Abstract
The transcription factor lymphoid enhancer factor 1 (LEF-1) is directed to the nucleus by a nine-amino-acid nuclear localization signal (NLS; KKKKRKREK) located in the high-mobility-group DNA binding domain. This NLS is recognized by two armadillo repeat proteins (pendulin/Rch1/α-P1/hSrp1α and Srp1/karyopherin-α/α-S1/NPI-1) which function in nuclear transport as the importin-α subunit of NLS receptors. T-cell factor 1 (TCF-1), a related transcription factor, contains a similar sequence (KKKRRSREK) in the identical position within its HMG DNA binding domain. We show that this sequence functions as an NLS in vivo but is not recognized by these two importin-α subtypes in a yeast two-hybrid assay and only weakly recognized in an in vitro binding assay. Transfer of the LEF-1 NLS to TCF-1 can confer pendulin/Rch1 binding, demonstrating that the NLS is the primary determinant for recognition. We have constructed a set of deletion mutations in pendulin/Rch1 to examine the differential NLS recognition more closely. We find that the entire armadillo repeat array of pendulin/Rch1 is necessary to maintain high affinity and specificity for the LEF-1 NLS versus the TCF-1 NLS. Importin-β, the second subunit of the NLS receptor complex, does not influence in vitro NLS binding affinity or specificity. To test whether this differential recognition is indicative of distinct mechanisms of nuclear transport, the subcellular localization of LEF-1 and TCF-1 fused to green fluorescent protein (GFP)) was examined in an in vitro nuclear transport assay. GFP–LEF-1 readily localizes to the nucleus, whereas GFP–TCF-1 remains in the cytoplasm. Thus, LEF-1 and TCF-1 differ in several aspects of nuclear localization.
A recent survey of DNA and RNA binding proteins with delimited nuclear localization signals (NLSs) found that these signals are often contained within or are near the domain involved in DNA or RNA binding (27). NLSs serve to target the protein to the nucleus through a direct binding of 60-kDa NLS receptors known collectively as importins or karyopherins (2, 15, 32, 35). These receptors are composed primarily of reiterated hydrophobic repeat motifs called armadillo repeats (named after Drosophila armadillo) (37). Although the armadillo repeat regions are known to be involved in NLS binding, the precise domain responsible for NLS recognition has not been defined. Understanding how NLS receptors bind and direct their ligands to the nucleus is important because in the case of RNA and DNA binding proteins, many are shuttled through nuclear pores by an NLS receptor that remains tightly bound to the nucleic acid binding domain for an undetermined length of time.
The DNA binding domain of the transcription factor lymphoid enhancer factor 1 (LEF-1) binds and bends specific DNA sequences within the promoters or enhancers of genes (11, 50, 54). LEF-1 regulates gene expression by engaging in protein-protein contact with enhancer and promoter binding proteins on these bent templates (4, 5, 12, 30, 45). The DNA binding and bending activities are carried out by an 88-amino-acid (aa) high-mobility-group (HMG) DNA binding domain in a region near the C terminus of the protein (6, 11). This HMG domain can bind and bend DNA as a separate independent protein fragment, and the three-dimensional structure of this fragment associated with DNA has been determined (29). The first 68 aa of the DNA binding/bending domain, termed the HMG box, carry DNA sequence specificity. The remaining 18-aa portion of the DNA binding/bending domain appears to play a role in DNA binding affinity. A stretch of 9 aa immediately C terminal to the HMG box acts as a flexible linker region for the final nine residues of the DNA binding domain to swing over the minor groove and back under to make specific contacts with the phosphate backbone in the bent major groove. Deletion or amino acid replacement in this region destroys DNA binding by lowering binding affinities up to 2 orders of magnitude. We refer to these last nine residues as the B box because the amino acid sequence is composed almost entirely of basic amino acids (KKKKRKREK). We have previously shown that the B box performs dual duties in DNA binding and in targeting LEF-1 protein to the nucleus by functioning as the NLS of the protein (39). The B box is the only NLS signal in LEF-1, being both necessary and sufficient for nuclear localization. LEF-1 is not the only HMG DNA binding protein that can recognize its specific DNA binding site. A highly related but distinct transcription factor named T-cell factor 1 (TCF-1) can also bind and bend the same DNA sequence via its HMG DNA binding domain located in a similar position near the C terminus of the protein (52, 53). Not surprisingly, the HMG DNA binding domain of TCF-1 is nearly identical in amino acid sequence (95%), differing at only six positions. Two of the differences (KKKRRSREK [Fig. 1]) are in the B box. In this study, we show that like the LEF-1 B box, the TCF-1 B box can function as an NLS to direct TCF-1 or heterologous proteins to the nucleus.
FIG. 1.
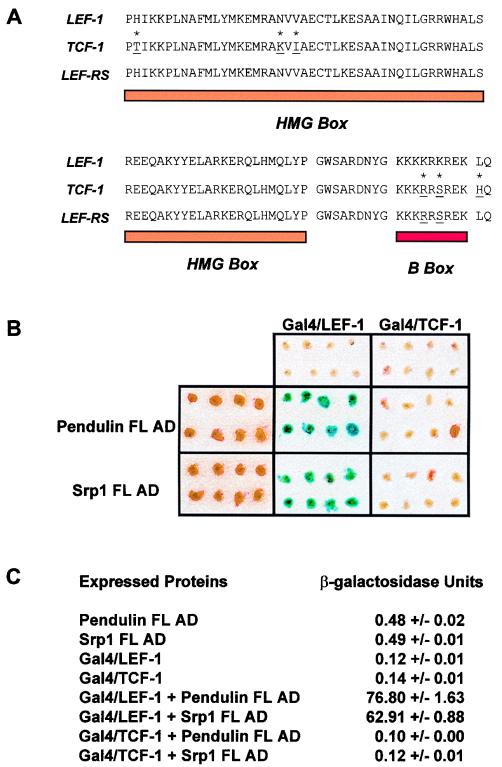
Yeast two-hybrid analysis of the interaction of pendulin and Srp1 with LEF-1 and TCF-1 HMG DNA binding domains. (A) The amino acid sequences of the LEF-1 (from aa 297 to 384) and TCF-1 (from aa 152 to 239) HMG DNA binding domains are shown. Asterisks indicate the six positions where amino acid sequences differ. The amino acid sequence for a LEF-1 HMG box–TCF-1 B-box hybrid DNA binding domain called LEF-RS is shown. The residues specific for TCF-1 are underlined. (B) FL pendulin and Srp1 were fused to the yeast Gal4 transcription activation domain (AD) and assayed in the yeast two-hybrid system for the ability to interact with LEF-1 and TCF-1 HMG DNA binding domains fused to the yeast Gal4 DNA binding domain. β-Galactosidase activity was assessed in eight independent colonies by the filter assay method. (C) β-Galactosidase activity was assessed by solution assay. Three independent yeast colonies were tested for each set of conditions.
We have reported the isolation of cDNA clones encoding two NLS receptor subtypes, using the LEF-1 HMG DNA binding domain as bait in a yeast two-hybrid screen (39). These two receptors, variously referred to as importin-α/pendulin/Rch1/hSrp1α/α-P1/PTAC58/karyopherin α2 (9, 16, 21, 25, 49, 56) and Srp1/karyopherin-α/NPI-1/α-S1 (8, 34, 36, 58), bind directly to NLSs and, along with a second, armadillo repeat-containing subunit (importin-β/p97/karyopherin-β) (8), dock the NLS-bearing substrate at the nuclear pore (7, 14, 21, 40, 56). Subsequent transport steps are GTP/GDP dependent and involve movement of the NLS receptor and NLS-bearing substrate together as a complex through the pore into the nucleoplasm (2, 15, 32, 35). At some point the NLS receptor dissociates to recycle back to the cytoplasm with the recently described export factor CAS, leaving the NLS-bearing substrate to perform its function in the nucleus (26). We have shown that both NLS receptor subtypes (which we refer to as pendulin and Srp1) bind directly and specifically to the B box in LEF-1 (39). Furthermore, we have shown that neither NLS receptor subtype binds to TCF-1 in vitro or in vivo in a yeast two-hybrid system. As described above, the B box in the TCF-1 DNA binding domain differs from LEF-1 by only two amino acids. Since both LEF-1 and TCF-1 are nuclear transcription factors, we are interested in exploring this difference in NLS receptor recognition. Furthermore, since the NLS/B box of LEF-1 is directly involved in DNA binding, we wish to study the structural determinants of the NLS receptor that bind to this important region of the DNA binding domain.
NLS receptors are part of the armadillo repeat family, a newly recognized family defined by multiple, tandem armadillo repeat motifs (37). Armadillo repeats are on average 42 aa in length, with a preponderence of conserved hydrophobic residues. Arm repeats are found in proteins that carry out a wide variety of cellular functions, including cell adhesion (beta-catenin/armadillo, plakoglobin, and p120), signal transduction (beta-catenin/armadillo), GTP exchange (smgGDS), nuclear import (importins/karyopherins and transportins), tumor suppression (adenomatous polypsis coli), and others (10, 15, 17, 22, 23, 31, 35, 42, 43, 46). Arm repeats do not appear as single modules but exist in tandem arrays of at least 4 repeats and up to as many as 13 repeats. They may comprise most of the protein or only a small portion of it; NLS receptors are made up of eight or nine armadillo repeats flanked by relatively small, hydrophilic N- and C-terminal domains.
Overall, NLS receptors appear to recognize highly variable NLSs (56). How is the high degree of sequence variability in NLSs accommodated, yet fine specificity like that observed between LEF-1 and TCF-1 maintained? Although arm repeats are highly variable along an array, they are each highly conserved in both sequence and position in homologs from yeast to humans, implying that each individual arm repeat has specialized to engage in specific contacts or carry out a specific function (37). Conservation of the order of the arm repeats within an array also suggests that this specialized function requires cooperation from neighboring arm repeats to fold correctly and that groups of arm repeats engage in specific interactions. Our results support this model. We find that high levels of LEF-1 NLS binding in an in vitro binding assay require an intact, full-length importin-α armadillo repeat array. Truncation mutants of importin-α that leave an open, naked end of the array exhibit nondiscriminant, equivalent levels of binding to LEF-1 and TCF-1. Furthermore, in the in vitro binding assay used in this study, NLS specificity and affinity were found to be carried by the arm repeat array of the importin-α subunit alone, independent of the interaction with the arm repeat-containing importin-β subunit, an observation that differs from previous reports.
To test whether the observed differential recognition by the importin receptors is indicative of different modes of nuclear import for LEF-1 and TCF-1, the subcellular localizations of LEF-1 and TCF-1 were examined in vivo and in a nuclear transport assay in digitonin-permeabilized cells. LEF-1 and TCF-1 enter the nucleus in intact cells; however, only LEF-1 can be imported to the nucleus in the permeabilized cell assay. This finding suggests that while LEF-1 and TCF-1 are both nuclear transcription factors, their modes of nuclear transport appear to be different.
MATERIALS AND METHODS
Plasmid construction.
Full-length mouse pendulin (FL pendulin) and mouse Srp1 were constructed from partial yeast two-hybrid clones of pendulin (aa 25 to 529) and Srp1 (aa 191 to 538) in pACT that had been transferred into pBluescript (39). To obtain the first 25 aa of pendulin, primers were used to amplify the first 348 nucleotides of the coding region, using PCR from a mouse T-cell cDNA library (N-terminal 5′-GCGGATCCGCATGTCCACGAACGAGAATGC-3′ and C-terminal 5′-GGATGATGTTGTCTATAGGAGG-3′). A BamHI-XbaI fragment of this amplified, fully sequenced product was inserted into the partial clone mentioned above in homologous sites in pBluescript. To obtain coding sequence for the first 190 aa of Srp1, a degenerate N-terminal primer (5′-GCGGATCCCGATGTCNACCCCYGGNAAGGAGAA-3′) and a nondegenerate C-terminal primer (5′-GGTCATGGTCAAGCGGTTTTGC-3′) were used to amplify a 697-bp fragment, using PCR from mouse heart cDNA. An internal BamHI/BsgI fragment of this PCR product was transferred into the partial Srp1 clone in pBluescript at corresponding sites.
The HMG DNA binding domains of human LEF-1 (aa 297 to 384) and human TCF-1A (aa 152 to 239) were cloned in frame into the 3′ end of the yeast Gal4 DNA binding domain in plasmid pAS1.
The HMG DNA binding domain proteins were all cloned into a modified bacterial expression vector, pGEMEX, that could be used both for in vitro transcription/translation and for protein expression (6). The HMG DNA binding domain of LEF-1 (aa 297 to 384) in the modified pGEMEX vector was previously constructed (6). The HMG DNA binding domains of TCF-1 (aa 152 to 239) and LEF-RS (aa 297 to 384 with K377R and K379S) were both PCR amplified with the following pairs of primers: TCF-1 N-terminal 5′-CCAACCATCAAGAAGCCCCTCA-3′ and C-terminal 5′-TCATTGGTGCTTTTCCCTCGACCGCCT-3′ and LEF-RS N-terminal 5′-CCTCACATTAAGAAGCCTCTGAATGC-3′ and C-terminal 5′-TCACTGTAGTTTCTCTCTCGACCTCCT-3′ containing the two mutations. The HMG DNA binding domain of TCF-KK (aa 152 to 239 with R232K and S234K) was PCR amplified with primers N-terminal 5′-CCAACCATCAAGAAGCCCCTCA-3′ and C-terminal 5′-TCATTGGTGCTTTTCCCTCTTCCGCTT-3′ containing the two mutations. LEF-RS S→D (aa 297 to 384 with K377R and K379D) and TCF S→D (aa 152 to 239 with S234D) were PCR amplified by using the following pairs of primers: LEF-RS S→D N-terminal 5′-CCTCACATTAAGAAGCCTCTGAATGC-3′ and C-terminal 5′-TCACTGGTGTTTCTCTCTGTCCCT-3′ containing the two mutations and TCF S→D N-terminal 5′-CCAACCATCAAGAAGCCCCTCA-3′ and C-terminal 5′-TCATTGGTGCTTTTCCCTGTCCCG-3′ containing the one mutation. The HMG DNA binding domain of KLEFR (aa 297 to 384 with K377R) and KLEFS (aa 297 to 384 with K379S) were both PCR amplified with the same N-terminal primer, 5′-CCTCACATTAAGAAGCCTCTGAATGC-3′, and different C-terminal primers: 5′-TCACTGTAGTTTCTCTCTCTTCCTGCTCTTCTTCTT-3′ containing the single mutation for KLEFR and 5′-TCACTGTAGTTTCTCTCTAGATCTCTTC-3′ containing the single mutation for KLEFS. These amplified products were inserted in frame into the modified pGEMEX vector. The HMG DNA binding domain of STCFA (TCF-1A aa 152 to 239 with S234A) was PCR amplified from a green fluorescent protein (GFP)–TCF-1 DNA binding domain construct (described below), using primers N-terminal 5′-TCACACAATGTATACATCATG-3′ (primer hybridizes to the coding region of GFP starting at aa 147) and C-terminal 5′-TCATTGGTGCTTTTCCCGGGCCCGCCT-3′ containing the single mutation for STCFA. An EcoRI digest of this PCR fragment containing only the TCF-1 DNA binding domain portion was inserted in frame into the modified pGEMEX vector. Δ27 TCF-1 (aa 171 to 254; created by MseI digestion) and Δ19 TCF-1 (aa 179 to 254; created by StyI digestion) were also cloned in frame into the modified pGEMEX vector.
For the in vitro binding assay, all clones and deleted forms of pendulin were inserted into the pET15b (Novagen) vector for in vitro transcription and translation. A clone encoding FL pendulin and the 3′ untranslated region was excised from pBluescript by using BamHI and XhoI restriction sites and inserted in frame at the XhoI site of pET15b. BamHI/BglII fragments of the original yeast two-hybrid clones encoding aa 25 to 529 (ΔN25), 62 to 529 (ΔN62), 140 to 529 (ΔN140), 179 to 529 (ΔN179), 232 to 529 (ΔN232), 240 to 529 (ΔN240), and 243 to 529 (ΔN243) of pendulin in pACT were cloned in frame at the BamHI site of pET15b (39). ΔN301 (aa 301 to 529) was constructed by digestion with HindIII in the coding region of pendulin and XhoI at the 3′ end. ΔN328 (aa 328 to 529) was constructed by PCR amplification with primers starting at aa 328 (5′-ACTCAGAAAGTGATCGATGCA-3′) and ending at the stop codon (5′-TTAGAAGTTAAAGGTCCCAGG-3′). ΔN400 (aa 400 to 529) was constructed by digesting pendulin with BglI in the coding region and XhoI at the 3′ end. ΔN452 (aa 452 to 529) was constructed by digesting pendulin with PstI in the coding region and XhoI at the 3′ end. ΔN301, ΔN328, ΔN400, and ΔN452 were all cloned in frame at the BamHI site in pET15b. ΔC449 (aa 1 to 449), ΔC397 (aa 1 to 397), ΔC358 (aa 1 to 358), and ΔC300 (aa 1 to 300) were constructed by digestion with BamHI and PstI, BamHI and BglI, BamHI and Tth111I, and BamHI and HindIII, respectively. All were inserted in frame into the XhoI site in pET15b.
For the yeast two-hybrid assay, all pendulin deletions in pET15b except those originally obtained from the yeast two-hybrid screen were transferred into pACTII (S. Elledge, Baylor College of Medicine). Pendulin deletions in pET15b were digested with NcoI and EcoRV and ligated into pACTII at the NcoI and SmaI sites. The pendulin deletions in pACTII and Gal4–LEF-1 (aa 297 to 399) in pAS1 were simultaneously transformed into the Y190 yeast strain, and colonies were selected on medium lacking leucine and tryptophan.
Plasmid constructs were verified by sequencing using specific oligonucleotide primers and a Sequenase kit and restriction mapping. Fragments generated by PCR were completely sequenced.
Purification of the HMG DNA binding domains of LEF-1, TCF-1, and LEF-RS.
Recombinant HMG DNA binding domain proteins used as competitors in the in vitro binding assay were expressed in the pLysS bacterial strain as previously described (54). The bacterial pellet was resuspended in lysis buffer (phosphate-buffered saline [PBS], 1% Triton X-100, phenylmethylsulfonyl fluoride [PMSF]), sonicated to lyse bacteria (two 30-s pulses with a 1-min, 0°C incubation between pulses), and centrifuged at 17,000 × g for 10 min. Glycerol was added to 10%, and the supernatant was stored at −100°C (until the next step). A 30% (NH4)2SO4 cut of the crude lysate served as an enrichment step for the HMG DNA binding domain proteins. Cleared supernatant was then raised to 60% (NH4)2SO4, and precipitated proteins from this fraction were isolated by centrifugation at 34,000 × g for 10 min at 4°C. The 60% (NH4)2SO4 pellet was resuspended in TM 0.1M buffer (50 mM Tris [pH 7.9], 12.5 mM MgCl2, 1 mM EDTA, 1 mM dithiothreitol, 10% glycerol, 0.1% Nonidet P-40, 0.1 M KCl) and dialyzed against 30% (NH4)2SO4 in TM 0.1M, followed by 10% (NH4)2SO4 in TM 0.1M and finally TM 0.1M buffer. The supernatant was cleared by centrifugation at 17,000 × g at 4°C for 15 min. The supernatant was loaded onto a 1-ml Econo S column (Bio-Rad) in TM 0.1M buffer. The column was washed with 3 column volumes of TM 0.125M (0.125 M KCl), and proteins were eluted with a 10-column volume KCl gradient of 0.125 to 0.6 M KCl, followed by a 1-column volume 1 M KCl elution. Fractions containing the purified HMG DNA binding domain protein were pooled and dialyzed against TM 0.1M buffer and then loaded onto a 6-ml calf thymus DNA cellulose column (Sigma). The column was developed with a 0.1 to 0.8 M KCl gradient, and peak fractions containing HMG protein were pooled and dialyzed against TM 0.15M buffer (containing 0.15 M KCl). Protein concentrations were determined by the Bradford method and comparison to a standard curve of bovine serum albumin (BSA) protein. Silver stain analysis of sodium dodecyl sulfate (SDS)-polyacrylamide gels confirmed that each protein was purified to >98% homogeneity. To determine the fraction of purified HMG protein that was active for DNA binding, DNA titration experiments with a radiolabeled oligonucleotide encoding a LEF-1 binding site were performed as described previously (38). Binding was performed with DNA concentrations low enough that free protein was approximately equal to total protein, and DNA and DNA-protein complexes were quantitated by scintillation counting of shifted and nonshifted 32P-labeled oligonucleotide.
Peptides for competitors in the in vitro binding assay.
LEF-1 B-box peptide (KKKKRKREK) and TCF-1 B-box peptide (KKKRRSREK) were synthesized by Biosynthesis, Inc., Lewisville, Tex. Simian virus 40 (SV40) T antigen (T Ag) NLS peptide (PKKKRKVED) was synthesized by Genosys Biotechnologies Inc., The Woodlands, Tex. SV40 T Ag reverse NLS peptide (GYGDEVKRKKKP) was obtained from the laboratory of Masayasu Nomura (University of California, Irvine). Peptides were >70% pure.
GST fusion protein purification.
FL pendulin was inserted in frame 3′ of the glutathione S-transferase (GST) gene in pGEX-3X (Pharmacia Biotech, Inc.). GST-FL pendulin, GST-pendulin (aa 25 to 529), and GST were purified as previously described (39). A eukaryotic GFP-C1 open reading frame (Clontech) was fused in frame to the 3′ end of GST in pGEX-2T (Pharmacia Biotech) (referred to as GST-GFP). LEF-1 (aa 297 to 399) and TCF-1A (aa 152 to 254) were each fused in frame to the 3′ end of GFP in the GST-GFP pGEX-2T construct prepared as described above. These proteins were expressed in bacteria and purified over a glutathione affinity column as previously described (39). GST-GFP, GST–GFP–LEF-1, and GST–GFP–TCF-1 were judged to be ∼85% to 90% pure by Coomassie blue staining of SDS-polyacrylamide gels. DNA fragments encoding the DNA binding domains of LEF-1 (aa 297 to 384) and TCF-1A (aa 152 to 254) were each inserted in frame 3′ of GST in pGEX-2T (Pharmacia Biotech), and recombinant protein was obtained by overexpression in isopropyl-1-thio-β-d-galactopyranoside-treated Escherichia coli BL21 cells (0.2 mg/ml, 4 h, 37°C). Cells from a 1-liter culture were lysed by sonication in 20 ml of lysis buffer (1× PBS, 1% Triton X-100, 1 mM PMSF) two times for 30 s and then centrifuged for 15 min at 17,000 × g. GST–LEF-1 and GST–TCF-1 proteins were recovered from the pellet by extraction with 20 ml of 4 M guanidine-HCl in column buffer (CB; 50 mM Tris-HCl [pH 8], 1 mM dithiothreitol, 1 mM EDTA, 5% glycerol, 50 mM NH4SO4, 1 mM PMSF) for 1 h at 4°C on a rocking platform. The extract was centrifuged at 17,000 × g for 15 min, and the resulting supernatant was dialyzed against two changes of 1 M guanidine-HCl in CB followed by dialysis in two changes of CB. A white precipitate formed during dialysis was removed by centrifugation at 17,000 × g for 15 min. The supernatant was applied to a 1-ml glutathione-Sepharose column, and the column was washed with 30 column volumes of CB. GST fusion proteins were eluted in 5 ml of CB supplemented with 5 mM glutathione. Protein was quantified by the Bradford assay or SDS-polyacrylamide gel electrophoresis (PAGE) and Coomassie blue staining.
In vitro binding assay.
Radiolabeled HMG DNA binding domain or pendulin protein fragments were generated by using a coupled in vitro transcription/translation system (Promega) and [35S]methionine (DuPont NEN). The in vitro binding assay was performed as previously described (39) except that TBST (200 mM NaCl, 0.2% Tween 20, 10 mM Tris [pH 8.0]) was used. Additionally, ethidium bromide (200 μg/ml) and RNase A (100 μg/ml) were added to the binding assay only during the 30-min room temperature incubation with GST fusion protein bound to glutathione beads, 35S-labeled in vitro-translated proteins, and the TBST–0.2% BSA buffer. We have found that the addition of ethidium bromide and RNase A raises specific binding levels fivefold. Since LEF-1 and TCF-1 are DNA binding proteins, competing nucleic acid in the plasmid-programmed TNT (Promega) translation preparations may interfere with importin/karyopherin binding (28). For competition experiments, GST fusion protein bound to glutathione beads, radiolabeled proteins, TBST–0.2% BSA, and either NLS peptides or purified HMG DNA binding domain proteins of LEF-1, TCF-1, or LEF-RS used as competitors were incubated for 2 h instead of 30 min.
β-Galactosidase assays.
For qualitative plate assays, yeast colonies were patched onto minimal medium lacking leucine and tryptophan and incubated for 3 days. Colonies were transferred onto Whatman no. 50 filter paper, lysed in liquid nitrogen, and placed onto Whatman 3mm paper soaked in Z buffer (0.113 M Na2HPO4 · 7H2O, 0.04 M NaH2PO4 · H2O, 0.01 M KCl, 1 mM MgSO4 · 7H2O, 0.03 M β-mercaptoethanol [pH 7.0]) containing 1 mg of 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal) per ml. Plates were incubated at 30°C from 1 h to overnight. For liquid quantitative assays, colonies were grown to stationary phase in minimal medium lacking leucine or tryptophan or both. Yeast cultures were prepared and assayed as described previously (47). Three independent colonies were assayed in triplicate for each strain.
Importin-β depletion binding assay.
Twenty microliters of in vitro-translated FL pendulin or ΔN62 was incubated with 20 μl of either a 50% slurry of anti-importin-β antibody or control antibody covalently attached to Sepharose beads (generous gifts from S. A. Adam) in 1× PBS–0.1% gelatin–0.1% azide. The binding reaction mixtures were incubated for 30 min on a rotating platform at room temperature and then centrifuged for 5 min at 2,000 rpm to collect the beads. The depleted supernatants were assayed for quantitative depletion of importin-β and saved for later use in a binding assay with GST–LEF-1, GST–TCF-1, and GST in the in vitro binding assay protocol described above. Beads were washed three times with 1 ml of TBST, and 15 μl of 2× SDS sample buffer was added. Samples were analyzed by SDS-PAGE in the same manner as in the in vitro binding assay.
Fluorescence microscopy.
DNA sequences for TCF-1A aa 152 to 239 (GFP TCF wt [wild type]), 152 to 228, and 229 to 239 and LEF-1 aa 297 to 384 (GFP LEF wt) were fused in frame at the PvuII site (near the 3′ end) of coding sequences for a variant of GFP (S65T; generous gift of R. Tsien, University of California, San Diego) in a eukaryotic expression vector containing an SV40 origin of replication. Two single-amino-acid mutants for TCF-1A (aa 152 to 239), S234A (GFP STCFA) and S234D (GFP STCFD), and two single-amino-acid mutants for LEF-1 (aa 297 to 384), K377R (GFP KLEFR) and K379S (GFP KLEFS), were fused in frame at the PvuII site of GFP in the eukaryotic expression vector described above. All mutants were PCR amplified from GFP TCF wt and GFP LEF wt DNA binding domain constructs by using the same N-terminal primer, 5′-TCACACAATGTATACATCATG-3′. This primer hybridizes to the coding region of GFP starting at aa 147. The C-terminal primers used for each mutant are the same as those listed above for the identical mutation cloned into the modified pGEMEX vector. Cos-1 transfection, the expression vector, and immunofluorescence protocols were described previously (39). Slides were examined with a Zeiss Axioskop and an Oncor charge-coupled device (CCD) camera at 80× and UV illumination through a fluorescein isothiocyanate (FITC) filter.
In vitro nuclear import assay.
HeLa S100 cytosol extract used to reconstitute nuclear transport was prepared as described by Adam et al. (3). Digitonin permeabilization of HeLa cells and the in vitro nuclear import assay procedure were performed exactly as described previously (1) except for the following: in the 50-μl import reaction, HeLa cytosol was added to a final concentration of 4 mg/ml in place of NLS receptor and p97/importin-β; 40 μg of purified GST-GFP, GST–GFP-LEF-1, or GST–GFP-TCF-1 per ml was added where indicated. In experiments with ATP, an ATP-regenerating system was added in amounts described previously (1). In experiments without ATP, the ATP-regenerating system was omitted and 50 U of apyrase (Sigma) per ml was added. GST-FL pendulin was added to 30 μg/ml in the import reaction where indicated. The coverslips were mounted on glass slides in import buffer and sealed with epoxy. Slides were examined with a Zeiss Axioskop and an Oncor CCD camera at 80× and UV illumination through an FITC filter.
RESULTS
Two importin/karyopherin-α subtypes bind to LEF-1 and not to TCF-1.
We have previously reported that two importin/karyopherin-α subtypes (referred to here as Srp1 and pendulin) interact with the LEF-1 HMG DNA binding domain in a yeast two-hybrid assay (39). In this assay, LEF-1 coding sequences were fused to the C terminus of a fragment encoding the DNA binding domain of yeast Gal4. An identical fusion of sequences encoding the TCF-1 HMG DNA binding domain to Gal4 did not produce a protein capable of interacting with either pendulin or Srp1 in the yeast two-hybrid assay. This is remarkable given that the 235-aa Gal4–LEF-1 and Gal4–TCF-1 fusion proteins differ at only six positions (Fig. 1A). In the reported assay, partial coding sequences of pendulin and Srp1 were fused to the C terminus of the Gal4 transcription activation domain. To test the possibility that sequences missing from each of the partial pendulin and Srp1 open reading frames were necessary for TCF-1 recognition, full-length open reading frames were built by PCR amplification and placed into appropriate vectors for testing in the yeast two-hybrid assay. β-Galactosidase activity was assessed by filter and solution assay (Fig. 1B and C), and consistent with our previous observations, FL pendulin and Srp1 interacted with Gal4–LEF-1 but not Gal4–TCF-1.
LEF-1 and TCF-1 are coexpressed in differentiating T lymphocytes in the thymus. Interestingly, pendulin mRNA is highly expressed in thymus (as well as spleen and heart), while Srp1 is expressed ubiquitously at a very low level (39). We observe a similar pattern of relative expression in T-cell lines. Since pendulin is abundant in cells that express LEF-1 and TCF-1, we have analyzed the interaction between LEF-1 and pendulin and the lack of interaction between TCF-1 and pendulin.
Specificity of an in vitro assay for NLS binding.
The specificity of the interaction between pendulin and the LEF-1 and TCF-1 HMG DNA binding domains was tested in an in vitro GST pull-down assay. This assay was used previously to map the amino acids within the LEF-1 B box/NLS necessary for a direct interaction with the NLS receptors (39). Modifications to the GST pull-down protocol were added to improve levels of binding above background up to fivefold (see Materials and Methods). To test that these modifications do not alter the interaction between pendulin and the HMG DNA binding domains, several versions of the modified pull-down assay were assessed for specificity. For one assay, GST coding sequence was fused to the pendulin open reading frame (GST-pendulin [aa 25 to 529]). This fusion protein was expressed and purified in recombinant form from bacteria and incubated with 35S-labeled, in vitro-translated HMG protein (Fig. 2A). Glutathione-linked Sepharose beads were added to bring down any 35S-labeled HMG DNA binding domain that associated specifically with GST-pendulin. Wild-type LEF-1 and TCF-1 HMG DNA binding domains were tested as well as a LEF-1 mutation in which two of the amino acids in the B box/NLS were changed to that of the TCF-1 B box (LEF-RS). Only the LEF-1 HMG DNA binding domain interacted specifically with GST-pendulin. No interaction was observed with TCF-1, which differs by six residues, nor LEF-RS which only differs by two residues. In a different version of the assay, GST was fused to sequences encoding the wild-type HMG DNA binding domain of LEF-1 (GST–LEF-1) or TCF-1 (GST–TCF-1). Purified, recombinant protein derived from these fusion constructs was incubated with in vitro-translated 35S-labeled FL pendulin (Fig. 2B), and again, FL pendulin interacted with GST–LEF-1 and only weakly with GST–TCF-1 at levels near background. Thus, interactions with GST-pendulin in vitro in these new conditions are highly specific and sensitive to small differences in amino acid sequence within the B box.
FIG. 2.
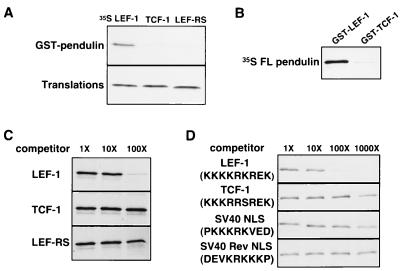
Specificity of an in vitro GST pull-down binding assay. (A) Sequences encoding the HMG DNA binding domains of LEF-1 (LEF (aa 297 to 384), TCF-1 (aa 152 to 239 of TCF-1A), and LEF-RS were in vitro translated and incubated with GST-pendulin fusion protein. 35S-labeled proteins that bound to the GST fusion proteins were sedimented with glutathione resin followed by SDS-PAGE of released proteins and autoradiography of the gel (GST pull-down assay described in the text and in Materials and Methods). Only the LEF-1 HMG DNA binding domain can bind to pendulin, and the 2-aa mutation (LEF-RS) destroys this interaction. (B) FL pendulin was generated by in vitro translation in reticulocyte lysates supplemented with [35S]methionine and then incubated with GST–LEF-1 or GST–TCF-1 in the GST pull-down assay. SDS-PAGE analysis shows that pendulin interacts with GST–LEF-1 but not GST–TCF-1. (C) The specificity of binding of in vitro-translated FL pendulin to GST–LEF-1 in the GST pull-down assay was analyzed by competition with 1-, 10-, and 100-fold molar excesses of purified, recombinant LEF-1, TCF-1, and LEF-RS HMG DNA binding domain proteins compared to the amount of GST-LEF-1 added. (D) As in panel C, the specificity of binding of in vitro-translated FL pendulin to GST–LEF-1 in the GST pull-down assay was analyzed by competition with 1-, 10-, 100-, and 1,000-fold molar excesses of peptides encoding the B boxes of LEF-1 and TCF-1, the SV40 T Ag NLS, and a mutant SV40 T Ag NLS which is the reverse sequence.
The specificity of the interaction was further explored by competition with purified LEF-1 HMG DNA binding domain, TCF-1 HMG DNA binding domain, or LEF-RS HMG DNA binding domain proteins (described above). These recombinant proteins were purified from bacterial lysates to near homogeneity (98%) by column chromatography, with a final purification on calf thymus DNA-cellulose to enrich the for properly folded HMG DNA binding domain. Gel shift analysis confirmed that each of these purified protein preparations was approximately 60 to 75% active for DNA binding (19). Each domain was added in the indicated molar equivalents relative to GST–LEF-1 affixed to glutathione beads (60 nM). Wild-type LEF-1 HMG DNA binding domain protein competes for binding at 100 molar excess (6 μM), whereas the same amount of TCF-1 HMG DNA binding domain or LEF-RS HMG DNA binding domain protein does not compete (Fig. 2C). Competition with peptides encoding the LEF-1 or TCF-1 B box show a similar pattern. The LEF-1 NLS peptide but not the TCF-1 B-box peptide competes for binding at 100-fold molar excess (down to 24% [Fig. 2D]). The LEF-1 NLS peptide competes even more for binding at 1,000-fold molar excess (60 μM), while at this concentration the TCF-1 NLS competes weakly for binding, lowering levels by 50%. At such high concentrations, it is possible that the amount of competition observed with the TCF-1 peptide is nonspecific. However, we also tested the wild-type and mutant SV40 NLS peptides (Fig. 2D). Whereas the wild-type SV40 peptide competes similarly to TCF-1 (48% at 1,000-fold molar excess), the Rev peptide does not compete for binding at 1,000-fold molar excess. Thus, the small amount of competition observed with the TCF-1 NLS may reflect low but significant binding to pendulin. It is interesting that the intact LEF-1 HMG DNA binding domain competes 5- to 10-fold better for binding to FL pendulin than the LEF-1 NLS alone, and the intact TCF-1 HMG DNA binding domain does not. These results imply that placement of the NLS within the HMG context further refines NLS recognition.
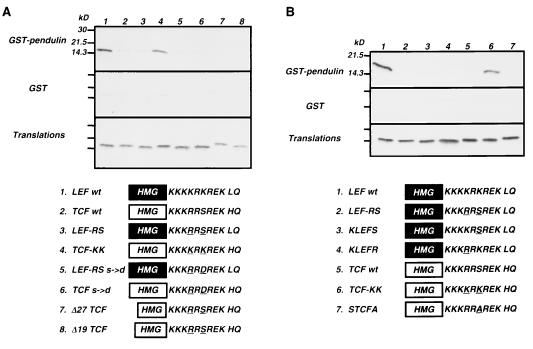
The contexts provided by the LEF-1 and TCF-1 HMG DNA binding domains are nearly identical (Fig. 1A). Two of six amino acid differences are within the B box; one difference is at the C terminus just outside the HMG DNA binding domain. The remaining three are in the N-terminal region of the HMG box; one of these, the threonine amino acid in TCF-1 (T299), is not conserved between humans and mice. The other residue differences are conserved. To test whether the context of LEF-1 enables better recognition of the TCF-1 B box, a swap experiment was performed in which sequences encoding the TCF-1 B box replaced the LEF-1 B box in the LEF-1 HMG DNA binding domain, and vice versa (Fig. 3). Each wild-type and modified DNA binding domain peptide was generated by in vitro translation and incubated with GST-pendulin beads in the GST pull-down assay. Context does not appear to play a major role in peptide recognition, because placement of the TCF-1 B box within the context of the LEF-1 HMG DNA binding domain did not enhance or enable recognition of the TCF-1 NLS (LEF-RS [Fig. 3A, lane 3; Fig. 3B, lane 2]). The context of the TCF-1 DNA binding domain is not entirely deleterious to recognition because the LEF-1 NLS, when placed within the TCF-1 context, is still recognized by GST-pendulin, albeit at levels that are approximately 30% lower (TCF-KK [Fig. 3A, lane 4; Fig. 3B, lane 6]). We also tested whether the TCF-1 B box was masked in any way by regions of the HMG box itself. Two different deletions which remove 19 and 27 aa from the N-terminal end of the HMG box do not enable TCF-1 B-box recognition. These two deletions appear to run aberrantly large on SDS-PAGE. The constructs encoding these deletion forms have been sequenced multiple times in both strands to confirm that these expression constructs are correct. Furthermore, LEF-1 antibody specifically recognizes these N-terminal truncation mutants in a Western analysis (data not shown). Therefore, the slower migration is not due to a frameshift mutation, but instead may reflect a disruption of the highly alpha-helical structure of this domain (53).
FIG. 3.
The B box is the primary site of importin/karyopherin-α recognition. Each of the wild type and mutant HMG DNA binding domains depicted at the bottom was generated by in vitro translation in reticulocyte lysates supplemented with [35S]methionine. Protein equivalent amounts of each translation product were incubated with either GST-pendulin or GST alone in the GST pull-down assay. A portion (10%) of the amount of each translation product added to the binding assay is shown in the bottom panels.
To determine whether the arginine or the serine residue in the TCF-1 B box was most deleterious for pendulin recognition, single amino acid changes within the LEF-1 NLS were made to match TCF-1 (Fig. 3B, lanes 3 and 4). Surprisingly, a single amino acid change from lysine to arginine (KLEFR) or lysine to serine (KLEFS) is enough to negate pendulin binding.
To mimic the negative charge of a phosphorylated serine, a mutation in the sequence encoding the serine residue in the TCF-1 B box was changed to encode an aspartate residue. Others have reported a functional role for phosphorylation near or within NLSs in other proteins, and it was possible that phosphorylation of the serine residue in the TCF-1 B box might enable importin/karyopherin-α recognition. However, this single amino acid change does not promote pendulin binding either within the context of TCF-1 or LEF-RS (Fig. 3A, lanes 5 and 6).
The sensitivity of pendulin binding to conservative single amino acid changes within an NLS is striking. NLS recognition is not thought to be so specific since bona fide NLS sequences differ greatly and consensus NLSs have been difficult to define. Arm repeats are thought to function directly as the NLS interaction domain, but it is not clear whether one arm repeat or a group of arm repeats are specific for one or more NLS sequences. Nor is it clear that it is always the arm repeats that recognize an NLS. Several groups have found the C-terminal hydrophilic domain of importin-α to interact with nuclear targeting signals. To explore the specificity of the LEF-1–pendulin interaction further, a deletion analysis of the arm repeat array of pendulin was performed. Since TCF-1 does not interact with pendulin in vitro, we reasoned that this protein could be used as a negative control for specificity to help identify which regions of the importin-α protein were involved in directing specificity of recognition.
NLS receptor deletion analysis in yeast.
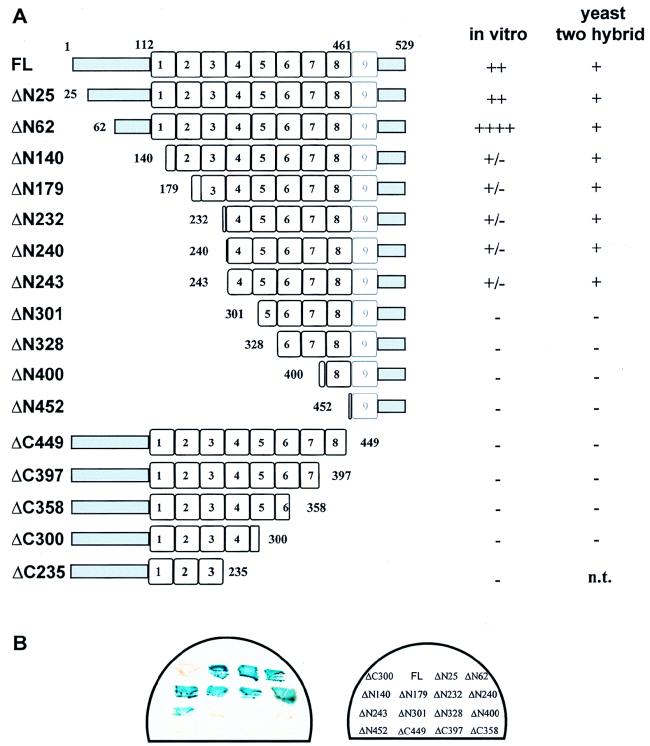
N- and C-terminal deletions of pendulin were constructed (Fig. 4A). Each deletion is shown relative to the borders of the armadillo repeats established by Yano et al. (59). While the true borders of armadillo repeats within the NLS receptor family have not been firmly established, the borders shown here are more congruent with those recently established for HEAT repeats, a more general type of repeat that includes armadillo repeats (29a). Each of the mutated NLS receptors was placed in a vector for yeast two-hybrid analysis with the HMG DNA binding domain of LEF-1 as bait (see Materials and Methods). Figure 4B shows the results of the yeast two-hybrid analysis in a filter assay format where one representative colony for each deletion was analyzed as a patch on the filter. FL pendulin readily associated with the DNA binding domain of LEF-1 in yeast to produce a strong increase in lacZ reporter gene expression. Deletion of the hydrophilic N-terminal region of pendulin had no obvious effect on β-galactosidase levels in the filter assay (ΔN25 and ΔN62), nor did further deletion into arm repeats 1 to 3 (ΔN140, ΔN179, ΔN232, ΔN240, and ΔN243). However, deletion of arm repeat 4 and beyond completely abrogated any interaction in yeast (ΔN301-ΔN452). None of the C-terminal deletions exhibited an interaction with LEF-1 in yeast (ΔC449-ΔC300). The results from this assay indicate that the first few armadillo repeats are not necessary for NLS recognition in yeast. It would also seem from our analysis of the C-terminal deletions that a complete C terminus is required; however, this may not be the case since these fusion proteins were expressed at much lower levels than the N-terminal deletion fusion proteins (10- to 15-fold lower, as judged by Western analysis [38a]). The yeast two-hybrid assay, although useful for detecting strong, specific interactions, may not be sensitive enough to detect weak but specific binding, and the variation in protein levels of the different deletions complicates any interpretation of the results.
FIG. 4.
Summary of pendulin deletions and corresponding LEF-1 binding activities. (A) The amino acid endpoints of the N- and C-terminal deletions are given for each construct and are shown relative to arm repeat borders that match those of Yano et al. (59). A summary of the activity of each deletion is given on the right for the in vitro binding assay and for the filter assay in the yeast two-hybrid system. n.t., not tested. (B) Filter assay for β-galactosidase activity in the yeast two-hybrid system. Each pendulin deletion (depicted in panel A) was cloned into a pACTI or pACTII yeast two-hybrid vector (gift of S. Elledge) in frame with sequences encoding the Gal4 transcription activation domain. The LEF-1 HMG DNA binding domain was inserted into a pAS vector to be expressed as a fusion protein with the Gal4 DNA binding domain. Both yeast two hybrid expression plasmids were cotransformed into strain Y190 and selected on Trp- and Leu-deficient selective medium for 4 days. An increase in the levels of β-galactosidase activity is an indication of the in vivo interaction between the two fusion proteins.
NLS receptor deletion analysis in vitro.
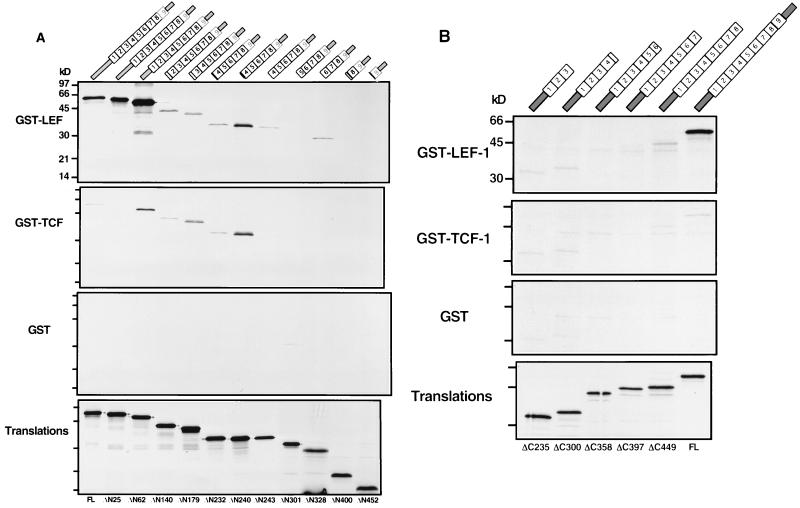
All of the N- and C-terminal deletions were tested for binding to GST–LEF-1, GST–TCF-1, and GST in vitro (Fig. 5). Deletion of the first N-terminal 24 aa does not appear to affect binding to GST-LEF-1 (Fig. 5A, ΔN25), but deletion to aa 62 (ΔN62) resulted in a dramatic and surprising 10-fold increase in GST–LEF-1 binding (when the specific activities of the translation products are accounted for, there is a 30-fold molar increase in binding). A small amount of GST–TCF-1 binding was detected with this protein but at levels greatly reduced compared to GST–LEF-1. No binding to GST alone was observed. Further deletion to aa 140 (ΔN140), a deletion predicted to disrupt the first armadillo repeat, resulted in a sharp decrease in GST–LEF-1 binding to levels that were more than 50-fold below the level of binding observed for ΔN62 and approximately 3- to 5-fold below the levels of binding observed with FL pendulin and ΔN25. Curiously, there was a slight increase in binding of ΔN240 to GST–TCF-1, reaching levels equivalent to that observed with GST–LEF-1. Further deletion into the arm repeat array continued this pattern—weak but equivalent recognition of GST–LEF-1 and GST–TCF-1 and not GST. We conclude from this pattern of binding that deletion into the arm repeat array not only causes a critical loss in NLS binding affinity but also damages specificity. The alternative hypothesis, that deletion to aa 140 has removed part of the LEF-1-specific binding domain, is unlikely given that there is no loss of binding or specificity with these deletions in the yeast two-hybrid assay.
FIG. 5.
In vitro analysis of the pendulin deletion constructs. Proteins encoded by N-terminal (A) and C-terminal (B) deletion constructs of pendulin were generated by translation in vitro in reticulocyte lysates supplemented with [35S]methionine. Translation products were tested for interaction with GST–LEF-1, GST–TCF-1, and GST alone in the GST pull-down assay. Protein equivalent amounts of each translation product were added to the binding reactions as shown in the bottom panel.
To define the C-terminal border of the LEF-1 recognition domain, we tested six truncation mutants (ΔC449 to ΔC235 [Fig. 5B]), with the shortest containing a translation stop codon in arm repeat 3. Each of these shortened proteins was generated by in vitro translation and tested in the GST pull-down assay with GST–LEF-1. In accordance with their lack of binding in the yeast two-hybrid assay, all of the C-terminal truncation mutants were incapable of interacting specifically with GST–LEF-1, and binding for all deletions was at least 15-fold lower than that for FL pendulin.
The first 55 aa of pendulin comprise an important domain referred to as the importin-β binding (IBB) domain (13, 57). The IBB domain binds directly to the 97 kDa co-NLS receptor called importin-β, and this interaction is essential for nuclear import. The IBB domain alone, when fused to a cytoplasmic reporter protein, can promote complete nuclear import, circumventing the usual requirement for an NLS-receptor interaction. Thus, the IBB domain is responsible for mediating the interaction of importin/karyopherin-α receptors to the protein import machinery. Our results indicate that removal of this domain (ΔN62) augments binding to GST–LEF-1 in vitro but not in the yeast two-hybrid assay. One possible difference between the yeast assay and the GST pull-down assay is that importin-β is present in reticulocyte lysates (∼100 ng/50 μl [1a]), whereas in yeast, KAP95 (yeast importin-β) may have limited access to the Gal4-pendulin bait in yeast nuclei.
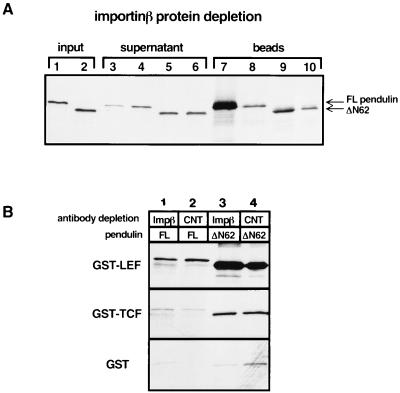
Binding of importin-β to importin/karyopherin-α has been reported to increase NLS binding (41). This report, coupled with the data presented above showing that the ΔN62 deletion mutant missing the IBB domain binds ∼30-fold better to the LEF-1 NLS, suggests either that removal of the IBB domain mimics importin-β binding or that importin-β binding in our GST pull-down assay is inhibitory or that ΔN62 creates an altered conformation of the protein, enabling better binding in vitro. Reticulocyte lysates contain endogenous importin-β protein, and therefore translation products of pendulin that contain an intact IBB domain are likely to interact with endogenous importin-β in the lysate. To determine whether the presence of endogenous importin-β influences pendulin recognition of GST–LEF-1 in our GST pull-down assay, importin-β protein was depleted from reticulocyte lysates that had been programmed with either FL pendulin or ΔN62 expression plasmid. Quantitative depletion of importin-β was achieved by using importin-β antibody covalently attached to beads (kindly provided by S. Adam), and complete depletion of the protein was confirmed by Western analysis with soluble importin-β antisera (data not shown). These depleted lysates were then used in a GST pull-down assay with GST–LEF-1, GST–TCF-1, and GST to examine both the specificity and affinity of pendulin for LEF-1 (Fig. 6B).
FIG. 6.
Importin-β does not influence the affinity or specificity of the pendulin–LEF-1 interaction. (A) Reticulocyte lysates programmed with constructs encoding either FL pendulin (lanes 1, 3, 4, 7, and 8) or ΔN62 pendulin (lanes 2, 5, 6, 9, and 10) were treated with importin-β antiserum (lanes 3, 5, 7, and 9) or control antiserum (lanes 4, 6, 8, and 10) covalently attached to beads. Lanes 1 and 2 depict 1.25% of the translation product in lysates prior to treatment with the antibody-coupled beads; lanes 3 through 6 show 1.25% of the translation product remaining after the antibody depletion; lanes 7 through 10 show 100% of the translation product that cosedimented with the antibody-coupled beads. (B) Analysis of the depleted lysates in the GST pull-down assay. Translation lysates treated with importin-β antisera (Impβ) or a control serum (CNT) were incubated with GST–LEF-1, GST–TCF-1, or GST alone in the GST pull-down assay. Depletion of importin-β from the reticulocyte lysates does not significantly affect the binding of pendulin to GST–LEF-1 or GST–TCF-1.
SDS-PAGE analysis of 35S-labeled pendulin remaining in the supernatant after depletion and products associated with the importin-β antibody beads shows that a significant amount (80%) of FL pendulin was depleted with the importin-β antibody (Fig. 6A). Depletion of FL pendulin with bead-bound control antibody was equal to the amount of 35S-labeled ΔN62 that was lost with either importin-β antibody beads or control antibody beads, indicating that nonspecific binding by antibody reduces the amount of any in vitro translation product by about 20%. This experiment demonstrates that most of the in vitro-translated FL pendulin associates with endogenous importin-β in the reticulocyte lysate (compare lanes 3 and 4 and lanes 7 and 8). Since FL pendulin binds the LEF-1 NLS poorly compared to ΔN62 in our standard GST pull-down assay, we can conclude either that ΔN62 assumes a conformation more favorable for LEF-1 interaction or that in this assay, importin-β binding is somewhat repressive for binding of FL pendulin to LEF-1. Clearly, removal of the IBB domain does not mimic importin-β binding.
As a further test, equal amounts of FL pendulin and ΔN62 from the various depleted lysates were tested for binding to GST–LEF-1, GST–TCF-1, and GST (Fig. 6B). The levels of ΔN62 binding to GST–LEF-1 and GST–TCF-1 from control and from importin-β-depleted extracts were identical. Depletion of importin-β did not affect the level of ΔN62 binding to GST–LEF-1, nor did it alter the specificity of binding for LEF-1 versus TCF-1. This is not surprising as ΔN62 is completely missing the IBB domain and therefore incapable of interacting with importin-β; importin-β depletion would be predicted to have no consequence. More importantly, a similar pattern of binding was observed with FL pendulin in the depleted extracts. That is, quantitative depletion of importin-β did not affect the specificity of FL pendulin for LEF-1 versus TCF-1, nor did it significantly affect the level of binding of LEF-1 (compare lanes 1 and 2). We conclude that the specificity of pendulin for LEF-1 versus TCF-1 and the 28- to 30-fold increase in LEF-1 NLS binding observed with the ΔN62 deletion is independent of importin-β binding, a result that is somewhat in contrast to reports from other groups.
The TCF-1 B box functions as an NLS in vivo.
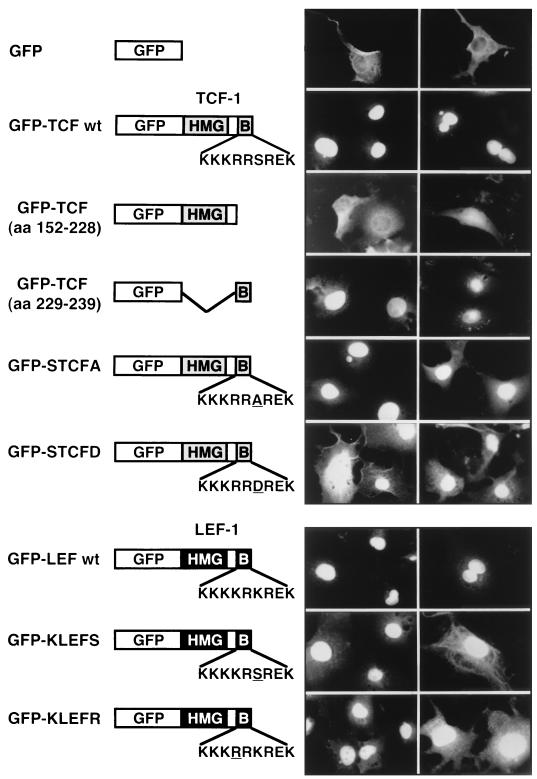
We wished to explore the functional consequences of the differential recognition of LEF-1 and TCF-1 by pendulin. Based on the high degree of amino acid sequence similarity between the TCF-1 B box and the LEF-1 B box/NLS, the TCF-1 B box has been proposed to be a functional NLS, although this has never been formally tested. Therefore, another possible explanation for the lack of interaction between TCF-1 and pendulin and Srp1 is that there is not a functional NLS within the TCF-1 HMG DNA binding domain. To test for this possibility, sequences encoding the wild-type TCF-1 HMG DNA binding domain were fused 3′ of sequences encoding GFP (GFP-TCF wt [Fig. 7]). A deletion mutant of TCF-1 missing the 9-aa B box (GFP-TCF [aa 152 to 228]) and a sequence encoding the 9-aa B box were also fused to GFP (GFP-TCF [aa 229 to 239]). These fusion proteins were introduced into Cos-1 cells by transient transfection, and green fluorescence was monitored to determine subcellular localization. Bright staining is observed in the nuclei of cells transfected with either the GFP–TCF wt or GFP-TCF (aa 229 to 239) expression plasmid. However, GFP–TCF-1 without the B box (GFP-TCF [aa 152 to 228]) remains in the cytoplasm. Thus, the TCF-1 B box can function as a nuclear targeting sequence in vivo both as an independent NLS and within the context of the HMG DNA binding domain.
FIG. 7.
The TCF-1 B box functions as an NLS. Mammalian expression plasmids encoding GFP fused to the TCF-1 HMG DNA binding domain (GFP-TCF wt; aa 152 to 239 of TCF-1A), the DNA binding domain missing the B box (aa 152 to 228), the B box alone (aa 229 to 239), and GFP alone were transiently transfected into Cos-1 cells; 48 h later, subcellular localization was assessed by immunofluorescence of formaldehyde-fixed cells, using a Zeiss Axioskop and an Oncor CCD camera with an FITC filter. Two examples of field views are shown for each transfected construct. Single amino acid changes in the TCF-1 (GFP-STCFA and GFP-STCFD) and LEF-1 B boxes (GFP-KLEFS and GFP-KLEFR) were also assessed for their effect on nuclear localization. Single amino acid substitutions are underlined.
To test whether phosphorylation of the serine residue within the TCF-1 NLS was involved in nuclear import, we examined the subcellular localization of a mutant TCF-1 NLS in which the serine was replaced by alanine (GFP-STCFA [Fig. 7]). GFP-STCFA was able to efficiently localize to the nucleus, demonstrating that phosphorylation of the TCF-1 NLS is not necessary for nuclear import. In fact, replacement of the serine with an aspartate residue to mimic phosphorylation (GFP-STCFD [Fig. 7]) appears to be somewhat deleterious to nuclear import, as more of this protein appears to remain in the cytoplasm.
Single amino acid substitutions within the LEF-1 NLS that were not recognized by pendulin in the in vitro GST pull-down assay (GFP-KLEFS and GFP-KLEFR) were tested for the ability to direct nuclear import in vivo. These mutant NLSs were able to direct import almost as efficiently as the wild-type LEF-1 NLS. We observed minor increases in cytoplasmic accumulation of these fusion proteins, suggesting that the single amino acid substitutions may have interfered slightly with nuclear import. Nevertheless, mutations in the LEF-1 NLS that disrupted pendulin interactions in vitro were not severely deleterious for nuclear transport in vivo. Likewise, wild-type TCF-1, STCFA, and STCFD proteins were all able to localize to the nucleus but did not show significant binding to pendulin in vitro (Fig. 3). Taken together, this discordance between lack of pendulin binding in vitro and positive nuclear import in vivo suggested that even weak NLS recognition by pendulin in vitro might be enough for nuclear import in vivo and therefore, although the TCF-1 NLS is not recognized well by pendulin in our assays, it might be a recognized target in vivo.
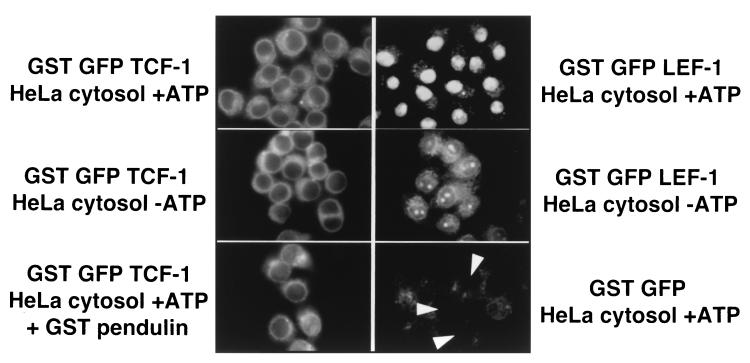
An in vitro nuclear transport assay using digitonin-permeabilized cells was performed to test whether this was the case. In this assay, nuclear transport is restored to cells depleted of cytoplasm and components of the nuclear transport pathway by addition of a cytoplasmic extract and an energy source. The extract contains pendulin, Srp1, importin-β, and other components necessary for NLS-driven nuclear import. Wild-type GFP–LEF-1 and GFP–TCF-1 coding sequences were fused in frame to the GST open reading frames, and the 70-kDa recombinant proteins were purified from bacteria. These purified preparations were added to digitonin-permeabilized HeLa cells in the presence of HeLa cytoplasmic extract and ATP, and nuclear import activity was monitored (Fig. 8). GST–GFP–LEF-1 efficiently localized to the nucleus whereas GST–GFP–TCF-1 did not localize to the nucleus at all but remained in the cytoplasm. Depletion of ATP was deleterious for GST–GFP–LEF-1 import, indicating that the observed nuclear import was energy dependent. The minor amount of GST–GFP–LEF-1 import in the absence of ATP is most likely due to incomplete depletion of endogenous ATP stores in the permeabilized cells. GST-GFP control protein was localized solely in the cytoplasm, indicating that the nuclei in these cells were intact and that nuclear import was a specific, NLS-dependent process. A large excess of purified GST-pendulin (0.8 μg, 180 nM) was added along with GST–GFP–TCF-1 to test whether weak pendulin interactions were enough to promote nuclear import. The bottom left panel of Fig. 8 shows that even a large excess of pendulin was unable to mediate any amount of TCF-1 nuclear import. These data provide strong evidence for differing nuclear transport mechanisms for LEF-1 and TCF-1.
FIG. 8.
LEF-1 but not TCF-1 can enter the nucleus in digitonin-permeabilized HeLa cells. Recombinant GST-GFP fusions of the LEF-1 and TCF-1 HMG DNA binding domains were purified and added to digitonin-permeabilized HeLa cells in the presence of a HeLa S100 cytosol extract with or without ATP and an energy regeneration system. Purified GST-pendulin protein (0.8 μg/180 nM) was added to the transport assay shown in the bottom right panel.
DISCUSSION
We have shown that FL pendulin and Srp1 bind to LEF-1 and not to TCF-1. At least for pendulin, the high affinity and discriminating specificity of this interaction require an intact armadillo repeat array because deletion into either the N- or C-terminal portion of the arm repeat region damages both aspects of pendulin binding. An exception is the N-terminal deletions of pendulin in the yeast two-hybrid assay, where these truncations maintain a preference for the LEF-1 NLS over the TCF-1 NLS. A major difference between the in vivo yeast assay and the in vitro GST assay is that in the yeast system, a heterologous transcription activation domain is fused onto the N-terminal end of the truncated arm repeat arrays. In the in vitro GST pull-down assay, the 35S-labeled N-terminal deletions have naked arm repeat ends. In both assays, any C-terminal truncation that leaves a naked arm repeat end destroys binding. One interpretation of the C-terminal truncation data might be that the NLS binding domain is located near the C-terminal end, an interpretation consistent with the model of Moroianu et al. (33). However, for reasons discussed below, we favor an alternative model. We propose, as have others, that the arm repeat region binds directly to NLSs. Furthermore, we propose that arm repeat arrays require anchoring domains to maintain a proper structure for NLS specificity.
The first crystal structure for an armadillo repeat region was solved recently by Huber and colleagues (20). Elegant structural analysis revealed that arm repeats are alpha helical and pack against one another to form an elongated superhelix of alpha helices. Neighboring arm repeats engage in extensive interactions giving rise to a protein core that is resistant to proteolysis and somewhat limited in flexibility. Thus, single arm repeats are unlikely to fold properly, and partial arm repeat arrays might be somewhat structurally distorted or denatured. In the yeast two-hybrid assay, the unanchored C-terminal truncations of pendulin accumulate to 10- to 15-fold-lower levels than the anchored N-terminal truncations, suggesting that these forms are not folded properly and are susceptible to degradation (38a).
In addition to the deletion mutants presented here, a set of excised arm repeat fragments has been constructed. Consistent with the loss of specificity observed with the N- and C-terminal deletions, portions of the pendulin arm repeat region bind weakly to both GST–LEF-1 and GST–TCF-1 but not to GST alone (38a). Arm repeats 4 to 8 retain a preference for the LEF-1 DNA binding domain, binding to TCF-1 at levels three- to fourfold lower. Although the overall levels of binding are much reduced, these arm repeat fragments bind preferentially to basic NLS-like sequences. We have found that even a 57-aa fragment of pendulin arm repeats 4 and 5 can transfer a preference for basic peptides when placed within the middle of the arm repeat array of beta-catenin (38a). These data may be consistent with the reported structure of the beta-catenin arm repeat array. In that structure, the alpha-helical superhelix creates a positively charged groove along the length of the arm repeat region (20). The authors propose that this basic groove is the site of interaction with beta-catenin substrates which are rich in acidic amino acids. Fragments of pendulin, while they are unlikely to be folded properly, might still assume a structure in such a way that a groove, or partially folded region rich in acidic side chains, retains a weak preference for basic amino acid sequences. The data presented in this report suggest that while the arm repeat array is the likely primary site of NLS recognition, perhaps through an arm repeat-formed groove, an intact arm repeat array flanked by anchoring domains is necessary for high-affinity and highly specific NLS binding.
What are the determinants of an NLS? Although there are two types of basic NLS sequence classes, single cluster and bipartite, it has not been possible to define a consensus NLS. Part of the reason may be that there is a family of importin/karyopherin-α subtypes, each of which may carry a set of distinct specificities. The experiments presented here approach the question of NLS specificity for the single subtype pendulin. Obviously, the presence of an arginine or serine in the TCF-1 B box is deleterious for pendulin recognition.
In addition to NLS specificity, the competition experiment shown in Fig. 2 suggests that context may play a role in NLS recognition. The 9-aa B box/NLS of LEF-1 competes with at least 10-fold-lower efficiency than the entire 88-aa LEF-1 HMG DNA binding domain. The structure of the LEF-1 HMG DNA binding domain alone and not complexed to DNA has not been determined. However, nuclear magnetic resonance analysis of several HMG boxes of highly divergent amino acid sequence shows that for each HMG box, three alpha helices fold back and pack against one another to form an L-shaped structure, a structure now considered to be a signature fold for HMG boxes (18, 53, 55). Therefore, this highly folded structure may play a role in promoting better NLS recognition. Such a role could be indirect, as in promoting a particular B box/NLS structure, or it could be direct by providing additional contacts for importin/karyopherin-α interaction. Preliminary evidence from a random mutagenesis screen in our laboratory shows that amino acid substitutions in the HMG box of TCF-1, a region far outside the B box, enables moderate levels of pendulin recognition. Nevertheless, the B-box exchange experiment demonstrates that the primary determinant for NLS recognition is the NLS itself (Fig. 3). The contexts provided by the LEF-1 and TCF-1 HMG boxes are virtually identical and not the primary factor in the differential NLS specificity described here.
We observe NLS specificity to be derived from the NLS sequence and the importin/karyopherin-α receptor. Importin-β, the 97-kDa coreceptor subunit for importin/karyopherin-α, does not appear to modulate importin-α specificity or affinity for LEF-1 NLS binding. Our conclusions differ from reports showing KAP60 and KAP95, the Saccharomyces cerevisiae homologs of importin-α and -β respectively, to exhibit enhanced NLS binding when present together as a complex (41). Significant differences between experimental systems, including the use of recombinant protein, the use of yeast homologs of the NLS receptor complex, and the use of a 12-aa NLS target rather than a larger highly folded domain such as the HMG DNA binding domain, may have contributed significantly to the contrasting observations.
More perplexing is our observation that deletion of the first 62 aa of pendulin causes greatly enhanced LEF-1 NLS binding in the GST pull-down assay. Weis et al. have constructed a similar deletion (to aa 66) of human pendulin and do not observe higher levels of binding to CBP80, a nuclear cap-binding protein that contains a bipartite NLS sequence (57). It is possible that the enhanced level of ΔN62 binding is specific for the LEF-1 NLS or similar targets. All deletion constructs were sequenced near the deletion endpoints to confirm that second-site mutations were not inadvertently created. That a second-site mutation far removed from the deletion endpoint is responsible for the increase in binding activity is possible but unlikely because ΔN62 coding sequences do not exhibit enhanced binding in the yeast two-hybrid assay. Although we are unable to explain the increased binding activity of ΔN62, analysis of the activity of this deletion mutant was useful in that it confirmed that importin-β binding does not play a role in NLS specific binding. Thus, differences in NLS specificity among different importin/karyopherin-α subtypes can be attributed to the unique armadillo repeat regions of each α subunit.
Pendulin has between 40 and 61% amino acid identity with all other known metazoan importin/karyopherin-α proteins, and it has 44% amino acid identity with the importin homolog Srp1 in S. cerevisiae (39, 51). Thus, pendulin is approximately as different from other subtypes as it is from the single yeast homolog. We and others have also shown that importin/karyopherins are widely but differentially expressed in mouse tissues to various levels. These overlapping patterns of expression coupled with the potential for distinct NLS specificities suggests that importin/karyopherin-α proteins do not merely subserve identical general housekeeping roles in nuclear import. Rather, each may impart a unique pattern of nuclear import activity. This subspecialization, combined with other coexpressed subtypes, would determine the overall pattern of nuclear import in cells.
How does TCF-1 reach the nucleus? Our in vivo nuclear transport assays demonstrate that the TCF-1 B box is an efficient NLS and able to target GFP to the nucleus as well as, if not better than, LEF-1 (Fig. 4). In contrast to these observations, our in vitro nuclear transport assays reveal that TCF-1 nuclear localization must differ from that of LEF-1. At least three formal possibilities require investigation. First, TCF-1 may be recognized by one of the other newly identified importin/karyopherins. If this is true, this alternative importin must be absent or inactive in the HeLa cytoplasmic extract used to reconstitute nuclear import in the digitonin-permeabilized cells. Second, the TCF-1 NLS may be modified in some way other than phosphorylation. This modification does not occur in reticulocyte lysates or in the import assay in digitonin-treated cells. Finally, the third possibility is that the TCF-1 NLS may direct import via a unique mechanism or via association with a heretofore unrecognized importin/karyopherin. There are at least three newly identified importins: importin-α3/α-Q1/Qip-1, importin-α4/α-Q2/karyopherin α3, and importin α6/α-S2. Importin α6/α-S2 is between 79 and 86% similar to mSrp1. Importin α-Q1/α3/Qip-1 and importin Q2/α4/karyopherin α3 are each 40 to 45% similar to Srp1 and pendulin and 85% identical to one another (24, 44, 48, 51). It is also possible that other importin-α receptors remain to be identified. In an attempt to identify one of these alternative importins as a receptor for the TCF-1 NLS, we have used the TCF-1 HMG DNA binding domain as bait in an extensive yeast two-hybrid screen. No importin receptor subtypes were identified. However, a negative result is not a definitive answer; it is possible that other subtypes were underrepresented in the yeast two-hybrid library or not inserted in frame in the two-hybrid vectors. Therefore it will be important to directly test these new importin receptors in the GST pull-down assay.
A difference in NLS receptor binding may have important functional consequences for LEF-1 and TCF-1. Both LEF-1 and TCF-1 are known to bind and cooperate with another armadillo repeat protein named beta-catenin to carry out Wnt/Wingless signal transduction into the nucleus. Identification of the pathway directing TCF-1 import will be an important step in determining whether different mechanisms of LEF-1 and TCF-1 nuclear transport promote different LEF-1, TCF-1, and beta-catenin function.
ACKNOWLEDGMENTS
We thank M. Nomura, H. Mangalam, and D. Guttridge for critical reading of the manuscript. We thank S. Adam for invaluable advice during the course of this work and for the importin-β reagents. We also acknowledge Karine Hovanes for valuable help and discussion with various aspects of this project.
This work was supported in part by grant CA 62079 from the NIH and in part by grant RPG-97-156-CSM from the American Cancer Society. M.L.W. is a member of the Developmental Biology Center at UCI.
REFERENCES
- 1.Adam E J H, Adam S A. Identification of cytosolic factors required for nuclear location sequence-mediated binding to the nuclear envelope. J Cell Biol. 1994;125:547–555. doi: 10.1083/jcb.125.3.547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1a.Adam, S. Personal communication.
- 2.Adam S A. The importance of importin. Trends Cell Biol. 1995;5:189–191. doi: 10.1016/s0962-8924(00)88991-6. [DOI] [PubMed] [Google Scholar]
- 3.Adam S A, Sterne-Marr R E, Gerace L. Nuclear protein import in permeabilized mammalian cells requires soluble cytoplasmic factors. J Cell Biol. 1990;111:807–816. doi: 10.1083/jcb.111.3.807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bagga R, Emerson B M. An HMG I/Y-containing repressor complex and supercoiled DNA topology are critical for long-range enhancer-dependent transcription in vitro. Genes Dev. 1997;11:629–639. doi: 10.1101/gad.11.5.629. [DOI] [PubMed] [Google Scholar]
- 5.Bruhn L, Munnerly A, Grosschedl R. ALY, a context-dependent coactivator of LEF-1 and AML-1, is required for TCR alpha enhancer function. Genes Dev. 1997;11:640–653. doi: 10.1101/gad.11.5.640. [DOI] [PubMed] [Google Scholar]
- 6.Carlsson P, Waterman M, Jones K. The hLEF/TCF-1α HMG protein contains a context-dependent transcriptional activation domain that induces the TCRα enhancer in T cells. Genes Dev. 1993;7:2418–2430. doi: 10.1101/gad.7.12a.2418. [DOI] [PubMed] [Google Scholar]
- 7.Chi N C, Adam E J H, Adam S A. Sequence and characterization of cytoplasmic nuclear protein import factor p97. J Cell Biol. 1995;130:265–274. doi: 10.1083/jcb.130.2.265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cortes P, Ye Z S, Baltimore D. Rag-1 interacts with the repeated amino acid motif of the human homologue to the yeast protein SRP1p. Proc Natl Acad Sci USA. 1994;91:7633–7637. doi: 10.1073/pnas.91.16.7633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cuomo C A, Irch S A, Gyuris J, Oettinger M A. Rch1, a protein that specifically interacts with the RAG-1 recombination activating protein. Proc Natl Acad Sci USA. 1994;91:6156–6160. doi: 10.1073/pnas.91.13.6156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Franke W W, Goldschmidt M D, Zimblemann R, Mueller H M, Schiller D L, Cowein P. Molecular cloning and amino acid sequence of human plakoglobin, the common junctional plaque protein. Proc Natl Acad Sci USA. 1989;86:4027–4031. doi: 10.1073/pnas.86.11.4027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Giese K, Cox J, Grosschedl R. The HMG domain of lymphoid enhancer factor 1 bends DNA and facilitates assembly of functional nucleoprotein structures. Cell. 1992;69:185–196. doi: 10.1016/0092-8674(92)90129-z. [DOI] [PubMed] [Google Scholar]
- 12.Giese K, Kingsley C, Kirshner J R, Grosschedl R. Assembly and function of a TCR alpha enhancer complex is dependent on LEF-1-induced DNA bending and multiple protein-protein interactions. Genes Dev. 1995;9:995–1008. doi: 10.1101/gad.9.8.995. [DOI] [PubMed] [Google Scholar]
- 13.Gorlich D, Henklein P, Laskey R A, Hartmann E. A 41 amino acid motif in importin-α confers binding to importin-β and hence transit into the nucleus. EMBO J. 1996;15:1810–1817. [PMC free article] [PubMed] [Google Scholar]
- 14.Gorlich D, Kostak S, Kraft R, Dingwall C, Laskey R A, Hartmann E, Prehn S. Two different subunits of importin cooperate to recognize nuclear localization signals and bind them to the nuclear envelope. Curr Biol. 1995;5:383–392. doi: 10.1016/s0960-9822(95)00079-0. [DOI] [PubMed] [Google Scholar]
- 15.Gorlich D, Mattaj I W. Nucleocytoplasmic transport. Science. 1996;271:1513–1518. doi: 10.1126/science.271.5255.1513. [DOI] [PubMed] [Google Scholar]
- 16.Gorlich D, Prehn S, Laskey R A, Hartmann E. Isolation of a protein that is essential for the first step of nuclear protein import. Cell. 1994;79:767–778. doi: 10.1016/0092-8674(94)90067-1. [DOI] [PubMed] [Google Scholar]
- 17.Groden J, Thilveris A, Samowitz W, Carlson M, Gelbart L, Albertsen H, Joslyn G, Stevens J, Spirio L, Robertson M, Sargeant L, Krapcho K, Wolff E, Burt R, Hughes J P, Warrington J, McPherson J, Wasmuth J, LePlaslier D, Abderrahim H, Cohen D, Leppert M. Identification and characterization of the familial adenomatous polyposis coli gene. Cell. 1991;66:589–600. doi: 10.1016/0092-8674(81)90021-0. [DOI] [PubMed] [Google Scholar]
- 18.Grosschedl R, Giese K, Pagel J. HMG domain proteins: architectural elements in the assembly of nucleoprotein structures. Trends Genet. 1994;10:94–100. doi: 10.1016/0168-9525(94)90232-1. [DOI] [PubMed] [Google Scholar]
- 19.Guttridge, K. Unpublished data.
- 20.Huber A H, Nelson W J, Weis W I. Three-dimensional structure of the armadillo repeat region of β-catenin. Cell. 1997;90:871–882. doi: 10.1016/s0092-8674(00)80352-9. [DOI] [PubMed] [Google Scholar]
- 21.Imamoto N, Shimamoto T, Takao T, Tachibana T, Kose S, Matsubae M, Sekimoto T, Shimonishi Y, Yoneda Y. In vivo evidence for involvement of a 58 kDa component of nuclear pore-targeting complex in nuclear protein import. EMBO J. 1995;14:3617–3626. doi: 10.1002/j.1460-2075.1995.tb00031.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kikuchi A, Kaibuchi K, Hori Y, Nonaka H, Sakoda T, Kawamura M, Mizuno T, Takai T. Molecular cloning of the human cDNA for a stimulatory GDP/GTP exchange protein for cKi-ras p21 and smg p21. Oncogene. 1992;7:289–293. [PubMed] [Google Scholar]
- 23.Kinzler K W, Nilbert N C, Su L K, Vogelstein B, Bryan T M, Levy D B, Smith K J, Presiinger A C, Hedge P, McKinechnie D, Finniear R, Markham A, Groeffen J, Boguski M S, Altschul S F, Horii A, Miyoshi Y, Miki Y, Nishisho I, Nakamura Y. Identification of the FAP locus genes from chromosome 5q21. Science. 1991;253:661–665. doi: 10.1126/science.1651562. [DOI] [PubMed] [Google Scholar]
- 24.Kohler M, Ansiequ S, Prehn S, Leutz A, Haller H, Hartmann E. Cloning of two novel human importin-α subunits and analysis of the expression pattern of the importin-α protein family. FEBS Lett. 1997;417:104–108. doi: 10.1016/s0014-5793(97)01265-9. [DOI] [PubMed] [Google Scholar]
- 25.Kussel P, Frasch M. Pendulin, a drosophila protein with cell cycle-dependent nuclear localization, is required for normal cell proliferation. J Cell Biol. 1995;129:1491–1507. doi: 10.1083/jcb.129.6.1491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kutay U, Bischoff F R, Kostka S, Kraft R, Gorlich D. Export of importin α from the nucleus if mediated by a specific nuclear transport factor. Cell. 1997;90:1061–1071. doi: 10.1016/s0092-8674(00)80372-4. [DOI] [PubMed] [Google Scholar]
- 27.LaCasse E C, Lefebvre Y A. Nuclear localization signals overlap DNA- or RNA-binding domains in nucleic acid-binding proteins. Nucleic Acids Res. 1995;23:1647–1656. doi: 10.1093/nar/23.10.1647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lai J-S, Herr W. Ethidium bromide provides a simple tool for identifying genuine DNA-independent protein associations. Proc Natl Acad Sci USA. 1992;89:6958–6962. doi: 10.1073/pnas.89.15.6958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Love J J, Li X, Case D A, Giese K, Grosschedl R, Wright P E. Structural basis for DNA bending by the architectural transcription factor LEF-1. Nature. 1995;376:791–795. doi: 10.1038/376791a0. [DOI] [PubMed] [Google Scholar]
- 29a.Malik H S, Eickbush T H, Goldfarb D S. Evolutionary specialization of the nuclear targeting apparatus. Proc Natl Acad Sci USA. 1997;94:13738–13742. doi: 10.1073/pnas.94.25.13738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mayall T P, Sheridan P L, Montminy M R, Jones K A. Distinct roles for P-CREB and LEF-1 in TCR alpha enhancer assembly and activation on chromatin templates in vitro. Genes Dev. 1997;11:887–899. doi: 10.1101/gad.11.7.887. [DOI] [PubMed] [Google Scholar]
- 31.McCrea P D, Turck C W, Gumbiner B. A homolog of the armadillo protein in Drosophila (plakoglobin) associated with E-cadherin. Science. 1991;254:1359–1361. doi: 10.1126/science.1962194. [DOI] [PubMed] [Google Scholar]
- 32.Melchior F, Gerace L. Mechanisms of nuclear protein import. Curr Biol. 1995;7:310–318. doi: 10.1016/0955-0674(95)80084-0. [DOI] [PubMed] [Google Scholar]
- 33.Moroianu J, Blobel G, Radu A. The binding site of karyopherin α for karyopherin β overlaps with a nuclear localization sequence. Proc Natl Acad Sci USA. 1996;93:6572–6576. doi: 10.1073/pnas.93.13.6572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Moroianu J, Blobel G, Radu A. Previously identified protein of uncertain function is karyopherin α together with karyopherin β docks import substrate at nuclear pore complexes. Proc Natl Acad Sci USA. 1995;92:2008–2011. doi: 10.1073/pnas.92.6.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nigg E. Nucleocytoplasmic transport: signals, mechanisms and regulation. Nature. 1997;386:779–787. doi: 10.1038/386779a0. [DOI] [PubMed] [Google Scholar]
- 36.O’Neill R E, Palese P. NPI-1, the human homolog of SRP-1, interacts with influenza virus nucleoprotein. Virology. 1995;206:116–125. doi: 10.1016/s0042-6822(95)80026-3. [DOI] [PubMed] [Google Scholar]
- 37.Peifer M, Berg S, Reynolds A B. A repeating amino acid motif shared by proteins with diverse cellular roles. Cell. 1994;76:789–791. doi: 10.1016/0092-8674(94)90353-0. [DOI] [PubMed] [Google Scholar]
- 38.Peterson J M, Skalicky J J, Donaldson L W, McIntosh L P, Alber T, Graves B J. Modulation of transcription factor ets-1 DNA binding: DNA-induced unfolding of an α helix. Science. 1995;269:1866–1869. doi: 10.1126/science.7569926. [DOI] [PubMed] [Google Scholar]
- 38a.Prieve, M. Unpublished data.
- 39.Prieve M, Guttridge K L, Munguia J E, Waterman M. The nuclear localization signal of lymphoid enhancer factor-1 is recognized by two differentially expressed Srp1-NLS receptor proteins. J Biol Chem. 1996;271:7654–7658. doi: 10.1074/jbc.271.13.7654. [DOI] [PubMed] [Google Scholar]
- 40.Radu A, Blobel G, Moore M S. Identification of a protein complex that is required for nuclear protein import and mediates docking of import substrate to distinct nucleoporins. Proc Natl Acad Sci USA. 1995;92:1769–1773. doi: 10.1073/pnas.92.5.1769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rexach M, Blobel G. Protein import into nuclei: association and dissociation reactions involving transport substrate, transport factors, and nucleoporins. Cell. 1995;83:683–692. doi: 10.1016/0092-8674(95)90181-7. [DOI] [PubMed] [Google Scholar]
- 42.Reynolds A B, Daniel J, McCrea P D, Wheelock M J, Wu J, Zhang Z. p120cas associates with E-cadherin complexes. Mol Cell Biol. 1992;14:8333–8342. doi: 10.1128/mcb.14.12.8333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Riggleman B, Schedl P, Wieschaus E. Spatial expression of the Drosophila segment polarity gene armadillo is post-transcriptionally regulated by wingless. Cell. 1990;63:549–560. doi: 10.1016/0092-8674(90)90451-j. [DOI] [PubMed] [Google Scholar]
- 44.Seki T, Tada S, Katada T, Enomoto T. Cloning of a cDNA encoding a novel importin-alpha homologue, Qip1: discrimination of Qip1 and Rch1 from hSrp1 by their ability to interact with DNA helicase Q1/RecQL. Biochem Biophys Res Commun. 1997;234:48–53. doi: 10.1006/bbrc.1997.6535. [DOI] [PubMed] [Google Scholar]
- 45.Sheridan P L, Sheline C T, Cannon K, Voz M L, Pazin M J, Kadonaga J T, Jones K A. Activation of the HIV-1 enhancer by the LEF-1 HMG protein on nucleosome-assembled DNA in vitro. Genes Dev. 1995;9:2090–2104. doi: 10.1101/gad.9.17.2090. [DOI] [PubMed] [Google Scholar]
- 46.Siomi H, Dreyfuss G. A nuclear localization domain in the hnRNP A1 protein. J Cell Biol. 1995;129:551–560. doi: 10.1083/jcb.129.3.551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Steffan J S, Keys D A, Dodd J A, Nomura M. The role of TBP in rDNA transcription by RNA polymerase I in Saccharomyces cerevisiae: TBP is required for upstream activation factor-dependent recruitment of core factor. Genes Dev. 1996;109:2551–2563. doi: 10.1101/gad.10.20.2551. [DOI] [PubMed] [Google Scholar]
- 48.Takeda S, Fujiwara T, Shimizu F, Kawai A, Shinomiya K, Okuno S, Ozaki K, Katagiri T, Shimada Y, Nagata M, Watanabe T, Takaichi A, Kuga Y, Suzuki M, Hishigaki H, Takahashi E, Shin S, Nakamura Y, Kirai Y. Isolation and mapping of karyopherin a3 (KPNA3), a human gene that is highly homologous to genes encoding Xenopus importin, yeast SRP1 and human RCH1. Cytogenet Cell Genet. 1997;76:87–93. doi: 10.1159/000134521. [DOI] [PubMed] [Google Scholar]
- 49.Torok I, Strand D, Schmitt R, Tick G, Torok T, Kiss I, Mechler B M. The overgrown hematopoietic organs-31 tumor suppressor gene of drosophila encodes an importin-like protein accumulating in the nucleus at the onset of mitosis. J Cell Biol. 1995;129:1473–1489. doi: 10.1083/jcb.129.6.1473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Travis A, Amsterdam A, Belanger C, Grosschedl R. LEF-1, a gene encoding a lymphoid-specific protein with an HMG domain, regulates T-cell receptor alpha enhancer function. Genes Dev. 1991;5:880–894. doi: 10.1101/gad.5.5.880. [DOI] [PubMed] [Google Scholar]
- 51.Tsuji L, Takumi T, Imamoto N, Yoneda Y. Identification of novel homologues of mouse importin α, the α subunit of the nuclear pore-targeting complex, and their tissue-specific expression. FEBS Lett. 1997;416:30–34. doi: 10.1016/s0014-5793(97)01092-2. [DOI] [PubMed] [Google Scholar]
- 52.van de Wetering M, Oosterwegel M, Dooijes D, Clevers H. Identification and cloning of TCF-1, a T lymphocyte specific transcription factor containing a sequence-specific HMG box. EMBO J. 1991;10:123–132. doi: 10.1002/j.1460-2075.1991.tb07928.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.van Houte L, van Oers A, van de Wetering M, Dooijes D, Kaptein R, Clevers H. The sequence-specific high mobility group 1 box of TCF-1 adopts a predominantly alpha-helical conformation in solution. J Biol Chem. 1993;268:18083–18087. [PubMed] [Google Scholar]
- 54.Waterman M L, Fischer W H, Jones K A. A thymus-specific member of the HMG protein family regulates the human T cell receptor C alpha enhancer. Genes Dev. 1991;5:656–669. doi: 10.1101/gad.5.4.656. [DOI] [PubMed] [Google Scholar]
- 55.Weir H, Kraulis P J, Hill C S, Raine A R C, Laue E D, Thomas J O. Structure of the HMG box motif in the B-domain of HMG1. EMBO J. 1993;12:1311–1319. doi: 10.1002/j.1460-2075.1993.tb05776.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Weis K, Mattaj I W, Lamond A I. Identification of hSRP1α as a functional receptor for nuclear localization sequences. Science. 1995;268:1049–1053. doi: 10.1126/science.7754385. [DOI] [PubMed] [Google Scholar]
- 57.Weis K, Ryder U, Lamond A I. The conserved amino-terminal domain of hSRP1α is essential for nuclear protein import. EMBO J. 1996;15:1818–1825. [PMC free article] [PubMed] [Google Scholar]
- 58.Yano R, Oakes M, Yamaghishi M, Dodd J A, Nomura M. Cloning and characterization of SRP1, a suppressor of temperature-sensitive RNA polymerase I mutations, in Saccharomyces cerevisiae. Mol Cell Biol. 1992;12:5640–5651. doi: 10.1128/mcb.12.12.5640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yano R, Oakes M L, Tabb M M, Nomura M. Yeast Srp1p has homology to armadillo/plakoglobin/β-catenin and participates in apparently multiple nuclear functions including the maintenance of the nucleolar structure. Proc Natl Acad Sci USA. 1994;91:6880–6884. doi: 10.1073/pnas.91.15.6880. [DOI] [PMC free article] [PubMed] [Google Scholar]