Abstract
Community health centers serve ethnically diverse populations that may pose challenges for record linkage based on name and date of birth. The objective was to identify an optimal deterministic algorithm to link patient encounters and laboratory results for hemoglobin A1c testing and examine its variability by health center site, patient ethnicity, and other variables. Based on data elements of last name, first name, date of birth, gender, and health center site, matches with ≥50% to < 100% of a maximum score were manually reviewed for true matches. Match keys based on combinations of name substrings, date of birth, gender, and health center were used to link encounter and laboratory files. The optimal match key was the first two letters of the last name and date of birth, which had a sensitivity of 92.7% and a positive predictive value of 99.5%. Sensitivity marginally varied by health center, age, gender, but not by ethnicity. An algorithm that was inexpensive, accurate, and easy to implement was found to be well suited for population-based measurement of clinical quality.
The objective of the current study was to apply an efficient, low-cost record linkage protocol for computerized data of ethnically diverse patients with diabetes served by seven community health centers in the United States. The protocol was originally developed to match Dutch cancer patients to a national population registry,1 and it was not known how the reported optimal match key would perform in a U.S. population and in subgroups of ethnicity and other demographic variables.
Background
The Community Health Center Network (CHCN) is a nonprofit organization founded in 1997 as a partnership of seven community health centers in Alameda County, CA. The CHCN provides operational support services such as claims processing and support for information technology. The health centers provide primary care for approximately 80,000 patients who are largely low-income Latinos, Asians, and African Americans. The CHCN has had an active clinical quality improvement program led by the medical directors at each community health center. For an annual audit of clinical quality, the medical directors have selected the percentage of patients with diabetes who are being monitored for glycemic control.2 Adapting criteria from the Health Plan Employer Data Information Set (HEDIS),3 this is defined by one or more laboratory tests of hemoglobin A1c during a measurement year in patients who have had two or more encounters with a diabetes diagnosis in the measurement year or preceding year.
Design Objectives
Computerized disease registries have emerged as an important tool for the clinical management of patients with chronic diseases during the primary care encounter and for population-based measurement of health care quality.4 Highly functional registries should be able to integrate information on encounters, laboratory results, pharmacy medications, visits to the emergency department, and hospitalizations. For measurement of clinical performance, record linkage can minimize or eliminate labor-intensive review of medical charts. Despite the availability of sophisticated and accurate record linkage software, there are significant technical, administrative, and financial barriers in linking clinical databases.
Most matching algorithms are probabilistic or deterministic. Probabilistic methods use scoring and the assigning of weights to each pairing of common data elements in two files. Weights based on exact odds further require that the files be initially analyzed to determine the frequencies of the individual elements in the files. Thus, probabilistic methods require advanced technical skills, sophisticated computing, and potentially expensive software.5,6,7,8 Deterministic matching uses a match key by joining all or some of the characters in names and dates and optionally appending codes for gender, postal office or address, race/ethnicity, marital status, or other personal characteristics. Compared with probabilistic methods, deterministic methods may not generate the highest possible yield of true matches nor minimize false positives. Because deterministic matching is simpler than other methods, a broader array of organizations may find this approach suited to the capabilities of their existing staff and computer resources.
Common enhancements to matching of names include standardizing phonetic or orthographic components and the use of nickname dictionaries. Recent research has demonstrated the utility of approximate string comparators to match names that contain typographical errors; letters that are inserted, deleted, or transposed; and permutated first, middle, or family names.9,10 These methods (e.g., Jaro-Winkler, longest common substring, Levenshtein edit distance) provide a measure of similarity between two strings and, when combined with deterministic matching, have been shown to increase the sensitivity of matching by as much as 10% compared with deterministic matching alone.9 Other enhancements include algorithms to break ties when several records in one file link to the same record in a different file.11 Some of these algorithms use information on the logical ordering of dates (e.g., date of birth, date of death) and other logical associations (e.g., a mother's residence is the same as her infant's), when such data are available.
A common practice for identifying the optimal key in deterministic matching employs trial-and-error combinations of first name, last name, middle name or initial; day, month, and year of birth; and other data elements that may be available. Progressively increasing and decreasing the first nth number of characters of the first name, last name, in combination with the day-month-year components of date of birth leads to hundreds of permutations. Additional data elements may be included or excluded if they improve the accuracy of matching.
Rather than embark on a time-consuming process of trial and error for matching patients with diabetes in the CHCN data warehouse with a file of laboratory results for hemoglobin A1c, the CHCN tested a protocol for matching of two files based on first name, last name, date of birth, gender, and postal code described by Van den Brandt et al.1 These researchers found an optimal match key composed of the date of birth, first four letters of the family name, and gender. This key had a sensitivity of 97.9% and a positive predictive value (PPV) of approximately 97.9%. Compared with other protocols identified in a literature review, the CHCN selected this one because it was simple and could be implemented with database programs and statistical applications already available at the CHCN. Robust testing of the protocol or match key has not been reported, and the CHCN explored its applicability in its patient population, which has a diverse racial and ethnic background.
System Description
Patient File
Each health center of the CHCN operates a computerized practice management system that generates a standardized computer file on patient encounters each quarter. These files include patient first name, last name, date of birth, gender, ethnicity, payer, and diagnosis code using the International Classification of Diseases, 9th edition, Clinical Modification (ICD-9-CM), treatment code using the Current Procedural Terminology (CPT-4), and date of service. The quarterly files are loaded into database tables of a data warehouse at the CHCN. For the 2002 annual audit of glycemic control, the encounter file consisted of 4,377 unique patient identification numbers associated with two or more dates of service with an ICD code (250) for diabetes from October 1, 2000, to September 30, 2002. This file was complete for identifiers with first and last name, date of birth, and gender, but had some missing data for race/ethnicity and payer of services at the last encounter. This file also had an unknown, but small percentage of duplicated patients. In the encounter file, patients with known race/ethnicity were mostly Asian, Latino, and other nonwhite groups (▶).
Table 1.
Demographic Characteristics and Missing Data of Patients in the Encounter File (N = 4,377) and Laboratory File (N = 3,806)
| Encounter |
Laboratory |
|||
|---|---|---|---|---|
| Item | No. | % | No. | % |
| Health center | ||||
| Missing | 0 | * | 1 | |
| A | 1,376 | 31 | 1,115 | 36 |
| B | 1,171 | 27 | 708 | 23 |
| C | 906 | 21 | 679 | 22 |
| D | 355 | 8 | 233 | 7 |
| E | 254 | 6 | 101 | 4 |
| F | 177 | 4 | 135 | 4 |
| G | 138 | 3 | 114 | 4 |
| Age (date of birth) | ||||
| Missing | 1 | 15 | ||
| <25 | 68 | 2 | 81 | 3 |
| 25–44 | 682 | 16 | 536 | 17 |
| 45–64 | 1,795 | 41 | 1,306 | 43 |
| 65+ | 1,831 | 42 | 1,148 | 37 |
| Gender | ||||
| Missing | 1 | 20 | ||
| Male | 1,569 | 36 | 1,076 | 35 |
| Female | 2,807 | 64 | 1,990 | 65 |
| Ethnicity/race | ||||
| Missing | 792 | |||
| Asian/Pacific Is. | 1,331 | 37 | ||
| Latino (Hispanic of any race) | 1,303 | 36 | ||
| African American | 537 | 15 | ||
| White (non-Hispanic) | 324 | 9 | ||
| Native American | 90 | 3 | ||
| Payer | ||||
| Missing | 452 | |||
| Medicare | 1,415 | 36 | ||
| Uninsured | 1,322 | 34 | ||
| Medicaid | 906 | 23 | ||
| Commercial | 192 | 5 | ||
| Other | 90 | 2 | ||
Missing data excluded from calculations of percentages.
Laboratory File
The community health centers contract with a commercial laboratory to provide specialized analytical diagnostic tests, including glycosylated hemoglobin. The laboratory's information system for requisition and notification of results cannot access unique identifiers of patients in practice management systems. Patient first name, last name, date of birth, and gender are entered onto paper requisition forms. Clinics can access the laboratory's database to look up the results of tests as soon as the results are known. The CHCN and each health center have also received electronic files that include first and last name, date of birth, and gender along with the numerical results of the test and date of requisition (service). The electronic file is loaded into database tables in the CHCN data warehouse. For the 2002 audit, the laboratory file consisted of all patients (n = 3,086) who had at least one hemoglobin A1c test from October 1, 2000, to September 30, 2002. This file was created using a patient key based on the first ten letters of the last name, first five letters of the first name, date of birth, and gender (e.g., SMITHSEBAS19501124M). This highly specific key was applied to aggregate laboratory data for the same patient into a single record that listed patient, multiple hemoglobin A1c test results, and dates. There were very few missing data for first and last name, date of birth, and gender. Ethnicity and payer were not collected as part of this file.
The laboratory file did not contain zip codes. However, because the community health centers serve distinct geographic areas, a health center identifier, which was available in both laboratory and encounter files, was used as an indicator of patients' residence.
Linkage Algorithm
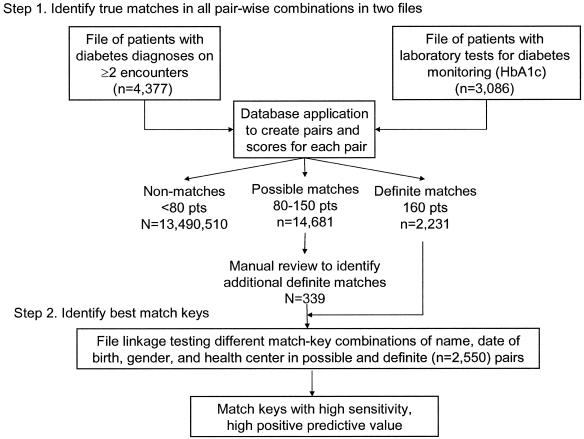
The linkage procedure was carried out in a multistep process (▶). In the first step, all true matches were identified from among all possible pairings of records between the encounter file and laboratory file. In the second step, combinations of match keys (one-way, two-way, three-way, etc.) were used to match records in the two files, and the sensitivity and PPV of each combination were calculated based on knowledge of true matches from step 1. Optimal keys were ones with very high sensitivity (percentage of all true matches) and very high PPV (low false-positive rate).
Figure 1.
Overview of the processing steps.
In step 1, to prioritize pairings that were definite, possible, or unlikely true matches, scores were assigned to particular identifiers and a total score for each pairing was calculated (▶). Pairings with less than half the maximum (< 80) points were considered to lack true matches. The following criteria were applied to identify the subgroup of pairings that had all the definite and possible matches: year of birth OR identical first four characters of the last name AND a score of ≥80 points based on all matching identifiers.
Table 2.
Identifiers and Their Score for Agreement in Computer Linkage
| Data Element | Abbreviation | Points |
|---|---|---|
| Year of birth | YOB | 20 |
| Month of birth | MOB | 20 |
| Day of birth | DYOB | 20 |
| First four letters of last name | F4 | 40 |
| Fifth and higher letters of last name | F5 | 20 |
| First initial of first name | I | 20 |
| Gender | G | 10 |
| Clinic | C | 10 |
| Total | 160 |
Pairings with the maximum number of points (160) were accepted as a true match (definite); that is, both files had exact matches on all possible identifiers: first four characters of last name (F4), fifth and subsequent letters of last name (F5), first initial of first name (I), year of birth (YOB), month of birth (MOB), day of birth (DYOB), gender (G), and health center (C). For possible matches (pairings with scores ≥80 and < 160 points) each identifier from the encounter file and laboratory file were visually compared, and a decision was made by one investigator (LH) whether the pair was a true match. In a few cases, the consensus of a second reviewer (NAM) was sought.
A standard database application (Microsoft Access®) was used to create all pairwise combinations of patients in the encounter file and the laboratory file and to assign scores for each matching identifier and a sum of points for each pairing. An MS Access data entry form was used to record whether a possible match was a true match and to classify as many as three reasons for lack of agreement per mismatched record pairs (inserted/deleted characters or names, typographic errors, name permutations, etc.).
Match Keys
To determine the optimal linkage key, 34 combinations of last name, first initial of first name, date of birth, gender, and health center were preplanned and tested as match keys in the pairings with ≥80 points. The combinations included the 17 one-, two-, and three-way combinations reported by Van den Brandt et al.1 An additional key, the first ten letters of the last name/first five letters of the first name/date of birth/gender, was examined because it was widely used for record linkage by database managers at the CHCN, but it had not been systematically evaluated for accuracy.
Several match keys not included by Van den Brandt et al.1 were created from the first two letters of the last name (F2). The CHCN staff who process health care bills have found F2 to be an effective “pocket” to search for true matches.
In the visual inspection of possible matches, last names and first names were occasionally transposed. This prompted us to add another match key post-analysis that combined a date of birth, the first two letters of the last name (F2), or a match on the first two letters of the last name (F2) in one file with the first two letters of the first name (I2) in the second file.
Statistical Analyses
The accuracy of each match key was determined by its sensitivity and PPV. To minimize redundancy, the results of several suboptimal keys were eliminated from this presentation.
An optimal key was defined as the one with a very high sensitivity and a very high PPV. To examine whether optimal keys varied by health center, patient ethnicity, age, gender, or payer, two by n contingency tables of sensitivity and the covariate of interest were constructed. Similar univariate analyses were carried out for PPV. Chi-square was used for the test of statistical differences of proportions and p < 0.05 was the criterion for statistical significance. No correction of the p-value was made due to multiple comparisons. Statistical analyses were carried out in STATA 7 (College Station, TX).
Results
The crossing of the two files resulted in 13,507,422 pairings (4,377 × 3,086) and 16,892 pairs scored ≥80 points. The number of pairs with the maximum of 160 points was 2,211. The visual inspection of the 14,681 pairings between 80 points (55%) and 150 (0.2%) points identified an additional 339 true matches for a total of 2,550 (▶). True matches concentrated in the pairings with the highest scores. More than 90% of true matches occurred in pairings with scores of ≥140. A small peak of true matches occurred at the lowest point range (80–100). False positives diminished as scores increased and comprised 0.17% of pairs scoring 140 to 160 points. The false positives in the 110- to 130-point range (N = 433) have a preponderance of individuals with common family names for the population (e.g., Lee, Nguyen, Hernandez, Rodriguez) but did not agree on other identifiers. The most common reasons for mismatching were lack of agreement on the day, year, and month of birth (▶). Compound names were also a prevalent cause of mismatches. Typically, one of the names in compound first names was either dropped or reversed. Compound last names were sometimes parsed between the first name and last name in the matching file.
Table 3.
Distribution of Matches Linking Patients with Diabetes (N = 4,377) and Patients with Laboratory Results (N = 3,806), Including Matches Identified by Visual Inspection
| Score | Total | False Pos. | True Pos. |
|---|---|---|---|
| 80–100 | 13,990 | 13,885 | 105 |
| 110–130 | 471 | 433 | 38 |
| 140–160 | 2,431 | 24 | 2,407 |
| Total | 16,892 | 14,342 | 2,550 |
Pos. = positive.
Table 4.
Common Discrepancies in Identifiers of True Matches in Record Pairs with 80 or More Points
| Identifier | Description of Error | Record Pairs with an Error, N | Mismatched Records per 100 True Matches |
|---|---|---|---|
| Total records with any error* | 339 | 13.3 | |
| I | Double first name (e.g., Mary Ann vs. Ann) | 69 | 2.7 |
| DOB | Day of birth transcription | 58 | 2.3 |
| DOB | Year of birth transcription | 43 | 1.7 |
| I, F | First part of compound last (family) name (e.g., Benton-Jones) written as first (given) name of second file | 37 | 1.5 |
| F4 | Misspelling or transposed characters | 37 | 1.5 |
| F5 | Misspelling or transposed characters | 30 | 1.2 |
| F | Interchange of compound last names (e.g., Perez Jones vs. Jones Perez) | 27 | 1.1 |
| DOB | Month of birth transcription | 21 | 0.8 |
| G | Transcription | 18 | 0.7 |
Multiple errors occurred in some record pairs, which is not reflected in total.
The match keys had a wide range of sensitivities and PPVs (▶). As expected, keys that used all identifiers were highly specific (PPV = 100%), but missed many true matches (sensitivity, ∼70%) (▶, keys 11 and 12). The key most similar to the optimal key of Van den Brandt et al.1 (▶, key 8) was also highly specific but had a low sensitivity (72.6%). Date of birth alone contributed to high sensitivity of matching (▶, key 3). Several keys based on F2/DOB alone or combined with gender, first name initial, or the interchange of the first two letters of the first and last name gave both high sensitivity (92.7%–93.2%) and high PPV (99.4%–99.5%) (▶, keys 1–2). Keys based on F2/DOB were optimal at three of the health centers (not shown). At the other health centers, similarly sensitive keys were F2/C/I (two centers), DOB/C, and DOB/G/I. Comparing the results of F2/DOB and F4/DOB (▶, key 2 vs. 7), it appears that errors in the third and fourth characters of the family name decreased sensitivity using the optimal key F4/DOB of Van den Brandt et al.
Table 5.
Matches, True Positives, False Negatives, Sensitivity, and Positive Predictive Value of Selected Keys Used to Link Patients with Diabetes and Patients with Laboratory Results
| No. | Key | Matches | True Positives | False Negatives | Sensitivity, % | PPV % |
|---|---|---|---|---|---|---|
| 1 | (F2 or F2=12)/DOB | 2,390 | 2,376 | 174 | 93.2 | 99.4 |
| 2 | F2/DOB | 2,377 | 2,364 | 186 | 92.7 | 99.5 |
| 3 | DOB | 2,583 | 2,408 | 142 | 94.4 | 93.2 |
| 4 | F2/C/I | 4,188 | 2,454 | 96 | 96.2 | 58.6 |
| 5 | F9 | 11,582 | 1,946 | 604 | 76.3 | 16.8 |
| 6 | All, except DOB | 2,671 | 1,896 | 654 | 74.4 | 71.0 |
| 7 | F4/DOB | 1,896 | 1,889 | 661 | 74.1 | 99.6 |
| 8 | DOB/F4/G/C* | 1,857 | 1,852 | 698 | 72.6 | 99.7 |
| 9 | DOB/F4/C/I | 1,863 | 1,861 | 689 | 73.0 | 99.9 |
| 10 | DOB/F4/G/I | 1,844 | 1,844 | 706 | 72.3 | 100.0 |
| 11 | F10/I5/DOB/G | 1,813 | 1,813 | 737 | 71.1 | 100.0 |
| 12 | F4/F5/I/DOB/G/C All identifiers | 1,791 | 1,791 | 759 | 70.2 | 100.0 |
PPV = positive predictive value.
An optimal key reported by Van den Brandt et al.1
Positive predictive value of the optimal key F2/DOB did not significantly vary by health center, ethnicity, age, gender, or payer (▶). However, sensitivity varied by health centers (79.2%–94.9%) and other demographic variables with the notable exception of ethnicity. The sensitivity of F2/DOB was greater in males than females and at ages younger than 45 years. There were no significant differences in sensitivity between known payers. However, the sensitivity in matches with unknown payer was lower than known payers.
Table 6.
Sensitivity and Positive Predictive Value of Matching Using an Optimal Match Key (F2/DOB) by Health Center, Patient Demographics, and Payer
| Item | True Matches, N | Sensitivity, % | PPV, % |
|---|---|---|---|
| Health Center* | |||
| A | 906 | 94.9† | 98.9 |
| B | 582 | 94.9 | 99.8 |
| C | 491 | 92.3 | 99.8 |
| D | 180 | 90.0 | 100.0 |
| E | 97 | 90.7 | 100.0 |
| F | 80 | 79.2 | 98.8 |
| G | 37 | 88.1 | 100.0 |
| Ethnicity | |||
| Asian | 750 | 93.9 | 98.8 |
| African American | 218 | 94.0 | 100.0 |
| Latino | 752 | 93.1 | 99.9 |
| Native American | 53 | 91.4 | 98.2 |
| White (non-Hispanic) | 120 | 87.0 | 100.0 |
| Missing | 480 | 93.2 | 99.6 |
| Age | |||
| < 25 | 29 | 96.7† | 100.0 |
| 25–44 | 323 | 95.9 | 99.4 |
| 45–64 | 1,014 | 91.1 | 99.7 |
| 65+ | 1,007 | 94.1 | 99.1 |
| Gender | |||
| Male | 868 | 94.2† | 99.7 |
| Female | 1,492 | 91.6 | 99.4 |
| Payer | |||
| Uninsured | 734 | 93.7† | 99.6 |
| Medicaid | 482 | 91.5 | 99.6 |
| Medicare | 781 | 94.3 | 99.1 |
| Commercial | 102 | 94.4 | 100.0 |
| Missing | 219 | 88.3 | 99.6 |
Discussion
Although we did not confirm that the optimal linkage key used by researchers for the Dutch cancer registry was optimal for patients with diabetes at community health centers, the application of their protocol, with information from the CHCN staff, who manually match claims, led to the identification of match keys with both very high sensitivity and PPV. An important difference between our results and those of Van den Brandt et al.1 appear to be related to lack of agreement on the three and more characters of the family name (F2 vs. F4). This may be due to differences in population characteristics (large proportion of ethnic family names), differences in training and supervision of data entry staff at the health centers or disease registry, or differences in data entry applications. This also confirms an observation that the experience of those in the trenches performing actual matching should inform the construction of accurate deterministic match keys. Researchers have commented on the capacity of humans to deploy a lifetime memory of name variants and error recognition strategies that are difficult to reproduce in computer linkage algorithms.5
The types of name and date errors that produce mismatches in general were also found in our setting. However, some of these errors might have been expected to occur with a high frequency because our patient population was mostly nonwhite and had a high percentage of recent immigrants. Both maternal and paternal last names are used throughout Mexico and Latin America, the countries of origin of most of our Latino patients. Many Asian ethnic groups present family name and given name in a different order than those with an Anglo-Saxon acculturation. Likewise, recent immigrants accustomed to the day-month-year order for dates may be confused by the month-day-year convention used in United States. Because the error rates varied by health center, this information can be used to help improve protocols and training for staff who fill out laboratory requisition forms. This targeted feedback may be more useful than a general message to improve data quality.
We observed a familiar trade-off between sensitivity and specificity when an increasing number of identifiers were combined for a key. For the purpose of statistical aggregation of data used in epidemiologic research and population-based measurement of clinical quality, the optimal keys produced a negligible rate of false positives (< 0.5%), and false negatives (8%), whose exclusion would not constitute an important source of bias. Probabilistic methods and deterministic methods combined with string comparators may have improved the sensitivity9 but would have required the CHCN to purchase additional software and substantially increased human resources for training during implementation and for maintenance. The manual review of 14,681 possible matches took one researcher two to three workdays, and the programming of the database application for data entry, scoring of match pairs, and statistical analysis required less than one person-week. This is a substantially smaller amount of resources than an alternative in which probabilistic methods are implemented in commercially available software for an enterprise as large as the CHCN.
Other researchers have reported that the accuracy of matching can be influenced by race/ethnicity.12 This may be the result of proportionately higher missing data for Social Security number (a common blocking variable) for Latinos and other minorities, compared with whites. We did not find that the sensitivity of optimal keys varied by ethnicity. One explanation is that the pattern of errors in name and date of birth was not unique to race/ethnicity. Another possibility is that ethnicity itself was misclassified in the administrative data. Race/ethnicity in administrative data often has poor agreement with self-reports.13,14 One recent study of patients at a community health center had a proportion of agreement of approximately 56%. Race/ethnicity from administrative data used in this study had better agreement (74%) with the race/ethnicity indicated by patients on registration forms in medical charts.15
Although there were significant differences in sensitivity of the optimal key by age, sex, and payer, many of these differences are not likely to create an important bias in studies of a statistical nature. The direction of the bias appears to be logical: Poorer matching was found in females, possibly due to changes in last name after marriage, and poor matching was found in older age groups, possibly due to errors in the year 2000 conversion or informant bias. For the purpose of clinical management of individual patients, even this low false-positive rate may not be acceptable. It may be practical to apply the two-step approach of Van den Brandt et al. Matches with the maximum number of points can be loaded into a patient care database without a case-by-case human review. Matches >80 and < 160 points could be manually reviewed before being loaded into database tables.
The implications of this work are greater than just for diabetes research. Record linkage involving different types of clinical information is an essential part of monitoring controller medications in patients with asthma, Pap screening in women, and Chlamydia testing in young women.3 We have repeated this study, matching patients from an encounter file with unrestricted diagnoses to patients in a laboratory file for any test result. The optimal match keys in this study were confirmed in these generic files.
Conclusion
Over the next decade, the adoption of electronic standards for the sharing of clinical information between different health care organizations, falling prices for technology, and removal of other technical barriers will facilitate database integration. In the meantime, a record linkage algorithm based on a few common patient identifiers that is inexpensive, accurate, and easy to implement is preferable to manual look-up and re-entry of laboratory results. A relatively simple protocol that uses widely accessible database applications can be adapted to meet this need.
Supported by the Agency for Healthcare Research and Quality (1 R21 HS013543-01).
The CHCN Claims Department is acknowledged for their contribution of the F2 key and Ray Otake for bringing this information to the authors' attention. Khati Hendry provided insightful comments during the development of this project. Dr. Joseph Selby (Kaiser Permanente) is acknowledged for technical assistance in carrying out the literature review.
Protection of human subjects in research was approved for this project by the IRB of Kaiser Permanente-North.
Neil Maizlish was responsible for the design, analysis, and writing of this report. Linda Herrera contributed to the design, data collection, analysis, and revision of this report.
References
- 1.Van den Brandt PA, Schouten LJ, Goldbohm RA, Dorant E, Hunen PMH. Development of a record linkage protocol for use in the Dutch cancer registry for epidemiologic research. Int J Epidemiol. 1990;19:553–8. [DOI] [PubMed] [Google Scholar]
- 2.Maizlish NA, Shaw B, Hendry K. Glycemic control in diabetic patients served by community health centers. Am J Med Qual. 2004;19:172–9. [DOI] [PubMed] [Google Scholar]
- 3.National Committee for Quality Assurance. HEDIS 2003 (Health plan employer data and information set.) Technical specifications Volume 2 Washington, DC: National Committee for Quality Assurance, 2002.
- 4.Institute of Medicine. Using information technology. In: Crossing the quality chasm: a new health system for the 21st century. Washington, DC: National Academy Press, 2001, 164–80.
- 5.Fair M, LaLonde P, Newcombe HB. Application of exact odds for partial agreements of names in record linkage. Comp Biomed Res. 1991;24:58–71. [DOI] [PubMed] [Google Scholar]
- 6.Gill L, Goldacre M, Simmons H, Bettley G, Griffith M. Computerized linking of medical records: methodological guidelines. J Epidemiol Community Health. 1993;47:316–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Roos LL, Wadja A. Record linkage strategies. Meth Inform Med. 1991;30:117–23. [PubMed] [Google Scholar]
- 8.Winkler WE. Record linkage software and methods for merging administrative data. Washington, DC: Statistical Research Division, U.S. Bureau of the Census, 2001.
- 9.Grannis SJ, Overhage JM, McDonald C. Real world performance of approximate string comparators for use in patient matching. Medinfo. 2004;2004:43–7. [PubMed] [Google Scholar]
- 10.Friedman C, Sideli R. Tolerating spelling errors during patient validation. Comput Biomed Res. 1992;25:486–509. [DOI] [PubMed] [Google Scholar]
- 11.Machado CJ, Hill K. Probabilistic record linkage and an automated procedure to minimize the undecided-matched pair problem. Cad Saude Publica. 2004;20:915–25. [DOI] [PubMed] [Google Scholar]
- 12.Zingmond DS, Ye Z, Ettner SL, Liu H. Linking hospital discharge and death records–accuracy and sources of bias. J Clin Epidemiol. 2004;57:21–9. [DOI] [PubMed] [Google Scholar]
- 13.Kressin NR, Chang B-H, Hendricks A, Kazis LE. Agreement between administrative data and patients' self-reports of race/ethnicity. Am J Public Health. 2003;93:1734–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Moscou S, Anderson MR, Kaplan JB, Valencia L. Validity of racial/ethnic classifications in medical records data: an exploratory study. Am J Public Health. 2003;93:1084–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Community Health Center Network. Assessment of data quality in medical charts, practice management systems, and CHCN data warehouse. Alameda, CA: Community Health Center Network, 2003.