Abstract
Cryptococcosis is an opportunistic fungal infection caused by members of the two sibling species complexes: Cryptococcus neoformans and Cryptococcus gattii. Flucytosine (5FC) is one of the most widely used antifungals against Cryptococcus spp., yet very few studies have looked at the molecular mechanisms responsible for 5FC resistance in this pathogen. In this study, we examined 11 C. gattii clinical isolates of the major molecular type VGIII based on differential 5FC susceptibility and asked whether there were genomic changes in the key genes involved in flucytosine metabolism. Susceptibility assays and sequencing analysis revealed an association between a point mutation in the cytosine deaminase gene (FCY1) and 5FC resistance in two of the studied 5FC resistant C. gattii VGIII clinical isolates, B9322 and JS5. This mutation results in the replacement of arginine for histidine at position 29 and occurs within a variable stretch of amino acids. Heterologous expression of FCY1 and spot sensitivity assays, however, demonstrated that this point mutation did not have any effect on FCY1 activities and was not responsible for 5FC resistance. Comparative sequence analysis further showed that no changes in the amino acid sequence and no genomic alterations were observed within 1 kb of the upstream and downstream sequences of either cytosine permeases (FCY2-4) or uracil phosphoribosyltransferase (FUR1) genes in 5FC resistant and 5FC susceptible C. gattii VGIII isolates. The herein obtained results suggest that the observed 5FC resistance in the isolates B9322 and JS5 is due to changes in unknown protein(s) or pathway(s) that regulate flucytosine metabolism.
Keywords: Cryptococcus gattii, flucytosine resistance, cytosine deaminase, FCY1
Introduction
Members of the Cryptococcus neoformans and Cryptococcus gattii species complexes are the etiological agents of cryptococcosis. While C. neoformans is responsible for most cases of infections globally, C. gattii continues to pose a serious public health threat, as witnessed by ongoing cases of infections in the Pacific Northwest region of Canada and the United States since 1999.1 Despite global concerns, a limited number of antifungals are effective against Cryptococcus spp., and resistance to these drugs is common in clinical settings.2 Current therapeutic options for the treatment of cryptococcosis include amphotericin B formulations, flucytosine, and the triazoles.2 Flucytosine (5-fluorocytosine, 5FC) is a fluorinated pyrimidine with activity against a number of fungal pathogens. It has no intrinsic antifungal capacity, but after uptake by susceptible fungi it is converted into 5-fluorouracil (5FU), which is further converted into metabolites that inhibit fungal RNA and DNA synthesis. 5FC combined with amphotericin B is currently the gold standard for the treatment of cryptococcal meningoencephalitis,3 and reduced mortality with the use of this regimen has been demonstrated in a randomized trial.4
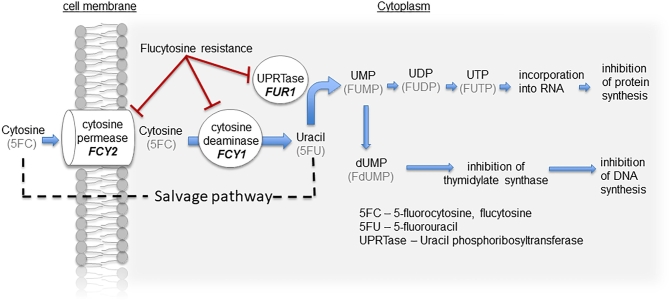
It is well known that flucytosine metabolism utilizes the pyrimidine salvage pathway that operates in fungal and bacterial species. Mutations in three key components of this pathway have been implicated in promoting flucytosine resistance in fungal species: cytosine permease (FCY2), cytosine deaminase (FCY1), and uracil phosphoribosyltransferase (UPRTase, encoded by FUR1).5,6 Flucytosine is transported into susceptible fungi by the membrane-bound cytosine permease and subsequently deaminated to 5-fluorouracil (5FU) by cytosine deaminase (Fig. 1). The absence of cytosine deaminase in mammalian cells allows selective toxicity on fungal organisms. Following deamination of 5FC, 5FU is converted into 5-fluorouridine monophosphate (FUMP) via the action of UPRTase. FUMP is then further reduced to 5-fluorodeoxyuridine monophosphate (FdUMP), which interferes with DNA synthesis. Inhibition of protein synthesis occurs through a separate mechanism where FUMP is phosphorylated to become 5-fluorouridine diphosphate (FUDP) and 5-fluorouridine triphosphate (FUTP), which is in turn incorporated into fungal RNA, thereby disrupting the amino acid pool and ultimately causing abnormal protein synthesis.
Figure 1.
The main enzymatic steps involved in the uptake, conversion, and mechanism of action of fluoropyrimidines. 5FC, flucytosine; 5FU, 5-fluorouracil; UPRTase/FUR1, uracil phosphoribosyltransferase; FUMP, 5-fluorouridine monophosphate; FUDP, 5-fluorouridine diphosphate; FUTP, 5-fluorouridine triphosphate; FdUMP, 5-fluorodeoxyuridine monophosphate. This Figure is reproduced in color in the online version of Medical Mycology.
Due to the high incidence of acquired or primary (intrinsic) resistance, the use of 5FC as monotherapy is not recommended except in selected cases of urinary candidiasis and chromoblastomycosis.3,7 Acquired resistance is often due to a decrease in UPRTase activity and is most frequently found following treatment with 5FC.8 This resistance is thought to be a result of either the failure of the organism to metabolize the drug or the loss of pyrimidine biosynthesis feedback control.8 On the other hand, primary resistance is thought to be the result of impaired cellular deamination or decreased 5FC uptake due to mutations in the FCY1 or FCY2 genes respectively.9 At the present time, there is a paucity of data on the regulatory and resistance mechanisms of 5FC in Cryptococcus spp. A recent genome-scale study on the regulatory and resistance mechanisms of 5FC in C. neoformans identified 177 5FC responsive genes.10 In particular, a C. neoformans strain lacking the transcription factor, Mbs1, displayed increased susceptibility to 5FC.10 On the other hand, the 5FC resistance mechanisms have not been previously explored in C. gattii at the molecular level, and it is generally thought that they are similar to those found in Saccharomyces cerevisiae and Candida albicans.8 In the herein presented work, we tested 11 C. gattii clinical and veterinary isolates belonging to the major molecular type VGIII/AFLP5 for 5FC susceptibility and identified two resistant isolates with 5FC MIC > 64 μg/ml. Our data surprisingly show that the observed 5FC resistance did not directly involve changes in either protein sequences or upstream and downstream genomic sequences of cytosine permeases, cytosine deaminase, or uracil phosphoribosyltransferase genes. We speculate that the 5FC resistance in the two examined C. gattii VGIII clinical isolates, B9322 and JS5, is due to changes in regulatory protein(s) or pathway(s) that control flucytosine metabolism.
Methods
Reagents, isolates, and culture conditions
Flucytosine (5FC) (Cat. F7129) and cytosine (Cat. C3506) were obtained from Sigma. All 11 C. gattii strains were grown and maintained on yeast-peptone-dextrose (YPD) medium. 5FC resistant C. gattii VGIII strains (B9322 and JS5) and susceptible strains (JS91 and those with MICs < 4 μg/ml shown in Table 1) were obtained from a collection of clinical and veterinary C. gattii isolates.11–15 The S. cerevisiae WT and mutant strain lacking cytosine deaminase (Sc fcy1Δ) were a gift from Joseph Heitman (Duke University).16
TABLE 1.
In vitro susceptibility testing and FCY1 profiles of 11 Cryptococcus gattii clinical and veterinery isolates used in this study.
| Strains | Other Collection | Year of | Mating | FCY1 | 5FC MIC | Original | ||||
|---|---|---|---|---|---|---|---|---|---|---|
| ID | numbers | Country | isolation | Source | Serotype | Type | Sequence (5΄ → 3΄) | Mutations | (ug/mL) | Reference |
| B9322 | WM1 1.949 | USA | 2011 | clin | B | alpha | TTGAAGAGTCATTCG | LKSHS | >64 | Ref 13 |
| JS5 | WM10.165 | USA | 2005 | vet | B | alpha | TTGAAGAGTCATTCG | LKSHS | >64 | Ref 12 |
| JS69 | WM10.186 | USA | 2006 | vet | B | alpha | TTGAAGAGTCGTTCG | LKSRS | 0.5 | Ref 14 |
| JS91 | WM11.40 | USA | 2011 | vet | C | a | TTGAAGAGCCGTTCG | LKSRS | 0.5 | Ref 14 |
| JS110 | WM11.139 | USA | 2011 | vet | B | a | TTGAAGAGCCGTTCG | LKSRS | 1 | Ref 12 |
| B8965 | WM11.941 | USA | 2010 | clin | B | a | TTGAAGAGCCGTTCG | LKS RS | 1 | Ref 13 |
| B9151 | WM1 1.945 | USA | 2011 | clin | B | alpha | TTGAAGAGCCGTTCG | LKSRS | 1 | Ref 12 |
| B8260 | WM1 1.937 | USA | 2009 | vet | C | alpha | TTGAAGAGCCGTTCG | LKSRS | 0.5 | Ref 13 |
| B8262 | WM1 1.938 | USA | 1992 | clin | B | alpha | TTGAAGAGICGTTCG | LKSRS | 1 | Ref 15 |
| 07-11763 | WM09.45 | USA | 2007 | vet | B | alpha | TTGAAGAGICGTTCG | LKS RS | 1 | Ref 12 |
| 08-7686 | WM09.47 | USA | 2008 | vet | B | alpha | TTGAAGAGICGTTCG | LKSRS | 0.5 | Ref 12 |
Genome sequencing and in vitro susceptibility testing of C. gattii clinical isolates
Isolates included in this study all belong to the C. gattii VGIII major molecular type and had previously undergone whole genome sequencing as part of a larger population genomics survey.11–15 Isolates with differential 5FC susceptibility were chosen for comparison, and genomic sequences of regions predicted to be involved in 5FC susceptibility were compared (see Table 2). Susceptibility testing was performed on 11 C. gattii VGIII isolates in accordance with the CLSI reference document M27-A3.17Candida krusei ATCC 6258 and Candida parapsilosis ATCC 22019 were used as controls for susceptibility testing. The strains JS5 and B9322 showing 5FC MICs >64 μg/ml were chosen for comparison to nine isolates with 5FC MICs <4 μg/ml (Table 1).
Table 2.
List of putative Cryptococcus gattii genetic loci involved in cytosine metabolism.
| C. gattii genes | Broad institute ID | Amino acid changes |
|---|---|---|
| Cytosine deaminase (FCY1) | CNAG_00613.2 | R29H mutation in two isolates |
| Cytosine permease (FCY2) | CNAG_01681.2 | No change |
| Cytosine-purine permease, (FCY3) | CNAG_04982.2 | No change |
| Cytosine permeases (FCY4) | CNAG_04276.2 | No change |
| Uracil phosphoribosyltransferase (FUR1) | CNAG_02337.2 | No change |
Spot sensitivity assays
5FC and 5FU resistance assays were carried out on minimal media (SD) supplemented with 2.5–10 μg/ml 5FC or 1–10 μg/ml 5FU, respectively. Cytosine assimilation assays were done on minimal media lacking a nitrogen source (without NH4SO4) and supplemented with 1–4 mM cytosine. Sterile-filtered stock solutions of 5FC (5.2 μg/ml), 5FU (5.0 μg/ml), and cytosine (40 mM) were prepared by dissolving the powder in ddH2O or DMSO (as the case for 5FU) and added to SD agar post-autoclave. Inocula from overnight cultures were used to start new cultures for 8–10 hours on the day of the experiment. Cultures were washed three times with 1X phosphate-buffered saline (PBS) and resuspended in 1X PBS. Serial 10-fold dilutions were prepared (101 cells/5 μl to 105 cells/5 μl) on 96-well plates, and 5 μl of each dilution was spotted on assay plates using a multichannel pipet.
Heterologous expression of C. gattii FCY1 in yeast
C. gattii FCY1 wild-type (CgFCY1wt) and FCY1 mutant (CgFCY1mut) complementary DNAs (cDNAs) were ordered from GenScript Inc., Piscataway, NJ, USA. The CgFCY1mut cDNA carried a point mutation at nucleotide position 86, resulting in the Arg29His substitution in the expressed protein. Both wild-type and mutant cDNAs were commercially synthesized and subcloned into the pUC57 plasmid backbone. To produce the yeast heterologous expression constructs expressing either CgFCY1wt or CgFCY1mut cDNA under the control of either a strong GDP promoter or a weak ADH promoter, pUC57 plasmids carrying CgFCY1 cDNAs were digested with BamHI and EcoRI, and the gel purified cDNAs were subcloned into the yeast shuttle expression plasmids p416GDP and p416ADH (Fig. S1).18 The resultant expression plasmids were then introduced into S. cerevisiae fcy1Δ via lithium acetate transformation using a standard method.19 Transformants were selected on uracil-deficient medium and tested for their ability to assimilate cytosine and grow on SD media in the presence of flucytosine.
Heterologous expression of C. gattii FCY1 in the KN99 fcy1Δ background
C. gattii FCY1 wild-type (CgFCY1wt) and FCY1 mutant (CgFCY1mut) cDNAs and C. neoformans FCY1 wild-type (CnFCY1wt) and FCY1 mutant (CnFCY1mut) cDNAs were subcloned into the Jmm180 Cryptococcus expression plasmid under the control of the constitutive actin promoter (Fig. S1). The resultant expression plasmids were then introduced into the KN99 FCY1 deletion strain (KN99 fcy1Δ) via biolistic transformation.20 Transformants were selected on uracil-deficient medium and tested on minimal media (SD) in the presence of flucytosine and assimilate cytosine as a sole nitrogen source.
Results
Mutations in the proteins that mediate the uptake or metabolism of flucytosine (5FC) are the basis for 5FC resistance.6,21 Primary resistance, the focus of this study, refers to the inherent resistance in the absence of prior drug exposure. We investigated primary resistance in C. gattii VGIII isolates to 5FC by identifying and analyzing genomic sequences of cytosine permease genes (FCY2, FCY3, FCY4 – 3 putative genes found in C. gattii), cytosine deaminase gene (FCY1), and uracil phosphoribosyltransferase (UPRTase, encoded by FUR1) in 11 clinical isolates (Table 2). These transporters and enzymes are the major components of the pyrimidine salvage pathway found in fungi and bacteria and are known to play essential roles in 5FC metabolism (Fig. 1). Susceptibility testing and sequence analysis of 11 C. gattii isolates revealed an association between a point mutation in the FCY1 gene and 5FC resistance in two clinical isolates (B9322 and JS5) where the substitution of guanine for adenine at nucleotide position 86 resulted in the exchange of arginine for histidine at the amino acid position 29 (Table 1). Comparative analysis of the open reading frames (ORF) and the 1 kb upstream and downstream sequences of the three putative cytosine permeases (FCY2-4) and UPRTase (FUR1) between the 5FC resistant and susceptible isolates indicated that these genes were either isogenic or displayed silent mutations (Table 2) (Fig. S2). Together, our data suggested that a single point mutation in FCY1 may have given rise to 5FC resistance in the isolates B9322 and JS5.
The enzyme structure of cytosine deaminase in S. cerevisiae has been resolved.22,23 The structure predicts that among the FCY1 active-site residues, a histidine and two cysteines (highlighted in yellow) coordinate zinc within the zinc-binding motif, while a glutamate residue serves as a proton shuttle (Fig. 2). The amino acid sequence alignment of FCY1 from S. cerevisiae and C. gattii revealed that these residues along with the amino acids that mediate substrate specificity (highlighted in green) are conserved in C. gattii. However, the Arg29His mutation found in the 5FC resistant isolates, B9322 and JS5, occurred outside of the active-site residues and within a string of amino acids that are not conserved (Fig. 2).
Figure 2.
Homology model of cytosine deaminase (FCY1) and ClustalW alignment of amino acid sequences from C. gattii FCY1 (CgFcy1) and S. cerevisiae FCY1 (ScFcy1). The amino acid residues highlighted in green and yellow represent the active site in Fcy1 based on the crystal structure of ScFCY1.22,23 The histidine and two cysteines (shaded in yellow) act as ligands in the zinc binding motif. Residues with a predicted role in substrate specificity are labeled in green color. The single amino acid residue that is mutated in 5FC resistant isolates (Table 1) is in bold-face and underlined.
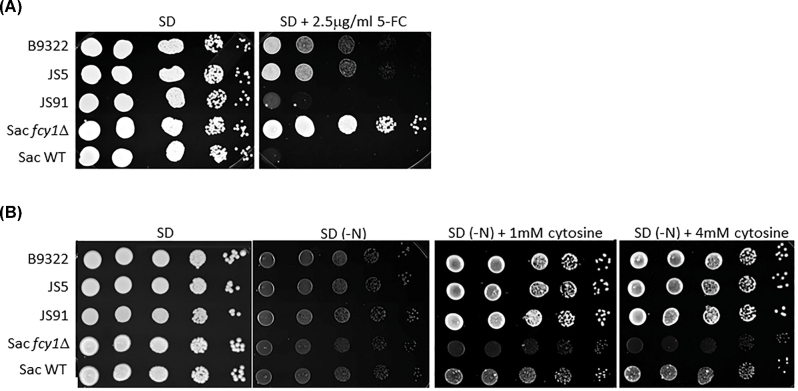
Consistent with the MIC data (5FC MIC >64 μg/ml, Table 1), the isolates B9322 and JS5 were able to grow on minimal media (SD) in the presence of 2.5 μg/ml 5FC (Fig. 3A). In contrast, the JS91 control strain, deemed susceptible according to its low MIC value (0.5 μg/ml, Table 1), was not viable (Fig. 3A). A few remaining colonies were observed, but they were likely the result of spontaneous 5FC resistance as previously reported.24 We also tested a strain of S. cerevisiae lacking the gene encoding cytosine deaminase (S. cerevisiae fcy1Δ) and showed that it was highly resistant to 5FC as predicted (Fig. 3A).
Figure 3.
Two C. gattii clinical isolates are resistant to flucytosine but retain the ability to assimilate cytosine. (A) Growth of 5FC resistant isolates (B9322 and JS5), 5FC susceptible isolate (JS91) and S. cerevisiae control strains (WT and fcy1Δ) on minimal media (SD) containing 2.5 μg/ml 5FC. Serial dilutions of cells (105 cells/5 μl to 101 cells/5 μl) were spotted on SD agar supplemented with 2.5 μg/ml 5FC. (B) B9322 and JS5 retain the ability to assimilate cytosine as a sole nitrogen source. Serial dilutions of cells (105 cells/5μl to 101 cells/5μl) were spotted on SD agar supplemented with 1–4 mM of cytosine as the sole nitrogen source. SD denotes synthetic dextrose minimal media. SD (-N) denotes synthetic dextrose media lacking a nitrogen source (NH4SO4).
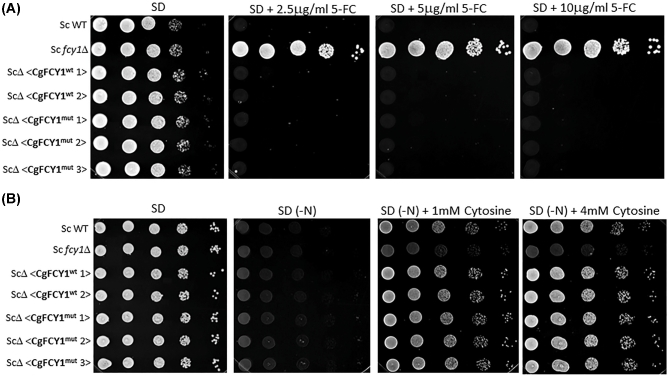
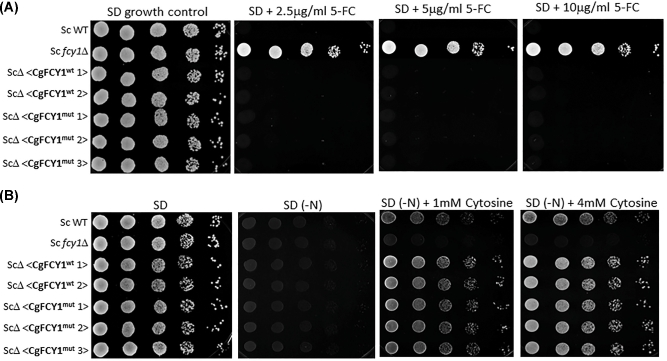
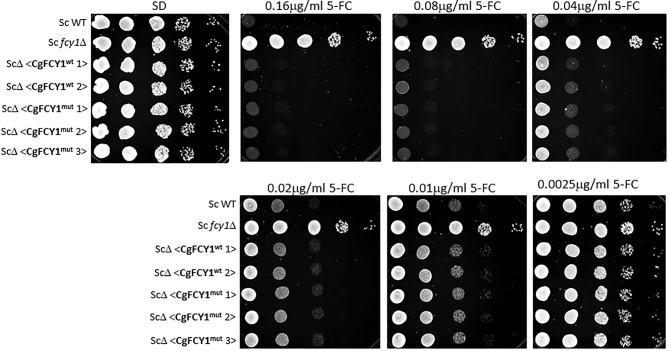
Since the deamination of cytosine to uracil results in the production of ammonia, the ability of fungi to assimilate cytosine as a sole nitrogen source has been used as a proxy method to quickly determine the presence of cytosine deaminase activity.21 Surprisingly, we found that despite the point mutation in the FCY1 gene and the observed 5FC resistance, both isolates, B9322 and JS5, grew robustly on media containing cytosine as the sole nitrogen source suggesting that the Arg29His substitution has little to no effect on cytosine deaminase activity (Fig. 3B). We suspected that the Arg29His substitution may alter the overall FCY1 structure in such a manner that the enzyme becomes defective at metabolizing 5FC (thus giving rise to 5FC resistance) while retaining its ability to assimilate cytosine. To test this idea, both wild-type and mutant FCY1 cDNAs from C. gattii were heterologously expressed in the S. cerevisiae fcy1Δ genetic background under the control of the strong GDP promoter, and the yeast transformants were tested for their susceptibility to flucytosine and their ability to assimilate cytosine (Fig. 4). Contrary to our expectation, S. cerevisiae fcy1Δ cells expressing the mutant cytosine deaminase (CgFCY1mut) were susceptible to flucytosine and grew on cytosine media to the same extent as those expressing the wildtype version (CgFCY1wt), suggesting that the Arg29His substitution has no effect on cytosine deaminase activity. While the heterologous expression data indicated that there was no causal link between the Arg29His substitution in FCY1 and the 5FC resistance observed in the isolates B9322 and JS5, we thought that expressing the mutant FCY1 protein (CgFCY1mut) under the control of a strong promoter might have masked any differences we observed between wild-type and mutant proteins. To rule out this possibility, CgFCY1wt and CgFCY1mut proteins were heterologously expressed in the S. cerevisiae fcy1Δ genetic background under the control of the weak ADH promoter (Fig. 5). The ADH promoter had previously been assayed to have activities that were 72 times less than the activities associated with the GDP promoter.18 Despite the low expression, S. cerevisiae fcy1Δ cells expressing the CgFCY1mut protein were susceptible to 5FC and able to assimilate cytosine to the same level as those expressing the wild-type version (CgFCY1wt) (Fig. 5). The similar 5FC susceptibility observed between transformants expressing either CgFCY1wt or CgFCY1mut protein held true even over a subtle range of 5FC concentrations (Fig. 6).
Figure 4.
Heterologous expression of C. gattii FCY1 wildtype (CgFCY1wt) and FCY1 mutant (CgFCY1mut) proteins under the control of the strong GDP promoter in the S. cerevisiae fcy1Δ genetic background (ScΔ). (A) Growth of S. cerevisiae wildtype, fcy1Δ, and heterologously expressed strains on minimal media (SD) in the presence of 2.5–10 μg/ml flucytosine (5FC). (B) Growth of S. cerevisiae strains on minimal media containing 1–4 mM cytosine as a sole nitrogen source. SD(-N) denotes synthetic dextrose agar lacking a nitrogen source.
Figure 5.
Heterologous expression of C. gattii FCY1 wildtype (CgFCY1wt) and FCY1 mutant (CgFCY1mut) proteins under the control of the weak ADH promoter in the S. cerevisiae fcy1Δ genetic background (ScΔ). (A) Growth of S. cerevisiae wildtype, S. cerevisiae fcy1Δ, and heterologously expressed strains on minimal media (SD) in the presence of 2.5–10 μg/ml flucytosine (5FC). (B) Growth of S. cerevisiae strains on minimal media containing 1–4 mM cytosine as a sole nitrogen source. SD(-N) denotes synthetic dextrose agar lacking a nitrogen source.
Figure 6.
Heterologous expression of C. gattii FCY1 wildtype (CgFCY1wt) and FCY1 mutant (CgFCY1mut) proteins under the control of the weak ADH promoter in the S. cerevisiae fcy1Δ genetic background (ScΔ). Growth of S. cerevisiae wild-type, S. cerevisiae fcy1Δ, and heterologously expressed strains on minimal media (SD) in the presence of 0.0025–0.16 μg/ml flucytosine (5FC).
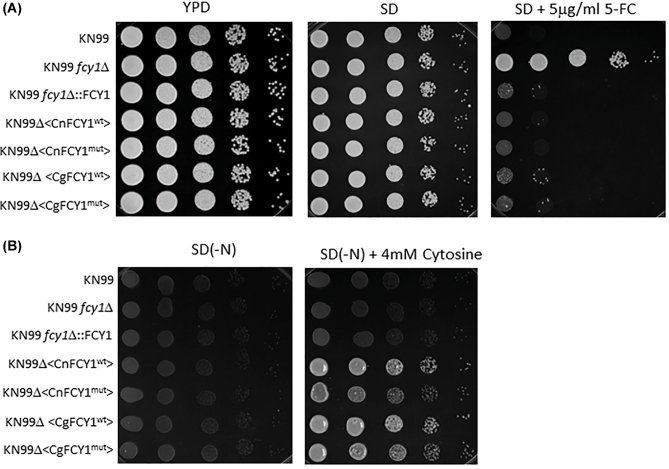
To further rule out the unlikely scenario that the CgFCY1mut protein may behave unpredictably when expressed in S. cerevisiae, both CgFCY1wt and CgFCY1mut proteins were expressed in the KN99 fcy1Δ genetic background and transformants were tested for flucytosine susceptibility and their ability to assimilate cytosine (Fig. 7). We decided to heterologously express the C. gattii FCY1 proteins in the KN99 fcy1Δ genetic background because the strain KN99 belongs to C. neoformans, a sibling species of C. gattii and the KN99 fcy1Δ null mutant had recently been made available to the scientific community.25 Similar to the results obtained from the S. cerevisiae heterologous system, no differences in flucytosine susceptibility and cytosine assimilation were observed between KN99 fcy1Δ transformants expressing either the CgFCY1wt or CgFCY1mut proteins (Fig. 7). In addition, the KN99 FCY1 mutant allele, CnFCY1mut, was also generated and expressed in the KN99 fcy1Δ genetic background. As expected, transformants expressing the mutant FCY1 protein from KN99 were found to restore flucytosine susceptibility and cytosine assimilation to wildtype level (Fig. 7).
Figure 7.
Expression of C. neoformans and C. gattii FCY1 wildtype and FCY1 mutant proteins under the control of the actin promoter in the KN99 fcy1Δ genetic background (KN99Δ). Growth of KN99 wildtype, KN99 fcy1Δ, and heterologously expressed strains on minimal media (SD) in the presence of 5 μg/ml flucytosine (5FC) or 4 mM cytosine as a sole nitrogen source. CgFCY1wt/CgFCY1mut and CnFCY1wt/CnFCY1mut denote FCY1 wild-type and mutant proteins from C. gattii and C. neoformans, respectively. SD(-N) denotes synthetic dextrose agar lacking a nitrogen source.
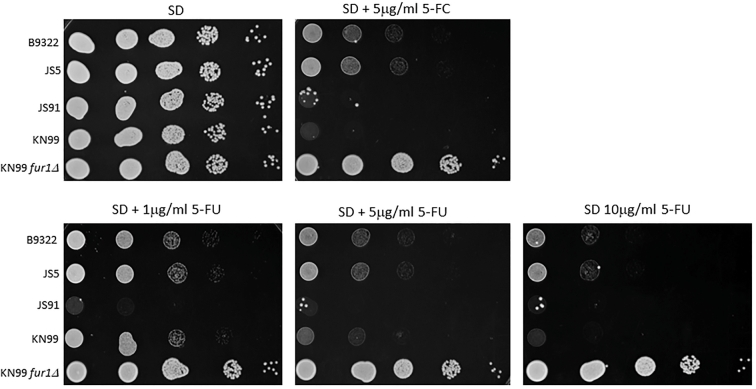
To assess whether the observed flucytosine resistance in B9322 and JS5 is due to pathway components downstream of FCY1, the isolates were tested for their ability to grow on 5-fluorouracil (5FU), a toxic metabolite that normally forms as a result of the deamination of 5FC (Fig. 1). Consistent with the contribution of FUR1 to flucytosine resistance, both isolates B9322 and JS5 were able to survive on 5FU over a wide concentration range (Fig. 8). Several colonies from the 5FC susceptible isolate, JS91, could be observed growing on media containing 5FU; however, these were likely resistant cells that spontaneously arise during prolonged drug exposure. Interestingly, sequencing analysis revealed that there were no amino acid changes in FUR1 among 5FC resistant and 5FC susceptible isolates (Fig. S2). In addition, analysis of 1 kb of the upstream and downstream sequences of the FUR1 gene did not show any genomic changes spanning the 2 kb genomic distance, except for the presence of several SNPs that are not unique to the B9322 and JS5 isolates (Fig. S3A,B).
Figure 8.
B9322 and JS5 are resistant to 5-fluorouracil (5FU). Growth of 5FC resistant isolates (B9322 and JS5), 5FC susceptible isolate (JS91) and KN99 control strains (KN99 WT and KN99 fur1Δ) on minimal media (SD) containing 5 μg/ml 5FC or 1–10 μg/ml 5FU. Serial dilutions of cells (105 cells/5 μl to 101 cells/5μl) were spotted on SD agar supplemented with either 5FC or 5FU.
Discussion
Flucytosine (5FC) remains an essential adjunct to amphotericin B during the treatment of severe cryptococcal infections.4 While the molecular mechanisms responsible for 5FC resistance have been well studied in Candida spp5,6,21,24,26,27 and S. cerevisiae28–31, little is known about the 5FC resistance mechanisms in Cryptococcus spp.8,10 5FC resistance has been previously described in C. neoformans and C. gattii VGIII isolates analyzed in epidemiological studies32,33; however, amino acid changes within key enzymes in the 5FC metabolic pathway have not yet been examined in these isolates. Primary 5FC resistance in C. neoformans has been reported to occur in 1–25% of isolates.5,34,35 Unfortunately, past reports examining the mechanism of resistance in these strains have been limited and to our knowledge an assessment of 5FC resistance mechanism in C. gattii has not been previously performed.
Genotyping of C. gattii at subspecies level identified four major molecular types (VGI/AFLP4, VGII/AFLP6, VGIII/AFLP5, and VGIV/AFLP7) among C. gattii isolates.36 The molecular type VGIII has received greater interest in recent years because it has been identified as the main cause of disease in otherwise healthy individuals.16 Based on multilocus sequencing typing (MLST) analysis, C. gattii VGIII is subdivided into VGIIIa and VGIIIb subgroups with the VGIIIa isolates showing more virulence.16 VGIIIa and VGIIIb have been shown to correspond to serotype B and serotype C, respectively.12 Nine of the C. gattii VGIII isolates used in this study belong to the VGIIIa subgroup (serotype B) and two isolates belong to the VGIIIb subgroup (serotype C) (Table 1). Based on the analysis of 55 isolates, C. gattii was recently proposed to be divided into five species.37 However, this new nomenclature remains very controversial, leading to the proposal of leading members of the Cryptococcus research community to apply the term “species complex” in combination with the major molecular type designation until all scientific aspects concerning the different major genetic groups within the C. gattii species complex are availble.38 As a result, the current study is referring to the isolates used herein as C. gattii VGIII.
In the herein presented work, we identified an association between 5FC resistance and the Arg29His mutation found within the cytosine deaminase gene (FCY1) of two C. gattii VGIII clinical isolates, B9322 and JS5. This association is consistent with the role of FCY1 as the enzyme responsible for the conversion of 5FC to 5FU (5-fluorouracil), a toxic metabolite that inhibits DNA and protein synthesis. Our finding is also in agreement with previous studies on 5FC resistance where deletion or mutations in the FCY1 gene (also known as FCA1 in Candida spp.) were found to contribute to 5FC resistance in both Candida spp.8,21,24,26 and S. cerevisiae.29 With the exception of the Arg29His mutation found in FCY1 in the isolates B9322 and JS5, no amino acid changes were observed in either cytosine permeases (FCY2-4) or FUR1, suggesting that the Arg29His mutation may be responsible for the observed 5FC resistance (Table 2) (data not shown).
To assess the impact of the Arg29His mutation on FCY1 activities, the isolates B9322 and JS5 were tested on media containing cytosine as the sole nitrogen source. Since ammonia is the byproduct produced from the deamination of cytosine to uracil, assessing the ability of C. gattii to assimilate cytosine would establish whether the Arg29His mutation is essential for FCY1 activity.16 Interestingly, both isolates B9322 and JS5 were found to grow robustly on cytosine media, suggesting the existence of active FCY1 in the resistant isolates (Fig. 3B). The finding that the isolates B9322 and JS5 can assimilate cytosine therefore nullified our hypothesis that the Arg29his mutation on FCY1 was responsible for their resistance to 5FC. An alternative interpretation of the cytosine assimilation data, however, could potentially rescue our hypothesis. We thought that the Arg29His mutation altered the overall FCY1 structure in such a manner that the enzyme was defective at metabolizing 5FC while retained its ability to assimilate cytosine. This notion is consistent with our finding that the Arg29His substitution occurs in a stretch of amino acids that is not conserved (Fig. 2). It is conceivable that a nonessential mutation reduces the affinity of cytosine deaminase to 5FC but not cytosine. To test this idea, we heterologously expressed the C. gattii FCY1 wild-type and mutant proteins (CgFCY1wt and CgFCY1mut) in both S. cerevisiae fcy1Δ and the C. neoformans KN99 fcy1Δ genetic backgrounds and assessed their resistance to 5FC and growth on cytosine media. Spot sensitivity data obtained from these heterologous expression studies unequivocally demonstrate that the Arg29His mutation has no effect on FCY1 activities towards either flucytosine or cytosine (Fig. 4–7). Thus, there is no causal link between the point mutation found in FCY1 and the observed 5FC resistance in the isolates B9322 and JS5.
Interestingly, the 5FC resistant isolates, B9322 and JS5, were viable on media containing 5-fluorouracil (5FU), a highly toxic metabolite that requires functional UPRTase (encoded by FUR1) for its antifungal activities. Analysis of FUR1 among 5FC resistant and 5FC susceptible isolates showed that no amino acid changes and no genomic alterations were observed within 1 kb of the upstream and downstream sequences of the FUR1 gene (Fig. S3A,B), suggesting that the observed 5FC resistance in C. gattii is likely due to changes in unknown protein(s) or pathway(s) that regulate FUR1 expression. Consistent with this notion, DNA microarray data suggests that FUR1 expression in S. cerevisiae is mediated by several positive regulators, including CAD1, CST6, and SSP1.8,39
While the data from this work did not yield any breakthrough in the understanding of the molecular mechanism responsible for 5FC resistance in two clinical isolates of C. gattii, they provide important insights into where we should direct our efforts in order to better understand 5FC resistance in this important human pathogen. The majority of work done in S. cerevisiae and other fungal pathogens, particularly Candida spp., have focused on the three key mediators of flucytosine resistance—namely, cytosine permease (FCY2), cytosine deaminase (FCY1), and UPRTase (FUR1).5,6,8,21,24,27,40 Mutations in either the coding sequence or the immediate upstream and downstream sequences flanking the FCY2, FCY1, and FUR1 genes have been shown to promote flucytosine resistance in the above studies. The herein presented data suggest that the 5FC resistance in the isolates B9322 and JS5 is not mediated by changes in either the genes’ coding or regulatory sequences but is likely be due to unknown factors or pathways that modulate flucytosine metabolism. Consistent with the notion that 5FC resistance can be mediated by changes in proteins that are not part of the pyrimidine salvage pathways is the deletion of the transcription factor MBS1 for which it has been shown that it confers increased susceptibility to 5FC in C. neoformans.10 A recent genome-wide screen for genes that promote flucytosine resistance in S. cerevisiae identified over 180 genes, particularly those involved in DNA repair, RNA and protein metabolism.41 These aforementioned studies and the herein presented work further underline the need to examine 5FC resistance mechanisms globally in order to arrive at a better understanding of 5FC resistance in fungal pathogens.
Supplementary Material
Acknowledgments
We are especially grateful to Joseph Heitman (Duke University) for providing the S. cerevisiae WT and fcy1Δ deletion strains and to the National Institute of Allergy and Infectious Diseases (NIAID) for funding.
The findings and conclusions in this report are those of the authors and do not necessarily represent the official position of the Centers for Disease Control and Prevention.
Contributor Information
Kiem Vu, Department of Pharmacology, University of California, Davis, California, USA.
George R Thompson, III, Department of Medical Microbiology and Immunology, University of California, Davis, California, USA; Department of Internal Medicine, Division of Infectious Diseases, University of California Davis Medical Center, Davis, California, USA.
Chandler C Roe, Translational Genomics Research Institute, Flagstaff, Arizona, USA.
Jane E Sykes, Department of Medicine and Epidemiology, School of Veterinary Medicine, University of California, Davis, California, USA.
Elizabeth M Dreibe, Translational Genomics Research Institute, Flagstaff, Arizona, USA.
Shawn R Lockhart, Mycotic Diseases Branch, Centers for Disease Control and Prevention, Atlanta, Georgia USA.
Wieland Meyer, Molecular Mycology Research Laboratory, Center for Infectious Diseases and Microbiology, Marie Bashir Institute for Emerging Infectious Diseases and Biosecurity, Westmead Clinical School, Sydney Medical School, Westmead Hospital, The University of Sydney, Westmead Institute for Medical Research, Sydney, Australia.
David M Engelthaler, Translational Genomics Research Institute, Flagstaff, Arizona, USA.
Angie Gelli, Department of Pharmacology, University of California, Davis, California, USA.
Supplementary material
Supplementary data are available at MMYCOL online.
Declaration of interest
The authors report no conflicts of interest. The authors alone are responsible for the content and the writing of the paper.
References
- 1. Datta K, Bartlett KH, Baer Ret al. Spread of Cryptococcus gattii into Pacific Northwest region of the United States. Emerg Infect Dis. 2009; 15: 1185–1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Coelho C, Casadevall A. Cryptococcal therapies and drug targets: the old, the new and the promising. Cell Microbiol. 2016; 18: 792–799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Perfect JR, Dismukes WE, Dromer Fet al. Clinical practice guidelines for the management of cryptococcal disease: 2010 update by the Infectious Diseases Society of America. Clin Infect Dis. 2010; 50: 291–322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Day JN, Chau TT, Wolbers Met al. Combination Antifungal Therapy for Cryptococcal Meningitis. N Engl J Med. 2013; 368: 1291–1302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Cuenca-Estrella M, Diaz-Guerra TM, Mellado E, Rodriguez-Tudela JL. Flucytosine primary resistance in Candida species and Cryptococcus neoformans. Eur J Clin Microbiol Infect Dis. 2001; 20: 276–279. [DOI] [PubMed] [Google Scholar]
- 6. Hope WW, Tabernero L, Denning DW, Anderson MJ. Molecular mechanisms of primary resistance to flucytosine in Candida albicans. Antimicrob Agents Chemother. 2004; 48: 4377–4386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Rex JH, Walsh TJ, Sobel JDet al. Practice guidelines for the treatment of candidiasis. Infectious Diseases Society of America. Clin Infect Dis. 2000; 30: 662–678. [DOI] [PubMed] [Google Scholar]
- 8. Whelan WL. The genetic basis of resistance to 5-fluorocytosine in Candida species and Cryptococcus neoformans. Crit Rev Microbiol. 1987; 15: 45–56. [DOI] [PubMed] [Google Scholar]
- 9. Block ER, Jennings AE, Bennett JE. 5-fluorocytosine resistance in Cryptococcus neoformans. Antimicrob Agents Chemother. 1973; 3: 649–656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Song MH, Lee JW, Kim MSet al. A flucytosine-responsive Mbp1/Swi4-like protein, Mbs1, plays pleiotropic roles in antifungal drug resistance, stress response, and virulence of Cryptococcus neoformans. Eukaryotic cell. 2012; 11: 53–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Engelthaler DM, Hicks ND, Gillece JDet al. Cryptococcus gattii in North American Pacific Northwest: whole-population genome analysis provides insights into species evolution and dispersal. MBio. 2014; 5: e01464–014714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Firacative C, Roe CC, Malik Ret al. MLST and whole-genome-based population analysis of Cryptococcus gattii VGIII links clinical, veterinary and environmental strains, and reveals divergent serotype specific sub-populations and distant ancestors. PLoS Negl Trop Dis. 2016; 10: e0004861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Lockhart SR, Iqbal N, Harris JRet al. Cryptococcus gattii in the United States: genotypic diversity of human and veterinary isolates. PLoS One. 2013; 8: e74737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Singer L.M., Meyer W., Firacative C.et al. Antifungal drug susceptibility and phylogenetic diversity among Cryptococcus isolates from dogs and cats in North America. J Clin Microbiol. 2014; 52: 2061–2070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Iqbal N, DeBess EE, Wohrle Ret al. Correlation of genotype and in vitro susceptibilities of Cryptococcus gattii strains from the Pacific Northwest of the United States. J Clin Microbiol. 2010; 48: 539–544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Byrnes EJ, 3rd Li W, Ren Pet al. A diverse population of Cryptococcus gattii molecular type VGIII in southern Californian HIV/AIDS patients. PLoS Pathog. 2011; 7: e1002205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. CLSI . Reference method for broth dilution antifungal susceptibility testing of yeasts; approved standard—third edition. CLSI document M27-A3. Clinical and Laboratory Standards Institute, Wayne, PA. 2008. [Google Scholar]
- 18. Mumberg D, Muller R, Funk M. Yeast vectors for the controlled expression of heterologous proteins in different genetic backgrounds. Gene. 1995; 156: 119–122. [DOI] [PubMed] [Google Scholar]
- 19. Goetz RD, Woods RA. Yeast transformation by the LiAc/SS carrier DNA/PEG method. Methods Mol Biol. 2006; 313: 107–120. [DOI] [PubMed] [Google Scholar]
- 20. Davidson RC, Cruz MC, Sia RAet al. Gene disruption by biolistic transformation in serotype D strains of Cryptococcus neoformans. Fungal Genet Biol. 2000; 29: 38–48. [DOI] [PubMed] [Google Scholar]
- 21. Edlind TD, Katiyar SK. Mutational analysis of flucytosine resistance in Candida glabrata. Antimicrob Agents Chemother. 2010; 54: 4733–4738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Ireton GC, Black ME, Stoddard BL. The 1.14 A crystal structure of yeast cytosine deaminase: evolution of nucleotide salvage enzymes and implications for genetic chemotherapy. Structure. 2003; 11: 961–972. [DOI] [PubMed] [Google Scholar]
- 23. Ko TP, Lin JJ, Hu CYet al. Crystal structure of yeast cytosine deaminase. Insights into enzyme mechanism and evolution. J Biol Chem. 2003; 278: 19111–19117. [DOI] [PubMed] [Google Scholar]
- 24. Florent M, Noel T, Ruprich-Robert Get al. Nonsense and missense mutations in FCY2 and FCY1 genes are responsible for flucytosine resistance and flucytosine-fluconazole cross-resistance in clinical isolates of Candida lusitaniae. Antimicrob Agents Chemother. 2009; 53: 2982–2990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Lee KT, So YS, Yang DHet al. Systematic functional analysis of kinases in the fungal pathogen Cryptococcus neoformans. Nat Commun. 2016; 7: 12766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Papon N, Noel T, Florent Met al. Molecular mechanism of flucytosine resistance in Candida lusitaniae: contribution of the FCY2, FCY1, and FUR1 genes to 5-fluorouracil and fluconazole cross-resistance. Antimicrob Agents Chemother. 2007; 51: 369–371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. McManus BA, Moran GP, Higgins JA, Sullivan DJ, Coleman DC. A Ser29Leu substitution in the cytosine deaminase Fca1p is responsible for clade-specific flucytosine resistance in Candida dubliniensis. Antimicrob Agents Chemother. 2009; 53: 4678–4685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Paluszynski JP, Klassen R, Rohe M, Meinhardt F. Various cytosine/adenine permease homologues are involved in the toxicity of 5-fluorocytosine in Saccharomyces cerevisiae. Yeast. 2006; 23: 707–715. [DOI] [PubMed] [Google Scholar]
- 29. Erbs P, Exinger F, Jund R. Characterization of the Saccharomyces cerevisiae FCY1 gene encoding cytosine deaminase and its homologue FCA1 of Candida albicans. Current Genetics. 1997; 31: 1–6. [DOI] [PubMed] [Google Scholar]
- 30. Weber E, Rodriguez C, Chevallier MR, Jund R. The purine-cytosine permease gene of Saccharomyces cerevisiae: primary structure and deduced protein sequence of the FCY2 gene product. Mol Microbiol. 1990; 4: 585–596. [DOI] [PubMed] [Google Scholar]
- 31. Kern L, de Montigny J, Jund R, Lacroute F. The FUR1 gene of Saccharomyces cerevisiae: cloning, structure and expression of wild-type and mutant alleles. Gene. 1990; 88: 149–157. [DOI] [PubMed] [Google Scholar]
- 32. Pfaller MA, Messer SA, Boyken Let al. Global trends in the antifungal susceptibility of Cryptococcus neoformans (1990 to 2004). J Clin Microbiol. 2005; 43: 2163–2167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Hagen F., Illnait-Zaragozi M-T., Bartlett K. H.et al. In vitro antifungal susceptibilities and amplified fragment length polymorphism genotyping of a worldwide collection of 350 clinical, veterinary, and environmental Cryptococcus gattii isolates. Antimicrob. Agents Chemother. 2010; 54: 5139–5149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Espinel-Ingroff A, Aller AI, Canton Eet al. Cryptococcus neoformans-Cryptococcus gattii species complex: an international study of wild-type susceptibility endpoint distributions and epidemiological cutoff values for fluconazole, itraconazole, posaconazole, and voriconazole. Antimicrob Agents Chemother. 2012; 56: 5898–5906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Govender NP, Patel J, van Wyk Met al. Trends in antifungal drug susceptibility of Cryptococcus neoformans isolates obtained through population-based surveillance in South Africa in 2002–2003 and 2007–2008. Antimicrob Agents Chemother. 2011; 55: 2606–2611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Cogliati M. Global molecular epidemiology of Cryptococcus neoformans and Cryptococcus gattii: an atlas of the molecular types. Scientifica (Cairo). 2013; 675213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Hagen F, Khayhan K, Theelen Bet al. Recognition of seven species in the Cryptococcus gattii/Cryptococcus neoformans species complex. Fungal Genet Biol. 2015; 78: 16–48. [DOI] [PubMed] [Google Scholar]
- 38. Kwon-Chung KJ, Bennett JE, Wickes BLet al. The case for adopting the “species complex” nomenclature for the etiologic agents of cryptococcosis. mSphere. 2017; 2: e00357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Cohen BA, Pilpel Y, Mitra RD, Church GM. Discrimination between paralogs using microarray analysis: application to the Yap1p and Yap2p transcriptional networks. Mol Biol Cell. 2002; 13: 1608–1614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Chen YN, Lo HJ, Wu CCet al. Loss of heterozygosity of FCY2 leading to the development of flucytosine resistance in Candida tropicalis. Antimicrob Agents Chemother. 2011; 55: 2506–2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Costa C, Ponte A, Pais Pet al. New mechanisms of flucytosine resistance in C. glabrata unveiled by a Chemogenomics analysis in S. cerevisiae. PloS one. 2015; 10: e0135110. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.