Abstract
Base pairing between U2 snRNA and the branchpoint sequence (BPS) is essential for pre-mRNA splicing. Because the metazoan BPS is short and highly degenerate, this interaction alone is insufficient for specific binding of U2 snRNP. The splicing factor U2AF binds to the pyrimidine tract at the 3′ splice site in the earliest spliceosomal complex, E, and is essential for U2 snRNP binding in the spliceosomal complex A. We show that the U2 snRNP protein SAP 155 UV cross-links to pre-mRNA on both sides of the BPS in the A complex. SAP 155’s downstream cross-linking site is immediately adjacent to the U2AF binding site, and the two proteins interact directly in protein-protein interaction assays. Using UV cross-linking, together with functional analyses of pre-mRNAs containing duplicated BPSs, we show a direct correlation between BPS selection and UV cross-linking of SAP 155 on both sides of the BPS. Together, our data are consistent with a model in which U2AF binds to the pyrimidine tract in the E complex and then interacts with SAP 155 to recruit U2 snRNP to the BPS.
The pre-mRNA splicing reaction is carried out with extreme precision in order to generate mRNAs that encode functional proteins. The accuracy of splicing depends on multiple sequence elements located at the 5′ and 3′ splice sites, at the branch site, and within exons. Networks of RNA-RNA, RNA-protein, and protein-protein interactions involving each of these sequence elements contribute to the specificity of splicing. Additional specificity is derived from the recognition of each sequence element multiple times prior to the two catalytic steps of splicing. These successive recognition events occur as the spliceosomal complexes E, A, B, and C assemble on pre-mRNA in a stepwise pathway (for reviews, see references 17, 25, and 32).
One of the critical sequence elements in the intron is the branchpoint sequence (BPS). This element contains an adenosine that functions as the nucleophile for catalytic step I of splicing. Despite its key role in splicing, the BPS is weakly conserved in metazoans, and additional elements are required for BPS recognition. The most important of these is the pyrimidine tract located immediately downstream from the BPS. The splicing factor U2AF, which is composed of two subunits (U2AF65 and U2AF35), binds to the pyrimidine tract in the E complex, with U2AF65 directly contacting the pre-mRNA (4, 33 [for reviews, see references 17, 25, and 32]). The essential splicing factor SF1 (also known as mBBP) interacts with U2AF65 and also has sequence specificity for the BPS (2, 5, 16). Thus, this network of interactions is thought to function in the initial recognition of the pyrimidine tract and BPS.
The BPS is recognized a second time during spliceosome assembly by formation of a duplex between the BPS and U2 snRNA (for review, see reference 19). This duplex is first established in the A complex and plays an essential role in splicing by specifying the branch-site adenosine as the nucleophile for catalytic step I (23). Two multisubunit splicing factors, SF3a and SF3b, are components of U2 snRNP and are required for binding to the branch site (for reviews, see references 17, 25, and 32). SF3a consists of three subunits (SAPs 61, 62, and 114), and SF3b is thought to consist of at least four subunits (SAPs 49, 130, 145, and 155). All of the SF3a and SF3b subunits, except SAP 130, UV cross-link to pre-mRNA in the A complex. These proteins bind sequence independently to a 25-nucleotide (nt) region immediately upstream of the branch site and are thought to function in part by anchoring U2 snRNP tightly to the pre-mRNA (for reviews, see references 17, 25, and 32).
The high level of degeneracy of the mammalian BPS indicates that specific mechanisms must exist for targeting U2 snRNP to the BPS. U2AF65 is thought to interact directly with the BPS and promote annealing of U2 snRNA (28). However, U2AF65 does not specifically recognize U2 snRNA sequences. Thus, other mechanisms must be responsible for targeting this snRNA to the BPS. The proteins that bind to pre-mRNA in the vicinity of the BPS early in spliceosome assembly (discussed above) are candidates for factors involved in this process. In addition, Fleckner and coworkers (13) identified a putative RNA helicase, UAP56, that interacts with U2AF65 and is required for U2 snRNP binding. Other proteins that interact with pre-mRNA near or at the BPS early in spliceosome assembly include BPS72, BPS70 (11), p80, and p14 (18, 24). None of these proteins has been shown to play a direct role in targeting U2 snRNP to the BPS.
In this study, we show that the SF3b and U2 snRNP component SAP 155 cross-links to pre-mRNA on both sides of the BPS in the A complex and also can interact directly with U2AF. Moreover, cross-linking of SAP 155 in the A complex correlates with BPS selection in a pre-mRNA containing duplicate BPSs. The SAP 155-U2AF interaction is conserved in Schizosaccharomyces pombe, suggesting that it is functionally important. In contrast to all other factors mentioned above, SAP 155 is the only example of a protein with the combined characteristics of (i) interacting with U2 snRNP, (ii) directly contacting U2AF, and (iii) UV cross-linking to pre-mRNA immediately next to U2AF (i.e., near the BPS). Furthermore, temporally, U2AF UV cross-linking to the pyrimidine tract in the E complex is followed by SAP 155 UV cross-linking on both sides of the BPS in the A complex. Thus, together, these observations raise the possibility that recruitment of U2 snRNP to the branch site involves binding of U2AF to the pyrimidine tract followed by an interaction with SAP 155 to position U2 snRNP at the BPS.
MATERIALS AND METHODS
Plasmids.
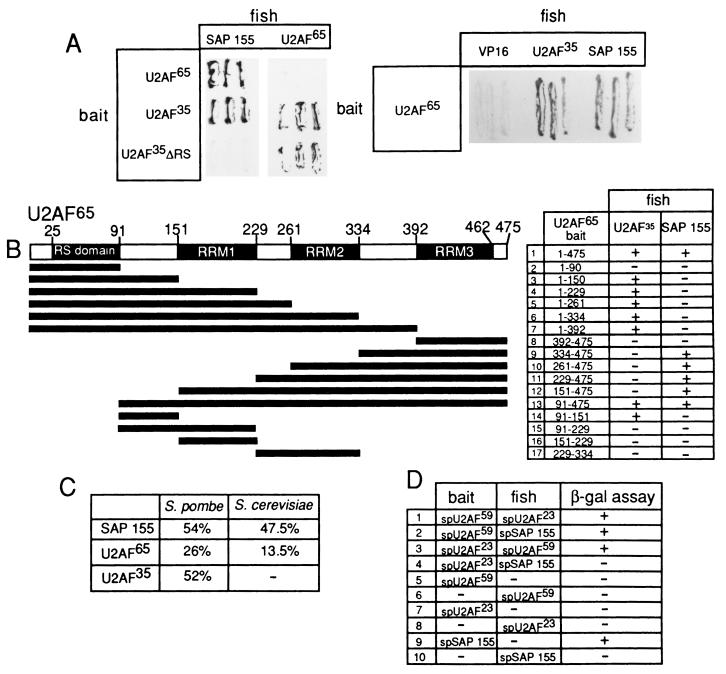
pAdML was described in reference 14. Derivatives of this pre-mRNA (see Fig. 6 and 7) are identical to pAdML, except where indicated in the figures. Glutathione S-transferase (GST) fusion constructs were made by inserting the indicated sequences (see Fig. 4) into the pGEX2TK vector (Pharmacia). S. pombe SAP 155 was cloned from an S. pombe DNA library by PCR with oligonucleotides 5′ CGCGGATCCATGTCAACTGGTACGTATCC 3′ and 5′ AAAACTGCAGTTAGATGCAAATATGTAAAG 3′ and subcloned into the BamHI and PstI sites of pGBT9 and pGAD424. The bait and fish plasmids used in Fig. 5A and B were constructed by insertion of the indicated sequences downstream of either the LexA DNA binding domain in the pEG202 bait vector or the VP16 activator domain in the fish vector pVP16. Vectors were transformed and cells were grown on selective media (HULL, a medium lacking histidine or leucine to select for bait and fish, respectively, and uracil and lysine to maintain selection). The bait and fish plasmids used in Fig. 5D were constructed by inserting the indicated sequences downstream of either the GAL4 DNA binding domain in the bait plasmid pGBT9 (Clontech) or the GAL4 activation domain in the fish plasmid pGAD424 (Clontech [30]).
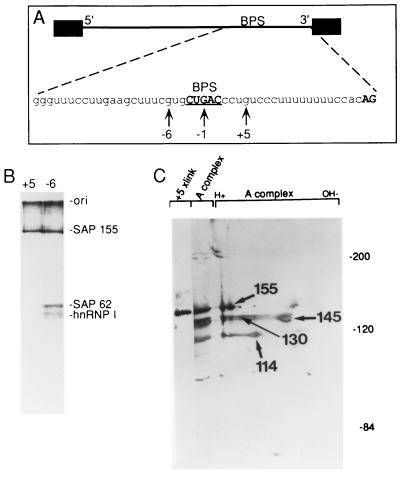
FIG. 6.
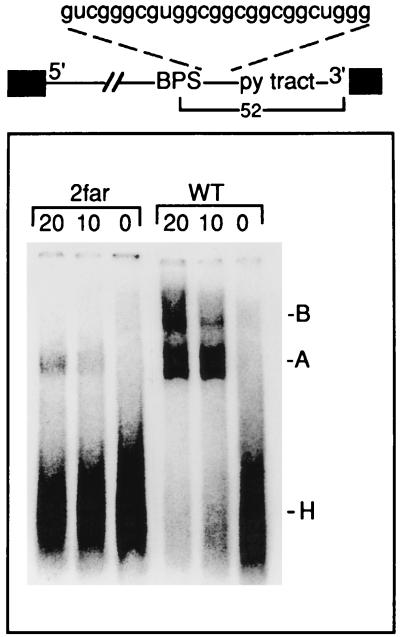
The distance between the BPS and pyrimidine (py) tract is critical for A complex assembly. A spacer of the sequence indicated was inserted immediately downstream of the BPS to generate the pre-mRNA 2far. The distance between the BPS and 3′ splice site is 52 nt. 2far or wild-type pre-mRNA was incubated under splicing conditions for the times indicated (in minutes) and then fractionated on a native gel. The H, A, and B complexes are indicated.
FIG. 7.
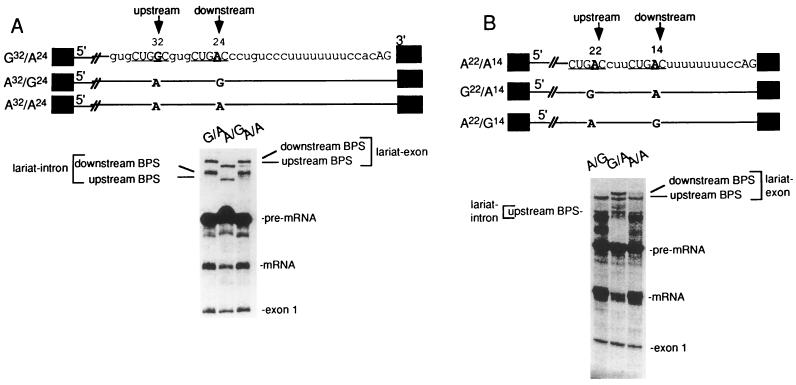
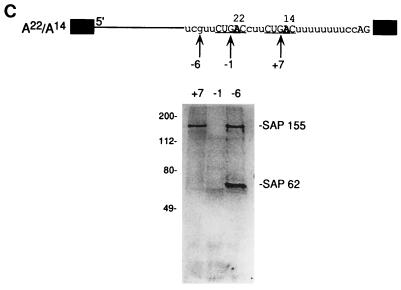
SAP 155 cross-linking to pre-mRNA correlates with BPS selection (A) The BPS closest to the 3′ splice site is selected. The structures of pre-mRNAs containing duplicated BPSs are shown. Pre-mRNAs were incubated under splicing conditions for 40 min, and total RNA was fractionated on a 15% polyacrylamide denaturing gel. The bands corresponding to spliced products and intermediates are indicated. (B) An optimal distance between the BPS and 3′ splice site is necessary for BPS selection. The structures of pre-mRNAs containing duplicated BPSs are indicated. The branch-site adenosine is in boldface, and the distance from this adenosine to the 3′ splice site is shown. The pre-mRNAs were incubated under splicing conditions for 45 min, and then total RNA was fractionated on a 13% polyacrylamide denaturing gel. The splicing intermediates and products are indicated. (C) SAP 155 UV cross-links on both sides of the functional BPS. A22/A14 pre-mRNA was 32P site-specifically labeled at the indicated guanosines, assembled into the A complex, isolated by gel filtration, and UV cross-linked. After RNase A treatment, cross-linked proteins were fractionated by SDS-polyacrylamide gel electrophoresis, and cross-linked proteins were detected by phosphorimager analysis. SAPs 155 and 62 are indicated.
FIG. 4.
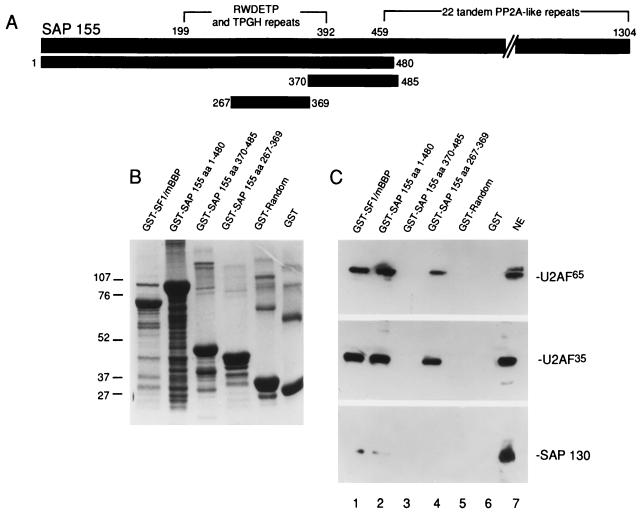
A specific amino-terminal region of SAP 155 is sufficient to mediate the interaction with U2AF. (A) Structure of SAP 155. The amino-terminal RWDETP and TPGH repeats and carboxy-terminal PP2A-like repeats are shown (29). Numbers indicate amino acids. The portions of SAP 155 that were used to make fusion proteins are indicated. (B) Coomassie stain of GST fusion proteins. Markers are indicated to the left in kilodaltons. (C) Lanes 1 to 6: GST fusion proteins bound to glutathione beads were incubated with a 25-μl aliquot of nuclear extract and washed, and then Western analysis of the bound proteins was carried out. Total nuclear extract (NE) (4 μl) is shown in lane 7. The blot was probed successively with U2AF65, U2AF35, and SAP 130 antibodies.
FIG. 5.
Analysis of the SAP 155-U2AF interaction by the yeast two-hybrid assay. (A) β-Galactosidase filter lift assays were done with cells transformed with the indicated bait and fish vectors. (B) Structure of U2AF65 and derivatives of U2AF65 that were used as baits for two-hybrid assays. U2AF35 and SAP 155 (aa 171 to 775) were used as fish. The interactions were assayed with the β-galactosidase filter assay. +, interactions between the fish and bait (blue color production on the filter). −, no interaction (relative to the VP16 negative control). (C) The percent identity between Homo sapiens, S. pombe, and S. cerevisiae SAP 155, U2AF35, U2AF65 homologs is shown. (D) Two-hybrid assays of S. pombe (sp) U2AF65, U2AF35, and SAP 155. The fish and baits are indicated. As with human SAP 155, S. pombe SAP 155 used as a bait transactivates the β-galactosidase (β-gal) promoter in the absence of a fish construct.
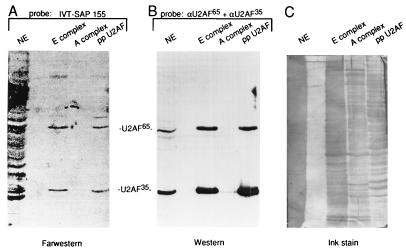
Far-Western analysis.
Partially purified U2AF (ppU2AF) was prepared by mixing nuclear extract with poly(U)-Sepharose and washing it five times in 250 mM NaCl–10 mM Tris (pH 8), followed by elution of bound proteins with sodium dodecyl sulfate (SDS)-sample buffer (33). Spliceosomal complexes E and A were affinity purified as described previously (4, 27). For far-Western blots, aliquots of nuclear extract, ppU2AF, or purified spliceosomes were fractionated on an SDS–9% polyacrylamide gel, blotted onto nitrocellulose or polyvinylidene difluoride membranes, and probed with in vitro-translated (IVT) SAP 155 (35). U2AF65 and SAP 155 were IVT by using a coupled transcription-translation system (Promega). Five micrograms of RNase A was added after the translation reaction, and this mixture was then incubated for 10 min prior to the probing of the blots.
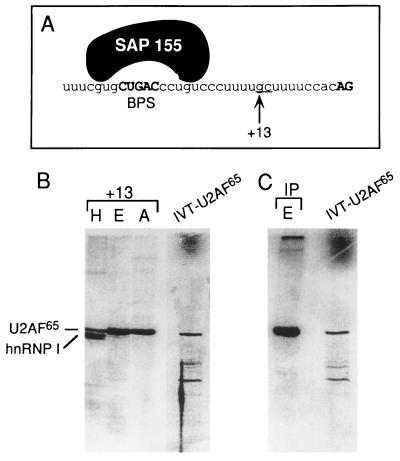
UV cross-linking of complexes assembled on site-specifically labeled pre-mRNA.
AdML pre-mRNAs containing a single 32P-labeled guanosine residue were synthesized by the technique of Moore and Sharp (20) with the modifications used by Chiara et al. (11). Spliceosomal complexes were assembled on the site-specifically labeled pre-mRNA, fractionated by gel filtration, and UV cross-linked (11). A 200-μl aliquot was treated with RNase A (10 μg). After acetone precipitation, proteins were fractionated on a SDS–polyacrylamide gel and detected by phosphorimager (11). For immunoprecipitation of cross-linked U2AF65, pre-mRNA site specifically labeled at the 3′ splice site (see Fig. 2, +13) was assembled into the E complex, isolated by gel filtration, and UV cross-linked. A 500-μl aliquot of a fraction containing the E complex was incubated with RNase A (10 μg) for 30 min at 37°C. The fraction was then mixed overnight at 4°C with U2AF65 polyclonal antibodies (36) immobilized on protein A-Trisacryl beads. After being washed with 500 mM NaCl and 1% Nonidet P-40, total bound protein was fractionated on a 9% SDS gel.
FIG. 2.
U2AF65 UV cross-linking next to SAP 155 in the A complex. (A) Nucleotide sequence of the 3′ portion of the AdML intron. The arrow indicates the G residue that was site-specifically labeled. The SAP 155 cross-linking sites in the A complex are indicated. At the +13 site, the UU was changed to GC in order to transcribe the 3′ RNA for site-specific labeling (see Materials and Methods). This alteration has no effect on A complex assembly or splicing. (B) The proteins that cross-link at the +13 site in the H, E, and A complexes are shown next to IVT U2AF65 (as a marker). (C) Pre-mRNA site-specifically labeled at +13 was assembled into the E complex and UV cross-linked, and then an immunoprecipitation (IP) with U2AF65 antibodies was carried out. IVT U2AF65 was run as a marker.
GST pull down assays.
GST fusion proteins were expressed in Escherichia coli, bound to glutathione-Sepharose beads, and incubated for 1 h at 4°C with 25 μl of nuclear extract. Each sample was washed six times with NETN (0.5% Nonidet P-40, 20 mM Tris [pH 8], 100 mM NaCl, 1 mM EDTA, 2 mM phenylmethylsulfonyl fluoride). Bound proteins were eluted in protein sample buffer, fractionated on SDS gels, transferred to polyvinylidene difluoride membranes, and probed with the indicated antibodies.
Yeast two-hybrid assays.
For Fig. 5A and B, β-galactosidase filter lift assays were used to test for two-hybrid interactions (12). All interactions that are denoted as positive turned blue in 10 or greater independent assays compared to the negative control at the same time point. For Fig. 5D, interactions were assayed as described by Wentz-Hunter and Potashkin (30).
Splicing assays.
Splicing reaction mixtures (25 μl) containing 32P-labeled pre-mRNAs (20 ng) were incubated under standard splicing conditions for 45 min. Total RNA was isolated and separated on polyacrylamide denaturing gels. For native gel analysis, sample dye containing 2.5 mg of heparin per ml was added to splicing reactions mixtures, which were then fractionated on 4% polyacrylamide nondenaturing gels.
RESULTS
SAP 155 interacts with pre-mRNA on both sides of the BPS.
To identify splicing factors involved in targeting U2 snRNP to the BPS, we analyzed proteins that UV cross-link to pre-mRNA in the vicinity of the BPS in spliceosomal complex A. Pre-mRNAs were site-specifically labeled with 32P at guanosines located 6 nt upstream (−6) or 5 nt downstream (+5) of the branch-site adenosine (Fig. 1A), assembled into the A complex, and then isolated by gel filtration (see Materials and Methods). As shown in Fig. 1B (lane −6), and consistent with previous work (15), the SF3a component SAP 62 and the SF3b component SAP 155 cross-link at the −6 site (low levels of hnRNP I from contaminating H complex are also detected at −6 [Fig. 1B, −6]). A single high-molecular-weight protein, comigrating with SAP 155 on SDS and two-dimensional gels, was detected at +5 (Fig. 1B and C, +5). This protein does not cross-link at −1 (11) (see Fig. 7C). In addition, the +5 and −6 cross-linked proteins are present in affinity-purified A complex and comigrate with SAP 155 on two-dimensional gels (reference 15 and data not shown). We conclude that SAP 155 UV cross-links to pre-mRNA on both sides of, but not directly at, the BPS in the A complex. Coomassie-stained gels show that SAP 155 is equimolar with the other high-molecular-weight U2 snRNP proteins in the A complex (data not shown). Assuming that these proteins all bind to pre-mRNA as monomers, a single SAP 155 monomer may contact both sides of the BPS. We cannot exclude the alternative possibility that there is a mixture of A complexes in which SAP 155 binds upstream of the BPS in one population and downstream of the BPS in another population. However, we have obtained no evidence for a temporal order of SAP 155 cross-linking at either the upstream or downstream site during spliceosome assembly (data not shown).
FIG. 1.
U2 snRNP protein SAP 155 UV cross-linking to pre-mRNA on both sides of the branch site. (A) Schematic of pre-mRNA. The nucleotide sequence of the 3′ portion of the AdML intron is shown. The BPS and AG dinucleotide at the 3′ splice site are in boldface, and the BPS is also underlined. The arrows indicate the G residues that were labeled, and the numbers are relative to the branch-site adenosine. (B) Pre-mRNAs labeled at −6 or +5 were assembled into the A complex, isolated by gel filtration, and UV cross-linked. After treatment with RNase A, proteins were fractionated on a 9% SDS gel (equal counts per minute of each pre-mRNA were loaded). The positions of SAPs 155 and 62 and hnRNP I are indicated. hnRNP I cross-linking is due to contamination of the A complex with the H complex in this experiment. (C) The +5 cross-linked sample was fractionated on a 6% SDS gel next to the A complex and next to the A complex separated in two dimensions. The A complex samples were detected by staining and the +5 cross-link (xlink) was detected by phosphorimager.
U2AF65 UV cross-links immediately downstream of SAP 155 in the A complex.
To identify proteins near SAP 155 on the pre-mRNA in the A complex, we site-specifically labeled pre-mRNA 10 nt downstream from the branch site. Only SAP 155 was detected at this site, and the level was extremely low relative to that detected at +5 and −6 (data not shown). In contrast, when pre-mRNA was labeled at a site 13 nt from the branch site (Fig. 2A, +13), one major band, which cofractionates with IVT U2AF65, was detected in both the E and A complexes (Fig. 2B, lanes E and A) (33, 34). As expected (33, 34), this cross-linked band is specifically immunoprecipitated by antibodies to U2AF65 (Fig. 2C, lane IP), but not by control antibodies (data not shown).
U2AF65 cross-linking was detected at similar levels in the E and A complexes (Fig. 2B, compare lanes E and A). This is consistent with previous work showing that U2AF65 is detected in both the E and A complexes when they are isolated by gel filtration alone and assayed by UV cross-linking (8). (Note that when the complexes are isolated by the more stringent two-step gel filtration-affinity chromatography procedure, U2AF65 is detected at much lower levels in the A complex than in the E complex [4]. Thus, together, these data indicate that U2AF65 undergoes a conformational change during the E-to-A complex transition.) We conclude that SAP 155 is the closest cross-linked protein upstream of U2AF65 on the pre-mRNA in the A complex.
U2AF and SAP 155 interact directly.
The observation that SAP 155 and U2AF65 UV cross-link to adjacent sites on the pre-mRNA in the A complex raised the possibility that these proteins may interact with each other directly. To test this possibility, we used IVT SAP 155 to probe a far-Western blot containing nuclear extract, purified E complex, purified A complex, or a partially purified preparation of U2AF (ppU2AF; see Materials and Methods and reference 33). (Note that RNase A was added to the probes to disrupt the potential for RNA-mediated interactions.) An ink stain of the blot shows that there are a large number of proteins in each lane (Fig. 3C). Significantly, the IVT SAP 155 probe detects only two main bands in the E complex and in ppU2AF (Fig. 3A). The same two proteins were also detected by antibodies to U2AF65 and U2AF35 (Fig. 3B). Neither the SAP 155 probe nor the U2AF antibodies detected bands in the A complex (Fig. 3A and B), consistent with studies showing that U2AF is largely dissociated from the affinity-purified A complex (4 [also see above]). The observation that SAP 155 interacts with U2AF65 and U2AF35 on the far-Western blot and does not interact with a very large number of other abundant proteins present on this blot indicates that the SAP 155-U2AF65 and the SAP 155-U2AF35 interactions are specific. In addition, when IVT-U2AF65 was used to probe a far-Western blot containing the purified A complex, only SAP 155 was detected (data not shown). Thus, SAP 155 is the only U2 snRNP protein detected by U2AF.
FIG. 3.
U2AF and SAP 155 interact directly. (A) IVT SAP 155 (aa 148 to 775) was used to probe a blot containing nuclear extract (NE), E complex, A complex, or partially purified U2AF (ppU2AF). U2AF65 and U2AF35 are indicated. (B) The blot in panel B was probed with U2AF65 and U2AF35 antibodies (α) (Western blotting). (C) An ink stain of the blot probed in panels A and B.
The SAP 155 probe detected several proteins when a large amount of total nuclear extract was used for the far-Western analysis (Fig. 3A and C, NE). However, it is unlikely that these interactions are specific. This conclusion is based on two-dimensional far-Western analysis, which revealed that the SAP 155 probe detects the proteins that are among the most abundant in the nuclear extract (data not shown). None of the proteins detected by SAP 155 in the total extract appears to correspond to known spliceosomal proteins, and Western analysis shows that they do not correspond to the SR family of splicing factors (data not shown). Other spliceosomal proteins, such as the components of SF3a and -b, detected only single proteins on far-Western blots of nuclear extract (3, 10). However, the proteins in the SF3a and SF3b complexes are tightly associated in stable complexes that copurify over multiple chromatographic steps and are not disrupted under very-high-salt conditions (reference 7 and unpublished observations). The observation that SAP 155 is a less-specific probe on far-Western blots than the SF3a and -b components may be mechanistically important, since the SAP 155-U2AF interaction is expected to be lower affinity so that it can be disrupted prior to catalytic step II of splicing (9).
To demonstrate SAP 155-U2AF interactions by another method and delimit a region of SAP 155 necessary for interactions with U2AF, GST fusion proteins containing different portions of SAP 155 were constructed (Fig. 4A). These proteins were coupled to glutathione beads and mixed with nuclear extract (containing RNase A). After extensive washing, the bound proteins were fractionated on a gel, and Western blot analyses were carried out. A Coomassie-stained gel of the SAP 155 and control GST fusion proteins is shown in Fig. 4B. As reported previously (2), GST-SF1/mBBP (amino acids [aa] 1 to 361) pulls down both U2AF65 and U2AF35 (Fig. 4C, lane 1). Similarly, aa 1 to 480 or 267 to 369 of SAP 155 pull down both U2AF65 and U2AF35 (Fig. 4C, lanes 2 and 4). In contrast, GST alone, GST fused to random amino acids, or a GST fusion protein containing SAP 155 aa 370 to 485 does not interact with either U2AF subunit (Fig. 4C, lanes 3, 5, and 6). A GST fusion protein containing the SAP 155 carboxy terminus also does not interact with U2AF (data not shown).
The blot shown in Fig. 4C was reprobed with antibodies to the U2 snRNP protein, SAP 130 (as a negative control). SAP 130, which was detected in total nuclear extract (Fig. 4C, lane 7), was not detected significantly with any of the GST fusion proteins (Fig. 4C, lanes 1 to 6). Furthermore, two additional spliceosomal proteins (hSLU7 and hPRP16) were also not detected with GST-SAP 155 (data not shown). Thus, these data indicate that SAP 155 and U2AF interact specifically with each other in total nuclear extract. Moreover, a distinct domain on SAP 155, extending from aa 267 to 369, is necessary and sufficient for this interaction with U2AF. Interestingly, this domain contains several repeats of the sequence RWDETP and a large number of TPGH repeats which are potential phosphorylation sites (29).
To determine whether U2AF and SAP 155 can also interact in vivo, yeast two-hybrid assays were carried out. An amino-terminal portion of SAP 155 (aa 171 to 775) was fused to the activation domain of VP16 and used as a fish construct. SAP 155 fused to the LexA DNA binding domain could not be used as a bait construct because it transactivates the β-galactosidase promoter in the absence of a fish construct (data not shown). By using the SAP 155 fish construct, interactions with full-length U2AF65 and U2AF35 baits were detected (Fig. 5A, left panel, column 1). No interaction was detected with U2AF35 lacking its RS domain (Fig. 5A, left panel, column 1). In contrast, U2AF65 interacts with both U2AF35 and U2AF35ΔRS, as previously reported (35) (Fig. 5A, left panel, column 2). With U2AF65 as a bait, interactions with SAP 155 and U2AF35 were detected, whereas no interactions were detected with the VP16 construct alone (Fig. 5A, right panel). Interactions between U2AF65 and U2AF35 were detected at 10 min, whereas SAP 155-U2AF interactions were detected at 15 min. In contrast, no interactions were detected in control experiments for at least 3 h (data not shown). We conclude that SAP 155 interacts with both subunits of U2AF in the two-hybrid assay, confirming the interactions detected by far-Western analysis (Fig. 3).
A specific region containing the third RRM of U2AF65 mediates the interaction with SAP 155.
An amino-terminal region (aa 64 to 182) of U2AF65 is required for the interaction with U2AF35 (35), the Arg-Ser-rich (RS) domain (aa 25 to 91) of U2AF65 promotes annealing of U2 snRNA to pre-mRNA (28), aa 138 to 183 of U2AF65 interact with UAP56 (13), and a region containing the third RRM (aa 334 to 475) of U2AF65 interacts with SF1/mBBP (6). To define the domain on U2AF65 required for the interaction with SAP 155, derivatives of U2AF65 were used as baits (Fig. 5B) to test interactions with three different fish containing U2AF35, SAP 155, or VP16 alone (as a negative control) (Fig. 5B). β-Galactosidase activity in liquid culture was quantified for a subset of these bait-fish combinations and confirmed the data obtained from assays on plates (data not shown). As shown in Fig. 5B, and consistent with previous work (35), aa 91 to 151 of U2AF65 are sufficient for the interaction with U2AF35, and derivatives of U2AF65 lacking this domain do not interact with U2AF35 (Fig. 5B, row 14, and rows 8 to 12, respectively; we note that a U2AF65 derivative containing aa 91 to 229 failed to interact with U2AF35 despite containing the U2AF35-interaction domain [row 15]. This protein is expressed in yeast, but could be improperly folded because of the additional sequences.)
Amino acids 334 to 475 of U2AF65 are sufficient for the interaction with SAP 155 (Fig. 5B, row 9). The region between aa 334 and 392 is required, since a construct containing only aa 392 to 475 fails to interact with SAP 155 (Fig. 5B, row 8). Similarly, aa 392 to 475 are required, since a construct containing aa 1 to 392 does not interact with SAP 155 but does still interact with U2AF35. We conclude that a specific domain of U2AF65, which contains the third RRM, is required for the interaction with SAP 155. Significantly, this region of U2AF65 also interacts with SF1/mBBP (6). Because SF1/mBBP binds to pre-mRNA in the E complex, whereas SAP 155 does not bind until the A complex, it is possible that there is an exchange of SF1/mBBP for SAP 155 during the E-to-A complex transition.
For U2AF35, the RS domain is required for the interaction with SAP 155. Because U2AF35 lacks an RRM and is not significantly similar to U2AF65, it is likely that structurally distinct domains of U2AF65 and U2AF35 interact with SAP 155.
SAP 155-U2AF65 interactions are conserved in S. pombe.
As shown in Fig. 5C, SAP 155, U2AF65, and U2AF35 are conserved in S. pombe (22, 29, 30). In addition, S. pombe SAP 155 contains two of the amino-terminal RWDETP motifs that are present in the U2AF interaction domain of human SAP 155 (see Fig. 4A). In contrast, the amino terminus of Saccharomyces cerevisiae SAP 155 is not conserved (29), and there is no homolog for U2AF35 in S. cerevisiae. The high level of sequence conservation of U2AF and SAP 155 between S. pombe and humans prompted us to test the possibility that SAP 155-U2AF interactions also occur in S. pombe. An S. pombe SAP 155 fish construct was tested with S. pombe U2AF65 (U2AF59) and U2AF35 (U2AF23) baits. As shown in Fig. 5D, the interaction between S. pombe SAP 155 and U2AF59 does indeed occur. In contrast, the U2AF35-SAP 155 interaction is not conserved (Fig. 5D). Interestingly, despite the high similarity between S. pombe U2AF23 and U2AF35 (Fig. 5C), U2AF23 lacks an RS domain. As suggested above, the RS domain of U2AF35 is required for U2AF35-SAP 155 interactions.
Parameters for U2 snRNP binding.
The data presented above show that U2AF and SAP 155 interact directly in both humans and S. pombe and that SAP 155 cross-links on both sides of the BPS (in humans). Because U2AF binding precedes SAP 155 (and U2 snRNP) binding during spliceosome assembly, one possible role for these RNA-protein and protein-protein interactions is in recruiting U2 snRNP to the BPS. In this recruitment step, U2AF bound to the pyrimidine tract may contact SAP 155 to position and/or stabilize U2 snRNP at the BPS. Because SAP 155 and SF1/mBBP both interact with the same domain on U2AF65 and because this domain is involved in U2AF65–pre-mRNA interactions (references 6 and 34 and this study), it is not possible to use mutant U2AF65 lacking the interaction domain to test the functional significance of the SAP 155-U2AF65 interaction. Thus, in order to gain further insight into the mechanism for recruiting U2 snRNP, we investigated the parameters in the pre-mRNA that are important for U2 snRNP binding.
Previous work indicated that the distance between the pyrimidine tract and BPS is critical for catalytic step I of the splicing reaction (26, 27a). To determine whether this distance is also important for U2 snRNP binding, we constructed a pre-mRNA, designated 2far, in which a 27-nt spacer was inserted between the BPS and U2AF65 binding sites (Fig. 6, see schematic). This spacer lacks adenosines because of their potential to function as cryptic branch sites. Comparison of spliceosome assembly with wild-type versus 2far pre-mRNA shows that this insertion dramatically decreases A complex assembly (Fig. 6). In contrast, E complex assembly on wild-type and 2far pre-mRNAs occurs with the same efficiency (data not shown). The observation that the E complex can assemble on 2far pre-mRNA indicates that the block to A complex assembly is unlikely to be due to a general negative effect of the insertion sequence. These data indicate that the distance between the BPS and pyrimidine tract is a critical parameter for U2 snRNP binding.
We next asked whether the proximity of the BPS to the pyrimidine tract is important for BPS selection. To test this parameter, pre-mRNAs containing tandemly duplicated BPSs were constructed (Fig. 7). In the first set of experiments, both BPSs were located within the BPS-to-AG distance that is normally found in metazoan pre-mRNAs (18 to 40 nt) (Fig. 7A). Pre-mRNAs containing a G substitution for the branch-site A were used to generate markers for the lariats. Only the downstream BPS was used when the upstream BPS contained a G (Fig. 7A, lane G/A), and only the upstream BPS was used when the downstream BPS contained a G (Fig. 7A, lane A/G). However, when both BPSs contained an A, and thus were in direct competition, lariat formation occurred primarily at the downstream BPS (Fig. 7A, lane A/A). These data show that the BPS closest to the pyrimidine tract is preferentially selected. This result, together with the observation that the distance between the BPS and pyrimidine tract is critical for U2 snRNP binding (Fig. 6), supports a model for U2 snRNP binding in which a factor(s) bound at the BPS interacts with a factor(s) bound at the pyrimidine tract. Moreover, there must be a mechanism for constraining the interaction linearly along the RNA, since the BPS and pyrimidine tract must be located adjacent to each other for efficient U2 snRNP binding and BPS selection. A model of U2 snRNP binding that incorporates these data is presented below (see Discussion).
An optimal distance between the BPS and pyrimidine tract for branch-site selection.
To determine whether the BPS nearest to the pyrimidine tract is always selected or whether there is an optimal distance between the BPS and pyrimidine tract, we constructed a pre-mRNA containing duplicated BPSs in which the downstream BPS is located directly adjacent to the run of U’s where U2AF binds and the upstream BPS is located 8 nt further upstream. The branch sites are located 14 and 22 nt upstream from the 3′ splice site in this construct (Fig. 7B, schematic, A22/A14). Pre-mRNAs containing G substitutions for either BPS were used to generate markers for the lariats (Fig. 7B, schematic, G22/A14 and A22/G14). Significantly, the upstream BPS is preferentially used in A22/A14 pre-mRNA (Fig. 7B, lane A/A). Moreover, when there is a G substitution in the upstream BPS, lariat formation occurs at the downstream BPS but is much less efficient (Fig. 7B, compare lanes G/A versus A/A). Thus, from the analyses shown in Fig. 7A and B, we conclude that the BPS nearest to the 3′ splice site is preferentially selected, but it cannot be located too close to the 3′ splice site. Although other explanations are possible, a reasonable interpretation of these data is that there are steric constraints on the factors bound at the pyrimidine tract and those bound at the BPS.
SAP 155 contacts pre-mRNA on both sides of the functional BPS in A22/A14 pre-mRNA.
SAP 155 cross-links on both sides of the BPS in functional A complex (Fig. 1). However, there is no direct evidence that this interaction is essential for splicing. To determine whether the cross-linking of SAP 155 to the pre-mRNA is likely to be functionally important, we took advantage of the observation that there are two potentially functional BPSs in A22/A14 pre-mRNA, yet only the upstream one is used for catalytic step I. We then asked where SAP 155 cross-links in the A complex assembled on this pre-mRNA. The pre-mRNA was 32P site-specifically labeled at −6, −1, or +7 relative to the A22 BPS and then assembled into the A complex (Fig. 7C). (Note that it was not possible to label downstream of the A14 BPS because the pyrimidine sequence in this region cannot be used as a transcription template for the site-specific labeling [see Materials and Methods].) Strikingly, the data reveal that SAP 155 cross-links on both sides of the functional A22 BPS, and no cross-linking of SAP 155 is detected on the upstream side of the nonfunctional A14 BPS (−1 site, Fig. 7C). Moreover, SAP 62, which cross-links at the −6 site on wild-type pre-mRNA, is also detected at the −6 site of the functional BPS in A22/A14 pre-mRNA (Fig. 7C). Thus, the wild-type pattern of cross-linked proteins is observed surrounding the functional, but not the nonfunctional, BPS in A22/A14 pre-mRNA. Furthermore, as observed with wild-type pre-mRNA (Fig. 1), the cross-linking efficiencies of SAP 155 are the same both upstream and downstream of the functional BPS in A22/A14 pre-mRNA. Finally, the sequences where SAP 155 cross-links are different in wild-type and A22/A14 pre-mRNAs (compare schematics, Fig. 1A and 7C). The latter observation argues that SAP 155–pre-mRNA interactions on both sides of the BPS occur generally and are not peculiar to one pre-mRNA sequence. Moreover, the observation that the interaction between SAP 155 and the functional BPS is detected at a time in the splicing reaction (the A complex) well prior to BPS use (catalytic step I) is consistent with the proposal that SAP 155–pre-mRNA interactions play a role in recruiting U2 snRNP to the BPS.
DISCUSSION
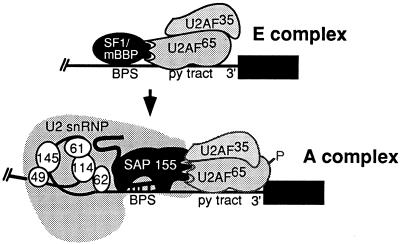
The BPS is highly degenerate in metazoan pre-mRNAs. Nevertheless, this sequence element plays a critical role in splicing by base pairing with U2 snRNA to specify the nucleophile for catalytic step I. Because the BPS is so degenerate, specific mechanisms must exist for targeting U2 snRNA to its binding site. Our data suggest a model for how U2 snRNP is recruited to the BPS (Fig. 8). In the E complex, U2AF binds to the pyrimidine tract and interacts directly with SF1/mBBP (2, 4–6, 33). During the transition to the A complex, U2AF recruits U2 snRNP through direct interactions with SAP 155 (Fig. 8). An amino-terminal region of SAP 155 and a carboxy-terminal portion of U2AF65 (which contains the third RRM) are necessary for this interaction. In addition, the amino terminus of SAP 155 interacts with U2AF35. In the A complex, SAP 155 contacts pre-mRNA on both sides of the BPS. SAP 155 and SF1/mBBP interact with the same region of U2AF65 (6). Thus, the SF1/mBBP-U2AF interaction in the E complex may be replaced by the SAP 155-U2AF interaction in the A complex. Annealing of U2 snRNA to the BPS may be achieved by a direct interaction between the RS domain of U2AF65 and the BPS, as recently proposed (Fig. 8) (28). Finally, the other SF3a and -b subunits (SAPs 49, 61, 62, 114, and 145) function to anchor U2 snRNP tightly to the pre-mRNA (15). The RNA-protein and protein-protein interactions of these SF3a and -b proteins are thought to fold the pre-mRNA into a distinct structure upstream of the branch site (15). As indicated in the model, U2AF65 is phosphorylated in the A complex and becomes less tightly bound to pre-mRNA (8). Ultimately, U2AF is replaced by U5 snRNP (9).
FIG. 8.
Model for recruitment of U2 snRNP. The 3′ portion of the intron containing the BPS and the pyrimidine (py) tract are shown. Only the factors discussed in this study are indicated. Proteins that interact directly are shown touching. P indicates the phosphorylation of U2AF65 that occurs in the A complex. Note that U2AF65 and U2AF35 are much less tightly bound in the A complex than in the E complex. SAPs 49, 61, 62, 114, 145, and 155 are shown.
Parameters for BPS selection.
In order to determine parameters important for U2 snRNP binding to the BPS and for BPS selection, we analyzed a series of mutant pre-mRNAs. These studies yielded three key results. First, insertion of a spacer between the BPS and pyrimidine tract inhibits U2 snRNP binding. Second, in a pre-mRNA in which a duplicate BPS is inserted upstream of the normal BPS, the one closer to the pyrimidine tract is used. Third, a BPS located too close to the pyrimidine tract is not selected. Together, these data indicate that recruitment of U2 snRNP to the BPS (and BPS selection) requires an interaction between a factor(s) bound to the pyrimidine tract and a factor(s) bound to the BPS. Moreover, the distance requirement indicates that this interaction is constrained linearly along the pre-mRNA. Specifically, the data are not consistent with a mechanism in which factors bound at the pyrimidine tract can interact with factors bound at a distant BPS, with the intervening RNA looped out. The data also indicate that there must be an optimal distance between the BPS and pyrimidine tract, most likely to accommodate the factors bound at each site. Our observations on the parameters for BPS selection (that the nearest BPS to the pyrimidine tract is selected and that there are constraints on the BPS-to-pyrimidine tract distance) could be explained by SF1/mBBP-U2AF interactions, SAP 155-U2AF interactions, or both (2, 5, 33, 34) (Fig. 8). At present, it is not possible to distinguish between these and other possibilities.
Several observations are consistent with the model that SAP 155-U2AF interactions function in recruiting U2 snRNP to the BPS. U2AF is necessary for A complex assembly, and SAP 155 is a component of the splicing factor SF3b, which is essential for A complex assembly (for reviews, see references 17, 25, and 32). Temporally, U2AF cross-linking to the pyrimidine tract precedes SAP 155 UV cross-linking on both sides of the BPS. Moreover, the RNA-protein and protein-protein interactions of SAP 155 and U2AF appear to be functionally important for U2 snRNP binding. First, in a pre-mRNA containing duplicated BPSs, SAP 155 interacts with pre-mRNA on both sides of the functional BPS only. Second, a specific domain on SAP 155 and a specific domain on U2AF are required for the SAP 155-U2AF interaction. Third, the SAP 155-U2AF interaction is conserved in S. pombe. Significantly, S. pombe SAP 155 and U2AF59 were recently shown to interact as synthetic lethal mutants (27b), providing genetic evidence for the importance of the SAP 155-U2AF65 interaction.
The amino terminus of SAP 155 is not conserved in S. cerevisiae, indicating that the same SAP 155-U2AF65 interactions that we have detected in S. pombe and humans are not likely to be involved in recruiting U2 snRNP in S. cerevisiae. The S. cerevisiae BPS is stringently conserved. Thus, it is possible that the U2 snRNA-BPS duplex and/or BBP plays a role in recruiting U2 snRNP in yeast (2 [see reference 19 for review]). It is also possible that interactions between MUD2 and the U2 snRNP protein PRP11 (the SAP 62 homolog) function in recruiting U2 snRNP in S. cerevisiae (1). The yeast splicing factor PRP5, which is a member of the DEAD box family of ATPases, is required for U2 snRNP binding and mediates a conformational change in U2 snRNP (21, 31). Thus, this protein is another candidate for recruiting U2 snRNP. These observations, together with our data, indicate that the specificity for U2 snRNP binding most likely involves the formation of a vast number of RNA-RNA, RNA-protein, and protein-protein contacts.
ACKNOWLEDGMENTS
We are grateful to Z. Zhou and K. Chua for useful discussions and critical reading of the manuscript and to M. Rosbash and N. Abovich for useful discussions and for the mBBP clone. We are grateful to Ron McKinney for excellent technical assistance with the two-hybrid assays of the S. pombe constructs.
This work was supported by an NIH grant to R.R. and an NIH grant (R01GM47487) and ACS grant (JFRA-545) to J.P.
REFERENCES
- 1.Abovich N, Liao X C, Rosbash M. The yeast MUD2 protein: an interaction with PRP11 defines a bridge between commitment complexes and U2 snRNP addition. Genes Dev. 1994;8:843–854. doi: 10.1101/gad.8.7.843. [DOI] [PubMed] [Google Scholar]
- 2.Abovich N, Rosbash M. Cross-intron bridging interactions in the yeast commitment complex are conserved in mammals. Cell. 1997;89:403–412. doi: 10.1016/s0092-8674(00)80221-4. [DOI] [PubMed] [Google Scholar]
- 3.Bennett M, Reed R. Correspondence between a mammalian spliceosome component and an essential yeast splicing factor. Science. 1993;262:105–108. doi: 10.1126/science.8211113. [DOI] [PubMed] [Google Scholar]
- 4.Bennett M, Michaud S, Kingston J, Reed R. Protein components specifically associated with prespliceosome and spliceosome complexes. Genes Dev. 1992;6:1986–2000. doi: 10.1101/gad.6.10.1986. [DOI] [PubMed] [Google Scholar]
- 5.Berglund J A, Chua K, Abovich N, Reed R, Rosbash M. The splicing factor BBP interacts specifically with the pre-mRNA branchpoint sequence UACUAAC. Cell. 1997;89:781–787. doi: 10.1016/s0092-8674(00)80261-5. [DOI] [PubMed] [Google Scholar]
- 6.Berglund J A, Abovich N, Rosbash N. A cooperative interaction between U2AF65 and mBBP/SF1 facilitates branchpoint region recognition. Genes Dev. 1998;12:858–867. doi: 10.1101/gad.12.6.858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brosi R, Hauri H P, Kramer A. Separation of splicing factor SF3 into two components and purification of SF3a activity. J Biol Chem. 1993;268:17640–17646. [PubMed] [Google Scholar]
- 8.Champion-Arnaud P, Gozani O, Palandjian L, Reed R. Accumulation of a novel spliceosomal complex on pre-mRNAs containing branch site mutations. Mol Cell Biol. 1995;15:5750–5756. doi: 10.1128/mcb.15.10.5750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chiara M, Palandjian L, Reed R. Evidence the U5 snRNP recognizes the 3′ splice site for catalytic step II in mammals. EMBO J. 1997;16:4746–4759. doi: 10.1093/emboj/16.15.4746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chiara M D, Champion-Arnaud P, Buvoli M, Nadal-Ginard B, Reed R. Specific protein-protein interactions between the essential mammalian spliceosome-associated proteins SAP 61 and SAP 114. Proc Natl Acad Sci USA. 1994;91:6403–6407. doi: 10.1073/pnas.91.14.6403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chiara M D, Gozani O, Bennett M, Champion-Arnaud P, Palandjian L, Reed R. Identification of proteins that interact with exon sequences, splice sites, and the branchpoint sequence during each stage of spliceosome assembly. Mol Cell Biol. 1996;16:3317–3326. doi: 10.1128/mcb.16.7.3317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fields S, Song O. A novel genetic system to detect protein-protein interactions. Nature. 1989;340:245–246. doi: 10.1038/340245a0. [DOI] [PubMed] [Google Scholar]
- 13.Fleckner J, Zhang M, Valcarcel J, Green M R. U2AF65 recruits a novel human DEAD box protein required for the U2 snRNP-branchpoint interaction. Genes Dev. 1997;11:1864–1872. doi: 10.1101/gad.11.14.1864. [DOI] [PubMed] [Google Scholar]
- 14.Gozani O, Patton J G, Reed R. A novel set of spliceosome-associated proteins and the essential splicing factor PSF bind stably to pre-mRNA prior to catalytic step II of the splicing reaction. EMBO J. 1994;13:3356–3367. doi: 10.1002/j.1460-2075.1994.tb06638.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gozani O, Feld R, Reed R. Evidence that sequence-independent binding of highly conserved U2 snRNP proteins upstream of the branch site is required for assembly of spliceosomal complex A. Genes Dev. 1996;10:233–243. doi: 10.1101/gad.10.2.233. [DOI] [PubMed] [Google Scholar]
- 16.Kramer A. Purification of splicing factor SF1, a heat-stable protein that functions in the assembly of a presplicing complex. Mol Cell Biol. 1992;12:4545–4552. doi: 10.1128/mcb.12.10.4545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kramer A. The structure and function of proteins involved in mammalian pre-mRNA splicing. Annu Rev Biochem. 1996;65:367–409. doi: 10.1146/annurev.bi.65.070196.002055. [DOI] [PubMed] [Google Scholar]
- 18.MacMillan A M, Query C C, Allerson C R, Chen S, Verdine G L, Sharp P A. Dynamic association of proteins with the pre-mRNA branch region. Genes Dev. 1994;8:3008–3020. doi: 10.1101/gad.8.24.3008. [DOI] [PubMed] [Google Scholar]
- 19.Madhani H D, Guthrie C. Dynamic RNA-RNA interactions in the spliceosome. Annu Rev Genet. 1994;28:1–26. doi: 10.1146/annurev.ge.28.120194.000245. [DOI] [PubMed] [Google Scholar]
- 20.Moore M J, Sharp P A. Site-specific modification of pre-mRNA: the 2′-hydroxyl groups at the splice sites. Science. 1992;256:992–997. doi: 10.1126/science.1589782. [DOI] [PubMed] [Google Scholar]
- 21.O’Day C L, Dalbadie-McFarland G, Abelson J. The Saccharomyces cerevisiae Prp5 protein has RNA-dependent ATPase activity with specificity for U2 small nuclear RNA. J Biol Chem. 1996;271:33261–33267. doi: 10.1074/jbc.271.52.33261. [DOI] [PubMed] [Google Scholar]
- 22.Potashkin J, Naik K, Wentz-Hunter K. U2AF homolog required for splicing in vivo. Science. 1993;262:573–575. doi: 10.1126/science.8211184. [DOI] [PubMed] [Google Scholar]
- 23.Query C C, Moore M J, Sharp P A. Branch nucleophile selection in pre-mRNA splicing: evidence for the bulged duplex model. Genes Dev. 1994;8:587–597. doi: 10.1101/gad.8.5.587. [DOI] [PubMed] [Google Scholar]
- 24.Query C C, Strobel S A, Sharp P A. Three recognition events at the branch-site adenine. EMBO J. 1996;15:1392–1402. [PMC free article] [PubMed] [Google Scholar]
- 25.Reed R. Initial splice-site recognition and pairing during pre-mRNA splicing. Curr Opin Genet Dev. 1996;6:215–220. doi: 10.1016/s0959-437x(96)80053-0. [DOI] [PubMed] [Google Scholar]
- 26.Reed R. The organization of 3′ splice-site sequences in mammalian introns. Genes Dev. 1989;3:2113–2123. doi: 10.1101/gad.3.12b.2113. [DOI] [PubMed] [Google Scholar]
- 27.Reed R. Protein composition of mammalian spliceosomes assembled in vitro. Proc Natl Acad Sci USA. 1990;87:8031–8035. doi: 10.1073/pnas.87.20.8031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27a.Smith C W J, Porro E B, Patton J G, Nadal-Ginard B. Scanning from an independently specified branchpoint defines the 3′ splice site of mammalian introns. Nature. 1989;342:243–247. doi: 10.1038/342243a0. [DOI] [PubMed] [Google Scholar]
- 27b.Tani, T. Personal communication.
- 28.Valcarcel J, Gaur R K, Singh R, Green M R. Interaction of U2AF65 RS region with pre-mRNA branch point and promotion of base pairing with U2 snRNA. Science. 1996;273:1706–1709. doi: 10.1126/science.273.5282.1706. [DOI] [PubMed] [Google Scholar]
- 29.Wang C, Chua K, Seghezzi W, Lees E, Reed R. Phosphorylation of the spliceosomal protein SAP 155 coupled with splicing catalysis. Genes Dev. 1998;12:1409–1414. doi: 10.1101/gad.12.10.1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wentz-Hunter K, Potashkin J. The small subunit of the splicing factor U2AF is conserved in fission yeast. Nucleic Acids Res. 1996;24:1849–1854. doi: 10.1093/nar/24.10.1849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wiest D K, O’Day C L, Abelson J. In vitro studies of the Prp9.Prp11.Prp21 complex indicate a pathway for U2 small nuclear ribonucleoprotein activation. J Biol Chem. 1996;271:33268–33276. doi: 10.1074/jbc.271.52.33268. [DOI] [PubMed] [Google Scholar]
- 32.Will C L, Luhrmann R. Protein functions in pre-mRNA splicing. Curr Opin Cell Biol. 1997;9:320–328. doi: 10.1016/s0955-0674(97)80003-8. [DOI] [PubMed] [Google Scholar]
- 33.Zamore P D, Green M R. Biochemical characterization of U2 snRNP auxiliary factor: an essential pre-mRNA splicing factor with a novel intranuclear distribution. EMBO J. 1991;10:207–214. doi: 10.1002/j.1460-2075.1991.tb07937.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zamore P D, Patton J G, Green M R. Cloning and domain structure of the mammalian splicing factor U2AF. Nature. 1992;355:609–614. doi: 10.1038/355609a0. [DOI] [PubMed] [Google Scholar]
- 35.Zhang M, Zamore P D, Carmo-Fonseca M, Lamond A I, Green M R. Cloning and intracellular localization of the U2 small nuclear ribonucleoprotein auxiliary factor small subunit. Proc Natl Acad Sci USA. 1992;89:8769–8773. doi: 10.1073/pnas.89.18.8769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zuo P, Maniatis T. The splicing factor U2AF35 mediates critical protein-protein interactions in constitutive and enhancer-dependent splicing. Genes Dev. 1996;10:1356–1368. doi: 10.1101/gad.10.11.1356. [DOI] [PubMed] [Google Scholar]