Abstract
DNA viruses are often used as vectors for foreign gene expression, but large DNA region from cloned or authentic viral genomes must usually be handled to generate viral vectors. Here, we present a unique system for generating adenoviral vectors by directly substituting a gene of interest in a small transfected plasmid with a replaced gene in a replicating viral genome in Cre-expressing 293 cells using the recombinase-mediated cassette exchange (RMCE) reaction. In combination with a positive selection of the viral cis-acting packaging signal connected with the gene of interest, the purpose vector was enriched to 97.5 and 99.8% after three and four cycles of infection, respectively. Our results also showed that the mutant loxP V (previously called loxP 2272), a variant target of Cre used in the RMCE reaction, was useful as a non-compatible mutant to wild-type loxP. This method could be useful for generating not only a large number of adenovirus vectors simultaneously, but also other DNA virus vectors including helper-dependent adenovirus vector.
INTRODUCTION
DNA viral vectors derived from adenoviruses, herpes viruses, vaccinia viruses, baculoviruses and so on have contributed to advances in both basic research and gene therapy. These vectors are particularly useful owing to their high expression level and the excellent stability of their viral genomes. However, the construction of these vectors, i.e. the introduction of a gene of interest to a specific site in the viral genome, has been hampered by the large sizes of viral genomes (∼36–200 kb). Consequently, viral vectors have been generated utilizing either authentic viral genome DNAs from adenoviruses (1), vaccinia viruses (2) or baculoviruses (3), or cloned full-length viral DNAs of adenoviruses (4,5), Epstein–Barr viruses (6) or baculoviruses (7). However, these large sections of DNA are difficult to handle, and the efficiency of obtaining viral vectors is low.
For first-generation adenovirus vector construction, the most efficient method available to date is the COS–TPC method (1), which utilizes viral genome DNA–terminal protein complex (DNA–TPC) instead of a cloned intact viral genome. This method is highly efficient because DNA–TPC yields ∼103-fold more infectious viruses than deproteinated viral genomes (8) or cloned intact viral DNA. DNA–TPC is usually prepared from purified virus particles but is, of course, present abundantly in infected host cells as part of the replicating viral genome. If a gene of interest could be transferred directly into the replicating viral DNA–TPC in these cells, vectors could be efficiently constructed without manipulating large sections of viral genome DNA. Therefore, we designed a novel approach where a gene of interest initially located in a small plasmid is transferred directly to a specific site in the replicating ‘recipient’ viral genome via a pathway mediated by recombinase-mediated cassette exchange (RMCE) and Cre recombinase.
The site-specific recombinase Cre, derived from bacteriophage P1, catalyzes the precise excision of DNA flanked by a pair of target sequences, loxP, in the same orientation. Cre also exhibits another recombination activity: double-reciprocal recombination or RMCE [(9) and for a review see (10)], in which two DNA regions on different molecules are both flanked by a wild type (wt) and a mutant loxP sequence. Cre exchanges the two regions flanked by the wt and mutant loxP sequences while maintaining the orientation of the regions. The mutant loxP 511 (11), which has a single-base substitution within the central eight-base region of loxP, has been used in the RMCE reactions (9,12–16). Ideally, such ‘exclusive’ mutants would not be able to recombine with wt loxP, but would exclusively and efficiently recombine only with identical mutant loxPs. We found a mutant loxP V (previously called loxP 2272) with two-base substitutions whose recombination efficiency was as high as that of loxP 511, but recombination with wt loxP was strictly undetectable in an in vitro assay (17), an Escherichia coli system using the M13 phagemid (18), or in mammalian chromosomes (19). And loxP V, together with wt loxP, has been successfully applied in the RMCE reactions, yielding a higher fidelity and efficiency than those obtained by loxP 511 together with wt loxP using embryonic stem (ES) cells with the selection of drug resistance (20,21). In the present study, we established a highly efficient method to produce adenoviral vectors by using the RMCE reaction mediated by the Cre and the mutant loxP V without the isolation of the viral clones. The method probably provides an efficient and a reliable tool not only for generating a large number of adenovirus vectors at once, but also for other viral vector construction and offers various applications in genetic manipulation of mammalian cell chromosomes.
MATERIALS AND METHODS
Cell lines
The human embryonic kidney cell line, 293 (22), and the monkey kidney cell line, CV1, were cultured in DMEM supplemented with 10% fetal calf serum (FCS). The 293 cells constitutively express adenoviral E1 genes and support the replication of E1-substituted recombinant adenoviruses. After infection with the recombinant adenoviruses, the cells were maintained in DMEM supplemented with 5% FCS. The 293 cell line that constitutively expressed nuclear localization signal (NLS)-tagged Cre recombinase under the control of the CAG promoter (23) (termed 293FNCre) was isolated and maintained with 10% FCS in DMEM containing 1.5 mg/ml of geneticin. The characterization of this cell line will be presented elsewhere.
Construction of recipient viruses
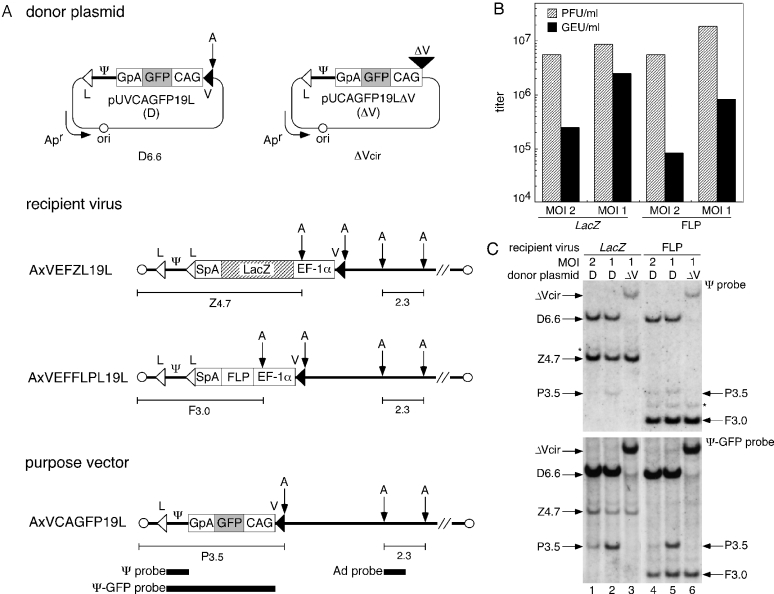
The mutant loxP V (V) used in this study was identical to loxP 2272 (17,19), with two transversion mutations within the eight-base central region (ATGTATGC) of wt loxP (L): T to A at the second base and G to C at the seventh base. To construct an expression unit flanked by L and V, we first cloned the ‘L sequence-SwaI site-V sequence’ at the SacI site of pUC19 (termed pUVwL). To construct the recipient viruses using the COS–TPC method (1), the left-end fragment of the adenovirus genome, including an L sequence at the end of the viral-packaging signal, was introduced into the cassette cosmid pAxcw (1) and termed pAx19Lcw (since the position where the L sequence was introduced was at the SgrAI site, 191 bp from the left end of the adenovirus genome, we included 19L in the name of the constructs). The ‘V-SwaI-L’ unit from pUVwL was then cloned at the SwaI site of pAx19Lcw; the resulting cassette cosmid for the recipient virus was named pAxVwL19L. The expression unit of the recipient viruses, which expresses LacZ or FLP recombinase under the control of a human polypeptide elongation factor 1α (EF-1α) promoter (24), was cloned at the SwaI site of pAxVwL19L. Each cosmid was mixed with adenovirus DNA–terminal protein complex and transfected to the 293 cells, resulting in the generation of recipient viruses (Figure 2A) expressing LacZ (AxVEFZL19L) or FLP (AxVEFFLPL19L) through homologous recombination.
Figure 2.
Production of a first-generation adenovirus vector by Cre-mediated RMCE. (A) Structure of the donor plasmids, recipient viruses and purpose vector. pUVCAGFP19L (top left), donor plasmid carrying a GFP-expression unit flanked by L and V. pUCAGFP19LΔV (top right), control plasmid lacking V. AxVEFZL19L and AxVEFFLPL19L, LacZ and FLP recipient viruses, respectively. AxVCAGFP19L, purpose vector. The position of the probes (closed bars, bottom) and the size (kb) of the specific bands corresponding to fragments from the recipient viruses (Z4.7 or F3.0), the purpose vector (P3.5) and the adenovirus constant region (2.3) detected by Southern blotting are shown. V, a mutant loxP V; CAG, CAG promoter; GFP, green fluorescent protein; GpA, rabbit β-globin poly(A) signal; Ψ, viral-packaging signal; L, wild-type loxP; Apr, ampicillin-resistance gene; ori, plasmid replication origin; EF-1α, EF-1α promoter; SpA, SV40 early poly(A) signal; and A, AflII site. (B) Virus titer of the produced purpose vector and residual recipient viruses in passage 1 stock. The indicated MOIs are those for the initial infection of the 293FNCre cells with the recipient viruses. PFU, plaque-forming units; GEU, GFP-expressing units. (C) Detection of the infectious viral genomes in passage 1 stock. 293FNCre cells were infected with LacZ (lanes 1–3) or FLP (lanes 4–6) recipient viruses at an MOI of 2 or 1 and cultured in a 6-well plate, followed by transfection with the donor plasmid (D) or the control plasmid (ΔV) at 3.0 μg/well. The passage 1 stock was prepared on day 5 and then used to infect CV1 cells to detect the infectious viral genomes. Specific bands corresponding to fragments from the purpose vector (P3.5) and each recipient virus (LacZ, Z4.7; FLP, F3.0) were detected using Southern blotting with Ψ (upper panel) or Ψ-GFP probes (lower panel) using the same nylon membrane. The bands representing D6.6 and ΔVcir, corresponding to AflII-cleaved 6.6 kb pUVCAGFP19L donor and circular pUCAGFP19LΔV, respectively, appear to be ‘carryover’ DNA initially transfected to the 293FNCre cells and then co-transferred to the CV1 cells, together with the adenoviral infection (36). These bands were not detected at and after the second cycle of infection (data not shown). Note that the DNA was detected as an AflII-digested linear form of the donor plasmid and a circular form of the control plasmid. The bands indicated by the asterisk (*) appear to have been caused by recipient viruses carrying two packaging signals generated by the re-integration of the excised circular Ψ into either loxP site on the recipient viral genomes.
Construction of the donor plasmid
The XhoI–SwaI fragment of pAxVwL19L containing the L sequence and the viral-packaging signal was inserted into pUVwL; the resulting cassette donor plasmid for the gene of interest was named pUVw19L. The expression unit of the GFP gene in the donor plasmid was derived from pxCAEGFP (25) [originally derived from pEGFP-C1 (EGFP vector; Clontech)], and cloned at the SwaI site of pUVw19L (termed pUVCAGFP19L or D; Figure 2A, left). Another plasmid lacking V (termed pUCAGFP19LΔV or ΔV; Figure 2A, right) was also constructed to serve as a negative control.
Production of first-generation adenovirus vector by RMCE
Under standard conditions, 293FNCre cells were infected with the recipient virus at an MOI of 1 and cultured in a 6-well plate for 1 h, followed immediately by the transfection of the donor plasmid at 3.0 μg/well using TransFast (Promega). When a viral cytopathic effect was clearly observed (normally 5 days after infection, under the above conditions), the cells were harvested together with the medium (∼1.5 ml/well). The cell suspension was then sonicated for 3 min (6 cycles, 30 s each) using a Bioruptor II sonicator (CosmoBio, Tokyo, Japan) at maximum power (200 W) and centrifuged at 1900 g (5000 r.p.m.; Tomy TMP 11 rotor) for 5 min at 4°C using a microcentrifuge. The supernatant was stored at −80°C as a virus stock (‘passage 1 stock’). To enrich the purpose vector by eliminating the residual recipient virus, 293FNCre cells were infected with 50 μl of the virus stock and cultured in a 6-well plate. A total of five passages were performed (including the initial infection step), and each virus stock was prepared as described above. The production of the purpose vector was monitored by green fluorescent protein (GFP) expression observed using a fluorescence microscope (IX70; Olympus) throughout the culture period.
Titration of the produced purpose vector
The total virus titer (PFU/ml) and the purpose vector titer [GFP-expressing units (GEU)/ml] of the passage 1 stock were measured using an end-point assay, as described previously (26). Briefly, 50 μl of DMEM with 5% FCS was dispensed into each well of a 96-well plate and 8 rows of 3-fold serial dilutions of the initial virus stock, starting at a dilution of 1:10 were prepared, followed by the addition of 293 cells (3 × 105 cells) in 50 μl of the medium to each well. Twelve days later, the end point of the cytopathic effect of the total viruses or GFP-expressing viruses was determined by using microscopy, and the 50% tissue culture infectious dose (TCID50) was calculated. In our laboratory, 1 TCID50/ml corresponded to ∼1 PFU/ml (26).
Southern blot analyses
To detect the ‘infectious virus’ DNA of purpose vector and the residual recipient viruses, CV1 cells were infected with the virus stocks at 100 μl per 6 cm dish. After 24 h, the total DNA was prepared from the dish according to the method described by Saito et al. (27). After AflII digestion, 20 μg of the DNA was electrophoresed on a 0.8% agarose gel at 35 V for 17 h. Before alkaline treatment, the gel was exposed to 0.1 N HCl for partial depurination (28), and the DNA was then transferred onto a Hybond-N nylon membrane (Amersham Biosciences) using the capillary-transfer method (29). Specific DNA was detected with a DIG DNA Labeling and Detection kit (Roche Diagnostics). A 0.3 kb XhoI–BspEI fragment of the Ψ gene, a 3.4 kb KpnI–XmnI fragment of the Ψ-GFP gene and a 0.3 kb AflII–BstEII fragment of the adenovirus genome (corresponding to the coding region of the viral protease gene) were labeled with digoxigenin-UTP as probes (Figure 2A), and specific DNAs were detected by autoradiography using chemiluminescence of CDP-Star (Roche Diagnostics).
Ratio of purpose vector to recipient viruses
To determine the ratio of produced purpose vector to residual recipient viruses, the total DNA was extracted from CV1 cells infected with each virus stock and was subjected to Southern blotting or quantitative real-time PCR. Since the ‘carryover’ donor plasmid DNA was transferred to the CV1 cells when the passage 1 stock was used (Figure 2C), the results of the real-time PCR were disturbed by this DNA. Hence, for passages 1 and 2 stocks, we quantified the density of the specific bands using Southern blot analysis, while for passage 3–5 stocks we measured the amount of each viral DNA using real-time PCR. TaqMan primers and probes specific for the GFP gene in the purpose vector (forward primer, 5′-CTG CTG CCC GAC AAC CA-3′; reverse primer, 5′-TGT GAT CGC GCT TCT CGT T-3′; and TaqMan probe, 5′-FAM-CTG AGC ACC CAG TCC GCC CTG-TAMRA-3′) and specific for the LacZ gene in the recipient virus (forward primer, 5′-GGC TTT CGC TAC CTG GAG AGA-3′; reverse primer, 5′-GTA TTT AGC GAA ACC GCC AAG AC-3′; and TaqMan probe, 5′-FAM-TGA TCC TTT GCG AAT ACG CCC ACG-TAMRA-3′) were designed using Primer Express software. The difference in the detection efficiency between the GFP and LacZ genes was corrected using the control plasmid pEFVGV-CALNLZ (19), containing both GFP and LacZ gene on the same plasmid.
RESULTS
Production of adenovirus vectors using RMCE
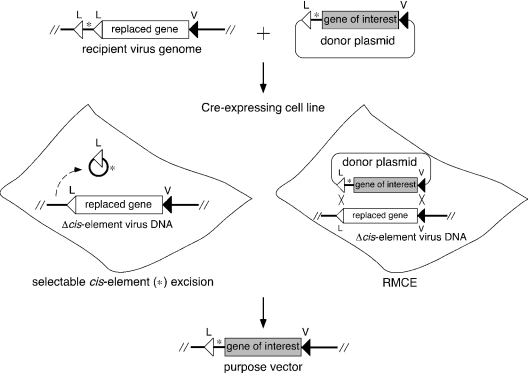
Using our novel method, a ‘gene of interest’ in a donor plasmid was substituted for a ‘replaced gene’ initially located in a replicating recipient virus genome using RMCE, together with a positive selection to eliminate residual recipient viruses, via viral cis-element excision (Figure 1). The ‘recipient virus’ genome contained a viral cis-element (denoted by *) flanked by a pair of wt loxP sequences (denoted by L) and a replaced gene flanked by wt loxP and the mutant loxP V (denoted by V), while a ‘donor plasmid’ contained a linked set of viral cis element and a gene of interest flanked by L and V. When the recipient virus and the donor plasmid were co-introduced into a Cre-expressing cell line, the purpose vector was produced as a result of the Cre-mediated excision of the viral cis-element in the recipient virus and the subsequent RMCE of the replaced gene in the Δcis-element recipient virus with the gene of interest from the donor plasmid. In the present study, we attempted to use this method to produce a first-generation adenovirus vector as a model.
Figure 1.
Strategy for the generation of viral vectors using the RMCE reaction. L, wt loxP and V, mutant loxP V.
To produce a first-generation adenovirus vector using RMCE, we first established a 293 cell line, 293FNCre, that constitutively expressed an NLS-tagged Cre (NCre). When the 293FNCre cells were infected with viruses bearing a pair of wt loxP sequences, AxCALNLZ (25), AxVEFZL19L and AxVEFFLPL19L described below, ∼80% of the replicating viral genome underwent Cre-mediated excision (data not shown). A similar level of Cre expression was reported by Parks et al. (30) We then prepared two different recipient viruses expressing LacZ and FLP (denoted as the LacZ recipient and the FLP recipient or AxVEFZL19L and AxVEFFLPL19L, respectively, in Figure 2A) and compared the efficiency of the production of a purpose vector containing the donor plasmid-derived GFP gene. Note that the function of the FLP gene per se was not significant. In these recipient viruses, the viral-packaging signal (Ψ) corresponded to the viral cis element (*) shown in Figure 1, while the LacZ- and FLP-expression units corresponded to the replaced gene. The 293FNCre cells were infected with one of the recipient viruses at a multiplicity of infection (MOI: PFU/cell) of 1 or 2, followed by the transfection of the donor plasmid (Figure 2A) at 3.0 μg/well on a 6-well plate. Under these conditions, a viral cytopathic effect was observed on day 5. The cells were harvested, and the virus stock (referred to as ‘passage 1 stock’) was prepared. The initial viral titer of the purpose vector produced by the RMCE was determined by infecting authentic 293 cells on 96-well plates with serially diluted passage 1 stock. The total virus titer, i.e. the residual recipient virus plus the purpose vector (PFU/ml), and the titer of the purpose vector only (GEU/ml) were then determined by observing the end point of the viral cytopathic effect and GFP-expressing fluorescent cells, respectively (Figure 2B). A significant titer (1 × 105 to 2 × 106 GEU/ml) of GFP-expressing purpose vector was detected in the passage 1 stock, showing that the purpose vector was efficiently generated by the RMCE. Moreover, the titer of purpose vector expressing GFP (closed bar) was higher when each recipient virus was infected at an MOI of 1, rather than at an MOI of 2. The titer was also higher when the LacZ recipient, rather than the FLP recipient was used; when both recipients were infected at an MOI of 1, the percentages of purpose vector in the initial viral stock were ∼30 and 4%, respectively, based on the GEU/PFU ratio (Figure 2B).
To confirm the efficient production of the purpose vector, the viral genomes of the infectious particles in the passage 1 stock were analyzed as follows. CV1 cells were infected with the passage 1 stock and then, 24 h later, completely washed with PBS (−) to remove any viral particles remaining on the surfaces of the cells. The total DNA, including the infectious viral genomes of the produced purpose vector and the residual recipient virus, was then extracted, digested using AflII, and subjected to Southern blotting using a Ψ or Ψ-GFP probe (Figure 2C, upper and lower panels, respectively; the positions of the probes are shown in Figure 2A, bottom). The Ψ probe detected the left-end fragments (denoted Z4.7, F3.0 and P3.5) of the two residual recipient viruses, the purpose vector and ‘carryover’ donor plasmid DNA (see Figure 2C legend) with uniform sensitivity, while the Ψ-GFP probe was designed to detect the purpose vector-derived P3.5 fragment ∼10-fold more strongly than the other fragments. When the cells were infected with one of the recipient viruses at an MOI of 1 (Figure 2C: LacZ, lane 2; FLP, lane 5), the P3.5 bands within the total DNA from the CV1 cells were faintly detected by the Ψ probe (upper panel), but were clearly detected by the Ψ-GFP probe (lower panel). When an MOI of 2 was used (LacZ, lane 1; FLP, lane 4), the P3.5 bands were only detected by the Ψ-GFP probe (lower panel). In contrast, the P3.5 bands were not detected when a control plasmid lacking V (denoted ΔV) was transfected instead of the donor plasmid (D) (lanes 3 and 6, both panels). These results showed that a purpose vector with the expected structure was indeed produced by the RMCE. The percentage of purpose vector to the residual recipient viruses expressing LacZ (P3.5 to Z4.7) or FLP (P3.5 to F3.0) was higher when the lower MOI was used (lane 1 versus lane 2 for LacZ and lane 4 versus lane 5 for FLP) and when the LacZ recipient, rather than the FLP recipient was used (lane 2 versus lane 5), confirming the result of the viral titration described above.
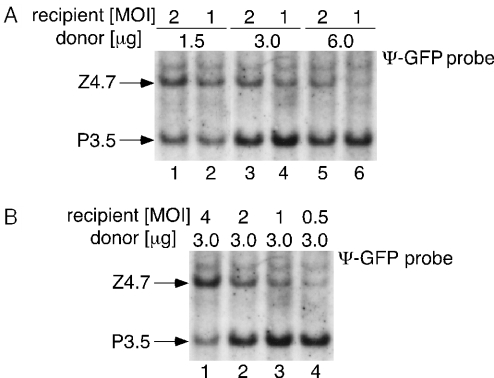
Optimization of vector production
To optimize the conditions for efficient production of the purpose vector, 293FNCre cells were infected with the LacZ recipient at an MOI of 2 or 1, transfected with various amounts of the donor plasmid, and the passage 1 stock was prepared. The viral genomes of the produced purpose vector and the residual recipient virus in infectious particles within the stock were then analyzed using the Ψ-GFP probe (Figure 3A) as described above. The ratio of the purpose vector to the residual recipient virus (Figure 3A, P3.5 to Z4.7) was clearly higher when the donor plasmid was transfected at 3.0 μg/well (lanes 3 and 4) than when it was transfected at 1.5 μg/well (lanes 1 and 2), irrespective of the MOI. No notable improvement was obtained when the amount of the donor plasmid was increased to 6.0 μg/well (lanes 5 and 6). Thus, we fixed the amount of the donor plasmid at 3.0 μg/well in subsequent experiments. We then optimized the MOI by infecting 293FNCre cells with the LacZ recipient at various MOIs (Figure 3B), followed by donor plasmid transfection. The purpose vector was produced more efficiently when an MOI of 1 or 0.5 (lanes 3 or 4) was used, compared with when an MOI of 4 or 2 (lanes 1 or 2) was used; thus, an MOI of ∼1 or 0.5 was selected as optimal, although the percentage of the purpose vector increased further when a lower MOI was used (see below). The percentage of purpose vector in the passage 1 stock was quantified as ∼25% when the LacZ recipient, an MOI of 0.5, and a donor plasmid amount of 3.0 μg/well of the donor plasmid were used. (Figure 3B, lane 4 and Table 1, first row, right. Note that the Ψ-GFP probe was used in Figure 3B.)
Figure 3.
Optimal conditions for producing adenovirus vector by Cre-mediated RMCE. The virus stock was prepared on day 5 and then used to infect CV1 cells for a Southern blot analysis. Note that the Ψ-GFP probe was used to detect the specific bands. The results were reproduced in three experiments, and a representative result is shown. (A) Optimal amount of the donor plasmid DNA. 293FNCre cells were infected with the LacZ-expressing recipient virus at an MOI of 2 or 1 and cultured in a 6-well plate, followed by transfection with the indicated amounts of donor plasmid (μg/well). (B) Optimal MOI of the recipient virus. 293FNCre cells were infected with the LacZ-recipient virus at the indicated MOIs, followed by transfection with the donor plasmid at 3.0 μg/well.
Table 1.
Ratio of produced purpose vector to residual recipient viruses during passages through 293FNCre cells
| Passage | Ratio (purpose vector:recipient virus) | |
|---|---|---|
| MOI 2a | MOI 0.5a | |
| 1b | 5:95 | 25:75 |
| 2b | 40:60 | 85:15 |
| 3c | 93.0:7.0 | 97.5:2.5 |
| 4c | 98.5:1.5 | 99.8:0.2 |
| 5c | 99.9:0.1 | 99.97:0.03 |
aThe initial MOIs used to infect the 293FNCre cells with the LacZ-recipient virus are shown.
bThe estimated percentages by quantifying the density of the Z4.7- and P3.5-bands specific to the purpose vector and the recipient virus, respectively.
cThe estimated percentages by measuring the amounts of the infectious viral genome of the purpose vector and the recipient virus using quantitative real-time PCR by detecting GFP and LacZ genes, respectively.
Vector purification by serial passage
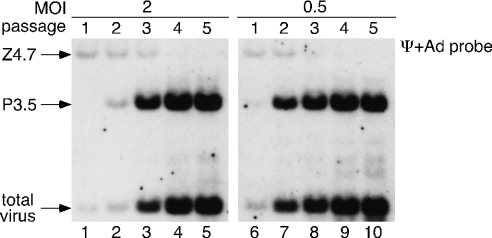
Since the produced purpose vector can replicate by itself and the residual recipient viruses are eventually eliminated because of the excision of a viral-packaging signal (Ψ), the vector stock can be purified by serial passages in Cre-expressing 293 cells or isolated by limiting dilutions in authentic 293 cells. To purify the purpose vector, 293FNCre cells were infected with 50 μl of the passage 1 stock and cultured in a 6-well plate; passage 2 stock was then prepared on day 5. A total of five passages were performed (including the initial infection step), and each prepared viral stock was used to infect CV1 cells for a Southern blot analysis of the infected viral genomes from infectious particles (Figure 4). The ratio of purpose vector to residual recipient virus (P3.5 to Z4.7) dramatically increased with each passage in the 293FNCre cells, especially when an MOI of 0.5 was initially used (right panel), and the Z4.7 band specific for the recipient virus was no longer detectable after the fourth passage at an MOI of 2 (lane 4) or after the third passage at an MOI of 0.5 (lane 8). When initiated at an MOI of 0.5, the percentage of purpose vector in the stock reached ∼80–90% after no more than two passages (Table 1), indicating that only 10 days are required to produce an adenovirus vector sufficient for practical screening applications. Moreover, a stock purity of 99.8% was obtained after four passages.
Figure 4.
Purification of the purpose vector by serial passage. CV1 cells infected with passages 1–5 stocks were used in a Southern blot analysis. The initial MOIs (MOI 2, left panel; 0.5, right panel) used to infect the 293FNCre cells with the LacZ-recipient virus are shown. The Ψ probe and Ad probe were simultaneously used to detect specific bands. The results were reproduced in three experiments, and a representative result is shown.
DISCUSSION
In the present study, a novel system for producing viral vectors was established based on an RMCE reaction mediated by Cre recombinase and the mutant loxP V. Here, we demonstrated the efficient production of a first-generation adenovirus vector by the replacement of a specific gene (replaced gene) in the replicating adenovirus genome with a gene of interest from plasmid DNA in Cre-expressing 293 cells (293FNCre cells). Although the first successful example of the RMCE was reported using FLP/FRT system (31), we chose Cre/loxP system because Cre is much more active, ∼90-fold more, than FLP in the recombination efficiency in mammalian cells (32).
Several factors were found to be important for the efficient construction of adenovirus vectors using the RMCE reaction. First, two different recipient viruses, a LacZ recipient and an FLP recipient, were used and the former recipient was found to produce the purpose vector much more efficiently than the latter, based on the viral titers (Figure 2B) and a Southern blot analysis of the infectious viral genomes (Figure 2C). The structures of both recipient viruses were identical, except for the inserted genes (LacZ and FLP). A possible reason for the difference in vector production efficiencies may be due to the differences in the genome sizes of the recipient viruses. Relative to the size of the wt adenovirus-5 genome (35 935 nt = 100%), the genome of the LacZ recipient was much larger (103.5%) than those of the FLP recipient (97.8%) and the GFP-expressing purpose vector (95.4%). In general, when two viral clones with different genome sizes are simultaneously replicated in the permissive cells, the virus with the shorter genome tends to become dominant, probably because shorter genomes replicate faster than longer ones (Y. Kanegae and I. Saito, unpublished data). A recipient virus with a genome larger than that of the LacZ recipient described here may be difficult to construct, since such a genome size would be near the upper limit of the adenovirus packaging (105%) (33). Therefore, the purpose vector has two different selective advantages against the LacZ recipient virus; the majority of the recipients were not packaged by excision of their packaging signal, and the purpose vector outgrew the recipients of much larger size.
On the other hand, if undesired recombination between wt loxP (L) and loxP V (V) should occur, viruses lacking either a replaced gene or a gene of interest would be generated (refer to Figure 2A), and such viruses would replicate much faster and might become majority of the viral population. Langer et al. (34) faced this problem in construction of adenoviral vector when using loxP 511 by RMCE and hence their trial was unsuccessful. Although the loxP 511 was the first mutant applied to the RMCE reaction as an ‘exclusive’ or ‘non-compatible’ mutant loxP, our in vitro analyses revealed that most of the mutant loxPs with single-base substitutions, including loxP 511, slightly but reproducibly recombined with wt loxP (17). In contrast, most of the mutant loxPs with two-base substitutions exhibited a high fidelity against wt loxP but a low recombination efficiency with identical mutant loxP, while the mutant loxP V showed both a high fidelity and high recombination efficiency not only in vitro analysis, but also in mammalian cells. In the present study, the viruses without an insert were not evident and did not disturb the process of the vector construction, even though thousands of copies of viral genomes per cell replicates in 293FNCre cells. Even after the fifth serial passage using the LacZ recipient, no putative virus genome lacking an insert was detected, showing such virus, if any, was present at most 20-fold less than the purpose vector (M. Ishimura and Y. Kanegae, unpublished data). In addition, the designs of the virus structures in the present study were chosen such that the recipient virus and the purpose vector would lose their viral packaging signal if L–V recombination occurred (Figure 2A).
Second, our data showed that a low initial MOI (1.0 or 0.5) of the recipient virus produced a much higher ratio of the purpose vector (Figure 3B), suggesting that only a small amount of recipient virus is required by using this method. This result may indicate that large amounts of recipient virus are difficult to eliminate and that a small amount of recipient virus is sufficient to generate purpose vectors. However, we do not recommend using an MOI of <0.5 because we observed undesired generation of rearranged viruses containing only part of the gene of interest, probably because uncontrolled multiple cycles of infection occurred within a single passage (data not shown).
Third, using the LacZ recipient, we observed that the purpose vector was purified and the recipient virus was eliminated during serial passages. The proportion of recipient virus in the stock constantly decreased by ∼5–10-fold per passage, reaching a percentage of no more than 0.03% after five cycles of infection (Table 1). This result shows that the residual recipient virus can be efficiently eliminated from the stock, irrespective of the amount of residual recipient virus, by excising the packaging signal. Suprisingly, the percentage of purpose vector in the stock after the initial infection cycle reached as high as 25% without any kind of external selection. One obvious reason for such high efficiency was that, in our construction design, the genome of the purpose virus grew faster than that of the recipient viruses depending on the genome size, even within the initial cycle. However, the efficiency of RMCE itself might be more important. Previous reports (20,21) using RMCE method and loxP V were concerned about drug-selected isolation of purpose cell clones in murine ES cell lines and hence the efficiency of RMCE was not examined. Our preliminary results, using CV1 cells, which is replication-non-permissive for adenovirus vector, indicated that without any selection, approximately no <1% of the infected cells did receive the GEU through the RMCE reaction (data not shown).
The method described here is the first successful example of RMCE applied to the construction of viral vectors in mammalian cells. For adenovirus vector construction, this method provides several unique features, compared with the conventional methods. First, this method only requires that a small donor plasmid containing cDNA be introduced into the vector, while other methods require very large plasmids or cosmids containing nearly the full-length of adenovirus genome (1,4,5). These large plasmids are often difficult to handle, especially for new users of adenovirus vectors. RMCE-related system using att of lambda phage have been commercially available recently (35). Because, however, lambda-phage recombination system can work only within E.coli, large viral genome should be handled before the generation of viral vector. Second, a purpose vector can be quickly obtained by transfection plus infection followed by more than three times serial passages. Although this virus stock contained a trace of remaining recipient virus, the pure purpose virus can be readily isolated using limiting-dilution, if desired.
Therefore, this simple and efficient method will probably be useful in genomic/proteomic approaches for expression screening using adenovirus vectors not only in culture cell but also in vivo directly when hundreds of cDNA candidates are to be examined. Using this method, a set of 24- or even 96-different recombinant adenoviruses could be obtained by transfecting 293FNCre cells with a large set of donor plasmids in a multiple-well plate and subsequently infecting the cells with the recipient virus for serial passages (using the same multi-well plate layout). The purpose vector would become dominant after about three cycles, without requiring the tedious isolation of each recombinant virus. After expression screening, the desired adenoviral clones could be readily isolated for further large-scale expression experiments or for use in animal experiments.
The present work shows that a given DNA plasmid sequence can be efficiently transferred into a DNA viral genome using a viral cis element, such as the packaging signal utilized in the present study. Therefore, this method could be used for the construction of large DNA vectors other than first-generation adenovirus vectors. The study of DNA viruses with large genomes, such as various herpes viruses, including Epstein–Barr virus, could probably have benefits from this technique. This method may also be applicable to the development of helper-dependent adenovirus vectors (30) generated by the substitution of ∼30 kb of foreign DNA. This possibility is now being studied in our laboratory.
Acknowledgments
The authors thank Dr J. Miyazaki for the CAG promoter, Ms K. Nishijima and Ms E. Kondo for their excellent secretarial support, and Ms Y. Nishiwaki for real-time PCR analysis. We also thank Ms S. Kondo for her continuous support and encouragement. This work was supported in part by grants-in-aid from the Ministry of Education, Culture, Sports, Science and Technology to M.N., Y.K. and I.S. Funding to pay the Open Access publication charges for this article was provided by the grant above.
Conflict of interest statement. None declared.
REFERENCES
- 1.Miyake S., Makimura M., Kanegae Y., Harada S., Sato Y., Takamori K., Tokuda C., Saito I. Efficient generation of recombinant adenoviruses using adenovirus DNA–terminal protein complex and a cosmid bearing the full-length virus genome. Proc. Natl Acad. Sci. USA. 1996;93:1320–1324. doi: 10.1073/pnas.93.3.1320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Merchlinsky M., Moss B. Introduction of foreign DNA into the vaccinia virus genome by in vitro ligation: recombination-independent selectable cloning vectors. Virology. 1992;190:522–526. doi: 10.1016/0042-6822(92)91246-q. [DOI] [PubMed] [Google Scholar]
- 3.Kitts P.A., Possee R.D. A method for producing recombinant baculovirus expression vectors at high frequency. Biotechniques. 1993;14:810–817. [PubMed] [Google Scholar]
- 4.Bett A.J., Haddara W., Prevec L., Graham F.L. An efficient and flexible system for construction of adenovirus vectors with insertions or deletions in early regions 1 and 3. Proc. Natl Acad. Sci. USA. 1994;91:8802–8806. doi: 10.1073/pnas.91.19.8802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mizuguchi H., Kay M.A. Efficient construction of a recombinant adenovirus vector by an improved in vitro ligation method. Hum. Gene Ther. 1998;9:2577–2583. doi: 10.1089/hum.1998.9.17-2577. [DOI] [PubMed] [Google Scholar]
- 6.Kanda T., Yajima M., Ahsan N., Tanaka M., Takada K. Production of high-titer Epstein–Barr virus recombinants derived from Akata cells by using a bacterial artificial chromosome system. J. Virol. 2004;78:7004–7015. doi: 10.1128/JVI.78.13.7004-7015.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ernst W.J., Grabherr R.M., Katinger H.W. Direct cloning into the Autographa californica nuclear polyhedrosis virus for generation of recombinant baculoviruses. Nucleic Acids Res. 1994;22:2855–2856. doi: 10.1093/nar/22.14.2855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sharp P.A., Moore C., Haverty J.L. The infectivity of adenovirus 5 DNA–protein complex. Virology. 1976;75:442–456. doi: 10.1016/0042-6822(76)90042-8. [DOI] [PubMed] [Google Scholar]
- 9.Feng Y.Q., Seibler J., Alami R., Eisen A., Westerman K.A., Leboulch P., Fiering S., Bouhassira E.E. Site-specific chromosomal integration in mammalian cells: highly efficient CRE recombinase-mediated cassette exchange. J. Mol. Biol. 1999;292:779–785. doi: 10.1006/jmbi.1999.3113. [DOI] [PubMed] [Google Scholar]
- 10.Baer A., Bode J. Coping with kinetic and thermodynamic barriers: RMCE, an efficient strategy for the targeted integration of transgenes. Curr. Opin. Biotechnol. 2001;12:473–480. doi: 10.1016/s0958-1669(00)00248-2. [DOI] [PubMed] [Google Scholar]
- 11.Hoess R.H., Wierzbicki A., Abremski K. The role of the loxP spacer region in P1 site-specific recombination. Nucleic Acids Res. 1986;14:2287–2300. doi: 10.1093/nar/14.5.2287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bouhassira E.E., Westerman K., Leboulch P. Transcriptional behavior of LCR enhancer elements integrated at the same chromosomal locus by recombinase-mediated cassette exchange. Blood. 1997;90:3332–3344. [PubMed] [Google Scholar]
- 13.Bethke B., Sauer B. Segmental genomic replacement by Cre-mediated recombination: genotoxic stress activation of the p53 promoter in single-copy transformants. Nucleic Acids Res. 1997;25:2828–2834. doi: 10.1093/nar/25.14.2828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Trinh K.R., Morrison S.L. Site-specific and directional gene replacement mediated by Cre recombinase. J. Immunol. Methods. 2000;244:185–193. doi: 10.1016/s0022-1759(00)00250-7. [DOI] [PubMed] [Google Scholar]
- 15.Lauth M., Moerl K., Barski J.J., Meyer M. Characterization of Cre-mediated cassette exchange after plasmid microinjection in fertilized mouse oocytes. Genesis. 2000;27:153–158. doi: 10.1002/1526-968x(200008)27:4<153::aid-gene40>3.0.co;2-b. [DOI] [PubMed] [Google Scholar]
- 16.Hong S.H., Lee M.S., Park S., Kim H.I., Shin H.J., Kwon M.H., Kim K.H. A phagemid system enabling easy estimation of the combinatorial antibody library size. Immunol. Lett. 2004;91:247–253. doi: 10.1016/j.imlet.2003.12.011. [DOI] [PubMed] [Google Scholar]
- 17.Lee G., Saito I. Role of nucleotide sequences of loxP spacer region in Cre-mediated recombination. Gene. 1998;216:55–65. doi: 10.1016/s0378-1119(98)00325-4. [DOI] [PubMed] [Google Scholar]
- 18.Siegel R.W., Jain R., Bradbury A. Using an in vivo phagemid system to identify non-compatible loxP sequences. FEBS Lett. 2001;505:467–473. doi: 10.1016/s0014-5793(01)02806-x. [DOI] [PubMed] [Google Scholar]
- 19.Kondo S., Okuda A., Sato H., Tachikawa N., Terashima M., Kanegae Y., Saito I. Simultaneous on/off regulation of transgenes located on a mammalian chromosome with Cre-expressing adenovirus and a mutant loxP. Nucleic Acids Res. 2003;31:e76. doi: 10.1093/nar/gng076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kolb A.F. Selection-marker-free modification of the murine beta-casein gene using a lox2272 [correction of lox2722] site. Anal. Biochem. 2001;290:260–271. doi: 10.1006/abio.2000.4984. [DOI] [PubMed] [Google Scholar]
- 21.Araki K., Araki M., Yamamura K. Site-directed integration of the cre gene mediated by Cre recombinase using a combination of mutant lox sites. Nucleic Acids Res. 2002;30:e103. doi: 10.1093/nar/gnf102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Graham F.L., Smiley J., Russell W.C., Nairn R. Characteristics of a human cell line transformed by DNA from human adenovirus type 5. J. Gen. Virol. 1977;36:59–74. doi: 10.1099/0022-1317-36-1-59. [DOI] [PubMed] [Google Scholar]
- 23.Niwa H., Yamamura K., Miyazaki J. Efficient selection for high-expression transfectants with a novel eukaryotic vector. Gene. 1991;108:193–199. doi: 10.1016/0378-1119(91)90434-d. [DOI] [PubMed] [Google Scholar]
- 24.Kim D.W., Uetsuki T., Kaziro Y., Yamaguchi N., Sugano S. Use of the human elongation factor 1 alpha promoter as a versatile and efficient expression system. Gene. 1990;91:217–223. doi: 10.1016/0378-1119(90)90091-5. [DOI] [PubMed] [Google Scholar]
- 25.Kanegae Y., Lee G., Sato Y., Tanaka M., Nakai M., Sakaki T., Sugano S., Saito I. Efficient gene activation in mammalian cells by using recombinant adenovirus expressing site-specific Cre recombinase. Nucleic Acids Res. 1995;23:3816–3821. doi: 10.1093/nar/23.19.3816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kanegae Y., Makimura M., Saito I. A simple and efficient method for purification of infectious recombinant adenovirus. Jpn. J. Med. Sci. Biol. 1994;47:157–166. doi: 10.7883/yoken1952.47.157. [DOI] [PubMed] [Google Scholar]
- 27.Saito I., Oya Y., Yamamoto K., Yuasa T., Shimojo H. Construction of nondefective adenovirus type 5 bearing a 2.8-kilobase hepatitis B virus DNA near the right end of its genome. J. Virol. 1985;54:711–719. doi: 10.1128/jvi.54.3.711-719.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Saito I., Groves R., Giulotto E., Rolfe M., Stark G.R. Evolution and stability of chromosomal DNA coamplified with the CAD gene. Mol. Cell. Biol. 1989;9:2445–2452. doi: 10.1128/mcb.9.6.2445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sambrook J., Fritsch E.F., Maniatis T. Molecular Cloning: A Laboratory Manual, 3rd edn. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1992. [Google Scholar]
- 30.Parks R.J., Chen L., Anton M., Sankar U., Rudnicki M.A., Graham F.L. A helper-dependent adenovirus vector system: removal of helper virus by Cre-mediated excision of the viral packaging signal. Proc. Natl Acad. Sci. USA. 1996;93:13565–13570. doi: 10.1073/pnas.93.24.13565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Seibler J., Bode J. Double-reciprocal crossover mediated by FLP-recombinase: a concept and an assay. Biochemistry. 1997;36:1740–1747. doi: 10.1021/bi962443e. [DOI] [PubMed] [Google Scholar]
- 32.Nakano M., Odaka K., Ishimura M., Kondo S., Tachikawa N., Chiba J., Kanegae Y., Saito I. Efficient gene activation in cultured mammalian cells mediated by FLP recombinase-expressing recombinant adenovirus. Nucleic Acids Res. 2001;29:e40. doi: 10.1093/nar/29.7.e40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Parks R.J., Graham F.L. A helper-dependent system for adenovirus vector production helps define a lower limit for efficient DNA packaging. J. Virol. 1997;71:3293–3298. doi: 10.1128/jvi.71.4.3293-3298.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Langer S.J., Ghafoori A.P., Byrd M., Leinwand L. A genetic screen identifies novel non-compatible loxP sites. Nucleic Acids Res. 2002;30:3067–3077. doi: 10.1093/nar/gkf421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hartley J.L., Temple G.F., Brasch M.A. DNA cloning using in vitro site-specific recombination. Genome Res. 2000;10:1788–1795. doi: 10.1101/gr.143000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yoshimura K., Rosenfeld M.A., Seth P., Crystal R.G. Adenovirus-mediated augmentation of cell transfection with unmodified plasmid vectors. J. Biol. Chem. 1993;268:2300–2303. [PubMed] [Google Scholar]