Abstract
A highly protective malaria vaccine would greatly facilitate the prevention and elimination of malaria and containment of drug-resistant parasites1. A high level (more than 90%) of protection against malaria in humans has previously been achieved only by immunization with radiation-attenuated Plasmodium falciparum (Pf) sporozoites (PfSPZ) inoculated by mosquitoes2–4; by intravenous injection of aseptic, purified, radiation-attenuated, cryopreserved PfSPZ (‘PfSPZ Vaccine’)5,6; or by infectious PfSPZ inoculated by mosquitoes to volunteers taking chloroquine7–10 or mefloquine11 (chemoprophylaxis with sporozoites). We assessed immunization by direct venous inoculation of aseptic, purified, cryopreserved, non-irradiated PfSPZ (‘PfSPZ Challenge’12,13) to malaria-naive, healthy adult volunteers taking chloroquine for antimalarial chemoprophylaxis (vaccine approach denoted as PfSPZ-CVac)14. Three doses of 5.12 × 104 PfSPZ of PfSPZ Challenge12,13 at 28-day intervals were well tolerated and safe, and prevented infection in 9 out of 9 (100%) volunteers who underwent controlled human malaria infection ten weeks after the last dose (group III). Protective efficacy was dependent on dose and regimen. Immunization with 3.2 × 103 (group I) or 1.28 × 104 (group II) PfSPZ protected 3 out of 9 (33%) or 6 out of 9 (67%) volunteers, respectively. Three doses of 5.12 × 104 PfSPZ at five-day intervals protected 5 out of 8 (63%) volunteers. The frequency of Pf-specific polyfunctional CD4 memory T cells was associated with protection. On a 7,455 peptide Pf proteome array, immune sera from at least 5 out of 9 group III vaccinees recognized each of 22 proteins. PfSPZ-CVac is a highly efficacious vaccine candidate; when we are able to optimize the immunization regimen (dose, interval between doses, and drug partner), this vaccine could be used for combination mass drug administration and a mass vaccination program approach to eliminate malaria from geographically defined areas.
PfSPZ-CVac was safe and well tolerated by volunteers and no serious adverse events occurred. The frequencies of grade 1–3 adverse events were similar in recipients of placebo and PfSPZ-CVac (Extended Data Fig. 1a, Supplementary Table 1), and with increasing PfSPZ dose (Extended Data Fig. 1a). Chloroquine was generally well tolerated; however, two volunteers dropped out of the trial after the loading dose because they experienced nausea and vomiting (Extended Data Fig. 2).
Controlled human malaria infection (CHMI) with PfSPZ Challenge (NF54) by direct venous inoculation (DVI)12,13 was conducted 8–10 weeks after the last vaccine doses. All placebo (normal saline plus chloroquine) recipients (n = 13) developed parasitaemia. Protective efficacy was 33% (3 out of 9; P = 0.08, log-rank test), 67% (6 out of 9; P = 0.0047) and 100% (9 out of 9; P = 0.0001) in the 3.2 × 103 (group I) 1.28 × 104 (group II) and 5.12 × 104 (group III) PfSPZ dose groups, respectively (Fig. 1a). The median time from inoculation until first positive thick blood smear (prepatent period) for volunteers that received placebo, 3.2 × 103 PfSPZ, and 1.28 × 104 PfSPZ were 11.0, 12.5, and 12.9 days, respectively (P = 0.18; Kruskal–Wallis rank-sum test). Quantitative real-time polymerase chain reaction (qPCR) detected parasitaemia between 6 and 11 days after DVI of PfSPZ for CHMI (Fig. 1b) and was negative in protected volunteers. Non-protected, PfSPZ-CVac vaccinees and controls had similar blood parasite multiplication rates (9.7-fold increase in parasitaemia per each ~48 hour asexual blood-stage cycle; 95% confidence interval: 7.6, 12.4; control versus group I or II: P = 0.52 or 0.66; linear mixed effect model); immunization did not induce detectable functional immunity against the Pf blood stage. This is consistent with data showing that mice15 and humans8 that are immunized with chemoprophylaxis with sporozoites (CPS) are not protected against blood-stage parasite challenge. CHMI was well tolerated (Extended Data Fig. 1b, Supplementary Table 1), and there were no cases of severe malaria.
Figure 1 |. Protective efficacy of PfSPZ-CVac.
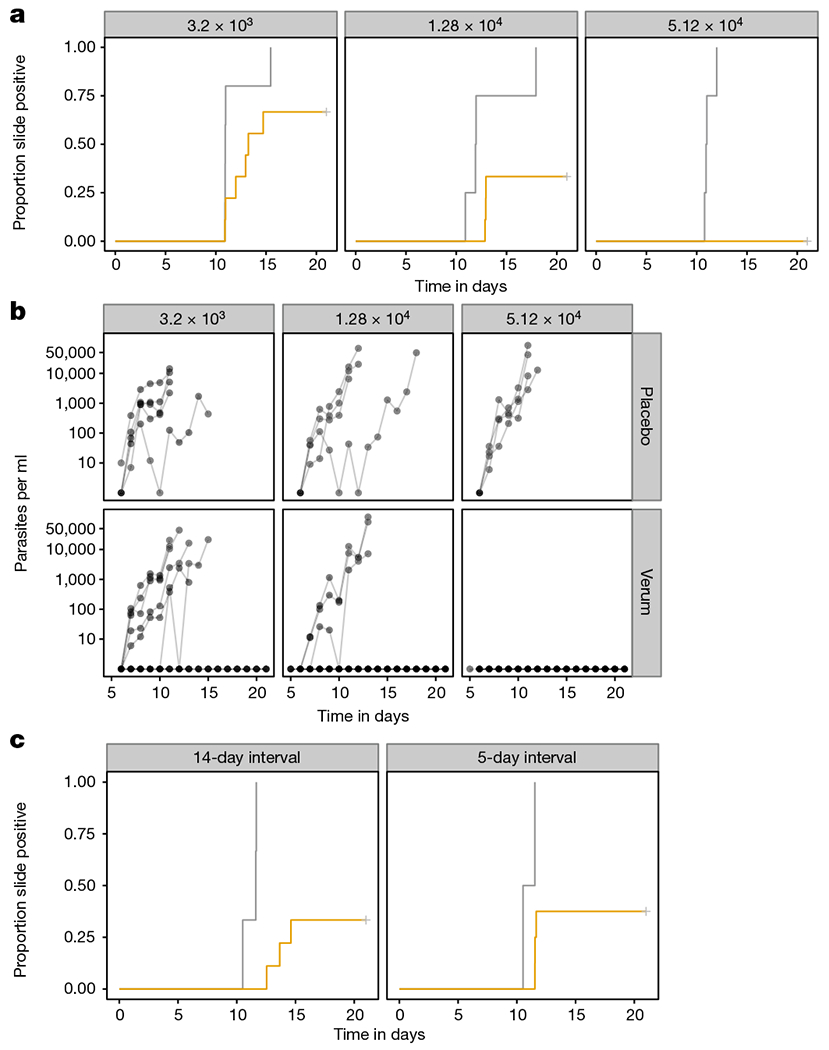
a, Proportion of controls and vaccinees who developed microscopically detectable parasitaemia after CHMI by DVI of 3.2 × 103 PfSPZ Challenge, 8–10 weeks after last immunization, and 7–9 weeks after last dose of chloroquine. Vaccinees received three doses of 3.2 × 103 (n = 9), 1.28 × 104 (n = 9), or 5.12 × 104 (n = 9) PfSPZ and controls (n = 13) received three doses of normal saline (vaccinees in yellow, placebo recipients in grey). b, Parasitaemia over time measured by qPCR. c, Proportion of controls and vaccinees who developed microscopically detectable parasitaemia after CHMI by DVI of 3.2 × 103 PfSPZ Challenge 70–72 days after last immunization, and 65–67 days after last dose of chloroquine. Vaccinees received three doses of 5.12 × 104 PfSPZ at 14-day (n = 9) and 5-day intervals (n = 9) PfSPZ and controls (n = 6) received three doses of normal saline (vaccinees in yellow, placebo recipients in grey). One vaccinee in the 5-day interval group and one control did not participate in the CHMI.
Transient parasitaemia by qPCR following first vaccination was dose-dependent (Fig. 2a, b); subjects in groups I, II, and III had median peak parasitaemias of 448, 939, and 15,755 parasites per ml, respectively. Parasitaemia decreased in most volunteers following subsequent immunizations. All 14 individuals with a ≥100-fold decrease in post-immunization parasitaemia between first and third immunization were protected. However, 4 out of 6 protected volunteers in group II had 5–67-fold decreases.
Figure 2 |. Transient parasitaemia following vaccination.
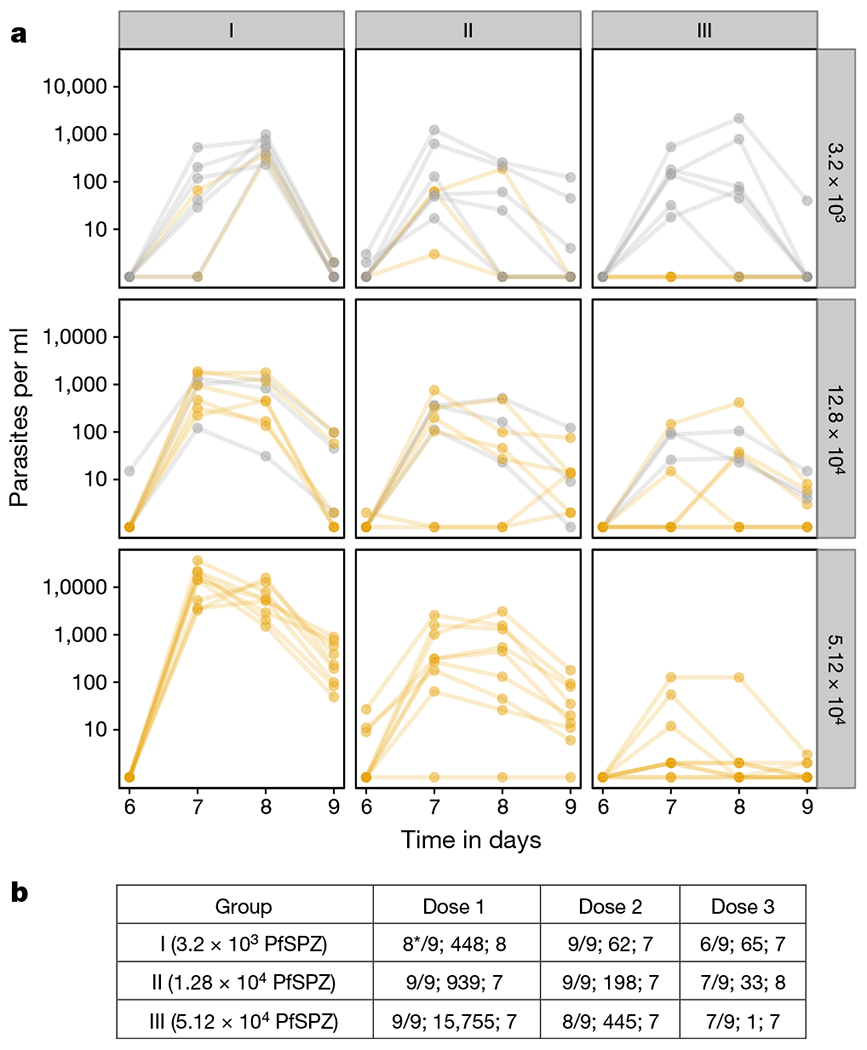
a, Parasitaemia measured by qPCR in the three dosage groups after each immunization. The subjects who were protected and not protected against CHMI are in yellow and grey, respectively. b, Number of subjects positive per number injected, median peak parasite density, and day of peak parasite density after each dose of PfSPZ-CVac. *Technical problem with day 7 and 8 samples of one volunteer.
There was a dose response of antibodies (Fig. 3a–c) against Pf circumsporozoite protein (PfCSP), as detected by ELISA, automated immunofluorescence assay, and inhibition of sporozoite invasion assay, and results of the three assays were highly correlated (Supplementary Table 2). In groups I and II, antibodies against PfCSP by ELISA, PfSPZ by automated immunofluorescence assay, or inhibition of sporozoite invasion did not correlate with protection (Fig. 3a–c, Supplementary Table 2). Levels of antibodies to PfCSP increased following each immunization (Supplementary Table 3), and decreased in all subjects in group III from two weeks after last dose until CHMI (Supplementary Table 4).
Figure 3 |. Anti-plasmodial antibody responses in vaccinated volunteers.
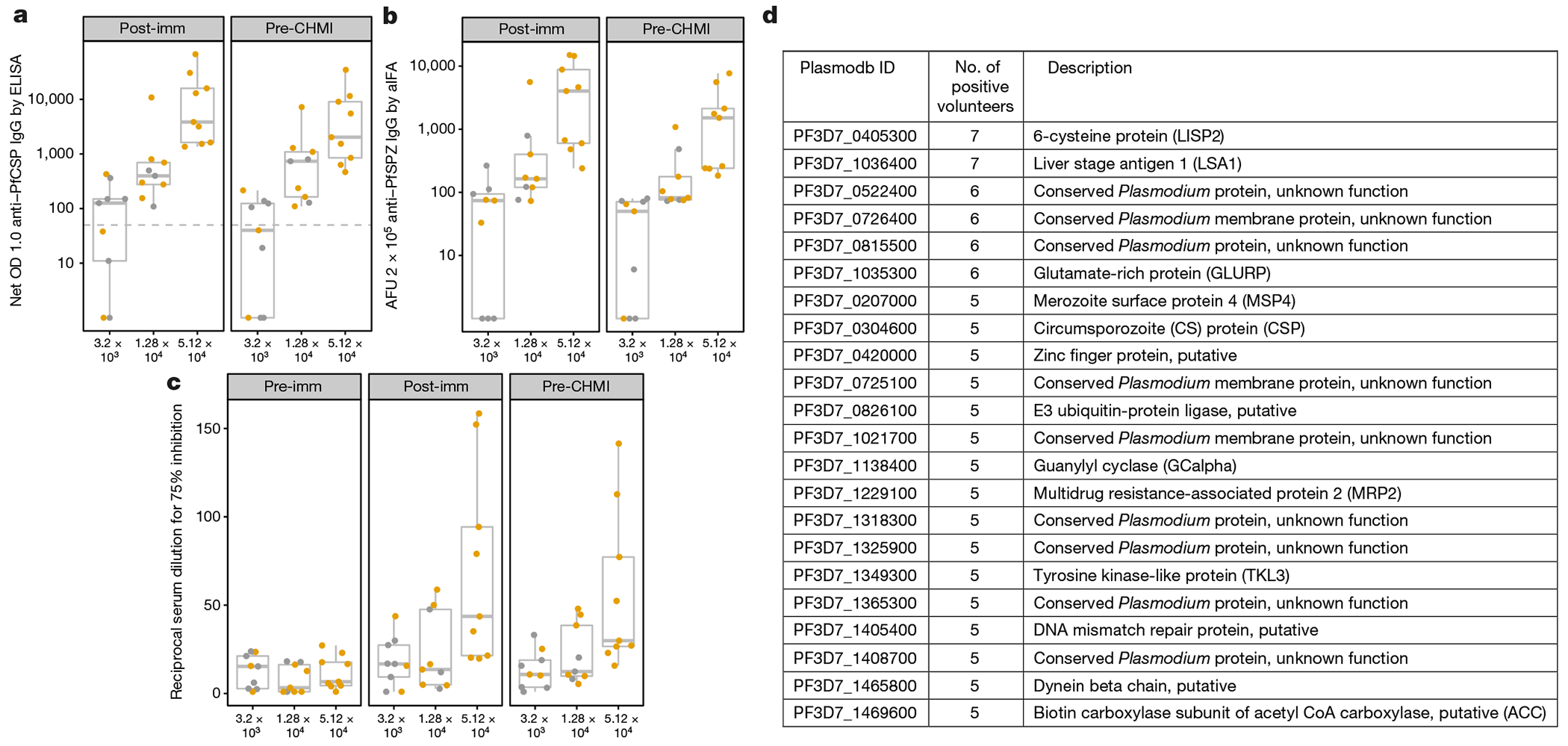
Antibodies were assessed in sera taken before any immunizations (pre-immunization), two weeks following last immunization (post-immunization) and one day before CHMI (pre-CHMI). a–d, Antibodies were assessed to PfCSP by ELISA (a); air-dried PfSPZ by automated immunofluorescence assay (b); live PfSPZ by inhibition of sporozoite invasion (c); and 7,455 Pf peptides on a proteome array (d). a, PfCSP ELISA results are reported as net optical density (OD) 1.0; reciprocal serum dilution at which the optical density was 1.0 in post-immunization or pre-CHMI sera minus the OD 1.0 in pre-immunization sera. All negative net values were assigned a value of 1. Values above the dashed line are considered positive. b, Automated immunofluorescence assay (aIFA) results are reported as arbitrary fluorescent units (AFU) 2 × 105; reciprocal serum dilution at which the AFU were 2 × 105 in post-immunization and pre-CHMI sera. c, Inhibition of sporozoite invasion values are reported as the reciprocal dilution of pre-immunization, post-immunization and pre-CHMI sera that inhibited by 75% the numbers of PfSPZ invading as compared to in negative controls without serum. d, The 22 proteins on the proteome array recognized by post-immunization sera from at least five volunteers from group III (highest dose) are delineated. The list is derived from bipartite graph analysis following normalization and background correction using values from sera taken before injection of PfSPZ Challenge in vaccinees and controls, and after injection of normal saline in controls. The threshold of positivity for the array studies was more conservative compared to the ELISA analyses; for example, for PfCSP, 5 out of 9 array-positive compared to 9 out of 9 ELISA-positive. In a–c, protected individuals are represented in yellow and unprotected ones in grey, and box plots display median (middle line), 25th (lower hinge) and 75th (upper hinge) quartile. Whiskers extend to values within 1.5× the inter-quartile ranges of the lower and upper hinges, respectively.
Post-immunization sera were assessed for antibodies to other proteins that are first expressed in (1) PfSPZ (PfSSP2/TRAP, PfCelTOS, PfMSP5, PfAMA1), (2) early liver stages (PfEXP1, PfLSA1), and (3) late liver stages (PfMSP1, PfEBA175) by ELISA. In group III, 9 out of 9 subjects had antibodies to PfCSP; 5 out of 9 to PfMSP5; 3 out of 9 to PfAMA1; 1 out of 9 to PfEXP1; 3 out of 9 to PfLSA1; and 4 out of 9 to PfMSP1 (Supplementary Table 5).
We assessed sera two weeks after the third dose for anti-Pf IgG antibodies using proteome arrays containing 7,455 in vitro translated complete or partial Pf peptides, representing 4,805 of the proteins that were predicted to be translated by the approximately 5,400 protein-coding genes in the Pf genome. More than 5 out of 9 (56%) group III volunteers developed antibodies to 22 proteins (Fig. 3d, Supplementary Fig. 1). The two most highly recognized proteins, PfLSA1 (ref. 16) and PfLISP2 (ref. 17), are expressed during the liver stage. Antibody responses in sera from 5 out of 9 immunized volunteers in groups I and II were detected for only four and two of these 22 proteins, respectively (Supplementary Fig. 1). There was no significant association of antibody responses against any protein with protection (Supplementary Table 6).
Peripheral blood mononuclear cells sampled before vaccination, two weeks after last vaccination, and one day before CHMI were assessed by multi-parameter flow cytometry following incubation with PfSPZ or Pf-infected red blood cells (PfRBC)6. PfRBC were used as a surrogate for late liver-stage parasites18. There was a dose-dependent increase in frequency of PfSPZ- and PfRBC-specific memory CD4 T cells expressing any combination of IFN-γ, IL-2, or TNFα (also known as TNF-α) (Fig. 4a, b), and in CD4 T-cell cytokine polyfunctionality, including IL-4 and IL-10 responses (Extended Data Figs 3a–f, 4a, b).
Figure 4 |. T-cell immunogenicity and correlates of protection.
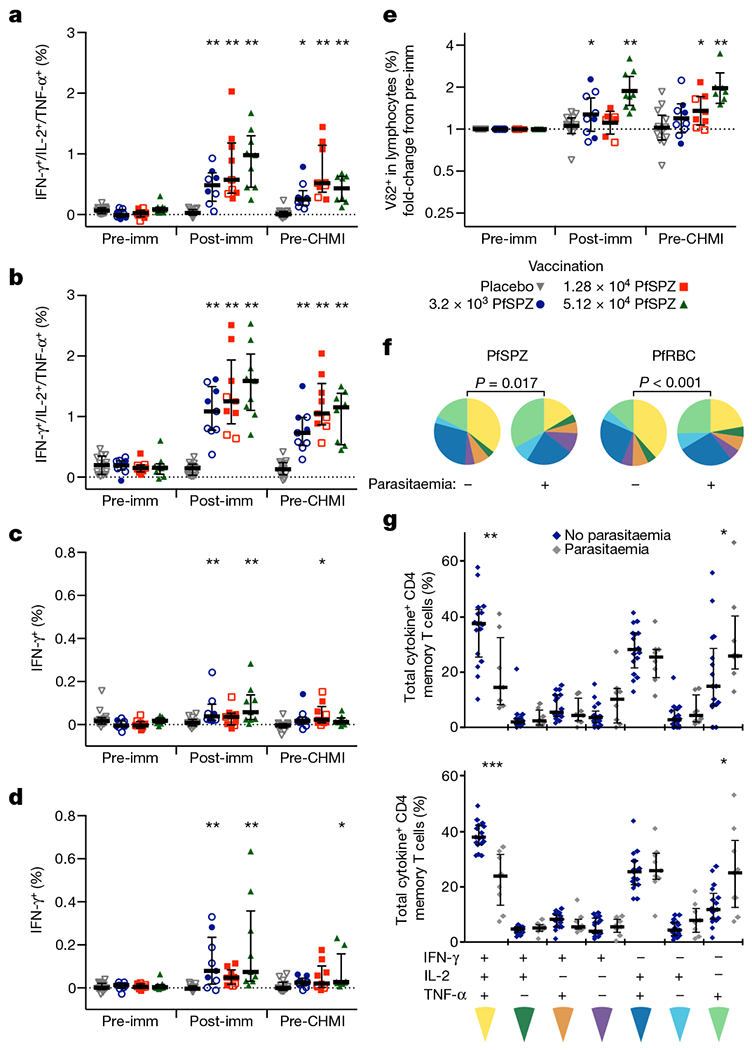
a, b, Memory CD4 T cells producing IFN-γ, IL-2, and/or TNF-α following PfSPZ (a) or PfRBC stimulation (b). c, d, Memory CD8 T cells producing IFN-γ following PfSPZ (c) or PfRBC stimulation (d). For a–d, results are the percentage of cytokine-producing cells after incubation with Pf antigen minus the percentage of cells after incubation with control antigen stimulation. e, Fold change compared to pre-immunization in the frequency of Vδ2+ T cells as a percentage of total lymphocytes. f, Cytokine polyfunctionality of PfSPZ- or PfRBC-specific memory CD4 T cells. Pie charts show the fraction of each cytokine combination out of the total cytokine response comparing subjects that were parasitaemic (+) or not parasitaemic (−) after CHMI. g, Individual data points for f, showing the composition of PfSPZ- (top) or PfRBC-specific (bottom) memory CD4 T cells producing any combination of IFN-γ, IL-2, and/or TNF-α at the time of CHMI. Subjects that remained without parasitaemia are shown in blue and subjects that developed parasitaemia are shown in grey. a–e, The difference from pre-vaccine within a dose group was assessed by two-way ANOVA with Bonferroni correction. Open symbols denote parasitaemic after CHMI; closed symbols denote not parasitaemic after CHMI. f, Comparison between pie graphs was by a non-parametric partial permutation test; g, comparison between parasitaemic and not parasitaemic responses was by Wilcoxon test. Bars are median ± interquartile range (a–d, g) or geometric mean and 95% confidence interval (e). Pre-imm, 3 days before first immunization; post-imm, 14 days after third immunization; pre-CHMI, 1 day before CHMI. *P < 0.05, **P < 0.01, *** P < 0.001.
PfSPZ- and PfRBC-specific CD8 T cells were detected 14 days after the last immunization (Fig. 4c, d), most consistently in the group treated with 5.12 × 104 PfSPZ, but were much lower in frequency than CD4 T cells. There was a dose-dependent increase in the frequency of circulating γδ T cells (Fig. 4e), as seen with the PfSPZ Vaccine5,6. Memory γδ T cells showed an increase in IFN-γ secretion and expression of the activation markers CD38 after vaccination (Extended Data Fig. 4c–f). γδ T-cell expansion was primarily restricted to the Vγ9+Vδ2+ sub-family (Extended Data Fig. 5a–f).
At the time of CHMI, there was a significant difference in the T-cell cytokine expression profiles of subjects who did develop parasitaemia, and those that did not (Fig. 4f). CD4 T cells that simultaneously produced IFN-γ, IL-2, and TNF-α were more frequent in subjects who did not develop parasitaemia, whereas those that produced only TNF-α were more frequent in those who became parasitaemic (Fig. 4g). When stratified by dose group, sterile protection was strongly associated with a higher frequency of PfRBC-specific polyfunctional IFN-γ-, IL-2-, and TNF-α-producing CD4 T cells; a weaker association with protection was also found for PfRBC-specific IFN-γ+ CD8 T-cell responses (Extended Data Table 1).
We used the 100% protective administered dose of 5.12 × 104 PfSPZ and assessed three-dose regimens at 14- and 5-day intervals together with 6 and 4 doses of chloroquine, respectively, and conducted CHMI 70 or 72 days after the last vaccination. All of the five placebo recipients developed parasitaemia, whereas we observed 67% (6 out of 9, P = 0.000045, log-rank test) and 63% (5 out of 8, P = 0.012) protective efficacy in subjects immunized at 14- and 5-day intervals (Fig. 1c). Antibodies to PfCSP 2 weeks and 10 weeks (pre-CHMI) after last dose were higher, but not significantly (P > 0.05), in vaccinees immunized at 5- or 14-day intervals than in those who received the vaccine at 28-day intervals (Extended Data Fig. 6).
Three doses of PfSPZ-CVac (5.12 × 104 PfSPZ) administered at 4-week intervals protected 9 out of 9 (100%) vaccinees against CHMI ten weeks after the last dose. The only other malaria vaccine that has induced such protection is the radiation-attenuated PfSPZ Vaccine. 6 out of 6 subjects were protected 3 weeks after administration of 6.75 × 105 PfSPZ of PfSPZ Vaccine divided into five doses. However, immunization with a lower total dosage of 1.2–1.8 × 105 PfSPZ of PfSPZ Vaccine protected only 9% (1 out of 11) of vaccinees at 3 weeks5,19. PfSPZ-CVac-immunized subjects received 1.536 × 105 PfSPZ in three doses, and had 100% protection at 10 weeks. The requirement for a higher dose of PfSPZ Vaccine as compared to PfSPZ-CVac is probably due to the fact that the PfSPZ in PfSPZ Vaccine do not replicate20,21 (Extended Data Fig. 7). The quantity and breadth of expressed proteins in hepatocytes are far less than in PfSPZ-CVac, where parasites develop into mature liver-stage schizonts with up to 4 × 104 nuclei and may express more than 4,000 proteins. This resulted in induction of antibodies against liver-stage antigens, including PfLISP2, PfLSA1, and PfMRP2 in proteome microarrays (Fig. 3d), and PfMSP1 in ELISA. Furthermore, PfSPZ- and PfRBC-specific memory CD4 T-cell responses two weeks after final vaccination reported here are the highest yet observed, including responses after a regimen of considerably higher doses of PfSPZ Vaccine6, indicating that PfSPZ-CVac is substantially more potent than PfSPZ Vaccine.
There was no significant correlation between antibody responses detected by ELISA, automated immunofluorescence assay, inhibition of sporozoite invasion, or proteome array and protection. Antibodies against PfSPZ induced by CPS and PfSPZ Vaccine have had significant functional activity in vivo in mice with human livers5,6,22, and we also report significant functional activity (Fig. 3c). We hypothesize that antibodies that inhibit PfSPZ invasion of hepatocytes contribute to protection, but the primary effectors of protection are cellular immune responses that eliminate parasite-infected hepatocytes during the 5–6 days of parasite maturation in the liver5–9,11,20,23–27. Consistent with this hypothesis, PfSPZ- and PfRBC-specific polyfunctional cytokine-producing memory CD4 T cells correlated with sterile protection in this study, as observed following CPS immunization7. At the time of CHMI, only 2 out of 9 protected volunteers in group III had antigen-specific IFNγ+ CD8 T cells, the cell type known to be essential for protection in model systems23–25. This is probably because PfSPZ-based vaccines induce tissue-resident T cells in the liver that do not recirculate in the peripheral blood5,6,20,28.
Immunization and CHMI were performed with the same Pf strain (PfNF54). When CPS-immunized subjects underwent CHMI with heterologous parasites at 14 months, efficacy was dose-dependent; one out of two who received the full dose of around 45 infected mosquitoes was protected29. However, studies using irradiated PfSPZ-containing mosquitoes showed protection against CHMI with heterologous strains of Pf4,30. Most importantly, increased doses of PfSPZ Vaccine have now been shown to induce short-term (3 weeks) and long-term (33 weeks) protection against heterologous CHMI in the US, and 24-week protection against intense natural transmission of malaria in Mali31–33. We hypothesize that higher doses of PfSPZ Challenge in PfSPZ-CVac will be required to achieve optimal long-term protection against heterogeneous parasites in the field. If this is not achieved with PfSPZ-CVac based on Pf, strain NF54, a vaccine including several strains of PfSPZ can be manufactured.
Although immunization with three doses of vaccine at 28-day intervals is feasible and similar to many other vaccine regimens (for example, three-dose infant primary immunizations for many vaccines are given at 28- to 56-day intervals), reducing time to complete immunization would increase use of the vaccine. We assessed three doses at 5-day intervals and achieved 63% protective efficacy against a first CHMI at 10 weeks after last vaccine dose (Fig. 1c) and observed a consistent decrease in liver to blood inoculum following second and third immunizations (Extended Data Fig. 8). Although this 10-day regimen was less protective than the 56-day regimen (63% versus 100%), given the dose response seen with the 56-day regimen (Fig. 1a) and based on the previous data with PfSPZ Vaccine, we hypothesize that increasing numbers of PfSPZ per dose will increase protective efficacy to 100% with the 10-day regimen.
Although this represents an important first step, in order to finalize a PfSPZ-CVac immunization regimen for elimination campaigns and for travellers, our next clinical trials will address: (1) duration of protection (CPS protects for at least 28 months10); (2) responses in malaria-exposed populations; (3) efficacy against heterogeneous natural exposure and heterologous CHMI; (4) immunization of all age groups, pregnant women and immunocompromised subjects; (5) other drugs for chemoprophylaxis; and (6) mass drug administration with PfSPZ Challenge to create PfSPZ-CVac for elimination campaigns, our ultimate goal.
METHODS
Participants.
The study was approved by the ethics committee of the medical faculty and the university clinics of the University of Tübingen and strictly adhered to Good Clinical Practice and the principles of the Declaration of Helsinki. The study is registered at ClinicalTrials.gov (https://clinicaltrials.gov/ct2/show/NCT02115516) and in the EudraCT database, number 2013-003900-38. The study was carried out under FDA IND 15862 and with approval of the Paul-Ehrlich-Institute. Volunteers were healthy, 18–45 years old, malaria-naive adults. The full list of inclusion and exclusion criteria is given in the clinical trial protocol (Supplementary Information). All volunteers, except those in the second part who received the every 5-day regimen, received 10 mg kg−1 or 620 mg chloroquine base (Resochin, Bayer) loading dose 2 days before the first dose of PfSPZ Challenge, whichever dose was less, followed by weekly doses of 5 mg kg−1 or 310 mg through 5 days after the last dose of PfSPZ Challenge. Volunteers who were immunized on days 0, 5, and 10 received chloroquine on days 0 (loading dose), 5, 10, and 15. Randomization was performed on the day of first immunization by an independent party through provision of sealed envelopes to the syringe preparation team members, who diluted PfSPZ Challenge12,13,34 or loaded placebo (normal saline) into syringes at an allocation ratio of 9:5, PfSPZ:placebo. Only the syringe preparation team was aware of allocation to the intervention and had no other role in the trial. The rest of the team remained blinded until completion of CHMI of group III. PfSPZ Challenge dose escalation for groups I, II, and III was staggered by at least four weeks and in each group three sentinel volunteers (PfSPZ:placebo, 2:1) received injections one to seven days before the main group. For group I, II, and III the three immunizations were given at 28-day intervals and CHMI by DVI of 3.2 × 103 PfSPZ was done 8–10 weeks following the last immunization. In the second part the three immunizations were administered at 14-day and 5-day intervals and CHMI was done at 10 weeks post immunization. Chloroquine concentrations were measured by mass spectrometry in the blood of selected volunteers on the day before CHMI to exclude carry-over from the immunization period. Following CHMI volunteers were regularly visited for 20 weeks. The primary efficacy endpoint was the proportion of volunteers with thick blood smears positive for Pf within 21 days of CHMI, primary safety endpoint was occurrence of related grade 3 or serious adverse events from first chloroquine dose until the end of the follow-up.
Study design and population.
We performed a randomized, placebo-controlled, double-blind trial in healthy, malaria-naive, 18–45 year olds (TÜCHMI-002; ClinicalTrials.gov ID: NCT02115516). Between 1 May and 4 July 2014, 42 volunteers were randomized to receive either three doses of normal saline (placebo) or 3.2 × 103 PfSPZ (PfSPZ Challenge12,13,34) (group I), 1.28 × 104 PfSPZ (group II), or 5.12 × 104 PfSPZ (group III) by DVI12,13 at four-week intervals (Extended Data Fig. 2). All volunteers received oral chemoprophylaxis with chloroquine starting with a loading dose (10 mg kg−1 chloroquine base or 620 mg, whichever dose was less) two days before first PfSPZ inoculation followed by weekly maintenance doses (5 mg kg−1) through five days after last PfSPZ inoculation; total of 10 doses. Chloroquine is not active against SPZ or liver-stage parasites35 and Pf asexual blood-stage parasites leave the liver between days 5 and 6 following inoculation36; hence dosing five days following inoculation ensured high drug levels upon liver egress. Dose-escalation was staggered in four-week intervals and at each dose escalation the ratio of placebo-immunized to PfSPZ-CVac-immunized subjects was 5:9. Following PfSPZ dose escalation, accelerated regimens (14- and 5-day intervals) were assessed using essentially the same procedures. A full report will be published separately. Eight to ten weeks after last vaccine or placebo dose (51–67 days after last chloroquine dose), protective efficacy was assessed by CHMI using DVI with PfSPZ Challenge12,13.
Procedures.
Daily thick blood smears were performed as previously described37 from day 6 to 21, following each DVI for immunization and CHMI, during antimalarial treatment and at each late follow-up visit. Slides were considered positive when at least two readers detected two unambiguous parasites, each. A negative slide was defined as no observed parasites in the volume of blood required to detect with 95% probability less than 10 parasites per μl (~0.5 μl). In case of discordance, a third reading was performed. In addition, 1.2 ml blood was sampled in EDTA tubes (Sarstedt) for nucleic acid extraction at the same time-points. DNA extraction control 610 (Bioline) was added and total nucleic acid (DNA and RNA) was isolated from 0.5 ml blood using the QIAamp DNA blood mini kit (Qiagen) according to the manufacturer’s specifications but without addition of RNase. Parasitaemia was calculated from a standard curve generated from serially diluted (2–20,000 parasites per ml) Pf 3D7 ring-stage synchronized cultured parasites, counted by microscopy and cytometry. All purified nucleic acid samples were stored at −20 °C until time of use. Reverse transcription quantitative polymerase chain reaction (RT–qPCR) was performed using TaqMan RNA-to-CT 1-Step Kit using published primers and probes38, with a different fluorophore and addition of a minor groove binder (probe: VIC-ATGGCCGTTTTTAGTTCGTG-NFQMGB; primers: 5′-GCTCTTTCTTGATTTCTTGGATG-3′ and 5′AGCAGGTTAAGATCTCGTTCG-3′). Reactions were done in 384 wells at 48 °C for 20 min (reverse transcription), 96 °C for 10 min (enzyme activation), and 45 cycles of 95 °C for 15 s, 62 °C 1 min. Samples were run as triplicates with no-template control, no-RT control and positive controls in the same plate. Amplification controls were assessed manually and cycle values (Cq) were calculated with the second derivative maximum method (LightCycler 480 software; version 1.5.1.62). The assay was validated in accordance to MIQE guidelines38,39 and had a lower limit of quantification of 3 parasites per ml. qPCR results were not reported to the clinical and microscopy teams during the study period to maintain blinding of the study.
Statistics.
Sample size was calculated with the intention to show, with a power of 80% and a two-tailed alpha of 5%, a difference in proportion of infected volunteers of 25% or less of immunized volunteers and 99% in controls, allocated in a 2:1 ratio (8 PfSPZ:4 controls), allowing for one dropout in each group (9:5). Clinical data were captured on paper case report forms and transferred to an electronic database (OpenClinica; version 3.2) by double data entry. Efficacy data were reported as proportions (primary) and time to parasitaemia. Safety and tolerability data were listed and reported as summary statistics. Results of immunological assays were explored by post hoc analyses and used to generate hypotheses about correlates and immunological mechanisms of protection. Analyses were coded in R (version 3.2.3)40 when not otherwise stated. Where possible, estimate and 95% confidence interval are given. Box plots display median (middle line), 25th (lower hinge) and 75th (upper hinge) quartile. Whiskers extend to values within 1.5× the inter-quartile ranges of the lower and upper hinges, respectively. A two-sided P value less than 5% was considered statistically significant.
Flow cytometry data were analysed with Pestle v1.7, SPICE v5.3 (ref. 41) and Prism 6 (GraphPad). Graphs were rendered in FlowJo, SPICE, and Prism. For vaccine immunogenicity, comparisons to pre-vaccine were performed using Wilcoxon signed rank test with Bonferroni correction for multiple comparisons or two-way ANOVA with Bonferroni correction, as specified in the text. Immune responses assessed at baseline, two weeks after final immunization, and at the time of CHMI were compared to outcome (parasitaemia or no parasitaemia) after CHMI. Assessment of immune responses that correlated with sterile protection was made using a stratified Wilcoxon test controlling for vaccine dose group as a covariate.
Antibody responses.
Sera were assessed for vaccine-induced antibodies by ELISA (enzyme-linked immunosorbent assay), immunofluorescence assay, inhibition of sporozoite invasion assay and protein arrays representing 91% of the Pf proteome.
ELISA procedure.
ELISAs were performed for antigens first expressed in PfSPZ (PfCSP, PfSSP2/TRAP, PfCelTOS, PfMSP5, PfAMA1), early liver stages (PfEXP1 and PfLSA1) and late liver stages (PfMSP1 and PfEBA175). The ELISA methods for each antigen are described below.
Recombinant (r) proteins used in ELISA assays are listed in Supplementary Table 7. 96-well plates (Nunc Maxisorp Immuno Plate) were coated overnight at 4 °C with 0.5 μg to 2.0 μg recombinant proteins (Supplementary Table 7) in 50 μl per well in coating buffer (KPL). Plates were washed three times with 2 mM imidazole, 160 mM NaCl, 0.02% Tween 20, 0.5 mM EDTA and blocked with 1% Bovine Serum Albumin (BSA) blocking buffer (KPL) containing 1% or 5% non-fat dry milk (Supplementary Table 7) for 1 h at 37 °C. Plates were washed three times and serially diluted serum samples (in triplicates) were added and incubated at 37 °C for 1 h. After three washes, peroxidase labelled goat anti-human IgG (KPL) was added at a dilution of 0.05 μg ml−1 to 0.2 μg ml−1 (Supplementary Table 7) and incubated at 37 °C for 1 h. Plates were washed three times, ABTS peroxidase substrate was added for plate development, and the plates were incubated for defined periods at 22 °C room temperature (Supplementary Table 7). The plates were read with a Spectramax Plus384 microplate reader (Molecular Devices) at 405 nm. The data were collected using Softmax Pro GXP v5 and fit to a 4-parameter logistic curve, to calculate the serum dilution at OD 1.0. A negative control (pooled serum from non-immune individuals from malaria free area) was included in all assays. The following positive control sera were used for the different antigens: serum from an individual with anti-PfCSP antibodies for PfCSP; pooled sera from individuals immunized with PfLSA-1 and PfEBA-175 subunit vaccines respectively for PfLSA1 and PfEBA175; pooled sera from volunteers from a malaria-endemic area (acquired from a blood bank in Kenya) for PfAMA1, PfEXP1, and PfMSP1. No positive control sera were available for PfMSP5, PfSSP2/TRAP or PfCelTOS.
Samples were considered positive if the difference between the post-immunization OD 1.0 and the pre-immunization OD 1.0 (net OD 1.0) was ≥50 and the ratio of post-immunization OD 1.0 to pre-immunization OD 1.0 (ratio) was ≥3.
Immunofluorescence assay.
Purified PfSPZ (NF54 strain) from aseptic Anopheles stephensi mosquitoes produced by Sanaria were resuspended in phosphate buffered saline (PBS (pH 7.4)) to obtain 5 × 103 PfSPZ per 40 μl 40 μl (5 × 103 PfSPZ) were added to each well of Greiner cellstar clear-bottom black 96-well plates (Sigma-Aldrich). After addition of the suspension, plates were left at room temperature for 12–18 h for air-drying.
Immunofluorescence procedure.
50 μl of sera diluted in PBS with 2% BSA were added to each well of the 96-well plate containing air-dried PfSPZ. Sera samples were added at twofold dilutions starting at 1:50. After adding samples, plates were incubated at 37 °C for 1 h. Plates were washed in PBS three times on an Aquamax Microplate washer. Alexa Fluor 488 conjugated goat anti-human IgG (Molecular Probes) was diluted to 1:250 in PBS with 2% BSA and 50 μl was added to each well. The plates were then incubated for 1 h at 37 °C. Plates were washed three times with PBS on an Aquamax Microplate washer. 100 μl PBS was added to each well. The plates were sealed using a plate sealer and stored in the dark at 4 °C until data acquisition. Samples were assessed by scanning the plates using an Acumen eX3 laser scanning imaging cytometer. The positive control was pooled human serum taken two weeks after the last immunization from 12 uninfected volunteers immunized 4 or 5 times with 1.35 × 105 PfSPZ Vaccine5. The Acumen image cytometer scans the entire surface area of each well in a 96-well plate and the fluorescence intensity values (arbitrary units) therefore represent the signal from all 5 × 103 PfSPZ that were seeded in each well.
Immunofluorescence analysis and readout.
The data obtained from the Acumen image cytometer were plotted to fit a 4-parameter sigmoidal curve in GraphPad Prism software using serum dilution (log transformed) as the x axis variable and arbitrary fluorescence units (AFU) on the y axis. Over many iterations during development of this assay we have determined that sera from naive volunteers in the USA and Europe, including pre-immune sera, always register an arbitrary fluorescence value less than 2 × 105 even at the highest concentration (1:50 dilution, see above) used in this assay. Moreover, for sera that react to PfSPZ, 2 × 105 AFU falls in the exponential portion of their sigmoidal curves. Therefore, 2 × 105 was chosen as a threshold in the automated immunofluorescence assay and the results for each volunteer for antibodies to PfSPZ are reported as the reciprocal serum dilution at which fluorescence intensity was equal to 2 × 105 AFU.
Inhibition of sporozoite invasion.
HC-04 (1F9) (ref. 42) cells (hepatocytes) were obtained from the Naval Medical Research Center. Master and working cell banks were produced, and after establishing they were free of mycoplasma, were quality control released. For the assay they were cultured in complete medium (10% FBS in DMEM/F12 with 100 units per ml penicillin and 100 μg per ml streptomycin; Gibco by Life Technologies) in Entactic-Collagen IV-Laminin (ECL)-coated 96-well clear bottom black well plates (Greiner Bio-One North America) at a density of 2.5 × 104 cells per well, and incubated for 24 h at 37 °C, 5% CO2 with 85% relative humidity. Twenty-four hours later cells were infected with 104 aseptic, purified, cryopreserved PfSPZ per well, without or with sera diluted in a 12-point series from subjects immunized with PfSPZ Vaccine. The assay control included PfSPZ added with media alone. All subjects were assessed at pre-immunization (baseline), post-immunization and pre-CHMI time points. Three hours later, PfSPZ that had not invaded the HC-04 cells were removed by washing three times with Dulbecco’s phosphate-buffered saline (DPBS), and the cultures were fixed using 4% paraformaldehyde for 15 min at room temperature. Differential immunostaining was performed to differentiate between PfSPZ inside the hepatocytes versus PfSPZ outside the hepatocytes. PfSPZ outside the hepatocytes were stained with an anti-PfCSP mAb (2A10, 6.86 μg ml−1) (Protein Potential LLC, with permission from New York University School of Medicine) conjugated with Alexa Fluor 633 (far-red) (1 μg ml−1; custom-conjugated at GenScript). The hepatocytes were then permeabilized with 0.1% Triton X-100 for 10 min at room temperature, and the PfSPZ inside the hepatocytes were stained with the anti-PfCSP mAb (2A10, 6.86 μg ml−1) conjugated with Alexa Fluor 488 (green; 1 μg ml−1, conjugated from Genscript). The numbers and intensity of infected hepatocytes (green only) were counted by scanning the plates using an Acumen eX3 laser scanning imaging cytometer. The data obtained from the Acumen image cytometer were plotted to fit a 4-parameter sigmoidal curve in GraphPad Prism software using serum dilution (log transformed) as the x axis variable and arbitrary fluorescence units on the y axis. 75% inhibition was interpolated from the sigmoidal curves as the reciprocal serum dilution at which the fluorescent intensity of infected wells with serum was 25% of the negative control without serum. The number of invaded PfSPZ scored in this assay in the absence of sera ranged from 400–600 (intensity of 1–3 × 106 fluorescence units) (4% to 6% of those added to the wells).
Proteome array.
A whole-proteome microarray with 91% coverage of the Pf proteome (PfWPM) was produced by Antigen Discovery, Inc. (ADI). Proteins were expressed as previously described43 from a library of Pf partial or complete open reading frames (ORFs) cloned into a T7 expression vector pXI using an in vitro transcription and translation (IVTT) system, the Escherichia coli cell-free Rapid Translation System (RTS) kit (5 Prime). The library was created through an in vivo recombination cloning process with PCR-amplified Pf ORFs, and a complementary linearized expressed vector transformed into chemically competent E. coli was amplified by PCR and cloned into pXI vector using a high-throughput PCR recombination cloning method described elsewhere44. Each expressed protein includes a 5′ polyhistidine (HIS) epitope and 3′ haemagglutinin (HA) epitope. After expressing the proteins according to manufacturer’s instructions, translated proteins were printed onto nitrocellulose-coated glass AVID slides (Grace Bio-Labs) using an Omni Grid Accent robotic microarray printer (Digilabs, Inc.). Microarray chip printing and protein expression were quality checked by probing random slides with anti-HIS and anti-HA monoclonal antibodies with fluorescent labelling. PfWPM chips contained 7,455 Pf peptide fragments, representing proteins from 4,805 unique genes, 302 IgG positive control spots and 192 spotted IVTT reactions without Pf ORFs (IVTT controls). For each PfWPM chip, 3 replicates were printed per microarray slide on 3 nitrocellulose pads. IgG-positive control spots were included as an assay control, whereas IVTT control spots were included as a sample-level normalization factor. Serum samples were diluted 1:100 in a 3 mg ml−1 E. coli lysate solution in protein arraying buffer (Maine Manufacturing) and incubated at room temperature for 30 min. Chips were rehydrated in blocking buffer for 30 min. Blocking buffer was removed, and chips were probed with pre-incubated serum samples using sealed, fitted slide chambers to ensure no cross-contamination of sample between pads. Chips were incubated overnight at 4 °C with agitation. Chips were washed five times with TBS-0.05% Tween 20, followed by incubation with biotin-conjugated goat anti-human IgG (Jackson ImmunoResearch) diluted 1:200 in blocking buffer at room temperature. Chips were washed three times with TBS-0.05% Tween 20, followed by incubation with streptavidin-conjugated SureLight P-3 (Columbia Biosciences) at room temperature protected from light. Chips were washed three times with TBS-0.05% Tween 20, three times with TBS, and once with water. Chips were air dried by centrifugation at 1,000g for 4 min and scanned on a ScanArray Express HT spectral scanner (Perkin-Elmer), and spot and background intensities were measured using an annotated grid file (.GAL). Data were exported and normalized using the IVTT control spots for statistical analysis in R40.
Proteome microarray data processing.
Raw spot and local background fluorescence intensities, spot annotations and sample phenotypes were imported and merged in R, where all subsequent procedures were performed40. Foreground spot intensities were adjusted by local background by subtraction, and negative values were converted to 1. Next, all foreground values were transformed using the base 2 logarithm (log2). The dataset was normalized to remove systematic effects by subtracting the median signal intensity of the IVTT controls for each sample. As the IVTT control spots carry the chip, sample and batch-level systematic effects, but also antibody background activity to the IVTT system, this procedure normalizes the data and provides a relative measure of the specific antibody binding to the non-specific antibody binding to the IVTT controls (a.k.a. background). With the normalized data, a value of 0.0 means that the intensity is no different than the background and a value of 1.0 indicates a doubling with respect to background. A seropositivity threshold was established for each protein on the chip using the top 2.5th percentile of the pre-immunization samples for each protein. Reactive antigens were defined as those that had seropositive responses after immunization and before CHMI, but which did not show seropositive responses in the mock-immunization group.
T-cell responses.
PBMCs were isolated by density-gradient centrifugation from heparinized whole blood. Assessment of cellular immune responses using multi-parameter flow cytometry was performed on PBMCs from cryopreserved samples at the completion of the study, as described6. In brief, PBMCs were thawed and rested in complete RPMI for 8 h followed by stimulation for 17 h with: (1) 1.5 × 105 viable, irradiated, aseptic, purified, cryopreserved PfSPZ from a single production lot; (2) PfSPZ Vaccine diluent (1% human serum albumin, HSA, CSL Behring); (3) 2 × 105 lysed RBC, >90% infected with late-stage schizonts (PfRBC) from a single production lot; or (4) 2 × 105 donor-matched uninfected erythrocytes from a single production lot. For the last 5 h of the stimulation, 10 μg ml−1 Brefeldin A (BD) was added to the culture.
After stimulation, cells were stained as previously described45. The staining panels are shown in Supplementary Table 8 and the antibody clones and manufacturers are shown in Supplementary Table 9. Briefly, cells were surface stained with CCR7 at 37 °C for 20 min. Dead cells were identified by Aqua Live-Dead dye (Invitrogen), as per manufacturer’s instructions. This was followed by 15 min surface staining at room temperature for CD4, CD8, CD14, CD38, CD45RA, CD56, CD57, CD127, CD161, TCR-γδ, TCR-Vδ1, TCR-Vδ2, TCR-Vγ9, TCR-Vα7.2, CXCR6, or PD-1. Cells were washed, fixed, and permeabilized using Cytofix/Cytoperm kit (BD) and stained intracellularly for CD3, IFN-γ, IL-2, TNF-α, IL-4, IL-10, perforin, or Ki-67. Cells were washed, fixed in 0.5% paraformaldehyde, and measured on a modified LSR II (BD).
Flow cytometry data were analysed using FlowJo v9.9 (Tree Star) blinded to vaccination group and CHMI outcome. All antigen-specific cytokine frequencies are reported after background subtraction of identical gates from the same sample incubated with the control antigen stimulation (HSA or uninfected erythrocytes).
Data availability.
The data that support the findings of these studies are available in part on request from the corresponding author (S.L.H.) subject to restrictions. Some data are not publicly available, as they contain information that could compromise research participant privacy/consent.
Extended Data
Extended Data Figure 1 |. Distribution of adverse events.
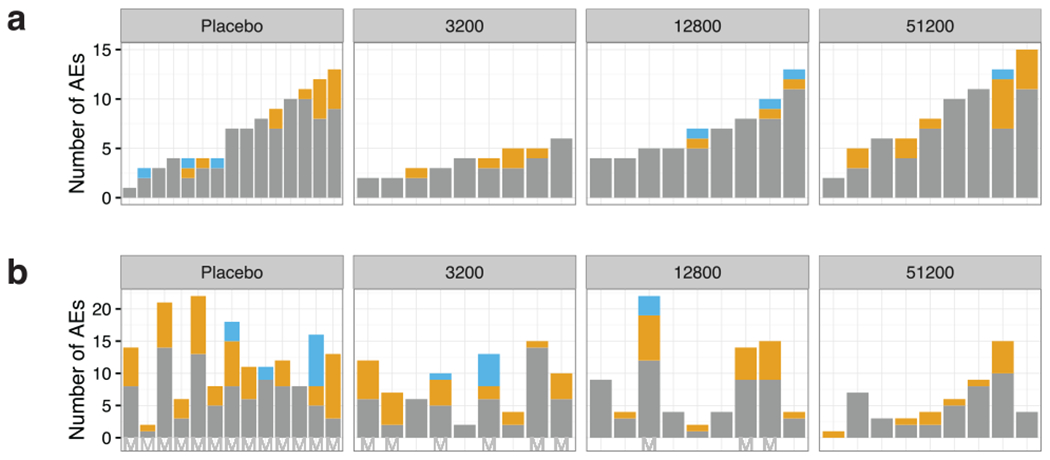
a, b, The number of adverse events (AEs) regardless of attribution to investigational product. Each bar represents one volunteer sorted on the number of adverse events from the time of first injection with normal saline (controls) or PfSPZ-CVac until the time of CHMI, approximately 17 weeks later (a) and adverse events in the same volunteers from initiation of CHMI until the end of follow-up (b). Mild (grade 1) adverse events are depicted in grey, moderate (grade 2) in yellow and severe (grade 3) in blue. Non-protected volunteers are marked with an ‘M’ on the x axis.
Extended Data Figure 2 |.
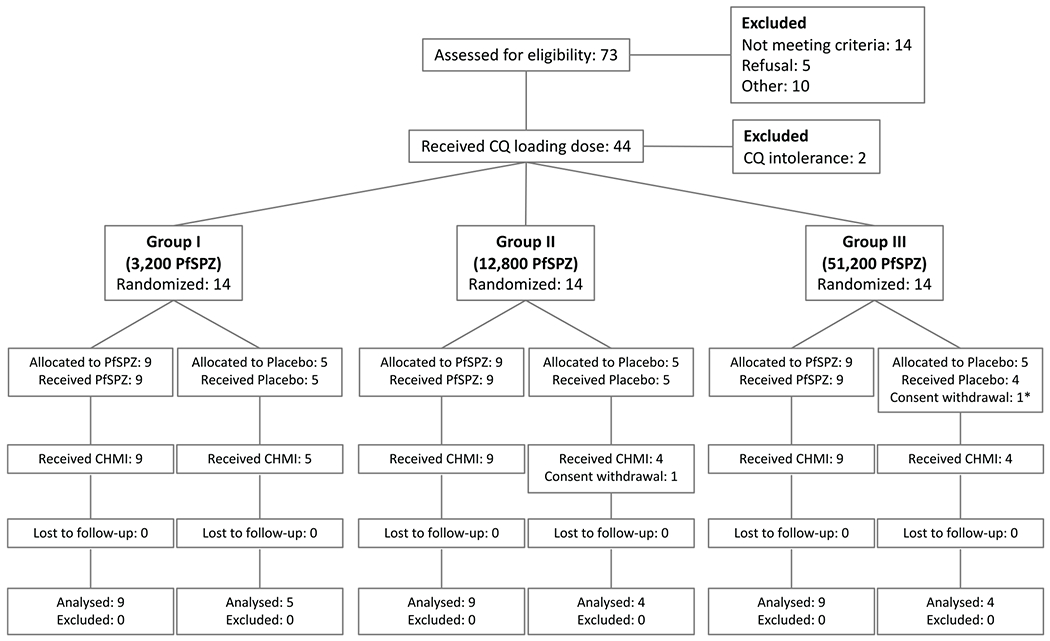
CONSORT study flow chart.
*Participant received only the first Placebo injection
Extended Data Figure 3 |. CD4 T-cell cytokine polyfunctionality.
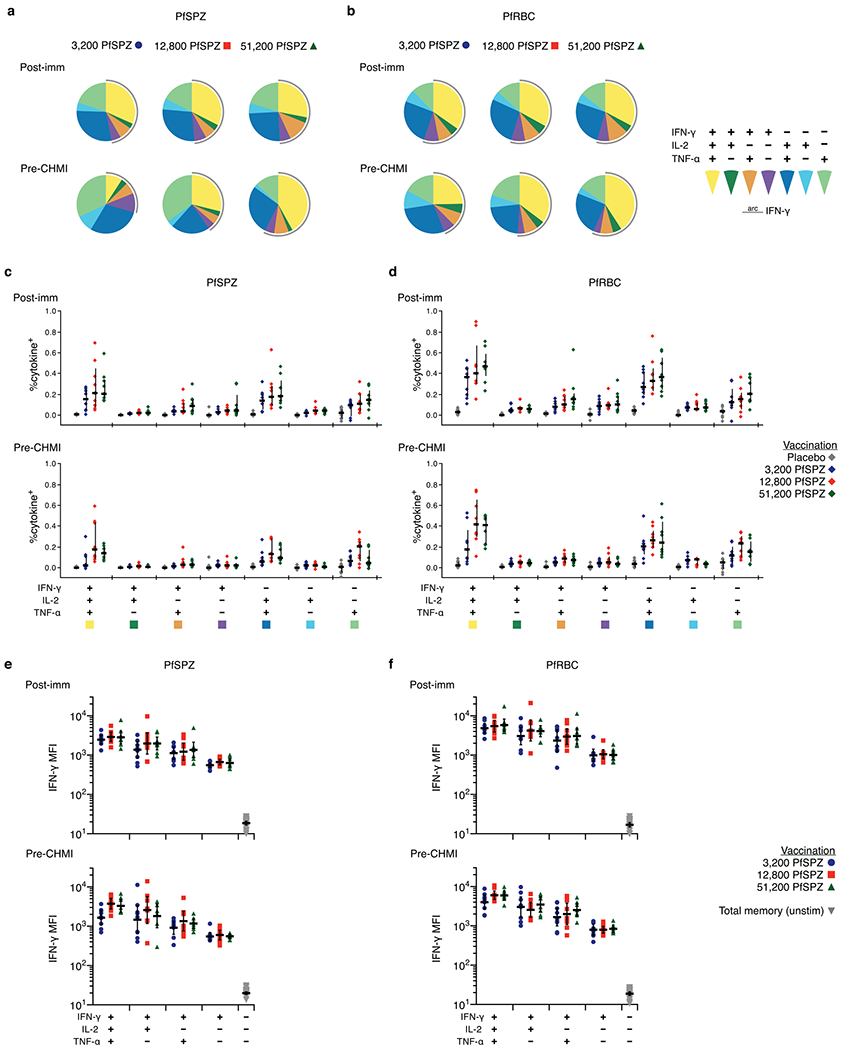
PBMCs from subjects were drawn 14 days after third immunization (post-imm) or 1 day before CHMI (pre-CHMI), stimulated with PfSPZ, PfRBC, or stimulation controls, and stained for intracellular cytokine expression. a, b, The pie charts show the proportion of memory CD4 T cells expressing any combination of IFN-γ, IL-2, or TNF-α for each dose group after stimulation with PfSPZ (a) or PfRBC (b). Responses are background subtracted from control antigen stimulations 1% HSA or uninfected erythrocytes. c, d, The magnitude of the memory CD4 T-cell response for each combination of cytokines is shown in c and d. There is a trend towards higher polyfunctionality as dose increases. e, f, The median fluorescence intensity (MFI) for IFN-γ is shown for the different combination of IFN-γ+ cells following PfSPZ or PfRBC stimulation. Cells that simultaneously produce IFN-γ, IL-2, and TNF-α have the highest IFN-γ MFI.
Extended Data Figure 4 |. T-cell immunogenicity.
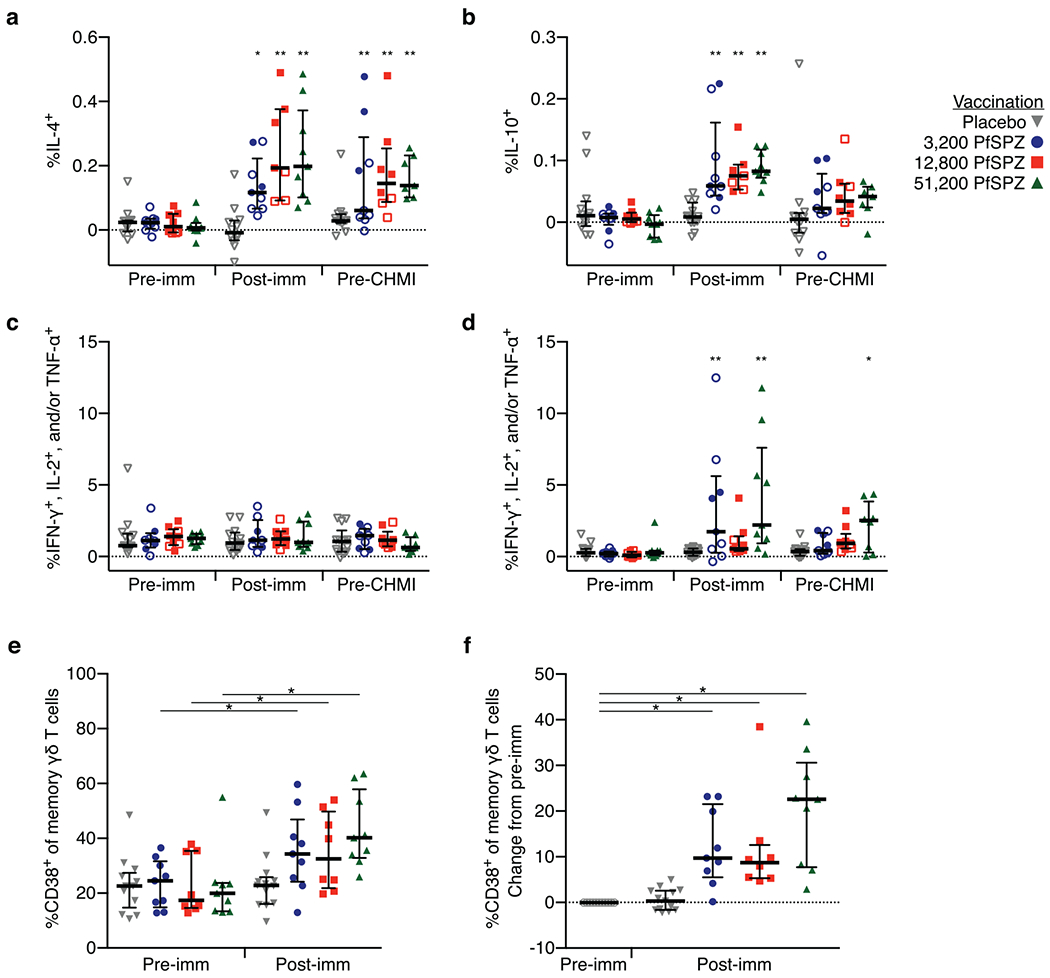
a, b, Memory CD4 T cells producing IL-4 (a) or IL-10 (b) after PfRBC stimulation. Memory γδ T cells producing IFN-γ, IL-2, or TNF-α following PfSPZ (c) or PfRBC stimulation (d). For a–d, results are the percentage of cytokine-producing cells after incubation with PfSPZ minus the percentage of cells after incubation with vaccine diluent (medium with 1% HSA) as control or percentage of cytokine-producing cells after incubation with asexual Pf-infected red blood cells (PfRBC) minus uninfected RBCs as control. e, f, Total memory γδ T cells assessed before immunization (pre-imm) and 14 days after third immunization (post-imm) for the percentage of cells expressing CD38. The absolute frequencies are shown in e and the change from pre-vaccination to post-vaccination is shown in f. For a–d, within a dose group, the difference from pre-vaccine was assessed by two-way ANOVA with Bonferroni correction. Data are median ± interquartile range. For e, f, difference from pre-vaccine was assessed by Wilcoxon signed rank test. P values were corrected for multiple comparisons by the Bonferroni method. *P < 0.05, *P < 0.01. Data are median ± interquartile range. Pre-imm, 3 days before first immunization; post-imm, 14 days after third immunization; pre-CHMI, 1 day before CHMI.
Extended Data Figure 5 |. Sub-family analysis of γδ T cells.
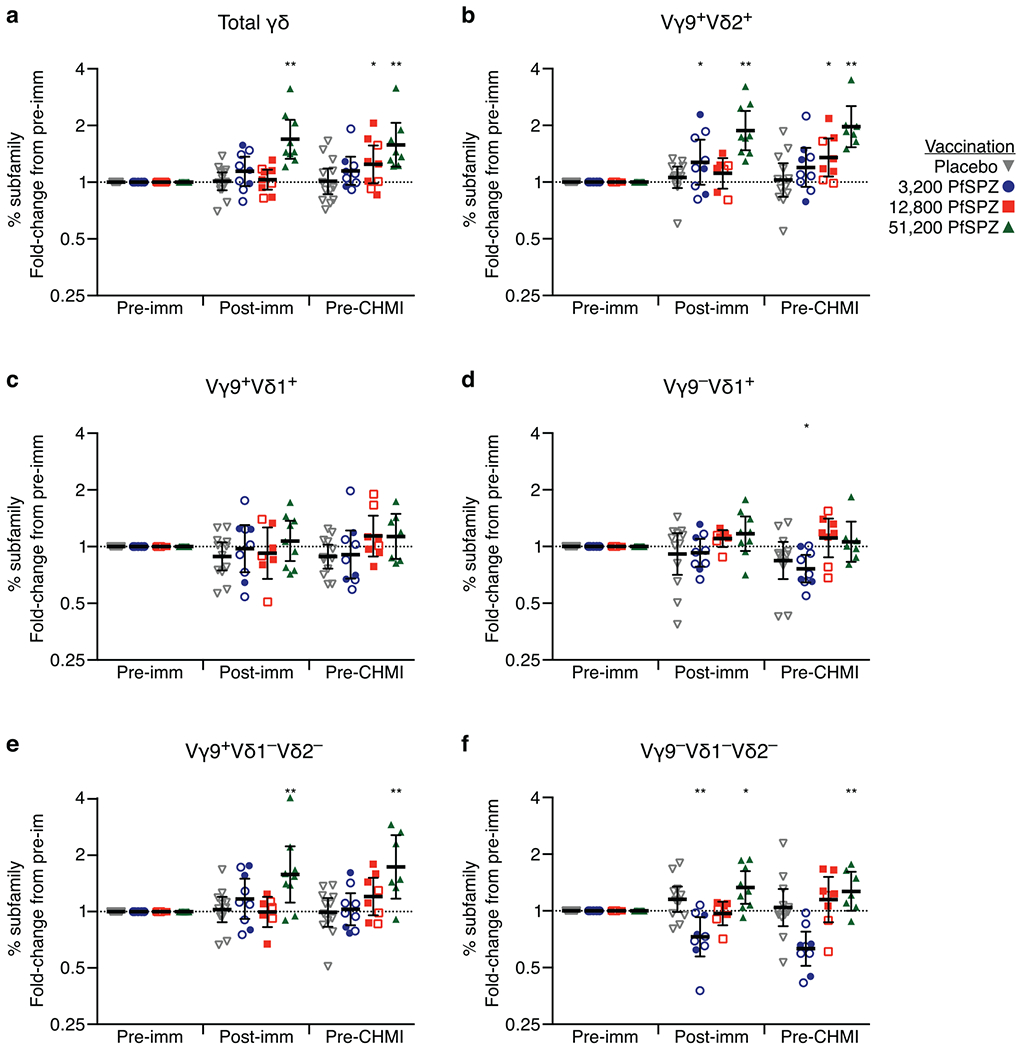
a–f, The frequency of the circulating γδ T-cell subsets as a percentage of total lymphocytes was assessed in unstimulated PBMCs before the first immunization (pre-imm), 2 weeks after final immunization (post-imm), and the day before CHMI (pre-CHMI). Fold change compared to pre-imm is shown for total memory γδ T cells (a), Vγ9+Vδ2+ (b), Vγ9+Vδ1+ (c), Vγ9−Vδ1+ (d), Vγ9+Vδ1−Vδ2− (e), and Vγ9−Vδ1−Vδ2− (f) subfamilies. The frequency of Vγ9−Vδ2+ subset is low to undetectable. Within a dose group, the difference from pre-imm was assessed by two-way ANOVA with Bonferroni correction. *P < 0.05, **P < 0.01. Data are geometric mean ± 95% CI.
Extended Data Figure 6 |. Anti-plasmodial antibody responses in vaccinated volunteers who were immunized with three doses of 5.12 × 104 PfSPZ at 28-day, 14-day, or 5-day intervals.
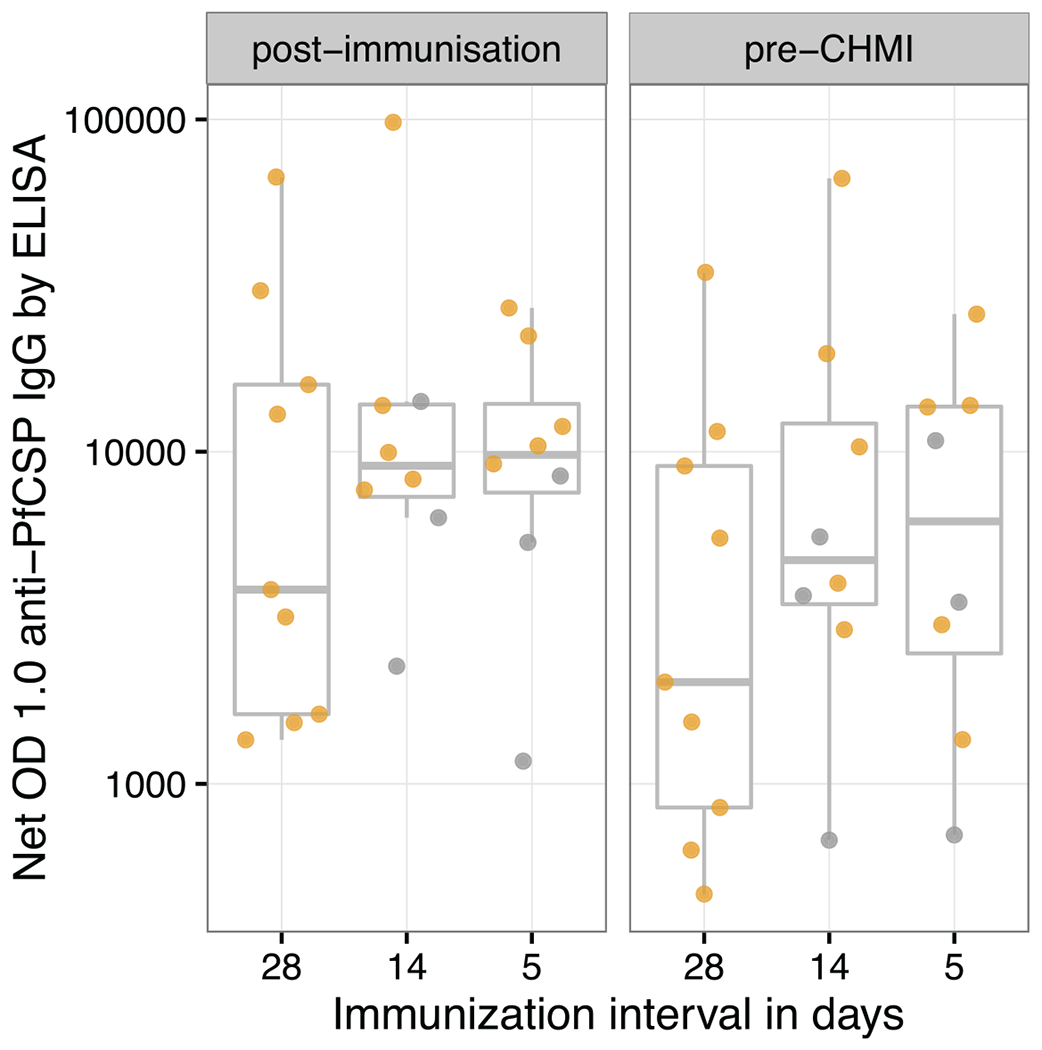
Antibodies to PfCSP by ELISA were assessed in sera taken before any immunizations (pre-immunization), two weeks following last immunization (post-immunization) and 10 weeks after last immunization, which was one day before CHMI (pre-CHMI). PfCSP ELISA results are reported as net OD 1.0; the reciprocal serum dilution at which the optical density was 1.0 in post-immunization or pre-CHMI sera minus the OD 1.0 in pre-immunization sera. All values met criteria for positivity. Protected volunteers are represented by yellow circles and unprotected volunteers by grey circles.
Extended Data Figure 7 |. Development of PfSPZ Vaccine and PfSPZ-CVac in hepatocytes.
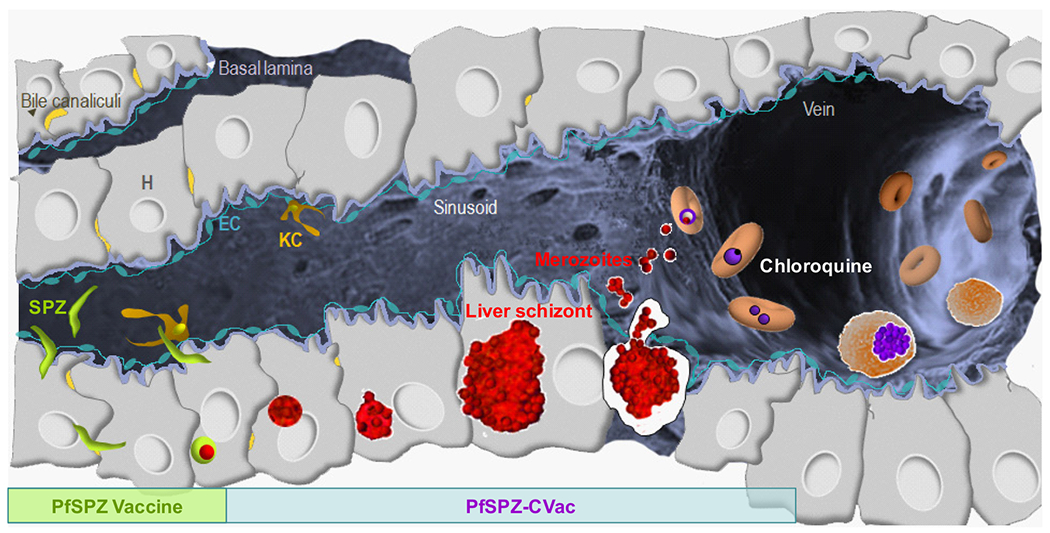
Radiation-attenuated PfSPZ in PfSPZ Vaccine invade hepatocytes and partially develop, but do not replicate. They are metabolically active and non-replicating. Infectious PfSPZ in PfSPZ-CVac invade hepatocytes and fully develop. A single PfSPZ replicates exponentially producing more than 104 merozoites. These merozoites are released into the circulation, and each merozoite can invade a different erythrocyte. Chloroquine prevents complete parasite development within erythrocytes, thereby preventing the development of merozoites that can invade new erythrocytes.
Extended Data Figure 8 |. Transient parasitaemia following vaccination at 5-day intervals.
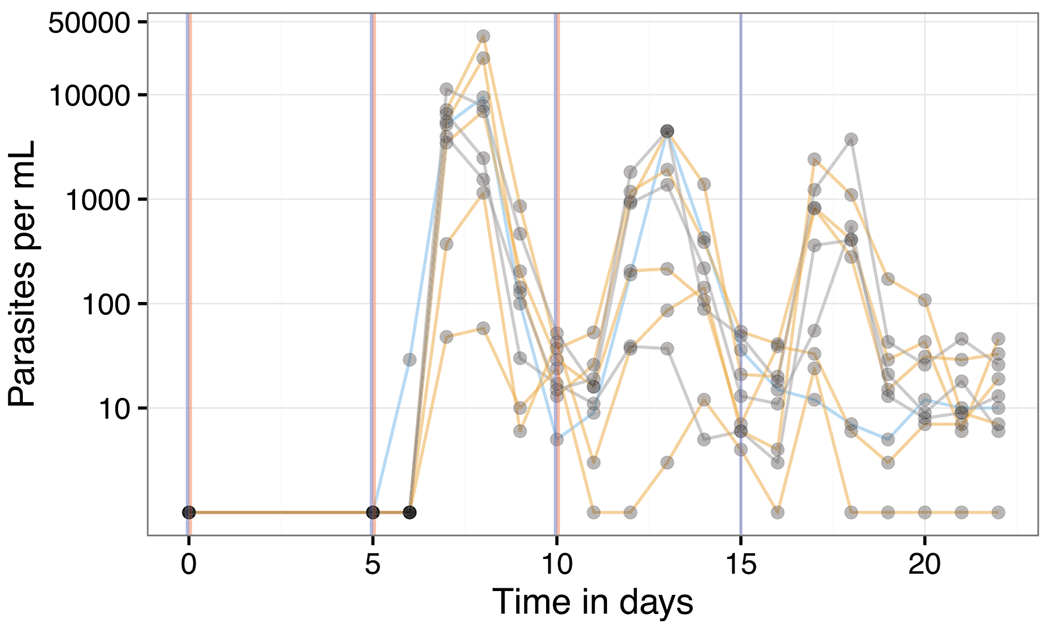
Parasitaemia measured by qPCR over 22 days. The subjects who were protected and not protected against CHMI are in yellow and grey, respectively. The times of PfSPZ inoculations are shown as vertical red lines and the time of last CQ administration as a vertical blue line. CQ was given as 10 mg kg−1 (maximum 620 mg) loading dose on day 0 followed by 5 mg kg−1 (maximum 310 mg) chloroquine base on days 5, 10, and 15.
Extended Data Table 1 |.
Pf-specific T-cell correlates of sterile protection, stratified by group
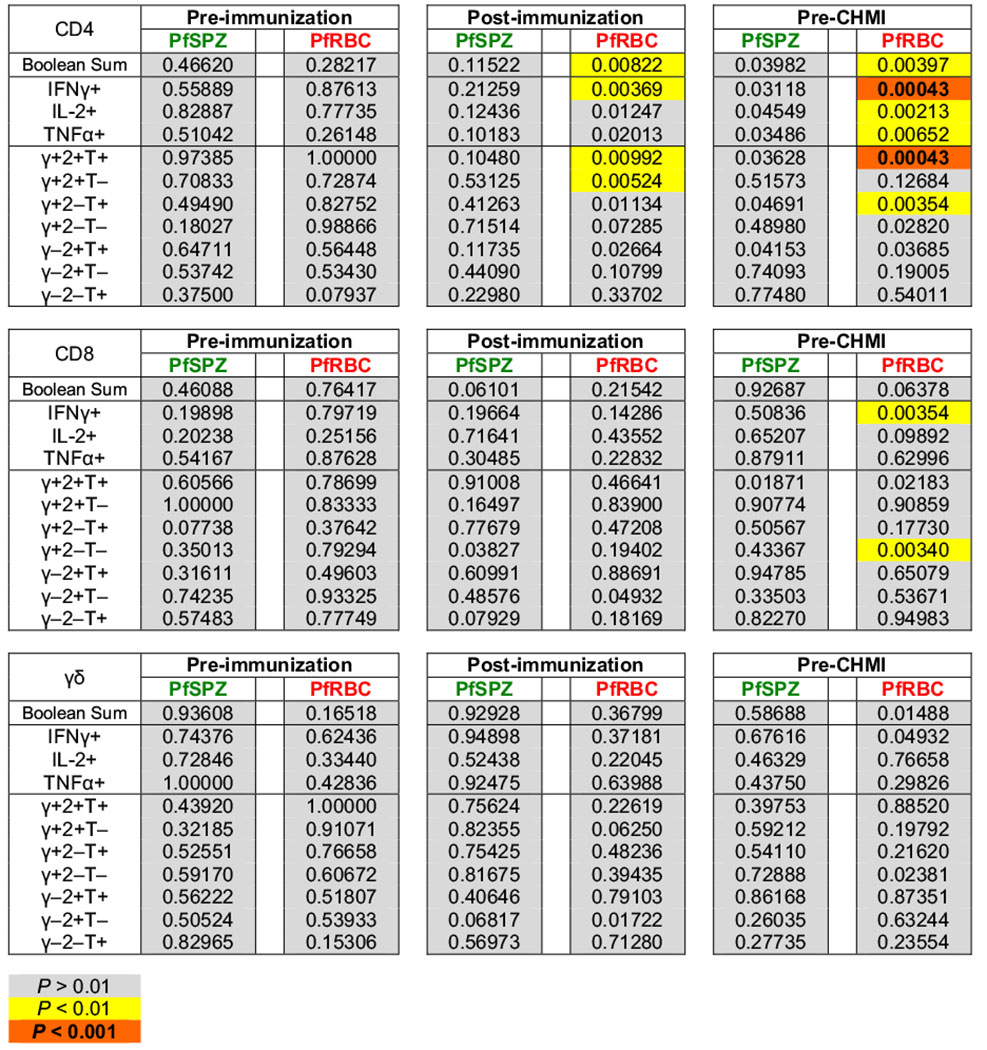
|
A complete table of P values is shown for the association of each individual immune parameter with sterile protection (parasitaemia versus no parasitaemia) after CHMI. P values were determined by stratified Wilcoxon test controlling for vaccine dose as a co-variate. Boolean sum is the total cytokine-positive response (IFN-γ, IL-2, or TNF-α). T cells positive for more than one cytokine are only counted once. γ = IFN-γ; 2 = IL-2; T = TNF-α. P values less than 0.01 are highlighted. In this exploratory analysis, P values are intentionally not corrected for multiple comparisons.
Supplementary Material
Acknowledgements
The authors thank the vaccine trial participants for their contribution and commitment to vaccine research. We thank F. Adomat, S. Adukpo, M. Aldejohann, S. Bolte, S. Borrmann, A. Bouyoukou Hounkpatin, S. Brückner, E. Bruske, J. Fernandes, P. Granados Bayün, J. Hass, S. Jeyaraj, J. Keim, A. Knoblich, R. Köllner, A. Kreidenweiss, D. N. Ndungu, R. Ritter, J. A. Selvaraj, Z. Sulyok, S. Theil, N. Theurer, and I. Westermann for support in conducting the trial, and P. Darrah and M. Roederer for assistance with the interpretation of the T-cell data. We thank the Sanaria and Protein Potential teams for manufacture and shipping of investigational products, PfSPZ Challenge and diluents, regulatory, quality, and clinical site activities, and legal and administrative support, including especially D. Cheney (née Padilla), Y. Abebe, E. Saverino, Y Wu, E. Fomumbod, A. Awe, M. King, M. Orozco, A. Patil, Y. Wen, K. Nelson, J. Overby, S. Matheny, V. Pitch, B. Jiang, L. Gao, R. Xu, T. T. Wai, S. Monsheimer, P. De La Vega, M. Laskowski, H. Huang, M. Marquette, J. Jackson, F. Beams, R. Douglas, R. C. Thompson, D. Dolberg and A. Hoffman. We thank J. Inglese and P. Dranchak of the National Center for Advancing Translational Sciences (NCATS), NIH for support with the automated immunofluorescence assay and inhibition of sporozoite invasion assays. We appreciate the expert reviews of the Safety Monitoring Committee (W. Chen, P. Coyne and P. Zanger). The clinical trial was funded by the German Federal Ministry of Education and Research (BMBF) through the German Center for Infection Research (DZIF). Manufacture of investigational product by Sanaria was supported in part by the National Institute of Allergy and Infectious Diseases of the National Institutes of Health under SBIR award numbers 5R44AI058375 and 5R44AI055229. T cell studies were supported by the intramural research program of the VRC, NIAID, NIH. Proteome microarray studies were supported by NIAID SBIR grant 5R44AI066791 and funding from the Bill & Melinda Gates Foundation.
Footnotes
Online Content Methods, along with any additional Extended Data display items and Source Data, are available in the online version of the paper; references unique to these sections appear only in the online paper.
Supplementary Information is available in the online version of the paper.
The authors declare competing financial interests: details are available in the online version of the paper. Readers are welcome to comment on the online version of the paper.
References
- 1.malERA Consultative Group on Vaccines. A research agenda for malaria eradication: vaccines. PLoS Med. 8, e1000398 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Clyde DF, Most H, McCarthy VC & Vanderberg JP Immunization of man against sporozite-induced falciparum malaria. Am. J. Med. Sci 266, 169–177 (1973). [DOI] [PubMed] [Google Scholar]
- 3.Rieckmann KH, Carson PE, Beaudoin RL, Cassells JS & Sell KW Letter: Sporozoite induced immunity in man against an Ethiopian strain of Plasmodium falciparum. Trans. R. Soc. Trop. Med. Hyg 68, 258–259 (1974). [DOI] [PubMed] [Google Scholar]
- 4.Hoffman SL et al. Protection of humans against malaria by immunization with radiation-attenuated Plasmodium falciparum sporozoites. J. Infect. Dis 185, 1155–1164 (2002). [DOI] [PubMed] [Google Scholar]
- 5.Seder RA et al. Protection against malaria by intravenous immunization with a nonreplicating sporozoite vaccine. Science 341, 1359–1365 (2013). [DOI] [PubMed] [Google Scholar]
- 6.Ishizuka AS et al. Protection against malaria at 1 year and immune correlates following PfSPZ vaccination. Nat. Med 22, 614–623 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bijker EM et al. Cytotoxic markers associate with protection against malaria in human volunteers immunized with Plasmodium falciparum sporozoites. J. Infect. Dis 210, 1605–1615 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bijker EM et al. Protection against malaria after immunization by chloroquine prophylaxis and sporozoites is mediated by preerythrocytic immunity. Proc. Natl Acad. Sci. USA 110, 7862–7867 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Roestenberg M. et al. Protection against a malaria challenge by sporozoite inoculation. N. Engl. J. Med 361, 468–477 (2009). [DOI] [PubMed] [Google Scholar]
- 10.Roestenberg M. et al. Long-term protection against malaria after experimental sporozoite inoculation: an open-label follow-up study. Lancet 377, 1770–1776 (2011). [DOI] [PubMed] [Google Scholar]
- 11.Bijker EM et al. Sporozoite immunization of human volunteers under mefloquine prophylaxis is safe, immunogenic and protective: a double-blind randomized controlled clinical trial. PLoS One 9, e112910 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gómez-Pérez GP et al. Controlled human malaria infection by intramuscular and direct venous inoculation of cryopreserved Plasmodium falciparum sporozoites in malaria-naive volunteers: effect of injection volume and dose on infectivity rates. Malar. J 14, 306 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mordmüller B. et al. Direct venous inoculation of Plasmodium falciparum sporozoites for controlled human malaria infection: a dose-finding trial in two centres. Malar. J 14, 117 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bastiaens GJ et al. Safety, immunogenicity, and protective efficacy of intradermal immunization with aseptic, purified, cryopreserved Plasmodium falciparum sporozoites in volunteers under chloroquine prophylaxis: a randomized controlled trial. Am. J. Trop. Med. Hyg 94, 663–673 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Beaudoin RL, Strome CPA, Mitchell F & Tubergen TA Plasmodium berghei: immunization of mice against the ANKA strain using the unaltered sporozoite as an antigen. Exp. Parasitol 42, 1–5 (1977). [DOI] [PubMed] [Google Scholar]
- 16.Guerin-Marchand C. et al. A liver-stage-specific antigen of Plasmodium falciparum characterized by gene cloning. Nature 329, 164–167 (1987). [DOI] [PubMed] [Google Scholar]
- 17.Orito Y. et al. Liver-specific protein 2: a Plasmodium protein exported to the hepatocyte cytoplasm and required for merozoite formation. Mol. Microbiol 87, 66–79 (2013). [DOI] [PubMed] [Google Scholar]
- 18.Tarun AS et al. A combined transcriptome and proteome survey of malaria parasite liver stages. Proc. Natl Acad. Sci. USA 105, 305–310 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Richie TL et al. Progress with Plasmodium falciparum sporozoite (PfSPZ)-based malaria vaccines. Vaccine 33, 7452–7461 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Epstein JE et al. Live attenuated malaria vaccine designed to protect through hepatic CD8+ T cell immunity. Science 334, 475–480 (2011). [DOI] [PubMed] [Google Scholar]
- 21.Suhrbier A, Winger LA, Castellano E & Sinden RE Survival and antigenic profile of irradiated malarial sporozoites in infected liver cells. Infect. Immun 58, 2834–2839 (1990). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Behet MC et al. Sporozoite immunization of human volunteers under chemoprophylaxis induces functional antibodies against pre-erythrocytic stages of Plasmodium falciparum. Malar. J 13, 136 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schofield L. et al. γ Interferon, CD8+ T cells and antibodies required for immunity to malaria sporozoites. Nature 330, 664–666 (1987). [DOI] [PubMed] [Google Scholar]
- 24.Weiss WR, Sedegah M, Beaudoin RL, Miller LH & Good MF CD8+ T cells (cytotoxic/suppressors) are required for protection in mice immunized with malaria sporozoites. Proc. Natl Acad. Sci. USA 85, 573–576 (1988). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Doolan DL & Hoffman SL The complexity of protective immunity against liver-stage malaria. J. Immunol 165, 1453–1462 (2000). [DOI] [PubMed] [Google Scholar]
- 26.Belnoue E. et al. Protective T cell immunity against malaria liver stage after vaccination with live sporozoites under chloroquine treatment. J. Immunol 172, 2487–2495 (2004). [DOI] [PubMed] [Google Scholar]
- 27.Weiss WR & Jiang CG Protective CD8+ T lymphocytes in primates immunized with malaria sporozoites. PLoS One 7, e31247 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fernandez-Ruiz D. et al. Liver-resident memory CD8+ T cells form a front-line defense against malaria liver-stage infection. Immunity 45, 889–902 (2016). [DOI] [PubMed] [Google Scholar]
- 29.Schats R. et al. Heterologous protection against malaria after immunization with Plasmodium falciparum sporozoites. PLoS One 10, e0124243 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Egan JE et al. Humoral immune responses in volunteers immunized with irradiated Plasmodium falciparum sporozoites. Am. J. Trop. Med. Hyg 49, 166–173 (1993). [DOI] [PubMed] [Google Scholar]
- 31.Epstein JE et al. Protection against Plasmodium falciparum malaria by PfSPZ Vaccine. JCI Insight 2, e89154 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lyke K. et al. PfSPZ Vaccine induces strain-transcending T cells and durable protection against heterologous malaria challenge. Proc. Natl Acad. Sci. USA (in the press). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sissoko MS et al. Safety and efficacy of PfSPZ Vaccine against Plasmodium falciparum via direct venous inoculation in healthy malaria-exposed Malian adults: a randomised, double-blind trial. Lancet Infect Dis. (in the press). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Roestenberg M. et al. Controlled human malaria infections by intradermal injection of cryopreserved Plasmodium falciparum sporozoites. Am. J. Trop. Med. Hyg 88, 5–13 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sahu T. et al. Chloroquine neither eliminates liver stage parasites nor delays their development in a murine chemoprophylaxis vaccination model. Front. Microbiol 6, 283 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fairley NH Chemotherapeutic suppression and prophylaxis in malaria. Trans. R. Soc. Trop. Med. Hyg 38, 311–365 (1945). [PubMed] [Google Scholar]
- 37.Planche T. et al. Comparison of methods for the rapid laboratory assessment of children with malaria. Am. J. Trop. Med. Hyg 65, 599–602 (2001). [DOI] [PubMed] [Google Scholar]
- 38.Kamau E, Alemayehu S, Feghali KC, Saunders D & Ockenhouse CF Multiplex qPCR for detection and absolute quantification of malaria. PLoS One 8, e71539 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bustin SA et al. The MIQE guidelines: minimum information for publication of quantitative real-time pCR experiments. Clin. Chem 55, 611–622 (2009). [DOI] [PubMed] [Google Scholar]
- 40.R Core Team. R: A Language and Environment for Statistical Computing. (2015).
- 41.Roederer M, Nozzi JL & Nason MC SPICE: exploration and analysis of post-cytometric complex multivariate datasets. Cytometry A 79, 167–174 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sattabongkot J. et al. Establishment of a human hepatocyte line that supports in vitro development of the exo-erythrocytic stages of the malaria parasites Plasmodium falciparum and P. vivax. Am. J. Trop. Med. Hyg 74, 708–715 (2006). [PubMed] [Google Scholar]
- 43.Felgner PL et al. Pre-erythrocytic antibody profiles induced by controlled human malaria infections in healthy volunteers under chloroquine prophylaxis. Sci. Rep 3, 3549 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Davies DH et al. Profiling the humoral immune response to infection by using proteome microarrays: high-throughput vaccine and diagnostic antigen discovery. Proc. Natl Acad. Sci. USA 102, 547–552 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lamoreaux L, Roederer M & Koup R Intracellular cytokine optimization and standard operating procedure. Nat. Protocols 1, 1507–1516 (2006). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that support the findings of these studies are available in part on request from the corresponding author (S.L.H.) subject to restrictions. Some data are not publicly available, as they contain information that could compromise research participant privacy/consent.


