Abstract
Chromatin assembly factor I (CAF-I) is a three-subunit histone-binding complex conserved from the yeast Saccharomyces cerevisiae to humans. Yeast cells lacking CAF-I (cacΔ mutants) have defects in heterochromatic gene silencing. In this study, we showed that deletion of HIR genes, which regulate histone gene expression, synergistically reduced gene silencing at telomeres and at the HM loci in cacΔ mutants, although hirΔ mutants had no silencing defects when CAF-I was intact. Therefore, Hir proteins are required for an alternative silencing pathway that becomes important in the absence of CAF-I. Because Hir proteins regulate expression of histone genes, we tested the effects of histone gene deletion and overexpression on telomeric silencing and found that alterations in histone H3 and H4 levels or in core histone stoichiometry reduced silencing in cacΔ mutants but not in wild-type cells. We therefore propose that Hir proteins contribute to silencing indirectly via regulation of histone synthesis. However, deletion of combinations of CAC and HIR genes also affected the growth rate and in some cases caused partial temperature sensitivity, suggesting that global aspects of chromosome function may be affected by the loss of members of both gene families.
Two molecules each of the core histones H2A, H2B, H3, and H4 tightly associate with 146 bp of DNA to form a nucleosome, the fundamental repeat structure of chromatin (38; for a review, see reference 73). Numerous genetic and biochemical studies have shown that core histones also contribute to gene activation (6, 16) and repression (18, 35) and have direct roles in position-dependent gene silencing (for reviews, see references 12 and 37). The latter point has been most clearly demonstrated in the budding yeast Saccharomyces cerevisiae, in which genes adjacent to telomeres and the mating type genes present at the silent HM loci are subject to position-dependent transcriptional repression. Mutations within the N-terminal residues of histones H3 and H4 disrupt this gene silencing (22, 41, 49, 71). Furthermore, these same N-terminal regions make direct contact with SIR gene products required for silencing in S. cerevisiae (14). Thus, chromatin structures responsible for gene silencing include nucleosomes as important components.
It is presently unknown how many proteins mediate assembly of histones on DNA in vivo or how such factors contribute to gene silencing. One factor involved in these processes is a three-subunit protein complex termed chromatin assembly factor I (CAF-I). CAF-I was initially purified from human somatic cell nuclear extracts on the basis of its ability to assemble histone octamers during DNA replication in vitro (62, 66; for reviews, see references 26 and 32). CAF-I-like activities have also been detected in extracts of Drosophila and Xenopus embryos (25). Purification of yeast CAF-I led to identification of the genes encoding the three yeast CAF-I subunits, each of which is homologous to its human counterpart (28). These genes have been termed CAC1, CAC2, and CAC3 (for chromosome assembly complex). Deletion of any of the three CAC genes results in decreased telomeric gene silencing (9, 28). cacΔ mutants display decreased stability of the transcriptionally silent state at both telomeres and the silent HML locus (8, 42). However, cacΔ mutants have no known growth defects. These data indicate that yeast CAF-I contributes to position-dependent gene silencing but is not the only cellular factor responsible for chromatin assembly.
Loss of SIR2, SIR3, or SIR4 gene function in S. cerevisiae completely abolishes gene silencing at both HM loci and telomeres (1, 55). These genes encode proteins that are structural components of silenced heterochromatin (15, 67) and are not known to be involved in the assembly of histones on DNA. Indeed, mutants specifically defective in histone deposition have not been described. There could be several reasons for this. Formation of nucleosomes from newly synthesized histones during DNA replication is an essential process because progression through S phase of the cell cycle in the absence of histone H2B or H4 synthesis causes lethality (13, 30). However, if the process of chromatin formation were performed by multiple, partially redundant factors, recovery of mutations in this process would be difficult in standard genetic screens because of the weak phenotypes of single mutants. In this case, disruption of multiple factors would be required to observe strong phenotypes resulting from chromatin malformation or malfunction.
We describe here a novel phenotype of cacΔ mutants: the residual gene silencing in these cells is sensitive to mutations in the HIR1, HIR2, and HIR3 genes and to changes in histone gene dosage. The three HIR-encoded proteins act to restrict transcription of three of the four histone gene pairs to the G1/S phase transition of the yeast cell cycle and to regulate the HTA1-HTB1 locus in response to altered levels of the gene products of this locus, histones H2A and H2B (43, 48, 60, 65). The HIR gene products regulate the histone HTA1-HTB1 promoter through a negative cis-acting site, although the Hir proteins themselves do not appear to directly contact DNA. hirΔ mutants display a Hir− phenotype: HTA1-HTB1 transcription is not repressed (i) outside of the G1/S phase; (ii) when cells are treated with hydroxyurea (HU), which inhibits DNA synthesis and causes accumulation of unassembled histone proteins; or (iii) when the HTA and HTB genes are present on high-copy-number plasmids. Although hirΔ mutants had no defects in gene silencing when CAF-I was intact, we observed a synergistic loss of telomeric and HM gene silencing in cacΔ hirΔ double mutants. These data suggest that pathways responsible for formation of heterochromatin in the absence of CAF-I are easily perturbed by changes in histone levels or histone stoichiometry. Furthermore, we provide evidence that the Hir proteins also have a role in cooperating with CAF-I to ensure proper cell growth and viability.
MATERIALS AND METHODS
Plasmids.
To make the cac3Δ::LEU2 deletion allele, a HindIII-BamHI fragment of pJJ283 (23) containing the LEU2 gene was inserted into ClaI- and BglII-cut pPK96 (28) to generate pPK112. pPK112 was digested with ApaI and SacI prior to transformation.
To make the cac1Δ::hisG-URA3-hisG deletion allele, a 5.4-kb BamHI-XbaI fragment containing the URA3 and kanamycin resistance (kan) genes flanked by direct repeats of bacterial hisG DNA was inserted into BglII- and NheI-digested pPK98 (28) to create pPK102. pPK102 was digested with BamHI for transformation.
To make the cac2Δ::hisG-URA3-hisG deletion allele, a 5.4-kb BamHI-BglII fragment containing the hisG-URA3-kan-hisG DNA was inserted into SnaBI- and SphI-digested, Klenow polymerase-treated pPK55 (28) to create pPK101. pPK101 was digested with EcoRI and KpnI prior to transformation.
To make pPK118 [YEp351-(HHT1-HHF1)], the 5.5-kb HindIII-BamHI fragment from pCC67 (3) was inserted into HindIII- and BamHI-digested YEp351. To make pPK119 [YEplac181-(HHT2-HHF2)], the 2.7-kb SmaI-EcoRI fragment from pCC66 (3) was inserted into SmaI- and EcoRI-digested YEplac181. To make pPK120 [YEplac181-(HTA2-HTB2)], the 3.55-kb SmaI-EcoRI fragment from pCC223 (3) was inserted into BamHI- and EcoRI-digested YEplac181. To make pPK128 [YEp351-(HHT1-HHF1)-(HTA1-HTB1)], the 6.4-kb BamHI fragment of pCC67 (3) was inserted into BamHI-digested pPK118.
To make the HTA1Δ-neg transplacement fragment, a 1.1-kb HindIII fragment from the HTA1-HTB1 promoter, containing a 54-bp deletion of the Hir-responsive negative site marked by a XhoI site (47), was inserted into a pUC18-based plasmid that contained a HindIII-SacI insert carrying the 3′ end of the HTB1 gene and the HIS3 gene inserted as a BamHI fragment. The resulting plasmid, pMA100, was digested with SalI and EcoRI to liberate an ∼4.0-kb fragment that contained the entire HTA1-HTB1 promoter with the negative site deleted, the entire HTB1 gene and 3′-flanking sequences, and the HIS3 gene.
Yeast strains.
The genotypes of yeast strains used in this study are shown in Table 1. All strains used were derived from strain W303 (70) by transformation or by crosses with other strains in this background, with the exception of the spt21Δ::HIS3 allele, which was backcrossed from strain GNX193-1B (44) to W303 strains six times prior to the construction of the strains used here. Previously described deletion alleles include the following: cac1Δ::LEU2, cac2Δ::TRP1, cac3Δ::URA3, cac3Δ::hisG-URA3-kan-hisG, and URA3-VIIL (28); hir1Δ::HIS3 and hir3Δ::HIS3 (53); sir1Δ::LEU2 (21); (hht1-hhf1)Δ::LEU2 (plasmid pUK192 [39]); (hht2-hhf2)Δ::HIS3 (pUK431 [39]); hhf2Δ::LEU2 (pPK21 [29]); (hta2-htb2)Δ::TRP1 (pJH21 [16]); and hmlΔ::LEU2 (pJH455 [76]).
TABLE 1.
S. cerevisiae strains used in this study
| Straina | Genotype | Reference or source |
|---|---|---|
| W303-1A | MATa | 70 |
| W303Δ1H | MATa hir1Δ::HIS3 | 53 |
| W303Δ2 | MATa hir2Δ::URA3 | 60 |
| W303Δ1Δ2(FOA) | MATa hir1Δ::HIS3 hir2Δ::ura3 (FOA)r) | 58 |
| YB 0152 | MATa cac2Δ::TRP1 | 28 |
| AJL387-5a Δsir2 | MATa rad1Δ::LEU2 sir2Δ::TRP1 LYS2::dam/HIS3::lys2 URA3-VIIL | 33 |
| JRY4470 | MATa sir2Δ::LEU2 ura3::LEU2 TRP1-URA3-VIIL | J. Rine (University of California, Berkeley) |
| PKY028 | MATa | 28 |
| PKY031 | MATa cac2Δ::hisG-URA3-kan-hisG | This work |
| PKY034 | MATa cac3Δ::hisG-URA3-kan-hisG | This work |
| PKY035 | MATa cac1Δ::hisG-URA3-kan-hisG | This work |
| PKY090 | MATa URA3-VIIL | 28 |
| PKY102 | MATα cac1Δ::LEU2 hir1Δ::HIS3 | This work |
| PKY103 | MATa cac2Δ::TRP1 hir1Δ::HIS3 | This work |
| PKY106 | MATa cac1Δ::LEU2 URA3-VIIL | 28 |
| PKY107 | MATa cac2Δ::TRP1 URA3-VIIL | 28 |
| PKY110 | MATα cac1Δ::LEU2 hir1Δ::HIS3 hir2Δ::URA3 | This work |
| PKY111 | MATa cac2Δ::TRP1 hir1Δ::HIS3 hir2Δ::URA3 | This work |
| PKY112 | MATa cac1Δ::LEU2 cac2Δ::TRP1 hir1Δ::HIS3 hir2Δ::URA3 | This work |
| PKY117 | MATa hir1Δ::HIS3 URA3-VIIL | This work |
| PKY132 | MATα cac3Δ::URA3 hir1Δ::HIS3 | This work |
| PKY136 | MATa cac1Δ::LEU2 hir2Δ::URA3 | This work |
| PKY137 | MATα cac2Δ::TRP1 hir2Δ::URA3 | This work |
| PKY149 | MATa sir1Δ::LEU2 cac2Δ::TRP1 | This work |
| PKY264 | MATa cac2Δ::TRP1 hir1Δ::HIS3 URA3-VIIL | This work |
| PKY268 | MATa HMR+::ADE2 | This work |
| PKY269 | MATa HMR+::ADE2 cac1Δ::LEU2 | This work |
| PKY299 | MATa cac1Δ::LEU2 LYS2::dam/HIS3::lys2 URA3-VIIL | This work |
| PKY300 | MATa LYS2::dam/HIS3::lys2 URA3-VIIL | This work |
| PKY302 | MATa cac1Δ::LEU2 hir1Δ::HIS3 URA3-VIIL | This work |
| PKY305 | MATa cac2Δ::TRP1 LYS2::dam/HIS3::lys2 URA3-VIIL | This work |
| PKY310 | MATa cac2Δ::TRP1 hir1Δ::HIS3 LYS2::dam/HIS3::lys2 URA3-VIIL | This work |
| PKY311 | MATa hir1Δ::HIS3 LYS2::dam/HIS3::lys2 URA3-VIIL | This work |
| PKY328 | MATa hir3Δ::HIS3 | This work |
| PKY329 | MATa hir1Δ::HIS3 spt21Δ::HIS3 URA3-VIIL | This work |
| PKY332 | MATa cac3Δ::LEU2 spt21Δ::HIS3 URA3-VIIL | This work |
| PKY352 | MATa hir3Δ::HIS3 URA3-VIIL | This work |
| PKY353 | MATa cac2Δ::TRP1 hir3Δ::HIS3 URA3-VIIL | This work |
| PKY354 | MATa hir3Δ::HIS3 spt21Δ::HIS3 URA3-VIIL | This work |
| PKY360 | MATa cac1Δ::LEU2 spt21Δ::HIS3 URA3-VIIL | This work |
| PKY361 | MATa spt21Δ::HIS3 URA3-VIIL | This work |
| PKY365 | MATa cac3Δ::LEU2 hir3Δ::HIS3 URA3-VIIL | This work |
| PKY375 | MATa HMR+::ADE2 cac1Δ::LEU2 hir1Δ::HIS3 | This work |
| PKY377 | MATa cac2Δ::TRP1 spt21Δ::HIS3 URA3-VIIL | This work |
| PKY378 | MATa sir1Δ::LEU2 | This work |
| PKY380 | MATa sir1Δ::LEU2 hir3Δ::HIS3 | This work |
| PKY382 | MATa cac2Δ::TRP1 hir3Δ::HIS3 | This work |
| PKY384 | MATa sir1Δ::LEU2 cac2Δ::TRP1 hir3Δ::HIS3 | This work |
| PKY408 | MATa (hht1-hhf1)Δ::LEU2 URA3-VIIL | This work |
| PKY409 | MATa cac2Δ::TRP1 (hht1-hhf1)Δ::LEU2 URA3-VIIL | This work |
| PKY410 | MATa (hht2-hhf2)Δ::HIS3 URA3-VIIL | This work |
| PKY411 | MATa cac2Δ::TRP1 (hht2-hhf2)Δ::HIS3 URA3-VIIL | This work |
| PKY412 | MATa hhf2Δ::LEU2 URA3-VIIL | This work |
| PKY413 | MATa cac2Δ::TRP1 hhf2Δ::LEU2 URA3-VIIL | This work |
| PKY499 | MATa (hta2-htb2)Δ::TRP1 URA3-VIIL | This work |
| PKY500 | MATa cac1Δ::LEU2 (hta2-htb2)Δ::TRP1 URA3-VIIL | This work |
| PKY512 | MATa cac1Δ::hisG-URA3-kan-hisG cac2Δ::TRP1 | This work |
| PKY581 | MATa HTA1Δneg::HIS3 URA3-VIIL | This work |
| PKY582 | MATa cac1Δ::LEU2 HTA1Δneg::HIS3 URA3-VIIL | This work |
| PKY730 | MATa hm1Δ::LEU2 sir1Δ::LEU2 cac2Δ::TRP1 hir3Δ::HIS3 | This work |
All strains are derived from the W303 (70) genetic background and have the genotype leu2-3,112 ura3-1 his3-11,15 trp1-1 ade2-1 can1-100.
Gene disruptions were performed by lithium acetate transformation of a wild-type diploid as described elsewhere (24). Selected colonies were purified twice after transformation and then sporulated. Correct gene disruptions were confirmed in all cases by DNA hybridization. The HTA1Δneg-HIS3 strains were further checked to ensure that they had a Hir− phenotype (48): total RNA isolated from log-phase cells treated with 0.2 M HU for 30 min prior to harvest was analyzed by Northern blot hybridization to show that the HTA1 mRNA was not down regulated (data not shown).
Genetic procedures and media.
Standard procedures were used for genetic crosses and tetrad analysis. Standard yeast media used for crosses and for scoring genetic marker segregations were as described elsewhere (24). YPAD is yeast extract-peptone-dextrose (YPD) medium supplemented with adenine at 50 mg/liter. 5-Fluoro-orotic acid (FOA) was added in all cases to synthetic media at a concentration of 1 mg/ml.
Viability and growth assays.
Viability tests were conducted by plating serial dilutions of cells growing exponentially at 30°C in YPD broth onto prewarmed YPD plates and incubating at 30 and 37°C. Samples (80 μl fixed in phosphate-buffered saline–3.7% formaldehyde) were also counted with a hemocytometer immediately before duplicate platings were performed to determine total cell numbers. Generation times were determined in YPD medium at 30 or 37°C by following the absorbance at 660 nm; for several cacΔ hirΔ double mutants, generation times were determined by counting cell numbers with a hemocytometer. Because the trp1-1 allele present in the W303 strain background causes slow growth at 16°C, we performed control experiments to show that cacΔ hirΔ double mutants had growth defects at this temperature that were independent of the status of the TRP1 gene (data not shown).
Telomeric silencing assays.
To measure telomeric silencing, the URA3 gene inserted next to the chromosome VIIL telomere was used as previously described (28); originally described in reference 11). We found that equivalent results were obtained by using log-phase cells (A600, ∼0.6) from liquid medium or cells taken from plates and adjusted to an A600 of ∼0.6 (see, e.g., reference 57). To assess quantitatively the proportion of silenced cells in a population, cells were plated on synthetic medium with or without FOA (either complete or selective for plasmids for the data in Table 4), and the number of FOA-resistant colonies per viable cell plated was determined after 8 days of incubation at 30°C. Small FOA-resistant microcolonies formed by the cacΔ strains were counted under a dissecting microscope. Colonies on synthetic complete (or selective)-medium plates were counted after 3 days to assess the number of viable plated cells. To correct for variations in the potency of the FOA in different batches of plates, the fraction of FOA-resistant cells was normalized to that of the wild-type strain for each repetition of the experiment. In experiments using strains carrying plasmids, duplicate transformants of each strain tested gave the same results. In some cases, visual, semiquantitative estimates of telomeric silencing were obtained by spotting 5-μl quantities of 10-fold serial dilutions of cells onto medium with or without FOA; however, data from these spot tests were not used to obtain the numerical values presented.
TABLE 4.
Effects of histone overexpression on telomeric silencing in wild-type and cac2Δ cells
| Strain | Relevant genotypea | FOA/total cell ratiob | n |
|---|---|---|---|
| PKY367 | wt + YEp351 | 0.51 ± 0.091 | 6 |
| PKY368 | wt + YEp351-(HTA1-HTB1) | 0.55 ± 0.14 | 3 |
| PKY451 | wt + YEp351-(HHT1-HHF1) | 0.54 ± 0.11 | 3 |
| PKY460 | wt + YEp351-(HHT1-HHF1 + HTA1-HTB1) | 0.72 ± 0.44 | 3 |
| PKY454 | wt + YEplac181 | 0.66 ± 0.22 | 3 |
| PKY452 | wt + YEplac181-(HHT2-HHF2) | 0.59 ± 0.22 | 3 |
| PKY453 | wt + YEplac181-(HTA2-HTB2) | 0.55 ± 0.095 | 3 |
| PKY369 | cac2Δ + YEp351 | 0.10 ± 0.047 | 6 |
| PKY370 | cac2Δ + YEp351-(HTA1-HTB1) | 0.013 ± 0.0095 | 3 |
| PKY455 | cac2Δ + YEp351-(HHT1-HHF1) | 0.31 ± 0.040 | 3 |
| PKY461 | cac2Δ + YEp351-(HHT1-HHF1 + HTA1-HTB1) | 0.46 ± 0.082 | 3 |
| PKY458 | cac2Δ + YEplac181 | 0.066 ± 0.012 | 3 |
| PKY456 | cac2Δ + YEplac181-(HHT2-HHF2) | 0.20 ± 0.053 | 3 |
| PKY457 | cac2Δ + YEplac181-(HTA2-HTB2) | 0.021 ± 0.011 | 3 |
wt, wild type.
Telomeric silencing of the URA3-VIIL reporter was assayed as described in the legend to Fig. 3, except that the media used were synthetic complete medium (SC) minus leucine and SC minus leucine plus FOA in order to maintain selection for the plasmids. For each strain, the fraction of Leu+ cells resistant to FOA was measured; the averages of data from n experiments ± standard deviations are shown.
Quantitative mating assays.
Quantitative assays were performed as described elsewhere (7), using strains 216 (MATa his1) and 217 (MATα his1) as the testers. Patch mating tests (see Fig. 3C) were performed by growing cell patches overnight on YPD medium at 30°C and then replica plating the patches either onto synthetic dextrose agar plates onto which had been spread a fresh culture of the appropriate mating tester or onto YPD plates as a control for cell growth. Patches were photographed after 2 days of growth at 30°C.
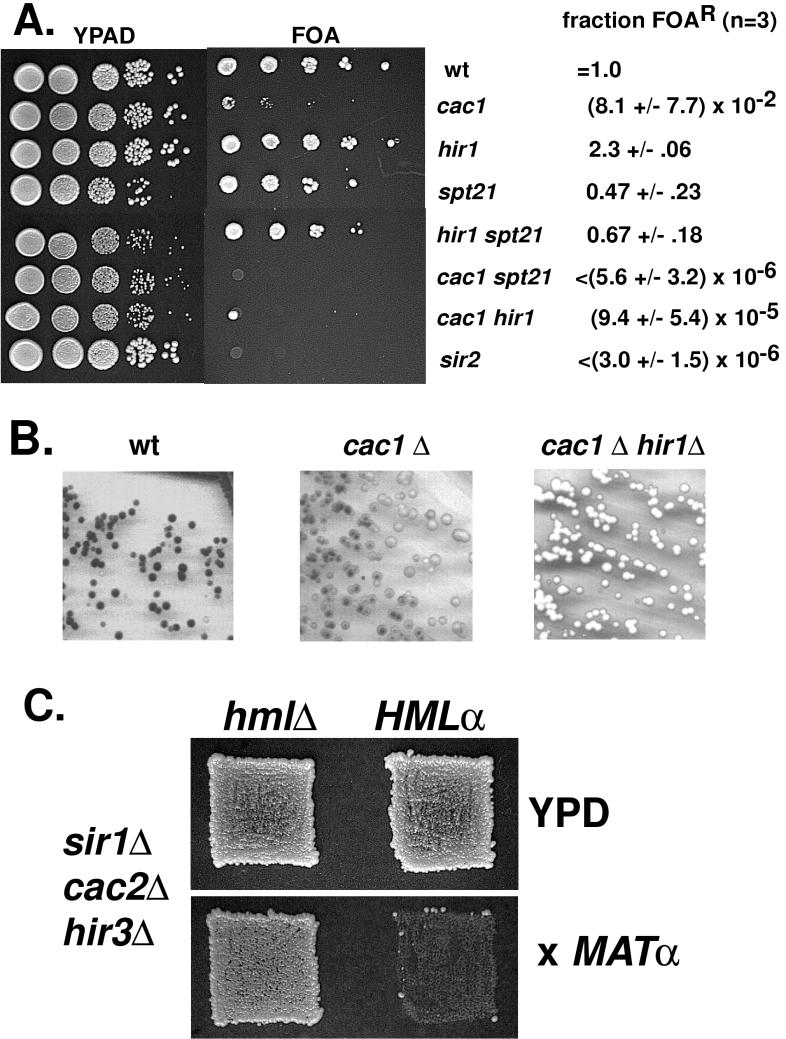
FIG. 3.
Deletion of both CAC and HIR genes synergistically reduces silencing of a telomere-proximal URA3 gene, the ADE2 gene at HMR, and the a genes at the natural HML locus. (A) Quantitation of silencing in representative strains. PKY090 (CAC+), PKY106 (cac1Δ), PKY117 (hir1Δ), PKY361 (spt21Δ), PKY329 (spt21Δ hir1Δ), PKY360 (cac1Δ spt21Δ), PKY302 (cac1Δ hir1Δ), and JRY4470 (sir2Δ) were used. Tenfold serial dilutions of each strain were spotted onto nonselective (YPAD) medium to observe the total number of cells and onto FOA medium to observe cells capable of telomeric silencing. For each independent experiment, the fraction of viable cells resistant to FOA (FOAR) was normalized to the value obtained for the wild-type (wt) strain; the averages of data from three experiments ± standard deviations are shown. <, no FOA-resistant colonies were observed (of 2 × 106 to 4 × 106 plated). (B) Strains containing the HMR+::ADE2 allele (68) were grown on YPD medium without additional adenine. A dark colony color indicates silencing of the ADE2 gene. Strains PKY268 (wild type [wt]), PKY269 (cac1Δ), and PKY375 (cac1Δ hir1Δ) were used. (C) Strains PKY730 (MATa hm1Δ sir1Δ cac2Δ hir3Δ) (left) and PKY364 (MATa HMLα sir1Δ cac2Δ hir3Δ) (right) were replica plated onto YPD medium to test for growth (upper panel) or onto synthetic dextrose medium spread with a lawn of mating tester strain 217 (MATα his1) to select for diploid formation (lower panel).
Dam methylase accessibility assays.
These assays were performed essentially as described elsewhere (33), except that the enzyme DpnII (New England Biolabs) was used instead of MboI. In this case, DNA was first digested with HindIII and then reaction mixtures were adjusted with the buffer recommended by the manufacturer for digestion with DpnII.
RESULTS
The CAC2/HIR1 subfamily of WD repeat proteins.
The CAC2 and CAC3 genes encode the smaller two subunits of yeast CAF-I (28). Cac2p and Cac3p contain WD repeat motifs found in many eukaryotic proteins (45). Cac3p (previously described in the literature as Msi1p [19, 28, 56]) is a member of the histone-binding p48 subfamily of WD proteins which includes the p48 subunit of human CAF-I, the yeast histone acetyltransferase-associated protein Hat2p, and the human histone acetyltransferase-associated protein p46 (28, 50, 74, 75). All members of the p48 subfamily share conserved residues outside the canonical WD repeat signature and are members of at least one multisubunit protein complex that either binds or modifies histone proteins (28, 50, 69, 72, 74, 75, 79). Cac2p is not a member of the p48 subfamily. However, examination of database entries showed that the yeast protein most similar to Cac2p is Hir1p, which contains WD motifs in its N-terminal half (34, 60). The yeast Cac2p and Hir1p proteins both have conserved human homologs, the CAF-I p60 subunit and the HIRA protein, respectively (27, 34). Comparison of CAC2, HIR1, and their human homologs showed that they encode a distinct subfamily of WD proteins (Fig. 1). Each of the seven WD repeats in these proteins includes conserved residues other than those that define a generic WD motif consensus, and each repeat contains distinctly conserved residues not shared by the other six repeats (Fig. 1). Furthermore, the N-to-C-terminal order of the WD repeats in these four proteins is conserved.
FIG. 1.
The CAC2/HIR1 subfamily of WD repeat proteins. The yeast CAC2 gene (28), the human CAF-I p60 gene (27), the human HIRA gene (34), and the yeast HIR1 gene (34, 60) were aligned by using the PILEUP program (Genetics Computer Group, Madison, Wis.). Each protein sequence is shown starting from the initiator methionine through the seven WD motifs depicted on each line. Amino acids identical in at least three proteins are shadowed in black; conservative changes are underlined. A WD motif consensus from a large number of proteins is illustrated between repeats 3 and 4 (45), with shadowed H’s representing hydrophobic residues and DPGN representing a region that often includes those amino acids.
Regulation of histone gene promoters by Hir proteins is thought to involve the formation of repressive chromatin structures by the histone-binding protein Hir2p (3a), suggesting that the homology shared by CAC2 and HIR1 may reflect the presence of a conserved domain involved in chromatin assembly. One phenotypic consequence of defects in chromatin assembly is the reduction of heterochromatic gene silencing (28). We expect that assembly of chromatin is accomplished by a large number of functionally overlapping factors since cacΔ mutants display only a partial loss of telomeric silencing. We therefore tested whether deletion of both CAC and HIR genes in the same cell would result in more-severe phenotypes with respect to heterochromatic gene silencing and histone gene regulation.
Growth defects in cacΔ hirΔ mutants.
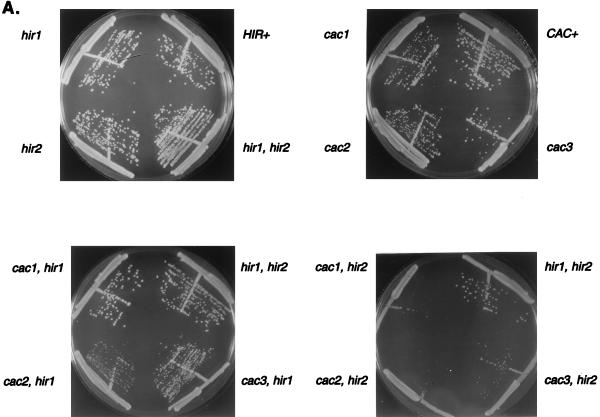
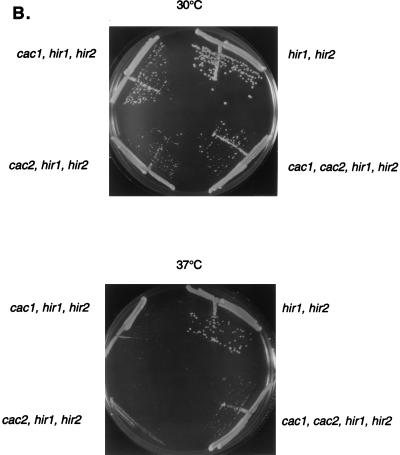
Loss of single CAC or HIR genes does not result in growth defects, nor does loss of multiple members within the same group (9, 28, 60) (Fig. 2). However, most cacΔ hirΔ double mutants grew somewhat more slowly than wild-type or single-mutant cells at 30°C, as reflected by decreased colony sizes on plates; this phenotype was exacerbated at low (16°C) (data not shown) and high (37°C) (Fig. 2A) temperatures, although there was no specific morphological defect (e.g., cell cycle arrest phenotype) associated with growth at 37°C. These colony phenotypes arose only when deletions of CAC and HIR genes were combined, and they were most pronounced in cac2Δ hir2Δ double mutants and in triple mutants with deletions of both HIR1 and HIR2, for which growth defects were noted at both 30 and 37°C (Fig. 2B). cacΔ hirΔ double-mutant cells also had longer generation times than cacΔ or hirΔ single-mutant cells in liquid YPD medium. For example, cac2Δ or hir2Δ single-mutant strains doubled every 90 to 100 min at both 30 and 37°C, whereas cac2Δ hir2Δ double mutants had a 125-min generation time at 30°C and a 195-min generation time at 37°C (Table 2). A cac2Δ hir1Δ hir2Δ triple mutant had an even longer generation time, doubling every 225 min at 30°C. Also, cacΔ hirΔ colonies were smaller after tetrad dissection, including those double mutants that do not have significantly decreased colony sizes or increased doubling times at 30°C in liquid culture (e.g., cac1Δ hir1Δ mutants [Fig. 2C]). In contrast, small tetrad colony phenotypes were never observed on deletion of multiple members of either the CAC or HIR gene family (data not shown).
FIG. 2.
Growth defects of cacΔ hirΔ mutants. Cultures were streaked onto YPD plates for distribution into single colonies, and the plates were incubated for 3 to 5 days. (A) cacΔ hirΔ strains grown at 37°C; (B) triple and quadruple cacΔ hirΔ deletion strains grown at 30 and 37°C. Strains used were W303 (HIR+ CAC+), W303Δ1 (hir1Δ), W303Δ2 (hir2Δ), W303Δ1Δ2(FOA) (hir1Δ hir2Δ), PKY035 (cac1Δ), PKY031 (cac2Δ), PKY034 (cac3Δ), PKY102 (cac1Δ hir1Δ), PKY103 (cac2Δ hir1Δ), PKY132 (cac3Δ hir1Δ), PKY136 (cac1Δ hir2Δ ), PKY137 (cac2Δ hir2Δ), PKY103 (cac3Δ hir2Δ), PKY110 (cac1Δ hir1Δ hir2Δ), PKY111 (cac2Δ hir1Δ hir2Δ), and PKY112 (cac1Δ cac2Δ hir1Δ hir2Δ). (C) Slow growth of cac1Δ hir1Δ spores after germination. Strain PKY102 (MATα cac1Δ::LEUZ hir1Δ::HIS3) was mated to PKY090 (MATa URA3-VIIL). The resulting diploid was sporulated, and tetrads were dissected. The viable progeny, shown here after 3 days of growth on YPAD medium at 30°C, are of two sizes. Scoring of markers revealed that all of the smaller progeny were His+ Leu+, indicating that they were cac1Δ hir1Δ double mutants (data not shown). Note that the number of complete tetrads with a 3:1 ratio of large to small colonies is below the expected 2:3 frequency for tetratypes; this is presumably because of the proximity of CAC1 to the chromosome XVI centromere.
TABLE 2.
Generation times and viabilities of yeast strains at 30 and 37°Ca
| Strain | Relevant genotype | Generation time (min, 30°C) | % Viability at 37°Cb |
|---|---|---|---|
| W303-1A | Wild type | 95 | 100 |
| W303Δ1 | hir1Δ | 90 | 100 |
| W303Δ2 | hir2Δ | 90 | 100 |
| W303Δ1Δ2(FOA) | hir1Δ hir2Δ | 105 | 100 |
| PKY080 | cac1Δ | 90 | 100 |
| PKY076 | cac2Δ | 90 | 100 |
| PKY512 | cac1Δ cac2Δ | 105 | 100 |
| PKY102 | cac1Δ hir1Δ | 95 | 100 |
| PKY103 | cac2Δ hir1Δ | 110 | 100 |
| PKY136 | cac1Δ hir2Δ | 125 | 100 |
| PKY137 | cac2Δ hir2Δ | 125 | 18 |
| PKY110 | cac1Δ hir1Δ hir2Δ | 130 | 56 |
| PKY111 | cac2Δ hir1Δ hir2Δ | 225 | <1.0 |
At 37°C, cac2Δ hir2Δ and cac1Δ hir1Δ hir2Δ strains produced colonies with two classes of colony sizes: the majority of colonies were small and a minority were large after 3 days. At 30°C, all of the colonies were large.
Percent viability values are the averages of data from two independent experiments, except for strain W303Δ1Δ2(FOA), which was assayed once. All strains tested were 100% viable at 30°C.
The slower growth rates of cacΔ hirΔ double mutants at 37°C did not generally result from cell inviability because, with one exception, the double mutants were equally viable when plated on YPD medium at 30 and 37°C (Table 2). The exception was the cac2Δ hir2Δ double mutant, which showed a 4- to 10-fold loss of viability at 37°C but not at 30°C. cac2Δ hir1Δ hir2Δ triple mutants and cac1Δ cac2Δ hir1Δ hir2Δ quadruple mutants, however, exhibited not only very reduced colony sizes but also reduced viability at 37°C (Fig. 2B). For example, >99% of cac2Δ hir1Δ hir2Δ cells were inviable when plated at 37°C, although the triple mutant showed no loss of viability when plated at 30°C (Table 2).
The synthetic growth phenotypes could occur because cacΔ hirΔ double mutants are more severely defective in histone synthesis and/or chromatin formation than single mutants. To test the first possibility, we examined whether cacΔ mutants displayed appropriate transcriptional regulation of the histone H2A1-encoding HTA1 gene (60). Using an HTA1-lacZ reporter gene, we found that transcription from the HTA1 promoter was unaltered from that of the wild type in both single and double cacΔ mutant cells: there was no observed effect on the overall levels of HTA1-lacZ transcripts or on the ability of the reporter gene to be repressed when DNA synthesis was blocked with HU or when extra copies of genes encoding histones H2A and H2B were present (data not shown and references 43 and 60). In addition, the HTA1 reporter gene was derepressed to the same extent in hirΔ single mutants and in cacΔ hirΔ double mutants (data not shown). Therefore, CAF-I does not appear to regulate HTA1 transcription.
Synergistic effects of cacΔ and hirΔ mutations on telomeric gene silencing.
To test whether the growth defects of cacΔ hirΔ double mutants were due to defective chromatin formation, we first analyzed telomeric chromatin structure and function. In S. cerevisiae, telomere-proximal genes are subject to position-dependent but gene-independent silencing, termed the telomeric position effect (TPE) (11). TPE can be quantitatively assayed by placing the URA3 gene adjacent to a telomere; the fraction of cells in a population that are resistant to the drug FOA, a metabolic poison for Ura+ cells, represents the level of silencing of the URA3 gene. In cacΔ mutants, TPE is reduced but not abolished (9, 28). We quantitated the levels of TPE in cacΔ mutants, hirΔ mutants, and cacΔ hirΔ double mutants. hir1Δ mutants had no defects in TPE; in fact, there was a slight increase in the TPE in these strains (Fig. 3A). hir1Δ mutants were indistinguishable from hir2Δ or hir3Δ mutants in this assay (data not shown). In contrast, cacΔ hirΔ double mutants had TPE levels approximately 3 orders of magnitude below the levels observed for the partially derepressed cacΔ mutants. These synergies were observed in multiple cacΔ hirΔ double-mutant combinations: deletion of cac1Δ or cac2Δ in combination with either hir1Δ, hir2Δ, or hir3Δ gave similar results, with the fraction of FOA-resistant colonies being in the range of 10−4 to 10−5 relative to wild-type cells (Fig. 3A and data not shown). cac3Δ mutants had a weaker TPE defect than cac1Δ or cac2Δ mutants (28), and cac3Δ hirΔ double mutants were also slightly less derepressed than other cacΔ hirΔ combinations (data not shown).
The HIR1, HIR2, and HIR3 genes do not encode the only regulators of histone gene transcription. Mutations of several genes alter the site of transcription initiation at gene promoters made nonfunctional by Ty transposon insertions (for a review, see reference 77). Mutants lacking these genes (termed SPT [for suppressor of Ty] genes) thus have an Spt− phenotype in which a functional transcription start site is restored to gene promoters with Ty insertions. A subset of spt mutants also have a Hir− phenotype because they are unable to repress HTA1 transcription upon treatment with HU; conversely, hirΔ mutations also alter Ty transcription and thus cause Spt− phenotypes (59, 64). SPT genes that are required for a normal Hir+ phenotype include SPT1, which is identical to HIR2 (59); SPT11 and SPT12, encoding histones H2A and H2B (3); and SPT10 and SPT21, which are required for transcription of the HTA2-HTB2 and HHT2-HHF2 histone gene pairs (5).
To test whether other genes that misregulate histone gene expression also affect the TPE, we measured telomeric silencing in spt21Δ, cacΔ spt21Δ, and hirΔ spt21Δ strains (Fig. 3A and data not shown). In all combinations, the spt21Δ deletion behaved like a hirΔ deletion with respect to telomeric gene silencing. spt21Δ cells had nearly wild-type levels of TPE (the approximately twofold reduction in TPE could be due to the fact that this mutation is pleiotropic, affecting expression at a large number of loci [44]). Combining spt21Δ with a hir1Δ or hir3Δ deletion did not result in a synergistic change in the TPE (Fig. 3A and data not shown), as observed for combinations of hirΔ deletions (data not shown). In contrast, cacΔ spt21Δ double mutants were as severely reduced for TPE as were cacΔ hirΔ double mutants (Fig. 3A and data not shown). Thus, these data extend our observation that mutations that cause misregulation of histone synthesis also cause a dramatic reduction of telomeric silencing in cacΔ mutants.
Synergistic effects of cacΔ and hirΔ mutations on HM gene silencing.
Position-dependent transcriptional repression at the HML and HMR loci of S. cerevisiae ensures that only the a or α genes at the MAT locus are expressed; defects in silencing at HML or HMR cause coexpression of a and α genes in the same cell, resulting in a reduced mating efficiency (for a review, see reference 37). Although cacΔ mutants mate as efficiently as wild-type cells (9, 28) (Table 3), we asked whether combinations of cacΔ and hirΔ mutations cause synergistic silencing defects at HM loci. We quantitated mating of MATa strains as an assay for silencing of the α genes at HML, and we measured mating of isogenic MATα strains as an assay for silencing of the a genes at HMR. As previously observed, cacΔ or hirΔ gene deletions by themselves did not confer a mating defect (Table 3). However, in MATa cacΔ hirΔ double mutants (e.g., cac2Δ hir3Δ [Table 3]), approximately a 10-fold reduction in mating was observed, indicative of HML derepression. This demonstrates that the synergistic loss of position-dependent gene silencing in cacΔ hirΔ mutants is not limited to telomere-proximal genes.
TABLE 3.
Mating efficiencies of strains with different combinations of sir1Δ, cac2Δ, and hir3Δ deletions
| Strain genotype | Mating efficiency in backgrounda:
|
|
|---|---|---|
| MATα | MATa | |
| Wild type | 1.0 (3) | 1.0 (5) |
| cac2Δ | 1.2 ± 0.34 (3) | 0.83 ± 0.34 (4) |
| hir3Δ | 0.69 ± 0.35 (3) | 0.91 ± 0.12 (4) |
| sir1Δ | 0.29 ± 0.15 (3) | 0.38 ± 0.13 (4) |
| sir1Δ cac2Δ | 0.50 ± 0.18 (3) | 3.3 × 10−2 ± 2.0 × 10−2 (4) |
| sir1Δ hir3Δ | 0.54 ± 0.12 (3) | 0.55 ± 0.18 (4) |
| cac2Δ hir3Δ | 1.0 (2) | 9.4 × 10−2 ± 3.6 × 10−2 (5) |
| sir1Δ cac2Δ hir3Δ | 6.2 × 10−2 ± 5.9 × 10−2 (3) | 1.9 × 10−4 ± 1.4 × 10−4 (5) |
In each independent experiment, data were normalized to values obtained simultaneously for the wild-type strain. The values are averages of data from multiple experiments (numbers in parentheses) ± standard deviations. The data for the MATα cac2Δ hir3Δ strain are averages of values from two experiments, presented without standard deviations.
Silencing at HMR is stronger than that at HML because of the more highly redundant nature of the cis-acting silencer HMR-E (37). For example, cac2Δ hir3Δ mutants did not display an HMR silencing defect as assayed by mating (Table 3). However, more-sensitive assays for reduction in silencing at HMR, which rely on the presence of mutations within the silencers or on different reporter genes, have been developed. In one case, it was found that replacing the a genes normally present at HMR with the ADE2 gene allows for very sensitive detection of changes in silencing by a colony color assay (68). Our strains carry an ade2-1 mutation at the ADE2 locus, which causes red colony color on adenine-limited media; the HMR::ADE2 allele is largely repressed, rendering cells pink. However, cis- or trans-acting mutations that reduce silencing lead to pink- or white-sectored or entirely white colonies because of increased expression of the ADE2 gene. We found that CAC gene deletions gave strains carrying the HMR::ADE2 allele a mottled pink and white appearance, in which a large variety of shades of pink are evident in a population (shown as a population of mixed dark and light colonies in a black-and-white photograph [Fig. 3B]). This suggests that cells are rapidly switching between the silenced and nonsilenced states at HMR::ADE2 in cacΔ mutants, consistent with recent data examining switching at telomeric and HML loci (8, 42). In contrast, hirΔ mutants showed the same uniformly pink colony color as wild-type cells (data not shown). cacΔ hirΔ HMR::ADE2 mutants appeared white (Fig. 3B), suggesting that the two mutations act synergistically at HMR to derepress transcription, as we had observed at telomeres and HML.
The SIR1-4 (for silent information regular) genes are important for efficient silencing at the HM loci (for a review, see reference 37). Deletion of either SIR2, SIR3, or SIR4 results in a complete loss of silencing at the HM loci (55). In contrast, in sir1 mutant cells, HML silencing is mitotically metastable, with transcription of the resident genes switching between the repressed and active states, resulting in a population of genetically identical cells, each of which exists in one of two epigenetic states (51). This observation is consistent with the idea that the SIR1 gene is required for efficient establishment of the silenced state at HM loci, which normally occurs in virtually every cell. sir1 cells have a mild mating defect (55) (Table 3). However, several mutations which individually cause no mating deficiency cause synergistic reductions in silencing when combined with sir1Δ mutations (see, e.g., reference 54). Thus, sir1Δ mutants provide a sensitized background for the detection of silencing defects at the HM loci.
To test whether cacΔ or hirΔ mutations enhance the sirΔ mating defect, we quantitated the mating efficiencies of strains that contained all possible combinations of SIR1, CAC2, and HIR3 gene deletions (Table 3). MATa sir1Δ hir3Δ mutants mated at the same levels as sir1Δ cells. In contrast, MATa sir1Δ cac2Δ cells mated approximately 10-fold less efficiently than MATa sir1Δ cells, indicative of a role for CAC2 in HM silencing that is different from that of SIR1. This defect was not observed in MATα sir1Δ cac2Δ cells, presumably due to the stronger silencing at the HMRa locus. A dramatic loss of silencing was observed at HMLα in MATa sir1Δ cac2Δ hir3Δ triple mutants, which mated approximately 100-fold less efficiently than MATa sir1Δ cac2Δ double mutants. The sir1Δ cac2Δ hir3Δ combination also weakened silencing at HMRa sufficiently to reduce mating in a MATα strain approximately eightfold compared to sir1Δ cac2Δ mutants. To confirm that the reduction in mating efficiency observed in the sir1Δ cac2Δ hir3Δ triple mutants was due to a loss of silencing at HM loci and not to inactivation of the pheromone signaling pathway, we tested whether deletion of HMLα would restore mating. Indeed, deletion of HML restored mating to a MATa sir1Δ cac2Δ hir3Δ strain (Fig. 3C), indicating that coexpression of a and α genes is the cause of the mating defects in the triple mutant. Together, these data demonstrate that loss of both CAC and HIR genes synergistically weakens silencing at both telomeres and the HM loci and that the role of CAC and HIR genes does not overlap with that of SIR1.
Defects in telomeric chromatin structure.
Position-dependent gene silencing at yeast telomeres and HM loci results in changes in chromatin structure as revealed by increased protection from nucleases and methylases (10, 36, 61). To test whether the reductions in telomeric gene silencing in cacΔ hirΔ mutants are related to chromatin structure, we expressed the bacterial dam methylase gene in various yeast strains and tested the DNA of these strains for digestion by methylation-sensitive restriction enzymes (Fig. 4). The DNA was hybridized with a URA3 DNA probe, which recognizes both the mutant ura3-1 gene at the normal chromosomal position and the URA3 gene inserted next to the chromosome VIIL telomere. Figure 4A illustrates the expected sizes of DNA fragments from both loci; previous work has established that the GATC sequence internal to the telomeric copy of the URA3 gene is largely protected from methylation in wild-type but not sir2Δ mutant cells, which lack telomeric silencing (10, 33). Methylase access at this sequence was assayed by digestion with the restriction enzyme DpnI, which requires methylation of the GATC recognition sequence for activity. Conversely, DpnII digestion of GATC sequences is inhibited by methylation. Sau3AI digestion of GATC sequences is insensitive to dam methylation and thus serves as a positive control for digestion.
FIG. 4.
Increased accessibility of dam methylase to telomeric chromatin in a cac2Δ hir1Δ double mutant. (A) Diagram of the URA3 gene adjacent to the chromosome VIIL telomere [(TG)n] and the ura3-1 gene at the natural locus on chromosome V (33, 78). Restriction sites for HindIII and 5′-GATC-3′ sequences are indicated by H3 and Sau, respectively. The GATC sequence that is more protected from methylation in wild-type cells than in sir cells is indicated by Sau∗. Expected restriction fragments (A, B, C, and the telomeric fragment T) are shown schematically. (B) Strains expressing bacterial dam methylase were PKY300 (CAC+), PKY299 (cac1Δ), PKY305 (cac2Δ), PKY310 (cac2Δ hir1Δ), PKY311 (hir1Δ), and AJL387-5aΔsir2 (sir2Δ). DNA from each strain was digested with four different combinations of restriction enzymes prior to agarose gel electrophoresis and DNA hybridization with a URA3 probe. Shown from left to right, for each strain: digestion with HindIII alone, digestion with HindIII plus DpnI, digestion with HindIII plus DpnII, digestion with HindIII plus Sau3AI. “DpnI protection” indicates the fraction of total counts in the HindIII plus DpnI lanes present in the B restriction fragment, indicating protection of the GATC site internal to the telomeric URA3 gene from dam methylase. The values presented are averages of data from two independent experiments.
A convenient metric for quantifying methylase accessibility at the telomere is the ratio of the band B signal to the total signal in the HindIII- and DpnI-digested samples (Fig. 4A, second lane in each set of four); complete resistance to digestion by the methylation-dependent enzyme DpnI at the GATC site within the telomeric URA3 gene would result in a value of 0.5, because the endogenous ura3-1 locus is known to be fully accessible to the methylase (33). We observed average ratios of 0.33 and 0.06 in wild-type and sir2Δ cells, respectively. In cac1Δ and cac2Δ cells, there was a decrease in this ratio compared to that of wild-type cells, to 0.22 and 0.21, respectively. In contrast, the ratio in hir1Δ cells was 0.40. Thus, the extent of methylase access in these strains has the same rank order as the strength of telomeric silencing. Consistent with this correlation, we observed a ratio of 0.12 for cac2Δ hir1Δ cells, well below the value observed for cacΔ cells but not as low as that for sir2Δ cells. We also digested DNA samples with the methylase-sensitive enzyme DpnII (third lane for each strain). Liberation of fragment C from the telomeric locus, reflecting protection from methylase activity, was greatly inhibited in cac2Δ hir1Δ and sir2Δ cells. We also noted that none of the mutants analyzed exhibited substantial changes in telomere length, as shown by the migration of the telomeric fragment labeled “T” in the HindIII-digested samples (first lane for each strain). Together, these data demonstrate that cac2Δ hir1Δ cells are defective in forming the telomeric chromatin structure required for gene silencing but are able to maintain wild-type telomere length.
Effects of changes in histone gene copy number on telomeric gene silencing.
We have considered two classes of models to explain the synergistic loss of chromatin-mediated gene silencing in cacΔ hirΔ and cacΔ spt21Δ double mutants. First, the HIR genes and SPT21 could encode chromosome assembly factors that act in a pathway partially redundant to CAF-I, so that severe silencing defects are not observed until both classes of genes are mutated. A second possibility is that cacΔ mutants are sensitive to the loss of these genes because nucleosome assembly pathways that operate in the absence of CAF-I are intrinsically sensitive to perturbations in histone levels. Both hirΔ and spt21Δ mutations cause misregulation of histone synthesis: three of the four histone gene loci are derepressed in hirΔ mutants (48, 65), while the histone HTA2-HTB2 and HHT2-HHF2 gene pairs are not fully activated in spt21Δ mutants (5). Thus, although hirΔ and spt21Δ mutations cause misregulation of histone gene transcription in opposite directions, both sets of mutations share the property of altering histone levels and, in some instances, histone stoichiometry (59). Note that the two models proposed are not mutually exclusive.
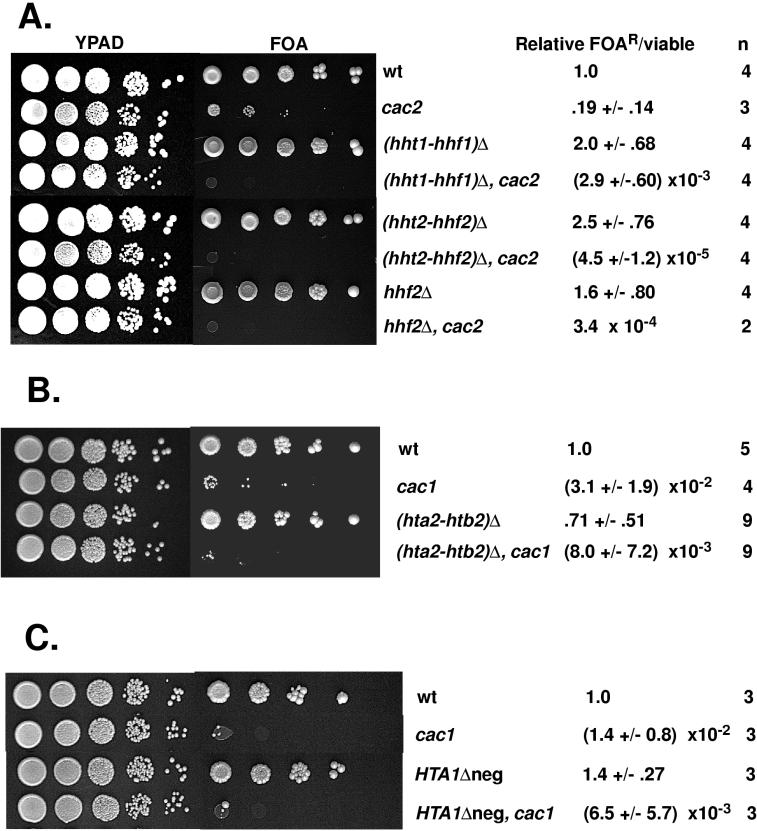
To test for effects of histone levels on the TPE in cacΔ mutants, we performed two types of experiments. We first overexpressed individual histone gene pairs (HTA-HTB or HHT-HHF) in wild-type and cac2Δ cells and measured the TPE in the transformants (Table 4). Northern blots showed that overexpressed histone mRNAs accumulated to similar levels in wild-type and cac2Δ cells (data not shown), again indicating that CAF-I does not regulate histone gene expression. In wild-type cells, overexpression of any of the four histone gene pairs resulted in TPE levels equivalent to those of control strains carrying the vectors alone. As previously observed, cac2Δ mutants displayed reduced TPE levels (28) (Fig. 5A). Unlike the case for wild-type cells, telomeric silencing in cac2Δ mutants was either increased or further reduced depending on the types of histones that were overexpressed. Overexpression of histones H2A and H2B resulted on average in a threefold (HTA2-HTB2) or an eightfold (HTA1-HTB1) reduction in telomeric silencing. In contrast, overexpression of histones H3 and H4 from either HHT-HHF gene pair resulted in a threefold stimulation of silencing. Furthermore, overexpression of all four core histones (using the HHT1-HHF1 and HTA1-HTB1 gene pairs) simultaneously led to a greater than fourfold enhancement of silencing in cac2Δ mutants, approaching the level of silencing in a wild-type strain in these experiments. These data suggest that TPE is more sensitive to changes in histone levels in the absence of CAF-I. In addition, the opposing effects of H2A-H2B versus H3-H4 overexpression on the TPE in cac2Δ mutants further suggest that CAF-I-independent pathways are selectively perturbed by an altered histone stoichiometry.
FIG. 5.
Changes in histone gene dosage and expression affect telomeric gene silencing in a cacΔ mutant but not in wild-type cells. Telomeric silencing of the URA3-VIIL reporter was assayed as described in the legend to Fig. 3. For each strain, the fraction of viable cells resistant to FOA (FOAR) was normalized to the value obtained for the wild-type strain; the averages of values from n experiments ± the standard deviations are shown, except for the hhf2Δ cac2Δ strain, for which the averages of data from two experiments are presented. (A) Deletion of histone H3 and H4 genes. Strains PKY090 (wild type [wt]), PKY107 (cac2Δ), PKY408 [(hht1-hhf1)Δ], PKY409 [(hht1-hhf1)Δ cac2Δ], PKY410 [(hht2-hhf2)Δ], PKY411 [(hht2-hhf2)Δ cac2Δ), PKY412 (hhf2Δ), and PKY413 (hhf2Δ cac2Δ) were used. (B) Deletion of the HTA2-HTB2 gene pair encoding histones H2A and H2B. Strains PKY090 (wt), PKY106 (cac1Δ), PKY499 [(hta2-htb2)Δ], and PKY500 (hta2-htb2)Δ cac1Δ] were used. (C) Deletion of the negative regulatory site in the HTA1-HTB1 promoter. Strains PKY090 (wt), PKY106 (cac1Δ), PKY581 (HTA1Δneg), and PKY582 (HTA1Δneg cac1Δ) were used.
We also tested the effects on TPE of a reduction in the histone gene copy number. Deletion of either gene pair encoding histones H3 and H4 or deletion of only the HHF2 gene, which encodes histone H4, did not reduce silencing in an otherwise wild-type strain (Fig. 5A). In contrast, any of these histone H3-H4 deletions caused a strong synergistic loss of telomeric silencing if combined with a cac2Δ deletion [approximately 100-fold in the case of (hht1-hhf1)Δ cac2Δ compared to cac2Δ]; other combinations resulted in larger effects (Fig. 5A), similar to the multiple-order-of-magnitude effects observed in cacΔ hirΔ mutants (Fig. 3). Although silencing was reduced to similar extents in cacΔ strains on deletion of either HIR or H3-H4 genes, no 37°C growth defects were associated with H3-H4 deletions, even when combined with cacΔ deletions (data not shown). We also determined the effects of deleting the gene pairs encoding histones H2A and H2B. However, deletion of the HTA1-HTB1 gene pair caused lethality in our strain background (data not shown), even though the same deletion allele has been used to construct viable haploid strains in other backgrounds (17). Deletion of the other gene pair, HTA2-HTB2, was not lethal and did not have a statistically significant effect on telomeric silencing in an otherwise wild-type cell (Fig. 5B). Telomeric silencing in an (hta2-htb2)Δ cac1Δ double mutant was slightly reduced, on average, relative to that of a cac1Δ mutant, although the standard deviations in these experiments were overlapping. However, this may be an underestimate of the silencing defect because of dosage compensation by the remaining HTA1-HTB1 gene pair (43). Together, these data suggest that telomeric silencing is most significantly perturbed by reduction of the level of histone H3-H4 expression in the absence, but not in the presence, of CAF-I.
Telomeric silencing is largely unaffected by misregulation of HTA1-HTB1 transcription.
The previous data do not rule out the possibility that the Hir proteins also directly function with CAF-I in formation of heterochromatin. To test this possibility, we constructed strains in which the cis-acting negative site within the HTA1-HTB1 promoter was deleted (HTA1Δ-neg mutants). The Hir proteins negatively regulate expression of this gene pair through this site (48); the resulting cells thus constitutively express the HTA1 and HTB1 genes, yet they contain functional Hir proteins. We measured the TPE in these strains in the presence and absence of the CAC1 gene to determine whether misregulation of the HTA1 and HTB1 genes in the presence of functional Hir proteins was sufficient to cause a synergistic loss of TPE in a cac1Δ mutant. We observed that the HTA1Δ-neg mutation had no effect on TPE itself (Fig. 5C), consistent with the observation that hirΔ mutants, which cannot negatively regulate HTA1 synthesis, also do not have TPE defects (Fig. 3). The HTA1Δ-neg cac1Δ double mutant displayed a TPE level approximately twofold lower than that of a cac1Δ strain, a decrease similar to that observed in a cacΔ mutant on overproduction of histones H2A and H2B (Table 4). However, the TPE in the HTA1Δ-neg cac1Δ double mutant was much stronger than that observed in cacΔ hirΔ strains (Fig. 3). We conclude that misregulation of the HTA1-HTB1 gene pair does not contribute in a major way to the synergistic loss of telomeric silencing observed in cacΔ hirΔ strains.
DISCUSSION
Multiple contributions to heterochromatin formation.
Yeast cells lacking CAF-I have reduced levels of telomeric gene silencing (9, 28). In this study, we found that cacΔ mutants also have subtle defects in silencing at the silent mating type loci HMR and HML that can be detected with sensitive assays (Fig. 3C; Table 3), consistent with other recent studies (8). The extensive residual silencing observed in cacΔ mutants suggests that CAF-I is partially redundant to other factors which participate in heterochromatin formation. The defect in silencing in cacΔ mutants appears to be related to the stability of the silenced chromatin once it is formed, because cacΔ mutants activate previously repressed telomere-proximal genes at a higher frequency (42) and more rapidly escape cell cycle arrest caused by exposure to alpha mating pheromone, a sensitive assay for the stability of HML silencing (8). These findings are consistent with the small colony size of FOA-resistant cacΔ mutants in TPE assays (Fig. 3) (9, 28) and the variegated appearance of cacΔ HMR::ADE2 strains on adenine-limited media (Fig. 3B). The synergistic reduction of HML silencing in sir1Δ cacΔ double mutants (Table 3) (8) thus may result from a loss of factors that contribute to two aspects of silencing: establishment of the silenced state (Sir1p [51]) and maintenance of heterochromatin (CAF-I). However, because the defect in HML silencing in sir1Δ cacΔ double mutants is partial, there must be other factors that contribute to silencing in the absence of Sir1p and CAF-I.
Several lines of evidence presented here suggest that the Hir proteins represent one set of factors that cooperate with CAF-I in the formation of heterochromatin. Combination of hirΔ and cacΔ gene deletions caused dramatic and synergistic reductions in silencing at both telomeres and HM loci (Fig. 3; Table 3). Moreover, these phenotypes correlated with changes in chromatin structure that have been associated with a loss of silencing (10, 33, 61): the telomeric chromatin of a cac2Δ hirΔ mutant displayed a large increase in accessibility to dam methylase relative to that of mutants lacking either gene alone (Fig. 4).
Disruption of HIR genes alone had little, if any, effect on gene silencing at telomeres or the silent HM loci (Fig. 3; Table 3). One interpretation of these data is that the role of Hir proteins in gene silencing is functionally redundant to other mechanisms. However, in the absence of Hir proteins, histone synthesis is misregulated, resulting in the constitutive transcription of both HHT-HHF loci and one HTA-HTB locus (48, 60, 65). It is therefore possible that hirΔ mutants do have defects in heterochromatin function but that these defects are phenotypically masked by the simultaneous overexpression of the four core histone proteins, such as we observed in a cac2Δ strain transformed with high-copy-number HTA-HTB plus HHT-HHF genes (Table 4).
Role of HIR genes in silencing through regulation of histone synthesis.
Why is silencing in cacΔ mutants sensitive to the loss of the HIR genes? One possible explanation is that Hir proteins and CAF-I play partially redundant roles in the formation of heterochromatin and that the loss of both factors is required to observe dramatic decreases in silencing. Because hirΔ mutants misregulate expression of a subset of the yeast histone genes (48, 60), a second hypothesis proposes that changing the absolute levels or the relative stoichiometry of the core histones would be detrimental to gene silencing in the absence of CAF-I.
Several lines of evidence suggest that the second hypothesis is correct. Telomeric silencing in cacΔ mutants, but not in wild-type cells, is strongly reduced by deletion of genes encoding histones H3 and H4 (Fig. 5A) and is moderately reduced by H2A-H2B overexpression (Table 4). In contrast, deletion of the HTA2-HTB2 gene pair encoding H2A-H2B had a slight effect on silencing in a cacΔ mutant (Fig. 5B), and the TPE in a cac2Δ strain was enhanced by overproduction of histones H3 and H4 (Table 4). Therefore, we also propose that deletion of SPT21, which is required for expression of both HTA2-HTB2 and HHT2-HHF2 (5), causes a synergistic loss of telomeric silencing in cacΔ mutants because of the loss of expression of the HHT2-HHF2 gene pair (Fig. 3).
Together, these data demonstrate that silencing in cacΔ mutants is sensitive to both the overall level and the relative stoichiometry of histone proteins. We suggest that this reflects the sensitivity of an alternative silencing pathway(s) to altered histone levels. When the CAC genes are intact, this alternative pathway is not required for telomeric or HM silencing, so changes in histone gene expression have little effect on the formation of heterochromatin. However, in cacΔ mutants, this pathway plays a substantial role in silencing, and it is easily perturbed by changes in histone levels or stoichiometry. Because human CAF-I is known to be a histone H3-H4 binding and deposition factor (63, 74, 75), we expect that the redundant yeast pathway also performs this function. When cellular levels of histones H3 and H4 fall below a certain threshold, as might occur in the case of an HHT-HHF or SPT21 gene deletion, this pathway appears to function inefficiently, analogous to the functioning of an enzyme when the substrate concentration is far below the Km value. Note that overexpression of histones H3 and H4 in a cacΔ mutant slightly improved the TPE, consistent with an improved function of a putative backup pathway (Table 4). In contrast, overexpression of histones H2A and H2B in a cacΔ mutant further reduced telomeric silencing (Table 4). We propose that this occurs by sequestration of unassembled (H3-H4)2 tetramers, because this phenotype is suppressed by co-overexpression of H3-H4. Changes in histone stoichiometry have also been shown to reduce mitotic chromosome stability (40). In that case, overproduction of either H2A-H2B or H3-H4 led to increased chromosome loss but simultaneous overproduction of all four core histone proteins did not. This suggests that histone stoichiometry, but not necessarily the absolute levels of histones, is important for preventing chromosome loss. In contrast, in the absence of CAF-I, heterochromatin formation is sensitive both to the levels of H3-H4 and to the stoichiometry of the four core histones. CAF-I ensures that heterochromatin formation occurs efficiently even in cells that exhibit abnormalities in histone gene expression.
Our data are also consistent with the possibility that the CAF-I-independent silencing pathway is not mediated by assembly proteins and that histones are able to assemble properly on DNA on their own provided that they are present in a sufficiently high concentration. Although we cannot presently rule out this possibility, we do not favor it because studies of several organisms have found that unassembled histone proteins are complexed with other proteins, including factors involved in the deposition of histones onto DNA (2, 4, 20, 31, 75).
Global defects in cacΔ hirΔ mutants.
In addition to causing reductions in heterochromatic gene silencing, simultaneous deletion of the CAC and HIR genes revealed other phenotypes that suggest more-global chromosomal roles for CAF-I and the Hir proteins. cacΔ hirΔ mutants displayed growth defects at 30°C that were exacerbated at both high (37°C [Fig. 2; Table 2) and low (16°C [data not shown]) temperatures. The histone H3-H4 gene pair deletions did not cause synergistic growth defects when combined with cacΔ mutations (data not shown), suggesting that the growth defects in cacΔ hirΔ mutants neither caused nor arose from a loss of heterochromatic silencing. Furthermore, deletion of both the HIR1 and HIR2 genes in cacΔ mutants increased the generation time at 30°C and reduced the viability at 37°C (Table 2). Because histone genes are no more derepressed in hir1Δ hir2Δ double mutants than in the corresponding single mutants (58), this suggests that the synthetic growth phenotypes do not solely result from changes in histone gene expression, although subtle effects on histone levels may not be detected.
Formation of heterochromatin is not required for yeast viability; sir2, sir3, and sir4 mutants completely lack telomeric and HM silencing (1, 55) and have wild-type growth rates (54a; see also reference 46). Therefore, the growth defects observed in cacΔ hirΔ mutants suggest the existence of a more global defect in chromosomes. It remains to be determined whether these growth phenotypes result from changes in gene expression, structural defects in chromosomes, or both. Global defects are also consistent with data showing that a combination of cacΔ and hirΔ mutations results in a synergistic stimulation of the rate of Ty transposition (52).
Do Hir proteins directly contribute to (hetero)chromatin formation?
It is unlikely that cacΔ and hirΔ mutations act synergistically to cause repression of genes required for silencing (e.g., SIR genes or assembly factor genes), because there exists no evidence that CAF-I or Hir proteins act as transcriptional activators; indeed, when tethered to heterologous promoters, Hir proteins are general repressors of transcription (65). Furthermore, cacΔ mutants display no phenotypes common to other mutants lacking transcriptional activation functions, such as failure to grow on alternative carbon sources or amino acid auxotrophies, nor do they have an Spt− phenotype (data not shown). Thus, the synergistic effect of cacΔ and hirΔ mutations on gene silencing is likely to result from a shared defect in chromosome function.
What overlapping role in chromosome function might be shared by CAF-I and Hir proteins? One possibility is that Hir proteins and CAF-I are both histone deposition factors. The Hir proteins are sensitive monitors of the intracellular levels of histones H2A and H2B and modulate HTA1-HTB1 transcription in response to altered levels of these histones (43). This suggests that they may be histone binding proteins. Indeed, Hir2p can bind directly to immobilized H3, H4, and H2B molecules (53a), suggesting that repression of the HTA1-HTB1 promoter could be mediated through histone deposition by Hir protein complexes (3a). We note that heterochromatic regions in S. cerevisiae contain all four core histones (15) and that human CAF-I directly binds and deposits histones H3 and H4 (63, 75). A simple hypothesis would be that Hir proteins are among several factors able to deposit histones H2A and H2B to complete nucleosome formation initiated by CAF-I or other activities. An alternative hypothesis is that Hir proteins are structural components of chromatin and that their presence in heterochromatin marks it for nucleosome assembly by another histone deposition factor when CAF-I is absent. In either model, the CAF-I and Hir protein complexes might function in partially overlapping ways to form repressive nucleosomal structures at some chromosomal locations, such that the loss of both complexes results in synergistic defects in chromatin formation.
ACKNOWLEDGMENTS
We thank members of the Kaufman and Osley laboratories and D. deBruin, R. Bacon, A. Dillin, and S. Okamura for critical reviews of the manuscript; S. Liebman and J. Berman for communication of results prior to publication; J. Boeke, J. Haber, M. Grunstein, A. Lustig, J. Rine, and F. Winston for strains and plasmids; and J. Chuang for excellent technical assistance.
This work was supported by Department of Energy funds awarded to P.D.K. and administered through the Lawrence Berkeley National Laboratory under contract DE-AC03-76SF00098 and by National Institutes of Health grants GM55712 (to P.D.K.) and GM40118 (to M.A.O.).
REFERENCES
- 1.Aparicio O M, Billington B L, Gottschling D E. Modifiers of position effect are shared between telomeric and silent mating-type loci in S. cerevisiae. Cell. 1991;66:1279–1287. doi: 10.1016/0092-8674(91)90049-5. [DOI] [PubMed] [Google Scholar]
- 2.Chang L, Loranger S S, Mizzen C, Ernst S G, Allis C D, Annunziato A T. Histones in transit: cytosolic histone complexes and diacetylation of H4 during nucleosome assembly in human cells. Biochemistry. 1997;36:469–480. doi: 10.1021/bi962069i. [DOI] [PubMed] [Google Scholar]
- 3.Clark-Adams C D, Norris D, Osley M A, Fassler J S, Winston F. Changes in histone gene dosage alter transcription in yeast. Genes Dev. 1988;2:150–159. doi: 10.1101/gad.2.2.150. [DOI] [PubMed] [Google Scholar]
- 3a.Compagnone-Post, P. A., J. Recht, and M. A. Osley. Unpublished data.
- 4.Dilworth S M, Black S J, Laskey R A. Two complexes that contain histones are required for nucleosome assembly in vitro: role of nucleoplasmin and N1 in Xenopus egg extracts. Cell. 1987;51:1009–1018. doi: 10.1016/0092-8674(87)90587-3. [DOI] [PubMed] [Google Scholar]
- 5.Dollard C, Ricupero-Hovasse S L, Natsoulis G, Boeke J D, Winston F. SPT10 and SPT21 are required for transcription of particular histone genes in Saccharomyces cerevisiae. Mol Cell Biol. 1994;14:5223–5228. doi: 10.1128/mcb.14.8.5223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Durrin L K, Mann R K, Kayne P S, Grunstein M. Yeast histone H4 N-terminal sequence is required for promoter activation in vivo. Cell. 1991;65:1023–1031. doi: 10.1016/0092-8674(91)90554-c. [DOI] [PubMed] [Google Scholar]
- 7.Ehrenhofer-Murray A E, Rivier D H, Rine J. The role of SAS2, an acetyltransferase homologue of Saccharomyces cerevisiae, in silencing and ORC function. Genetics. 1997;145:923–934. doi: 10.1093/genetics/145.4.923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Enomoto S, Berman J. Chromatin assembly factor I contributes to the maintenance, but not the reestablishment, of silencing at the yeast silent mating loci. Genes Dev. 1998;12:219–232. doi: 10.1101/gad.12.2.219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Enomoto S, McCune-Zierath P D, Gerami-Nejad M, Sanders M, Berman J. RLF2, a subunit of yeast chromatin assembly factor I, is required for telomeric chromatin function in vivo. Genes Dev. 1997;11:358–370. doi: 10.1101/gad.11.3.358. [DOI] [PubMed] [Google Scholar]
- 10.Gottschling D E. Telomere-proximal DNA in Saccharomyces cerevisiae is refractory to methyltransferase activity in vivo. Proc Natl Acad Sci USA. 1992;89:4062–4065. doi: 10.1073/pnas.89.9.4062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gottschling D E, Aparicio O M, Billington B L, Zakian V A. Position effect at S. cerevisiae telomeres: reversible repression of PolII transcription. Cell. 1990;63:751–762. doi: 10.1016/0092-8674(90)90141-z. [DOI] [PubMed] [Google Scholar]
- 12.Grunstein M. Histone acetylation in chromatin structure and transcription. Nature. 1997;389:349–352. doi: 10.1038/38664. [DOI] [PubMed] [Google Scholar]
- 13.Han M, Chang M, Kim U-J, Grunstein M. Histone H2B repression causes cell-cycle-specific arrest in yeast: effects on chromosomal segregation, replication, and transcription. Cell. 1987;48:589–597. doi: 10.1016/0092-8674(87)90237-6. [DOI] [PubMed] [Google Scholar]
- 14.Hecht A, Laroche T, Strahl-Bolsinger S, Gasser S M, Grunstein M. Histone H3 and H4 N-termini interact with SIR3 and SIR4 proteins: a molecular model for the formation of heterochromatin in yeast. Cell. 1995;80:583–592. doi: 10.1016/0092-8674(95)90512-x. [DOI] [PubMed] [Google Scholar]
- 15.Hecht A, Strahl-Bolsinger S, Grunstein M. Spreading of transcriptional repressor SIR3 from telomeric heterochromatin. Nature. 1996;383:92–96. doi: 10.1038/383092a0. [DOI] [PubMed] [Google Scholar]
- 16.Hirschhorn J N, Bortvin A L, Ricupero-Hovasse S L, Winston F. A new class of histone H2A mutations in Saccharomyces cerevisiae causes specific transcriptional defects in vivo. Mol Cell Biol. 1995;15:1999–2009. doi: 10.1128/mcb.15.4.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hirschhorn J N, Brown S A, Clark C D, Winston F. Evidence that SNF2/SWI2 and SNF5 activate transcription in yeast by altering chromatin structure. Genes Dev. 1992;6:2288–2298. doi: 10.1101/gad.6.12a.2288. [DOI] [PubMed] [Google Scholar]
- 18.Huang L, Zhang W, Roth S Y. Amino termini of histones H3 and H4 are required for a1-α2 repression in yeast. Mol Cell Biol. 1997;17:6555–6562. doi: 10.1128/mcb.17.11.6555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hubbard E J, Yang X L, Carlson M. Relationship of the cAMP-dependent protein kinase pathway to the SNF1 protein kinase and invertase expression in Saccharomyces cerevisiae. Genetics. 1992;130:71–80. doi: 10.1093/genetics/130.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ito T, Bulger M, Kobayashi R, Kadonaga J T. Drosophila NAP-1 is a core histone chaperone that functions in ATP-facilitated assembly of regularly spaced nucleosomal arrays. Mol Cell Biol. 1996;16:3112–3124. doi: 10.1128/mcb.16.6.3112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ivy J M, Klar A J S, Hicks J B. Cloning and characterization of four SIR genes of Saccharomyces cerevisiae. Mol Cell Biol. 1986;6:688–702. doi: 10.1128/mcb.6.2.688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Johnson L M, Kayne P S, Kahn E S, Grunstein M. Genetic evidence for an interaction between SIR3 and histone H4 in the repression of the silent mating loci in Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 1990;87:6286–6290. doi: 10.1073/pnas.87.16.6286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jones J S, Prakash L. Yeast Saccharomyces cerevisiae selectable markers in pUC18 polylinkers. Yeast. 1990;6:363–366. doi: 10.1002/yea.320060502. [DOI] [PubMed] [Google Scholar]
- 24.Kaiser C, Michaelis S, Mitchell A. Methods in yeast genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1994. [Google Scholar]
- 25.Kamakaka R T, Bulger M, Kaufman P D, Stillman B, Kadonaga J T. Postreplicative chromatin assembly by Drosophila and human chromatin assembly factor 1. Mol Cell Biol. 1996;16:810–817. doi: 10.1128/mcb.16.3.810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kaufman P D. Nucleosome assembly: the CAF and the HAT. Curr Opin Cell Biol. 1996;8:369–373. doi: 10.1016/s0955-0674(96)80012-3. [DOI] [PubMed] [Google Scholar]
- 27.Kaufman P D, Kobayashi R, Kessler N, Stillman B. The p150 and p60 subunits of chromatin assembly factor 1: a molecular link between newly synthesized histones and DNA replication. Cell. 1995;81:1105–1114. doi: 10.1016/s0092-8674(05)80015-7. [DOI] [PubMed] [Google Scholar]
- 28.Kaufman P D, Kobayashi R, Stillman B. Ultraviolet radiation sensitivity and reduction of telomeric silencing in Saccharomyces cerevisiae cells lacking chromatin assembly factor I. Genes Dev. 1997;11:345–357. doi: 10.1101/gad.11.3.345. [DOI] [PubMed] [Google Scholar]
- 29.Kayne P S, Kim U J, Han M, Mullen J R, Yoshizaki F, Grunstein M. Extremely conserved histone H4 N terminus is dispensable for growth but essential for repressing the silent mating loci in yeast. Cell. 1988;55:27–39. doi: 10.1016/0092-8674(88)90006-2. [DOI] [PubMed] [Google Scholar]
- 30.Kim U-J, Han M, Kayne P, Grunstein M. Effects of histone H4 depletion on the cell cycle and transcription of S. cerevisiae. EMBO J. 1988;7:2211–2219. doi: 10.1002/j.1460-2075.1988.tb03060.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kleinschmidt J A, Fortkamp E, Krohne G, Zentgraf H, Franke W W. Co-existence of two different types of soluble histone complexes in nuclei of Xenopus laevis oocytes. J Biol Chem. 1985;260:1166–1176. [PubMed] [Google Scholar]
- 32.Krude T. Nucleosome assembly during DNA replication. Curr Biol. 1995;5:1232–1234. doi: 10.1016/s0960-9822(95)00245-4. [DOI] [PubMed] [Google Scholar]
- 33.Kyrion G, Liu K, Liu C, Lustig A J. RAP1 and telomere structure regulate telomere position effects in Saccharomyces cerevisiae. Genes Dev. 1993;7:1146–1159. doi: 10.1101/gad.7.7a.1146. [DOI] [PubMed] [Google Scholar]
- 34.Lamour V, Lecluse Y, Desmaze C, Spector M, Bodescot M, Aurias A, Osley M A, Lipinski M. A human homolog of the S. cerevisiae HIR1 and HIR2 transcriptional repressors cloned from the DiGeorge syndrome critical region. Hum Mol Genet. 1995;4:791–799. doi: 10.1093/hmg/4.5.791. [DOI] [PubMed] [Google Scholar]
- 35.Lenfant F, Mann R K, Thomsen B, Ling X, Grunstein M. All four core histone N-termini contain sequences required for the repression of basal transcription in yeast. EMBO J. 1996;15:3974–3985. [PMC free article] [PubMed] [Google Scholar]
- 36.Loo S, Rine J. Silencers and domains of generalized repression. Science. 1994;264:1768–1771. doi: 10.1126/science.8209257. [DOI] [PubMed] [Google Scholar]
- 37.Loo S, Rine J. Silencing and heritable domains of gene expression. Annu Rev Cell Dev Biol. 1995;11:519–548. doi: 10.1146/annurev.cb.11.110195.002511. [DOI] [PubMed] [Google Scholar]
- 38.Luger K, Mäder A W, Richmond R K, Sargent D F, Richmond T J. Crystal structure of the nucleosome core particle at 2.8Å resolution. Nature. 1997;389:251–260. doi: 10.1038/38444. [DOI] [PubMed] [Google Scholar]
- 39.Mann R K, Grunstein M. Histone H3 N-terminal mutations allow hyperactivation of the yeast GAL1 gene in vivo. EMBO J. 1992;11:3297–3306. doi: 10.1002/j.1460-2075.1992.tb05408.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Meeks-Wagner D, Hartwell L H. Normal stoichiometry of histone dimer sets is necessary for high fidelity of mitotic chromosome transmission. Cell. 1986;44:43–52. doi: 10.1016/0092-8674(86)90483-6. [DOI] [PubMed] [Google Scholar]
- 41.Megee P C, Morgan B A, Mittman B A, Smith M M. Genetic analysis of histone H4: essential role of lysines subject to reversible acetylation. Science. 1990;247:841–845. doi: 10.1126/science.2106160. [DOI] [PubMed] [Google Scholar]
- 42.Monson E K, de Bruin D, Zakian V A. The yeast Cac1 protein is required for the stable inheritance of transcriptionally repressed chromatin at telomeres. Proc Natl Acad Sci USA. 1997;94:13081–13086. doi: 10.1073/pnas.94.24.13081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Moran L, Norris D, Osley M A. A yeast H2A-H2B promoter can be regulated by changes in histone gene copy number. Genes Dev. 1990;4:752–763. doi: 10.1101/gad.4.5.752. [DOI] [PubMed] [Google Scholar]
- 44.Natsoulis G, Dollard C, Winston F, Boeke J D. The products of the SPT10 and SPT21 genes of Saccharomyces cerevisiae increase the amplitude of transcriptional regulation at a large number of unlinked loci. New Biol. 1991;3:1249–1259. [PubMed] [Google Scholar]
- 45.Neer E J, Schmidt C J, Nambudripad R, Smith T F. The ancient regulatory-protein family of WD-repeat proteins. Nature. 1994;371:297–300. doi: 10.1038/371297a0. [DOI] [PubMed] [Google Scholar]
- 46.Nislow C, Ray E, Pillus L. SET1, a yeast member of the Trithorax family, functions in transcriptional silencing and diverse cellular processes. Mol Biol Cell. 1997;8:2421–2436. doi: 10.1091/mbc.8.12.2421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Osley M A, Gould J, Kim S, Kane M Y, Hereford L. Identification of sequences in a yeast histone promoter involved in periodic transcription. Cell. 1986;23:537–544. doi: 10.1016/0092-8674(86)90285-0. [DOI] [PubMed] [Google Scholar]
- 48.Osley M A, Lycan D. trans-acting regulatory mutations that alter transcription of Saccharomyces cerevisiae histone genes. Mol Cell Biol. 1987;7:4204–4210. doi: 10.1128/mcb.7.12.4204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Park E-C, Szostak J W. Point mutations in the yeast histone H4 gene prevent silencing of the silent mating type locus HML. Mol Cell Biol. 1990;10:4932–4934. doi: 10.1128/mcb.10.9.4932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Parthun M, Widom J, Gottschling D E. The major cytoplasmic histone acetyltransferase in yeast: links to chromatin replication and histone metabolism. Cell. 1996;87:85–94. doi: 10.1016/s0092-8674(00)81325-2. [DOI] [PubMed] [Google Scholar]
- 51.Pillus L, Rine J. Epigenetic inheritance of transcriptional states in S. cerevisiae. Cell. 1989;59:637–647. doi: 10.1016/0092-8674(89)90009-3. [DOI] [PubMed] [Google Scholar]
- 52.Qian Z, Huang H, Hong J Y, Burck C L, Johnston S D, Berman J, Carol A, Liebman S W. Yeast Ty1 retrotransposition is stimulated by a synergistic interaction between mutations in chromatin assembly factor I and histone regulatory proteins. Mol Cell Biol. 1998;18:4783–4792. doi: 10.1128/mcb.18.8.4783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Recht J, Dunn B, Raff A, Osley M A. Functional analysis of histones H2A and H2B in transcriptional repression in Saccharomyces cerevisiae. Mol Cell Biol. 1996;16:2545–2553. doi: 10.1128/mcb.16.6.2545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53a.Recht, J., and M. A. Osley. Unpublished data.
- 54.Reifsnyder C, Lowell J, Clarke A, Pillus L. Yeast SAS silencing genes and human genes associated with AML and HIV-1 Tat interactions are homologous with acetyltransferases. Nat Genet. 1996;14:42–49. doi: 10.1038/ng0996-42. [DOI] [PubMed] [Google Scholar]
- 54a.Rine, J. Personal communication.
- 55.Rine J, Herskowitz I. Four genes responsible for a position effect on expression from HML and HMR in Saccharomyces cerevisiae. Genetics. 1987;116:9–22. doi: 10.1093/genetics/116.1.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ruggieri R, Tanaka K, Nakafuku M, Kaziro Y, Toh-e A, Matsumoto K. MSI1, a negative regulator of the RAS-cAMP pathway in Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 1989;86:8778–8782. doi: 10.1073/pnas.86.22.8778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Rundlett S E, Carmen A A, Kobayashi R, Bavykin S, Turner B M, Grunstein M. HDA1 and RPD3 are members of distinct yeast histone deacetylase complexes that regulate silencing and transcription. Proc Natl Acad Sci USA. 1996;93:14503–14508. doi: 10.1073/pnas.93.25.14503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sherwood P. Ph.D. thesis. New York, New York: Cornell University; 1993. [Google Scholar]
- 59.Sherwood P W, Osley M A. Histone regulatory (hir) mutations suppress delta insertion alleles in Saccharomyces cerevisiae. Genetics. 1991;128:729–738. doi: 10.1093/genetics/128.4.729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sherwood P W, Tsang S V-M, Osley M A. Characterization of HIR1 and HIR2, two genes required for regulation of histone gene transcription in Saccharomyces cerevisiae. Mol Cell Biol. 1993;13:28–38. doi: 10.1128/mcb.13.1.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Singh J, Klar A J. Active genes in budding yeast display enhanced in vivo accessibility to foreign DNA methylases: a novel in vivo probe for chromatin structure of yeast. Genes Dev. 1992;6:186–196. doi: 10.1101/gad.6.2.186. [DOI] [PubMed] [Google Scholar]
- 62.Smith S, Stillman B. Purification and characterization of CAF-I, a human cell factor required for chromatin assembly during DNA replication. Cell. 1989;58:15–25. doi: 10.1016/0092-8674(89)90398-x. [DOI] [PubMed] [Google Scholar]
- 63.Smith S, Stillman B. Stepwise assembly of chromatin during DNA replication in vitro. EMBO J. 1991;10:971–980. doi: 10.1002/j.1460-2075.1991.tb08031.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Spector M. Ph.D. thesis. New York, New York: Cornell University; 1994. [Google Scholar]
- 65.Spector M S, Raff A, DeSilva H, Lee K, Osley M A. Hir1p and Hir2p function as transcriptional corepressors to regulate histone gene transcription in the Saccharomyces cerevisiae cell cycle. Mol Cell Biol. 1997;17:545–552. doi: 10.1128/mcb.17.2.545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Stillman B. Chromatin assembly during SV40 DNA replication in vitro. Cell. 1986;45:555–565. doi: 10.1016/0092-8674(86)90287-4. [DOI] [PubMed] [Google Scholar]
- 67.Strahl-Bolsinger S, Hecht A, Luo K, Grunstein M. SIR2 and SIR4 interactions differ in core and extended telomeric heterochromatin in yeast. Genes Dev. 1997;11:83–93. doi: 10.1101/gad.11.1.83. [DOI] [PubMed] [Google Scholar]
- 68.Sussel L, Vannier D, Shore D. Epigenetic switching of transcriptional states: cis- and trans-acting factors affecting establishment of silencing at the HMR locus in Saccharomyces cerevisiae. Mol Cell Biol. 1993;13:3919–3928. doi: 10.1128/mcb.13.7.3919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Taunton J, Hassig C A, Schreiber S L. A mammalian histone deacetylase related to the yeast transcriptional regulator Rpd3p. Science. 1996;272:408–411. doi: 10.1126/science.272.5260.408. [DOI] [PubMed] [Google Scholar]
- 70.Thomas B, Rothstein R. Elevated recombination rates in transcriptionally active DNA. Cell. 1989;58:619–630. doi: 10.1016/0092-8674(89)90584-9. [DOI] [PubMed] [Google Scholar]
- 71.Thompson J S, Ling X, Grunstein M. Histone H3 amino terminus is required for telomeric and silent mating locus repression in yeast. Nature. 1994;369:245–247. doi: 10.1038/369245a0. [DOI] [PubMed] [Google Scholar]
- 72.Tyler J K, Bulger M, Kamakaka R T, Kobayashi R, Kadonaga J T. The p55 subunit of Drosophila chromatin assembly factor 1 is homologous to a histone deacetylase-associated protein. Mol Cell Biol. 1996;16:6149–6159. doi: 10.1128/mcb.16.11.6149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.van Holde K E. Chromatin. Berlin, Germany: Springer-Verlag; 1989. [Google Scholar]
- 74.Verrault A, Kaufman P D, Kobayashi R, Stillman B. Nucleosomal DNA regulates the core-histone-binding subunit of the human Hat1 acetyltransferase. Curr Biol. 1998;8:96–108. doi: 10.1016/s0960-9822(98)70040-5. [DOI] [PubMed] [Google Scholar]
- 75.Verreault A, Kaufman P D, Kobayashi R, Stillman B. Nucleosome assembly by a complex of CAF-I and acetylated histones H3/H4. Cell. 1996;87:95–104. doi: 10.1016/s0092-8674(00)81326-4. [DOI] [PubMed] [Google Scholar]
- 76.White C I, Haber J E. Intermediates of recombination during mating type switching in Saccharomyces cerevisiae. EMBO J. 1990;9:663–673. doi: 10.1002/j.1460-2075.1990.tb08158.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Winston F, Carlson M. Yeast SNF/SWI transcriptional activators and the SPT/SIN chromatin connection. Trends Genet. 1992;8:387–391. doi: 10.1016/0168-9525(92)90300-s. [DOI] [PubMed] [Google Scholar]
- 78.Wright J H, Gottschling D E, Zakian V A. Saccharomyces telomeres assume a non-nucleosomal chromatin structure. Genes Dev. 1992;6:197–210. doi: 10.1101/gad.6.2.197. [DOI] [PubMed] [Google Scholar]
- 79.Zhang Y, Iratni R, Erdjument-Bromage H, Tempst P, Reinberg D. Histone deacetylases and SAP18, a novel polypeptide, are components of a human Sin3 complex. Cell. 1997;89:357–364. doi: 10.1016/s0092-8674(00)80216-0. [DOI] [PubMed] [Google Scholar]