Abstract
Here the authors present a Grid-aware middleware system, called GridPACS, that enables management and analysis of images in a massive scale, leveraging distributed software components coupled with interconnected computation and storage platforms. The need for this infrastructure is driven by the increasing biomedical role played by complex datasets obtained through a variety of imaging modalities. The GridPACS architecture is designed to support a wide range of biomedical applications encountered in basic and clinical research, which make use of large collections of images. Imaging data yield a wealth of metabolic and anatomic information from macroscopic (e.g., radiology) to microscopic (e.g., digitized slides) scale. Whereas this information can significantly improve understanding of disease pathophysiology as well as the noninvasive diagnosis of disease in patients, the need to process, analyze, and store large amounts of image data presents a great challenge.
The use of biomedical imaging is growing in prevalence and has become a key component in both basic research and clinical practice. Although advances in data acquisition technologies have improved the resolution and speed at which we can collect image data, most researchers have access to a limited research repository mainly due to lack of efficient software for managing, manipulating, and sharing large volumes of image data. For instance, a single digitized microscopy image can reach up to several tens of gigabytes in size, and a research study may require access to hundreds of such images. Querying and retrieving the data of interest so that the analysis can be completed quickly is a challenging step in large datasets. Formulation and execution of effective image analysis workflows are also crucial. An image analysis workflow can consist of many steps of simple and complex operations on image data as well as interactive visualization. There is also a need to support the information service needs of collaborative studies in which datasets may reside on various heterogeneous distributed resources including a wide range of networks, computation platforms, and data storage systems.
In this work we present a software system that is designed to address these data management and processing challenges. The contributions of this report include: (1) an efficient client frontend that implements the functionality to submit queries in a uniform way against distributed image databases consisting of multiple studies and modalities; (2) extensible metadata schema for images that can be used to represent two-dimensional (2D), three-dimensional (3D), and time-dependent images with optional application-specific metadata; and (3) an integrated runtime system that provides support for storage and management of distributed image databases, defined by metadata schemas, and for processing large numbers of images quickly. GridPACS extends the traditional image archival systems and our previous work1,2 in the following ways:
Virtualized and Federated Data Access. An image dataset can be partitioned and accessed at multiple distributed sites. Multiple image servers can be grouped to form a collective that can be queried as if it were a single, centralized server entity.
Scalability. The infrastructure makes it possible to employ compute and storage clusters to store and manipulate large image datasets. An image server can be set up on any number of nodes of a cluster. New image server nodes can be added to or deleted from the system with little overhead. A single image can be partitioned and stored on multiple server nodes.
Active Storage. The architecture supports invocation of procedures on ensembles of images. These procedures can be used to, for example, carry out subsetting, error correction, image processing, feature analysis, and rendering. Processing of data can be split across storage nodes (where image data are stored) and compute nodes without requiring the client to download the data to a local machine. The system is extensible in that application developers can define and deploy their own procedures.
On-demand Database Creation. An application-specific data type (e.g., results of an image analysis workflow) can be registered in the system as a new schema. New versions of existing schemas may also be created by altering various aspects such as the existence, cardinality, ordering, and value constraints of attributes and elements. Moreover, the schema can be a composition of new attributes and/or elements and references to existing schemas. Application-defined metadata types and image datasets conforming to a given schema can be automatically manifested as custom databases at runtime, and image data adhering to these data types can be stored in these databases.
Our previous work3,4,5 developed middleware frameworks for distributed data management and data processing. GridPACS is an application of these middleware frameworks optimized to support distributed image datasets. This work also complements our earlier work on digitized microscopy,1 which focused on efficient storage and retrieval of large digitized microscopy images and distributed image processing,2 which developed a runtime system for distributed execution of image processing operations. GridPACS implements support for images from multiple imaging modalities and builds image management support on a generic XML-based data management system. In GridPACS, image datasets and data processing workflows are modeled by XML schemas. These schemas and their instances are stored, retrieved, and managed by distributed data management services. Images might be organized in grids of 2D tiles, 3D volumes, and 2D/3D time-dependent datasets. Through the use of XML schemas, multiple attributes of arbitrary complexity can be defined and associated with an image. Examples of such attributes include image type, unique identifier of the study for which the image was acquired, and acquisition date. Distributed execution services carry out instantiation of data processing operations (components) on distributed platforms, management of data flow between operations, and data retrieval from and storage to distributed collections of data servers.
Background
The growth of radiology imaging data led to the development of Picture Archiving and Communication Systems (PACS)6 that store images in the DICOM standard format.7 PACS-based systems support storage, management, and retrieval of image datasets and metadata associated with the image data. A client using these systems typically retrieves and downloads the data of interest, often specified through a graphical user interface, to the local workstation for application-specific processing and analysis. The effectiveness of imaging studies with such configurations is severely limited by the capabilities of the client's workstation, while the server's compute capabilities are underutilized. Advanced analysis techniques and exploration of collections of datasets can easily overwhelm the capacity of most advanced desktop workstations as well as premium clinical review stations. A number of projects targeted systems for federation of multiple PACS instances.8,9,10 GridPACS extends such systems by not only enabling federation at the Grid scale but also making it possible to set up image databases on storage clusters and integrate data processing and data retrieval in a distributed server environment.
The Open Microscopy Environment project (<http://openmicroscopy.org>) supports a database-driven system for analysis of biological images. The system consists of a relational database that stores image data and metadata. Images in the database can be processed using a series of modular programs. These programs are connected to the database; a module in the processing sequence reads its input data from the database and writes its output back to the database so that the next module in the sequence can work on it. The Virtual Slidebox project (<http://www.path.uiowa.edu/virtualslidebox>) at the University of Iowa is a web-based portal to a database of digitized microscopy slides used in education. The users can search for virtual slides and view them through the portal. The Visible Mouse project (<http://tvmouse.compmed.ucdavis.edu>) provides access to basic information about the mouse and mouse pathology through a web-based portal. The Visible Mouse system can be used for educational and research applications.
Manolakos and Funk11 describe a Java-based tool for rapid prototyping of image processing operations. This tool uses a component-based framework, called JavaPorts, and implements a master-worker mechanism. Oberhuber12 presents an infrastructure for remote execution of image processing applications using SGI ImageVision library, which is developed to run on SGI machines, and NetSolve.13 Andrade et al.14 developed a distributed semantic caching system, called ActiveProxy-G, which allows proxies interspersed between application clients and application servers to speed up execution of related queries by exploiting cached aggregates. ActiveProxy-G requires integration of user-defined operators in the system to enable caching of application-specific intermediate results and searching of cached results.
In our work, we address the metadata and data management issues using a generic, distributed, XML-based data management system. This system, called Mobius,3,4 is designed as a set of loosely coupled services with well-defined protocols. Building on Mobius, GridPACS allows for modeling and storage of image data and workflows as XML schemas and XML documents, enabling use of well-defined protocols for storing and querying data. A client can define and publish data models to efficiently store, query, reference, and create virtualized XML views into distributed and interconnected databases. Support for image data processing is an integral part of GridPACS. The image processing component builds on a component-based middleware5 that provides combined task- and data-parallelism through pipelining and transparent component copies. This enables combined use of distributed storage and compute clusters. Data operations that carry out data subsetting and filtering operations can be pushed to storage nodes to reduce the volume of network traffic, whereas compute-intensive operations can be instantiated on high-performance compute clusters. Complex image analysis workflows can be formed from networks of individual data processing components.2
The Distributed Parallel Storage Server (DPSS)15 project developed tools to use distributed storage servers to supply data streams to multiuser applications in an Internet environment. The DPSS was used in the implementation of a remote, large scale data visualization system prototype.16 Our middleware has similarities to DPSS in that it is used for storing and retrieving data. However, DPSS is designed as a block-server, whereas data in our system is managed as semistructured documents conforming to schemas. This allows for more complex data querying capabilities, schema validation, and database creation from complex schemas.
There are a number of projects that target development of infrastructures for shared access to data and computation in various areas of medicine, science, and engineering. The Biomedical Informatics Research Network (BIRN) (<http://www.nbirn.net>)17,18 initiative focuses on support for collaborative access to and analysis of datasets generated by neuroimaging studies. The BIRN project uses the Storage Resource Broker (SRB),19 which provides a distributed file system infrastructure as a distributed data management middleware layer. MammoGrid20,21,22 is a multi-institutional project funded by the European Union (EU). The objective of this project is to apply Grid middleware and tools to build a distributed database of mammograms and investigate how it can be used to facilitate collaboration between researchers and clinicians across the EU. eDiamond23,24 targets deployment of Grid infrastructure to manage, share, and analyze annotated mammograms captured and stored at multiple sites. One of the goals of MammoGrid and eDiamond is to develop and promote standardization in medical image databases for mammography and other cancer diseases. MEDIGRID25,26,27 is another project initiated recently to investigate application of Grid technologies for manipulating large medical image databases. These large scale and multi-institutional projects share the same goal of deploying an infrastructure, building on Grid technologies, to facilitate sharing of medical image data across institutions, even countries. A major requirement of these projects is the capability to support storage, management, integration, and analysis of large image datasets and associated metadata. Our system is designed to address these needs through customization of two generic middleware tools in an integrated package; a distributed, XML-based data and metadata management system and a distributed data processing engine. In that respect, GridPACS provides a core, Grid-enabled technology that can be employed by those projects.
Design Objectives
The GridPACS architecture is designed to support a wide range of biomedical imaging applications encountered in translational research projects, basic science efforts, and clinical medicine. These applications have in common the need to store, query, process, and visualize large collections of related images. In this section, we briefly describe two biomedical imaging applications with which our group has direct experience and how these application scenarios motivate the requirement for GridPACS.
Dynamic Contrast Enhanced MRI Studies
Dynamic Contrast Enhanced Magnetic Resonance Imaging (DCE-MRI)28,29 is a powerful method for cancer diagnosis and staging and for monitoring cancer therapy. It aims to detect differences in the microvasculature and permeability of abnormal tissue as compared with healthy tissue. After an extracellular contrast agent is injected in the subject, a sequence of images are obtained over some period of time. The time-dependent signal changes acquired in these images are quantified by a pharmacokinetic model to map out the differences between tissues. State-of-the-art research studies in DCE-MRI make use of large datasets, which consist of time-dependent, multidimensional collections of data from multiple imaging sessions. Although single images are relatively small (2D images consists of 1282 to 5122 pixels, 3D volumes of 64 to 512 2D images), a single study carried out on a patient can result in an ensemble of hundreds of 3D image datasets. A study that involves time-dependent studies from different patients can involve thousands of images.
There are a wide variety of computational techniques used to quantitatively characterize DCE-MRI results.28,30 These studies can be used to aid tumor identification and staging and to assess efficacy of cancer treatments. Systematic development and assessment of image analysis techniques require an ability to efficiently invoke candidate image quantification methods on large collections of image data. A researcher might use GridPACS to iteratively apply several different image analysis methods on data from multiple different studies to assess ability to predict outcome or effectiveness of a treatment across patient groups. Such a study would likely involve multiple image datasets containing thousands of 2D and 3D images at different spatial and temporal resolutions.
Digital Microscopy
Digital microscopy1 is being increasingly used in basic research studies as well as in cooperative studies that involve anatomic pathology. For instance, both the Cancer and Leukemia Group B (CALGB) and the Children's Oncology Group are making use of fully digitized slides to support slide review. Researchers in both groups are developing computer-based methods to assist anatomic pathologists in quantifying histologic features for disease grading. Digital microscopy is also used for basic science applications. One such example involves the characterization of the effects of gene knock-outs through phenotypical and microanatomic change, including differences in cellular and tissue organization, in mouse placenta. These studies involve digitizing roughly 1,000 sequential thin sections of mouse placenta that are stained with H & E.
These applications target different goals and make use of different types of datasets. Nevertheless, they exhibit several common requirements that motivate the need for a system like GridPACS. First, in all of these application scenarios, it is anticipated that data will be generated by distributed groups of researchers and clinicians. For instance, researchers from different institutions can generate various portions of mouse placenta datasets. In most cases it is not practical to allocate a single, very large storage system for a project. Instead, several sites associated with a project will contribute storage. These requirements motivate distributed storage and virtualized data access. Second, they all involve significant processing on large volumes of data. This characteristic requires the ability to push data processing near data sources. Third, we also expect each such application to be in a position to make use of computing capacity located at multiple sites. Hence, a system should support combined use of distributed storage and compute resources to rapidly access data and pass it to relevant processing functions.
System Description
We have designed GridPACS to support distributed storage, retrieval, and querying of image data and descriptive metadata. Queries can include extraction of spatial subregions, defined by a bounding box and other user-defined predicates; requests to execute user-defined image analysis operations on data subsets; and queries against metadata. These queries may span datasets across multiple sites. For example, a researcher may want to perform a statistical comparison of images from two datasets located at two different institutions.
GridPACS can use clusters of storage and compute machines to store and serve image data. It manages the distributed storage resources on the server and encapsulates efficient storage methods for image datasets. In addition, GridPACS can maintain metadata describing image analysis workflows and can checkpoint intermediate results during execution of a workflow. It supports efficient execution of image analysis as a network of image processing components on commodity clusters and multiprocessor machines. The network of components can consist of multiple stages of pipelined operations, parameter studies, and interactive visualization.
Descriptive metadata, workflows, and system metadata are modeled as XML schemas. The close integration of image data and metadata makes it straightforward to maintain detailed provenance31 information about how imagery has been acquired and processed; the framework manages annotations and data generated as result of data analysis. In the rest of this section we provide a more detailed description of the GridPACS architecture. We first present the two middleware systems, Mobius3,4 and DataCutter,5 that provide the underlying runtime support.
Mobius and DataCutter Middleware
Mobius
Mobius provides a set of generic services and protocols to support distributed creation, versioning, and management of data models and data instances; on-demand creation of databases; federation of existing databases; and querying of data in a distributed environment. Its design is motivated by the requirements of Grid-wide data access and integration32,33 and by earlier work done at General Electric's Global Research Center.34 Mobius services employ XML schemas to represent metadata definitions (data models) and XML documents to represent and exchange data instances. A more detailed description of Mobius can be found in Hastings et al.3 and Langella et al.4 We briefly describe the functionality of core services used for GridPACS implementation.
Mobius GME: global model exchange
For any application it is essential to have models for the data and metadata. By creating and publishing a common schema for data captured and referenced in a collaborative study, research groups can ensure that applications developed by each group can correctly interact with the shared data sets and interoperate. The Global Model Exchange (GME) is a distributed service that provides a protocol for publishing, versioning, and discovering XML schemas. Because the GME is a global service, it is implemented as an architecture, in which there are multiple GMEs, each of which is an authority for managing schemas defined within a set of namespaces. Namespaces provide a mechanism for differentiating common terminology between multiple markup vocabularies. Other services such as storage services can use the GME to match instance data with their data type definitions.
Mobius Mako: distributed data storage and retrieval
The Mako is a distributed data storage service that provides users the ability to create on-demand databases; store, retrieve, and query instance data; and organize instance data into collections. Mako exposes data resources as XML data services through a set of well-defined interfaces based on the Mako protocol. The initial Mako distribution contains an implementation to expose XML databases that support the XMLDB API (<http://www.xmldb.org>). It also contains the implementation of MakoDB, an XML database optimized for interacting in the Mako framework. To meet its data storage demands, the GridPACS system is composed of several clusters, with each machine in a cluster running a MakoDB implementation of a Mako. Mako client interfaces enable clients to communicate with Makos over a network. Databases that store XML instance documents can be created by using the client interfaces to submit the XML schema that models the instance data to a Mako. Once submitted, the client interfaces can be used to perform operations on instance data, such as submit, retrieve, delete, and query using XPath.
DataCutter
DataCutter5 supports a filter-stream programming model for developing data-intensive applications. In this model, the application processing structure is implemented as a set of components (referred to as filters) that exchange data through a stream abstraction. Filters are connected via logical streams, which denote a unidirectional data flow from one filter (i.e., the producer) to another (i.e., the consumer). The overall processing structure of an application is realized by a filter group, which is a set of filters connected through logical streams. Processing of data through a filter group can be pipelined. Multiple filter groups can be instantiated simultaneously and executed concurrently. In GridPACS, DataCutter streams are strongly typed with types defined by XML schemas managed by GMEs. The current runtime implementation provides a multithreaded execution environment and uses TCP for point-to-point stream communication between two filters placed on different machines.
GridPACS Implementation
GridPACS has been realized as four main components in an integrated system; frontend client, backend metadata and data management, image storage, and distributed execution. The client application can be used by a user to upload, query, and inspect the data at the server. The backend metadata and data management service is implemented as a collection of federated Mobius Mako servers to manage distributed image databases.
▶ shows a sample environment of frontend clients and backend image data servers. The image storage component allows declustered data storage and parallel access of image chunks. The distributed execution component allows image processing operations to be performed on the images by multiple compute servers simultaneously.
Figure 1.
A sample GridPACS instance.
GridPACS Frontend and Client Component
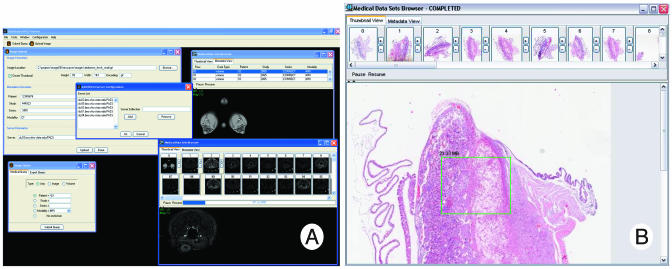
The GridPACS client frontend, shown in ▶, provides an intuitive, centralized view of the GridPACS backend. It creates a local workspace for the user by hiding the details of retrieving and locating datasets within the backend. The results of queries are displayed in a browser, which provides mechanisms for traversing both volumetric and single image datasets via thumbnails and a metadata viewer.
Figure 2.
(A) GridPACS client screen shot. (B) Medical datasets browser screen shot.
Accessing and displaying large datasets over a network of distributed servers can quickly become problematic for clients because of issues such as network delays, large data sizes and quantities, and multiple servers. The GridPACS client application uses several techniques to overcome these adversities. The first important feature is the intelligent use of background threading for tasks that communicate with network resources. The client application uses an active thread pool that allows sequences of tasks to be performed in the background, whereas the main application thread continues to process user inputs and refresh the display. Another powerful feature is the use of on-demand, lazy data loading. All data within datasets are left on the backend until it is absolutely needed. Only a reference to the data is stored in the client. The dataset object model abstracts this concept by presenting get methods that cache and immediately return data if it is present and transparently load the needed data from the backend when it is not. This allows the application components to work with the datasets as though they are all locally present, and the object model handles the requests and local storage when they are not. The dataset browser takes full advantage of this feature and is able to present a large amount of data quickly.
Distributed Metadata and Data Management
Image data are modeled using XML schemas that are managed by the GME. An instance of the schema corresponds to an image dataset with images and associated image attributes stored across multiple storage systems running data management servers (i.e., Makos). In a cluster environment, multiple Mako servers can be instantiated on each node, and one or more GMEs can be instantiated to manage schemas.
Our image model is made up of two components: the image metadata and the image data. The image metadata component contains a few basic attributes and a detail set entity that allows a collection of user-defined key value pair metadata. This allows application-specific metadata to be attached to an image while maintaining a common generic model. The image data component contains either a Base64 encoded version of the image data or a reference to a binary submitted with the XML instance. The Mako protocol supports the attachment of binary objects to XML documents, which is much more efficient than the Base64 encoding of binary objects.
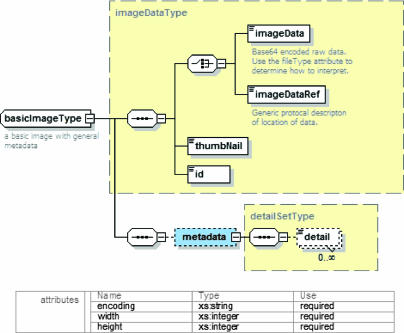
We have designed a number of core models that any user can extend with specific versions that are more relevant to their domain. ▶ shows the basic single image type. The basicImageType is a simple data model that has four main elements: the image data or a pointer to it, a thumbnail, an id, and a generic set of key-value metadata. It also defines three required attributes: the image data encoding type, the height, and the width. This model is the basic starting point for representing an image in storage system. All images in the storage system are of this type or its extensions.
Figure 3.
Basic image model.
A simple extension to the 2D image model, shown in ▶, is the volumeImageType shown in ▶a. The volumeImageType extends the basicImageType and adds a few more descriptive attributes that are relevant to the image if it belongs to a volume. The new attributes xloc, yloc, and zloc are attributes that describe the image starting location with respect to the volume container. ▶b shows the volumeType, which is the container for a set of volumeImageTypes. This type contains a set of volumeImageTypes as well as some required metadata, which describe the dimensions of the volume.
Figure 4.
(A) Volume image model. (B) Volume model.
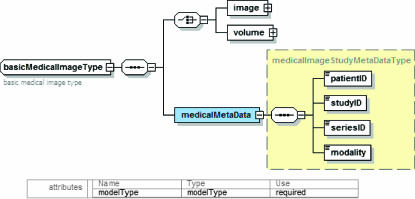
To handle medical image data, the basic image and volume models described above are used in a new model, which is more specific to medical image data sets. This model, shown in ▶, contains two basic elements, the image as defined by basicImageType or a volume as defined by volumeType and a set of medical image data set specific metadata. This model can be used to describe medical images or volumes and can be queried using the attached medical metadata and the metadata of the image data.
Figure 5.
Medical image dataset model.
Image Data Storage
To process data, a number of XML instances must be created and stored that adhere to the models defined by users and registered in the system. When input data sets are ingested, they are registered and indexed so that they can be referenced by clients in queries. To maximize the efficiency of parallel storage and data accesses for image data, GridPACS uses the Mako/MakoDB framework of Mobius and data declustering techniques. The goal of declustering35,36 is to distribute the data across as many storage units as possible so that data elements that satisfy a query can be retrieved from many sources in parallel. An application programming interface (API) is provided by the system that can be used by an application to partition a large byte array into smaller pieces (chunks) and associate metadata description with each piece (represented by a schema). For example, if the byte array corresponds to a large 2D image, each chunk can be a rectangular subsection of the image. The metadata associated with a chunk can be the bounding box of the chunk, its location within the image, etc.
The system currently supports several data distribution policies. These policies take into account the storage capacity and I/O performance of the storage systems that host the backend Mako databases, as well as the network performance. An application can specify a preferred declustering mechanism when it requests a distribution plan.
Distributed Execution
The distributed execution service builds on the runtime system developed by Hastings et al.2 This runtime system consists of four pieces: (1) an image processing toolkit, (2) a visualization toolkit, (3) a runtime system for execution in a distributed environment, and (4) an XML-based schema for process and workflow description. An image analysis application is represented as a filter group consisting of filters that process the data in a pipelined fashion. That is, the functions constituting the processing structure of the application are implemented as application components using DataCutter filter support.5 The system implements a simple abstraction layer for executing image processing operations implemented using functions from the Insight Segmentation and Registration Toolkit (ITK)37 and the Visualization Toolkit (VTK).38
In this version of GridPACS, we have adapted the workflow schema developed by Hastings et al.,2 which was based on the process modeling of the Distributed Process Management System (DPM).39 The Grid Service communities are defining standards to describe, register, discover, and execute workflows. We ultimately plan to adopt a model based on these emerging workflow standards. GridPACS describes workflows in a way that is comparable to systems such as Pegasus40,41 and Condor's DAGMan.42 The execution of an application is modeled by a directed acyclic task graph of filters. We have extended the earlier implementation2 to use the Mako service structure for managing workflow descriptions, querying input datasets, and storing both intermediate results and image analysis output. As discussed earlier, the Mako allows a document to be comprised of other document pieces stored in the data grid. We use this functionality to support nested workflows. That is, a workflow can consist of filters and other workflows. A process in a workflow document can be a reference to another process description enabling one researcher's process model to be comprised of references to other process model instances. GridPACS currently supports internal and external filter definitions. An internal filter represents a process or function that adheres to an interface, DataCutter in this case, and is implemented as an application-specific filter in the system. An external filter represents a self-contained executable that exists outside the system and communicates with other filters through files.
To use the distributed metadata and data management service, the runtime system has two system-level filters, MakoReader and MakoWriter. These filters provide interfaces between Mako servers and other filters in the workflow. The MakoReader filter retrieves images from Mako servers, converts them into VTK/ITK data structures, and passes them to the first stage filters in the workflow. The MakoWriter filter can be placed between two application filters, which are connected to each other in the workflow graph, when the output of a filter needs to be check-pointed. The last-stage filters in the workflow also connect to MakoWriter filters to store output in Mako servers. The execution of a workflow and check-pointing can be done stage by stage (i.e., all the data are processed by one stage and stored on Mako servers before the next stage is executed) or pipelined (i.e., all stages execute concurrently; the MakoWriter filters interspersed between stages both send data to Mako servers and pass it to the next filter in the workflow).
Status Report
In this section, we present a performance evaluation of GridPACS. In this evaluation we used datasets generated by time-dependent magnetic resonance imaging (MRI) scans and digitized microscopes. A group of three PC clusters with a total of 35 nodes were used to run GridPACS servers. One cluster (OSUMED) consists of 24 Pentium III nodes, with 512 MB memory and 300 GB disk space per node. The nodes of this cluster are connected to each other over a Fast Ethernet Switch. The second cluster (DC) has five dual-processor nodes, each with Xeon 2.4GHz CPUs, 2GB memory, and 250GB disk space. The third cluster (MOB) consists of 6 nodes with dual AMD Opteron CPUs. Each node of this cluster has 8GB memory and 1.5TB RAID disk array. The OSUMED cluster is connected to the MOB and DC clusters via a shared 100-Mbps wide-area network, whereas MOB and DC are connected to each other via a 1-Gbps network.
To obtain insight into how GridPACS performs with regard to creating an image database, we grouped five nodes of the MOB cluster into a federated database server using a virtual Mako instance running on one of these nodes. The client communicates with the virtual Mako, which delegates requests to the Mako servers running on the individual MOB nodes. The virtual Mako also served as a metadata server. The client machine was connected to the image database server over a 100-Mbps network. When submitting an image, the ingestion client first creates an XML document, based on the XML schema that contains the metadata information about the image, which includes patient name, study id, imaging modality, and date of acquisition. This document is then submitted with the actual image data as a binary attachment. Upon receiving the document, the virtual Mako server redirects the document to a Mako server based on a predefined distribution strategy. The Mako server stores the attachment locally on the file system, and ingests the metadata into the metadata server for registration and indexing.
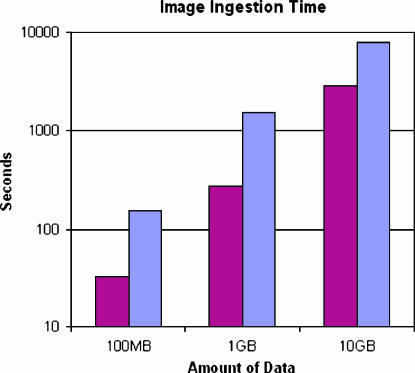
In our experimental setup, the size of each radiology image was on average 100 KB, whereas digitized microscopy images ranged from 400 MB to 5 GB in size. The client partitioned each microscopy image into 512 × 512 pixel tiles and submitted each tile separately. Again, an XML document was created for each image. However, this document contained pointers to all the tiles as well as other metadata information. Each tile was associated with a bounding box and location within the original image. This information is encoded in an XML file, to which the tile is attached. ▶ shows the timing results for creating image databases of 100 MB, 1 GB, and 10 GB in size. For both the small images (i.e., radiology images) and large images (i.e., digitized microscopy images), as the size of the datasets increases, the time to create the databases increases linearly.
Figure 6.
Performance numbers for creation of an image database. Red bars represent large images, and blue bars represent small images.
The results show that the time to insert the same amount of image data is significantly less for the larger images than the smaller ones. This is expected, because approximately the same amount of metadata is used to describe both large and small images. Radiology images typically are small; therefore, they have higher metadata density per megabyte than digitized microscopy images. For the same amount of image data, radiology images may result in 300 times the metadata overhead compared with a microscopy image dataset.
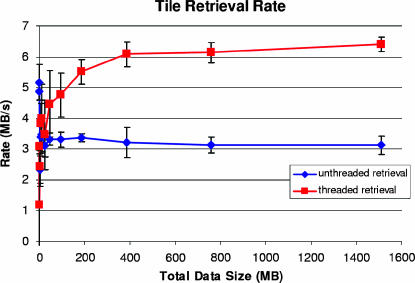
A query into the GridPACS system is executed in two phases. In the first phase, the list of images (or image tiles) that satisfies the query is retrieved from the metadata server. Then, the image tiles and their metadata are retrieved from the image servers. To evaluate the querying capabilities, we have uploaded 1,200 digitized microscopy images, equivalent to 1 Terabyte of data, to the system. Each image was divided into 512 × 512 pixel tiles, which were stored across 30 nodes, 24 of which were grouped into a single Virtual Mako server. The metadata associated with images is stored on a cluster of five Mako servers. The experiments are carried out using a client program connected to GridPACS via a 100-Mbps Ethernet connection. The client submitted queries that selected rectangular regions of a 40,000 × 40,000 pixel image in the database. The tests were run with and without multithreading at the client side. For the multithreaded test, eight concurrent active threads were used. ▶ shows that the throughput for large dataset retrieval is twice as high for the multithreaded version (threaded retrieval in the figure) compared with the unthreaded version and achieves better usage of available network bandwidth.
Figure 7.
Multithreaded tile retrieval (threaded retrieval) outperforms single-threaded (unthreaded) retrieval.
To evaluate the basic functionality of data retrieval paired with on-demand image analysis, we implemented an image analysis scenario that classifies and segments nuclei in digitized microscopy images. The image analysis algorithm assumes that each pixel represents a mixture of actual materials, nucleus, cytoplasm, red blood cells, and background.43 The complete image is then processed in chunks, and pixels are classified based on shortest distance to each of the cluster centers in feature space. The resulting classification is then segmented into components that represent individual nuclei. This image analysis scenario is implemented as a set of two filters: a data retrieval filter that interacts with Mako servers to query for images and retrieve image chunks and a processing filter that implements the image analysis function. The client query specifies an image and a set of computation nodes for image processing. The data retrieval filters and processing filters execute on these nodes, retrieve portions of the image dataset (as defined in the query), apply the image analysis operation, and send the results to the client. The performance results are shown in ▶, where 10 parallel nodes are used to process image chunks. In this experiment, the size of the image was varied (as shown in the x-axis as “Data Chunks”). The Mako servers run on five of these nodes, and each image is distributed across these nodes. The graph also shows the timing results for data retrieval only. As expected, the execution time increases when data analysis is performed. However, the increase is not substantial, mainly due to the capability of GridPACS to pipeline the processing of image chunks.
Figure 8.
Performance numbers when image analysis is performed as part of data retrieval in GridPACS. The image size is varied in these experiments so that the number of chunks for the image ranged from 1 to 4,096.1. Blue bars represent retrieval results with analysis, and red bars represent retrieval results without analysis.
Conclusions
We presented a distributed image archival and processing system, called GridPACS. The salient features of GridPACS are the following. (1) Manifested databases can be stored and accessed at distributed sites. (2) GridPACS infrastructure allows image servers to be added to or deleted from the system easily and efficiently and can distribute individual images across multiple image servers. (3) GridPACS provides support for active storage. Remote invocation of procedures on ensembles of images is supported. These procedures can be executed on a distributed collection of storage and compute platforms. New procedures can be added to the system. (4) Application-defined metadata types and image datasets can be manifested automatically as custom databases at runtime, and image data adhering to these data types can be stored in these databases. These features extend traditional PACSs, which use centralized repositories for image archival, to a Grid-enabled system that addresses the distributed storage, querying, and processing requirements of large biomedical image projects in an integrated system.
Supported in part by the National Science Foundation under Grants #ACI-9619020 (UC Subcontract #10152408), #EIA-0121177, #ACI-0203846, #ACI-0130437, #ANI-0330612, #ACI-9982087, Lawrence Livermore National Laboratory under Grant #B517095 (UC Subcontract #10184497), NIH NIBIB BISTI #P20EB000591, Ohio Board of Regents BRTTC #BRTT02-0003.
References
- 1.Catalyurek U, Beynon MD, Chang C, Kurc T, Sussman A, Saltz J. The virtual microscope. IEEE Trans Inf Technol Biomed. 2003;7:230–48. [DOI] [PubMed] [Google Scholar]
- 2.Hastings S, Kurc T, Langella S, Catalyurek U, Pan T, Saltz J. Image processing for the grid: A toolkit for building grid-enabled image processing applications. CCGrid: IEEE Int Symp on Cluster Comput and the Grid. Tokyo, Japan: IEEE Press, 2003.
- 3.Hastings S, Langella S, Oster S, Saltz J. Distributed data management and integration: The Mobius project. GGF Semantic Grid Workshop. 2004;2004:20–38. [Google Scholar]
- 4.Langella S, Hastings S, Oster S, Kurc T, Catalyurek U, Saltz J. A distributed data management middleware for data-driven application systems. Proc 2004 IEEE Int Conf on Cluster Comput. San Diego, CA: September, 2004.
- 5.Beynon MD, Kurc T, Catalyurek U, Chang C, Sussman A, Saltz J. Distributed processing of very large datasets with DataCutter. Parallel Comput. 2001;27:1457–78. [Google Scholar]
- 6.Huang H, Witte G, Ratib O, Bakker A, Chuang K. Picture Archiving and Communication Systems (PACS) in Medicine. Berlin, Germany: Springer-Verlag, 1991.
- 7.N.E.M.A. American College of Radiology. Digital imaging and communications in medicine (DICOM): Version 3.0. ACR-NEMA Committee, Working Group VI, 1993.
- 8.Martinez R, Alsafadi Y, Kim J. Application of the distributed computing environment to global PACS. Proc SPIE Med Imaging Conf. February, 1994.
- 9.Sheng OL, Garcia CH-M. The design of medical image databases: A distributed approach. Proc 9th Int Phoenix Conf on Comp and Comm. 1990:296–303.
- 10.Reijns G. A distributed picture archiving and communications system for hospitals using image pre-fetching. 28th Hawaii Int Conf on System Sciences (HICSS'95). 1995:470–81.
- 11.Manolakos E, Funk A. Rapid prototyping of component-based distributed image processing applications using javaports. Workshop on Computer-Aided Medical Image Analysis, CenSSIS Research and Industrial Collaboration Conf. 2002.
- 12.Oberhuber M. Distributed high-performance image processing on the internet. Master's thesis, Technische Universitat Graz. 2002.
- 13.Casanova H, Dongarra J. NetSolve: A network enabled server for solving computational science problems. Int J Supercomput Appl. 1997;11(3):212–23. [Google Scholar]
- 14.Andrade A, Kurc T, Sussman A, Saltz J. Active Proxy-G: Optimizing the query execution process in the grid. Proc 2002 ACM/IEEE SC Conference (SC2002). ACM Press, 2002.
- 15.Johnston W, Guojun J, Hoo G, et al. Distributed environments for large data-objects: Broadband networks and a new view of high performance, large scale storage-based applications. Proc Internetworking'96. Nara, Japan. September, 1996.
- 16.Bethel W, Tierney B, Lee J, Gunter D, Lau S. Using high-speed WANs and network data caches to enable remote and distributed visualization. Proc 2000 ACM/IEEE SC Conference (SC 2000). Dallas, TX. November; 2000.
- 17.Peltier S, Ellisman M. The Biomedical Informatics Research Network. The Grid, Blueprint for a New Comput Infrastructure. , ed 2 Philadelphia: Elsevier, 2003.
- 18.Santini S, Gupta A. The role of internet imaging in the biomedical informatics research network. Proc SPIE, Vol. 5018. San Jose, CA: Internet Imaging IV, January, 2003.
- 19.Rajasekar A, Wan M, Moore R. MySRB & SRB—Components of a data grid. 11th Int Symp on High Performance Distributed Comput (HPDC-11). July, 2002.
- 20.Amendolia R, Estrella F, Hauer T, et al. Grid databases for shared image analysis in the mammogrid project. Eighth Int Database Engineering & Applications Symp (Ideas'04). July, 2004.
- 21.Amendolia SR, Brady M, McClatchey R, Mulet-Parada M, Odeh M, Solomonides T. Mammogrid: Large-scale distributed mammogram analysis. Stud Health Technol Inform. 2003;95:194–9. [PubMed] [Google Scholar]
- 22.Rogulin D, Estrella F, Hauer T, McClatchey R, Amendolia SR, Solomonides A. A grid information infrastructure for medical image analysis. Distributed Databases and Processing in Med Img Comput Workshop (DiDaMIC-2004). September, 2004.
- 23.Brady M, Gavaghan D, Simpson A, Highnam R, Mulet M. eDiamond: A grid-enabled federated database of annotated mammograms. In: Grid Computing: Making the Global Infrastructure a Reality. New York, NY: John Wiley & Sons, Ltd., 2003.
- 24.Solomonides A, McClatchey R, Odeh M, et al. Mammogrid and eDiamond: Grids applications in mammogram analysis. Proc IADIS Int Conf: e-Society. 2003:1032–3.
- 25.Duque H, Montagnat J, Pierson JM, Brunie L, Magnin I. DM2: A distributed medical data manager for grids. BioGrid'03, the 3rd Int Symp on Cluster Comput and the Grid (CCGrid 2003). May 2003:606–11.
- 26.Montagnat J, Breton V, Magnin IE. Using grid technologies to face medical image analysis challenges. In: BioGrid'03, 3rd Int Symp on Cluster Comput and the Grid (CCGrid 2003). May 2003:588–93.
- 27.Tweed T, Miguet S. Medical image database on the grid: Strategies for data distribution. HealthGrid'03. Jan 2003:152–62.
- 28.Knopp MV, Giesel F, Marcos H, von Tengg-Kobligk H, Choyke P. Dynamic contrast-enhanced magnetic resonance imaging in oncology. Top Magn Reson Imaging. 2001;12(2):301–8. [DOI] [PubMed] [Google Scholar]
- 29.Padhani AR. Dynamic contrast-enhanced MRI in clinical oncology: Current status and future directions. J Magn Reson Imaging. 2002;16:407–22. [DOI] [PubMed] [Google Scholar]
- 30.Lucht RE, Knopp MV, Brix G. Classification of signal-time curves from dynamic MR mammography by neural networks. J Magn Reson Imaging. 2001;19(1):51–7. [DOI] [PubMed] [Google Scholar]
- 31.Foster I, Voeckler J, Wilde M, Zhao Y. Chimera: A virtual data system for representing, querying, and automating data derivation. Proc 14th Conf on Scientific and Statistical Database Mgmt. 2002.
- 32.Antonioletti M, Atkinson M, Malaika S, et al. Grid Data Service Specification. Technical report. Global Grid Forum. 2003.
- 33.Antonioletti M, Krause A, Hastings S, et al. Grid Data Service Specification: The XML Realisation. Technical report. Global Grid Forum. 2003.
- 34.Langella S. Intellegent data management system. Master's thesis, Troy, NY: Computer Science Department, Rensselear Polytechnic Institute, 2002.
- 35.Faloutsos C, Bhagwat P. Declustering using fractals. 2nd Int Conf on Parallel and Distributed Information Systems. San Diego, CA, 1993:18–25.
- 36.Moon B, Saltz J. Scalability analysis of declustering methods for multidimensional range queries. IEEE Trans Knowl and Data Eng. 1998;10(2):310–27. [Google Scholar]
- 37.National Library of Medicine. Insight Segmentation and Registration Toolkit (ITK). [cited 2002 October]. Available at: http://www.itk.org/.
- 38.Schroeder W, Martin K, Lorensen B. The Visualization Toolkit: An Object-Oriented Approach to 3D Graphics. , ed 2 Upper Saddle River, NJ: Prentice Hall, 1997.
- 39.Hastings S. Distributed architectures: A java-based process management system. Master's thesis, Troy, NY: Computer Science Department, Rensselear Polytechnic Institute, 2002.
- 40.Deelman E, Blythe J, Gil Y, Kesselman C. Pegasus: Planning for execution in grids. GriPhyN Technical Report 2002-20. 2002.
- 41.Deelman E, Blythe J, Gil Y, et al. Mapping abstract complex workflows onto grid environments. J Grid Comput. 2003;1(1):25–39. [Google Scholar]
- 42.Frey J, Tannenbaum T, Foster I, Livny M, Tuecke S. Condor-G: A computation management agent for multi-institutional grids. Proc Tenth IEEE Symp on High Performance Distributed Comput (HPDC10). IEEE Press. August 2001.
- 43.Pan T, Mosaliganti K, Machiraju R, Cowden D, Saltz J. Large scale image analysis and nuclear segmentation for digital microscopy images. Ninth Ann Conf on Adv Practice, Instruction and Innovation through Inform (APIII 2004). 2004.