Abstract
To perform multiplex profiling of single cells and eliminate the risk of potential sample loss caused by centrifugation, we developed a microfluidic flow cytometry and mass spectrometry system (μCytoMS) to evaluate the drug uptake and induced protein expression at the single cell level. It involves a microfluidic chip for the alignment and purification of single cells followed by detection with laser-induced fluorescence (LIF) and inductively coupled plasma mass spectrometry (ICP-MS). Biofunctionalized nanoprobes (BioNPs), conjugating ∼3000 6-FAM-Sgc8 aptamers on a single gold nanoparticle (AuNP) (Kd = 0.23 nM), were engineered to selectively bind with protein tyrosine kinase 7 (PTK7) on target cells. PTK7 expression induced by oxaliplatin (OXA) uptake was assayed with LIF, while ICP-MS measurement of 195Pt revealed OXA uptake of the drug in individual cells, which provided further in-depth information about the drug in relation to PTK7 expression. At an ultralow flow of ∼0.043 dyn/cm2 (20 μL/min), the chip facilitates the extremely fast focusing of BioNPs labeled single cells without the need for centrifugal purification. It ensures multiplex profiling of single cells at a throughput speed of 500 cells/min as compared to 40 cells/min in previous studies. Using a machine learning algorithm to initially profile drug uptake and marker expression in tumor cell lines, μCytoMS was able to perform in situ profiling of the PTK7 response to the OXA at single-cell resolution for tests done on clinical samples from 10 breast cancer patients. It offers great potential for multiplex single-cell phenotypic analysis and clinical diagnosis.
Keywords: single-cell heterogeneity, multiplex profiling, microfluidic chip, biofunctionalized nanoprobes, flow cytometry, mass spectrometry
Single cell analysis of cancer has received much attention in recent years, and significant progress has been made.1,2 For example, further understanding on drug-resistance mechanisms,3 tumor cell metastasis,4 and personalized prognosis5 have been achieved through studying and observing the heterogeneity in drug uptake and accompanying protein expressions on individual tumor cells. Nevertheless, there are still significant challenges with regard to the throughput, multiplex biomarker detection, low retention rate of precious samples, and accuracy of current single-cell assays. Also, these assays normally involve sample preparation, i.e., centrifugation or mixing by pipet, that can result in cell loss or cell damage.6−8
Recent advances in mass spectrometry (MS) have enabled metabonomic and proteomic analyses at the single-cell resolution, through employing matrix-assisted laser desorption/ionization, secondary ion technique and electrospray ionization.9−11 MS coupling with microfluidic chip can minimize reaction volumes, reduce sample transfer steps, stabilize single-cell stream, and improve detection sensitivity.12 In addition, microfluidic systems can enable sample preprocessing before analysis of individual cells, e.g., secondary flow effect for high-throughput single-cell separation/alignment13,14 and hydrodynamic filtration for staining/labeling various probes on target cells.15,16 Robust microfluidic platforms were developed to measure cellular phenotyping characteristics.17 Using a series of mass probes prepared by assembling specific organic tags or antibodies on nanoparticles (NPs) or magnetic beads, amplified and simplified mass signals can enable the study of expression of multiple proteins and metabolites on single cells.11,18,19 Unfortunately, direct monitoring of drug uptake along with protein expression in multiple single cells remains a significant challenge due to low-throughput and matrix interfering effects.
Owing to its low matrix effect, high sensitivity, and wide dynamic range,20 time-resolved inductively coupled plasma mass spectrometry (ICP-MS) has great potential for single-cell analysis and has been extensively used to phenotype at the single-cell resolution.21−23 Further, extensive efforts were made in single-cell profiling using cytometry (CyTOF). It combines the merits of flow cytometry and ICP-MS, which can explore multiparameters of individual cells with high sensitivity and resolution.11,24 However, CyTOF is very costly, and it is not readily available in most general laboratories. Also, the highly integrated interface between flow cytometry and time-of-flight mass spectrometry means that cells cannot be easily recovered and recultured after cytofluorescence data acquisition and counting. The cells are directly ionized and then introduced to TOF-MS. Therefore, the more economical approach of coupling laser-induced fluorescence (LIF) with ICP-MS was proposed for acquiring high-throughput multidimensional data from single cells. The multiple MS fluorescent probes, e.g., aptamer nanoparticle probe and Mito-Tracker Green, were used to label target biomarkers or organelles of cells to enable monitoring and displaying information on single cells.25 Aptamers Sgc8 and SYL3C were able to identify PTK7 and Epithelial Cell Adhesion Molecule (EpCAM), respectively.26 Probes may be readily achieved by attaching these aptamers with modified fluorescent groups onto metal nanoparticles. These probes facilitate simultaneous collection and analysis of information on cell counting, fluorescence sensing and MS detection of individual cells.16,27 Fluorescence/MS signals of probes enable assaying the expression level of target proteins on single cells and have been widely applied as binding ligand in tumor cell analysis.16,26,28 In addition, machine learning algorithms have been demonstrated to be able to perform functional phenotype profiling of bioparticles.29 However, during flow, specific binding of recognition-ligands on cells may be adversely affected by larger shearing stress at high flow rate,30 and massive cell sampling may result in cell loss due to the centrifugal operation employed in sample purification.31 Thus, integration of a microfluidic chip with MS to conduct simultaneous sample preprocessing and detection under low flow rates will yield better single-cell information so as to enable more effective and comprehensive multiplex profiling of cell phenotype and heterogeneity.
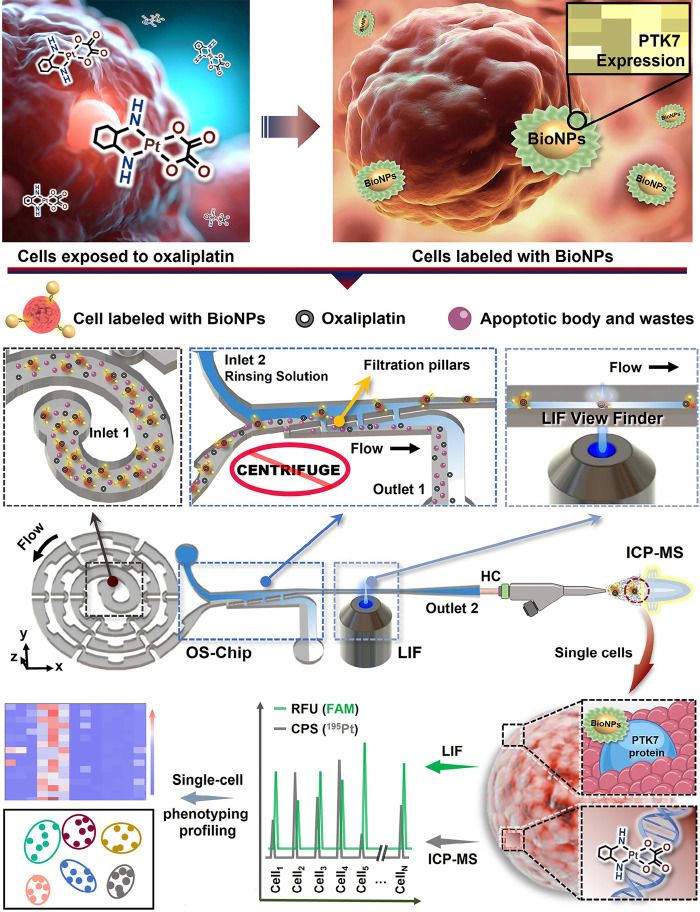
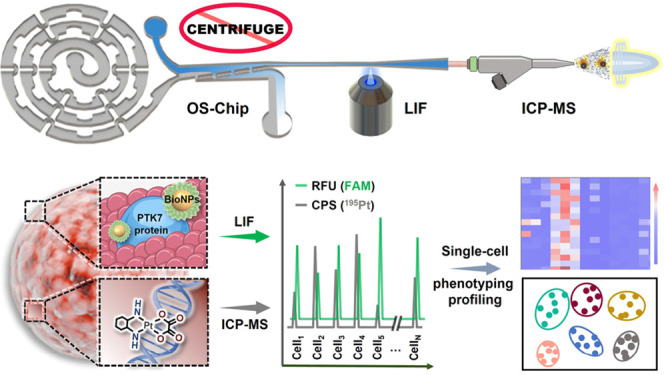
Here, we proposed a microfluidic flow cytometry/mass spectrometry system (μCytoMS) consisting of a single cell focusing microfluidic chip coupled with LIF and ICP-MS detection as illustrated in Scheme 1 and Figure S1. Our μCytoMS enables simultaneous high-throughput multiparameter detection of biomarker expression and agent uptake at the single-cell resolution. A specially designed microfluidic chip was connected horizontally to ICP-MS for aligning and sampling single cells with no background interference. The chip enables excellent single-cell alignment and purification under ultralow flow condition, which ensures unicellular profiling with high throughput, temporal, and signal/noise ratio and is fully compatible with downstream analysis of living cells. This chip does not require tedious centrifugation and, thus, avoid sample loss while reducing the time for purification step. Furthermore, accurate fluorescence signal collection was achieved for the analysis of protein heterogeneity by using high-resolution LIF View Finder. For effective labeling of PTK7 biomarkers, which is a sensitive mediator of oxaliplatin (OXA) on the surface of single cells,32 a panel of BioNPs probes were fabricated by self-assembling ca. 3000 Sgc8 aptamers onto each single AuNP. Based on LIF detection of the probes, high-throughput evaluation of PTK7 expressed on individual cell could be achieved from the amplified fluorescence arising from 6-FAM conjugating at 3′ end of Sgc8. At the same time, the MS signal of the 195Pt isotope provided information on the presence of OXA in single cells, thus facilitating sensitive and high-throughput protein-drug evaluation at the single cell level.
Scheme 1. Working Principles of the Microfluidic System for Single Cell Alignment/Purification in the LIF/ICP-MS Assays.
After OXA exposure, the trypsinized cell suspension was incubated with BioNPs to enable the capture of these cells by the probe. The cells conjugated with BioNPs were then aligned and purified in the microfluidic chip. Finally, the single cells were detected with LIF and introduced to TRA-ICP-MS to perform efficient background-free multiplex single-cell analysis.
Finally, we performed clinical validation by evaluating OXA uptake and agent-protein interaction in circulating tumor cells (CTCs) obtained from cancer patient’s blood after treatment with platinum drugs by monitoring signal (cps) of 195Pt at the single-cell resolution. μCytoMS was demonstrated to be able to evaluate OXA uptake and PTK7 expression at the single-cell level based on ten breast cancer patients’ CTCs. In addition, machine learning algorithms, i.e., linear discriminant analysis (LDA) and receiver operating characteristic (ROC) curves, were performed and proven reliable for cell phenotyping and heterogeneity profiling.
Results and Discussion
Flow of Single Cell after Purification
It is widely recognized that Dean-like secondary flow effect and hydrodynamic filtration (HDF) effect were employed for particle alignment and sample medium exchange, respectively.16 The inertial alignment arises from secondary flow acting on bioparticles, which boosts migration laterally to equilibrium positions in the spiral channel under combination of inertial lift force (FL) and Dean drag force (FD).33 Periodic dimensional confinement in the spiral microchannel has proven to boost Dean flow effect, thus enabling particle alignment for subsequent application for single cell analysis.30 In addition, noticeable flow rate gradients can be generated near the filtration pillars arising from the HDF structure where large particles stay in the low flow rate region in the main channel, while small particles and carrier-medium of large particles pass through the filter to the high flow rate region. Thus, HDF structure is regularly used for exchanging the carrier-medium of large particles.15 Based on the above principle, the device was designed and fabricated as an integrated multiparameter single-cell sampling/analyzing system with LIF and ICP-MS detection.30,31 In this work, a one-stop sampling chip (OS-Chip) was designed, as shown in Scheme 1. It consisted of a series of components: (1) two inlets for introducing sample exposed to OXA and incubated with BioNPs (Inlet 1) and rinsing solution (Inlet 2); (2) an enhanced spiral channel consisting of periodic high-curvature curves and dimensional-confinement pillars for the generation of single-cell stream; (3) an omnipotent purifier for sample purification; (4) a LIF View Finder for fluorescence data collection; (5) a horizontal capillary to connect the microchip to ICP-MS. Low aspect ratio (i.e., AR = H/W = 30 μm/300 μm = 1/10, where H and W represent the height and the width of the spiral channel, respectively) was applied for enhancing focusing and alignment of cells.34 This configuration strengthens the secondary-flow and HDF effects to promote single-cell alignment and purification. In addition, the LIF View Finder enabled the LIF detector to precisely collect the fluorescence signal from fluorescent aptamer probes tagged on the tumor cells. Subsequently, these cells were transported horizontally via the horizontal capillary to the ICP-MS for high-temporal resolution elemental analysis for isotopes of 197Au or 195Pt. This provided a more stable single-cell stream when compared to the conventional vertical configuration. Here, a series of signals from individual cells may provide detailed information on cellular heterogeneity in terms of cell counting, phenotyping, protein expression, and dosing monitoring/controlling. Further description on tumor recognizing probes, single-cell sampling device, and parametric evaluation of LIF and ICP-MS will be elaborated in the following sections.
Fabrication and Characterization of BioNPs
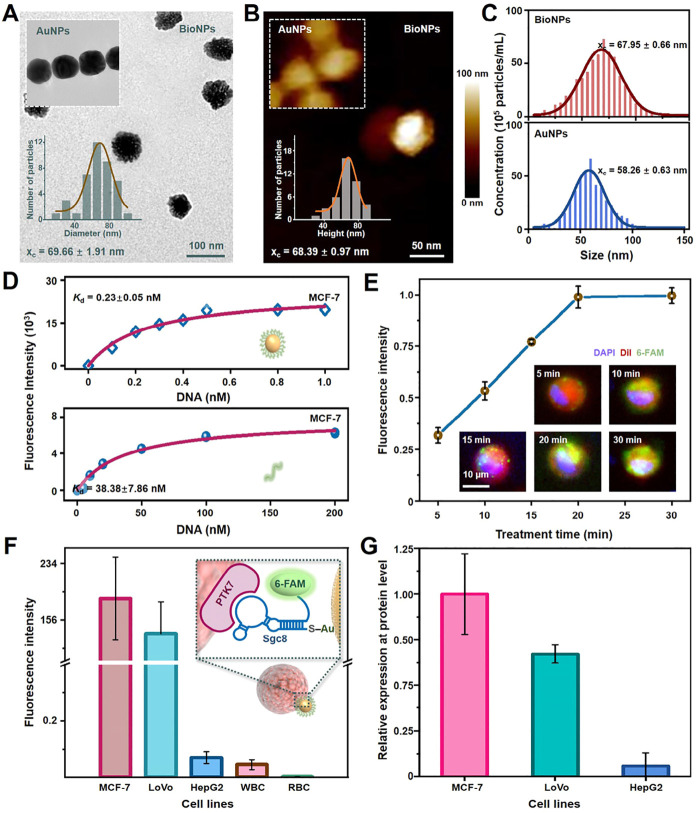
BioNPs nanoprobes were functionalized with thiolated aptamers (Sgc8) using classical method (Figure 1A-C).26 The size change of the gold nanoparticles before and after modification was evaluated by three methods (TEM, AFM, and NTA), which indicated that the aptamers were successfully conjugated to the gold nanoparticles. Compared with the standard gold nanoparticles, the average diameter of the modified probes increased by about 8 nm. Aptamers Sgc8 can selectively bind with the biomarker protein PTK7 expressed on the surface of tumor cells. The conjugate of nanoprobe-tumor cell was produced via the multivalent effect, due to the sufficient affinity by ca. 3000 Sgc8 aptamers loaded on each single AuNP of 60.8 nm (Figures S1A-C). The Kd value of Sgc8 in the BioNPs was 0.23 ± 0.050 nM against MCF-7 cells (Figure 1D), which exhibited ca. 170-fold promotion of the binding affinity with respect to that of the monovalent aptamers of Sgc8 (38.38 ± 7.68 nM). Competition analysis shows that the BioNPs could bind more stably to the biomarker compared with the aptamer (Figure S1D). In comparison with previous study,16,26 BioNPs exhibited 100% capture efficiency with even greater binding capacity against the previously reported Sea urchin-DMA-AuNPs (Kd=∼0.35 nM, 30 min, Figure 1E). A reduction of treatment time for PTK7+ cells was also achieved (20 min) due to more aptamers being labeled on the surface of larger nanoparticles.16 Furthermore, fluorescence intensities of MCF-7 cells were negatively correlated with OXA concentration, which illustrated the decrease expression level of PTK7 with increased OXA concentration (Figure S1C). In Figures 1F and S1, the fluorescence intensities of various cell lines were different when incubated with the same concentration of oxaliplatin. The intensity of MCF-7 cells (PTK7+) was higher than that of other biomarker protein-positive cell lines (LoVo cells) conjugated with BioNPs nanoprobes. In addition, minimal fluorescence was observed from PTK7-negative cells (white blood cells and red blood cells). Furthermore, the results of the real-time PCR (qPCR) analysis were consistent with the conclusions obtained from the fluorescence imaging analysis (Figure 1G). These observations indicated that fluorescence variation of specific BioNPs probes may provide information for distinguishing the cell subgroups and related protein expression level.
Figure 1.
Fabrication and characterization of BioNPs. (A) TEM images and (B) AFM images of the BioNPs nanoprobes along with their size distribution. (C) Nanoparticle tracking analysis (NTA) confirmed that AuNPs were conjugated with Sgc8 aptamers. (D) The evaluation of the dissociation constants of BioNPs and Sgc8-AuNPs to MCF-7 cells. (E) Scrutinizing the treatment time of BioNPs nanoprobes conjugating on MCF-7 cells. Incubation of the probe with the cells can be completed in 20 min. DAPI, DiI, and 6-FAM represent the nucleus, cell membrane and BioNPs probe, respectively. (F) The relative fluorescence intensity of BioNPs conjugated on various cell lines. (G) The relative expressions of PTK7 in various cell lines by real-time PCR (qPCR). The data indicate mean ± standard deviation (SD). Three independent groups tested by using biological replicates (n = 3) were used for each data point.
One-Stop Horizontal Single-Cell Sampling
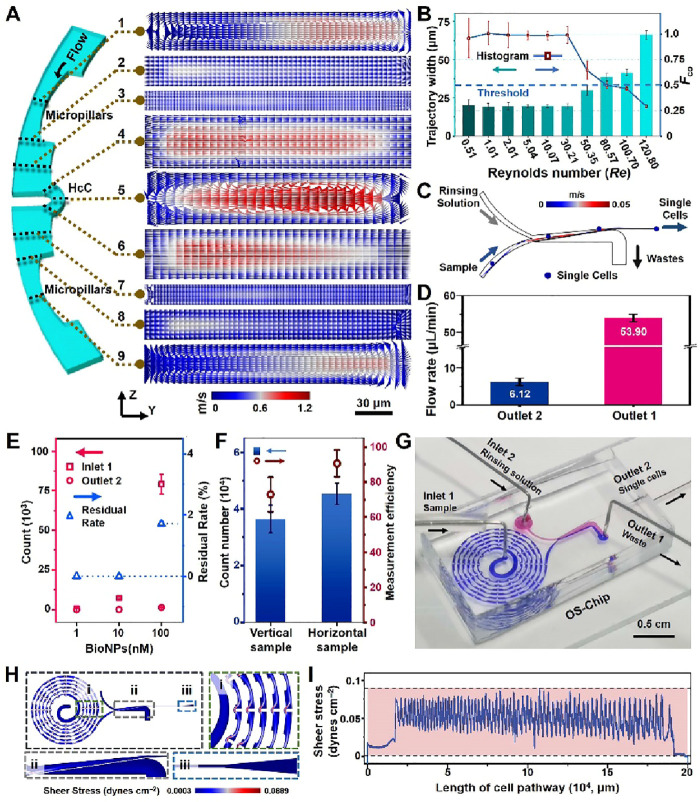
To analyze single cells, it is imperative to construct a platform for single-cell sampling and fluorescent-signal acquisition. For this purpose, the cell focusing and alignment capability and efficiency were evaluated for five different spiral chip configurations. Full details of the performances and discussion can be found in the Supporting Information (Figures S3–S7). Arising from the high-curvature channel (HcC) in the D3-ESC chip configuration, particles were found to undergo alignment in the periodic confinement space, which then resulted in significantly enhanced Dean-like secondary flow effect to facilitate generation of single particle stream (Figure 2A, Re = 1.01, 10 μL/min).13,35 To validate the profound vortex strengthening and fluid motion, a numerical model on the D3-ESC was developed and simulated with a flow rate of 10 μL/min. For cross-section of 1/9 by 5, corresponding to central region of the two wider micropillars and HcC, obvious Dean vortices were generated (Figure S8). This led to Dean-like secondary flow train and boosted progressive alignment to continuous single particle stream.30 Significantly, periodic HcCs resulted in sustained focusing of particles, as compared to a conventional spiral microdevice that reduces the curvature in the outer loops/channels, which may disrupt single-cell alignment. For further evaluation of the capability of D3-ESC in generating single stream of particles, fluorescence microbeads with diameter of 19.3 μm were used to perform focusing experiments under various flow conditions (Re = 0.51–201.42, 5–2000 μL/min). Figures 2B, S9, and S10 illustrated that particles were stably concentrated to generate single stream near the inner side of the channel within a wide range of flow rates (Re = 0.51–50.35, 5–500 μL/min). The average width of the particle trajectory was 21.04 ± 3.94 μm, and Fco value was found to be 0.94 ± 0.13. However, with the increase in Re, the position of particle focusing will induce outward movement within the range of Re = 80.57–120.79, and the width of trajectory will then broaden or even split into two streams for Re > 150.10. This observation is in accordance with the results reported in a previous study.30 It is probable that excessive lateral flow velocity in y-axis has led to formation of two pairs of Dean vortices or two equilibrium positions.33 It is noteworthy that the present study has achieved high-throughput cell focusing at an extremely low flow rate of 5 μL/min with Re = 0.51 and a cell density of 106 cells/mL (Figure S11). This benefits from the specially designed D3-ESC configuration of the microfluidic chip, which significantly promotes excellent generation of Dean-like secondary flow.
Figure 2.
Evaluation of OS-Chip. (A) Fluid velocity field of nine cross sections (1–9) in the y-axis along the micropillar-HcC-micropillars at a flow rate of 10 μL/min and Re = 1.01. (B) Fco values in the D3-ESC configuration under various flow rates of 5, 10, 20, 50, 100, 300, 500, 800, 1000, and 1200 μL/min (Re = 0.51, 1.01, 2.01, 5.04, 10.07, 30.21, 50.35, 80.57, 100.70, and 120.80). (C) Simulation results of the single-cell trajectory achieved in the purifier with P2 configuration. (D) The actual flow rates in Outlets 1 and 2. An independent t-test is performed to obtain statistical pairwise differences between Outlets 1 and 2 (***p < 0.001). (E) Evaluation of the purification performance for the P2 by using different concentrations of AuNPs suspension collected from the Outlet 2. Residual Rate represents residual quantity of AuNPs in the collected medium from Outlet 2. (F) The comparison of transport efficiency or measurement efficiency for the single cells between the vertical and horizontal sampling modes. (G) Photograph of the OS-Chip with two dyes (blue and red) loading in two inlets (Inlet 1 and Inlet 2). Shear stress gradient (H) and shear stress of the whole cell path (I) in the microfluidic chip. Three independent chips prepared by using biological replicates (n = 3) were used for each data point.
Huge amounts of interferents in single-cell samples may seriously affect the accuracy of subsequent fluorescence/ICP-MS detection. These can include the committed free or unbound BioNPs, cell debris produced after digestion as well as complex organic/inorganic salt matrix.36 Thus, six model purification chips were designed and fabricated, among which purification chip P2 was chosen for rinsing the interferents after performing the comparison tests. Full details were discussed in the Supporting Information and illustrated in Figures S12–S17. Here, the fluorescent particles and staining MCF-7 cells were sifted out from the particle/cell suspension to the Outlet 2 with sample loss of 0.69 ± 0.089% (Figures S17C and S18). This observation is in agreement with the result of particle-tracing simulation as illustrated in Figure 2C. While centrifugation has traditionally been considered the most common method for washing samples, the centrifugation process has been known to cause sample loss as well as reduced sample viability.31 To verify this, we performed multiple centrifugations on the cells. As shown in Figure S17D, after five centrifugations, the loss of cells was nearly 60%, while cell viability decreased from 93.6% to 82.3%. This is the reason why there are a growing number of centrifugation-free techniques being developed to directly process and assay samples. In addition, the flow rates at Outlet 1 and Outlet 2 were 53.90 and 6.12 μL/min, respectively (Figures 2D and S19), which ensure a >3-fold concentration of the cell and match the requirement for LIF detection of the single cells and their subsequent sampling into ICP-MS by the micronebulizer. Thereafter, a range of concentrations of BioNPs probe suspension, OXA-containing medium, trypsinized medium containing massive vesicles, and whole blood with various hematocrit (Hct) were tested to determine the purification capability of the microfluidic chip for downstream single cell evaluation in these complex matrices. The results presented in Figures 2E and S19–S23 show removal of probes as high as 10 nM with a 0% residual rate. In addition, massive vesicles-containing trypsinized medium, OXA-containing medium, and diluted whole blood with model cells (Hct = 2.5%–15%) were all effectively removed. The above observations clearly indicated that the P2 microfluidic device is able to depurate the excessive interferents including nanoscale contaminants, complex matrix in biological samples, as well as ionic species or drug molecules.
Numerical simulations were also performed to investigate the fluid motions at the interface of vertical and horizontal sampling modes to ICP-MS (Figures S24A,B). For conventional microchip-ICP-MS interfacing, a tubing is generally inserted vertically at the outlet of the chip for sampling the aligned single-cell stream. In such a case, it can inevitably cause sudden changes in the flow field and disrupt the aligned single-cell stream. This may present obstacle for achieving high-efficiency single-cell transportation and analysis.37,38 In contrast, the stable flow field distribution in a horizontal sampling mode enables a reliable aligned single-cell stream, as shown in Movie S1, and significant promotion for the single-cell transport efficiency has been observed (Figure 2F). As such, the OS-Chip was constructed as characterized by the dye experiments (Figures 2G and S24C). With optimized low flow conditions (Inlet 1:Inlet 2 = 20:40 μL/min), the cells were hardly damaged in the alignment/purification process under an average shear stress of 0.043 dyn/cm2 (Figures 2H–I and S25). The μCytoMS system was constructed by coupling the OS-Chip to LIF and ICP-MS (Figure 3A). MCF-7 cells after exposure to 105 mL–1 of AuNPs suspension (60.8 ± 7.5 nm in diameter) were detected by measuring the isotope of 197Au (Figure S26). A cell transport efficiency of 90.6 ± 7.6% was derived (n = 5, confidence limit of 95%) by adopting an Enya Mist nebulizer with a 30-folds higher sampling efficiency than the conventional concentric nebulizer. Surprisingly, the cell transport efficiency may even be improved up to 96.0 ± 10.3% (n = 3, confidence limit of 95%) by reducing the flow rate of Inlet 1 to 5 μL/min. Considering that the sampling throughput is a vital parameter in single-cell processing and assaying, the inlet flow rates may be adjusted based on the specific requirements of the study. For fluorescence monitoring of the single cells, the LIF View Finder was connected to the end of Outlet 1. The narrow channel ensured a stable single-cell flow path and reliable single-cell fluorescence signal acquisition. The present OS-Chip mode enabled single-cell alignment and purification at 500 cells/min under ultralow flow conditions, which is 20-fold higher in comparison with previous studies (Table S2) involving multidimensional unicellular profiling strategies.11 It achieved a more concise configuration of the purifying unit with higher purification efficiency. LIF View Finder and appropriate interface further ensured almost-lossless measurement efficiency for single-cell sampling and profiling.
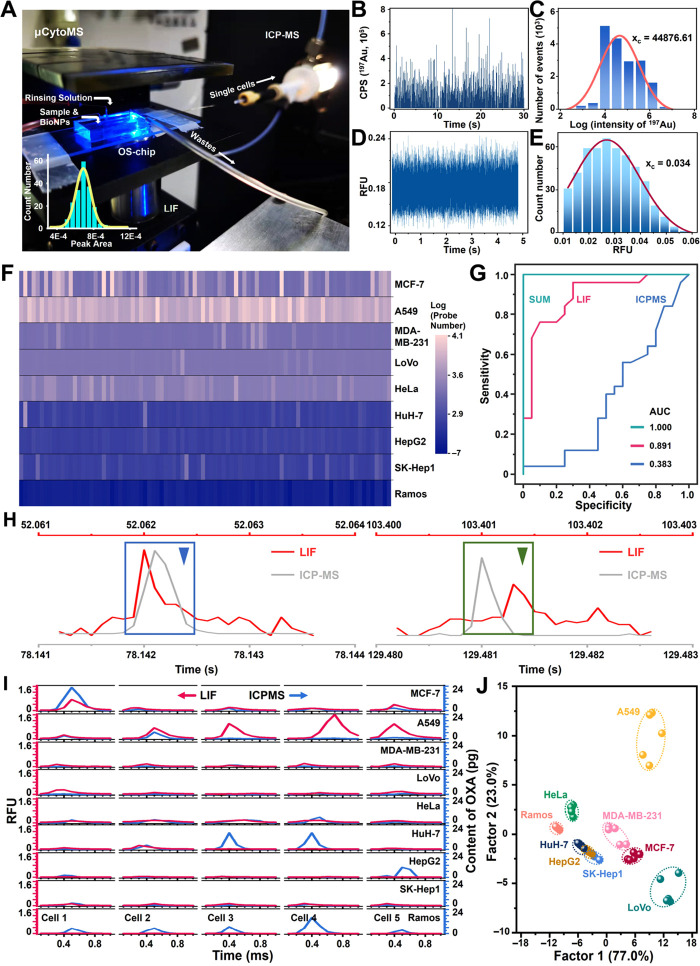
Figure 3.
Analytical performance of μCytoMS. (A) Photograph of the μCytoMS system. The inset represents the frequency histogram and Gauss fit of the peak for single fluorescent particle signals. It indicates that the system is able to quantify fluorescent response of individual particles. ICP-MS temporal profile (B) and intensity distribution (C) of 197Au spikes of AuNPs solution for the detection of single BioNPs probes (n = 5, the confidence limit is 95%). (D) Fluorescence burst data of single Apt-AuNPs probes during a 5 s time window achieved from LIF. (E) Frequency histogram of peak height of single BioNPs probe Gauss fit from LIF. (F) Heatmap of the number of probe labeling on 90 single-cells from nine cell lines. (G) ROC curves showing the high accuracy (AUC = 1) of the SUM signature in cell phenotyping. SUM means integrated dual-mode detection of LIF and ICP-MS with higher accuracy than LIF (AUC = 0.891) or ICP-MS (AUC = 0.383) alone for cell analysis. (H) Close-up of the green and blue triangles marked signals in Figure S30A,B, along with LIF (red line) and ICP-MS (gray line) data coincidence after correction with the Correction time. (I) Content of the OXA and the fluorescence burst data of the same 5 single-cells from nine cell lines. (J) LDA canonical score plots for the response of the BioNPs assemblies to single cells from nine cell lines. Three independent groups to be tested by using biological replicates (n = 3) were used for each data point.
Evaluation of μCytoMS in Analytical Performance
It is important to maintain a steady spike intensity when operating the μCytoMS system. For this purpose, fluorescence/MS signal of single-probe (Figure 3B–E), cell density, and Fluorescence/MS correction time (26.080 ± 0.449 s) were scrutinized with full details given in Figures S27–S29 and Table S1. Briefly, dwell time (tdwell) of 0.1 ms, cell number density of 2.5 × 104 cell/mL, and laser strobing of 10 kHz were employed for our studies.25 These ensured consistent matching of the fluorescence/MS data collection with that of single-cell events.
For the purpose of evaluating the performance of single-cell detection and phenotypic analysis of the μCytoMS system, five PTK7-highly expressed cancer cell lines (MCF-7, A-549, MDA-MB-231, LoVo, and HeLa cells), three PTK7-low expressed cancer cell lines (HepG2, HuH-7, and SK-Hep-1 cells), and one PTK7-negative cell line (Ramos cells) were chosen as the model cells. The cells were trypsinized and ca. 1500 cells were ushered into the microchannel at 20 μL/min, followed by flowing through the LIF View Finder and to ICP-MS for measureing the expression levels of PTK7 and the dosage of OXA in various single cells (Figures S30 and S31). By using the optimized data filtering based on iterative algorithm as reported in our previous work,25,30 1389/1483, 1185/1336, 1405/1392, 1319/1425, 1352/1023, 1440/1483, 1432/1097, 1485/1182, and 968/968 effective single-cell fluorescence bursts/195Pt spike signals corresponding to MCF-7, A549, MDA-MB-231, LoVo, HeLa, HepG2, HuH-7, SK-Hep-1, and Ramos cells, respectively, were acquired within a sampling time interval of 180 s. It is directly related to the total mass of the OXA at the single cell level and the number of BioNPs probes labeled with PTK7 being expressed on the cell surface (Figure S31C). Figure 3F shows that the number of probes labeling on PTK7-highly expressed cell lines was much more than those on PTK7-low expressed cell lines, and much less was observed on PTK7-negative cell lines. The receiver operating characteristic (ROC) curve was adopted for evaluating the reliability of μCytoMS. The results indicated that with respect to detection either by ICP-MS or LIF only, the present detection strategy by both LIF and ICP-MS exhibited better accuracy and resolution for cellular phenotyping (AUC = 1 in total ROC, Figure 3G). The single-cell LIF-ICP-MS signal after screening against the correction time is given in Figures 3H and S29, which ensures subsequent fluorescence and MS data analysis for the corresponding LIF and ICP-MS detection for the same single cell. The compatible fluorescence/MS spectrum matching from nine cell lines further revealed the validation of protein expression and drug uptake (Figure 3I). The results clearly illustrated that PTK7 expression varied significantly even among the same cell line, indicating that cell-to-cell heterogeneity and analysis stochasticity are always present in profiling any cell population.39,40 For single LoVo cells, the number of probes was fewer than other PTK7+ cell lines, while the contents of platinum were found to be more than others. On the other hand for single MCF-7 cells, the number of probes were more than other PTK7+ cell lines, while the contents of platinum were found to be less than others. It is probably because that the variations in the proteins and transport mechanisms of oxaliplatin in the different cell lines, or there are more OXA targets in DNA of colon cancer cells than of the other cancers such as breast cancer, so that OXA would accumulate, in turn, to lead to decreased expression level for PTK7.41 Few probes were labeled on HepG2 cells due to low PTK7 expression, but the accumulation of OXA was highest, probably due to the drug amassing in the liver after the medication was metabolized within the body.42 However, the PTK7 expression on some individual cells was opposite to the overall trend. This indicates that the system can detect rare cells and provide more detailed and evidence-based specific information for personalized clinical diagnosis and treatment. Furthermore, LDA was applied to the μCytoMS response matrix comprising nine fluorescence units of BioNPs probe × nine masses of OXA.43Figure 3J illustrated that all the cell lines were clearly categorized into six individual clusters, which illustrated the competency of the sensitive μCytoMS for phenotyping various tumor cell lines at the single-cell resolution. It is noteworthy that two breast cancer cell lines instead of three hepatoma cell lines were separated into two independent clusters. It suggested that the multiplex strategy has potential for subgroup analysis of homologous tumor cell lines with highly expressed biotarget, which can be promising for future applications for cancer screening.
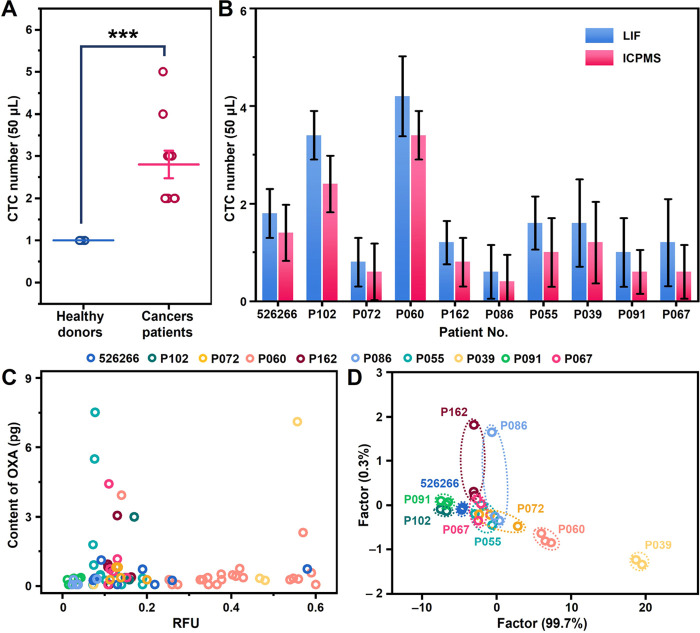
To assess the clinical profiling feasibility using μCytoMS, blood samples from ten breast cancer patients and five healthy volunteers were treated and analyzed (Table S3). The treatment processes are given in the “Blood Processing” section and the Supporting Information under the section of immunofluorescence staining. It was illustrated in Figures 4A and S32 that CTCs were identified in all the patients with 1–4 CTCs per 50 μL of blood (with a mean of 1.24 ± 0.96), while no CTCs were identified in the healthy controls. It is very interesting to see that LIF identified relatively more CTCs than ICP-MS probably due to the direct recognition strategy with LIF (Figure 4B). Thus, lossless single-cell sampling of ICP-MS is extremely critical for future development in analyzing rare cells to collect as much MS data as possible. Figure 4C indicates that the obtained plot of MS data reflects the heterogeneous signaling states of the CTCs identified. In P055’s solitary CTC, a pronounced concentration of oxaliplatin was observed, coupled with a diminished expression of PTK7. This phenomenon could potentially be attributed to the advanced stage of the patient’s breast cancer. The extended duration of chemotherapy might have facilitated an increased accumulation of the drug, thereby exhibiting a degree of therapeutic effectiveness. These data provided important and critical information that will be needed to facilitate individual diagnosis of cancer patients. Furthermore, it was seen that the LDA plots categorized the ten cancer patients into three individual clusters (Figure 4D), which verified that the μCytoMS provided differentiation in the clinical exploration. It is noticed that the clusters of P091 and P102 were very close, which may be related to the potential bone metastases in these three patients.44,45 On the other hand, the widest cluster ranges of P162, P086, and P072 were probably due to the extremely high degree and extent of lymph node involvement. The μCytoMS performed simultaneous quantification of PTK7 expression and platinum-based drug content in individual CTCs, providing nuanced insights into tumor cell metastasis. Elevated platinum levels alongside sustained PTK7 expression implied limited efficacy, contrasting with reduced PTK7 expression, signaling a positive therapeutic response. Variations in PTK7 and drug content among single CTCs from the same blood sample may indicate patient improvement or deterioration, necessitating a comprehensive evaluation. While clinical implementation is a future endeavor, our system’s contribution to patient-specific CTC analysis at the single-cell level establishes a fundamental framework for informing personalized treatment strategies.
Figure 4.
Evaluation of performance of μCytoMS on clinical samples. (A) Quantification of CTCs in 50 μL of blood samples from patients and healthy controls. (B) The numbers of CTCs were detected respectively from 50 μL of cancer patient blood with detection by LIF and ICP-MS. (C) The scatter plot for μCytoMS profiling of individual patient CTCs. (D) LDA canonical score plots for the response of the BioNPs assemblies to single CTCs from ten patients. The data indicate mean ± standard deviation (SD). Three independent groups to be tested by using biological replicates (n = 5) were used for each data point. The Student’s t test was used to compare the means of two groups (A).
Conclusions
A microfluidic cytometry/mass spectrometry system (μCytoMS) was constructed for enabling in situ high-throughput analysis of drug uptake and associated protein expression on single cells by the multiscale integration of BioNPs engineering, microfluidic single-cell alignment, and fluorescence/ICP-MS dual mode detections. The high-affinity BioNPs probes bound selectively with biomarker PTK7 on tumor cells, which enabled LIF sensing of PTK7 expression level induced by OXA uptake, and the dosage of OXA uptake was evaluated by the measurement of 195Pt isotope with ICP-MS. The expression of PTK7 may be further elucidated by correlating with AuNPs conjugated on a single cell. The microchip facilitated purification and alignment of single cells at ultralow flow conditions, and the horizontal interface ensures superefficient matching of the single-cell focusing microfluidic unit to ICP-MS. The system efficiently eliminated the drawbacks of cell damage, sample loss, and time-consuming process of conventional centrifugation pretreatment, and it provides extremely high cell transport efficiency. The system assisted deciphering and phenotyping of the heterogeneity of nine cell lines and CTCs from 10 breast cancer patients at single-cell resolution with free background interference. Taken together, our single-cell analysis will not only enhance characterization of cancer heterogeneity, but also potentially lead to better assessment of treatment efficacy and hopefully to better clinical outcomes.
Methods
Cell Culture and Sample Preparation
The cell lines employed for the present study were acquired from the Cancer Institute and Hospital of Chinese Academy of Medical Science (Beijing, China). These cell lines include two living breast cancer cell lines, MCF-7 and MDA-MB-231, three hepatoma cell lines, HepG2, HuH-7, and SK-Hep-1, one lung carcinoma cancer cell, A549, one colorectal cancer cell, LoVo, one cervical cancer cell, HeLa, and one lymphocytoma cell, Ramos. The cells were cultured in the DMEM and 1640 medium containing 10% fetal bovine serum (FBS) and 100 U/mL penicillin and 100 U/mL streptomycin. The culturing was performed in an incubator at 37 °C under 5% CO2, and the cells were trypsinized and reseeded every 2 days. The cell number was counted by a cell counter (Countess II, Thermofisher, USA) to obtain optimal cell density for ICP-MS analysis (n = 3). For single-cell analysis, the cell suspension was appropriately diluted with a PBS solution containing 1% (w/v) Tween-20.
For exploring the effect of oxaliplatin (OXA) on the expression of PTK7 on the surface of cells, OXA solutions (50 μg/L, incubation for 2 h) in DMEM media were used for cell culture. After culturing, the cells were trypsinized without any centrifuging and washing process and divided into two parts. One part was centrifuged at 900 rpm for 4 min and washed four times with PBS solution to prepare cell suspension for control testing. Another part was introduced into the microfluidic chip directly after treatment with BioNPs for 20 min (10 μL of probes:105 cells). Afterward, the cell suspension was sprayed into aerosol by using an Enya Mist nebulizer and horizontal micro spray chamber after aligning from the OS-Chip, and introduced into the ICP torch for time-resolved ICP-MS measurement.
Blood Processing
Whole blood samples from volunteers and cancer patients (pathologically or clinically confirmed) were obtained from Liaoning Cancer Hospital and Institute (#1) and General Hospital of the Northern Theater Command (#2–#10) (Table S1). This study was approved by the Animal and Medical Ethics Committee of Northeastern University (NEU-EC-2021B006S). The blood samples were drawn into evacuated EDTA-coated collection tubes. All the blood samples were diluted 4 times with PBS, treated with BioNPs and processed by the microchip within 24 h by injecting 200 μL of diluted whole blood into the chip at a flow rate 20 μL/min. In addition, sufficient BioNPs ensure that every artificial CTC in blood is identified and labeled (10 μL of probes/200 μL of diluted whole blood).
Acknowledgments
Financial support from National Natural Science Foundation of China (22274017, 22334003), the Fundamental Research Funds for the Central Universities (N232410019), and the National University of Singapore (NUS) Start-Up Grant (A-8001301-00-00) are highly appreciated. Support provided by the Institute for Health Innovation and Technology at NUS is also acknowledged.
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acsnano.3c12803.
Additional information on materials and chemicals, apparatus and imaging analysis, preparation and characterization of BioNPs conjugates, binding affinities analysis, numerical simulations, the channel Reynolds number and the Dean number, microfluidic device design and fabrication, LIF detector construction, coefficient of focusing (Fco), evaluation of the purifier, the shear stress, operating procedure, cell viability, data processing, different detection parameters, cell transport efficiency, immunofluorescence staining, additional discussions of competition assays for BioNPs and aptamer, optimization of the single-cell alignment unit, optimization of purification unit and comparison with centrifugation, optimization of cell number density and laser strobing, unification of single-cell LIF-ICP-MS signals, applications in real samples, Table S1: ICP-MS instrumental and operational parameters, comparison of current status of single-cell analysis to μCytoMS in this work, patients’ information, and additional figures as noted in the text (PDF)
Single cells in the Purifier and LIF View Finder (MP4)
Author Contributions
X.Z., C.T.L., M.-L.C., and J.-H.W. designed the experiments. X.Z. performed the majority of the experiments, analyzed the data and wrote the draft of the manuscript. X.W. and C.-X.W. prepared the experiments and collected part of the data. X.M. and J.W. analyzed parts of the data. J.-J.B., X.-Y.S., and Y.W. performed part of the experiment and collected data. Ting Yang edited the draft manuscript. C.T.L., M.-L.C., and J.-H.W., edited the final manuscript.
The authors declare no competing financial interest.
Supplementary Material
References
- Zenobi R. Introduction: Frontiers of Analytical Science. Chem. Rev. 2021, 121 (19), 11699–11700. 10.1021/acs.chemrev.1c00652. [DOI] [PubMed] [Google Scholar]
- Chatzimichail S.; Supramaniam P.; Salehi-Reyhani A. Absolute Quantification of Protein Copy Number in Single Cells With Immunofluorescence Microscopy Calibrated Using Single-Molecule Microarrays. Anal. Chem. 2021, 93 (17), 6656–6664. 10.1021/acs.analchem.0c05177. [DOI] [PubMed] [Google Scholar]
- Reza K. K.; Dey S.; Wuethrich A.; Wang J.; Behren A.; Antaw F.; Wang Y. L.; Sina A. I.; Trau M. In Situ Single Cell Proteomics Reveals Circulating Tumor Cell Heterogeneity during Treatment. ACS Nano 2021, 15 (7), 11231–11243. 10.1021/acsnano.0c10008. [DOI] [PubMed] [Google Scholar]
- Zhuang J. L.; Wu Y. J.; Chen L.; Liang S. P.; Wu M. H.; Zhou L. D.; Fan C. H.; Zhang Y. Q. Single-Cell Mobility Analysis of Metastatic Breast Cancer Cells. Adv. Sci. 2018, 5 (12), 1801158. 10.1002/advs.201801158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi L.; Zhang W. C.; Ren X. L.; Xu R. L.; Yang Z. M.; Chen R. Q.; Tu C.; Li Z. H. Cross-Talk of Multiple Types of RNA Modification Regulators Uncovers the Tumor Microenvironment and Immune Infiltrates in Soft Tissue Sarcoma. Frontiers in Immunology 2022, 13, 921223. 10.3389/fimmu.2022.921223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu L. L.; Ding H. M.; Qu X.; Shi X. N.; Yang J. M.; Huang M. J.; Zhang J. L.; Zhang H. M.; Song J.; Zhu L.; Song Y. L.; Ma Y. Q.; Yang C. Y. Fluidic Multivalent Membrane Nanointerface Enables Synergetic Enrichment of Circulating Tumor Cells with High Efficiency and Viability. J. Am. Chem. Soc. 2020, 142 (10), 4800–4806. 10.1021/jacs.9b13782. [DOI] [PubMed] [Google Scholar]
- Zhang H.; Kong S.; Booth A.; Boushaba R.; Levy M. S.; Hoare M. Prediction of shear damage of plasmid DNA in pump and centrifuge operations using an ultra scale-down device. Biotechnol. Prog. 2007, 23 (4), 858–865. 10.1002/bp070066z. [DOI] [PubMed] [Google Scholar]
- Ehrenfest D. M. D.; Pinto N. R.; Pereda A.; Jimenez P.; Del Corso M.; Kang B. S.; Nally M.; Lanata N.; Wang H. L.; Quirynen M. The impact of the centrifuge characteristics and centrifugation protocols on the cells, growth factors, and fibrin architecture of a leukocyte- and platelet-rich fibrin (L-PRF) clot and membrane. Platelets 2018, 29 (2), 171–184. 10.1080/09537104.2017.1293812. [DOI] [PubMed] [Google Scholar]
- Amantonico A.; Oh J. Y.; Sobek J.; Heinemann M.; Zenobi R. Mass spectrometric method for analyzing metabolites in yeast with single cell sensitivity. Angew. Chem.-Int. Ed. 2008, 47 (29), 5382–5385. 10.1002/anie.200705923. [DOI] [PubMed] [Google Scholar]
- Do T. D.; Comi T. J.; Dunham S. J. B.; Rubakhin S. S.; Sweedler J. V. Single Cell Profiling Using Ionic Liquid Matrix-Enhanced Secondary Ion Mass Spectrometry for Neuronal Cell Type Differentiation. Anal. Chem. 2017, 89 (5), 3078–3086. 10.1021/acs.analchem.6b04819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu S. T.; Liu M. X.; Bai Y.; Liu H. W. Multi-Dimensional Organic Mass Cytometry: Simultaneous Analysis of Proteins and Metabolites on Single Cells. Angew. Chem.-Int. Ed. 2021, 60 (4), 1806–1812. 10.1002/anie.202009682. [DOI] [PubMed] [Google Scholar]
- Zhu Y.; Clair G.; Chrisler W. B.; Shen Y. F.; Zhao R.; Shukla A. K.; Moore R. J.; Misra R. S.; Pryhuber G. S.; Smith R. D.; Ansong C.; Kelly R. T. Proteomic Analysis of Single Mammalian Cells Enabled by Microfluidic Nanodroplet Sample Preparation and Ultrasensitive NanoLC-MS. Angew. Chem.-Int. Ed. 2018, 57 (38), 12370–12374. 10.1002/anie.201802843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Carlo D.; Irimia D.; Tompkins R. G.; Toner M. Continuous inertial focusing, ordering, and separation of particles in microchannels. Proc. Natl. Acad. Sci. U.S.A. 2007, 104 (48), 18892–18897. 10.1073/pnas.0704958104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pei H.; Li L.; Han Z.; Wang Y.; Tang B. Recent advances in microfluidic technologies for circulating tumor cells: enrichment, single-cell analysis, and liquid biopsy for clinical applications. Lab Chip 2020, 20 (21), 3854–3875. 10.1039/D0LC00577K. [DOI] [PubMed] [Google Scholar]
- Yamada M.; Seki M. Hydrodynamic filtration for on-chip particle concentration and classification utilizing microfluidics. Lab Chip 2005, 5 (11), 1233–1239. 10.1039/b509386d. [DOI] [PubMed] [Google Scholar]
- Zhang X.; Wei X.; Men X.; Wu C.-X.; Bai J.-J.; Li W.-T.; Yang T.; Chen M.-L.; Wang J.-H. Dual-Multivalent-Aptamer-Conjugated Nanoprobes for Superefficient Discerning of Single Circulating Tumor Cells in a Microfluidic Chip with Inductively Coupled Plasma Mass Spectrometry Detection. ACS Appl. Mater. Interfaces 2021, 13 (36), 43668–43675. 10.1021/acsami.1c11953. [DOI] [PubMed] [Google Scholar]
- Bounab Y.; Eyer K.; Dixneuf S.; Rybczynska M.; Chauvel C.; Mistretta M.; Tran T.; Aymerich N.; Chenon G.; Llitjos J.-F.; Venet F.; Monneret G.; Gillespie I. A.; Cortez P.; Moucadel V.; Pachot A.; Troesch A.; Leissner P.; Textoris J.; Bibette J.; Guyard C.; Baudry J.; Griffiths A. D.; Védrine C. Dynamic single-cell phenotyping of immune cells using the microfluidic platform DropMap. Nat. Protoc. 2020, 15 (9), 2920–2955. 10.1038/s41596-020-0354-0. [DOI] [PubMed] [Google Scholar]
- Xu S.; Ma W.; Bai Y.; Liu H. Ultrasensitive Ambient Mass Spectrometry Immunoassays: Multiplexed Detection of Proteins in Serum and on Cell Surfaces. J. Am. Chem. Soc. 2019, 141 (1), 72–75. 10.1021/jacs.8b10853. [DOI] [PubMed] [Google Scholar]
- Pichaandi J.; Zhao G.; Bouzekri A.; Lu E.; Ornatsky O.; Baranov V.; Nitz M.; Winnik M. A. Lanthanide nanoparticles for high sensitivity multiparameter single cell analysis. Chemical Science 2019, 10 (10), 2965–2974. 10.1039/C8SC04407D. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y.; Chao Y.; Miao Y.; Li Y.; Xu T.; Li S.; Peng J. Time-resolved ICP-MS analysis of mineral element contents and distribution patterns in spermatogenic cells of different types. Anal. Chim. Acta 2023, 1255, 341054. 10.1016/j.aca.2023.341054. [DOI] [PubMed] [Google Scholar]
- Zhou Y.; Li H.; Sun H. Cytotoxicity of arsenic trioxide in single leukemia cells by time-resolved ICP-MS together with lanthanide tags. Chem. Commun. 2017, 53 (20), 2970–2973. 10.1039/C7CC00143F. [DOI] [PubMed] [Google Scholar]
- Tsang C.-N.; Ho K.-S.; Sun H.; Chan W.-T. Tracking Bismuth Antiulcer Drug Uptake in Single Helicobacter pylori Cells. J. Am. Chem. Soc. 2011, 133 (19), 7355–7357. 10.1021/ja2013278. [DOI] [PubMed] [Google Scholar]
- Yang Y.-S. S.; Atukorale P. U.; Moynihan K. D.; Bekdemir A.; Rakhra K.; Tang L.; Stellacci F.; Irvine D. J. High-throughput quantitation of inorganic nanoparticle biodistribution at the single-cell level using mass cytometry. Nat. Commun. 2017, 8 (1), 14069. 10.1038/ncomms14069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Q.; Ge W.; Wang T.; Lan J.; Martínez-Jarquín S.; Wolfrum C.; Stoffel M.; Zenobi R. High-Throughput Single-Cell Mass Spectrometry Reveals Abnormal Lipid Metabolism in Pancreatic Ductal Adenocarcinoma. Angew. Chem., Int. Ed. 2021, 60 (46), 24534–24542. 10.1002/anie.202107223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C.; Wei X.; Men X.; Zhang X.; Yu Y.-L.; Xu Z.-R.; Chen M.-L.; Wang J.-H. Two-Dimensional Cytometry Platform for Single-Particle/Cell Analysis with Laser-Induced Fluorescence and ICP-MS. Anal. Chem. 2021, 93 (23), 8203–8209. 10.1021/acs.analchem.1c00484. [DOI] [PubMed] [Google Scholar]
- Song Y.; Shi Y.; Huang M.; Wang W.; Wang Y.; Cheng J.; Lei Z.; Zhu Z.; Yang C. Bioinspired Engineering of a Multivalent Aptamer-Functionalized Nanointerface to Enhance the Capture and Release of Circulating Tumor Cells. Angew. Chem.-Int. Ed. 2019, 58 (8), 2236–2240. 10.1002/anie.201809337. [DOI] [PubMed] [Google Scholar]
- Bendall S. C.; Simonds E. F.; Qiu P.; Amir E.-a. D.; Krutzik P. O.; Finck R.; Bruggner R. V.; Melamed R.; Trejo A.; Ornatsky O. I.; Balderas R. S.; Plevritis S. K.; Sachs K.; Pe’er D.; Tanner S. D.; Nolan G. P. Single-Cell Mass Cytometry of Differential Immune and Drug Responses Across a Human Hematopoietic Continuum. Science 2011, 332 (6030), 687–696. 10.1126/science.1198704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu L.-L.; Zhang Z.-L.; Tang M.; Zhu D.-L.; Dong X.-J.; Hu J.; Qi C.-B.; Tang H.-W.; Pang D.-W. Spectrally Combined Encoding for Profiling Heterogeneous Circulating Tumor Cells Using a Multifunctional Nanosphere-Mediated Microfluidic Platform. Angew. Chem.-Int. Ed. 2020, 59 (28), 11240–11244. 10.1002/anie.201914468. [DOI] [PubMed] [Google Scholar]
- Liu C.; Zhao J.; Tian F.; Chang J.; Zhang W.; Sun J. λ-DNA- and Aptamer-Mediated Sorting and Analysis of Extracellular Vesicles. J. Am. Chem. Soc. 2019, 141 (9), 3817–3821. 10.1021/jacs.9b00007. [DOI] [PubMed] [Google Scholar]
- Zhang X.; Wei X.; Men X.; Jiang Z.; Ye W.-Q.; Chen M.-L.; Yang T.; Xu Z.-R.; Wang J.-H. Inertial-Force-Assisted, High-Throughput, Droplet-Free, Single-Cell Sampling Coupled with ICP-MS for Real-Time Cell Analysis. Anal. Chem. 2020, 92 (9), 6604–6612. 10.1021/acs.analchem.0c00376. [DOI] [PubMed] [Google Scholar]
- Mu T.; Toyoda H.; Kimura Y.; Yamada M.; Utoh R.; Umeno D.; Seki M. Laborless, Automated Microfluidic Tandem Cell Processor for Visualizing Intracellular Molecules of Mammalian Cells. Anal. Chem. 2020, 92 (3), 2580–2588. 10.1021/acs.analchem.9b04288. [DOI] [PubMed] [Google Scholar]
- Kopetz S.; Lesslie D. P.; Dallas N. A.; Park S. I.; Johnson M.; Parikh N. U.; Kim M. P.; Abbruzzese J. L.; Ellis L. M.; Chandra J.; Gallick G. E. Synergistic Activity of the Src Family Kinase Inhibitor Dasatinib and Oxaliplatin in Colon Carcinoma Cells Is Mediated by Oxidative Stress. Cancer Res. 2009, 69 (9), 3842–3849. 10.1158/0008-5472.CAN-08-2246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Carlo D. Inertial microfluidics. Lab Chip 2009, 9 (21), 3038–3046. 10.1039/b912547g. [DOI] [PubMed] [Google Scholar]
- Nivedita N.; Papautsky I. Continuous separation of blood cells in spiral microfluidic devices. Biomicrofluidics 2013, 7 (5), 14. 10.1063/1.4819275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gou Y. X.; Zhang S.; Sun C. K.; Wang P.; You Z.; Yalikun Y.; Tanaka Y.; Ren D. H. Sheathless Inertial Focusing Chip Combining a Spiral Channel with Periodic Expansion Structures for Efficient and Stable Particle Sorting. Anal. Chem. 2020, 92 (2), 1833–1841. 10.1021/acs.analchem.9b03692. [DOI] [PubMed] [Google Scholar]
- Moore T. L.; Rodriguez-Lorenzo L.; Hirsch V.; Balog S.; Urban D.; Jud C.; Rothen-Rutishauser B.; Lattuada M.; Petri-Fink A. Nanoparticle colloidal stability in cell culture media and impact on cellular interactions. Chem. Soc. Rev. 2015, 44 (17), 6287–6305. 10.1039/C4CS00487F. [DOI] [PubMed] [Google Scholar]
- Wei X.; Zhang X.; Guo R.; Chen M.-L.; Yang T.; Xu Z.-R.; Wang J.-H. A Spiral-Helix (3D) Tubing Array That Ensures Ultrahigh-Throughput Single-Cell Sampling. Anal. Chem. 2019, 91 (24), 15826–15832. 10.1021/acs.analchem.9b04122. [DOI] [PubMed] [Google Scholar]
- Cao Y.; Deng B.; Yan L.; Huang H. An environmentally-friendly, highly efficient, gas pressure-assisted sample introduction system for ICP-MS and its application to detection of cadmium and lead in human plasma. Talanta 2017, 167, 520–525. 10.1016/j.talanta.2017.02.057. [DOI] [PubMed] [Google Scholar]
- Altschuler S. J.; Wu L. F. Cellular Heterogeneity: Do Differences Make a Difference?. Cell 2010, 141 (4), 559–563. 10.1016/j.cell.2010.04.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Losick R.; Desplan C. Stochasticity and cell fate. Science 2008, 320 (5872), 65–8. 10.1126/science.1147888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misset J. L.; Bleiberg H.; Sutherland W.; Bekradda M.; Cvitkovic E. Oxaliplatin clinical activity: a review. Critical Reviews in Oncology/Hematology 2000, 35 (2), 75–93. 10.1016/S1040-8428(00)00070-6. [DOI] [PubMed] [Google Scholar]
- Almazroo O. A.; Miah M. K.; Venkataramanan R. Drug Metabolism in the Liver. Clinics in Liver Disease 2017, 21 (1), 1. 10.1016/j.cld.2016.08.001. [DOI] [PubMed] [Google Scholar]
- Zhang X.-W.; Liu M.-X.; He M.-Q.; Chen S.; Yu Y.-L.; Wang J.-H. Integral Multielement Signals by DNA-Programmed UCNP-AuNP Nanosatellite Assemblies for Ultrasensitive ICP-MS Detection of Exosomal Proteins and Cancer Identification. Anal. Chem. 2021, 93 (16), 6437–6445. 10.1021/acs.analchem.1c00152. [DOI] [PubMed] [Google Scholar]
- Jiang G.; Zhang M.; Yue B.; Yang M.; Carter C.; Al-Quran S. Z.; Li B.; Li Y. PTK7: A new biomarker for immunophenotypic characterization of maturing T cells and T cell acute lymphoblastic leukemia. Leuk. Res. 2012, 36 (11), 1347–1353. 10.1016/j.leukres.2012.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiao H.; Cui Z. W.; Yang S. B.; Ji D. K.; Wang Y. G.; Yang Y.; Han X. G.; Fan Q. M.; Qin A.; Wang T. Y.; He X. P.; Bu W. B.; Tang T. T. Targeting Osteocytes to Attenuate Early Breast Cancer Bone Metastasis by Theranostic Upconversion Nanoparticles with Responsive Plumbagin Release (vol 11, pg 7259, 2017). ACS Nano 2019, 13 (4), 4857–4857. 10.1021/acsnano.9b02283. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.