Abstract
Differentiation is a coordinated process of irreversible cell cycle exit and tissue-specific gene expression. To probe the functions of the retinoblastoma protein (RB) family in cell differentiation, we isolated HBP1 as a specific target of RB and p130. Our previous work showed that HBP1 was a transcriptional repressor and a cell cycle inhibitor. The induction of HBP1, RB, and p130 upon differentiation in the muscle C2C12 cells suggested a coordinated role. Here we report that the expression of HBP1 unexpectedly blocked muscle cell differentiation without interfering with cell cycle exit. Moreover, the expression of MyoD and myogenin, but not Myf5, was inhibited in HBP1-expressing cells. HBP1 inhibited transcriptional activation by the MyoD family members. The inhibition of MyoD family function by HBP1 required binding to RB and/or p130. Since Myf5 might function upstream of MyoD, our data suggested that HBP1 probably blocked differentiation by disrupting Myf5 function, thus preventing expression of MyoD and myogenin. Consistent with this, the expression of each MyoD family member could reverse the inhibition of differentiation by HBP1. Further investigation implicated the relative ratio of RB to HBP1 as a determinant of whether cell cycle exit or full differentiation occurred. At a low RB/HBP1 ratio cell cycle exit occurred but there was no tissue-specific gene expression. At elevated RB/HBP1 ratios full differentiation occurred. Similar changes in the RB/HBP1 ratio have been observed in normal C2 differentiation. Thus, we postulate that the relative ratio of RB to HBP1 may be one signal for activation of the MyoD family. We propose a model in which a checkpoint of positive and negative regulation may coordinate cell cycle exit with MyoD family activation to give fidelity and progression in differentiation.
During terminal differentiation, cells undergo an irreversible cell cycle withdrawal followed by the expression of tissue-specific markers to specify the final phenotype. The progression of differentiation is a precisely coordinated event in which the irreversible cell cycle arrest is tightly coupled to the expression of the tissue-specific genes. This communication of general and tissue-specific pathways guarantees fidelity by ensuring that appropriately arrested and viable cells proceed to the last steps in tissue biogenesis. A lapse in this coordination can give uncontrolled proliferation of otherwise-differentiated cells, a hallmark of preneoplastic changes that may eventually result in cancer. Alternatively, this lapse can yield defects in development, in which the balance of proliferation and differentiation must be tightly maintained. Despite the importance of these effects, the cellular and molecular mechanisms underlying the coordination of general and tissue-specific events in differentiation have not been extensively addressed.
Muscle cells represent the best-characterized differentiation system, with the landmark discovery of the MyoD transcription factor family (MyoD, myogenin, Myf5, and Mrf4) as critical regulators of muscle determination and differentiation (reviewed in reference 32). The major function of this family of basic helix-loop-helix proteins is to form heterodimers with ubiquitous basic helix-loop-helix E proteins which then activate muscle-specific genes. The transcriptional activation is achieved through direct binding to E-box elements (CANNTG), which are present in the promoters of numerous muscle-specific genes and of each MyoD family member (54, 57). There is considerable complexity with extensive functional redundancy and autoactivation among MyoD family members. Gene knockout studies have demonstrated that Myf5, MyoD, and myogenin are critical for normal muscle differentiation during development. Either Myf5 or MyoD is required, and recent studies have suggested that Myf5 may be upstream of MyoD (reviewed in reference 36). The elegant characterization of the MyoD family function provides a necessary backdrop for the elucidation of the important mechanisms that activate the tissue-specific differentiation pathways. Additionally, the C2C12 muscle cell line represents a feasible model system for probing fundamental mechanisms of differentiation.
The retinoblastoma family of growth suppressor proteins (RB, p130, and p107) are critical players in general cell cycle regulation (see reference 11 and reviews within). A major paradigm is that RB blocks G1 progression by inhibiting the E2F family of transcription factors. E2Fs are required for the activation of numerous genes that are necessary for G1-to-S progression (see reference 11 and reviews within). However, the functions of RB, p130, and p107 are not limited to G1 regulation and are important in other cellular processes, such as differentiation and apoptosis protection (reviewed in reference 53). These diverse functions suggest a critical and broad role in cellular regulation.
During differentiation in muscles and other tissues, RB and p130 expression is increased but p107 expression declines markedly. In quiescent and differentiated cells, the major E2F complex contains p130 (7, 8, 25, 43). We have shown that this p130-E2F complex coincides with transcriptional repression through E2F elements (43). This E2F-p130 complex is distinctive for G0 but not G1 (45). These observations are also consistent with the view that terminal differentiation may be an “irreversible G0” state. In addition, complexes of RB and E2F4 have been observed in certain differentiated cells (17). RB appears to be critical for maintaining the irreversible cell cycle arrest that occurs with differentiation, since RB−/− differentiated muscle cells could reenter the cell cycle (30, 41, 56). Recent studies with transgenic mice have demonstrated that a threshold level of RB is necessary for the characteristic irreversible cell cycle arrest during muscle development in animals (56).
Recent studies have indicated a surprising function for RB in tissue-specific gene expression in adipocyte and muscle differentiation (4, 30). The expression of tissue-specific genes in adipocyte and muscle differentiation is dictated by the functions of C/EBP and MyoD family transcription factors, respectively (reviewed in references 32 and 55). RB is necessary for adipocyte differentiation, as RB−/− cells fail to undergo adipogenesis. One molecular mechanism is a direct physical interaction of RB with C/EBP-α, resulting in the transcriptional activation of adipocyte-specific genes (4). In muscle tissue, RB augments the transcriptional ability of MyoD in the expression of muscle-specific genes (30). In muscle tissue, the molecular mechanism probably does not occur through a direct physical interaction since efforts to demonstrate a direct interaction have had mixed results (14, 30). Recent work by Kaelin and Lee has provided new evidence that the function of RB in MyoD activation and E2F regulation can be uncoupled by selective mutations. Both the N-terminal and pocket regions are required for the activation of differentiation in the cellular and animal models (37, 42).
Because of their dual roles in both cell cycle control and differentiation, RB and its targets are excellent candidates for studying the mechanisms that coordinate general cell cycle and tissue-specific regulation during differentiation. While E2F is certainly one RB and p130 target in differentiation, other cellular targets may also be necessary for cell cycle arrest and differentiation. A simple argument is that the concentration of RB family members is vastly greater than that of the E2Fs. To address the complex role of RB in differentiation, we and others have recently isolated HBP1 as a novel target of p130 and RB from differentiated muscle cells and from developing murine tissues (19, 49). HBP1 is a new member of the sequence-specific HMG box proteins, which include LEF1, SRY, and TCF and which have all been linked to differentiation and signaling (for reviews, see references 13, 15, and 31). HBP1 is a sequence-specific transcriptional repressor that contains the consensus LXCXE, or RB interaction, motifs. However, HBP1 interacts only with RB and p130 and not with p107 (49). Three independent observations suggest potential functions of HBP1 in muscle differentiation. First, HBP1, RB, and p130 are all increased with differentiation (8, 23, 49). Second, expression of the HBP1 protein represses the promoter of the N-MYC gene, a protooncogene that becomes downregulated in differentiating cells. Third, HBP1 expression can elicit cell cycle arrest, which is a necessary feature of terminal differentiation. Thus, our previous work suggests that HBP1 may promote the early stages of differentiation by facilitating cell cycle arrest through transcriptional repression of key cell cycle genes (49).
In the present study we have uncovered an additional and unexpected function of HBP1 in differentiation. Because overexpression of HBP1 gave efficient cell cycle arrest and HBP1 was normally induced with differentiation, we expected that HBP1 expression would enhance differentiation. Surprisingly, the expression of HBP1 blocked full terminal differentiation in muscle cells without blocking cell cycle exit. The HBP1 phenotype was distinct from the findings of studies with oncogenes that inhibited differentiation by preventing the required initial step of cell cycle exit. The HBP1 block was selective, as the expression of myogenin and MyoD was abolished. Yet, Myf5 expression was normal, which suggested that HBP1 might inhibit Myf5 functions to prevent expression of MyoD. Consistently, HBP1 was able to block transcriptional activation of the MyoD family and the reexpression of MyoD family members did restore differentiation in HBP1-expressing lines. Further investigation revealed that the relative ratio of RB to HBP1 appeared to dictate the progression of differentiation. Low RB/HBP1 ratios yielded cell cycle exit but inhibited tissue-specific gene expression. In contrast, high RB/HBP1 ratios yielded full differentiation. Similar changes were manifested in endogenous C2 muscle cell differentiation. Our experimental observations are consistent with a new model in which the relative ratio of RB to HBP1 may constitute a signal for MyoD family regulation. These and other new studies suggest that the coordination of cell cycle arrest with tissue-specific gene expression in differentiation may involve positive and negative regulation by HBP1 and RB.
MATERIALS AND METHODS
Plasmids. (i) Mammalian expression constructs.
pEF-BOS HBP1 wild-type and mutant constructs were as described previously (49) (Fig. 1). pEMSV-MyoD and pEMSV-myogenin were kindly provided by E. Olson. pCMV-HA-Myf5 was subcloned from EMSV-Myf5 (provided by E. Olson). pCMV-HA-RB was a gift from Q. Sheng and B. Schaffhausen.
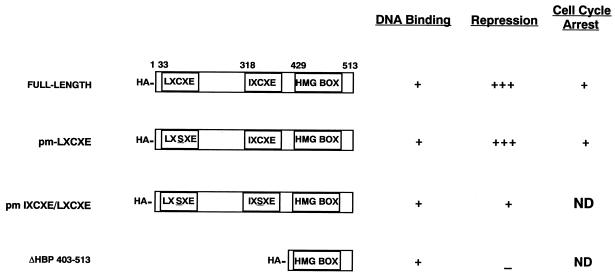
FIG. 1.
Schematic diagram of HBP1 mutants. The data presented here represent a summary of previous studies (49).
(ii) CAT reporter constructs.
MCK 4800-CAT was a generous gift from E. Olson. 4R-CAT was a generous gift from the late Harold Weintraub (51).
Cell culture, establishment of stable cell lines, and transfections.
C2C12 myogenic cells were cultured in Dulbecco’s modified Eagle medium (DMEM) supplemented with 15% (vol/vol) fetal calf serum (FCS). Myogenic differentiation was induced by growing cells in DMEM supplemented with 2% FCS.
To establish stable HBP1 cell lines, C2C12 cells were transfected by the calcium phosphate precipitation method with 30 μg of pEF-BOS HBP1 and 3 μg of TK-hygro. Stably transfected cells were selected in culture medium containing 250 μg of hygromycin B per ml (Calbiochem). Colonies of hygromycin-resistant cells were isolated approximately 10 days after selection and were propagated. HBP1 stable cell lines were screened by immunoprecipitation-Western analysis. Two HBP1 cell lines, designated B1-C2 and B2-C2, were further analyzed.
To reverse the nondifferentiation phenotype, 10 μg of pEMSV-MyoD, pEMSV-Myf5, or pEMSV-myogenin was transfected into either B1-C2 or B2-C2 together with 5 μg of Rous sarcoma virus–β-galactosidase (β-Gal) as a cotransfection marker. The cells were exposed to the DNA precipitates for 24 h in DMEM plus 15% FCS and were then grown in DMEM plus 2% FCS for an additional 40 h. Immunofluorescence staining was subsequently performed on the transfected cells.
To establish stable RB and HBP1 cell lines, B1-C2 cells were transfected by the calcium phosphate precipitation method with 30 μg of pCMV-RB and 3 μg of pEF-1α-puro. Stably transfected cells were selected in culture medium containing 6 μg of puromycin per ml (Calbiochem). Colonies of puromycin-resistant cells were isolated approximately 7 days after selection and were propagated. RB cell lines were screened by Western analysis by using an anti-RB monoclonal antibody (Pharmingen). One RB cell line, designated B1-B9, and three control puromycin-resistant lines were analyzed for differentiation.
Transient assay for differentiation.
The effect of overexpression of HBP1 on C2C12 differentiation was evaluated in a transient-transfection experiment. C2C12 cells grown on coverslips were transfected with plasmids encoding wild-type or mutant HBP1, Myf5, and β-Gal. Within 24 h of transfection, the cells were grown in medium supplemented with 2% FCS for another 40 h. Cells were fixed and immunostained for β-Gal and myosin heavy chain (MHC) to visualize transfected and differentiated cells, respectively. The percentage of differentiated cells was determined as a ratio of double-positive (MHC+ [differentiated]; β-Gal+ [transfected]) to total β-Gal-positive (transfected) cells. Each transfection was repeated three times, and cells on two different coverslips from each experiment were counted. Generally, 200 to 300 cells were counted for each experiment.
Immunoprecipitation and Western blotting.
Cells were lysed on plates in TNN buffer (50 mM Tris-HCl, pH 7.4; 100 mM NaCl; 5 mM EDTA; 0.5% Nonidet P-40; 1 μg pepstatin per ml; 0.5 mM EGTA; 200 μM phenylmethylsulfonyl fluoride [PMSF]; 0.5 mM dithiothreitol [DTT]; 1 μg of leupeptin per ml). Cell lysates were precleared with protein A-Sepharose beads before antibodies were added. The immune complexes were formed for 1 h at 4°C with gentle agitation and then collected onto protein A-Sepharose beads by gentle agitation at 4°C for another hour. After washes with TNN buffer, the beads were boiled in sodium dodecyl sulfate (SDS) sample buffer for 10 min, and the supernatant was analyzed by SDS-polyacrylamide gel electrophoresis (PAGE) and Western blot analysis enhanced chemiluminescence (with ECL; Renaissance).
For detection of HBP1 transgene product or transfected HBP1, anti-HBP1 rabbit antisera were used for immunoprecipitation, and monoclonal antihemagglutinin (anti-HA) antibody 12CA5 was used to detect the HA-tagged HBP1 transgene product by Western blot analysis. For detection of HA-Myf5, anti-Myf5 rabbit antisera were used for immunoprecipitation and 12CA5 was used in the Western analysis. For detection of the RB transgene product, cell lysates were immunoprecipitated with anti-RB antibodies (Santa Cruz), and RB was detected with a monoclonal anti-RB antibody (Pharmingen) by Western blot analysis.
For Western blotting of myogenin and MHC, antimyogenin monoclonal antibody F5D and anti-MHC monoclonal antibody FS9 were used, respectively (generous gifts of Woody Wright and Frank Stockdale, respectively).
Immunofluorescence staining.
C2C12 cells, grown on coverslips (Fisher Scientific), were fixed in 30% methanol–70% acetone for 20 min at −20°C. The coverslips were air dried and were rehydrated in phosphate-buffered saline (PBS) for 3 min. Each coverslip was covered with 50 μl of appropriately diluted primary antibodies and incubated at 37°C for 1 h in a humidified chamber. After several washes with PBS, each coverslip was covered with 50 μl of appropriately diluted secondary antibodies and then incubated at 37°C for 1 h in a humidified chamber. The coverslips were then washed three times with PBS, and the cells were counterstained with 50 μl of Hoechst dye for 5 min at room temperature. The coverslips were washed once in PBS and once in distilled water and mounted on slides with 50% glycerol–50% distilled water. Immunofluorescent cells were visualized under a microscope and counted.
For detection of transfected cells, rabbit anti-β-Gal (final concentration, 36 μg/ml; 5 Prime→3 Prime) was used as the primary antibody combined with rhodamine-conjugated goat anti-rabbit secondary antibodies (Jackson). For detection of differentiated cells, anti-myosin heavy-chain monoclonal antibody FS9 was used as primary antibody combined with fluorescein isothiocyanate-conjugated donkey anti-mouse secondary antibodies (Jackson).
For detection of cells that incorporated bromodeoxyuridine (BrdU), cells on coverslips were first treated with 2 N HCl for 1 h at 37°C before incubation with primary mouse anti-BrdU antibody (Boehringer Mannheim). BrdU-positive cells were stained with fluorescein-conjugated donkey anti-mouse secondary antibodies.
T2 RNase protection assay.
Total cellular RNA was isolated with Trizol reagent (Sigma) according to the manufacturer’s directions and treated with RNase-free DNase (Amersham) to remove residual DNA. T2 RNase protection assays were performed as previously described (48). The probes were derived from murine MyoD (gift of the late H. Weintraub [51]), murine Myf5 (gift of Yukang Wang, murine HBP1 cDNA (49), or pTRI-GAPDH-mouse template (Ambion).
CAT assays.
C2C12 cells were scraped off the plates 48 h after transfection and were lysed in lysis buffer (250 mM Tris-HCl, pH 7.5; 1 mM EDTA; 1 μg of pepstatin per ml; 0.5 mM EGTA; 200 μM PMSF; 1 μg of leupeptin per ml) by four cycles of freezing and thawing. The amounts of CAT protein in cell extracts were determined with a chloramphenicol acetyltransferase (CAT) enzyme-linked immunosorbent assay kit (Boehringer Mannheim) according to the manufacturer’s specifications and with a linear standard curve. All transcription data were normalized for transfection efficiency with β-Gal, whose activity was quantitated by an ONPG (o-nitrophenyl-β-d-galactopyranoside) assay and by using a linear standard curve. The normalized reporter activity was expressed as a ratio of nanograms of CAT to units of β-Gal.
Cell labeling and immunoprecipitations.
C2C12 cells that were grown in 100-mm-diameter tissue culture plates were differentiated for 4 days in DMEM complemented with 2% FCS. After cells were washed twice with methionine-free DMEM (GIBCO), the cells were preincubated in methionine-free DMEM for 20 min. The cells were then labeled for 4 h with 1 mCi of 35S-methionine label per 100-mm dish in methionine-free DMEM supplemented with 2% FCS. The cells were washed with cold PBS and lysed in TNN buffer on ice for 20 min. The cell lysates were collected and were briefly centrifuged at 6,000 rpm at 4°C. The supernatants were pooled, precleared by incubation with protein A-Sepharose beads, and divided into three portions. One-twelfth of the extract was used as a control for total RB expression by a double immunoprecipitation with anti-RB antibodies (Santa Cruz). The remaining 11/12 of the extract were divided into two portions and immunoprecipitated with anti-HBP1 antibodies for 1 h at 4°C. The immune complexes were collected onto protein A-Sepharose beads over a 1-h period at 4°C, and the beads were washed three times with TNN buffer. The beads were then boiled in release buffer (50 mM Tris-HCl, pH 7.4; 1% SDS; 5 mM DTT) for 15 min and then chilled on ice. The supernatants were collected, and TNN buffer was added to each sample to a total volume of 1 ml. The second immunoprecipitations were carried out overnight at 4°C with anti-RB or control (anti-β-Gal) antibodies, and the immune complexes were again collected onto protein A-Sepharose beads. After the beads were washed four times with TNN buffer, they were boiled in SDS sample buffer and the proteins were resolved by SDS–7% PAGE and visualized with a phosphorimager after a 3-month exposure.
RESULTS
HBP1 inhibits differentiation by altering the MyoD family expression pattern.
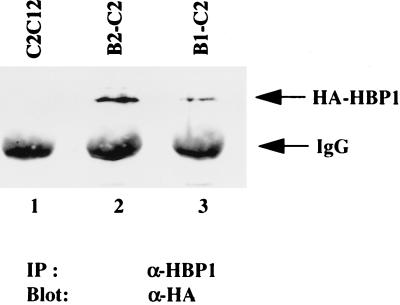
Differentiation can be divided into two stages: initiation of an irreversible cell cycle arrest and expression of tissue-specific genes. Our studies have implicated a role for HBP1 in cell cycle control during C2C12 muscle cell differentiation (49). To directly investigate the potential role of HBP1 in the full muscle differentiation program, we established two stable C2 cell lines that constitutively expressed HBP1. The HA-tagged rat HBP1 transgene products in these two mouse lines, designated B1-C2 and B2-C2, were detected by immunoprecipitation with anti-HBP1 antibodies followed by Western analysis with an anti-HA antibody (Fig. 2). Because rat and mouse HBP1 could be distinguished in RNase protection assays, we determined that the relative expression of exogenous rat HA-HBP1 was modestly increased over the endogenous levels in the stable lines (3.7-fold in B1-C2 and 5.7-fold in B2-C2 [data not shown]). The overall levels of HBP1 in the cell lines were similar to the expression of HBP1 in fully differentiated C2 myotubes (49).
FIG. 2.
Detection of HBP1 transgene product in B1-C2 and B2-C2 cells. Cell lysates from each cell line were prepared, and the level of HBP1 transgene product in each cell line was detected by immunoprecipitation with anti-HBP1 antibodies followed by Western analysis with anti-HA (12CA5) antibody as described in Materials and Methods. The analyzed lysates are depicted in the figure as follows: lane 1, normal C2 cells; lane 2, B1-C2 HBP1 expressing line; lane 3, B2-C2 HBP1 expressing line.
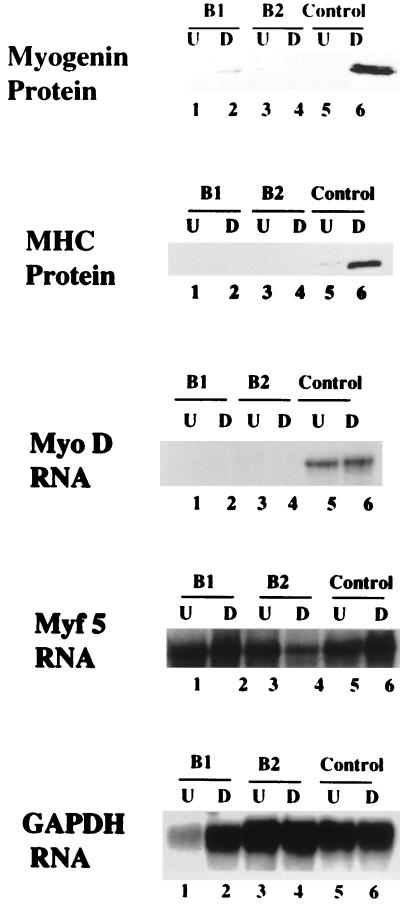
When these two cell lines were subjected to differentiation conditions, a surprising result was that terminal differentiation was completely inhibited. As shown in Fig. 3, the expression of several molecular markers of differentiation—MyoD, myogenin, and MHC—was undetectable in B1-C2 and B2-C2, suggesting that the expression of HBP1 had blocked terminal differentiation. In contrast, the differentiation markers were easily detected in a control hygromycin-resistant line, verifying the expected efficient differentiation under the same conditions. In addition to defective expression of differentiation markers, B1-C2 and B2-C2 cells did not undergo the differentiation-specific event of myotube formation, even after prolonged incubation (8 days) in differentiation media. Myotube formation began at 72 h in normal C2C12 cells and three control hygromycin-resistant lines (data not shown).
FIG. 3.
Expression levels of terminal differentiation markers. The levels of MHC, myogenin, MyoD, and Myf5 were scored in B1-C2, B2-C2, and control cell lines. For myogenin and MHC, the protein levels were detected by Western blot analysis with monoclonal antimyogenin (F5D) and monoclonal anti-MHC (FS9) antibodies, respectively, in cell lysates prepared from each line. For MyoD and Myf5, the RNA levels were quantitated in a T2 RNase protection assay with total RNA that was isolated from each cell line. GAPDH (glyceraldehyde-3-phosphate dehydrogenase) was employed as an RNA loading control. The tested cell lines are indicated: lanes 1 and 2, B1-C2; and lanes 3 and 4, B2-C2; and lanes 5 and 6, hygromycin-resistant control line without HA-HBP1. The odd numbers represent undifferentiated conditions (15% serum) and are denoted “U.”. The even numbers represent differentiated conditions (2% serum) and are denoted “D.” A representative experiment is shown here, and identical results were obtained in two independent analyses.
Although differentiation was clearly blocked in the HBP1-expressing cell lines, this did not reflect a global inhibition in the expression of all MyoD family members. While MyoD and myogenin expression was clearly absent, Myf5 expression was surprisingly unaffected in the HBP1 cell lines (Fig. 3). This apparently normal Myf5 expression also indicated that the inhibition of muscle differentiation by HBP1 expression was not due to reversal of the myogenic phenotype, since Myf5 served as a marker of the myogenic lineage. The expression of Mrf4 was not tested, since its expression is limited to mature muscle fiber and is not generally manifested in tissue culture (reviewed in reference 32). Thus, we conclude that HBP1 blocks differentiation by interfering with the expression of MyoD and myogenin but not the expression of Myf5. Consistent with our finding, genetic studies in mice have indicated that Myf5 is functionally upstream of MyoD during muscle development (36, 47).
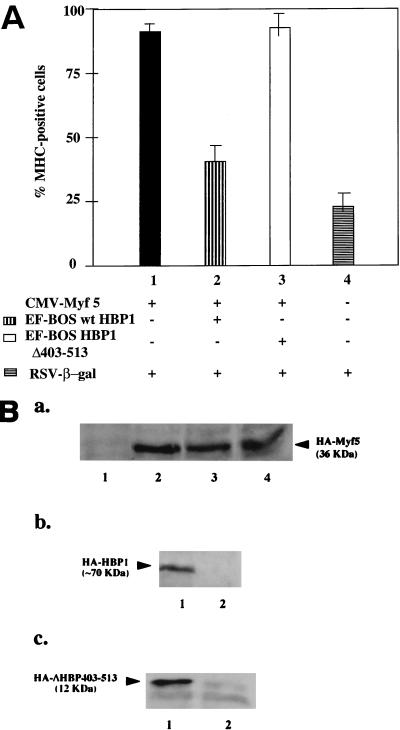
To confirm that the differentiation block in B1-C2 and B2-C2 was not due to aberrant clonal selection of cell lines, we verified our results in an experiment involving transient expression of HBP1. The design of the transient differentiation is based on the observation that complete differentiation in normal C2 cells was a relatively slow process (∼96 h). The expression of Myf5 (or other MyoD family members) could accelerate the terminal differentiation program to give complete differentiation at ∼48 h. Thus, the time line of the experiment was transfection of undifferentiated cells followed by a 40-h incubation in differentiation medium. The transfected cells were marked by the coexpression of β-Gal. By immunofluorescent staining, we then counted the cells that were doubly positive for the expression of β-Gal (transfected cells) and for MHC (differentiated cells). The percentage of differentiation was determined as the portion of MHC-positive cells in the transfected cell population.
As shown in Fig. 4A, Myf5 did induce nearly complete differentiation in the transfected population (lane 1). Coexpression of HBP1 and Myf5 decreased the percentage of differentiated cells (lane 2, 40 ± 10%), whereas coexpression of the DNA binding domain of HBP1 (ΔHBP403-513) caused little inhibition of differentiation (lane 3, 87.6%). Under the assay conditions, 20 ± 8% of control transfected cells exhibited differentiation in the absence of exogenous Myf5 expression, and this level of differentiation constituted the baseline of the assay (lane 4). As shown in Fig. 4B, the inhibition of differentiation by HBP1 was not due to loss of exogenous Myf5 expression, which was similar irrespective of HBP1 coexpression (Fig. 4B, lanes 2 to 4). This experiment demonstrated that the differentiation efficiency in the presence of HBP1 approached that of the control in which there was no exogenous Myf5 expression (compare columns 2 and 4 in Fig. 4A). Therefore, we conclude that HBP1 can efficiently inhibit terminal differentiation when expressed either stably or transiently and provide independent support for a functional role of HBP1 as a dominant inhibitor in C2C12 differentiation.
FIG. 4.
Overexpression of HBP1 inhibited C2C12 differentiation in a transient-differentiation assay. A transient-differentiation assay was devised to score the effects of HBP1 on C2C12 differentiation. The basis of the assay was that expression of MyoD family members could accelerate C2 cell differentiation. (A) C2C12 cells grown on coverslips were transfected with plasmids encoding wild-type or mutant HBP1, Myf5, and β-Gal. After 24 h, the transfected cells were cultured in medium supplemented with 2% FCS for another 40 h. Cells were fixed and immunostained for β-Gal and MHC. The percentage of differentiated cells (MHC positive) among the transfected cells (β-Gal positive) was determined. Each transfection was repeated three times, and cells on two different coverslips from each experiment were counted. The total number of cells counted for each experiment was 200 to 300. All lanes contain β-Gal and the following additions: lane 1, Myf5 (filled column); lane 2, Myf5 and wild-type HBP1 (vertical stripes); lane 3, Myf5 and HA-ΔHBP403-513 (open column); and lane 4, no addition (horizontal stripes). (B) To measure protein expression levels, the lysates from a parallel experiment were analyzed for the expression of HA-Myf5, HA-ΔHBP403-513, and HA-HBP1 by using immunoprecipitation with anti-Myf5 or anti-HBP1 antibodies, respectively, followed by Western blot analyses of immune complexes with the anti-HA antibody. The constructs and antibodies are described in Materials and Methods. A representative experiment is shown. (Ba) Western blot showing Myf5 expression. The assay was performed as described for panel B. Cells were transfected with β-Gal and Myf5 (lane 4), Myf5 and HBP1 (lane 3), Myf5 and ΔHBP403-513 (lane 2), or no other expression vector (lane 1). (Bb) Western blot showing HBP1 expression. Cells were transfected with β-Gal, Myf5 and HBP1 (lane 1) or Myf5 (lane 2). (BC) Cells were transfected with β-Gal and Myf5 and ΔHBP403-513 (lane 1) or Myf5 (lane 2).
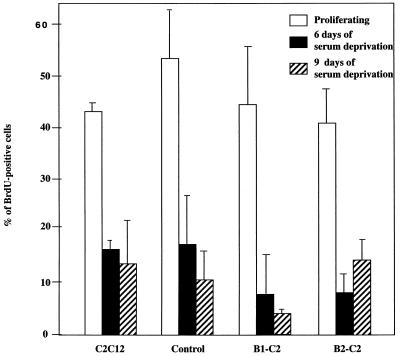
To understand the molecular basis for the block in differentiation, we first sought to determine whether HBP1-expressing cells could exit the cell cycle. Many oncogenes could inhibit differentiation by preventing cell cycle exit (for a review, see reference 22). Our existing evidence already strongly suggested that HBP1 enhanced, rather than prevented, cell cycle exit. First, in contrast to oncogenes that could stimulate S-phase entry, the expression of HBP1 directly inhibited cell cycle progression in C2 cells (49). Second, in a growth suppression assay, HBP1 led to a threefold inhibition of colony formation, suggesting that HBP1 had a moderate growth suppression effect rather than enhancing proliferation (unpublished data). In contrast, expression of oncogenes that could inhibit differentiation by preventing cell cycle arrest (e.g., RAS [18, 33]) generally gave increased colony formation in these same assays. Third, the B1-C2 and B2-C2 HBP1-expressing lines exhibited somewhat reduced growth rates (data not shown), a finding again inconsistent with oncogenic transformation. Lastly, we used BrdU incorporation as an assay for S phase. As shown in Fig. 5, the efficiencies of cell cycle exit in response to serum deprivation were equivalent in normal C2C12, control hygromycin-resistant, and HBP1-expressing B1-C2 and B2-C2 lines. Thus, we conclude that the block of differentiation by HBP1 is not due to defects in cell cycle exit. These data further suggest that HBP1 affects events downstream of cell cycle exit in the full differentiation pathway.
FIG. 5.
Suppression of S phase in HBP1-expressing cells upon serum deprivation. The HBP1-expressing cell lines (B1-C2 and B2-C2) and the control lines were assayed for the ability to exit the cell cycle upon serum deprivation. BrdU incorporation was used as a measure of the S phase, and quiescent cells should exhibit a reduction in BrdU-positive cells. All cell lines were grown on coverslips in differentiation medium for the indicated time period followed by a 1-h pulse of BrdU labeling. Cells were subsequently fixed and immunostained for BrdU. The percentages of BrdU-positive cells were determined by counting the cells from several random fields; approximately 200 to 300 cells were counted for each column. As controls, normal C2 and control hygromycin-resistant C2 cell lines were utilized. The open, filled, and diagonal striped bars represent the proliferating, 6-day-deprived, and 9-day-deprived cell populations for each cell line, respectively.
Expression of MyoD family members restores full differentiation.
The previous experiments suggested that HBP1 uncoupled the tightly coordinated processes of cell cycle exit and of tissue-specific gene expression. We next asked if the inhibition of differentiation by HBP1 could be reversed by expression of MyoD family members. On a practical level, a positive outcome would predict the location of the differentiation step that was affected by HBP1 and would further argue that HBP1 did not block differentiation by nonspecific and pleiotropic means. Instead, ectopic expression of HBP1 probably interfered with “appropriate” signals to activate tissue-specific genes. Mechanistically, any restoration of differentiation by MyoD family members would be informative. A positive result might suggest that HBP1 blocked the transcriptional function of Myf5 and the subsequent activation of the MyoD and myogenin promoter via E-box elements (5, 50, 54, 57). Thus, we hypothesized that reexpression of Myf5, MyoD, or myogenin should complement the differentiation defect imposed by HBP1. A positive result would further suggest that these MyoD family members could be downstream of HBP1.
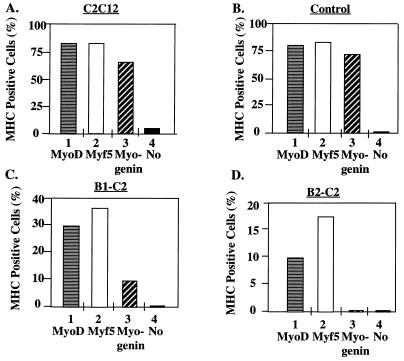
We transiently expressed Myf5, MyoD, and myogenin into control hygromycin-resistant or HBP1-expressing cells and scored MHC expression as a marker for differentiation. As described above, a β-Gal expression vector was used as a cotransfection marker, and a double immunostaining assay was used to quantitate the percentage of transfected cells (β-Gal positive) that were also differentiated (MHC positive). As controls, the expression of Myf5, MyoD, or myogenin led to efficient differentiation (Fig. 6A and B), whereas β-Gal alone gave little differentiation (5 to 10%; indicated by “No” in Fig. 6). Together, these data verified that differentiation was controlled by the exogenous MyoD family member.
FIG. 6.
Expression of MyoD family members can overcome inhibition of differentiation in the B1-C2 and B2-C2 lines. The purpose of this experiment was to determine whether MyoD family members could rescue the differentiation defect imposed by HBP1. A modification of the transient-differentiation assay in Fig. 4 was used. Expression vectors encoding MyoD, Myf5, or myogenin were transiently transfected into either control or HBP1-expressing cell lines (B1-C2 or B2-C2). A β-Gal expression vector was cotransfected to identify the transfected cells in each experiment. At 24 h posttransfection, cells were grown in differentiation medium for an additional 40 h before they were stained with antibodies. Double immunostaining for differentiated and transfected cells was performed with anti-MHC monoclonal antibody and anti-β-Gal antisera, followed by staining with fluorescein-conjugated anti-mouse immunoglobulin G and rhodamine-conjugated anti-rabbit immunoglobulin G, respectively. The percentages of differentiated and transfected cells were quantitated from approximately 200 to 300 cells for each experimental point. The indicated cell lines are as described in the legend to Fig. 5. The percentage of MHC-positive cells was determined for MyoD (horizontal stripes), Myf5 (open column), myogenin (diagonal stripes), or β-Gal (filled; denoted “No”).
As shown in Fig. 6C and D, the reexpression of Myf5, MyoD, or myogenin could partially restore differentiation in both of the HBP1-expressing lines, as determined by an increase in the number of MHC-positive cells. In the absence of any MyoD family member expression, there was no detectable differentiation in the HBP1-expressing lines, consistent with the initial analysis in Fig. 2. Myogenin was the least effective for unknown reasons. From Fig. 6 we conclude that the HBP1 differentiation block is not a result of a nonspecific impairment in the intrinsic differentiation program generated during the selection of the cell lines. Furthermore, these results suggest that MyoD family may act downstream of HBP1 in a muscle differentiation pathway.
HBP1 blocks transcriptional activation by the MyoD family.
If HBP1 blocked differentiation upstream of the MyoD family, a logical mechanism would be interference with the transcriptional activation functions of the MyoD family. This potential mechanism would abolish not only expression of downstream muscle-specific genes but also autoactivation among MyoD family members (5, 50, 54, 57). To assess potential regulation by HBP1 of transcriptional activation by MyoD family members, both natural differentiation-specific (muscle creatine kinase [MCK]) and synthetic E-box promoters (4R-CAT) were utilized in standard transcriptional assays. The natural promoter provided information on transcriptional regulation in the context of a complex promoter regulated by differentiation. The synthetic promoter measured effects directly through the isolated E-box element that was specific for MyoD family members. While the roles of individual MyoD family members have been unraveled in genetic and developmental studies, all members were functionally equivalent in tissue culture with respect to transcriptional activation (reviewed in reference 32). Thus, our experiments could only address whether HBP1 could block the general transcription function of the MyoD family and could not evaluate the functions of any specific member. Therefore, we used Myf5, MyoD, and myogenin interchangeably in our transcription assays and obtained similar results.
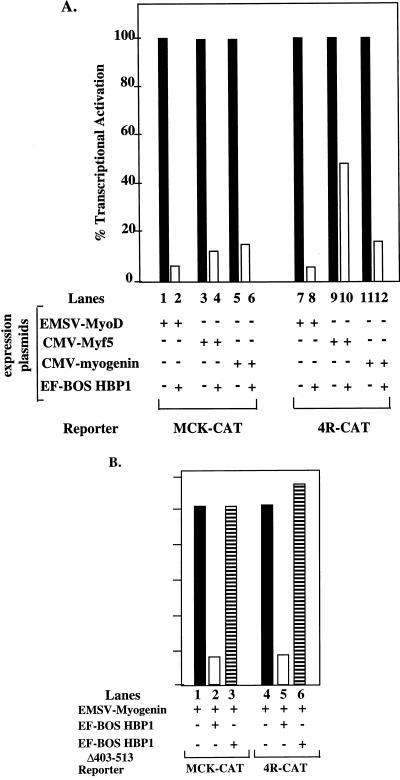
As expected, the expression of MyoD, Myf5, and myogenin led to significant activation of the synthetic E-box (4R-CAT) and of the natural MCK promoters. Noticeably, the coexpression of HBP1 resulted in a large inhibition of activation in each case (Fig. 7A). By Western analysis, the transfected MyoD family members were expressed at the same levels regardless of HBP1 expression (data not shown). Thus, the inhibition of HBP1 was not a result of defective MyoD family expression. The N-terminal repression domain of HBP1 was required for inhibition of the MyoD family transactivation (as represented by myogenin). Despite efficient expression (Fig. 4B), the HBP1 mutant ΔHBP403-513 was not functional (Fig. 7B). Thus, we conclude that expression of HBP1 can elicit efficient inhibition of transcriptional activation by MyoD family members and that this function depends upon the N-terminal domain of HBP1.
FIG. 7.
Inhibition by HBP1 of MyoD family transcriptional activation. (A) HBP1 can inhibit MyoD family activation of a natural differentiation-specific MCK-CAT or a simplified muscle-specific 4R-CAT reporter constructs. MCK-CAT denotes a CAT reporter construct driven by the ∼3 kb of the differentiation-specific muscle creatine kinase promoter (46). 4R-CAT denotes a simplified and muscle-specific reporter in which CAT expression is driven by four reiterated MyoD family binding sites (E-box elements) upstream of a minimal thymidine kinase promoter (51). The effect of HBP1 on transcriptional activation by either MyoD (lanes 1 and 2), Myf5 (lanes 3 and 4), or myogenin (lanes 5 and 6) was quantitated in C2 cells as described in Materials and Methods. In each set, transcriptional activities in the presence or absence of HBP1 were denoted by open or filled columns, respectively. (B) Inhibition of MyoD family transcriptional activation requires the N-terminal region of HBP1. Myogenin was used as a representative member of the MyoD family, and the relative inhibition by wild-type HBP1 (open column) and by ΔHBP403-513 (horizontal stripes) (see Fig. 1 for description) was compared by using assays similar to those described for panel A.
While HBP1 functioned as a sequence-specific transcriptional repressor (49), the apparent inhibition of MyoD transcriptional activation was probably not a result of DNA binding. First, HBP1 did not bind to the E-box element in an in vitro gel shift assay (data not shown). Second, it was important to note that the inhibition by HBP1 was specific for transcriptional activation by MyoD family members and did not reflect an inhibition of general transcriptional function. Other control promoters such as B-MYB were not repressed upon HBP1 expression (49). Similarly, the basal activity of the E-box reporter was also not directly affected by HBP1 expression (see Fig. 8C, lane 2). Third, our preliminary evidence on direct physical interactions of HBP1 and MyoD family members was mixed. We could demonstrate a specific physical interaction of HBP1 with all MyoD family members in glutathione S-transferase (GST) capture assays, but specific in vivo interactions were not detected (data not shown). While the block in transcriptional activation was specific, HBP1 might target other cofactors necessary for MyoD transcriptional activation (see Discussion), but the precise physical interactions were not yet clear. While functionally important, the physical interactions might also be transient and impossible to detect by immunoprecipitations that required stable contacts. However, these and previous studies suggest that HBP1 may elicit repression in different contexts but that not all promoters are equally affected.
FIG. 8.
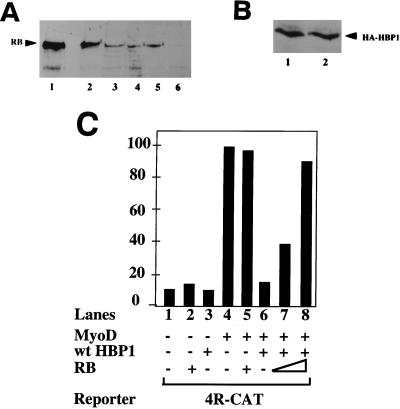
Expression of RB reverses the HBP1-mediated inhibition of differentiation and of MyoD transcriptional activation. Whereas Table 1 represents a direct quantitation of differentiation in the test cell lines, this figure depicts the relative expression levels of exogenous and endogenous RB protein. The presence of HA-RB transgene product was detected in lysates from each cell line by immunoprecipitation with anti-RB antibodies followed by Western analysis of immune complexes with a monoclonal anti-RB antibody (as described in Materials and Methods). This protocol allowed a direct comparison of “overexpressed” RB levels relative to endogenous RB. C2 cell line B1-B9 represents a line coexpressing HBP1 and RB; the C2 cell line B1-B2 represents a line expressing HBP1 only, but it was isolated with selection conditions identical to those for B1-B9. (A) Expression of RB and HA-RB in cell lines. Lanes: 1, C2C12 transiently transfected with HA-RB expression vector; 2, B1-B9 (HBP1+RB); 3, B1-B2 (control hygromycin- and puromycin-resistant line; HBP1 only); 4, B1-C2 (HBP1 only); 5, C2C12; 6, C2C12 cell extracts immunoprecipitated with anti-β-Gal antibodies as a negative control. The position of the RB protein is indicated and was determined in the positive control (lane 1). (B) Expression of HBP1 in cell lines. The levels were quantitated by Western blotting with an anti-HA antibody of an anti-HBP1 immunoprecipitation. This control experiment was performed to ensure that the reversal of the differentiation phenotype by RB was not due to the loss of HBP1 expression. Lanes: 1, B1-B9 (HBP1+RB); 2, B1-B2 (HBP1 only). (C) Effect of RB on HBP1-mediated inhibition of MyoD activation of 4R-CAT. The transcriptional activities were determined by transient-transfection assays in C2 cells by using specific promoter constructs together with wild-type or mutant HBP1 and RB expression vectors. Rous sarcoma virus–β-Gal was used as an internal transfection control to normalize transfection efficiency. The transfection output is expressed as a normalized ratio of CAT protein to β-Gal activity, and the combinations of expressed proteins are indicated. One representative experiment is shown in each graph, and each quantitation represents duplicate transfections that varied by <10%. Each experiment was repeated three to five times. Protein expression levels were equivalent in all transfections by Western blotting or immunoprecipitation followed by Western analyses (data not shown).
Expression of RB reverses the HBP1 inhibition of MyoD family transcriptional activation and of cell differentiation.
The observations in Fig. 2 to 7 suggested that HBP1, a target for RB and p130, had an apparent negative role in differentiation. Since previous work suggested a cooperative role of RB and MyoD in promoting muscle differentiation (30), we sought to determine whether RB could overcome the block in differentiation and in MyoD family activation by HBP1. A positive outcome would suggest that the activation of terminal differentiation genes might result from both positive activation by MyoD family members and RB-mediated relief of negative repression by HBP1.
To address whether RB could activate the tissue-specific aspects of differentiation, we generated RB cell lines in the background of one HBP1 cell line, B1-C2. We reasoned that the relatively low level of HBP1 expressed in this line could be more easily counteracted by RB expression. Using a second antibiotic (puromycin) selection marker, we isolated 10 lines from B1-C2 cells transfected with CMV-HA-RB. Only one (B1-B9) contained a detectable amount of RB transgene as determined by anti-HA immunoblot analysis (data not shown), a finding consistent with the known potent growth suppression ability of RB. This line contained a higher overall level of RB compared to control and C2C12 lines (Fig. 8A). The ability of this line to differentiate was examined by detection of MHC-expressing cells by using the immunostaining assay. We found that when subjected to differentiation conditions this cell line could now partially differentiate, containing ∼15% of MHC-positive cells (Table 1). Three control sister cell lines were isolated by double antibiotic selection, but they had no detectable RB transgene expression. None of these control lines was able to differentiate appreciably (Table 1; Fig. 8A, lane 3; and data not shown). The expression of the HA-HBP1 transgene was intact in all of these cell lines as shown by immunoprecipitation-Western analysis (Fig. 8B, lanes 1 and 2, and data not shown). This indicated that the partial reversal of differentiation in the HA-HBP1/RB cell line was not due to the loss of HBP1. Thus, the elevated RB expression level shown in Fig. 8A correlated well with the reversal of the HBP1-imposed differentiation defect in the B1-B9 line.
TABLE 1.
MHC-positive cellsa
| Expt | No. MHC positive/total no. (%) in cell line:
|
||||
|---|---|---|---|---|---|
| C2C12 | B1-B9 (RB-HBP1) | Con 1 | Con 2 | Con 3 | |
| 1 | 33/127 (26.0) | 32/225 (14.2) | 1/207 (0.48) | 2/321 (0.62) | 7/217 (3.2) |
| 2 | 30/96 (31.3) | 34/219 (15.5) | 1/194 (0.52) | 0/426 (0) | 5/207 (2.4) |
RB-expressing B1-B9 and three individual control lines with both selection markers were grown for 5 days in differentiation media. MHC-positive cells were detected by immunofluorescence and were quantitated from duplicate plates. Con, control.
Consistent with these findings, while HBP1 blocked MyoD-dependent transcription (Fig. 8C, lane 6), the reexpression of RB could also restore MyoD-dependent activation of transcription (Fig. 8C, lanes 7 and 8). Neither RB nor HBP1 alone affected the basal activity of 4R-CAT (Fig. 8C, lanes 2 and 3). It should be noted that the RB-positive C2 cells differed from previous studies in which RB−/− fibroblasts were used to show the dependence of RB on MyoD transcription. No exogenous effects of RB on MyoD activity were observed in the RB-positive C2C12 line (30) (Fig. 8C, compare lanes 4 and 5). We also verified that MyoD transcriptional activation did not occur in the RB-negative C33A cervical carcinoma line (data not shown); however, this would not provide a feasible test system for muscle differentiation. In Fig. 8, the use of the C2C12 system allowed a direct and concurrent test of HBP1 regulatory functions on MyoD transcriptional activation and on overall cell differentiation.
To further explore the mechanisms by which RB opposed HBP1 inhibition of differentiation, we examined functions of HBP1 mutants that were deficient in RB and p130 binding. For reference, HBP1 contained both LXCXE and IXCXE RB interaction motifs; mutation of either one retained interaction with RB family members. Mutation in both motifs abolished binding to RB and p130. In the context of sequence-specific repression of the N-MYC promoter, both motifs were necessary for full repression (49; see Fig. 1 for a summary).
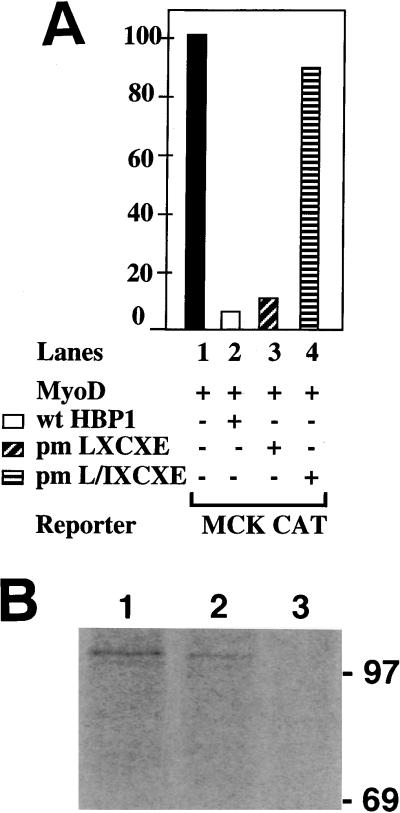
Intriguingly, the inhibition of MyoD family transcriptional activation by HBP1 depended on the ability to bind RB and p130 (Fig. 9). This function was retained in an HBP1 mutant with a single LXCXE mutation (pmLXCXE) (Fig. 9A, lane 3) but was abolished by mutation of both RB interaction motifs (pmL/IXCXE) (Fig. 9A, lane 4). The expression levels of wild-type and mutant HBP1 were similar as shown by anti-HBP1 immunoprecipitation following anti-HA Western analysis (data not shown).
FIG. 9.
Regulation of MyoD family transcriptional activation by HBP1 and RB. (A) Effect of HBP1 mutants on MyoD activation of a MCK-CAT reporter construct. The role of RB binding in the inhibition of MyoD family activation was tested by using the indicated mutants of HBP1. As described in Fig. 1 and reference 49, the wild-type HBP1 and pm-LXCXE are functional in RB binding and in repression of the N-MYC promoter, but the pm-L/IXCXE is defective in both functions. The indicated proteins were expressed in conjunction with MyoD: lane 1, no HBP expression vector (filled bar); lane 2, wild-type HBP1 (open bar); lane 3, pm-LXCXE (diagonal stripes); and lane 4, pm-L/I XCXE (horizontal stripes). (B) In vivo association of RB and HBP1 in differentiated C2C12 myotubes. HBP1 was shown to interact with RB in differentiated C2C12 myotubes. C2C12 were completely differentiated for 4 days in DMEM supplemented with 2% FCS. Cells were metabolically labeled with 35S-methionine, and cell lysates were collected for double immunoprecipitations as described in Materials and Methods. The first immunoprecipitations were carried out with anti-RB antibodies (lane 1) or anti-HBP1 antibodies (lanes 2 and 3), and the second immunoprecipitations were carried out with anti-RB antibodies (lanes 1 and 2) and anti-β-Gal antibodies (lane 3). Note that the amount of extracts used in lane 1 was approximately one-sixth of that used in lane 2 or 3.
We have already shown that HBP1 could specifically interact with RB and p130 in transfected cells (49). However, the demonstration of a physical interaction between endogenous HBP1 and RB or p130 in muscle cells would support the proposed functional connection between HBP1 and RB (and/or p130) in terminal differentiation. While simple in design, the low abundance of HBP1 posed a significant technical hurdle. HBP1 was present at an ∼10-fold-lower level than E2F, which was already a rare protein (47a, 52). Because the expression of endogenous RB and HBP1 was maximal in differentiated myotubes, we reasoned that this would be an optimal cell type for detecting HBP1-RB complexes.
Fully differentiated C2 cells were labeled with 35S-methionine, and coimmunoprecipitation was used to assess the endogenous interaction of RB and HBP1. As shown in Fig. 9B, the position of authentic RB was determined by immunoprecipitation with anti-RB antibodies in C2 extracts (lane 1). A double immunoprecipitation with anti-HBP1 followed by anti-RB was performed. As shown in Fig. 9B (lane 2), full-length RB was present in the test immunoprecipitation, suggesting a specific association of HBP1 with RB. No RB-specific band was evident in the control immunoprecipitation in which an irrelevant antibody (anti-β-Gal antibody) was used (Fig. 9B, lane 3). A similar strategy was attempted with p130, but the poorer quality of the anti-p130 antibodies precluded definitive analysis and the detection of endogenous complexes with HBP1. From Fig. 9, we conclude that endogenous HBP1 and RB complexes do exist, although at a rare abundance. This experiment extends and confirms previous results and suggests that our experimental system may mimic endogenous conditions.
DISCUSSION
This study focuses on the roles of RB and HBP1 in the coordination of cell cycle exit and tissue-specific gene expression in full muscle differentiation. We have shown that the expression of HBP1 inhibited muscle cell differentiation without blocking cell cycle exit. This study differs significantly from previous studies that have utilized oncogenes to block cell differentiation (e.g., E1A, RAS, mdm-2, and cyclin D1 [12, 35, 44]). In these cases, oncogenes prevented cell cycle exit, a necessary first step in differentiation. Thus, HBP1 and RB must be involved in steps that coordinate cell cycle exit and tissue-specific gene expression.
Because this study revealed unexpected results, we tried to eliminate several concerns. The first involved the use of cell lines. While ectopic expression of HBP1 in cell lines was necessary for our studies, we emphasize that exogenous HBP1 levels were modest. The level in the HBP1-expressing line was approximately the endogenous level obtained upon differentiation induction. Additionally, our experiments excluded the possibility that the observations in the HBP1-expressing lines were an artifact of stable cell line selection. We showed that the inhibition of differentiation was corroborated under both stable and transient expression, which argued against unique aspects of either experimental system. A second concern was that the HBP1-expressing cell lines were inherently defective in differentiation due to multiple, uninteresting, pleiotropic mutation. However, reexpression of MyoD and RB resulted in the expression of muscle differentiation markers and indicated that the HBP1-expressing lines retained the inherent ability to differentiate. A third concern was that the HBP1 protein was simply sequestering RB in an E1A-like manner. E1A and other viral oncogenes bind RB and enhance cell cycle entry (reviewed in reference 9). In contrast, the increased expression of HBP1 gave cell cycle exit, but not reentry. However, HBP1 did block tissue-specific gene expression in differentiation. Since RB is involved in both cell cycle and tissue-specific regulation during differentiation (reviewed in reference 53), the selective effect of HBP1 is incompatible with a general E1A-like sequestration of RB.
Regulation of specific MyoD family members.
Both cellular differentiation and transcriptional activity assays indicated that ectopic HBP1 expression blocked differentiation by interfering with tissue-specific gene expression. Our transcriptional experiments indicated that HBP1 abrogated the transcriptional activation by Myf5 and other MyoD family members. Consistent with this, ectopic expression of HBP1 inhibited muscle differentiation with a loss of MyoD and myogenin expression. Strikingly, Myf5 expression remained intact. A feature of the MyoD family is autoactivation. For example, MyoD can activate the myogenin promoter (54). Thus, HBP1 expression probably blocked differentiation by inhibiting Myf5 function and preventing the subsequent activation of the MyoD and myogenin promoters (Fig. 10B). Consistently, reexpression of MyoD or myogenin did rescue the differentiation-defective phenotype of HBP1 expression.
FIG. 10.
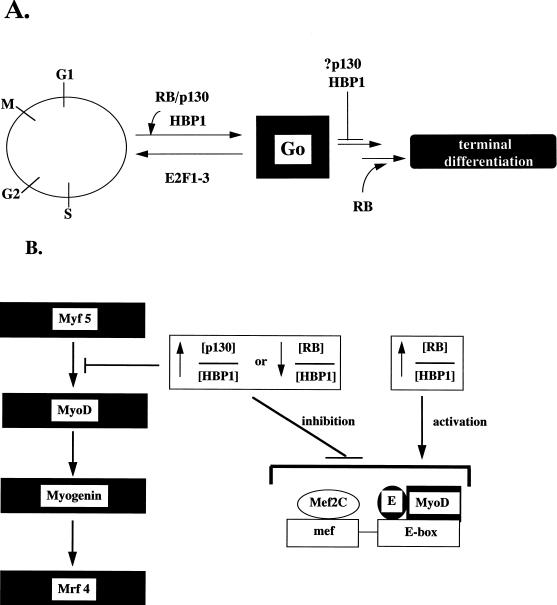
Summary model of HBP1 and RB functions in differentiation. We postulate that muscle differentiation can be divided into general cell cycle exit (G0) and tissue-specific gene expression (terminal differentiation) coordinated by the RB family (RB and p130) and their targets, such as HBP1. A high p130/HBP1 or low RB/HBP1 ratio may favor the cell cycle exit but act as a negative signal for terminal differentiation. This transient suspension of differentiation may eventually be relieved by activation of RB, resulting in a high RB/HBP1 ratio. This complex regulatory mechanism may be an effective means for ensuring fidelity in differentiation by ensuring that only viable and arrested cells proceed to the irreversible expression of genes that specify individual tissue phenotypes.
Is there evidence supporting differential functions for the highly homologous Myf5 and MyoD transcription factors? Recent studies have suggested a distinction in myogenesis, and Myf5 may lie functionally upstream of MyoD. First, Myf5 expression precedes MyoD in murine development, and MyoD expression is delayed in the absence of Myf5 (3, 39). However, either Myf5 or MyoD is required for full muscle development, since deletion of both abolishes muscle formation (40). Second, recent data now suggest that two parallel pathways governed by Myf5 and Pax3 may function upstream of MyoD to activate its expression. A Pax3−/− Myf5−/− mouse not only lacks muscle but, importantly, lacks MyoD expression, while MyoD expression is retained when either gene alone is deleted (24, 47; reviewed in reference 36). A direct investigation on the location of the HBP1 block will require the use of animal systems, since tissue culture systems do not recapitulate the subtle differences between MyoD and Myf5.
The unusual pattern of MyoD family members has also been observed in studies with CDK5, a differentiation-specific cyclin-dependent kinase (CDK) family member. The expression of a dominant negative CDK5 in Xenopus sp. inhibited muscle differentiation by blocking MyoD expression, but Myf5 expression was normal. Myogenin was not tested (34). Direct CDK5 expression enhanced muscle differentiation in C2 cells, which nicely complemented the Xenopus studies (20). Remarkably, CDK5 and HBP1 are both induced within 24 h of C2 cell differentiation (34, 49). The induction kinetics and unusual phenotype in differentiation inhibition suggest that CDK5 and HBP1 may conceivably target a step that resides between Myf5 and MyoD. However, the outcomes of CDK5 and HBP1 activity have opposite predictions, but this intriguing scenario remains to be tested.
Although our study has demonstrated a clear inhibition of Myf5 function by HBP1, an open question is whether a direct physical interaction occurs between HBP1 and Myf5. We have detected a physical complex of HBP1 and Myf5 in vitro, but in vivo interaction experiments have been uninformative (data not shown). Alternatively, HBP1 may inhibit Myf5 function by targeting Mef2c or E proteins, which are cofactors for Myf5 transcriptional activation (26, 27, 29).
Coordination of differentiation by HBP1 and RB.
We have shown that RB can reverse the HBP1-mediated inhibition of MyoD family transcriptional activation and differentiation. The simplest explanation is that the relative ratio of RB to HBP1 dictates whether only cell cycle arrest or full differentiation occurs. At a lowered RB/HBP1 ratio cell cycle exit, but not tissue-specific gene expression, persists. The interaction with RB and p130 is central to HBP1-mediated inhibition and suggests that the active inhibitor may be a complex of HBP1 with p130 or RB. Additionally, experimentally increasing the ratio of RB to HBP1 could partially rescue the HBP1 inhibition of differentiation and of MyoD transcriptional activity. In normal C2 cell differentiation, the accumulation of the underphosphorylated form of RB occurs just prior to the onset of tissue-specific gene expression. This suggests that the accumulation of the underphosphorylated RB may be one signal for activation of MyoD and of tissue-specific gene expression. Thus, our experimental system mimics the normal C2 cell differentiation. Because the RB regulation of MyoD family members is probably indirect, the elevated ratio of RB to HBP1 may provide the signal for activation of the MyoD family and of tissue-specific gene expression (see model Fig. 10). The importance of RB has also been underscored by recent work in which the functions of RB in MyoD activation and in E2F regulation could be uncoupled (37, 42). Activation of E2F does not require the N-terminal region, yet it is necessary for full muscle differentiation. Additionally, specific pocket mutants that were defective in E2F regulation still supported differentiation. Thus, both the N-terminal and pocket regions are required for the activation of differentiation in both cellular and animal models.
Our data do not exclude the possible involvement of another RB family member, p130, in the negative signaling pathway during differentiation. Indeed, we speculate that HBP1 and p130 may act as active inhibitors of MyoD-like master regulators and block expression of the differentiation-specific genes. In the adipocyte system, Classon et al. have recently demonstrated that loss of p130 and p107 results in an unexpected increase in differentiation. Due to functional compensation, both p130 and p107 must be deleted to yield this phenotype. Their results suggest opposite functions for RB and p130 and are consistent with an inhibitory function for p130 in differentiation (6). Preliminary data obtained in our laboratory suggest that the expression of p130 alone cannot support muscle differentiation in C2 cells, despite efficient cell cycle arrest (42a).
In the present study, HBP1 inhibited MyoD family transcriptional functions; this negative function also required binding to either p130 or RB. The collective data do raise the novel possibility that HBP1 and p130 are active inhibitors of tissue-specific gene expression during differentiation. Yet, both HBP1 and p130 are active inducers of cell cycle exit. These dual functions of HBP1 and p130 may ensure that cell cycle exit is complete prior to expression of tissue-specific genes. Perhaps p130 is involved in negative regulation but RB is necessary for positive activation of tissue-specific gene expression. Thus, RB and p130 could conceivably have opposite roles in the progression of differentiation.
While the least complex model for RB family member function has been provided, further investigation is clearly needed to firmly establish these opposite roles for RB and for p130 or p107. A considerable complication is that there is extensive functional compensation by other RB family members when one is mutated (16, 28). HBP1 binds both p130 and RB, and we cannot distinguish the functions of these related proteins in the C2 cell line, in which all three proteins are upregulated during differentiation. The ideal reagents for testing the functions in differentiation will be RB−/− p107−/− p130−/− and/or HBP1−/− cells or mice, but neither currently exists.
A differentiation checkpoint?
The paradoxical induction of proteins that induce cell cycle arrest but block differentiation may be a general feature, since three distinct examples have now been described. In adipocytes, the apparent negative inhibitor induced upon differentiation is GADD153/CHOP, and this protein shows striking functional similarity to HBP1. GADD153/CHOP was originally isolated as a gene that was induced with growth arrest and DNA damage and was later shown to be an inhibitor of the C/EBP transcription factor family (38). Like MyoD in muscle, members of the C/EBP family are critical factors in adipocyte differentiation and are positively regulated by RB (4). First, like HBP1, CHOP expression is normally induced in adipocyte differentiation, but ectopic expression paradoxically blocks adipocyte differentiation. Similar to our studies, the reexpression of the master regulator C/EBP-α can also restore differentiation in CHOP-expressing cells, suggesting that CHOP also blocks differentiation by blocking the master regulator (2). Second, like HBP1, direct expression of CHOP leads to cell cycle arrest (1, 49). Like HBP1, CHOP also has ubiquitous tissue distribution, which suggests that negative regulation of differentiation may not be limited to adipocyte or muscle, respectively (1, 21, 47a). While HBP1 is regulated by RB and p130, it is not known whether this is true for CHOP. Recently, the p202 protein has been described as another inducible yet negative inhibitor of differentiation. Like HBP1 and CHOP, expression of p202 elicits cell cycle arrest but blocks differentiation. Intriguingly, p202 also binds RB (10).
To explain the observations in the current work and in the literature, we propose the existence of a differentiation checkpoint that regulates coordination of cell cycle exit and tissue-specific gene expression. This hypothetical checkpoint would consist of both positive and negative regulation involving RB, p130, and their target proteins. In the early phase of differentiation (about 24 h), cell cycle exit predominates but there is no tissue-specific differentiation. Both p130 and HBP1 levels are high in this early phase. We propose that p130 and HBP1 simultaneously elicit cell cycle arrest but actively block the activation of MyoD and tissue-specific gene expression. The lower ratio of RB to HBP1 may signal that the environment for tissue-specific gene expression is inappropriate until cell cycle exit is complete. The functional similarities of three distinct proteins (HBP1, CHOP, and p202) predict the existence of a general inhibitory pathway that provides cell cycle arrest but may transiently suspend tissue-specific gene expression during differentiation. Additionally, the repressor complex E2F4-p130 may also contribute to cell cycle exit in this early phase (see, for example, references 8 and 43).
When cell cycle exit is completed, there would be activation of MyoD and of tissue-specific genes at about h 48 of C2 differentiation. We hypothesize that a positive signal is generated by the higher ratio of RB to HBP1. The accumulation of the underphosphorylated form of RB may contribute to the activation of MyoD, C/EBP, and other regulators of tissue-specific gene expression. In this way, both negative and positive regulation of tissue-specific gene expression would ensure that cell cycle exit is complete prior to activation of MyoD and other global regulators.
We emphasize that this differentiation checkpoint is meant as a framework for generating future studies. The results presented in the current study and in the literature do support the notion of negative and positive regulation in the coordination of cell cycle exit and tissue-specific gene expression in a full differentiation pathway. While the mechanics of MyoD family members are well understood, the mechanisms underlying their activation are still unclear. A component of the regulatory signal must be the completion of cell cycle exit, since this necessarily precedes tissue-specific gene expression. Thus, the dual functions in regulating cell cycle exit and tissue-specific gene expression suggest that proteins such as HBP1 and RB might be excellent candidates in a checkpoint that coordinates the progression and fidelity during differentiation. How other MyoD cofactors such as Mef2C and p300 fit into an RB-p130-mediated pathway is unclear. In any case, the current studies do provide a new view on the role of RB family members in cell differentiation. An important future goal is the elucidation of the precise mechanisms by which cell cycle exit signals to activate MyoD family transcription.
ACKNOWLEDGMENTS
We thank the following colleagues for their generosity: Eric Olson (myogenin, MyoD, Myf5, and MCK CAT), Woody Wright (antimyogenin F5D), Frank Stockdale (continuous supply of anti-MHC), and Yukang Wang (murine Myf5). We thank Eric Paulson, Andrew Leiter, Brian Schaffhausen, and Larry Feig for many helpful discussions. We especially thank Marie Classon, Ed Harlow, Bill Kaelin, and Wen-Hwa Lee for providing information prior to publication.
The work was supported by grants to A.S.Y. from the AHA, the NIH (GM44634), and the Digestive Disease Center at New England Medical Center (NIDDK, P30 DK-34928). A.S.Y. is an Established Investigator of the American Heart Association.
REFERENCES
- 1.Barone M V, Crozat A, Tabaee A, Philipson L, Ron D. CHOP (GADD153) and its oncogenic variant, TLS-CHOP, have opposing effects on the induction of G1/S arrest. Genes Dev. 1994;8:453–464. doi: 10.1101/gad.8.4.453. [DOI] [PubMed] [Google Scholar]
- 2.Batchvarova N, Wang X Z, Ron D. Inhibition of adipogenesis by the stress-induced protein CHOP (Gadd153) EMBO J. 1995;14:4654–4661. doi: 10.1002/j.1460-2075.1995.tb00147.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Braun T, Rudnicki M, Arnold H-H, Jaenisch R. Targeted inactivation of the muscle regulatory gene Myf-5 results in abnormal rib development and perinatal death. Cell. 1992;71:369–382. doi: 10.1016/0092-8674(92)90507-9. [DOI] [PubMed] [Google Scholar]
- 4.Chen P L, Riley D J, Chen Y, Lee W H. Retinoblastoma protein positively regulates terminal adipocyte differentiation through direct interaction with C/EBPs. Genes Dev. 1996;10:2794–2804. doi: 10.1101/gad.10.21.2794. [DOI] [PubMed] [Google Scholar]
- 5.Cheng T C, Tseng B S, Merlie J P, Klein W H, Olson E N. Activation of the myogenin promoter during mouse embryogenesis in the absence of positive autoregulation. Proc Natl Acad Sci USA. 1995;92:561–565. doi: 10.1073/pnas.92.2.561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Classon, M., C. Gorka, B. K. Kennedy, N. Dyson, and E. Harlow. Opposing roles for pRB and p107/p130 in adipocyte differentiation. Submitted for publication.
- 7.Cobrinik D, Whyte P, Peeper D, Jacks T, Weinberg R. Cell cycle-specific association of E2F with the p130 E1A-binding protein. Genes Dev. 1993;7:2392–2404. doi: 10.1101/gad.7.12a.2392. [DOI] [PubMed] [Google Scholar]
- 8.Corbeil H, Whyte P, Branton P E. Characterization of transcription factor E2F complexes during muscle and neuronal differentiation. Oncogene. 1995;11:909–920. [PubMed] [Google Scholar]
- 9.Cress W D, Nevins J R. Use of the E2F transcription factor by DNA tumor virus regulatory proteins. In: Farnham P J, editor. Transcriptional control of cell growth: the E2F gene family. Vol. 208. New York, N.Y: Springer-Verlag; 1996. pp. 63–78. [DOI] [PubMed] [Google Scholar]
- 10.Datta B, Min W, Burma S, Lengyel P. Increase in p202 expression during skeletal muscle differentiation: inhibition of MyoD protein expression and activity of p202. Mol Cell Biol. 1998;18:1074–1083. doi: 10.1128/mcb.18.2.1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Farnham P J. Transcriptional control of cell growth: the E2F family. Vol. 208. New York, N.Y: Springer-Verlag; 1996. [Google Scholar]
- 12.Fiddler T A, Smith L, Tapscott S J, Thayer M J. Amplification of MDM2 inhibits MyoD-mediated myogenesis. Mol Cell Biol. 1996;16:5048–5057. doi: 10.1128/mcb.16.9.5048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Grosschedl R. Higher order nucleoprotein complexes in transcription: analogies with site-specific recombination. Curr Opin Cell Biol. 1995;7:362–370. doi: 10.1016/0955-0674(95)80091-3. [DOI] [PubMed] [Google Scholar]
- 14.Gu W, Schneider J W, Condorelli G, Kaushal S, Mahdavi V, Nadal-Ginard B. Interaction of myogenic factors and the retinoblastoma protein mediates muscle cell commitment and differentiation. Cell. 1993;72:309–324. doi: 10.1016/0092-8674(93)90110-c. [DOI] [PubMed] [Google Scholar]
- 15.Hunter T. Oncoprotein networks. Cell. 1997;88:333–346. doi: 10.1016/s0092-8674(00)81872-3. [DOI] [PubMed] [Google Scholar]
- 16.Hurford R K, Cobrinik D, Lee M-H, Dyson N. pRb and p107/p130 are required for the regulated expression of different sets of E2F responsive genes. Genes Dev. 1997;11:1447–1463. doi: 10.1101/gad.11.11.1447. [DOI] [PubMed] [Google Scholar]
- 17.Ikeda M A, Jakoi L, Nevins J R. A unique role for the Rb protein in controlling E2F accumulation during cell growth and differentiation. Proc Natl Acad Sci USA. 1996;93:3215–3220. doi: 10.1073/pnas.93.8.3215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lassar A, Thayer M, Overell R, Weintraub H. Transformation by activated ras or fos prevents myogenesis by inhibiting expression of MyoD1. Cell. 1989;58:659–667. doi: 10.1016/0092-8674(89)90101-3. [DOI] [PubMed] [Google Scholar]
- 19.Lavender P, Vandel L, Bannister A J, Kouzarides T. The HMG-box transcription factor HBP1 is targeted by the pocket proteins and E1A. Oncogene. 1997;14:2721–2728. doi: 10.1038/sj.onc.1201243. [DOI] [PubMed] [Google Scholar]
- 20.Lazaro J B, Kitzmann M, Poul M A, Vandromme M, Lamb N J, Fernandez A. Cyclin dependent kinase 5, cdk5, is a positive regulator of myogenesis in mouse C2 cells. J Cell Sci. 1997;110:1251–1260. doi: 10.1242/jcs.110.10.1251. [DOI] [PubMed] [Google Scholar]
- 21.Lesage F, Hugnot J P, Amri E Z, Grimaldi P, Barhanin J, Lazdunski M. Expression cloning in the K+ transport-defective yeast and regulation of HBP1, a new putative HMG transcriptional regulator. Nucleic Acids Res. 1994;22:3685–3688. doi: 10.1093/nar/22.18.3685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Maione R, Amati P. Interdependence between muscle differentiation and cell-cycle control. Biochim Biophys Acta. 1997;1332:M19–M30. doi: 10.1016/s0304-419x(96)00036-4. [DOI] [PubMed] [Google Scholar]
- 23.Maione R, Fimia G, Holman P, Schaffhausen B, Amati P. Retinoblastoma antioncogene is involved in the inhibition of myogenesis by polyomavirus large T antigen. Cell Growth Differ. 1994;5:231–237. [PubMed] [Google Scholar]
- 24.Maroto M, Reshef R, Munsterberger E, Koester S, Goulding M, Lassar A B. Ectopic Pax-3 activates MyoD and Myf-5 expression in embryonic mesoderm and neural tissue. Cell. 1997;89:139–148. doi: 10.1016/s0092-8674(00)80190-7. [DOI] [PubMed] [Google Scholar]
- 25.Moberg K, Starz M A, Lees J A. E2F-4 switches from p130 to p107 and pRB in response to cell cycle reentry. Mol Cell Biol. 1996;16:1436–1449. doi: 10.1128/mcb.16.4.1436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Molkentin J D, Olson E N. Defining the regulatory networks for muscle development. Curr Opin Genet Dev. 1996;6:445–453. doi: 10.1016/s0959-437x(96)80066-9. [DOI] [PubMed] [Google Scholar]
- 27.Molkentin J D, Olson E N. Combinatorial control of muscle development by basic helix-loop-helix and MADS-box transcription factors. Proc Natl Acad Sci USA. 1996;93:9366–9373. doi: 10.1073/pnas.93.18.9366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mulligan G J, Wong J, Jacks T. p130 is dispensable in peripheral T lymphocytes: evidence for functional compensation by p107 and pRB. Mol Cell Biol. 1998;18:206–220. doi: 10.1128/mcb.18.1.206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Murre C, McCaw P S, Vaessin H, Caudy M, Jan L Y, Jan Y N, Cabrera C V, Buskin J N, Hauschka S D, Lassar A B, Weintraub H, Baltimore D. Interactions between heterologous helix-loop-helix proteins generate complexes that bind specifically to a common DNA sequence. Cell. 1989;58:537–544. doi: 10.1016/0092-8674(89)90434-0. [DOI] [PubMed] [Google Scholar]
- 30.Novitch B G, Mulligan G J, Jacks T, Lassar A B. Skeletal muscle cells lacking the retinoblastoma protein display defects in muscle gene expression and accumulate in S and G2 phases of the cell cycle. J Cell Biol. 1996;135:441–456. doi: 10.1083/jcb.135.2.441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nusse R. A versatile transcriptional effector of wingless signaling. Cell. 1997;89:321–323. doi: 10.1016/s0092-8674(00)80210-x. [DOI] [PubMed] [Google Scholar]
- 32.Olson E N, Klein W H. bHLH factors in muscle development: deadlines and commitments, what to leave in and what to leave out. Genes Dev. 1994;8:1–8. doi: 10.1101/gad.8.1.1. [DOI] [PubMed] [Google Scholar]
- 33.Olson E N, Spizz G, Tainsky M. The oncogenic forms of N-ras or H-ras prevent skeletal myoblast differentiation. Mol Cell Biol. 1987;7:2104–2111. doi: 10.1128/mcb.7.6.2104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Phillipot A, Porro E, Kirschner M W, Tsai L-H. The role of cyclin-dependent kinase 5 and a novel regulatory subunit in regulating muscle differentiation and patterning. Genes Dev. 1997;11:1409–1421. doi: 10.1101/gad.11.11.1409. [DOI] [PubMed] [Google Scholar]
- 35.Rao S, Chu C, Kohtz S. Ectopic expression of cyclin D1 prevents activation of gene transcription by myogenic basic helix-loop-helix proteins. Mol Cell Biol. 1994;14:5259–5267. doi: 10.1128/mcb.14.8.5259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rawls A, Olson E N. MyoD meets its maker. Cell. 1997;89:5–8. doi: 10.1016/s0092-8674(00)80175-0. [DOI] [PubMed] [Google Scholar]
- 37.Riley D J, Liu C Y, Lee W H. Mutations of N-terminal regions render the retinoblastoma protein insufficient for functions in development and tumor suppression. Mol Cell Biol. 1997;17:7342–7352. doi: 10.1128/mcb.17.12.7342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ron D, Habener J F. CHOP, a novel developmentally regulated nuclear protein that dimerizes with transcription factors C/EBP and LAP and functions as a dominant-negative inhibitor of gene transcription. Genes Dev. 1992;6:439–453. doi: 10.1101/gad.6.3.439. [DOI] [PubMed] [Google Scholar]
- 39.Rudnicki M, Braun T, Hinuma S, Jaenisch R. Inactivation of MyoD in mice leads to up-regulation of the myogenic HLH gene Myf-5 and results in apparently normal muscle development. Cell. 1992;71:383–390. doi: 10.1016/0092-8674(92)90508-a. [DOI] [PubMed] [Google Scholar]
- 40.Rudnicki M, Braun T, Hinuma S, Jaenisch R. MyoD or Myf 5 is required for formation of skeletal muscle. Cell. 1993;75:1351–1359. doi: 10.1016/0092-8674(93)90621-v. [DOI] [PubMed] [Google Scholar]
- 41.Schneider J W, Gu W, Zhu L, Mahdavi V, Nadal-Ginard B. Reversal of terminal differentiation mediated by p107 in Rb−/− muscle cells. Science. 1994;264:1467–1470. doi: 10.1126/science.8197461. [DOI] [PubMed] [Google Scholar]
- 42.Sellers W R, Novitch B G, Miyake S, Heith A, Otterson G A, Kaye F J, Lassar A B, Kaelin W G J. Stable binding to E2F is not required for the retinoblastoma protein to activate transcription, promote differentiation, and suppress tumor cell growth. Genes Dev. 1998;12:95–106. doi: 10.1101/gad.12.1.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42a.Sheppard, K. A., and A. S. Yee. Unpublished observations.
- 43.Shin E, Shin A, Paulding C, Schaffhausen B, Yee A S. Multiple changes in E2F transcription factor function occur upon muscle differentiation. Mol Cell Biol. 1995;15:2252–2262. doi: 10.1128/mcb.15.4.2252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Skapek S X, Rhee J, Spicer D B, Lassar A B. Inhibition of myogenic differentiation in proliferating myoblast by cyclin D1-dependent kinase. Science. 1995;267:1022–1023. doi: 10.1126/science.7863328. [DOI] [PubMed] [Google Scholar]
- 45.Smith E J, Leone G, DeGregori J, Jakoi L, Nevins J R. The accumulation of an E2F-p130 transcriptional repressor distinguishes a G0 cell state from a G1 cell state. Mol Cell Biol. 1996;16:6965–6976. doi: 10.1128/mcb.16.12.6965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sternberg E A, Spizz G, Perry M W, Vizard D, Weil T, Olson E N. Identification of upstream and intragenic regulatory elements that confer cell type-restricted and differentiation-specific expression on the muscle creatine kinase gene. Mol Cell Biol. 1988;8:2896–2909. doi: 10.1128/mcb.8.7.2896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tajbakhsh S, Rocancourt D, Cossu G, Buckingham M. Redefining the genetic hierarchies controlling skeletal myogenesis: Pax-3 and Myf-5 act upstream of MyoD. Cell. 1997;89:127–138. doi: 10.1016/s0092-8674(00)80189-0. [DOI] [PubMed] [Google Scholar]
- 47a.Tevosian, S. Unpublished data.
- 48.Tevosian S, Paulson K E, Bronson R, Yee A S. Expression of the E2F-1/DP-1 transcription factor during murine development. Cell Growth Differ. 1996;7:43–52. [PubMed] [Google Scholar]
- 49.Tevosian S G, Shih H, Mendelson K G, Sheppard K A, Paulson K E, Yee A S. HBP1: an HMG box transcriptional repressor that is targeted by the retinoblastoma family. Genes Dev. 1997;11:383–396. doi: 10.1101/gad.11.3.383. [DOI] [PubMed] [Google Scholar]
- 50.Thayer M J, Tapscott S J, Davis R L, Wright W, Lassar A B, Weintraub H. Positive auto-regulation of the myogenic determination gene MyoD1. Cell. 1989;58:241–248. doi: 10.1016/0092-8674(89)90838-6. [DOI] [PubMed] [Google Scholar]
- 51.Weintraub H, Davis R, Lockshon D, Lassar A. MyoD binds cooperatively to two sites in a target enhancer sequence: occupancy of two sites is required for activation. Proc Natl Acad Sci USA. 1990;87:5623–5627. doi: 10.1073/pnas.87.15.5623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yee A S, Raychaudhuri P, Jakoi L, Nevins J R. The adenovirus-inducible factor E2F stimulates transcription following specific DNA binding. Mol Cell Biol. 1989;9:578–585. doi: 10.1128/mcb.9.2.578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yee A S, Shih H S, Tevosian S G. New perspectives on retinoblastoma family functions in differentiation. 1998. Front. Biosci., in press. [DOI] [PubMed] [Google Scholar]
- 54.Yee S P, Rigby P W. The regulation of myogenin gene expression during the embryonic development of the mouse. Genes Dev. 1993;7:1277–1289. doi: 10.1101/gad.7.7a.1277. [DOI] [PubMed] [Google Scholar]
- 55.Yeh W C, Cao Z, Classon M, McKnight S L. Cascade regulation of terminal adipocyte differentiation by three members of the C/EBP family of leucine zipper proteins. Genes Dev. 1995;9:168–181. doi: 10.1101/gad.9.2.168. [DOI] [PubMed] [Google Scholar]
- 56.Zackenhaus E, Jiang Z, Chung D, Marth J D, Phillips R A, Gallie B L. pRB controls proliferation, differentiation, and death of skeletal muscle cells and other lineages during embryogenesis. Genes Dev. 1996;10:3051–3064. doi: 10.1101/gad.10.23.3051. [DOI] [PubMed] [Google Scholar]
- 57.Zingg J M, Pedrazza-Alva G, Jost J P. MyoD1 promoter autoregulation is mediated by two proximal E-boxes. Nucleic Acids Res. 1994;22:2234–2241. doi: 10.1093/nar/22.12.2234. [DOI] [PMC free article] [PubMed] [Google Scholar]