Summary
Only a small number of avian species inhabit salty environments. To understand how they adapted, we examined the evolution of kidney sizes, supraorbital salt glands (SSGs), and the utilization of salty habitats across 230 species spanning 25 avian orders. Phylogenetic analysis indicates that SSGs, large kidneys, and thriving in salty habitats emerged convergently in birds. Transition rate analysis reveals that species possessing SSGs and large kidneys tended to move from low-to high-salinity environments, while others moved in the opposite direction. However, habitat salinity also influenced kidney evolution; lineages residing in high-salinity environments tended to develop larger kidneys than those in low-salinity environments. Our findings suggest that SSGs and large kidneys may have evolved through adaptation to high salinity. Overall, habitat conditions and physiological traits influenced avian adaptation to salty environments in a reciprocal manner. These results shed the new light on the evolutionary mechanisms underlying functional diversity in birds.
Subject areas: Ornithology, Evolutionary biology, Phylogeny
Graphical abstract

Highlights
-
•
Birds have independently evolved larger kidneys and salt glands multiple times
-
•
Kidney sizes and salt glands in birds correlate positively with habitat salinity
-
•
Habitat salinity constrains the evolution of bird kidney sizes and salt glands
Ornithology; Evolutionary biology; Phylogeny;
Introduction
Oceans provide vast space and rich resources for organisms. However, organisms must overcome the osmotic challenge of saltwater intake to live in such environments. Therefore, less than 3% of avian species (i.e., seabirds) live entirely or partially in high-osmolarity environments.1
Supraorbital salt glands (SSGs, salt glands) in birds, also known as glandulae nasalis,2,3 produce concentrated solutions primarily consisting of NaCl or KCl, which help maintain proper internal solute levels while minimizing water loss.4,5,6 Glands with a similar function might have evolved, independently and convergently in reptiles such as turtles, sea snakes, lizards, and crocodiles.7,8 These glands are widespread and found in various parts of the skull across different evolutionary lineages, although their homology remains to be examined. Moreover, the unique positioning of the salt glands above the eye is a distinctive feature of birds.3,4 Supraorbital salt glands have been discovered in at least 40 bird families, which cover almost all traditional bird orders except for the Passeriformes.8 These glands are particularly common in marine birds such as gulls, petrels, albatrosses, auks, and penguins but can also be found in some freshwater species, such as dabbling ducks, mallards, and rails.9 Desert-dwelling birds such as ostriches and North African partridges1,8 and some carnivorous birds with high-protein diets such as Tawny Eagles10 also possess supraorbital salt glands. Avian supraorbital salt glands display significant levels of plasticity in both their structure and function,1,6,11,12,44 but their size and excretory capacity largely depend on the species’ environmental salinity and diet.13,14
Birds also use their kidneys to maintain the osmotic balance of their blood.15 The avian kidney comprises units of lobule with a renal medulla containing loops of Henle able to extract water from urine with higher salt concentrations than in the plasma. It has been shown that the mass of the kidney is positively correlated with the number of medullary cones, the smaller units of a renal medulla, which are associated with its ability to regulate osmotic pressure.15 How birds physiologically maintain their osmostasis has been studied intensively.14 However, we know little about how the avian physiological traits associated with osmoregulation, such as the SSGs and large kidneys, have evolved in response to the types of habitats they use.
Non-passerine birds with SSGs tend to have larger kidneys (compared with their body sizes) than others.1 Hughes, therefore, proposed that birds with high-salinity tolerance have larger kidneys for better osmoregulation. However, shared ancestry among species can lead to a significant association between two traits evolving independently.19 Without controlling for such “phylogenetic signals” (as Hughes did not1), we do not know whether high-salinity environments could have driven these two physiological traits to evolve in birds. Independent evolution of similar traits, convergent evolution, in the same environmental conditions, has long been considered convincing evidence of adaptation.56 If a salty environment could drive the evolution of a larger kidney and the presence of an SSG, could it lead to the convergent evolution of the larger kidneys and SSGs in different avian lineages? It is also unclear whether the evolution of the two physiological traits was correlated or independent in birds.
Results
Convergent evolution of larger kidney sizes, presence of supraorbital salt glands, and the use of high salinity habitat
In our dataset, species in completely or mainly saline habitats usually have large kidneys (residual kidney weight ≧ 0.13, 67%) and SSGs (91%); in contrast, few species in mainly and completely freshwater habitats have “large kidneys” (88% had residual kidney weight <0.13), and most (80%) do not have SSGs. Ancestral characteristics reconstruction suggested that the ancestor of birds probably lived in freshwater environments, had no SSG, and had medium-sized kidneys (the probabilities of having “non-large kidneys” and of having “large-kidneys” are roughly equal) (Figure 1).
Figure 1.
Estimating ancestral states and reconstructing traits in avian species: habitat salinity, presence of SSGs, and kidney sizes
Estimated ancestral states for the level of habitat salinity (A) and the presence of SSGs (B) for the 230 avian species studied and kidney sizes for the 167 avian species studied (C). The characteristics of each species are shown at the tips of the phylogenetic tree. The posterior probability of the reconstructed characteristic state of each trait is shown in the pie on each ancestral node. The gray and black bars next to the phylogenetic tree indicate the order of the species. The color patterns at the rightmost indicate the characteristic state of the extant species.
Interestingly, our results suggest that the birds’ utilization of high-salinity habitats evolved together with the growth of their kidneys and the development of SSGs. The use of high-salinity habitats (sum of posterior probability for completely or mainly saline habitats >0.5) occurred independently at least 14 times in 12 orders of birds (Anseriformes, Charadriiformes, Coraciiformes, Gaviiformes, Gruiformes, Phoenicopteriformes, Podicipediformes, Pelecaniformes, Passeriformes, Suliformes and Sphenisciformes); SSGs evolved independently 8 times in 14 orders of birds (Accipitriformes, Anseriformes, Charadriiformes, Falconiformes, Galliformes, Gaviiformes, Gruiformes, Pelecaniformes, Phoenicopteriformes, Podicipediformes, Procellariiformes, Sphenisciformes, Struthioniformes and Suliformes) (Figure 1). Intriguingly, both the utilization of high-salinity habitats and the presence of SSGs evolved in the same nodes for at least four avian lineages (Columbea, Aequornithes, and part of Galloanserae and Charadriiformes). Furthermore, SSGs are present in the common ostrich (Struthio camelu), the 13 Accipitriformes species, and 5 Falconiformes species that do not utilize high-salinity habitats. In contrast, four species in Passeriformes, the ash-throated flycatcher (Myiarchus cinerascens), the marsh wren (Cistothorus palustris), the snow bunting (Plectrophenax nivalis), and the Savannah sparrow (Passerculus sandwichensis) and one species in Coraciiformes, the belted kingfisher (Megaceryle alcyon), have evolved to utilize high-salinity habitats without having SSGs.
The use of high-salinity habitats and the presence of SSGs are evolutionarily inert in birds. Only five lineages, the pin-tailed duck (Anas acuta), the redhead (Aythya Americana), the greater scaup (Aythya marila), the upland sandpiper (Bartramia longicauda), and the long-billed dowitcher (Limnodromus scolopaceus), have evolved from ancestors utilizing high salinity habitats to descendants using freshwater habitats. Only four lineages, the white stork (Ciconia Ciconia), the African fish eagle (Haliaeetus vocifer), the American kestrel (Falco sparverius), and the Robert Falco (Falco jugger) have lost SSGs possessed by their ancestors.
By contrast, the evolution of birds’ kidney size has been relatively labile. Large kidneys evolved at least 25 times in twelve orders of birds (Struthioniformes, Anseriformes, Galliformes, Phoenicopteriformes, Podicipediformes, Gaviiformes, Gruiformes, Pelecaniformes, Suliformes, Charadriiformes, Coraciiformes, and Passeriformes). In addition, lineages derived from those with large kidneys have 16 times reverted to having residual kidney weight <0.13, namely the mallard duck (Anus platyrhynchos), the pin-tailed duck (A. acuta), the redhead (Aythya Americana) in the non-Passeriformes, and the horned lark (Eremophila alpestris), the American bushtit (Psaltriparus minimus), the cedar waxwing (Bombycilla cedrorum), the cactus wren (Campylorhynchus brunneicapillus), the northern mockingbird (Mimus polyglottos), the Bendire’s thrasher (Toxostoma bendirei), the zebra finch (Taeniopygia guttata), the chipping sparrow (Spizella passerina), the grey-cheeked thrush (Catharus minimus), the russet nightingale-thrush (C. occidentalis), the hermit thrush (C. guttatus), the ancestors of corvoidea, and all species of passeroidea in Passeriformes. However, birds in other avian lineages re-acquired large kidneys. These include the Eurasian magpie (Pica pica) in corvoidea, the painted redstart (Myioborus pictus), Bullock’s oriole (Icterus bullockii), Brewer’s blackbird (Euphagus cyanocephalus), the rufous-crowned sparrow (Aimophila ruficeps), the Savannah sparrow, and Lincoln’s sparrow (Melospiza lincolnii) in passeroidea, Carduelinae and a “masked” clade (Piranga, Cardinalis, and Caryothraustes) in Cardinalidae. It is worth noting that birds adapted to high-salinity habitats have all evolved at least one characteristic assumed to be related to osmostasis except the snow bunting (Plectrophenax nivalis).
In addition, the results of search.conv clarify that when birds inhabit saline environments, the kidney size and SSGs in different lineages tend to be similar (found highly significant under “state,” p = 0.001). Furthermore, birds' habitat diversity within each clade is diverse. Different clades do not exhibit convergent evolution toward the same direction (found no significance under “clade,” p = 0.117).
Correlated evolution of large kidneys and functional supraorbital salt glands
Our results show high phylogenetic signals in levels of habitat salinity (λ = 0.78, p ≪ 0.001; K = 0.26, p = 0.001; null hypothesis λ and K = 0), the presence of SSGs (λ = 1, p ≪ 0.001; K = 1.373, p = 0.001; null hypothesis λ and K = 0), and residual kidney weight (λ = 0.7, p ≪ 0.001; K = 0.2, p value = 0.012; null hypothesis λ and K = 0). After controlling for the phylogenetic signal, the PGLS results show that birds in high-salinity environments tend to have larger kidneys; the residual kidney weight is significantly positively correlated with levels of habitat salinity (λ = 0.54, K = 0, δ = 0.26, estimate = 0.07, s.e. = 0.01, t = 7.38, p ≪ 0.001, R2 = 0.25). The evolutionary dependency between residual kidney weight and habitat types is also supported by the results of the correlated evolution analysis, except at the 25th percentile cut-off point (logBF for the set of cut-off points: 25th = 0.67; 50th = 10.872; 75th = 23.041).
The results of our PGLM analysis show that the presence of SSGs is significantly positively correlated with levels of habitat salinity (λ = 0.91, K = 0.42, δ = 2.09, estimate = 0.488, s.e. = 0.226, t = 2.16, p = 0.03, R2 = 0.099) (Figure 2). The results of the correlated evolutionary analyses also support such an association; the salt gland characteristic and habitat types are highly evolutionarily dependent on each other (logBF = 16.012). Furthermore, the presence of SSGs is positively correlated with kidney size (λ = 0.46, K = 0, δ = 0.3, estimate = 0.21, s.e. = 0.03, t = 6.4, p ≪ 0.001, R2 = 0.19) (Figure 3). The results of the correlated evolutionary analyses show that the residual kidney weight and salt gland characteristics are strongly dependent on each other (logBF in the set of the cut-offs point 25th = 8.85; 50th = 16.618; 75th = 16.305).
Figure 2.
The proportion of avian species living in completely saline, mainly saline, mainly freshwater, and completely freshwater habitats with and without supraorbital salt glands
Figure 3.
The allometric effect of body weight on kidney weights
Linear regression between the log(body weight) and log(kidney size) is log(kidney size) = 1.164994 ± 0.072970 + (0.928436 ± 0.019287) log(body weight) (p < 2.2e-16; adjusted r2 = 0.933).
Supraorbital salt glands and larger kidneys drove the evolution of salinity adaptation
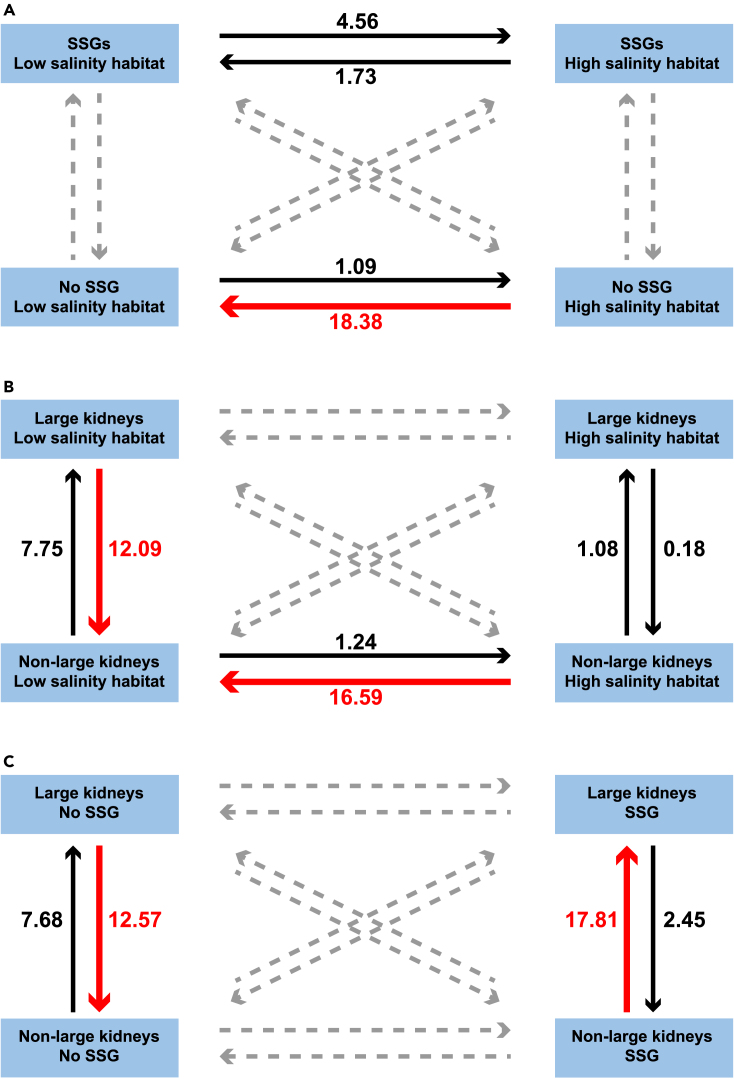
Transition rate analysis indicates that the presence of SSGs drove birds to shift from low-to high-salinity habitats, while the lack of SSGs drove the evolution of habitat use in the other direction. For species with SSGs, the transition rate from low to high-salinity habitats was 4.56, while the reverse was 1.73; in contrast, for species without SSGs, the transition rate was biased toward low-salinity habitats (18.38 vs. 1.09 for low-to high-salinity habitats transition; Figure 4A). Similarly, kidney size probably drove the evolution of birds’ habitat use; for species with large kidneys, the transition rate from high-to low-salinity habitats was about 13.4 times higher than that from low-to high-salinity habitats (16.59 vs. 1.24 for low-to high-salinity habitats transition, Figure 4B).
Figure 4.
Evolutionary transition rates of salinity adaptation traits in avian species
Evolutionary transition rates between the use of high salinity habitats and (A) the presence of SSGs, (B) larger kidneys, and (C) between the larger kidneys and the presence of SSGs. For each characteristic, the transition rates between the three different combinations of characteristic categories were estimated by the dependent model in BayesTraits13,14 with the assumption that the two characteristics evolved interdependently. The dashed arrows are the estimated minimum transition rates (≒0). The thin black arrow indicates the estimated lower transition rates (<10). The thick red arrows indicate the estimated higher transition rates (>10).
Results of the transition rate analysis also support correlated evolution between the presence of SSGs and kidney sizes. For species without SSGs, the transition rate from large to non-large kidneys was 12.57, and the reverse was 7.68, whereas for species with SSGs, the transition rate from non-large to large kidneys was 17.81 and the reverse was 2.45 (Figure 4C).
Salinity is a force to fine-tune the evolution of avian kidney size
Our results suggest that different levels of environmental salinity might have modulated the evolution of kidney size (Figure 4B). For species living in low-salinity habitats, the transition rate from large to non-large kidneys was 12.09, and the reverse was 7.75, whereas, for species in high-salinity habitats, the transition rate from non-large to large kidneys (1.08) was about five times higher than that from large to non-large kidneys (0.18).
Discussion
Our ancestral state reconstruction suggests that high-salinity habitats evolved independently in different lineages of birds (Figure 1), as it did with sea snakes.29 Furthermore, we found that this ecological trait was quite evolutionarily inert; among the 230 species we sampled, only five species whose ancestors had used saline habitats reverted to using freshwater habitats. Similarly, we found that the two physiological traits related to salt tolerance, namely the presence of SSGs and large kidneys, also evolved convergently in birds. Of these, the presence of SSGs was also highly evolutionarily conserved (Figure 1). Therefore, our results suggest that the evolutionary potential of organisms might be constrained by the cost of niche shift.30,31 The “cost" of niche shift may arise because of antagonistic pleiotropy, in which a mutation in a single gene controlling multiple traits increases fitness in one trait while also reducing fitness in another one.33 Alternatively, such “cost" may be rendered by a reduction in evolvability, the capacity to generate variation useful for adaptive change, after shifting to a new ecological niche.34,35
Shifting to high salinity habitats might have allowed these avian lineages to escape from the intensive competition with other species using freshwater habitats and paved the way for further adaptive radiations.36 It allowed avian lineages (such as Charadriiformes37) to radiate by filling various empty niches in such habitats. However, ecological niche shift may constrain changes in other directions and thus limit niche diversity and evolvability38,39 later. Therefore, birds using high-salinity habitats might become an "evolutionary dead-end"40,41 after the empty salty niche is filled. The speciation rate for these birds could be reduced or even followed by an increasing extinction rate.
Convergent evolution (the independent evolution of similar phenotypes8) to use high-salinity habitats also implies that different lineages of birds might have adopted different genetic routes to develop complex traits when adapting to high-salinity habitats.42,43 For instance, the supraorbital salt glands of birds have been considered to be derived from the nasal gland, which can also be found in reptiles and most mammals.8 The convergent evolution of salt glands in the tetrapod has been suggested to be the result of the cooption of an existing nasal gland in the ancestor of tetrapods.8 The convergent evolution of SSGs we found in this study implies that the development of this complex phenotype could have been modulated by the same set of developmental toolkits for the same existing gland in different lineages of birds independently. Therefore, the evolution of salt tolerance could have been driven by mutations in different compartments of the same genetic pathways, or genetic toolkit, that regulates the developmental process of SSGs, kidney size, and other physiological traits related to salt tolerance or osmostasis. Convergent phenotypic evolution with divergent genetic causes has been shown in the cases of four Passerellidae sparrows,45 the Savannah sparrow (P. sandwichensis beldingi), Nelson’s sparrow (Ammospiza nelsoni subvirgatus), the song sparrow (Melospiza melodia pusillula), and the swamp sparrow (M. georgiana nigrescens); fewer than 50% of genes that were considered to contribute to adaptation to salty coastal habitats were shared among them. Hence the convergent evolution of salt-tolerance phenotypes suggests that birds should provide a rich opportunity to investigate the diverse genetic mechanisms underpinning their salt-tolerant phenotype.
In 1970, Hughes1 found that species in avian orders composed mainly of non-saline species have relatively smaller kidneys whereas species associated with saline habitats have larger kidneys. However, he neither tested for the positive associations between kidney size and environmental salinity nor controlled for the effect of phylogenetic constraints on the evolution of kidney size and habitat use. Therefore, our results provide the first modern evolutionary test to demonstrate that avian species living in high-salinity environments have larger kidneys. Such an evolutionary trend can be illustrated by avian orders whose species mainly use freshwater habitats. For example, species specialized to high-salt habitats, such as the black-faced spoonbill of Pelecaniformes, the American coot (Fulica Americana) of Gruiformes, and the Savannah sparrow of Passeriformes, all have the largest kidneys recorded in their order (Table 1). The importance of kidney size in osmoregulation is also supported by the result that lineages without large kidneys tend to shift their habitats from high-to low-salinity habitats (Figure 4B).
Table 1.
List of avian species with information on the sample size, presence/absence (+/−) of supraorbital salt glands (SSGs), kidney weight in grams, body weight in grams, the residual of kidney weight/body weight (K/W), and habitat type (completely freshwater, mainly freshwater, mainly saline, completely saline)
| Species | Sample size | SSG | Kidney (g) | Body (g) | K/W residuals | Habitat type |
|---|---|---|---|---|---|---|
| Accipitriformes | ||||||
| Accipiter gentilis10 | 1 | + | NA | NA | NA | Complete freshwater |
| Accipiter striatus10 | 1 | + | NA | NA | NA | Complete freshwater |
| Aquila rapax2 | 3 | + | 13.9 | 2532 | −0.187 | Completely freshwater |
| Aquila verreauxii10 | 1 | + | NA | NA | NA | Complete freshwater |
| Buteo rufofuscus10 | 1 | + | NA | NA | NA | Mainly freshwater |
| Buteo jamaicensis15 | 1 | – | 7.6 | 1225 | −0.155 | Completely freshwater |
| Gyps africanus2 | 1 | – | 35.8 | 5270 | −0.073 | Completely freshwater |
| Haliaeetus vocifer2 | 1 | – | 18.4 | 3500 | −0.197 | Completely freshwater |
| Gyps coprotheres10 | 1 | + | NA | NA | NA | Complete freshwater |
| Polemaetus bellicosus10 | 1 | + | NA | NA | NA | Complete freshwater |
| Terathopius ecaudatus10 | 1 | + | NA | NA | NA | Complete freshwater |
| Torgos tracheliotos10 | 1 | + | NA | NA | NA | Complete freshwater |
| Anseriformes | ||||||
| Anas platyrhynchos1 | 1 | + | 6.5 | 1305 | −0.249 | Mainly saline |
| Anas acuta15 | 1 | + | 5.1 | 862 | −0.186 | Mainly freshwater |
| Anas rubripes54 | 1 | + | NA | NA | NA | Mainly freshwater |
| Anas crecca55 | 1 | + | NA | NA | NA | Mainly saline |
| Anas clypeata55 | 1 | + | NA | NA | NA | Mainly saline |
| Aythya valisineria55 | 1 | + | NA | NA | NA | Mainly saline |
| Anser anser57 | 1 | + | NA | NA | NA | Mainly saline |
| Cygnus atratus58 | 1 | + | NA | NA | NA | Mainly saline |
| Aythya americana15 | 2 | + | 8.4 | 1055 | −0.051 | Mainly freshwater |
| Aythya marila2 | 1 | + | 9.1 | 787 | 0.102 | Mainly freshwater |
| Aythya affinis2 | 1 | + | 18.1 | 1041 | 0.288 | Mainly saline |
| Oxyura jamaicensis1 | 1 | + | 5.4 | 411 | 0.139 | Mainly saline |
| Mergus serrator2 | 1 | + | 9.7 | 700 | 0.178 | Mainly saline |
| Somateria mollissima59 | 1 | + | NA | NA | NA | Completely saline |
| Caprimulgiformes | ||||||
| Chordeiles minor15 | 3 | – | 0.6 | 80 | −0.152 | Completely freshwater |
| Amazilia tzacatl2 | 1 | – | 0.04 | 5 | −0.204 | Completely freshwater |
| Selasphorus platycercus15 | 2 | – | 0.03 | 3 | −0.129 | Completely freshwater |
| Selasphorus rufus15 | 1 | – | 0.04 | 4 | −0.114 | Completely freshwater |
| Cathartiformes | ||||||
| Cathartes aura1 | 1 | – | 12.6 | 1761 | −0.083 | Completely freshwater |
| Charadriiformes | ||||||
| Charadrius alexandrinus1 | 1 | + | 0.6 | 37 | 0.161 | Completely saline |
| Charadrius vociferus15 | 3 | + | 1.3 | 88 | 0.145 | Mainly saline |
| Arenaria melanocephala1 | 1 | + | 2 | 118 | 0.214 | Completely saline |
| Bartramia longicauda15 | 1 | + | 1.6 | 141 | 0.045 | Completely freshwater |
| Actitis macularius15 | 2 | + | 0.7 | 41 | 0.186 | Mainly saline |
| Tringa melanoleuca15 | 2 | + | 2.9 | 212 | 0.137 | Mainly saline |
| Calidris melanotos15 | 2 | + | 1.1 | 85 | 0.087 | Mainly saline |
| Calidris minutilla1 | 1 | + | 0.3 | 19 | 0.13 | Mainly saline |
| Calidris ruficollis1 | 1 | + | 0.8 | 46 | 0.198 | Mainly saline |
| Limnodromus scolopaceus15 | 2 | + | 1.8 | 130 | 0.129 | Mainly freshwater |
| Calidris pusilla15 | 2 | + | 0.4 | 23 | 0.177 | Mainly saline |
| Limosa fedoa1 | 1 | + | 6.4 | 388 | 0.236 | Mainly saline |
| Larus californicus60 | 1 | + | NA | NA | NA | Mainly saline |
| Larus marinus59 | 1 | + | NA | NA | NA | Completely saline |
| Larus hyperboreus61 | 1 | + | NA | NA | NA | Completely saline |
| Larus delawarensis15 | 4 | + | 7.2 | 488 | 0.194 | Mainly saline |
| Larus heermanni1 | 1 | + | 7.8 | 544 | 0.185 | Completely saline |
| Larus glaucescens1 | 1 | + | 7.4 | 618 | 0.111 | Mainly saline |
| Larus occidentalis1 | 2 | + | 15.5 | 1283 | 0.136 | Completely saline |
| Chlidonias niger15 | 3 | + | 1 | 60 | 0.187 | Mainly saline |
| Aethia cristatella1 | 2 | + | 6 | 289 | 0.328 | Completely saline |
| Aethia pusilla1 | 2 | + | 2.3 | 96 | 0.358 | Completely saline |
| Fratercula cirrhata1 | 1 | + | 11.7 | 734 | 0.24 | Completely saline |
| Uria aalge1 | 1 | + | 17.5 | 1031 | 0.277 | Completely saline |
| Alle alle2 | 2 | + | 2.1 | 103 | 0.29 | Completely saline |
| Cepphus grylle61 | 1 | + | NA | NA | NA | Completely saline |
| Uria lomvia61 | 1 | + | NA | NA | NA | Completely saline |
| Alca torda61 | 1 | + | NA | NA | NA | Completely saline |
| Recurvirostra americana62 | 1 | + | NA | NA | NA | Mainly saline |
| Rynchops niger59 | 1 | + | NA | NA | NA | Mainly saline |
| Fratercula corniculata1 | 2 | + | 9.7 | 572 | 0.26 | Completely saline |
| Fratercula arctica61 | 1 | + | NA | NA | NA | Completely saline |
| Charadrius hiaticula61 | 1 | + | NA | NA | NA | Mainly saline |
| Pluvialis apricaria61 | 1 | + | NA | NA | NA | Mainly saline |
| Rissa tridactyla61 | 1 | + | NA | NA | NA | Completely saline |
| Actitis hypoleucos61 | 1 | + | NA | NA | NA | Mainly saline |
| Calidris alpina61 | 1 | + | NA | NA | NA | Mainly saline |
| Calidris canutus61 | 1 | + | NA | NA | NA | Completely saline |
| Calidris minuta61 | 1 | + | NA | NA | NA | Mainly saline |
| Calidris alba61 | 1 | + | NA | NA | NA | Mainly saline |
| Gallinago gallinago61 | 1 | + | NA | NA | NA | Mainly saline |
| Limosa lapponica61 | 1 | + | NA | NA | NA | Mainly saline |
| Tringa glareola61 | 1 | + | NA | NA | NA | Completely freshwater |
| Tringa ochropus61 | 1 | + | NA | NA | NA | Completely freshwater |
| Ciconiiformes | ||||||
| Ciconia ciconia2 | 3 | – | 23 | 3350 | −0.082 | Completely freshwater |
| Columbiformes | ||||||
| Columba livia15 | 1 | – | 2.8 | 286 | 0.001 | Completely freshwater |
| Zenaida asiatica15 | 3 | – | 0.9 | 141 | −0.205 | Mainly freshwater |
| Zenaida macroura1 | 1 | – | 0.7 | 110 | −0.214 | Completely freshwater |
| Columbina inca15 | 2 | – | 0.3 | 43 | −0.201 | Completely freshwater |
| Coraciiformes | ||||||
| Megaceryle alcyon15 | 2 | – | 2.2 | 173 | 0.1 | Mainly saline |
| Cuculiformes | ||||||
| Geococcyx californianus15 | 2 | – | 1.7 | 190 | −0.05 | Completely freshwater |
| Falconiformes | ||||||
| Falco sparverius2 | 1 | – | 1.1 | 112 | −0.025 | Completely freshwater |
| Falco biarmicus10 | 1 | + | NA | NA | NA | Complete freshwater |
| Falco cherrug10 | 1 | + | NA | NA | NA | Mainly freshwater |
| Falco chicquera10 | 1 | + | NA | NA | NA | Complete freshwater |
| Falco jugger10 | 1 | – | NA | NA | NA | Complete freshwater |
| Falco peregrinus10 | 1 | + | NA | NA | NA | Mainly saline |
| Polihierax semitorquatus10 | 1 | + | NA | NA | NA | Complete freshwater |
| Galliformes | ||||||
| Bonasa umbellus15 | 2 | – | 4.9 | 577 | −0.041 | Completely freshwater |
| Lagopus lagopus15 | 5 | – | 6 | 602 | 0.03 | Completely freshwater |
| Tympanuchus phasianellus15 | 1 | – | 5.1 | 791 | −0.151 | Completely freshwater |
| Centrocercus urophasianus15 | 2 | – | 22.3 | 2013 | 0.111 | Completely freshwater |
| Callipepla squamata15 | 2 | – | 1.4 | 168 | −0.084 | Completely freshwater |
| Callipepla gambelii15 | 3 | – | 1.2 | 150 | −0.105 | Completely freshwater |
| Cyrtonyx montezumae15 | 2 | – | 1.3 | 212 | −0.211 | Completely freshwater |
| Phasianus colchicus15 | 4 | – | 8.2 | 1283 | −0.14 | Completely freshwater |
| Perdix perdix15 | 2 | – | 3.8 | 521 | −0.11 | Completely freshwater |
| Numida meleagris2 | 1 | – | 7.3 | 1620 | −0.286 | Completely freshwater |
| Coturnix coturnix63 | 18 | + | NA | NA | NA | Mainly freshwater |
| Ammoperdix heyi49 | 1 | + | NA | NA | NA | Completely freshwater |
| Gaviiformes | ||||||
| Gavia stellata2 | 3 | + | 25.2 | 1549 | 0.27 | Mainly saline |
| Gruiformes | ||||||
| Gallirallus owstoni64 | 4 | + | NA | NA | NA | Completely freshwater |
| Balearica pavonina2 | 2 | – | 26 | 4448 | −0.144 | Completely freshwater |
| Fulica americana1 | 1 | + | 9 | 699 | 0.146 | Mainly saline |
| Otidiformes | ||||||
| Ardeotis kori2 | 2 | – | 44.3 | 7770 | −0.138 | Completely freshwater |
| Pelecaniformes | ||||||
| Pelecanus erythrorhynchos15 | 2 | + | 66 | 6777 | 0.09 | Mainly saline |
| Ardea herodias15 | 1 | – | 15.8 | 1840 | −0.002 | Mainly saline |
| Nycticorax nycticorax15 | 4 | – | 4.6 | 623 | −0.099 | Mainly freshwater |
| Botaurus lentiginosus15 | 2 | – | 4.7 | 625 | −0.091 | Mainly saline |
| Platalea minorc | 4 | + | 21.52 | 1573 | 0.196 | Completely saline |
| Threskiornis aethiopicusc | 3 | + | 9.08 | 1068 | −0.022 | Mainly saline |
| Gorsachius melanolophusc | 3 | – | 2.38 | 397 | −0.203 | Completely freshwater |
| Bubulcus ibisc | 3 | – | 1.26 | 238 | −0.271 | Completely freshwater |
| Pelecanus occidentalis59 | + | NA | NA | NA | Completely saline | |
| Phoenicopteriformes | ||||||
| Phoeniconaias minor2 | 5 | + | 18 | 1541 | 0.126 | Completely saline |
| Piciformes | ||||||
| Colaptes auratus15 | 1 | – | 1 | 126 | −0.114 | Completely freshwater |
| Colaptes chrysoides15 | 2 | – | 1 | 116 | −0.08 | Completely freshwater |
| Melanerpes uropygialis15 | 3 | – | 0.7 | 68 | −0.019 | Completely freshwater |
| Melanerpes formicivorus15 | 2 | – | 0.5 | 63 | −0.134 | Completely freshwater |
| Sphyrapicus varius15 | 2 | – | 0.5 | 45 | 0.002 | Completely freshwater |
| Sphyrapicus thyroideus15 | 3 | – | 0.5 | 43 | 0.021 | Completely freshwater |
| Podicipediformes | ||||||
| Podiceps nigricollis15 | 2 | + | 3.7 | 330 | 0.064 | Mainly saline |
| Podilymbus podiceps15 | 2 | + | 5.9 | 496 | 0.101 | Mainly saline |
| Procellariiformes | ||||||
| Phoebastria immutabilis65 | 1 | + | NA | NA | NA | Completely saline |
| Phoebastria nigripes65 | 1 | + | NA | NA | NA | Completely saline |
| Phoenicopterus ruber65 | 1 | + | NA | NA | NA | Completely saline |
| Phoebastria irrorata66 | 1 | + | NA | NA | NA | Completely saline |
| Psittaciformes | ||||||
| Melopsittacus undulatus15 | 6 | – | 0.3 | 39 | −0.162 | Completely freshwater |
| Sphenisciformes | ||||||
| Eudyptula minor65 | 1 | + | NA | NA | NA | Completely saline |
| Pygoscelis adeliae65 | 1 | + | NA | NA | NA | Completely saline |
| Pygoscelis papua65 | 1 | + | NA | NA | NA | Completely saline |
| Spheniscus mendiculus44 | 1 | + | NA | NA | NA | Completely saline |
| Spheniscus humboldti65 | 1 | + | NA | NA | NA | Completely saline |
| Strigiformes | ||||||
| Bubo virginianus1 | 1 | – | 3.7 | 635 | −0.201 | Completely freshwater |
| Micrathene whitneyi15 | 2 | – | 0.4 | 37 | −0.015 | Completely freshwater |
| Struthioniformes | ||||||
| Struthio camelus2 | 1 | + | 920 | 123000 | 0.06 | Completely freshwater |
| Suliformes | ||||||
| Urile pelagicus1 | 1 | + | 16.9 | 1300 | 0.168 | Completely saline |
| Fregata minor65 | 1 | + | NA | NA | NA | Completely saline |
| Aptenodytes patagonicus65 | 1 | + | NA | NA | NA | Completely saline |
| Sula nebouxii65 | 1 | + | NA | NA | NA | Completely saline |
| Sula sula65 | 1 | + | NA | NA | NA | Completely saline |
| Passeriformes | ||||||
| Pachyramphus aglaiae15 | 1 | – | 0.295 | 29 | −0.049 | Completely freshwater |
| Tyrannus tyrannus15 | 3 | – | 0.604 | 49 | 0.05 | Completely freshwater |
| Tyrannus verticalis15 | 1 | – | 0.388 | 38.8 | −0.048 | Completely freshwater |
| Tyrannus vociferans15 | 2 | – | 0.557 | 46.4 | 0.037 | Completely freshwater |
| Pitangus sulphuratus15 | 1 | – | 0.657 | 79 | −0.107 | Completely freshwater |
| Contopus sordidulus15 | 3 | – | 0.157 | 14.3 | −0.036 | Completely freshwater |
| Contopus cooperi15 | 1 | – | 0.515 | 36.5 | 0.1 | Completely freshwater |
| Pyrocephalus rubinus15 | 2 | – | 0.211 | 13.9 | 0.104 | Completely freshwater |
| Empidonax oberholseri15 | 1 | – | 0.204 | 12.1 | 0.145 | Completely freshwater |
| Myiarchus cinerascens15 | 4 | – | 0.299 | 26.3 | −0.003 | Mainly freshwater |
| Eremophila alpestris15 | 4 | – | 0.284 | 27.5 | −0.044 | Mainly freshwater |
| Tachycineta thalassina15 | 2 | – | 0.282 | 15.7 | 0.18 | Mainly freshwater |
| Tachycineta bicolor15 | 3 | – | 0.315 | 18 | 0.173 | Mainly freshwater |
| Hirundo rustica15 | 3 | – | 0.325 | 18 | 0.187 | Completely freshwater |
| Petrochelidon pyrrhonota15 | 5 | – | 0.374 | 22.2 | 0.163 | Completely freshwater |
| Progne subis15 | 2 | – | 0.544 | 46.6 | 0.025 | Mainly freshwater |
| Cyanocitta stelleri15 | 2 | – | 1.101 | 103.7 | 0.007 | Completely freshwater |
| Aphelocoma ultramarina15 | 2 | – | 1.274 | 120.2 | 0.01 | Completely freshwater |
| Pica pica15 | 4 | – | 2.556 | 198.2 | 0.11 | Completely freshwater |
| Baeolophus wollweberi15 | 4 | – | 0.144 | 10.4 | 0.055 | Completely freshwater |
| Auriparus flaviceps15 | 4 | – | 0.107 | 7 | 0.087 | Mainly freshwater |
| Psaltriparus minimus15 | 1 | – | 0.102 | 8 | 0.012 | Mainly freshwater |
| Sitta carolinensis15 | 1 | – | 0.225 | 15.8 | 0.08 | Completely freshwater |
| Certhia familiaris15 | 2 | – | 0.145 | 8.4 | 0.145 | Mainly freshwater |
| Troglodytes aedon15 | 3 | – | 0.175 | 11 | 0.117 | Completely freshwater |
| Cistothorus palustris15 | 2 | – | 0.278 | 13 | 0.255 | Mainly saline |
| Thryomanes bewickii15 | 1 | – | 0.212 | 12.2 | 0.159 | Completely freshwater |
| Campylorhynchus brunneicapillus15 | 5 | – | 0.424 | 38.4 | −0.005 | Completely freshwater |
| Catherpes mexicanus15 | 1 | – | 0.18 | 11.2 | 0.122 | Completely freshwater |
| Salpinctes obsoletus15 | 2 | – | 0.249 | 15.5 | 0.132 | Mainly freshwater |
| Mimus polyglottos15 | 3 | – | 0.508 | 49.1 | −0.026 | Completely freshwater |
| Dumetella carolinensis15 | 1 | – | 0.452 | 36.8 | 0.04 | Completely freshwater |
| Toxostoma bendirei15 | 1 | – | 0.66 | 63.6 | −0.017 | Completely freshwater |
| Toxostoma curvirostre15 | 3 | – | 0.875 | 72.4 | 0.053 | Completely freshwater |
| Toxostoma crissale15 | 2 | – | 0.677 | 58.8 | 0.026 | Completely freshwater |
| Oreoscoptes montanus15 | 3 | – | 0.57 | 40 | 0.107 | Completely freshwater |
| Turdus migratorius15 | 3 | – | 1.172 | 86.6 | 0.107 | Completely freshwater |
| Catharus guttatus15 | 1 | – | 0.359 | 31 | 0.009 | Completely freshwater |
| Catharus ustulatus15 | 1 | – | 0.461 | 29.8 | 0.134 | Mainly freshwater |
| Catharus minimus15 | 1 | – | 0.396 | 33.2 | 0.024 | Completely freshwater |
| Catharus occidentalis15 | 1 | – | 0.252 | 31.2 | −0.147 | Completely freshwater |
| Catharus aurantiirostris15 | 1 | – | 0.405 | 33.8 | 0.027 | Completely freshwater |
| Sialia mexicana15 | 4 | – | 0.408 | 29.3 | 0.088 | Mainly freshwater |
| Polioptila caerulea15 | 2 | – | 0.078 | 5.1 | 0.078 | Completely freshwater |
| Polioptila melanura15 | 3 | – | 0.086 | 5.4 | 0.097 | Completely freshwater |
| Bombycilla cedrorum15 | 4 | – | 0.471 | 40.1 | 0.023 | Completely freshwater |
| Phainopepla nitens15 | 3 | – | 0.368 | 25.3 | 0.103 | Completely freshwater |
| Sturnus vulgaris15 | 4 | – | 1.133 | 82.2 | 0.113 | Mainly freshwater |
| Vireo solitarius15 | 1 | – | 0.169 | 16 | −0.05 | Completely freshwater |
| Setophaga ruticilla15 | 1 | – | 0.1 | 9.5 | −0.066 | Mainly freshwater |
| Myioborus pictus15 | 1 | – | 0.163 | 9.1 | 0.163 | Completely freshwater |
| Myioborus miniatus15 | 1 | – | 0.097 | 9.5 | −0.079 | Completely freshwater |
| Passer domesticus15 | 5 | – | 0.301 | 29.3 | −0.044 | Completely freshwater |
| Taeniopygia guttata15 | 2 | – | 0.09 | 11.9 | −0.203 | Completely freshwater |
| Dolichonyx oryzivorus15 | 3 | – | 0.396 | 35.2 | 0.001 | Mainly freshwater |
| Sturnella neglecta15 | 2 | 1.247 | 113.4 | 0.025 | Completely freshwater | |
| Agelaius phoeniceus15 | 4 | – | 0.668 | 67.5 | −0.036 | Mainly freshwater |
| Icterus cucullatus15 | 2 | – | 0.294 | 25.2 | 0.007 | Completely freshwater |
| Icterus bullockii15 | 3 | – | 0.41 | 33.1 | 0.041 | Completely freshwater |
| Euphagus carolinus15 | 1 | – | 0.687 | 66.2 | −0.016 | Mainly freshwater |
| Euphagus cyanocephalus15 | 3 | – | 0.863 | 70.9 | 0.055 | Mainly freshwater |
| Quiscalus quiscula15 | 3 | – | 1.037 | 94.3 | 0.019 | Completely freshwater |
| Molothrus ater15 | 2 | – | 0.398 | 49.7 | −0.137 | Completely freshwater |
| Piranga ludoviciana15 | 2 | – | 0.514 | 33.2 | 0.138 | Completely freshwater |
| Cardinalis cardinalis15 | 3 | – | 0.59 | 47 | 0.057 | Completely freshwater |
| Cardinalis sinuatus15 | 1 | – | 0.574 | 35.1 | 0.163 | Completely freshwater |
| Passerina caerulea15 | 1 | – | 0.273 | 28.1 | −0.07 | Completely freshwater |
| Coccothraustes vespertinus15 | 1 | – | 0.86 | 52.3 | 0.177 | Completely freshwater |
| Pinicola enucleator15 | 1 | – | 0.833 | 53.5 | 0.154 | Completely freshwater |
| Passerculus sandwichensis15 | 3 | – | 0.261 | 17.7 | 0.254 | Completely saline |
| Pipilo erythrophthalmus15 | 1 | – | 0.397 | 39.6 | −0.046 | Mainly freshwater |
| Pooecetes gramineus15 | 2 | – | 0.285 | 26.3 | −0.024 | Completely freshwater |
| Chondestes grammacus15 | 1 | – | 0.166 | 26.2 | −0.257 | Completely freshwater |
| Aimophila ruficeps15 | 1 | – | 0.264 | 17.9 | 0.099 | Completely freshwater |
| Amphispiza bilineata15 | 6 | – | 0.154 | 12.9 | −0.003 | Completely freshwater |
| Junco hyemalis15 | 2 | – | 0.216 | 21.7 | −0.067 | Completely freshwater |
| Spizella passerina15 | 1 | – | 0.125 | 13.3 | −0.106 | Completely freshwater |
| Spizella pallida15 | 1 | – | 0.17 | 13 | 0.037 | Completely freshwater |
| Spizella breweri15 | 4 | – | 0.178 | 10.6 | 0.14 | Completely freshwater |
| Zonotrichia albicollis15 | 3 | – | 0.279 | 26.5 | −0.036 | Completely freshwater |
| Melospiza lincolnii15 | 5 | – | 0.228 | 18.1 | 0.03 | Mainly freshwater |
| Melospiza melodia15 | 2 | – | 0.247 | 21.1 | 0.003 | Completely freshwater |
| Calcarius lapponicus15 | 2 | – | 0.261 | 26.7 | −0.068 | Mainly freshwater |
| Plectrophenax nivalis15 | 1 | – | 0.337 | 31.1 | −0.019 | Mainly saline |
Data sources are shown in superscript after the species name; a superscript C indicates data collected in the current study.
In addition to kidney size, our results suggest that the presence of SSGs in species also correlates with their habitats' salinity. We found that species with SSGs tend to become high-salinity specialists; in contrast, species without SSGs shift from high-salinity habitats to low-salinity habitats (Figure 4). Therefore, the transition rate between the SSG and environmental salinity (from low-to high-salinity habitats was 16.47, while the reverse was 3.35). This not only supports the important role of the SSG in osmoregulation in high-salinity environments13,46 but also suggests that the presence of SSGs could be a physiological prerequisite for birds in some lineages to use the harsh high-salinity environment.
Hughes proposed that SSGs should be associated with increased renal mass.1 According to our PGLS results, the presence of SSGs is positively correlated with kidney size; our evolutionary dependency analysis results also suggest that SSGs tend to be highly correlated with the evolution of kidney sizes. This is also supported by the results of transition rate analysis; kidney size tended to decrease in species without SSGs and increase in species with SSGs (Figure 4). Our results support the intimate interactions between the kidneys and the salt glands for the osmoregulation suggested by Hughes.1
The correlated evolution of two traits could be caused either by the linkage between the genes underpinning the two traits (genetic correlation) or because the two traits respond to the same selection pressure.47 Genetic correlation is the proportion of variance that two traits share due to genetic causes.48 Such correlations may arise from linkage disequilibrium (i.e., the non-random association of alleles at two or more loci) or pleiotropy, which occurs when a single gene influences two or more characteristics.48 However, if the correlated evolution between the kidney size and functional salt glands had been caused by genetic linkage, we would have found a perfect (or nearly perfect) correlation between these two traits. Although the result of the correlated evolutionary analysis was highly significant (logBF >8.0) for the kidney size and presence of SSGs, the r2 between relative kidney size and the functional salt gland was only 0.19. Therefore, our results suggest that the association between the large kidneys and the presence of SSGs was probably driven by the selection force of environmental salinity influencing both of them independently. This result implies that birds may need large kidneys and salt glands working synergistically to regulate the osmotic pressure in high-salinity habitats effectively. Or if multiple genes control each of the two traits, and some of the associated genes are linked, the correlations between the traits could be imperfectly correlated. Or perhaps the evolution of the traits was genetically correlated, and initially led to a perfect or near-perfect correlation between them which has to some extent broken down more recently., but has not they could still drift apart recently.
We also found that the correlation between kidney size and habitat type used (r2 = 0.25) is much higher than that between the presence of supraorbital salt glands and habitat type used (r2 = 0.02). The lower correlation between the presence of SSGs and habitat use might be because birds that can produce hypertonic salt secretions from SSGs are not restricted to species living in high-osmotic environments. For instance, the salt glands of raptor species (e.g., the tawny eagle), were found to produce secretions in response to a high protein diet.10 The common ostrich also produces salt secretions from the salt gland when facing heat stress.49,50
Our analysis suggests that larger kidneys and the presence of SSGs are highly associated with the level of habitat salinity in birds. Almost all birds adapted to high-salinity habitats have evolved at least one characteristic that is assumed to be related to osmostasis. There are two lines of evidence to support the conclusion that salty environments drive the evolution of SSG and large kidneys. (1) Transition rate analysis indicates that species living in high salinity habitats tend to develop larger kidneys and SSGs; in contrast, species living in low-salinity habitats tend to develop smaller kidneys and to lose their SSGs (Figures 4B and 4C). (2) The results of our investigation of morphological convergence reveal that birds’ kidney size and SSGs tend to undergo convergent evolution when they occupy saline water environments. Therefore, the salty environment might not only play a role in facilitating the emergence of physiological traits that participate in osmoregulation but could also further fine-tune the evolutionary dynamics of these physiological traits.
Our results suggest that environmental salinity could be a powerful driving force for large kidneys' convergent and correlated evolution and the presence of SSGs in birds. The strong association between the presence of SSGs and large kidneys suggests that birds might need both to handle the strong selection pressure posed by high-salinity environments. Because it is highly energetically demanding to maintain osmotic balance through SSGs and large kidneys in high-salinity environments,51,52 evolution to utilize salty habitats may be constrained in birds. Such physiological constraints might explain why less than 3% of avian species live entirely or partially in such high-osmolarity environments.53 Our results provide tantalizing evidence of correlated evolution between different physiological traits to handle high osmotic pressure in high-salinity environments and suggest that such osmotic pressure has independently contributed to the expansion of avian physiological and functional diversity.
Limitations of the study
Since kidney weight measurements require fresh samples, they are not easy to obtain. Only about 56% of the orders of birds were included in this study. However, such limitations probably only affect the estimation of the numbers of species having adapted to high-salinity habitats, changed kidney size, or developed or lost SSGs in the evolutionary process, rather than the trend of the results from the phylogenetic analysis, the ancestral state reconstruction, and the correlated evolution analysis.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Biological samples | ||
| Pelicaniforme samples | Taiwan Biodiversity Research Institute | SPBorgan001 SPBorgan002 SPBorgan003 SPBorgan004 ASibis001 ASibis002 ASibis003 BcNH001 BcNH002 MNH001 MNH002 MNH003 Bubibis001 Bubibis002 Bubibis003 GE001 GE002 |
| Deposited data | ||
| Analyzed data | This paper | https://doi.org/10.17632/bkvjx52nsr.1 |
| Software and algorithms | ||
| R software | Team, R.C.67 | https://www.r-project.org/ |
| phytools | Revell19 | https://github.com/liamrevell/phytools |
| caper | Orme et al.23 | https://github.com/cran/caper/blob/master/R/brunch.R |
| phylolm | Ho et al.24 | https://github.com/lamho86/phylolm |
| RRphylo | Castiglione et al.32 | https://github.com/cran/RRphylo |
| BayesTraits 3.0 | Pagel and Meade26 | http://www.evolution.reading.ac.uk/BayesTraitsV3/BayesTraitsV3.html |
Resource availability
Lead contact
Further information and requests for resources should be directed to and will be fulfilled by the lead contact, Shou-Hsien Li (e-mail: t43028@ntnu.edu.tw).
Materials availability
This study did not generate new unique reagents.
Data and code availability
-
•
Data: All the raw data enrolled in this study have been deposited to the Digital Commons Data and are publicly available as of the date of publication. The accession number is listed in the key resources table.
-
•
Code: This paper does not report original code.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
Method details
Collection of characteristics
Data on kidney and body weight were collected for 161 avian species and on SSGs for 224 species, both featuring known phylogenetic information from the literature.1,2,10,15,44,54,55,56,57,58,59,60,61,62,63,64,65,66 In 2023, we conducted searches using Google Scholar, utilizing various combinations of search terms such as 'salt gland', 'kidney size', and 'osmotic regulation' to obtain the most pertinent data. Because information on kidney weight was only published for two species of Pelicaniformes, we measured kidney and body weights from six additional Pelicaniformes species, namely the great egret (Ardea alba; n= 2), cattle egret (Bubulcus ibis; n= 3), black-crowned night heron (Nycticorax nycticorax; n= 2), Malayan night heron (Gorsachius melanolophus; n= 3), black-faced spoonbill (Platalea minor; n= 4), and African sacred ibis (Threskiornis aethiopicus; n= 3) which live in habitats with various levels of salinity (Table 1). All Pelicaniforme samples measured in this study were from carcasses of roadkill or of rescued individuals archived in the Taiwan Biodiversity Research Institute (TBRI). Before measurement, carcasses were stored in sealed plastic bags to prevent desiccation, and frozen at -20°C. After thawing, we followed the method of Maryanne (1970) to weigh the birds to the nearest 0.1 g; then, we took out the kidneys, dried them with absorbent paper, and weighed them to the nearest 0.001 g. Our subsequent analyses related to kidney size included 167 species and those relating to SSGs covered 230 species representing 25 avian orders.
We categorized the habitat type used by these birds into different levels of salinity: (1) completely freshwater (completely living in freshwater habitats, such as forests, grasslands, streams, lakes, and reservoirs.), (2) mainly freshwater (almost exclusively living in freshwater habitats; only a few records appear in saline habitats such as saltmarshes, saltpans, mangroves, seacoasts, beaches, and marine environments), (3) mainly saline (including the ocean and habitats associated with high salinity; sometimes appearing in freshwater habitats or just for parts of the year), and (4) completely saline habitats (exclusively living in habitats with high salinity) by assessing the habitat information on these species described in Birds of the World.16 The completely saline habitat has the highest level of environmental salinity, followed by mainly saline, mainly freshwater, and then completely freshwater.
Standardization of kidney sizes
Since the data obtained from the literature mostly only provides the average values of multiple individuals for each species, this study uses the average values of each species for subsequent analysis. Because both the body and kidney sizes of the avian species included in this study differ by up to five orders of magnitude (3 ∼ 123,000 g for body weight; 0.03 ∼ 920 g for kidney weight), we log-transformed body and kidney weights to better meet the statistical assumptions of a regression test. We used the residual from linear regressions (Log(kidney size) = 1.165 ± 0.073 + (0.928 ± 0.0193) log(body weight) (p < 2.2e-16; adjusted r2: 0.933; Figure 3).) of kidney weight on body weight as a proxy for relative kidney sizes to control the allometric effect.
Ancestral state reconstruction
To infer the macroevolutionary history of the ecological and physiological traits associated with salt tolerance, we extracted 10,000 avian phylogenetic trees of all 230 species based on the complete Bayesian maximum clade credibility (MCC) species-level avian phylogeny from http://birdtree.org 17 with the Hackett constraint.18 Using the R package, phytools,19 we constructed a consensus tree based on the majority rule of the 10,000 trees we extracted. We estimated the branch length with a least square method. In the consensus tree of the 167 avian species the values were: Minimum: 0.188, 1st Quartile: 3.565, Median: 7.831, 3rd Quartile: 17.514, Maximum: 108.376. And in the consensus tree of the 230 avian species the values were: Minimum: 0.12, 1st Quartile: 3.252, Median: 7.337, 3rd Quartile: 15.979, Maximum: 108.376. Then, we used a Bayesian inference method (stochastic characteristic mapping with the "make.simmap" function) implemented in the phytools package for R26 to reconstruct the ancestral state of the kidney size of the 167 avian species and SSG and habitat type of the 230 avian species. To compare the continuous data of residual kidney weight with the discrete data of SSGs and habitat type, we recoded the residual kidney weight data as a binary variable using the fourth quartile (the 75th percentile) of residual kidney weights as a reference. Species with residual kidney weights equal to or greater than 0.13 (the 75th percentile) were considered to have ‘large kidneys’, while those with residual kidney weights less than 0.13 were considered to have ‘non-large’ kidneys. The parameters were set with 1,000 MCMC generations and the ARD (“All Rates Different”) model, which assumes that rates of trait evolution were not the same among all branch. The transition rates between different characteristic states were calculated by simulations, and posterior probabilities were mapped to the phylogeny using the "densityMap" function in phytools.19 The simulation involves two steps: first, simulating ancestral states at each internal node by sampling the posterior distribution of states; second, generating a substitution (mutational) history by sampling the posterior distribution conditioned on the reconstructions and observed states at the tips of the topology. Posterior probability values ≥ 50% indicated a characteristic state being ancestral to a clade.
Phylogenetic signals in kidney sizes, possession of SSGs, and habitat salinity
After using the Shapiro-Wilk test to check the normality of the data, we used the function ‘phylosig’ in the package ‘phytools19’ to calculate the Pagel’s λ20 and Blomberg et al.’s K21 for the phylogenetic signal in the variation of relative kidney sizes, the presence of SSGs, and the category of habitat. Assuming a Brownian motion model of trait evolution, a λ of 0 indicates that trait correlation between species is independent of their shared evolutionary history; by contrast, a λ of 1 suggests that trait correlation between species is constrained by their shared evolutionary history.20 Also, assuming a Brownian motion model of trait evolution, K values greater than 1 indicate a higher trait variance among clades than expected, whereas K values smaller than 1 imply a lower trait variance within clades than expected.22 Therefore, low K values suggest that their shared evolutionary history constrains the trait variance. We used phylogenetic generalized least-squares (PGLS) in the R package ‘caper’23 to control the phylogenetic effect and infer the association between variations in relative kidney size and habitat types with different levels of salinity and between variations in relative kidney size and the presence of SSGs. We used the phylogenetic generalized linear model (PGLM) in R package ‘phylolm’24 to infer the linear association between the possession of SSGs and habitat salinity.
Evolutionary transitions in kidney size, SSGs and levels of habitat salinity
We used the Discrete module of BayesTraits 3.025,26 to estimate the transition rates of correlated evolution between habitat salinity, relative kidney sizes, and the possession of SSGs on a phylogeny. We took the dependent model approach, which assumes that the rate of change in one trait depends on the state of the other trait. We estimated the log Bayes factor (logBF) for the dependent model (allowing correlation between variables) against the independent model (null model, which fixes all correlations to be zero) as twice the difference between the estimated log marginal likelihoods using the formula logBF = 2∗(log marginal likelihood dependent model – log marginal likelihood, independent model). Then, we interpreted comparisons where logBF > 2 as having weak support, logBF > 5 as having moderate support, and logBF > 10 as having strong support to reject the null model.27
Because the transition rate of correlated evolutionary analyses can only be calculated from binary data, residual kidney weight was transformed into a binary variable by reference to the fourth quartile of residual kidney weights; species with residual kidney weights equal to or larger than 0.13 (the 75th percentile) were considered to have ‘large kidneys,’ and those with residual kidney weights less than 0.13 were supposed to have ‘non-large kidneys.’ To test the sensibility of this cutoff value for our analysis, we repeated these analyses setting the cut-off points to the 25th and 50th percentiles of residual kidney weight, as suggested by Fristoe et al.28 We similarly re-grouped the habitat categories into two by combining the entirely freshwater habitat and the mainly freshwater habitat to be 0, and the mainly saline habitat and the completely saline habitat to be 1. We ran an MCMC chain with 5.05 million iterations and a burn-in of 50,000 iterations and sampled every 1000 iterations. We scaled the branch length of the phylogenetic trees by 0.001 and used an exponential prior with a mean of 10 for all parameters.
Searching for morphological convergence among species in diverse saline habitat types
We used the RRphylo32 method to perform phylogenetic ridge regression on trees and data, yielding branch-wise evolutionary rates and ancestral character estimates (ACEs) at each node. This process is applied independently to each phenotype component for multivariate data, using a normalization factor to prevent extreme rate values and multicollinearity while assuming minimized rate variation within clades. We also employed search.conv,32 a fast and effective method for identifying phenotypic convergence among clades or species groups within specific categories. With search.conv, the phenotypic distance between species is quantified as the angle between their phenotypic vectors (i.e., multivariate phenotypes for each species). Under a Brownian motion model of evolution, this angle should increase proportionally to the patristic distance, the sum of the lengths of the branches that link two nodes in a tree, between species. However, when morphological convergence is present, the angle (per unit time) becomes smaller than the expected.
When comparing clades, the function calculates the mean angle over all possible combinations of species pairs, taking one species per clade, and divides this value by the patristic distance between the nodes subtending the clades (i.e., the phylogenetic distance between the most recent common ancestors, MRCAs, of the clades). This value is contrasted with a random distribution of 1,000 angle-by-time values to assess significance. When comparing species, the function randomly samples two species within the tree, computes the angle between them, and divides it by the patristic distance between their immediate ancestors.
Given two clades presumed to evolve under convergence, search.conv derives the ACEs for the MRCAs for both clades from the RRphylo results and calculates the angle between them. This angle is added to the mean angle between species and divided by the patristic distance between the MRCAs. If the "summed" angle is smaller than expected by chance, it indicates that the clades converged since their origin and subsequently followed parallel phenotypic evolutionary trajectories. The significance level is assessed as described above by randomizing phenotypes across the tree tips. Finally, if no specific hypothesis about converging clades is available, the function automatically scans the phylogeny to identify instances of convergence.
Under the "state case", search.conv computes the mean angle over all possible combinations of species pairs using one species per state. Each angle is divided by the patristic distance between the species. Significance is assessed by contrasting this value with a family of 1,000 random angles obtained by shuffling the state across the species.
In this study, we employed search.conv for both "clade" and "state" scenarios, by categorizing species based on their usage of saline and non-saline habitats. We further conducted the analyses by functional variable values (kidney sizes and presence of SSGs) for each species, utilizing these vectors as input for search.conv.
Quantification and statistical analysis
Statistical Analysis Methods and Software: Used R v 4.2.3 language and related packages (phytools, caper, phylolm, RRphylo) and BayesTraits V3.0, to perform various statistical analyses, including ancestral state reconstruction, correlated evolutionary transition rate estimation, phylogenetic signal testing, phylogenetically adjusted least squares regression, phylogenetically adjusted generalized linear models, phylogenetic ridge regression, and detection of morphological convergence.
Location of Statistical Details: All statistical details of this research, such as the types of statistical tests, the exact value and meaning of sample size n, the definition and measurement of central tendency and dispersion, the setting of significance level, etc., were found in Table 1 and Method details.
Premises and Strategies of Statistical Analysis: Before performing statistical analysis, we tested the normality of the data and log-transformed or binarized the data to meet the assumptions of the statistical model. This study obtained all species with available characteristic values and phylogenetic information, stratified them based on their habitat types and physiological characteristics, and did not exclude any data or species during the analysis process.
Acknowledgments
We greatly thank two anonymous reviewers who provide numerous insightful comments to improve the quality of this article. We thank the Taiwan Biodiversity Research Institute for providing the specimens they archived. We are grateful to Yao-Hua Chen, Jun-Ling Yao, and Yi-Hua Guo who assisted with sample preparation and anatomy. We are in debt to Chih-Ming Hung who had provided many insightful comments to the early draft of this article. We thank Alan Watson for greatly improving the readability of this article. We thank the Ministry of Science and Technology of Taiwan (MOST 111-2621-B-003 -002 -MY3) for their financial support.
Author contributions
Shou-Hsien Li, Chi-Cheng Chiu, and Ben-Yang Liao designed the research; Chi-Cheng Chiu, and Cheng-Te Yao performed the research; Chi-Cheng Chiu analyzed data; and Shou-Hsien Li, Chi-Cheng Chiu, Ben-Yang Liao, and Cheng-Te Yao wrote the article. All authors approved the final submission.
Declaration of interests
None of the authors of this article have any affiliations with or involvement in any organization or entity with any financial or non-financial interest (such as patent or stock ownership, membership of a company board of directors, membership of an advisory board or committee for a company, and consultancy for or receipt of speaker’s fees from a company) in the subject matter or materials discussed in this article.
Published: February 9, 2024
Contributor Information
Ben-Yang Liao, Email: liaoby@nhri.edu.tw.
Shou-Hsien Li, Email: t43028@ntnu.edu.tw.
References
- 1.Hughes M.R. Relative Kidney Size in Nonpasserine Birds with Functional Salt Glands. Condor. 1970;72:164–168. doi: 10.2307/1366626. [DOI] [Google Scholar]
- 2.Quiring D.P. McGraw-Hill; 1950. Functional Anatomy of the Vertebrates. [Google Scholar]
- 3.Baumel J.J. Publications of the Nuttall Ornithological Club; 1993. Handbook of Avian Anatomy: Nomina Anatomica Avium. [Google Scholar]
- 4.Schmidtnielsen K., Jorgensen C.B., Osaki H. Extrarenal salt excretion in birds. Am. J. Physiol. 1958;193:101–107. doi: 10.1152/ajplegacy.1958.193.1.101. [DOI] [PubMed] [Google Scholar]
- 5.Hughes M.R. Regulation of salt gland, gut and kidney interactions. Comp. Biochem. Physiol. Mol. Integr. Physiol. 2003;136:507–524. doi: 10.1016/j.cbpb.2003.09.005. [DOI] [PubMed] [Google Scholar]
- 6.Gutiérrez J.S., Dietz M.W., Masero J.A., Gill Jr R.E., Dekinga A., Battley P.F., Sánchez-Guzmán J.M., Piersma T. Functional ecology of saltglands in shorebirds: flexible responses to variable environmental conditions. Funct. Ecol. 2012;26:236–244. doi: 10.1111/j.1365-2435.2011.01929.x. [DOI] [Google Scholar]
- 7.Vidal N., Hedges S.B. The molecular evolutionary tree of lizards, snakes, and amphisbaenians. C. R. Biol. 2009;332:129–139. doi: 10.1016/j.crvi.2008.07.010. [DOI] [PubMed] [Google Scholar]
- 8.Babonis L.S., Brischoux F. Perspectives on the Convergent Evolution of Tetrapod Salt Glands. Integr. Comp. Biol. 2012;52:245–256. doi: 10.1093/icb/ics073. [DOI] [PubMed] [Google Scholar]
- 9.Carpenter R.E., Stafford M.A. The secretory rates and the chemical stimulus for secretion of the nasal salt glands in the Rallidae. Condor. 1970;72:316–324. doi: 10.2307/1366010. [DOI] [Google Scholar]
- 10.Cade T.J., Greenwald L. Nasal Salt Secretion in Falconiform Birds. Condor. 1966;68:338–350. doi: 10.2307/1365449. [DOI] [Google Scholar]
- 11.Schmidt-Nielsen K., Kim Y.T. The effect of salt intake on the size and function of the salt gland of ducks. Auk. 1964;81:160–172. doi: 10.2307/4082766. [DOI] [Google Scholar]
- 12.Shuttleworth T.J., Hildebrandt J.P. Vertebrate salt glands: Short-and long-term regulation of function. J. Exp. Zool. 1999;283:689–701. doi: 10.1002/(sici)1097-010x(19990601)283:7<689::aid-jez7>3.0.co;2-t. [DOI] [PubMed] [Google Scholar]
- 13.Peaker M., Linzell J.L. Cambridge University Press; 1975. Salt Glands in Birds and Reptiles. [PubMed] [Google Scholar]
- 14.Skadhauge E. A quantitative survey of salt and water excretion. Comp. Biochem. Physiol. A Comp. Physiol. 1982;71:481–483. doi: 10.1016/0300-9629(82)90196-7. [DOI] [PubMed] [Google Scholar]
- 15.Johnson O.W. Some Morphological Features of Avian Kidneys. Auk. 1968;85:216–228. doi: 10.2307/4083582. [DOI] [Google Scholar]
- 16.Billerman S.M., Keeney B.K., Rodewald P.G., Schulenberg T.S., editors. Birds of the World. Cornell Laboratory of Ornithology; 2020. [Google Scholar]
- 17.Jetz W., Thomas G.H., Joy J.B., Redding D.W., Hartmann K., Mooers A.O. Global Distribution and Conservation of Evolutionary Distinctness in Birds. Curr. Biol. 2014;24:919–930. doi: 10.1016/j.cub.2014.03.011. [DOI] [PubMed] [Google Scholar]
- 18.Hackett S.J., Kimball R.T., Reddy S., Bowie R.C.K., Braun E.L., Braun M.J., Chojnowski J.L., Cox W.A., Han K.-L., Harshman J., et al. A Phylogenomic Study of Birds Reveals Their Evolutionary History. Science. 2008;320:1763–1768. doi: 10.1126/science.1157704. [DOI] [PubMed] [Google Scholar]
- 19.Revell L.J. phytools: an R package for phylogenetic comparative biology (and other things) Methods Ecol. Evol. 2012;3:217–223. doi: 10.1111/j.2041-210X.2011.00169.x. [DOI] [Google Scholar]
- 20.Pagel M. The Maximum Likelihood Approach to Reconstructing Ancestral Character States of Discrete Characters on Phylogenies. Syst. Biol. 1999;48:612–622. [Google Scholar]
- 21.Pagel M. Detecting correlated evolution on phylogenies: a general method for the comparative analysis of discrete characters. Proc. R. Soc. B. Biol. Sci. 1994;255:37–45. doi: 10.1098/rspb.1994.0006. [DOI] [Google Scholar]
- 22.Pagel M., Lutzoni F. In: Biological Evolution and Statistical Physics Lecture Notes in Physics. Lässig M., Valleriani A., editors. Springer; 2002. Accounting for phylogenetic uncertainty in comparative studies of evolution and adaptation; pp. 148–161. [DOI] [Google Scholar]
- 23.Orme D., Freckleton R., Thomas G., Petzoldt T., Fritz S., Isaac N., Pearse W. 2018. Caper: Comparative Analyses of Phylogenetics and Evolution in R. [Google Scholar]
- 24.Ho L.S.T., Ane C., Lachlan R., Tarpinian K., Feldman R., Yu Q., van der Bijl W. 2016. Phylolm. [Google Scholar]
- 25.Pagel M., Meade A., Barker D. Bayesian Estimation of Ancestral Character States on Phylogenies. Syst. Biol. 2004;53:673–684. doi: 10.1080/10635150490522232. [DOI] [PubMed] [Google Scholar]
- 26.Pagel M., Meade A. Bayesian Analysis of Correlated Evolution of Discrete Characters by Reversible-Jump Markov Chain Monte Carlo. Am. Nat. 2006;167:808–825. doi: 10.1086/503444. [DOI] [PubMed] [Google Scholar]
- 27.Meade A., Pagel M. 2014. BayesTraits. [Google Scholar]
- 28.Fristoe T.S., Iwaniuk A.N., Botero C.A. Big brains stabilize populations and facilitate colonization of variable habitats in birds. Nat. Ecol. Evol. 2017;1:1706–1715. doi: 10.1038/s41559-017-0316-2. [DOI] [PubMed] [Google Scholar]
- 29.Gearty W., Carrillo E., Payne J.L. Ecological Filtering and Exaptation in the Evolution of Marine Snakes. Am. Nat. 2021;198:506–521. doi: 10.1086/716015. [DOI] [PubMed] [Google Scholar]
- 30.Futuyma D.J., Moreno G. The Evolution of Ecological Specialization. Annu. Rev. Ecol. Syst. 1988;19:207–233. [Google Scholar]
- 31.Schluter D. Ecological Character Displacement in Adaptive Radiation. Am. Nat. 2000;156:S4–S16. doi: 10.1086/303412. [DOI] [Google Scholar]
- 32.Castiglione S., Tesone G., Piccolo M., Melchionna M., Mondanaro A., Serio C., Di Febbraro M., Raia P. A new method for testing evolutionary rate variation and shifts in phenotypic evolution. Methods Ecol. Evol. 2018;9:974–983. [Google Scholar]
- 33.MacArthur R.H. Princeton University Press; 1984. Geographical Ecology: Patterns in the Distribution of Species. [Google Scholar]
- 34.Colegrave N., Collins S. Experimental evolution: experimental evolution and evolvability. Heredity. 2008;100:464–470. doi: 10.1038/sj.hdy.6801095. [DOI] [PubMed] [Google Scholar]
- 35.Woods R.J., Barrick J.E., Cooper T.F., Shrestha U., Kauth M.R., Lenski R.E. Second-Order Selection for Evolvability in a Large Escherichia coli Population. Science. 2011;331:1433–1436. doi: 10.1126/science.1198914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ackerly D.D., Schwilk D.W., Webb C.O. Niche Evolution and Adaptive Radiation: Testing the Order of Trait Divergence. Ecology. 2006;87:S50–S61. doi: 10.1890/0012-9658(2006)87[50:NEAART]2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- 37.Baker A.J., Pereira S.L., Paton T.A. Phylogenetic relationships and divergence times of Charadriiformes genera: multigene evidence for the Cretaceous origin of at least 14 clades of shorebirds. Biol. Lett. 2007;3:205–209. doi: 10.1098/rsbl.2006.0606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Buckling A., Wills M.A., Colegrave N. Adaptation Limits Diversification of Experimental Bacterial Populations. Science. 2003;302:2107–2109. doi: 10.1126/science.1088848. [DOI] [PubMed] [Google Scholar]
- 39.Collar D.C., O’Meara B.C., Wainwright P.C., Near T.J. Piscivory Limits Diversification of Feeding Morphology in Centrarchid Fishes. Evolution. 2009;63:1557–1573. doi: 10.1111/j.1558-5646.2009.00626.x. [DOI] [PubMed] [Google Scholar]
- 40.Colles A., Liow L.H., Prinzing A. Are specialists at risk under environmental change? Neoecological, paleoecological and phylogenetic approaches. Ecol. Lett. 2009;12:849–863. doi: 10.1111/j.1461-0248.2009.01336.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Simpson G.G. Columbia University Press; 1944. Tempo and Mode in Evolution. [Google Scholar]
- 42.Stern D.L. The genetic causes of convergent evolution. Nat. Rev. Genet. 2013;14:751–764. doi: 10.1038/nrg3483. [DOI] [PubMed] [Google Scholar]
- 43.Arendt J., Reznick D. Convergence and parallelism reconsidered: what have we learned about the genetics of adaptation? Trends Ecol. Evol. 2008;23:26–32. doi: 10.1016/j.tree.2007.09.011. [DOI] [PubMed] [Google Scholar]
- 44.Fänge R., Schmidt-Nielsen K., Robinson M. Control of Secretion From the Avian Salt Gland. Am. J. Physiol. 1958;195:321–326. doi: 10.1152/ajplegacy.1958.195.2.321. [DOI] [PubMed] [Google Scholar]
- 45.Walsh J., Benham P.M., Deane-Coe P.E., Arcese P., Butcher B.G., Chan Y.L., Cheviron Z.A., Elphick C.S., Kovach A.I., Olsen B.J., et al. Genomics of rapid ecological divergence and parallel adaptation in four tidal marsh sparrows. Evol. Lett. 2019;3:324–338. doi: 10.1002/evl3.126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Holmes W.N., Phillips J.G. The Avian Salt Gland. Biol. Rev. 1985;60:213–256. doi: 10.1111/j.1469-185X.1985.tb00715.x. [DOI] [Google Scholar]
- 47.Price T., Langen T. Evolution of correlated characters. Trends Ecol. Evol. 1992;7:307–310. doi: 10.1016/0169-5347(92)90229-5. [DOI] [PubMed] [Google Scholar]
- 48.Falconer D.S. Pearson Education India; 1996. Introduction to Quantitative Genetics. [Google Scholar]
- 49.Schmidt-Nielson K., Borut A., Lee P., Crawford E. Nasal Salt Excretion and the Possible Function of the Cloaca in Water Conservation. Science. 1963;142:1300–1301. doi: 10.1126/science.142.3597.1300. [DOI] [PubMed] [Google Scholar]
- 50.Sheldon F.H., Whittingham L.A. In: Avian Molecular Evolution and Systematics. Mindell D.P., editor. Academic Press; 1997. Phylogeny in studies of bird ecology, behavior, and morphology. [Google Scholar]
- 51.Peña-Villalobos I., Valdés-Ferranty F., Sabat P. Osmoregulatory and metabolic costs of salt excretion in the Rufous-collared sparrow Zonotrichia capensis. Comp. Biochem. Physiol. Mol. Integr. Physiol. 2013;164:314–318. doi: 10.1016/j.cbpa.2012.10.027. [DOI] [PubMed] [Google Scholar]
- 52.Sabat P., Narváez C., Peña-Villalobos I., Contreras C., Maldonado K., Sanchez-Hernandez J.C., Newsome S.D., Nespolo R., Bozinovic F. Coping with Salt Water Habitats: Metabolic and Oxidative Responses to Salt Intake in the Rufous-Collared Sparrow. Front. Physiol. 2017;8 doi: 10.3389/fphys.2017.00654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rajpar M.N., Ozdemir I., Zakaria M., Sheryar S., Rab A. In: Intechopen. Seabirds H.M., editor. 2018. Seabirds as bioindicators of marine ecosystems; pp. 47–65. [Google Scholar]
- 54.Barnes G.G., Nudds T.D. Salt tolerance in American black ducks, mallards, and their F1-hybrids. Auk. 1991;108:89–98. doi: 10.1093/auk/108.1.89. [DOI] [Google Scholar]
- 55.Cooch F.G. A preliminary study of the survival value of a functional salt gland in prairie Anatidae. Auk. 1964;81:380–393. doi: 10.2307/4082692. [DOI] [Google Scholar]
- 56.Losos J.B. Convergence, adaptation, and constraint. Evolution. 2011;65:1827–1840. doi: 10.1111/j.1558-5646.2011.01289.x. [DOI] [PubMed] [Google Scholar]
- 57.Hanwell A., Linzell J.L., Peaker M. Salt-gland secretion and blood flow in the goose. J. Physiol. 1971;213:373–387. doi: 10.1113/jphysiol.1971.sp009387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hughes M.R. The effects of salt water adaptation on the Australian black swan, Cygnus atratus (Latham) Comp. Biochem. Physiol. A Comp. Physiol. 1976;55:271–277. doi: 10.1016/0300-9629(76)90143-2. [DOI] [PubMed] [Google Scholar]
- 59.Schmidt-Nielsen K. The salt-secreting gland of marine birds. Circulation. 1960;21:955–967. doi: 10.1161/01.CIR.21.5.955. [DOI] [PubMed] [Google Scholar]
- 60.McFARLAND L.Z. Minimal salt load required to induce secretion from the nasal salt glands of sea gulls. Nature. 1964;204:1202–1203. doi: 10.1038/2041202a0. [DOI] [PubMed] [Google Scholar]
- 61.Staaland H. Anatomical and physiological adaptations of the nasal glands in Charadriiformes birds. Comp. Biochem. Physiol. 1967;23:933–944. doi: 10.1016/0010-406X(67)90354-4. [DOI] [PubMed] [Google Scholar]
- 62.Rubega M.A., Oring L.W. Excretory organ growth and implications for salt tolerance in hatchling American avocets Recurvirostra americana. J. Avian Biol. 2004;35:13–15. doi: 10.1111/j.0908-8857.2004.03126.x. [DOI] [Google Scholar]
- 63.Dunson W.A., Dunson M.K., Ohmart R.D. Evidence for the presence of nasal salt glands in the roadrunner and the coturnix quail. J. Exp. Zool. 1976;198:209–216. doi: 10.1002/jez.1401980210. [DOI] [PubMed] [Google Scholar]
- 64.Carpenter R.E., Stafford M.A. The secretory rates and the chemical stimulus for secretion of the nasal salt glands in the Rallidae. Condor. 1970;72:316–324. doi: 10.2307/1366010. [DOI] [Google Scholar]
- 65.McFARLAND L.Z. Captive marine birds possessing a functional lateral nasal gland (salt gland) Nature. 1959;184:2030–2031. [Google Scholar]
- 66.Watson G.E., Divoky G.J. Identification of Diomedea leptorhyncha Coues 1866, an albatross with remarkably small salt glands. Condor. 1971;73:487–489. [Google Scholar]
- 67.Team, R.C. 2022. R: A language and environment for statistical computing. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
-
•
Data: All the raw data enrolled in this study have been deposited to the Digital Commons Data and are publicly available as of the date of publication. The accession number is listed in the key resources table.
-
•
Code: This paper does not report original code.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.