Abstract
Acute myeloid leukemia (AML) is a genetically extremely heterogeneous disease. Drug resistance after induction therapy is a very frequent event resulting in poor medium survival times. Therefore, the identification of new targets and treatment modalities is a medical high priority issue. We addressed our attention to circular RNAs (circRNAs), which can act as oncogenes or tumor suppressors in AML. We searched the literature (PubMed) and identified eight up-regulated and two down-regulated circ-RNAs with activity in preclinical in vivo models. In addition, we identified twenty-two up-regulated and four down-regulated circRNAs with activity in preclinical in vitro systems, but pending in vivo activity. Up-regulated RNAs are potential targets for si- or shRNA-based approaches, and down-regulated circRNAs can be reconstituted by replacement therapy to achieve a therapeutic benefit in preclinical systems. The up-regulated targets can be tackled with small molecules, antibody-based entities, or other modes of intervention. For down-regulated targets, up-regulators must be identified. The ranking of the identified circRNAs with respect to therapy of AML will depend on further target validation experiments.
Keywords: Driver mutations, genetic heterogeneity, preclinical validation, reconstitution therapy, shRNA, siRNA, review
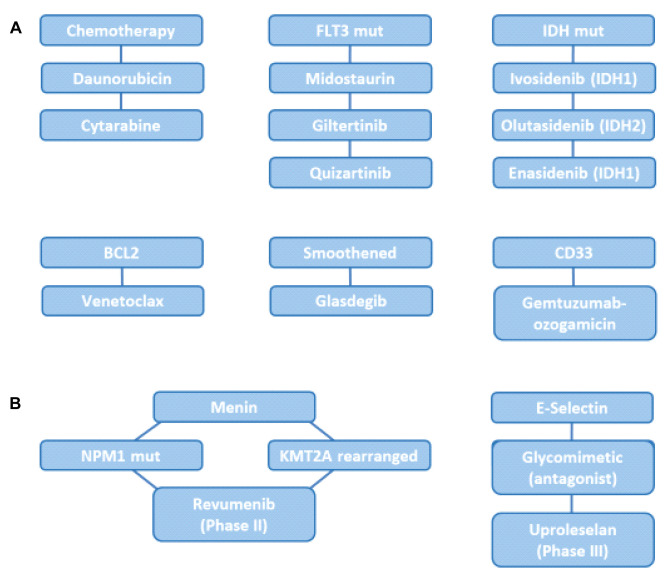
Acute myeloid leukemia (AML) is a clinically heterogeneous disease with clonal proliferation of undifferentiated myeloid progenitors (1). Presently the annual incidence is 20,000 cases in the US and 12,000 in Europe (2). Over the years Cytarabine plus Daunorubicine has remained the standard therapy of AML (3,4). Chemotherapy plus hematopoietic stem cell transplantation enables a five-year overall survival of 40% of patients younger than 60 years and 10-20% of patients older than 60 years (3,4). The exception is acute promyelocytic leukemia (APL), which is caused by a fusion protein derived from the retinoic acid receptor and can be cured by treatment with all-trans retinoic acid (ATRA) and arsenic trioxide (5). Recently, several agents, such as Daunorubicin, Cytarabine, inhibitors of mutant FLT3 (mutFLT3), such as Midostaurin, Giltertinib and Quizartinib, as well as inhibitors of mutant IDH (mutIDH), such as Ivosidenib, Enasidenib and Olutasidenib, BCL2 (Venetoclax), Smoothened (Glasdegib) and CD33 (Gemtuzumab-ozogamicin) are approved for treatment of AML (Figure 1) (3-10). Agents such as Revumenib, which targets mutant nucleophosmin 1 (mutNPM1) and rearranged histone-lysine-N-methyltransferase 2A (KMT2A) (11) are in Phase II and E-selectin antagonist Uproleselan (12,13) is undergoing Phase III clinical studies (Figure 1).
Figure 1. Drugs and targets for treatment of AML. A) Approved drugs and targets. B) Selected ongoing clinical trials. BCL2: B-cell lymphoma 2; CD33, CD47: cluster of differentiation 33 and 47; FLT3 mut: mutant fms-like tyrosine kinase; IDH1 mut: mutant isocitrate dehydrogenase 1; NPM mut: mutant nucleophosmin 1; KMT2A: lysine-specific methyltransferase 2A.
In addition to mutNPM1 and mutFLT3, several driver mutations have been identified which reveal defined molecular subgroups of AML. These mutations affect signaling pathways, DNA methylation enzymes, chromatin-modifying enzymes, transcription factors, components of the spliceosome complex and tumor suppressor genes. Mutations in transcription factor CCAAT/enhancer binding protein α (CEBPA), mixed lineage leukemia or myeloid lymphoid leukemia (MLL), neuroblastoma ras oncogene homolog (NRAS) and runt-related transcription factor 1 (RUNX1) have been identified (6,10). It remains to be seen whether some of these targets can be validated for treatment of AML especially in the context of combination therapy. Taken together, the identification of additional validated targets and the design of new treatment entities for AML is an important medical issue.
In the past few years, unprecedented clinical results have been achieved by treatment of lymphomas and leukemias with chimeric antigen receptor (CAR)-based T-cell therapy (14). However, treatment of AML with CAR-based therapy targeting cluster of differentiation 33 (CD33) and CD123 has been hampered by toxicities due to expression of these antigens on endothelial cells, hematopoietic stem, and progenitor cells, resulting in a critical toxicity profile (15). Recently, colony-stimulating factor 1 receptor (CSF-1R) and CD86 have been identified as antigens, which are over-expressed in AML and show limited expression in normal tissues (15). CARs directed against these antigens mediated activity in AML-based in vivo models. It remains to be seen whether these approaches will be game-changers in the field.
Circular RNAs and Their Role in Oncology
CircRNAs are transcribed by RNA Pol II and can be generated by RNA binding protein induced circularization, intron pairing and spliceosome-dependent lariat-driven circularization, creating new backsplicing junctions (16,17). circRNAs are preferentially located in the cytosol and are expressed in a cell-, tissue- and developmental stage-specific manner (16,17). CircRNAs were described first in viroids (18) and later in mammalian cells (19). A crucial finding was the co-localization of circRNA ciRS-7 and micro RNA-7 (miR-7) in neocortical and hippocampal neurons suggesting interactions and the identification of more than seventy conserved binding sites for miR-7 on ciRS-7 involved in sponging of miR-7 (20,21). Subsequently it was shown that circRNA Cdr1 is involved in brain function in the mouse through interaction with miR-671 by mediating synaptic transmission (22).
circRNAs are dysregulated in cancer and their landscape has been investigated in 40 types of cancer (23). They can exert tumor-suppressive as well as oncogenic functions (23). In addition to sponging of miRs, circRNAs can mediate transcriptional regulation by recruiting transcription factors to promoter regions of DNA methylating as well as histone methylating and acetylating enzymes (24). circRNAs are involved in tethering or sequestering proteins, translocation, and redistribution of proteins and in modulating translation (24). Some circRNAs can encode internal peptides or proteins, which can be translated via internal ribosome entry sites (IRES) or m6A-induced ribosome engagement sites (MIRES) (25). These proteins can regulate yes-associated protein (YAP)-Hippo and WNT/β-catenin signaling pathways and malignant progression by phosphorylation and ubiquitinylation of specific proteins (25). circRNAs can affect all aspects of cancer from dormancy to proliferation, invasion, and metastasis (26).
Knowledge on circRNA in AML with respect to their role as diagnostic and therapeutic biomarkers and from the point of view of mechanistic insights has been summarized in (2,27,28). In our review, we focus on their role as possible new therapeutic entities and as tools for target identification.
Up-regulated circRNAs With In Vivo Activity in Preclinical Models
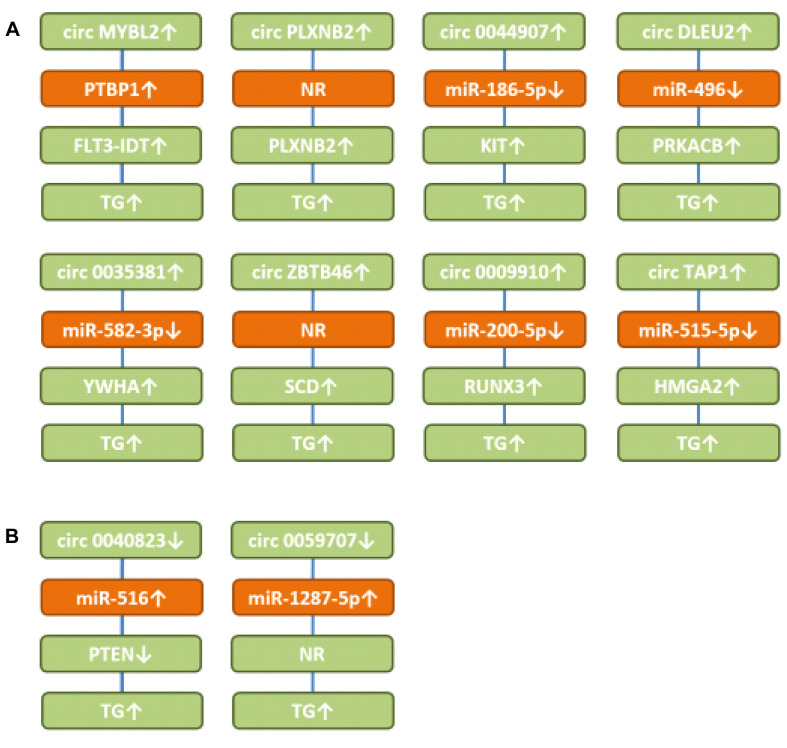
Circ MYB proto-oncogene like 2 (circMYBL2) up-regulates fms-like tyrosine kinase 3-internal duplication domain (FLT3-ITD). circMYBL2 (Figure 2A) is over-expressed in FLT3-ITD+ AML (29). Its knockdown in MOLM13 cells (FLT3-ITD+ AML) impaired proliferation (29). After tail vein injection, circMYBL2 promoted infiltration of MOLM13 cells into the bone marrow, spleen and liver and its knockdown extended survival of immuno-deficient mice (29). circMYBL2 targeted mutFLT3 kinase and the corresponding pathway for myeloid differentiation and progression. It has been shown that circMYBL2 recruited polypyrimidine tract binding protein1 (PTBP1) resulting in improved translation of FLT3-ITD (1). PTBP1 has been shown to enhance proliferation of FLT3-ITD AML cells and has been identified as a potential target for AML therapy (30). It should be emphasized that FLT3 mutations are present in 30% of AML cases with internal domain duplication (ITD) representing the most common mutation (31). Identification and clinical evaluation of mutant FLT3 inhibitors are ongoing approaches (32,33).
Figure 2. Circular RNAs with efficacy in AML-related preclinical in vivo systems. A) Up-regulated circular RNAs. B) Down-regulated circular RNAs. circDLEU2: circ deleted in lymphocytic leukemia 2; circMYBL2: circMYB proto-oncogene like 2; circZBTB46: circ zinc finger and BTB domain containing 46; circTAP1: circ threonine-aspartase 1; FLT3: fms-like tyrosine kinase 3; FLT-ITD: fms-like tyrosine kinase internal duplication domain; KIT: receptor tyrosine kinase KIT; HMGA2: high-mobility group A2; miR: microRNA; NR: not resolved; PLXNB2: plexin B2; PTBP1: polypyrimidine tract binding protein1; PTEN: phosphatase and tensin homolog; PRKACB: cAMP-dependent protein kinase A catalytic subunit β; RUNX3: runt-related transcription factor 3; SCD: stearoyl-CoA-desaturase; TG: tumor growth; YWHAZ: tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein zeta.
Circ PlexinB2 (circPLXNB2) up-regulates PlexinB2 (PLXNB2). Circ PLXNB2 (Figure 2A) correlated with worse prognosis in AML patients (34). Over-expression of circ PLXNB2 promoted proliferation and inhibited apoptosis in OCI-AML3 cells. circPLXNB2 increased leukemic burden of transfected HL60 AML cells after subcutaneous implantation into immuno-compromised mice (34). It up-regulated the expression of PLXNB2, B-cell lymphoma 2 (BCL2) and cyclin D1 by a yet unresolved mechanism. Plexins bind to semaphorins and are involved in axon guidance and signal transduction, such as activation of GTPase RHOA, mitogen-activated protein kinase A (MAPK) and phosphatidyl-inositol-3-kinase (PI3K) signaling (35,36). PLXNB2 acts as a receptor for angiogenin and regulates angiogenesis in tumors including AML (37). PLXNB2 has been shown to be involved in growth and invasion of ovarian carcinoma (38).
circ0044907 up-regulates transmembrane tyrosine kinase KIT. High expression of KIT in the bone marrow of AML patients correlated with poor prognosis (39). In AML cell lines HL60 and TIB-202, loss of circ0044907 (Figure 2A) reduced proliferation and induced apoptosis. This was due to sponging of miR-186-5p and subsequent up-regulation of KIT. Silencing of circ0044907 in HL60 AML cells reduced their growth after subcutaneous implantation into nude mice. KIT contributes to signal transduction in hematopoietic stem cells, mast cells and Cajal cells of the gastrointestinal tract by interaction with stem cell factor (SCF) as a ligand and stimulating MAPK-janus kinase – signal transducer and activators of transcription (JAK-STAT) and PI3K-signaling (40,41). KIT mutations are associated with a poor prognosis in AML patients (42,43). Inhibition of KIT has emerged as an important strategy for treatment of AML (44).
Circ deleted in lymphocytic leukemia 2 (circDLEU2) up-regulates cAMP-dependent protein kinase A catalytic subunit β (PRKACB). circDLEU2 (Figure 2A) was up-regulated in AML, promoted proliferation of MOLM-13 and MV-4-11 AML cells and inhibited autophagy (45). After subcutaneous injection of circDLEU2-transfected cells, it promoted tumor formation in nude mice. circDLEU2 sponged miR-496 and up-regulated PRKACB, a cyclic AMP-dependent ser/thr kinase (46). However, its function seems to be context-dependent, because it inhibits proliferation of non-small cell lung carcinoma cells (47). Furthermore, in cholangiocarcinoma, a fusion protein involving PRKACB plays an oncogenic role (48,49).
circ0035381 up-regulates tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein zeta (YWHAZ). circ0035381 (Figure 2A) was up-regulated in AML bone marrow samples and AML cells (50). Knockdown of circ0035381 repressed proliferation of AML cells and induced apoptosis and mitochondrial damage in vitro. circ0035381 sponged miR-582-3p and up-regulated YWHAZ. The latter belongs to the family of 14-3-3 proteins, which are over-expressed in a wide range of cancers and are involved in cell growth, cell-cycle, apoptosis, migration, and invasion (51). Down-regulation of circ0035381 also decreased growth of AML cells in nude mice. It has been shown that targeting YWHAZ by siRNA, shRNA or miRs can suppress malignant properties of cancer cells (51). YWHAZ can interact with a diverse range of signaling proteins by binding to an amphipathic helix and activating it by phosphorylation (52). It has been shown that YWHAZ is increased in AML and that its knockdown by siRNA reduced cell growth and proliferation of AML cells (53,54).
Circ zinc finger and BTB domain containing 46 (circZBTB46) up-regulates stearoyl-CoA desaturase (SCD). circZBTB46 (Figure 2A) was up-regulated in AML patients and expression correlated with disease progression (55). circZBTB46 promoted cell proliferation and cell progression in HL-60 and K562 AML cells in vitro. Its knockdown impaired tumorigenesis of HL-60 AML cells in nude mice. It up-regulated SCD probably by a miR-sponge mechanism, but this issue needs to be resolved in further detail. SCD is an endoplasmic reticulum localized enzyme involved in the biosynthesis of mono-unsaturated fatty acids. It has been shown that inhibition of SCD induces lipid oxidation and cell death by ferroptosis (56). The latter represents a reactive-oxygen species-dependent form of cell death with the characteristics of iron accumulation and lipid peroxidation (57,58). In AML, it has been shown that a ferroptosis-related gene signature together with an immune infiltration pattern predicts the overall survival of patients (59). Induction of ferroptosis in hematological diseases including AML is an actively pursued approach (60).
circ0009910 up-regulates runt-related transcription factor 3 (RUNX3). circ0009910 (Figure 2A) was up-regulated in AML patients and predicted risk of poor outcome (61). It promoted proliferation and inhibited apoptosis in AML cell lines in vitro and in xenografts by sponging miR-200-5p (61). It has been shown by other groups that miR-200-5p targets RUNX3, a promoter of AML progression (62,63).
Circ threonine aspartase 1 (circTAP1) up-regulates high mobility group AT-hook fold 2 (HMGA2). circTAP1(Figure 2A) was found up-regulated in AML patients and cells (64). Its knockdown in HL-60 and THP-1 AML cells inhibited proliferation and induced apoptosis in vitro. It sponged miR-515-5p and up-regulated HMGA2. Knockdown of circTAP1 inhibited growth of HL-60 AML xenografts after subcutaneous implantation into nude mice. HMGA2 is a member of the high mobility group proteins, which are critical regulators in cancer development. HMGA2 binds to AT-rich regions of DNA, increases proliferation by promoting cell-cycle entry and inhibits apoptosis. HMGA2 mediates epithelial mesenchymal transition (EMT), MAPK/extracellularly regulated kinase (ERK), TGFβ/SMAD, PI3K/AKT, nuclear factor ĸB (NFĸB) and STAT signaling (65). It has been shown that high expression of HMGA2 in AML patients correlates with poor outcome (66). Amplified HMGA2 can promote growth of AML cells by regulating the AKT pathway (67). Furthermore, it has been shown that HMGA2 can promote AML progression by activating WNT/β-catenin signaling (68).
Down-regulated circRNAs With In Vivo Efficacy in Preclinical Models
circ0040823 up-regulates phosphatase and tensin homolog (PTEN). circ0040823 (Figure 2B) was down-regulated in AML patient leukemic cells, inhibited AML cell proliferation and induced apoptosis and cell-cycle arrest (69). It sponged miR-516 resulting in up-regulation of PTEN. circ0040823 suppressed the growth of AML-xenograft tumors in nude mice. PTEN acts as a tumor suppressor with phosphatase-dependent and -independent (scaffold) activities affecting survival, migration, proliferation, and metabolism of tumor cells (70). PTEN can be down-regulated in AML (71) and PTEN deletion drives AML resistance to mitogen-activated protein kinase kinase (MEK) inhibitors (72).
circ0059707 up-regulates miR-1287-5p. Down-regulation of circ0059707 (Figure 2B) predicted a poor prognosis in AML patients (73). circ0059707 inhibited growth of K562 and THP1 AML cells by up-regulating miR-1287-5p in vitro and growth of K562 xenografts in nude mice after subcutaneous implantation. The mode of action was not resolved in further detail.
Up-regulated Circular RNAs With In Vitro Efficacy in AML-related Cellular Systems. Circular RNAs Targeting Transmembrane Proteins
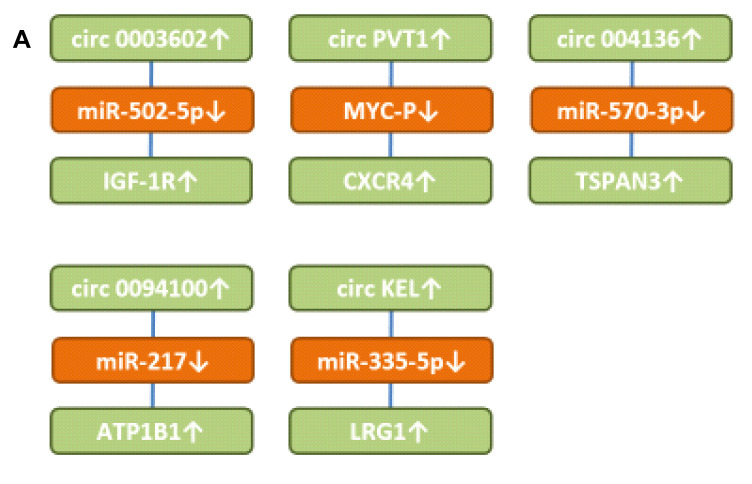
circ0003602 up-regulates insulin-like growth factor 1 receptor (IGF-1R). circ0003602 (Figure 3A) was up-regulated in AML patients and predicted poor prognosis (74). Knockdown of circ0003602 induced inhibition of proliferation, migration, invasion and caused apoptosis. circ0003602 sequestered miR-502-5p resulting in up-regulation of IGF-1R. It was shown that sumoylation of the IGF-1R promoted proliferation of AML cells (75). An IGF-1R inhibitor has been shown to suppress cell survival and resistance to chemotherapy in AML cells (76). Targeting IGF-1R has yielded disappointing results in Phase III studies in several types of cancer (77,78).
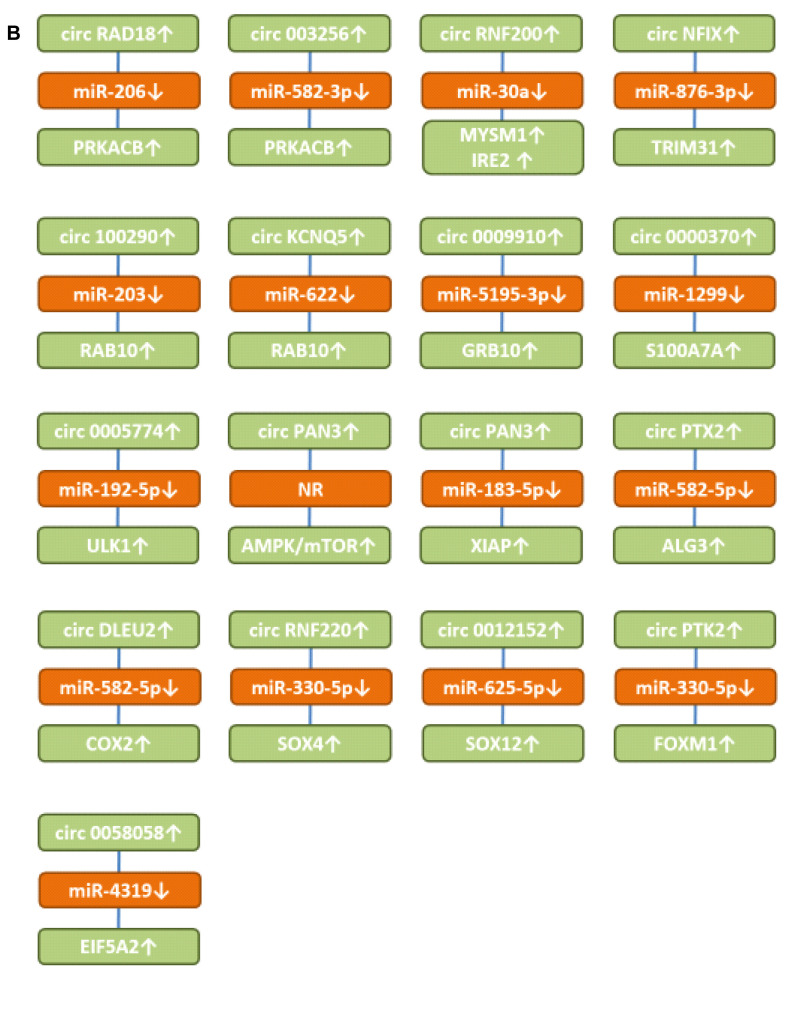
Figure 3. Up-regulated circRNAs with in vitro efficacy in preclinical AML-related cellular systems. A) Circular RNAs targeting transmembrane receptors and secreted proteins. B) Circular RNAs involved in signaling and up-regulating transcription factors. ALG3: α-1,3 mannosyltransferase; AMPK: AMP-activated protein kinase; circATP1B1: circ Na, K ATPase β subunit1; circDLEU2: circ deleted in lymphocytic leukemia 2; circKCNQ5: circ potassium voltage-gated KQT member; circKEL: Circ KELL blood group locus; circPAN3: circ pan domain 3; circNFIX: circ nuclear factor IX; circPTK2: circ protein tyrosine kinase 2; circPVT1: circ plasmacytoma variant translocation 1; circRAD18: circ E3 ubiquitin-protein ligase 18; circRNF220: circ ring finger 220; COX2: cyclooxygenase 2; CXCR4: CXC motif chemokine receptor 4; EIF5A2: eukaryotic translation initiation factor 5A2; FOXM1: forkhead box M1; GRB10: growth factor receptor bound protein 10; IGF-1R: insulin growth factor 1 receptor; IRES2: immediate early response 2; LRG1: leucine rich α2-glycoprotein; miR: microRNA; mTOR: mammalian target of rapamycin; NR: not resolved; MYSMI: deubiquitinase MYSMI; RAB10: ras-related protein 10; PRKACB: cAMP-dependent protein kinase A catalytic subunit β; S100A7A: S100 protein S100A7; SOX4 and SOX12: sex-determining region 4 or 12 related high mobility group; TSPAN3: tetraspanin 3; ULK1: unc 51-like kinase; TRIM31: tripartite motif 31; XIAP: X-linked inhibitor of apoptosis.
Circ plasmacytoma variant translocation 1 (circPVT1) up-regulates CXC motif chemokine receptor 4 (CXCR4). circPVT1 (Figure 3A) was up-regulated in AML patients and correlated with poor prognosis (79). In THP-1 AML cells, knockdown of circPVT1 inhibited cell viability, migration, and induced apoptosis. circPVT1 stabilized MYC protein by decreasing phosphorylation of Thr 58 to prevent its degradation and up-regulated its downstream effector CXCR4 (79). The latter and its ligand chemokine ligand 12 (CXCL12) play a key role in survival and migration of normal and malignant stem cells in the bone marrow (80). CXCR4 is a driver of pathogenesis of AML and high expression of AML predicts a poor prognosis (81,82). Small molecules, peptides, monoclonal antibodies (mAbs) and mAb-drug conjugates have been developed to target CXCR4 expressing malignant cells (83).
circ004136 up-regulates tetraspanin 3 (TSPAN3). circ004136 (Figure 3A) knockdown hampered cell viability, cell-cycle progression, migration, invasion and promoted cell apoptosis (84). It up-regulated TSPAN3 by sponging miR-570-3p. TSPAN3 is required for development and progression of AML (85). TSPAN3 deficiency disables responses to CXCL12/stromal cell derived factor 1 (SDF1) and leads to defects of AML localization within the bone marrow niche (85). TSPAN3 is a target of the RNA binding protein Musashi2, which is a predictor of poor outcome in AML patients and plays an important role in the pathogenesis of AML (86-88).
circ0094100 up-regulates Na, K-ATPase β subunit1 (ATP1B1). circ0094100 (Figure 3A) was elevated in AML tissues and cells (89). circ0094100 knockdown inhibited AML cell viability and cell-cycle progression and promoted apoptosis. It sponged miR-217 and up-regulated ATP1B1. Over-expression of transmembrane protein ATP1B1 predicts an adverse prognosis in cytogenetically normal AML patients (90). ATP1B1 encodes the Na, K ATPase β subunit, a key regulator of Na+ and K+ electrochemical gradients across the plasma membrane and an essential regulator of cellular activity. Its impairment causes apoptosis (91,92).
Circ Kell blood group locus (circKEL) up-regulates leucine-rich-alpha2-glycoprotein 1 (LRG1). circKEL (Figure 3A) was elevated in patients with AML compared to healthy controls and correlated with worse overall survival (93). It sponged miR-335-5p and up-regulated LRG1. The latter is a secreted glycoprotein which inhibits apoptosis in Kasumi leukemia cells (94). Down-regulation of LRG1 inhibits cell-cycle related proteins and JAK/STAT signaling in Kasumi cells (94). LRG1 promotes angiogenesis by binding to accessory receptor endoglin and mediates signaling through the anaplastic lymphoma kinase 1 (ALK1)/Smad 1,5,8 pathway by modulating transforming growth factor β (TFGβ) (95). LRG1 also functions as a mediator of vascular niche formation in metastasis (96). An antibody-drug conjugate directed against LRG1 has been constructed and is presently evaluated as an anticancer agent (97).
Up-regulated Circular RNA Regulating Signaling-related Targets in AML-related Cellular Systems In Vitro
Circ E3 ubiquitin-protein ligase RAD18 (circRAD18) up-regulates cyclic AMP-dependent protein kinase catalytic subunit β (PRKACB). circRAD18 (Figure 3B) was over-expressed in AML patients and AML cell lines (98). In THP1 and HL60 AML cells knockdown of circ RAD18 inhibited cell-cycle progression, migration and invasion and facilitated apoptosis. circRAD18 sponged miR-206 resulting in up-regulation of PRKACB. Over-expression of miR-206 repressed cell-cycle repression of AML cells by binding to PRKACB.
circ0003256 up-regulates cyclic AMP-dependent protein kinase catalytic subunit β (PRKACB). circ0003256 (Figure 3B) was highly expressed in pediatric AML patients and cells (99). Suppression of circ0003256 hindered proliferation and induced apoptosis in THP1 and MV4-11 AML cells. circ0003256 sponged miR-582-3p resulting in up-regulation of PRKACB.
circRNF220 up-regulates deubiquitinase MYSMI and immediate early response 2 (IRE2). circRNF200 (Figure 3B) was highly expressed in pediatric AML patients and was an independent and reliable predictor of relapse in these patients (100). In THP1 and HL60 AML cells, circRNF220 enhanced proliferation and impaired apoptosis. It sponged miR-30a and up-regulated MYSMI and IRE2. MYSMI functions as a regulator of hematopoietic stem cell function, blood cell production, immune response and plays an important role in myeloid cell development (101). IRE2 promotes tumor cell motility and metastasis and predicts poor survival in CRC patients (102). In HCC, IRE2 regulates cell adhesion and motility via integrin β1-mediated signaling pathway (103) and the activity of RHO GTPases (104).
Circ nuclear factor IX (circNFIX) up-regulates tripartite motif 31 (TRIM31). circNFIX (Figure 3B) was found up-regulated in AML tissues and cell lines (105). Knockdown of circNFIX in HL60 and MOLM13 AML cells restrained proliferation and induced apoptosis in vitro. circNFIX sponged miR-876-3p and up-regulated TRIM31. Silencing of miR-876-3p attenuated the effect of NFIX depletion on the progression of AML cells. Ectopic expression of TRIM31 rescued the effect of circNFIX silencing on AML cell proliferation. TRIM31 plays a dual role in progression and suppression of cancer (106). It acts as an E3-ubiquitin ligase which can regulate p53, PI3K-AKT, NFĸB and WNT/β catenin pathways (106). The TRIM family contributes to tumorigenesis, cancer development and drug resistance (107). It has been shown that TRIM31 promotes AML progression through WNT/β catenin signaling (108). In breast cancer, TRIM31 mediates tumor progression through regulation of ubiquitinylation of p53 (109).
circ100290 up-regulates ras-related protein 10 (RAB10). circ100290 (Figure 3B) was found increased in AML samples and cell lines (110). Its down-regulation suppressed proliferation and induced cell-cycle arrest and apoptosis of AML cells. circ100290 sponged miR-203 resulting in up-regulation of RAB10 (111). A miR-203 inhibitor could reverse the effect of circ100290 knockdown on proliferation and apoptosis in AML cells.
Circ potassium voltage-gated subfamily KQT member 5 (circKCNQ5) up-regulates ras-related protein 10 (RAB10). circKCNQ5 (Figure 3B) was increased in bone marrow samples of childhood AML and AML cell lines (112). Its knockdown suppressed AML cell proliferation and promoted apoptosis. circKCNQ5 sponged miR-622 and up-regulated RAB10. RAB GTPases act as coordinators of vesicle traffic (111,113). RAB10 can be found in the endoplasmic reticulum, Golgi apparatus, and endosomes/phagosomes. It is involved in endosomal sorting, exocytosis, axonal growth in neurons and phagocytosis in macrophages (114). In gastric cancer, RAB10 has been shown to regulate apoptosis and autophagy (115).
circ0009910 up-regulates growth factor receptor bound protein 10 (GRB10). circ0009910 (Figure 3B) was up-regulated in AML bone marrow and AML cell lines (116). In HL-60 and MOLM-13 AML cells, blocking of circ0009910 with siRNA suppressed proliferation, cell-cycle progression and facilitated apoptosis. It sponged miR-5195-3p and up-regulated GRB10. The latter is an adapter protein containing a pleckstrin homology domain as well as a src-homology 2 (SH2) domain. It plays a role in insulin-growth factor 1 mediated signaling and ligand-induced ubiquitination, internalization, and stability of IGR-1R (117-119). GRB10 is involved in autocrine proliferation of AML cells via the insulin-growth factor (IGF)/IGF-1R pathway (120).
circ0000370 up-regulates S100 protein A7A (S100A7A). circ0000370 (Figure 3B) was found increased in FLT3-ITD+ AML patients (121). Its knockdown decreased viability of MV4-11 and MOLM-14 AML cells. circ0000370 over-expression inhibited apoptosis in U937 and H60 AML cells. It sponged miR-1299 and increased S100A7A expression (121). At least twenty-four S100 proteins are known. They have two calcium binding sites and contain a helix-loop-helix. They exert intracellular functions and act as secreted proteins. In cancer, they are involved in proliferation, cytoskeleton dynamics, invasion, migration, calcium homeostasis, energy metabolism and inflammation (122). In breast cancer, S100A7A promotes epidermal growth factor (EGF) signaling (123) and increases mammary tumorigenesis through up-regulation of inflammatory pathways (124). S100A7A is markedly increased in epidermal hyperproliferation disorders, acts as a cytokine, and is involved in autoimmune conditions such as psoriasis (125).
circ0005774 up-regulates unc-51 like kinase (ULK1). circ0005774 (Figure 3B) was highly expressed in pediatric AML and AML cells (126). In HL-60 and NB4 AML cells, blocking of circ000574 enhanced apoptosis, suppressed cell viability and cell-cycle entry, and inhibited proliferation markers, such as proliferation nuclear antigen (PCNA). It sponged miR-192-5p and up-regulated ULK1. The latter acts as a ser/thr kinase that participates in the initiation of autophagy (127). Autophagy represents an essential degradation process in the cellular response to stress and its inhibition is evaluated as a therapy for AML and FLT3-ITD AML (128-130).
Circ pan domain protein 3 (circPAN3) up-regulates mammalian target of rapamycin (mTOR) signaling. circPAN3 (Figure 3B) was increased in Adriamycin (ADM)-resistant AML cell lines and bone marrow samples from relapsed patients (131). Down-regulation of circPAN3 restored ADM sensitivity in ADM-resistant cell lines K562 and THP-1, whereas lentivirus-mediated over-expression of circPAN3 had the opposite effect. circPAN3 facilitates drug resistance through autophagy and influences regulation of apoptosis-related proteins via the AMPK/mTOR pathway. Down-regulation of adenosine-monophosphate (AMP)-activated protein kinase (AMPK) stimulated mTOR (131,132). The mechanistic underpinnings were not resolved in further detail. It has been shown that AMPK enhances chemotherapy effects in AML (133). The PI3K/AKT/mTOR pathway is constitutively activated in AML patients and drugs which inhibit mTOR and activate AMPK promote differentiation and block blast proliferation in AML (134,135). Disappointing clinical results with inhibitors of this pathway have been obtained with monotherapy and in combination with induction therapy. Combination with other agents will be an important issue for future studies.
Circ pan domain protein 3 (circPAN3) up-regulates X-linked inhibitor of apoptosis (XIAP). circPAN3 (Figure 3B) conferred ADM-resistance of THP-1 AML cells by sponging miR-183-5p and subsequent up-regulation of XIAP (136). The latter belongs to the family of inhibitor of apoptosis proteins (IAPs), binds and inhibits caspases 3, 7 and 9 (137,138). It has been shown that XIAP inhibitors induce differentiation and impair clonogenic capacity of AML stem cells (139). AML patients with over-expression of both survivin and XIAP showed unfavorable response to induction therapy in 100% of the patients and short survival (30 days) (140). Combined inhibition of XIAP and BCL2 gave rise to maximal therapeutic efficacy in AML patients (141).
Circ protein tyrosine kinase 2 (PTK2) up-regulates α(1-3)-mannosyltransferase (ALG3). circPTK2 (Figure 3B) was found over-expressed in AML peripheral blood samples and cell lines (142). Its knockdown restrained proliferation and glycolysis of AML cells. circPTK2 sponged miR-582-3p, resulting in up-regulation of ALG3. Glycosyltransferases have an impact on malignancy in diverse experimental models and are potential prognostic biomarkers (143). ALG3 is involved in the first steps of N-glycosylation and contributes to the malignancy of NSCLC (144) and oral squamous cell carcinoma by regulating the CDK-cyclin pathway (145). In AML, the function of ALG3 remains to be explored in further detail.
Circ deleted in lymphocytic leukemia 2 (circDLEU2) up-regulates cyclo-oxygenase 2 (COX2). circDLEU2 (Figure 3B) was up-regulated in AML marrow samples and cells (146). In HL-60 and THP1 AML cells, knockdown of circDLEU2 repressed proliferation, whereas it induced apoptosis and cell arrest in G0/G1 phase. circDLEU2 acted as a sponge for miR-582-5p and up-regulated COX2. In cancer, COX2 can induce cancer stem cell-like activity, apoptosis resistance, angiogenesis, inflammation, and metastasis through its metabolite prostaglandin E2 (147). COX2 inhibitor celecoxib exerted antitumor effects in HL60 AML cells and inhibited autophagy by affecting lysosome function (148,149). Combination of celecoxib and doxorubicin increased growth inhibition and apoptosis in AML cells (149). Furthermore, a celecoxib derivative inhibited focal adhesion and induces caspase-dependent apoptosis in AML cells (150). Vascular endothelial growth factor (VEGF) has been shown to modulate proliferation and chemoresistance of AML cells through endothelin-1 dependent induction of COX2 (151).
Circ ring finger 200 (circRNF200) up-regulates sex-determining region (SRY)-related high mobility group box4 (SOX4). circRNF200 (Figure 3B) was found to be highly expressed in AML patients (152). In HL-60 and THP-1 AML cells, knockdown of circRNF200 inhibited cell growth, cell-cycle progression, invasion, and glycolytic metabolism in vitro. circRNF200 sponged miR-330-5p resulting in up-regulation of SOX4. Knockdown of circRNF200 down-regulated SOX4 expression by increasing miR-330-5p. Inhibition of miR-330-5p neutralized the impairment of circRNF200 in AML cell development. SOX proteins are a family of transcription factors with more than twenty members with functions in development, cell-fate decision, differentiation, tumor formation and metastasis (153). Over-expression of SOX4 correlated with poor prognosis in AML patients (154). It has been shown that SOX4 induces proliferation of AML cells (155). In AML, SOX4 is a direct target and crucial mediator of transcription factor C/EBP (156). SOX4 also promotes progression of AML by inducing RHO GTPase activating protein 9 (ARHGAP9) (157).
circ0012152 up-regulates sex-determining region Y-related high mobility group box 12 (SOX12). circ0012152 (Figure 3B) was found increased in AML tissues and cells (158). Its knockdown suppressed proliferation and promoted cell death in AML cells. circ0012152 inhibited miR-625-5p expression and up-regulated SOX12. The latter was preferentially expressed in CD34 cells in AML and promoted expression of β-catenin and WNT signaling (159). In K61 and MO7e AML cells, knockdown of SOX12 decreased transcription factor TWIST1 and β-catenin (160). SOX12 has been identified as a promoter of AML progression (161).
circPTK2 up-regulates forkhead box protein M1 (FOXM1). circPTK2 (Figure 3B) was highly expressed in AML patients (162). Interference with circPTK2 suppressed proliferation and induced apoptosis in AML cells. circPTK2 sponged miR-330-5p and up-regulated FOXM1. Si-circPTK2 mediated inhibition of malignant behavior of AML cells and was partly counteracted by addition of anti-miR-330-5p. FOXM1 is a proliferation-associated transcription factor expressed during the cell-cycle and involved in self-renewal and tumorigenesis (163,164). FOXM1 promotes proliferation of AML cells through modulation of cell-cycle progression (165). Over-expression of FOXM1 is associated with adverse prognosis in FLT3-ITD AML (166). Blast cell survivors after induction therapy correlated with a signature of FOXM1 activity (167). Furthermore, it has been shown that nuclear FOXM1 drives chemo-resistance of AML (168). NPM1 binds to FOXM1, and this interaction is required for the level and nuclear localization of FOXM1 (169,170).
circ0058058 up-regulates eucaryotic translation initiation factor 5A2 (EIF5A2). circ0058058 (Figure 3B) was up-regulated in AML patients and its knockdown inhibited proliferation, migration, and invasion, but accelerated apoptosis (171). It sponged miR-4319 and up-regulated EIF5A2. The latter acts as a translation initiation factor and is also involved in promoting translation of reiterated proline sequences (172). It plays an important role in tumor growth, invasion, metastasis and in the tumor microenvironment (173). EIF5A emerges as two isoforms, EIF5A1 and EIF5A2. EIF5A1 is constitutively expressed, EIF5A2 is found in a few tissues, highly expressed in several types of cancer, and is considered as a possible oncogene. Its function depends on post-translational modification of one of the two lysine residues of EIF5A2, creating amino acid hypusine, making it susceptible to the modification pathway (173). In AML, it has been shown that miR-9 and miR-33b increase sensitivity to Daunorubicin by targeting EIF5A2 (174).
Down-regulated Circular RNAs With In Vitro Efficacy in AML-related Cellular Systems
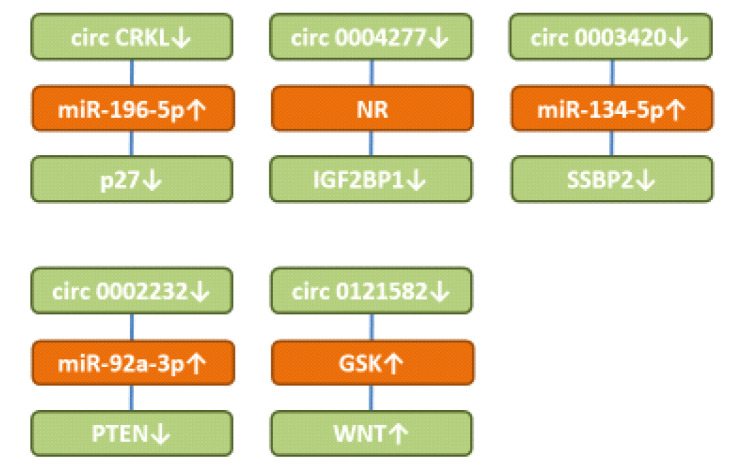
circCKRL up-regulates protein p27. circCKRL (Figure 4) was found down-regulated in AML samples and cell lines (175). circCRKL suppressed cell proliferation in THP-1 and MOLM13 ALM cells. It sponged miR-196-5a and miR-196-5b resulting in up-regulation of p27. The latter is a member of the kinase inhibitory family (KIP) and acts as a CDK inhibitor that mediates cell-cycle inhibition (176). It has been shown that AML patients with high level of p27 had a significantly increased disease-free survival (177). It has been observed that the KIP cell cycle pathway can be inactivated epigenetically in AML patients (178). FLT3 and FLT3-ITD can inhibit p27 by phosphorylation in AML patients (179).
Figure 4. Down-regulated circular RNA with activity in preclinical AML-related cellular systems. circCRKL: circ CRK-like protein; GSK-3β: glycogen synthase kinase 3β; IGF2BP1: insulin-like growth factor mRNA binding protein 1; miR: microRNA; p27: protein p27; PTEN: phosphatase and tensin homolog; SSBP2: single-stranded DNA binding protein 2; WNT: WNT signaling.
circ0003420 suppresses expression of insulin-like growth factor mRNA binding protein 1 (IGF2BP1). Expression of circ0003420 (Figure 4) correlated with poor clinical results and impaired therapeutic effects in AML (180). Its down-regulation leads to up-regulation of IGF2BP1. Restoration of IGF2BP1 diminished the effects of circ 0003420 on replication, apoptosis, and the leukemic stem cell phenotype of KG-1a AML cells. IGF2BP1 has six canonical RNA binding sites and acts as a fine tuner of expression of genes involved in proliferation, growth and chemoresistance (181). It is regarded as an oncogene, which can orchestrate stem cell properties. However, tumor-suppressive roles of this protein have also been observed (181). IGF2BP1 maintains leukemia cell properties by regulating genes, such as homeobox B4 (HOXB4), proto-oncogene myeloblastosis (MYB) and alcohol dehydrogenase 1A1 (ALDH1A1) (182). IGF2BP1 also has been shown to mediate the tumorigenic functions of LIN288 in AML cells (183).
circ0004277 down-regulates single-stranded DNA binding protein 2 (SSBP2). circ0004277 (Figure 4) was found to be reduced in AML patients (184). It inhibited viability, migration, and invasion of U937 and KG-1a AML cells. circ0004237 absorbs miR-134-5p and up-regulates SSBP2. The latter interacts with single-stranded DNA, mediating DNA damage response and maintenance of genomic stability and acts as a tumor suppressor gene in AML (184). circ0004277 is frequently deleted in AML and its restoration leads to cell-cycle arrest in AML cell lines (184). Loss of nuclear expression of SSBP2 is associated with poor prognostic factors in colorectal cancer (185). It has been observed that the SSBP2 gene is frequently deleted in prostate cancer (186).
circ0121582 inhibits glycogen synthase kinase 3β (GSK3β). circ0121582 (Figure 4) was down-regulated in AML (187). It sponged miR-224 resulting in up-regulation of GSK3β. It also bound to the GSK promoter in the nucleus where it recruited the DNA methylase TET1 to inhibit its transcription. GSKβ1 inhibits WNT/β catenin signaling by phosphorylation of GSK3β, which is subsequently ubiquitinated by βTrCP and targeted for proteosomal degradation (188). WNT signaling is required for the development of leukemic stem cells in AML (189-191). In addition, it has been shown that WNT antagonists can be epigenetically inactivated in AML (192). Inhibition of WNT signaling is pursued as a strategy to eliminate AML cells (193).
Technical issues. Identification of up- and down-regulated circRNAs allows identification of potential targets and new ways of therapeutic intervention in patients with AML by manipulating the expression levels of the corresponding circRNA. The first step of drug development is the evaluation of the identified targets and circRNA in standard preclinical in vitro and in vivo models. Up-regulated circRNAs reveal targets, which can be inhibited by small molecules, antibody-derived entities, or other types of intervention. circRNA by itself can be inhibited with siRNA or shRNA or antisense oligos (194,195). Down-regulated circRNAs reveal targets, which functionally can be reconstituted with small molecules. This approach is hampered by druggability issues and the specificity of the identified compounds. The other option is reconstitution of circRNA expression by transduction of plasmid- or retrovirus-based expression vectors into corresponding recipient cells (196,197).
Regarding manipulation of up-regulated circRNAs with siRNA or shRNA, specificity issues, immunogenicity, and optimization of pharmacokinetic and pharmacodynamic properties are critical parameters. These issues will not be discussed in further detail in this review. In case of hematological malignancies, delivery of siRNA or shRNA is facilitated by easier accessibility in comparison to solid tumors. Several lipid-based delivery systems for these agents have been developed in the past few years (198-200). Also, the targeting of lipid nanoparticles (LNP) to specific cells is a critical issue (200). The first clinically approved product (siRNA-LNP) in this field was Onpattro for the treatment of transerythrin amyloidosis (201).
Regarding identified targets, such as transcription factors, druggability issues might arise (202). The new compound class of proteolysis targeting chimeras (PROTACS) might lead to unprecedented breakthroughs. PROTACS are divalent compounds composed of a targeting moiety, a linker and an E3 ligase binding entity leading to proteosomal degradation of the corresponding target (203-205). In addition to undruggable targets, resistance mutations, protein aggregates, protein isoforms, protein scaffolds and over-expressed proteins in general are targets for PROTACS (205). At least twenty clinical degraders have been approved for clinical investigation, and fourteen are intended for oral administration (205).
Conclusion
The vast majority of the identified circRNAs act by sponging miRs. In four cases, the MOA is not resolved. In addition, circMYBL2 acts by recruiting PTBP1 (Figure 2A) and circ0121582 sponges miR-224 and binds to the promoter of GSK3β (Figure 4). Three different circRNAs have PRKCAB as a target (Figure 2A and Figure 3B), RAB10 was up-regulated by two different circRNAs (Figure 3B). Furthermore, we observed that circPAN3 targets mTOR as well as XIAP (Figure 3A). CircDLEU2 up-regulates PRKCAB as well as COX2. Inhibition of miR-330-5p by two different circRNAs up-regulates SOX4 as well as FoxM1 (Figure 3B), whereas miR-582-3p inhibits expression of YWHAZ and ALG3 (Figure 2A and Figure 3B).
mutFLT3 was revealed as a clinically validated target of up-regulated cirRNA MYBL2 with efficacy in preclinical in vivo models (Figure 2A). In addition, transmembrane proteins KIT and PLXNB2, enzymes PRKCAB and SCD, adapter protein YHWA, transcription factor RUNX31 and multiple function protein HMGA2 were identified as targets of up-regulated circRNAs showing efficacy in preclinical in vivo models (Figure 2A). PTEN has emerged as a target for reconstitution therapy (Figure 2B).
Transmembrane proteins IGFR1, CXCR4, TSPAN3, ATP1B1 and secreted protein LRG1 (Figure 3A) have been identified as targets of up-regulated circRNA with in vitro, but pending in vivo activity in preclinical systems. Furthermore, fifteen up-regulated targets have been identified (Figure 3B). In addition to miR-4961 (Figure 2A), miR-206 and -582-3p target PRKACB (Figure 3B). RAB10 was identified as a target of two circRNAs (Figure 3B). Enzymes, such as ULK1, mTOR, ALG3 and COX2 as well as transcription factors SOX4, SOX12 and FOXM1 have been identified as potential targets for treatment of AML. As targets for replacement therapy, p27, SSBP2 and IGF2BP1 and their corresponding down-regulated circRNAs have been revealed (Figure 4).
Ranking of the identified circRNAs and their corresponding targets with respect to treatment of AML will depend on the outcome of further target validation experiments.
Conflicts of Interest
AN is and UHW was an employee of Roche.
Authors’ Contributions
AN and UHW contributed equally to all aspects of the review.
References
- 1.Papaemmanuil E, Gerstung M, Bullinger L, Gaidzik VI, Paschka P, Roberts ND, Potter NE, Heuser M, Thol F, Bolli N, Gundem G, Van Loo P, Martincorena I, Ganly P, Mudie L, McLaren S, O'Meara S, Raine K, Jones DR, Teague JW, Butler AP, Greaves MF, Ganser A, Döhner K, Schlenk RF, Döhner H, Campbell PJ. Genomic classification and prognosis in acute myeloid leukemia. N Engl J Med. 2016;374(23):2209–2221. doi: 10.1056/NEJMoa1516192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jamal M, Song T, Chen B, Faisal M, Hong Z, Xie T, Wu Y, Pan S, Yin Q, Shao L, Zhang Q. Recent progress on circular RNA research in acute myeloid leukemia. Front Oncol. 2019;9:1108. doi: 10.3389/fonc.2019.01108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Halik A, Arends CM, Bullinger L, Damm F, Frick M. Refining AML treatment: the role of genetics in response and resistance evaluation to new agents. Cancers (Basel) 2022;14(7):1689. doi: 10.3390/cancers14071689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Liu H. Emerging agents and regimens for AML. J Hematol Oncol. 2021;14(1):49. doi: 10.1186/s13045-021-01062-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ravandi F, Stone R. Acute promyelocytic leukemia: a perspective. Clin Lymphoma Myeloma Leuk. 2017;17(9):543–544. doi: 10.1016/j.clml.2017.06.037. [DOI] [PubMed] [Google Scholar]
- 6.Döhner K, Döhner H. Molecular characterization of acute myeloid leukemia. Haematologica. 2008;93(7):976–982. doi: 10.3324/haematol.13345. [DOI] [PubMed] [Google Scholar]
- 7.Döhner H, Wei AH, Löwenberg B. Towards precision medicine for AML. Nat Rev Clin Oncol. 2021;18(9):577–590. doi: 10.1038/s41571-021-00509-w. [DOI] [PubMed] [Google Scholar]
- 8.Bazinet A, Kantarjian H. Moving toward individualized target-based therapies in acute myeloid leukemia. Ann Oncol. 2023;34(2):141–151. doi: 10.1016/j.annonc.2022.11.004. [DOI] [PubMed] [Google Scholar]
- 9.Bouligny IM, Maher KR, Grant S. Mechanisms of myeloid leukemogenesis: Current perspectives and therapeutic objectives. Blood Rev. 2023;57:100996. doi: 10.1016/j.blre.2022.100996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kayser S, Levis MJ. The clinical impact of the molecular landscape of acute myeloid leukemia. Haematologica. 2023;108(2):308–320. doi: 10.3324/haematol.2022.280801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Perner F, Stein EM, Wenge DV, Singh S, Kim J, Apazidis A, Rahnamoun H, Anand D, Marinaccio C, Hatton C, Wen Y, Stone RM, Schaller D, Mowla S, Xiao W, Gamlen HA, Stonestrom AJ, Persaud S, Ener E, Cutler JA, Doench JG, McGeehan GM, Volkamer A, Chodera JD, Nowak RP, Fischer ES, Levine RL, Armstrong SA, Cai SF. MEN1 mutations mediate clinical resistance to menin inhibition. Nature. 2023;615(7954):913–919. doi: 10.1038/s41586-023-05755-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Muz B, Abdelghafer A, Markovic M, Yavner J, Melam A, Salama NN, Azab AK. Targeting E-selectin to tackle cancer using uproleselan. Cancers (Basel) 2021;13(2):335. doi: 10.3390/cancers13020335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.DeAngelo DJ, Jonas BA, Liesveld JL, Bixby DL, Advani AS, Marlton P, Magnani JL, Thackray HM, Feldman EJ, O'Dwyer ME, Becker PS. Phase 1/2 study of uproleselan added to chemotherapy in patients with relapsed or refractory acute myeloid leukemia. Blood. 2022;139(8):1135–1146. doi: 10.1182/blood.2021010721. [DOI] [PubMed] [Google Scholar]
- 14.Marvin-Peek J, Savani BN, Olalekan OO, Dholaria B. Challenges and advances in chimeric antigen receptor therapy for acute myeloid leukemia. Cancers (Basel) 2022;14(3):497. doi: 10.3390/cancers14030497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gottschlich A, Thomas M, Grünmeier R, Lesch S, Rohrbacher L, Igl V, Briukhovetska D, Benmebarek MR, Vick B, Dede S, Müller K, Xu T, Dhoqina D, Märkl F, Robinson S, Sendelhofert A, Schulz H, Umut Ö, Kavaka V, Tsiverioti CA, Carlini E, Nandi S, Strzalkowski T, Lorenzini T, Stock S, Müller PJ, Dörr J, Seifert M, Cadilha BL, Brabenec R, Röder N, Rataj F, Nüesch M, Modemann F, Wellbrock J, Fiedler W, Kellner C, Beltrán E, Herold T, Paquet D, Jeremias I, von Baumgarten L, Endres S, Subklewe M, Marr C, Kobold S. Single-cell transcriptomic atlas-guided development of CAR-T cells for the treatment of acute myeloid leukemia. Nat Biotechnol. 2023;41(11):1618–1632. doi: 10.1038/s41587-023-01684-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kristensen LS, Hansen TB, Venø MT, Kjems J. Circular RNAs in cancer: opportunities and challenges in the field. Oncogene. 2018;37(5):555–565. doi: 10.1038/onc.2017.361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kristensen LS, Jakobsen T, Hager H, Kjems J. The emerging roles of circRNAs in cancer and oncology. Nat Rev Clin Oncol. 2022;19(3):188–206. doi: 10.1038/s41571-021-00585-y. [DOI] [PubMed] [Google Scholar]
- 18.Sanger HL, Klotz G, Riesner D, Gross HJ, Kleinschmidt AK. Viroids are single-stranded covalently closed circular RNA molecules existing as highly base-paired rod-like structures. Proc Natl Acad Sci USA. 1976;73(11):3852–3856. doi: 10.1073/pnas.73.11.3852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hsu M, Coca-Prados M. Electron microscopic evidence for the circular form of RNA in the cytoplasm of eukaryotic cells. Nature. 1979;280(5720):339–340. doi: 10.1038/280339a0. [DOI] [PubMed] [Google Scholar]
- 20.Hansen TB, Jensen TI, Clausen BH, Bramsen JB, Finsen B, Damgaard CK, Kjems J. Natural RNA circles function as efficient microRNA sponges. Nature. 2013;495(7441):384–388. doi: 10.1038/nature11993. [DOI] [PubMed] [Google Scholar]
- 21.Hansen TB, Kjems J, Damgaard CK. Circular RNA and miR-7 in cancer. Cancer Res. 2013;73(18):5609–5612. doi: 10.1158/0008-5472.CAN-13-1568. [DOI] [PubMed] [Google Scholar]
- 22.Piwecka M, Glažar P, Hernandez-Miranda LR, Memczak S, Wolf SA, Rybak-Wolf A, Filipchyk A, Klironomos F, Cerda Jara CA, Fenske P, Trimbuch T, Zywitza V, Plass M, Schreyer L, Ayoub S, Kocks C, Kühn R, Rosenmund C, Birchmeier C, Rajewsky N. Loss of a mammalian circular RNA locus causes miRNA deregulation and affects brain function. Science. 2017;357(6357):eaam8526. doi: 10.1126/science.aam8526. [DOI] [PubMed] [Google Scholar]
- 23.Vo JN, Cieslik M, Zhang Y, Shukla S, Xiao L, Zhang Y, Wu YM, Dhanasekaran SM, Engelke CG, Cao X, Robinson DR, Nesvizhskii AI, Chinnaiyan AM. The landscape of circular RNA in cancer. Cell. 2019;176(4):869–881.e13. doi: 10.1016/j.cell.2018.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhou WY, Cai ZR, Liu J, Wang DS, Ju HQ, Xu RH. Circular RNA: metabolism, functions and interactions with proteins. Mol Cancer. 2020;19(1):172. doi: 10.1186/s12943-020-01286-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Prats AC, David F, Diallo LH, Roussel E, Tatin F, Garmy-Susini B, Lacazette E. Circular RNA, the key for translation. Int J Mol Sci. 2020;21(22):8591. doi: 10.3390/ijms21228591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li J, Sun D, Pu W, Wang J, Peng Y. Circular RNAs in cancer: Biogenesis, function, and clinical significance. Trends Cancer. 2020;6(4):319–336. doi: 10.1016/j.trecan.2020.01.012. [DOI] [PubMed] [Google Scholar]
- 27.Singh V, Uddin MH, Zonder JA, Azmi AS, Balasubramanian SK. Circular RNAs in acute myeloid leukemia. Mol Cancer. 2021;20(1):149. doi: 10.1186/s12943-021-01446-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhou M, Gao X, Zheng X, Luo J. Functions and clinical significance of circular RNAs in acute myeloid leukemia. Front Pharmacol. 2022;13:1010579. doi: 10.3389/fphar.2022.1010579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sun YM, Wang WT, Zeng ZC, Chen TQ, Han C, Pan Q, Huang W, Fang K, Sun LY, Zhou YF, Luo XQ, Luo C, Du X, Chen YQ. circMYBL2, a circRNA from MYBL2, regulates FLT3 translation by recruiting PTBP1 to promote FLT3-ITD AML progression. Blood. 2019;134(18):1533–1546. doi: 10.1182/blood.2019000802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang QX, Pan YM, Xiao HL, An N, Deng SS, Du X. Alternative splicing analysis showed the splicing factor polypyrimidine tract-binding protein 1 as a potential target in acute myeloid leukemia therapy. Neoplasma. 2022;69(05):1198–1208. doi: 10.4149/neo_2022_220314N279. [DOI] [PubMed] [Google Scholar]
- 31.Daver N, Schlenk RF, Russell NH, Levis MJ. Targeting FLT3 mutations in AML: review of current knowledge and evidence. Leukemia. 2019;33(2):299–312. doi: 10.1038/s41375-018-0357-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhao JC, Agarwal S, Ahmad H, Amin K, Bewersdorf JP, Zeidan AM. A review of FLT3 inhibitors in acute myeloid leukemia. Blood Rev. 2022;52:100905. doi: 10.1016/j.blre.2021.100905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wu M, Li C, Zhu X. FLT3 inhibitors in acute myeloid leukemia. J Hematol Oncol. 2018;11(1):133. doi: 10.1186/s13045-018-0675-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lin L, Wang Y, Bian S, Sun L, Guo Z, Kong D, Zhao L, Guo D, Li Q, Wu M, Wang Y, Wang Y, Li Y. A circular RNA derived from PLXNB2 as a valuable predictor of the prognosis of patients with acute myeloid leukaemia. J Transl Med. 2021;19(1):123. doi: 10.1186/s12967-021-02793-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Worzfeld T, Offermanns S. Semaphorins and plexins as therapeutic targets. Nat Rev Drug Discov. 2014;13(8):603–621. doi: 10.1038/nrd4337. [DOI] [PubMed] [Google Scholar]
- 36.Negishi M, Oinuma I, Katoh H. Plexins: axon guidance and signal transduction. Cell Mol Life Sci. 2005;62(12):1363–1371. doi: 10.1007/s00018-005-5018-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yu W, Goncalves KA, Li S, Kishikawa H, Sun G, Yang H, Vanli N, Wu Y, Jiang Y, Hu MG, Friedel RH, Hu GF. Plexin-B2 mediates physiologic and pathologic functions of angiogenin. Cell. 2017;171(4):849–864.e25. doi: 10.1016/j.cell.2017.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liang Y, Meng K, Qiu R. Circular RNA Circ_0013958 functions as a tumor promoter in ovarian cancer by regulating miR-637/PLXNB2 axis. Front Genet. 2021;12:644451. doi: 10.3389/fgene.2021.644451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liu L, Qiang X. Hsa_circ_0044907 promotes acute myeloid leukemia progression through upregulating oncogene KIT via sequestering miR-186-5p. Hematology. 2022;27(1):960–970. doi: 10.1080/16078454.2022.2113574. [DOI] [PubMed] [Google Scholar]
- 40.Roskoski R Jr. The role of small molecule Kit protein-tyrosine kinase inhibitors in the treatment of neoplastic disorders. Pharmacol Res. 2018;133:35–52. doi: 10.1016/j.phrs.2018.04.020. [DOI] [PubMed] [Google Scholar]
- 41.Katagiri S, Chi S, Minami Y, Fukushima K, Shibayama H, Hosono N, Yamauchi T, Morishita T, Kondo T, Yanada M, Yamamoto K, Kuroda J, Usuki K, Akahane D, Gotoh A. Mutated KIT tyrosine kinase as a novel molecular target in acute myeloid leukemia. Int J Mol Sci. 2022;23(9):4694. doi: 10.3390/ijms23094694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ayatollahi H, Shajiei A, Sadeghian MH, Sheikhi M, Yazdandoust E, Ghazanfarpour M, Shams SF, Shakeri S. Prognostic importance of C-KIT mutations in core binding factor acute myeloid leukemia: a systematic review. Hematol Oncol Stem Cell Ther. 2017;10(1):1–7. doi: 10.1016/j.hemonc.2016.08.005. [DOI] [PubMed] [Google Scholar]
- 43.Miettinen M, Lasota J. KIT (CD117): a review on expression in normal and neoplastic tissues, and mutations and their clinicopathologic correlation. Appl Immunohistochem Mol Morphol. 2005;13(3):205–220. doi: 10.1097/01.pai.0000173054.83414.22. [DOI] [PubMed] [Google Scholar]
- 44.Chi SG, Minami Y. Emerging targeted therapy for specific genomic abnormalities in acute myeloid leukemia. Int J Mol Sci. 2022;23(4):2362. doi: 10.3390/ijms23042362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wu DM, Wen X, Han XR, Wang S, Wang YJ, Shen M, Fan SH, Zhang ZF, Shan Q, Li MQ, Hu B, Chen GQ, Lu J, Zheng YL. Role of circular RNA DLEU2 in human acute myeloid leukemia. Mol Cell Biol. 2018;38(20):e00259–18. doi: 10.1128/MCB.00259-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Taylor SS, Wallbott M, Machal EMF, Søberg K, Ahmed F, Bruystens J, Vu L, Baker B, Wu J, Raimondi F, Ongeri EM, Herberg FW, Skålhegg BS. PKA Cβ: a forgotten catalytic subunit of cAMP-dependent protein kinase opens new windows for PKA signaling and disease pathologies. Biochem J. 2021;478(11):2101–2119. doi: 10.1042/BCJ20200867. [DOI] [PubMed] [Google Scholar]
- 47.Chen Y, Gao Y, Tian Y, Tian DL. PRKACB is downregulated in non-small cell lung cancer and exogenous PRKACB inhibits proliferation and invasion of LTEP-A2 cells. Oncol Lett. 2013;5(6):1803–1808. doi: 10.3892/ol.2013.1294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mertens JC, Rizvi S, Gores GJ. Targeting cholangiocarcinoma. Biochim Biophys Acta Mol Basis Dis. 2018;1864(4 Pt B):1454–1460. doi: 10.1016/j.bbadis.2017.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nakamura H, Arai Y, Totoki Y, Shirota T, Elzawahry A, Kato M, Hama N, Hosoda F, Urushidate T, Ohashi S, Hiraoka N, Ojima H, Shimada K, Okusaka T, Kosuge T, Miyagawa S, Shibata T. Genomic spectra of biliary tract cancer. Nat Genet. 2015;47(9):1003–1010. doi: 10.1038/ng.3375. [DOI] [PubMed] [Google Scholar]
- 50.Xue F, Li M, Liu Y, Xu C, Li H, Liu H. Circ_0035381 regulates acute myeloid leukemia development by modulating YWHAZ expression via adsorbing miR-582-3p. Biochem Genet. 2023;61(1):354–371. doi: 10.1007/s10528-022-10244-1. [DOI] [PubMed] [Google Scholar]
- 51.Gan Y, Ye F, He XX. The role of YWHAZ in cancer: A maze of opportunities and challenges. J Cancer. 2020;11(8):2252–2264. doi: 10.7150/jca.41316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Muslin AJ, Tanner JW, Allen PM, Shaw AS. Interaction of 14-3-3 with signaling proteins is mediated by the recognition of phosphoserine. Cell. 1996;84(6):889–897. doi: 10.1016/s0092-8674(00)81067-3. [DOI] [PubMed] [Google Scholar]
- 53.Su R, Gong JN, Chen MT, Song L, Shen C, Zhang XH, Yin XL, Ning HM, Liu B, Wang F, Ma YN, Zhao HL, Yu J, Zhang JW. c-Myc suppresses miR-451⊣YWTAZ/AKT axis via recruiting HDAC3 in acute myeloid leukemia. Oncotarget. 2016;7(47):77430–77443. doi: 10.18632/oncotarget.12679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Liang R, Chen XQ, Bai QX, Wang Z, Zhang T, Yang L, Dong BX, Gao GX, Gu HT, Zhu HF. Increased 14-3-3ζ expression in the multidrug-resistant leukemia cell line HL-60/VCR as compared to the parental line mediates cell growth and apoptosis in part through modification of gene expression. Acta Haematol. 2014;132(2):177–186. doi: 10.1159/000357377. [DOI] [PubMed] [Google Scholar]
- 55.Long F, Lin Z, Long Q, Lu Z, Zhu K, Zhao M, Yang M. CircZBTB46 protects acute myeloid leukemia cells from ferroptotic cell death by upregulating SCD. Cancers (Basel) 2023;15(2):459. doi: 10.3390/cancers15020459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tesfay L, Paul BT, Konstorum A, Deng Z, Cox AO, Lee J, Furdui CM, Hegde P, Torti FM, Torti SV. Stearoyl-CoA desaturase 1 protects ovarian cancer cells from ferroptotic cell death. Cancer Res. 2019;79(20):5355–5366. doi: 10.1158/0008-5472.CAN-19-0369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tang D, Chen X, Kang R, Kroemer G. Ferroptosis: molecular mechanisms and health implications. Cell Res. 2021;31(2):107–125. doi: 10.1038/s41422-020-00441-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lei G, Zhuang L, Gan B. Targeting ferroptosis as a vulnerability in cancer. Nat Rev Cancer. 2022;22(7):381–396. doi: 10.1038/s41568-022-00459-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yin Z, Li F, Zhou Q, Zhu J, Liu Z, Huang J, Shen H, Ou R, Zhu Y, Zhang Q, Liu S. A ferroptosis-related gene signature and immune infiltration patterns predict the overall survival in acute myeloid leukemia patients. Front Mol Biosci. 2022;9:959738. doi: 10.3389/fmolb.2022.959738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhang J, Liu Y, Li Q, Xu A, Hu Y, Sun C. Ferroptosis in hematological malignancies and its potential network with abnormal tumor metabolism. Biomed Pharmacother. 2022;148:112747. doi: 10.1016/j.biopha.2022.112747. [DOI] [PubMed] [Google Scholar]
- 61.Ping L, Jian-Jun C, Chu-Shu L, Guang-Hua L, Ming Z. Silencing of circ_0009910 inhibits acute myeloid leukemia cell growth through increasing miR-20a-5p. Blood Cells Mol Dis. 2019;75:41–47. doi: 10.1016/j.bcmd.2018.12.006. [DOI] [PubMed] [Google Scholar]
- 62.Chen Y, Wang X, Cheng J, Wang Z, Jiang T, Hou N, Liu N, Song T, Huang C. MicroRNA-20a-5p targets RUNX3 to regulate proliferation and migration of human hepatocellular cancer cells. Oncol Rep. 2016;36(6):3379–3386. doi: 10.3892/or.2016.5144. [DOI] [PubMed] [Google Scholar]
- 63.Zhang W, Ma Q, Long B, Sun Z, Liu L, Lin D, Zhao M. Runt-related transcription factor 3 promotes acute myeloid leukemia progression. Front Oncol. 2021;11:725336. doi: 10.3389/fonc.2021.725336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lin Y, Huang Y, Liang C, Xie S, Xie A. Silencing of circTASP1 inhibits proliferation and induces apoptosis of acute myeloid leukaemia cells through modulating miR-515-5p/HMGA2 axis. J Cell Mol Med. 2021;25(15):7367–7380. doi: 10.1111/jcmm.16765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Mansoori B, Mohammadi A, Ditzel HJ, Duijf PHG, Khaze V, Gjerstorff MF, Baradaran B. HMGA2 as a critical regulator in cancer development. Genes (Basel) 2021;12(2):269. doi: 10.3390/genes12020269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Marquis M, Beaubois C, Lavallée VP, Abrahamowicz M, Danieli C, Lemieux S, Ahmad I, Wei A, Ting SB, Fleming S, Schwarer A, Grimwade D, Grey W, Hills RK, Vyas P, Russell N, Sauvageau G, Hébert J. High expression of HMGA2 independently predicts poor clinical outcomes in acute myeloid leukemia. Blood Cancer J. 2018;8(8):68. doi: 10.1038/s41408-018-0103-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Tan L, Wei X, Zheng L, Zeng J, Liu H, Yang S, Tan H. Amplified HMGA2 promotes cell growth by regulating Akt pathway in AML. J Cancer Res Clin Oncol. 2016;142(2):389–399. doi: 10.1007/s00432-015-2036-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Yang S, Gu Y, Wang G, Hu Q, Chen S, Wang Y, Zhao M. HMGA2 regulates acute myeloid leukemia progression and sensitivity to daunorubicin via Wnt/β-catenin signaling. Int J Mol Med. 2019;44(2):427–436. doi: 10.3892/ijmm.2019.4229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wang N, Yang B, Jin J, He Y, Wu X, Yang Y, Zhou W, He Z. Circular RNA circ_0040823 inhibits the proliferation of acute myeloid leukemia cells and induces apoptosis by regulating miR-516b/PTEN. J Gene Med. 2022;24(3):e3404. doi: 10.1002/jgm.3404. [DOI] [PubMed] [Google Scholar]
- 70.Lee YR, Chen M, Pandolfi PP. The functions and regulation of the PTEN tumour suppressor: new modes and prospects. Nat Rev Mol Cell Biol. 2018;19(9):547–562. doi: 10.1038/s41580-018-0015-0. [DOI] [PubMed] [Google Scholar]
- 71.Morotti A, Panuzzo C, Crivellaro S, Carrà G, Torti D, Guerrasio A, Saglio G. The role of PTEN in myeloid malignancies. Hematol Rep. 2015;7(4):5844. doi: 10.4081/hr.2015.6027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Smith AM, Zhang CRC, Cristino AS, Grady JP, Fink JL, Moore AS. PTEN deletion drives acute myeloid leukemia resistance to MEK inhibitors. Oncotarget. 2019;10(56):5755–5767. doi: 10.18632/oncotarget.27206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ma J, Wen X, Xu Z, Xia P, Jin Y, Lin J, Qian J. The down-regulation of Circ_0059707 in acute myeloid leukemia promotes cell growth and inhibits apoptosis by regulating miR-1287-5p. Curr Oncol. 2022;29(9):6688–6699. doi: 10.3390/curroncol29090525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Ye Q, Li N, Zhou K, Liao C. Homo sapiens circular RNA 0003602 (Hsa_circ_0003602) accelerates the tumorigenicity of acute myeloid leukemia by modulating miR-502-5p/IGF1R axis. Mol Cell Biochem. 2022;477(2):635–644. doi: 10.1007/s11010-021-04277-0. [DOI] [PubMed] [Google Scholar]
- 75.Zhang J, Huang FF, Wu DS, Li WJ, Zhan HE, Peng MY, Fang P, Cao PF, Zhang MM, Zeng H, Chen FP. SUMOylation of insulin-like growth factor 1 receptor, promotes proliferation in acute myeloid leukemia. Cancer Lett. 2015;357(1):297–306. doi: 10.1016/j.canlet.2014.11.052. [DOI] [PubMed] [Google Scholar]
- 76.He Y, Zhang J, Zheng J, Du W, Xiao H, Liu W, Li X, Chen X, Yang L, Huang S. The insulin-like growth factor-1 receptor kinase inhibitor, NVP-ADW742, suppresses survival and resistance to chemotherapy in acute myeloid leukemia cells. Oncol Res. 2010;19(1):35–43. doi: 10.3727/096504010x12828372551821. [DOI] [PubMed] [Google Scholar]
- 77.Hua H, Kong Q, Yin J, Zhang J, Jiang Y. Insulin-like growth factor receptor signaling in tumorigenesis and drug resistance: a challenge for cancer therapy. J Hematol Oncol. 2020;13(1):64. doi: 10.1186/s13045-020-00904-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Werner H, Sarfstein R, Bruchim I. Investigational IGF1R inhibitors in early stage clinical trials for cancer therapy. Expert Opin Investig Drugs. 2019;28(12):1101–1112. doi: 10.1080/13543784.2019.1694660. [DOI] [PubMed] [Google Scholar]
- 79.Sheng XF, Hong LL, Fan L, Zhang Y, Chen KL, Mu J, Shen SY, Zhuang HF. Circular RNA PVT1 regulates cell proliferation, migration, and apoptosis by stabilizing c-Myc and downstream target CXCR4 expression in acute myeloid leukemia. Turk J Haematol. 2023;40(2):82–91. doi: 10.4274/tjh.galenos.2023.2022.0435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Cancilla D, Rettig MP, DiPersio JF. Targeting CXCR4 in AML and ALL. Front Oncol. 2020;10:1672. doi: 10.3389/fonc.2020.01672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Ladikou EE, Chevassut T, Pepper CJ, Pepper AG. Dissecting the role of the CXCL12/CXCR4 axis in acute myeloid leukaemia. Br J Haematol. 2020;189(5):815–825. doi: 10.1111/bjh.16456. [DOI] [PubMed] [Google Scholar]
- 82.Calandra G, Bridger G, Fricker S. CXCR4 in clinical hematology. Curr Top Microbiol Immunol. 2010:173–191. doi: 10.1007/82_2010_26. [DOI] [PubMed] [Google Scholar]
- 83.Peled A, Tavor S. Role of CXCR4 in the pathogenesis of acute myeloid leukemia. Theranostics. 2013;3(1):34–39. doi: 10.7150/thno.5150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Bi J, Pu Y, Yu X. Exosomal circ_0004136 enhances the progression of pediatric acute myeloid leukemia depending on the regulation of miR-570-3p/TSPAN3 axis. Anticancer Drugs. 2021;32(8):802–811. doi: 10.1097/CAD.0000000000001068. [DOI] [PubMed] [Google Scholar]
- 85.Kwon HY, Bajaj J, Ito T, Blevins A, Konuma T, Weeks J, Lytle NK, Koechlein CS, Rizzieri D, Chuah C, Oehler VG, Sasik R, Hardiman G, Reya T. Tetraspanin 3 is required for the development and propagation of acute myelogenous leukemia. Cell Stem Cell. 2015;17(2):152–164. doi: 10.1016/j.stem.2015.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Byers RJ, Currie T, Tholouli E, Rodig SJ, Kutok JL. MSI2 protein expression predicts unfavorable outcome in acute myeloid leukemia. Blood. 2011;118(10):2857–2867. doi: 10.1182/blood-2011-04-346767. [DOI] [PubMed] [Google Scholar]
- 87.Kharas MG, Lengner CJ, Al-Shahrour F, Bullinger L, Ball B, Zaidi S, Morgan K, Tam W, Paktinat M, Okabe R, Gozo M, Einhorn W, Lane SW, Scholl C, Fröhling S, Fleming M, Ebert BL, Gilliland DG, Jaenisch R, Daley GQ. Musashi-2 regulates normal hematopoiesis and promotes aggressive myeloid leukemia. Nat Med. 2010;16(8):903–908. doi: 10.1038/nm.2187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Griner LN, Reuther GW. Aggressive myeloid leukemia formation is directed by the Musashi 2/Numb pathway. Cancer Biol Ther. 2010;10(10):979–982. doi: 10.4161/cbt.10.10.14010. [DOI] [PubMed] [Google Scholar]
- 89.Cao J, Huang S, Li X. Rapamycin inhibits the progression of human acute myeloid leukemia by regulating the circ_0094100/miR-217/ATP1B1 axis. Exp Hematol. 2022;112-113:60–69.e2. doi: 10.1016/j.exphem.2022.07.298. [DOI] [PubMed] [Google Scholar]
- 90.Shi JL, Fu L, Ang Q, Wang GJ, Zhu J, Wang WD. Overexpression of ATP1B1 predicts an adverse prognosis in cytogenetically normal acute myeloid leukemia. Oncotarget. 2016;7(3):2585–2595. doi: 10.18632/oncotarget.6226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Yin W, Cheng W, Shen W, Shu L, Zhao J, Zhang J, Hua ZC. Impairment of Na+,K+-ATPase in CD95(APO-1)-induced human T-cell leukemia cell apoptosis mediated by glutathione depletion and generation of hydrogen peroxide. Leukemia. 2007;21(8):1669–1678. doi: 10.1038/sj.leu.2404791. [DOI] [PubMed] [Google Scholar]
- 92.Yin W, Li X, Feng S, Cheng W, Tang B, Shi Y, Hua ZC. Plasma membrane depolarization and Na,K-ATPase impairment induced by mitochondrial toxins augment leukemia cell apoptosis via a novel mitochondrial amplification mechanism. Biochem Pharmacol. 2009;78(2):191–202. doi: 10.1016/j.bcp.2009.03.025. [DOI] [PubMed] [Google Scholar]
- 93.Wu ZJ, Sun Q, Gu DL, Wang LQ, Li JY, Jin H. Expression of circ-KEL in acute myeloid leukemia and its regulatory mechanisms in leukemic cells. Zhonghua Xue Ye Xue Za Zhi. 2021;42(3):230–237. doi: 10.3760/cma.j.issn.0253-2727.2021.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Xiao S, Zhu H. Leucine-rich alpha-2-glycoprotein1 gene interferes with regulation of apoptosis in leukemia KASUMI-1 cells. Med Sci Monit. 2018;24:8348–8356. doi: 10.12659/MSM.911249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Wang X, Abraham S, McKenzie JAG, Jeffs N, Swire M, Tripathi VB, Luhmann UFO, Lange CAK, Zhai Z, Arthur HM, Bainbridge J, Moss SE, Greenwood J. LRG1 promotes angiogenesis by modulating endothelial TGF-β signalling. Nature. 2013;499(7458):306–311. doi: 10.1038/nature12345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Singhal M, Gengenbacher N, Abdul Pari AA, Kamiyama M, Hai L, Kuhn BJ, Kallenberg DM, Kulkarni SR, Camilli C, Preuß SF, Leuchs B, Mogler C, Espinet E, Besemfelder E, Heide D, Heikenwalder M, Sprick MR, Trumpp A, Krijgsveld J, Schlesner M, Hu J, Moss SE, Greenwood J, Augustin HG. Temporal multi-omics identifies LRG1 as a vascular niche instructor of metastasis. Sci Transl Med. 2021;13(609):eabe6805. doi: 10.1126/scitranslmed.abe6805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Javaid F, Pilotti C, Camilli C, Kallenberg D, Bahou C, Blackburn J, R Baker J, Greenwood J, Moss SE, Chudasama V. Leucine-rich alpha-2-glycoprotein 1 (LRG1) as a novel ADC target. RSC Chem Biol. 2021;2(4):1206–1220. doi: 10.1039/d1cb00104c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Wang Y, Guo T, Liu Q, Xie X. CircRAD18 accelerates the progression of acute myeloid leukemia by modulation of miR-206/PRKACB axis. Cancer Manag Res. 2020;12:10887–10896. doi: 10.2147/CMAR.S277432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Liu J, Qiu B. Identification of circular RNA Circ_0003256 as a novel player in pediatric acute myeloid leukemia. J Pediatr Hematol Oncol. 2023;45(1):29–37. doi: 10.1097/MPH.0000000000002372. [DOI] [PubMed] [Google Scholar]
- 100.Liu X, Liu X, Cai M, Luo A, He Y, Liu S, Zhang X, Yang X, Xu L, Jiang H. CircRNF220, not its linear cognate gene RNF220, regulates cell growth and is associated with relapse in pediatric acute myeloid leukemia. Mol Cancer. 2021;20(1):139. doi: 10.1186/s12943-021-01395-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Fiore A, Liang Y, Lin YH, Tung J, Wang H, Langlais D, Nijnik A. Deubiquitinase MYSM1 in the hematopoietic system and beyond: a current review. Int J Mol Sci. 2020;21(8):3007. doi: 10.3390/ijms21083007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Neeb A, Wallbaum S, Novac N, Dukovic-Schulze S, Scholl I, Schreiber C, Schlag P, Moll J, Stein U, Sleeman JP. The immediate early gene Ier2 promotes tumor cell motility and metastasis, and predicts poor survival of colorectal cancer patients. Oncogene. 2012;31(33):3796–3806. doi: 10.1038/onc.2011.535. [DOI] [PubMed] [Google Scholar]
- 103.Xu Z, Zhu L, Wu W, Liao Y, Zhang W, Deng Z, Shen J, Yuan Q, Zheng L, Zhang Y, Shen W. Immediate early response protein 2 regulates hepatocellular carcinoma cell adhesion and motility via integrin β1-mediated signaling pathway. Oncol Rep. 2017;37(1):259–272. doi: 10.3892/or.2016.5215. [DOI] [PubMed] [Google Scholar]
- 104.Xu X, Zhou W, Chen Y, Wu K, Wang H, Zhang J, Zhou Y, Zeng J, Yang J, Deng Z, Zhang Y, Shen W. Immediate early response protein 2 promotes the migration and invasion of hepatocellular carcinoma cells via regulating the activity of Rho GTPases. Neoplasma. 2020;67(03):614–622. doi: 10.4149/neo_2020_190818N781. [DOI] [PubMed] [Google Scholar]
- 105.Huang L, Huang L, Ming X, Wu J, Liu W, Xiao Y. CircNFIX knockdown inhibited AML tumorigenicity by the miR-876-3p/TRIM31 axis. Hematology. 2022;27(1):1046–1055. doi: 10.1080/16078454.2022.2115699. [DOI] [PubMed] [Google Scholar]
- 106.Guo Y, Lin P, Hua Y, Wang C. TRIM31: A molecule with a dual role in cancer. Front Oncol. 2022;12:1047177. doi: 10.3389/fonc.2022.1047177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Huang N, Sun X, Li P, Liu X, Zhang X, Chen Q, Xin H. TRIM family contribute to tumorigenesis, cancer development, and drug resistance. Exp Hematol Oncol. 2022;11(1):75. doi: 10.1186/s40164-022-00322-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Xiao Y, Deng T, Ming X, Xu J. TRIM31 promotes acute myeloid leukemia progression and sensitivity to daunorubicin through the Wnt/β-catenin signaling. Biosci Rep. 2020;40(4):BSR20194334. doi: 10.1042/BSR20194334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Guo Y, Li Q, Zhao G, Zhang J, Yuan H, Feng T, Ou D, Gu R, Li S, Li K, Lin P. Loss of TRIM31 promotes breast cancer progression through regulating K48- and K63-linked ubiquitination of p53. Cell Death Dis. 2021;12(10):945. doi: 10.1038/s41419-021-04208-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Fan H, Li Y, Liu C, Liu Y, Bai J, Li W. Circular RNA-100290 promotes cell proliferation and inhibits apoptosis in acute myeloid leukemia cells via sponging miR-203. Biochem Biophys Res Commun. 2018;507(1-4):178–184. doi: 10.1016/j.bbrc.2018.11.002. [DOI] [PubMed] [Google Scholar]
- 111.Li G, Marlin MC. Rab family of GTPases. Methods Mol Biol. 2015;1298:1–15. doi: 10.1007/978-1-4939-2569-8_1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Hu X, Yin J, He R, Chao R, Zhu S. Circ_KCNQ5 participates in the progression of childhood acute myeloid leukemia by enhancing the expression of RAB10 via binding to miR-622. Hematology. 2022;27(1):431–440. doi: 10.1080/16078454.2022.2056983. [DOI] [PubMed] [Google Scholar]
- 113.Stenmark H. Rab GTPases as coordinators of vesicle traffic. Nat Rev Mol Cell Biol. 2009;10(8):513–525. doi: 10.1038/nrm2728. [DOI] [PubMed] [Google Scholar]
- 114.Chua CEL, Tang BL. Rab 10-a traffic controller in multiple cellular pathways and locations. J Cell Physiol. 2018;233(9):6483–6494. doi: 10.1002/jcp.26503. [DOI] [PubMed] [Google Scholar]
- 115.Duan X, Yu X, Li Z. Circular RNA hsa_circ_0001658 regulates apoptosis and autophagy in gastric cancer through microRNA-182/Ras-related protein Rab-10 signaling axis. Bioengineered. 2022;13(2):2387–2397. doi: 10.1080/21655979.2021.2024637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Wang D, Ming X, Xu J, Xiao Y. Circ_0009910 shuttled by exosomes regulates proliferation, cell cycle and apoptosis of acute myeloid leukemia cells by regulating miR-5195-3p/GRB10 axis. Hematol Oncol. 2021;39(3):390–400. doi: 10.1002/hon.2874. [DOI] [PubMed] [Google Scholar]
- 117.Morrione A. Grb10 proteins in insulin-like growth factor and insulin receptor signaling (review) Int J Mol Med. 2000;5(2):151–154. doi: 10.3892/ijmm.5.2.151. [DOI] [PubMed] [Google Scholar]
- 118.Morrione A. Grb10 adapter protein as regulator of insulin-like growth factor receptor signaling. J Cell Physiol. 2003;197(3):307–311. doi: 10.1002/jcp.10363. [DOI] [PubMed] [Google Scholar]
- 119.Lim MA, Riedel H, Liu F. Grb10: more than a simple adaptor protein. Front Biosci. 2004;9(1-3):387–403. doi: 10.2741/1226. [DOI] [PubMed] [Google Scholar]
- 120.Doepfner KT, Spertini O, Arcaro A. Autocrine insulin-like growth factor-I signaling promotes growth and survival of human acute myeloid leukemia cells via the phosphoinositide 3-kinase/Akt pathway. Leukemia. 2007;21(9):1921–1930. doi: 10.1038/sj.leu.2404813. [DOI] [PubMed] [Google Scholar]
- 121.Zhang L, Bu Z, Shen J, Shang L, Chen Y, Wang Y. A novel circular RNA (hsa_circ_0000370) increases cell viability and inhibits apoptosis of FLT3-ITD-positive acute myeloid leukemia cells by regulating miR-1299 and S100A7A. Biomed Pharmacother. 2020;122:109619. doi: 10.1016/j.biopha.2019.109619. [DOI] [PubMed] [Google Scholar]
- 122.Donato R, Cannon BR, Sorci G, Riuzzi F, Hsu K, Weber DJ, Geczy CL. Functions of S100 proteins. Curr Mol Med. 2013;13(1):24–57. [PMC free article] [PubMed] [Google Scholar]
- 123.Paruchuri V, Prasad A, McHugh K, Bhat HK, Polyak K, Ganju RK. S100A7-downregulation inhibits epidermal growth factor-induced signaling in breast cancer cells and blocks osteoclast formation. PLoS One. 2008;3(3):e1741. doi: 10.1371/journal.pone.0001741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Nasser MW, Qamri Z, Deol YS, Ravi J, Powell CA, Trikha P, Schwendener RA, Bai XF, Shilo K, Zou X, Leone G, Wolf R, Yuspa SH, Ganju RK. S100A7 enhances mammary tumorigenesis through upregulation of inflammatory pathways. Cancer Res. 2012;72(3):604–615. doi: 10.1158/0008-5472.CAN-11-0669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Eckert RL, Lee KC. S100A7 (Psoriasin): a story of mice and men. J Invest Dermatol. 2006;126(7):1442–1444. doi: 10.1038/sj.jid.5700265. [DOI] [PubMed] [Google Scholar]
- 126.Li Q, Luan Q, Zhu H, Zhao Y, Ji J, Wu F, Yan J. Circular RNA circ_0005774 contributes to proliferation and suppresses apoptosis of acute myeloid leukemia cells via circ_0005774/miR-192–5p/ULK1 ceRNA pathway. Biochem Biophys Res Commun. 2021;551:78–85. doi: 10.1016/j.bbrc.2021.02.058. [DOI] [PubMed] [Google Scholar]
- 127.Liu L, Yan L, Liao N, Wu WQ, Shi JL. A review of ULK1-mediated autophagy in drug resistance of cancer. Cancers (Basel) 2020;12(2):352. doi: 10.3390/cancers12020352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Du W, Xu A, Huang Y, Cao J, Zhu H, Yang B, Shao X, He Q, Ying M. The role of autophagy in targeted therapy for acute myeloid leukemia. Autophagy. 2021;17(10):2665–2679. doi: 10.1080/15548627.2020.1822628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Hwang DY, Eom JI, Jang JE, Jeung HK, Chung H, Kim JS, Cheong JW, Min YH. ULK1 inhibition as a targeted therapeutic strategy for FLT3-ITD-mutated acute myeloid leukemia. J Exp Clin Cancer Res. 2020;39(1):85. doi: 10.1186/s13046-020-01580-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Xiang H, Zhang J, Lin C, Zhang L, Liu B, Ouyang L. Targeting autophagy-related protein kinases for potential therapeutic purpose. Acta Pharm Sin B. 2020;10(4):569–581. doi: 10.1016/j.apsb.2019.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Shang J, Chen WM, Liu S, Wang ZH, Wei TN, Chen ZZ, Wu WB. CircPAN3 contributes to drug resistance in acute myeloid leukemia through regulation of autophagy. Leuk Res. 2019;85:106198. doi: 10.1016/j.leukres.2019.106198. [DOI] [PubMed] [Google Scholar]
- 132.Chomanicova N, Gazova A, Adamickova A, Valaskova S, Kyselovic J. The role of AMPK/mTOR signaling pathway in anticancer activity of metformin. Physiol Res. 2021;70(4):501–508. doi: 10.33549/physiolres.934618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Ghiraldeli L, Anderson R, Pladna K, Pardee TS. Adenosine Monophosphate Activated Protein Kinase (AMPK) enhances chemotherapy response in Acute Myeloid Leukemia (AML) Cancer Lett. 2022;535:215659. doi: 10.1016/j.canlet.2022.215659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Visnjic D, Dembitz V, Lalic H. The role of AMPK/mTOR modulators in the therapy of acute myeloid leukemia. Curr Med Chem. 2019;26(12):2208–2229. doi: 10.2174/0929867325666180117105522. [DOI] [PubMed] [Google Scholar]
- 135.Nepstad I, Hatfield KJ, Grønningsæter IS, Reikvam H. The PI3K-Akt-mTOR signaling pathway in human acute myeloid leukemia (AML) cells. Int J Mol Sci. 2020;21(8):2907. doi: 10.3390/ijms21082907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Shang J, Chen WM, Wang ZH, Wei TN, Chen ZZ, Wu WB. CircPAN3 mediates drug resistance in acute myeloid leukemia through the miR-153-5p/miR-183-5p–XIAP axis. Exp Hematol. 2019;70:42–54.e3. doi: 10.1016/j.exphem.2018.10.011. [DOI] [PubMed] [Google Scholar]
- 137.Tu H, Costa M. XIAP's profile in human cancer. Biomolecules. 2020;10(11):1493. doi: 10.3390/biom10111493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Hanifeh M, Ataei F. XIAP as a multifaceted molecule in Cellular Signaling. Apoptosis. 2022;27(7-8):441–453. doi: 10.1007/s10495-022-01734-z. [DOI] [PubMed] [Google Scholar]
- 139.Moreno-Martínez D, Nomdedeu M, Lara-Castillo MC, Etxabe A, Pratcorona M, Tesi N, Díaz-Beyá M, Rozman M, Montserrat E, Urbano-Ispizua A, Esteve J, Risueño RM. XIAP inhibitors induce differentiation and impair clonogenic capacity of acute myeloid leukemia stem cells. Oncotarget. 2014;5(12):4337–4346. doi: 10.18632/oncotarget.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Ibrahim AM, Mansour IM, Wilson MM, Mokhtar DA, Helal AM, Al Wakeel HM. Study of survivin and X-linked inhibitor of apoptosis protein (XIAP) genes in acute myeloid leukemia (AML) Lab Hematol. 2012;18(1):1–10. doi: 10.1532/LH96.11005. [DOI] [PubMed] [Google Scholar]
- 141.Hashimoto M, Saito Y, Nakagawa R, Ogahara I, Takagi S, Takata S, Amitani H, Endo M, Yuki H, Ramilowski JA, Severin J, Manabe RI, Watanabe T, Ozaki K, Kaneko A, Kajita H, Fujiki S, Sato K, Honma T, Uchida N, Fukami T, Okazaki Y, Ohara O, Shultz LD, Yamada M, Taniguchi S, Vyas P, de Hoon M, Momozawa Y, Ishikawa F. Combined inhibition of XIAP and BCL2 drives maximal therapeutic efficacy in genetically diverse aggressive acute myeloid leukemia. Nat Cancer. 2021;2(3):340–356. doi: 10.1038/s43018-021-00177-w. [DOI] [PubMed] [Google Scholar]
- 142.Shang Z, Ming X, Wu J, Liu W, Xiao Y. CircPTK2 promotes cell viability, cell cycle process, and glycolysis and inhibits cell apoptosis in acute myeloid leukemia by regulating miR-582-3p/ALG3 axis. Expert Rev Hematol. 2022;15(12):1073–1083. doi: 10.1080/17474086.2022.2114894. [DOI] [PubMed] [Google Scholar]
- 143.Pucci M, Duca M, Malagolini N, Dall'Olio F. Glycosyltransferases in cancer: Prognostic biomarkers of survival in patient cohorts and impact on malignancy in experimental models. Cancers (Basel) 2022;14(9):2128. doi: 10.3390/cancers14092128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Ke SB, Qiu H, Chen JM, Shi W, Han C, Gong Y, Chen YS. ALG3 contributes to the malignancy of non-small cell lung cancer and is negatively regulated by MiR-98-5p. Pathol Res Pract. 2020;216(3):152761. doi: 10.1016/j.prp.2019.152761. [DOI] [PubMed] [Google Scholar]
- 145.Shao P, Wei C, Wang Y. ALG3 contributes to the malignant properties of OSCC cells by regulating CDK-Cyclin pathway. Oral Dis. 2021;27(6):1426–1434. doi: 10.1111/odi.13687. [DOI] [PubMed] [Google Scholar]
- 146.Dong S, Zhong H, Li L. Circ_DLEU2 knockdown represses cell proliferation, migration and invasion, and induces cell apoptosis through the miR-582-5p/COX2 pathway in acute myeloid leukemia. Histol Histopathol. 2023;38(2):171–183. doi: 10.14670/HH-18-507. [DOI] [PubMed] [Google Scholar]
- 147.Hashemi Goradel N, Najafi M, Salehi E, Farhood B, Mortezaee K. Cyclooxygenase-2 in cancer: A review. J Cell Physiol. 2019;234(5):5683–5699. doi: 10.1002/jcp.27411. [DOI] [PubMed] [Google Scholar]
- 148.Lu Y, Liu XF, Liu TR, Fan RF, Xu YC, Zhang XZ, Liu LL. Celecoxib exerts antitumor effects in HL-60 acute leukemia cells and inhibits autophagy by affecting lysosome function. Biomed Pharmacother. 2016;84:1551–1557. doi: 10.1016/j.biopha.2016.11.026. [DOI] [PubMed] [Google Scholar]
- 149.Chen C, Xu W, Wang CM. Combination of celecoxib and doxorubicin increases growth inhibition and apoptosis in acute myeloid leukemia cells. Leuk Lymphoma. 2013;54(11):2517–2522. doi: 10.3109/10428194.2013.781170. [DOI] [PubMed] [Google Scholar]
- 150.Casanova I, Bosch R, Lasa A, Parreño M, Céspedes MV, Brunet S, Nomdedéu JF, Mangues MA, Sierra J, Mangues R. A celecoxib derivative inhibits focal adhesion signaling and induces caspase-8-dependent apoptosis in human acute myeloid leukemia cells. Int J Cancer. 2008;123(1):217–226. doi: 10.1002/ijc.23516. [DOI] [PubMed] [Google Scholar]
- 151.Hua KT, Lee WJ, Yang SF, Chen CK, Hsiao M, Ku CC, Wei LH, Kuo ML, Chien MH. Vascular endothelial growth factor-C modulates proliferation and chemoresistance in acute myeloid leukemic cells through an endothelin-1-dependent induction of cyclooxygenase-2. Biochim Biophys Acta. 2014;1843(2):387–397. doi: 10.1016/j.bbamcr.2013.10.015. [DOI] [PubMed] [Google Scholar]
- 152.Zhang Z, Lin S, Yin J, Yu W, Xu C. CircRNF220 plays a pathogenic role to facilitate cell progression of AML in vitro via sponging miR-330-5p to induce upregulation of SOX4. Histol Histopathol. 2022;37(10):1019–1030. doi: 10.14670/HH-18-472. [DOI] [PubMed] [Google Scholar]
- 153.Grimm D, Bauer J, Wise P, Krüger M, Simonsen U, Wehland M, Infanger M, Corydon TJ. The role of SOX family members in solid tumours and metastasis. Semin Cancer Biol. 2020;67(Pt 1):122–153. doi: 10.1016/j.semcancer.2019.03.004. [DOI] [PubMed] [Google Scholar]
- 154.Lu JW, Hsieh MS, Hou HA, Chen CY, Tien HF, Lin LI. Overexpression of SOX4 correlates with poor prognosis of acute myeloid leukemia and is leukemogenic in zebrafish. Blood Cancer J. 2017;7(8):e593. doi: 10.1038/bcj.2017.74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Sun X, Liu H, Li T, Qin L. MicroRNA 339 5p inhibits cell proliferation of acute myeloid leukaemia by directly targeting SOX4. Mol Med Rep. 2018;18(6):5261–5269. doi: 10.3892/mmr.2018.9552. [DOI] [PubMed] [Google Scholar]
- 156.Fung TK, Leung AY, So CW. Sox4you: a new player in C/EBPα leukemia. Cancer Cell. 2013;24(5):557–559. doi: 10.1016/j.ccr.2013.10.016. [DOI] [PubMed] [Google Scholar]
- 157.He X, Zou H, Wang F. SOX4-induced upregulation of ARHGAP9 promotes the progression of acute myeloid leukemia. Drug Dev Res. 2021;82(8):1227–1234. doi: 10.1002/ddr.21837. [DOI] [PubMed] [Google Scholar]
- 158.Shang Z, Ming X, Wu J, Xiao Y. Downregulation of circ_0012152 inhibits proliferation and induces apoptosis in acute myeloid leukemia cells through the miR-625-5p/SOX12 axis. Hematol Oncol. 2021;39(4):539–548. doi: 10.1002/hon.2895. [DOI] [PubMed] [Google Scholar]
- 159.Wan H, Cai J, Chen F, Zhu J, Zhong J, Zhong H. SOX12: a novel potential target for acute myeloid leukaemia. Br J Haematol. 2017;176(3):421–430. doi: 10.1111/bjh.14425. [DOI] [PubMed] [Google Scholar]
- 160.Dabiri A, Sharifi M, Sarmadi A. Knockdown of SOX12 expression induced apoptotic factors is associated with TWIST1 and CTNNB1 expression in human acute myeloid leukemia cells. Int J Mol Cell Med. 2021;10(4):249–258. doi: 10.22088/IJMCM.BUMS.10.4.249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Wang Y, Guo X, Wang L, Xing L, Zhang X, Ren J. miR-342-3p inhibits acute myeloid leukemia progression by targeting SOX12. Oxid Med Cell Longev. 2022;2022:1275141. doi: 10.1155/2022/1275141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Yi L, Zhou L, Luo J, Yang Q. Circ-PTK2 promotes the proliferation and suppressed the apoptosis of acute myeloid leukemia cells through targeting miR-330-5p/FOXM1 axis. Blood Cells Mol Dis. 2021;86:102506. doi: 10.1016/j.bcmd.2020.102506. [DOI] [PubMed] [Google Scholar]
- 163.Liao GB, Li XZ, Zeng S, Liu C, Yang SM, Yang L, Hu CJ, Bai JY. Regulation of the master regulator FOXM1 in cancer. Cell Commun Signal. 2018;16(1):57. doi: 10.1186/s12964-018-0266-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Gartel AL. FOXM1 in cancer: Interactions and vulnerabilities. Cancer Res. 2017;77(12):3135–3139. doi: 10.1158/0008-5472.CAN-16-3566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Nakamura S, Hirano I, Okinaka K, Takemura T, Yokota D, Ono T, Shigeno K, Shibata K, Fujisawa S, Ohnishi K. The FOXM1 transcriptional factor promotes the proliferation of leukemia cells through modulation of cell cycle progression in acute myeloid leukemia. Carcinogenesis. 2010;31(11):2012–2021. doi: 10.1093/carcin/bgq185. [DOI] [PubMed] [Google Scholar]
- 166.Liu LL, Zhang DH, Mao X, Zhang XH, Zhang B. Over-expression of FoxM1 is associated with adverse prognosis and FLT3-ITD in acute myeloid leukemia. Biochem Biophys Res Commun. 2014;446(1):280–285. doi: 10.1016/j.bbrc.2014.02.094. [DOI] [PubMed] [Google Scholar]
- 167.Williams MS, Basma NJ, Amaral FMR, Wiseman DH, Somervaille TCP. Blast cells surviving acute myeloid leukemia induction therapy are in cycle with a signature of FOXM1 activity. BMC Cancer. 2021;21(1):1153. doi: 10.1186/s12885-021-08839-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Khan I, Halasi M, Zia MF, Gann P, Gaitonde S, Mahmud N, Gartel AL. Nuclear FOXM1 drives chemoresistance in AML. Leukemia. 2017;31(1):251–255. doi: 10.1038/leu.2016.270. [DOI] [PubMed] [Google Scholar]
- 169.Khan I, Eklund EE, Gartel AL. Therapeutic vulnerabilities of transcription factors in AML. Mol Cancer Ther. 2021;20(2):229–237. doi: 10.1158/1535-7163.MCT-20-0115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Bhat UG, Jagadeeswaran R, Halasi M, Gartel AL. Nucleophosmin interacts with FOXM1 and modulates the level and localization of FOXM1 in human cancer cells. J Biol Chem. 2011;286(48):41425–41433. doi: 10.1074/jbc.M111.270843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Zhang T, Zhou Y, Guan J, Cheng H. Circ_0058058 knockdown inhibits acute myeloid leukemia progression by sponging miR-4319 to regulate EIF5A2 expression. Cancer Biother Radiopharm. 2023;38(10):738–748. doi: 10.1089/cbr.2020.4170. [DOI] [PubMed] [Google Scholar]
- 172.Ning L, Wang L, Zhang H, Jiao X, Chen D. Eukaryotic translation initiation factor 5A in the pathogenesis of cancers. Oncol Lett. 2020;20(4):81. doi: 10.3892/ol.2020.11942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Mathews MB, Hershey JW. The translation factor eIF5A and human cancer. Biochim Biophys Acta. 2015;1849(7):836–844. doi: 10.1016/j.bbagrm.2015.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Liu Y, Lei P, Qiao H, Sun K, Lu X, Bao F, Yu R, Lian C, Li Y, Chen W, Xue F. miR-9 enhances the chemosensitivity of AML cells to daunorubicin by targeting the EIF5A2/MCL-1 axis. Int J Biol Sci. 2019;15(3):579–586. doi: 10.7150/ijbs.29775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Liu W, Cheng F. Circular RNA circCRKL inhibits the proliferation of acute myeloid leukemia cells via the miR-196a-5p/miR-196b-5p/p27 axis. Bioengineered. 2021;12(1):7704–7713. doi: 10.1080/21655979.2021.1982310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Bencivenga D, Caldarelli I, Stampone E, Mancini FP, Balestrieri ML, Della Ragione F, Borriello A. p27 Kip1 and human cancers: A reappraisal of a still enigmatic protein. Cancer Lett. 2017;403:354–365. doi: 10.1016/j.canlet.2017.06.031. [DOI] [PubMed] [Google Scholar]
- 177.Yokozawa T, Towatari M, Iida H, Takeya K, Tanimoto M, Kiyoi H, Motoji T, Asou N, Saito K, Takeuchi M, Kobayashi Y, Miyawaki S, Kodera Y, Ohno R, Saito H, Naoe T. Prognostic significance of the cell cycle inhibitor p27Kip1 in acute myeloid leukemia. Leukemia. 2000;14(1):28–33. doi: 10.1038/sj.leu.2401640. [DOI] [PubMed] [Google Scholar]
- 178.Chim CS, Wong AS, Kwong YL. Epigenetic inactivation of the CIP/KIP cell-cycle control pathway in acute leukemias. Am J Hematol. 2005;80(4):282–287. doi: 10.1002/ajh.20503. [DOI] [PubMed] [Google Scholar]
- 179.Peschel I, Podmirseg SR, Taschler M, Duyster J, Götze KS, Sill H, Nachbaur D, Jäkel H, Hengst L. FLT3 and FLT3-ITD phosphorylate and inactivate the cyclin-dependent kinase inhibitor p27(Kip1) in acute myeloid leukemia. Haematologica. 2017;102(8):1378–1389. doi: 10.3324/haematol.2016.160101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Lin G, Fei Y, Zhang Y. Hsa-circ_0003420 induces apoptosis in acute myeloid leukemia stem cells and impairs stem cell properties. Immunopharmacol Immunotoxicol. 2021;43(5):622–631. doi: 10.1080/08923973.2021.1963272. [DOI] [PubMed] [Google Scholar]
- 181.Elcheva IA, Wood T, Chiarolanzio K, Chim B, Wong M, Singh V, Gowda CP, Lu Q, Hafner M, Dovat S, Liu Z, Muljo SA, Spiegelman VS. RNA-binding protein IGF2BP1 maintains leukemia stem cell properties by regulating HOXB4, MYB, and ALDH1A1. Leukemia. 2020;34(5):1354–1363. doi: 10.1038/s41375-019-0656-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Zhou J, Bi C, Ching YQ, Chooi JY, Lu X, Quah JY, Toh SH, Chan ZL, Tan TZ, Chong PS, Chng WJ. Inhibition of LIN28B impairs leukemia cell growth and metabolism in acute myeloid leukemia. J Hematol Oncol. 2017;10(1):138. doi: 10.1186/s13045-017-0507-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Liu Y, Chen X, Liu J, Jin Y, Wang W. Circular RNA circ_0004277 inhibits acute myeloid leukemia progression through microRNA-134-5p/single stranded DNA binding protein 2. Bioengineered. 2022;13(4):9662–9673. doi: 10.1080/21655979.2022.2059609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Liang H, Samanta S, Nagarajan L. SSBP2, a candidate tumor suppressor gene, induces growth arrest and differentiation of myeloid leukemia cells. Oncogene. 2005;24(16):2625–2634. doi: 10.1038/sj.onc.1208167. [DOI] [PubMed] [Google Scholar]
- 185.Chung Y, Kim H, Bang S, Jang K, Paik SS, Shin SJ. Nuclear expression loss of SSBP2 is associated with poor prognostic factors in colorectal adenocarcinoma. Diagnostics (Basel) 2020;10(12):1097. doi: 10.3390/diagnostics10121097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Liu JW, Nagpal JK, Sun W, Lee J, Kim MS, Ostrow KL, Zhou S, Jeronimo C, Henrique R, Van Criekinge W, Moon CS, Califano JA, Trink B, Sidransky D. ssDNA-binding protein 2 is frequently hypermethylated and suppresses cell growth in human prostate cancer. Clin Cancer Res. 2008;14(12):3754–3760. doi: 10.1158/1078-0432.CCR-07-4763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Chen JJ, Lei P, Zhou M. hsa_circ_0121582 inhibits leukemia growth by dampening Wnt/β-catenin signaling. Clin Transl Oncol. 2020;22(12):2293–2302. doi: 10.1007/s12094-020-02377-9. [DOI] [PubMed] [Google Scholar]
- 188.Liu J, Xiao Q, Xiao J, Niu C, Li Y, Zhang X, Zhou Z, Shu G, Yin G. Wnt/β-catenin signalling: function, biological mechanisms, and therapeutic opportunities. Signal Transduct Target Ther. 2022;7(1):3. doi: 10.1038/s41392-021-00762-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Wang Y, Krivtsov AV, Sinha AU, North TE, Goessling W, Feng Z, Zon LI, Armstrong SA. The Wnt/beta-catenin pathway is required for the development of leukemia stem cells in AML. Science. 2010;327(5973):1650–1653. doi: 10.1126/science.1186624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Gruszka AM, Valli D, Alcalay M. Wnt signalling in acute myeloid leukaemia. Cells. 2019;8(11):1403. doi: 10.3390/cells8111403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Staal FJ, Famili F, Garcia Perez L, Pike-Overzet K. Aberrant Wnt signaling in leukemia. Cancers (Basel) 2016;8(9):78. doi: 10.3390/cancers8090078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Valencia A, Román-Gómez J, Cervera J, Such E, Barragán E, Bolufer P, Moscardó F, Sanz GF, Sanz MA. Wnt signaling pathway is epigenetically regulated by methylation of Wnt antagonists in acute myeloid leukemia. Leukemia. 2009;23(9):1658–1666. doi: 10.1038/leu.2009.86. [DOI] [PubMed] [Google Scholar]
- 193.Rodrigues ACBDC, Costa RGA, Silva SLR, Dias IRSB, Dias RB, Bezerra DP. Cell signaling pathways as molecular targets to eliminate AML stem cells. Crit Rev Oncol Hematol. 2021;160:103277. doi: 10.1016/j.critrevonc.2021.103277. [DOI] [PubMed] [Google Scholar]
- 194.Kara G, Calin GA, Ozpolat B. RNAi-based therapeutics and tumor targeted delivery in cancer. Adv Drug Deliv Rev. 2022;182:114113. doi: 10.1016/j.addr.2022.114113. [DOI] [PubMed] [Google Scholar]
- 195.Ranasinghe P, Addison ML, Dear JW, Webb DJ. Small interfering RNA: Discovery, pharmacology and clinical development—An introductory review. Br J Pharmacol. 2023;180(21):2697–2720. doi: 10.1111/bph.15972. [DOI] [PubMed] [Google Scholar]
- 196.Pahle J, Walther W. Vectors and strategies for nonviral cancer gene therapy. Expert Opin Biol Ther. 2016;16(4):443–461. doi: 10.1517/14712598.2016.1134480. [DOI] [PubMed] [Google Scholar]
- 197.Schambach A, Morgan M. Retroviral vectors for cancer gene therapy. Recent Results Cancer Res. 2016;209:17–35. doi: 10.1007/978-3-319-42934-2_2. [DOI] [PubMed] [Google Scholar]
- 198.Dong Y, Siegwart DJ, Anderson DG. Strategies, design, and chemistry in siRNA delivery systems. Adv Drug Deliv Rev. 2019;144:133–147. doi: 10.1016/j.addr.2019.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Eygeris Y, Gupta M, Kim J, Sahay G. Chemistry of lipid nanoparticles for RNA delivery. Acc Chem Res. 2022;55(1):2–12. doi: 10.1021/acs.accounts.1c00544. [DOI] [PubMed] [Google Scholar]
- 200.Semple SC, Leone R, Barbosa CJ, Tam YK, Lin PJC. Lipid nanoparticle delivery systems to enable mRNA-based therapeutics. Pharmaceutics. 2022;14(2):398. doi: 10.3390/pharmaceutics14020398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Akinc A, Maier MA, Manoharan M, Fitzgerald K, Jayaraman M, Barros S, Ansell S, Du X, Hope MJ, Madden TD, Mui BL, Semple SC, Tam YK, Ciufolini M, Witzigmann D, Kulkarni JA, van der Meel R, Cullis PR. The Onpattro story and the clinical translation of nanomedicines containing nucleic acid-based drugs. Nat Nanotechnol. 2019;14(12):1084–1087. doi: 10.1038/s41565-019-0591-y. [DOI] [PubMed] [Google Scholar]
- 202.Bushweller JH. Targeting transcription factors in cancer - from undruggable to reality. Nat Rev Cancer. 2019;19(11):611–624. doi: 10.1038/s41568-019-0196-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Li X, Pu W, Zheng Q, Ai M, Chen S, Peng Y. Proteolysis-targeting chimeras (PROTACs) in cancer therapy. Mol Cancer. 2022;21(1):99. doi: 10.1186/s12943-021-01434-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Arora P, Singh M, Singh V, Bhatia S, Arora S. PROTACs in treatment of cancer: a review. Mini Rev Med Chem. 2021;21(16):2347–2360. doi: 10.2174/1389557521666210226150740. [DOI] [PubMed] [Google Scholar]
- 205.Békés M, Langley DR, Crews CM. PROTAC targeted protein degraders: the past is prologue. Nat Rev Drug Discov. 2022;21(3):181–200. doi: 10.1038/s41573-021-00371-6. [DOI] [PMC free article] [PubMed] [Google Scholar]