Abstract
Purpose
A variable number of tandem repeats (VNTR) in the insulin gene (INS) control region may be involved in type 2 diabetes (T2D). The TH01 microsatellite is near INS and may regulate it. We investigated whether the TH01 microsatellite and INS VNTR, assessed via the surrogate marker single nucleotide polymorphism rs689, are associated with T2D and serum insulin levels in a Mexican population.
Methods
We analyzed a main case–control study (n = 1986) that used univariate and multivariate logistic regression models to calculate the risk conferred by TH01 and rs689 loci for T2D development; rs689 results were replicated in other case–control (n = 1188) and cross-sectional (n = 1914) studies.
Results
TH01 alleles 6, 8, 9, and 9.3 and allele A of rs689 were independently associated with T2D, with differences between sex and age at diagnosis. TH01 alleles with ≥ 8 repeats conferred an increased risk for T2D in males compared with ≤ 7 repeats (odds ratio, ≥ 1.46; 95% confidence interval, 1.1–1.95). In females, larger alleles conferred a 1.5-fold higher risk for T2D when diagnosed ≥ 46 years but conferred protection when diagnosed ≤ 45 years. Similarly, rs689 allele A was associated with T2D in these groups. In males, larger TH01 alleles and the rs689 A allele were associated with a significant decrease in median fasting plasma insulin concentration with age in T2D cases; the reverse occurred in controls.
Conclusion
Larger TH01 alleles and rs689 A allele may potentiate insulin synthesis in males without T2D, a process disabled in those with T2D.
Supplementary Information
The online version contains supplementary material available at 10.1007/s40618-023-02175-4.
Keywords: Diabetes, INS, Insulin, rs689, TH01
Introduction
In the control region of the INS gene, there is a variation in the number of tandem repeats (VNTR) polymorphism at 390 base pairs (bp) from the start of INS transcription (Fig. 1A) that is associated with type 1 diabetes [1–3]. The INS VNTR consists of a sequence of 14–15 nucleotides (5′-ACAGGGGTGTGGGG-3′) repeated in tandem [4] and has been shown to participate in the regulation of INS in healthy individuals [5–7]. INS VNTR alleles are categorized according to the number of repeats in class I (26–63), class II (64–140), and class III (141–209). While class I INS VNTR alleles are considered to confer a risk of developing type 1 diabetes, class III alleles are generally associated with protection [4]. Unlike class III alleles, class I alleles have been associated with increased insulin production [8, 9].
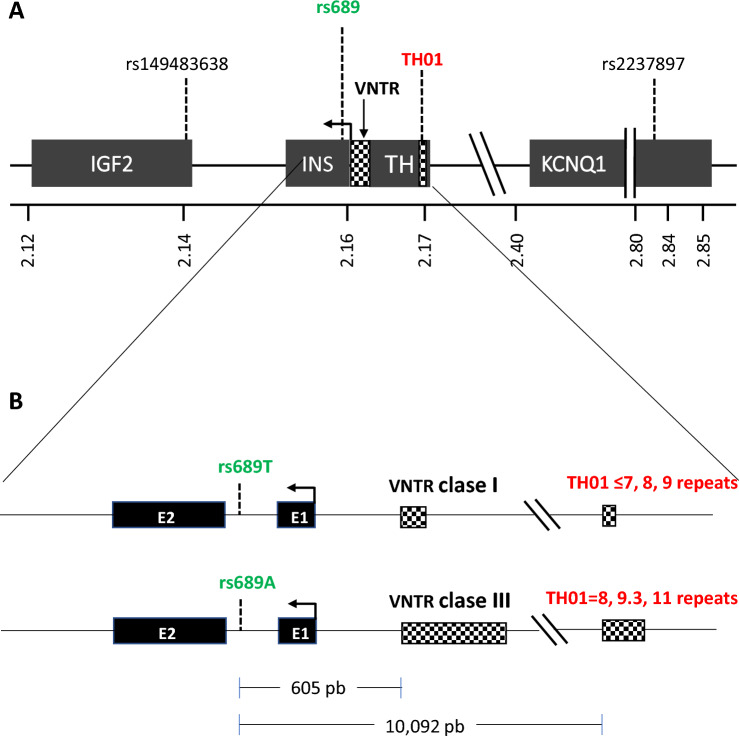
Fig. 1.
Genomic map of region 11p15.5. Panel A shows the locations of genes IGF2, INS, TH, and KCNQ1 and the markers SNP rs689, VNTR INS, and TH01 microsatellite. The positions are based on the human genome version GRCh38. The square arrow indicates the direction and site of initiation of gene transcription. Panel B shows a zoom of the INS-TH region indicating the distance between the rs689 and VNTR and TH01 markers and the alleles of rs689, INS VNTR and TH01 linked in European and Japanese populations (rs689/INS VNTR) and found in this report (rs689/TH01). bp base pairs, SNP single nucleotide polymorphism, VNTR variable number of tandem repeats
Although INS VNTR has been associated with type 2 diabetes (T2D) [10–13], the results remain controversial. Moreover, because most studies were conducted in European populations, there is little evidence in Latin American populations. There could be different associations between INS VNTR and T2D in the latter population, as polymorphisms in SLC16A11, INS-IGF2, and HNF1A genes have previously been reported to have strong associations with T2D in Mexicans but not in European populations [14, 15].
The INS VNTR is complex; therefore, a surrogate marker, the single nucleotide polymorphism (SNP) rs689, has been frequently used [13]. This marker is found within intron 1 of INS and is in complete linkage disequilibrium with the INS VNTR in European populations (allele A is associated with class III alleles; allele T, with class I alleles; Fig. 1B). In addition, the tyrosine hydroxylase (TH)01 microsatellite, a tetranucleotide (AATG) repeated 3–11 times in tandem, is also located relatively close to INS (approximately 9800 bp; Fig. 1A). It is located within the TH gene that is in partial linkage imbalance with INS VNTR in European [1, 16] and Japanese [17] populations. In addition, in vitro experiments have demonstrated that TH01 has enhancer functions, i.e., it can regulate gene expression at a distance, a common attribute for microsatellites [18], and could potentially influence INS expression [19].
Interestingly, although TH01 is associated with obesity [20], hypertension [21, 22], coronary heart disease [23], metabolic syndrome, and high triglyceride levels [24, 25], its association with T2D has not been explored. Therefore, this study was conducted to investigate whether the TH01 microsatellite and INS VNTR (through SNP rs689) are associated with T2D and fasting plasma insulin concentration in the Mexican population. Since in a previous study in Mexican population [26] we found important differences in the association of genes with T2D between males and females and between individuals diagnosed early (≤ 45 years) and late (≥ 46 years), in this paper we also explore whether TH01 and rs689 have a different influence on the development of T2D between sexes and age of presentation. The main study was a case–control study that used logistic regression models to calculate the risk (odds ratio; OR) conferred of each locus for T2D in a Mexican population; a clinical replica case–control study and a cross-sectional study were also conducted.
Materials and methods
Sample selection and study design
Both TH01 and rs689 were analyzed in the main case–control study, which included individuals of the Diabetes in Mexico Study (DMS), the details of which have been previously described [14, 15]. Briefly, participants were recruited between November 2009 and August 2013 from two tertiary-level hospitals in Mexico City; T2D was diagnosed based on the American Diabetes Association (ADA) [27] criteria. A total of 988 cases (unrelated individuals aged > 20 years, with a previous diagnosis of T2D or fasting blood glucose > 125 mg/dL) and 998 controls (healthy individuals aged > 50 years with fasting blood glucose < 100 mg/dL); all participants were self-recognized as Mexican mestizo.
For the replication of rs689, a case–control study included 593 cases of T2D and 595 controls (recruited between January 2014 and January 2015 from a tertiary care hospital in Mexico City). Cases consisted of individuals previously diagnosed with T2D according to the ADA criteria, who agreed to participate and were continuously recruited during routine medical visits. Controls were individuals aged ≥ 50 years, who attended the same clinics for reasons other than T2D, had fasting blood glucose < 100 mg/dL, and agreed to participate in the study.
Another replica of rs689, a population-based cross-sectional study, was conducted on 1172 from 1914 patients recruited between July and December 2017 from a hospital in Puebla, Mexico. Healthy individuals were invited to participate through flyers distributed in the hospital’s neighborhood. The procedures used for TH01 and rs689 SNP genotyping are in Online Resource 1.
Ethics
The protocol for the main study was approved by the local ethics committee of each study site and the Federal Commission for the Protection Against Health Risks (COFEPRIS) (CAS/OR/CMN/113300410D0027-0577/2012). The clinical replica study was approved by the Ethics and Research Committees of the Comisión Nacional de Investigación Científica of the Instituto Mexicano del Seguro Social (IMSS R-2014-785-005), while the cross-sectional study was approved by the Ethics and Research Committees of the Hospital General de Puebla “Ignacio Romero Vargas” (68/ENS/INV/REV/2017). All protocols complied with the Declaration of Helsinki and local ethical guidelines for clinical studies in Mexico. Written informed consent was obtained from all participants.
Statistical analyses
For the sample size calculation, multivariate logistic regression (MLR) models were considered to have good performance when a baseline of 100 cases (i.e., the univariate logistic regression models) and 15 additional cases for each variable were introduced into the model [28]. Given that only two variables were to be introduced in the MLR models, we calculated that a minimum number of 115 cases were needed in the comparison groups of the main case–control study in which both loci were studied.
Logistic regression models were used to calculate the risk or protection conferred by the TH01 and rs689 loci for T2D development in the whole sample and stratified by sex and the median (45 years) age of T2D presentation (≤ 45 and ≥ 46 years). Cases diagnosed with T2D at ≤ 45 years were compared with controls aged ≤ 54 years, and cases diagnosed with T2D at ≥ 46 years were compared with controls aged ≥ 55 years. The statistical significance of differences in the distribution of genotypes between the observed and expected results was calculated using the chi-square test according to the Hardy–Weinberg law, and the linkage disequilibrium between TH01 and rs689 SNP alleles, using Arlequin (version 3.5.2.2; http://cmpg.unibe.ch/software/arlequin35/) [29].
A post-hoc power analysis was performed for each model using the G*Power software version 3.1.9.7 (https://www.psychologie.hhu.de/arbeitsgruppen/allgemeine-psychologie-und-arbeitspsychologie/gpower), considering the sample size, odds ratio (OR), probability of the event in the control group, and an α = 0.05 [30]. All statistical tests were two-sided, and the significance level was set at P < 0.05. The statistical analyses were conducted using SPSS software version 25 (IBM Corp., Armonk, NY, USA). Additional information on the statistical methods is provided in Online Resource 1.
Results
Participants and demographic characteristics
The 53.3%, 60.6%, and 73.5% of participants were female in the main case–control (n = 1986), replica case–control (n = 1188), and cross-sectional (n = 1172) studies, respectively. In both case–control studies, nearly 50% of participants were T2D cases, whereas in the cross-sectional study, the prevalence of T2D was 5.4%. At enrollment, the mean (SD) age of controls vs. cases and the mean age at the time of T2D diagnosis were similar across the studies (Table 1).
Table 1.
Participant demographic characteristics (n = 4346)
| Variable | Female | Male | Both sexes | |||
|---|---|---|---|---|---|---|
| Control | Cases | Control | Cases | Control | Cases | |
| Main case–control study (n = 1986): mean ± SD (n) | ||||||
| Age (years) | 59.7 ± 11.2 (535) | 55.4 ± 12.1 (524)¶ | 58.6 ± 11.4 (463) | 55.8 ± 11.4 (464)§ | 59.2 ± 11.3 (998) | 55.6 ± 11.7 (988)¶ |
| BMI (kg/m2) | 28 ± 4.9 (531) | 29.6 ± 5.5 (524)¶ | 26.9 ± 4.1 (457) | 28.7 ± 5 (459)¶ | 27.5 ± 4.6 (988) | 29.2 ± 5.3 (983)¶ |
| Waist (cm) | 93.3 ± 11.3 (348) | 97.7 ± 11.6 (409)¶ | 93.4 ± 10.5 (310) | 98.7 ± 12.7 (279)¶ | 93.4 ± 10.9 (658) | 98.1 ± 12 (688)¶ |
| Hip (cm) | 103.6 ± 11 (327) | 106.5 ± 11.5 (408)§ | 98.9 ± 7.9 (298) | 101.7 ± 10.8 (275)§ | 101.4 ± 9.9 (625) | 104.6 ± 11.5 (683)¶ |
| WHR | 0.9 ± 0.07 (327) | 0.92 ± 0.06 (407)§ | 0.94 ± 0.06 (298) | 0.97 ± 0.06 (275)¶ | 0.92 ± 0.07 (625) | 0.94 ± 0.07 (682)¶ |
| Age at T2D diagnosis (year) | 45.9 ± 10.6 (524) | 46.2 ± 10.9 (464) | 46 ± 10.8 (988) | |||
| Years with the disease | 9.4 ± 8.4 (524) | 9.6 ± 9 (464) | 9.5 ± 8.7 (988) | |||
| Replica case–control study (n = 1188): mean ± SD (n) | ||||||
| Age (years) | 53.4 ± 9.6 (423) | 57 ± 8.8 (297)¶ | 55.8 ± 11 (172) | 57.3 ± 10.2 (296) | 54.1 ± 10.1 (595) | 57.1 ± 9.5 (593)¶ |
| BMI (kg/m2) | 28.9 + 5.1 (423) | 30.6 + 5.1 (297)¶ | 28.9 + 4.3 (172) | 28.3 + 4.3 (296) | 28.9 + 4.9 (595) | 29.5 + 4.9 (593)† |
| Waist (cm) | 92 + 11.2 (423) | 96.4 + 10.9 (297)¶ | 98.3 + 10.3 (172) | 98.5 + 11 (296) | 93.8 + 11.3 (595) | 97.4 + 11 (593)¶ |
| Hip (cm) | 105.2 + 11.9 (423) | 107 + 11.5 (297)† | 102.7 + 8 (172) | 101.1 + 8.2 (296)* | 104.5 + 11 (595) | 104.1 + 10.4 (593) |
| WHR | 0.89 + 0.4 (423) | 0.9 + 0.1 (297) | 0.96 + 0.1 (172) | 0.97 + 0.1 (296)‡ | 0.91 + 0.3 (595) | 0.94 + 0.1 (593)* |
| Age at T2D diagnosis (year) | 45.3 ± 7 (297) | 44.3 ± 7.5 (296) | 44.8 ± 7.3 (593) | |||
| Years with the disease | 11.7 ± 7.8 (297) | 12.9 ± 7.5 (296) | 12.3 ± 7.7 (593) | |||
| Cross-sectional study (n = 1172a): mean ± SD (n) | ||||||
| Age (years) | 49.3 ± 10.7 (789) | 49 ± 11.7 (72) | 48.8 ± 11.1 (279) | 49.5 ± 10.9 (32) | 49.1 ± 10.8 (1068d) | 49.2 ± 11.4 (104) |
| BMI (kg/m2) | 28.5 ± 4.7 (789) | 31.1 ± 4.6 (72)§ | 28.6 ± 4 (279) | 30.7 ± 7.3 (32)§ | 28.5 ± 4.5 (1068) | 31 ± 5.5 (104)¶ |
| Waist (cm) | 91.2 ± 15.4 (789) | 98.1 ± 15.4 (72)§ | 95.3 ± 14.8 (279) | 101.8 ± 12.2 (32)§ | 92.3 ± 15.3 (1068) | 99.2 ± 14.6 (104)¶ |
| Age at T2D diagnosis (year)b | 49.8 ± 11.7 (63) | 48.8 ± 10.6 (29) | 49.5 ± 11.3 (92) | |||
| Years with the diseasec | ≈0 | ≈0.7 | ≈0 | |||
SD standard deviation
*p < 0.1; †p < 0.05, ‡p < 0.01, §p < 0.001, ¶p < 0.000
a1914 individuals were recruited but only 1172 were explored for rs689 locus
b92 newly diagnosed cases were included and age at recruitment was used. The age at diabetes diagnosis of the 12 individuals with type 2 diabetes diagnosed before recruitment was not collected
cDifference between mean age of all 104 cases less than the mean age of 92 newly diagnosed cases
dThis control group includes individuals with pre-diabetes (n = 529; A1c ≥ 5.7 to < 6.5), and non-diabetic individuals aged ≥ 40 years (n = 539; A1c < 5.7)
Frequency and association of alleles and genotypes of TH01 with T2D using regression models
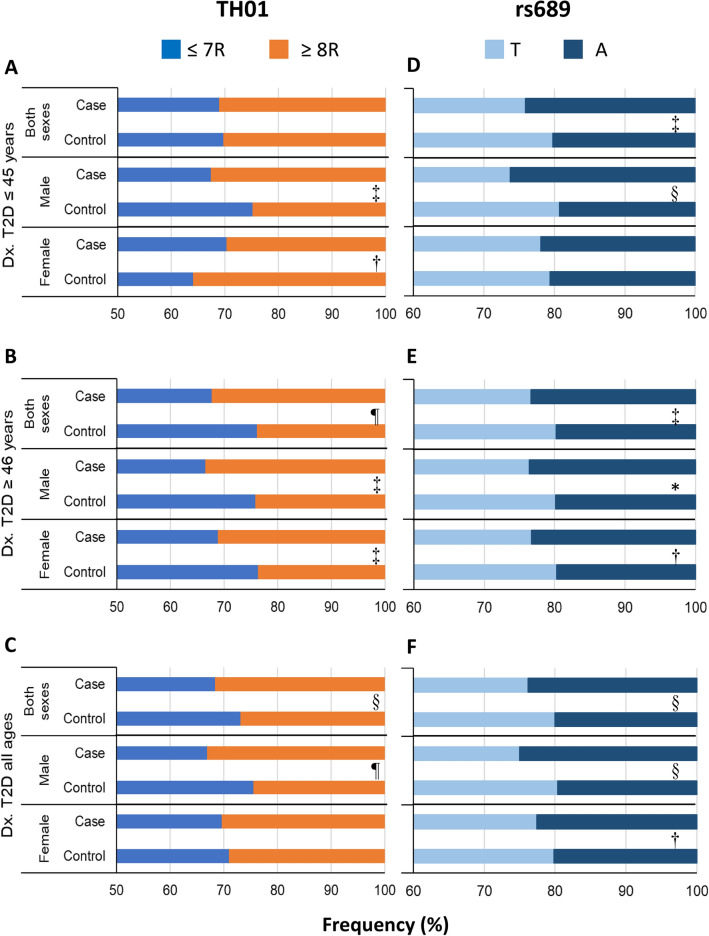
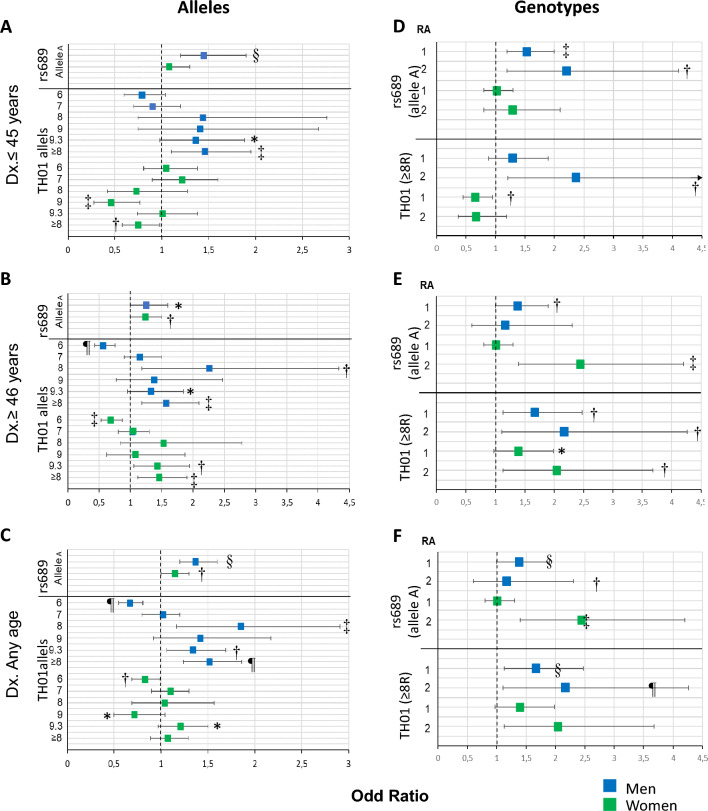
The frequency of alleles and genotypes of TH01 was only assessed in the main study and summarized in Fig. 2A–C and Online Resource 2. Though we identified 9 out of 11 alleles reported for the TH01 microsatellite [31], only five (6, 7, 8, 9, and 9.3) had a frequency ≥ 5% in cases or controls. Allele 6 had a significant protective effect towards T2D diagnosed at ≥ 46 years, with a 32.0% reduced risk in females and 44.0% in males (Fig. 3B); in addition, allele 9 had a protective effect for T2D in females diagnosed at ≤ 45 years (54.0%) (Fig. 3A). The opposite effect was observed for allele 8, which conferred a 2.26-times greater risk for T2D diagnosed at ≥ 46 years in males, and allele 9.3, which conferred a 1.43-times increased risk in females diagnosed at ≥ 46 years (Fig. 3B) and a 1.34-times increased risk in males diagnosed at either age cutoff (Figs. 3C). The differences between alleles were seen more clearly when pooled by size; large alleles (L) with ≥ 8 repeats (R; 8, 9, 9.3, and 11) conferred a similarly increased risk for T2D diagnosed at ≥ 46 years in males (1.57-times) and females (1.46-times) than small alleles (S) with ≤ 7 R (3, 4, 5, 6, 7; as the reference; Fig. 3B). In contrast, L alleles conferred an opposite risk for T2D diagnosed at ≤ 45 years in males and females: while they conferred a 1.46-times increased risk in males, they decreased 25% the risk in females (Fig. 3A).
Fig. 2.
Comparison of allelic frequencies of TH01 and rs689 markers between cases and controls by sex and age of T2D presentation. For TH01, only the main case–control study was explored (n = 1986; 3972 chromosomes), whereas, for rs689, the main and replica case–control and cross-sectional studies were explored as a pool (n = 4341; 8682 chromosomes). Cases diagnosed with T2D at ≤ 45 years were compared with controls aged ≤ 54 years, and cases diagnosed with T2D at ≥ 46 years were compared with controls aged ≥ 55 years. ≤ 7R = alleles with 3, 4, 5, 6, and 7 repeats, and ≥ 8R = alleles with 8, 9, 9.3, and 11 repeats. Dx diagnosis, OR odds ratio, SNP single nucleotide polymorphism, T2D type 2 diabetes. The chi-square test was used to assess the statistical significance; statistically significant values are labelled as *p < 0.1; †p < 0.05, ‡p < 0.01, §p < 0.001, ¶p < 0.0001
Fig. 3.
Association study with alleles and genotypes of TH01 and rs689 markers stratified by sex and age of T2D presentation. See the legend of Fig. 2. Univariate logistic regressions were performed in the whole sample and stratified by sex and age of T2D presentation. The Wald test was used to assess the statistical significance using the Enter method; statistically significant values are labelled with as follows: *p < 0.1; †p < 0.05, ‡p < 0.01, §p < 0.001, ¶p < 0.0001. CI confidence interval, OR odds ratio. The power (1 − β error probability) > 0.99 for both markers, with p < 0.05 in all groups
The associations of T2D with genotypes were greater than those observed with alleles (Online Resource 3); in males diagnosed at any age (Figs. 3D, E, F) and in women diagnosed at ≥ 46 years (Fig. 3E) the homozygous L/L genotypes conferred a much higher risk than heterozygous L/S genotypes. For example, the risk in males increased from an OR of 1.48 (95% CI 1.13–1.95; p = 0.005) when heterozygous to 2.26 (95% CI 1.41–3.64; p = 0.0007) when homozygous (Fig. 3F), indicating an additive effect with the number of alleles with ≥ 8R.
Frequency of alleles and genotypes of SNP rs689 and its association with T2D
The frequency of alleles and genotypes of rs689 was assessed in the main and replica studies (n = 4346). Allele A was significantly more frequent in cases than in controls (23.8% vs. 20.0%; p < 0.0001); although this difference was observed in females (22.6% vs. 20.2%; p < 0.05), it was markedly greater in males (25% vs. 19.6%; p < 0.001) (Fig. 2F). Notoriously, when the analysis was performed stratifying the sample by age of T2D presentation, the difference in men was statistically significant only in those diagnosed at ≤ 45 years (Fig. 2D), while the difference in women was only significant in those diagnosed at ≥ 46 years (Fig. 2E). Similar differences were found in the frequency of genotypes AT and AA between all cases and controls (Online Resource 4).
Logistic regression analysis demonstrated that allele A confers a significantly higher risk for T2D (OR 1.25; 95% CI 1.1–1.4; p < 0.0001), with a comparatively higher risk in males (OR 1.37; 95% CI 1.2–1.6; p < 0.001) than in females (OR 1.15; 95% CI 1–1.3; p = 0.043) (Fig. 3C). Likewise, as in the frequency comparison, the allele A confers a high risk for T2D only in men diagnosed early (OR 1.45; 95% CI 1.2–1.9, p < 0.001) and in women diagnosed late (OR 1.24; 95% CI 1–1.5; p = 0.032). (Figs. 3A, B).
For T2D diagnosed early, both genotypes (AT/AA) were found to confer a risk only in males, with an additive effect; the risk was much greater for AA (OR 2.2; 95% CI 1.2–4.1; p = 0.013) than AT (OR 1.53; 95% CI 1.2–2; p = 0.002) (Fig. 3D). For T2D diagnosed at ≥ 46 years (Fig. 3E), a higher risk was conferred only by AA in females (OR 2.44; 95% CI 1.4–4.2; p = 0.0015) and AT in males (OR 1.38; 95% CI 1–1.9; p = 0.036). Despite the similar differences across the studies in the allelic and genotypic frequencies, the risk conferred by allele A, or genotypes AT and AA, towards T2D diagnosis was observed mainly in males diagnosed at an earlier age in the two replica studies (Online Resources 5–10).
Linkage disequilibrium analysis
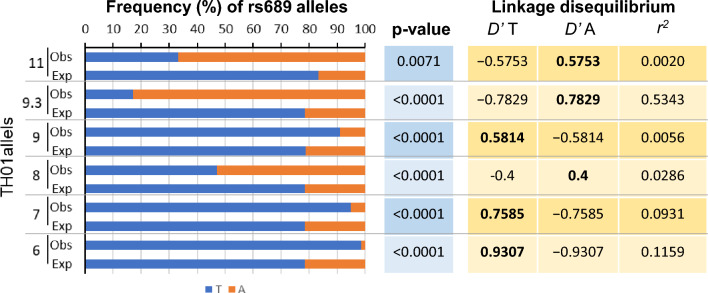
Our analyses showed that TH01 alleles with 3–7 repeats were in linkage disequilibrium with allele T of SNP rs689 (Fig. 4). Alleles 6 and 7 of TH01 were inherited with allele T of SNP rs689 98.5% (D’ = 0.9307) and 94.8% (D’ = 0.7585) of the time. The differences in the frequency of haplotypes 6/T vs. 6/A and 7/T vs. 7/A completely departed from the expected random distribution if they were in linkage equilibrium (p < 0.00001). In contrast, TH01 alleles with 8–11 repeats, except allele 9, were in partial linkage disequilibrium with allele A of SNP rs689. Allele 9.3 of TH01 was highly linked and inherited with allele A for 83.0% of the time (D’ = 0.7829). In contrast, alleles 8 and 11 were partially linked to allele A and segregated together for 52.9% and 66.7% of the time, respectively, and their respective D’ values were lower (0.3882 and 0.5753). Similarly, the frequencies of haplotypes 9.3/A, 8/A, and 11/A were much more frequent than haplotypes with a T than if they were in equilibrium and randomly distributed (p < 0.01). Allele 9 was in linkage disequilibrium with allele T, and they were inherited together 90.1% of the time (D’ = 0.5814).
Fig. 4.
Linkage disequilibrium between TH01 and SNP rs689 alleles. The analysis was performed only in the main case–control study (n = 1986; 3972 chromosomes). The graph shows the frequency (observed) with which each A and T allele of the rs689 SNP is inherited in conjunction with each of the TH01 alleles in phased data. The expected frequency was calculated assuming that alleles A and T are not linked to alleles of TH01. The significance of the difference between the two frequencies was calculated with the chi-square test (p value). D’ linkage disequilibrium statistic, r2 correlation between allelic values at two loci
Multivariate logistic regression of TH01 and SNP rs689
TH01 and SNP rs689 alleles were both associated with T2D in the same groups as reported in univariate logistic regression models; the association remained significant for both loci in MLR models, which is explained by the partial linkage disequilibrium between some alleles of these two markers, suggesting that both loci contribute independently to T2D in the MLR model (Online Resource 11). In fact, the value of R2 increased from the first (TH01) to the second (rs689) block introduced in this model, supporting the additive contribution of the second marker, which could be indicative of a combined effect by which both loci contribute to T2D.
In males, both ≥ 8R haplotypes were associated with T2D, with haplotype ≥ 8R/T conferring a higher risk than haplotype ≥ 8R/A (Online Resource 12). This finding indicates a predominant effect of the microsatellite TH01 over that of rs689. When explored individually, the T allele of the SNP rs689 confers protection, although, in females, the effect differed between age groups. In females, ≥ 8R/T was protective for T2D development at ≤ 45 years, while ≥ 8R/A had no effect; the opposite effect was observed among the groups diagnosed with T2D at ≥ 46 years of age.
Association of insulin concentration with age and the alleles of TH01 and rs689
The median fasting plasma insulin concentration (mIU/mL) was significantly higher among cases than in controls (9.5 [5.5–16.2] vs 6.8 [4.8–10]; p < 0.0001) (Online Resource 13). Interestingly, insulin concentrations were higher in cases diagnosed at ≤ 45 years than at ≥ 46 years; this difference was greater in males than in females. In fact, insulin concentrations decreased with age at T2D diagnosis (r = − 0.111; p < 0.0001), which was much greater in males (r = − 0.189; p < 0.0001) than in females (r = − 0.051; p > 0.05) and independent of allele type, suggesting the effect was likely related to age.
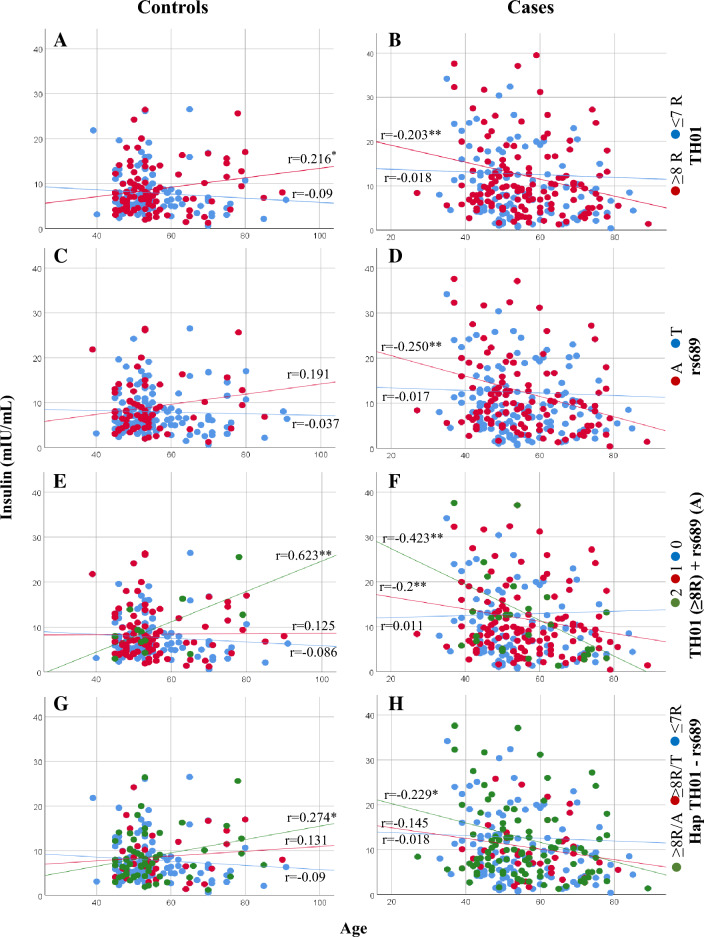
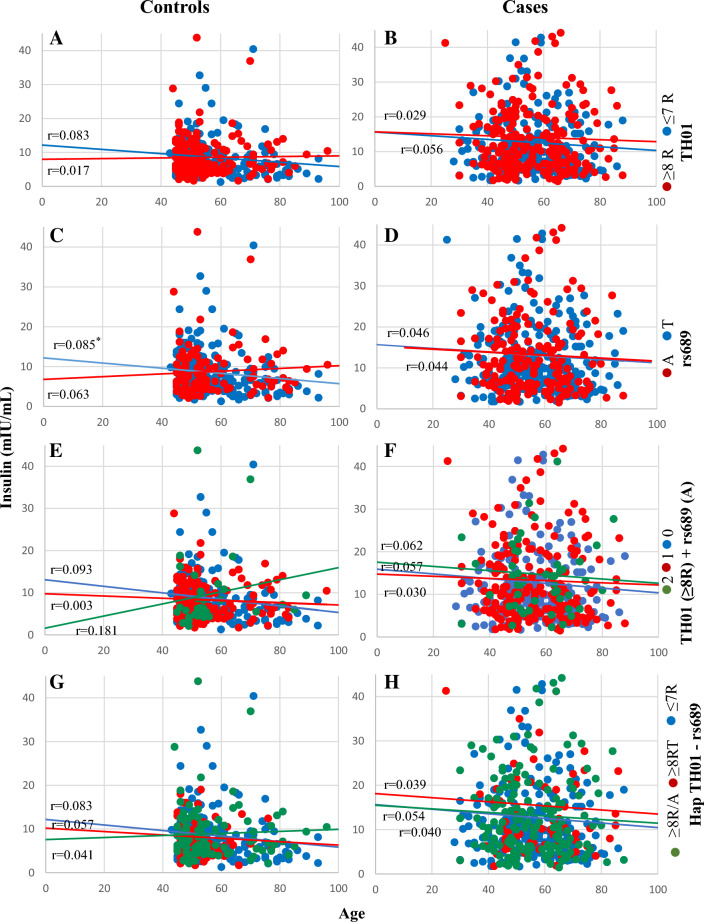
Insulin concentration significantly decreased with participant age for the entire main study population (r = − 0.046; p = 0.02). In males with T2D, insulin decreased significantly with age in those with alleles that had ≥ 8R of TH01 (r = − 0.203; p = 0.009; Fig. 5B) or the rs689 allele A (r = − 0.250; p = 0.004; Fig. 5D). When both alleles were present, there was a significant increase in the negative correlation (r = − 0.423; p = 0.007; Fig. 5F), which occurred only with ≥ 8R/A and not with ≥ 8R/T. In contrast, in males in the control group, an inverse correlation was observed for those with these alleles, whereby the fasting plasma insulin concentration increased with age (Fig. 5A–G). However, no significant correlations between insulin concentration and age were observed in females with T2D or in the control group for the alleles of the two markers (Fig. 6A–H).
Fig. 5.
Correlation between fasting plasma insulin concentration and age of controls and cases in males according to TH01 and rs689 alleles. A, B TH01 alleles C, D SNP rs689 alleles E, F TH01 ≥ 8R alleles + /or rs689 A allele and G, H Hap TH01—rs689. In panels E, F, the numbers 2 (green circle), 1 (red circle), and 0 (blue circle) represent TH01 (≥ 8R) + rs689 A allele, TH01 (≥ 8R) or rs689 A allele, and neither, respectively. *p < 0.05, **p < 0.01. R repeats, SNP single nucleotide polymorphism
Fig. 6.
Correlation between fasting plasma insulin concentration and age of controls and cases in females according to TH01 and rs689 alleles. A, B TH01 alleles C, D SNP rs689 alleles E, F TH01 ≥ 8R alleles + /or rs689 A allele and G, H Hap TH01—rs689. In panels E, F, the numbers 2 (green circle), 1 (red circle), and 0 (blue circle) represent TH01 (≥ 8R) + rs689 A allele, TH01 (≥ 8R) or rs689 A allele, and neither, respectively. *p < 0.05, **p < 0.01. R repeats, SNP single nucleotide polymorphism
Discussion
Our results indicate an association between TH01 microsatellite and the SNP rs689 with T2D and fasting plasma insulin concentrations. The degree of association varied with age at T2D diagnosis and sex. TH01 alleles with ≥ 8R and the rs689 A allele conferred an increased risk of developing T2D at any age in males and at ≥ 46 years in females, and a protective (≥ 8R) or neutral (A) effect towards developing T2D at ≤ 45 years among females. TH01 alleles with ≤ 7R have either an inverse (allele 6) or a neutral (allele 7) effect. Fasting plasma insulin decreased linearly with age in male cases who had alleles with ≥ 8R or the rs689 A allele but increased slightly in controls with those alleles. In contrast, fasting plasma insulin remained constant in male cases and controls who had alleles with ≤ 7R or rs689 T allele. Contrarily, the decrease of insulin concentration with age in females is not influenced by TH01 or rs689. Linkage disequilibrium and multivariate analysis suggest that TH01 is partially in linkage imbalance with rs689 and that the risk or protection conferred by both loci for T2D appear to be independent of each other, even though there may be an additive effect.
Although inconsistent, most studies indicate that class I alleles stimulate the INS gene 1.5- to 3 times more than class III alleles [1, 5, 8, 9, 32–35]. While Le Stunff et al. noted an association between T/T genotypes (VNTR I/I) with a higher fasting plasma insulin level [32], other studies have found greater stimulation of allele III for insulin gene expression in in vitro experiments [6] or equal to allele I for plasma insulin secretion [7]. Allele A of rs689, linked with allele III, influences alternative splicing of intron 1 of INS through differential recognition of its 3′ splice site, resulting in an increased production of mature transcripts and more proinsulin in culture supernatants than transcripts from allele T [7].
The influence of VNTR on gene activity or plasma insulin secretion in T2D remains unknown. However, studies have demonstrated the involvement of VNTR in INS expression, a mechanism that could be altered or affected in T2D in some populations [2, 7]. Our results with SNP rs689 support this hypothesis, as insulin concentration significantly decreased with age in male participants with T2D who were positive for allele A (class III), while it remained constant in those positive for allele T (class I).
The association of ≥ 8R with a slight increase in fasting plasma insulin concentrations with age in controls suggests a probable involvement of TH01 in regulating INS expression. However, because this association was also observed with allele A of rs689, the association between TH01 microsatellite alleles and fasting plasma insulin levels could be due to a linkage effect with allele A. In cases with these alleles or with the ≥ 8R/A haplotype, fasting plasma insulin levels decreased with age, which could suggest that the regulation of INS expression could be progressively affected by age in the presence of class III (of VNTR INS) and/or ≥ 8R (of TH01) alleles. Because allele 6, which is completely linked to allele T of rs689, was found to be a protector for T2D and its frequency increased with age, it could have an inverse effect compared with the ≥ 8R/A haplotype on INS gene expression during aging.
Alleles with ≥ 8R are protective for T2D development at ≤ 45 years and confer a greater risk at a later age in females, indicating the crucial role of estrogen in this association. Epidemiological studies have demonstrated that estrogens are protective against T2D. First, hormone therapy has been shown to reduce the incidence of diabetes by 35% in postmenopausal females with coronary heart disease [36], and secondly, women with early menopause have a higher risk of T2D [37]. There is evidence that estrogen represses the expression of insulin mRNA in pancreatic β-cells through indirect genomic signaling [38] and can stimulate the degradation of misfolded proinsulin, thereby protecting the production of insulin and delaying the onset of diabetes [39]. Our analysis is limited by including only Mexican participants and, therefore, not necessarily generalizable to other populations.
In conclusion, insulin increases with age in males when TH01 alleles with ≥ 8R or allele A of rs689 are present, suggesting an involvement in a mechanism that maintains insulin synthesis in individuals without T2D. In contrast, this is somehow disabled in patients with T2D, thus warranting further research.
Supplementary Information
Below is the link to the electronic supplementary material.
Acknowledgements
The authors wish to thank Aafreen Saiyed of Edanz (www.edanz.com) for providing editorial support for this manuscript, in accordance with Good Publication Practice (GPP 2022) guidelines (http://www.ismpp.org/gpp-2022).
Author contributions
Conceived and/or designed the work: JB, HG-R, JA-D, PK-M, and RT-C. Acquired data: LO, EJ-T, EB, MC-L, REB, ER-M, MM-M, AA-S, AV-S, JJP-R, HG-O, LAM-J, AM, and DAA-H. Interpreting the results: JB, HG-R, JA-D, PK-M, and RT-C. Drafted the manuscript: JB. All authors have critically revised the manuscript, approved the final version of the manuscript, and agree to be held accountable for all aspects of the work.
Funding
This work was supported by the Carlos Slim Foundation, the Laboratorio Huella Génica, and the Faculty of Medicine of the National Autonomous University of Mexico (UNAM).
Data availability
The datasets generated during and/or analyzed during the current study are available from the corresponding author on reasonable request.
Declarations
Conflict of interest
All authors have nothing to disclose.
Research involving human participants and/or animals
The protocol for the main study was approved by the local ethics committee of each study site and the Federal Commission for the Protection Against Health Risks (COFEPRIS) (CAS/OR/CMN/113300410D0027-0577/2012). The clinical replica study was approved by the Ethics and Research Committees of the Comisión Nacional de Investigación Científica of the Instituto Mexicano del Seguro Social (IMSS R-2014-785-005), while the cross-sectional study was approved by the Ethics and Research Committees of the Hospital General de Puebla “Ignacio Romero Vargas” (68/ENS/INV/REV/2017). All protocols complied with the Declaration of Helsinki and local ethical guidelines for clinical studies in Mexico.
Informed consent
Written informed consent was obtained from all participants.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
J. Berumen, Email: jaimeberumen47@gmail.com
H. Gallardo-Rincón, Email: hgallardo@fundacioncarlosslim.org
References
- 1.Bennett ST, Lucassen AM, Gough SCL, et al. Susceptibility to human type 1 diabetes at IDDM2 is determined by tandem repeat variation at the insulin gene minisatellite locus. Nat Genet. 1995;9:284–292. doi: 10.1038/ng0395-284. [DOI] [PubMed] [Google Scholar]
- 2.Pugliese A, Zeller M, Fernandez A, et al. The insulin gene is transcribed in the human thymus and transcription levels correlated with allelic variation at the INS VNTR-IDDM2 susceptibility locus for type 1 diabetes. Nat Genet. 1997;15:293–297. doi: 10.1038/ng0397-293. [DOI] [PubMed] [Google Scholar]
- 3.Stead JD, Buard J, Todd JA, Jeffreys AJ. Influence of allele lineage on the role of the insulin minisatellite in susceptibility to type 1 diabetes. Hum Mol Genet. 2000;9:2929–2935. doi: 10.1093/hmg/9.20.2929. [DOI] [PubMed] [Google Scholar]
- 4.Coleta RD, Alexander AL, Alva CBD, et al. Insulin-like growth factor 1 gene (CA)n repeats and a variable number of tandem repeats of the insulin gene in Brazilian children born small for gestational age. Clinics (Sao Paulo) 2013;68:785–791. doi: 10.6061/clinics/2013(06)10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cocozza S, Riccardi G, Monticelli A, et al. Polymorphism at the 5' end flanking region of the insulin gene is associated with reduced insulin secretion in healthy individuals. Eur J Clin Invest. 1988;18:582–586. doi: 10.1111/j.1365-2362.1988.tb01271.x. [DOI] [PubMed] [Google Scholar]
- 6.Kennedy GC, German MS, Rutter WJ. The minisatellite in the diabetes susceptibility locus IDDM2 regulates insulin transcription. Nat Genet. 1995;9:293–298. doi: 10.1038/ng0395-293. [DOI] [PubMed] [Google Scholar]
- 7.Ahmed S, Bennett ST, Huxtable SJ, Todd JA, Matthews DR, Gough SCL. INS VNTR allelic variation and dynamic insulin secretion in healthy adult non-diabetic Caucasian subjects. Diabet Med. 1999;16:910–917. doi: 10.1046/j.1464-5491.1999.00169.x. [DOI] [PubMed] [Google Scholar]
- 8.Lucassen AM, Screaton GR, Julier C, Elliott TJ, Lathrop M, Bell JI. Regulation of insulin gene expression by the IDDM associated, insulin locus haplotype. Hum Mol Genet. 1995;4:501–506. doi: 10.1093/hmg/4.4.501. [DOI] [PubMed] [Google Scholar]
- 9.Vafiadis P, Bennett ST, Colle E, Grabs R, Goodyer CG, Polychronakos C. Imprinted and genotype-specific expression of genes at the IDDM2 locus in pancreas and leucocytes. J Autoimmun. 1996;9:397–403. doi: 10.1006/jaut.1996.0054. [DOI] [PubMed] [Google Scholar]
- 10.Knowler WC, Pettitt DJ, Vasquez B, Rotwein PS, Andreone TL, Permutt MA. Polymorphism in the 5' flanking region of the human insulin gene. Relationships with noninsulin-dependent diabetes mellitus, glucose and insulin concentrations, and diabetes treatment in the Pima Indians. J Clin Invest. 1984;74:2129–2135. doi: 10.1172/JCI111638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Huxtable SJ, Saker PJ, Haddad L, et al. Analysis of parent-offspring trios provides evidence for linkage and association between the insulin gene and type 2 diabetes mediated exclusively through paternally transmitted class III variable number tandem repeat alleles. Diabetes. 2000;49:126–130. doi: 10.2337/diabetes.49.1.126. [DOI] [PubMed] [Google Scholar]
- 12.Lindsay RS, Walker JD, Halsall I, et al. Insulin and insulin propeptides at birth in offspring of diabetic mothers. J Clin Endocrinol Metab. 2003;88:1664–1671. doi: 10.1210/jc.2002-021018. [DOI] [PubMed] [Google Scholar]
- 13.Hansen SK, Gjesing AP, Rasmussen SK, et al. Large-scale studies of the HphI insulin gene variable-number-of-tandem-repeats polymorphism in relation to Type 2 diabetes mellitus and insulin release. Diabetologia. 2004;47:1079–1087. doi: 10.1007/s00125-004-1418-3. [DOI] [PubMed] [Google Scholar]
- 14.Estrada K, Aukrust I, Bjørkhaug L, et al. Association of a low-frequency variant in HNF1A with type 2 diabetes in a Latino population. JAMA. 2014;311:2305–2314. doi: 10.1001/jama.2014.6511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Williams AL, Jacobs SB, Moreno-Macías H, et al. Sequence variants in SLC16A11 are a common risk factor for type 2 diabetes in Mexico. Nature. 2014;506:97–101. doi: 10.1038/nature12828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.McGinnis RE, Spielman RS. Insulin expression: is VNTR allele 698 really anomalous? Nat Genet. 1995;10:378–380. doi: 10.1038/ng0895-378. [DOI] [PubMed] [Google Scholar]
- 17.Awata T. IDDM and variable number of tandem repeats (VNTR) in the 5'region of the insulin gene: a review. Nihon Rinsho. 1997;55:376–381. [PubMed] [Google Scholar]
- 18.Gymrek M. A genomic view of short tandem repeats. Curr Opin Genet Dev. 2017;44:9–16. doi: 10.1016/j.gde.2017.01.012. [DOI] [PubMed] [Google Scholar]
- 19.Rodriguez S, Gaunt TR, Day IN. Molecular genetics of human growth hormone, insulin-like growth factors and their pathways in common disease. Hum Genet. 2007;122(1):1–27. doi: 10.1007/s00439-007-0378-3. [DOI] [PubMed] [Google Scholar]
- 20.Gu D, O'Dell SD, Chen XH, Miller GJ, Day IN. Evidence of multiple causal sites affecting weight in the IGF2-INS-TH region of human chromosome 11. Hum Genet. 2002;110(2):173–181. doi: 10.1007/s00439-001-0663-5. [DOI] [PubMed] [Google Scholar]
- 21.Sharma P, Hingorani A, Jia H, Ashby M, Hopper R, Clayton D, et al. Positive association of tyrosine hydroxylase microsatellite marker to essential hypertension. Hypertension. 1998;32(4):676–682. doi: 10.1161/01.HYP.32.4.676. [DOI] [PubMed] [Google Scholar]
- 22.Jindra A. Association analysis of two tyrosine hydroxylase gene polymorphisms in normotensive offspring from hypertensive families. Blood Press. 2000;9(5):250–254. doi: 10.1080/080370500448623. [DOI] [PubMed] [Google Scholar]
- 23.Klintschar M, Immel UD, Stiller D, Kleiber M. TH01, a tetrameric short tandem repeat locus in the tyrosine hydroxylase gene: Association with myocardial hypertrophy and death from myocardial infarction? Dis Markers. 2005;21(1):9–13. doi: 10.1155/2005/256254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rodriguez S, Gaunt T, O´Dell S, Chen X, Gu D, Hawe E, et al. Haplotypic analyses of the IGF2-INS-TH gene cluster in relation to cardiovascular risk traits. Hum Mol Genet. 2004;13(7):715–725. doi: 10.1093/hmg/ddh070. [DOI] [PubMed] [Google Scholar]
- 25.Rodriguez S, Gaunt T, Dennison E, Chen X, Syddall E, Phyllips D, et al. Replication of IGF2-INS-TH*5 haplotype effect on obesity in older men and study of related phenotypes. Eur J Hum Gen. 2006;14(1):109–116. doi: 10.1038/sj.ejhg.5201505. [DOI] [PubMed] [Google Scholar]
- 26.Berumen J, Orozco L, Betancourt-Cravioto M, Gallardo H, Zulueta M, Mendizabal L, et al. Influence of obesity, parental history of diabetes, and genes in type 2 diabetes: a case–control study. Sci Rep. 2019;9(1):2748. doi: 10.1038/s41598-019-39145-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.American Diabetes Association Standards of medical care in diabetes-2022 Abridged for primary care providers. Clin Diabetes. 2022;40(1):10–38. doi: 10.2337/cd22-as01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Babyak MA. What you see may not be what you get: a brief, nontechnical introduction to overfitting in regression-type models. Psychosom Med. 2004;66:411–421. doi: 10.1097/01.psy.0000127692.23278.a9. [DOI] [PubMed] [Google Scholar]
- 29.Excoffier L, Lischer HEL. Arlequin suite ver 3.5: A new series of programs to perform population genetics analyses under Linux and Windows. Mol Ecol Resour. 2010;10:564–567. doi: 10.1111/j.1755-0998.2010.02847. [DOI] [PubMed] [Google Scholar]
- 30.Faul F, Erdfelder E, Lang AG, Buchner A. G*Power 3: a flexible statistical power analysis program for the social, behavioral, and biomedical sciences. Behav Res Methods. 2007;39:175–191. doi: 10.3758/bf03193146. [DOI] [PubMed] [Google Scholar]
- 31.Puers C, Hammond HA, Jin L, Caskey T, Schumm JW. Identification of repeat sequence heterogeneity at the polymorphic short tandem repeat locus HUMTH01 (AATG) n and reassignment of alleles in population analysis by using a locus-specific allelic ladder. Am J Hum Genet. 1993;53:953–958. [PMC free article] [PubMed] [Google Scholar]
- 32.Le Stunff C, Fallin D, Schork NJ, Bougnères P. The insulin gene VNTR is associated with fasting insulin levels and development of juvenile obesity. Nat Genet. 2000;26:444–446. doi: 10.1038/82579. [DOI] [PubMed] [Google Scholar]
- 33.Owerbach D, Gabbay KH. The search for IDDM susceptibility genes: the next generation. Diabetes. 1996;45:544–551. doi: 10.2337/diab.45.5.544. [DOI] [PubMed] [Google Scholar]
- 34.Bennett ST, Wilson AJ, Esposito L, et al. Insulin VNTR allele-specific effect in type 1 diabetes depends on identity of untransmitted paternal allele. Nat Genet. 1997;17:350–352. doi: 10.1038/ng1197-350. [DOI] [PubMed] [Google Scholar]
- 35.Luthman H, Söderling-Barros J, Persson B, et al. Human insulin-like growth-factor-binding protein. Low-molecular-mass form: protein sequence and cDNA cloning. Eur J Biochem. 1989;180:259–265. doi: 10.1111/j.1432-1033.1989.tb14641.x. [DOI] [PubMed] [Google Scholar]
- 36.Kanaya AM, Herrington D, Vittinghoff E, et al. Glycemic effects of postmenopausal hormone therapy: the heart and estrogen/progestin replacement study. A randomized, double-blind, placebo-controlled trial. Ann Intern Med. 2003;138:1–9. doi: 10.7326/0003-4819-138-1-200301070-00005. [DOI] [PubMed] [Google Scholar]
- 37.Brand JS, Van Der Schouw YT, Onland-Moret NC, et al. Age at menopause, reproductive life span, and type 2 diabetes risk: results from the EPIC-InterAct study. Diabetes Care. 2013;36:1012–1019. doi: 10.2337/dc12-1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sekido T, Nishio SI, Ohkubo Y, et al. Repression of insulin gene transcription by indirect genomic signaling via the estrogen receptor in pancreatic beta cells. In Vitro Cell Dev Biol Anim. 2019;55:226–236. doi: 10.1007/s11626-019-00328-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Xu B, Allard C, Alvarez-Mercado AI, et al. Estrogens promote misfolded proinsulin degradation to protect insulin production and delay diabetes. Cell Rep. 2018;24:181–196. doi: 10.1016/j.celrep.2018.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets generated during and/or analyzed during the current study are available from the corresponding author on reasonable request.