Abstract
In order to further understand how DNA polymerases discriminate against incorrect dNTPs, we synthesized two sets of dNTP analogues and tested them as substrates for DNA polymerase α (pol α) and Klenow fragment (exo−) of DNA polymerase I (Escherichia coli). One set of analogues was designed to test the importance of the electronic nature of the base. The bases consisted of a benzimidazole ring with one or two exocyclic substituent(s) that are either electron-donating (methyl and methoxy) or electron-withdrawing (trifluoromethyl and dinitro). Both pol α and Klenow fragment exhibit a remarkable inability to discriminate against these analogues as compared to their ability to discriminate against incorrect natural dNTPs. Neither polymerase shows any distinct electronic or steric preferences for analogue incorporation. The other set of analogues, designed to examine the importance of hydrophobicity in dNTP incorporation, consists of a set of four regioisomers of trifluoromethyl benzimidazole. Whereas pol α and Klenow fragment exhibited minimal discrimination against the 5- and 6-regioisomers, they discriminated much more effectively against the 4- and 7-regioisomers. Since all four of these analogues will have similar hydrophobicity and stacking ability, these data indicate that hydrophobicity and stacking ability alone cannot account for the inability of pol α and Klenow fragment to discriminate against unnatural bases. After incorporation, however, both sets of analogues were not efficiently elongated. These results suggest that factors other than hydrophobicity, sterics and electronics govern the incorporation of dNTPs into DNA by pol α and Klenow fragment.
INTRODUCTION
Fidelity of base pairing during DNA replication is the foundation upon which a stable genetic code is built. It is therefore not surprising that replicative DNA polymerases have evolved to make very few errors [one in 104–>105 dNTPs polymerized, (1–3)] when selecting which of the four natural dNTPs to insert opposite a given template base. The manner in which DNA polymerases do this, however, is not yet well understood. Nor is it clear if all polymerases use the same mechanism, or if several different mechanisms have evolved. The earliest theories on polymerase fidelity proposed that polymerase discrimination arose from the hydrogen bonds between the nascent base pair (4,5). Although it is logical to assume that the proper match of the hydrogen bonds between a correct base pair is the basis for the high fidelity of DNA replication, the difference in thermodynamic stability between a matched and mismatched base pair cannot solely account for the low error rates (6–8).
It has thus been argued that polymerases amplify the small thermodynamic advantage of correct base pairing through other factors, such as the geometry of a Watson–Crick pair (9,10). Because both a G–C and an A–T pair have roughly the same shape, it has been proposed that the active sites of DNA polymerases contain a pocket of this geometry. According to this hypothesis, the distortion caused by a mismatch does not allow the polymerase to adopt the conformation necessary to catalyze phosphodiester bond formation. On the other hand, a proper geometric fit in the active site promotes catalysis and thus proper nucleotide insertion. The ability of dNTPs to diffuse in and out of the active site easily allows for the sampling necessary in this type of nucleotide selection.
In support of this idea, some DNA polymerases incorporate nucleotide analogues that are isosteric to the natural dNTPs but lack hydrogen-bonding capability (11,12). However, this only truly supports the idea that hydrogen bonds are not necessary for the polymerization reaction. It does not preclude the possibility that other nucleotides, which lack hydrogen bonds and are also of altered geometry, can also serve as good substrates for DNA polymerases. Indeed, we recently showed that some DNA polymerases efficiently incorporate nucleotide analogues that form base pairs with geometry that is most likely severely distorted from that of a Watson–Crick pair. For example, both pol α and Klenow fragment (KF) of Escherichia coli DNA polymerase I, exonuclease deficient polymerize dNTP analogues containing the bases benzimidizole, 5- or 6-nitrobenzimidazole, and 5-nitroindole orders of magnitude more efficiently than they misincorporate a natural dNTP (13). Both enzymes incorporated the nitrated derivatives more efficiently than they incorporated the parent benzimidazole. Likewise, the Romesberg and Schultz groups showed that KF will polymerize a variety of large, hydrophobic purine and pyrimidine analogues at rates that occasionally approach those for a natural, cognate dNTP (14–20).
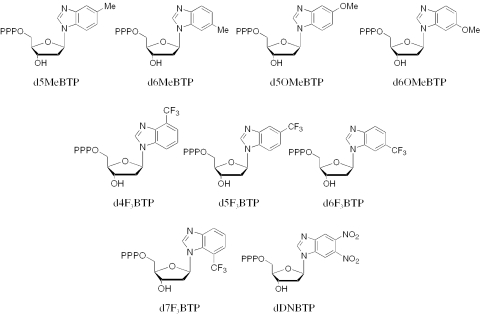
To better understand the ability of pol α and KF to polymerize dNTP analogues containing bases whose shape does not closely resemble the canonical dNTPs, we synthesized a series of dNTP analogues containing benzimidazole derivatives (Figure 1). The results of these studies indicate that the inability of pol α and KF to discriminate against unnatural bases does not result from either the hydrophobicity of the bases or the electronic nature of the aromatic ring (electron rich or deficient).
Figure 1.
Structures of the dNTP analogues discussed.
MATERIALS AND METHODS
All reagents were of the highest quality commercially available. Unlabeled dNTPs were from Sigma and radiolabeled dNTPs from New England Nuclear. Synthetic DNA oligonucleotides were purchased from Oligos, etc. or BioSearch, and their concentrations determined spectrally. Klenow fragment (exo−) was purchased from New England BioLabs, and human DNA pol α (4-subunit complex) was expressed and purified as previously described (21). 5-Nitrobenzimidazole was purchased from Lancaster and 5-methylbenzimidazole and 5-methoxybenzimidazole were purchased from Sigma. 4-Trifluoromethyl-1H-benzimidazole, 5-trifluoromethyl-1H-benzimidazole (22) and 1-β-D-2′-deoxyribofuranosyl-(4-methylbenzimidazole)-5′-triphosphate (dZTP) (11) were synthesized as previously described.
5,6-Dinitrobenzimidazole
To a stirred solution of 2 g 5-nitrobenzimidazole in 30 ml fuming H2SO4 at 0°C was added 15 ml of 1:1 fuming H2SO4/KNO3. After stirring 6 h at 110°C, the temperature was lowered to 0°C and 37% ammonia water was added dropwise to neutralize the solution. The neutralized crude product was extracted into EtOAc and purified by silica column chromatography (EtOAc/toluene, 7:3) to give the title compound as a creamy white powder in 70% yield.
1H NMR (300 MHz, d6-DMSO): δ 8.46 (s, 2H, ArH), 8.74 (s, 1H, ArH).
3′,5′-O-toluyl protected nucleosides
The bases were glycosylated using the procedure of Kazimierczuk et al. (23). Briefly, 1-methyl-2-deoxy-3,5-bis-O-p-toluoyl-α-D-erythro-pentofuranose (Aldrich) was dissolved in AcOH saturated with HCl. HCl(g) was bubbled through the reaction solution for 5 min. After ∼10 min, a solid white precipitate developed, which was isolated, washed with Et2O and dried briefly. The substituted benzimidazole (1.5 eq) was dissolved in dry CH3CN and treated with NaH (2 eq). After 45 min, 1-chloro-2-deoxy-3,5-bis-O-p-toluoyl-α-D-erythro-pentofuranose (1 eq) was added and the reaction allowed to stir for 1.5 h at room temperature. The reaction was then quenched with aqueous NH4Cl and partitioned between water and EtOAc. The organic layer was dried, filtered and concentrated. Flash chromatography of the resulting solid (silica, EtOAc) yielded the protected nucleoside. Regioisomer separation was achieved by a combination of flash chromatography and preparative HPLC (silica) using EtOAc/MeOH, 9:1.
Deprotection of the nucleosides
The separate isomers were dissolved in MeOH and treated with NaOMe (1 eq). After stirring at room temperature for 1.5 h, the reaction was quenched with solid ammonium bicarbonate and partitioned between water and EtOAc. The organic layer was then dried, filtered, concentrated and chromatographed on silica (EtOAc/MeOH, 9:1), giving deprotected nucleosides in generally good yield (80–90%). Regioisomer assignments were determined via GOESY NMR experiments performed on either the protected or deprotected nucleosides. Interactions between the C7 proton on the benzimidazole ring and the anomeric protons on the sugar were observed. Individual proton assignments were determined using COSY experiments.
5/6-Methylbenzimidazole deoxyriboside
1H NMR (400 MHz, CDCl3): δ 2.37 (s, 3H, Me), 2.38 (s, 3H, Me), 2.41-2.48 (m, 4H, 2′, 2′), 3.70-3.84 (dd, 4H, 5′, 5′, J1 = 41.7, J2 = 12.2), 4.02 (m, 2H, 4′), 4.60-4.62 (m, 2H, 3′), 6.17 (t, 2H, 1′, J = 5.9 Hz), 7.0 (d, 2H, Ar, J = 8.4 Hz), 7.15 (s, 1H, Ar), 7.23 (d, 1H, Ar, J = 8.3 Hz), 7.44 (s, 1H, Ar), 7.51 (d, 1H, Ar, J = 8.3 Hz), 8.2 (br s, 2H, Ar); HRMS (ESI+): 249.1237 ([M•H]+ calc. 249.1234).
5-Methoxybenzimidazole deoxyriboside
1H NMR (500 MHz, CDCl3): δ 2.49 (m, 1H 2′), 2.51 (m, 1H, 2′), 3.81-3.90 (m, 5H, OMe, 5′, 5′), 4.08 (dd, 1H, 4′, J1 = 7.5 Hz, J2 = 4 Hz), 4.71 (m, 1H, 3′), 6.29 (t, 1H, 1′, J = 6 Hz), 6.95 (d, 1H, ArH, J = 9 Hz), 7.39 (d, 1H, ArH, J = 9.5 Hz), 8.13 (s, 1H, ArH); one proton obscured by solvent peak; HRMS (EI+): 264.1111 (M+ calc. 264.1110).
6-Methoxybenzimidazole deoxyriboside
1H NMR (500 MHz, CDCl3): δ 2.48 (m, 1H, 2′), 2.59 (m, 1H, 2′), 3.77 (dd, 1H, 5′, J1 = 12 Hz, J2 = 3 Hz), 3.82 (s, 3H, OMe), 3.87 (dd, 1H, 5′, J1 = 12 Hz, J2 = 3 Hz), 4.05 (dd, 1H, 4′, J1 = 7Hz, J2 = 3 Hz), 4.69 (dd, 1H, 3′, J1 = 11 Hz, J2 = 5 Hz), 6.21 (t, 1H, 1′, J = 6 Hz), 6.86 (dd, 1H, ArH, J1 = 8.5 Hz, J2 = 2.5 Hz), 6.89 (s, 1H, ArH), 7.56 (d, 1H, ArH, J = 8.5 Hz), 8.15 (br s, 1H, ArH); HRMS (EI+): 264.1108 (M+ calc. 264.1110).
4-Trifluoromethylbenzimidazole deoxyriboside
1H NMR (500 MHz, d6-acetone): δ 2.56 (m, 1H, 2′), 2.77 (m, 1H, 2′), 3.81 (m, 2H, 5′), 4.09 (m, 1H, 4′), 4.30 (br, 1H, 5′-OH), 4.62 (br, 1H, 3′-OH), 4.70 (m, 1H, 3′), 6.53 (dd, 1H, 1′, J1 = 6 Hz, J2 = 7.5 Hz), 7.45 (t, 1H, ArH, J = 8 Hz), 7.61 (d, 1H, ArH, J = 8 Hz), 8.09 (d, 1H, ArH, J = 8 Hz), 8.58 (s, 1H, ArH); (MS) (MALDI+): 303 ([M•H]+calc. 303).
5-Trifluoromethylbenzimidazole deoxyriboside
1H NMR (400 MHz, d6-acetone): δ 2.52 (m, 1H, 2′), 2.74 (m, 1H, 2′), 3.79 (m, 2H, 5′), 4.05 (dd, 1H, 4′, J1 = J2 = 4.8 Hz), 4.24 (t, 1H, 5′-OH, J = 8 Hz), 4.58 (d, 1H, 3′-OH, J = 5.2 Hz), 4.66 (m, 1H, 3′), 6.50 (t, 1H, 1′, J = 6 Hz), 7.58 (dd, 1H, ArH, J1 = 8.8 Hz, J2 = 1.2 Hz), 7.97 (d, 1H, ArH, J = 8.4 Hz), 8.00 (s, 1H, ArH), 8.58 (s, 1H, ArH); HRMS (EI+): 302.0882 (M+ calc. 302.0878).
6-Trifluoromethylbenzimidazole deoxyriboside
1H NMR (400 MHz, d6-acetone): δ 2.52 (m, 1H, 2′), 2.75 (m, 1H, 2′), 3.80 (m, 2H, 5′), 4.07 (dd, 1H, 4′, J1 = 4.4 Hz, J2 = 4.8 Hz), 4.27 (br s, 1H, 5′-OH), 4.58 (br s, 1H, 3′-OH), 4.67 (m, 1H, 3′), 6.55 (dd, 1H, 1′, J1 = J2 = 8 Hz), 7.56 (dd, 1H, ArH, J1 = 9.6 Hz, J2 = 1.6 Hz), 7.85 (d, 1H, ArH, J = 11.2 Hz), 8.25 (s, 1H, ArH), 8.61 (s, 1H, ArH); HRMS (EI+): 302.0882 (M+ calc. 302.0878).
7-Trifluoromethylbenzimidazole deoxyriboside
1H NMR (500 MHz, d6-acetone): δ 2.572 (m, 2H, 2′), 3.94 (m, 2H, 5′), 4.08 (m, 1H, 4′), 4.45 (br, 1H, 5′-OH), 4.65 (br, 1H, 3′-OH), 4.71 (m, 1H, 3′), 6.62 (t, 1H, 1′, J = 6 Hz), 7,44 (t, 1H, ArH, J = 8 Hz), 7.72 (d, 1H, ArH, J = 8 Hz), 8.01 (d, 1H, ArH, J = 8 Hz), 8.93 (s, 1H, ArH); MS (MALDI) : 303 ([M•H]+ calc. 303).
5,6-Dinitrobenzimidazole deoxyriboside
1H NMR (400 MHz, d6-acetone): δ 2.55 (m, 1H, 2′), 2.78 (m, 1H, 2′), 3.85 (m, 2H, 5′), 4.15 (dd, 1H, 4′, J1 = J2 = 3.6 Hz), 4.40 (t, 1H, 5′-OH, J = 6 Hz), 4.62 (d, 1H, 3′-OH, J = 3.9 Hz), 4.71 (m, 1H, 3′), 6.63 (t, 1H, 1′, J = 5.4 Hz), 8.41 (s, 1H, ArH), 8.81 (s, 1H, ArH), 8.90 (s, 1H, ArH).
Nucleoside phosphorylation
The nucleosides were phosporylated using the method of Ludwig (24). Briefly, the nucleosides (1 eq) were prepared by co-evaporation from pyridine and dried in vacuo overnight. They were then dissolved in freshly and carefully distilled PO(OMe)3. The mixture was cooled to 0°C and POCl3 (1.5 eq) added. After stirring overnight at 4°C, the reaction was warmed to room temperature and both tributylammoniumpyrophosphate (5 eq) and tributylamine (5 eq) were added simultaneously. After stirring for 0.5 h, the reaction was quenched with 2 mL 1 M triethylammonium bicarbonate (TEAB). The mixture was diluted to 50 mL and applied directly to an ion exchange column (DE-52 resin). The column was washed with water to remove unreacted starting materials, and the product eluted with a gradient of 0–1 M TEAB. Fractions containing the dNTP were identified by UV activity and concentrated. The nucleotide was then desalted (BioGel P-2 resin) prior to use in enzymatic assays. Some nucleotides also required further purification by preparative HPLC (C18 resin, 0–100% CH3CN in 20 mM triethylammonium acetate).
1-β-D-2′-Deoxyribofuranosyl-(5/6-methylbenzimidazole)-5′-triphosphate (d5/6MeBTP): 31P NMR (400 MHz, D2O): δ –9.61(br s, γ-P), –10.40 (br s, α-P), –22.18 (br s, β-P), MS (ESI+): 489 ([M•H+] calc. 489), 590 ([M•N(Et)3 H]+ calc. 590).
1-β-D-2′-Deoxyribofuranosyl-(5-methoxybenzimidazole)-5′-triphosphate (d5OMeBTP): 31P NMR (400 MHz, D2O): δ –9.13(br s, γ-P), –10.26 (br s, α-P), –22.01 (br s, β-P), MS (ESI+): 505 ([M•H]+ calc. 505).
1-β-D-2′-Deoxyribofuranosyl-(6-methoxybenzimidazole)-5′-triphosphate (d6OMeBTP): 31P NMR (400 MHz, D2O): δ –9.19 (br s, γ-P), –10.27 (br s, α-P), –22.10 (br s, β-P), MS (ESI+): 505 ([M•H]+ calc. 505).
1-β-D-2′-Deoxyribofuranosyl-(4-trifluoromethylbenzimidazole)-5′-triphosphate (d4F3BTP): 31P NMR (400 MHz, D2O): δ –9.80 (d, γ-P, J = 42 Hz), –10.40 (d, α-P, J = 50 Hz), –22.16 (br t, β-P, J1 = 50 Hz, J2 = 42 Hz); 19F NMR (400 MHz, D2O): δ –61.2, MS (MALDI−): 541 ([M-H]− calc. 541).
1-β-D-2′-Deoxyribofuranosyl-(5-trifluoromethylbenzimidazole)-5′-triphosphate (d5F3BTP): 31P NMR (400 MHz, D2O): δ –4.27 (br m, γ-P), –9.45 (br m, α-P), –18.2 (br s, β-P); 19F NMR (400 MHz, D2O): δ –61.0 (s); MS (MALDI–): 541 ([M-H]− calc. 541).
1-β-D-2′-Deoxyribofuranosyl-(6-trifluoromethylbenzimidazole)-5′-triphosphate (d6F3BTP): 31P NMR (400 MHz, D2O): δ –5.02 (br m, γ-P), −10.0 (br s, α-P), −20.4 (br s, β-P); 19F NMR (400 MHz, D2O): δ –61.1 (s); MS (MALDI–): 541 ([M-H]− calc. 541).
1-β-D-2′-Deoxyribofuranosyl-(7-trifluoromethylbenzimidazole)-5′-triphosphate (d7F3BTP): 31P NMR (400 MHz, D2O): δ −9.25 (br d, γ-P, J = 46 Hz), −10.20 (d, α-P, J = 47.6 Hz), –21.85 (br t, β-P, J1 = 46 Hz, J2 = 47.6 Hz); 19F NMR (400 MHz, D2O): δ −57.8; MS (MALDI−): 541 ([M-H]− calc. 541).
1-β-D-2′-Deoxyribofuranosyl-(5,6-dinitrobenzimidazole)-5′-triphosphate (dDNBTP): 31P NMR (400 MHz, D2O): δ –5.01 (br s, γ-P), −9.94 (br s, α-P), –20.5 (br s, β-P); MS (MALDI–): 563 ([M-H]− calc. 563).
5′-End labeling of primers and annealing of primer/template pairs
DNA primers were 5′-[32P]-labeled using polynucleotide kinase and [γ-32P]ATP, gel purified and annealed to the appropriate template as described previously (25,26).
Polymerization assays with pol α and KF
All kinetic data were determined under steady-state conditions. Assays contained 1 μM 5′-[32P]-primer/template, 50 mM Tris–HCl (pH 7.6), 10 mM MgCl2, 1 mM dithiothreitol, 0.05 mg/mL bovine serum albumin, and various concentrations of dNTPs and/or analogues, in a total volume of 5 or 10 μL. Polymerization reactions were initiated by the addition of enzyme, incubated at 37°C for 5 min, and quenched by adding an equal volume of gel loading buffer (90% formamide in 1× Tris/Borate/EDTA buffer, 0.05% xylene cyanol and bromophenol blue). Products were separated by denaturing gel electrophoresis (20% acrylamide, 8 M urea) and analyzed by phosphorimagery (Molecular Dynamics). Kinetic parameters were determined by fitting the data to the Michaelis–Menten equation using KaleidaGraph software. Although enzyme concentration was adjusted to keep product formation <20%, all reported Vmax values were normalized to the same final enzyme concentration (2 nM for pol α; 3.3 unit/mL for KF). A Vmax of 1% extension corresponds to a kcat of 1 min−1 for pol α, and 0.83 min−1 for KF.
Polymerase read-through assays
Since both polymerases inserted the analogues much more efficiently than they did an incorrect dNTP, elongation past an incorporated analogue could be measured by simply including both the analogue triphosphate and the next correct dNTP in the reactions (i.e. TTP for DNAG,C,T and dATP for DNAA). In order to investigate the possibility of polymerase read-through, reaction conditions were adjusted such that a large amount of the primer was converted to primer +1 to provide a reasonable amount of substrate for the extension reaction. Thus, enzyme concentrations were 4 to 10 times higher than those used in the assays described above, and the dNTP concentrations were generally 100 μM. These conditions typically result in 50–100% of the primers being elongated via polymerization of an analogue triphosphate.
RESULTS
Our aim was to further explore DNA polymerase selectivity as it directly relates to the electronic character and hydrophobicity of the aromatic ring of the base. Therefore, we synthesized the nine dNTP analogues in Figure 1 and tested them as substrates for pol α and KF. Incorporation of d5OMeBTP, d6OMeBTP, d4F3BTP, d5F3BTP, d6F3BTP, d7F3BTP and dDNBTP was measured under steady-state conditions to determine how effectively these enzymes discriminate against them. In the case of compounds d5MeBTP and d6MeBTP, they were tested as a mixture of the two regioisomers—it proved impossible to separate them from one another at any stage in the synthesis, precluding the measurement of any accurate kinetic parameters for the individual regioisomers. (A variety of chromatographic techniques, including normal and reverse phase silica flash columns and HPLC, as well as crystallization were attempted on the protected and deprotected nucleosides and the dNTPs. In every case the two regioisomers were inseparable.) Comparison of the 5- and 6-substituted series was used to assess the importance of the electronic character of the purine ring for both KF and pol α. Incorporation of the four regioisomers of dCF3TP was compared to test the importance of hydrophobicity and stacking potential. In order to minimize the possibility that the DNA sequence around the template base being copied influenced the results, primer:templates were designed to monitor the polymerization of analogues from all four natural bases in essentially the same sequence context (Figure 2). For one of the four templates (DNAA), one additional base in the single-stranded template had to be altered in order to prevent insertion of two consecutive dTTPs.
Figure 2.
Sequences of the DNA primer/template pairs used. Primers are oriented 5′ to 3′ and templates 3′ to 5′ so that the complementary regions are easily visualized. The template base from which the incoming dNTP will be incorporated is underlined.
Incorporation by polymerase α
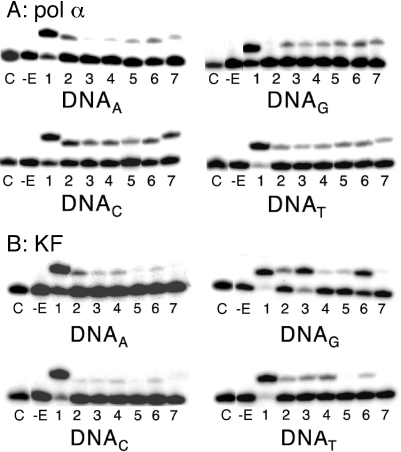
Pol α used all of the analogues as substrates (Table 1; Figure 3A). Two sets of control experiments indicated that the products generated were not due to contaminating normal dNTPs in the analogue triphosphates. First, using high percentage acrylamide gels (30 or 40% with 4 M urea), the products due to analogue incorporation had different electrophoretic mobilities than the products due to incorporation of the natural dNTPs. On 30% acrylamide gels, the products due to analogue incorporation had different electrophoretic mobilities than the products generated by incorporation of dATP, dCTP or dTTP, although the products generated by analogue and dG incorporation often comigrated. On 40% gels, however, the products due to analogue incorporation had different mobilities than the products due to dG incorporation. Second, none of the products due to analogue incorporation were readily elongated upon addition of the next correct dNTP. If the analogue incorporation had actually resulted from a contaminating natural dNTP, then the product of analogue incorporation should have been rapidly elongated on at least one of the templates since it would have actually been a correct base pair.
Table 1.
Kinetic parameters for the analogues, determined as described in Materials and Methods
| Template | Analog | Pol α | Klenow fragment | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Vmax (% ext) | KM (μM) | V/K (% ext/μM) | Discrimination | Vmax (% ext) | KM (μM) | V/K (% ext/μM) | Discrimination | ||
| DNAA | d5OMeBTP | 1.7 (0.8) | 132 (79) | 0.013 | 396 | 25.8 (2.1) | 247 (38) | 0.104 | 703 |
| DNAG | d5OMeBTP | 1.0 (0.6) | 38 (29) | 0.026 | 322 | 89.0 (23.8) | 11 (3) | 8.0 | 17 |
| DNAC | d5OMeBTP | 6.4 (3.2) | 25 (4) | 0.26 | 88 | 14.4 (0.7) | 562 (179) | 0.0257 | 5770 |
| DNAT | d5OMeBTP | 2.0 (1.0) | 40 (17) | 0.050 | 187 | 39.2 (17) | 145 (65) | 0.271 | 480 |
| DNAA | d6OMeBTP | 0.85 (0.4) | 20 (12) | 0.044 | 119 | 21.9 (12.9) | 60 (38) | 0.37 | 200 |
| DNAG | d6OMeBTP | 0.85 (0.2) | 29 (10) | 0.029 | 279 | 10.0 (4.6) | 26 (14) | 0.39 | 351 |
| DNAC | d6OMeBTP | 8.2 (5.1) | 11 (6) | 0.72 | 32 | 13.2 (11.5) | 242 (242) | 0.0545 | 2720 |
| DNAT | d6OMeBTP | 3.3 (1.9) | 21 (15) | 0.15 | 62 | 25.4 (11.5) | 41 (12) | 0.62 | 211 |
| DNAA | d5F3BTP | 2.1 (1.1) | 27 (24) | 0.077 | 68 | 6.4 (1.1) | 79 (18) | 0.080 | 913 |
| DNAG | d5F3BTP | 1.9 (0.3) | 9 (4) | 0.21 | 39 | 61.5 (17.3) | 160 (68) | 0.386 | 355 |
| DNAC | d5F3BTP | 8.1 (2.5) | 9 (0.4) | 0.87 | 26 | 0.9 (0.3) | 67 (48) | 0.014 | 10500 |
| DNAT | d5F3BTP | 11.3 (3) | 17 (10) | 0.66 | 14 | 1.4 (0.2) | 37 (15) | 0.037 | 3530 |
| DNAA | d6F3BTP | 3.2 (2.5) | 31 (23) | 0.10 | 50 | 10.1 (1.3) | 75 (12) | 0.13 | 546 |
| DNAG | d6F3BTP | 2.7 (1.8) | 17 (15) | 0.15 | 53 | 89.5 (23.5) | 58 (33) | 1.5 | 89 |
| DNAC | d6F3BTP | 13 (4.2) | 19 (10) | 0.69 | 33 | 10.4 (2.7) | 263 (61) | 0.0394 | 3760 |
| DNAT | d6F3BTP | 15.9 (12.2) | 35 (28) | 0.45 | 21 | 24.3 (36.4) | 33 (13) | 0.73 | 179 |
| DNAA | dDNBTP | 3.4 (0.2) | 29 (1) | 0.12 | 45 | 1.4 (0.5) | 39 (15) | 0.036 | 2030 |
| DNAG | dDNBTP | 1.6 (0.3) | 3 (0.4) | 0.48 | 17 | 6.7 (3.6) | 81 (13) | 0.083 | 1650 |
| DNAC | dDNBTP | 15.6 (2.3) | 6 (5) | 2.5 | 9 | 1.6 (0.3) | 250 (15) | 0.0064 | 23100 |
| DNAT | dDNBTP | 5.7 (1.1) | 16 (12) | 0.36 | 26 | 1.6 (0.1) | 130 (5) | 0.012 | 10500 |
| DNAA | d5/6MeBTP | nd | nd | nd | nd | 16.8 | 169 | 0.099 | 740 |
| DNAG | d5/6MeBTP | nd | nd | nd | nd | 87.5 | 109 | 0.805 | 170 |
| DNAC | d5/6MeBTP | nd | nd | nd | nd | 20.5 | 1570 | 0.0131 | 11300 |
| DNAT | d5/6MeBTP | nd | nd | nd | nd | 13.8 | 111 | 0.124 | 1050 |
Discrimination is defined as the ratio of Vmax/KM of the cognate dNTP to that of the analogue. A Vmax of 1% extension corresponds to a kcat of 1 min−1 for pol α, and 0.83 min−1 for KF. Values are averages of at least three experiments; standard deviations are in parentheses. nd: not determined.
Figure 3.
Representative incorporation of analogues. The template is indicated in the figure. C: no dNTP control; -E: no enzyme control; 1: cognate dNTP; 2: d5/6MeBTP; 3: d5OMeBTP; 4: d6OMeBTP; 5: d5F3BTP; 6: d6F3BTP; 7: dDNBTP. All assays were performed as described in Materials and Methods, and contained 10 μM dNTP analogue and either (A) 4 nM pol α or (B) 6.6 U/mL KF.
Remarkably, most of the analogues had Vmax/KM values only one order of magnitude lower than that for a cognate base with at least one of the templates tested. In general, pol α showed little preference for pairing a given analogue with a specific template base. This is shown most dramatically with d6F3BTP, in which the difference in Vmax/KM values for the best and worst templates is only a factor of 2.5. Even though the analogues are approximately the size of a purine, pol α showed only a slight preference to polymerize the analogs opposite pyrimidines. Rather, pol α exhibits a general permissiveness towards incorporation of all of the analogues, regardless of template base partner. In fact, the difference between the most efficient incorporation of an analogue across from a given template base (template C:dDNBTP) and the least (template A:d5OMeBTP) is only 44-fold. In contrast, previous work using these templates showed that on average, pol α discriminates against polymerizing natural, non-cognate dNTPs by 4 orders of magnitude [(13) and data not shown]. Thus, pol α incorporated all of these compounds 100–1000-fold more efficiently than natural mismatches.
Pol α discriminates much more effectively against 4- and 7-CF3-benzimidazole relative to the 5- and 6-substituted analogues (Figure 4A). Indeed, the discrimination against 7-CF3-benzimidazole approaches that seen for an incorrect natural dNTP. All four of these regioisomers have similar hydrophobicity (J. W. Engels, unpublished data). Since previous studies have shown that the stacking ability of a base closely matches the amount of buried surface area (27), these four bases should also have similar stacking ability. If stacking ability and/or hydrophobicity were the primary determinants for the incorporation of unnatural bases, then all four of these bases should have been similarly incorporated. This was not observed, however, indicating that other factors likely dominate incorporation of these unnatural bases.
Figure 4.
Differential discrimination of four regioisomers. (A) Discrimination by pol α; (B) Discrimination by KF. The analogues and templates used are indicated in the figure; the numerical value given above a bar is the discrimination factor. Discrimination is defined as the ratio of Vmax/KM of the cognate dNTP to that of the analogue. Vmax/KM were determined as described in Materials and Methods.
Incorporation by Klenow fragment
KF shows significantly more discrimination against the analogues than pol α (Table 1; Figure 3B). For example, KF discriminates against dDNBTP, which is the best substrate for pol α, by a factor of 103–104. This level of discrimination is similar to that observed for discrimination against an incorrect, natural dNTP. Conversely, the best base pair formed by KF (template G:d5OMeBTP) has a Vmax/KM only 17-fold worse than a canonical G:C base pair. Thus, the efficiency of incorporation by KF varied by 1400-fold between the best and worst incorporation events, a marked contrast to the lack of differentiation shown by pol α. Additionally, for a given analogue, KF shows a greater tendency to prefer pairing it with a specific template base. For four of the five analogues studied [as well as the mixture of d5/6MeBTP], this results in a surprising preference for pairing opposite a template guanosine. The reasons for this are not clear at this time.
As with pol α, KF discriminates against 5- or 6-substituted analogues less efficiently than it discriminates against natural non-cognate dNTPs (10–200-fold). However discrimination against d4CF3TP and d7CF3TP was significantly greater than that observed for pol α (Figure 4B). KF incorporated d7CF3TP so poorly that detectable incorporation only occurred with a 10-fold higher enzyme concentration and high concentrations of nucleotide (500 μM). This extremely low level of incorporation only allows us to provide a lower limit for discrimination with d7CF3TP. Again, the variation in discrimination against the four regioisomers of trifluoromethylbenzimidazole demonstrates that increased hydrophobicity alone cannot account for the incorporation of these analogues.
Polymerase read-through of the analogs
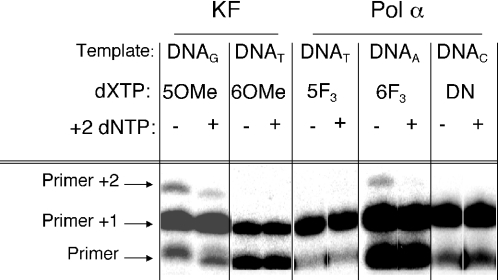
Both enzymes were tested for the ability to continue elongation of a primer once an analogue has been incorporated. In most cases, no elongation occurred (<0.2%, Figure 5). In a few instances, however, KF or pol α extended past an incorporated analogue (Table 2). Up to 3.1% of the product generated via incorporation of a single analogue was further elongated via incorporation of a second analogue. If either the enzyme or dNTP concentration was reduced, the analogue read-through was likewise reduced. Curiously, the enzymes tended to polymerize two consecutive analogues more efficiently than an analogue followed by a cognate dNTP. In cases where two consecutive analogues were incorporated, inclusion of the cognate dNTP for the 2 position did not increase elongation of primer +1 product, but rather decreased it. Presumably, this occurred due to the next correct dNTP binding in the active site and preventing binding of another molecule of analogue triphosphate [e.g. d6F3BTP on DNAA with pol α (Figure 5)]. Based on the amount of analogue read-through when it was seen, extension past an analogue is at best 104 times less efficient than extension past a natural cognate base pair.
Figure 5.
After incorporation of a dNTP analogue, pol α and KF do not readily polymerize the next correct dNTP. Assays were performed as described under Materials and Methods. The DNA polymerase, dNTP analogue, and template examined are shown in the figure. For DNAA, ‘+2 dNTP’ is dATP; for all other templates, it is TTP. When present, the +2 dNTP was at 100 μM. For the dXTP, 5OMe = d5OMeBTP, 6OMe = d6OMeBTP, etc. The data in the first and fourth set of lanes (d5OMeBTP and d6F3BTP, respectively) are significantly darkened in order to highlight the +2 product generated in the presence of only the analogue.
Table 2.
Extension past incorporated analogues
| Template | Analogue | +2 dNTP | Polymerase | %Elongation |
|---|---|---|---|---|
| DNAG | d5OMeBTP | d5OMeBTP | KF | 3.1 |
| DNAA | d6OMeBTP | d6OMeBTP | KF | 1.3 |
| DNAG | dOMeBTP | d6OMeBTP | KF | 1.9 |
| DNAA | d6F3BTP | d6F3BTP | Pol α | 0.9 |
| DNAT | d5OMeBTP | TTP | KF | 2.5 |
| DNAT | d6OMeBTP | TTP | KF | 2.5 |
Percent elongation indicates the amount of ‘primer +1’ DNA that was extended to ‘primer + 2’. Reactions contained 100 μM analogue dNTP, 100 μM TTP if indicated, and a 10-fold increase in polymerase relative to standard kinetic assays, with no correction in the resultant % extension.
DISCUSSION
We examined the ability of pol α and KF to polymerize a series of dNTP analogues whose bases consist of benzimidazole derivatives containing either electron-withdrawing or -donating groups. With the exception of the 7-trifluoromethylbenzimidazole base, pol α demonstrated a remarkable inability to discriminate against polymerization of these analogues, even though their shape varies substantially from the natural dNTPs. KF also polymerized the analogues, albeit somewhat less efficiently than pol α. Indeed, KF discriminated against polymerizing the most highly modified base, 5,6-dinitrobenzimidazole, almost as effectively as it discriminates against incorrect, natural dNTPs. On the other hand, pol α discriminated against this analogue 2–3 orders of magnitude less effectively than an incorrect, natural dNTP, incorporating it with an efficiency approaching that of a cognate dNTP.
Stacking ability and/or hydrophobicity cannot account for the inability of pol α to discriminate against these unnatural bases. Previous work has suggested that incorporation of dNTPs containing unnatural bases by T4 DNA polymerase, a B family polymerase like pol α, is primarily driven by the enhanced stacking ability of the unnatural bases relative to a natural base (28). The 4-, 5-, 6-, and 7-trifluoromethylbenzimidazole dNTPs have similar hydrophobicity and should have virtually identical stacking abilities (J. W. Engels, unpublished data). Importantly, pol α and KF incorporate these analogs with very different efficiencies. Thus, enhanced stacking ability and/or hydrophobicity of these bases relative to the natural bases cannot be the primary factor that causes either polymerase to exhibit minimal discrimination against most of the hydrophobic bases we have tested.
For the most part, pol α appears to lack the machinery to discriminate against benzimidazole derivatives bearing substituents at the 5 and 6 positions. The similarity of the data for all of the analogues reported herein, as well as previously described compounds (13), reinforces the idea that pol α has no specific interactions with any of the analogues. The shape and chemical properties of a methoxy, trifluoromethyl and nitro group vary significantly. If pol α made specific interactions with these compounds, then one would have expected to observe significant differences among the compounds. Similarly, if dNTP (and thus base pair) geometry played a dominant role in nucleotide incorporation by pol α, then a preference for either 5- or 6-substituted benzimidazoles would be expected. However, no such differences are seen. Interestingly, pol α discriminates best against the 4- and 7-CF3TPs, even though these two analogues, particularly d4CF3TP, could potentially form a more geometrically pleasing base pair with the natural pyrimidines.
KF also incorporates many of the analogues, but to a lesser extent than pol α and with greater discrimination. A simple comparison of the Vmax/KM values relative to an average value for natural mismatches by KF shows a preference for incorporating these compounds over natural mismatches by approximately two orders of magnitude. However, if these analogues are considered only as purine analogues, and the data compared only to the misincorporation of purines on a specific template, then the difference is not as great—only a factor of 10. This is because on the template sequence examined, KF misincorporates purines more effectively than it misincorporates pyrimidines [(13) and data not shown)].
The electronic character of the aromatic ring system likely does not greatly impact the ability of pol α and KF to polymerize these modified derivatives. Varying the nature of the substituents from electron-donating (methyl and methoxy) to electron-withdrawing (trifluoromethyl and nitro) did not significantly alter the ability of these enzymes to polymerize the analogues. Therefore, it is doubtful that the electronic character of the aromatic ring dominates the selection mechanism of these polymerases.
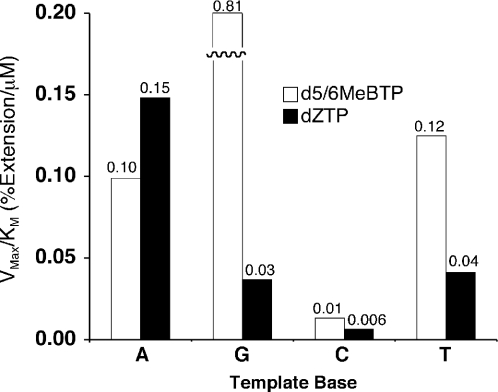
These data also provide further evidence that the shape of the base pair between the incoming dNTP and the template base being replicated is not a critical factor in determining incorporation of a dNTP. A comparison of the data obtained from the mixture of d5MeBTP and d6MeBTP with the incorporation of dZTP (11), a close isostere of dATP, provides a direct estimation of the effect of shape. These three compounds contain the same structural moieties, benzimidazole and a methyl group, but in different orientations. As shown in Figure 6, KF polymerizes d5/6MeBTP with similar or greater efficiency relative to dZTP for all template bases. Most importantly, incorporation of d5/6MeBTP across from a thymidylate residue is three times more efficient than that of dZTP, even though dZTP is indeed an adenine isostere. This comparison provides direct evidence that nascent base pair shape is not a primary principle governing polymerase fidelity.
Figure 6.
Comparison of incorporation of analogues containing 4- and 5/6-methylbenzimidazole. Kinetic data were determined as described in Materials and Methods. The numerical value above a bar is the specificity constant for that dNTP on the indicated template. d5/6MeBTP refers to the mixture of 5- and 6-methylbenzimidazole nucleoside triphosphate; dZTP refers to 4-methylbenzimidazole nucleoside triphosphate.
It may seem counterintuitive for a DNA polymerase to incorporate nucleotide analogues that differ so greatly from the natural bases. However, a lack of similarity to the natural substrate may in fact give rise to the inability of an enzyme to discriminate against an analogue. Having had no exposure to molecules such as the analogues presented here, polymerases have had no evolutionary pressure to develop mechanisms to discriminate against their chemical features. On the other hand, polymerases have had significant pressure to develop mechanisms to discriminate specifically against the three natural dNTPs that do not match a given template nucleotide. In such a mechanism, the enzyme recognizes a specific component of a non-cognate base in such a way as to prevent nucleotide insertion. Unnatural bases that lack these specific components would therefore be incorporated relatively easily, as these compounds are. The similar levels of discrimination against a variety of base analogues further argues that the incorporation of the analogues result from a lack of discrimination against, rather than a selection for the analogues.
While geometry does not appear to play an important role in determining whether or not pol α and KF polymerize a dNTP, the geometry of a newly synthesized base pair may very well play a critical role in determining whether or not the polymerase adds the next dNTP. Any base pair formed between one of the analogues and a template base almost certainly has a geometry very different than that of a canonical base pair. Elongation past these new pairs was either very inefficient or absent altogether. Thus, these results are consistent with previous work showing that correct geometry of the base pair at the 3′-terminus of the primer is critical to allow rapid addition of the next correct dNTP (11,18–20,29,30).
Surprisingly, we found several cases in which an incorporated analogue is more efficiently elongated by polymerization of a second analogue rather than the next correct dNTP. A priori, one might have expected the polymerase to more rapidly incorporate the next correct dNTP since it can form a correctly shaped and hydrogen bonded base pair, whereas a second analogue cannot. This result raises the possibility that DNA polymerases may recognize specific features found on a natural base to help prevent elongation of a misshapen base-pair (e.g. a mismatch). Experiments to test this hypothesis are in progress.
In total, our data indicate that neither hydrogen bonding, nor base pair geometry, nor electronic character play a dominant role in the fidelity mechanism of pol α and KF for single nucleotide insertion. The pol α data are most consistent with a negative selection model, wherein specific features of the natural bases allow the enzyme to discriminate against mismatches. The results with KF are less straightforward, and perhaps indicate the existence of yet another mechanism or a combination of mechanisms (e.g. a combination of negative selection and base pair geometry). The fact that KF normally has an intrinsic exonuclease activity may provide an explanation for why it would have a different mechanism for the fidelity of single nucleotide insertion than pol α. Alternatively, these slightly different discrimination mechanisms may be the evolutionary result of pol α and KF belonging to different polymerase families. Ultimately, what is clear for both enzymes is that DNA polymerase fidelity is a process that is much more complicated than that previously thought.
Acknowledgments
This work was supported by National Institutes of Health Grant GM54194 to R.D.K. and a Howard Hughes Medical Institute predoctoral fellowship to K.K. Funding to pay the Open Access publication charges for this article was provided by GM54194.
Conflict of interest statement. None declared.
REFERENCES
- 1.Kunkel T.A., Bebenek K. Recent studies of the fidelity of DNA synthesis. Biochim. Biophys. Acta. 1988;951:1–15. doi: 10.1016/0167-4781(88)90020-6. [DOI] [PubMed] [Google Scholar]
- 2.Bebenek K., Kunkel T. The fidelity of retroviral reverse transcriptases. In: Skalka A.M., Goff S., editors. Reverse Transcriptase. Plainview, NY: Cold Spring Harbor Laboratory Press; 1993. pp. 85–102. [Google Scholar]
- 3.Roberts J.D., Kunkel T.A. Fidelity of DNA polymerases. In: DePamphilis M., editor. DNA Replication in Eukaryotic Cells: Concepts, Enzymes and Systems. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1996. pp. 217–247. [Google Scholar]
- 4.Watson J.D., Crick F.H. Molecular structure of nucleic acids: a structure for deoxyribose nucleic acid. Nature. 1953;171:737–738. doi: 10.1038/171737a0. [DOI] [PubMed] [Google Scholar]
- 5.Watson J.D., Crick F.H. Genetical implications of the structure of deoxyribonucleic acid. Nature. 1953;171:964–967. doi: 10.1038/171964b0. [DOI] [PubMed] [Google Scholar]
- 6.Raszka M., Kaplan N.O. Association by hydrogen bonding of mononucleotides in aqueous solution. Proc. Natl Acad. Sci. USA. 1972;69:2025–2029. doi: 10.1073/pnas.69.8.2025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mildvan A.S. Mechanism of enzyme action. Annu. Rev. Bio. 1974;43:357–399. doi: 10.1146/annurev.bi.43.070174.002041. [DOI] [PubMed] [Google Scholar]
- 8.Loeb L.A., Kunkel T.A. Fidelity of DNA synthesis. Annu. Rev. Bio. 1982;51:429–457. doi: 10.1146/annurev.bi.51.070182.002241. [DOI] [PubMed] [Google Scholar]
- 9.Kool E.T., Morales J.C., Guckian G.M. Mimicking the structure and function of DNA: insights into DNA stability and replication. Angew. Chem. Int. Ed. Engl. 2000;39:990–1009. doi: 10.1002/(sici)1521-3773(20000317)39:6<990::aid-anie990>3.0.co;2-0. [DOI] [PubMed] [Google Scholar]
- 10.Kool E.T. Active site tightness and substrate fit in DNA replication. Annu. Rev. Bio. 2002;71:191–219. doi: 10.1146/annurev.biochem.71.110601.135453. [DOI] [PubMed] [Google Scholar]
- 11.Morales J.C., Kool E.T. Efficient replication between non-hydrogen bonded nucleoside shape analogs. Nat. Struct. Biol. 1998;5:950–954. doi: 10.1038/2925. [DOI] [PubMed] [Google Scholar]
- 12.Morales J.C., Kool E.T. Varied molecular interactions at the active sites of several DNA Polymerases: nonpolar nucleoside isosteres as probes. J. Am. Chem. Soc. 2000;122:1001–1007. doi: 10.1021/ja993464+. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chiaramonte M., Moore C.L., Kincaid K., Kuchta R.D. Facile polymerization of dNTPs bearing unatural base analogues by DNA polymerase alpha and Klenow Fragment (DNA Polymerase I) Biochemistry. 2003;42:10472–10481. doi: 10.1021/bi034763l. [DOI] [PubMed] [Google Scholar]
- 14.Ogawa A.K., Wu Y., McMinn D.L., Liu J., Schultz P.G., Romesberg F.E. Efforts toward the expansion of the genetic alphabet: information storage and replication with unnatural hydrophobic base pairs. J. Am. Chem. Soc. 2000;122:3274–3287. [Google Scholar]
- 15.Berger M., Wu Y., Ogawa A.K., McMinn D.L., Schultz P.G., Romesberg F.E. Universal bases for hybridization, replication and chain termination. Nucleic Acids Res. 2000;28:2911–2914. doi: 10.1093/nar/28.15.2911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wu Y., Ogawa A.K., Berger M., McMinn D.L., Schultz P.G., Romesberg F.E. Efforts toward expansion of the genetic alphabet: optimization of interbase hydrophobic interactions. J. Am. Chem. Soc. 2000;122:7621–7632. [Google Scholar]
- 17.Ogawa A.K., Wu Y., Berger M., Schultz P.G., Romesberg F.E. Rational Design of an unnatural base pair with increased kinetic selectivity. J. Am. Chem. Soc. 2000;122:8803–8804. [Google Scholar]
- 18.Yu C., Henry A.A., Romesberg F.E., Schultz P.G. Polymerase recognition of unnatural base pairs. Angew. Chem. Int. Ed. Engl. 2002;41:3841–3844. doi: 10.1002/1521-3773(20021018)41:20<3841::AID-ANIE3841>3.0.CO;2-Q. [DOI] [PubMed] [Google Scholar]
- 19.Matsuda S., Henry A.A., Schultz P.G., Romesberg F.E. The effect of minor-groove hydrogen-bond acceptors and donors on the stability and replication of four unnatural base pairs. J. Am. Chem. Soc. 2003;125:6134–6139. doi: 10.1021/ja034099w. [DOI] [PubMed] [Google Scholar]
- 20.Henry A.A., Yu C., Romesberg F.E. Determinants of unnatural nucleobase stability and polymerase recognition. J. Am. Chem. Soc. 2003;125:9638–9646. doi: 10.1021/ja035398o. [DOI] [PubMed] [Google Scholar]
- 21.Zerbe L.K., Goodman M.F., Efrati E., Kuchta R.D. Abasic template lesions are strong chain terminators for DNA primase but not for DNA polymerase alpha during the synthesis of new DNA strands. Biochemistry. 1999;38:12908–12914. doi: 10.1021/bi991075m. [DOI] [PubMed] [Google Scholar]
- 22.Moore C.L., Zivkovic A., Engels J.W., Kuchta R.D. Human DNA primase uses Watson-Crick hydrogen bonds to distinguish between correct and incorrect nucleoside triphosphates. Biochemistry. 2004;43:12367–12374. doi: 10.1021/bi0490791. [DOI] [PubMed] [Google Scholar]
- 23.Kazimierczuk Z., Cottam H.B., Revankar G.R., Robins R.K. Synthesis of 2′-deoxytubercidin, 2′-deoxyadenosine, and related 2′-deoxynucleosides via a novel direct stereospecific sodium salt glycosylation procedure. J. Am. Chem. Soc. 1984;106:6379–6382. [Google Scholar]
- 24.Ludwig J. A new route to nucleoside 5′-triphosphates. Acta Biochim. Biophys. Acad. Sci. Hung. 1981;16:131–135. [PubMed] [Google Scholar]
- 25.Kuchta R.D., Mizrahi V., Benkovic P.A., Johnson K.A., Benkovic S.J. Kinetic mechanism of DNA polymerase I (Klenow) Biochemistry. 1987;26:8410–8417. doi: 10.1021/bi00399a057. [DOI] [PubMed] [Google Scholar]
- 26.Sambrook J., Fritsch E.F., Maniatis T. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratories; 1989. [Google Scholar]
- 27.Guckian K.M., Schweitzer B.A., Ren R.X.F., Sheils C.J., Tahmassebi D.C., Kool E.T. Factors contributing to aromatic stacking in water: evaluation in the context of DNA. J. Am. Chem. Soc. 2000;122:2213–2222. doi: 10.1021/ja9934854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Reineks E.Z., Berdis A.J. Evaluating the contribution of base stacking during translesion DNA replication. Biochemistry. 2004;43:393–404. doi: 10.1021/bi034948s. [DOI] [PubMed] [Google Scholar]
- 29.Morales J.C., Kool E.T. Minor groove interactions between polymerase and DNA: more essential to replication than Watson–Crick hydrogen bonds? J. Am. Chem. Soc. 1999;121:2323–2324. doi: 10.1021/ja983502+. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kool E.T. Synthetically modified DNAs as substrates for polymerases. Curr. Opin. Chem. Biol. 2000;4:602–608. doi: 10.1016/s1367-5931(00)00141-1. [DOI] [PubMed] [Google Scholar]