Searches of the GenBank and EMBL databases were performed with the FASTA program. Identity matrices were calculated with pileup and distances in the GCG Wisconsin package.
Preparation of RNA from organs of C3H/HeJ mice.
Several organs were dissected from male and female C3/HeJ mice and immediately frozen in liquid nitrogen. Organs were homogenized in a mortar with 6 M guanidinium isothiocyanate with sea sand as previously described (4, 21). After centrifugation of the debris, the RNA was pelleted by ultracentrifugation (SW-28 rotor, 113,000 × g) and solubilized in 200 μl of diethylpyrocarbonate-treated water.
RT-PCR.
Samples (1 μg) of RNA were taken from the different tissues, and first-strand cDNA was prepared by the use of Moloney leukemia murine virus reverse transcriptase (RT; SuperscriptII; Gibco Bethesda Research Laboratories [BRL]) and oligo(dT)12-18 as a primer. Half of the final volume (10 μl) was used for PCR. The sequences of the primers (5′tacatcacggagaaagtggctcagataaagcag3′ and 5′ctcgtgcccttgcttgatgatgacaat3′) correspond to sequences 1185 and 2171, respectively, in adseverin. In adseverin (D5), they correspond to sequences 1185 and 1871, respectively. The amplified fragments were analyzed on a 1% agarose gel. They had sizes of 986 and 686 bp, respectively. The amplified fragments were blotted onto nitrocellulose, and Southern blotting was performed as described by Ausubel et al. (4). The probe was derived from the SstI fragment of mouse adseverin that was isolated and randomly primed in accordance with the manufacturer’s (Boehringer Mannheim) instructions. The probes for the dynamitin control experiment were dyn01 (5′gtagaactgttgcaagccaaagtga3′) and dyn02 (5′ctttcccagcctcttcatccgag3′) and were designed for the expressed sequence tag of dynamitin, vi64g01.r1.
Selective amplification of adseverin (D5).
A 0.5-μg sample of IL-9-stimulated TS2 cell RNA was taken, and first-strand cDNA was prepared as described above. Half of the final volume (10 μl) was used for PCR. The backward primers corresponding to the flanking region of the spliced-out fragment in adseverin (D5) had the following sequences: D3, 5′aatctc3′; D5, 5′tcaatctcca3′; D7, 5′cttcaatctccaca3′; D10, 5′cttcttcaatctccacaatt3′; D15, 5′cggaacttcttcaatctccacaattctggt3′; D20, 5′tctcccggaacttcttcaatctccacaattctggtgatag3′; d5/25, 5′gtgaactctcccggaacttcttcaatctcca3′; d10/20, 5′tctcccggaacttcttcaatctccacaatt3′. pDpcr (5′cccacagg agaggaagactgccatgaagacagctgaggag3′) was used as the forward primer. The control backward primers were Dcontr1 (5′gcatcaacgtcaacctccacaattctggtg3′) and Dcontr2 (5′cccggaacttcttcaataatgaatcttccag3′), which correspond to the sequence of adseverin. PCRs were performed with Taq DNA polymerase in accordance with the manufacturer’s (Gibco BRL) instructions, although the number of reaction cycles was restricted to 15. The amplified fragments were analyzed on a 1.5% agarose gel.
Cloning and expression of mouse adseverin and adseverin (D5) in E. coli.
Plasmid p9016 was digested with NcoI/SstI and SstI/NsiI, and the adseverin-encoding fragments were ligated into NcoI/PstI-opened vector pSE380 (1), resulting in plasmid pSEGLP.
The Eco0109/BsmI fragment of plasmid p9034, encoding mouse adseverin (D5), was exchanged with the Eco0109/BsmI fragment of plasmid pSEGLP, resulting in plasmid pSEGLP (D5). E. coli MC1061 [F− araΔ139 Δ(ara-leu)7697 ΔlacX74 galU galK hsdR2(rK−mK+) mcrB1 rpsL Str+] was used as the transformation host.
Purification of recombinant mouse adseverin and adseverin (D5) from E. coli.
A single colony of E. coli MC1061 harboring plasmid pSEGLP or pSEGLP(D5) was picked from a freshly transformed plate. A preculture was grown overnight and diluted 50 times in 1.8 liters of Luria-Bertani medium containing the antibiotic triacillin (Gibco-BRL) at a concentration of 100 μg/ml. Upon a cell density of 2.5 × 108/ml, isopropyl-β-d-thiogalactopyranoside was added to a final concentration of 1 mM, after which the bacteria were grown for another 4 h. Cells were collected by centrifugation and resuspended in 40 ml of buffer A (25 mM Tris-HCl [pH 7.5], 0.5 mM CaCl2, 50 mM NaCl) plus 1 mM phenylmethylsulfonyl fluoride (PMSF) or a protease inhibitor mixture (300-μg/ml leupeptin, 100-μg/ml pepstatin, 100 mM benzamidine, 50-μg/ml antipain) in the case of mouse adseverin (D5). The cells were opened by being passed twice through a French press (Aminco, Danvers, Mass.). Mouse adseverin was purified after removal of cell debris by centrifugation; the proteins in the supernatant were precipitated with 40% (wt/vol) ammonium sulfate. The protein pellet was solubilized in 40 ml of buffer A–1 mM PMSF and dialyzed against 1 liter of the same buffer (with three changes). Dialyzed proteins were loaded on a DEAE column that had previously been equilibrated with buffer A. The flowthrough of the column, containing adseverin, was dialyzed against buffer B (25 mM Tris-HCl [pH 8.5], 50 mM NaCl, 1 mM EGTA) plus 1 mM PMSF and loaded onto a second DEAE column that had been equilibrated with this buffer. Here, too, adseverin was found in the flowthrough, and the corresponding fraction was passed on to a Mono Q column (HR5/5; Pharmacia) that had previously been equilibrated with buffer B and was eluted with a gradient of NaCl. Adseverin eluted at around 190 mM NaCl and was more than 95% pure. During purification, activity was tested with the falling-ball assay (24).
E. coli bacteria containing pSEGLP(D5) were grown and lysed as described above, except that the bacteria were passed four times through a French press. Mouse adseverin (D5) was purified from inclusion bodies. These were washed twice in each of the following solutions: buffer C (25 mM Tris-HCl [pH 6.5], 1 mM EGTA, 25 mM NaCl), buffer C plus 0.475 M NaCl, buffer C plus 0.975 M NaCl, buffer C, buffer C plus 0.5 M urea, and buffer C. The washed inclusion bodies, enriched for adseverin (D5), were solubilized in buffer C containing 10 mM dithiothreitol, protease inhibitor (see above), and 8 M urea. After overnight dialysis against buffer C with 6 M urea, the proteins were loaded onto a DEAE column (2.5 by 10 cm) that had previously been equilibrated with the same buffer. The flowthrough of the column, which contained adseverin (D5), was dialyzed against the same buffer at pH 8.0 and loaded onto a gel filtration column (Superdex 200; Pharmacia). This procedure yielded a protein more than 95% pure, as estimated by SDS-polyacrylamide gel electrophoresis (PAGE). The purified protein was renatured by stepwise dialysis against 4, 2, and 0 M urea (in buffer C), yielding soluble and biologically active mouse adseverin (D5).
Amino-terminal amino acid sequence determination.
Bacterially expressed proteins were separated by SDS-PAGE, and blotted onto Problot membranes as described by Bauw et al. (5). The appropriate band was cut out, and the amino-terminal amino acid sequence was determined by Edman degradation on an A470 gas-phase sequenator equipped with a 120A on-line phenylthiohydantoin amino acid analyzer (Applied Biosystems, Foster City, Calif.) in accordance with the instructions of the manufacturer.
Actin preparation.
Actin was prepared from cow muscle as described by Spudich and Watt (34). Actin was kept in G buffer (2 mM Tris-HCl [pH 7.6], 0.2 mM ATP, 0.5 mM 2-mercaptoethanol, 0.2 mM CaCl2). Labelling of actin with N-pyrenyliodoacetamide was performed as described by Brenner and Korn (6).
Nucleating, severing, and capping activities of mouse adseverin and adseverin (D5).
All fluorescence measurements with pyrene-labeled actin were performed at room temperature with an SFM25 fluorimeter (Kontron Instruments, Zurich, Switzerland). The excitation and emission wavelengths were 365 and 388 nm, respectively. Human plasma gelsolin was purified as described by Bryan (7) and used as a control. Assays were performed as follows. For the nucleation assay, 6 μM actin (10% pyrene labelled) was preincubated with mouse adseverin, mouse adseverin (D5), or human gelsolin at a 1:1,000 molar ratio at room temperature for 10 min. The total volume was adjusted to 675 μl with G buffer. Polymerization was initiated by addition of 75 μl 10-fold-concentrated F buffer (final concentrations, 100 mM KCl and 2 mM MgCl2), and the increase in fluorescence was measured. Assays were performed in the presence of Ca2+ or EGTA at a concentration of 0.2 or 2 mM, respectively.
For the severing assay, 8 μM actin (25% pyrene labelled) in 675 μl of G buffer was allowed to polymerize by addition of 75 μl of 10-fold-concentrated F buffer. After 15 min, human gelsolin was added at a molar ratio of 1:300 (final concentration, 26 nM) and the mixture was kept on ice overnight in order to equilibrate. Subsequently, these precapped filaments were diluted 20 times in 750 μl of G buffer with Ca2+ or EGTA in the absence or presence (5 nM final concentration) of adseverin or adseverin (D5), and the fluorescence was measured. For the capping assay, F actin (final concentration, 1.2 μM) nuclei were prepared as follows. A 40 μM actin stock (150 μl) was mixed with 4.5 ml of G buffer. A 0.5-ml sample of 10-fold-concentrated F buffer was added to initiate polymerization, and the mixture was kept on ice overnight. A 262-μl volume of these nuclei was mixed with 52.5 μl of actin (25% pyrene labelled) to a final concentration of 3 μM. Polymerization was initiated by adding 35 μl of 10-fold-concentrated F buffer in the absence or presence (3 nM final concentration) of adseverin or adseverin (D5), and the fluorescence was measured.
Mouse adseverin and adseverin (D5) complex formation with G actin.
A 182-μl volume of mouse adseverin (concentration, 0.5 mg/ml) or 1 ml of mouse adseverin (D5) (concentration, 0.1 mg/ml) was mixed with 78 μl of G actin (concentration, 1.28 mg/ml) in a collodion bag (Sartorius AG) and dialyzed overnight against a buffer containing 2 mM Tris, 10 mM CaCl2, 50 mM NaCl, 0.2 mM dithiothreitol, and 0.2 mM ATP. The mixture was loaded onto a Superdex 200 gel filtration column (Pharmacia) equilibrated and run with the same buffer, and the fractions were analyzed by SDS-PAGE. To determine the role of Ca2+ in complex formation, we performed the same experiment but with a buffer containing 2 mM Tris, 30 mM EGTA, 50 mM NaCl, 0.2 mM DTT, and 0.2 mM ATP.
RESULTS
IL-9 upregulates the expression of a member of the gelsolin family in T cells.
Based on specific or upregulated expression in IL-9-stimulated T cells, we previously identified and isolated four genes. Three of them corresponded to granzyme A, granzyme B, and the α chain of the high-affinity receptor for IgE (FcɛRIα) (22). The sequence of the fourth IL-9-induced gene did not show a perfect match to any of the DNA sequences in the GenBank and EMBL databases but, interestingly, displayed similarity to gelsolin and adseverin. Initially, we identified two independent cDNA clones by differential screening methods (clones 9016 and 9034). We used these clones for further screening of the same library and isolated seven cDNAs independently; the 5′ part was obtained by rapid amplification of 5′ cDNA ends. Four of these clones contained the same 2,144-nucleotide open reading frame, encoding a protein of 715 amino acid residues (calculated mass, 80,293 Da) (Fig. 1). Comparison of the complete sequence with the information stored in the SwissProt database showed high degrees of identity with bovine adseverin (90%) and bovine scinderin (88%), both members of the gelsolin family. These proteins are known to be important regulators of the subcortical microfilament network (31, 32). We propose that these clones encode murine adseverin, although we cannot exclude the possibility that we identified a novel variant of adseverin. Identity scores with adseverin, scinderin, and other members of the gelsolin family are shown in Table 1.
FIG. 1.
Amino acid sequence of mouse adseverin (GenBank accession no. U04354). The sequence missing in mouse adseverin (D5) (GenBank accession no. Y13971) corresponds to amino acids 528 to 628 and is underlined. Superscripts indicate the different domains in accordance with the plasma gelsolin crystal structure (8).
TABLE 1.
Identity scores of different members of the gelsolin family with mouse adseverina
| Proteinb | Relative identity
score
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9c | 10c | |
| 1 | 1.00 | 0.96 | 0.94 | 0.60 | 0.59 | 0.60 | 0.39 | 0.36 | 0.50 | 0.49 |
| 2 | 1.00 | 0.94 | 0.60 | 0.59 | 0.60 | 0.39 | 0.36 | 0.49 | 0.49 | |
| 3 | 1.00 | 0.60 | 0.59 | 0.59 | 0.39 | 0.36 | 0.49 | 0.48 | ||
| 4 | 1.00 | 0.98 | 0.90 | 0.36 | 0.34 | 0.46 | 0.47 | |||
| 5 | 1.00 | 0.88 | 0.35 | 0.33 | 0.45 | 0.46 | ||||
| 6 | 1.00 | 0.37 | 0.34 | 0.46 | 0.46 | |||||
| 7 | 1.00 | 0.43 | 0.35 | 0.35 | ||||||
| 8 | 1.00 | 0.32 | 0.32 | |||||||
| 9 | 1.00 | 0.90 | ||||||||
| 10 | 1.00 | |||||||||
The proteins were compared with the pileup and distances programs from the GCG Wisconsin software package with default parameter settings.
Proteins (EMBL accession no.): 1, human gelsolin (A07400); 2, pig plasma gelsolin (M36927); 3, mouse gelsolin (J04953); 4, bovine adseverin (D26549); 5, bovine scinderin (X78479); 6, mouse adseverin (U04354); 7, lobster gelsolin (Z29534); 8, drosophila gelsolin (X75629); 9, mouse villin (M98454); 10, human villin (A26237).
Villin headpiece was omitted in the identity calculation.
Like several members of the gelsolin family, mouse adseverin displays the characteristic repeat of the six homologous domains (19), each containing the three conserved sequence motifs initially defined by Ampe and Vandekerckhove (2) in fragmin and by Way and Weeds (40) in porcine gelsolin. The two putative polyphosphoinositide binding sites described for gelsolin (46) and villin (17) are also present in the corresponding regions of mouse adseverin albeit that the second one is less conserved (Fig. 2).
FIG. 2.
Comparison of the sequences of the PIP2 binding sites of different actin binding proteins (17, 45, 46). PIP2 binding sites are underlined.
Interestingly, in three of the seven independently isolated cDNA clones we observed the same internal deletion of 300 nucleotides in the coding region. This deleted segment corresponds to most of the fifth domain and a short amino-terminal part of the sixth segment of mouse adseverin (amino acids 528 to 628 of adseverin) (Fig. 1). The corresponding protein is hereafter referred to as mouse adseverin (D5). It contains 615 amino acids and has a molecular weight of 69,150. Given the fact that three of the seven clones, each unique isolates, encode this variant, it is unlikely that we observed a cloning artifact, but rather, we believe that the shorter mRNA results from an alternative splicing event and represents the first naturally occurring five-segmented member of the gelsolin family (see also the Discussion).
Mouse adseverin is expressed in various cell lines and tissues but at low levels; mouse adseverin (D5) is present only in blood cells.
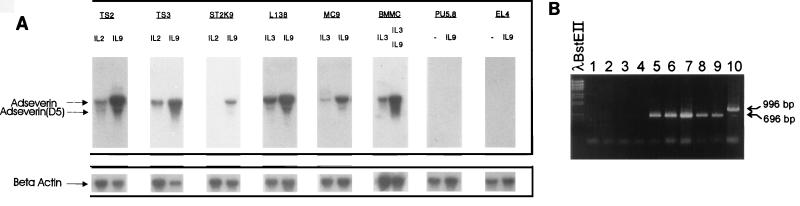
We first measured mouse adseverin RNA transcript expression in various IL-9-responsive cell lines by Northern blot analysis (Fig. 3A) and found that in T-cell clones (TS2 and TS3) and mast cell lines (L138 and MC9) adseverin was significantly upregulated by IL-9. We observed the same effect with freshly derived mast cells in the presence of IL-3 and IL-9 but not with mast cells in the presence of IL-3 alone. In contrast, we observed no expression of mouse adseverin in the PU5.8 murine macrophage cell line or in the EL4 lymphoma cell line, even after IL-9 stimulation. Macrophages are a good source from which to purify cytoplasmic gelsolin (44, 45); thus, there is no cross-hybridization between the cytoplasmic gelsolin mRNA and the mouse adseverin probe. Closer inspection of the Northern blots of the T cells (TS2) and mast cells (L138 and MC9) reveals a fainter signal of a band with a smaller size, which may represent mouse adseverin (D5) mRNA (Fig. 3A). By using different primers with sequences corresponding to the regions flanking the spliced-out fragment and applying short PCR conditions, we were able to selectively amplify adseverin (D5) cDNA from IL-9-induced TS2 cells in an RT-PCR experiment (Fig. 3B).
FIG. 3.
(A) The IL-9-responsive cell lines indicated were cultured in the presence of saturating concentrations of the indicated cytokines for at least 3 days. After electrophoresis of 10 μg of total RNA and transfer to nitrocellulose, the filter was hybridized with a specific 32P-labelled mouse adseverin cDNA probe. Hybridization with a β-actin probe was used as a control to compare the amounts of RNA in the lanes. (B) Selective amplification of adseverin (D5) cDNA. RT-PCR of total RNA of IL-9-induced TS2 cells. pDpcr was used as the forward primer. The sequences of the different primers are described in Materials and Methods. Lanes: 1, primer D3; 2, primer D5; 3, primer D7; 4, primer D10; 5, primer D15; 6, primer D20; 7, primer d5/25; 8, primer d10/20; 9, primer Dcontr1; 10, primer Dcontr2.
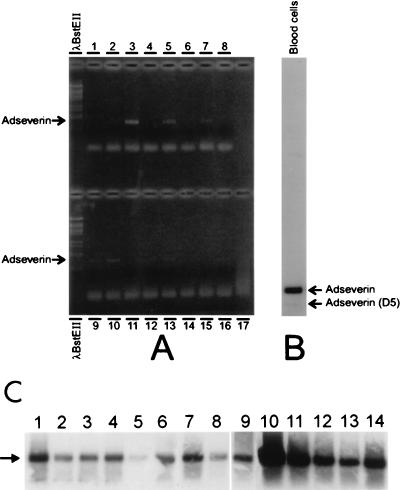
We also prepared RNAs from several organs or tissues from male and female mice but failed to detect any positive signal for either adseverin or adseverin (D5) in a Northern blot (data not shown). With the more sensitive RT-PCR technique, we were able to amplify a 986-bp adseverin fragment in liver, kidney, spleen, intestine, and muscle samples (Fig. 4A). Via Southern blotting of the same amplified PCR fragments, we detected a signal in all of the tissues examined (Fig. 4C). The absence of a signal in conventional Northern blots and the positive results obtained by RT-PCR indicate that the adseverin message is only present at low levels in the tissues analyzed. In the case of the adseverin (D5) variant, we detected only a faint band of the amplified fragment in blood cells by Southern blotting (Fig. 4B), which is in accordance with the fact that we picked up both forms from a T-lymphocyte library. All other tissues were negative (Fig. 4C). This further supports the notion that the message encoding mouse adseverin (D5) is indeed present in vivo.
FIG. 4.
(A) RT-PCR of RNAs prepared from different organs and tissues. The methods and linkers used are described in Materials and Methods. (A) Upper lanes: 1, blood; 2, heart; 3, liver; 4, lung; 5, kidney; 6, pancreas; 7, intestine; 8, thymus. Lower lanes: 9, muscle; 10, spleen; 11, brain; 12, testis; 13, ovary; 14, skin; 15, uterus; 16, pancreas; 17, negative control (linkers). (B) Southern blot of RT-PCR fragment of RNA from blood cells. (C) Southern blot of RT-PCR fragments of different tissues. Lanes: 1, heart; 2, lung; 3, pancreas; 4, thymus; 5, brain; 6, ovary; 7, skin; 8, uterus; 9, pancreas; 10, liver; 11, kidney; 12, intestine; 13, muscle; 14, spleen. The arrow indicates the amplified adseverin band.
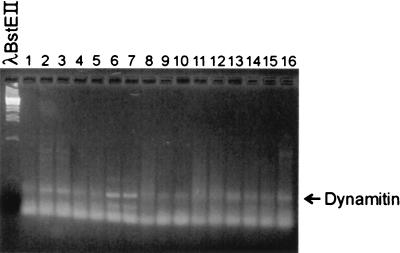
A control RT-PCR experiment was performed with primers against a dynamitin expressed sequence tag. Dynamitin is a low-abundance household protein. We were able to amplify a band of 423 bp in all tissues (Fig. 5).
FIG. 5.
Agarose (2.5%) gel of RT-PCR fragments of different tissues. Lanes: 1, blood; 2, heart; 3, liver; 4, lung; 5, kidney; 6, pancreas; 7, intestine; 8, thymus; 9, muscle; 10, spleen; 11, brain; 12, testis; 13, ovary; 14, skin; 15, uterus; 16, pancreas.
Expression and purification of recombinant mouse adseverin and adseverin (D5) in E. coli.
Upon expression of adseverin in E. coli about half of the recombinant protein is in the insoluble fraction. In addition, this fraction contained another induced protein with a mass of about 49,000 Dal resulting from internal initiation at amino acid 307, as determined by amino-terminal amino acid sequencing of electroblotted protein. For adseverin (D5), a larger amount of the protein was present in the insoluble fraction, as was a similar initiation form.
We tried to purify adseverin by the procedure described for gelsolin by Bryan (7), in which extracts are successively passed over a DEAE column in the presence or absence of Ca2+. Under these conditions, gelsolin is not retained by the first column but is by the second. In contrast, adseverin is present in the flowthrough of a DEAE column regardless of whether Ca2+ is present or not. Nevertheless, these purification steps proved very efficient because most of the contaminating proteins were removed. Mouse adseverin was further purified on a Mono Q column.
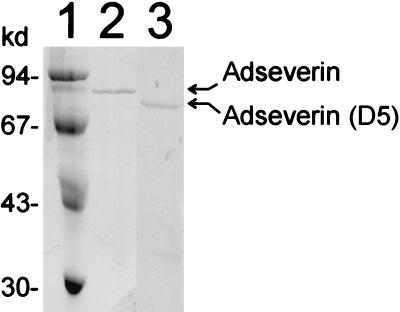
Since recombinant mouse adseverin (D5) was deposited mainly in inclusion bodies, we used these as starting material. We solubilized them in 8 M urea and purified the protein under denaturating conditions by using ion-exchange chromatography and gel filtration. After purification in the presence of urea, we renatured adseverin (D5) in a stepwise fashion, yielding soluble and active protein (Fig. 6).
FIG. 6.
SDS-PAGE of purified proteins. Lanes: 1, molecular size markers; 2, adseverin; 3, adseverin (D5). kd, kilodaltons.
Characterization of the actin binding properties of mouse adseverin and adseverin (D5); the latter does not nucleate actin polymerization in vitro.
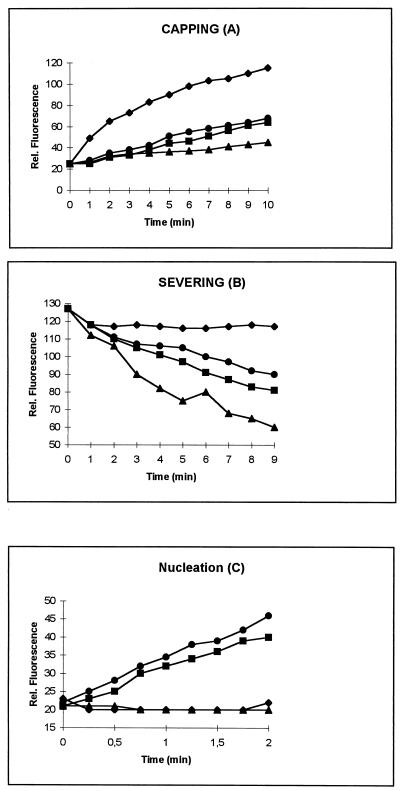
Initially, we assayed the different proteins in a falling-ball experiment indicating that they are all active (24) (Table 2). In addition, we examined the effects of both proteins on actin polymerization by using pyrenyl-labelled actin (Fig. 7). In all experiments, we employed human plasma gelsolin as a reference. We first investigated the ability of adseverin and adseverin (D5) to cap actin filaments. We prepared F-actin nuclei with a final concentration of 1.2 μM. These nuclei were added to 1.8 μM pyrenyl-actin (25% labelled) in the presence of the different actin binding proteins at 3 nM under polymerization conditions. Filament capping results in slower polymerization and a slower increase in the associated fluorescence, which is what we observed for both mouse adseverin and adseverin (D5), but only in the presence of Ca2+ (Fig. 7A).
TABLE 2.
Falling times for different actin binding proteins in a falling-ball experimenta
| Sample | Falling time(s) |
|---|---|
| Negative control | ∞ (>60 min) |
| Mouse adseverin | 4 |
| Mouse adseverin (D5) | 12 |
| Gelsolin | 5 |
The total volume was 200 μl. The actin concentration was 1.2 mg/ml. The molar ratio of actin to actin binding protein was 1:500. Actin was polymerized in a capillary in the presence or absence of actin binding protein. After 30 min, the time necessary for a ball to move 3 cm was measured.
FIG. 7.
Characterization of the different actin binding properties of mouse adseverin and adseverin (D5) in the presence of Ca2+. Assays were done as described in Materials and Methods. Symbols: •, adseverin; , gelsolin; ▴, adseverin (D5) ⧫, negative control (contains only actin). Rel., relative.
For the severing assay, we used actin filaments precapped with gelsolin. After reaching equilibrium, the samples were diluted below the critical monomer concentration of the pointed end in low-salt buffer containing either adseverin or adseverin (D5) in the presence or absence of Ca2+. Severing results in the generation of new pointed ends which will depolymerize under the conditions used while no polymerization or depolymerization will take place at the capped plus end. We only observed a faster decrease in fluorescence, indicative of depolymerization, in samples containing adseverin or adseverin (D5) and Ca2+, and the latter protein seems to sever slightly more efficiently (Fig. 7B).
It is known that gelsolin nucleates actin polymerization. This results in the reduction of the lag phase for actin polymerization typically observed in vitro. Likewise, we assayed the nucleating capacities of adseverin and adseverin (D5) and observed little difference between the actin-nucleating activities of gelsolin and adseverin. Interestingly, adseverin (D5) was completely inactive with respect to the in vitro induction of actin polymerization (Fig. 7C).
Characterization of the complexes of mouse adseverin and adseverin (D5) formed with G actin.
We determined the complexes formed by adseverin and adseverin (D5) with G actin via gel filtration. In the case of adseverin, after mixing the protein with G actin, we detected a protein peak of 170 kDa corresponding to the 1:2 adseverin-actin complex. After addition of EGTA, we detected only two protein peaks, one of approximately 80 kDa and another of approximately 45 kDa, containing adseverin and actin, respectively. This indicates that complete dissociation of the complex occurred and no 1:1 EGTA-resistant complex was found, which is in contrast with most members of the gelsolin family. In the case of adseverin (D5), we did not detect a stable complex with G actin in a gel filtration experiment, regardless of the presence or absence of Ca2+.
DISCUSSION
In a search for proteins that are induced by IL-9 treatment of murine T-helper cells, we identified two members of the gelsolin family: mouse adseverin and adseverin (D5). Mouse adseverin is characterized by a typical six-domain organization and is likely to be the murine homolog of bovine adseverin (also called scinderin), given its high level of identity with this protein. Although the expression level of mouse adseverin is significantly upregulated in T-helper cells, hence its detection, it is expressed at a low level but constitutively in other cells. We demonstrated its presence in all of the tissues and most of the cells examined, but expression levels may be very low since only a more sensitive technique indicated that mRNA was present. Previously, bovine adseverin had only been purified from adrenal and exocytotic cells (31). It was also immunologically detected in pituitary, brain, and kidney tissues after partial enrichment (25, 28, 37), again pointing at the low abundance of the protein. It is important to mention here that mouse adseverin is strongly upregulated in mast cells, which contain many granules and are known to be exocytotically active. Adseverin has been suggested to be involved in secretory processes (28, 31). Mouse adseverin (D5) appears to be a completely novel member of the gelsolin family. It lacks most of the fifth domain and a small part of the sixth segment and is the first such member of this family known. Previously, only members with six or three segments had been isolated. We detected adseverin (D5) in blood cells only, which suggests very narrow tissue specificity. This fact, combined with its low expression level, may explain why the five-segmented form had previously escaped detection, unless the 74-kDa protein called bovine adseverin in some early reports corresponds to adseverin (D5). Indeed, the predicted molecular mass of mouse adseverin (D5) (69,150 Da) is in reasonable agreement with this (25, 32). In a report published after cloning of the gene, a molecular mass of 80 kDa was assigned (closer to the 80,293 Da of mouse adseverin) (28). However, nucleating activity was observed in those studies.
The fact that the nucleotide sequences of the cDNA clones, with the exception of the spliced-out part, are exactly the same suggests that adseverin and adseverin (D5) are encoded by a single gene and likely result from an alternative splicing event. Given the different tissue expression patterns of the two forms, splicing does not occur haphazardly and therefore must be regulated, possibly as a response to IL-9 activation.
From our biochemical characterization of adseverin and adseverin (D5), it is clear that both, like gelsolin, show Ca2+-dependent actin binding activity. The curves we obtained for adseverin in capping, severing, and nucleation experiments resemble those we obtained for (plasma) gelsolin, suggesting that they modulate actin dynamics similarly. In contrast, adseverin (D5) seems to have slightly increased capping and severing activity and is completely deficient in nucleation of actin polymerization. The carboxy-terminal half of gelsolin is known to be important for Ca2+ regulation of the amino-terminal half (20), and the lack of the fifth domain apparently does not influence Ca2+ regulation of severing and capping. Therefore, the inability of adseverin (D5) to nucleate actin polymerization is probably not due to disturbed Ca2+ regulation. It is generally believed that for efficient nucleation, two actin monomers need to be brought into close proximity to each other in the correct orientation (13). The fragment containing domains S1 to S3 caps and severs F actin but does not nucleate actin polymerization, indicating that domains S4 to S6 are essential for nucleation (41). Since mouse adseverin (D5) is not able to nucleate actin polymerization, it is clear that intact S5 is required for nucleation. This is in agreement with a recent study showing that the S5 and S6 domains of scinderin are required for this activity (26). However, this contrasts with a mutagenesis study of gelsolin suggesting that an actin binding site is present in S4 (30). Other scenarios may be possible. One possibility is that the fifth segment is a functionally necessary spacer between the actin binding site in S4 and the Ca2+ binding site in S6 (30, 41). Alternatively, the fifth domain can enhance or determine the binding activity of the neighboring fourth G-actin binding segment. A similar phenomenon, although not so extreme, has been observed for the third domain of gelsolin with respect to severing and capping (35). This domain, although not strictly required for severing, is necessary for the strong capping of S1 to S3.
The observation that adseverin, but not adseverin (D5), formed a stable complex with G actin in a gel filtration experiment is in accordance with the observed effects of both proteins on nucleation. The facts that a 2:1 G-actin–adseverin complex was completely dissociated upon chelation of Ca2+ and no EGTA-resistant 1:1 complex was observed prove its extreme Ca2+ sensitivity and regulation.
We showed that PU5.8 macrophage (the primary source of cytoplasmic gelsolin [45]) express no mouse adseverin as judged by Northern blotting. This may be indicative of a regulatory mechanism in which the expression of mouse adseverin and that of gelsolin are complementary. In this respect, it is of interest that gelsolin knockout mice are relatively healthy, showing normal embryonic development and longevity (42). It is clear from these experiments that gelsolin is not important for embryogenesis. However, taking into account our in vitro actin binding data and given the ubiquitous presence of adseverin in tissues, the latter could take over the role of gelsolin. Comparison of the adseverin mRNA levels in gelsolin knockout mice and wild-type mice and/or the construction of adseverin knockout and adseverin-gelsolin double-knockout mice would be informative. Besides the potentially complementary role, one can wonder about the significance of the fact that adseverin, adseverin (D5), and gelsolin are all present in the same cell. From our results, it is clear that adseverin and adseverin (D5) can be regulated at the transcriptional level by cytokines (here, IL-9). In this respect, we should point out that IL-9 is both a factor of activation and an inducer of differentiation, suggesting that the level of adseverin and adseverin (D5) is increased during differentiation and/or an immune response.
ACKNOWLEDGMENTS
We are grateful to Caroline-Aurore Seghers for help with fluorescence experiments.
J.C.R. is a Research Associate and J.L. is a Scientific Associate (Televie) with the Fonds National de la Recherche Scientifique, Belgium. C.A. is a Research Associate of the Flanders Fund for Scientific Research (FWO). This work was supported in part by the Flanders Action for Biotechnology (VLAB-COT), the Action Levenslijn 7.0040.94, GOA 91/96-3 to J.V., and in part by the Belgian Federal Service for Scientific, Technical and Cultural Affairs and the Operation Televie.
REFERENCES
- 1.Amann E, Brosius J, Ptashne M. Vectors bearing a hybrid trp-lac promoter useful for regulated expression of cloned genes in Escherichia coli. Gene. 1983;25:167–178. doi: 10.1016/0378-1119(83)90222-6. [DOI] [PubMed] [Google Scholar]
- 2.Ampe C, Vandekerckhove J. The F-actin capping proteins of Physarum polycephalum: cap42(a) is very similar, if not identical, to fragmin and is structurally and functionally very homologous to gelsolin; cap42(b) is Physarum actin. EMBO J. 1987;6:4149–4157. doi: 10.1002/j.1460-2075.1987.tb02761.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Andre E, Lottspeich F, Schleicher M, Noegel A. Severin, gelsolin, and villin share a homologous sequence in regions presumed to contain F-actin severing domains. J Biol Chem. 1988;263:722–727. [PubMed] [Google Scholar]
- 4.Ausubel F, Brent R, Kingston R, Moore D, Seidman J, Smith J, Struhl K. Current protocols in molecular biology 1993. New York, N.Y: Greene and Wiley-Interscience; 1993. [Google Scholar]
- 5.Bauw G, De Loose M, Inzé D, Van Montagu M, Vandekerckhove J. Alterations in the phenotype of plant cells studied by NH2-terminal amino-acid-sequence analysis of proteins electroblotted from two-dimensional gel-separated total extracts. Proc Natl Acad Sci USA. 1987;84:4806–4810. doi: 10.1073/pnas.84.14.4806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brenner S L, Korn E D. On the mechanism of actin monomer-polymer subunit exchange at steady state. J Biol Chem. 1983;258:5013–5020. [PubMed] [Google Scholar]
- 7.Bryan J. Gelsolin has three actin-binding sites. J Cell Biol. 1988;106:1553–1562. doi: 10.1083/jcb.106.5.1553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Burtnick L D, Koepf E D, Grimes J, Jones E Y, Stuart D I, McLaughin P J, Robinson R C. The crystal structure of plasma gelsolin; implication for actin severing, capping and nucleation. Cell. 1997;90:661–670. doi: 10.1016/s0092-8674(00)80527-9. [DOI] [PubMed] [Google Scholar]
- 9.Dieffenbach C W, SenGupta D N, Krause D, Sawzak D, Silverman R H. Cloning of murine gelsolin and its regulation during differentiation of embryonal carcinoma cells. J Biol Chem. 1989;264:13281–13288. [PubMed] [Google Scholar]
- 10.Gettemans J, De Ville Y, Waelkens E, Vandekerckhove J. The actin-binding properties of the Physarumactin-fragmin complex. Regulation by calcium, phospholipids, and phosphorylation. J Biol Chem. 1995;270:2644–2651. doi: 10.1074/jbc.270.6.2644. [DOI] [PubMed] [Google Scholar]
- 11.Giebing T, Hinssen H, D’Haese J. The complete sequence of a 40-kDa actin-modulating protein from the earthworm Lumbricus terrestris. Eur J Biochem. 1994;225:773–779. doi: 10.1111/j.1432-1033.1994.0773b.x. [DOI] [PubMed] [Google Scholar]
- 12.Gips S J, Kandzari D E, Goldschmidt-Clermont P J. Growth factor receptors, phospholipases, phospholipid kinases and actin reorganization. Semin Cell Biol. 1994;5:201–208. doi: 10.1006/scel.1994.1025. [DOI] [PubMed] [Google Scholar]
- 13.Hesterkamp T, Weeds A G, Mannherz H G. The actin monomers in the ternary gelsolin: 2 actin complexes are in an antiparallel orientation. Eur J Biochem. 1993;218:507–513. doi: 10.1111/j.1432-1033.1993.tb18403.x. [DOI] [PubMed] [Google Scholar]
- 14.Hültner L, Druez C, Moeller J, Schmitt E, Uyttenhove C, Rüde E, Dörmer P, Van Snick J. Mast cell growth-enhancing activity (MEA) is structurally related and functionally identical to the novel mouse T cell growth factor P40/TCGFIII (interleukin 9) Eur J Immunol. 1990;20:1413–1416. doi: 10.1002/eji.1830200632. [DOI] [PubMed] [Google Scholar]
- 15.Janmey P A. Phosphoinositides and calcium as regulators of cellular actin assembly and disassembly. Annu Rev Physiol. 1994;56:169–191. doi: 10.1146/annurev.ph.56.030194.001125. [DOI] [PubMed] [Google Scholar]
- 16.Janmey P A, Stossel T P. Modulation of gelsolin function by phosphatidylinositol 4,5-bisphosphate. Nature. 1987;325:362–364. doi: 10.1038/325362a0. [DOI] [PubMed] [Google Scholar]
- 17.Janmey P A, Lamb J, Allen P G, Matsudaira P T. Phosphoinositide-binding peptides derived from the sequences of gelsolin and villin. J Biol Chem. 1992;267:11818–11823. [PubMed] [Google Scholar]
- 18.Kraus-Friedman N. Signal transduction and calcium: a suggested role for the cytoskeleton in inositol 1,4,5-triphosphate action. Cell Motil Cytoskelet. 1994;28:279–284. doi: 10.1002/cm.970280402. [DOI] [PubMed] [Google Scholar]
- 19.Kwiatkowski D J, Stossel T P, Orkin S H, Mole J E, Colten H R, Yin H L. Plasma and cytoplasmic gelsolins are encoded by a single gene and contain a duplicated actin-binding domain. Nature. 1986;227:455–458. doi: 10.1038/323455a0. [DOI] [PubMed] [Google Scholar]
- 20.Kwiatkowski D J, Janmey P A, Yin H L. Identification of critical functional and regulatory domains in gelsolin. J Cell Biol. 1989;108:1717–1726. doi: 10.1083/jcb.108.5.1717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liou G I, Matragon S. A simple method to homogenize multiple tissue samples in small sizes without cross contamination. BioTechniques. 1992;13:719. [PubMed] [Google Scholar]
- 22.Louahed Y, Kermouni N, Van Snick J, Renauld J C. IL-9 induces expression of granzymes and high-affinity IgE receptor in murine T helper clones. J Immunol. 1995;154:5061–5070. [PubMed] [Google Scholar]
- 23.Lück A, D’Haese J, Hinssen H. A gelsolin-related protein from lobster muscle: cloning, sequence analysis and expression. Biochem J. 1995;305:767–775. doi: 10.1042/bj3050767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.MacLean-Fletcher S, Pollard T. Viscometric analysis of the gelation of Acanthamoebaextracts and purification of two gelation factors. J Cell Biol. 1980;85:414–428. doi: 10.1083/jcb.85.2.414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Maekawa S, Sakai H. Inhibition of actin regulatory activity of the 74-kDa protein from bovine adrenal medulla (adseverin) by some phospholipids. J Biol Chem. 1990;265:10940–10942. [PubMed] [Google Scholar]
- 26.Marcu M G, Zhang L, Elzagallaai A, Trifaro J M. Localization by segmental deletion analysis and functional characterization of a third actin-binding site in domain 5 of scinderin. J Biol Chem. 1998;273:3661–3668. doi: 10.1074/jbc.273.6.3661. [DOI] [PubMed] [Google Scholar]
- 27.Müllauer L, Fujita H, Ishizaki A, Kuzumaki N. Tumor-suppressive function of mutated gelsolin in ras-transformed cells. Oncogene. 1993;8:2531–2536. [PubMed] [Google Scholar]
- 28.Nakamura S, Sakurai T, Nonomura Y. Differential expression of bovine adseverin in adrenal gland revealed by in situ hybridization. Cloning of a cDNA for adseverin. J Biol Chem. 1994;269:5890–5896. [PubMed] [Google Scholar]
- 29.Paunio T, Kangas H, Kalkkinen N, Haltia M, Palo J, Peltonen L. Toward understanding the pathogenic mechanisms in gelsolin-related amyloidosis: in vitro expression reveals an abnormal gelsolin fragment. Hum Mol Genet. 1994;12:2223–2229. doi: 10.1093/hmg/3.12.2223. [DOI] [PubMed] [Google Scholar]
- 30.Pope B, Maclver S, Weeds A. Localization of the calcium-sensitive actin monomer binding site in gelsolin to segment 4 and identification of calcium binding sites. Biochemistry. 1995;34:1583–1588. doi: 10.1021/bi00005a014. [DOI] [PubMed] [Google Scholar]
- 31.Rodriguez Del Castillo A, Lemaire S, Tchakarov L, Jeyapragrasan M, Doucet J P, Vitale M L, Trifaro J M. Chromaffin cell scinderin, a novel calcium-dependent actin filament-severing protein. EMBO J. 1990;9:43–52. doi: 10.1002/j.1460-2075.1990.tb08078.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sakurai T, Kurokawa H, Nonomura Y. Comparison between the gelsolin and adseverin domain structure. J Biol Chem. 1991;166:15979–15983. [PubMed] [Google Scholar]
- 33.Schmitt E, van Brandwijk R, Van Snick J, Siebold B, Rüde E. TCGF III/P40 is produced by naive murine CD4+T cells but is not a general T cell growth factor. Eur J Immunol. 1989;19:2167–2170. doi: 10.1002/eji.1830191130. [DOI] [PubMed] [Google Scholar]
- 34.Spudich J A, Watt S. The regulation of rabbit skeletal muscle contraction. J Biol Chem. 1971;246:4866–4871. [PubMed] [Google Scholar]
- 35.Sun H Q, Wooten D C, Janmey P A, Yin H L. The actin side-binding domain of gelsolin also caps actin filaments. Implications for actin filament severing. J Biol Chem. 1994;269:9473–9479. [PubMed] [Google Scholar]
- 36.Tanaka M, Mullauer L, Ogiso Y, Fujita H, Moriya S, Furuuchi K, Harabayashi T, Shinohara N, Koyanagi T, Kuzumaki N. Gelsolin: a candidate for suppressor of human bladder cancer. Cancer Res. 1995;55:3228–3232. [PubMed] [Google Scholar]
- 37.Tchakarov L, Vitale M L, Jeyapragrasan M, Rodriguez Del Castillo A, Trifaro J M. Expression of scinderin, an actin filament-severing protein, in different tissues. FEBS Lett. 1990;268:209–212. doi: 10.1016/0014-5793(90)81010-l. [DOI] [PubMed] [Google Scholar]
- 38.T’Jampens D, Meerschaert K, Constantin B, Bailey J, Cook L J, De Corte V, De Mol H, Goethals M, Van Damme J, Vanderkerckhove J, Gettemans J. Molecular cloning, over-expression, developmental regulation and immunolocalization of fragmin P, a gelsolin-related actin-binding protein from Physarum polycephalumplasmodia. J Cell Sci. 1997;110:1215–1226. doi: 10.1242/jcs.110.10.1215. [DOI] [PubMed] [Google Scholar]
- 39.Uyttenhove C, Simpson J, Van Snick J. Functional and structural characterization of P40, a mouse glycoprotein with T-cell growth factor activity. Proc Natl Acad Sci USA. 1988;85:6934–6938. doi: 10.1073/pnas.85.18.6934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Way M, Weeds A G. Nucleotide sequence of pig plasma gelsolin. Comparison of protein sequence with human gelsolin and other actin-severing proteins shows strong homologies and evidence for large internal repeats. J Mol Biol. 1988;203:1127–1133. doi: 10.1016/0022-2836(88)90132-5. [DOI] [PubMed] [Google Scholar]
- 41.Way M, Gooch J, Pope B, Weeds A G. Expression of human plasma gelsolin in Escherichia coliand dissection of actin binding sites by segmental deletion mutagenesis. J Cell Biol. 1989;109:593–605. doi: 10.1083/jcb.109.2.593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Witke W, Sharpe A H, Hartwig J H, Aruma T, Stossel T, Kwiatkowski D I. Hemostatic, inflammatory, and fibroblast responses are blunted in mice lacking gelsolin. Nature. 1995;81:41–51. doi: 10.1016/0092-8674(95)90369-0. [DOI] [PubMed] [Google Scholar]
- 43.Yamamoto K, Pardee J D, Reidler J, Stryer L, Spudich J A. Mechanism of interaction of Dictyosteliumseverin with actin filaments. J Cell Biol. 1982;95:711–719. doi: 10.1083/jcb.95.3.711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yin H L, Stossel T P. Control of cytoplasmic actin gel-sol transformation by gelsolin, a calcium-dependent regulatory protein. Nature. 1979;281:583–586. doi: 10.1038/281583a0. [DOI] [PubMed] [Google Scholar]
- 45.Yin H L, Stossel T P. Purification and structural properties of gelsolin, a Ca2+-activated regulatory protein of macrophages. J Biol Chem. 1980;255:9490–9493. [PubMed] [Google Scholar]
- 46.Yu F X, Sun H Q, Janmey P A, Yin H. Identification of a polyphosphoinositide-binding sequence in an actin monomer-binding domain of gelsolin. J Biol Chem. 1992;267:14616–14621. [PubMed] [Google Scholar]