Abstract
Previous studies have identified a conserved AG dinucleotide at the 3′ splice site (3′SS) and a polypyrimidine (pPy) tract that are required for trans splicing of polycistronic pre-mRNAs in trypanosomatids. Furthermore, the pPy tract of the Trypanosoma brucei α-tubulin 3′SS region is required to specify accurate 3′-end formation of the upstream β-tubulin gene and trans splicing of the downstream α-tubulin gene. Here, we employed an in vivo cis competition assay to determine whether sequences other than those of the AG dinucleotide and the pPy tract were required for 3′SS identification. Our results indicate that a minimal α-tubulin 3′SS, from the putative branch site region to the AG dinucleotide, is not sufficient for recognition by the trans-splicing machinery and that polyadenylation is strictly dependent on downstream trans splicing. We show that efficient use of the α-tubulin 3′SS is dependent upon the presence of exon sequences. Furthermore, β-tubulin, but not actin exon sequences or unrelated plasmid sequences, can replace α-tubulin exon sequences for accurate trans-splice-site selection. Taken together, these results support a model in which the informational content required for efficient trans splicing of the α-tubulin pre-mRNA includes exon sequences which are involved in modulation of trans-splicing efficiency. Sequences that positively regulate trans splicing might be similar to cis-splicing enhancers described in other systems.
In trypanosomatid protozoa, transcription of protein-encoding genes is polycistronic and pre-mRNAs are colinear with the corresponding chromosomal gene arrangements. mRNA-encoding sequences are separated by short intergenic regions, ranging in size from 100 to 500 nucleotides (nt). Monocistronic mRNAs are generated from polycistronic pre-mRNAs by two RNA-processing reactions, namely, trans splicing and 3′ cleavage-polyadenylation (for a review, see reference 23). Trans splicing entails the addition of a 39-nt leader sequence from the spliced leader (SL) RNA to the 5′ ends of mature mRNAs, whereas the 3′ end cleavage-polyadenylation process of trypanosome pre-mRNAs is thought to be akin to the 3′ cleavage-polyadenylation process occurring in higher eukaryotes. As a result of these reactions, the intergenic regions of trypanosome pre-mRNAs are cut off and discarded. Sequence comparisons and mutagenesis experiments have identified two essential elements of the 3′ splice site (3′SS) region required for trans splicing in trypanosomatids, namely, the conserved AG dinucleotide at the 3′SS and a polypyrimidine (pPy) tract of various lengths located just upstream of the 3′SS (8, 9, 14, 19, 25). However, no sequence analogous to the mammalian or yeast branch site consensus has yet been identified. Furthermore, no specific sequences for 3′-end formation and polyadenylation, like the AAUAAA sequence in higher eukaryotes, seem to be present in trypanosomatid mRNA (10, 12, 14, 19, 25). The only common features among trypanosome poly(A) sites are the presence of an adenosine residue before or after the poly(A) addition site(s) and the fact that 3′-end cleavage of pre-mRNA generates several closely spaced 3′ ends, rather than a unique end, as seen in vertebrate mRNAs. On the other hand, accurate choice of the poly(A) site requires a downstream 3′SS. Several lines of evidence indicate that in polycistronic pre-mRNAs, poly(A) site selection is coupled to downstream trans splicing. Inhibition of trans splicing by destruction of U2 small nuclear RNA (snRNA) in permeabilized cells of Trypanosoma brucei inhibits 3′-end formation of tubulin mRNA, as well as that of the majority of mRNAs (21). In addition, in the Leishmania dihydrofolate reductase-thymidine synthetase pre-mRNA, poly(A) site selection is specified by sequences spanning the 3′SS region (12). For the T. brucei α-tubulin 3′SS region, we determined that the pPy tract governs both trans splicing and polyadenylation (14). Similarly, a pPy tract associated with a cryptic 3′SS is required for polyadenylation of procyclin mRNA (10, 19). The former observation supported a model in which trans-splice-site selection and poly(A)-site selection were functionally coupled via recognition of the pPy tract, but it was not clear whether this element was independently recognized by the trans-splicing and 3′-end cleavage-polyadenylation machineries or whether the pPy tract functioned solely as part of the 3′SS. Mechanistic coupling of polyadenylation and trans splicing has also been described for Caenorhabditis elegans in the case of SL2 trans-spliced mRNAs (11). However, in this system the situation appears to be reversed in that trans splicing of the downstream mRNA is dependent upon a functional 3′-end-formation signal upstream, namely, the AAUAAA hexanucleotide.
There are several major gaps in our understanding of pre-mRNA processing in trypanosomatids. How are intergenic regions of trypanosome pre-mRNAs accurately recognized by the pre-mRNA-processing machinery? What distinguishes these regions from other regions in the pre-mRNA? In particular, why are mRNA-encoding regions not substrates for trans splicing and polyadenylation? At present there is no evidence of transcriptional regulation in trypanosomes at the level of transcription initiation, although in T. brucei regulation of transcription at the level of transcript elongation seems possible (18). Therefore, it is hypothesized that modulation of gene expression in terms of mRNA output on a per gene basis is achieved primarily by (i) regulatory loops that involve pre-mRNA turnover in combination with differential rates of trans splicing and polyadenylation and (ii) mRNA turnover. In this scenario, the pre-mRNA cis-acting signals for RNA processing, namely, the 3′ splice acceptor site and the poly(A) site, by virtue of their interactions with the RNA-processing machineries, would be major determinants for regulating gene expression in trypanosomes.
In the experiments reported here, we sought to answer the question of whether sequences other than those of the conserved AG dinucleotide and the pPy tract play a role in trans-splice-site selection. To this end we developed a cis competition assay using as a substrate a pre-mRNA containing tandem duplications of the α-tubulin 3′SS region. We show that the identification of the α-tubulin 3′SS requires downstream exon sequences, located in the 5′ untranslated region (5′UTR) of α-tubulin mRNA. The informational content of the 5′UTR is complex, consisting of sequences which are essential for trans-splice-site choice, as well as sequences which appear to exert a negative effect. Furthermore, our results demonstrate that use of the wild-type (wt) poly(A) site of β-tubulin mRNA strictly depends on active trans splicing at the downstream 3′SS and that the pPy tract functions primarily for 3′SS identification but has no role per se in polyadenylation.
MATERIALS AND METHODS
Plasmid constructs.
A dicistronic expression vector was assembled from gene cassettes obtained by PCR amplification with specific oligonucleotides carrying unique restriction sites at their ends. The starting plasmid was pGFPΔBX (20a), which contains inserted into pSP72 (Stratagene) downstream from the SP6 promoter the ribosomal promoter, the α-β-tubulin intergenic region, the green fluorescent protein (GFP)-coding region, and the PARP 3′UTR adjacent to the T7 promoter. A unique BamHI site between GFP and the PARP 3′UTR was used to insert the β-α-tubulin intergenic region followed by the chloramphenicol acetyltransferase (CAT)-coding region to give the construct pWT.
For mutagenesis, the β-α-tubulin intergenic region was transferred into pBluescriptII KS− (Stratagene) by using the flanking XbaI and XhoI sites at the 5′ and 3′ ends, respectively. Mutagenesis was performed by two sequential PCRs, and the introduced mutations were verified by DNA sequencing. To construct AS1 and AS2, a unique SmaI site was introduced at position 474 of the β-α-tubulin intergenic region. The SmaI-containing intergenic region was put back into the dicistronic construct to give pWTS+, and PCR fragments with SmaI sites at either end were inserted. In all the other constructs the SmaI site was changed to a SalI site by the insertion of a linker. The UTR and SM mutants were generated by three sequential PCRs with pWTS+ as a template. Two PCRs were performed with the mutagenic oligonucleotides and either GFPOUT (5′-GACCACATGGTCCTTCTTGAG-3′) or CAT-5 (5′-GCCATTGGGATATATCAACGGTGG-3′) as an external oligonucleotide. Sequences to be duplicated were PCR amplified with oligonucleotides containing SalI sites at either end.
β-Tubulin and actin 5′UTR sequences were generated by annealing two partially overlapping oligonucleotides. After conversion to double-stranded DNA with Klenow polymerase and digestion with BglII and XhoI, the fragments were put in place of the α-tubulin 5′UTR between the BglII site at the 3′SS (AGATCT; the AG dinucleotide is underlined) and the XhoI site immediately upstream of the CAT initiation codon.
DNA transfections and nucleic acid analysis.
Transient transfection of procyclic forms of T. brucei rhodesiense, RNA isolation, and primer extension analysis were done essentially as described previously (5, 14). U6 snRNA was primer extended with oligonucleotide U6-D, complementary to nt 66 to 83 of U6 snRNA, and CAT mRNA was primed with CAT-5, complementary to nt 26 to 39 of the CAT-coding region. Northern blotting was performed by standard procedures after the RNA was separated by electrophoresis through a 1.7% agarose–formaldehyde gel. Blots were hybridized to radiolabeled antisense PCR probes complementary to the coding regions of GFP or CAT mRNAs. Hybridizations were carried out at 50°C in a solution containing 50% formamide, 5× SET (1× SET is 150 mM NaCl, 10 mM Tris-HCl at pH 7.5, and 1 mM EDTA), 10× Denhardt’s solution, 100 μg of Saccharomyces cerevisiae RNA per ml, and 1% sodium dodecyl sulfate, and blots were washed at 60 to 65°C in 2× SSC (1× SSC is 150 mM NaCl plus 15 mM Na citrate)–0.1% sodium dodecyl sulfate.
3′- and 5′-end analysis by rapid amplification of cDNA ends (RACE) was carried out according to the protocol of the manufacturer (Gibco BRL). The cDNA for GFP 3′-end RACE was amplified with the gene-specific oligonucleotide GFPOUT, 48 nt upstream from the GFP termination codon. For 5′-end analysis of CAT mRNA, the first-strand synthesis was carried out with the CATIN oligonucleotide (5′-CCCATATCACCAGCTCACCG-3′), 233 nt downstream from the CAT initiation codon. This was followed by amplification with the SL-specific oligonucleotide Eco-SL (5′-GGGAATTCCGCTATTATTAGAACAGTTTCT-3′) and with CAT-5, located 22 nt downstream from the CAT initiation codon. 3′- and 5′-end PCR products were analyzed by agarose gel electrophoresis and sequenced after purification with a QIAquick PCR purification kit (Qiagen).
RESULTS
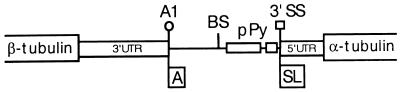
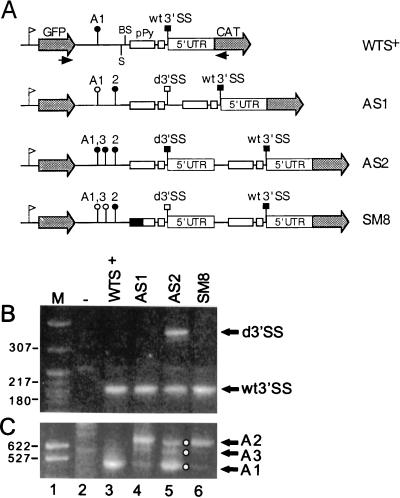
In T. brucei, transcription of the α- and β-tubulin genes gives rise to polycistronic pre-mRNAs consisting of alternating head-to-tail α and β repeating units separated by short intergenic regions (16). Figure 1 shows a schematic representation of the various elements present in the β-α-tubulin intergenic region: the β-tubulin poly(A) addition region of about 10 nt (A1), a putative branch site region, and a long and a short pPy tract followed by the α-tubulin 3′SS. In the experiments reported here, we sought to identify sequence elements contributing to the selection of the α-tubulin 3′SS. To do this, we established an in vivo competition assay based on the premise that a tandem duplication of all the sequences necessary for 3′SS selection will result in equal levels of use of both 3′SSs. This construct then sets the stage for introducing mutations in one of the two 3′SS regions and testing their effects under the stringent conditions of the competition assay. To simulate the chromosomal arrangement, we used a dicistronic expression vector where sequences extending from the nucleotide after the β-tubulin termination codon to the nucleotide preceding the α-tubulin initiation codon (14) were placed between the GFP gene and the CAT gene. Since no RNA polymerase II promoters are known in trypanosomes, expression of this plasmid was driven by the ribosomal promoter, which has been shown to direct pre-mRNA synthesis in T. brucei (27). In order to be able to duplicate various portions of the α-tubulin 3′SS region, we engineered by site-directed mutagenesis an SmaI site just upstream of the putative branch sites to generate WTS+ (Fig. 2A). This location was chosen, since block substitution mutagenesis revealed that this region does not harbor sequences involved in poly(A)- and/or 3′SS-site selection (data not shown). In our first test construct (AS1), we duplicated the minimal 3′SS region, comprising sequences from 4 nt upstream of the branch site region to the 3′SS that includes the AG dinucleotide (Fig. 2A). WTS+ and AS1 were transfected into T. brucei procyclic cultured cells by electroporation, and RNA was prepared 4 h after transfection, that is, at the time when RNA accumulation is maximal. In each experiment reported here, RNA samples were equalized by monitoring the expression of a cotransfected U6 snRNA gene (data not shown). To test for trans-splice-site and poly(A)-site selection, we employed 5′- and 3′-end RACE, respectively, using CAT- and GFP-specific oligonucleotide primers as described in Materials and Methods. As expected, when WTS+ was transfected into trypanosome cells, trans splicing occurred at the wt 3′SS, as was illustrated by the amplification of a PCR fragment of the predicted size of 200 nt (Fig. 2B, lane 3). Sequence analysis of this PCR product further confirmed that addition of the SL sequence was accurate to the nucleotide (data not shown). 3′-end RACE analysis of WTS+-transfected RNA generated a major fragment of about 500 nt (Fig. 2C, lane 3), which upon sequence analysis demonstrated correct polyadenylation at the wt β-tubulin polyadenylation site region, or A1 site (Fig. 1). Thus, the 200- and 500-nt PCR fragment are diagnostic for accurate use of the wt trans splice and poly(A) sites, respectively.
FIG. 1.
Schematic representation of a portion of the β-tubulin–α-tubulin gene cluster of T. brucei. Open boxes indicate the β- and α-tubulin-coding regions, which are not drawn to scale. The thin line indicates the 145-nt-long intergenic region between the β-tubulin poly(A) sites or A1 sites and the AG dinucleotide at the α-tubulin 3′SS. The positions of the long and short pPy tracts are indicated, as is the position of the putative branch sites (BS). SL and A flags mark the positions of SL and poly(A)-site additions, respectively.
FIG. 2.
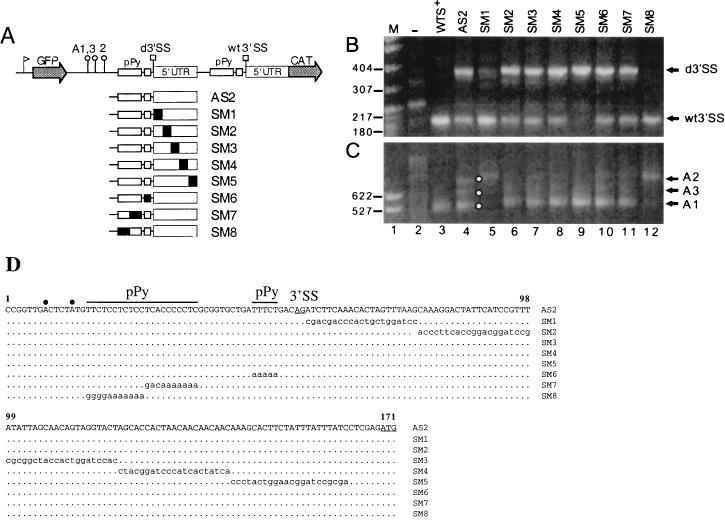
cis competition between duplicated α-tubulin 3′SSs. (A) Schematic representation of plasmid constructs used for transfection. WTS+ contains the β- and α-tubulin sequences from the nucleotide after the β-tubulin termination codon to the nucleotide preceding the α-tubulin ATG. These sequences are sandwiched between the GFP- and CAT-coding regions. The ribosomal promoter, indicated by a flag upstream, directs synthesis of the pre-mRNA. An SmaI site (S) was introduced by site-directed mutagenesis at position 474 of the β-tubulin–α-tubulin intergenic region (14) of the parent plasmid to generate WTS+. In AS1 the minimal 3′SS of α-tubulin mRNA, from 4 nt upstream from the branch sites (BS) to and including the AG dinucleotide, was duplicated at the SmaI site of WTS+. In AS2 the duplicated region included the 5′UTR of α-tubulin mRNA to the nucleotide preceding the ATG translation initiation codon. SM8 is a mutant derivative of AS2 in which the 5′ half of the long pPy tract of the duplicated 3′SS was mutagenized as described in Materials and Methods. wt3′SS and d3′SS represent the wt and duplicated 3′SSs, respectively. Filled squares and circles indicate the usage of the 3′SS and poly(A) sites, respectively, whereas open squares and circles indicate that the 3′SS and poly(A) sites, respectively, are not used. The identities of the other symbols are as described in the legend to Fig. 1. Arrows below the GFP- and CAT-coding regions indicate the approximate positions of gene-specific oligonucleotides which were used for 5′- and 3′-end RACE analyses. Results of 5′-end RACE (B) and 3′-end RACE (C) analyses of transcripts produced by transient expression of the constructs diagrammed in panel A are shown. Arrows indicate the positions of the amplified DNA fragments diagnostic of usage of the duplicated or wt 3′SS and of the A1 to 3 polyadenylation sites. Lane M, MspI digest of pBR322 DNA as a marker. Representative molecular sizes (in base pairs) are shown. Lane −, RACE products obtained with RNA from mock-transfected cells.
5′-end RACE analysis of RNA isolated from cells transfected with AS1, which contains a duplication of the minimal 3′SS region, produced the 200-nt fragment diagnostic of trans splicing at the wt 3′SS (Fig. 2B, lane 4) but revealed only trace amounts of a 5′-end RACE product of 263 nt, which is the expected size for trans splicing at the duplicated site. This result was confirmed by Southern hybridization of 5′-end RACE products and also by primer extension analysis (data not shown). 3′-end RACE analysis of AS1-derived RNA generated a prominent fragment of about 650 nt and a few lower-molecular-weight fragments that are barely visible in the reproduction shown (Fig. 2C, lane 4). Direct sequence analysis of the 650-nt fragment revealed that poly(A) addition occurred 113 nt downstream of the wt A1 site, at a site we termed A2, which is just upstream of the duplicated-branch-site sequence (Fig. 2A). Thus, in summary, the duplicated minimal α-tubulin 3′SS region in the context of plasmid AS1 was not efficiently recognized by the trans-splicing machinery and polyadenylation of GFP mRNA ignored the wt A1 site, indicating that the mere presence of a downstream pPy tract, even when it was placed at the same position as in the β-α-tubulin intergenic region, is not sufficient to specify efficient 3′-end cleavage and polyadenylation.
We can put forward two hypothesis that lead to the skipping of the duplicated 3′SS. First, it is possible that sequences downstream of the AG dinucleotide, namely α-tubulin exon sequences, are required for 3′SS selection. Alternatively, sequences located downstream of the AG dinucleotide at the duplicated site, which are not present in the wt configuration, may exert a negative effect on 3′SS choice. To address this issue, we constructed plasmid AS2, in which the duplicated 3′SS region was extended in the 3′ direction to include 106 nt of α-tubulin exon sequences encompassing the entire 5′UTR. When AS2-derived RNA was analyzed by 5′-end RACE, two size classes of PCR fragments were obtained: the shorter one corresponded to use of the wt3′SS and the longer one, whose size was 369 nt, corresponded to use of the duplicated 3′SS (Fig. 2B, lane 5). Sequence analysis of the PCR fragments confirmed that this was indeed the case. 3′-end RACE analysis revealed a more complex pattern with three size classes of PCR fragments (Fig. 2C, lane 5). The shortest product was diagnostic of molecules with 3′ ends at the A1 site, whereas the longest product indicated that in AS2 RNA there exist GFP mRNA molecules polyadenylated at the A2 site. The third product defined a new poly(A) addition site, termed A3, and positioned the 3′ ends of a small proportion of GFP mRNAs at a site located between the A1 and A2 sites. The intensities of the three 3′-end RACE products varied from experiment to experiment, and in some experiments the three size classes were much less defined than those shown in Fig. 2C, resulting in almost a smear of bands. We think that this is because, in general, polyadenylation in trypanosomes occurs at several sites within a short region and that the greater the heterogeneity is at the 3′ end, the more difficult it is to obtain specific 3′-end RACE products.
We have previously proposed a model where trans-splice-site selection and poly(A)-site selection are functionally coupled via recognition of a pPy tract just upstream of the 3′SS (14). To test whether the three distinct poly(A) sites in AS2-derived RNA are guided by the same or different pPy tracts, we impaired the function of the duplicated 3′SS by mutating the first half of the long pPy tract in plasmid AS2 to generate construct SM8. As expected, this mutation drastically reduced the appearance of the 369-nt 5′-end RACE fragment diagnostic of usage of the duplicated 3′SS (Fig. 2B, lane 6). As a consequence, polyadenylation at the A1 and A3 sites was also reduced (Fig. 2C, lane 6), indicating that it is the duplicated 3′SS that directs 3′-end cleavage and polyadenylation at these two sites and that the wt 3′SS is coupled to polyadenylation at the A2 site.
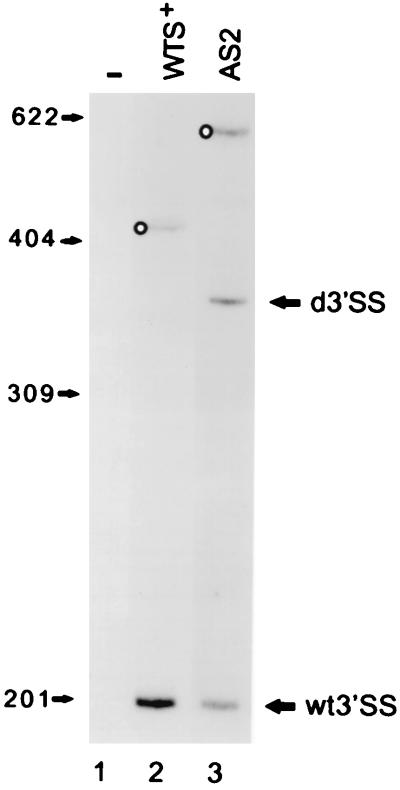
To quantitate more precisely the abundance of CAT mRNAs trans spliced at the wt or at the duplicated 3′SS, we performed primer extension analysis on AS2-derived RNA using a 5′-end-labeled oligonucleotide primer. Figure 3 shows that the two types of CAT mRNA (lane 3) were represented in approximately equal amounts. Both in AS1-derived (lane 2) and in AS2-derived (lane 3) RNAs we observed an additional primer extension product. The origin of these products is uncertain because we could not detect them by 5′-end RACE, and therefore they most likely do not represent trans-spliced RNAs. One possible explanation is that they are derived by cleavage of the pre-mRNA. These presumptive sites of cleavage map upstream from the wt poly(A) site, and they do not seem to correlate with the presence of consensus 3′ splice acceptor sequences.
FIG. 3.
Analysis of 3′SS use by primer extension analysis. RNAs from cells transfected with the construct indicated above each lane were reverse transcribed by using as a primer a 32P-labeled oligonucleotide complementary to nt 26 to 39 of the CAT-coding region. The positions of the primer extension products diagnostic of use of the duplicated 3′SS (d3′SS) or wt 3′SS are indicated. Open circles indicate primer extension products whose origins are uncertain (see the text for details). Sizes are indicated in nucleotides.
We also considered the possibility that CAT mRNA trans spliced at the duplicated site is preferentially degraded due to its primary structure, which consists of tandemly repeated sequences at the 5′ end that are not present in CAT mRNA trans spliced at the wt 3′SS. To exclude this possibility, we measured the stability of these two RNA molecules by using the methyltransferase inhibitor sinefungin, which blocks trans splicing in T. brucei cells (15, 22). These experiments showed that CAT mRNA trans spliced at the duplicated 3′SS was as stable as CAT mRNA trans spliced at the wt 3′SS (data not shown).
Taken together, the experiments described above demonstrated that in the pre-mRNA derived from the AS2 construct both the duplicated and the wt 3′SS were used and that the usage of the duplicated and wt 3′SS correlated directly with the usage of the A1-A3 and A2 poly(A) sites, respectively. However, it appeared that the duplicated 3′SS behaved somewhat differently from the wt 3′SS in poly(A)-site selection, in that it directed 3′-end formation not only at the A1 site but also, albeit to a lesser extent, at the A3 site. Although at present we do not fully understand the reason for this difference in selectivity, one possible explanation is that the structure of the pre-mRNA with a duplicated 3′SS is different from that of wt pre-mRNA, and this difference in structure might affect to some extent poly(A)-site choice. Notwithstanding this difference, we concluded that the AS2 pre-mRNA substrate fulfilled the criteria for the cis competition assay, since both 3′SSs appeared to be used to similar extents.
Specific sequences in the α-tubulin 5′UTR and in the 3′SS region positively and negatively affect trans-splice- and poly(A)-site selection.
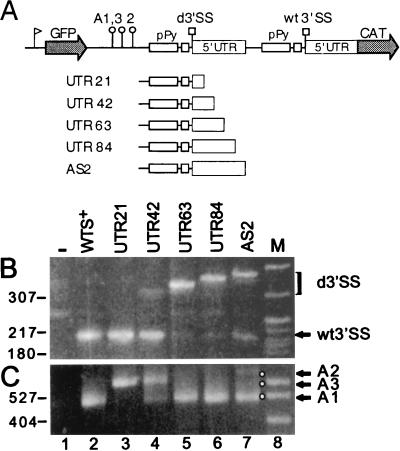
To investigate in detail what sequences in the α-tubulin 5′UTR are required for fully competitive 3′SS selection, we used the AS2 construct as our baseline construct and introduced a series of mutations. The first set of constructs had the same minimal duplicated 3′SS region but differed by increments of 21 nt in the lengths of their 5′UTR sequences, which were appended to their duplicated 3′SS regions (Fig. 4A). Panels B and C of Fig. 4 show, respectively, the results of the 5′- and 3′-end RACE analyses of RNAs from transfected cells, and the results are summarized in Table 1. In our PCR analysis, in which two different cDNAs were amplified with the same set of oligonucleotides, we focused on detecting any reproducible change in the proportions of the fragments diagnostic of usage of the duplicated and wt 3′SSs rather than on determining absolute amounts. To control for potential PCR artifacts, amplification conditions were monitored with different cDNA dilutions and various numbers of amplification cycles. From these experiments it was evident that the first 42 nucleotides of the α-tubulin 5′UTR were sufficient to provide competitive ability to the duplicated 3′SS region, albeit at a lower level than that which we observed with AS2-derived RNA. Addition of further UTR sequences (constructs UTR63 and UTR84) resulted in almost exclusive usage of the duplicated 3′SS, and we could barely detect use of the wt 3′SS (lanes 5 and 6 in Fig. 4B). Interestingly, when the α-tubulin 5′UTR was further increased to include the last 22 nucleotides (construct AS2), both the duplicated and wt 3′SSs were used (lane 7). 3′-end RACE analysis of the same RNA samples corroborated these observations, in that usage of the duplicated 3′SS correlated with the appearance of polyadenylation at the A1 site (Fig. 4C).
FIG. 4.
Effects of 5′UTR truncations on the relative use of duplicated and wt3′SSs. (A) Schematic representations of the AS2 parent construct and of the 5′UTR deletion derivatives containing either 21 (UTR21), 42 (UTR42), 63 (UTR63), or 84 (UTR84) nt of the 106-nt-long α-tubulin 5′UTR. Results of 5′-end RACE (B) and 3′-end RACE (C) analyses of RNAs from transfected cells are shown. The sizes of the RACE products diagnostic of duplicated 3′SS use varied because of the various sizes of the 5′UTR portion included in each construct. Lane M, MspI digest of pBR322 DNA as a marker. Representative molecular sizes (in base pairs) are shown. Lane −, RACE products obtained with RNA from mock-transfected cells. d3′SS, duplicated 3′SS.
TABLE 1.
Summary of 3′SS and poly(A)-site selection
| Mutant | wt 3′SS | Duplicated 3′SS | Poly(A) site(s) |
|---|---|---|---|
| WTS+ | + | NAa | 1 |
| AS1 | + | − | 2 |
| AS2 | + | + | 1, 2, 3 |
| UTR21 | + | − | 2 |
| UTR42 | + | + | 1, 2, 3 |
| UTR63 | + | + | 1, 2, 3 |
| UTR84 | + | + | 1, 2, 3 |
| SM1 | + | +/− | 2 |
| SM2 | + | + | 1, 2, 3 |
| SM3 | + | + | 1, 2, 3 |
| SM4 | + | + | 1, 2, 3 |
| SM5 | − | − | 1, 2, 3 |
| SM6 | + | + | 1, 2, 3 |
| SM7 | + | + | 1, 2, 3 |
| SM8 | + | − | 2 |
| ACTsub | − | NA | 1 |
| βTUBsub | + | NA | 1 |
| RADsub | − | NA | 1 |
| ACTsubD | + | − | 2 |
| βTUBsubD | + | + | 1, 2, 3 |
NA, not applicable.
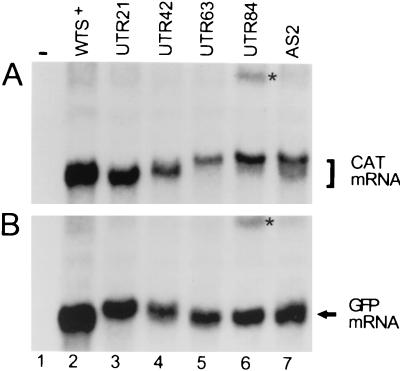
Because the 5′ ends of CAT mRNAs derived from the various UTR constructs differed from each other and from those generated with AS2, we analyzed the steady-state amounts of CAT mRNAs by Northern blot analysis (Fig. 5A). In agreement with the results of the 5′-end RACE (Fig. 4B), the size increase of CAT mRNA was comparable to the progressive increase of the duplicated 5′UTR (Fig. 5A). In the AS2-derived RNA (lane 7), two CAT mRNA bands can be discerned, and these bands most likely correspond to CAT mRNA trans spliced at the wt and duplicated 3′SSs. In quantitative terms there seemed to be less accumulation of CAT mRNA in RNAs derived from the AS2 and the UTR constructs relative to that in the control (WTS+) (lane 2). A similar trend was seen in the analysis of GFP mRNA (Fig. 5B). Whether this behavior reflects instability of the mRNAs, of the pre-mRNAs, or of both remains to be determined.
FIG. 5.
Northern analysis of CAT and GFP transcripts in cells transfected with the constructs shown in Fig. 4A. The asterisks indicate the positions of putative dicistronic transcripts.
In a second set of constructs we introduced a series of block substitutions, covering the region from the nucleotide after the AG dinucleotide to the nucleotide preceding the ATG initiation codon (mutants SM1 to -5) (Fig. 6A). The various constructs were transfected into trypanosome cells, and the resulting RNAs were analyzed by 5′- and 3′-end RACE analyses (Fig. 6B and C); a summary of the results is shown in Table 1. Mutations in SM2, SM3, and SM4 did not affect trans-splice-site choice (compare lanes 6 to 8 in Fig. 6B with lane 4). The mutation in SM1 greatly decreased the amount of CAT mRNA trans spliced at the duplicated 3′SS (Fig. 6B, lane 5). On the other hand, the mutation in SM5 appeared to direct almost exclusive use of the duplicated 3′SS (Fig. 6B, lane 9). Analysis of the corresponding 3′-end RACE products (Fig. 6C, lanes 5 and 9) indicated agreement with the predominant use of the wt 3′SS in mutant SM1 and with the almost exclusive use of the duplicated 3′SS in mutant SM5.
FIG. 6.
Effects of block substitutions on the relative use of the duplicated and wt α-tubulin 3′SSs. (A) Schematic representations of the parent AS2 construct and of the mutagenized SM derivatives. Substituted nucleotides are indicated by filled boxes. Results of 5′-end RACE (B) and 3′-end RACE (C) analyses of RNA from transfected cells are shown. The identity of each construct is indicated above each lane. Lane M, MspI digest of pBR322 DNA as a marker. Representative molecular sizes (in base pairs) are shown. Lane −, RACE products obtained with RNA from mock-transfected cells. (D) Sequences of the α-tubulin 3′SS acceptor and 5′UTR, which was duplicated in the AS2 mutant. The nucleotide changes in mutants SM1 to -8 are indicated in lowercase letters. Dots represent unchanged nucleotides. Filled circles above the sequence indicate the putative branch site adenosines. d3′SS, duplicated 3′SS.
A second series of block substitution mutants (SM6 to SM8) (Fig. 6A) was generated to determine the effects of some of our previously described pPy tract mutations by the more stringent competitive assay. The phenotypes of mutants SM6 and SM7 in terms of use of the duplicated 3′SS (Fig. 6B and C) closely resembled the results obtained with our previously described mutants BM5 and BM3 (14). On the other hand, mutant SM8, in which only the first half of the large pPy tract was mutated, had a severe effect on use of the duplicated 3′SS site, whereas a similar mutant (BM2, in our previous nomenclature) had no effect when it was assayed in the context of a single α-tubulin 3′SS region (14).
In conclusion, the last set of mutants confirmed our previous results establishing that the pPy tract is an essential element for 3′SS identification in trypanosomes and also highlighted the fact that not all residues of the large pPy tract contribute equally to 3′SS selection. Furthermore, the results obtained with the SM1 mutation strongly argue that α-tubulin exon sequences immediately downstream of the AG dinucleotide contribute essential information for the identification of the 3′SS. Finally, it seems likely that the last 20 nucleotides of the α-tubulin 5′UTR harbor an inhibitory element for identification of the α-tubulin 3′SS.
Can other 5′UTRs substitute for the α-tubulin 5′UTR?
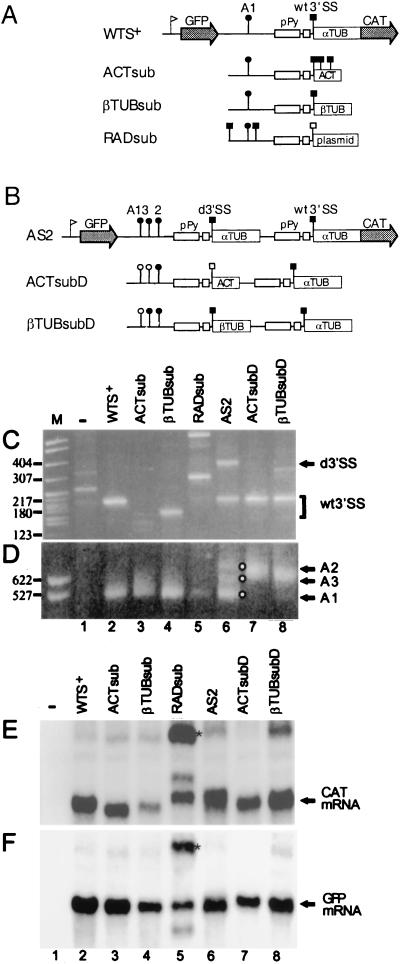
To test whether the function provided by the α-tubulin 5′UTR could be replaced by other sequences, we constructed three plasmids (Fig. 7A) in which the α-tubulin 5′UTR was replaced by the β-tubulin 5′UTR (βTUBsub), by the actin 5′UTR (ACTsub), or by random plasmid sequences of similar lengths (RADsub). These three constructs contained a single α-tubulin 3′SS region and differed only in the sequences between the 3′SS and the ATG initiation codon. 5′-end RACE and primer extension analysis (Fig. 7C and data not shown) of transfected RNAs showed that the ACTsub construct produced CAT mRNA with heterogeneous 5′ ends (lane 3), with trans splicing occurring at the α-tubulin 3′SS, as well as at two other closely spaced AG dinucleotides within the actin 5′UTR. In contrast, the βTUBsub-derived RNA showed one prominent band, whose size was consistent with use of the α-tubulin 3′SS (lane 4). In the RADsub contruct, use of the α-tubulin 3′SS was undetectable (lane 5). However, we detected two prominent bands whose sizes indicated that they were derived from trans-splicing events which had occurred about 100 and 400 nt upstream from the wt α-tubulin 3′SS. 3′-end RACE indicated that both ACTsub and βTUBsub directed polyadenylation at the A1 site, and this was also true for the little material that could be amplified from the RADsub RNA sample (Fig. 7D). Northern blot analysis of the same RNA samples revealed that in all cases there was substantial accumulation of both CAT and GFP mRNAs (Fig. 7E and F). This was even true when trans-splice-site selection became completely erratic, as it was with RADsub-derived RNA (lanes 5). Nevertheless, trans splicing of RADsub-derived RNA must have been severely affected because substantial amounts of putative dicistronic transcripts accumulated in these cells (Fig. 7E and F).
FIG. 7.
Effects of replacement of the α-tubulin 5′UTR with the actin or β-tubulin 5′UTR or unrelated sequences. (A and B) Schematic representations of the various constructs used for transfection. ACTsub contains the 55-nt-long actin 5′UTR in place of the α-tubulin 5′UTR; βTUBsub contains the 59-nt-long β-tubulin 5′UTR. In RADsub the α-tubulin 5′UTR was replaced with a 100-nt-long fragment derived from the plasmid vector pCRTMII. In ACTsubD and βTUBsubD the actin and β-tubulin 5′UTR substitutions of ACTsub and βTUBsub replaced the corresponding regions of the duplicated 3′SS of plasmid AS2. The positions of the SL addition and poly(A) sites for the RADsub constructs are only indicative and were not precisely determined. Symbols are as defined in the legend of Fig. 2A. ACT, actin; αTUB, α-tubulin; βTUB, β-tubulin; d3′SS, duplicated 3′SS. Results of 5′-end RACE (C) and 3′-end RACE (D) analyses of RNA from transfected cells are shown. Lane M, MspI digest of pBR322 DNA as a marker. Representative molecular sizes (in base pairs) are shown. Lane −, RACE products obtained with RNA from mock-transfected cells. (E and F) Northern blot analysis of CAT and GFP transcripts in cells transfected with the constructs shown in Fig. 7A and B. Asterisks indicate the positions of putative dicistronic transcripts.
The behavior of the ACTsub and βTUBsub chimeras was also investigated in the competition assay by constructing plasmids ACTsubD and βTUBsubD (Fig. 7B). When the actin 5′UTR function was tested, it failed to compete for the wt α-tubulin 3′SS, as can be deduced from the results of the 5′- and 3′-end RACE analyses (Fig. 7C and D, lanes 7). On the other hand, the β-tubulin 5′UTR directed some trans splicing at the duplicated 3′SS (lanes 8). In summary, (i) random sequences cannot substitute for the function provided by the α-tubulin 5′UTR for recognition of the pre-mRNA by the trans-splicing machinery, (ii) actin 5′UTR sequences can probably replace the α-tubulin 5′UTR only in the direct-type assay and not in the competition assay, and (iii) the β-tubulin 5′UTR can function both in the direct assay and, to a lesser extent, in the competition assay. From these observations it appears that the α-tubulin 5′UTR contributes a function that can be provided to some degree by other trypanosome 5′UTRs.
DISCUSSION
The goal of the work described in this paper was to analyze in detail the contributions of the various elements of the α-tubulin 3′SS region to trans-splice- and poly(A)-site selection. The strategy we chose is based on a competition assay using a substrate containing two alternative α-tubulin 3′SSs. This type of assay has been used many times in the past, for instance, for examining the role of U-rich tracts and other sequence elements for cis splicing of yeast introns in vivo (17). The competition assay is more stringent than a direct assay and is suitable to detect the contribution of minor sequence elements and/or structural determinants present in pre-mRNA.
The AS2 pre-mRNA substrate, which we used for our experiments, fulfilled the criteria for a cis competition assay in that both 3′SSs were used to similar extents. Importantly, the concomitant use of the A1-A3 and A2 poly(A) sites, which were coupled to use of the duplicated and wt 3′SSs, respectively, provided independent internal controls for our analyses. Thus, the alternate use of the two competing 3′SSs was corroborated by analysis of the 3′-end cleavage and polyadenylation products which were simultaneously formed by processing of the pre-mRNA.
3′-end cleavage and polyadenylation cannot be uncoupled from trans splicing at the α-tubulin 3′SS.
In our initial experiments we discovered that the minimal α-tubulin 3′SS region was not recognized by the trans-splicing machinery (construct AS1 in Fig. 2), because the duplicated minimal 3′SS was not used to a detectable level. In the context of the AS1 substrate, the wt A1 polyadenylation site was almost completely ignored and the majority of GFP mRNA was polyadenylated at a new downstream poly(A) addition site (A2). Thus, the pPy tract of the duplicated 3′SS region, which is located downstream of the A1 site in the same configuration as in the wt pre-mRNA, did not function to direct polyadenylation at the wt A1 site. This result suggests that the pPy tract of the α-tubulin 3′SS, which we had previously shown was essential for accurate and efficient poly(A)-site selection, is recognized only in the context of its function in trans splicing. These observations, together with previous work from our own and other laboratories, further strengthen the concept that polyadenylation cannot be uncoupled from trans splicing during pre-mRNA processing in trypanosomes (10, 12, 14, 19, 25). It is tempting to speculate that in these organisms, which separated very early from the main branch of eukaryotic evolution, there exists a unique pre-mRNA processing body which is endowed with both trans-splicing and polyadenylation functions. The factors required for 3′-end cleavage and polyadenylation might associate with the pre-mRNA after the 3′SS region has been earmarked by association with components of the trans spliceosome, or they might be in a complex with some components of the trans spliceosome. In trypanosomatid protozoa in vivo, 3′-end cleavage and polyadenylation always takes place a short distance upstream from a 3′SS, i.e., within 100 to a few hundred nucleotides. Our findings with the AS1 and AS2 constructs and earlier work by LeBowitz et al. (12), which showed that poly(A)-site selection moves in concert with the 3′SS in a Leishmania pre-mRNA intergenic region, support a model in which the factor(s) responsible for cleaving the pre-mRNA at the poly(A) site can reach only a certain short distance away from the 3′SS. Perhaps in trypanosomes the 3′SS substitutes for the AAUAAA signal found in pre-mRNAs of higher eukaryotes which directs 3′-end cleavage and polyadenylation at a short distance downstream from itself.
α-Tubulin 5′UTR modulates 3′SS usage.
Among the block substitution mutations of the α-tubulin 5′UTR, mutant SM1 had the strongest phenotype, in that we observed a drastic reduction in use of the duplicated 3′SS as assessed by 5′- and 3′-end RACE analyses (Fig. 6). We interpret the phenotype of the SM1 mutant to indicate that the first 20 nucleotides immediately downstream of the AG dinucleotide of the 3′SS contain a sequence element which plays an important positive role in 3′SS identification. However, the first 20 nucleotides of the 5′UTR by themselves were not sufficient to direct trans splicing to the duplicated 3′SS, as we observed with the UTR21 construct. Indeed, there was a clear increase in the use of the duplicated 3′SS in UTR42, and this trend reached a maximum with UTR63, when the use of the duplicated site predominated. Thus, these data can be interpreted to indicate that the sequences between 42 and 63 nt downstream from the 3′SS gave a competitive advantage to the duplicated 3′SS relative to that of the wt 3′SS. There are several possibilities to explain these results. One possibility we favor is that the information provided by the 5′UTR is complex and redundant. The SM1 mutation gave the most severe phenotype, possibly because the sequences downstream of the AG dinucleotide might be engaged in direct interactions with trans-spliceosomal components, like the newly discovered U5 snRNA (4, 26). The lack of detectable phenotypes for the SM2 to SM4 substitutions can be explained by assuming that the function provided by each block of nucleotides is additive and that therefore when one block is changed, the others take over. The alternate and not mutually exclusive possibility is that there needs to be some spacing between the duplicated 3′SSs because of steric hindrance between the complexes assembling on the two sites.
The 5′UTR of α-tubulin also contains an element which reduces use of the 3′SS. This element coincides with the last 20 nucleotides of the sequence, just before the ATG initiation codon. The existence of this inhibitory sequence was brought to light by the SM5 mutation (Fig. 6), in which mutation of this block of nucleotides led to almost exclusive use of the duplicated 3′SS, and by the deletion of this element as seen in UTR84 (Fig. 4). Thus, the informational content of the α-tubulin 5′UTR for trans-splice-site selection is complex.
Potential functions of α-tubulin exon sequences in 3′SS selection.
The α-tubulin 5′UTR sequences between positions 1 and 70 after the 3′SS could be considered a trans-splicing enhancer, since they modulate the use of the α-tubulin 3′SS. Exonic splicing enhancers have been identified in many pre-mRNAs of higher eukaryotes and are defined as positive cis-acting regulatory sequences that promote use of upstream splice sites (for a review, see reference 7). Interestingly, enhancer-dependent interactions dramatically stimulate trans splicing of synthetic pre-mRNAs molecules in vitro (1, 2). The best-characterized splicing enhancers consist of short purine-rich sequences which bind to specific members of the SR protein family, a class of proteins that modulate splice-site choice (for a review, see reference 24). More recently, a new family of exonic splicing enhancers, termed the A/C-rich splicing enhancer or ACE family, has been identified by in vivo selection (3). Also, ACE activity is thought to be mediated by interaction with a subset of SR proteins. We inspected the α-tubulin 5′UTR for the presence of purine and A/C-rich sequences and found a number of A/C-rich sequence elements distributed throughout. By assuming that these motifs act synergistically, one can rationalize the observation that the 5′UTR sequences 20 to 60 nt downstream from the 3′SS seem to have an additive effect on enhancing recognition of the duplicated 3′SS. Similar A/C-rich elements can be identified in the β-tubulin 5′UTR but not in the actin 5′UTR, which could not efficiently substitute for the α-tubulin 5′UTR in the cis competition assay. These observations are consistent with the possibility that the A/C-rich sequences are important for the modulatory activity of the α-tubulin 5′UTR, but further experiments are required to validate this possibility. The enhancer function of α-tubulin 5′UTR sequences on trans splicing may be mediated by protein factors akin to the aforementioned SR proteins. At present, however, there is no evidence for the existence of SR-like proteins in trypanosomes. Finally, the fact that the actin 5′UTR was unable to substitute for the α-tubulin 5′UTR might suggest that in trypanosomes, different classes of 3′SS which differ in their requirements for adjacent exon sequences exist.
Another possibility to explain the requirement of exon sequences for 3′SS recognition in trypanosomes is to imagine that the folding of the pre-mRNA is essential to present the correct 3′SS to components of the trans spliceosome. Although other interpretations are possible, perhaps mutation and removal of the last 20 nucleotides of the 5′UTR, as in mutants SM5 and UTR84, respectively, relieve some structural constraint from the pre-mRNA. Indeed, RNA secondary structure has been shown to have profound effects on splice-site choice and splicing efficiency (6). An RNA-based splicing enhancer, consisting of two short complementary sequences which base pair with each other, has been proposed as the basis for the increased splicing efficiency of the yeast intron rp51b by Libri and colleagues (13). Computer-aided folding of the α-tubulin intergenic region and 5′UTR, however, was not informative.
Potential role of 5′UTR sequences in regulation of gene expression.
Gene regulation is one of the most intriguing aspects of trypanosomatid biology. Since trypanosomes do not seem to use as a regulatory mechanism the process of transcription initiation, one needs to postulate that other regulatory mechanisms are operational in these organisms. Other steps downstream in the cascade of events leading from the primary transcript to the generation of mature mRNA, such as trans splicing and polyadenylation, pre-mRNA turnover, transport, mRNA turnover, etc., might be regulated. For each of these steps regulation has been amply documented for other eukaryotic systems. It is our working hypothesis that regulation of pre-mRNA processing in trypanosomes plays a major role in determining the output of mature mRNA molecules per gene. Our study of trans splicing in a permeable cell system has revealed that trans splicing takes place on nascent pre-mRNA chains, at least for those pre-mRNAs, like tubulin and actin, which are efficiently trans spliced in permeable cells and in vivo (21). In contrast, calmodulin pre-mRNA does not seem to be efficiently trans spliced in permeable cells (21) or in vivo, where under steady-state conditions more than 10% of calmodulin RNA is found in the form of polycistronic transcripts (20). This result argues that, at least for calmodulin pre-mRNA, 3′SSs are often skipped and that trans-splicing efficiency might be important in regulating the final amount of mature mRNA produced by the cell. Skipping of 3′SS was also observed in our transfection experiments whenever trans splicing became erratic, as with the RADsub construct (Fig. 7), where the α-tubulin 5′UTR was substituted by unrelated plasmid sequences and abundant dicistronic transcripts were easily detected by Northern blot analysis. Thus, we propose that 5′UTR sequences play an important role in determining the rate of trans splicing at a given 3′SS. Our in vivo competition assay will prove very useful for understanding in more detail the architecture of trypanosome 3′SS regions and whether there exist different classes of pre-mRNAs with different processing rates, as our observations with the calmodulin pre-mRNA seem to suggest.
ACKNOWLEDGMENTS
We thank the class of 1997 of the Biology of Parasitism Course in Woods Hole, Mass., for their enthusiasm, hard work, and critical comments and for initiating some of the experiments; Anna Polotsky and Helen Kwon for continuous and excellent technical support; and Susan Baserga, Joan Steitz, and Sandra Wolin for critical reading of the manuscript.
This work was supported by grant AI28798 from the National Institutes of Health to E.U. and by a Burroughs Wellcome Scholar Award in Molecular Parasitology to E.U. C.L.-E. was partially supported by the Consejo Nacional de Investigaciones Científicas y Tecnológicas (CONICIT) of Venezuela.
REFERENCES
- 1.Bruzik J P, Maniatis T. Enhancer-dependent interaction between 5′ and 3′ splice sites in trans. Proc Natl Acad Sci USA. 1995;92:7056–7059. doi: 10.1073/pnas.92.15.7056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chiara M D, Reed R. A two-step mechanism for 5′ and 3′ splice-site pairing. Nature. 1995;375:510–513. doi: 10.1038/375510a0. [DOI] [PubMed] [Google Scholar]
- 3.Coulter L R, Landree M A, Cooper T A. Identification of a new class of exonic splicing enhancers by in vivo selection. Mol Cell Biol. 1997;17:2143–2150. doi: 10.1128/mcb.17.4.2143. . (Erratum, 17:3468.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dungan J M, Watkins K P, Agabian N. Evidence for the presence of a small U5-like RNA in active trans-spliceosomes of Trypanosoma brucei. EMBO J. 1996;15:4016–4029. [PMC free article] [PubMed] [Google Scholar]
- 5.Fantoni A, Dare A O, Tschudi C. RNA polymerase III-mediated transcription of the trypanosome U2 small nuclear RNA gene is controlled by both intragenic and extragenic regulatory elements. Mol Cell Biol. 1994;14:2021–2028. doi: 10.1128/mcb.14.3.2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Goguel V, Rosbash M. Splice site choice and splicing efficiency are positively influenced by pre-mRNA intramolecular base pairing in yeast. Cell. 1993;72:893–901. doi: 10.1016/0092-8674(93)90578-e. [DOI] [PubMed] [Google Scholar]
- 7.Hertel K J, Lynch K W, Maniatis T. Common themes in the function of transcription and splicing enhancers. Curr Opin Cell Biol. 1997;9:350–357. doi: 10.1016/s0955-0674(97)80007-5. [DOI] [PubMed] [Google Scholar]
- 8.Hotz H R, Hartmann C, Huober K, Hug M, Clayton C. Mechanisms of developmental regulation in Trypanosoma brucei: a polypyrimidine tract in the 3′-untranslated region of a surface protein mRNA affects RNA abundance and translation. Nucleic Acids Res. 1997;25:3017–3026. doi: 10.1093/nar/25.15.3017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Huang J, Van der Ploeg L H. Requirement of a polypyrimidine tract for trans-splicing in trypanosomes: discriminating the PARP promoter from the immediately adjacent 3′ splice acceptor site. EMBO J. 1991;10:3877–3885. doi: 10.1002/j.1460-2075.1991.tb04957.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hug M, Hotz H R, Hartmann C, Clayton C. Hierarchies of RNA-processing signals in a trypanosome surface antigen mRNA precursor. Mol Cell Biol. 1994;14:7428–7435. doi: 10.1128/mcb.14.11.7428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kuersten S, Lea K, MacMorris M, Spieth J, Blumenthal T. Relationship between 3′ end formation and SL2-specific trans-splicing in polycistronic Caenorhabditis elegans pre-mRNA processing. RNA. 1997;3:269–278. [PMC free article] [PubMed] [Google Scholar]
- 12.LeBowitz J H, Smith H Q, Rusche L, Beverley S M. Coupling of poly(A) site selection and trans-splicing in Leishmania. Genes Dev. 1993;7:996–1007. doi: 10.1101/gad.7.6.996. [DOI] [PubMed] [Google Scholar]
- 13.Libri D, Stutz F, McCarthy T, Rosbash M. RNA structural patterns and splicing: molecular basis for an RNA-based enhancer. RNA. 1995;1:425–436. [PMC free article] [PubMed] [Google Scholar]
- 14.Matthews K R, Tschudi C, Ullu E. A common pyrimidine-rich motif governs trans-splicing and polyadenylation of tubulin polycistronic pre-mRNA in trypanosomes. Genes Dev. 1994;8:491–501. doi: 10.1101/gad.8.4.491. [DOI] [PubMed] [Google Scholar]
- 15.McNally K P, Agabian N. Trypanosoma brucei spliced-leader RNA methylations are required for trans splicing in vivo. Mol Cell Biol. 1992;12:4844–4851. doi: 10.1128/mcb.12.11.4844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Muhich M L, Boothroyd J C. Polycistronic transcripts in trypanosomes and their accumulation during heat shock: evidence for a precursor role in mRNA synthesis. Mol Cell Biol. 1988;8:3837–3846. doi: 10.1128/mcb.8.9.3837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Patterson B, Guthrie C. A U-rich tract enhances usage of an alternative 3′ splice site in yeast. Cell. 1991;64:181–187. doi: 10.1016/0092-8674(91)90219-o. [DOI] [PubMed] [Google Scholar]
- 18.Pays E, Vanhamme L, Berberof M. Genetic controls for the expression of surface antigens in African trypanosomes. Annu Rev Microbiol. 1994;48:25–52. doi: 10.1146/annurev.mi.48.100194.000325. [DOI] [PubMed] [Google Scholar]
- 19.Schurch N, Hehl A, Vassella E, Braun R, Roditi I. Accurate polyadenylation of procyclin mRNAs in Trypanosoma brucei is determined by pyrimidine-rich elements in the intergenic regions. Mol Cell Biol. 1994;14:3668–3675. doi: 10.1128/mcb.14.6.3668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tschudi C, Ullu E. Polygene transcripts are precursors to calmodulin mRNAs in trypanosomes. EMBO J. 1988;7:455–463. doi: 10.1002/j.1460-2075.1988.tb02833.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20a.Ullu, E. Unpublished data.
- 21.Ullu E, Matthews K R, Tschudi C. Temporal order of RNA-processing reactions in trypanosomes: rapid trans splicing precedes polyadenylation of newly synthesized tubulin transcripts. Mol Cell Biol. 1993;13:720–725. doi: 10.1128/mcb.13.1.720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ullu E, Tschudi C. Trans splicing in trypanosomes requires methylation of the 5′ end of the spliced leader RNA. Proc Natl Acad Sci USA. 1991;88:10074–10078. doi: 10.1073/pnas.88.22.10074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ullu E, Tschudi C, Günzl A. Trans-splicing in trypanosomatid protozoa. In: Smith D F, Parsons M, editors. Molecular biology of parasitic protozoa. Oxford, United Kingdom: Oxford University Press; 1996. pp. 115–133. [Google Scholar]
- 24.Valcarcel J, Green M R. The SR protein family: pleiotropic functions in pre-mRNA splicing. Trends Biochem Sci. 1996;21:296–301. [PubMed] [Google Scholar]
- 25.Vassella E, Braun R, Roditi I. Control of polyadenylation and alternative splicing of transcripts from adjacent genes in a procyclin expression site: a dual role for polypyrimidine tracts in trypanosomes? Nucleic Acids Res. 1994;22:1359–1364. doi: 10.1093/nar/22.8.1359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Xu Y, Ben-Shlomo H, Michaeli S. The U5 RNA of trypanosomes deviates from the canonical U5 RNA: the Leptomonas collosoma U5 RNA and its coding gene. Proc Natl Acad Sci USA. 1997;94:8473–8478. doi: 10.1073/pnas.94.16.8473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zomerdijk J C, Kieft R, Borst P. Efficient production of functional mRNA mediated by RNA polymerase I in Trypanosoma brucei. Nature. 1991;353:772–775. doi: 10.1038/353772a0. [DOI] [PubMed] [Google Scholar]