Abstract
Reorganization of the actin cytoskeleton is an early cellular response to a variety of extracellular signals. Dissection of pathways leading to actin rearrangement has focused largely on those initiated by growth factor receptors or integrins, although stimulation of G protein-coupled receptors also leads to cytoskeletal changes. In transfected Cos-7SH cells, activation of the chemoattractant formyl peptide receptor induces cortical actin polymerization and a decrease in the number of central actin bundles. In this report, we show that cytoskeletal reorganization can be transduced by G protein βγ heterodimers (Gβγ), phosphoinositide 3-kinase γ (PI3-Kγ), a guanosine exchange factor (GEF) for Rac, and Rac. Expression of inactive variants of either PI3-Kγ, the Rac GEF Vav, or Rac blocked the actin rearrangement. Neither wortmannin nor LY294002, pharmacologic inhibitors of PI3-K, could inhibit the actin rearrangement induced by a constitutively active Rac. The inhibition of cytoskeletal reorganization by the dominant negative Vav variants could be rescued by coexpression of a constitutively active form of Rac. In contrast, a Vav variant with its pleckstrin homology (PH) domain missing constitutively induced JNK activation and led to cytoskeletal reorganization, even without stimulation by PI3-Kγ. This suggests that the PH domain of Vav controls the guanosine exchange activity of Vav, perhaps by a mechanism regulated by D3 phosphoinositides generated by PI3-K. Taken together, these findings delineate a pathway leading from activation of a G protein-coupled receptor to actin reorganization which sequentially involves Gβγ, PI3-Kγ, a Rac GEF, and Rac.
Multiple signal transduction pathways converge to induce rearrangements of the actin cytoskeleton in order to mediate motility, shape change, and attachment to substrate. These pathways are initiated by a variety of extracellular stimuli which activate transmembrane receptors of different classes. Most studies have focused on pathways which are triggered by activation of growth factor receptors. These pathways are postulated to involve a variety of signaling molecules, including small GTP binding proteins, protein kinases, and lipid kinases.
Ras was the first small GTP binding protein to be implicated in reorganization of the actin cytoskeleton (2). Microinjection of Ras proteins was shown to induce cytoskeletal changes leading to the formation of ruffles in quiescent fibroblasts. Later, small GTP binding proteins of the Rho family were demonstrated to be critical in regulating specialized actin cytoskeletal structures (9). Microinjection of active forms of Cdc42 leads to the formation of filopodia, active Rac variants induce lamellipodia and membrane ruffles, and active forms of Rho cause stress fiber and focal contact assembly (25). The formation of these actin structures by extracellular stimuli can be blocked by dominant negative variants of the corresponding Rho family members. Further, at least in Swiss 3T3 fibroblasts, there is a hierarchical order to these proteins, with Cdc42 able to activate Rac, which, in turn, can activate Rho.
Phosphoinositide 3-kinases (PI3-K enzymes) are enzymes which phosphorylate phosphatidylinositide lipids at the D3 position of the inositol ring, leading to the formation of lipid second messengers which are critical in the transduction of a variety of signals. PI3-K enzymes are known to be involved in the regulation of actin polymerization. The cortical actin assembly initiated by platelet-derived growth factor or insulin requires PI3-Kα/β isoforms (12, 18, 32). G protein-coupled receptors also induce actin rearrangement; however, no published reports specifically address the role of PI3-Kγ, the isoform of PI3-K activated by G protein βγ heterodimers (Gβγ) (28). Like the growth factor-activated PI3-K enzymes (p85/p110α/β), this G protein-activated PI3-K also exists as a heterodimer composed of a catalytic subunit, p110γ, and an adapter subunit, p101 (29, 30).
Since relatively little is known about the signaling pathways leading from activation of G protein-coupled receptors to cytoskeletal reorganization, we studied the events occurring after stimulation of the chemoattractant formyl peptide receptor (fPR), a seven-transmembrane-domain receptor coupled to a pertussis toxin-sensitive, heterotrimeric G protein. In this report, we show that stimulation of the fPR leads to a loss of central F-actin cables and reappearance of the F-actin in a cortical pattern in transfected Cos-7SH cells. We demonstrate that the fPR-stimulated cytoskeletal change is pertussis toxin sensitive, implying that it is mediated by the release of Gβγ. Signaling downstream of the fPR involves the activation of PI3-Kγ. This pathway is dependent on Rac-1 but is independent of Cdc42. It also requires a guanosine exchange factor (GEF) for Rac, such as Vav, situated between PI3-Kγ and Rac in this signaling pathway. Lastly, we show that a variant of the Rac GEF, Vav, missing its pleckstrin homology (PH) domain, is constitutively active in leading to Rac activation as measured biochemically by JNK activation and assayed functionally by the ability to induce cytoskeletal changes in the absence of other upstream stimulatory signals. Taken together, these data delineate a signaling pathway beginning with activation of a G protein-coupled receptor and culminating in cytoskeletal rearrangement that sequentially involves PI3-Kγ, a Rac GEF, and Rac. Moreover, the data suggest a mechanism by which the PH domain of a GEF may regulate its exchange activity.
MATERIALS AND METHODS
Mammalian expression vectors.
The cDNA clone of p110γ was obtained by reverse transcription-PCR from HL-60 mRNA, and several individual isolates were fully sequenced. Any region with ambiguity or discrepancy with the published clone was sequenced in both directions. The sequences of all of the full-length clones agreed with each other but disagreed with the sequence currently in the data bank at the following points: (i) an additional 3-base, in-frame insertion corresponding to an alanine at nucleotides 408 to 410 and (ii) a missense mutation at amino acid 459 corresponding to an Arg-to-Gln substitution. All of these sequence discrepancies were found in multiple independent isolates. All p110γ variants were engineered to contain an additional 9-amino-acid hemagglutinin (HA) epitope tag, YPYDVPDYA, recognized by the monoclonal antibody 12CA5 (16). The plasmid that directs the expression of an EE epitope (EEEEYMPME)-tagged variant of p101 was a gift from Len Stephens (Babraham Institute, London, United Kingdom). Plasmids that direct the synthesis of Gβ1 and Gγ2 were a generous gift from Janet Robishaw (Geisinger Institute, Danville, Pa.). pcDNA3-1 oncogenic vav (a gift from Linda Van Aelst, Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.), and pCMV oncogenic dbl were created by cloning the BamHI fragment from pc11dbl (a gift from Alessandra Eva, Cornell University, Ithaca, N.Y.) into pCMV6. The generation of pcDNA3Jnk1, which encodes FLAG-tagged Jnk1, has been described previously (1). The catalytically inactive variants of p110γ, Δ948–981 p110γ-HA (His948 to Arg981) and K833R p110γ-HA, and the constitutively active variant, myr-p110γ-HA (which contains a fusion of the myristoylation sequence MGQSLT of the Rasheed sarcoma virus to the alternative start site of p110γ), were generated by PCR mutagenesis by the techniques of Landt et al. (19) and Ho et al. (13). Plasmids directing the expression of the Vav variants Δ342–348 Vav, L213Q Vav, Y174F Vav, and Δ407–510 Vav (Vav ΔPH) were generated by PCR mutagenesis and were also engineered to contain the HA epitope at their carboxyl terminus. Plasmids encoding myc-tagged V12N17 Rac and myc-tagged N17 Cdc42 were a gift from Alan Hall (University College, London, United Kingdom). The plasmid directing the synthesis of myc-V12 Rac was a gift from Judy Meinkoth (Department of Pharmacology, University of Pennsylvania, Philadelphia, Pa.). The plasmid encoding the D7 variant of Lsc was a gift from Ian Whitehead (University of North Carolina, Chapel Hill, N.C.). The plasmid directing the expression of Δ568–574 Lsc-HA was generated by PCR mutagenesis. All p110γ, Vav, and Lsc mutants were cloned into pCMV5, and all sequences were fully confirmed.
Cell culture, immunoblotting, and indirect immunofluorescence.
Cos-7SH or Cos-7 cells grown in Dulbecco’s minimal essential medium (Gibco BRL, Gaithersburg, Md.) with 10% fetal bovine serum (HyClone, Logan, Utah) and 1% penicillin-streptomycin (Gibco BRL) were transiently transfected by calcium phosphate-DNA coprecipitation. For immunoblotting, the cells were lysed in boiling 1% sodium dodecyl (SDS), normalized for their protein concentration, fractionated by SDS-polyacrylamide gel electrophoresis (PAGE), and immunoblotted with anti-HA (HA.11) or anti-myc (9E10) (Babco, Berkeley, Calif.). Fixation, permeabilization, staining, and photography were done as previously described (22). Rhodamine-labeled phalloidin (Molecular Probes, Eugene, Oreg.) was used to stain the cells for the presence of F-actin, as specified by the manufacturer’s protocol.
JNK assay.
Cos-7 cells in 60-mm plates were transfected with a total of 2 to 5 μg of DNA by using Lipofectamine reagent (Life Technologies, Gaithersburg, Md.). The cells were lysed in 0.5 ml of JLB (25 mM HEPES [pH 7.5], 150 mM NaCl, 1% Nonidet P-40, 0.25% sodium deoxycholate, 1 mM EDTA, 25 mM NaF, 10 mM β-glycerol phosphate, 1 mM sodium vanadate, 10 μg of leupeptin per ml, 10 μg of aprotinin per ml) per plate, clarified by centrifugation, and then precleared with ∼20 μl of protein G-Sepharose beads (Pharmacia Biotech, Piscataway, N.J.). The lysates were incubated for 2 h on ice with 2 to 3 μg of M2 anti-FLAG monoclonal antibody (Eastman Kodak, New Haven, Conn.). Immune complexes were collected on 20 μl of protein G-Sepharose beads for 1 to 2 h. The beads were washed three times with JLB and once with JKB (40 mM HEPES [pH 7.5], 10 mM MgCl2, 1 mM dithiothreitol) and resuspended in 60 μl of JKB. Then 30 μl of beads was assayed for phosphorylation of glutathione S-transferase–Jun (amino acids 1 to 79) in a 40-μl final volume with 50 μM ATP and 5 to 10 μCi of [32P]ATP. The reaction was stopped by the addition of SDS-PAGE loading buffer, and samples were then subjected to SDS-PAGE (10% polyacrylamide) and stained with Coomassie blue. FLAG-JNK expression levels were checked by anti-FLAG Western blotting of immune complexes and examination of the stained kinase gel (JNK visible).
RESULTS
Effect of the chemoattractant fPR and PI3-Kγ on the actin cytoskeleton.
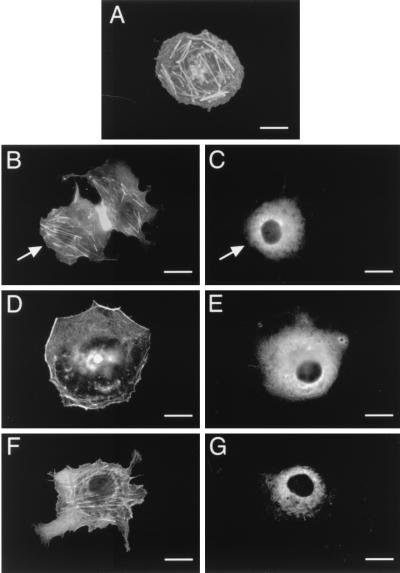
We wished to determine whether the actin reorganization initiated by stimulation of a G protein-coupled receptor is dependent on PI3-Kγ. To do this, we transiently expressed PI3-Kγ (p110γ-HA and p101) with the fPR in Cos-7SH cells (which express neither PI3-Kγ nor fPR endogenously) and analyzed the transfectants for actin rearrangement before and after stimulation with the agonist peptide formylmethionine-leucine-phenylalanine (fMLP). Cells which were transfected with empty pCMV5 vector had thick central actin fibers and no peripheral actin staining (Fig. 1A). Cells expressing PI3-Kγ (p110γ-HA and p101) and the fPR appeared the same as mock-transfected cells prior to fMLP stimulation (Fig. 1B). However, after a 30-min stimulation with 300 nM fMLP, these cells showed a dramatic cortical actin polymerization and a decrease in the number of central actin bundles (Fig. 1D). The fPR-stimulated, PI3-Kγ-mediated actin rearrangements were blocked by overnight pretreatment with 100 μM pertussis toxin (Fig. 1F) and could be mimicked by coexpression of PI3-Kγ with Gβγ subunits (Fig. 2A), but not Gαi subunits (Fig. 2C). This is consistent with a model in which the stimulated fPR initiates Gβγ release from Gαi, thus activating PI3-Kγ and triggering cytoskeletal changes.
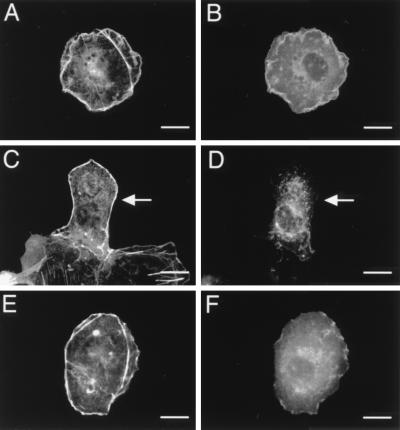
FIG. 1.
Stimulation of the fPR, acting via PI3-Kγ, leads to reorganization of the actin cytoskeleton. Cos-7SH cells transiently expressing either empty vector (pCMV5) or the chemoattractant fPR with PI3-Kγ (p110γ-HA and p101) were fixed and stained with monoclonal anti-HA antibody 12CA5, followed by fluorescein isothiocyanate (FITC)-conjugated goat anti-mouse and rhodamine-labeled phalloidin. Transfected cells were identified by the presence of FITC staining (C, E, and G), and their pattern of F-actin polymerization was assessed by phalloidin staining (A, B, D, and F). (A) Mock-transfected cells. (B and C) Cells expressing fPR and PI3-Kγ in the resting state. (D and E) Cells expressing fPR and PI3-Kγ after a 30-min stimulation with 300 nM fMLP. (F and G) Cells expressing fPR and PI3-Kγ after overnight exposure to 100 μM pertussis toxin followed by a 30-min stimulation with 300 nM fMLP. Cells shown are representative of cells from at least three experiments. Bars, 25 μm. When more than one cell is shown in a field, transfected cells are indicated by arrows.
FIG. 2.
Overexpression of Gβγ subunits, but not Gαi subunits, stimulates PI3-Kγ to reorganize the actin cytoskeleton. Cos-7SH cells transiently expressing PI3-Kγ (p110γ-HA and p101) along with the Gβ1 and Gγ2 (A and B) or Gαi (C and D) subunits of heterotrimeric G proteins were fixed and stained as described in the legend to Fig. 1. Transfected cells were identified by the presence of FITC staining (B and D), and their pattern of F-actin polymerization was assessed by phalloidin staining (A and C). (A and B) Cells expressing Gβγ and PI3-Kγ. (C and D) Cells expressing Gαi and PI3-Kγ. Cells shown are representative of cells from at least three experiments. Bars, 25 μm. The bottom panel is a representative anti-HA Western blot showing equivalent levels of p110γ-HA expression. Lanes: 1, Gβγ and PI3-Kγ; 2, Gαi and PI3-Kγ.
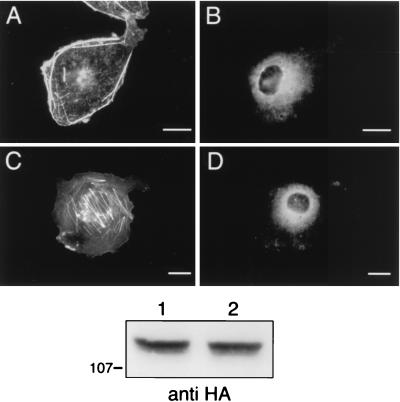
Treatment of cells with 100 nM wortmannin, a pharmacologic inhibitor of PI3-K, blocked the Gβγ- and PI3-Kγ-mediated effects on the actin cytoskeleton, suggesting that these effects required D3 phosphoinositide production (Fig. 3A). LY294002 (50 μM), another PI3-K inhibitor, also blocked the actin rearrangements (Fig. 3C). As further evidence for the second-messenger requirement, we generated two catalytically inactive p110γ variants. Based on a report showing that deletion of the putative ATP binding site within p110α destroyed lipid kinase activity (14), we generated a plasmid which encodes an HA-tagged p110γ variant with the analogous mutation (Δ948–981 p110γ-HA) and found that the expressed protein also fails to phosphorylate phosphatidylinositol to form phosphatidylinositol 3-phosphate (22a). A second HA-tagged p110γ variant, K833R p110γ-HA, was recently shown to also lack lipid kinase activity (21). Both of these catalytically inactive p110γ variants were incapable of stimulating actin reorganization when expressed with p101 and Gβγ, again supporting the need for D3 phosphoinositide production to carry signals from PI3-Kγ to the cytoskeleton (Fig. 3E and G). Although the expression of the catalytically inactive mutants was variable (Fig. 3, bottom panel), both Δ948–981 p110γ-HA and K833R p110γ-HA consistently failed to induce cortical actin reorganization.
FIG. 3.
Inhibition of PI3-Kγ activity blocks the Gβγ-mediated actin rearrangements. Cos-7SH cells transfected with PI3-Kγ (p110γ-HA and p101) along with Gβ1 and Gγ2 were split and then treated overnight with either 100 nM wortmannin (A and B), 50 μM LY294002 (C and D), or dimethyl sulfoxide (DMSO) diluent alone as a control (not shown). Cos-7SH cells transiently expressing p101 and Gβ1/Gγ2 along with HA-tagged, catalytically inactive p110γ variants Δ948–981 p110γ (E and F) and K833R p110γ (originally defined as K799R) (G and H) were fixed and stained as described in the legend to Fig. 1. Transfected cells were identified by the presence of FITC staining (B, D, F, and H), and their pattern of F-actin polymerization was assessed by phalloidin staining (A, C, E, and G). Cells shown are representative of cells from at least three experiments. Bars, 25 μm. When more than one cell is shown in a field, transfected cells are indicated by arrows. The bottom panel is an anti-HA Western blot showing expression of HA-tagged p110γ variants. Lanes: 1, Gβγ, p101 and wild-type p110γ; 2, Gβγ, p101 and Δ948–981 p110γ; 3, Gβγ, p101 and K833R p110γ.
A p110γ variant with a myristoyl group at its N terminus has been shown to be constitutively active biochemically (21). When expressed in Cos-7SH cells, some of the myr-p110γ-HA was membrane associated (Fig. 4B) and caused cytoskeletal reorganization, even in the absence of p101 and Gβγ (Fig. 4A). This suggests a potential role for Gβγ in recruiting p110γ to the membrane and further argues that this membrane localization is sufficient to lead to cytoskeletal changes.
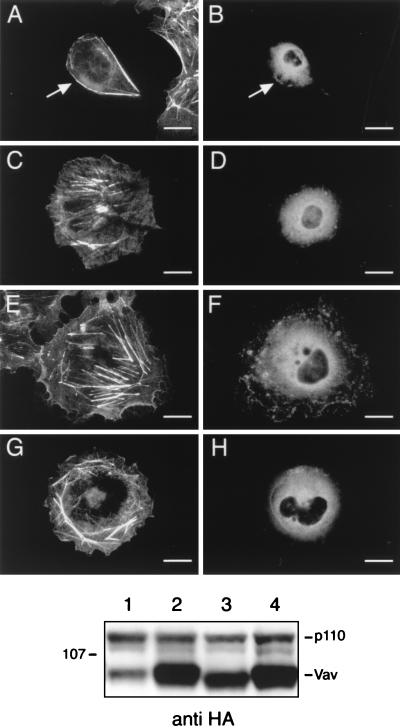
FIG. 4.
Coexpression of a dominant negative Rac, but not dominant negative Cdc42, blocks the myr-p110γ-HA-induced cytoskeletal reorganization. Cos-7SH cells transiently expressing a constitutively active, HA-tagged p110γ variant myristoylated at its N terminus (myr-p110γ-HA) alone (A and B) or along with dominant negative variants of either Rac (myc-V12N17 Rac) (C and D) or Cdc42 (myc-N17 Cdc42) (E and F) or cells transfected with fPR, PI3-Kγ, and the dominant negative Rac followed by a 30 min stimulation with 300 nM fMLP (G and H) were fixed and stained as described in the legend for Fig. 1. Transfected cells were identified by the presence of FITC staining (B, D, F, and H), and their pattern of F-actin polymerization was assessed by phalloidin staining (A, C, E, and G). Cells shown are representative of cells from at least five experiments. Bars, 25 μm. When more than one cell is shown in a field, transfected cells are indicated by arrows. The middle panel is an anti-HA Western blot showing equivalent levels of myr-p110γ or wild-type p110γ expression. Lanes: 1, myr-p110γ-HA; 2, myr-p110γ-HA and myc-V12N17 Rac; 3, myr-p110γ-HA and myc-N17 Cdc42; 4, fPR, PI3-Kγ (p110γ-HA and p101), and myc-V12N17 Rac. The bottom panel is an anti-myc Western blot showing approximately equivalent levels of expression of myc-V12N17 Rac and mycN17 Cdc42. The lanes are as described for the anti-HA blot in the middle panel.
fPR and PI3-Kγ-mediated cell ruffling requires Rac1 but not Cdc42.
Small GTP binding proteins of the Rho family have been demonstrated to play a critical role in the transduction of extracellular signals to the cytoskeleton (9). Several investigators have shown that Rac, a member of this family of GTPases, acting downstream of PI3-Kα/β, is required for the formation of actin ruffles seen in response to growth factors or insulin (12, 18). We wished to determine if Rac was similarly involved in the mediation of PI3-Kγ-dependent effects on the cytoskeleton. We therefore coexpressed a myc epitope-tagged, dominant negative variant of Rac (myc-V12N17 Rac) with myr-p110γ and found that it completely inhibited the ability of myr-p110γ to stimulate actin reorganization (Fig. 4C). By contrast, a myc-tagged, dominant negative variant of Cdc42 (myc-N17 Cdc42) had minimal impact on the myr-p110γ-initiated actin effects (Fig. 4E), thus implying that the pathway between PI3-Kγ and the actin cytoskeleton is dependent on Rac but not Cdc42. The actin changes induced by fMLP stimulation of the fPR were also blocked by the dominant negative Rac variant (Fig. 4G). Anti-HA Western blotting demonstrated approximately equal levels of myr-p110γ-HA and p110γ-HA (Fig. 4, middle panel). Anti-myc epitope Western blotting demonstrated approximately equal expression of myc-N17 Cdc42 and myc-V12N17 Rac (Fig. 4, bottom panel).
Neither wortmannin nor LY294002 could inhibit the cortical actin polymerization produced by the constitutively active Rac variant L61 Rac (Fig. 5A, C, and E). Moreover, neither of the catalytically inactive p110γ mutants (Δ948–981 p110γ-HA and K833R p110γ-HA) could inhibit the cytoskeletal changes induced by the constitutively active Rac variant V12 Rac (data not shown). These data imply that in these cells, Rac functions downstream of PI3-Kγ.
FIG. 5.
Pharmacologic inhibitors of PI3-K do not inhibit the cytoskeletal changes induced by a constitutively active Rac. Cos-7SH cells transiently expressing a constitutively active, HA-tagged Rac variant (HA-L61Rac) were treated overnight with either DMSO diluent alone (A and B), 100 nM wortmannin (C and D), or 50 μM LY294002 (E and F). The cells were fixed and stained as described in the legend to Fig. 1. Transfected cells were identified by the presence of FITC staining (B, D, and F), and their pattern of F-actin polymerization was assessed by phalloidin staining (A, C, and E). Cells shown are representative of cells from at least three experiments. Bars, 25 μm. When more than one cell is shown in a field, transfected cells are indicated by arrows.
Signals between PI3-Kγ and Rac are mediated by a GEF for Rac.
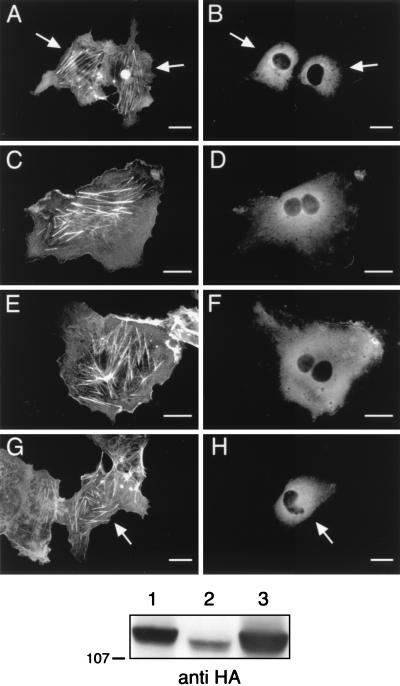
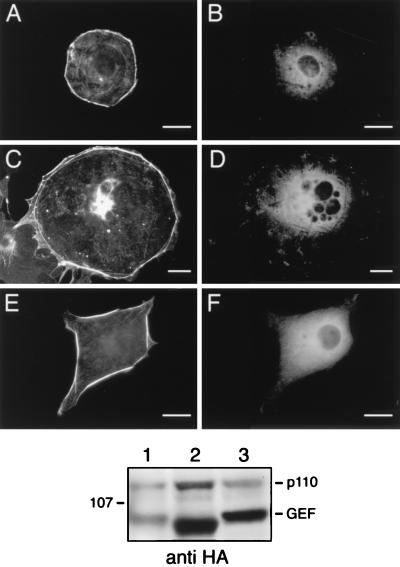
Rho family GTPases are regulated by proteins related to the product of the dbl oncogene. Dbl family members act as guanine nucleotide exchange factors (GEFs) for small GTP binding proteins, activating these proteins by catalyzing the exchange of GDP for GTP on Rho family members (3). Recently, a member of this family, Vav, has been shown to stimulate GDP/GTP exchange for Rac-1 (5). We wished to determine if the p110γ-mediated cytoskeletal changes required the action of a Rac GEF. To do this, we compared the cytoskeletal effects of coexpressing either wild-type Vav or catalytically inactive Vav variants along with myr-p110γ-HA. Coexpression of wild-type Vav had little influence on the ability of myr-p110γ-HA to induce cortical actin rearrangement (Fig. 6A). It has been shown that an inactive Vav variant (L213Q Vav) is unable to induce Rac activation (4). In contrast to wild-type Vav, the inactive L213Q Vav completely inhibited the ability of myr-p110γ-HA to lead to cytoskeletal reorganization (Fig. 6C). We constructed another Vav variant, missing 6 amino acids within the Dbl homology (DH) domain (Δ342–348 Vav), analogous to a known inactivating mutation within Dbl (11). We found that this Vav variant (Δ342–348 Vav) also blocked myr-p110γ-induced actin rearrangement (Fig. 6E). Lastly, another inactive Vav variant which cannot be tyrosine phosphorylated (Y174F Vav) was recently described by Han et al. (10). This variant also blocked the myr-p110γ-induced cortical actin rearrangements (Fig. 6G). These data support the concept that exchange activity on Rac is necessary to transduce signals from PI3-Kγ to Rac. Anti-HA immunoblotting of cell lysates demonstrated that HA–wild-type Vav, HA-L213Q Vav, HA-Δ342–348 Vav, and HA-Y174F Vav were expressed at approximately equal levels and that the Vav variants did not influence the expression of myr-p110γ (Fig. 6, bottom panel).
FIG. 6.
Coexpression of inactive variants of Vav inhibits myr-p110γ-induced cytoskeletal reorganization. Cos-7SH cells were transiently transfected with plasmids encoding myr-p110γ-HA and either wild-type Vav or inactive Vav variants and then fixed and stained as described in the legend to Fig. 1. Transfected cells were identified by the presence of FITC staining (B, D, F, and H), and their pattern of F-actin polymerization was assessed by phalloidin staining (A, C, E, and G). The cells were transfected with myr-p110γ-HA and either wild-type Vav (A and B), L213Q Vav (C and D), Δ342–348 Vav (E and F), or Y174F Vav (G and H). Cells shown are representative of cells from at least four experiments. Bars, 25 μm. When more than one cell is shown in a field, transfected cells are indicated by arrows. The bottom panel is an anti-HA Western blot showing approximately equivalent levels of myr-p110γ-HA and HA-tagged Vav and Vav variants. Lanes: 1, myr-p110γ-HA and HA–wild-type Vav; 2, myr-p110γ-HA and HA-L213Q Vav; 3, myr-p110γ-HA and HA-Δ342–348 Vav; 4, myr-p110γ-HA and HA-Y174F Vav.
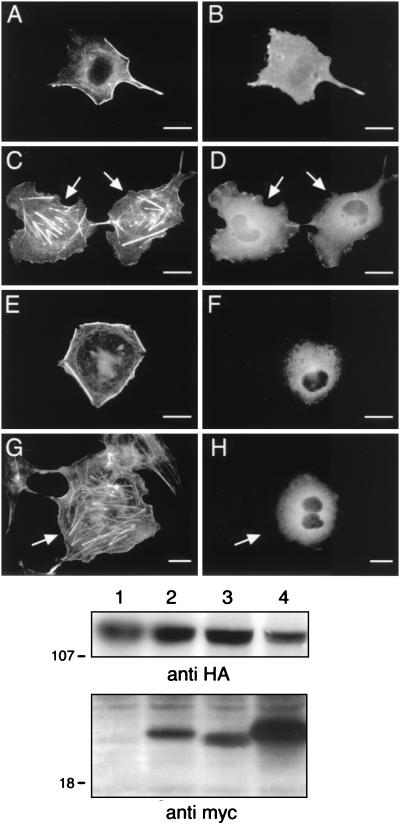
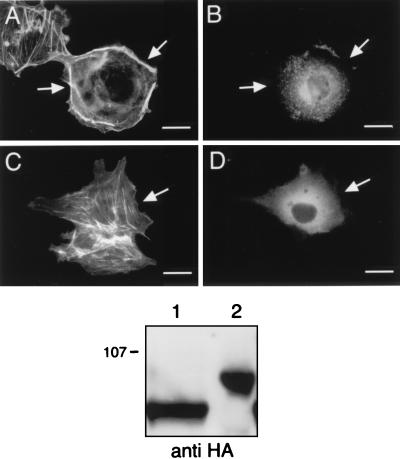
To demonstrate that Vav was truly acting upstream of Rac, we coexpressed a constitutively active Rac variant (myc-V12 Rac) along with dominant negative Vav variants. As shown in Fig. 7A and 7C, the constitutively active Rac rescued the block of cytoskeletal reorganization induced by two different dominant negative Vav variants, showing that Vav was situated upstream of Rac. As another control, we constructed an inactive Lsc variant with the analogous 6-amino-acid deletion (Δ568–574 Lsc) and showed that it had no effect on the myr-p110γ-initiated cytoskeletal changes (Fig. 7E). Since Lsc is a GEF that is known to be active on Rho and not Rac (8), this provides further evidence for the necessity of Rac-specific exchange activity in this pathway. Anti-HA immunoblotting of cell lysates demonstrated approximately equal expression of the mutant GEFs and myr-p110γ (Fig. 7, bottom panel).
FIG. 7.
Coexpression of a constitutively active Rac variant rescues the block produced by the inactive Vav variants on myr-p110γ-HA-induced cytoskeletal reorganization, and an inactive variant of Lsc fails to block this pathway. Cos-7SH cells were transiently transfected with plasmids encoding myr-p110γ-HA and either inactive Vav variants along with a constitutively active Rac variant or an inactive Lsc variant and then fixed and stained as described in the legend to Fig. 1. Transfected cells were identified by the presence of FITC staining (B, D, and F), and their pattern of F-actin polymerization was assessed by phalloidin staining (A, C, and E). (A and B) Cells expressing a constitutively active Rac variant, myc-V12 Rac, along with myr-p110γ-HA and the inactive L213Q Vav variant. (C and D) Cells expressing a constitutively active Rac variant, myc-V12 Rac, along with myr-p110γ-HA and the inactive Δ342–348 Vav variant. (E and F) Cells coexpressing myr-p110γ-HA along with an inactive variant of Lsc, a GEF for Rho and not Rac, with the analogous 6-amino-acid deletion within the DH domain (Δ568–574 Lsc-HA). Cells shown are representative of cells from at least four experiments. Bars, 25 μm. The bottom panel is an anti-HA Western blot demonstrating equivalent levels of expression of myr-p110γ-HA and HA-tagged Vav and Lsc variants. Lanes: 1, myr-p110γ-HA, L213Q Vav, and V12 Rac; 2, myr-p110γ-HA, Δ342–348 Vav, and V12 Rac; 3, myr-p110γ-HA and Δ568–574 Lsc.
The PH domain of Vav plays an inhibitory role in Vav function.
All GEFs in the Dbl family have PH domains immediately adjacent to their DH domains (3). PH domains are sequences of approximately 100 amino acids which are postulated to recruit molecules to membranes by specific interactions with polyphosphoinositide lipids (7, 20, 23). We wished to determine the role of the PH domain of Vav in the function of this molecule. We therefore constructed a Vav variant with its PH domain deleted (Vav ΔPH, missing residues 407 to 510) and tested its ability to affect cytoskeletal reorganization mediated by myr-p110γ. In contrast to the three inactive variants (L213Q Vav, Δ342–348 Vav, and Y174F Vav), Vav ΔPH did not block the ability of myr-p110γ-HA to cause actin rearrangement (data not shown). Moreover, this Vav variant was able to induce Rac activation and lead to cytoskeletal reorganization, even in the absence of transfected PI3-Kγ or other upstream stimulatory molecules (Fig. 8A). By contrast, expression of wild-type Vav alone had no effect on the actin cytoskeleton (Fig. 8C). Anti-HA immunoblotting of cell lysates demonstrated equal expression of Vav ΔPH and wild-type Vav (Fig. 8, bottom panel).
FIG. 8.
A Vav variant missing its PH domain is constitutively active in leading to cytoskeletal reorganization. Cos-7SH cells transiently expressing Vav ΔPH alone (A and B) or wild-type Vav alone (C and D) were fixed and stained as described in the legend to Fig. 1. Transfected cells were identified by the presence of FITC staining (B and D), and their pattern of F-actin polymerization was assessed by phalloidin staining (A and C). Cells shown are representative of cells from at least five experiments. Bars, 25 μm. Transfected cells are indicated by arrows. The bottom panel is an anti-HA Western blot demonstrating equivalent levels of expression of HA-tagged Vav ΔPH and wild-type Vav. Lanes: 1, HA-Vav ΔPH; 2, HA–wild-type Vav.
As another test of the constitutive activity of the Vav ΔPH variant, we compared the ability of this variant with that of the wild-type and oncogenic Vav to stimulate JNK activity (Fig. 9). Previously, Vav-induced activation of JNK was shown to occur through a Rac-dependent pathway (4). As shown in Fig. 9, overexpression of wild-type Vav had little influence on JNK activity, but the Vav ΔPH variant was essentially as active as oncogenic Vav. Western blot analysis demonstrated that Vav ΔPH consistently expressed at dramatically lower levels than wild-type Vav or oncogenic variants of Vav (data not shown). Despite lower levels of expression, ΔPH Vav always led to an increase in Rac activation roughly equivalent to that of oncogenic Vav. Deletion of the PH domain within Vav thus appears to create a constitutively active variant, arguing that the PH domain plays an autoinhibitory role within this protein in the resting state.
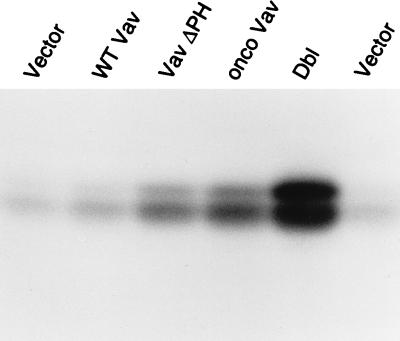
FIG. 9.
Effect of Vav variants on Rac activation. Cos-7 cells were transiently transfected with plasmids encoding Vav variants and FLAG-tagged JNK. Rac activation was measured as the ability to activate JNK. JNK kinase assays were performed after the anti-FLAG immunoprecipitations. Lane vector contains empty vector and shows the background level of JNK activity. Expression of wild-type Vav leads to no, or minimal, increases in JNK activity (lane WT Vav), while the Vav variant with its PH domain deleted markedly stimulates JNK activity (lane Vav ΔPH), to a level similar to that of oncogenic Vav (lane onco Vav), although not as powerfully as oncogenic Dbl (lane Dbl).
DISCUSSION
Taken together, the above findings delineate a pathway leading from activation of a G protein-coupled receptor to actin cytoskeletal reorganization which sequentially involves Gβγ, PI3-Kγ, a Rac GEF, and Rac. The involvement of PI3-Kγ and Rac in the mediation of cytoskeletal changes initiated by G protein-coupled receptors might have been inferred by analogy to the pathway used by growth factor receptors. However, the interaction between PI3-Kγ and Vav in this assay system suggests that Rac GEFs may be activated by D3 phosphoinositides. These observations raise several issues, including (i) the relative positions of PI3-Kγ and Rac within this signaling pathway, (ii) the mechanism by which the inactive Vav variants are able to block signaling, (iii) the mechanism by which D3 phosphoinositides activate Rac GEFs, (iv) the role of tyrosine phosphorylation in Vav activation, and (v) the identity of the endogenous Rac GEF present in Cos-7SH cells.
The first issue is whether Rac lies upstream or downstream of PI3-K in this pathway. Several investigators have found that Rho family members can bind to the p85 subunit of PI3-Kα/β isoforms and thereby lead to enzyme activation (31, 33). Recently, Keely et al. have shown that Rac and Cdc42 induce cell motility and invasion in T47D mammary epithelial cells (15). Since the Rac-induced effects were blocked by treatment with the PI3-K inhibitors wortmannin and LY294002, these investigators argued that PI3-K activation was downstream of Cdc42 and Rac. In contrast to these data, Hawkins et al. demonstrated that the platelet-derived growth factor-stimulated activation of Rac in porcine aortic endothelial cells occurred downstream of PI3-K, since neither wortmannin nor a dominant negative p85 (Δp85) could inhibit ruffle formation induced by expression of a constitutively active Rac, V12 Rac-1 (12). Our data show that neither treatment with pharmacologic inhibitors of PI3-K (wortmannin or LY294002) nor coexpression of catalytically inactive variants of p110γ had an effect on the cytoskeletal changes induced by V12 Rac. This suggests that in Cos-7SH cells, as in porcine aortic endothelial cells, Rac lies downstream of a PI3-K. This positioning is further supported by our finding that a dominant negative Rac blocks both PI3-Kγ-mediated actin changes induced by fPR stimulation or Gβγ overexpression and the actin rearrangement induced by a constitutively active PI3-Kγ variant.
The next question is the mechanism by which the inactive Vav variants are able to block this signaling pathway. This could be explained by two hypotheses. First, the dominant negative Vav variants might bind and sequester either PI3-Kγ or the active lipid products of PI3-K. Second, they might bind and sequester Rac, their downstream target. Since coexpression of the active Rac can rescue the block conferred by the inactive Vav molecules, the first hypothesis seems more likely. Preliminary experiments have failed to show a direct association between p110γ and Vav (21a), but experiments designed to explore this issue are under way in our laboratory.
The next issue is the mechanism of activation of a Rac GEF by PI3-Kγ. The role of the PH domain within the Dbl family members has been a source of speculation since they were first described. As has been shown for other PH domains, it is possible that the PH domain of the Rac GEF plays a role in targeting the GEF to specific intracellular locations (27, 34). It is also possible that the PH domain regulates Rac GEF function through binding to D3 phosphoinositide second messengers produced by PI3-Kγ. Consistent with the latter possibility is work by Han et al., who showed that, in vitro, the exchange activity of Vav is inhibited when its PH domain is bound to phosphatidylinositol 4,5-bisphosphate and is stimulated when bound to phosphatidylinositol 3,4,5-trisphosphate (10). Our data show that deletion of the PH domain allows Vav to activate JNK and to mediate cytoskeletal changes in the absence of other stimuli. Our observations could be explained by either hypothesis, i.e., that D3 phosphoinositides and the PH domain act cooperatively either to affect the intracellular localization of Vav or to merely release its intrinsic exchange activity. The latter hypothesis is supported by (i) the observation that the boundaries of Sos PH and DH domains are overlapping and hence interdependent (17), (ii) our indirect immunofluorescence data for HA-tagged Vav which indicate that the intracellular localization of Vav is not grossly influenced by deletion of its PH domain (Fig. 8D and F), and (iii) the recently published work from the Bar-Sagi laboratory showing that the ability of the DH domain of Sos to activate Rac was inhibited by the PH domain (24). Our data and the studies by the Bar-Sagi (24) and Broek (10) groups support the concept that the PH domains of Rac GEFs regulate guanosine exchange activity in a fashion determined by binding to products of PI3-K enzymes.
Tyrosine phosphorylation by Src family members is critical for Vav function (5), and others have used tyrosine phosphorylation of Vav as a marker for its activation (6). Han et al. have shown that a Y-to-F mutation at position 174 on Vav abolishes phosphorylation by Lck in vitro (10). Our data and those of others have shown that the PH domain of Vav also regulates Vav activity (10). This raises the question of the relative contribution of phosphorylation or lipid binding to Vav activation. Han et al. have shown that phosphatidylinositol 3,4,5-trisphosphate binding to the PH domain of Vav increases the tyrosine phosphorylation on Vav, thus arguing that tyrosine phosphorylation is the critical step in Vav activation. An alternative hypothesis is that phosphorylation at Y174 is required to allow the proper phosphoinositide to bind to the PH domain and that the phosphoinositide binding to the PH domain then activates Vav. Preliminary experiments from our laboratory suggest that this latter hypothesis may be correct, since a Vav variant with both the phosphorylation site mutation and PH domain truncation (Y174F ΔPH Vav) appears to constitutively induce cytoskeletal reorganization (21a). This suggests that if the inhibitory PH domain is deleted, tyrosine phosphorylation is no longer required for Vav activation. Experiments are under way in our laboratory to further explore the steps required in Vav activation.
The last issue is the identity of the endogenous Rac GEF in our transfected cells. Since Vav-1 expression is restricted to hematopoietic cells, it cannot account for the activation of Rac in Cos-7SH cells when activated PI3-Kγ is expressed alone. A nonhematopoietic Vav (Vav-2 or Vav-T) has recently been identified, although it is not known whether this has exchange activity for Rac (26). Certainly, the striking inhibition of actin reorganization seen when the inactive Vav variants are expressed implies that these variants are competing with an endogenous exchange factor with similar binding characteristics. However, at this point, the identity of that endogenous Rac exchange factor is unknown.
In conclusion, our observations show a potentially linear signaling pathway that is initiated by a G protein-coupled receptor and leads to cytoskeletal reorganization via the sequential action of Gβγ, PI3-Kγ, a Rac GEF, and Rac. The data also suggest that the Rac GEF, Vav, is activated, directly or indirectly, by D3 phosphoinositide products of PI3-Kγ. Lastly, our findings suggest that the PH domain located carboxyl to the Dbl motif in Vav has an inhibitory effect on Vav activity. The mechanism by which D3 phosphoinositides and the Vav PH domain regulate exchange activity is an area of current study.
ACKNOWLEDGMENT
Alice D. Ma and Ara Metjian contributed equally to this work.
REFERENCES
- 1.Bagrodia S, Derijard B, Davis R J, Cerione R A. Cdc42 and PAK-mediated signaling leads to Jun kinase and p38 mitogen-activated protein kinase activation. J Biol Chem. 1995;270:27995–27998. doi: 10.1074/jbc.270.47.27995. [DOI] [PubMed] [Google Scholar]
- 2.Bar-Sagi D, Feramisco J R. Induction of membrane ruffling and fluid phase pinocytosis in quiescent fibroblasts by ras proteins. Science. 1986;233:1061–1068. doi: 10.1126/science.3090687. [DOI] [PubMed] [Google Scholar]
- 3.Cerione R A, Zheng Y. The Dbl family of oncogenes. Curr Opin Cell Biol. 1996;8:216–222. doi: 10.1016/s0955-0674(96)80068-8. [DOI] [PubMed] [Google Scholar]
- 4.Crespo P, Bustelo X R, Aaronson D S, Coso O A, Lopez-Barahona M, Barbacid M, Gutkind J S. Rac-1 dependent stimulation of the JNK/SAPK signaling pathway by Vav. Oncogene. 1996;13:455–460. [PubMed] [Google Scholar]
- 5.Crespo P, Schuebel K E, Ostrom A A, Gutkind J S, Bustelo X R. Phosphotyrosine-dependent activation of Rac-1 GDP/GTP exchange by the vav proto-oncogene product. Nature. 1997;385:169–172. doi: 10.1038/385169a0. [DOI] [PubMed] [Google Scholar]
- 6.Deckert M, Tartare-Deckert S, Couture C, Mustelin T, Altman A. Functional and physical interactions of Syk family kinases with the Vav proto-oncogene product. Immunity. 1996;5:591–604. doi: 10.1016/s1074-7613(00)80273-3. [DOI] [PubMed] [Google Scholar]
- 7.Gibson T J, Hyvönen M, Musacchio A, Saraste M, Birney E. PH domain: the first anniversary. Trends Biochem Sci. 1994;19:349–353. doi: 10.1016/0968-0004(94)90108-2. [DOI] [PubMed] [Google Scholar]
- 8.Glaven J A, Whitehead I P, Nomanbhoy T, Kay R, Cerione R A. Lfc and Lsc oncoproteins represent two new guanine nucleotide exchange factors for the Rho GTP-binding protein. J Biol Chem. 1996;271:27374–27381. doi: 10.1074/jbc.271.44.27374. [DOI] [PubMed] [Google Scholar]
- 9.Hall A. Small GTP-binding proteins and the regulation of the actin cytoskeleton. Annu Rev Cell Biol. 1994;10:31–54. doi: 10.1146/annurev.cb.10.110194.000335. [DOI] [PubMed] [Google Scholar]
- 10.Han J, Luby-Phelps K, Das B, Shu X, Xia Y, Mosteller R D, Krishna U M, Falck J R, White M A, Broek D. Role of substrates and products of PI 3-kinase in regulating activation of Rac-related guanosine triphosphatases by Vav. Science. 1998;279:558–560. doi: 10.1126/science.279.5350.558. [DOI] [PubMed] [Google Scholar]
- 11.Hart M J, Eva A, Zangrilli D, Aaronson S A, Evans T, Cerione R A, Zheng Y. Cellular transformation and guanine nucleotide exchange activity are catalyzed by a common domain on the dbl oncogene gene product. J Biol Chem. 1994;269:62–65. [PubMed] [Google Scholar]
- 12.Hawkins P T, Eguinoa A, Qiu R-G, Stokoe D, Cooke F T, Walters R, Wennstrom S, Claesson-Welsh L, Evans T, Symons M, Stephens L. PDGF stimulates an increase in GTP-Rac via activation of phosphoinositide 3-kinase. Curr Biol. 1995;5:393–403. doi: 10.1016/s0960-9822(95)00080-7. [DOI] [PubMed] [Google Scholar]
- 13.Ho S N, Hunt H D, Horton R M, Pullen J K, Pease L R. Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene. 1989;7:51–59. doi: 10.1016/0378-1119(89)90358-2. [DOI] [PubMed] [Google Scholar]
- 14.Hu Q, Klippel A, Muslin A J, Fantl W J, Williams L T. Ras-dependent induction of cellular responses by constitutively active phosphatidylinositol 3-kinase. Science. 1995;268:100–103. doi: 10.1126/science.7701328. [DOI] [PubMed] [Google Scholar]
- 15.Keely P J, Westwick J K, Whitehead I P, Der C J, Parise L V. Cdc42 and Rac1 induce integrin mediated cell motility and invasiveness through PI(3)K. Nature. 1997;390:632–636. doi: 10.1038/37656. [DOI] [PubMed] [Google Scholar]
- 16.Kolodziej P A, Young R A. Epitope tagging and protein surveillance. Methods Enzymol. 1991;194:508–519. doi: 10.1016/0076-6879(91)94038-e. [DOI] [PubMed] [Google Scholar]
- 17.Koshiba S, Kigawa T, Kim J-H, Shirouzu M, Bowtell D, Yokoyama S. The solution structure of the pleckstrin homology domain of mouse Son-of sevenless 1 (mSos1) J Mol Biol. 1997;269:579–591. doi: 10.1006/jmbi.1997.1041. [DOI] [PubMed] [Google Scholar]
- 18.Kotani K, Yonezawa K, Hara K, Ueda H, Kitamura Y, Sakaue H, Ando A, Chavanieu A, Calas B, Grigorescu F, Nishiyama M, Waterfield M D, Kasuga M. Involvement of phosphoinositide 3-kinase in insulin- or IGF-1-induced membrane ruffling. EMBO J. 1994;13:2313–2321. doi: 10.1002/j.1460-2075.1994.tb06515.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Landt O, Grunert H-P, Hahn U. A general method for rapid site-directed mutagenesis using the polymerase chain reaction. Gene. 1990;96:125–128. doi: 10.1016/0378-1119(90)90351-q. [DOI] [PubMed] [Google Scholar]
- 20.Lemmon M A, Ferguson K M, Schlessinger J. PH domains: diverse sequences with a common fold recruit signaling molecules to the cell surface. Cell. 1996;85:621–624. doi: 10.1016/s0092-8674(00)81022-3. [DOI] [PubMed] [Google Scholar]
- 21.Lopez-Ilasca M, Crespo P, Pellici P G, Gutkind J S, Wetzker R. Linkage of G protein-coupled receptors to the MAPK signaling pathway through PI 3-kinase gamma. Science. 1997;275:394–397. doi: 10.1126/science.275.5298.394. [DOI] [PubMed] [Google Scholar]
- 21a.Ma, A. D., and C. S. Abrams. Unpublished data.
- 22.Ma A D, Brass L F, Abrams C S. Pleckstrin associates with plasma membranes and induces the formation of membrane projections. J Cell Biol. 1997;136:1071–1079. doi: 10.1083/jcb.136.5.1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22a.Metjian, A., et al. Unpublished data.
- 23.Musacchio A, Gibson T, Rice P, Thompson J, Saraste M. The PH domain: a common piece in the structural patchwork of signalling proteins. Trends Biochem Sci. 1993;18:343. doi: 10.1016/0968-0004(93)90071-t. [DOI] [PubMed] [Google Scholar]
- 24.Nimnual A S, Yatsula B A, Bar-Sagi D. Coupling of Ras and Rac guanosine triphosphatases through the Ras exchanger Sos. Science. 1998;279:560–563. doi: 10.1126/science.279.5350.560. [DOI] [PubMed] [Google Scholar]
- 25.Nobes C D, Hall A. Rho, Rac, and Cdc42 GTPases regulate the assembly of multimolecular focal complexes associated with actin stress fibers, lamellipodia, and filopodia. Cell. 1995;81:53–62. doi: 10.1016/0092-8674(95)90370-4. [DOI] [PubMed] [Google Scholar]
- 26.Okumura K, Kaneko Y, Nonoguchi K, Nishiyama H, Yokoi H, Higuchi T, Itoh K, Yoshida O, Miki T, Fujita J. Expression of a novel isoform of Vav, Vav-T, containing a single Src homology 3 domain in murine testicular germ cells. Oncogene. 1997;14:713–720. doi: 10.1038/sj.onc.1200878. [DOI] [PubMed] [Google Scholar]
- 27.Olson M F, Sterpetti P, Nagata K, Toksoz D, Hall A. Distinct roles for DH and PH domains in the LBC oncogene. Oncogene. 1997;15:2827–2831. doi: 10.1038/sj.onc.1201594. [DOI] [PubMed] [Google Scholar]
- 28.Stephens L, Smrcka A, Cooke F T, Jackson T R, Sternweis P C, Hawkins P T. A novel phosphoinositide 3 kinase activity in myeloid-derived cells is activated by G protein βγ subunits. Cell. 1994;77:83–93. doi: 10.1016/0092-8674(94)90237-2. [DOI] [PubMed] [Google Scholar]
- 29.Stephens L R, Eguinoa A, Erdjument-Bromage H, Lui M, Cooke F, Coadwell J, Smrcka A S, Thelen M, Cadwallader K, Tempst P, Hawkins P T. The Gβγ sensitivity of a PI3K is dependent upon a tightly associated adaptor p101. Cell. 1997;89:105–114. doi: 10.1016/s0092-8674(00)80187-7. [DOI] [PubMed] [Google Scholar]
- 30.Stoyanov B, Volinia S, Hanck T, Rubio I, Loubtchenkov M, Malek D, Stoyanova S, Vanheasebroek B, Dhand R, Nurnberg B, Gierschik P, Seedorf K, Hsuan J J, Waterfield M D, Wetzker R. Cloning and characterization of a G protein-activated human phosphoinositide-3 kinase. Science. 1995;269:690–693. doi: 10.1126/science.7624799. [DOI] [PubMed] [Google Scholar]
- 31.Tolias K F, Cantley L C, Carpenter C L. Rho family GTPases bind to phosphoinositide kinases. J Biol Chem. 1995;270:17656–17659. doi: 10.1074/jbc.270.30.17656. [DOI] [PubMed] [Google Scholar]
- 32.Wennstrom S, Hawkins P, Cooke F, Hara K, Yonezawa K, Kasuga M, Jackson T, Claesson-Welsh L, Stephens L. Activation of phosphoinositide 3-kinase is required for PDGF-stimulated membrane ruffling. Curr Biol. 1994;4:385–393. doi: 10.1016/s0960-9822(00)00087-7. [DOI] [PubMed] [Google Scholar]
- 33.Zheng Y, Bagrodia S, Cerione R A. Activation of phosphoinositide 3-kinase activity by Cdc42Hs binding to p85. J Biol Chem. 1994;269:18727–18730. [PubMed] [Google Scholar]
- 34.Zheng Y, Zangrilli D, Cerione R A, Eva A. The pleckstrin homology domain mediates transformation by oncogenic Dbl through specific intracellular targeting. J Biol Chem. 1996;271:10917–10920. doi: 10.1074/jbc.271.32.19017. [DOI] [PubMed] [Google Scholar]