Abstract
The T box transcription antitermination regulatory system, found in Gram-positive bacteria, is dependent on a complex set of interactions between uncharged tRNA and the 5′-untranslated mRNA leader region of the regulated gene. One of these interactions involves the base pairing of the acceptor end of cognate tRNA with four bases in a 7 nt bulge of the antiterminator RNA. In vitro selection of randomized tRNA binding to Bacillus subtilis tyrS antiterminator model RNAs was used to determine what, if any, sequence trends there are for binding beyond the known base pair complementarity. The model antiterminator RNAs were selected for the wild-type tertiary fold of tRNA. While there were no obvious sequence correlations between the selected tRNAs, there were correlations between certain tertiary structural elements and binding efficiency to different antiterminator model RNAs. In addition, one antiterminator model selected primarily for a kissing tRNA T loop–antiterminator bulge interaction, while another antiterminator model resulted in no such selection. The selection results indicate that, at the level of tertiary structure, there are ideal matches between tRNAs and antiterminator model RNAs consistent with in vivo observations and that additional recognition features, beyond base pair complementarity, may play a role in the formation of the complex.
INTRODUCTION
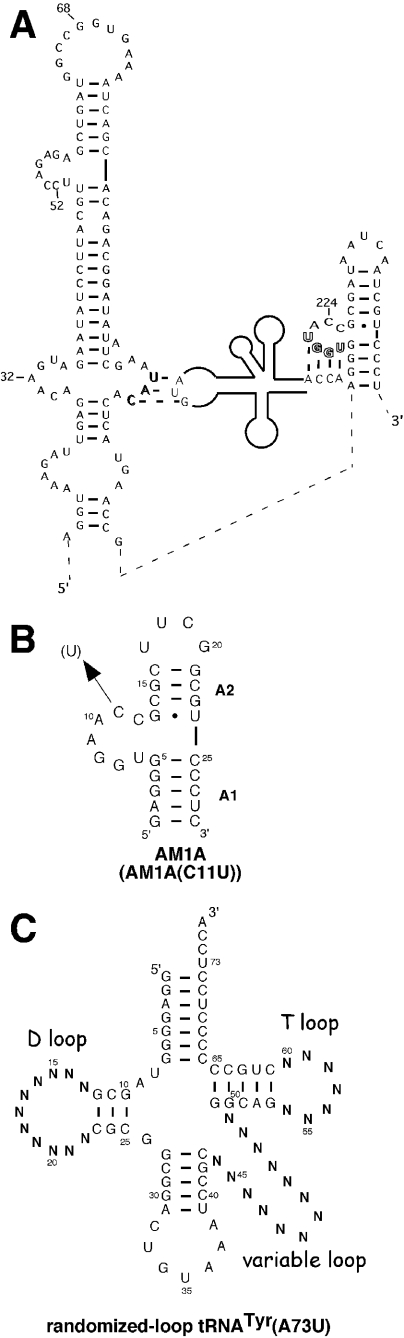
The T box transcription termination control system, primarily found in Gram-positive bacteria, utilizes a novel tRNA–mRNA molecular interaction to effect transcription antitermination (1–3). This regulatory mechanism has been identified in a variety of aminoacyl-tRNA synthetase, amino acid biosynthesis and amino acid transport genes. Phylogenetic and mutational data (4) and structural mapping (5) have identified a complex secondary structure for the 5′-untranslated leader region of the mRNA of T box genes as well as base pairing interactions between the 5′ leader mRNA and the appropriate cognate tRNA (Figure 1A). The recognition involves the tRNA anticodon loop base pairing with a portion of the 5′ leader termed the specifier sequence (5,6) along with the uncharged tRNA amino acid acceptor end base pairing with four of seven bulged nucleotides in the antiterminator (5,7). This latter tRNA–mRNA base pairing is presumed to stabilize the formation of the antiterminator over a competing, mutually exclusive, terminator stem–loop structure (5,8,9) in order to effect antitermination. The specificity of the regulatory response is directed primarily by these two tRNA–mRNA interactions (3). In vitro tRNA–antiterminator binding studies (8) as well as in vitro transcription studies (10,11) indicate that this tRNA–mRNA interaction occurs in the absence of additional factors. In addition, limited studies with model tRNAs illustrate that the full tRNA is important for optimal in vivo transcription antitermination (12) as well as in vitro tRNA binding to the antiterminator (8).
Figure 1.
(A) Secondary structure model of B.subtilis tyrS leader region and the interaction with tRNA. A Tyr (UAC) specifier sequence in a side bulge of stem–loop I (shown in shadow) base pairs with the anticodon of the cognate tRNA (2,4). Also of importance is the base pairing of the uncharged acceptor end of tRNA with the first four bases (shown in outline) of the antiterminator bulge. This latter interaction also involves covariation of the variable base of the antiterminator (position 222) with the discriminator base of the tRNA (7). (B) Secondary structure of antiterminator model RNA AM1A. The variant model, AM1A(C11U), contains a U at position 11 (corresponding to position 224 in the full leader). These antiterminator model RNAs bind B.subtilis tRNATyr(A73U) in vitro (8) and are functionally relevant in vivo (4). (C) Structure model of B.subtilis tRNATyr(A73U) randomized in D, T and variable loop regions.
In vitro evolutionary selection techniques have been extensively used to identify RNA and DNA aptamers to proteins and small molecules as well as to identify novel nucleic acid sequences for ribozymes and other biologically functional nucleic acid systems (13,14). Few studies, however, have focused on selection involving intermolecular association of two or more nucleic acids. While G-quartet, triplex and antisense interactions can involve intermolecular association of two or more nucleic acid strands, they necessitate recognition and binding at the primary sequence level as opposed to the recognition of tertiary structure of the nucleic acid. The few in vitro selection studies that have utilized binding to a nucleic acid with a tertiary fold as the selection event have illustrated the propensity for RNA to form complementary base pairs between loops. Examples include aptamers to HIV-1 TAR RNA (15), 16S rRNA decoding region (16) and tRNA (17). The resulting kissing complexes, however, do not always involve standard antisense recognition as is illustrated by the non-canonical base pairs identified in selection studies of the HIV dimerization initiation site (18). Counter selection methods to preclude kissing interactions can aid the selection of alternate interactions, but even then, the resulting sequences appear to involve extensive recognition of the primary sequence instead of the tertiary fold of the nucleic acid (19).
In vitro selections of aptamer analogs of tRNA have also been investigated. Some studies involve selecting for small RNAs that mimic tRNA in a cellular process, such as binding tRNA synthetase (20) or receptor-mediated mitochondrial tRNA import (21). Other studies have investigated randomized regions within otherwise native tRNA, such as tRNA synthetase binding by tRNA randomized in the loop regions (22) and autolytic cleavage with Pb2+ of tRNA randomized at selected positions (23). While in vitro selection techniques have been applied to select for RNAs that bind tRNA (17), few, if any, studies have been done to select for tRNAs that bind to a specific RNA molecule.
This work investigates the factors controlling selection of tRNAs randomized in the D, T and variable loop regions for binding to Bacillus subtilis tyrS T box antiterminator model RNAs. The antiterminator model RNA AM1A is based on the antiterminator sequence of B.subtilis tyrS with minor modifications. The loop region and initial base pair in stem A1 (Figure 1B) were modified to facilitate structure elucidation and preparation via in vitro transcription (8,24). In addition, the variable base in the antiterminator of B.subtilis tyrS (corresponding to position 9 in AM1A) was changed from the wild-type sequence of U to an A in order to avoid homodimerization (8). A functionally relevant interaction with tRNA is achieved by covarying the tRNA discriminator base (position 73) to complement the antiterminator variable base (8). These modifications have all been shown to be functionally relevant in vivo (4). Consequently, for these model studies the ‘wild-type’ tRNA is B.subtilis tRNATyr(A73U). The model RNAs [AM1A and the reduced function variant AM1A(C11U)] bind B.subtilis tRNATyr(A73U) with differing affinities (8); have different antitermination efficiencies in vivo (2); have subtle structural/dynamic differences (M. Gerdeman, T. Henkin and J. Hines, unpublished data); and are functionally relevant in vitro (8) and in vivo (4), thus, making them excellent models for probing further tertiary interactions with tRNA via in vitro evolutionary selection.
MATERIALS AND METHODS
Oligonucleotides
Synthetic single-stranded DNAs (ssDNAs) were purchased from Oligos Etc., Inc. Biotinylated RNAs were purchased from Dharmacon Research, Inc. All oligonucleotides were purified by 20% denaturing (7 M Urea) PAGE (19:1 acrylamide:bisacrylamide) unless otherwise specified.
Pool preparation
The tRNATyr(A73U) was randomized in the D, T and variable loop regions (Figure 1C). Based on the starting amount of purified DNA, the complexity of the randomized pool of tRNA initially used was ∼9–12 × 1014 molecules comparable with other selections involving large randomized regions (≥30) for selecting aptamers of small molecules (25) or for investigating RNA–RNA interactions (26). The DNA used was as follows:
Template: 5′-TGGAGGAGGGGGGCAG(N7)CTGCC(N13)GCGGATTTACAGTCCGCCGCG(N11)CGCTACCCCTCC-3′, where N is any base.
PCR primers: Primer 1, 5′-GATAATACGACTCACTATAGGAGGGGTAGC-3′, where the underlined sequence is the T7 promoter region for T7 RNA polymerase reaction; Primer 2, 5′-TGGAGGAGGGGGGCAG-3′.
The initial DNA pool was generated from the synthetic ssDNA template by elongation of the primer complementary to the 5′ fixed region with Klenow fragment DNA polymerase I (27) (Roche) and purified by non-denaturing polyacrylamide gel (10%, 29:1 acrylamide:bisacrylamide). Double-stranded DNA products were transcribed using a T7 Ampliscribe Kit (Epicentre Technology).
In vitro selection
The in vitro selection was performed at room temperature. Model antiterminator RNAs (Figure 1B) were immobilized via a 5′-biotinylated-RNA–streptavidin complex. Two forms of streptavidin were investigated: UltraLink™ and MagnaBind™ Streptavidin beads (Pierce). UltraLink involves the separation of complexes with the beads from the supernatant by centrifugation while Magnabind utilizes a magnetic holder.
The RNA pool was renatured by heating in 50 mM Tris–HCl, pH 7.6, 250 mM NaCl, and 5 mM MgCl2 at 90°C for 2 min, followed by cooling to room temperature for 10 min. To eliminate non-specific binding, the RNA pool was mixed with the streptavidin beads in Ultrafree MC filter units (Millipore) in binding buffer containing 50 mM Tris–HCl, pH 7.6, 250 mM NaCl and 5 mM MgCl2 at room temperature. The RNAs, which did not bind to UltraLink™ or MagnaBind™ streptavidin beads, were separated by centrifuging or by magnetic separation, respectively. The unbound RNA was mixed with 30–60 pmol of either 5′ biotinylated AM1A or 5′ biotinylated AM1A(C11U) in 360–400 μl of binding buffer for 15–30 min. The RNA solution was then mixed with 200 μl of streptavidin beads that had been previously equilibrated with (200 μl) binding buffer by washing (3×). The resulting mixture was incubated for 15–30 min at room temperature on a rotating shaker. The beads were then separated from the supernatant either by centrifuging or by magnetic separation as appropriate. RNAs that were retained by the beads were eluted twice with 200 μl elution buffer (0.3 M sodium acetate, pH 5.2, 5 mM EDTA and 7 M urea) at 90°C over the course of 5 min. The eluted fractions were combined and the RNA was precipitated by ethanol with the addition of glycogen as a carrier (1 μl of a 20 μg/μl solution).
The recovered RNAs were amplified by RT–PCR. RNAs were reverse-transcribed at 42°C for 1 h in 20 μl of reaction mixture that contained 50 mM Tris (pH 8.3), 50 mM KCl, 5 mM MgCl2, 5 mM DTT, 2.5 μM 3′ primer, 0.5 mM dNTP mixture and 10 U reverse transcriptase (Promega). The dNTP mixture and enzyme were added after an annealing step (2 min at 90°C followed by incubation at room temperature for 10 min). The non A-tailing Pfu turbo polymerase (Stratagene) was used for amplification by PCR. The 20 μl of cDNA reaction mixture produced after reverse-transcription was diluted in 80 μl of 20 mM Tris–HCl, pH 8.8, 2 mM MgSO4, 10 mM KCl, 10 mM (NH4)2SO4, 0.1% Triton X-100, 0.1 mg/ml nuclease-free BSA, 2.5 U Pfu Turbo DNA polymerase and 0.4 μM each of 5′ and 3′ primers. The reaction mixture was cycled at 94°C for 1.15 min, 50°C for 1.15 min and 72°C for 1.15 min for 8–12 cycles (fewer cycles were used for later rounds of selection in order to increase stringency). The PCR product was confirmed by electrophoresis of 3 μl of the PCR mixture on 2% agarose gel and visualized using ethidium bromide.
The PCR product was precipitated in ethanol and used directly in the in vitro transcription with the T7 Ampliscribe Kit (Epicentre Technologies). Transcription was performed according to the manufacturer's protocol, at 37°C for 3 h. To remove remaining DNA template, the mixture was incubated with 1 μl of DNase supplied with the kit at 37°C for 30 min. Loading buffer (20 mM EDTA, 0.02% bromophenol blue and xylene cyanol marker dyes) was added directly to the mixture and the RNAs were purified by denaturing (7 M urea) 10% PAGE (19:1 acrylamide:bisacrylamide). Following crush and soak elution and ethanol precipitation, the isolated RNA was used for the next cycle of selection and amplification.
Cloning and sequencing
Cloning was performed initially after the sixth in vitro selection cycle and again following completion of the seventh cycle. DNA from the RT–PCR was directly cloned using the TA Cloning Kit (Invitrogen) following the supplied protocol. DNA was isolated and purified from individual colonies using the QIAprep Spin Miniprep Kit (Qiagen) according to the manufacturer's protocol. Plasmids were prepared for sequencing using the BigDye terminator Cycle Sequencing Ready Reaction DNA sequencing Kit (Applied Biosystem, CA) following the supplied protocol and sequenced using an Applied Biosystem 3700 DNA Analyzer.
Gel mobility shift assay
Gel mobility shift assay conditions that were previously used with the antiterminator model RNAs (8) were adapted to a smaller gel electrophoresis system using Novex 20% native TBE gels (38:1 acrylamide/bis-acrylamide) (Invitrogen). The running buffer was 0.5× TBE, 50 mM NaCl, 5 mM MgCl2 (where 1× TBE is 50 mM Tris-borate, pH 8.3 and 1 mM EDTA). The sample binding buffer was 0.5× TBE, 50 mM NaCl, 5 mM MgCl2 and 10% glycerol and was added to the reaction mixture as a 5× solution. RNA was monitored via either chemiluminescence or 5′-32P-end-labeling.
For the chemiluminescence detected gel shifts, 5′ biotinylated RNA [AM1A or AM1A(C11U)] was present in the 10 μl reactions mixtures at a concentration of 4 nM. The mixtures also contained a range of selected tRNA concentrations (e.g. 0–250 μM). The biotinylated RNA and tRNAs were renatured separately by heating in binding buffer at 90°C for 2 min and cooling down to room temperature for 10 min and then mixed. Binding mixtures were incubated at 4°C for 30–40 min prior to loading on the gel. The gels were run at 100 V for 4–5 h at room temperature. After electrophoresis, the gel was blotted onto a Nylon positively charged membrane (Roche). Membranes having the biotin-labeled RNA were developed using the LightShift Chemiluminescent EMSA Kit (Pierce) and conducted according to the manufacturer's protocol. Visualization and quantification of the chemiluminescent signal on the membrane was achieved using a Gel Doc/Chemi Doc system (Bio-Rad).
For the radioactively labeled gel shift experiments, stock solutions of tRNAs and 5′-32P-end-labeled antiterminator model RNAs in binding buffer were separately prepared and heated at 90°C for 2 min and allowed to cool slowly to room temperature for 10 min. The equal volume of tRNA and 32P-end-labeled antiterminator model RNA solution were mixed and incubated at 4°C for 30–40 min. The mixture was then loaded onto a 15% acrylamide gel (29:1 acrylamide: bisacrylamide) and run between 6–10 W for 4–5 h at 4°C. Binding buffer was 0.5× TBE, 5 mM MgCl2 and 50 mM NaCl. The gels were exposed on X-ray film for overnight at −80°C followed by visualization and quantification of the developed films on a Gel Doc/Chemi Doc system (Bio-Rad).
Evaluating tRNA tertiary interactions
In order to evaluate selected sequences with non-canonical base pairings at sites normally involved in the tertiary fold of tRNA (28), the selected base pairings at these positions were examined in the context of the crystal structure of yeast tRNAPhe (PDB entry 1EHZ) (29). The appropriate base changes were made using Biopolymer and InsightII (Accelrys). The ability of the new bases to base pair without any further structural change was then evaluated. The only base pairs that were considered viable for retaining the tertiary interaction [color coded in Figure 2 for B.subtilis tRNATyr(A73U)] were those that, when directly substituted, readily fit with no obvious steric interactions and were oriented to favor the formation of at least one hydrogen bond (≤2.5 Å H-bond distance; 120° < bond angle < 180°). The stringent criterion for an isosteric replacement within the context of the structurally well-characterized yeast tRNAPhe was used in order to have a conservative analysis of the sequence data. It does not preclude the possibility of alternate or novel tertiary interactions that can only be definitively determined through detailed structural studies of each tRNA sequence isolated.
Figure 2.
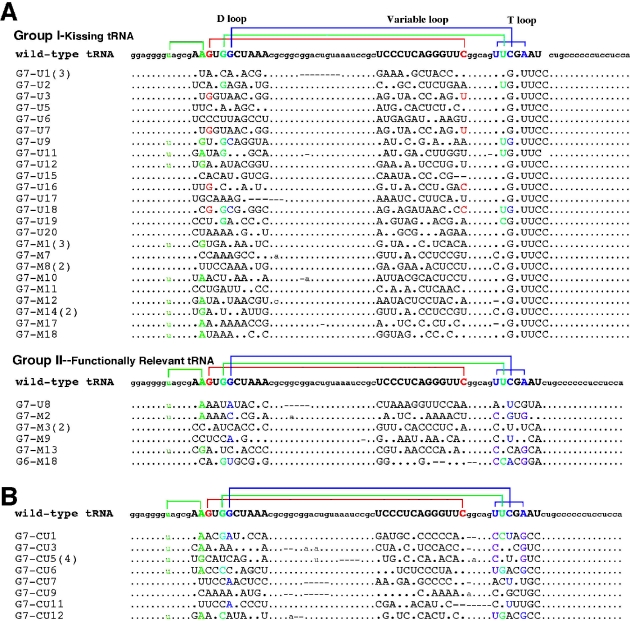
(A) Nucleotide sequences of selected clones of randomized tRNA after sixth and seventh in vitro selection cycles against antiterminator model RNA AM1A. Group I corresponds to the kissing tRNA sequences, while Group II corresponds to the functionally relevant tRNA sequences. (B) Nucleotide sequences of selected clones of randomized tRNA after the sixth and seventh in vitro selection cycles against antiterminator model RNA AM1A(C11U). For both (A) and (B), the parentheses next to the sequence number indicate the number of identical sequences. Sequences from randomized regions are shown in capital. Dots indicate the sequences identical to the wild-type B.subtilis tRNATyr(A73U). Lines indicate base deletion. Tertiary base pairing interactions are represented by color coding as shown for the wild-type B.subtilis tRNATyr(A73U) sequence. (See Materials and Methods for evaluation criteria and details regarding selection). Base pair color coding is as follows: 8:14, green; 15:48, red; 18:55, light blue; 19:56, dark blue; 54:58, pink.
Kd determination
Kd values were determined by quantifying the amount of complex formed (i.e. fraction bound) in the gel mobility shift assay at each tRNA concentration, and the resulting concentration-dependent data fit to a standard single site binding curve using Prism (GraphPad). Two-site binding as well as linear curve fits were also evaluated and in each case, the single-site binding provided to be the statistically best fit for the data.
RESULTS
In vitro selection techniques were used to investigate the sequence trends involved in tRNA binding to two T box antiterminator model RNAs. The two models [AM1A and AM1A(C11U), Figure 1B] differ in their functional activity in vivo (2): tRNA binding affinity in vitro (8) as well as in their structure and flexibility (M. Gerdeman, T. Henkin and J. Hines, unpublished data). The tRNA sequences selected to bind the antiterminator model RNAs are shown in Figure 2. Color coding of the bases reflects retention of base pairings involved in the tertiary fold of tRNA (for evaluation criteria see Materials and Methods). Nomenclature of the clones was determined by the number of selection cycles involved (G6 or G7) and the type of selection, UltraLink™ (U) or MagnaBind™ (M) streptavidin beads. The sequences have been grouped to illustrate general trends as well as to illustrate differences between the sequences selected by the functional antiterminator model RNA, AM1A, compared with those selected by the reduced function variant, AM1A(C11U). In general, the tRNA sequences selected to bind AM1A (Figure 2A) fell into two groups, kissing (Group I) and functionally relevant (Group II). The sequences selected to bind AM1A(C11U) (Figure 2B) using identical starting pool and selection conditions resulted in only one group of tRNA binding in a functionally relevant manner. The relative numbers of each type of tertiary base pairing for each selected group are schematically summarized in Figure 3. Details are discussed below.
Figure 3.
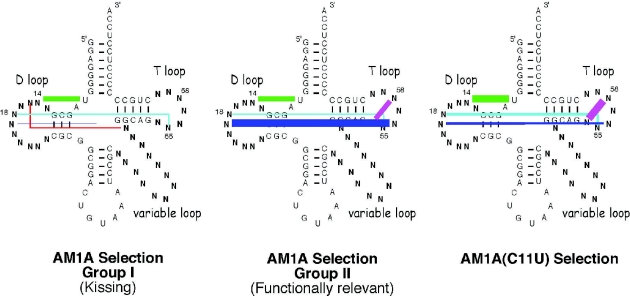
Schematic representation of the relative percentage of native tRNA tertiary base pairings observed for each of the selection groups shown in Figure 2. Tertiary base pairings are color coded as in Figure 2. Relative percentage of selected tRNA sequences with designated base pairing represented by line widths as follows: <10%, hairline; 10–19%, 1 pt line; 20–39%, 2 pt line; 39–50%, 4 pt line; >50%, 6 pt line.
Tertiary fold of tRNA selected
Many of the selected tRNAs had complementarity at positions where long-range base pairing is found in native tRNA (28). This tertiary folding was especially prevalent in the functionally relevant, non-kissing tRNA sequences (Group II) selected for binding AM1A and, in most of the tRNA sequences selected for binding AM1A(C11U) (Figures 2 and 3). While the complementary base pairs were often canonical, the primary sequence was not conserved; in particular, none of the selected tRNA sequences contained the contiguous GG seen at positions 18 and 19 in the D loop of canonical tRNA. While the sequences and relative importance of tertiary interactions often differed significantly from that found in the wild-type B.subtilis tRNATyr, there is precedence for alternate tRNA forms in nature as exhibited by the mitochondrial tRNAs (30).
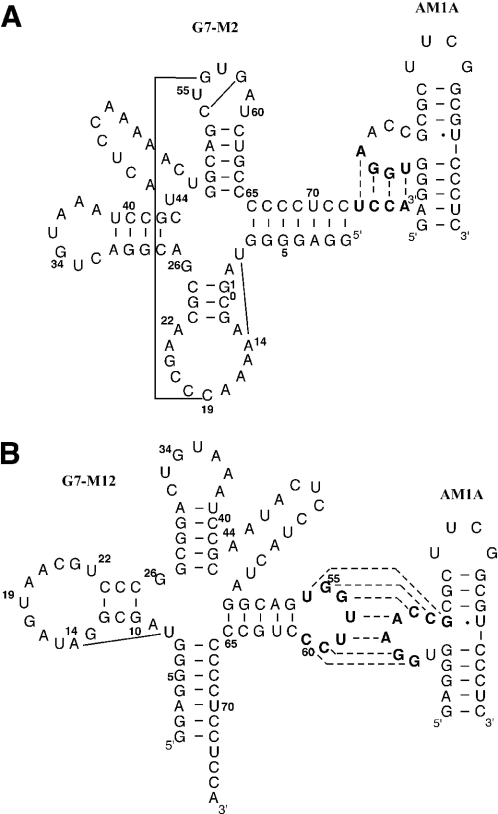
The most notable difference between the non-kissing, functionally relevant tRNA sequences (Group II) selected to bind AM1A and the sequences selected to bind AM1A(C11U) was the variation in which of the tertiary interactions were dominant in the selected sequences. In the case of the AM1A selected functional tRNAs, the tertiary interaction between the D and T loops was the dominant feature retained (e.g. G7–M2, Figure 4A) with 57% (4/7 sequences) having the 19:56 bp (Figure 3). Of somewhat less selective importance were the 8:14 (43%, 3/7) and 54:58 (43%, 3/7) bp. In contrast, for the AM1A(C11U) selection, the 8:14 (72%, 8/11) and 54:58 (72%, 8/11) base pairings were the dominant tertiary interactions retained. In addition, both base pairs between the D and T loop, 18:55 (27%, 3/11) and 19:56 (27%, 3/11), were of equal importance in the AM1A(C11U) selection, but for AM1A the 19:56 bp was preferentially selected ∼4-fold more often than the 18:55 (14%, 1/7) bp. The difference in the relative importance of the various tertiary base pairs in the selected tRNAs is consistent with AM1A and AM1A(C11U) having subtle differing recognition features for binding tRNA and relates to the structural and dynamic differences within the bulge region of the model antiterminator RNAs (M. Gerdeman, T. Henkin and J. Hines, unpublished data). These different recognition features most likely play a role in the decreased B.subtilis tRNATyr(A73U) affinity for AM1A(C11U) compared with AM1A (8) and in the reduced antitermination efficiency in vivo for the corresponding C224U base change in the context of the full leader mRNA (2).
Figure 4.
Secondary structure of representative selected tRNA/antiterminator complexes involving (A) functionally relevant acceptor end-bulge nucleotide pairing and (B) the kissing tRNA T loop–bulge nucleotide complex. For both, solid lines indicate complementary nucleotides involved in tRNA tertiary structure interactions (for evaluation criteria see Materials and Methods). Dashed lines indicate complementary nucleotides involved in tRNA–antiterminator interaction.
Kissing tRNA loop–antiterminator bulge interaction selected
Of the tRNA sequences selected for binding AM1A 81% (30/37) of the selected sequences showed a kissing loop–bulge complementarity between the T loop of the tRNA and the bulge of the antiterminator (e.g. G7–M12, Figure 4B). None of the other randomized tRNA loops exhibited this complementary relationship to the antiterminator bulge nucleotides. In addition, none of the tRNA selected to bind AM1A(C11U) had a kissing loop–bulge interaction. As summarized schematically in Figure 3, only 14% (5/37) of the kissing tRNA sequences retained any tertiary fold complementarity between the D and T loop and none had the T-loop base pair. In contrast 43% (16/37) retained other aspects of tRNA tertiary structure (e.g. 8:14 and 15:48 bp). The lack of tertiary structure involving the T loop presumably facilitates the base pairing of all the T-loop nucleotides with nucleotides in the bulge of the antiterminator.
Another interesting observation with the AM1A selection studies is that when the separation step (separating free from bound tRNA) involved centrifugation (i.e. Ultralink streptavidin beads), all but one of the 18 selected tRNA sequences were the tighter binding kissing tRNAs (Figure 2). In contrast, the use of a magnetic separation (Magnabind streptavidin beads) resulted in an increase in the number of non-kissing sequences presumably owing to the weaker binding tRNAs surviving the separation method. However, even with the magnetic separation, the kissing tRNAs still dominated the cloned sequences for AM1A.
Selected tRNA/antiterminator binding rivals wild-type tRNA binding
Binding of selected non-kissing, functionally relevant tRNA (Group II) sequences with model antiterminator RNAs was comparable with or better than that seen with wild-type tRNA (Table 1). This was especially pronounced for tRNAs selected to bind AM1A(C11U). In addition, specificity for a fully matched acceptor end to the antiterminator was observed [e.g. G7–M2 versus G7–M2(ACCA)] and was comparable with the specificity seen with wild-type tRNA [B.subtilis tRNATyr(A73U) versus B.subtilis tRNATyr] and AM1A (8). However, the extent of the specificity for a complementary base pair between the discriminator base of the tRNA and the variable base of the antiterminator (position 9 in AM1A) appears to be affected by the tRNA structure. In the case of G6–M18 there was only a modest reduction in binding affinity upon introduction of a discriminator base-variable base mismatch [G6–M18(ACCA)]. It is possible that the different tertiary base pairs in G6–M18 (18:55, 19:56 and 54:58) compared with G7–M2 (8:14, 19:56 and 54:58) may play a role in how well a mismatch at the discriminator base of tRNA is tolerated when binding antiterminator model RNA.
Table 1.
Kd (μM) of representative selected tRNAs and mutants for binding antiterminator model RNAs
| tRNA | AM1Aa | AM1A(C11U)a |
|---|---|---|
| B.subtilis tRNATyr(A73U) | 63b | 200b |
| AM1A Group I (kissing) | ||
| G7–U12 | 0.3 | – |
| G7–U12(ACCA)c | 0.5 | – |
| G7–M12 | 0.3 | – |
| G7–M12(mTL)d | 134 ± 30 | – |
| G7–M12(mTL)(AGGU)c,d | n.d. | – |
| G7–M14 | 0.5 | – |
| G7–M14(ACCA)c | 0.5 | – |
| AM1A Group II (functionally relevant) | ||
| G7–M2 | 36 ± 4 | – |
| G7–M2(ACCA)c | n.d. | – |
| G6–M18 | 7 | – |
| G6–M18(ACCA)c | 11 ± 5 | – |
| AM1A(C11U) | ||
| G7–CU1 | 53 ± 22 | 5 |
| G7–CU1(ACCA)c | – | 7 |
| G7–CU5 | – | 26 ± 6 |
n.d., no binding detected; ‘—’, not applicable. All values ± 1.0 (unless otherwise noted) based on replicate measurements.
aValues (except as noted) determined by gel shift as described in Materials and Methods.
bValues from (8).
cSequence of acceptor end 5′–3′ introducing one (ACCA) or complete (AGGU) mismatch with antiterminator model RNAs.
dSequence in the T loop 5′-AAGGAAC-3′.
Binding studies of the kissing T loop–antiterminator model RNA (Table 1) indicated a low micromolar to nanomolar affinity, i.e. approximately two orders of magnitude tighter binding than the native binding of the acceptor end of tRNA with AM1A. When the kissing base pairing was disrupted [e.g. G7–M12(mTL)], binding was significantly reduced even compared with that found with the wild-type tRNATyr(A73U). When the tRNA sequence was further mutated to disrupt the acceptor end complementarity to the antiterminator bulge nucleotides G7–M12(mTL)(AGGU), no binding was observed. These data confirm the kissing T loop–antiterminator bulge interaction for the Group I tRNA sequences.
DISCUSSION
The most significant and intriguing result of these studies is that antiterminator model RNAs select for the correct tertiary fold of tRNA presumably because the correct tRNA tertiary fold presents the acceptor end for optimal binding to the bulge nucleotides of the antiterminator. The details of the fold [i.e. which of the tRNA tertiary base pairings (28) are preferentially selected] varies with the antiterminator model RNA (Figures 2 and 3). These variations indicate a selective pressure for optimal tRNA–antiterminator matches consistent with in vivo studies highlighting the existence of optimal matches (6). No other tRNA sequence conservation was observed consistent with few, if any, specific base pairing interactions between other regions of the tRNA and the antiterminator. These observations are consistent with in vivo transcription antitermination results that indicated an overall requirement for the tertiary fold of the tRNA rather than the primary sequence (12).
The functionally relevant, non-kissing tRNA sequences selected to bind AM1A (Group II) had binding affinities and sequence specificity rivaling that seen in the wild-type tRNA model, B.subtilis tRNATyr(A73U), binding AM1A (Table 1), indicating that the antiterminator alone can select for a tertiary fold of tRNA that binds optimally. It is intriguing that the 19:56 bp along with the 18:55 and 54:58 bp were the dominant tertiary interactions retained in the AM1A functionally relevant (Figure 2A, Group II) selected tRNA sequences, whereas only the 8:14 and 54:58 bp were dominant in the AM1A(C11U) selected sequences (Figure 3). Since the randomized pool and selection conditions were identical for the individual selections to each of the two antiterminator model RNAs, the selection differences most likely reflect structural and/or dynamic differences in how each antiterminator model interacts with tRNA. This is consistent with the spectroscopic data, which indicate that antiterminator model RNA AM1A(C11U) is less thermodynamically stable than AM1A and has less stacking of the bases presumably in the region of the bulge (M. Gerdeman, T. Henkin and J. Hines unpublished data). These differences could affect how AM1A(C11U) binds tRNA and result in the selection of a different class of tRNAs from that seen with AM1A, namely, tRNAs that are optimally suited for the altered flexibility of AM1A(C11U). The preferential binding of the selected sequence G7–CU1 to AM1A(C11U) compared with AM1A (Table 1) indicates that the tRNAs selected were optimized for binding AM1A(C11U). In the native context, this C to U mutation results in reduced tRNA binding (8) and antitermination function (2,4). The tRNA selection differences between the two antiterminator model RNAs are consistent with the antiterminator structure playing a role in recognition of the tRNA acceptor end beyond the known base pairing interaction. Previous in vivo T box transcription antitermination studies have indicated the importance of optimal tRNA-antiterminator matches (6,12) and in vitro transcription data highlight the functional role of the antiterminator in dynamically sampling the tRNA acceptor end (31).
While the kissing tRNAs (Figure 2A, Group I) are not functionally relevant to the T box transcription control mechanism, the fact that only AM1A selected for these tRNAs again illustrates differences between the two antiterminator model RNAs. The selection differences are consistent with in vivo antitermination efficiency (2) and in vitro tRNA affinity differences (8) observed with this single C to U nucleotide change. The observed selection differences are most likely due to functionally significant structure and dynamic differences between the two antiterminator model RNAs. Surprisingly, all of the kissing loop–bulge sequences selected for binding AM1A involved the T loop, indicating that the tertiary fold of the selected tRNA presents the T loop in a favorable manner for interaction with the antiterminator RNA more so than do the other loops in tertiary folded tRNA. Aptamers selected to bind tRNA can readily participate in complementary base pairing with the D as well as the T loop (17), indicating that the specificity for the T loop, in the case of the antiterminator, may be due to additional structural constraints imposed by the tRNA–antiterminator complex. The selection of tRNA sequences, where the T loop is complementary to the bulge nucleotides of AM1A, indicates that this antiterminator model RNA is predisposed to readily bind RNA leading to a strong selection force for complementary RNA sequences. This is consistent with previous observations that the B.subtilis tyrS wild-type sequence in the bulge of the antiterminator had such a propensity for binding RNA that the corresponding model RNA readily formed homodimers (8).
The model antiterminator RNAs investigated differ in their ability to bind wild-type B.subtilis tRNATyr(A73U) (8), in their in vivo antitermination efficiency (2), and have subtle structural differences (M. Gerdeman, T. Henkin and J. Hines, unpublished data). The fact that the two model antiterminator RNAs selected for different tRNA tertiary interactions is consistent with these other observed functional differences. In the case of AM1A, tRNA sequences were selected that bound, via the functionally relevant acceptor end–bulge nucleotide base pairing, with affinity comparable with the wild-type tRNA sequence. In the case of AM1A(C11U) a tRNA was selected that bound the antiterminator model RNA significantly better than the wild-type tRNA consistent with in vivo data, indicating that optimal tRNA–antiterminator matches may exist (6). The dominant tertiary base pairings within the selected tRNAs differed depending on the antiterminator model. The differing selection results with the two model antiterminator RNAs further support the concept that binding between the T box antiterminator RNA and tRNA involves additional recognition features (e.g. structure and dynamics) beyond the base pairing complementarity of the tRNA acceptor end with the first four bases in the bulge of the antiterminator.
Acknowledgments
This work was supported by National Institutes of Health Grant GM61048. Funding to pay the Open Access publication charges for this article was provided by the Department of Chemistry & Biochemistry Ohio University.
Conflict of interest statement. None declared.
REFERENCES
- 1.Grundy F.J., Henkin T.M. tRNA as a positive regulator of transcription antitermination in B.subtilis. Cell. 1993;74:475–482. doi: 10.1016/0092-8674(93)80049-k. [DOI] [PubMed] [Google Scholar]
- 2.Rollins S.M., Grundy F.J., Henkin T.M. Analysis of cis-acting sequence and structural elements required for antitermination of the Bacillus subtilis tyrS gene. Mol. Microbiol. 1997;25:411–421. doi: 10.1046/j.1365-2958.1997.4851839.x. [DOI] [PubMed] [Google Scholar]
- 3.Grundy F.J., Henkin T.M. The T box and S box transcription termination control systems. Front. Biosci. 2003;8:d20–d31. doi: 10.2741/908. [DOI] [PubMed] [Google Scholar]
- 4.Grundy F.J., Moir T.R., Haldeman M.T., Henkin T.M. Sequence requirements for terminators and antiterminators in the T box transcription antitermination system: disparity between conservation and functional requirements. Nucleic Acids Res. 2002;30:1646–1655. doi: 10.1093/nar/30.7.1646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yousef M.R., Grundy F.J., Henkin T.M. Structural transitions induced by the interaction between tRNAGly and the Bacillus subtilis glyQS T box leader RNA. J. Mol. Biol. 2005 doi: 10.1016/j.jmb.2005.03.061. In press. [DOI] [PubMed] [Google Scholar]
- 6.Grundy F.J., Hodil S.E., Rollins S.M., Henkin T.M. Specificity of tRNA–mRNA interactions in Bacillus subtilis tyrS antitermination. J. Bacteriol. 1997;179:2587–2594. doi: 10.1128/jb.179.8.2587-2594.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Grundy F.J., Rollins S.M., Henkin T.M. Interaction between the acceptor end of tRNA and the T box stimulates antitermination in the Bacillus subtilis tyrS gene: a new role for the discriminator base. J. Bacteriol. 1994;176:4518–4526. doi: 10.1128/jb.176.15.4518-4526.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gerdeman M.S., Henkin T.M., Hines J.V. In vitro structure-function studies of the Bacillus subtilis tyrS mRNA antiterminator: evidence for factor independent tRNA acceptor stem binding specificity. Nucleic Acids Res. 2002;30:1065–1072. doi: 10.1093/nar/30.4.1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Henkin T.M. tRNA-directed transcription antitermination. Mol. Microbiol. 1994;13:381–387. doi: 10.1111/j.1365-2958.1994.tb00432.x. [DOI] [PubMed] [Google Scholar]
- 10.Grundy F.J., Winkler W.C., Henkin T.M. tRNA-mediated transcription antitermination in vitro: codon–anticodon pairing independent of the ribosome. Proc. Natl Acad. Sci. USA. 2002;99:11121–11126. doi: 10.1073/pnas.162366799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Putzer H., Condon C., Brechemeier-Baey D., Brito R., Grunberg-Manago D. Transfer RNA-mediated antitermination in vitro. Nucleic Acids Res. 2002;30:3026–3033. doi: 10.1093/nar/gkf415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Grundy F.J., Collins J.A., Rollins S.M., Henkin T.M. tRNA determinants for transcription antitermination of the Bacillus subtilis tyrS gene. RNA. 2000;6:1131–1141. doi: 10.1017/s1355838200992100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Feigon J., Dieckmann T., Smith F.W. Aptamer structures from A to Z. Chem. Biol. 1996;3:611–617. doi: 10.1016/s1074-5521(96)90127-1. [DOI] [PubMed] [Google Scholar]
- 14.Famulok M., Jenne A. Oligonucleotide libraries—variatio delectat. Curr. Opin. Chem. Biol. 1998;2:320–327. doi: 10.1016/s1367-5931(98)80004-5. [DOI] [PubMed] [Google Scholar]
- 15.Ducongé F., Toulmé J.J. In vitro selection identifies key determinants for loop–loop interactions: RNA aptamers selective for the TAR RNA element of HIV-1. RNA. 1999;5:1605–1614. doi: 10.1017/s1355838299991318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tok J.B., Cho J., Rando R.R. RNA aptamers that specifically bind to a 16S ribosomal RNA decoding region construct. Nucleic Acids Res. 2000;28:2902–2910. doi: 10.1093/nar/28.15.2902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Scarabino D., Crisari A., Lorenzini S., Williams K., Tocchini-Valentini G.P. tRNA prefers to kiss. EMBO J. 1999;18:4571–4578. doi: 10.1093/emboj/18.16.4571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lodmell J.S., Ehresmann C., Ehresmann B., Marquet R. Convergence of natural and artificial evolution on an RNA loop–loop interaction: the HIV-1 dimerization initiation site. RNA. 2000;6:1267–1276. doi: 10.1017/s1355838200000698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sekkai D., Dausse E., Di Primo C., Darfeuille F., Boiziau C., Toulmé J.-J. In vitro selection of DNA aptamers against the HIV-1 TAR RNA hairpin. Antisense Nucleic Acid Drug Dev. 2002;12:265–274. doi: 10.1089/108729002320351584. [DOI] [PubMed] [Google Scholar]
- 20.Peterson E.T., Pan T., Coleman J., Uhlenbeck O.C. In vitro selection of small RNAs that bind to Escherichia coli phenylalanyl-tRNA synthetase. J. Mol. Biol. 1994;242:186–192. doi: 10.1006/jmbi.1994.1571. [DOI] [PubMed] [Google Scholar]
- 21.Bhattacharyya S.N., Chatterjee S., Adhya S. Mitochondrial RNA import in Leishmania tropica: aptamers homologous to multiple tRNA domains that interact cooperatively or antagonistically at the inner membrane. Mol. Cell. Biol. 2002;22:4372–4382. doi: 10.1128/MCB.22.12.4372-4382.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Asahara H., Nameki N., Hasegawa T. In vitro selection of RNAs aminoacylated by Escherichia coli leucyl-tRNA synthetase. J. Mol. Biol. 1998;283:605–618. doi: 10.1006/jmbi.1998.2111. [DOI] [PubMed] [Google Scholar]
- 23.Pan T., Uhlenbeck O.C. In vitro selection of RNAs that undergo autolytic cleavage with Pb2+ Biochemistry. 1992;31:3887–3895. doi: 10.1021/bi00131a001. [DOI] [PubMed] [Google Scholar]
- 24.Gerdeman M.S., Henkin T.M., Hines J.V. Solution structure of B.subtilis T box antiterminator RNA: seven nucleotide bulge characterized by stacking and flexibility. J. Mol. Biol. 2003;326:189–201. doi: 10.1016/s0022-2836(02)01339-6. [DOI] [PubMed] [Google Scholar]
- 25.Tao J., Frankel A.D. Arginine-binding RNAs resembling TAR identified by in vitro selection. Biochemistry. 1996;35:2229–2238. doi: 10.1021/bi951844b. [DOI] [PubMed] [Google Scholar]
- 26.Aldaz-Carroll L., Tallet A., Dausse E., Yurchenko L., Toulmé J.J. Apical loop-internal loop interactions: a new RNA–RNA recognition motif identified through in vitro selection against RNA hairpins of the hepatitis C virus mRNA. Biochemistry. 2002;41:5883–5893. doi: 10.1021/bi0121508. [DOI] [PubMed] [Google Scholar]
- 27.Green M.R., Maniatis T., Melton D.A. Human beta-globulin pre-mRNA synthesized in vitro is accurately spliced in Xenopus oocyte nuclei. Cell. 1983;32:681–694. doi: 10.1016/0092-8674(83)90054-5. [DOI] [PubMed] [Google Scholar]
- 28.Arnez J.G., Moras D. Transfer RNA. In: Neidle S., editor. Oxford Handbook of Nucleic Acid Structure. Oxford: Oxford University Press; 1999. pp. 603–651. [Google Scholar]
- 29.Shi H., Moore P.B. The crystal structure of yeast phenylalanine tRNA at 1.93 Å resolution: a classic structure revisited. RNA. 2000;6:1091–1105. doi: 10.1017/s1355838200000364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Helm M., Brulé H., Friede D., Giegé R., Pütz D., Florentz C. Search for characteristic structural features of mammalian mitochondrial tRNAs. RNA. 2000;6:1356–1379. doi: 10.1017/s1355838200001047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Grundy F.J., Yousef M.R., Henkin T.M. Monitoring uncharged tRNA during transcription of the Bacillus subtilis glyQS gene. J. Mol. Biol. 2005;346:73–81. doi: 10.1016/j.jmb.2004.11.051. [DOI] [PubMed] [Google Scholar]