Abstract
Two different approaches for introducing pathogenic mutations into the β-amyloid precursor protein gene in mouse embryonic stem cells were compared. Both approaches require two sequential modifications of the targeting locus by homologous recombinations. One approach was a “targeting-in-out” procedure that is based on a double-replacement strategy, and the other was a “hit-and-run” procedure that makes use of an unstable genomic duplication after vector integration. Both approaches showed similar targeting frequencies for the first step. In the targeting-in-out procedure, targeted-in embryonic stem cell clones with the desired mutation and an intron-located selection cassette were obtained at a high frequency after the first step. Targeting out, however, resulted not only in the expected loss of the intron-located selection cassette but also in unavoidable reversion to wild type. In contrast, pure mutants, i.e., those without additional genomic changes, were generated by the hit-and-run procedure. Although targeted-in embryonic stem cells might be used to generate animals with modified β-amyloid precursor protein, the hit-and-run procedure appears to be the superior way to target gene modifications in vivo, leading to pure, correct mutants. For further improvements, optimization of the homologous recombination efficiency could be envisaged.
Few homologous recombination methods have thus far been described and applied to generate subtle modifications in the mammalian genome. Two procedures have common features, namely, modification of the targeting locus in a first step and then allowing for selection for the modification in a second step. The “targeting-in-out” procedure requiring two subsequent replacements at the locus of interest had been described for introduction of subtle mutations into the α2-Na,K-ATPase (2) and Col1a-1 (22) genes of mouse embryonic stem (ES) cells. Another procedure, termed “hit and run,” which makes use of an intermediate genomic duplication was described for subtle changes at the Hox-2.6 locus (11) and for a directly selectable insertion into the hypoxanthine phosphoribosyltransferase gene of mouse ES cells (21). Both procedures depend entirely on two homologous recombination steps. Another system, the Cre-loxP system (reviewed by Marth [14]), can also be used for subtle gene targeting, yet after introduction of loxP sequences into the desired genomic locus by homologous recombination, Cre-recombinase then induces a site-specific recombination between the loxP sequences.
In the present study, pathogenic β-amyloid precursor protein (β-APP) mutations known to segregate with a familial form of early-onset Alzheimer’s disease (AD) in Sweden (SFAD) (15) or with hereditary cerebral hemorrhage with amyloidosis in a Dutch family (HCHWA-D) (12) were introduced into mouse ES cells. These two mutations are located on exons 16 and 17 in the β-APP gene like all other pathogenic β-APP mutations known so far (for an overview, see the report of Schellenberg [17]). Gene targeting was approached in the present study by homologous recombination between the cellular β-APP gene and mutant targeting vectors by using a targeting-in-out and a hit-and-run procedure. The efficiencies and characteristics of both homologous recombination steps of both methods were evaluated and compared. The targeted mutated ES cells may be used to establish mice with a mutant β-APP gene under the control of endogenous genomic regulatory sequences without background wild-type gene expression.
MATERIALS AND METHODS
Isolation and characterization of β-APP mouse genomic clones.
All molecular biological methods not explained in detail here were performed by standard techniques (6, 16). Lambda phage clones containing exons 16 and 17 of the β-APP gene were obtained by screening a C57BL/6 mouse brain genomic library in λEMBL3a, a kind gift from Wolfgang Wille (Universität Köln, Cologne, Germany). Phage plaques were transferred in duplicate to nylon membranes (GeneScreen Plus; DuPont NEN), and denatured DNA was tested with two different probes, i.e., one on each filter. Probe 1, an oligonucleotide complementary to the 3′ end of human β-APP exon 18 (5′-GGCGGGGGTCTAGTTCTGCATCTGCTCAAAGAACTTGTAGGTTGG-3′) was end labeled with [α-32P]dATP by using terminal deoxynucleotidyl transferase and hybridized to membrane-bound DNA for 16 to 30 h at 60°C in 6× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate)–1× Denhardt’s solution with 10 μg of carrier DNA per ml. Membranes were washed with 0.5× SSC–0.1% sodium dodecyl sulfate (SDS) at 52°C prior to autoradiography. As a second probe, a 241-bp HincII rat cDNA fragment covering exons 16 and 17 was random prime labeled with [α-32P]dCTP by using the T7QuickPrime kit (Pharmacia). Hybridization for the second probe was done in 4.8× SSC–20 mM Tris-HCl (pH 7.4 at 20°C)–45% formamide–8% dextran sulfate–1× Denhardt’s solution–0.1% SDS for 16 to 30 h at 42°C. Membranes exposed to this probe were washed with 0.2× SSC–0.1% SDS at 58°C prior to autoradiography. Double-positive phage plaques were enriched and purified, and inserts were subcloned into plasmid pBluescriptII SK− (Stratagene). These clones were further analyzed by restriction mapping and double-strand sequencing by using the dideoxy chain termination method with Sequenase (U.S. Biochemicals) and the Bst sequencing kit (Bio-Rad).
Construction of targeting vectors.
Site-directed mutagenesis was performed with the Transformer kit (Clontech). Desired mutations within exons 16 and 17 of β-APP were introduced into mouse genomic subclones in pBluescriptII by using the mutator oligonucleotide Mut-1 (5′-GATCTCGGAAGTGAATCTAGACGCGGAGTTCGGACATGATTCAG-3′) for the SFAD mutation on exon 16 or Mut-2 (5′-GAAGGTGTTCTTTGCGCAGGACGTCGGATCGAACAAAGGCGC-3′) for the HCHWA-D mutation on exon 17 (underlined letters indicate nucleotide exchanges). Simultaneously, the unique NotI restriction site in the polylinker region was deleted for selection with the oligonucleotide Sel-1 (5′-CCGCGGTGGCAGCTGCTCTAGAAC-3′). Targeting vectors were constructed by using an 11-kb genomic β-APP fragment containing modified exons 16 and 17 starting 1.5 kb upstream of exon 16 and ending at the 5′ splice site of exon 18 (Fig. 1). Genes for neomycin resistance (Neo) and thymidine kinase from herpes simplex virus (TK) (3) were joined to form a selection cassette and attached to the genomic β-APP fragment. The Neo gene of this cassette, derived from transposon Tn5 (4), is under control of the mouse phosphoglycerate kinase promoter and polyadenylation signal (1). It was originally cloned by Michael McBurney (Ottawa, Ontario, Canada) and provided by Colin L. Stewart (Nutley, N.J.). The TK gene was excised from the vector plC19R/MC1-TK (constructed by Suzi Mansour, Salt Lake City, Utah) (13). For the replacement vector pBL-RV(in), the selection cassette was cloned into the HindIII site of the genomic β-APP fragment (1.0 kb downstream of exon 17) in pBluescriptII (Fig. 1A). No selection genes were put into the replacement vector pBL-RV(out) designed for a second transfection of ES cells that already contained TK and Neo after a preceding transformation (Fig. 1A). The insertion vector pGEM-IV(hit-run) was constructed by recircularization of the SalI-excised genomic β-APP fragment followed by linearization with KpnI (2.0 kb downstream of exon 17) and subcloning it into pGEM7 ZF− (Promega). The selection cassette was then inserted at the reopened SalI site. Prior to transfection, all targeting vectors were linearized and excised from pBluescriptII or pGEM7 with SalI or KpnI, respectively.
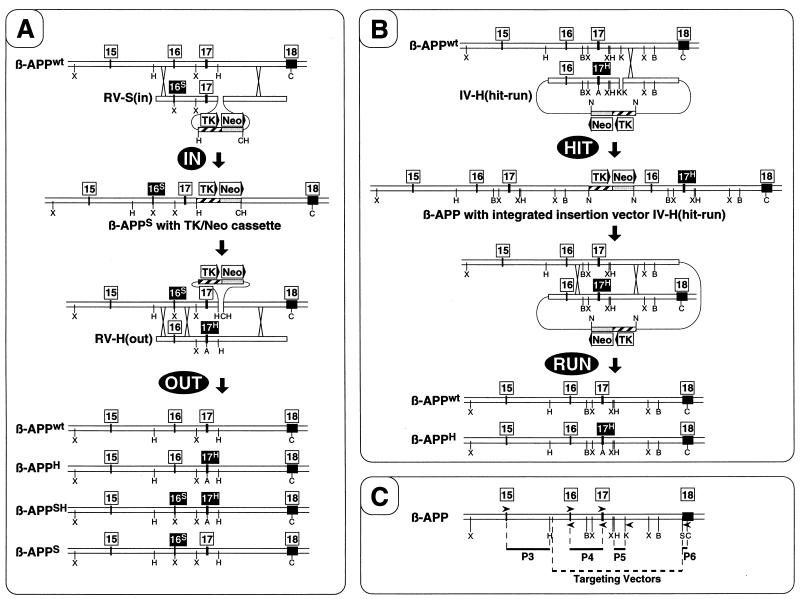
FIG. 1.
Approaches for introduction of subtle mutations into ES cells by homologous recombination. (A) The targeting-in-out procedure. Boxes with numbers indicate the positions of exons. Those exons carrying the SFAD (16S) or HCHWA-D (17H) mutation are highlighted, and mutated β-APP alleles are labeled by the same superscript letters. Boxes with text and arrowheads indicate the positions and orientations of selection genes. The locations of crossovers during homologous recombination are indicated by crosses. After the first homologous recombination (targeting IN), ES cells having replaced the targeted locus by the replacement vector RV-S(in) are enriched by selection for neomycin resistance (Neo) with G418. Isolated targeted-in ES cells are subsequently targeted OUT again by homologous recombination with RV-H(out) and selection with FIAU against the viral TK. Four possible end arrangements are shown, three of which contain mutations but lack the selection cassette. (B) The hit-and-run procedure. Selection of hit clones with a partial genomic duplication due to targeted integration of the vector IV-H(hit-run) by homologous recombination is performed as in the targeting-in step. After the run, ES cells that had undergone spontaneous intrachromosomal recombination resulting in loss of the duplication and selection cassette are counterselected with FIAU against TK. This process ends up either in reversion to wild-type (wt) or in expected targeted mutated ES cells without a selection cassette. (C) Primers and probes. Probes 3 to 6 (P3 to P6) for genomic Southern blot analysis are indicated by dark solid lines, and the genomic fragment used for construction of targeting vectors is shown as a dotted line. The binding sites and 5′-to-3′ orientation of primers used for PCR are shown by arrowheads. The positions of recognition sites for restriction endonucleases are indicated by single letters: A, AatII; B, BamHI; C, ClaI; H, HindIII; K, KpnI; N, NotI; S, SalI; X, XbaI.
Cell culture.
E14 ES cells derived from mouse strain 129/Sv with a karyotype of 40XY were provided by Horst Blüthmann (Basel, Switzerland). To prevent totipotent ES cells from differentiating, they were cultivated permanently on a feeder layer of irradiated primary mouse embryonic fibroblasts (PMEFs) transgenic for the Neo gene. PMEFs were prepared from 13-day-old ICR-M-TKneo2 embryos (19) and kept for a maximum of seven passages in high-glucose Dulbecco’s modified Eagle’s medium (DMEM) containing 10% fetal calf serum, 2 mM l-glutamine, 1× minimal essential medium nonessential amino acids (Gibco), 100 U of penicillin-streptomycin (Gibco) per ml, and 100 μM β-mercaptoethanol at 37°C under 10% CO2. Prior to serving as feeders for ES cells, PMEFs were allowed to build a nearly confluent monolayer and were inactivated by irradiation with 3,000 rads by using a cesium source. E14 cells were routinely passaged every 2 to 3 days and cultured in the same medium as that used for PMEFs alone, but supplemented with 103 U of leukemia inhibitory factor (ESGRO; Gibco) per ml and with 15% fetal calf serum.
Transfection and selection of ES cells.
For transfection, 2 × 107 ES cells were resuspended in 800 μl of ice-cold phosphate-buffered saline with 20 to 25 μg of linearized targeting vector and pulsed in cuvettes with a 0.4-cm electrode gap at 280 V and 500 μF with Genepulser electroporation equipment (Bio-Rad). After electroporation, cells were taken up in culture medium without selective drugs and seeded at a density of 2 × 106 to 4 × 106 treated cells per 6-cm-diameter culture dish on an irradiated feeder. For positive selection, i.e., for cells expressing the Neo gene, culture medium was replaced 24 h after electroporation with selection medium containing in addition 175 μg of G418 (Geneticin; Gibco) per ml. E14 cells were kept for 9 days under G418 selection conditions with fresh medium supplied every day before single ES colonies were picked by using glass capillary mouth pipettes. Counterselection for ES cells having lost the viral TK gene previously cointroduced by positive selection was performed by supplementation of medium with 0.2 μM fialuridine (FIAU; Oclassen) after a period of 1 to 5 days in nonselective medium. This negative selection was done for 7 days either directly for cells targeted with insertion vectors or when using replacement vectors after electroporation with a second replacement vector lacking the TK gene. Medium was also renewed every day, and single ES colonies were picked as described above.
Analysis of targeted ES cells by PCR.
DNA was extracted from 1 × 103 to 2 × 103 cells (ES cells cultured on PMEFs) in 10 μl of distilled water by using 100 μl of InstaGene purification matrix (Bio-Rad) as described in the manufacturer’s instructions. Lysate aliquots of 20 μl were used as a template for each 50-μl PCR mixture corresponding to 200 to 400 cell equivalents. All PCRs were performed by using the Expand Long Template PCR system with buffers 1 and 2 (Boehringer Mannheim) under modified conditions. Reactions were performed in thin-walled 500-μl tubes by using a TRIO Thermoblock Cycler (Biometra), and the enzyme mix was added after the initial DNA denaturation at 92°C for 2 min. Fifteen cycles of 92°C for 10 s, 65°C for 30 s, and 68°C for 2 to 6 min (depending on length of the expected fragment) were followed by another 25 cycles, with elongation times increasing for 20 s each round. All primers used for PCR are listed in Table 1, and binding sites are indicated in Fig. 1C. Mutation-specific fragments were selectively amplified and visualized directly by agarose gel electrophoresis of the PCR mixtures.
TABLE 1.
Oligonucleotides used as PCR primers for detection of targeted mutated ES cells
| Primer | Binding site | Orientationa | Specificityb | Sequencec |
|---|---|---|---|---|
| 15F | Exon 15 | → | 5′-TGCTGACCGAGGACTGACCACTCGACCAG-3′ | |
| 16F-wt | Exon 16 | → | wt | 5′-GAGATCTCGGAAGTGAAGATGGATGCAGAA-3′ |
| 16F-S | Exon 16 | → | SFAD | 5′-GAGATCTCGGAAGTGAATCTAGACGCGGAG-3′ |
| 16R-wt | Exon 16 | ← | wt | 5′-CCTGAATCATGTCCGAATTCTGCATCCATC-3′ |
| 16R-S | Exon 16 | ← | SFAD | 5′-CCTGAATCATGTCCGAACTCCGCGTCTAGA-3′ |
| 17F-wt | Exon 17 | → | wt | 5′-GGCGTTGTCATAGCAACCGTGATTG-3′ |
| 17F-H | Exon 17 | → | HCHWA-D | 5′-TGTTCTTTGCGCAGGACGTCGGA-3′ |
| 17R | Exon 17 | ← | 5′-ATCACCAGGGTGATGACAATCACGGTTGCT-3′ | |
| 17R-H | Exon 17 | ← | HCHWA-D | 5′-ATGGCGCCTTTGTTCGATCCGACGTCCTGC-3′ |
| 17+R | Intron 17d | ← | 5′-TGGTTCTCTCTGTGGTAGCCTTGG-3′ | |
| NeoF | Neo gene | → | 5′-ATATTGCTGAAGAGCTTGGCGGC-3′ |
Arrows indicate the 5′-to-3′ orientation of primers binding to genomic DNA with respect to the transcriptional direction of the β-APP gene.
If specificity is not indicated, primers bind unspecifically to wild-type (wt) and mutant DNA.
Underlined letters represent mutant-specific nucleotide exchanges.
The binding site of primer 17+R is located 2.0 kb upstream of exon 17.
Genomic Southern blot analysis.
After extraction of genomic DNA from cells (ES cells cultured on PMEFs) in accordance with standard procedures, samples containing 5 μg of DNA were digested for 5 to 10 h with different combinations of restriction enzymes as described in the manufacturer’s instructions (Boehringer Mannheim). After agarose gel electrophoresis, DNA was denatured and blotted by capillary transfer to nitrocellulose membranes (BA85, 0.45-μm pore size; Schleicher & Schuell). For hybridization, four genomic mouse β-APP restriction fragments specific for intron 15 (probe 3), intron 16 (probe 4), intron 17 (probe 5), and exon 18 (probe 6) were used (Fig. 1C). These probes were random prime radiolabeled and hybridized to the immobilized DNA as described above for probe 2. Autoradiography was performed after washing the membranes with 0.2× SSC–0.1% SDS at 50 to 70°C depending on the background radioactivity.
RESULTS
Genomic structure of the 3′ end of the mouse β-APP gene.
A genomic clone which contains an insert of 12.5 kb starting 1.5 kb upstream of exon 16 and ending 200 bp downstream of the 3′ noncoding region of exon 18 was isolated. Sequencing of exons 16 to 18 showed 100% nucleotide identity to the previously reported cDNA sequence from mouse strain BALB/c, including the large 3′ noncoding region of exon 18 (9). The exon-intron boundaries of exons 15 to 18 were found at the same positions as in humans (23) and rabbits (5). The position of exon 15, which was absent on the characterized genomic clone, was determined to be 5.6 kb upstream of exon 16 (Fig. 1) by PCR by using primers 15F and 16R-wt (Table 1).
Introduction of mutations into ES cells by targeting in-out.
For the first step of this approach (Fig. 1A), ES cells were transfected with the replacement vector RV-S(in), an 11-kb genomic β-APP fragment carrying the SFAD mutation and a selection cassette with Neo and TK genes. Forty to 60% of the 2 × 107 ES cells electroporated with 20 μg of targeting vector at 280 V and 500 μF survived this procedure. Selection for cells that had integrated the replacement vector was started after 24 h by addition of the antibiotic G418. While mock-transfected ES cells were eliminated completely within 5 days of selection, G418-resistant (G418r) ES cell colonies became visible after 4 to 5 days. Counting of G418r colonies after a total selection time of 9 days showed a frequency of 1.4 × 10−4 to 2.0 × 10−4 per cell surviving electroporation for integration of the Neo gene located on the replacement vector into the genome.
Single colonies were picked, expanded, and analyzed for homologous recombination events by PCR by using primers 15F and 16R-S (Table 1). This detected specifically the SFAD mutation at the expected position on exon 16, i.e., 5.7 kb downstream of exon 15 (Fig. 1). By agarose gel electrophoresis, 17 to 21% of the G418r cells were found to be PCR positive (Fig. 2A, left panel). After PCR with primers 16F-S and 17R (Table 1), the expected mutant-specific band at 2.8 kb was obtained from all clones previously identified; this band would, however, also be obtained from random vector integrations (Fig. 2A, right panel). For verification of these results, Southern analysis of XbaI-restricted genomic DNA from isolated ES cell clones was performed by using intron 15-specific probe 3, which is unable to bind directly to the targeting vector (Fig. 1C). Thus, the correctly positioned SFAD mutation would be detected by probe 3 hybridizing to an additional mutation-specific restriction fragment. This fragment is shorter by 1.8 kb than the wild-type fragment because of the new XbaI site on exon 16 of the targeted allele. All clones identified in the first PCR screen were verified by this method (Fig. 2B, left panel), excluding the possibility of false-positive signals caused by random integrations of the targeting vector and primer extension during PCR (10). However, the intensities of bands representing wild-type and mutant alleles are not equal. The genomic DNA was extracted from cocultures of ES cells with inactivated feeder cells (mouse embryonic fibroblasts), and the upper wild-type band is thus generally expected to be more intense; it derives from the wild-type allele of ES cells and from both alleles of feeder cells. Indeed, Southern blot analysis of tail biopsy samples from chimeric mice generated by using these targeted-in ES cell clones showed mainly equal ratios of wild-type to mutant bands (unpublished data). The possibility of additional randomly integrated targeting vectors was also tested by Southern analysis. HindIII-restricted genomic DNA from targeted-in ES cell clones was hybridized with the intron 16-specific probe 4 (Fig. 1), which detected the HindIII site 1.8 kb upstream of exon 16 that is not present on the targeting vector that begins just 300 bp upstream of this restriction site. A single band at 5.6 kb, corresponding to the wild type and the SFAD mutant, was detected. Additional bands of different lengths, as consequences of random integrations or vector repeats, were lacking in all clones tested (Fig. 2B, right panel). Southern blot analysis was also used to verify the correct position of the selection cassette by using probe 6, which is specific for exon 18, on ClaI-restricted DNA (Fig. 1). As expected, an additional 7.2-kb band was found for the mutant allele while the wild-type signal was larger than 15 kb (data not shown). Thus, with the vector RV-S(in), the frequency for targeted homologous recombination, coupled with mutagenesis of exon 16 and integration of the selection cassette, was 2.9 × 10−5 to 3.3 × 10−5 per viable cell after electroporation under the conditions used.
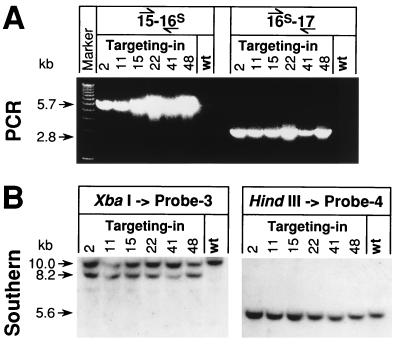
FIG. 2.
Characterization of targeted-in ES cells (clones 2, 11, 15, 22, 41, and 48). (A) Identification of positive ES cell clones by screening with SFAD-specific PCR by using primer pairs 15F–16R-S and 16F-S–17R (left and right panels, respectively). PCR products were analyzed directly by electrophoresis of 20-μl aliquots of the reaction mix on 1% agarose gels. The mutant-specific bands at 5.7 and 2.8 kb indicate the distance of the SFAD mutation, located on exon 16, from the neighboring exons 15 and 17. (B) Verification of positive ES cell clones by Southern blot analysis. Genomic DNA isolated from positive ES cell clones (cultured on PMEFs) was digested with either XbaI or HindIII, separated by electrophoresis on 1% agarose gels, and hybridized with intron 15-specific probe 3 or with intron 16-specific probe 4 after transfer to a nitrocellulose membrane. An additional 8.2-kb band detected with probe 3 on XbaI-restricted DNA indicated the targeted mutation by homologous recombination with RV-S(in) in PCR-positive cells (left panel). Because the preparations also contained DNA from feeder cells, the upper wild-type band appeared generally more intense than the 8.2-kb mutant band. For exclusion of additional random integrations of targeting vectors, HindIII-restricted DNA was tested with probe 4. Except for the 5.6-kb band for the wild type (wt) and SFAD, no additional bands indicative of random integrations of RV-S(in) were found (right panel).
Clones of “hit” ES cells with the targeted mutation were further expanded and kept under G418 selection to prevent spontaneous reversion to wild type. For targeting out, mutated ES cells were transfected with the replacement vector RV-H(out), a simple genomic fragment modified by the HCHWA-D mutation on exon 17. The purpose of this second transfection was to introduce an additional mutation (HCHWA-D) and to replace the nearby selection cassette with the original intron sequence (Fig. 1A). For selection against the viral TK gene located on the selection cassette, the culture medium of transfected ES cells was supplemented with 0.2 μM FIAU 3 to 4 days after electroporation. While growth of wild-type ES cells was not affected by this treatment, TK-positive clones were killed within 2 to 4 days of selection. After 5 days, single FIAU-resistant (FIAUr) colonies became visible and were picked after 7 days of selection. With 0.8 × 10−3 to 4.4 × 10−3 resistant colonies per viable cell after electroporation, FIAUr colonies appeared up to 25 times more frequently after the targeting-out step than did G418r colonies after the targeting-in step. Nevertheless, the frequency still indicates a stringent selection for TK-negative cells. If the detected stronger intensities of the 10-kb wild-type bands for targeted-in clones in Fig. 2 had not originated from inactivated feeder cells but from a large fraction of wild-type ES cells in targeted-in clones, the observed selection would not have occurred. No correlation between a higher intensity of the wild-type band and the rate of FIAUr clones after targeting out was found. All FIAUr clones subjected to another round of G418 selection were killed, and no surviving subclones grew, verifying the loss of the complete selection cassette by the targeting-out step.
After the selection, FIAUr clones were screened by PCR with primers 17F-H and 17+R (Table 1) for integration of RV-H(out) containing the HCHWA-D mutation (Fig. 1A). Random integrations would be detected as well, but only homologous recombination with the correct locus and replacement of the targeted-in TK gene should make targeted-out cells FIAUr, for which they were preselected. In testing 384 FIAUr ES cell clones, derived from 12 different targeted-in clones, not a single PCR-positive HCHWA-D clone was detected. PCR conditions were sensitive enough because addition of RV-H(out) directly to negative cell lysates prior to PCR gave strong 2.1-kb signals, even for a copy number 10 times smaller than expected for a targeted clone (data not shown). In addition, equally strong PCR signals were obtained from cell lysates for wild-type control with primers 17F-wt and 17+R (Table 1; Fig. 1), indicating sufficient quality of the DNA templates (data not shown). However, after the targeting out, FIAUr clones could still have retained the SFAD mutation and lost only the selection cassette. This possibility was tested by SFAD-specific PCR with primers 15F and 16R-S as in the targeting-in step. Again no positive clones were found, suggesting that all clones have reverted to wild type. Therefore, if all intermediate clones obtained by the targeting-in step were equally capable to undergo targeting out, the frequency of the second recombination step was less than 1.1 × 10−5 per viable cell after electroporation. Moreover, identical frequencies of FIAUr colonies were observed for ES cell clones targeted out with RV-H(out) and for mock-transfected clones, indicating a process independent of a homologous recombination with RV-H(out).
Targeting mutations by use of the hit-and-run technique.
The hit-and-run approach represents a different strategy for introduction of subtle mutations. Although this procedure requires two sequential recombinations as does the targeting in-out method, only one transfection, at the beginning, needs to be done (Fig. 1B). For this hit step, the ES cells were electroporated with the targeting vector IV-H(hit-run) as described above. The insertion vector contained the same 11-kb genomic DNA fragment as the replacement vectors, with the HCHWA-D mutation on exon 17. However, the ends of the fragment were connected by the selection cassette and the targeting vector was linearized within the genomic region by introduction of a double-strand break 2.0 kb upstream of exon 17 (Fig. 1B). Selection with G418 for 9 days resulted in 1.2 × 10−4 to 4.5 × 10−4 resistant ES colonies per viable cell after electroporation, which is similar to the frequency obtained with the corresponding replacement vector RV-S(in) (1.4 × 10−4 to 2.0 × 10−4).
Single G418r clones were screened for the correctly positioned HCHWA-D mutation on exon 17 by PCR by using primers 17F-H and 17+R (Fig. 1). Both primers could also bind to the targeting vector but in the wrong orientation for direct PCR. Of the tested cells, 23 to 25% gave a correct 2.1-kb PCR fragment (Fig. 3A). Misleading signals could possibly arise by primer extension (10) on randomly integrated insertion vectors or by amplification from randomly integrated head-to-tail tandem repeats of IV-H(hit-run) as frequently reported in transgenic animals with multiple integrations of the transgene (20). This was evaluated by Southern analysis with intron 17-specific probe 5 (Fig. 1) on XbaI-restricted DNA from potential hit clones. Only a single strong band at 3.2 kb was observed for all clones tested (Fig. 4C). The same pattern (single band at 6.8 kb) was detected by probe 5 hybridization to DNA cut with BamHI (data not shown). Since no additional fragments were found, random integrations can be excluded. However, the intensities of the 3.2- and 6.8-kb signals were clearly elevated in some clones relative to others, indicating a higher copy number of β-APP intron 17. Indeed, the presence of vector repeats, integrated site specifically at the targeted locus, was confirmed in these clones by Southern analysis of NotI-restricted DNA with intron 16-specific probe 4 (Fig. 4B). Only from the clones with tandem repeats of IV-H(hit-run) could an intron 16-containing 11-kb genomic fragment be excised between two neighboring selection cassettes, since those were flanked by restriction sites for the rare-cutting enzyme (Fig. 1). Finally, the exact position of the HCHWA-D mutation on the hit clones was determined. The mutation on exon 17 could be located on both sites of the partial genomic duplication (Fig. 1B) or even at several positions in those clones with a targeted integrated tandem repeat of IV-H(hit-run). A PCR with primers 15F and 17R-H was used to detect the HCHWA-D mutation at the 5′ site of the duplication, and another reaction with primers NeoF and 17R-H detected the mutation downstream of the selection cassette (Fig. 1B). The signal for either the 5′-located mutation (8.5 kb) or the 3′-located mutation (5.3 kb) or both were found in all clones tested (Fig. 4A). Thus, all 12 clones analyzed were hits and contained targeted integrations of IV-H(hit-run). Five of them contained targeted integrated tandem repeats with the HCHWA-D mutation at different positions. The others had integrated a single copy of the targeting vector, i.e., six clones with the mutation only at the 3′ site of the duplication and one with HCHWA-D only at the 5′ site. Considering all hit clones (including those with insertion vector repeats), the frequency for targeted integration of IV-H(hit-run) together with the HCHWA-D mutation by homologous recombination was 3.0 × 10−5 to 5.0 × 10−5 per viable cell after electroporation. This indicates similar efficiencies for the first steps of both targeting approaches.
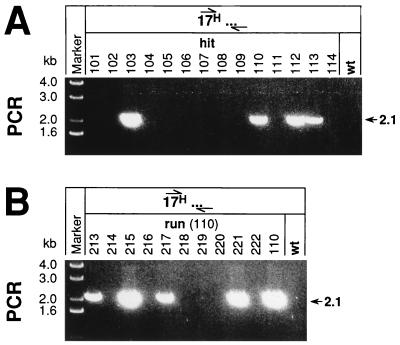
FIG. 3.
Screening for hit-and-run ES cells with the HCHWA-D mutation. (A) Identification of potential hit ES cell clones by screening with an HCHWA-D-specific PCR by using primers 17F-H and 17+R. PCR products were analyzed directly by electrophoresis of 20-μl aliquots of the reaction mix on 1% agarose gels. The mutant-specific band at 2.1 kb indicates the distance between the HCHWA-D mutation on exon 17 and the binding site of primer 17+R (17H…) located on the other side of the double-strand break in the 11-kb genomic fragment of the insertion vector IV-H(hit-run). (B) Detection of ES cell clones retaining the HCHWA-D mutation in their genome after the run of the hit clone 110. The same PCR conditions and analysis used for detection of hit clones were used for screening of run clones. wt, wild type. Numbers at tops of lanes are clone designations.
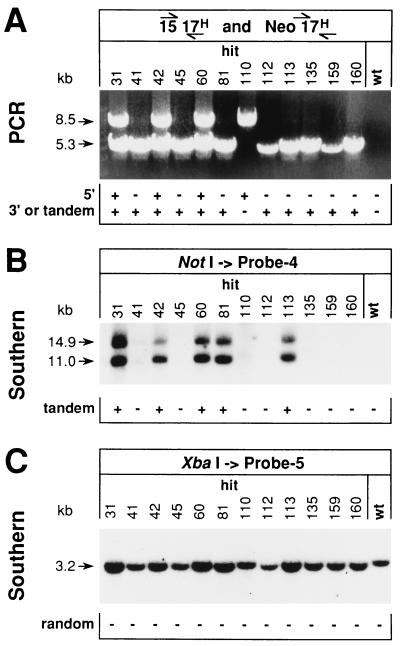
FIG. 4.
Characterization of potential hit ES cells. (A) Analysis of the targeted locus of hit clones. The position of the HCHWA-D mutation on the partial genomic duplication of hit clones was investigated by two mutation-specific PCRs by using primer pairs 15F–17R-H for detection of the 5′ location and NeoF–17R-H for the 3′ location. A combination of 15-μl aliquots from both reaction mixtures was analyzed directly by electrophoresis on 0.6% agarose gels. The band at 8.5 kb indicates the site-specific 5′ position of the HCHWA-D mutation, and the 5.3-kb band indicates the mutation when located 3′ of the selection cassette. (B) Detection of possible repetitive integrations of the targeting vector by Southern blot analysis. Genomic DNA isolated from positive ES cell clones was digested with NotI, separated by electrophoresis on 0.6% agarose gels, and hybridized with intron 16-specific probe 4 after transfer to a nitrocellulose membrane. The appearance of a band at 11 kb indicated the presence of head-to-tail tandem repeats of IV-H(hit-run) in some clones. Because of incomplete NotI restriction, a 14.9-kb band was detected together with the expected 11-kb band, showing elongated fragments, i.e., products with an attached selection cassette. (C) Exclusion of additional random integrations of targeting vectors. XbaI-restricted DNA was run on 0.9% agarose gels and tested with intron 17-specific probe 5. Except for the 3.2-kb band for the wild type and for alleles with site-specific integrations of IV-H(hit-run), no additional bands indicative of random integrations were detected. However, because of the higher copy number, stronger 3.2-kb signals were obtained from those clones with specifically integrated vector repeats. wt, wild type. Numbers at tops of lanes are clone designations.
Hit ES cell clones were expanded and kept under G418 selection before the second homologous recombination step, the run, was initiated. Hit clones were replated, and withdrawal of G418 allowed survival of cells losing their selection cassette with the Neo gene. After 1 to 5 days in nonselective medium, the counterselection against ES cells still retaining the selection cassette with the TK gene was started by the addition of FIAU to the culture medium and was maintained for 7 days. As expected, the longer that ES cells were kept in nonselective medium, the more FIAUr colonies were obtained. A constantly low number of clones survived the FIAU selection after a selection-free period of 1 to 3 days (1.8 × 10−4 to 3.9 × 10−4 per plated cell). After 4 days without selection, the number of FIAUr clones per plated cell increased drastically (5.5 × 10−2), and after 5 days, it was ∼1.2 × 10−1. This indicates the presence of residual TK activity that kills cells up to 3 days after loss of the TK gene. These observations have to be considered in calculating the rate of de novo-appearing FIAUr clones per cell per day. Based on a 24-h generation time of the cells, the observed frequency reflects a 3-day delay. Thus, the relevant cell number was eight times smaller, and consequently the rate of de novo-appearing FIAUr clones per cell per day was 3.6 × 10−2 to 3.8 × 10−2.
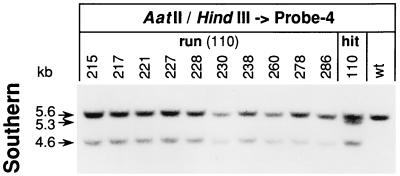
After the “run” procedure, FIAUr ES colonies were again screened by PCR (primers 17F-H and 17+R) for the presence of the HCHWA-D mutation. Revertants that had lost the selection cassette but retained the mutation were obtained from only 1 of the 12 hit clones subjected to counterselection. But, 24 to 31% of the FIAUr colonies which derived from this single clone (no. 110) were PCR positive (Fig. 3B). Such positive ES colonies, as well as the precursor clone, were further characterized by Southern analysis. Cellular genomic DNA, restricted with endonucleases HindIII and AatII, was hybridized with the intron 16-specific probe 4 (Fig. 1). While a single band at 5.6 kb was detected on wild-type DNA, an additional 4.6 kb band was found on DNA from targeted mutated hit-and-run clones, indicating the presence of the HCHWA-D allele by the mutant-specific AatII site on exon 17 (Fig. 5). Unequal intensities of wild-type and mutant bands are found, as in the targeted-in cell analysis, due to the presence of various amounts of feeder cell DNA. By Southern blot analysis, the 5′ position of the HCHWA-D mutation on the partial genomic duplication of clone 110 was confirmed, as had already been shown by PCR, making this clone unique among the hit clones tested (Fig. 4A). Together with the same two bands as those for the hit-and-run clones, representing the wild-type allele and the HCHWA-D mutation on the 5′ site of the partial duplication, a third band at 5.3 kb was detected, indicating that exon 17 on the 3′ site is of the wild type (Fig. 5). Since all PCR-positive FIAUr clones were verified as pure HCHWA-D mutants, the frequency of intrachromosomal recombination leading to de novo-generated mutants per day and cultured cell was 1.1 × 10−2 to 1.2 × 10−2 for the run of clone 110.
FIG. 5.
Characterization of ES cells produced by the hit-and-run procedure. Verification of PCR-positive clones of the hit-and-run procedure by Southern blot analysis. Aliquots containing 5 μg of genomic DNA from ES cell clones (cultured on PMEFs) were double digested with AatII/HindIII and hybridized with intron 16-specific probe 4 after electrophoretic separation through 2% agarose gels and transfer to a nitrocellulose membrane. A single band at 5.6 kb was obtained from wild-type ES cells (wt) while an additional band at 4.6 kb was obtained from hit-and-run clones for the targeted allele carrying the HCHWA-D mutation. From the hit clone 110, which served as precursor for these positive clones, a third band at 5.3 kb was detected, indicating the absence of an HCHWA-D mutation on the 3′ site of the partial genomic duplication. Numbers at tops of lanes are clone designations.
DISCUSSION
Introduction of subtle mutations into the mammalian genome is still a difficult task. Especially these days, however, it has become increasingly interesting to subtly modify proteins in animals for functional analysis. In this study, an isolated mouse genomic β-APP fragment was used for introduction of SFAD and HCHWA-D mutations into the genome of mouse ES cells by homologous recombination by two different methods. Both procedures had several common features. (i) All targeting vectors contained the same 11-kb-long homology to the cellular β-APP gene. (ii) The same selection cassette was used to control the homologous recombinations by G418 selection for the presence of the neomycin gene in the first step and by FIAU selection for the absence of the TK gene in the second step. Finally, (iii) the experimental conditions for transfection and selection of ES cells were standardized. These fixed parameters, together with defined differences between the two methods, allowed interpretation of the results with respect to some general mechanisms related to gene targeting by truly homologous recombination.
Genomic DNA and the selection cassette were arranged differently on the two types of vectors. Whereas the replacement vectors for the in-out approach were linearized at the ends of the β-APP region, the insertion vector for the hit-and-run procedure contained a double-strand break within the genomic fragment. Independent of the vector types, the frequencies for ES cells that were either targeted in or hit and had integrated the desired mutation into their genome after the first homologous recombination were nearly identical. Also the ratios of homologous recombination to random integration of targeting vectors of the two methods were comparable. These results confirm that mainly the overall extent of homology (11 kb long in these experiments) and not the arrangement of the homologous regions determines the probability of recombination in mammalian cells, and the frequencies fit well with predicted values (7).
Characterization of targeted-in ES cells revealed correctly replaced β-APP loci with the targeted mutation and the intron-located selection cassette; no vector repeats were found at the modified β-APP locus. In contrast, hit ES cell clones had frequently integrated not a single copy of the insertion vector, but also actually contained site-directed integrations of tandemly arranged (head-to-tail) vector repeats (42% of all hit clones). One explanation for this difference between the hit and targeting-in steps might be a different capacity for self-assembly of insertion and replacement vectors. Only head-to-tail arrangements of insertion vectors, as found in several hit clones, resemble at the junction between two vectors the naturally occurring chromosomal structure of the β-APP gene. Such a structure might be more stable than artificial combinations of other vector arrangements. The observed multicopy inserts might thus originate from homologous recombinations between the targeted locus and preassembled insertion vector repeats.
The second part of both procedures, i.e., the run or targeting out, was more complex than the first step. A major issue was a high spontaneous wild-type reversion rate of targeted-in or hit clones when they were subjected to FIAU counterselection. The number of FIAUr clones was more than 2 orders of magnitude greater than expected for targeting out, and ES cells which were mock transfected for targeting out showed the same high frequency of wild-type reversion. For the run procedure as well, high frequencies of spontaneous wild-type reversion were found. These reversion rates were even 10 times higher than for targeted-in clones. Within each group, however, the reversion frequencies of individual targeted-in or hit clones were similar. The higher reversion frequency of hit clones compared to that of targeted-in clones suggests different mechanisms for the two procedures. In both cases, dominating wild-type reversion cannot be explained by a defective TK gene, silencing of the targeted locus, or insufficient FIAU selection, since the TK gene was physically lost together with the targeted mutation. Loss of the entire targeted allele cannot be excluded. However, this might at best explain the less-frequent wild-type reversion in the targeting-in-out procedure, since the frequency of random chromosomal aberrations should not depend on the targeting procedure. Similarly unlikely is the existence of few wild-type ES cells, which might have survived the initial neomycin selection by metabolic cooperation with mutant cells and then have reappeared as wild-type clones after FIAU selection. Otherwise, a broader distribution of the reversion frequencies for individual clones would be expected, regardless of the method. If the high reversion rate was caused by metabolic cooperation, the number of wild-type cells surviving the first selection step might be reduced by simple changes of the experimental procedures like culturing ES cells at lower densities. This is unlikely, and thus a reduction of the wild-type reversion rate is probably hard to achieve. Finally, in the targeting-out step, the electroporation itself may have directly induced the reversion to wild-type cells, possibly by an SOS response, as was suggested previously (18).
Despite the observed high frequency of wild-type reversion, pure mutant ES cells without a selection cassette were generated by the hit-and-run procedure. However, only 1 of 12 hit clones resulted after the run in desired targeted mutant cells. For the other hit clones, only wild-type reversion was found although the rates of cells surviving FIAU selection were similar for all hit clones. A high percentage (24 to 31%) of FIAUr cells originating from this particular clone contained the desired mutation, suggesting that the extremely frequent spontaneous wild-type reversion in the hit-and-run procedure (3.6 × 10−2 to 3.8 × 10−2 per cell per day) happens because of intrachromosomal recombination between the duplicated β-APP sequences in the genome of hit ES cells. The genomic structure of the targeted locus of this particular hit clone was different from that of the rest. Among the seven hit clones with a single-copy integration of the targeting vector, it was the only one which contained the HCHWA-D mutation at the 5′ site of the partial β-APP duplication. All other hit clones either had the HCHWA-D mutation at the 3′ site or contained a multiple vector integration. An increased probability for crossovers close to the double-strand break has been reported for integration of insertion vectors by homologous recombination (8). This could have triggered generation of more hit clones with the HCHWA-D mutation at the 3′ site of the duplication in the present study. Yet, why after intrachromosomal recombination the mutation preferentially persisted in the genome when located at the 5′ site of the duplication is unclear. One reason might be a translocation of the HCHWA-D mutation from 3′ to 5′ after the integration, bringing the mutation into an environment of correctly methylated DNA sequences which might be less susceptible to loss through intrachromosomal recombination. Alternatively, the recombination machinery might be coupled to DNA replication and induce an orientation-specific preference for maintenance of the 5′ part of the duplication in the genome.
The location of the mutation on the intermediate genomic duplication in hit clones is very crucial for keeping the mutation in the genome after loss of the duplication by intrachromosomal homologous recombination and thus should be considered in optimization strategies of the hit-and-run procedure. Screening a broad range of hit clones with different genomic patterns, i.e., with the mutation at either side of the duplication, should be preferred over intense screening of clones from the same hit origin. A high frequency of intrachromosomal recombination resulting in mutant run clones might then be expected for some hit clones. Moreover, both approaches can be optimized by increasing the efficiency for homologous recombination itself. According to Deng and Capecchi (7), the maximal frequency of homologous recombination obtained with isogenic DNA of more than 14 kb is 10 times higher than for the 11-kb nonisogenic genomic β-APP fragment used in this study. Such an increased frequency might then be sufficient for identification of, e.g., targeted-out cells with subtle mutations among the large number of wild-type revertants.
In conclusion, by use of the targeting-in-out approach, only SFAD mutant ES cells with a remaining selection cassette in intron 17 were obtained, while pure HCHWA-D mutant ES cells were isolated after the hit-and-run procedure. This second approach turned out to be very efficient. From one intermediate clone containing the partial genomic duplication, ∼30% of the ES cell clones analyzed after counterselection were pure HCHWA-D mutants. Finally, the hit-and-run technique, with such high efficiency, has the advantage of being independent of a second transfection and leads to pure mutants without leaving behind any sequence in the genome.
ACKNOWLEDGMENTS
The mouse genomic library used for cloning the β-APP fragment was a kind gift of W. Wille (Institut für Genetik, Universität Köln). We thank H. Blüthmann for providing mouse E14 ES cells and PMEFs and Y. Lang for technical assistance with cell culture and transfection techniques (both of F. Hoffmann-La Roche, Basel, Switzerland). We are also very grateful to P. Malherbe and J. Kemp for critical reading of the manuscript.
REFERENCES
- 1.Adra C N, Boer P H, McBurney M W. Cloning and expression of the mouse pgk-1 gene and the nucleotide sequence of its promoter. Gene. 1987;60:65–74. doi: 10.1016/0378-1119(87)90214-9. [DOI] [PubMed] [Google Scholar]
- 2.Askew G R, Doetschman T, Lingrel J B. Site-directed point mutations in embryonic stem cells: a gene targeting tag-and-exchange strategy. Mol Cell Biol. 1993;13:4115–4124. doi: 10.1128/mcb.13.7.4115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Colbère-Garapin F, Chousterman S, Horodniceaunu F, Kourilsky P, Garapin A C. Cloning of the active thymidine kinase gene of herpes simplex virus type 1 in Escherichia coli K-12. Proc Natl Acad Sci USA. 1979;76:3755–3759. doi: 10.1073/pnas.76.8.3755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Colbère-Garapin F, Horodniceaunu F, Kourilsky P, Garapin A. A new dominant hybrid selective marker for higher eukaryotic cells. J Mol Biol. 1981;150:1–14. doi: 10.1016/0022-2836(81)90321-1. [DOI] [PubMed] [Google Scholar]
- 5.Davidson J S, West R L, Kotikalapudi P, Maroun L E. Sequence and methylation in the β/A4 region of the rabbit amyloid precursor protein gene. Biochem Biophys Res Commun. 1992;188:905–911. doi: 10.1016/0006-291x(92)91141-c. [DOI] [PubMed] [Google Scholar]
- 6.Davis L G, Dibner M D, Battey J F. Basic methods in molecular biology. 1st ed. New York, N.Y: Elsevier; 1986. [Google Scholar]
- 7.Deng C, Capecchi M R. Reexamination of gene targeting frequency as a function of the extent of homology between the targeting vector and the targeting locus. Mol Cell Biol. 1992;12:3365–3371. doi: 10.1128/mcb.12.8.3365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Deng C, Thomas D R, Capecchi M R. Location of crossovers during gene targeting with insertion and replacement vectors. Mol Cell Biol. 1993;13:2134–2140. doi: 10.1128/mcb.13.4.2134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.De Strooper B, Van Leuven F, Van Den Berghe H. The amyloid β protein precursor or proteinase nexin II from mouse is closer related to its human homolog than previously reported. Biochim Biophys Acta. 1991;1129:141–143. doi: 10.1016/0167-4781(91)90231-a. [DOI] [PubMed] [Google Scholar]
- 10.Frohman M A, Martin G R. Detection of homologous recombinants. In: Innis M A, Gelfand D H, Sninsky J J, White T J, editors. PCR protocols: a guide to methods and applications. San Diego, Calif: Academic Press; 1990. pp. 228–236. [Google Scholar]
- 11.Hasty P, Ramirez-Solis R, Krumlauf R, Bradley A. Introduction of a subtle mutation into the Hox-2.6 locus in embryonic stem cells. Nature. 1991;350:243–246. doi: 10.1038/350243a0. [DOI] [PubMed] [Google Scholar]
- 12.Levy E, Carman M D, Fernandez-Madrid I J, Power M D, Lieberburg I, van Duinen S G, Bots G T, Luyendijk W, Frangione B. Mutation of the Alzheimer’s disease amyloid gene in hereditary cerebral hemorrhage, Dutch type. Science. 1990;248:1124–1126. doi: 10.1126/science.2111584. [DOI] [PubMed] [Google Scholar]
- 13.Mansour S L, Thomas K R, Cappecchi M R. Disruption of the proto-oncogene int-2 in mouse embryo-derived stem cells: a general strategy for targeting mutations to non-selectable genes. Nature. 1988;336:348–352. doi: 10.1038/336348a0. [DOI] [PubMed] [Google Scholar]
- 14.Marth J D. Recent advances in gene mutagenesis by site-directed recombination. J Clin Invest. 1996;98:S47–S50. doi: 10.1172/JCI118634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mullan M, Crawford F, Axelman K, Houlden H, Lilius L, Winbald B, Lannfelt L. A pathogenic mutation for probable Alzheimer’s disease in the APP gene at the N-terminus of β-amyloid. Nat Genet. 1992;1:345–347. doi: 10.1038/ng0892-345. [DOI] [PubMed] [Google Scholar]
- 16.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 17.Schellenberg G D. Genetic dissection of Alzheimer’s disease, a heterogeneous disorder. Proc Natl Acad Sci USA. 1995;1995:8552–8559. doi: 10.1073/pnas.92.19.8552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Stacey A, Schnieke A, McWhir J, Cooper J, Colman A, Melton D W. Use of double-replacement gene targeting to replace the murine α-lactalbumin gene with its human counterpart in embryonic stem cells and mice. Mol Cell Biol. 1994;14:1009–1016. doi: 10.1128/mcb.14.2.1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Stewart C L, Schütze S, Vanek M, Wagner E F. Expression of retroviral vectors in transgenic mice obtained by embryo infection. EMBO J. 1987;6:383–388. doi: 10.1002/j.1460-2075.1987.tb04766.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Storb U, Denis K A, Brinster R L, Witte O N. Pre-B cells in kappa-transgenic mice. Nature. 1985;316:356–358. doi: 10.1038/316356a0. [DOI] [PubMed] [Google Scholar]
- 21.Valancius V, Smithies O. Testing an “in-out”-targeting procedure for making subtle genomic modifications in mouse embryonic stem cells. Mol Cell Biol. 1991;11:1402–1408. doi: 10.1128/mcb.11.3.1402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wu H, Liu X, Jaenisch R. Double replacement: strategy for efficient introduction of subtle mutations into the murine Col1a-1 gene by homologous recombination in embryonic stem cells. Proc Natl Acad Sci USA. 1994;91:2819–2823. doi: 10.1073/pnas.91.7.2819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yoshikai S, Sasaki H, Doh-ura K, Furuya H, Sakaki Y. Genomic organization of the human beta-protein precursor gene. Gene. 1990;87:257–263. doi: 10.1016/0378-1119(90)90310-n. [DOI] [PubMed] [Google Scholar]