Abstract
V(D)J recombination in vivo requires a pair of signals with distinct spacer elements of 12 and 23 bp that separate conserved heptamer and nonamer motifs. Cleavage in vitro by the RAG1 and RAG2 proteins can occur at individual signals when the reaction buffer contains Mn2+, but cleavage is restricted to substrates containing two signals when Mg2+ is the divalent cation. By using a novel V(D)J cleavage substrate, we show that while the RAG proteins alone establish a moderate preference for a 12/23 pair versus a 12/12 pair, a much stricter dependence of cleavage on the 12/23 signal pair is produced by the inclusion of HMG1 and competitor double-stranded DNA. The competitor DNA serves to inhibit the cleavage of substrates carrying a 12/12 or 23/23 pair, as well as the cutting at individual signals in 12/23 substrates. We show that a 23/33 pair is more efficiently recombined than a 12/33 pair, suggesting that the 12/23 rule can be generalized to a requirement for spacers that differ from each other by a single helical turn. Furthermore, we suggest that a fixed spatial orientation of signals is required for cleavage. In general, the same signal variants that can be cleaved singly can function under conditions in which a signal pair is required. However, a chemically modified substrate with one noncleavable signal enables us to show that formation of a functional cleavage complex is mechanistically separable from the cleavage reaction itself and that although cleavage requires a pair of signals, cutting does not have to occur simultaneously at both. The implications of these results are discussed with respect to the mechanism of V(D)J recombination and the generation of chromosomal translocations.
Immunoglobulin and T-cell-receptor genes are assembled during lymphoid development from component gene segments by a series of site-specific genomic rearrangement events collectively termed V(D)J recombination (for review, see reference 13). All recombinationally active gene segments are flanked by recombination signal sequences (RSSs) which serve as recognition elements for the V(D)J recombinase. The consensus RSS consists of heptamer (5′-CACAGTG-3′) and nonamer (5′-ACAAAAACC-3′) elements separated by a nonconserved spacer of 12 bp (12-signal) or 23 bp (23-signal).
V(D)J recombination occurs in two well-defined stages. In the first stage, a double-strand break (DSB) is introduced between the RSS and the flanking coding sequence (20, 21, 26). The resulting cleavage intermediates, a covalently closed hairpinned coding end and a blunt signal end, have been observed both in vivo (20, 21, 26) and in vitro (16, 30). In the second stage, the broken signal ends are ligated head-to-head to form signal joints, and the coding ends are sealed together in coding joints. Formation of the initial DSB requires the combined action of the two lymphoid-specific recombinase proteins RAG1 and RAG2 (16, 17, 25). No breaks are observed in cells lacking either one of these proteins (26). Subsequent resolution of these broken molecules relies on a series of steps that have yet to be defined, but which appear to involve a number of general DNA repair factors (11).
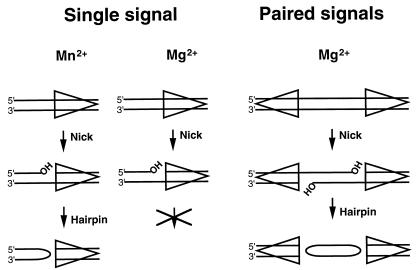
The initial cleavage steps can be reproduced by using purified RAG1 and RAG2 proteins and an oligonucleotide substrate containing an RSS (16). Together the RAG proteins specifically recognize the RSS and introduce a DSB between the RSS and flanking coding sequence. Cleavage occurs in two steps (16). In the first step, a nick is introduced at the signal/coding border on the top strand (see Fig. 1, single signal). In a second, separable step, the free 3′ hydroxyl carries out a nucleophilic attack on the phosphodiester bond on the bottom strand, leaving a hairpinned coding flank and a blunt, 5′ phosphorylated signal end, the same intermediates observed in vivo. Throughout this paper, “cleavage” refers to this hairpin and DSB formation.
FIG. 1.
Cation dependence of V(D)J cleavage. The steps of V(D)J cleavage that occur at a single site or at a signal pair are shown. In Mg2+, hairpinning does not readily occur at a single site, but is robust when the substrate contains a signal pair. All reactions require both RAG1 and RAG2 proteins. The top and bottom strands referred to in the text are with respect to the single-signal substrate as shown. The triangle indicates the RSS with the nonamer at the point.
Efficient recombination requires a pair of RSSs, one 12-signal and one 23-signal (13). Because all immunoglobulin and T-cell receptor gene segments of a given type are flanked by signals of the same spacer length, this 12/23 rule serves to ensure that rearrangements occur only between elements that could create a functional receptor gene. While there is a requirement for a pair of signals for cleavage in vivo (28), in vitro conditions can be manipulated to permit a comparison of the requirements for cleavage at a single site versus cleavage at a signal pair (coupled cleavage). Single-site or coupled cleavage depends on the divalent metal ion (6, 32). With a single site, Mn2+ permits hairpin formation, whereas only nicking proceeds in Mg2+. However, hairpin formation does occur in Mg2+ when two signals are present (Fig. 1). Thus, cleavage is coupled in Mg2+, requiring two signals for efficient DSB formation.
The requirement for two signals is separable in vitro from the requirement for a proper 12/23 pair. When cleavage is carried out in Mg2+ with purified RAG1 and RAG2 proteins, the preference for a 12/23 pair over a 12/12 pair is only three- to fivefold (24, 32). In vivo, the preference is closer to 100-fold (9, 28). In vitro cleavage in the presence of nuclear extract more readily approximates this preference, which has led to the suggestion that additional factors may be required to enforce the full extent of the 12/23 rule observed in vivo (6, 24).
The requirement for a 12/23 signal pair for cleavage would serve to decrease the chance of the RAG proteins introducing inappropriate and deleterious DSBs within the genome. It is therefore of considerable interest to understand the nature of the constraints on coupled cleavage: whether a synaptic complex containing both signals must be formed prior to cleavage, whether cleavage then occurs simultaneously at both RSSs or asynchronously, and what the sequence and structural requirements are for coupled cleavage compared to cleavage at an individual RSS.
The requirement for a pair of signals has been reproduced in vitro on a large DNA fragment in the presence of the purified RAG1 and RAG2 proteins (32) or with cell extracts containing the RAG proteins (6). When purified RAG proteins are used, cleavage is impaired when the recombination signals are closer than 1 kb, and it becomes undetectable when the signals are separated by only 200 bp (11a, 32). In the presence of cellular extracts, a 200-bp fragment can still serve as a substrate (6). This length constraint is likely to reflect the limitation of flexibility of naked DNA and makes analysis of the individual steps and requirements for coupled cleavage difficult.
Here we have developed a novel synthetic DNA substrate that facilitates this analysis. We show that the two cleavage events of coupled cleavage can be uncoupled in time and that the 12/23 rule can be generalized to a requirement for a signal pair with the spacer lengths differing by one helical turn. In addition, we define conditions in which the dependence on a 12/23 signal pair is as strict as is observed in vivo. These conditions require only the RAG1 and RAG2 proteins augmented by HMG1, demonstrating that no additional nuclear protein factors are required for this restriction in vitro.
MATERIALS AND METHODS
Proteins.
The core domains of RAG1 (amino acids 384 to 1008) and RAG2 (amino acids 1 to 383) were purified as described previously (16, 30). The purified proteins were dialyzed against 500 volumes of 20 mM HEPES (pH 7.5), 100 mM potassium glutamate, 20% glycerol, and 2 mM dithiothreitol for 4 h and immediately frozen in liquid nitrogen. T4 polynucleotide kinase and ligase were purchased from New England Biolabs. HMG1 was kindly provided by Reid Johnson (UCLA).
Preparation of oligonucleotide substrates.
All oligonucleotides were synthesized by the core facility of the department of Molecular Biology at Massachusetts General Hospital and gel purified as described previously (33). The large oligonucleotides (over 100 nucleotides [nt]) were constructed by the ligation of two oligonucleotides (one of them phosphorylated) by using a bridge oligonucleotide. A standard 12/23 deletion substrate for coupled cleavage, shown in Fig. 2A, has three oligonucleotides: DRK124, 5′-GAACGCGTGGTTTTTGTACA GCCAGACAGTGGAGTACTACCACTGTGCAGGTGGATCCCCGGGGA TCAGCAGGGATGGAGTTCTGAGGTCATTACTGCTGCAGGACGACCT GCACAGTGCTACAGACTGGAACAAAAACCCAGGTCTC-3′ (148-mer, bottom strand); DRK125, 5′-GAGACCTGGGTTTTTGTTCCAGTCTGTAGCACTGTCCAGGTCGTCCTGCAG-3′ (51-mer, 12-signal top strand); and DRK126, 5′-GATCCCCGGGGATCCACCTGCACAGTGGTAGTACTCC ACTGTCTGGCTGTACAAAAACCACGCGTTC-3′ (67-mer, 23-signal top strand). Two substrates with identical signals (12/12 and 23/23) were made by insertion of the same signal sequence twice. To construct substrates containing a bad flank, we replaced 2 nt (5′TG3′) with the dinucleotide 5′AC3′. All other substrates were made by inserting or deleting nucleotides to the desired size, and their sequences are as described by Cuomo et al. (4). To make substrates containing an Rp phosphorothioate linkage, we synthesized two oligonucleotides, DRK200 [5′-G(s)CAGGTCGTCCTGCAG-3′] and DRK209 [G(s)CAGGTGGATCCCCGGGGGATCAGCAGGG-3′], which both have a phosphorothioate linkage (indicated by “s”) between the first two nucleotides. The Rp stereoisomer was isolated by using C4 reverse-phase high-performance liquid chromatography, as described in reference 31, and was used to make a complete oligonucleotide as shown above.
FIG. 2.
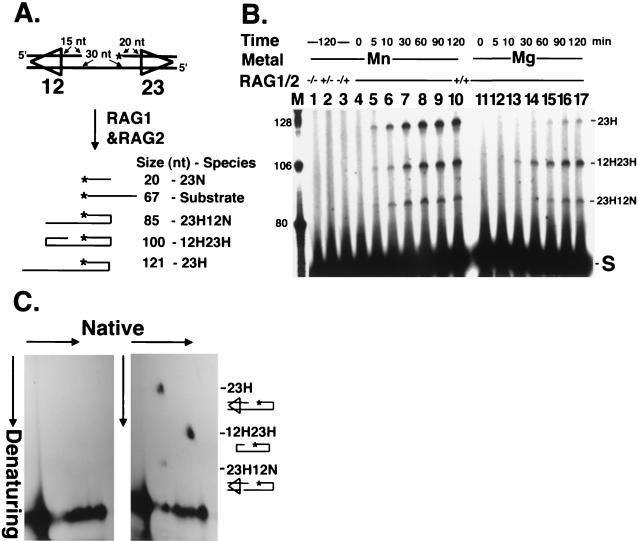
Coupled cleavage on an oligonucleotide substrate. (A) The standard tethered substrate with a 12/23 signal pair. The asterisk marks the position of the 32P label. The sizes of labeled species expected on a denaturing gel and their corresponding structures are shown. 23N migrates below the substrate and is not visible on the gels presented. (B) Time course of cleavage. Cleavage reactions were carried out in Mn2+ or Mg2+ as indicated for the times shown. The presence (+) or absence (−) of each RAG protein is indicated. Lane M contains labeled synthetic oligonucleotide markers of the sizes shown. S, substrate. (C) Identification of coupled cleavage products by two-dimensional gel electrophoresis. DNA was electrophoresed under native conditions in the first dimension and under denaturing conditions in the second. Coupled-cleavage reactions were carried out in Mg2+ with the substrate diagrammed in Fig. 2A without (left panel) or with (right panel) RAG1 and RAG2 proteins. The three species migrating off the diagonal are labeled.
Oligonucleotides were phosphorylated under standard conditions (New England Biolabs) in a 20-μl volume at 37°C for 90 min, with 25 pmol of oligonucleotide and 16.6 pmol of [γ-32P]ATP (100 μCi; purchased from NEN) in the presence of 20 U of T4 polynucleotide kinase. Phosphorylated oligonucleotides were separated from free [γ-32P]ATP by using a G-50 spin column and then were annealed with other oligonucleotides in 20 mM HEPES (pH 7.5)–100 mM potassium glutamate, after being heated at 94°C for 5 min, at a cooling rate of −1.5°C per min to 4°C (with a PCR program from MJ Research, Inc.). For labeling at the 3′ end, we annealed all oligonucleotides (each 25 pmol) under the conditions described above and incorporated [α-32P]dCMP in a reaction containing 5 U of Sequenase and [α-32P]dCTP (9 pmol) and cold dCTP (15 pmol) at 37°C for 5 min. The labeled substrates were purified and annealed as described above.
Oligonucleotide cleavage assay.
The oligonucleotide cleavage assay was initiated by the addition of RAG1 (100 ng [1.3 pmol]) and RAG2 (30 ng [0.71 pmol]) to a 10-μl reaction mixture containing 25 mM HEPES (pH 7.5), 2 mM dithiothreitol, 60 mM potassium glutamate, 1 mM MnCl2 or MgCl2, and 0.25 pmol of the indicated substrate DNA. Proteins were diluted into enzyme dilution buffer (25 mM HEPES [pH 7.5], 20% glycerol, 2 mM dithiothreitol, 0.05% Nonidet P-40). Each reaction mixture was incubated at 30°C for 2 h and stopped by the addition of a mixture of 10 μl of 94% formamide, 20 mM EDTA, 0.05% bromophenol blue, and 0.05% xylene cyanol. Fragments were separated by gel electrophoresis in 7 M urea–8% polyacrylamide–30% formamide at 75 W for 3 to 4 h in 0.67× Tris-borate-EDTA containing 12.5 mM HEPES (pH 7.5) and visualized by autoradiography.
For Fig. 3, reactions were carried out as described above, except that 100 ng of HMG1 (4 pmol as a monomer) or 50 pmol of double-stranded nonspecific competitor DNA (5′-TTAAGACTAGAGAGGAGGTAAGGTTC-3′ and its complementary strand), or both, was included in the reaction mixture. All other experiments were carried out without HMG1 or competitor DNA.
FIG. 3.
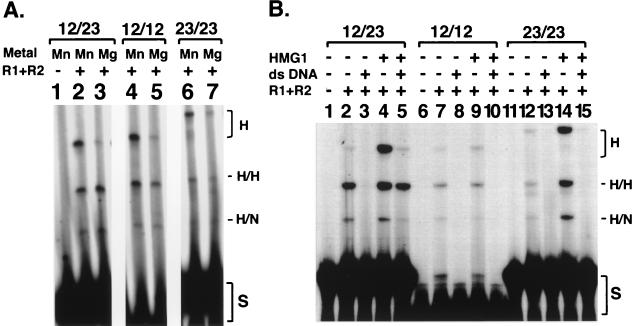
Strict 12/23 dependence with purified factors. (A) Effect of signal pairs on coupled cleavage. Cleavage of substrates with 12/23, 12/12, or 23/23 signal pairs in Mn2+ or Mg2+ is shown. All lanes are from the same gel with the same exposure. (B) Effect of HMG1 and added nonspecific double-stranded DNA (ds DNA) on coupled cleavage in Mg2+. H, single hairpin at labeled right-side signal; H/H, hairpin at both signals; H/N, hairpin at right signal, nick at left signal; S, substrate.
In vivo recombination assay.
To construct recombinational plasmid substrates containing a 33-bp spacer, pPS8 containing only a 12-signal (12) was linearized by HincII digestion and dephosphorylated by treatment with alkaline phosphatase. The linearized plasmid was ligated to a double-stranded oligonucleotide containing a 33-signal derived by annealing two complementary oligonucleotides (5′-GATCCCCGGGGATCCACCTGCACAGTGGTAGTACTCCACTGTCT GGCTGTGTCTGGCTGTACAAAAACCACGCGTTC-3′) phosphorylated at both 5′ ends. The resulting plasmid is pDRK512, which can form a signal joint by deletional recombination. The 33/23 deletional substrate pDRK513 was constructed by replacing the 12-signal with a 23-signal (two complementary oligonucleotides annealed with BamHI overhangs at both ends; 5′-GATCCCACAGTGGTAGTACTCCACTGTCTGGCTGTACAAAAACCCTCGG-3′) at the BamHI site of pDRK512. The DNA sequence and signal orientation of both plasmids were verified by DNA sequencing. The 12/12 substrate (pPS10) is as previously described (12).
All substrates form signal joints following V(D)J deletional recombination. The frequency of recombination was determined by the ratio of (Ampr + Camr)/Ampr colonies derived from transformation of Escherichia coli MC1061 transformed with plasmids recovered from HeLa cells 8 h posttransfection. Signal joint formation was verified by hybridization as described previously (12). The averages of six independent transfections are presented. An 8- to 10-fold difference in recombination for pDRK512 and pDRK513 was observed in all six transfections.
RESULTS
A novel substrate permits analysis of coupled cleavage on synthetic DNA.
The study of the coupling mechanism that operates in V(D)J cleavage would be simplified if an easily manipulable synthetic DNA substrate exhibited coupled cleavage. We therefore constructed a novel oligonucleotide substrate containing two duplex segments (each containing an RSS and coding flank sequence) separated from each other by a flexible tether of 30 nt of single-stranded DNA (Fig. 2A). This tether allows the two RSSs to contact each other without the large length of intervening DNA that would otherwise be necessary. With this substrate, all possible V(D)J cleavage products can be observed, depending on the site of 32P labeling. Thus, both nick and hairpin formation can be monitored and the product of cleavage at each individual RSS can be distinguished from the product of a cleavage at both RSSs. Figure 2A shows the cleaved fragments expected in our standard reaction when the 5′ end of the 23-signal sequence is labeled with 32P. Under these conditions, three species larger than the starting substrate can be readily distinguished by denaturing gel electrophoresis.
Coupled cleavage of this substrate was evident and revealed the same divalent cation preferences previously described for large DNA fragments. As shown in Fig. 2B (lanes 11 to 17), cleavage is coupled in the presence of Mg2+, and the major product, cleaved at both signals, contains two hairpinned coding ends (see below). Seventy-five percent of the cleavage products are from cleavage at both signals (12H23H), while two other fragments, 23H (hairpinned only at the 23-signal) and 23H12N (hairpinned at the 23-signal, nicked at the 12-signal), represent 9% and 16% of the product, respectively. Approximately 10% of the total substrate is cleaved. The coupled cleavage product appeared within 10 min and reached a maximum at about 90 min. Single-site cleavage, in contrast, is increased when Mn2+ is the divalent cation (Fig. 2B, lanes 4 to 10). In that situation, 52% of the product is singly cut at the 23-RSS (23H), 36% is doubly cut (12H23H), and 12% is 23H12N. The identity of the fragments produced by cleavage of the tethered substrate was confirmed by two-dimensional gel electrophoresis (Fig. 2C). As expected, the two fragments (indicated 23H and 23H12N) in the right panel have the same mobility under native conditions, but are separated under denaturing conditions, because the denatured 23H species is 36 nt longer than 23H12N. Migration of the 12H23H species was also consistent with its identification as a doubly hairpinned molecule. The 12H23H fragment migrates faster than the other species under native conditions, but it is intermediate in size in the second dimension as shown.
Strict observation of the 12/23 rule in vitro with purified components.
V(D)J recombination shows a marked preference for a 12/23 signal pair, although recombination of a 12/12 substrate can be detected at a low frequency (100-fold lower than a 12/23 pair) (9, 12). Similarly, in vivo cleavage greatly favors the 12/23 pair of RSSs (28). To test the selectivity of two signals in our cleavage assay, we compared coupled cleavage of similar substrates containing two identical signal sequences (12/12 or 23/23) with that of the 12/23 combination shown in Fig. 2A. The densitometric scan showed that the amount of coupled cleavage with a 12/12 substrate was three- to fourfold less than that observed with the 12/23 pair (Fig. 3A, compare lanes 3 and 5). Cleavage was barely detectable when two 23-signals were used in combination (Fig. 3A, lane 7). These results are similar to those of a previous report (32) analyzing cleavage with purified RAG proteins. The similarity between those experiments with an 8-kb DNA fragment as a substrate and the experiments presented here underscores that the tethered substrate behaves substantially the same as a much larger duplex fragment.
Other workers have reported a more dramatic 12/23 preference when cleavage was carried out in the presence of nuclear extract (6, 24). We sought to find conditions that could reproduce this greater 12/23 preference with purified components. One possible missing component is the ubiquitous nuclear HMG1 protein, which has been shown to preferentially stimulate cleavage at 23-signals and to improve coupled cleavage (29). An additional reason that 12/12 signals may be used excessively in our cleavage reactions (and in other reactions with purified components [24, 32]) might be that the high effective concentration of the two signals within the substrate permits the formation of a less favorable cleavage complex. The addition of an excess of nonspecific competitor DNA might be expected to diminish this effect.
We compared the effects of inclusion of HMG1 (4 pmol), separately or together with a 200-fold excess of double-strand nonspecific competitor (50 pmol), on cleavage of 12/23, 12/12, or 23/23 substrates (0.25 pmol) (Fig. 3B). With the addition of HMG1 alone, little effect on the 12/12 substrate was seen (Fig. 3B, compare lanes 7 and 9), but both coupled cleavage and single-site cleavage (at the 23 signal) were increased for the 12/23 and 23/23 substrates (Fig. 3B, compare lanes 2 and 4 and lanes 12 and 14). The addition of competitor DNA without HMG1 abolished cleavage for all three substrates (Fig. 3B, lanes 3, 8, and 13). Strikingly, the combination of the HMG1 and competitor DNA selectively enhanced coupled cleavage of the 12/23 substrate (Fig. 3B, compare lane 5 with lanes 10 and 15). Whereas no cleavage was detected for the 12/12 or 23/23 substrate, the coupled cleavage product for the 12/23 substrate was as robust as with HMG1 alone. Under these more stringent conditions, we observed at least a 60-fold preference for coupled cleavage of a 12/23 pair over a 12/12 or 23/23 pair. Furthermore, there was 20- to 30-fold-more coupled (12H23H) product than with single-site (23H) cleavage. This residual single-site cleavage is in keeping with the level of single-site cleavage previously detected in vivo (28). Thus, faithful restoration in vitro of the full extent of 12/23 coupling found in vivo requires no other proteins besides RAG1, RAG2, and HMG1.
A generalization of the 12/23 rule: spacer lengths must be offset by a single helical turn.
It is not understood why a pair of signals is required for cleavage and why a 12/23 pair is so greatly preferred. The naturally occurring spacer lengths are offset from each other by approximately one helical turn, suggesting that this spacing permits the proper interaction between RAG proteins on the helical surface of the DNA. A single RSS containing a spacer with one additional helical turn (a 33- or 34-RSS) can also be cleaved, albeit weakly, by the RAG proteins (in Mn2+) (4, 18), whereas RSSs with spacers of nonintegral lengths (e.g., 18 bp) are not cleaved (references 4 and 18 and see below). Coupled cleavage with tethered substrates containing a 33/23 or 33/12 pair was too faint to be reliably measured (data not shown).
We therefore asked if a 33-signal could function in vivo and whether it would pair preferentially with a 12- or 23-signal. Recombination frequencies were measured in a standard transfection assay in which RAG1 and RAG2 expression plasmids were cotransfected into HeLa cells in combination with a recombination substrate (5, 8). Substrates containing 12/23, 12/33, and 23/33 signal pairs were compared. While the 12/23 pair was by far the most efficient substrate (a recombination frequency of 3.3%), the recombination frequencies measured for the 23/33 substrate (0.06%) were reproducibly about 8- to 10-fold higher than that for the 12/33 pair (0.008%) (Table 1) and slightly higher than that for a 12/12 substrate (0.048%). Thus, although the 33-signal is intrinsically a poor substrate (as seen in vitro), it can serve to mediate recombination in vivo and preferentially pairs with a 23-signal. In vivo, 12-signals are cleaved equivalently to 23-signals (28). Thus, the preferred pairing with the 23-signal rather than the 12-RSS is not simply a reflection of the strength of an individual signal, but rather of the ability of particular signals to cooperate to coordinate cleavage.
TABLE 1.
Effect of 33-bp spacer signal on V(D)J recombination in vivo
| Substrate | Signal configuration | % Recombination |
|---|---|---|
| pJH200 | 12/23 | 3.3 |
| pDRK512 | 33/12 | 0.008 |
| pDRK513 | 33/23 | 0.060 |
| pPS10 | 12/12 | 0.048 |
Coupled cleavage can occur on prenicked substrates.
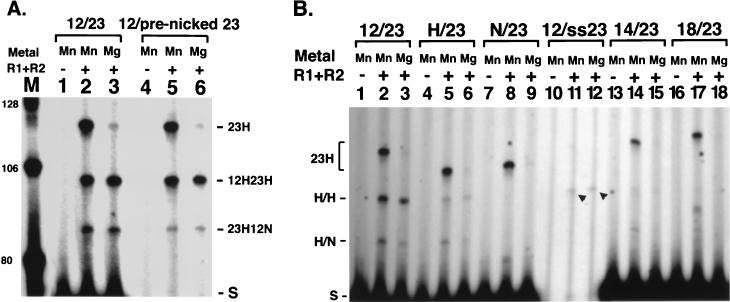
In Mg2+, generation of a DSB requires a pair of signals, but nicks are efficiently generated on substrates containing a single RSS. Therefore, such nicking could in principle occur at an individual RSS in vivo, prior to synapsis. We asked if a coupled-cleavage complex could still form on DNA that already contained a nick, or if the prenicked species would be a dead-end product (Fig. 4A). A substrate identical to that shown in Fig. 2A was constructed, except that two separate oligonucleotides were annealed to generate the top strand at the 23-signal, thereby forming a prenicked substrate. Coupled cleavage of this substrate was very similar to that with a covalently sealed top strand (Fig. 4A, compare lanes 3 and 6). Thus, nicking is a separable step from the formation of the complex that leads to coupled cleavage. Furthermore, RAG1 and RAG2 can bind a prenicked substrate in Mg2+ while remaining competent to assemble the proper structure for coupled cleavage.
FIG. 4.
Signal sequence alterations affect coupled cleavage. (A) Prenicked signal undergoes coupled cleavage. Lanes 1 to 3, standard deletional substrate; lanes 4 to 6, substrate with nick at 23-signal. All other labeling is as in Fig. 2. (B) Noncanonical signals in coupled cleavage. All alterations are in the 12-signal, and 32P label is at the 23 side as in Fig. 1. Arrowheads indicate the coupled cleavage product from the single-stranded signal substrate. H, heptamer intact, nonamer replaced; N, nonamer intact, heptamer replaced; ss23, single-stranded 23-signal, coding flank is duplex, bottom strand (with tether) is retained; 14, 14-spacer RSS; 18, 18-spacer RSS. Other labeling is as in Fig. 3.
A single-stranded RSS sequence supports coupled cleavage.
Previous work yielded the surprising observation that an RSS containing a single-stranded signal sequence can be recognized and cleaved by the RAG proteins in Mn2+ (4, 18). Interestingly, we found that a substrate can undergo coupled cleavage when it carries an intact 12-signal and a single-stranded 23-signal (with duplex coding flank intact) (Fig. 4B, lanes 11 and 12). While cleavage is not robust, it is clearly higher than that seen for the 18/23 pair (see below). Thus, a single-stranded signal can be specifically recognized and cleaved in the presence of Mg2+, indicating that the previous observations were not simply an artifact of single-site cleavage in Mn2+. Moreover, because the single-stranded signal is capable of directing coordinated cleavage, it must participate in the assembly of a coupled-cleavage complex. These observations further support the idea that unpairing of the DNA around the signal/coding border is a significant parameter in V(D)J cleavage (4, 18) and suggest the opening of the RSS and binding of the RAG proteins to single-stranded DNA may be an important aspect of the reaction.
Effects of signal and coding sequence alterations on coupled cleavage.
RAG-mediated cleavage of a substrate containing a single RSS in Mn2+ is surprisingly tolerant of variations in the signal sequence (4, 18). As discussed below, we found that similar variations are tolerated under coupled cleavage conditions. However, in keeping with the coordinated nature of the cleavage reaction in Mg2+, cleavage at a wild-type signal is depressed by the presence of an impaired signal and the overall coupled-cleavage rate is dictated by the cleavage potential of the impaired signal.
We examined cleavage in substrates lacking a heptamer or nonamer at one RSS or with altered spacer lengths between these elements. Coupled cleavage occurs at 10% of the wild-type level when the substrate contains an intact 23-RSS paired with a signal containing only a heptamer sequence (hep/23) (Fig. 4B, lane 3 versus lane 6), in keeping with observations on cleavage at a single RSS (4, 18). Thus, the absence of the nonamer at one RSS does not have a more significant effect on coupled cleavage and synapsis than on single-site cleavage. While very weak nonamer-directed cleavage has been detected at a single site, no coupled cleavage was detected in the non/23 substrate, in which the heptamer of the 12-RSS was replaced, leaving only an intact nonamer (Fig. 4B, lane 9). Either an intact heptamer at both signals is required for coupled cleavage, or any residual cleavage that occurred was below the level of detection. Substitution of a 14-bp spacer for the 12-bp spacer resulted in a 20-fold decrease in cleavage (Fig. 4B, lane 15). As expected, an 18/23 substrate failed to support any coupled cleavage, in keeping with the lack of function of a single RSS with an 18-bp spacer (Fig. 4B, lane 18).
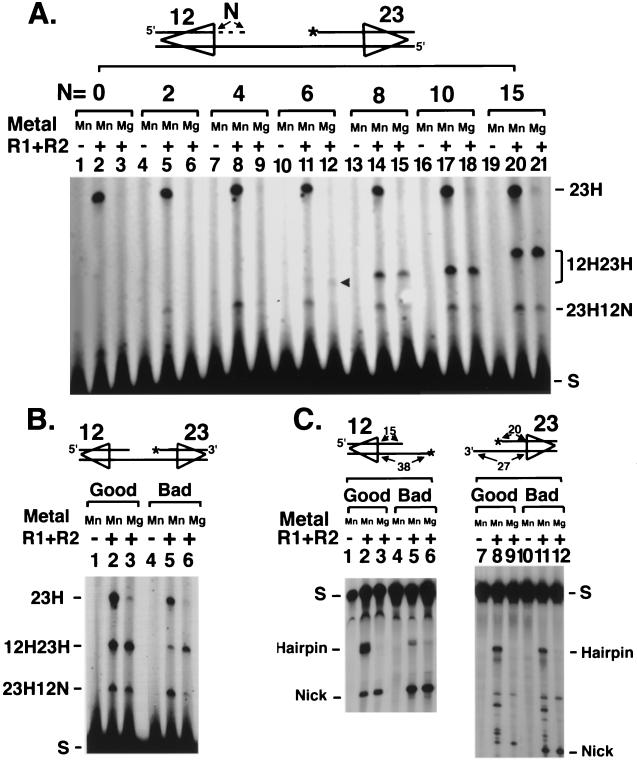
We also considered the effect of alterations in the length or sequence of the coding flank on coupled cleavage compared to that of cleavage at a single site. Tethered substrates containing different lengths of duplex coding sequence at the 12-signal while retaining the standard 20-nt coding flank at the 23-RSS were compared (Fig. 5A). All substrates retained a single-stranded coding flank on one strand, which is required to maintain the tether. Coupled cleavage (hairpins at both signals) required at least 6 bp of duplex DNA in the coding flank (Fig. 5A, lane 12), and the efficiency of coupled cleavage increased as coding length increased up to 15 nt. Nearly identical results were obtained when coding flank length was varied at a single signal in Mn2+. While some coding flank sequence is required for cleavage (as is apparent from the requirement for coding flank sequence for binding [1, 10]), no additional coding flank sequence is required to form a stable coupled-cleavage complex.
FIG. 5.
Effect of coding flank DNA on cleavage. (A) Six base pairs of duplex coding flank DNA is required for hairpinning. N, length of coding flank included at the position indicated. The arrow indicates the position of the double-hairpin product. (B) Coupled cleavage rescues bad flanks. 12/23 deletion substrate with good (lanes 1 to 3) or bad (lanes 4 to 6) dinucleotides flanking both RSSs is shown. See text for description of good and bad flanks. (C) The 23-signals do not exhibit bad flank effects in Mn2+. Single-site cleavage reactions on 12- or 23-signals with positions of hairpin and nicked products are indicated. Positions labeled with 32P are indicated with asterisks. All other labeling is as in Fig. 2.
Particular coding sequences flanking the RSS have been classified as “good flanks” (e.g., 5′TA3′ and 5′TG3′) or “bad flanks” (e.g., 5′TC3′ and 5′AC3′) based on their ability to undergo V(D)J recombination in vivo mediated by RAG1 mutants in a specific small region (19, 22). The same good flank or bad flank preferences are seen when wild-type RAG1 is used to cleave a single signal sequence in vitro (4, 18). Bad flank effects were shown to be overcome by unpairing the flanking dinucleotide, suggesting that bad flanks were more refractory to unpairing during the cleavage process (4, 18). Here we find that the bad flank effect is suppressed in coupled cleavage, where a substrate with two bad flanks behaves indistinguishably from one with two good flanks (Fig. 5B, compare lane 6 with lanes 3 and 5). Similar results have been observed with linearized plasmid substrates (32) and suggest that interactions between the two signals might aid unpairing sufficiently to overcome the bad flank effect.
The effect of coding flank sequence is seen at 12- but not 23-signals.
In the course of the experiments discussed above, we made the surprising observation that bad flank effects only occur in vitro at 12-signals and not at 23-signals. While cleavage at a 12-signal with a bad flank in the coupled substrate was inefficient in Mn2+, as expected, cleavage at the 23-signal with a bad flank was close to normal (Fig. 5B, lane 5). The lack of a bad flank effect at the 23-signal did not result from the coupling occurring in the tethered substrate in Mn2+. A comparison of four separate single RSS substrates containing either a 12- or 23-signal with good or bad flanks showed that hairpin formation in Mn2+ at the bad flank 12-signal was less than 10% of that seen with a 12-RSS with a good flank (Fig. 5C, lane 2 versus lane 5). However, hairpin formation at the 23-signal was not affected by coding flank sequence at all. 23-RSS substrates with either good or bad flank sequence were efficient in hairpin formation (Fig. 5C, lanes 8 and 11), suggesting there is some mechanistic difference between the recognition or cleavage properties of the two signals.
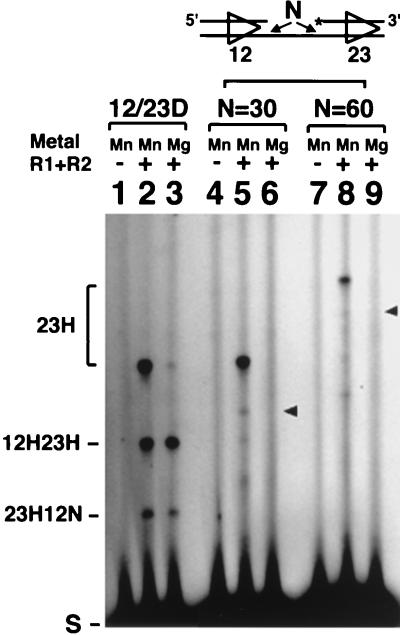
Inversion versus deletion substrates in coupled cleavage.
Joining of gene segments at the endogenous antigen receptor loci occurs primarily via deletional rearrangement, but inversional rearrangements also occur (13). The orientation of the two signals with respect to each other determines which type of rearrangement event takes place. Because the same RAG proteins and same RSSs are involved in both inversional and deletional rearrangement, it has seemed likely that cleavage will require the same structural alignment of the two signal sequences. An inversional tethered substrate was generated by flipping the direction of the 12-signal in the standard tethered molecule. RAG-mediated cleavage of this substrate is expected to generate two large fragments: a 106-nt fragment from cleavage at both signals and a 121-nt fragment arising from either a DSB at the 23-signal or a DSB at the 23-signal and a nick at the 12-RSS. In the presence of Mn2+, a 121-nt fragment was generated that was indicative of independent cleavage at the 23-signal (Fig. 6, lane 5). 32P labeling at the 5′ end of the 12-signal revealed that the 12-RSS was also efficiently cleaved in this construct (data not shown). However, although both RSSs could be cleaved independently in Mn2+, no coupled cleavage occurred in the presence of Mg2+ (Fig. 6, lane 6). Even a longer tether of 60 nt (Fig. 6, lanes 7 to 9) or 90 nt (data not shown) failed to allow coupled cleavage. The expected positions for a coupled-cleavage product are indicated by arrows. The failure of this substrate to undergo coupled cleavage is likely to reflect its inability to assemble into the proper configuration.
FIG. 6.
Inversion substrate cannot undergo coordinated cleavage. 12/23D, standard deletion substrate (lanes 1 to 3). The inversion substrate contains 30 nt (lanes 4 to 6) or 60 nt (lanes 7 to 9) of tether. Arrowheads indicate the expected position for doubly cut products. Other labeling is as in Fig. 2.
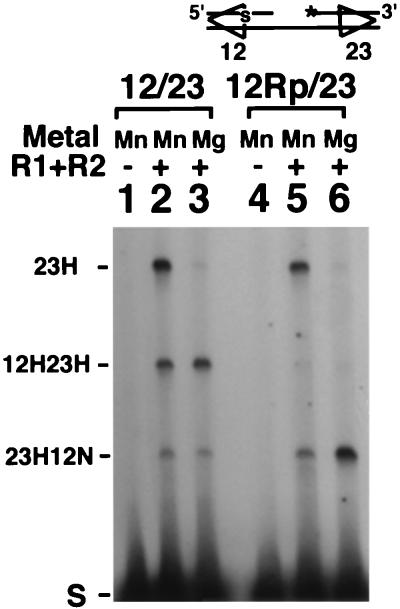
Cleavage can be uncoupled from signal pair recognition.
It has been unclear whether both signals are cleaved simultaneously during coupled cleavage and whether both signals must be cleavable for cutting to occur at either signal in Mg2+. The Rp stereoisomeric form of a phosphorothioate linkage is uncleavable by RAG proteins (31). We took advantage of this observation, placing an Rp phosphorothioate linkage at the signal/coding border of the 12-RSS to make a tethered substrate that could be bound but not cleaved at the 12-signal. Cleavage of this Rp substrate in Mn2+ and Mg2+ was compared to cleavage of a standard tethered substrate (Fig. 7). In the uncoupled (Mn2+) reaction, cleavage results in the formation of two fragments, 23H and 23H12N (Fig. 7, lane 5). As expected, no 23H12H species is observed, because the 12-signal can undergo nicking but not hairpinning. When cleavage was carried out under conditions requiring a signal pair, cleavage still proceeded, with the 23H12N product predominating (Fig. 7, lane 6). Production of the 23H12N species indicates that coupled cleavage can still occur in this substrate, even though DSB formation is impeded. Thus, an impediment to cleavage does not block proper communication between the two signals, and coupled-cleavage conditions do not demand simultaneous cleavage at both signals. A particular order of site cleavage is also not required, because coupled cleavage can also occur when the Rp linkage is placed at the 23-signal (data not shown).
FIG. 7.
An uncleavable linkage does not prevent coupling between signals. Analysis of cleavage of the standard 12/23 substrate (lanes 1 to 3) and a substrate (12Rp/23) containing a noncleavable phosphorothioate linkage at the 12-signal at the position indicated by S. Other labeling is as in Fig. 2.
DISCUSSION
The tethered oligonucleotide substrate described here displays all the properties previously described for coupled cleavage on larger DNA fragments and allows a more detailed description of the process. RAG1 and RAG2 alone are sufficient to carry out coordinated cleavage, so that analysis of coupled cleavage can be greatly simplified. The assembly of the substrate from short synthetic oligonucleotides made it possible to study the effects of different structural features, such as nicks or single-stranded sequence, and different chemical variations (e.g., phosphorothioate substitutions). In addition, the analysis of the cleavage reaction was greatly facilitated because both nick and hairpin products were detected directly, and by changing the position of the radioactive label, cleavage at any position was monitored. In the future, the simplicity of the reaction system could facilitate isolation of a synaptic complex by gel shift or identification of contact sites between the RAG proteins and DNA by UV cross-linking.
Evidence of a functional synaptic complex.
While there is no direct physical evidence as yet of a synaptic complex, it is highly likely that such a complex exists. Other biochemically related recombination reactions, such as bacteriophage Mu transposition, assemble the active proteins and DNA substrates into a synaptic complex (3, 23). The requirement for a pair of signals, the decrease in cleavage and recombination that occurs as those signals are brought closer together (2, 14, 27), and the implication that both sets of broken signal and coding ends must exist together in a complex to permit hybrid joint formation (7, 15) all argue for the assembly of a synaptic complex during V(D)J cleavage. However, these experiments do not distinguish between whether cutting must occur before pairing or vice versa.
The evidence presented here provides further strong support that RSSs are held in a synaptic complex with the RAG proteins and that this complex must form prior to cleavage. First, the lack of coupled cleavage on an inversion substrate that is identical to a deletion substrate except for RSS orientation suggests that a unique configuration of the signals and RAG proteins must exist in order for cleavage to occur. Second, the decrease in cleavage at a consensus signal that results from its pairing with a mutated signal argues for the direct communication between the signals that would occur in a synaptic complex. Third, the phosphorothioate experiments show that one signal can be cleaved separately from the other, but the second signal is still required. Thus a structure that includes both RSSs must be assembled prior to cleavage. Once this structure has been generated, either site can be cleaved regardless of cleavage at its partner. Thus, the transition to an active cleavage complex occurs as a result of pairing, but separately from hairpin formation. Similar communication between two sites has been found in other site-specific recombination systems.
As expected, cleavage can occur independently at each RSS when Mn2+ is used as the divalent metal ion, but the inclusion of Mg2+ constrains the system to require two signals. Nevertheless, we show that coordinated cleavage and synapsis can also occur in Mn2+. The generation of doubly cut DNA in Mn2+ is impaired in the inversional substrate, in parallel with the blocking of coupled cleavage in Mg2+. Rather than producing doubly cut DNA at a level equivalent to that seen for single 23-RSS hairpinning (as is seen in the deletion substrate), double cuts in Mn2+ are barely detectable. In fact, the amount of double cuts is consistent with the expected frequency for completely independent cleavage events at the two signals. The apparent decrease in double cuts in Mn2+ (as well as coupled cleavage in Mg2+) with this substrate compared to its deletional counterpart probably results from its failure to assemble the proper synaptic architecture.
Experiments with the standard deletion substrate also support the conclusion that synapsis can occur in Mn2+. The yield of 12H23H doubly cut product in Mn2+ (2 to 3%) is 15-fold higher than would be predicted for the random occurrence of two independent cleavage events on the same molecule. The amounts of the 23H and 12H species are typically 3% and 5%, respectively (for example, see Fig. 2B and 4A [also data not shown]), and therefore independent double cuts would be expected at a level of only 0.15% (the product of independent events at each signal). The kinetics of hairpin formation are also consistent with the double-hairpin products arising largely independently of the single-site cleavage pathway. In Mn2+, 23H and 23H12H species arise with essentially the same kinetics (Fig. 2B). Were the 23H12H species to arise from a second independent event, this product should accumulate later than the 23H species. Thus, while two signals and a synaptic complex are not required for cleavage in Mn2+, such complexes can form, and when they do, coupled cleavage results.
Requirements for 12/23 coupling: protein factors.
The 12/23 rule has two elements. The first is that a pair of signals is required. The second is that a 12/23 pair is strongly preferred over a 12/12 or 23/23 pair. The requirement for two signals is faithfully reproduced in vitro in the presence of Mg2+. However, in cleavage reactions carried out only with purified RAG proteins, the 12/23 preference is only three- to fivefold over a 12/12 pair (references 24 and 32 and this work). Cleavage reactions in the presence of nuclear extract exhibited a far more dramatic preference for a 12/23 pair, suggesting that there were additional factors in vivo that restricted cleavage to a 12/23 pair (6, 24).
We have now obtained at least a 60-fold preference for the 12/23 pair by using purified components: RAG1, RAG2, HMG1, and double-stranded DNA. This level of preference is in keeping with that observed in vivo (28), where a 50-fold preference for cleavage of a 12/23 substrate over that of a 12/12 substrate was observed. While we cannot rule out that additional factors are involved in vivo, our results suggest that 12/23 restriction can be accomplished by the RAG proteins themselves, aided in binding and complex assembly by HMG1 or another HMG-like family member. No additional protein factors would appear to be necessary in vitro, and it is likely that the stronger 12/23 preference that was observed with nuclear extracts simply reflected the presence of DNA and other factors that could nonspecifically interfere with assembly of aberrant complexes. Under our stringent 12/23 reaction conditions, the amount of single hairpin (e.g., 23H) is also significantly decreased (Fig. 7) to a level 20- to 30-fold lower than the amount of coupled product. In vivo, rare cleavage at a single site is observed at a level estimated to be 30- to 50-fold lower than when a 12/23 pair is present, suggesting that our reaction conditions are faithfully replicating the in vivo situation. Because the aberrant single-site cleavages are inhibited under conditions which channel the system into the biologically relevant reaction, it would appear that single-site cleavage in Mg2+ may occur via a separate pathway from that involved in coupled cleavage.
Requirements for 12/23 coupling: DNA signals.
What are the RSS requirements for 12/23 coupling? It is clear that coupled cleavage requires two functional RSSs, and much of the restriction on substrates occurs at the level of the individual RSS. If a signal can function for cleavage at a single site, it will serve for coupled cleavage. Also, in addition to the nucleotide sequence of the heptamer and nonamer, a strict spatial relationship between the heptamer and nonamer sequence is clearly important. Not only do RSSs with altered spacer lengths fail to support coupled cleavage (e.g., 18-bp spacer RSS), but an RSS with a single-stranded spacer region is also ineffective (less efficient than a heptamer alone [11a]). Previous work showed that the spacer length for a functional RSS must be offset by one or more full helical turns (12,23,33/34) (4, 18). Here we show that signal pairing occurs preferentially between two signals offset by a single helical turn (12/23 or 23/33). Two functional RSSs of different spacer length (such as a 12/33 pair) are not sufficient.
Thus, there must be a mechanism for detecting the presence of a pair of signals with proper spacer length. However, few differences between the properties of each signal have been noted. Direct binding assays have shown that both a 12-RSS and a 23-RSS can be bound by the RAG proteins, although binding to the 23-RSS is somewhat less efficient (1, 10). The gel mobility of each bound complex is the same, suggesting that the same number of RAG proteins bind each site (1, 10). Moreover, the effects of mutations in 12-signals closely parallel those in 23-signals, suggesting that site recognition by the two signals is also the same (4, 18).
The two signals do differ in their response to the addition of HMG1 protein (references 24 and 29 and this paper), and the increased cleavage at 23-signals has been taken to suggest that the HMG1 effect results from structural distortion in the 23-signal that permits the heptamer and nonamer to come into better register with each other, improving the binding of the RAG proteins. While HMG1 strongly stimulates cleavage at a 23-signal, very little effect is seen at 12-signals (29) or at 33-signals (11a). The failure to affect 33-signals, where HMG1 might have been expected to help in bringing the heptamer and nonamer into closer alignment, may mean that HMG1 cannot sufficiently distort the DNA of the 33-signal to facilitate cleavage. Alternatively, there may be some specific feature about HMG1 stimulation of the 23-RSS cleavage. This distinction may also relate to the observed preference for a 23/33 pair over a 12/33 pair for both recombination and cleavage. It appears that the 33-RSS behaves like the 12-RSS, both in pairing with a 23-RSS and in the absence of an HMG1 effect.
We have also observed one additional difference in the behaviors of the 12- and 23-signals. While cleavage at a 12-RSS displays great sensitivity to coding flank sequence, 23-signals appear largely unaffected by bad flank sequences. Since the effect of bad flanks at a 12-RSS can be overcome either by unpairing two bases of coding flank DNA or by the addition of a second signal to allow coupled cleavage, it appears that the bad flanks are a reflection of the need to unpair the DNA to facilitate hairpin formation (4, 18, 32). Perhaps binding of the RAG proteins to a 23-RSS already introduces a degree of distortion in the DNA that facilitates the unpairing of the coding flank, making the 23-signal immune to bad flank effects.
Implications for the mechanism of V(D)J recombination and the generation of genomic rearrangements.
The dependence on a pair of signals for cleavage in Mg2+ and the results of time course experiments suggested that cleavage was synchronized, occurring simultaneously (or nearly so) at each signal (6, 32). By introducing a noncleavable linkage, we were able to explore the requirements for coordinated cleavage more precisely. We found that while two signals are required to organize the cleavable structure, the two signals do not both have to be cleaved. The presence of two signals that can be bound by the RAG proteins is sufficient for DSB formation in Mg2+. Furthermore, there is no specific order to signal cleavage (e.g., the 12-RSS before the 23-RSS), because introduction of the noncleavable linkage at the signal/coding border of either the 12-signal or the 23-signal still permits cleavage at the other signal.
The results of the phosphorothioate experiments suggest a potential mechanism for how V(D)J-mediated translocations might occur despite strict 12/23 control. Because cleavage need not occur simultaneously, it should be possible for an active complex to assemble and cleave at only one site before falling apart. Any alterations that destabilize the synaptic complex (e.g., signals with lower affinity for the RAG proteins, mutations in the RAG proteins which decrease protein-protein interactions in the synaptic complex, or the absence of additional (unidentified) factors that might stabilize a complex formed over large chromosomal distances) could cause an increase in the occurrence of chromosomal breaks. Such breaks could be resolved by non-V(D)J pathways leading to large deletions and chromosomal translocations. If destabilizing mutations in the RAG proteins or accessory factors can be identified, it will be of interest to see if they do predispose pre-B or pre-T cells to the generation of deleterious chromosome breaks.
ACKNOWLEDGMENTS
We are grateful to Reid Johnson (UCLA) for providing HMG1 protein and Dan Herschlag (Stanford) for suggesting the phosphorothioate experiment. We also thank the members of our laboratory for valuable discussions.
This work was supported by a postdoctoral fellowship from the Cancer Research Institute (D.R.K.) and by National Institutes of Health grant GM58026 (M.A.O.), the Leukemia Society Scholars Program (M.A.O.), the Pew Scholars Program (M.A.O.), and Hoechst AG (M.A.O.).
REFERENCES
- 1.Akamatsu Y, Oettinger M A. Distinct roles of RAG1 and RAG2 in binding the V(D)J recombination signal sequences. Mol Cell Biol. 1998;18:4670–4678. doi: 10.1128/mcb.18.8.4670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Akamatsu Y, Tsurushita N, Nagawa F, Matsuoka M, Okazaki K, Imai M, Sakano H. Essential residues in V(D)J recombination signals. J Immunol. 1994;153:4520–4529. [PubMed] [Google Scholar]
- 3.Aldaz H, Schuster E, Baker T A. The interwoven architecture of the Mu transposase couples DNA synapsis to catalysis. Cell. 1996;85:257–269. doi: 10.1016/s0092-8674(00)81102-2. [DOI] [PubMed] [Google Scholar]
- 4.Cuomo C A, Mundy C L, Oettinger M A. DNA sequence and structure requirements for cleavage of V(D)J recombination signal sequences. Mol Cell Biol. 1996;16:5683–5690. doi: 10.1128/mcb.16.10.5683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cuomo C A, Oettinger M A. Analysis of regions of RAG-2 important for V(D)J recombination. Nucleic Acids Res. 1994;22:1810–1814. doi: 10.1093/nar/22.10.1810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Eastman Q M, Leu T M, Schatz D G. Initiation of V(D)J recombination in vitro obeying the 12/23 rule. Nature. 1996;380:85–88. doi: 10.1038/380085a0. [DOI] [PubMed] [Google Scholar]
- 7.Han J-O, Steen S B, Roth D B. Ku86 is not required for protection of signal ends or for formation of nonstandard V(D)J recombination products. Mol Cell Biol. 1997;17:2226–2234. doi: 10.1128/mcb.17.4.2226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hesse J E, Lieber M R, Gellert M, Mizuuchi K. Extrachromosomal DNA substrates in pre-B cells undergo inversion or deletion at immunoglobulin V-(D)-J joining signals. Cell. 1987;49:775–783. doi: 10.1016/0092-8674(87)90615-5. [DOI] [PubMed] [Google Scholar]
- 9.Hesse J E, Lieber M R, Mizuuchi K, Gellert M. V(D)J recombination: a functional definition of the joining signals. Genes Dev. 1989;3:1053–1061. doi: 10.1101/gad.3.7.1053. [DOI] [PubMed] [Google Scholar]
- 10.Hiom K, Gellert M. A stable RAG1-RAG2-DNA complex that is active in V(D)J cleavage. Cell. 1997;88:65–72. doi: 10.1016/s0092-8674(00)81859-0. [DOI] [PubMed] [Google Scholar]
- 11.Jackson S P, Jeggo P A. DNA double-strand break repair and V(D)J recombination: involvement of DNA-PK. Trends Biochem Sci. 1995;20:412–415. doi: 10.1016/s0968-0004(00)89090-8. [DOI] [PubMed] [Google Scholar]
- 11a.Kim, D. R. Unpublished observations.
- 12.Kirch S A, Sudarsanam P, Oettinger M A. Regions of RAG1 protein critical for V(D)J recombination. Eur J Immunol. 1996;26:886–891. doi: 10.1002/eji.1830260425. [DOI] [PubMed] [Google Scholar]
- 13.Lewis S M. The mechanism of V(D)J joining: lessons from molecular, immunological, and comparative analyses. Adv Immunol. 1994;56:27–150. doi: 10.1016/s0065-2776(08)60450-2. [DOI] [PubMed] [Google Scholar]
- 14.Lewis S M, Hesse J E. Cutting and closing without recombination in V(D)J joining. EMBO J. 1991;10:3631–3639. doi: 10.1002/j.1460-2075.1991.tb04929.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lewis S M, Hesse J E, Mizuuchi K, Gellert M. Novel strand exchanges in V(D)J recombination. Cell. 1988;55:1099–1107. doi: 10.1016/0092-8674(88)90254-1. [DOI] [PubMed] [Google Scholar]
- 16.McBlane J F, van Gent D C, Ramsden D A, Romeo C, Cuomo C A, Gellert M, Oettinger M A. Cleavage at a V(D)J recombination signal requires only RAG1 and RAG2 proteins and occurs in two steps. Cell. 1995;83:387–395. doi: 10.1016/0092-8674(95)90116-7. [DOI] [PubMed] [Google Scholar]
- 17.Oettinger M A, Schatz D G, Gorka C, Baltimore D. RAG-1 and RAG-2, adjacent genes that synergistically activate V(D)J recombination. Science. 1990;248:1517–1523. doi: 10.1126/science.2360047. [DOI] [PubMed] [Google Scholar]
- 18.Ramsden D A, McBlane J F, van Gent D C, Gellert M. Distinct DNA sequence and structure requirements for the two steps of V(D)J recombination signal cleavage. EMBO J. 1996;15:3197–3206. [PMC free article] [PubMed] [Google Scholar]
- 19.Roman C A, Baltimore D. Genetic evidence that the RAG1 protein directly participates in V(D)J recombination through substrate recognition. Proc Natl Acad Sci USA. 1996;93:2333–2338. doi: 10.1073/pnas.93.6.2333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Roth D B, Menetski J P, Nakajima P B, Bosma M J, Gellert M. V(D)J recombination: broken DNA molecules with covalently sealed (hairpin) coding ends in scid mouse thymocytes. Cell. 1992;70:983–991. doi: 10.1016/0092-8674(92)90248-b. [DOI] [PubMed] [Google Scholar]
- 21.Roth D B, Nakajima P B, Menetski J P, Bosma M J, Gellert M. V(D)J recombination in mouse thymocytes: double-strand breaks near T cell receptor δ rearrangement signals. Cell. 1992;69:41–53. doi: 10.1016/0092-8674(92)90117-u. [DOI] [PubMed] [Google Scholar]
- 22.Sadofsky M, Hesse J E, van Gent D C, Gellert M. RAG-1 mutations that affect the target specificity of V(D)J recombination: a possible direct role of RAG-1 in site recognition. Genes Dev. 1995;9:2193–2199. doi: 10.1101/gad.9.17.2193. [DOI] [PubMed] [Google Scholar]
- 23.Savilhati H, Mizuuchi K. Mu transpositional recombination:donor DNA cleavage and strand transfer in trans by the Mu transposase. Cell. 1996;85:271–280. doi: 10.1016/s0092-8674(00)81103-4. [DOI] [PubMed] [Google Scholar]
- 24.Sawchuk D J, Weis-Garcia F, Malik S, Besmer E, Bustin M, Nussenzweig M C, Cortes P. V(D)J recombination: modulation of RAG1 and RAG2 cleavage activity on 12/23 substrates by whole cell extract and DNA-bending proteins. J Exp Med. 1997;185:2025–2032. doi: 10.1084/jem.185.11.2025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schatz D G, Oettinger M A, Baltimore D. The V(D)J recombination activating gene (RAG-1) Cell. 1989;59:1035–1048. doi: 10.1016/0092-8674(89)90760-5. [DOI] [PubMed] [Google Scholar]
- 26.Schlissel M, Constantinescu A, Morrow T, Baxter M, Peng A. Double-strand signal sequence breaks in V(D)J recombination are blunt, 5′ phosphorylated, RAG-dependent, and cell-cycle-regulated. Genes Dev. 1993;7:2520–2532. doi: 10.1101/gad.7.12b.2520. [DOI] [PubMed] [Google Scholar]
- 27.Sheehan K M, Lieber M R. V(D)J recombination: signal and coding joint resolution are uncoupled and depend on parallel synapsis of the sites. Mol Cell Biol. 1993;13:1363–1370. doi: 10.1128/mcb.13.3.1363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Steen S B, Gomelsky L, Roth D B. The 12/23 rule is enforced at the cleavage step of V(D)J recombination in vivo. Genes Cells. 1996;1:543–553. doi: 10.1046/j.1365-2443.1996.d01-259.x. [DOI] [PubMed] [Google Scholar]
- 29.van Gent D C, Hiom K, Paull T T, Gellert M. Stimulation of V(D)J cleavage by high mobility group proteins. EMBO J. 1997;16:2665–2670. doi: 10.1093/emboj/16.10.2665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.van Gent D C, McBlane J F, Ramsden D A, Sadofsky M J, Hesse J E, Gellert M. Initiation of V(D)J recombination in a cell-free system. Cell. 1995;81:925–934. doi: 10.1016/0092-8674(95)90012-8. [DOI] [PubMed] [Google Scholar]
- 31.van Gent D C, Mizuuchi K, Gellert M. Similarities between initiation of V(D)J recombination and retroviral integration. Science. 1996;271:1592–1594. doi: 10.1126/science.271.5255.1592. [DOI] [PubMed] [Google Scholar]
- 32.van Gent D C, Ramsden D A, Gellert M. The RAG1 and RAG2 proteins establish the 12/23 rule in V(D)J recombination. Cell. 1996;85:107–113. doi: 10.1016/s0092-8674(00)81086-7. [DOI] [PubMed] [Google Scholar]
- 33.Vorndam A V, Kerschner J. Purification of small oligonucleotides by polyacrylamide gel electrophoresis and transfer to diethylaminoethyl paper. Anal Biochem. 1986;152:221–225. doi: 10.1016/0003-2697(86)90401-x. [DOI] [PubMed] [Google Scholar]