Abstract
3′-end processing of nucleus-encoded mRNAs includes the addition of a poly(A) tail that is important for translation initiation. Since the vast majority of chloroplast mRNAs acquire their 3′ termini by processing yet are not polyadenylated, we asked whether 3′ end maturation plays a role in chloroplast translation. A general characteristic of the 3′ untranslated regions of chloroplast mRNAs is an inverted repeat (IR) sequence that can fold into a stem-loop structure. These stem-loops and their flanking sequences serve as RNA 3′-end formation signals. Deletion of the Chlamydomonas chloroplast atpB 3′ IR in strain Δ26 results in reduced accumulation of atpB transcripts and the chloroplast ATPase β-subunit, leading to weakly photosynthetic growth. Of the residual atpB mRNA in Δ26, approximately 1% accumulates as a discrete RNA of wild-type size, while the remainder is heterogeneous in length due to the lack of normal 3′ end maturation. In this work, we have analyzed whether these unprocessed atpB transcripts are actively translated in vivo. We found that only the minority population of discrete transcripts of wild-type size is associated with polysomes and thus accounts for the ATPase β-subunit which accumulates in Δ26. Analysis of chloroplast rbcL mRNA revealed that transcripts extending beyond the mature 3′ end were not polysome associated. These results suggest that 3′-end processing of chloroplast mRNA is required for or strongly stimulates its translation.
Chloroplast genes are often organized into operons and gene clusters, which are transcribed into precursor transcripts that undergo complex processing events including splicing and intercistronic cleavages (reviewed in references 35 and 49). While intercistronic cleavages form some mRNA 5′ and 3′ termini, these can also be formed by other types of events. For example, 5′ ends are often formed by endonucleolytic processing of primary transcripts, and this may be the exclusive mode of 5′ end formation in chloroplasts in the green alga Chlamydomonas reinhardtii (reviewed in reference 12). Most plastid mRNAs contain inverted-repeat (IR) sequences in their 3′ untranslated regions, which are believed to fold into stem-loop structures. These IR sequences do not function as efficient transcription terminators but instead are thought to stabilize upstream sequences and mediate correct 3′-end processing (36, 37, 44, 47, 48). In most cases, the 3′ termini of mature transcripts lie immediately downstream of the IR.
Plastid 3′ IR sequences act to stabilize upstream mRNA segments in vitro and in vivo. When RNA molecules containing the IR sequences were incubated in spinach chloroplast protein extracts, they were correctly processed at their 3′ ends and the products were stable for several hours. However, when the IR sequences were deleted from the same RNA molecules and incubated in an identical protein extract, the RNA molecules were rapidly degraded (17, 37, 44, 46). The ability to introduce altered genes into the chloroplast of the green alga C. reinhardtii presented the opportunity to test the in vitro results in an in vivo context. When the 3′ IR of the chloroplast atpB gene was deleted in strain Δ26, atpB mRNA became heterogeneous and unstable, and the resulting decrease in protein accumulation limited photosynthetic growth (48). In addition, the nucleotide sequence of the 3′ untranslated regions (UTRs) can influence the accumulation of a correctly 3′-end-processed transcript, since the functionality of some Chlamydomonas 3′ IRs is orientation dependent in vivo (5, 37).
Evidence for possible involvement of the 3′ UTR in the initiation of translation has accumulated from studies of the poly(A) binding protein in the yeast Saccharomyces cerevisiae as well as in other systems (21, 39). In yeasts and plants, this protein was found to stimulate binding of the 40S ribosomal subunit to mRNA by association with the translation initiation factor eIF-4G, which also binds to eIF-4E and the 5′ cap of the mRNA (16, 40, 50). In mammalian cells, a protein called PAIP, an eIF4G homolog, binds the poly(A) binding protein and enhances translation (10). A model invoking mRNA circularization has been proposed, in which the mRNA 5′ and 3′ ends can interact via this association, which in turn is required for the initiation of translation (10, 16, 18, 40, 50). Although polyadenylation of mRNA has recently been described for spinach chloroplasts, it occurs primarily on degradation products and not at the 3′ end of the intact transcript (27, 30–32). As in Escherichia coli (41), chloroplast polyadenylation is transient and seems to target mRNA for rapid degradation.
The generally accepted finding that the poly(A) tails of eukaryotic, nucleus-encoded mRNAs stimulate translation initiation prompted us to look for a related phenomenon in chloroplasts, which share many features of prokaryotic mRNA metabolism (45). We used the green alga C. reinhardtii as a model system, since the chloroplast genome can be modified by biolistic transformation. Several strains in which the 3′ end processing elements of the atpB gene differed or were lacking were utilized. Analysis of these strains revealed that correctly processed atpB mRNA was highly enriched in polysomal fractions, whereas heterogeneous mRNA was poorly associated with polysomes regardless of its size. These results suggest that 3′ end processing of mRNA in the chloroplast may stimulate its translational activation.
MATERIALS AND METHODS
Plasmids and strains.
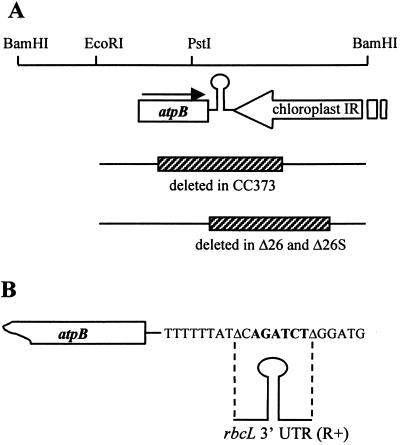
Construction of the plasmids pB17BS (atpB [wild-type]) and pΔ26 and of the corresponding Chlamydomonas strains P17 and Δ26 has been described previously (48). pΔ26 carries a deletion immediately downstream of the atpB coding region, extending from position 1490 (the stop codon is at position 1474) to position 3807, and the deleted bases are replaced by a 7-bp linker that includes a BglII site (Fig. 1B) (48). A fragment containing the 3′ UTR and flanking sequences of rbcL was previously described (36); this sequence was inserted in the BglII site of pΔ26, creating strain R+ (37).
FIG. 1.
The Chlamydomonas chloroplast atpB region and constructs used in this work. (A) Map of the 7.6-kb BamHI fragment of the Chlamydomonas chloroplast genome. A portion of the large IR of the chloroplast genome is shown as an open arrow. The inverted repeat downstream of the atpB coding region is shown as a stem-loop structure. The extents of the deletions in the chloroplast genomes of strains CC373, Δ26 and Δ26S are indicated by hatched boxes. (B) Detailed view of the sequence of the atpB 3′ UTR in Δ26 and Δ26S. The BglII site into which the rbcL 3′ UTR was inserted in strain R+ is shown in boldface type. The endpoints of the deletion in Δ26 and Δ26S are indicated by triangles.
Strain Δ26S was isolated during transformations used to create Δ26. Unlike Δ26, Δ26S grows robustly under photoautotrophic conditions and survives under high light (23). Δ26S was found to contain a recessive nuclear suppressor mutation, resulting in the accumulation of discrete 1.9- and 2.1-kb atpB transcripts that are 3′ end processed at cryptic sites and wild-type levels of the ATPase β-subunit (29).
Isolation of nucleic acids, filter hybridization, and PCR.
For nucleic acid preparations, cells were grown in 50 ml of HSA (high-salt medium containing acetate) to midlog phase. RNA and DNA were isolated as described previously (13, 36). For RNA filter hybridizations, 10 μg of total RNA was fractionated in 0.8% agarose–2.2 M formaldehyde gels, transferred to Amersham Hybond-N nylon membranes, and cross-linked by UV radiation. Prehybridization and hybridization were conducted in 50% formamide–5× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate)–10× Denhardt’s solution, 0.1% sodium dodecyl sulfate (SDS), and 0.1 mg of salmon sperm DNA per ml at 42°C. The blots were washed in 0.1× SSC–0.1% SDS at 65°C. Hybridization probes were generated by random priming in the presence of [α-32P]dATP, or with both [α-32P]dATP and [α-32P]dCTP for the experiment shown in Fig. 7. The BglII/EcoRI fragment of pΔ26 was used as an atpB probe, and a 5.8-kb EcoRI fragment was used to identify the psbA transcript (48). For rbcL, a PCR product covering nucleotides 2407 to 2620 (14) located in the coding region was used as a probe to detect 3′ end-processed mRNA. A PCR product extending from nucleotides 2775 to 2930 was used to identify unprocessed rbcL mRNA. The 3′ end of the mature mRNA is located at nucleotide 2677 (14). All quantification of 32P-labeled blots was carried out with a Fuji-Imaging Analyzer.
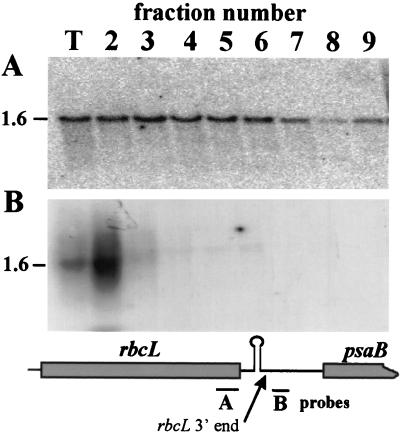
FIG. 7.
Distribution of rbcL mRNA in polysome gradient fractions. Lysates from wild-type cells were fractionated and analyzed as described in the legend to Fig. 4. The probes used in panels A and B are indicated on the map of the rbcL region shown at the bottom. Arrow, the 3′ end of mature rbcL mRNA.
Protein isolation and immunoblots.
Total proteins were resuspended in SDS denaturing sample buffer, fractionated in SDS-12% polyacrylamide gels, transferred to nitrocellulose membranes, and decorated with antibodies as described elsewhere (48). Antibodies directed against the chloroplast ATPase β-subunit (38) and the D1 protein of photosystem II (42) were used. Antigenic proteins were visualized by chemiluminescence and were quantitated by densitometric analysis (33).
Polysome fractionation.
To isolate total polysomes (4, 24) from Chlamydomonas cells, 30 ml of log-phase cells (2 × 106 cells/ml) was broken in a buffer containing 0.2 M Tris-HCl (pH 9)–0.2 M KCl–35 mM MgCl2–25 mM EGTA–0.2 M sucrose–1% Triton X-100–2% polyoxyethylene-10-tridecyl ether–0.5-mg/ml heparin–0.1-mg/ml chloramphenicol with a French press cell. Following centrifugation at 3,000 × g for 5 min, the supernatant was adjusted to 0.5% sodium-deoxycholate, incubated for 5 min on ice, and centrifuged for 15 min at 10,000 × g. Aliquots (1 ml) were layered onto 4 ml of 15 to 55% sucrose gradients in 40 mM Tris-HCl (pH 8.0)–20 mM KCl–10 mM MgCl2–0.5-mg/ml heparin–0.1-mg/ml chloramphenicol and centrifuged for 65 min at 45,000 rpm in a Beckman SW50 rotor. Ten fractions of 0.5 ml each were collected. The RNA of each fraction was purified by the addition of SDS to 0.5%, EDTA to 20 mM, phenol extraction, and precipitation with ethanol. Aliquots of each fraction were subjected to RNA blot analysis as described above. Since fractions 1 and 2 represent the buffer remaining from the sample loaded onto the gradient and are thus identical, only fraction 2 and subsequent fractions are presented in the figures. In control samples, polysomes were dissociated by the addition of EDTA (20 mM) to the algal lysates prior to gradient loading. In these gradients, 1 mM EDTA was substituted for 10 mM MgCl2.
RESULTS
Chlamydomonas strains containing altered atpB 3′ UTR sequences.
In order to test the relationship between 3′ end processing of mRNA and translational efficiency, we took advantage of four strains with unique 3′ processing properties for the atpB gene. The generation of these strains has been described previously (see Materials and Methods). The wild-type control strain was P17, which was created by transformation of the atpB deletion mutant CC373 (Fig. 1A) with a wild-type atpB gene. Strain Δ26 lacks nearly the entire atpB 3′ UTR and downstream sequences; these were replaced by a BglII linker, as shown in Fig. 1. This led to the accumulation of a heterogeneous and unstable set of atpB transcripts, weak photosynthetic growth, and sensitivity to high light. When the rbcL 3′ UTR was inserted into the BglII site of Δ26, normal photosynthetic growth as well as the accumulation of the atpB transcript was restored (36). Compared to the 1.9-kb length of the atpB transcript in wild-type cells, the atpB transcript harboring the 3′ UTR of the rbcL mRNA is 2.1 kb long (36). This strain was used to determine whether atpB sequences per se were required for 3′ UTR function or whether those of another gene could function equally well for the assays described in this paper. Finally, strain Δ26S was used. This strain is a derivative of Δ26 in which the chloroplast genome is unaltered, but there is a single, recessive nuclear mutation that permits the accumulation of reduced amounts (compared to wild-type cells) of a discrete atpB transcript, in spite of the deletion of the atpB 3′ UTR. We have hypothesized (29) that the gene mutated in Δ26S, CRP3, encodes a general chloroplast mRNA processing factor.
Variation of atpB transcript and ATPase β-subunit accumulation with 3′ UTR structure and function.
To measure the accumulation of all atpB transcripts in each strain described above, total RNA was fixed to filters by using a slot blot apparatus, and identical filters were hybridized with probes for atpB or with psbA as a loading control. Results from a typical hybridization are shown in Fig. 2A, and averaged results from several such experiments are shown in Fig. 3 (hatched bars). In agreement with previously obtained results (37, 47), atpB transcript accumulations relative to the wild-type strain were approximately 30% in Δ26, 45% in Δ26S, and 100% in R+. However, because of the technique used, it should be noted that hybridizing transcripts might not contain the entire atpB coding region or other parts of the message.
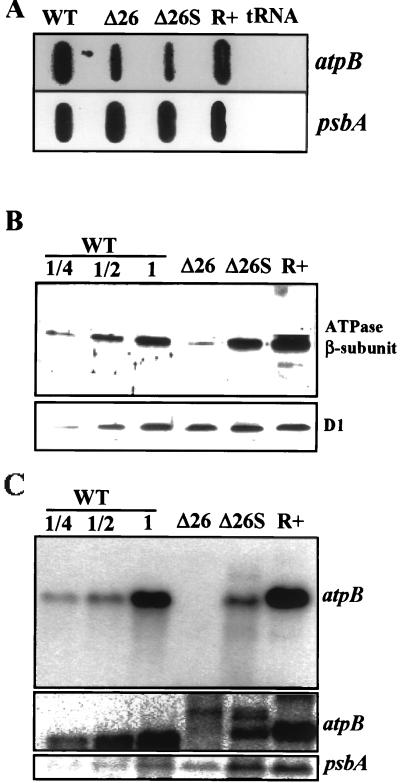
FIG. 2.
(A) Accumulation of total atpB transcripts. A 15-μg amount of total RNA from the indicated strains or yeast tRNA as a control was fixed to nylon filters with a slot blot apparatus, and identical filters were hybridized with 32P-labeled atpB and psbA probes. (B) Accumulation of the ATPase β-subunit. Total proteins from the indicated strains were fractionated by SDS-polyacrylamide gel electrophoresis, transferred to a nitrocellulose membrane, and incubated sequentially with antibodies directed against the ATPase β-subunit and the D1 protein of photosystem II. Antigenic proteins were visualized by chemiluminescence. (C) Accumulating discrete atpB transcripts. A 15-μg amount of total RNA from the indicated strains was analyzed by filter hybridization with sequential probing for atpB and psbA transcripts. The middle panel is a longer exposure (reproduced from a Fuji imager scan) which reveals the discrete processed transcripts in Δ26. Note that the size of atpB mRNA in the R+ transformant is 2.1 kb, compared to 1.9 kb in wild-type cells (36). WT, wild type.
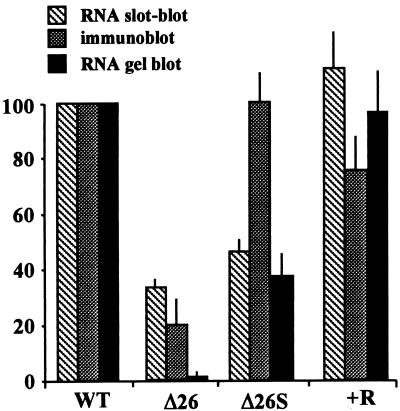
FIG. 3.
Quantification of atpB transcript and ATPase β-subunit protein levels. The total (slot blot) or discrete (gel blot) atpB transcript levels and the ATPase β-subunit levels were calculated from analyzing multiple (at least four) experiments as shown in Fig. 2. The signals were quantified with a Fuji-Imaging analyzer, and in both cases the level of the atpB transcript was normalized to that of the psbA RNA and is presented as a fraction of the amount in wild-type (WT) cells.
Δ26S was isolated as a spontaneous mutant that allowed rapid and high-light-tolerant photosynthetic growth (29). Since photosynthetic growth in Δ26 was limited by the synthesis of the ATPase β-subunit, we postulated that the restoration of rapid photosynthetic growth in Δ26S resulted from increased accumulation of the ATPase β-subunit. To measure the accumulation of the β-subunit in Δ26, Δ26S, and R+, total proteins were isolated from the same number of logarithmically growing cells and were subjected to immunoblot analysis with antibodies raised against the ATPase β-subunit or the D1 protein of the photosystem II reaction center as a loading control. As shown in Fig. 2B and 3 (stippled bars) and as reported previously (37, 48), the ATPase β-subunit accumulated to approximately 20% of the wild-type level in Δ26, fluctuating between 10 and 30%, depending on growth conditions (data not shown). However, in Δ26S and R+, the protein accumulated at or near the wild-type level. These higher levels of the ATPase β-subunit account for the wild-type photosynthetic growth characteristics of these strains.
Given these data and the fact that the two strains Δ26 and Δ26S have an identical atpB gene structure, it was of interest to determine the mechanism by which β-subunit accumulation was augmented in Δ26S relative to Δ26. Since we knew already that Δ26S, as opposed to Δ26, accumulates a population of homogeneous discrete transcripts (29), we used RNA gel blots to measure the relative amounts of these processed atpB transcripts in Δ26S. We use the term processed in this paper to indicate accumulating atpB transcripts of an approximately wild-type size; we infer that these transcripts are similarly 3′ end processed, since all atpB mRNA in each strain used in this study has no alterations at the atpB 5′ end.
Figures 2C and 3 (filled bars) show that the accumulation of processed atpB transcripts in Δ26S is 35 to 40% of the wild-type level, but only 1% or less in Δ26. This processed atpB RNA is of a length similar to that of the wild-type atpB transcript, although the 3′ end is located at a cryptic processing site inside the large chloroplast genome IR, since the normal atpB 3′ UTR has been deleted (Fig. 1) (29). A longer exposure of this gel (middle panel of Fig. 2C) shows that for Δ26, some discrete transcripts do accumulate, as well as a smear of transcripts both shorter and longer than wild-type atpB mRNA (this result is also observed with Δ26 cells suppressed by chloroplast gene amplification [23]). All of the data of Fig. 2 and 3 taken together suggest that the formation of atpB transcript with a distinct 3′ end is important for the accumulation of the ATPase-β subunit, which in turn facilitates photosynthetic growth. A hypothesis for the molecular mechanism of this phenomenon supported by these data is that 3′-end-processed atpB transcripts are more efficiently translated than their unprocessed and heterogeneous counterparts. An alternative explanation is that the formation of the mRNA 3′ end is not related to the translation efficiency and that the formation of approximately 35% more 3′-end-processed atpB transcript in Δ26S (Fig. 2 and 3) (28) is correlative rather than causal. In the second explanation, the suppression acts by increasing the translational efficiency of processed and unprocessed atpB transcripts compared to both wild-type and Δ26 cells. In order to distinguish between these two possibilities, we decided to determine the degree to which chloroplast polysomes are loaded with heterogeneous atpB transcripts versus 3′-end-processed transcripts.
Preferential polysome association of normally 3′-processed atpB transcripts.
The central question at this point of the work was that Δ26 and Δ26S accumulated similar amounts of atpB mRNA as measured by slot blots, yet Δ26S accumulated 4- to 10-fold more protein. While Δ26S does accumulate a greater amount of correctly processed atpB mRNA, as judged by RNA gel blots, Δ26 also accumulates a significant level of heterogeneous transcripts longer than the wild-type size, and these transcripts presumably include the entire atpB coding region. Thus, it was possible that certain types of atpB mRNA were preferentially translated.
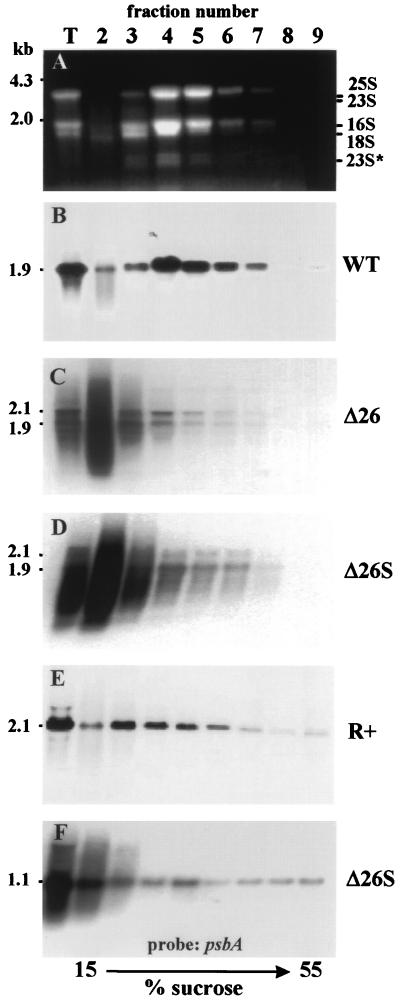
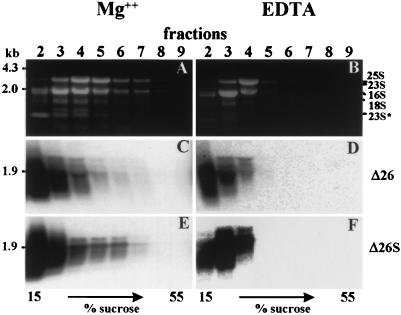
To measure the polysome association of atpB mRNAs in different strains, polysomal fractions derived from total cell lysates made in the presence of heparin, nonionic detergents, and chloramphenicol (to prevent runoff chloroplast translation) were sedimented in analytical sucrose gradients. RNA was isolated from 10 fractions and analyzed with RNA gel blots. Figure 4A shows the distribution of various rRNAs as revealed by ethidium bromide staining of each fraction and the total lysate (T). Since fractions 1 and 2 are identical and correspond to the 1 ml of cell lysate that was loaded on the gradient, results are presented only for fractions 2 to 9. The plastidic rRNAs (23S, 16S, and 23S*, an in vivo breakdown product of 23S rRNA [2, 28]) peak in fractions 4 and 5, indicating that a majority of polysomes are in these fractions. To verify that the rRNAs observed in fractions 4 to 7 are derived from polysomes, cell lysates were treated with EDTA and fractionated through EDTA-containing gradients. EDTA causes dissociation of polysomes into monosomes (2, 24). Indeed, when treated with EDTA, the rRNAs were no longer observed in fractions 5 to 7. Instead, they were concentrated in fractions near the top of the gradient (compare Fig. 5A and B).
FIG. 4.
Distribution of atpB transcripts in polysome gradients. Lysates from the strains indicated at the right were sedimented through analytical 15 to 55% sucrose gradients. Ten fractions were collected, and the RNA purified from the lysate of whole cells (T) as well as fractions 2 to 9 was assayed for atpB mRNA by RNA gel blot hybridization. (A) Ethidium bromide staining of the gel for wild-type cells to reveal the distribution of the rRNAs (similar gels were obtained for other strains but are not shown). 23S* is an in vivo breakdown product of plastid 23S rRNA (28). The migrations of DNA molecular weight markers in panel A and the atpB transcript (1.9 kb in the wild type [WT] and 2.1 kb in R+ [36]) are indicated at the left. (F) Gradient fractions derived from Δ26S (the ones shown in panel D) probed with a psbA-specific fragment. The relative exposure times for the panels were as follows: C > D > B = E = F.
FIG. 5.
Distribution of atpB transcripts in polysome gradients with or without EDTA. Lysates from Δ26 (A to D) and Δ26S (E and F) were sedimented and analyzed as described in the legend to Fig. 4. EDTA treatment is described in Materials and Methods.
We then analyzed the distribution of atpB transcripts between the nonpolysomal (fractions 2 to 4) and polysomal (fractions 4 to 7) fractions. In wild-type cells (Fig. 4B), most of the atpB transcripts were in fractions 4 to 6 and therefore polysome associated. In contrast, the majority of Δ26 atpB transcripts, irrespective of size, appeared as a smear in fraction 2 and were therefore nonpolysomal (Fig. 4C). The prolonged exposure of the blot in this panel (see also Fig. 2C) revealed the presence of two processed atpB transcripts of 1.9 and 2.1 kb in the polysomal fractions. These two 3′-end-processed atpB transcripts in Δ26 and Δ26S have been recently characterized (29). This result suggested that the ATPase β-subunit is translated in Δ26 cells from the 1.9- and 2.1-kb transcripts, rather than from the ones of variable size. As a control, EDTA treatment was used, and this treatment resulted in all atpB transcripts migrating in the nonpolysomal region of the gradient (Fig. 5C and D).
It was formally possible that the reduced amount of ATPase β-subunit in Δ26 cells resulted from the deletion of 2 kb downstream of the atpB gene, rather than from the low level of processed transcripts. To address this issue, we analyzed the polysome distribution of atpB transcripts in Δ26S and R+. Δ26S contains the same deletion as Δ26 but an increased amount of processed RNA, while R+ contains the deletion but also the inserted 3′ UTR of rbcL. Due to the high instability of the atpB transcripts in Δ26S cells, we repeatedly obtained a poor yield of polysomal transcripts. Figure 4D shows that in Δ26S, only processed (and some degraded) atpB transcripts were detected in the polysomal fractions, much as in Δ26. EDTA treatment caused slower sedimentation of these transcripts (Fig. 5E and F). In these two related strains, therefore, processed atpB transcripts are preferentially associated with polysomes. We conclude that although the total amounts of atpB transcript are similar in Δ26 and Δ26S, the increased amount of processed transcript in Δ26S is responsible for its wild-type protein level and normal photosynthetic growth.
R+ was used to see whether the presence of 3′ end processing signals from another gene could fully restore polysomal localization of atpB mRNA. Figure 4E shows that most atpB transcripts (which are 2.1 kb long in R+ [36]) are polysome associated in R+, similar to the distribution in wild-type cells (Fig. 4B), although slightly skewed toward more slowly migrating fractions. However, the nonpolysomal fraction 2 contained very little atpB mRNA. This result suggests that 3′ end processing strongly enhances polysome association of atpB transcripts.
Lack of nonspecific RNA degradation in polysomal fractions.
In order to verify that the heterogeneity of atpB transcripts in cell lysates of Δ26 and Δ26S is due to the lack of a stem-loop structure in the 3′ UTR and not because of general degradation activity, the gradient of Δ26S shown in Fig. 4D was reprobed with a psbA-specific fragment (Fig. 4F). As described before, the 1.1-kb psbA transcript is distributed between polysomal and nonpolysomal fractions (2, 11, 24, 25). A very small amount of RNA degradation was observed for the psbA transcript in the lysates obtained from Δ26S (Fig. 4F) or Δ26 (not shown) cells. Therefore, we concluded that transcripts other than the atpB are not subject to increased degradation in these strains relative to wild-type cells. However, some of the low-molecular-weight atpB transcripts observed in fraction 2 resulted from degradation during preparation of the cell lysates. In Δ26, atpB transcripts are highly labile in vivo because of the lack of a stem-loop structure (48). In addition, polysome fractionation subjects the RNA to additional manipulations as compared to direct phenol extraction (used, for example, for Fig. 2C). The appearance of Δ26 and Δ26S atpB transcripts longer than 2 kb in fraction 2 and not in the polysomal fractions indicates that at least some of the atpB transcripts observed in fraction 2 are unprocessed rather than degradation intermediates. Moreover, while probing the polysome gradient blots such as those presented in Fig. 4 with gene-specific probes, we often obtained transcripts longer than the mature size only in the nonpolysomal fractions 2 and 3 (see, for example, Fig. 4F). This hybridization signal could result from longer unprocessed transcripts that are not associated with ribosomes. No such hybridization signals were obtained in polysomal fractions 4 and 5 (Fig. 4F).
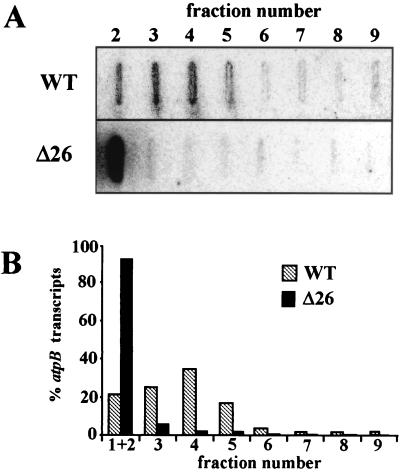
To ascertain independently that unprocessed transcripts not detected by RNA blot analysis were localized at the top of the gradient, total RNA from the polysome gradient fractions was extracted and analyzed by slot blot hybridization, as shown in Fig. 6A. These data clearly show that the vast majority of Δ26 atpB mRNA is found at the top of the gradient, while for wild-type cells it is primarily in the faster-sedimenting fractions. To quantify these results, the total amount of atpB mRNA in all fractions was set at 100%, and the distribution was calculated accordingly. The results presented in Figure 6B demonstrate that more than 98% of the atpB transcripts in Δ26 were found in fractions 1 to 3, with the remaining 1% in the polysomal fractions 4 to 6. In contrast, in wild-type cells more than half of the transcripts were found in fractions 4 and higher. These results further confirm our interpretations derived from the data shown in Fig. 4 and 5.
FIG. 6.
Quantification of total atpB transcripts in polysome gradient fractions. (A) Distribution of total atpB transcripts in polysome gradient fractions of the indicated strains was analyzed as described in the legend to Fig. 4, except that slot blot rather than gel blot analysis was used. (B) The signals were quantified with a Fuji-Imaging analyzer, and in both cases the level of the atpB transcript in each fraction is presented as a fraction of the total amount of atpB transcript, which was set to 100% (B). Fraction 1 was omitted for the reasons described in the text. WT, wild type.
Polysomal distribution of processed and unprocessed rbcL transcripts.
To determine whether the preferential polysomal association of 3′-end-processed mRNA also occurs with other chloroplast transcripts in Chlamydomonas, we examined the distribution of wild-type rbcL mRNA. To detect possible unprocessed transcripts, hybridization probes were prepared either from the 3′ end of the coding region, which should detect all rbcL transcripts, or from the sequences immediately downstream of the mature 3′ end, which should detect only unprocessed RNA. These probes are indicated as A and B, respectively, at the bottom of Fig. 7. Probe B lies between the 3′ end of the mature rbcL mRNA and that of psaB, which is downstream and on the opposite strand from rbcL (14) and encodes a 2.6-kb transcript (26). When polysome gradient fractions were analyzed for the presence of rbcL transcripts with probe A, the 1.6-kb mature mRNA was detected in all fractions, as shown in Fig. 7A, with the majority in the polysomal fractions. In contrast, probe B identified a heterogeneous set of transcripts of an average size slightly larger than that of the mature rbcL message, as shown in Fig. 7B. This hybridization occurred almost entirely in the nonpolysomal fraction 2. In order to ascertain that this hybridization signal was indeed obtained from unprocessed rbcL transcripts, a similar blot was probed with labeled antisense RNA corresponding to the sequence of probe B. The data obtained with this probe were identical to those shown in Fig. 7B. These results suggest that the lack of association of incompletely 3′-end-processed transcripts with polysomes may be a general phenomenon in Chlamydomonas chloroplasts.
DISCUSSION
The data presented here suggest that 3′ end processing may be required for translation of atpB and rbcL mRNAs in Chlamydomonas chloroplasts. Unprocessed atpB transcripts, defined as those that do not accumulate as an abundant size class of approximately 2 kb, were only present in nonpolysomal fractions. Processed mRNAs were present in both polysomal and nonpolysomal fractions. Since the 3′ ends of most chloroplast transcripts are generated from longer pre-mRNAs by exo- and/or endonucleolytic mechanisms (17, 36, 37, 44, 47), this 3′ processing apparatus may interact with or signal the translational machinery.
Our ability to detect a heterogeneous collection of putative processing intermediates or incorrectly processed transcripts for atpB and rbcL suggests that these molecules are relatively stable in the chloroplast. When they were analyzed by RNase protection, it was possible to detect partially processed transcripts in the Chlamydomonas chloroplast petD-trnR region (29), and in certain mutant backgrounds, intermediates in psaA and psbD processing, including psbD 3′ end processing, can also be readily detected (7). In land plants and Euglena, many intron-containing transcripts accumulate in chloroplasts (3, 9, 53), as well as ribosomal operon processing intermediates (1, 2, 34, 52). 16S rRNA processing intermediates are highly abundant in the maize nuclear mutant hcf7 (2), again indicating that such molecules are not necessarily unstable in chloroplasts. Partially processed mRNAs can also be loaded onto ribosomes, as has been shown for psbB operon mRNAs in maize (3).
The partitioning of mature chloroplast transcripts between polysomal and nonpolysomal fractions has been noted previously and can vary with growth conditions and the gene which is analyzed (25). In particular, psbA mRNA can be present to a large degree in nonpolysomal fractions. In the soluble phase of barley plastids, for example, nearly all psbA mRNA is nonpolysomal, with a higher proportion on polysomes isolated from membrane fractions (25). Spinach amyloplast ribosomes discriminate among mRNAs, with psbA being among those remaining nonpolysomal (11). In Chlamydomonas, the distribution of the psbA and rbcL mRNAs between thylakoids and stroma was found to fluctuate during the cell cycle (6). Thus, ribosome loading of mRNAs in chloroplasts appears to be a tightly regulated process.
The interaction of 3′ end processing with the translation machinery could ensure translation of only mature and full-length transcripts. Indeed, it is now well-established for nucleus-encoded transcripts that the poly(A) tail together with the poly(A)-binding protein is essential for translation initiation on 80S ribosomes (10, 16, 18, 40, 43). To explain this phenomenon, recent models have been presented in which the mRNA is drawn as a circle with the poly(A)-tail connected via the poly(A) binding protein to the translation initiation complex (10, 16, 18, 40). In fact, electron micrographs of cells actively synthesizing secreted peptide hormones show that the great majority of membrane-bound polysomes are circular (8).
In prokaryotes, transcription and translation are often coupled. The chloroplast translation apparatus in many respects resembles the prokaryotic system but also has eukaryotic characteristics (45). If an equivalent to the poly(A) tail-poly(A)-binding protein-mediated translation activation mechanism exists in chloroplasts, it must involve elements other than the poly(A) tail, which actually destabilizes chloroplast transcripts (27, 30–32). One candidate element would be the 3′-end stem-loop structure and/or proteins which bind in this region. However, the results presented here do not favor such an hypothesis, since in Δ26S cells, which produce wild-type levels of the ATPase β-subunit, the atpB gene lacks 2 kb of the 3′ UTR, including the stem-loop-forming sequences and the authentic 3′-end processing site (29). This observation can be reconciled in two ways: either the nuclear crp3 mutation in Δ26S cells overcomes the need for the wild-type 3′ UTR in terms of translation, or it is transcript length and/or the 3′ processing mechanism per se that confers translatability to atpB transcripts. In vitro chloroplast translation systems (19, 20) may help in resolving the role of 3′ end processing in atpB translation.
At least one case in which 3′-end processing is required for translation has been documented in prokaryotes. The E. coli R1 plasmid hok mRNA, which mediates plasmid stabilization by killing of plasmid-free segregants, is translated only following 3′ end processing (51). hok mRNA is folded in such a way that the unprocessed 3′ end and the 5′ end hybridize, inhibiting ribosome binding. Following 3′ end processing, the 5′ end becomes available to ribosomes (15). Whether this is a special or a more general mechanism in bacteria remains to be determined, and no information is available on whether long-range intramolecular interactions occur in chloroplast mRNAs. Chloroplast transformation in Chlamydomonas or tobacco offers a promising methodology for testing these and related possibilities.
ACKNOWLEDGMENTS
This work was supported by United States-Israel Binational Agricultural Research and Development Fund grant nos. US-2207-92 and US-2746-96 and by United States-Israel Binational Science Foundation grant no. 96-00418. H.L. was supported by an NSF grant to D.B.S.
Footnotes
This paper is dedicated to Robert Drager, who passed away on 30 March 1998.
REFERENCES
- 1.Audren H, Bisanz-Seyer C, Briat J F, Mache R. Structure and transcription of the 5S ribosomal RNA gene from spinach chloroplasts. Curr Genet. 1987;12:263–270. doi: 10.1007/BF00435288. [DOI] [PubMed] [Google Scholar]
- 2.Barkan A. Nuclear mutants of maize with defects in chloroplast polysome assembly have altered chloroplast RNA metabolism. Plant Cell. 1993;5:389–402. doi: 10.1105/tpc.5.4.389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barkan A. Proteins encoded by a complex chloroplast transcription unit are each translated from both monocistronic and polycistronic RNAs. EMBO J. 1988;7:2637–2644. doi: 10.1002/j.1460-2075.1988.tb03116.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barkan A. Tissue-dependent plastid RNA splicing in maize: transcripts from four plastid genes are predominantly unspliced in leaf meristems and roots. Plant Cell. 1989;1:437–446. doi: 10.1105/tpc.1.4.437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Blowers A D, Klein U, Ellmore G S, Bogorad L. Functional in vivo analyses of the 3′ flanking sequences of the Chlamydomonas chloroplast rbcL and psaB genes. Mol Gen Genet. 1993;238:339–349. doi: 10.1007/BF00291992. [DOI] [PubMed] [Google Scholar]
- 6.Breidenbach E, Jenni E, Boschetti A. Synthesis of two proteins in chloroplasts and mRNA distribution between thylakoids and stroma during the cell cycle of Chlamydomonas reinhardtii. Eur J Biochem. 1988;177:225–232. doi: 10.1111/j.1432-1033.1988.tb14366.x. [DOI] [PubMed] [Google Scholar]
- 7.Choquet Y, Goldschmidt-Clermont M, Girard-Bascou J, Kueck U, Bennoun P, Rochaix J D. Mutant phenotypes support a trans-splicing mechanism for the expression of the tripartite psaA gene in the Chlamydomonas reinhardtii chloroplast. Cell. 1988;52:903–914. doi: 10.1016/0092-8674(88)90432-1. [DOI] [PubMed] [Google Scholar]
- 8.Christensen A K, Kahn L E, Bourne C M. Circular polysomes predominate on the rough endoplasmic reticulum of somatotropes and mammotropes in the rat anterior pituitary. Am J Anat. 1987;178:1–10. doi: 10.1002/aja.1001780102. [DOI] [PubMed] [Google Scholar]
- 9.Copertino D W, Hallick R B. Group II twintron: an intron within an intron in a chloroplast cytochrome b-559 gene. EMBO J. 1991;10:433–442. doi: 10.1002/j.1460-2075.1991.tb07965.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Craig A, Haghighat A, Yu A, Sonenberg N. Interaction of polyadenylate-binding protein with eIF4G homologue PAIP enhances translation. Nature. 1998;392:520–523. doi: 10.1038/33198. [DOI] [PubMed] [Google Scholar]
- 11.Deng X, Gruissem W. Constitutive transcription and regulation of gene expression in non-photosynthetic plastids of higher plants. EMBO J. 1988;7:3301–3308. doi: 10.1002/j.1460-2075.1988.tb03200.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Drager, R. G., and D. B. Stern. Chloroplast RNA synthesis and processing. In J.-D. Rochaix, M. Goldschmidt-Clermont, and S. Merchant (ed.), Molecular biology of chlamydomonas: chloroplasts and mitochondria, in press. Kluwer Academic Publishers, Dordrecht, The Netherlands.
- 13.Drager R G, Zeidler M, Simpson C L, Stern D B. A chloroplast transcript lacking the 3′ inverted repeat is degraded by 3′→5′ exoribonuclease activity. RNA. 1996;2:652–663. [PMC free article] [PubMed] [Google Scholar]
- 14.Dron M, Rahire M, Rochaix J-D. Sequence of the chloroplast DNA region of Chlamydomonas reinhardtii containing the large subunit of riboluse bisphosphate carboxylase and part of its flanking genes. J Mol Biol. 1982;162:775–793. doi: 10.1016/0022-2836(82)90547-2. [DOI] [PubMed] [Google Scholar]
- 15.Franch T, Gerdes K. Programmed cell death in bacteria: translational repression by mRNA end-pairing. Mol Microbiol. 1996;21:1049–1060. doi: 10.1046/j.1365-2958.1996.771431.x. [DOI] [PubMed] [Google Scholar]
- 16.Gallie D R. Translation control of cellular and viral mRNAs. Plant Mol Biol. 1996;32:145–158. doi: 10.1007/BF00039381. [DOI] [PubMed] [Google Scholar]
- 17.Hayes R, Kudla J, Schuster G, Gabay L, Maliga P, Gruissem W. Chloroplast mRNA 3′-end processing by a high molecular weight protein complex is regulated by nuclear encoded RNA binding proteins. EMBO J. 1996;15:1132–1141. [PMC free article] [PubMed] [Google Scholar]
- 18.Hentze M W. eIF4G: a multipurpose ribosome adapter? Science. 1997;275:500–501. doi: 10.1126/science.275.5299.500. [DOI] [PubMed] [Google Scholar]
- 19.Hirose T, Sugiura M. Both RNA editing and RNA cleavage are required for translation of tobacco ndhD mRNA: a possible regulatory mechanism for the expression of chloroplast operon consisting of functionally unrelated genes. EMBO J. 1997;16:6804–6811. doi: 10.1093/emboj/16.22.6804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hirose T, Sugiura M. cis-acting elements and trans-acting factors for accurate translation of chloroplast psbA mRNAs: development of an in vitro translation system from tobacco chloroplasts. EMBO J. 1996;15:1687–1695. [PMC free article] [PubMed] [Google Scholar]
- 21.Jackson R J, Standart N. Do the poly(A) tail and 3′ untranslated region control mRNA translation? Cell. 1990;62:15–24. doi: 10.1016/0092-8674(90)90235-7. [DOI] [PubMed] [Google Scholar]
- 22.Keus R J A, Dekker A F, Kreuk K C J, Groot G S P. Transcription of ribosomal DNA in chloroplasts of Spirodela oligorhiza. Curr Genet. 1986;9:91–98. doi: 10.1007/BF00396209. [DOI] [PubMed] [Google Scholar]
- 23.Kindle K L, Suzuki H, Stern D B. Gene amplification can correct a photosynthetic growth defect caused by mRNA instability in Chlamydomonas chloroplasts. Plant Cell. 1994;6:187–200. doi: 10.1105/tpc.6.2.187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Klaff P, Gruissem W. Changes in chloroplast mRNA stability during leaf development. Plant Cell. 1991;3:517–530. doi: 10.1105/tpc.3.5.517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Klein R R, Mason H S, Mullet J E. Light-regulated translation of chloroplast proteins. I. Transcripts of psaA-psaB, psbA, and rbcL are associated with polysomes in dark-grown and illuminated barley seedlings. J Cell Biol. 1988;106:289–301. doi: 10.1083/jcb.106.2.289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kuck U, Choquet Y, Schneider M, Dron M, Bennoun P. Structural and transcription analysis of two homologous genes for the p700 chlorophyll a-apoproteins in Chlamydomonas reinhardtii: evidence for in vivo trans-splicing. EMBO J. 1987;6:2185–2196. doi: 10.1002/j.1460-2075.1987.tb02489.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kudla J, Hayes R, Gruissem W. Polyadenylation accelerates degradation of chloroplast mRNA. EMBO J. 1996;15:7137–7146. [PMC free article] [PubMed] [Google Scholar]
- 28.Leaver C J. Molecular integrity of chloroplast ribosomal ribonucleic acid. Biochem J. 1973;135:237–240. doi: 10.1042/bj1350237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Levy H, Kindle K L, Stern D B. A nuclear mutation that affects the 3′ processing of several mRNAs in Chlamydomonas reinhardtii chloroplasts. Plant Cell. 1997;9:825–836. doi: 10.1105/tpc.9.5.825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lisitsky I, Klaff P, Schuster G. Addition of poly(A)-rich sequences to endonucleolytic cleavage sites in the degradation of spinach chloroplast mRNA. Proc Natl Acad Sci USA. 1996;93:13398–13403. doi: 10.1073/pnas.93.23.13398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lisitsky I, Klaff P, Schuster G. Blocking polyadenylation of mRNA in the chloroplast inhibits its degradation. Plant J. 1997;12:1173–1178. [Google Scholar]
- 32.Lisitsky I, Kotler A, Schuster G. The mechanism of preferential degradation of polyadenylated RNA in the chloroplast: the exoribonuclease 100RNP/PNPase displays high binding affinity for poly(A) sequence. J Biol Chem. 1997;272:17648–17653. doi: 10.1074/jbc.272.28.17648. [DOI] [PubMed] [Google Scholar]
- 33.Lisitsky I, Liveanu V, Schuster G. RNA-binding characteristics of a ribonucleoprotein from spinach chloroplast. Plant Physiol. 1995;107:933–941. doi: 10.1104/pp.107.3.933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.McGarvey P, Helling R B. Processing of chloroplast ribosomal RNA transcripts in Euglena gracilis bacillaris. Curr Genet. 1989;15:363–370. doi: 10.1007/BF00419917. [DOI] [PubMed] [Google Scholar]
- 35.Rochaix J-D. Post-transcriptional regulation of chloroplast gene expression in Chlamydomonas reinhardtii. Plant Mol Biol. 1996;32:327–341. doi: 10.1007/BF00039389. [DOI] [PubMed] [Google Scholar]
- 36.Rott R, Drager R G, Stern D B, Schuster G. The 3′ untranslated regions of chloroplast genes in Chlamydomonas reinhardtii do not serve as efficient transcriptional terminators. Mol Gen Genet. 1996;252:676–683. doi: 10.1007/BF02173973. [DOI] [PubMed] [Google Scholar]
- 37.Rott R, Liveanu V, Drager R G, Stern D B, Schuster G. The sequence and structure of the 3′ untranslated regions of chloroplast transcripts are important determinants of mRNA accumulation and stability. Plant Mol Biol. 1998;36:307–314. doi: 10.1023/a:1005943701253. [DOI] [PubMed] [Google Scholar]
- 38.Rott R, Nelson N. Purification and immunological properties of proton-ATPase complexes from yeast and rat liver mitochondria. J Biol Chem. 1981;256:9224–9228. [PubMed] [Google Scholar]
- 39.Sachs A B, Davis R W. The polyadenylic acid binding protein is required for polyadenylic acid shortening and 60S ribosomal subunit-dependent translation initiation. Cell. 1989;58:857–868. doi: 10.1016/0092-8674(89)90938-0. [DOI] [PubMed] [Google Scholar]
- 40.Sachs A B, Sarnow P, Hentze M W. Starting at the beginning, middle, and end: translation initiation in eukaryotes. Cell. 1997;89:831–838. doi: 10.1016/s0092-8674(00)80268-8. [DOI] [PubMed] [Google Scholar]
- 41.Sarkar N. Polyadenylation of mRNA in prokaryotes. Annu Rev Biochem. 1997;66:173–197. doi: 10.1146/annurev.biochem.66.1.173. [DOI] [PubMed] [Google Scholar]
- 42.Schuster G, Timberg R, Ohad I. Turnover of photosystem II proteins during photoinhibition of Chlamydomonas reinhardtii. Eur J Biochem. 1988;177:403–410. doi: 10.1111/j.1432-1033.1988.tb14389.x. [DOI] [PubMed] [Google Scholar]
- 43.Standart N, Jackson R J. Regulation of translation by specific protein/mRNA interactions. Biochimie. 1994;76:867–879. doi: 10.1016/0300-9084(94)90189-9. [DOI] [PubMed] [Google Scholar]
- 44.Stern D B, Gruissem W. Control of plastid gene expression: 3′ inverted repeats act as mRNA processing and stabilizing elements, but do not terminate transcription. Cell. 1987;51:1145–1157. doi: 10.1016/0092-8674(87)90600-3. [DOI] [PubMed] [Google Scholar]
- 45.Stern D B, Higgs D C, Yang J. Transcription and translation in chloroplasts. Trends Plant Sci. 1997;2:308–315. [Google Scholar]
- 46.Stern D B, Jones H, Gruissem W. Function of plastid mRNA 3′ inverted repeats: RNA stabilization and gene-specific protein binding. J Biol Chem. 1989;264:18742–18750. [PubMed] [Google Scholar]
- 47.Stern D B, Kindle K L. 3′ end maturation of the Chlamydomonas reinhardtii chloroplast atpB mRNA is a two-step process. Mol Cell Biol. 1993;13:2277–2285. doi: 10.1128/mcb.13.4.2277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Stern D B, Radwanski E R, Kindle K L. A 3′ stem/loop structure of the Chlamydomonas chloroplast atpB gene regulates mRNA accumulation in vivo. Plant Cell. 1991;3:285–297. doi: 10.1105/tpc.3.3.285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sugita M, Sugiura M. Regulation of gene expression in chloroplasts of higher plants. Plant Mol Biol. 1996;32:315–326. doi: 10.1007/BF00039388. [DOI] [PubMed] [Google Scholar]
- 50.Tarum S Z, Sachs A B. Association of yeast poly(A) tail binding protein with translation initiation factor eIF-4G. EMBO J. 1996;15:7168–7177. [PMC free article] [PubMed] [Google Scholar]
- 51.Thisted T, Nielsen A K, Gerdes K. Mechanism of postsegregational killing: translation of hok, srnB, and pnd mRNAs of plasmids R1, F and R483 is activated by 3′ end processing. EMBO J. 1994;13:1950–1959. doi: 10.1002/j.1460-2075.1994.tb06464.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Vera A, Yokoi F, Sugiura M. The existence of pre-mature 16S rRNA species in plastid ribosomes. FEBS Lett. 1993;327:29–31. doi: 10.1016/0014-5793(93)81032-u. [DOI] [PubMed] [Google Scholar]
- 53.Westhoff P, Herrmann R G. Complex RNA maturation in chloroplasts: the psbB operon from spinach. Eur J Biochem. 1988;171:551–564. doi: 10.1111/j.1432-1033.1988.tb13824.x. [DOI] [PubMed] [Google Scholar]