Abstract
Objective
This paper introduces a privacy-preserving federated machine learning (ML) architecture built upon Findable, Accessible, Interoperable, and Reusable (FAIR) health data. It aims to devise an architecture for executing classification algorithms in a federated manner, enabling collaborative model-building among health data owners without sharing their datasets.
Materials and methods
Utilizing an agent-based architecture, a privacy-preserving federated ML algorithm was developed to create a global predictive model from various local models. This involved formally defining the algorithm in two steps: data preparation and federated model training on FAIR health data and constructing the architecture with multiple components facilitating algorithm execution. The solution was validated by five healthcare organizations using their specific health datasets.
Results
Five organizations transformed their datasets into Health Level 7 Fast Healthcare Interoperability Resources via a common FAIRification workflow and software set, thereby generating FAIR datasets. Each organization deployed a Federated ML Agent within its secure network, connected to a cloud-based Federated ML Manager. System testing was conducted on a use case aiming to predict 30-day readmission risk for chronic obstructive pulmonary disease patients and the federated model achieved an accuracy rate of 87%.
Discussion
The paper demonstrated a practical application of privacy-preserving federated ML among five distinct healthcare entities, highlighting the value of FAIR health data in machine learning when utilized in a federated manner that ensures privacy protection without sharing data.
Conclusion
This solution effectively leverages FAIR datasets from multiple healthcare organizations for federated ML while safeguarding sensitive health datasets, meeting legislative privacy and security requirements.
Keywords: Privacy-preserving machine learning, Federated machine learning, FAIR data, Distributed datasets
Graphical Abstract
1. Introduction
1.1. Background
Health data is accumulated through extensive use of digital technologies in healthcare, offering diverse opportunities for applications [1], [2], [3]. However, stringent privacy and security measures, such as the Health Insurance Portability and Accountability Act (HIPAA) [4] in the United States and the General Data Protection Regulation (GDPR) [5] in the European Union, and equivalent regulations in other countries [6], [7], [8], [9], have been established.
Machine learning algorithms greatly benefit from high-quality health data [10]. However, conventional methods necessitate centralizing data, posing conflicts with privacy regulations [11]. Federated machine learning emerged to tackle these challenges by enabling model training without the need to exchange or transfer sensitive data [12], [13]. This field stands as a distinct research area, marked by its advancements and ongoing challenges [14], [15], [16].
Data preparation stands as one of the most challenging and labor-intensive phases in machine learning [17]. For this reason, achieving health data interoperability is a significant step towards successful machine learning applications beyond health data sharing and other purposes of clinical informatics. The FAIR (Findable, Accessible, Interoperable, Reusable) guiding principles were introduced to formally outline guidelines for achieving machine-accessible and actionable interoperability [18], [19]. The health informatics community leads the adoption of FAIR principles to facilitate health data interoperability [20], [21], presenting numerous valuable opportunities for healthcare [22]. In a prior study, the authors introduced a FAIRification workflow [21] to formalize the steps for creating FAIR health datasets and developed an open-source software toolset to implement these steps [23].
1.2. Objective
This paper aims to outline the design of a privacy-preserving federated machine learning architecture built upon FAIR health datasets. This includes detailing the implementation decisions, deployment, and validation across five distinct hospital and health research institute settings spread throughout Europe. Following the FAIRification workflow [21], datasets of various healthcare and health research organizations were transformed into FHIR resources, adhering to the FAIR principles, using open-source FAIRification software tools [23]. Upon achieving FAIR compliance and conformance to HL7 FHIR, the data harmonization and preparation phase for machine learning purposes was nearly completed. However, due to restrictions on dataset exchange or transfer among organizations, a novel approach was necessary. Hence, the authors designed and implemented a federated machine learning architecture that maintains datasets within their respective locations, trains models locally on each FAIR dataset, and transfers only the trained local models to a trusted third party acting as the manager. This process facilitates the merging of local models into a global model capable of making predictions.
This paper introduces an architecture featuring a federated machine learning agent constructed and deployed on top of the FHIR repositories, housing FAIRified datasets within each participating organization. Additionally, a trusted federated machine learning manager was developed and deployed in the cloud to communicate with the agents and orchestrate the federated machine learning process. A browser-based graphical user interface (GUI) was developed on top of the manager to facilitate user interaction. This interface empowers data scientists to seamlessly design and execute federated machine learning algorithms. Users can access the platform through the GUI to perform the following actions:
-
•
Design their features/variables and create corresponding feature sets.
-
•
Create federated datasets based on these feature sets by defining eligibility criteria.
-
•
Choose from available machine learning algorithms and fine-tune various parameters.
-
•
Train machine learning models in a federated manner.
-
•
Utilize the trained models for future predictions.
2. Methods
2.1. Federated ML architecture
The federated ML architecture presented in this study comprises two main components: the Federated ML Manager and the Federated ML Agent. The Federated ML Manager (the “manager”) encompasses a set of services responsible for managing the projects, feature sets, and datasets. It orchestrates federated machine learning operations by connecting to the Federated ML Agents, querying results, aggregating local machine learning models to generate a global model, and conducting predictions in prospective studies using the global model. On the other hand, the Federated ML Agent (the “agent”) consists of components that enable health institutions to perform machine learning operations on FAIR health data.
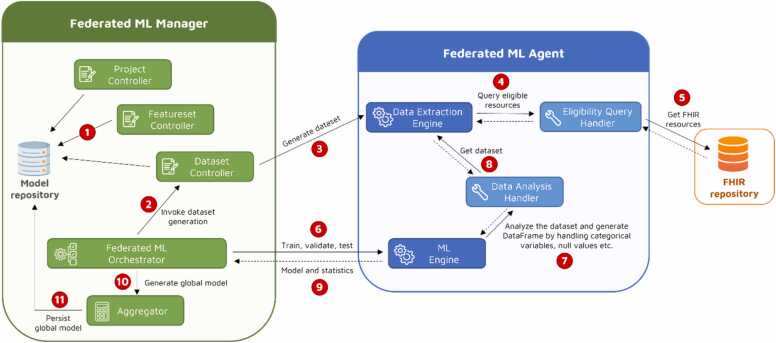
Fig. 1 illustrates the components of the manager and the agent, depicting the high-level communication between them. The numbers in the figure outline the general execution flow within the architecture. The flow commences with the project concept, overseen by the Project Controller, which holds machine learning resources like features, datasets, and models within a unified framework. Each distinct use case can be represented by a separate project.
Fig. 1.
Graphical representation of the Federated ML Architecture: components, subcomponents, and main interactions among them.
The Featureset Controller functions as a feature registry, facilitating the creation and management of features. A feature is defined by a combination of a FHIR search query and FHIR path scripts. These individual features are then organized into groups known as feature sets, enhancing the reusability of these features. This process is indicated by number 1 in Fig. 1. The subsequent numbers in the figure show the sequence in which the components interact with each other. The manager maintains a database known as the Model repository, responsible for persisting resources such as projects, feature sets, datasets, and ML models. Upon defining a feature set, the architecture allows for the creating datasets in a federated manner with respect to a feature set definition.
2.1.1. Data preparation
The methodology of this study is based on the utilization of FAIR data, achieving FAIRness through the adoption of the HL7 FHIR standard. In the preparation of FAIR data for a federated learning pipeline, we rely on standard data access mechanisms within HL7 FHIR, such as FHIR search and FHIR path. These mechanisms provide a standardized approach to data preparation, ensuring verifiability and repeatability across different settings. This standardized approach to data preparation enables the transfer of trained models to various settings, facilitating validation and utilization for online predictions. These models can be effectively transferred and utilized in settings where the same methodology is employed for data preparation, ensuring consistency and reliability in predictions across different environments.
The Dataset Controller functions by interfacing with the Data Extraction Engine of each agent and submitting the dataset generation request (represented as number 3 in the process flow). This request prompts the extraction of datasets from the FHIR repositories independently at each agent. Within the agent, the Data Extraction Engine receives the request, containing eligibility criteria along with a specified feature set. Eligibility criteria consist of a set of FHIR search query scripts allowing both inclusion and exclusion criteria to be specified. Table 1 showcases three example criteria along with their descriptions, illustrating their relationship with the FAIR data.
Table 1.
Three example eligibility criteria defined using a set of FHIR search query statements aligned with the semantics of the FAIR data stored in the FHIR repository.
| Criterion | FHIR search query | Description |
|---|---|---|
| Patients older than or equal to 18 years old | /Patient?birthdate=le2004 | Actual criterion is on the birth date of patients as of the execution of this query which was year 2022. |
| Patients diagnosed with chronic obstructive pulmonary disease (COPD) | /Condition?code=J44,J44.0,J44.1,J44.9 | FAIRified datasets encoded the conditions with ICD-10 and indicated COPD with given list of ICD-10 codes. |
| Patients without a psychiatric disease | /Condition?code:not=I60,I61,I63,G20,G30,F01,F02 | This query is semantically an exclusion criterion to exclude the patients who has conditions coded with the given ICD-10 codes. |
The eligible FHIR resources are queried from the FHIR repository through the Eligibility Query Handler (number 4 & 5). Subsequently, these results undergo conversion into a machine learning (ML)-ready format based on predefined feature definitions. The conversion process involves extracting the value of each feature from eligible resources using the corresponding FHIR search query and FHIR path script pair associated with the feature. This pair serves as an extraction specification [24] that transforms the FHIR resources into a tabular format suitable for ML algorithms, aligning with the predefined feature definitions. For example, the following pair selectively retrieves Patient resources and further extracts the gender value by evaluating the associated FHIR path script.
Each FHIR resource resulting from the search query corresponds to a row. The features represent the columns, and each feature yields a single value, populating the respective cell in this tabular conversion. Algorithm 1 shows the steps of the data preparation process, which are executed by different components as depicted in Fig. 1.
Algorithm 1
Data Preparation on FAIR Health Data.
 |
2.1.2. Model training
Once the datasets are generated at each agent, the orchestrator coordinates the federated model training process using the ML Engine and Data Analysis Handler components of the agents (denoted as numbers 6 to 8). Within the Data Analysis Handler, categorical values undergo encoding, and various strategies are employed to handle empty values through different imputing methods. Following the local models’ training and cross-validation, the manager collects and builds a boosted global model using the Aggregator. Subsequently, this model is persisted in the manager for future predictions (indicated by numbers 9 to 11). All communication between the manager and agents is asynchronous, allowing the manager to submit requests to agents without being blocked for a response. Requests are concurrently sent to agents, and the manager periodically polls the agents to check whether the results are ready.
2.2. Federated ML algorithm
2.2.1. Data preparation
The authors propose a novel data preparation methodology that utilizes FHIR search and FHIR path statements on top of an HL7 FHIR endpoint. This methodology allows the same data preparation pipeline to be employed across various settings without necessitating additional custom development efforts. The Dataset Controller within the manager triggers data preparation at each agent by interfacing with Data Extraction Engine endpoints. Within each agent, the Eligibility Query Handler queries eligible records from the FHIR repository using the provided FHIR search statements within dataset definitions. Following that, the Data Extraction Engine executes the FHIR search and FHIR path statements of each feature to extract the corresponding from each eligible FHIR resource. This process culminates in the generation of an ML-ready dataset in a tabular format, where each column represents the value of a feature, and the rows correspond to eligible FHIR resources.
2.2.2. Model training
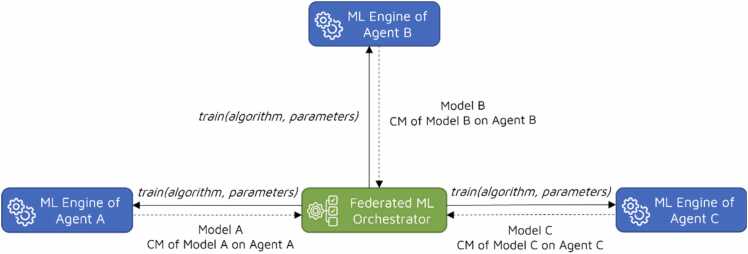
The privacy-preserving federated ML algorithm proposed in this study generates a global predictive model in four steps, utilizing the ML-ready tabular datasets prepared by each agent. Unlike existing federated learning algorithms such as Federated Averaging (FedAvg) [25], which update model parameters over several communication rounds between participants and a central server, the proposed algorithm creates a federated model in only two communication rounds by calculating weights for each local model. In the first step, the training phase begins with the Federated ML Orchestrator sending a training request to each agent. This request includes the name of the classification algorithm to be executed and the user-defined parameter values for the algorithm. Upon receiving this training request, the ML Engine within an agent first splits the data into training and test sets, allocating 80% for training and 20% for testing. Next, it trains a local model on the training set. It sends both the local model and its statistics, encompassing the confusion matrix (CM) containing true positive (TP), false positive (FP), true negative (TN) and false negative (FN) values, back to the Federated ML Orchestrator. Fig. 2 visually represents the training step within a federated setting involving three agents.
Fig. 2.
Training phase of the federated ML algorithm.
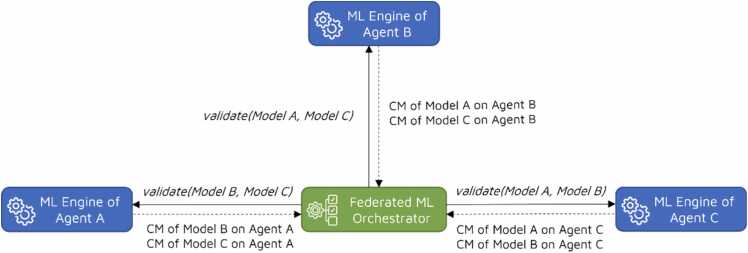
After receiving the local models and confusion matrices from all agents, the Federated ML Orchestrator distributes these models and matrices to every other agent. This enables each agent to independently validate the local models of other agents using their own training data and calculate the corresponding confusion matrices. Consequently, the agents forward these calculated matrices back to the Federated ML Orchestrator, as depicted in Fig. 3.
Fig. 3.
Validation phase of the federated ML algorithm.
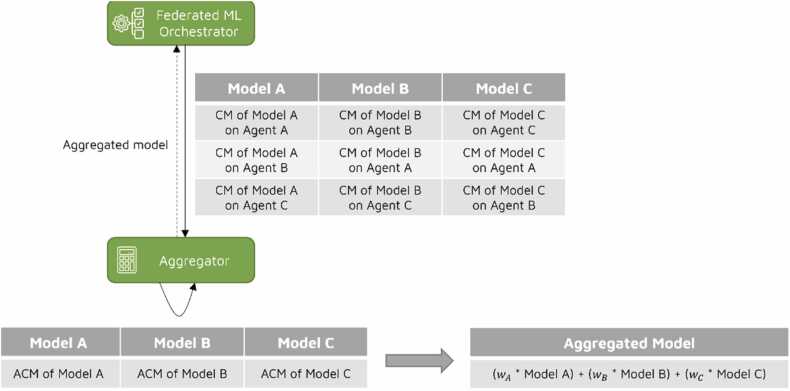
Following the training and validation process involving N agents, the Federated ML Orchestrator possesses N ML models and N * N confusion matrices. In the third step, all models and confusion matrices are forwarded to the Aggregator for generation of the global federated model. During the aggregation phase, illustrated in Fig. 4, the Aggregator combines the numbers within confusion matrices by summing up the TP, FP, TN, and FN values from each model. This process results in the generation of an aggregated confusion matrix (ACM) for each local model, simulating the scenario where all the data is centralized in a single data store.
Fig. 4.
Aggregation phase of the federated ML algorithm.
Once the ACMs are calculated for each model, the Aggregator proceeds to generate the global model. To do this, in this algorithm, the boosting approach is adapted to create a strong model from multiple local models by assigning a weight to each of these local models. In contrast to the traditional boosting approach, where weak classifiers are assigned weights in several iterations based on their performance on the weighted data, in the proposed approach weights are assigned based on the accuracy metric derived from the aggregated TP, FP, TN, FN values within the corresponding ACM. In this way, if a local model performs well in all participating sites, it gets greater weight. Likewise, if a local model performs well in one site, but it underperforms in other sites, it gets lower weight.
The calculated weights are normalized before assignment. The global model, or aggregated model, is then constituted by the weighted sum of the local models. This process can be mathematically expressed as follows:
When the Federated ML Orchestrator receives the aggregated model, it initiates the fourth and last step, the testing phase. The primary objective of this phase is to assess the performance of the global model on the data of agents, in comparison to their respective local models. This phase does not influence the global model itself. During this phase, the aggregated model, comprising various local models and their assigned weights, is disseminated to all agents for testing. Unlike conventional machine learning applications, where positive predictions are labeled as + 1 and negative predictions as 0, this algorithm uses + 1 for positive predictions and − 1 for negative predictions. Employing this approach, the algorithm computes the weighted sum based on the formula. If the result is a positive value, it predicts 1; otherwise, it predicts 0. For instance, consider an aggregated model with the formula , where the predictions of A, B, and C are negative, negative, and positive, respectively. The algorithm computes the formula as the following and generates an output of 0.
However, if the predictions for A, B, and C are negative, positive, and negative, respectively, the resulting output prediction would be 1. This calculation is derived as follows:
The pseudocode of the proposed privacy-preserving federated machine learning algorithm on FAIR health data is presented in Algorithm 2.
Algorithm 2
Privacy-Preserving Federated Machine Learning Algorithm on FAIR Health Data.
 |
3. Application & results
3.1. Datasets & FAIRification
The existing healthcare and health research datasets from the following five organizations have been FAIRified and prepared for use in this study. Ethical Board (EB) approvals were obtained in all countries, and the protocol numbers corresponding to each organization are listed below:
-
1.
Andalusian Health Service – Virgen del Rocío University Hospital (SAS) from Spain: 1269-M1-20
-
2.
Health Sciences Institute of Aragón (IACS) from Spain: 1269-M1-20
-
3.
Geneva University Hospital (UNIGE) from Switzerland: 2020-02683
-
4.
University of Porto (UP) from Portugal: PARECER A-13/2020
-
5.
Catholic University of the Sacred Heart (UCSC) from Italy: 1066/20-12/05/2020
The FAIRification workflow [21] provides a step-by-step guide tailored to making existing health data sets FAIR, specifically designed to meet the unique needs and requirements of health data. The Data Curation Tool, an open-source standalone software [23], addresses data curation and validation within this FAIRification workflow. This tool aids data owners in transforming their health datasets into HL7 FHIR resources. It accomplishes this through a user-friendly graphical user interface (GUI), providing features for terminology translations [26]. Moreover, onFHIR is an open-source implementation of the HL7 FHIR standard, serving as a versatile FHIR repository [27].
The datasets from the five distinct health organizations underwent transformation into HL7 FHIR resources through the same FAIRification workflow and the same set of software, resulting in the creation of FAIR datasets. Notably, this transformation process was independently conducted at each deployment site. To establish a FHIR repository, an instance of onFHIR [27] was deployed, serving as the platform for FHIR profiles constituting a common data model [23]. Utilizing the Data Curation Tool, corresponding FHIR resources were created, and stored in the FHIR repository. Table 2 provides insight into the quantity of FHIR resources available at the FHIR repository instances across participating organizations. These resources served as the foundational data used for training federated machine learning models in this study.
Table 2.
Total number of FHIR resources by resource type at each FAIR dataset.
| FHIR resource type | Total number of FHIR resources |
||||
|---|---|---|---|---|---|
| SAS | IACS | UNIGE | UP | UCSC | |
| Patient | 7873 | 7622 | 398 | 1313 | 1050 |
| Encounter | 11,954 | 12,106 | 410 | 0 | 0 |
| Observation | 73,817 | 11,999 | 2107 | 11,817 | 3150 |
| Condition | 118,616 | 78,040 | 1113 | 0 | 1081 |
| MedicationStatement | 42,164 | 70,880 | 2395 | 0 | 28,127 |
3.2. Deployment
The FHIR repository endpoints of the organizations were exclusively accessible within their local networks, solely by authenticated applications. As depicted in Fig. 5, a Federated ML Agent was deployed within the secure networks of each organization. This allowed the agent to access the FHIR repository and conduct the training of local models using the FAIRified data. In addition, a single trusted Federated ML Manager was deployed in the cloud. This manager was authorized by each organization to access its respective agent. Furthermore, each Federated ML Agent was accessible solely from the trusted Federated ML Manager, and the communication between the manager and the agents was encrypted to ensure security. Moreover, a web-based GUI was provided to the data scientists, enabling them to be authenticated by the manager and authorized to use the system via their web browsers.
Fig. 5.
The deployment setup of the federated machine learning architecture.
3.3. Experiment setup
Having the FAIR datasets and software deployed as per the experiment setup, data scientists from participating organizations conducted system testing based on a specific use case: predicting the risk of readmission among COPD patients within 30 days following hospital discharge. To represent this use case, a single project was established, ensuring every user worked within the same project through the web GUI of the Federated ML Manager. For this predictive analysis, 60 distinct features were defined using FHIR search queries and FHIR Path scripts to represent the independent variables. Additionally, one feature was created as the dependent variable to indicate whether readmission occurred within the 30-day period after discharge.
The users responsible for defining the features by providing FHIR search query and FHIR path scripts were required to possess a fundamental understanding of the FHIR standard. They needed to comprehend how the FAIRified datasets were represented by the FHIR resources and code systems specified in the FHIR profiles. Table 3 presents a few examples of the features defined in the experiments. For instance, in the Gender variable, the value is extracted directly from the gender field within the FHIR Patient resource. In the Smoking Status, Coronary heart disease, and Corticosteroids, the FHIR Path of “value:exists” examines the existence of a resource to be queried by the given FHIR search query. Similarly, in the Hemoglobin, the value is derived from the value field of valueQuantity field within the FHIR Observation resource. In these queries, standardized code systems such as LOINC (Logical Observation Identifiers Names and Codes) for smoking status (72166–2) and laboratory tests like hemoglobin (718−7), SNOMED-CT (Systematized Nomenclature of Medicine -- Clinical Terms) for concepts such as Yes (373066001), No (373067005) and Unknown (261665006), ICD-10 (International Classification of Diseases 10th Revision) for conditions, and ATC (Anatomical Therapeutic Chemical) for medications were utilized. These code systems were semantically encoded within the FAIR datasets.
Table 3.
A subset of the variable/feature definitions used in our experiments.
| Variable/ Feature | FHIR search query | FHIR path |
|---|---|---|
| Gender | /Patient | value:Patient.gender |
| Smoking Status | /Observation?code= 72166-2&value-concept= 373066001 | value:exists |
| Coronary heart disease | /Condition?code=I20,I21,I24,I25 | value:exists |
| Corticosteroids | /MedicationStatement?code= R03AK06,R03AK07,R03AK08, R03AK10,R03AK11 | value:exists |
| Hemoglobin | /Observation?code= 718-7 | value:Observation.valueQuantity.value |
For this specific use case, a dataset was generated utilizing the aforementioned featureset in a federated ML environment, engaging SAS, IACS, UNIGE, UP, and UCSC, as depicted in Fig. 5. The use case was constrained to patients aged older than 18 years and diagnosed with COPD. Consequently, two eligibility criteria were established: “/Patient?birthdate=le2004″ and “/Condition?code=J44,J44.0,J44.1,J44.9″. The former criteria in the FHIR Search Query identifies patients with a birthdate on or before 2004, while the latter filters patients whose condition records include at least one of the specified ICD-10 codes for COPD.
After creating the dataset in the federated environment, multiple federated machine learning models were generated using different base algorithms, including Logistic Regression, Support Vector Machine (SVM), Decision Trees, and Random Forest. These models employed various algorithm parameters such as threshold, regularization parameter, number of trees, maximum depth of a tree, and feature subset strategy. Among them, Random Forest yielded the most favorable outcome, which was somewhat anticipated due to significant data imbalance in certain agents (e.g., IACS, 98%), because tree-based algorithms, like Random Forest, are known to be resilient to data imbalances, outliers, and noise [28]. During the training phase, diverse values were provided for the maximum depth of a tree (5, 10, 15), minimum information gain (0.0, 0.2, 0.5), impurity metrics (gini, entropy) and number of trees (25, 50, 100) to the agents for testing while generating their respective local Random Forest models. Each agent’s ML Engine explored all combinations of these values and created the best-performing local model. Subsequently, the Aggregator aggregated these local models to generate a global federated model.
3.4. Results
The performance evaluation of the federated model took place in a prospective study involving patients from Andalusian Health Service – Virgen del Rocío University Hospital, Spain (SAS) (EB approval: 1269-M1–20) and the Institute for Pulmonary Diseases of Vojvodina, Serbia (IPBV) (EB approval: 110-V/10). This study focused on predicting the 30-day risk of readmission due to COPD following discharge for 100 recruited patients, comprising 22 from SAS and 78 from IPBV. The accuracy of the federated model's predictions was assessed by comparing its forecasts to the actual outcomes observed 30 days post-discharge. The federated model's predictions were accurate in 87% of cases within this prospective study.
It is important to note that this study primarily emphasizes the design and implementation of the federated architecture on FAIR health data. As such, a clinical evaluation was not conducted as part of this study. The authors presented a detailed analysis of the results following a clinical evaluation using the proposed architecture in another study [29], [30].
4. Discussion
The primary advantage of this study lies in demonstrating the practical application of a privacy-preserving federated machine learning architecture across five distinct healthcare and health research organizations. With the proposed solution, no organization needs to transfer its datasets to an off-site location, yet they can still train ML models by leveraging others' datasets. The training of ML models involves numerous prerequisites in terms of data preparation, many of which were already addressed by having FAIR datasets, despite their diverse locations. The proposed methodology capitalizes on these FAIR datasets, showcasing the significant promise of FAIR health data for machine learning, particularly in a federated execution while ensuring privacy preservation with no data sharing.
One major strength of the proposed methodology is its capacity to be applied in different settings given that the data adheres to FAIR through HL7 FHIR. In this study’s context, the Institute for Pulmonary Diseases of Vojvodina, Serbia (IPBV) effectively generated predictions using a federated model trained on datasets from entirely different healthcare organizations, and IPBV did not contribute to training this model due to the absence of retrospective data for that specific study.
The architecture developed in this study presents a versatile solution akin to a simple, no-code, GUI-based version of data science notebooks. It is not confined to a particular use case or set of variables; instead, it accommodates various classification scenarios that can be implemented by defining and/or reusing features, executing diverse algorithms, and adjusting several algorithm-specific parameters and missing data handling strategies. This adaptability ensures the reproducibility of the study. All agents and the manager operate within a secure environment, employing encrypted communication between the agent and manager. The agent is equipped with a highly restricted set of services, preventing the data from leaving its boundaries. Working in such a secure federated environment, both data scientists and clinicians can leverage datasets from other organizations to develop their ML models without direct access to those datasets. It is reported that studies on ML-based prediction models often exhibit a high risk of bias, primarily attributed to small study sizes, inadequate handling of missing data, and susceptibility to overfitting [31]. The proposed approach inherently addresses bias risk by employing local models from various datasets, potentially expanding the study size and mitigating overfitting while preserving privacy.
The presented methodology is privacy-preserving by design because the agents do not share any data, including the models, with each other. Instead, the confusion matrices are sent to a trusted orchestrator operated outside the agents’ domain. Importantly, this orchestrator remains unaware of the agents and their datasets. An alternative approach could have been an architecture without a trusted third party (the manager), where the agents share the confusion matrices with each other using secure multi-party computation techniques. However, this approach falls outside the scope of this work, as the authors’ design decision focused on achieving privacy through a trusted manager.
In the literature, only a few studies addressed the challenges of federated machine learning with health datasets while preserving privacy. The Personal Health Train (PHT) aims to establish a framework that integrates access-restricted data from multiple parties with different data governance policies in a privacy-preserving manner [16]. Despite applying strict de-identification and encryption measures within a secure environment, the PHT necessitates the transfer of datasets out of owner organizations into a central data store. In contrast, the proposed approach prohibits dataset transfers and implements federated versions of classification algorithms within a highly secure and trusted environment. Another approach, DataSHIELD (Data Aggregation Through Anonymous Summary-statistics from Harmonized Individual levEL Databases) [32], focuses on analyzing distributed data without granting the analyzer unrestricted access. Although DataSHIELD is a proven and widely used approach in epidemiological studies, it has limitations due to its reliance on the Opal data warehouse and a modified R statistical environment. The proposed methodology centers on FAIR datasets using the HL7 FHIR standard, a well-established standard widely used by various software vendors and organizations. Despite enabling the entire process through a GUI, the authors’ design does not impose library-related restrictions, allowing for the incorporation of new classification or other ML algorithms. Two other studies apply federated learning to healthcare datasets, but each concentrate on specific problems and the results of trained models for those issues [33], [34]. Notably, they do not rely on a standardized approach for data preparation; instead, they design their data transformation mechanisms and custom data models, which lack detailed explanations in their methods. In contrast, the proposed solution adheres to the standards of a FAIRification workflow centered around HL7 FHIR.
The presented methodology does not offer a specific framework tailored explicitly for time-series data. Feature definitions within the proposed approach are constrained by the availability of values in the FHIR repository and are subject to limitations based on FHIR path evaluation capabilities. Consequently, creating time-series for feature definitions becomes restricted within these confines. Additionally, the federated ML approach presented in this paper is limited to binary classification algorithms. Extensions to the presented methodology would be required to incorporate correlation or deep learning methods.
5. Conclusion
This paper presents the design for a privacy-preserving federated ML architecture and the algorithm for executing the federated learning process within this architecture. The system was deployed on top of already-FAIRified health data from five distinct healthcare and health research organizations across Europe, and an experimental evaluation was conducted in real-life settings. Data scientists defined several features and utilized them to extract datasets and train ML models employing various strategies and algorithm-specific parameters. The best-performing classification model was employed in a prospective study aimed at predicting the 30-day readmission possibilities of patients with COPD.
Irrespective of the positive results observed in the prospective study or the test statistics derived from the trained ML models, the experiments showed that the proposed solution can successfully utilize FAIR datasets from multiple health organizations for ML processes while preserving privacy within a trusted environment. Adherence to FAIR principles and adopting standards like HL7 FHIR in the health domain create new prospects for secondary use, such as federated ML. This approach assists healthcare and health research organizations in safely leveraging datasets from other entities to build more accurate models for classification problems. The utilization of FAIR datasets, which are generated through the FAIRification workflow, simplifies the process of data extraction and preparation for clinical study analyses for data teams of health data controllers, including hospitals. Datasets can be prepared for machine learning applications through a standardized and machine-processable pipeline.
Code availability
The source code for the FAIRification of health data and federated machine learning platform on top of the FAIR health data is provided as open source on GitHub. The same software was deployed during the evaluation after completing formal bilateral agreements for data access.
-
•
The agents and the central orchestration server backend of the federated learning platform software can be found at https://github.com/fair4health/ppddm
-
•
The frontend software of the federated learning platform can be found at https://github.com/fair4health/f4h-portal-ui
-
•
The FAIRification tools can be found at https://github.com/fair4health/common-data-model, https://github.com/fair4health/data-curation-tool, https://github.com/fair4health/data-privacy-tool
Funding
This work was supported by the FAIR4Health project [35], which received funding from the European Union’s Horizon 2020 research and innovation programme under grant agreement number 824666. Additionally, this study has been funded by Instituto de Salud Carlos III (ISCIII) through the project PT20/00088 and IMP/00019 and co-funded by the European Union.
CRediT authorship contribution statement
AAS: Conceptualization, Methodology, Software, Investigation, Writing – original draft. MG: Conceptualization, Methodology, Software, Writing – original draft, Visualization. CAR: Validation, Investigation, Resources, Writing – review & editing. GBLE: Conceptualization, Methodology, Writing – review & editing. AMG: Validation, Resources, Writing – review & editing. MJEC: Supervision. CLPC: Conceptualization, Supervision, Project administration, Writing – review & editing.
Declaration of Competing Interest
The authors declare the following financial interests/personal relationships which may be considered as potential competing interests: A. Anil Sinaci reports financial support was provided by EU Framework Programme for Research and Innovation Science with and for Society. Celia Alvarez-Romero reports financial support was provided by European Regional Development Fund. If there are other authors, they declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
We extend our sincere gratitude to the clinical researchers representing the health research organizations within the FAIR4Health consortium: Universite De Geneve (Switzerland), University Hospitals of Geneva (Switzerland), Universita Cattolica Del Sacro Cuore (Italy), Universidade Do Porto (Portugal), Instituto Aragonés de Ciencias de la Salud (Spain), Institut Za Plucne Bolesti Vojvodine (Serbia), and Servicio Andaluz de Salud (Spain).
References
- 1.Vayena E. Value from health data: European opportunity to catalyse progress in digital health. Lancet (Lond, Engl) 2021;397:652–653. doi: 10.1016/S0140-6736(21)00203-8. [DOI] [PubMed] [Google Scholar]
- 2.Alami H., Gagnon M.-P., Fortin J.-P. Digital health and the challenge of health systems transformation. MHealth. 2017;3:31. doi: 10.21037/mhealth.2017.07.02. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pashazadeh A., Navimipour N.J. Big data handling mechanisms in the healthcare applications: a comprehensive and systematic literature review. J Biomed Inform. 2018;82:47–62. doi: 10.1016/J.JBI.2018.03.014. [DOI] [PubMed] [Google Scholar]
- 4.Health Insurance Portability and Accountability Act of 1996 (HIPAA) n.d. https://www.cdc.gov/phlp/publications/topic/hipaa.html (accessed November 22, 2023).
- 5.General Data Protection Regulation (GDPR) n.d. 〈https://gdpr.eu/〉 (Accessed November 22, 2023).
- 6.The Data Protection Act n.d. 〈https://www.gov.uk/data-protection〉 (Accessed November 22, 2023).
- 7.Office of the Privacy Commissioner of Canada. The Personal Information Protection and Electronic Documents Act (PIPEDA) n.d. https://www.priv.gc.ca/en/privacy-topics/privacy-laws-in-canada/the-personal-information-protection-and-electronic-documents-act-pipeda/ (Accessed November 22, 2023).
- 8.Personal Information Protection Law of the People’s Republic of China n.d. 〈http://www.npc.gov.cn/npc/c30834/202108/a8c4e3672c74491a80b53a172bb753fe.shtml〉 (Accessed November 22, 2023).
- 9.Ministry of Electronics and Information Technology G of I. Information Technology Act n.d. 〈https://www.meity.gov.in/content/information-technology-act〉 (Accessed November 22, 2023).
- 10.Triantafyllidis A.K., Tsanas A. Applications of machine learning in real-life digital health interventions: review of the literature. J Med Internet Res. 2019;21(4) doi: 10.2196/12286. Https://WwwJmirOrg/2019/4/E12286 2019;21:e12286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Holzinger A. Machine learning for health informatics. Lect Notes Comput Sci Incl Subser Lect Notes Artif Intell Lect Notes Bioinforma. 2016:1–24. doi: 10.1007/978-3-319-50478-0_1. 9605 LNCS. [DOI] [Google Scholar]
- 12.Yang Q., Liu Y., Chen T., Tong Y. Federated machine learning. ACM Trans Intell Syst Technol (TIST) 2019;10 doi: 10.1145/3298981. [DOI] [Google Scholar]
- 13.Chamikara M.A.P., Bertok P., Khalil I., Liu D., Camtepe S. Privacy preserving distributed machine learning with federated learning. Comput Commun. 2021;171:112–125. doi: 10.1016/J.COMCOM.2021.02.014. [DOI] [Google Scholar]
- 14.Kairouz P., McMahan H.B., Avent B., Bellet A., Bennis M., Bhagoji A.N., et al. Advances and open problems in federated learning. Found Trends® Mach Learn. 2021;14:1–210. doi: 10.1561/2200000083. [DOI] [Google Scholar]
- 15.Cui J., Zhu H., Deng H., Chen Z., Liu D. FeARH: Federated machine learning with anonymous random hybridization on electronic medical records. J Biomed Inform. 2021;117 doi: 10.1016/J.JBI.2021.103735. [DOI] [PubMed] [Google Scholar]
- 16.Beyan O., Choudhury A., van Soest J., Kohlbacher O., Zimmermann L., Stenzhorn H., et al. Distributed analytics on sensitive medical data: the personal health train. Data Intell. 2020;2:96–107. doi: 10.1162/dint_a_00032. [DOI] [Google Scholar]
- 17.Brownlee J. Data preparation for machine learning: data cleaning, feature selection, and data transforms in Python. Mach Learn Master. 2020 [Google Scholar]
- 18.Wilkinson M.D., Dumontier M., Aalbersberg I.J.J., Appleton G., Axton M., Baak A., et al. The FAIR guiding principles for scientific data management and stewardship. Sci Data. 2016;3 doi: 10.1038/sdata.2016.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.FAIR Principles - GO FAIR n.d. https://www.go-fair.org/fair-principles/ (Accessed November 22, 2023).
- 20.van Reisen M., Stokmans M., Basajja M., Ong’ayo A.O., Kirkpatrick C., Mons B. Towards the tipping point for FAIR implementation. Data Intell. 2020;2:264–275. doi: 10.1162/dint_a_00049. [DOI] [Google Scholar]
- 21.Sinaci A.A., Núñez-Benjumea F.J., Gencturk M., Jauer M.L., Deserno T., Chronaki C., et al. From raw data to FAIR data: the FAIRification workflow for health research. Methods Inf Med. 2020;59:E21–E32. doi: 10.1055/S-0040-1713684. [DOI] [PubMed] [Google Scholar]
- 22.Queralt-Rosinach N., Kaliyaperumal R., Bernabé C.H., Long Q., Joosten S.A., van der Wijk H.J., et al. Applying the FAIR principles to data in a hospital: challenges and opportunities in a pandemic. J Biomed Semant. 2022;13(1):12. doi: 10.1186/s13326-022-00263-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.FAIR4Health Software n.d. https://github.com/fair4health (Accessed November 22, 2023).
- 24.Sinaci A.A., Laleci Erturkmen G.B. A federated semantic metadata registry framework for enabling interoperability across clinical research and care domains. J Biomed Inform. 2013;46:784–794. doi: 10.1016/J.JBI.2013.05.009. [DOI] [PubMed] [Google Scholar]
- 25.McMahan H.B., Moore E., Ramage D., y Arcas B.A. Federated learning of deep networks using model averaging. arXiv Prepr arXiv:1602. 2016;2(05629):2. doi: 10.48550/arxiv.1602.05629. [DOI] [Google Scholar]
- 26.Sinaci A.A., Gencturk M., Teoman H.A., Laleci Erturkmen G.B., Alvarez-Romero C., Martinez-Garcia A., et al. A data transformation methodology to create findable, accessible, interoperable, and reusable health data: software design, development, and evaluation study. J Med Internet Res. 2023;25 doi: 10.2196/42822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.onFHIR.io n.d. https://onfhir.io/ (Accessed November 22, 2023).
- 28.Drummond C., Holte R.C. Exploiting the cost (in) sensitivity of decision tree splitting criteria. InICML. 2000:1. [Google Scholar]
- 29.Alvarez-Romero C., Martinez-Garcia A., Vega J.T., Díaz-Jimènez P., Jimènez-Juan C., Nieto-Martín M.D., et al. Predicting 30-day readmission risk for patients with chronic obstructive pulmonary disease through a federated machine learning architecture on findable, accessible, interoperable, and reusable (FAIR) data: development and validation study. JMIR Med Inform. 2022;10(6) doi: 10.2196/35307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Carmona-Pírez J., Poblador-Plou B., Poncel-Falcó A., Rochat J., Alvarez-Romero C., Martínez-García A., et al. Applying the FAIR4Health solution to identify multimorbidity patterns and their association with mortality through a frequent pattern growth association algorithm. Int J Environ Res Public Health. 2022;19(4):2040. doi: 10.3390/ijerph19042040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Andaur Navarro C.L., Damen J.A.A., Takada T., Nijman S.W.J., Dhiman P., Ma J., et al. Risk of bias in studies on prediction models developed using supervised machine learning techniques: systematic review. BMJ. 2021;375:2281. doi: 10.1136/BMJ.N2281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gaye A., Marcon Y., Isaeva J., Laflamme P., Turner A., Jones E.M., et al. DataSHIELD: taking the analysis to the data, not the data to the analysis. Int J Epidemiol. 2014;43:1929–1944. doi: 10.1093/IJE/DYU188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vaid A., Jaladanki S.K., Xu J., Teng S., Kumar A., Lee S., et al. Federated learning of electronic health records to improve mortality prediction in hospitalized patients with COVID-19: machine learning approach. JMIR Med Inf. 2021;9(1) doi: 10.2196/24207. Https://MedinformJmirOrg/2021/1/E24207 2021;9:e24207. https://doi.org/10.2196/24207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sadilek A., Liu L., Nguyen D., Kamruzzaman M., Serghiou S., Rader B., et al. Privacy-first health research with federated learning. Npj Digit Med 2021 4:1. 2021;4:1–8. doi: 10.1038/s41746-021-00489-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Alvarez-Romero C., Martínez-García A., Sinaci A.A., Gencturk M., Méndez E., Hernández-Pérez T., et al. FAIR4Health: findable, accessible, interoperable and reusable data to foster health research. Open Res Eur. 2022;2 doi: 10.12688/openreseurope.14349.2. https://doi.org/10.12688%2Fopenreseurope.14349.2. [DOI] [PMC free article] [PubMed] [Google Scholar]