Abstract
Objective: To develop a model to store information in an electronic medical record (EMR) for the management of transplant patients. The model for storing donor information must be designed to allow clinicians to access donor information from the transplant recipient's record and to allow donor data to be stored without needlessly proliferating new Logical Observation Identifier Names and Codes (LOINC) codes for already-coded laboratory tests.
Design: Information required to manage transplant patients requires the use of a donor's medical information while caring for the transplant patient. Three strategies were considered: (1) link the transplant patient's EMR to the donor's EMR; (2) use pre-coordinated observation identifiers (i.e., LOINC codes with *∧DONOR specified in the system axes) to identify donor data stored in the transplant patient's EMR; and (3) use an information model that allows donor information to be stored in the transplant patient's record by allowing the “source” of the data (donor) and the “name” of the result (e.g., blood type) to be post-coordinated in the transplant patient's EMR.
Results: We selected the third strategy and implemented a flexible post-coordinated information model. There was no need to create new LOINC codes for already-coded laboratory tests. The model required that the data structure in the EMR allow for the storage of the “subject” of the test.
Conclusion: The selected strategy met our design requirements and provided an extendable information model to store donor data. This model can be used whenever it is necessary to refer to one patient's data from another patient's EMR.
During 2003, more than 25,000 persons in the United States underwent organ transplantation surgery.1 Nationwide and at LDS Hospital in Salt Lake City, UT, the number of transplantations increases each year. Transplant patients generate large quantities of data, predominantly laboratory results, which must be interpreted by the clinical team. In addition, clinicians must consider information about the organ donor in the management of a transplant patient. Many of the data related to transplants are stored electronically, but the Transplant Program at LDS Hospital used a paper-based medical record for lifelong monitoring of transplant patients. The paper record was not always accessible nor could it be used for automated decision support. A paper flowchart was used to document inpatient and outpatient laboratory results, medication doses, problems, procedures, and comments. The flowchart allowed clinicians to view trends in the data and provided context for the individual results and clinical observations that clinicians needed to interpret.
Clinicians need information about the organ donor to manage the care of the organ recipient, particularly the donor's history of infections and the quality of the donor organ. Several issues complicate the process of storing information about an organ donor in a transplant patient's record. First, there is no single standard for storing information about one person (e.g., donor, mother, newborn) in another person's record. Second, most donors are deceased and may not be registered in the electronic medical record (EMR) system. Registration in the system is required before unique system identifiers can be created so information can be linked within the EMR. Third, even if a donor has been previously registered in the system, the procurement agency may use its own patient identifier when final blood specimens are sent to the laboratory. Finally, privacy and security issues must be addressed to ensure that the donor's identifying information is only accessible to authorized clinicians accessing the EMR.
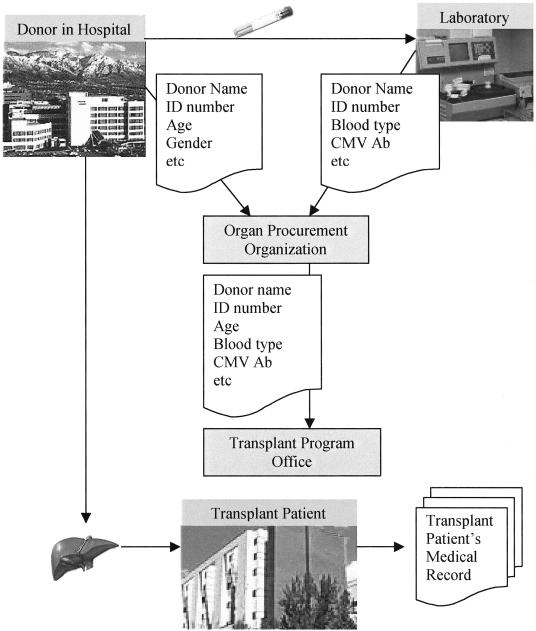
The typical process for transferring an organ and communicating information about the donor is illustrated in ▶. Clinical information about the donor is transferred to an organ procurement organization (OPO) and then relayed to a transplant program coordinator. Blood specimens are sent to a laboratory, and the results are relayed to an OPO and finally to the transplant coordinator. The organ may be harvested at the transplant center or may come from another institution that is local or out of state. Currently, in our facility, the donor's paper records are not included in the transplant patient's inpatient or outpatient medical records that are routinely used by the clinical team.
Figure 1.
Typical process for transferring an organ and communicating information about the donor at the time of transplantation.
As the patient population has increased and information technology has advanced, we recognized the need and opportunity to store all the transplantation-related information in the EMR and to use the information to improve decision making and the process and outcome of care.2 Strategies for storing transplantation-related information in an EMR have been briefly described in a few recent technical documents3,4,5,6 and publications.7 Earlier publications describe stand-alone transplantation information systems developed for single transplantation centers.8,9,10,11,12,13 To our knowledge, there is no published literature that addresses the requirements for developing a transplantation information system and implementing the storage of donor-related information in an EMR using an object-oriented data model.
While developing a model to store information in an EMR for the management of transplant patients, we identified several requirements and options for storing organ donor data. In the subsequent sections of this article, we describe the primary requirements that influenced the design of the information model and the options that were considered and how the model was developed and validated. Our approach has been implemented and may be useful for others to consider when developing information models for storing organ donor data in an EMR or when storing information about persons other than the patient (e.g., family history information) in a patient's record.
Background
Logical Observation Identifier Names and Codes (LOINC) codes can be used to report and store all standard laboratory results and many clinical observations in an EMR.14 LOINC codes provide a standard set of universal names and codes for identifying individual laboratory and clinical results, which allows users to merge clinical results from many sources into one database for patient care, clinical research, or management.5 They are intended to specify the “name” (observation identifier) for a name/value pair data element. Since LOINC codes are used by our institution and have been adopted by major laboratories and other organizations,5 it is important to evaluate how a proposed information model would affect the LOINC database.
The LOINC database currently contains about 34,000 different laboratory tests and clinical observations.5 Among 430 unique test names associated with blood typing and human leukocyte antigen (HLA) testing, approximately 70% have been repeated and new LOINC codes have been created that specify that the test result is applicable to a donor, not the patient. This pre-coordinated style is implemented by specifying “∧DONOR” in the system axis of the LOINC code. For example, the LOINC code for a donor's blood type is the following: ABO GROUP:TYPE:PT:BLD∧DONOR:NOM. To date, only ten laboratory tests other than HLA and blood bank codes have been repeated and pre-coordinated when they apply to a person other than the patient, including the patient's donor (n = 7), newborn (n = 1), and fetus (n = 2). For example, the LOINC code for a donor's hepatitis B core antibody result is the following: HEPATITIS B VIRUS CORE AB:ACNC:PT:SER∧DONOR:QN.
Given the interest in documenting laboratory results on organ donors and the presence of more than 10,000 LOINC codes for chemistry and microbiology tests alone, there is a potential for creating many new donor-specific codes for test names that already exist. Unbounded proliferation (combinatorial explosion) will occur if this approach is applied to other types of persons relevant to the patient (i.e., mother, father, sibling, other family member, household contact, sexual contact).
Since storing donor data in another persons record involves the transfer of data from one system to another (▶), it is important to consider the impact of an information model on information messaging and system interoperability. Health Level Seven (HL7) is an American National Standards Institute–accredited Standards Development Organization that focuses on interoperability standards for health care organizations.6 The mission of HL7 is to “provide standards for the exchange, management and integration of data that support clinical patient care and the management, delivery and evaluation of healthcare services. Specifically, to create flexible, cost effective approaches, standards, guidelines, methodologies, and related services for interoperability between healthcare information systems.”6 Version 3 of HL7 is based on a standard object-oriented information model, the HL7 Reference Information Model (HL7 RIM). The most important classes in the model (the so-called backbone classes) include (a) “Act” (e.g., procedure, observation, medication); (b) “Act_relationship” that specifies how acts are related to one another; (c) “Participation” that defines the context for the act (e.g., patient, subject, location); (d) “Roles” played by participants (e.g., patient, practitioner, specimen); and (e) “Entities” that play roles (e.g., persons, organizations, devices). Within this model, there is an “Observation” class for storing observations such as laboratory data. Within the observation class, there is an attribute that identifies the person on whom the test was performed (subject) and an attribute that identifies the person in whose record the observation is stored (recordTarget). Interoperability may be enhanced if the solution that we use can be implemented using the “subject” and “recordTarget” attributes of the HL7 RIM Observation class.
Formulation Process
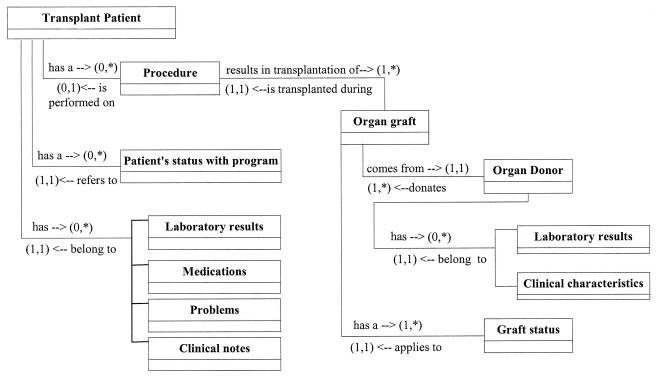
Initially, we assessed the clinician's data needs by reviewing flowcharts used in the transplant office, reviewing data that are currently captured in our EMR system, and by interviewing transplant physicians and nurses. We analyzed the data and information and created a data model that captures the relationship between entities required for the management of transplant patients (▶). It is necessary to store information about the patient's status with the transplant program, the surgical event, and the organ transplanted. This information must be incorporated into the existing model linking laboratory results, medications, problems, procedures, notes, and other clinical events reported for the patient (▶). It is also necessary to store information about the organ donor (▶).
Figure 2.
Relationship between entities required for the management of a transplant patient.
We identified two primary requirements for the design of an information model for storing information about an organ donor. First, the clinician must have the ability to access donor information while reviewing the transplant patient's medical record because donor information may affect the management of patients after transplantation surgery. Second, storage of donor data must not result in unbounded proliferation (combinatorial explosion) of new LOINC codes for already-coded laboratory tests.
We identified three options for storing donor information in the EMR (▶). The first strategy for storing donor information linked the transplant patient's EMR to data in the donor's EMR. This design is conceptually simple but has limitations. It is not practical to register a deceased person in the EMR system for several reasons. First, it is illogical to have deceased persons registered in the system because they may be mistaken for living patients. Second, the demographic data needed to accurately and uniquely identify the donor may not be available. This is particularly true if the organ is recovered from out of state or another hospital and the donor has never had contact with the transplant patient's hospital. In addition, one cannot guarantee that the records will remain linked and accessible over time. Record system managers may archive the electronic records of deceased persons. While archiving of a deceased patient's records may improve the performance of the EMR system, the archived data are no longer available when caring for the transplant patient.
Table 1.
Three Strategies for Storing Donor Information in an Electronic Medical Record
| Strategy | Patient Record | Structure of the Data: Test Name, Value, and Subject | ||
|---|---|---|---|---|
| 1. Create a donor record and link it to the transplant recipient's record | Transplant patient | Name |
Value |
|
| Blood type | A | |||
| CMV | Positive | |||
| Donor | Name |
Value |
||
| Blood type | A | |||
| CMV |
Positive |
|||
| 2. Pre-coordinate donor information in the transplant recipient's record | Transplant patient | Name |
Value |
|
| Donor blood type | A | |||
| Donor CMV |
Positive |
|||
| 3. Post-coordinate donor information in the transplant recipient's record | Transplant patient | Subject |
Name |
Value |
| Donor | Blood type | A | ||
| Donor | CMV | Positive | ||
CMV = cytomegalovirus.
The second strategy (▶) used pre-coordinated observation identifiers (i.e., LOINC codes with *∧DONOR specified in the system axes) to identify donor data that are stored in the transplant patient's EMR. A separate donor record is not created. This vocabulary model has the advantage of being a simple, physical data model. Each concept is expressed using a single data element (e.g., donor blood type). Use of this model may simplify queries for decision logic and data analysis. However, there are limitations of this model that concern the impact on LOINC codes. First, to implement this model, new LOINC codes for all the standard tests performed on donors would need to be created, and new LOINC codes would need to continue to be created as the tests of interest increase over time. Second, if this strategy were applied to models for storing data about a mother, newborn, or other family members relevant to a patient, a combinatorial explosion of LOINC codes would occur. For example, separate LOINC codes would need to be created for a mother's blood type, a newborn's blood type, and a father's blood type because each of these data elements may be relevant in the management of a given patient. Third, if this approach were implemented, the coded test name in the donor's laboratory record would need to be changed when the test results are stored in the transplant patient's record. In other words, it would require a map or crosswalk that correlates the name of the data element in the donor's record (patient's blood type) to the name of the stored data element in the transplant patient's record (donor's blood type). This process would require maintenance to ensure that the test codes were properly mapped between the laboratory system testing the donor's blood and the EMR system storing the donor's data in the transplant record.
A third strategy (▶) used an information model to allow donor data to be stored in the transplant patient's record. With this strategy, the “source” of the data (donor) and the “name” of the result (e.g., blood type) would be post-coordinated in the transplant patient's EMR as data were stored. The subject of the observation would be stored as a separate piece of data along with the observation and its value. Thus, a separate donor record would not be created. There are several advantages to this information model. First, there would be no need to create new LOINC codes for already-coded laboratory tests. Second, the model provides flexibility for clinicians to add new donor laboratory tests to the transplant patient's record using existing codes. This may be particularly beneficial over time if the types of tests performed on a donor expand. Third, there is no need to transform LOINC codes when the test result moves from a donor record to the transplant patient's record. The same LOINC code for a particular test name could be used in both the donor's laboratory result file and the transplant patient's record. Finally, this strategy can be implemented using the “subject” and “recordTarget” attributes of the HL7 RIM Observation class. Since this strategy is consistent with how subject and patient are modeled in the HL7 RIM, this strategy will be easier to use for persons already familiar with the HL7 strategy. Two important limitations of this model are that (1) it would require the data structure in the EMR to allow for the storage of the “subject” of the test and (2) users of the data would need to be aware that data are tagged by subject so they do not inadvertently mistake donor data for transplant patient data.
Model Description
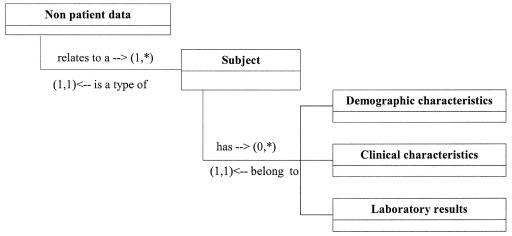
We selected the third strategy and implemented a post-coordinated information model to store donor information in the EMR. The laboratory and clinical observations included in the model are dependent on the context of the information (donor). We created an observation of “nonpatient” data, specified that the subject of the observations is an organ donor, and added the information about the donor that was relevant for the management of the transplant patient (▶). We requested only three new LOINC codes (▶) to store donor information in the transplant patient's record. We are able to use existing LOINC codes to name the laboratory test results and to specify the donor's relation to the patient (e.g., self, mother, sibling 1, grandmother, nonrelative).
Figure 3.
Relationship between entities required for storing information about someone other than the patient.
Table 2.
LOINC Codes Requested to Store Donor Data in the Information Model
| Component | System | Sample of Values | Suggested Short Name |
|---|---|---|---|
| Organ donor ID number | ∧DONOR | Donor identification number | |
| Subject of observation | ∧PATIENT | Organ donor, mother, newborn, sibling, household contact, sexual contact | Subject |
| Organ donor | ∧PATIENT | Cadaveric donor, living donor | Organ donor |
Logical Observation Identifier Names and Codes (LOINC) already existed for “Donor relation to patient.” The answer list for this code included terms such as “self,” “mother,” ”father,” “sibling 1,” “grandmother,” “nonrelative,” and so forth.
Validation through Example
We implemented the post-coordinated information model to store donor information in the EMR of liver transplant patients. We grouped the information about the donor, specified that the subject of the observations is an organ donor, and stored the following observations in the liver transplant patient's record: the donor's blood type; serologic status for CMV, hepatitis B, and hepatitis C; the final and peak results of selected laboratory tests that indicate kidney and liver function; and the unique donor identifier reported by the procurement organization and the United Network for Organ Sharing (UNOS). This donor information is directly linked with the coded data describing the operative procedure associated with transplantation of the graft. All this information is further linked to the laboratory and other clinical information for the transplant patient that already exists in the EMR. We were able to store and retrieve data accurately in the EMR, and clinicians were able to use and interpret the data accurately in caring for transplant patients.
Discussion
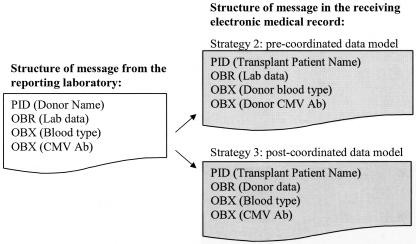
Our strategy for storing donor data has implications for the storage and transmission of data in the EMR. This new information model required a LOINC code to specify the subject (mother, donor, newborn) of the observation. The subject of the observation needs to be specified when the data are stored or transmitted in an HL7 message. The exchange of data between facilities or laboratories will be simplified, however, because there is no need to map codes and translate the donors' laboratory results from those of a “person” to those of a “donor” when the information is moved from the laboratory system to the transplant patient's record. The mapping that would be required to implement the second and third strategies is illustrated in ▶. The standard HL7 message for moving the data from one system to another would differ depending on which model is used: a pre-coordinated vocabulary model (strategy 2) or a post-coordinated information model (strategy 3). If strategy 3 is used and the laboratory data are directly interfaced from the laboratory performing the donor's tests or from the procurement agency, the mapping of the codes from one system to another is simple. If the transplant patient's identity is specified for the interface, the laboratory data could be “cut and pasted” into the donor data segment of the transplant patient's record. Similarly, if the transplant program uses living donors, it still may be important to store a core set of donor data in the transplant patient's record to facilitate access to data for decision support applications and future data analysis. Using the third strategy would facilitate the transfer of information within the system's EMR from the record of the donor to the record of the transplant patient.
Figure 4.
Comparison of the mapping required for data exchange using a pre-coordinated versus a post-coordinated data model.
The strategy that we propose has implications for maintenance of the LOINC terminology. First, our information model did not require creation of new LOINC codes for already-coded standard laboratory tests. This simplifies the addition of new laboratory tests to the data entry screen and database when the clinicians need to add new tests. Second, if our strategy was adopted universally, then hundreds of subject-specific LOINC codes could be inactivated, and no new LOINC codes would need to be created for subject specific tests. While this is a desirable outcome, many systems are using the existing LOINC codes, so it is unlikely that a change in creation of LOINC codes can be accomplished anytime soon.
There are limitations to the implementation of this model. First, if the donor provides organs for two different recipients, the donor information will need to be entered into the EMR twice. Since multiple organ transplantations are not common in our institution, this limitation is not burdensome. Second, legitimate security and privacy issues limit the extent of the donor information that can be stored in a transplant patient's record. The information stored in the transplant patient's record does not directly identify the donor nor does it provide information that would allow access to donor data in the OPO or UNOS secure databases.15 Both the procurement organization and UNOS would require authorization and authentication (and UNOS requires a password) before allowing access to donor information. Therefore, the information stored meets national standards for the privacy of individually identifiable health information,16 and it is unnecessary to control access to the record beyond the usual security measures used to control access to the EMR. We did not store donor data that would require long-term control of access privileges to a limited set of users, such as members of the transplant team.
The strategy that we propose also has implications for the various technical working groups who are discussing the issue of describing the context of an observation in the scope of an EMR. Specifying the “subject” of an observation is similar to the problem with specifying the timing of a result in relation to other symptoms or other test results. Our case example of storing information about one person in another person's record may be important to consider while designing and refining reference information models6 and data structures that require specifying the “subject” and context of an observation.
Conclusion
We have described how laboratory and other clinical data for both the transplant patient and the donor can be integrated in the transplant patient's EMR. This information can be accessed directly by the clinicians or used by decision support applications to enhance the management of transplant patients after surgery. In addition, the information can be used for research, and it provides linkage to extensive donor data located in other source files. We only stored data that were determined by the clinicians to be useful for post-transplantation management of the transplant patient. We met our objective to create an information model that allowed access of donor data from the transplant patient's record and did not require creation of new LOINC codes for already-coded standard laboratory tests.
Portions of this manuscript were presented as a poster at the annual meeting of the American Medical Informatics Association, November 2003, in Washington, DC. The abstract was published in the proceedings of the meeting: Staes CJ, Huff SM, Tilley C, Narus SP, Sorensen JB, Evans S. Development of an Information Model for Solid Organ Transplantation. Proc AMIA Symp. 2003:1015.
Supported by funding from a Medical Informatics Training Grant from the National Library of Medicine (CJS).
This project was approved by Institutional Review Boards from both the University of Utah and Intermountain Health Care.
References
- 1.Transplants by donor type. U.S. transplants performed: January 1, 1988–March 31, 2004. [online] 2004 [cited 2004 Jul 24]. Available from: http://www.OPTN.org/.
- 2.Dick R, Steen E, Detmer D, editors. The computer-based patient record: an essential technology for health care, Rev ed. Washington, DC: National Academy Press, 1997. [PubMed]
- 3.Purves I, Booth N. Durham and Darlington ERDIP Demonstrator Project: information models for the electronic health record [online]. 2002 [cited 2004 Aug 7]. Available from: http://www.nhsia.nhs.uk/erdip/pages/demonstrator/demonsites.asp?site=durham/.
- 4.O'Neil M. Using clinical terms in context in the clinical record. [online]. 1999 [cited 2004 Aug 8]. Available from: http://www.google.co.uk/search?q=cache:g8WOOT6X5ZsJ:www.nhsia.nhs.uk/context/pages/docs/relating.doc+%22Using+clinical+terms+in+context+in+the+clinical+record%22&hl=en/.
- 5.McDonald C, Huff SM, Suico J, Mercer K, editors. Logical Observation Identifiers Names and Codes (LOINCR) users' guide. Indianapolis: Regenstrief Institute, 2004.
- 6.HL7 Reference Information Model version 2.02. [online]. 2003 [cited 2004 Aug 8]. Available from: http://www.hl7.org/.
- 7.Coyle JF, Mori AR, Huff SM. Standards for detailed clinical models as the basis for medical data exchange and decision support. Int J Med Inf. 2003;69:157–74. [DOI] [PubMed] [Google Scholar]
- 8.Gordon RD, Markus B, Mitchell S. A liver transplant center information management system. Gastroenterol Clin North Am. 1988;17:61–9. [PubMed] [Google Scholar]
- 9.Kurtz M, Bennett T, Garvin P, Manuel F, Williams M, Langreder S. Demonstration of SLUMIS: a clinical database and management information system for a multi organ transplant program. Proc Annu Symp Comput Appl Med Care. 1991:889–90. [PMC free article] [PubMed]
- 10.Markus BH, Mitchell S, Gordon RD, Gillquist B, Tzakis AG, Starzl TE. TIMY—a center-oriented transplant information management system. Transplant Proc. 1988;20(1 Suppl 1):385–90. [PMC free article] [PubMed] [Google Scholar]
- 11.Mitchell SH, Zaltash P, Miller AL, Gordon RD. Transplant information management system: a center-oriented approach to transplant data management. Top Health Rec Manage. 1990;11:20–4. [PubMed] [Google Scholar]
- 12.Merion R. Iterative cooperative prototyping in the design of web-based transplant information systems. Transplant Proc. 1998;30:1634–6. [DOI] [PubMed] [Google Scholar]
- 13.Girardet RE, Oser AB, Klein EJ, Lansing AM, Lusk R, Nicholson CH. Computer tracking of multiple clinical variables in long-term treatment of heart transplant recipients. J Heart Lung Transplant. 1994;13:221–3. [PubMed] [Google Scholar]
- 14.McDonald CJ, Huff SM, Suico JG, Hill G, Leavelle D, Aller R, et al. LOINC, a universal standard for identifying laboratory observations: a 5-year update. Clin Chem. 2003;49:624–33. [DOI] [PubMed] [Google Scholar]
- 15.Dickinson DM, Ellison MD, Webb RL. Data sources and structure. Am J Transplant. 2003;3(Suppl 4):13–28. [DOI] [PubMed] [Google Scholar]
- 16.“Other requirements relating to uses and disclosures of protected health information.” Federal Register 45 CFR Part 164-514 (26 Feb 2001) p. 718–9. [online]. 2001 [cited 2004 Jul 25]; 10-1-02. Available from: http://www.gpoaccess.gov/fr/index.html/.