Abstract
The ubiquitous m7G cap of eukaryotic mRNAs and of precursors to the spliceosomal small nuclear RNAs (snRNAs) is the result of an essential RNA modification acquired during transcript elongation. In trypanosomes, the m7G cap is restricted to the spliced leader (SL) RNA and the precursors of U2, U3, and U4 snRNAs. mRNA capping in these organisms occurs posttranscriptionally by trans splicing, which transfers the capped SL sequence to the 5′ ends of all mRNAs. The SL cap is the most elaborate cap structure known in nature and has been shown to consist of an m7G residue followed by four methylated nucleotides. Using Crithidia fasciculata, we have characterized and purified the guanylyltransferase (capping enzyme), which transfers GMP from GTP to the diphosphate end of RNA. The corresponding gene codes for a protein of 697 amino acids, with the carboxy-terminal half of the C. fasciculata guanylyltransferase containing the six signature motifs previously identified in yeast capping enzymes. The amino-terminal half contains a domain that displays no resemblance to any other domain associated with capping enzymes. Intriguingly, this region harbors a consensus sequence for a phosphate-binding loop which is found in ATP- and GTP-binding proteins. This two-domain structure is also present in the Trypanosoma brucei capping enzyme, which shows 44% overall identity with the C. fasciculata capping enzyme. Thus, this structure appears to be common to all trypanosomatid protozoa and defines a novel class of capping enzymes.
Capping of RNA molecules with m7G is an essential modification which plays several roles in RNA metabolism and processing. In the nucleus it directs pre-mRNAs to the processing pathway and mRNAs and U small nuclear RNAs (U snRNAs) to the export pathway. In the cytoplasm, it regulates both mRNA translation initiation and mRNA turnover. In most eukaryotic organisms m7G capping is restricted to nascent RNA polymerase II (PolII) transcripts, namely, pre- mRNAs and precursors to U snRNAs. In contrast, in trypanosomatid protozoa mRNA capping occurs by a fundamentally different mechanism. It is well established that the m7G cap of mature mRNA is acquired posttranscriptionally by an RNA processing reaction, namely, trans splicing. In this process, the capped spliced leader (SL) sequence is transferred from the SL RNA to the 5′ ends of all trypanosome mRNAs (14, 23, 36). Thus, trans splicing can also be considered a trans-capping reaction. The cap structure of the Trypanosoma brucei and Crithidia fasciculata SL RNA is quite elaborate in that the m7G moiety is linked via a 5′-5′ triphosphate bridge to four methylated nucleotides, resulting in an unusual cap 4 structure (2) which is essential for utilization of the SL RNA in trans splicing (18, 43). At present the identity of the RNA polymerase transcribing the SL RNA genes is not defined, because the standard classification based on transcription inhibitors gave conflicting results as to whether PolII or PolIII is the responsible polymerase (8, 25, 26, 42). In addition to the SL RNA, a subset of trypanosome PolIII transcripts, namely, the U2, U3, and U4 snRNAs, is also initially m7G capped (7, 22, 24, 39). Despite the uncertainty about the polymerase transcribing the SL RNA genes, the fact that some PolIII transcripts are capped suggests that the mechanism of transcript selection by the capping enzyme is different in trypanosomes from that in other eukaryotes.
m7G capping is mediated by the stepwise action of three enzymatic activities (for a review, see reference 33). First, the γ-phosphate of a primary transcript is removed by RNA 5′-triphosphatase followed by GTP:RNA guanylyltransferase, or capping enzyme, which caps the RNA by the addition of GMP. Finally, the newly attached guanine residue is methylated at the N-7 position by the action of RNA (guanine-7-)methyltransferase. So far, only the reaction mechanism of guanylyltransferase has been examined in some detail. This enzyme catalyzes two sequential nucleotidyl transfer reactions, with a covalent enzyme-guanylate intermediate (19, 30). In this reaction, nucleophilic attack on the α-phosphate of GTP by guanylyltransferase results in the release of pyrophosphate and the formation of a covalent adduct in which GMP is linked to the guanylyltransferase through a phosphoamide bond to the ɛ-amino group of the catalytic lysine residue (30, 34, 38). To complete the reaction, GMP is transferred to the 5′ end of substrate RNA to yield an inverted 5′-5′ triphosphate bond.
The vaccinia virus capping enzyme is a multifunctional protein that carries out all three steps of the capping reaction (21, 40, 44). The protein is organized as a heterodimer, with subunits of 95 and 33 kDa (4, 10, 16, 29, 32). The RNA 5′-triphosphatase and guanylyltransferase have been mapped to the amino-terminal 60-kDa domain of the large subunit, while full (guanine-7-)methyltransferase activity requires both subunits. The subunit structure of the vaccinia virus capping enzyme is only partially maintained in cellular counterparts. In particular, cellular capping enzymes are distinct from their viral counterparts in that there is so far no example of a physical association between the capping and methyltransferase functions. In Saccharomyces cerevisiae, the purified capping enzyme consists of two monofunctional polypeptides: a 52-kDa guanylyltransferase and an 80-kDa triphosphatase (12, 13, 28). The guanylyltransferase is the product of the CEG1 gene, which is essential for cell viability (28). As in higher eukaryotes, the yeast (guanine-7-)methyltransferase is purified as a separate entity and is encoded by the ABD1 gene (15). The monofunctional domain structure of the guanylyltransferase is also present in other fungi, such as Schizosaccharomyces pombe (31) and Candida albicans (49), and in Chlorella virus PBCV-1 (11).
In higher eukaryotes, biochemical fractionation has shown that the guanylyltransferase from rat liver copurifies with an RNA triphosphatase activity but that the (guanine-7-)methyltransferase readily separates in early chromatography steps and purifies as an unassociated enzyme (48). Recent cloning of the Caenorhabditis elegans (37, 46) and mammalian (17, 50) capping enzymes showed that these enzymes consist of a single bifunctional polypeptide with two domains: a carboxy-terminal guanylyltransferase domain and an amino-terminal domain with RNA triphosphatase activity. Even though the guanylyltransferase domain is strictly conserved with respect to amino acids that are essential for in vivo function, the RNA triphosphatase domain does not resemble the vaccinia virus triphosphatase domain but rather has significant sequence and mechanistic similarities to the family of protein tyrosine phosphatases. Thus, it would appear from the above examples that unicellular and multicellular organisms differ with respect to the physical organization of enzymatic activities of the capping machinery; whereas unicellular organisms have a monofunctional guanylyltransferase, multicellular organisms have a bifunctional triphosphatase-guanylyltransferase.
As a first step toward understanding the process of RNA capping in trypanosomes, we have purified the capping enzyme from C. fasciculata and cloned the corresponding genes from C. fasciculata and T. brucei. Comparison of the predicted amino acid sequences of the trypanosome proteins with those of the available eukaryotic and viral capping enzymes revealed several unique structural features. In particular, the trypanosome capping enzymes are remarkable in that they include a novel amino-terminal domain that displays no resemblance to any other domain associated with capping enzymes.
MATERIALS AND METHODS
Purification of C. fasciculata guanylyltransferase.
C. fasciculata (ATCC 12858) was grown at 28°C in brain heart infusion broth (Difco) supplemented with 10 μg of hemin per ml, 2× BME vitamin solution (Gibco BRL), 10 U of penicillin G (Gibco BRL) per ml, and 10 μg of streptomycin sulfate (Gibco BRL) per ml. After being harvested and washed, the cells were stored at −70°C. Cell yields were approximately 1 to 2 ml of packed cells per liter.
All subsequent manipulations were performed at 4°C with a fast protein liquid chromatography system (Pharmacia Biotech) for chromatography steps. C. fasciculata cells (60 ml of packed-cell volume) were resuspended in breaking buffer (10 mM HEPES [pH 7.9], 10 mM KCl, 1.5 mM MgCl2, 1 mM dithiothreitol [DTT], and 10 μg of leupeptin per ml) at a total volume of 360 ml and passed through a Microfluidizer (Microfluidics). The crude cell extract was centrifuged at 4,000 × g for 15 min, and to the supernatant a 1/10 volume of 0.3 M HEPES (pH 7.9)–1.3 M KCl–30 mM MgCl2 was added. After centrifugation at 100,000 × g for 45 min, solid ammonium sulfate was added to the supernatant to give 40% saturation. The precipitate was collected by centrifugation at 10,000 × g for 20 min, dissolved in a minimal volume of buffer T (20 mM Tris-HCl [pH 7.5], 0.1 mM EDTA, 10% glycerol, 1 mM DTT, 10 μg of leupeptin per ml), and dialyzed overnight against the same buffer containing 100 mM KCl. After centrifugation, the supernatant was applied to an 80-ml DEAE–Sepharose CL-6B column equilibrated with buffer T containing 100 mM KCl. Bound proteins were step eluted with 400 mM KCl in buffer T, adjusted to pH 6.5 by the addition of a 1/50 volume of 1 M 2-(N-morpholino)ethanesulfonic acid (MES) (without pH adjustment), diluted fourfold with buffer M (20 mM MES [pH 6.5], 0.1 mM EDTA, 10% glycerol, 1 mM DTT, 10 μg of leupeptin per ml), and applied to an 8-ml Mono S column (Pharmacia Biotech) equilibrated with buffer M containing 100 mM KCl. A 120-ml linear gradient of 100 to 400 mM KCl was applied, and active fractions eluting between 260 and 340 mM KCl were pooled and concentrated with a Centricon-10 concentrator (Amicon). After buffer exchange to buffer T containing 100 mM KCl by use of a Hitrap desalting column (Pharmacia Biotech), the sample was loaded onto a 5-ml heparin-Sepharose high-performance column (Pharmacia Biotech), and bound proteins were step eluted with 400 mM KCl in buffer T. The eluate from the heparin column was diluted with 1 volume of buffer T and applied to a 5-ml GTP-agarose column (Sigma) equilibrated with buffer T containing 200 mM KCl. After extensive washing, the column was developed with a 20-ml gradient of 200 to 600 mM KCl. The enzyme eluted between 400 and 500 mM KCl.
Peptide sequencing.
Purified protein was separated by polyacrylamide gel electrophoresis (PAGE) and stained with Coomassie brilliant blue G (Sigma). Peptide sequences were obtained as previously described, except that Tween 20 was used at a concentration of 0.5% instead of 0.1% (47).
Assay for protein-guanylate complex formation.
Standard reaction mixtures (10 μl) contained 50 mM Tris-HCl (pH 7.5), 5 mM MgCl2, 2 mM DTT, [α-32P]GTP, and enzyme. After 15 min at 28°C, the reaction was terminated by the addition of sodium dodecyl sulfate (SDS) sample buffer, and the sample was boiled for 5 min and analyzed directly by SDS-PAGE on 10% acrylamide gels. Protein-guanylate complex formation was assessed by autoradiography of the gels.
Assay for RNA guanylyltransferase.
To measure RNA capping by guanylyltransferase, yeast low-molecular-weight RNA was used as a substrate as described previously (20). Yeast soluble RNA (type III; Sigma) was fractionated over a Superose 12 sizing column (Pharmacia Biotech), and RNA in the size range of 20 to 40 nucleotides was incubated at 28°C for 60 min in assay buffer (50 mM Tris-HCl [pH 7.5], 5 mM MgCl2, 2 mM DTT) with [α-32P]GTP and enzyme. RNA samples were then separated on a 10% denaturing polyacrylamide gel, and labeled RNA was isolated by soaking gel pieces in H2O. After ethanol precipitation in the presence of 30 μg of tRNA, samples were digested with nuclease P1 and nucleotide pyrophosphatase as described previously (41). The digestion products were chromatographed on polyethyleneimine-cellulose plates with 0.6 M potassium phosphate (pH 3.5) as a solvent.
Cloning of the C. fasciculata and T. brucei guanylyltransferase genes.
The following peptide sequences obtained from the purified C. fasciculata capping enzyme were used to design degenerate oligonucleotides: KKTQAVNVAEIYHLLHL (peptide 75K30) and KEVHSAIGNTVGGTDSA (peptide 75K32), corresponding to amino acids 149 to 165 and 340 to 356, respectively (amino acids that deviate from the predicted product of the open reading frame are underlined) (see Fig. 3). From the 75K30 sequence we designed three oligonucleotides: CAP1 (5′-ATYTCIGCNACRTTIACNGCYTG-3′), CAP2 (5′-AGIAGRTGRTADATYTCIGCNACRTT-3′), and CAP5 (5′-AARAARACICARGCNGTIAAYGTNGC-3′). CAP3 (5′-GCIATHGGIAAYACNGTNGG-3′) and CAP4 (5′-CCNACIGTRTTICCDATNGC-3′) correspond to DNA sequences predicted from the 75K32 peptide (I represents deoxyinosine). A C. fasciculata genomic library in lambda EMBL3 (kindly provided by D. MacMahon-Pratt) was screened with degenerate oligonucleotides labeled at the 5′ end with T4 polynucleotide kinase and [γ-32P]ATP. Filters were hybridized at 42°C for 2 days in buffer containing 5× SSPE (1× SSPE is 0.18 M NaCl, 10 mM NaH2PO4, and 1 mM EDTA [pH 7.7]), 5× Denhardt’s solution, 0.1% SDS, 100 μg of yeast soluble RNA (type III; Sigma) per ml, and 25 ng of oligonucleotides per ml. Filters were washed at 50°C in 5× SSPE–0.1% SDS for 2 hours with one change of solution. Positive phages were characterized by restriction enzyme mapping and Southern blotting. Convenient restriction fragments were subcloned into the pT3T7 vector and sequenced.
FIG. 3.
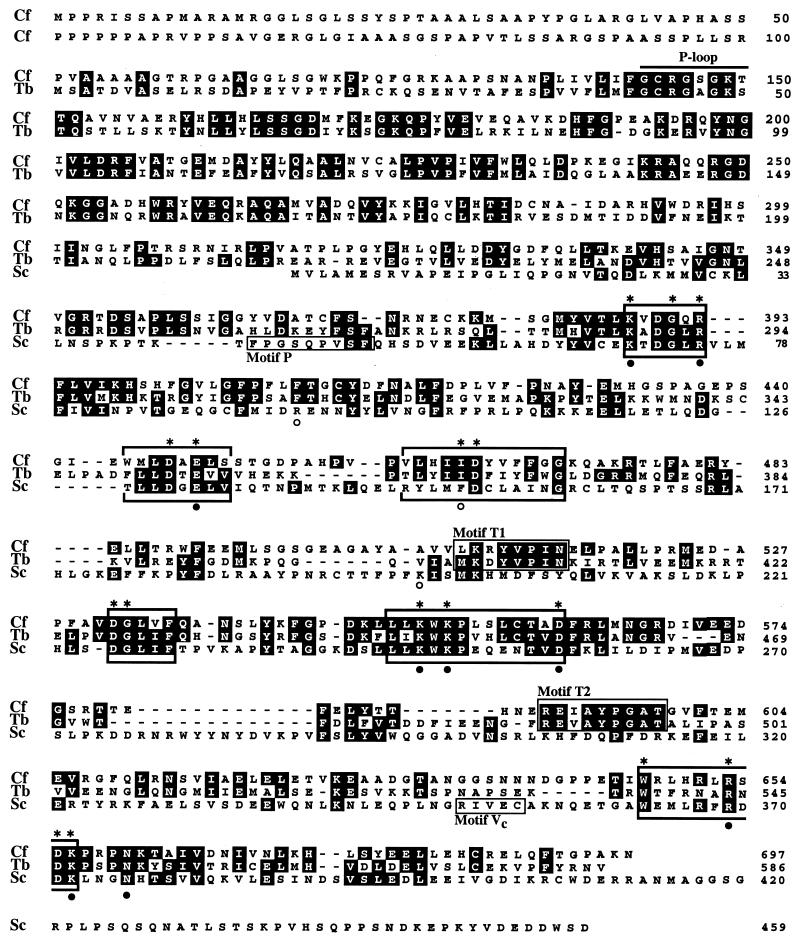
Alignment of the amino acid sequences of the C. fasciculata (Cf), T. brucei (Tb), and S. cerevisiae (Sc) capping enzymes. Amino acid identity is denoted by shaded residues. The six motifs characteristic of the nucleotidyltransferase superfamily (I, III, IIIa, IV, V, and VI) are indicated by brackets above and below the sequences (33, 46). Residues in these motifs that were shown to be essential for activity of the S. cerevisiae capping enzyme in vivo are indicated by asterisks above the sequences. Amino acids in physical proximity to the GTP moiety in the Chlorella virus capping enzyme-GTP cocrystal structure (9) are shown by filled circles (conserved in the trypanosome sequence) or open circles (not conserved in the trypanosome sequence). The phosphate-binding loop, or P-loop (27), with the consensus sequence GXXXXGK[T/S] is indicated toward the amino termini of the trypanosome proteins.
A T. brucei rhodesiense unidirectional oligo(dT)-primed cDNA library was constructed in lambda ZAP II (Stratagene), and clones for random sequencing were selected on the basis of the criterion of low reactivity with a total cDNA probe (43a). With this approach, an expressed sequence tag with homology to the C. fasciculata guanylyltransferase was isolated and used as a probe to screen a T. brucei genomic library. Positive phages were characterized as described above.
In vitro transcription and translation.
The T. brucei guanylyltransferase was cloned in frame into pET-28b (Novagen), and the TNT-coupled reticulocyte lysate system (Promega) was used to synthesize proteins from recombinant plasmids. The truncated construct expressing only the guanylyltransferase domain was constructed by PCR and contained amino acids 212 to 586. The lysine residue in motif I was mutated to an arginine residue by two sequential PCRs as described previously (7).
Nucleotide sequence accession number.
The GenBank nucleotide and protein sequence accession numbers for the T. brucei and C. fasciculata capping enzymes are AF059246 and AF059247, respectively.
RESULTS
Protein-guanylate complex formation in C. fasciculata extracts.
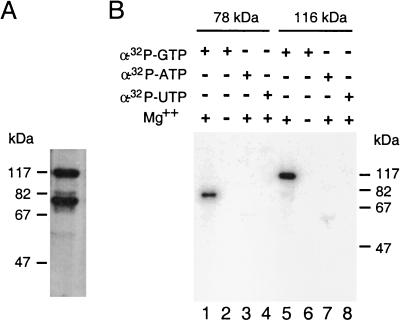
In T. brucei and C. fasciculata, the cap structure of the SL RNA consists of m7G linked via a 5′-5′ triphosphate bridge to four methylated nucleotides (2). In order to identify the trypanosome GTP:RNA guanylyltransferase, or capping enzyme, we took advantage of the fact that the transfer of GMP from GTP to the diphosphate termini of RNA proceeds through two reversible steps and that the intermediate in this reaction, an enzyme-GMP complex, can be conveniently identified by radioactive labeling of the protein with [α-32P]GTP. To determine whether trypanosome protein extracts have the ability to form such a complex, we chose C. fasciculata, which is readily accessible for extensive biochemical purifications. Aliquots of cell extracts were incubated in the presence of [α-32P]GTP for 10 min, denatured in the presence of SDS, and analyzed directly on an SDS-polyacrylamide gel. Autoradiography of the gel revealed the formation of two SDS-stable nucleotide-protein adducts, which migrated with apparent molecular masses of 116,000 and 78,000 daltons (Fig. 1A). Similarly, when a T. brucei whole-cell extract was used, two polypeptides, of approximately 116,000 and 70,000 daltons, were labeled with [α-32P]GTP (see Fig. 4, lane 1).
FIG. 1.
Formation of covalent protein-GMP complex by C. fasciculata extracts. (A) A C. fasciculata whole-cell extract was incubated with [α-32P]GTP, and complex formation was analyzed directly by SDS-PAGE followed by autoradiography. (B) Nucleotide and magnesium specificity for the formation of covalent protein complexes. Aliquots from Mono S fractions containing either the 78- or the 116-kDa polypeptide were incubated with the indicated α-32P-labeled nucleoside triphosphates in the presence (lanes 1, 3 to 5, 7, and 8) or absence (lanes 2 and 6) of 5 mM MgCl2. Samples were analyzed by SDS-PAGE and visualized by autoradiography.
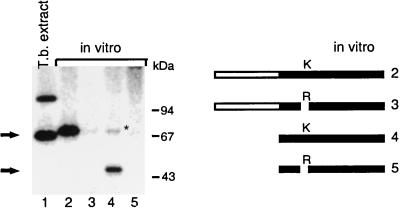
FIG. 4.
Formation of protein-GMP complex by recombinant T. brucei capping enzyme. Proteins were incubated in 50 mM Tris-HCl (pH 7.5)–5 mM MgCl2–5 mM DTT–[α-32P]GTP for 10 min at 28°C, denatured, and analyzed by SDS-PAGE. An autoradiograph of the gel is shown. The sources of proteins were as follows: the reaction mixture in lane 1 contained an aliquot (about 5 μg) of a T. brucei whole-cell extract, and the reaction mixtures in lanes 2 to 5 were incubated with an aliquot of a coupled in vitro transcription-translation reaction mixture that generated the proteins illustrated graphically to the right of the autoradiograph. The asterisk indicates a background band whose intensity varied from experiment to experiment.
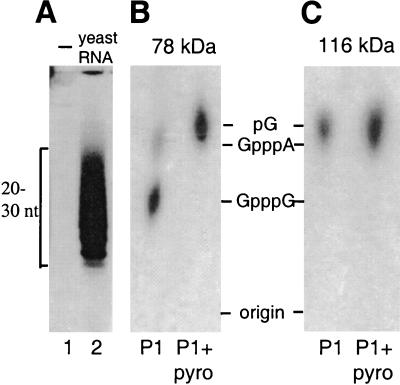
To determine whether any of these labeled polypeptides might be involved in RNA capping by endogenous RNA guanylyltransferase, the C. fasciculata cell extract was fractionated over a Mono S column. This chromatography resulted in the separation of the two polypeptides into distinct fractions, which were then used for further characterization. Both polypeptides required magnesium to bind GTP, and they were unable to bind either ATP or UTP in a covalent fashion (Fig. 1B). To directly look at cap formation, low-molecular-weight yeast RNA was incubated with an aliquot of the Mono S fraction containing either the 78- or the 116-kDa polypeptide in the presence of [α-32P]GTP (Fig. 2A). The labeled RNA was then gel purified and analyzed by digestion with different RNases and subsequent separation by thin-layer chromatography. The result of such an analysis showed that incubation of yeast RNA with the 78-kDa polypeptide fraction and subsequent digestion with nuclease P1 generated products which comigrated with the markers for GpppG and GpppA dinucleotides and that both products were susceptible to digestion with pyrophosphatase (Fig. 2B). These results were consistent with the action of guanylyltransferase, namely, to add GMP to the 5′ end of RNA by forming a pyrophosphate bridge. In contrast, a similar analysis with the 116-kDa polypeptide fraction did not show P1-resistant structures (Fig. 2C); therefore, characterization of this polypeptide was not pursued further. We concluded from these data that the enzymatic activity associated with the 78-kDa polypeptide was clearly distinct from a previously described 5′-end-labeling activity in T. cruzi extracts (51) and was most likely the C. fasciculata guanylyltransferase.
FIG. 2.
RNA capping by the Mono S fraction containing the 78-kDa polypeptide from C. fasciculata. An aliquot of the Mono S fraction containing either the 78- or the 116-kDa polypeptide was incubated with low-molecular-weight yeast RNA and [α-32P]GTP in 50 mM Tris-HCl (pH 7.5)–5 mM MgCl2–5 mM DTT. After phenol-chloroform extraction, labeled RNA was fractionated on a denaturing 25% urea-polyacrylamide gel. (A) Lane 2 shows labeling of exogenous yeast RNA with the 78-kDa polypeptide-containing fraction, which is indistinguishable from that obtained with the 116-kDa polypeptide-containing fraction. Lane 1 shows that no appreciable labeling was observed in the absence of exogenous yeast RNA. (B and C) Labeled RNA species were eluted from gel slices; one aliquot was digested with nuclease P1 (P1), and the other aliquot was digested with nuclease P1 and pyrophosphatase (P1+pyro). The digestion products were separated by thin-layer chromatography. The positions of marker nucleotides (nt) were determined by UV illumination.
Purification and cloning of the C. fasciculata guanylyltransferase.
Five different steps, including ammonium sulfate precipitation and chromatography on DEAE-Sepharose, Mono S, heparin-agarose, and GTP-agarose, were used for the purification of the 78-kDa polypeptide from a C. fasciculata whole-cell extract. In the final fraction, approximately five polypeptides were present, and the 78-kDa polypeptide was excised from a preparative SDS-polyacrylamide gel and subjected to digestion in situ with lysylendopeptidase. The obtained peptide sequences were then used to design degenerate DNA oligonucleotides, and the corresponding gene was isolated by screening of a C. fasciculata genomic library. Using this approach, we isolated a C. fasciculata gene with a predicted open reading frame of 697 amino acids, resulting in a protein with a molecular mass of 76 kDa.
The C. fasciculata capping enzyme consists of two domains.
The predicted open reading frame for the C. fasciculata capping enzymes is considerably larger than that for the S. cerevisiae guanylyltransferase, which is 459 amino acids long (26). A comparison between the trypanosome protein and the yeast protein revealed that the S. cerevisiae guanylyltransferase aligns with the carboxy-terminal half of the C. fasciculata capping enzyme (Fig. 3). The similarity is manifested by the presence of six motifs that were previously noted to be characteristic of eukaryotic and viral capping enzymes (33). These are relatively short stretches of amino acids that are found in the same order and with similar spacing in different members of this family of nucleotidyltransferases. So far, Shuman and collaborators have described six motifs (I, III, IIIa, IV, V, and VI) that are absolutely conserved in the superfamily of covalent nucleotidyltransferases (33, 46). Within these motifs, 16 residues in the yeast capping enzyme were found by mutational analysis to be essential for in vivo function, and all but 1 of these residues are conserved in the Crithidia protein (Fig. 3). The active site of guanylyltransferase is located in motif I, where GMP binds through a lysine residue, and in the C. fasciculata capping enzyme this residue is found at position 389 (KVDGQR). Except for the six motifs, there is little similarity between the trypanosome and yeast enzymes, namely, 25% identity and 38% similarity. Taken together, these structural similarities convinced us that with respect to the carboxy-terminal domain of the molecule, the isolated C. fasciculata protein was the capping enzyme.
There are several structural differences worth noting. The crystal structure of the Chlorella virus capping enzyme with bound GTP revealed 12 amino acids in proximity to GTP (9). Of these 12 residues, only 9 are conserved in the C. fasciculata protein (Fig. 3). Furthermore, Shuman and colleagues recently postulated two more conserved sequence motifs (P and Vc; Fig. 3) that define a new subgroup of capping enzymes, comprising the S. cerevisiae, S. pombe, C. albicans, C. elegans, and Chlorella virus enzymes (46). The C. fasciculata protein does not appear to be part of this group, since both motifs are absent.
What distinguishes the trypanosome capping enzymes even more from the S. cerevisiae guanylyltransferase, as well as from other fungal guanylyltransferases, is the presence of an additional domain at the amino terminus, comprising 316 amino acids in the C. fasciculata protein. A computer-assisted search with this amino-terminal domain against the available protein databases led to the finding that an area centering around position 146 in the C. fasciculata protein is most closely related to adenylate kinases from a variety of organisms. Closer inspection revealed that this similarity was confined to a phosphate-binding loop, or P-loop (27). The primary structure of this motif typically consists of a glycine-rich sequence followed by a conserved lysine and a serine or threonine: GXXXXGK[T/S]. The P-loop is one motif commonly found in ATP- and GTP-binding proteins, including, among others, adenylate kinases, elongation factors, myosin heavy chains, RecA protein, and Ras proteins (27, 45). This motif matched residues 143 to 150 of the C. fasciculata capping enzyme (Fig. 3).
The two-domain structure of the capping enzyme appears to be conserved in trypanosomatid protozoa.
Through an ongoing sequencing project in our laboratory of random T. brucei EST clones, we isolated an EST that showed considerable homology to the C. fasciculata guanylyltransferase. In particular, the predicted amino acid sequence of this EST revealed several of the signature motifs that are characteristic of guanylyltransferases. Using the EST as a probe, we isolated genomic clones from a T. brucei library, and further sequence analysis identified an open reading frame for a protein of 586 amino acids. Based on the primary structure of this protein as well as on functional studies (see below), we concluded that the isolated gene was the T. brucei homolog of the capping enzyme gene. The analysis of several independent genomic clones and genomic Southern hybridizations (data not shown) are consistent with the T. brucei capping enzyme being encoded by a single-copy gene.
Overall, the structure of the T. brucei capping enzyme is highly similar to that of the corresponding C. fasciculata protein (Fig. 3): the two-domain structure is conserved, with the guanylyltransferase domain again being located in the carboxy-terminal half of the molecule and with an amino-terminal domain that contains a P-loop consensus sequence. Between the two trypanosome sequences there is 44% identity at the amino acid level, with the highest degree of conservation being clustered around the signature motifs and toward the amino terminus of the T. brucei protein. This finding is quite similar to what is found for the S. cerevisiae and S. pombe enzymes, which are 38% identical.
As mentioned above, the recently postulated conserved sequence motifs P and Vc do not appear to be part of the trypanosome capping enzymes, since they are also absent from the T. brucei protein. Instead, we identified two trypanosome-specific motifs (T1 and T2; Fig. 3) that are not found in any other guanylyltransferase.
The recombinant T. brucei capping enzyme binds GMP.
Although the primary structure of the trypanosome capping enzymes revealed a guanylyltransferase domain, it was important to test whether the cloned genes actually encode a protein that can bind GMP in a covalent fashion. To do this, we initially expressed both the T. brucei and the C. fasciculata capping enzymes in Escherichia coli with several different expression vectors. Since this process did not result in the production of sufficient quantities of recombinant proteins in a soluble form to perform enzymatic tests, we used a coupled reticulocyte transcription-translation system to generate the T. brucei capping enzyme in vitro. Enzyme-GMP complex formation was then assayed with [α-32P]GTP. This assay showed that the full-length recombinant T. brucei capping enzyme formed an SDS-stable nucleotide-protein adduct that migrated slightly slower than the endogenous protein (Fig. 4, lane 1) due to the presence of an N-terminal histidine tag (lane 2). Furthermore, a truncated construct expressing only the guanylyltransferase domain also bound GMP (Fig. 4, lane 4). Both proteins lost the ability to bind GMP when the lysine residue of motif I (Fig. 3) was mutated to an arginine residue (Fig. 4, lanes 3 and 5). Taken together, these data further corroborated our conclusion that the cloned T. brucei protein has a guanylyltransferase domain.
DISCUSSION
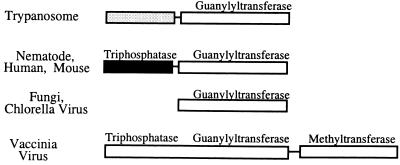
We have succeeded in purifying the C. fasciculata capping enzyme and in cloning the corresponding gene both from C. fasciculata and from T. brucei. We have based our identification on the presence of conserved signature motifs characteristic of nucleotidyltransferases and on the detection of guanylyltransferase activity in the biochemically purified C. fasciculata protein and in the T. brucei recombinant protein. The structure of the capping enzymes from these ancient protozoa complements the recently described structures of the C. elegans and mammalian capping enzymes (3, 17, 37, 46, 50). Figure 5 shows a diagram of the structure of the available eukaryotic capping enzymes. In terms of their overall structural arrangement, it is evident that the trypanosome capping enzymes are more akin to the C. elegans and mammalian capping enzymes than to the fungal capping enzymes. Relative to their fungal counterparts, the trypanosome, nematode, and mammalian enzymes contain an additional amino-terminal domain. In the C. elegans and mouse capping enzymes, this domain is approximately 230 amino acids long and has been shown to possess nucleotide triphosphatase activity, thus demonstrating that higher eukaryotic capping enzymes are bifunctional. Although the primary sequences of the trypanosomatid amino-terminal domains are different from the sequences of the latter, it is tempting to speculate that the trypanosome proteins are also bifunctional polypeptides, containing a triphosphatase domain fused to the guanylyltransferase domain. As pointed out above, we identified a P-loop or adenylate kinase homology region (Fig. 3) within the trypanosome-specific amino-terminal domain. This homology region is suggestive of triphosphatase activity, because adenylate kinase is known to transfer the γ-phosphate from ATP to AMP to generate two molecules of ADP. Thus, the amino-terminal domain of the trypanosome capping enzymes could conceivably remove the γ-phosphate from triphosphate-terminated RNA by a reaction mechanism that resembles the phosphotransferase reaction of adenylate kinases. Unfortunately, experimental proof of this enzymatic activity is not yet available, because we have not succeeded in producing either the C. fasciculata or the T. brucei full-length protein or the corresponding amino-terminal domain in sufficient quantities in soluble form in bacteria. In addition, we have not been able to detect triphosphatase activity in partially purified capping enzyme preparations.
FIG. 5.
Organization of functional domains in capping enzymes. Adopted from Wang et al. (46).
In most eukaryotic organisms, guanylyltransferase-mediated RNA capping is restricted to transcripts synthesized by PolII, namely, mRNAs and the majority of U snRNAs. Very recent data have demonstrated that the specificity of this capping reaction is brought about by an interaction of the phosphorylated form of the PolII carboxy-terminal domain with guanylyltransferase (3, 17, 50). However, in trypanosomatid protozoa, the mechanism of interaction between the capping enzyme and the transcriptional machinery must be fundamentally different for the following reasons. First, the large subunit of trypanosomatid PolII does not have a carboxy-terminal-like domain consisting of heptad repeats (6, 35), as has been described for the yeast and vertebrate enzymes (1, 5). Second, in trypanosomes capping is not restricted to PolII transcripts. In these organisms the capping enzyme acts upon the SL RNA, which is transcribed by an as-yet-undetermined RNA polymerase, and upon a specific subset of PolIII transcripts, namely, U2, U3, and U4 snRNAs. In contrast, the most abundant trypanosome PolIII transcripts, namely, tRNAs, 5S RNA, and 7SL RNA, are not capped. Thus, in principle, the trypanosome capping enzyme is capable of even greater selectivity than its higher eukaryotic counterpart, in that it discriminates between transcripts synthesized by the same RNA polymerase. Furthermore, as mentioned earlier, mRNA caps in trypanosomes are first formed on the SL RNA and then transferred to mRNAs by trans splicing. Once the RNA polymerase responsible for transcription of the SL RNA is unequivocally identified, it will become clear whether the trypanosome capping enzyme can cap RNAs other than PolIII transcripts. Notwithstanding this uncertainty, in the case of the U snRNAs it is unlikely that the specificity of the trypanosome capping enzyme can solely be accounted for by an interaction with the large subunit of PolIII. There are several alternate possibilities. For instance, it is possible that an interaction between the capping enzyme and the PolIII transcriptional machinery is mediated by factors which are gene specific and which assemble on gene-specific promoter elements. Indeed, our analysis of PolIII promoters of the U6 and U2 snRNA genes has demonstrated the existence of extragenic and intragenic control regions (7, 24). Whereas the extragenic control regions coincide with the A and B blocks of tRNA or tRNA-like gene promoters for the U6 and U2 genes, respectively, the intragenic promoter elements are clearly different in sequence between the two genes and may provide information on the assembly of gene-specific and possibly capping enzyme-specific transcription complexes. In this situation, capping would still occur cotranscriptionally, similar to the capping mechanism in higher eukaryotes. Another possibility is that the selection of RNA transcripts to be capped is based upon RNA determinants, which may be provided either by nascent or perhaps full-length transcripts.
Aside from the overall structural similarity, there is very little primary sequence conservation between the amino-terminal domains of the trypanosomatid proteins and of the nematode or mammalian proteins. Thus, the trypanosomatid capping enzymes define a new family or subgroup. At present, there are three distinct eukaryotic capping enzyme types, namely, the trypanosomal, the fungal, and the nematode-mammalian types, and among these three types there is little primary sequence conservation overall. We find it intriguing that cellular enzymes which are essential for cell viability display so little conservation in primary sequence. This finding could be taken as an indication that capping enzymes are in general quite tolerant to amino acid substitutions and are therefore relatively free to evolve and/or to coevolve with other cellular components with which they may interact. It is possible that the distinct structural features of each capping enzyme type reflect differences in substrate selection, differences in potential interactions with components of the transcription apparatus, or perhaps differences in enzymatic mechanisms. The possibility that the two-domain structure of the trypanosomal and nematode-mammalian enzymes was derived from different ancestral genes and that capping enzymes have a polyphyletic origin should also be considered. However, only further studies on the phylogeny of capping enzymes will clarify this point.
ACKNOWLEDGMENTS
We thank Diane MacMahon-Pratt for generously providing the C. fasciculata genomic library and Helen Kwon for excellent technical assistance. We thank David Bermudes, Christopher Yoo, and members of our laboratory for valuable criticism on the manuscript.
This investigation received financial support from National Institutes of Health grants AI28798 to E.U. and CA45508 to R.K. and from the UNDP/World Bank/WHO Special Programme for Research and Training in Tropical Diseases (TDR) (grant to C.T.).
REFERENCES
- 1.Allison L A, Moyle M, Shales M, Ingles C J. Extensive homology among the largest subunits of eukaryotic and prokaryotic RNA polymerases. Cell. 1985;42:599–610. doi: 10.1016/0092-8674(85)90117-5. [DOI] [PubMed] [Google Scholar]
- 2.Bangs J D, Crain P F, Hashizume T, McCloskey J A, Boothroyd J C. Mass spectrometry of mRNA cap 4 from trypanosomatids reveals two novel nucleosides. J Biol Chem. 1992;267:9805–9815. [PubMed] [Google Scholar]
- 3.Cho E J, Takagi T, Moore C R, Buratowski S. mRNA capping enzyme is recruited to the transcription complex by phosphorylation of the RNA polymerase II carboxy-terminal domain. Genes Dev. 1997;11:3319–3326. doi: 10.1101/gad.11.24.3319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cong P, Shuman S. Methyltransferase and subunit association domains of vaccinia virus mRNA capping enzyme. J Biol Chem. 1992;267:16424–16429. [PubMed] [Google Scholar]
- 5.Corden J L, Cadena D L, Ahearn J M, Jr, Dahmus M E. A unique structure at the carboxyl terminus of the largest subunit of eukaryotic RNA polymerase II. Proc Natl Acad Sci USA. 1985;82:7934–7938. doi: 10.1073/pnas.82.23.7934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Evers R, Hammer A, Kock J, Jess W, Borst P, Memet S, Cornelissen A W. Trypanosoma brucei contains two RNA polymerase II largest subunit genes with an altered C-terminal domain. Cell. 1989;56:585–597. doi: 10.1016/0092-8674(89)90581-3. [DOI] [PubMed] [Google Scholar]
- 7.Fantoni A, Dare A O, Tschudi C. RNA polymerase III-mediated transcription of the trypanosome U2 small nuclear RNA gene is controlled by both intragenic and extragenic regulatory elements. Mol Cell Biol. 1994;14:2021–2028. doi: 10.1128/mcb.14.3.2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gunzl A, Ullu E, Dorner M, Fragoso S P, Hoffmann K F, Milner J D, Morita Y, Nguu E K, Vanacova S, Wunsch S, Dare A O, Kwon H, Tschudi C. Transcription of the Trypanosoma brucei spliced leader RNA gene is dependent only on the presence of upstream regulatory elements. Mol Biochem Parasitol. 1997;85:67–76. doi: 10.1016/s0166-6851(96)02816-2. [DOI] [PubMed] [Google Scholar]
- 9.Hakansson K, Doherty A J, Shuman S, Wigley D B. X-ray crystallography reveals a large conformational change during guanyl transfer by mRNA capping enzymes. Cell. 1997;89:545–553. doi: 10.1016/s0092-8674(00)80236-6. [DOI] [PubMed] [Google Scholar]
- 10.Higman M A, Bourgeois N, Niles E G. The vaccinia virus mRNA (guanine-N7-)-methyltransferase requires both subunits of the mRNA capping enzyme for activity. J Biol Chem. 1992;267:16430–16437. [PubMed] [Google Scholar]
- 11.Ho C K, Van Etten J L, Shuman S. Expression and characterization of an RNA capping enzyme encoded by Chlorella virus PBCV-1. J Virol. 1996;70:6658–6664. doi: 10.1128/jvi.70.10.6658-6664.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Itoh N, Mizumoto K, Kaziro Y. Messenger RNA guanylyltransferase from Saccharomyces cerevisiae. I. Purification and subunit structure. J Biol Chem. 1984;259:13923–13929. [PubMed] [Google Scholar]
- 13.Itoh N, Yamada H, Kaziro Y, Mizumoto K. Messenger RNA guanylyltransferase from Saccharomyces cerevisiae. Large scale purification, subunit functions, and subcellular localization. J Biol Chem. 1987;262:1989–1995. [PubMed] [Google Scholar]
- 14.Laird P W, Zomerdijk J C, de Korte D, Borst P. In vivo labelling of intermediates in the discontinuous synthesis of mRNAs in Trypanosoma brucei. EMBO J. 1987;6:1055–1062. doi: 10.1002/j.1460-2075.1987.tb04858.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mao X, Schwer B, Shuman S. Yeast mRNA cap methyltransferase is a 50-kilodalton protein encoded by an essential gene. Mol Cell Biol. 1995;15:4167–4174. doi: 10.1128/mcb.15.8.4167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mao X, Shuman S. Intrinsic RNA (guanine-7) methyltransferase activity of the vaccinia virus capping enzyme D1 subunit is stimulated by the D12 subunit. Identification of amino acid residues in the D1 protein required for subunit association and methyl group transfer. J Biol Chem. 1994;269:24472–24479. [PubMed] [Google Scholar]
- 17.McCracken S, Fong N, Rosonina E, Yankulov K, Brothers G, Siderovski D, Hessel A, Foster S, Program A E, Shuman S, Bentley D L. 5′-Capping enzymes are targeted to pre-mRNA by binding to the phosphorylated carboxy-terminal domain of RNA polymerase II. Genes Dev. 1997;11:3306–3318. doi: 10.1101/gad.11.24.3306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McNally K P, Agabian N. Trypanosoma brucei spliced-leader RNA methylations are required for trans splicing in vivo. Mol Cell Biol. 1992;12:4844–4851. doi: 10.1128/mcb.12.11.4844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mizumoto K, Kaziro Y. Messenger RNA capping enzymes from eukaryotic cells. Prog Nucleic Acid Res Mol Biol. 1987;34:1–28. doi: 10.1016/s0079-6603(08)60491-2. [DOI] [PubMed] [Google Scholar]
- 20.Mizumoto K, Kaziro Y, Lipmann F. Reaction mechanism of mRNA guanylyltransferase from rat liver: isolation and characterization of a guanylyl-enzyme intermediate. Proc Natl Acad Sci USA. 1982;79:1693–1697. doi: 10.1073/pnas.79.6.1693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Moss B, Ensinger M J, Martin S A, Wei C M. Modification of the 5′-terminus of mRNA by guanylyl and methyl transferases from vaccinia virus. In: Haenni A L, Beaud G, editors. In vitro transcription and translation of viral genomes. Paris, France: Institut National de la Santé et de la Recherche Médicale; 1975. pp. 161–168. [Google Scholar]
- 22.Mottram J, Perry K L, Lizardi P M, Luhrmann R, Agabian N, Nelson R G. Isolation and sequence of four small nuclear U RNA genes of Trypanosoma brucei subsp. brucei: identification of the U2, U4, and U6 RNA analogs. Mol Cell Biol. 1989;9:1212–1223. doi: 10.1128/mcb.9.3.1212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Murphy W J, Watkins K P, Agabian N. Identification of a novel Y branch structure as an intermediate in trypanosome mRNA processing: evidence for trans splicing. Cell. 1986;47:517–525. doi: 10.1016/0092-8674(86)90616-1. [DOI] [PubMed] [Google Scholar]
- 24.Nakaar V, Dare A O, Hong D, Ullu E, Tschudi C. Upstream tRNA genes are essential for expression of small nuclear and cytoplasmic RNA genes in trypanosomes. Mol Cell Biol. 1994;14:6736–6742. doi: 10.1128/mcb.14.10.6736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rudenko G, Bishop D, Gottesdiener K, Van der Ploeg L H. Alpha-amanitin resistant transcription of protein coding genes in insect and bloodstream form Trypanosoma brucei. EMBO J. 1989;8:4259–4263. doi: 10.1002/j.1460-2075.1989.tb08611.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rudenko G, Lee M G, Van der Ploeg L H. The PARP and VSG genes of Trypanosoma brucei do not resemble RNA polymerase II transcription units in sensitivity to Sarkosyl in nuclear run-on assays. Nucleic Acids Res. 1992;20:303–306. doi: 10.1093/nar/20.2.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Saraste M, Sibbald P R, Wittinghofer A. The P-loop—a common motif in ATP- and GTP-binding proteins. Trends Biochem Sci. 1990;15:430–434. doi: 10.1016/0968-0004(90)90281-f. [DOI] [PubMed] [Google Scholar]
- 28.Shibagaki Y, Itoh N, Yamada H, Nagata S, Mizumoto K. mRNA capping enzyme. Isolation and characterization of the gene encoding mRNA guanyly[l]transferase subunit from Saccharomyces cerevisiae. J Biol Chem. 1992;267:9521–9528. [PubMed] [Google Scholar]
- 29.Shuman S. Functional domains of vaccinia virus mRNA capping enzyme. Analysis by limited tryptic digestion. J Biol Chem. 1989;264:9690–9695. [PubMed] [Google Scholar]
- 30.Shuman S, Hurwitz J. Mechanism of mRNA capping by vaccinia virus guanylyltransferase: characterization of an enzyme-guanylate intermediate. Proc Natl Acad Sci USA. 1981;78:187–191. doi: 10.1073/pnas.78.1.187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shuman S, Liu Y, Schwer B. Covalent catalysis in nucleotidyl transfer reactions: essential motifs in Saccharomyces cerevisiae RNA capping enzyme are conserved in Schizosaccharomyces pombe and viral capping enzymes and among polynucleotide ligases. Proc Natl Acad Sci USA. 1994;91:12046–12050. doi: 10.1073/pnas.91.25.12046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shuman S, Morham S G. Domain structure of vaccinia virus mRNA capping enzyme. Activity of the Mr 95,000 subunit expressed in Escherichia coli. J Biol Chem. 1990;265:11967–11972. [PubMed] [Google Scholar]
- 33.Shuman S, Schwer B. RNA capping enzyme and DNA ligase: a superfamily of covalent nucleotidyl transferases. Mol Microbiol. 1995;17:405–410. doi: 10.1111/j.1365-2958.1995.mmi_17030405.x. [DOI] [PubMed] [Google Scholar]
- 34.Shuman S, Surks M, Furneaux H, Hurwitz J. Purification and characterization of a GTP-pyrophosphate exchange activity from vaccinia virions. Association of the GTP-pyrophosphate exchange activity with vaccinia mRNA guanylyltransferase. RNA (guanine-7-)methyltransferase complex (capping enzyme) J Biol Chem. 1980;255:11588–11598. [PubMed] [Google Scholar]
- 35.Smith J L, Levin J R, Ingles C J, Agabian N. In trypanosomes the homolog of the largest subunit of RNA polymerase II is encoded by two genes and has a highly unusual C-terminal domain structure. Cell. 1989;56:815–827. doi: 10.1016/0092-8674(89)90686-7. [DOI] [PubMed] [Google Scholar]
- 36.Sutton R E, Boothroyd J C. Evidence for trans splicing in trypanosomes. Cell. 1986;47:527–535. doi: 10.1016/0092-8674(86)90617-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Takagi T, Moore C R, Diehn F, Buratowski S. An RNA 5′-triphosphatase related to the protein tyrosine phosphatases. Cell. 1997;89:867–873. doi: 10.1016/s0092-8674(00)80272-x. [DOI] [PubMed] [Google Scholar]
- 38.Toyama R, Mizumoto K, Nakahara Y, Tatsuno T, Kaziro Y. Mechanism of the mRNA guanylyltransferase reaction: isolation of N epsilon-phospholysine and GMP (5′ leads to N epsilon) lysine from the guanylyl-enzyme intermediate. EMBO J. 1983;2:2195–2201. doi: 10.1002/j.1460-2075.1983.tb01723.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tschudi C, Richards F F, Ullu E. The U2 RNA analogue of Trypanosoma brucei gambiense: implications for a splicing mechanism in trypanosomes. Nucleic Acids Res. 1986;14:8893–8903. doi: 10.1093/nar/14.22.8893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tutas D J, Paoletti E. Purification and characterization of core-associated polynucleotide 5′-triphosphatase from vaccinia virus. J Biol Chem. 1977;252:3092–3098. [PubMed] [Google Scholar]
- 41.Ullu E, Tschudi C. Accurate modification of the trypanosome spliced leader cap structure in a homologous cell-free system. J Biol Chem. 1995;270:20365–20369. doi: 10.1074/jbc.270.35.20365. [DOI] [PubMed] [Google Scholar]
- 42.Ullu E, Tschudi C. Permeable trypanosome cells as a model system for transcription and trans-splicing. Nucleic Acids Res. 1990;18:3319–3326. doi: 10.1093/nar/18.11.3319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ullu E, Tschudi C. Trans splicing in trypanosomes requires methylation of the 5′ end of the spliced leader RNA. Proc Natl Acad Sci USA. 1991;88:10074–10078. doi: 10.1073/pnas.88.22.10074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43a.Ullu, E., and C. Tschudi. Unpublished data.
- 44.Venkatesan S, Gershowitz A, Moss B. Modification of the 5′ end of mRNA. Association of RNA triphosphatase with the RNA guanylyltransferase-RNA (guanine-7-)methyltransferase complex from vaccinia virus. J Biol Chem. 1980;255:903–908. [PubMed] [Google Scholar]
- 45.Walker J E, Saraste M, Runswick M J, Gay N J. Distantly related sequences in the alpha- and beta-subunits of ATP synthase, myosin, kinases and other ATP-requiring enzymes and a common nucleotide binding fold. EMBO J. 1982;1:945–951. doi: 10.1002/j.1460-2075.1982.tb01276.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang G P, Deng L, Ho C K, Shuman S. Phylogeny of mRNA capping enzymes. Proc Natl Acad Sci USA. 1997;94:9573–9578. doi: 10.1073/pnas.94.18.9573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang R, Kobayashi R, Bishop J M. Cellular adherence elicits ligand-independent activation of the Met cell-surface receptor. Proc Natl Acad Sci USA. 1996;93:8425–8430. doi: 10.1073/pnas.93.16.8425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yagi Y, Mizumoto K, Kaziro Y. Association of an RNA 5′-triphosphatase activity with RNA guanylyltransferase partially purified from rat liver nuclei. EMBO J. 1983;2:611–615. doi: 10.1002/j.1460-2075.1983.tb01471.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yamada-Okabe T, Shimmi O, Doi R, Mizumoto K, Arisawa M, Yamada-Okabe H. Isolation of the mRNA-capping enzyme and ferric-reductase-related genes from Candida albicans. Microbiology. 1996;142:2515–2523. doi: 10.1099/00221287-142-9-2515. [DOI] [PubMed] [Google Scholar]
- 50.Yue Z, Maldonado E, Pillutla R, Cho H, Reinberg D, Shatkin A J. Mammalian capping enzyme complements mutant Saccharomyces cerevisiae lacking mRNA guanylyltransferase and selectively binds the elongating form of RNA polymerase II. Proc Natl Acad Sci USA. 1997;94:12898–12903. doi: 10.1073/pnas.94.24.12898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zwierzynski T A, Buck G A. In vitro capping in Trypanosoma cruzi identifies and shows specificity for the spliced leader RNA and U-RNAs. Nucleic Acids Res. 1990;18:4197–4206. doi: 10.1093/nar/18.14.4197. [DOI] [PMC free article] [PubMed] [Google Scholar]