Abstract
To explore the mechanisms by which CAG trinucleotide repeat tracts undergo length changes in yeast cells, we examined the polarity of alterations with respect to an interrupting CAT trinucleotide near the center of the tract. In wild-type cells, in which most tract changes are large contractions, the changes that retain the interruption are biased toward the 3′ end of the repeat tract (in reference to the direction of lagging-strand synthesis). In rth1/rad27 mutant cells that are defective in Okazaki fragment maturation, the tract expansions are biased to the 5′ end of the repeat tract, while the tract contractions that do not remove the interruption occur randomly on either side of the interruption. In msh2 mutant cells that are defective in the mismatch repair machinery, neither the small changes of one or two repeat units nor the larger contractions attributable to this mutation are biased to either side of the interruption. The results of this study are discussed in terms of the molecular paths leading to expansions and contractions of repeat tracts.
Repetitions of the trinucleotide CAG appear in the human genome and are the cause of more than 10 dominant hereditary neurological and neuromuscular diseases (19, 24). Disease alleles are distinguishable from normal alleles by their increased tract lengths. Furthermore, examination of tract lengths in affected parents and their children shows that tracts frequently change in length during parental transmission. In some cases, there is a distinct bias toward changes to longer tract lengths that lead to a condition more severe in the children than that in their affected parent. This phenomenon is the underlying cause of the genetic anticipation in which the disease exhibits an earlier onset as it passes through a pedigree.
The reasons for the instability of repetitive CAG tracts are becoming clearer. Like all repetitive sequences, they have an inherent instability based on the ability of the two strands of DNA to misalign. In addition, the trinucleotide repeats are able to form hairpin-like structures (5, 20, 29). The potential to form secondary structures in vivo imparts additional properties to them that contribute to their instability. One area that remains to be more fully illuminated is how the CAG repeat tracts are disruptive to the cellular complexes that replicate, transcribe, repair, and recombine DNA.
To understand the underlying causes for the instability of CAG repeat tracts, we and others have placed CAG repeat tracts in a yeast chromosome and observed their behavior in wild-type and mutant cells (3, 4, 14, 15, 25, 26). Our studies (14) and those of Freudenreich et al. (4) have shown that when CTG, the complement of CAG, is the lagging-strand template during replication, repeat tracts are approximately 10 times more unstable than when CAG serves as the lagging-strand template. In wild-type yeast cells, the overwhelming majority of tract length changes are contractions of 10 or more repeat units. This pattern is altered by the introduction of an rth1/rad27 mutation that is defective in Okazaki fragment maturation (22, 23). In an rth1/rad27 mutant, tracts become more unstable and approximately half of the events are tract expansions (3, 26). Repeat tracts also exhibit more changes in mismatch repair mutants. In these mutants, many of the events are small changes of one or two repeat units, most of which are losses of repeat units but some of which are gains (25).
In this study, we have characterized further the CAG tract length changes that occur in rth1/rad27 and msh2 mutant cells. These two mutants are of particular interest, because we believe that the tract length changes that arise in these two mutant backgrounds occur for different reasons. The absence of the flap endonuclease in rth1/rad27 mutant cells is likely the cause of the excess changes recovered in this strain. In particular, flaps of nucleotides at the 5′ ends of Okazaki fragments that give rise to tract expansions in the mutant either do not form or are efficiently removed in wild-type cells. In contrast, the phenotypic manifestations of the msh2 mutation likely arise because of the absence of a corrective activity. Thus, in this case the small loops that are the substrate for the mismatch repair machinery occur because of strand slippage during replication equally in both wild-type and mutant cells. They are normally invisible because they are efficiently removed by the wild-type mismatch repair system.
An indication of the independent paths leading to the events that arise in msh2 and rth1/rad27 mutant cells is the observation that the double mutant exhibits a spectrum of mutational events that is a composite of the events that occur in each single mutant. The clearest example of the composite pattern has been observed in the reversion pattern of a lys2 frameshift mutation (31). Most of the changes in the rth1/rad27 mutant are duplications of a 32-bp sequence, while most changes in the msh2 mutant are deletions of a single A residue in a run of six A residues. In the double mutant, both types of changes occur roughly in proportion to the individual contribution each mutation makes to the overall reversion rate. Similarly, in the case of GT dinucleotide tracts, expansions are the sole product of the rth1/rad27 mutant, while two-thirds of the events in msh2 mutant cells are contractions (8). The double mutant yields a composite that is two-thirds expansions and one-third contractions. We also have observed for CAG repeat tracts that the double mutant yields both the small tract changes indicative of the msh2 mutant and the tract expansions indicative of the rth1/rad27 mutant (27). For both the GT dinucleotide repeat tracts and the CAG trinucleotide repeat tracts, the results may be somewhat ambiguous because the mismatch repair system may be responsible for correcting some, but not all, of the changes created by the absence of the flap endonuclease encoded by RTH1/RAD27.
This study describes the results of mapping the polarity of the tract length changes occurring in wild-type and mutant cells employing long CAG repeat tracts with a single variant repeat near their centers. These studies provide insight into the pathways by which CAG tracts expand and contract.
MATERIALS AND METHODS
Construction of interrupted repeat tracts.
Repeat tracts containing interruptions of a single CAT repeat were made by a PCR scheme with primers that change one CAG repeat unit into a CAT triplet. In this scheme, two short CAG tracts were mutagenized to contain a CAT at the opposite ends of the tracts, and the resulting mutagenized tracts were joined by their CAT interruptions to produce one longer tract with a single CAT interruption near its middle. In the first step, two pairs of primers were used to copy a relatively short repeat tract (approximately 45 repeat units). One set of primers was DMLAde2L (5′-AGCGCTAGCCCGGGACACAAGGCTGAGCAG) and DMLAde2o (5′-GGAGCCCTGCTGAGGTGCTGCTGCTGATGCTG), and the other set of primers was DMLAde2m (5′-CCGGGACACAAGGCTGAGCATCAGCAGCAG) and DMLAde2R (5′-ATGGCTAGCGGAGCCCTGCTGAGGTGCTG). The bases that substitute the CAT for the CAG are in boldface. Each of the four primers contained either the 5′ (primers m and L) or the 3′ (primers o and R) unique sequence that flanks the human ataxin1 gene repeat tract (18). In addition, primers DMLAde2L and DMLAde2R included an NheI recognition site and a 3-bp clamp at their 5′ ends. In the second step of the scheme, the two products were digested with SfaNI [5′-GCATC(N)5], and the longer of the digestion products was ligated. This created a tract that was nearly twice as long as the template and had a single CAT interruption that was displaced by two CAG repeat units from the exact center.
The NheI recognition sites were used to clone the repeat into a HindIII site of ADE2 as previously described (14). Two derivatives were created with either CAG (tract CI) or CTG (tract DI) in the ADE2 coding strand. These disrupted copies of ADE2 were cloned into ARO2 in the same orientation. At this point, the tracts were sequenced. Tract CI has the sequence (CAG)43CAT(CAG)46, while tract DI has the sequence (CTG)48ATG(CTG)48. The length difference between the two tracts likely arose either during the PCR scheme or during propagation of the tracts in the cloning host Escherichia coli.
Strain construction.
Disruption of the ARO2 locus on chromosome VII was carried out with strain SSL204 as previously described (14). These strains were then mated to isogenic derivatives of SSL204A containing either the msh2 or the rth1/rad27 mutation (25, 26). Sporulation of the resulting diploids resulted in isogenic segregants with both the embedded repeat tracts and the desired mutation.
Measurement of tract length changes.
The general scheme of detecting tract length changes by PCR and measuring the frequency of tract changes has been previously described (14). In summary, template DNA is purified from sibling colonies arising from the dispersal of a parental colony into single cells, and tract lengths are measured by displaying on sequencing gels the PCR products created with primers to unique flanking sequences in ADE2. Sibling colonies lacking the parental band and containing a band of smaller (contraction) or larger (expansion) size arise from changes during growth of the parental colony (14). Approximately 30 sibling colonies from multiple parental colonies were analyzed (Table 1).
TABLE 1.
Compilation of tract length changes
| Genotype | Tract and no. of repeat units | No. of parental colonies | No. of sibling colonies | No. lacking parental band | No. of expansionsa | No. of contractions with the following no. of repeat units lost:
|
|
|---|---|---|---|---|---|---|---|
| ≤2 | ≥3 | ||||||
| Wild type | CI90 | 4 | 182 | 3 | 0 | 0 | 3 |
| rth1/rad27 | CI90 | 6 | 173 | 64 | 29 | 0 | 35 |
| msh2 | CI90 | 5 | 141 | 18 | 5 | 11 | 2 |
| Wild type | DI97 | 5 | 154 | 34 | 0 | 0 | 34 |
| rth1/rad27 | DI97 | 17 | 493 | 358 | 29 | 0 | 329 |
| msh2 | DI97 | 9 | 274 | 139 | 4 | 22 | 113 |
The tract expansions in the msh2 mutant ranged in size from 1 to 3 repeat units, while those in the rth1/rad27 mutant ranged from 1 to 40 repeat units in size.
Mapping of the positions of tract length changes.
The positions of the tract length changes were mapped relative to the CAT interruptions. PCR products were prepared with primers DMLAde2c (5′ primer) and DMLAde2b (3′ primer) that recognize sequences in ADE2 that flank the repeat tract. One of the two primers was 5′-end labeled with 32P. The PCR products were purified with Prep-a-gene (BioRad), digested with SfaNI (New England BioLabs), and displayed by electrophoresis on sequencing gels.
In some cases, e.g., when changes of a single repeat unit occurred or when the size of the digestion product was discordant with the magnitude of the size change, the analysis was repeated with an end label on the second PCR primer. In a few cases (7 of 136), changes were found to have occurred on both sides of the interruption. These rare events are likely the culmination of two events.
RESULTS
Creating strains with interrupted tracts.
Long CAG repeat tracts containing a single CAT interruption were constructed by a PCR scheme (see Materials and Methods) and were placed within yeast chromosome VII. Tracts were oriented with either CAG (tract CI) or CTG (tract DI) in the ADE2 coding strand. Isogenic derivatives of the wild-type strains carrying the interrupted tracts were created with either a deletion/disruption of rth1/rad27 encoding the flap endonuclease (22, 23) or a deletion/disruption of msh2 encoding a component of the mismatch repair system (10).
Patterns of changes in wild-type and mutant cells.
The relative stabilities and distinctive patterns of the tract length changes that occurred in the interrupted tracts in wild-type and mutant cells (Table 1) were in accord with our previous results (14, 25, 26). Because an ARS element (replication origin) is located 5′ to ADE2 (30), tract CI uses CAG as the lagging-strand template and tract DI uses CTG as the lagging-strand template. Consequently, tract DI is less stable than tract CI in wild-type cells as well as in mutant cells (Table 1) (4, 14). Furthermore, as previously observed for uninterrupted tracts, the overwhelming majority of tract length changes in wild-type cells are contractions of three or more repeat units (Table 1). In the rth1/rad27 mutant, the interrupted tracts are very unstable and exhibit the more frequent occurrence of tract expansions (Table 1), as previously found for uninterrupted tracts (3, 26). Also in accord with previous results, tract length changes in the interrupted repeat tract are more frequent in the msh2 mutant, and a new class of events that includes tract contractions of one or two repeat units and tract expansions of one to three repeat units is unique to this mutant (Table 1) (25).
Mapping the positions of tract length changes.
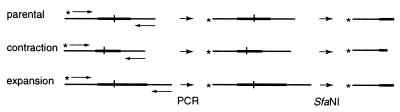
Because the CAT interruption introduced an SfaNI site into the repeat tract and because one of the two PCR primers used to measure tract length was end labeled, tract length changes could be mapped relative to the SfaNI site (Fig. 1). To map the position, the type of event—expansion or contraction—and the size of the tract length change were first determined by comparison of the PCR product copied from the altered tract to that from the parental colony. PCR products were then digested with SfaNI and run on a gel with appropriate standards. Depending on whether the change had occurred distal or proximal to the interruption in relation to the end label of the PCR primer, the change results in either the retention of the parental size SfaNI digestion product or in the appearance of an SfaNI fragment of altered length, respectively (Fig. 1). An example of some of the analyses are shown in Fig. 2. When large contractions occur, they often remove the interruption. Such changes result in the loss of the SfaNI site and consequently were recorded as the absence of SfaNI digestion products (Fig. 2).
FIG. 1.
Mapping scheme for repeat tract changes. The repeat tract (thickened bars), interruption (vertical tics), PCR primers (arrowhead lines), and 5′-end label (asterisks) are shown. To map changes, the sizes of the undigested PCR products were compared first, and then the products were digested with SfaNI to observe whether the labeled digestion product is the same as or smaller or larger than that of the parental size fragment. In the examples, the contraction occurred proximal to the interruption and the expansion distal to the interruption with respect to the label.
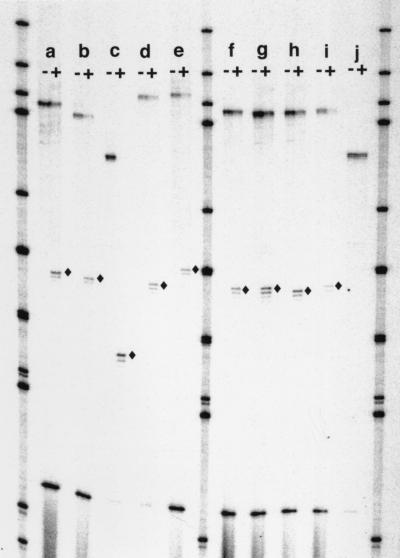
FIG. 2.
Examples of mapping. The PCR products displayed were derived from an end label placed on primer DMLAde2c (the 5′ primer), so that the digestion products correspond to the examples given in Fig. 1. PCR products (top bands) from various events are shown with the undigested (−) and SfaNI-digested (+) products run side by side. Diamonds, labeled digestion products. Because SfaNI digestion is incomplete, some undigested PCR product remains. The band near the bottom derives from the copy of ADE2 at its normal location on chromosome XV and serves as a control. It does not contain an SfaNI site. Also, because of its size, it does not elute well from the Prep-a-gene (see Materials and Methods) and appears in quantities lower than those expected. Size standards of end-labeled HpaII digestion products of KS+ DNA are run in the three lanes that are not marked. All bands shown are from tract CI. Lanes: a, control (parental); b, rth1/rad27 5′ contraction; c, rth1/rad27 3′ contraction; d, rth1/rad27 5′ expansion; e, rth1/rad27 3′ expansion; f, control (parental); g, msh2 5′ contraction; h, msh2 3′ contraction; i, msh2 3′ expansion; and j, msh2 contraction and loss of interruption.
Examples of changes.
Examples of tract length changes were selected from the siblings of multiple parental colonies (Table 2). Because more than a single example per parental colony was sometimes used, some examples were not strictly independent. Nevertheless, we attempted to minimize the possibility of dependence by using multiple parental colonies. More importantly, when multiple examples originating from a single parental colony were used, the examples of tract length changes that were chosen were different in size. The exception to our selection scheme was the events of the special class of small changes that occur only in the msh2 mutant (Table 1). In this case, we did not discard duplicate events of the same size, since most of the events were contractions of a single repeat unit (Table 2).
TABLE 2.
Analysis of tract length changes
| Strain and tract | Parent no. | Total no. of sib colonies | Lacking original size band | No. of increases | No. of decreases | Size changes and polaritya |
|---|---|---|---|---|---|---|
| Wild type | ||||||
| CI90 | 1 | 57 | 2 | 0 | 2 | −40!, −58! |
| CI90 | 2 | 36 | 1 | 0 | 1 | −10# |
| CI90 | 3 | 35 | 0 | 0 | 0 | NA |
| CI90 | 4 | 54 | 0 | 0 | 0 | NA |
| msh2 | ||||||
| CI90 | 1 | 28 | 3 | 0 | 3 | −1#, −2#, −10$ |
| CI90 | 2 | 20 | 3 | 0 | 3 | −1#, −1$, −2$ |
| CI90 | 3 | 32 | 4 | 4 | 0 | +1#, +1#, +1#, +2$ |
| CI90 | 4 | 31 | 2 | 1 | 1 | +1$; −1$ |
| CI90 | 5 | 30 | 6 | 0 | 6 | −1#, −1#, −1#, −1#, −1$, −25! |
| rth1/rad27 | ||||||
| CI90 | 1 | 30 | 10 | 3 | 7 | +15?, +20$, +40$; −3$, −15#(2), −15$, −35!, −47!, −75! |
| CI90 | 2 | 32 | 15 | 8 | 7 | +2$(2), +5$, +15#, +20?(2), +25$, +60?; −5$, −10$, −35!, −38!, −65!, −70!, −80! |
| CI90 | 3 | 20 | 8 | 3 | 5 | +2?, +13$, +25?; −15#, −20$, −25#, −47!, −76! |
| CI90 | 4 | 29 | 16 | 6 | 10 | +4?, +5$, +10$, +20$, +25$, +30?; −3?, −10?, −18?, −32?, −41$, −43?, −57!, −60!, −67!, −68! |
| CI90 | 5 | 32 | 12 | 8 | 4 | +5#, +8#, +12$, +15$, +20#, +25?, +30$, +35$; −5?, −10?, −25?, −71? |
| CI90 | 6 | 30 | 3 | 1 | 2 | +12$; −28?, −47? |
| Wild type | ||||||
| DI97 | 1 | 30 | 2 | 0 | 2 | −45!, −65! |
| DI97 | 2 | 31 | 8 | 0 | 8 | −8#, −20$, −41!, −42!, −45!, −51!, −54!, −65! |
| DI97 | 3 | 32 | 5 | 0 | 5 | −10#, −15#, −20#, −22#, −58! |
| DI97 | 4 | 31 | 14 | 0 | 14 | −12#, −24#, −30#, −35#(2), −40!, −45!, −46!(2), −48!(2), −49!, −50!, −60! |
| DI97 | 5 | 32 | 5 | 0 | 5 | −10#, −15#, −18#, −26!, −62#$ |
| msh2 | ||||||
| DI97 | 1 | 32 | 12 | 1 | 11 | +3$; −11$, −35#, −35$, −38#, −42!, −44!, −48!, −52!, −60!, −69!, −72! |
| DI97 | 2 | 32 | 23 | 1 | 22 | +1# −5#, −7#, −32!, −33#, −34$(2), −35$, −37$, −42#$, −41!, −43!(7), −64!, −65!, −67!, −76!(2) |
| DI97 | 3 | 31 | 15 | 1 | 14 | +1# −1#, −1$, −2#, −14$, −30#(4), −34!, −44!, −52!(2), −66!, −82! |
| DI97 | 4 | 30 | 13 | 0 | 13 | −1#, −1$, −5$, −13#, −34!, −42!, −49!, −50!(2), −61!, −65!, −73!(2) |
| DI97 | 5 | 32 | 14 | 1 | 13 | +1$; −1$, −1#, −20!(6), −30!, −35!, −73!, −86!(2) |
| DI97 | 6 | 28 | 18 | 0 | 18 | −1$, −1$, −1?, −5#(2), −8#, −14?, −40?, −46?(2), −52?, −70?, −72?(6) |
| DI97 | 7 | 32 | 18 | 0 | 18 | −1$, −5?, −12?, −25?(2), −34?, −36?, −40?(2), −44?, −47?, −52?(3), −57?(3), −67? |
| DI97 | 8 | 31 | 14 | 0 | 14 | −1$, −1$, −1#, −1#, −1$, −1?, −2$, −2$, −12?, −15?, −20?, −40?, −51?, −72? |
| DI97 | 9 | 26 | 12 | 0 | 12 | −1#, −1#, −15?, −20?, −30?, −38?, −42?(2), −44?, −53?, −54?, −73? |
| rth1/rad27 | ||||||
| DI97 | 1 | 32 | 24 | 3 | 21 | +8#$, +15$, +25#$; −10$(2), −13#(4), −18?, −22?, −24?, −25?, −37?, −41?, −43?(3), −47?, −50?, −61?, −63?, −65?(2) |
| DI97 | 2 | 28 | 27 | 1 | 26 | +5$; −12?, −19?, −22?, −25?, −30?(2), −32?(2), −38?, −39?(5), −45?, −47?, −49?(2), −56?(2), −63?, −65?, −67?(2), −70?, −79? |
| DI97 | 3 | 28 | 26 | 2 | 24 | +2$, +8#$; −3#, −7#(3), −11#(2), −22#(3), −24?, −34?(2), −38?, −42?, −44?(5), −53?, −56?, −58?(2), −74? |
| DI97 | 4 | 28 | 14 | 2 | 12 | +6$, +10$; −7$, −10?, −30?, −38?, −45?, −48?, −50?, −51?, −53?(2), −61?, −82? |
| DI97 | 5 | 28 | 18 | 0 | 18 | −3$(2), −11#(2), −20?, −22?(2), −42?, −45?, −46?, −48?, −51?, −53?, −58?(3), −75?, −84? |
| DI97 | 6 | 27 | 17 | 4 | 13 | +1#, +1?, +5$, +30$; −5$(2), −10$, −13!, −22?, −25?, −32?(2), −35?, −49?, −56?, −75?, −78? |
| DI97 | 7 | 29 | 16 | 2 | 14 | +5?, +28$; −6?, −13#, −22?, −28?(3), −38?, −50?(3), −58?, −71?(2), −78? |
| DI97 | 8 | 24 | 22 | 2 | 20 | +4#, +28$; −17#(4), −28?, −30?, −32?, −40?, −50?(2), −53?, −65?(4), −70?, −75?, −80?(2), −86? |
| DI97 | 9 | 26 | 18 | 1 | 17 | +5$; −6#, −22!, −26?, −27?(2), −30?, −33?, −36?(2), −40?, −44?, −53(3), −56?, −58?, −75? |
| DI97 | 10 | 32 | 20 | 4 | 16 | +4$, +25#(2), +40$; −4?, −8?(2), −13?(2), −18?, −22?, −25?, −28?(2), −38?, −42?, −44?(2), −76? |
| DI97 | 11 | 30 | 17 | 0 | 17 | −5$, −7?, −20?(2), −28?(2), −30?, −40?, −45?, −48?(2), −50?(2), −52?, −56?, −61?, −66? |
| DI97 | 12 | 31 | 25 | 1 | 24 | +5# −9?, −13?, −16?, −21?(2), −25?, −27?(2), −28?, −29?(3), −30?, −38?(2), −40?(2), −46?, −56?, −59?(2), −60?, −61?, −64? |
| DI97 | 13 | 32 | 21 | 3 | 18 | +3$, +10$, +15#$; −5?, −8?, −12?(4), −20?, −27?, −36?, −41?(3), −42?, −43?, −62?, −63?, −65?, −77? |
| DI97 | 14 | 26 | 21 | 1 | 20 | +5# −7?, −9?, −12?, −28?, −30?(2), −32?(2), −34?, −40?, −43?, −49?, −52?, −54?, −56?, −64?(5) |
| DI97 | 15 | 32 | 26 | 1 | 25 | +4$; −6?, −9?, −26?(2), −34?, −40?, −42?(2), −43?(2), −45?, −48?, −50?, −52?, −53?, −54?, −58?, −60?, −63?, −67?, −70?, −71?, −73?, −77?(2) |
| DI97 | 16 | 29 | 24 | 1 | 23 | +10$; −3?, −6?, −12?(3), −16?(2), −20?(3), −30?(2), −35?, −37?(2), −48?, −62?, −66?, −67?(2), −74?, −76?(2) |
| DI97 | 17 | 31 | 22 | 1 | 21 | +25#$; −3?, −10?, −12?, −15?, −20?, −22?, −30?, −34?, −39?, −42?, −45?, −50?, −52?, −54?, −62?, −63?, −65?(2), −68?, −78?(2) |
Each number with a plus or minus sign indicates the length of the tract expansion or tract contraction, respectively. Following the number are symbols representing the polarity of the changes in relation to the direction of Okazaki fragment synthesis. $, 5′ event; #, 3′ event; !, loss of interruption; ?, not tested. The listing of both $ and # following an event indicates that changes occurred on both sides of the interruption. Numbers in parentheses signify the numbers of multiple examples that were observed. As described in the text, except for the small changes that occurred in the msh2 mutant, only one of the examples was tested. NA, not applicable.
Although mechanisms by which examples of tract length changes of different sizes could have originated dependently on one another might be feasible, the most likely possibility is that they arose independently of each other. An indication of the likelihood that events from the same parental colony were independent was the observation that events from a parental colony often contained examples with different polarities (Table 2). Examples of different polarities were found even among the examples of small changes recovered from most msh2 parental colonies that contained multiple events of this class (Table 2). Although sequential tract length changes occurring during successive generations of cell growth could produce changes of different lengths, that would not be independent, an experimental determination of their incidence rules out a major contribution of sequential events. Seven of 136 tract length changes were found in which changes had occurred on both sides of the interruption (Table 2). From this value, we calculated that the probability of sequential events occurring on the same side of the interruption was approximately 5%. Thus, while we could not adhere to the strictest criteria of independence for technical reasons, we are confident that we achieved a high degree of independence.
Biases in the polarity of changes.
The results of the mapping analyses are given in Table 3, in which the polarities are broken down when possible by the classes of events—expansions, small contractions and large contractions—in wild-type cells and in the isogenic rth1/rad27 and msh2 mutant cells. The results show two significant examples of biases in polarity.
TABLE 3.
Polarity of changes in interrupted CAG tracts
| Genotype and tract | Expansion or contraction | No. of 5′ events | No. of 3′ events |
|---|---|---|---|
| Wild type | |||
| CI90 | Expansions | NAb | NA |
| CI90 | Contractions | 0 | 1 |
| rth1/rad27 | |||
| CI90 | Expansions | 15 | 4 |
| CI90 | Contractions | 6 | 3 |
| msh2 | |||
| CI90 | Expansions | 2 | 3 |
| CI90 | Contractions (≤2a) | 4 | 7 |
| CI90 | Contractions (≥3a) | 1 | 0 |
| Wild type | |||
| DI97 | Expansions | NA | NA |
| DI97 | Contractions | 1 | 12 |
| rth1/rad27 | |||
| DI97 | Expansions | 16 | 5 |
| DI97 | Contractions | 6 | 9 |
| msh2 | |||
| DI97 | Expansions | 2 | 2 |
| DI97 | Contractions (≤2a) | 11 | 8 |
| DI97 | Contractions (≥3a) | 7 | 9 |
Number of repeat units lost.
NA, not applicable.
First, in wild-type cells the contractions in tract DI that retain the interruption are strongly biased to have occurred on the 3′ side of the interruption (in relation to the direction of lagging-strand synthesis) (Table 3). The ratio of 12 3′ events to 1 5′ event is significantly different from random by χ2 analysis to P < 0.025 (and by the Fisher’s exact test to P < 0.05). Furthermore, neither the contractions that occur in the rth1/rad27 mutant nor those that occur in the msh2 mutant exhibit the same bias in polarity, i.e., in each case, the distributions are not significantly skewed from random.
Second, the expansions in tracts CI and DI in an rth1/rad27 mutant occur approximately three times as frequently to the 5′ side of the interruption as to the 3′ side (Table 3). (The ratio for each tract taken separately approaches significance by a χ2 analysis, yielding a P value of between 0.05 and 0.1. The combined ratio of 31 to 9 is significant to P = 0.01.) The bias in polarity of the expansions is striking, because the tract contractions in this mutant are distributed more randomly.
The data were also examined to see whether the sizes of the expansions differed between the 5′ and 3′ events. While the average sizes of the expansions occurring to the 3′ side was slightly less than the mean of those occurring to the 5′ side for both tracts CI and DI (12.0 versus 18.6 for tract CI and 8.0 versus 12.8 for tract DI), the ranges were overlapping (Table 2).
The behavior of tract DI in mutant cells.
We previously showed that trinucleotide tracts undergo more tract length changes in msh2 mutant cells than in wild-type cells (25). The increased frequency of tract length changes was accompanied by the appearance of a unique class of events containing expansions and contractions of one or two repeat units. Although this unique class accounted for almost all of the changes found in the msh2 mutant for the tracts of the stable orientation, the new class of small changes did not account for the increased frequency of changes occurring in tracts of the unstable orientation (25). The datum sets for tracts CI and DI corroborate this disparity (Table 1). For tract DI in the msh2 mutant, 139 of 274 sibling colonies showed a tract length change, while in the wild-type control for tract DI, 34 of 154 sibling colonies showed a tract length change. Thus, of the 139 events in the msh2 mutant, approximately 60 (34/154 × 274) could be attributed to the mechanism operating in wild-type cells and the remaining 79 could be regarded as resulting from the msh2 mutation. Only 22 of these 79 events in the msh2 cells are of the unique class of small contractions, meaning that a substantial portion of the excess events (approximately two-thirds in this analysis) are large contractions of three or more repeat units. Further scrutiny of the data shows that while the large contractions that occur in wild-type cells show a bias in their polarity, contractions of this class that occur in the msh2 mutant are not significantly biased (Table 3). These results suggest that the class of large contractions observed in the msh2 mutant arise both by the mechanism that occurs in wild-type cells and by another mechanism peculiar to msh2-deficient cells.
A similar pattern of behavior is also evident in the rth1/rad27 data. While half of the events recorded in tracts of the C orientation are expansions (Table 1) (26), fewer than half of the events of the D orientation that are attributable to this mutation are expansions. Instead, in the D orientation, additional large contractions are evident (Table 1) (26). As in the case of the msh2 mutant, these additional large contractions do not exhibit a bias in polarity (Table 3).
Loss of interruption and the absence of an effect of the interruption on the sizes of tract length changes.
Many of the contracted tracts lost the interrupted repeat, as evidenced by the failure of SfaNI digestion. (The selected examples in which the loss was authenticated are shown in Table 2. Most large contractions were not analyzed.) The mean size of the contractions retaining the interruption (taken from the data including wild-type and mutant cells for both tracts CI and DI but excluding contractions of two repeat units or fewer in the msh2 mutant) was 15.0 (n = 58; range, 3 to 38), while the value for contractions that were authenticated to have lost the interruption was 51.5 (n = 66; range, 13 to 86). Not surprisingly, considering the nature of the substrate, shorter contractions were more likely to retain the interruption than were longer contractions. Of interest is the result that the ranges of contractions retaining and losing the interruption overlap in the range of 13 to 38 repeat units.
We also examined the data to determine whether the interruption might act as an impediment to the contractions, as might be evidenced by a decrease in their sizes. No decrease in size was apparent when the data in the current study for contractions in wild-type cells were compared to data from our previous studies of uninterrupted D tracts in wild-type cells (14, 25, 26). In previous studies of uninterrupted D tracts of 60 to 78 repeat units in length, we recorded 40 contractions of which 8 were smaller than 20 repeat units. In this study, 9 of the 34 contractions that occurred in tract DI of 97 repeat units in length were of 20 repeat units or less (Table 2). Because these distributions are not significantly different (P = 0.5) by the χ2 test, we conclude that the interruption does not have a major effect on contraction size.
We also note that the mean sizes of expansions found for tracts CI and DI in the rth1/rad27 mutant are 17.2 and 11.7 repeat units, respectively. In our previous study (26) of an uninterrupted C tract of 78 repeat units, the mean size of expansions in the rth1/rad27 mutant was 16.6 repeat units. The mean value for the expansions of an uninterrupted D tract of 71 repeat units was 8.9 repeat units. Thus, as in the case of the contractions, the presence of an interruption does not appear to affect the size of tract expansions.
Finally, a comparison of the frequency of changes occurring in tracts DI and CI with uninterrupted repeat tracts of similar lengths shows that the interruptions do not appreciably stabilize these long tracts (25, 26). An additional study of an uninterrupted and an interrupted tract of approximately 30 repeat units found stabilization by the interruption (13). The results suggest that the ratio of interruptions to uninterrupted repeat units may influence the stabilizing effect of interruptions.
DISCUSSION
We have examined changes that occur in CAG repeat tracts interrupted with a single CAT triplet in wild-type cells and in rth1/rad27 and msh2 mutant cells. We observed polarity biases among some classes of events and random occurrences among other classes. In particular, the tract contractions that occur in wild-type cells carrying tract DI are biased toward the 3′ end (using the direction of synthesis along the lagging strand as the reference), while expansions that occur in rth1/rad27 mutant cells are biased toward the 5′ end. While the contractions that occur in wild-type cells are biased, the contractions that occur in rth1/rad27 mutant cells are not biased. Furthermore, both the small contractions of two or fewer repeat units as well as the longer contractions that occur in msh2 mutant cells are not biased.
The polarity distributions do not appear to be dependent on the orientation of the tracts. Although tracts CI and DI differ in their overall stability, both in wild-type and in mutant cells, the polarities of the changes are similar. In both tracts, the rth1/rad27-effected expansions show the same bias, whereas the contractions in the two tracts do not show a bias. Similarly, the changes that occur in either tract within the msh2 mutant are not biased. Although we were prevented because of their low incidence from examining certain classes of tract alterations, e.g., contractions in tract CI in a wild-type strain, our data suggest that tract orientation has little effect on the polarization of the changes.
Our results for the polarity of trinucleotide repeat changes in wild-type and msh2 mutant cells can be compared to the results for changes in interrupted GT dinucleotide tracts taking into account the fundamental differences in behavior between the two types of repeat tracts (21). Qualitatively, the two types of tracts appear to behave similarly in wild-type and msh2 mutant cells. Small expansions and contractions that occur in a dinucleotide repeat tract in wild-type cells are biased. The bias was designated as being to the 5′ side of the interruption with the GT strand as a reference (21). Changes that occur in trinucleotide tracts in wild-type cells, all of which involve contractions of many repeat units, are also biased. We designate our bias as being to the 3′ side of the interruption using the direction of synthesis of the Okazaki fragments in relation to the ARS element as the reference point. Whether the polarities are really the same and whether they both result from the same underlying cause, i.e., the movement of the replication fork, will require further investigation. The behaviors of di- and trinucleotide tracts are also similar in the msh2 mutant (21). In this mutant, the changes that occur are not biased in either di- or trinucleotide tracts.
Mechanism of tract expansion.
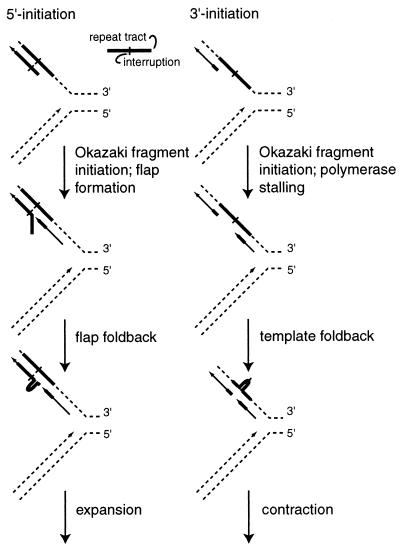
To account for the polarity bias of the expansions occurring in the rth1/rad27 mutant, we propose a model (Fig. 3) that differentiates between forks where the penultimate Okazaki fragments initiate either 5′ to the interruption (left column) or 3′ to the interruption (right column) using the direction of synthesis as the reference for the designation. In the case in which initiation of the penultimate Okazaki fragment occurs 5′ to the interruption, we envision that the newest Okazaki fragment is synthesized up to the penultimate fragment, generating a single-stranded polynucleotide flap. In the absence of the flap endonuclease (rth1/rad27), the flap is not removed and is fixed as an expansion. In the case in which initiation of the penultimate fragment occurs 3′ to the interruption, the polymerase making the newest Okazaki fragment may become stalled or slowed before it reaches the penultimate Okazaki fragment (described below). This provides the time for the template, in single-strand form, to fold back, forming a hairpin structure. Subsequent fixation makes these into tract contractions. In essence, deletions would be favored by stalling or slowing of the polymerase no matter where the fragment was initiated, and this may account for why the contractions in the rth1/rad27 mutant are randomly distributed. In contrast, expansions might occur only when two adjoining Okazaki fragments meet within the repeat tract, and this may be most probable when the most nascent fragment initiates outside the repeat tract and need only polymerize a short distance into the repeat tract before encountering the penultimate fragment. This would account for the three-to-one bias in the polarity of the expansions in the rth1/rad27 mutant.
FIG. 3.
Scheme for tract expansions and contractions in the rth1/rad27 mutant. The representation is of replication forks opening from left to right. The top template strand runs 5′ to 3′ and is the lagging strand. In this scheme, flaps that create foldback loops on the Okazaki fragment (left path) form only when two fragments meet within the repeat tract, which is a more likely event when the newest fragment must reach only partially into the repeat tract. Contractions (right path) occur when the polymerase stalls before reaching the penultimate fragment, giving time for the template to collapse. The example shown eliminates the interruption. Both Tishkoff et al. (31) and Gordenin et al. (6) have also provided molecular models that account for tract expansions and in some cases tract contractions.
The model could also account for why we previously observed that the ratio of expansions to contractions decreases as tract lengths become longer (26). Assuming that the newest Okazaki fragment initiates outside the CAG repeat tract, the chances of the newest and the penultimate Okazaki fragments meeting to produce an expansion will decrease because the possibility of the polymerase making it through the repeat tract decreases with the length of the repeat tract. More-advanced studies will be needed to determine whether CAG repeat tracts influence the initiation of Okazaki fragments.
We note that our results on the polarity of expansions in a yeast mutant are consistent with a study of CAG tract expansions in an E. coli plasmid (9). In the latter study, the expansions were described as occurring more frequently distal to the replication origin, i.e., to what would be the 5′ end of the newly synthesized lagging strand. Also, we note that an analysis of the CGG repeats at the fragile X locus in normal human chromosomes showed that most of the length differences occurred at one end of the repeat tract in reference to interrupting (cryptic) repeat units (11). Our model offers one explanation for how this bias among fragile X alleles might have occurred in human chromosomes, assuming that the opening of the replication fork is toward the expanded end and that the human flap endonuclease is hindered in its ability to process flaps containing CGG repeats (6, 16).
Mechanism of tract contractions.
The bias seen for the contractions that preserve the interruption within tract DI in wild-type cells may result from the foldback of the template strand before the polymerase copies it. Because the fork opens from 5′ to 3′ with respect to the lagging-strand template, the 5′ end of the template will become single stranded before the 3′ end of the template, giving more time for the 5′ end of the template to fold back on itself to create the alignment needed for a contraction (Fig. 3). Contractions caused by folding back of the 5′ end of the template are recorded as 3′ contractions in our scheme, in which the direction of synthesis is used as the reference direction. What should be recognized is that hairpins giving rise to contractions could also occur 5′ to the interruption but may be more likely to lead to the large events that eliminate the interruption.
A class of large contractions also occurs in tracts of the D orientation in both msh2 and rth1/rad27 mutants (Table 1) (25, 26). Although they are indistinguishable in size from the contractions that occur in wild-type cells, the additional large contractions found in the two mutants are different in that they do not have the decided polarity of those that take place in wild-type cells. These additional contractions might be accounted for by a general hindrance in the movement of the replication fork through repeat tracts in mutant cells coupled with the propensity of the CTG strand to fold back more readily than the CAG strand (5, 20, 29). The combination of these two factors could account for why there is no bias in the polarity of these events and why they are more evident in tracts of the D orientation than in those of the C orientation. While no direct evidence for difficulties in fork movement through the repeat tract are yet evident in yeast, the possibility that both mutations could lead to polymerase stalling is plausible. The proteins encoded by RTH1/RAD27 and MSH2 bind to proliferating cell nuclear antigen (PCNA) (7, 12, 33) and the absence of either protein or the failure of the action of either protein may signal the polymerases (bound directly or indirectly) to PCNA to pause.
In the case of the msh2 mutant, another formal possibility exists. Possibly, small loops containing one or two repeat units on the template strand that are not repaired during one round of replication collapse during the next round of replication into larger hairpins that are too large for the mismatch machinery to recognize (1, 17, 28, 32). Because the small changes that occur in this mutant background do not exhibit a bias in polarity, neither should the large changes resulting from their collapse. Furthermore, the differential stability of CTG and CAG hairpins could mean that the D orientation is more prone than the C orientation to this phenomenon.
Contractions that eliminate the interruption.
The largest contractions that eliminate the interruption also comment on the mechanism. Because they do not appear to be impeded by the interruption, their occurrence suggests that loop formation on the template as depicted in Fig. 3 involves interaction between distant repeat units rather than nucleation by adjacent units. Nucleation by adjacent units followed by zippering to yield a hairpin-like loop would be expected to be impeded by the interruption. The loss of an interrupting repeat concomitant with a shortening of the tract has also been observed in alleles of the human SCA1 gene (2). While most normal alleles of the human SCA1 gene contain CAT interruptions within a repeat tract of approximately 30 repeat units, rare chromosomes carry uninterrupted tracts of 20 repeat units. These rare alleles may have occurred by the process we observe for yeast.
These results begin to show that CAG repeat tracts may take more than one molecular path when they undergo expansions and contractions.
ACKNOWLEDGMENTS
This work was supported by grant PO1NS33718 from the National Institutes of Health.
We thank Shanda Reinke for help with sample preparation.
REFERENCES
- 1.Bishop D K, Kolodner R D. Repair of heteroduplex plasmid DNA after transformation into Saccharomyces cerevisiae. Mol Cell Biol. 1986;6:3401–3409. doi: 10.1128/mcb.6.10.3401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chung M Y, Ranum L P, Duvick L A, Servadio A, Zoghbi H Y, Orr H T. Evidence for a mechanism predisposing to intergenerational CAG repeat instability in spinocerebellar ataxia type I. Nature Genet. 1993;5:254–258. doi: 10.1038/ng1193-254. [DOI] [PubMed] [Google Scholar]
- 3.Freudenreich C H, Kantrow S M, Zakian V A. Expansion and length-dependent fragility of CTG repeats in yeast. Science. 1998;279:853–856. doi: 10.1126/science.279.5352.853. [DOI] [PubMed] [Google Scholar]
- 4.Freudenreich C H, Stavenhagen J B, Zakian V A. Stability of a CTG/CAG trinucleotide repeat in yeast is dependent on its orientation in the genome. Mol Cell Biol. 1997;17:2090–2098. doi: 10.1128/mcb.17.4.2090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gacy A M, Goellner G, Juranic N, Macura S, McMurray C T. Trinucleotide repeats that expand in human disease form hairpin structures in vitro. Cell. 1995;81:533–540. doi: 10.1016/0092-8674(95)90074-8. [DOI] [PubMed] [Google Scholar]
- 6.Gordenin D A, Kunkel T A, Resnick M A. Repeat expansion—all in a flap? Nature Genet. 1997;16:116–118. doi: 10.1038/ng0697-116. [DOI] [PubMed] [Google Scholar]
- 7.Johnson R E, Kovvali G K, Prakash L, Prakash S. Requirement of the yeast MSH3 and MSH6 genes for MSH2-dependent genomic stability. J Biol Chem. 1996;271:7285–7288. doi: 10.1074/jbc.271.13.7285. [DOI] [PubMed] [Google Scholar]
- 8.Johnson R E, Kovvali G K, Prakash L, Prakash S. Requirement of the yeast RTH1 5′ to 3′ exonuclease for the stability of simple repetitive DNA. Science. 1995;269:238–240. doi: 10.1126/science.7618086. [DOI] [PubMed] [Google Scholar]
- 9.Kang S, Ohshima K, Jaworski A, Wells R D. CTG triplet repeats from the myotonic dystrophy gene are expanded in Escherichia coli distal to the replication origin as a single large event. J Mol Biol. 1995;258:543–547. doi: 10.1006/jmbi.1996.0266. [DOI] [PubMed] [Google Scholar]
- 10.Kolodner R. Biochemistry and genetics of eukaryotic mismatch repair. Genes Dev. 1996;10:1433–1442. doi: 10.1101/gad.10.12.1433. [DOI] [PubMed] [Google Scholar]
- 11.Kunst C B, Warren S T. Cryptic and polar variation of the fragile X repeat could result in predisposing normal alleles. Cell. 1994;77:853–861. doi: 10.1016/0092-8674(94)90134-1. [DOI] [PubMed] [Google Scholar]
- 12.Li X, Li J, Harrington J, Lieber M R, Burgers P M. Lagging strand DNA synthesis at the eukaryotic replication fork involves binding and stimulation of FEN-1 by PCNA. J Biol Chem. 1995;270:22109–22112. doi: 10.1074/jbc.270.38.22109. [DOI] [PubMed] [Google Scholar]
- 13.Maurer, D. J., and D. M. Livingston. Unpublished observation.
- 14.Maurer D J, O’Callaghan B L, Livingston D M. Orientation dependence of trinucleotide CAG repeat instability in yeast. Mol Cell Biol. 1996;16:6617–6622. doi: 10.1128/mcb.16.12.6617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Miret J J, Pessoa-Brandao L, Lahue R S. Instability of CAG and CTG trinucleotide repeats in Saccharomyces cerevisiae. Mol Cell Biol. 1997;17:3382–3387. doi: 10.1128/mcb.17.6.3382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Murante R, Rust L, Bambara R A. Calf 5′ to 3′ exo/endonuclease must slide from a 5′ end of the substrate to perform structure-specific cleavage. J Biol Chem. 1995;270:30377–30383. doi: 10.1074/jbc.270.51.30377. [DOI] [PubMed] [Google Scholar]
- 17.Muster-Nassal C, Kolodner R D. Mismatch correction catalyzed by cell-free extracts of S. cerevisiae. Proc Natl Acad Sci USA. 1986;83:7618–7622. doi: 10.1073/pnas.83.20.7618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Orr H T, Chung M Y, Banfi S, Kwiatkowski T J, Jr, Servadio A, Beaudet A L, McCall A E, Duvick L A, Ranum L P, Zoghbi H Y. Expansion of an unstable trinucleotide CAG repeat in spinocerebellar ataxia type 1. Nature Genet. 1993;4:221–226. doi: 10.1038/ng0793-221. [DOI] [PubMed] [Google Scholar]
- 19.Paulson H L, Fischbeck K H. Trinucleotide repeats in neurogenetic disorders. Annu Rev Neurosci. 1996;19:79–107. doi: 10.1146/annurev.ne.19.030196.000455. [DOI] [PubMed] [Google Scholar]
- 20.Pearson C E, Sinden R R. Alternative structures in duplex DNA formed within trinucleotide repeats of the myotonic dystrophy and fragile X loci. Biochemistry. 1996;35:5041–5053. doi: 10.1021/bi9601013. [DOI] [PubMed] [Google Scholar]
- 21.Petes T D, Greenwell P W, Dominska M. Stabilization of microsatellite sequences by variant repeats in the yeast Saccharomyces cerevisiae. Genetics. 1997;146:491–498. doi: 10.1093/genetics/146.2.491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Prakash S, Sung P, Prakash L. DNA repair genes and proteins of Saccharomyces cerevisiae. Annu Rev Genet. 1993;27:33–70. doi: 10.1146/annurev.ge.27.120193.000341. [DOI] [PubMed] [Google Scholar]
- 23.Reagan M S, Pittenger C, Siede W, Friedberg E C. Characterization of a mutant strain of Saccharomyces cerevisiae with a deletion of the RAD27 gene, a structural homolog of the RAD2 nucleotide excision repair gene. J Bacteriol. 1995;177:364–371. doi: 10.1128/jb.177.2.364-371.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Reddy P S, Housman D E. The complex pathology of trinucleotide repeats. Curr Opin Cell Biol. 1997;9:364–372. doi: 10.1016/s0955-0674(97)80009-9. [DOI] [PubMed] [Google Scholar]
- 25.Schweitzer J K, Livingston D M. Destabilization of CAG trinucleotide repeat tracts by mismatch repair mutations in yeast. Hum Mol Genet. 1997;6:349–355. doi: 10.1093/hmg/6.3.349. [DOI] [PubMed] [Google Scholar]
- 26.Schweitzer J K, Livingston D M. Expansions of CAG repeat tracts are frequent in a yeast mutant defective in Okazaki fragment maturation. Hum Mol Genet. 1998;7:69–74. doi: 10.1093/hmg/7.1.69. [DOI] [PubMed] [Google Scholar]
- 27.Schweitzer, J. K., and D. M. Livingston. Unpublished observation.
- 28.Sia E A, Kokoska R J, Dominska M, Greenwell P, Petes T D. Microsatellite instability in yeast: dependence on repeat unit size and DNA mismatch repair genes. Mol Cell Biol. 1997;17:2851–2858. doi: 10.1128/mcb.17.5.2851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Smith G K, Jie J, Fox G E, Gao X. DNA CTG triplet repeats involved in dynamic mutations of neurological related gene-sequences form stable duplexes. Nucleic Acids Res. 1995;23:4303–4311. doi: 10.1093/nar/23.21.4303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stotz A, Linder P. The ADE2 gene from Saccharomyces cerevisiae: sequence and new vectors. Gene. 1990;95:91–98. doi: 10.1016/0378-1119(90)90418-q. [DOI] [PubMed] [Google Scholar]
- 31.Tishkoff D X, Filosi N, Gaida G M, Kolodner R D. A novel mutation avoidance mechanism dependent on S. cerevisiae RAD27 is distinct from DNA mismatch repair. Cell. 1997;88:253–263. doi: 10.1016/s0092-8674(00)81846-2. [DOI] [PubMed] [Google Scholar]
- 32.Tran H T, Gordenin D A, Resnick M A. The prevention of repeat-associated deletions in Saccharomyces cerevisiae by mismatch repair depends on the size and origin of deletions. Genetics. 1996;143:1579–1587. doi: 10.1093/genetics/143.4.1579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Umar A, Buermeyer A B, Simon J A, Thomas D C, Clark A B, Liskay R M, Kunkel T A. Requirement for PCNA in DNA mismatch repair at a step preceding DNA resynthesis. Cell. 1996;87:65–73. doi: 10.1016/s0092-8674(00)81323-9. [DOI] [PubMed] [Google Scholar]