Abstract
DRK, the Drosophila homolog of the SH2-SH3 domain adaptor protein Grb2, is required during signaling by the sevenless receptor tyrosine kinase (SEV). One role of DRK is to provide a link between activated SEV and the Ras1 activator SOS. We have investigated the possibility that DRK performs other functions by identifying additional DRK-binding proteins. We show that the phosphotyrosine-binding (PTB) domain-containing protein Disabled (DAB) binds to the DRK SH3 domains. DAB is expressed in the ommatidial clusters, and loss of DAB function disrupts ommatidial development. Moreover, reduction of DAB function attenuates signaling by a constitutively activated SEV. Our biochemical analysis suggests that DAB binds SEV directly via its PTB domain, becomes tyrosine phosphorylated upon SEV activation, and then serves as an adaptor protein for SH2 domain-containing proteins. Taken together, these results indicate that DAB is a novel component of the SEV signaling pathway.
Receptor tyrosine kinases (RTKs) play essential roles in the control of cell growth, specification of cell fate, and pattern formation during the development of multicellular organisms. Binding of a ligand to the extracellular domain of a RTK induces receptor dimerization, activation of the cytoplasmic kinase domain, and autophosphorylation on tyrosine residues (50). The activated receptor then initiates intracellular signaling in part by recruiting a number of cytoplasmic proteins (12). Although these proteins have diverse biochemical activities and biological functions, they often contain SRC homology 2 (SH2) or phosphotyrosine-binding (PTB) domains that can bind specific phosphotyrosine residues on the activated RTK. One important consequence of RTK stimulation is the activation of the Ras/mitogen-activated protein kinase (MAPK) pathway (19, 41, 42). For many RTKs, Ras activation is dependent on the binding of Grb2 directly to the tyrosine-phosphorylated RTK. The central SH2 domain of Grb2 is flanked by two SRC homology 3 (SH3) domains that interact with proline-rich sequences of the guanine nucleotide exchange factor SOS. Binding of the Grb2-SOS complex to an RTK brings SOS close to Ras and allows SOS to catalyze Ras activation. The activated GTP-bound Ras then stimulates a conserved protein kinase cascade consisting of Raf, MAPK kinase (MEK), and MAPK (3, 41). Once activated, MAPK modulates the activity of various transcription factors that control cellular responses to extracellular signals.
An extensively studied example of an RTK-mediated signaling process occurs during R7 photoreceptor cell specification in the Drosophila compound eye (26, 52, 64). The decision of the R7 precursor cell to adopt the photoreceptor cell fate is dependent on the stimulation of the sevenless RTK (SEV) by its ligand, the Bride of Sevenless protein, which is expressed on the surface of the adjacent R8 cell (39). By using sensitized genetic screens and epistasis experiments, numerous proteins that participate in SEV signal transduction have been identified. These include components of the Ras/MAPK pathway such as DRK (the Drosophila homolog of Grb2) (44, 55), SOS (8, 53), Ras1 (53), Raf (16), MEK (33, 60), and MAPK (6, 11).
The ability of activated Ras1 to bypass the requirement for SEV function during R7 development has suggested that the primary function of SEV is to activate Ras (20). However, the model that the sole function of activated SEV is to bind DRK-SOS has been questioned by genetic studies that suggest the existence of multiple intracellular signaling pathways downstream of SEV (1, 28, 45). For example, although the association of DRK and SOS does not depend on the carboxy (C)-terminal SH3 domain of DRK, mutations that affect this domain partially compromise SEV signaling. Furthermore, a C-terminal SH3 domain-truncated DRK cannot rescue the lethality associated with homozygous drk mutations (45). These data suggest that DRK-binding proteins besides SOS may play important roles in signaling by SEV and other RTKs. Biochemical studies performed with mammalian systems have provided evidence that such Grb2-binding partners do exist (21, 30, 38, 43). These include Cbl, a proto-oncogene product, and GAB1, a downstream component of the insulin and epidermal growth factor receptors.
Here we report that the PTB domain-containing protein Disabled (DAB) binds to the SH3 domains of DRK. DAB and DRK form a complex in vivo that appears to be distinct from the DRK-SOS complex. We demonstrate that DAB can bind SEV directly via its PTB domain. Moreover, the expression of activated SEV leads to increased DAB tyrosine phosphorylation that may provide docking sites for SH2 domain-containing proteins. We provide evidence that DAB is an important component of the SEV signaling pathway in vivo by showing that DAB is expressed in developing photoreceptor cells, that the removal of DAB function disrupts ommatidial development, and that either a reduction of DAB dosage or the expression of a PTB domain-mutated DAB suppresses the effects of excessive SEV signaling. Interestingly, a truncated DAB that lacks the DRK SH3 domain-binding region shows enhanced function during SEV signaling. This finding raises the possibility that either inhibition of DAB function by this region is alleviated upon DRK binding to activated SEV or DRK binding to this region serves normally to inhibit DAB function.
MATERIALS AND METHODS
Genetics.
Fly culture, crosses, and germ line transformation were performed by using standard procedures. The dab mutant flies were generously provided by S. M. Ahern and F. M. Hoffmann (University of Wisconsin, Madison). Marked clones of cells homozygous for either dab100, dab221, or dabM54 mutation were induced by X-ray irradiation of w, Tn-abl, abl1dab/+ first-instar larvae as previously described (53).
Histology.
Fixation and sectioning (1 to 2 μm) of adult eyes was performed essentially as described previously (59). Scanning electron microscopy (SEM) was performed as described elsewhere (37). Antibody staining of eye imaginal discs was essentially as described previously (22). The primary antibodies were rabbit affinity-purified anti-DAB antibodies (gift of A. R. Comer and F. M. Hoffmann) and mouse anti-ELAV monoclonal antibody. The primary antibodies were detected with either fluorescein isothiocyanate- or Cy5-conjugated secondary antibodies (Jackson Immunoresearch) that were diluted 1/200. The discs were then washed, mounted in Citifluor (Ted Pella, Inc.), and observed with an MRC 1024 confocal laser microscope (Bio-Rad).
DNA sequencing.
Double-stranded DNA sequencing was performed with a Sequenase 2.0 kit (U.S. Biochemical) with the protocol provided by the manufacturer.
Generation of 32P-labeled wild-type and mutant GTK-DRK fusion proteins.
Mutations in drk were generated by site-directed mutagenesis using reagents and protocols provided in the Mutagene kit (Bio-Rad). To construct the template for making the drk mutations, the entire coding sequence of the drk cDNA (∼750 bp) was released from the plasmid pGEX-drk (55) by BamHI and EcoRI digestion and subcloned into pBluescript (pBS) KS+, creating the plasmid pBS-drk. The mutagenic primers were 5′-GCCTCATACTTAGTAATTA-3′, 5′-CCTTCTTGATAAAAATCAGCGAATC-3′, and 5′-AATAGGAAACGCATCTTCC-3′. The presence of the point mutations was verified by sequencing using forward and reverse primers (Pharmacia). The resulting plasmids were designated pBS-drkP49L, pBS-drkR85K, and pBS-drkG199R, respectively. The wild-type and mutant drk cDNAs were released from pBS and subcloned into the pGEX-2TK vector (Pharmacia) at BamHI and EcoRI sites. The double-mutant construct was created by ligating the BamHI-BglI fragment (∼350 bp) of pBS-drkP49L and the BglI-EcoRI fragment of pBS-drkG199R (∼400 bp) into pGEX-2TK. The final plasmids were named pGTK-drkP49L, pGTK-drkR85K, pGTK-drkG199R, and pGTK-drkP49L/G199R.
Glutathione S-transferase (GST) fusion proteins were expressed in Escherichia coli and purified by glutathione affinity chromatography as described previously (57). The fusion proteins that contain a protein kinase A (PKA) phosphorylation site in the linker region between GST and DRK were named GTK-DRK. They were labeled with 32P by in vitro kinase reaction using PKA (Pharmacia) and [γ-32P]ATP (7,000 Ci/mmol; ICN Biomedicals, Inc.) according to the protocol provided by the manufacturer (Pharmacia). Fusions of this protein with DRK were termed GTK-DRK fusions.
Expression library screening.
A Drosophila 0- to 18-h embryonic λgt11 cDNA library was used (gift of A. Dingwall and M. Scott). Expression library plaque lifts were performed as described previously (56). Nitrocellulose filters were processed through a denaturation-renaturation cycle, blocked, and then hybridized to [32P]GTK-DRK probes as described previously (7). The filters were washed, dried, covered with Saran Wrap, and exposed to film at −70°C with an intensifying screen.
Construction of dabS119A mutant plasmid and fusion proteins.
The EcoRI-EcoRI fragment of dab cDNA was released from pPAC-dab (kindly provided by A. R. Comer and F. M. Hoffmann) and subcloned into pBS. The EcoRI-XhoI fragment of pBS-RI-dab-RI was subcloned into pBS. Site-directed mutagenesis using the primer 5′-GTGCATAAGATCGCCTTCATCGCG-3′ was performed to generate pBS-RI-dabS119A-XhoI. The EcoRI-XhoI fragment of pBS-RI-dab-RI was then replaced with EcoRI-dabS119A-XhoI.
The DNA coding sequence for the PTB domain of DAB was generated by PCR using wild-type dab cDNA as the template and the primers 5′-CGCGGATCCATGGTCAAGTCCCTGGTC-3′ and 5′-GCGAATTCCCGCCATCTCAATCTCCTT-3′. The PCR product was subcloned into pZero (Invitrogen) at the EcoRV site. It was then released from pZero by BamHI-EcoRI digestion and subcloned into pGEX-KG (25) to give plasmid pGEX-dabPTB.
A similar procedure was used to generate plasmid pGEX-dabPTBS119A except that the EcoRI-dabS119A-EcoRI fragment replaced wild-type dab cDNA as the template in the PCR experiment.
P-element transformation.
dab cDNA coding sequence which does not contain the alternatively spliced exon was released from pPAC-dab by NotI digestion, filled in with Klenow enzyme, and gel eluted. This fragment was then blunt-end ligated into the transformation vector pKB267 (5) which had been cut with KpnI plus EcoRI and filled in with Klenow enzyme. The direction of insert was verified by restriction enzyme digestion mapping. The resulting plasmid was called P[sE-dab].
To delete the proline-rich region coding fragment from P[sE-dab], the EcoRI-EcoRI fragment of P[sE-dab] was subcloned into a modified pBS vector in which the EcoRV site had been eliminated. The plasmid was then digested to completion with HpaI plus EcoRV and religated to generate pBS-RI-dabΔPro-RI. The EcoRI-EcoRI fragment of P[sE-dab] was subsequently replaced with the EcoRI-dabΔPro-EcoRI fragment to give P[sE-dabΔPro].
P[sE-dabS119A] was generated by replacing the EcoRI-EcoRI fragment of P[sE-dab] with that of pBS-RI-dabS119A-RI. The direction of insert and the presence of the point mutation were verified by restriction enzyme digestion mapping.
Production of rat anti-DAB antibodies.
A C-terminal peptide of DAB with the sequence CSNDFSDREKREQFE was made (Research Genetics) and coupled to keyhole limpet hemocyanin according to the manufacturer’s protocol. It was then injected into rats to raise polyclonal sera against DAB (BAbCO).
Pull-down and overlay experiments.
For pull-down experiments, the cells were lysed (5 × 107 cells per ml) in lysis buffer (50 mM HEPES [pH 7.5], 150 mM NaCl, 1 mM EDTA, 1% Triton X-100, 10% glycerol, 1 mM phenylmethylsulfonyl fluoride, 1 μg of aprotinin per ml, 1 μg of leupeptin per ml) with or without phosphatase inhibitors, and the extracts were cleared three times by centrifugation at 12,000 × g for 10 min; 200 μl of cell extract was then added to approximately 2 μg of fusion protein coupled to glutathione-agarose beads and incubated at 4°C for 4 h on a rotator. The beads were washed five times with washing buffer (lysis buffer with only 0.1% Triton X-100), resuspended in Laemmli buffer, and separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) on 9% gels.
For overlay experiments, the cells were lysed as described above, separated by SDS-PAGE on 5 to 12% gradient gels, and transferred onto nitrocellulose filters. The filters were then overlaid with fusion proteins at the concentration of 0.5 μg/ml and incubated at 4°C overnight (10 to 14 h). After the filters were washed extensively with TBS-T buffer, binding of fusion proteins to proteins on the filters was detected by immunoblotting as described below.
Immunoprecipitations and immunoblot analyses.
Immunoblotting and immunoprecipitation were performed as described previously (29). For Western blot analyses, the proteins were separated by SDS-PAGE on 9% gels except for anti-DAB immunoblotting, in which case the proteins were separated on 5 to 12% gradient gels. The following antibodies were used: rabbit polyclonal anti-DAB antibodies raised against a GST fusion protein containing amino acids 1600 to 2022 of DAB (24); rat anti-DAB, produced as described above; hemagglutinin (HA)-specific monoclonal antibody raised against the 12-amino-acid HA epitope CYPYDVPDYASL (BAbCO); rabbit anti-DRK antisera raised against a GST fusion protein containing the SH2 and C-terminal SH3 domains of DRK; monoclonal antibody 78C10, which recognizes the catalytic domain of SEV, used in anti-SEV immunoblotting; monoclonal antibody G11, raised against the C-terminal 13 amino acids of SEV, used in anti-SEV immunoprecipitation; and antiphosphotyrosine monoclonal antibody PY20 (Transduction Laboratories).
Tissue culture.
Drosophila Schneider cell line 2 (SL2) was maintained and transfected as previously described (54). Selection of polyclonal cell lines was in G418 (1 mg/ml) or hygromycin B (0.2 mg/ml). For heat shock induction, cells were treated for 30 min at 37°C and then returned to 23°C. The SEVS11 constructs were described previously (55).
The HA tag which contains the 10 amino acids YPYDVPDYAS was added to wild-type and mutant drk cDNA coding sequences by PCR using the primers 5′-GTCGGTACCATGGAAGCGATTGCC-3′ and 5′-CAGGGTACCTTAACTGGCGTAGTCGGGGACGTCGTAAGGATAGTTAACTGAATCATATG GCGT-3′. The resulting PCR products were subcloned into the pTA vector at the KpnI site, and the presence of the HA tag was verified by sequencing. The drk-HA fragments were then subcloned as KpnI fragments into the expression vector pAT-Hygro (1) for transfection into SL2 cells.
dab cDNA was released from pPAC-dab with NotI digestion, filled in with T4 polymerase, and blunt-end ligated into pAT-Hygro which had been cut with KpnI and filled with T4 polymerase. The resulting plasmid was named pAT-dab-Hygro. Plasmid pAT-dabΔPro-Hygro was generated by replacing the XhoI-XhoI fragment of pAT-dab-Hygro with the corresponding fragment from P[sE-dabΔPro].
RESULTS
DAB is a DRK-binding protein.
To identify Drosophila proteins that are capable of interacting with DRK, we produced DRK as a GTK-DRK fusion protein containing a PKA phosphorylation site in the linker region between GST and DRK (34). The fusion protein was expressed in bacteria, purified, 32P-labeled by using PKA and [γ-32P]ATP, and used to screen a Drosophila embryonic λgt11 cDNA expression library. From 500,000 plaques screened, 28 positive clones were identified, plaque purified, and subjected to further analysis. We found that two of the cDNA inserts were derived from Sos. In agreement with previous reports (45), their products bound with high affinity to the amino (N)-terminal SH3 domain of DRK (data not shown).
Sequence analysis of one insert indicated that it was derived from the previously described gene called disabled (dab) (24). The Drosophila dab gene encodes two alternatively spliced products of 220 and 250 kDa. DAB has three notable characteristics: (i) it has an N-terminally located PTB domain that may allow it to interact with tyrosine-phosphorylated proteins, (ii) it has a centrally located proline-rich region that contains several potential SH3 domain-binding motifs (PXXP) (47), and (iii) it is tyrosine phosphorylated at low levels when expressed in Drosophila tissue culture (SL2) cells and contains potential consensus binding sites for the SH2 domains of DRK, Src, Abl, and Nck (58).
The SH3 domains of DRK bind to the proline-rich region of DAB.
To characterize the nature of the in vitro DAB-DRK interaction, we determined which domains of DRK are required for binding to DAB. To answer this question, we made use of well-characterized mutations that had been shown to inactivate the function of either the SH2 or SH3 domain of Grb2 (13, 15, 18, 49). For example, changing the proline 49 residue to leucine (P49L) inactivates the N-terminal SH3 domain, while the arginine 86-to-lysine (R86K) mutation disrupts the SH2 domain and the glycine 203-to-arginine (G203R) mutation affects the C-terminal SH3 domain. We introduced the corresponding mutations, individually or in combination (P49L, R85K, G199R, P49L/G199R), into the [32P]GTK-DRK fusion protein and tested the ability of the mutant proteins to interact with the λgt11-encoded β-galactosidase–DAB fusion protein. As shown in Fig. 1A, mutation of the SH2 domain did not affect binding, indicating that the in vitro DAB-DRK interaction does not require a functional DRK SH2 domain. The DAB-DRK interaction, however, is dependent on the function of the SH3 domains because simultaneous mutations of both SH3 domains abolished binding. Moreover, while DAB binds to both SH3 domains, it appears to interact more strongly with the C-terminal domain.
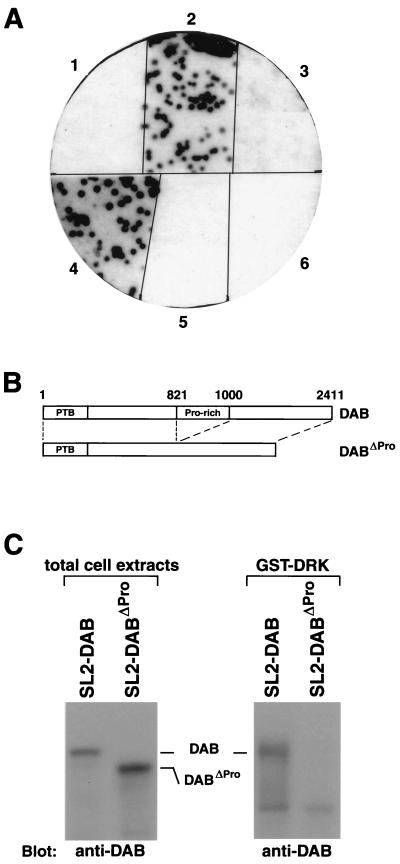
FIG. 1.
The in vitro interaction between DRK and DAB requires the SH3 domains of DRK and the proline-rich region of DAB. (A) Binding of [32P]GTK-DRK probes to λgt11-encoded β-galactosidase–DAB fusion protein. Plating of λgt11 clones, induction of β-galactosidase fusion protein expression, and protein transfer onto nitrocellulose filters were performed as described in Materials and Methods. Filters were hybridized in the presence of [32P]GTK, [32P]GTK-DRK, [32P]GTK-DRKP49L (N-terminal SH3 domain inactivated), [32P]GTK-DRKR85K (SH2 domain inactivated), [32P]GTK-DRKG199R (C-terminal SH3 domain inactivated), and [32P]GTK-DRKP49L/G199R (both SH3 domains inactivated), (sectors 1 to 6, respectively). Binding of GTK-DRK to β-galactosidase–DAB is dependent on the function of the DRK SH3 domains. (B) Schematic diagrams of DAB and DABΔPro. The numbers indicate amino acid residues. The region from asparagine 821 to aspartate 1000 is deleted in DABΔPro. Pro-rich, proline-rich region. (C) Interaction of GST-DRK with DAB and DABΔPro. Extracts from SL2-DAB and SL2-DABΔPro cells were incubated with GST-DRK fusion protein coupled to glutathione-agarose beads. The immunoblots both of total cell extracts (left) and of GST-DRK precipitates (right) were probed with rabbit anti-DAB polyclonal antibodies. Wild-type DAB and DABΔPro were expressed in SL2 cells under the control of the Actin5C promoter. The cell line used to prepare the extract is indicated above each lane. The positions of DAB and DABΔPro are indicated. Although DAB and DABΔPro were expressed at approximately equivalent levels, only wild-type DAB was precipitated by GST-DRK fusion protein.
The dependence of in vitro DAB-DRK binding on the function of the DRK SH3 domains suggests that the proline-rich region of DAB might be required for the interaction. To test this possibility, we expressed either wild-type DAB or a DAB protein lacking the proline-rich region (DABΔPro) in SL2 cells (Fig. 1B). Extracts from the resulting cell lines, SL2-DAB and SL2-DABΔPro, were incubated with GST-DRK fusion protein coupled to glutathione-agarose beads. Binding of full-length or truncated DAB to GST-DRK was monitored by immunoblotting with anti-DAB antibodies that recognize both forms of the protein. Full-length DAB, but not DABΔPro, was precipitated by the GST-DRK fusion protein (Fig. 1C). These results indicate that the in vitro interaction between DRK and DAB requires the presence of the proline-rich region of DAB and suggest that the SH3 domains of DRK bind directly to sequences within DAB proline-rich core.
DRK and DAB associate in vivo.
To determine that the binding of DAB and DRK occurs in vivo, we examined whether DRK and DAB could be coimmunoprecipitated from SL2 cell extracts. Although DRK was precipitated by anti-DAB antibodies, DAB was not detected in anti-DRK immunoprecipitates (data not shown). The reason for this failure may have been that the anti-DRK antibodies were raised against the C-terminal SH3 domain of DRK, which is required for interaction with DAB. To overcome this problem, we expressed an HA-tagged DRK in SL2 cells and immunoprecipitated the DRK-HA protein with an anti-HA antibody. We also performed the reciprocal experiment using anti-DAB antibodies. In each case, the immunoprecipitates were then immunoblotted with anti-DRK and anti-DAB antibodies. Our analysis showed that DRK-HA and DAB were coprecipitated by either anti-HA or anti-DAB antibodies (Fig. 2A). Furthermore, consistent with our in vitro binding analysis, the complex of DRK-HA with DAB was not detected when the HA-tagged DRK protein carried mutations inactivating both SH3 domains (P49L/G199R), while inactivation of the DRK SH2 domain did not affect binding (Fig. 2B). These results demonstrate that DRK and DAB associate in vivo in a DRK SH3 domain-dependent manner.
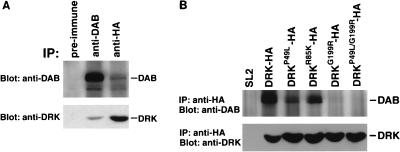
FIG. 2.
DAB and DRK associate in vivo. (A) Coimmunoprecipitation of DAB with DRK-HA. Extracts were prepared from SL2 cells that expressed an HA-tagged DRK protein. Immunoprecipitation was performed with rat preimmune serum, rat anti-DAB antiserum, or mouse anti-HA monoclonal antibody as indicated above each lane. Top, anti-DAB immunoblot; bottom, anti-DRK immunoblot of an identical gel. (B) The in vivo interaction between DRK and DAB requires the SH3 domains of DRK. Extracts were prepared from SL2 cells or SL2 cells expressing either wild-type or mutant DRK-HA protein (as indicated above each lane). They were then subjected to immunoprecipitation (IP) with an anti-HA antibody. Immunoblot analyses were performed with anti-DAB antibodies (top; two-thirds of the immunoprecipitates) and anti-DRK antibodies (bottom; one-third of the immunoprecipitates). Inactivation of the DRK SH2 domain (R85K) had no effect on binding, while inactivation of either SH3 domain (P49L or G199R) reduced binding and inactivation of both SH3 domains (P49L/G199R) abolished binding.
It has been reported that SOS binds DRK in vivo and that this interaction is mediated primarily through the N-terminal SH3 domain of DRK (45). Since DAB can bind to the C-terminal SH3 domain of DRK, we investigated whether a ternary complex of DAB, DRK, and SOS exists in vivo. While DRK was present in the anti-SOS immunoprecipitates, we were unable to coimmunoprecipitate DAB and SOS (data not shown). This finding suggests that DRK forms two distinct complexes in vivo, one with SOS and another with DAB.
DAB functions during ommatidial development and SEV signaling.
Since DRK plays an important role during photoreceptor development (44, 55), we investigated whether DAB might also participate in this process. We began by staining third-larval-instar eye imaginal discs with affinity-purified anti-DAB antibodies (Fig. 3A). Intense anti-DAB staining was observed both in the morphogenetic furrow and in developing ommatidial clusters posterior to the furrow. An apical-to-basal cross section revealed that DAB is localized to a small region just below the apical surface of the retinal epithelium (Fig. 3B). To determine which cells express DAB, the discs were costained with an antibody to ELAV, a neuronal marker present in the nuclei of developing and mature photoreceptors (Fig. 3C) (48). The results from these experiments showed that DAB is accumulated at the apical membrane of the developing photoreceptor cells. However, it was not possible to assign DAB expression to particular photoreceptors due to the apical constriction of these cells. The subcellular localization of DAB is similar to that of DRK (44), consistent with its role as a DRK-binding partner.
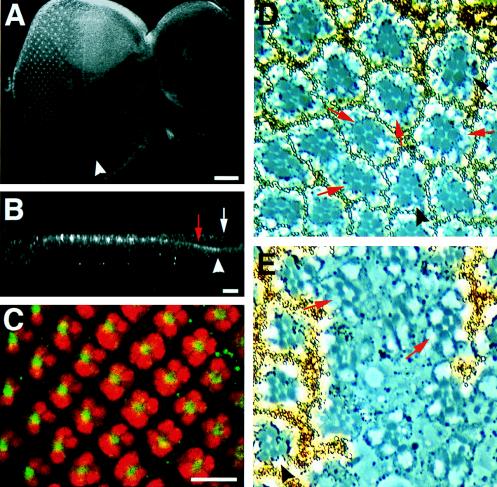
FIG. 3.
DAB is expressed in the ommatidial clusters posterior to the morphogenetic furrow, and loss of DAB function disrupts ommatidial development. (A to C) Confocal images from third-larval-instar eye antennal imaginal discs stained with affinity-purified anti-DAB antibodies only (A and B) or costained with anti-ELAV antibody (red) and anti-DAB antibodies (green) (C). (A) DAB expression is detected in the morphogenetic furrow (arrowhead), and there is a significant accumulation of DAB in the ommatidial clusters posterior to the furrow. (B) Apical-to-basal cross section. DAB expression is restricted to a small region just below the apical surface of the retinal epithelium. The red arrow indicates the retinal epithelium; the white arrow points at the peripodial membrane. The position of the morphogenetic furrow is marked by the arrowhead. In this panel, apical side is up and basal side is down. (C) Anti-ELAV antibody stained the nuclei of the developing and mature photoreceptors. DAB appeared to be located at the constricted apical surfaces of these cells. We were unable to identify unambiguously which photoreceptor cells contain the highest level of DAB. Scale bars indicate 100 μm (A) and 10 μm (B and C). (D and E) Photomicrographs of an eye section of a Tn-abl, abl1dab100, w1118/+ fly in which a clone of cells that are homozygous for both the w1118 and the dab100 mutations had been generated by X-ray induced mitotic recombination. Since all existing dab alleles were generated on abl− chromosomes and the two genes are closely linked, we used the Tn-abl transposon to provide functional Abl protein. The cells of the clones were genetically marked by the absence of a white gene and thus lack pigment granules. The pigment granules of the photoreceptors are small dark structures at the base of each rhabdomere; the pigment granules within the pigment cells, surrounding each ommatidium, are orange. (D) The most prominent phenotype is the presence of ommatidia which lack unpigmented R7 cells. Such ommatidia are indicated by the red arrows. Occasionally, ommatidia also lack unpigmented outer photoreceptor cells in addition to missing inner R7 photoreceptor (black arrow). The arrowhead points at a wild-type ommatidium. (E) Again, the most obvious phenotype is the lack of unpigmented, centrally located R7 cells in the ommatidia (red arrows). In this clone, many ommatidial clusters are also missing outer photoreceptors. The ommatidia are disorganized, and there are regions in which no photoreceptors are present. Overall, unpigmented photoreceptor cells are greatly underrepresented. An example of a wild-type ommatidium is indicated by the arrowhead. We also generated clones of Tn-abl;abl−/− cells as a control. As expected, the Tn-abl transposon is able to provide functional Abl protein, and thus all of the examined Tn-abl;abl−/− clones have wild-type ommatidia (data not shown).
We next examined whether dab function is required during the development of the photoreceptor cells. Since homozygous dab animals do not survive to adulthood, X-ray-induced mitotic recombination was used to generate marked clones of homozygous mutant dab cells in the eyes of dab/+ animals. Because all existing dab alleles were generated on chromosomes carrying tightly linked abelson (abl) mutations, a P-element transposon (Tn-abl) was used to provide abl function in the X-irradiated flies. The Tn-abl transposon has been shown to rescue the abl mutant phenotype (27). Numerous abnormalities were observed in dab homozygous mutant clones. The most common defects were the absence of the R7 cell and the lack of one or more outer photoreceptors (R1 to R6) in mosaic ommatidia (Fig. 3D and E). In addition, large dab mutant clones showed extensive ommatidial disorganization including regions in which no photoreceptors were present (Fig. 3E). This phenotype was observed with three different alleles of dab and resembles those observed in clones of cells homozygous for weak alleles of either Sos or Ras1 (51a). These results indicate that DAB has an important function during photoreceptor and ommatidial development.
The most extensively characterized step in ommatidial development is the specification of the R7 photoreceptor cell fate in response to the activation of SEV (26, 52, 64). The similarity of the dab clone phenotype to those of Sos and Ras1 suggests that DAB might also participate in SEV signal transduction. To investigate this possibility, we examined whether the level of DAB function is a limiting factor in determining the strength of SEV signaling. For this assay, we used flies carrying a P element expressing a constitutively activated SEV (SEVS11) under the control of the sev enhancer and hsp70 promoter sequences (sE) (4, 5). These transcriptional control elements direct expression in a subset of cells in the Drosophila eye, including the precursors of photoreceptors R7, R3, and R4 and all four of the cone cells. The expression SEVS11 in this pattern results in the generation of extra R7 photoreceptors due to the transformation of cone cell precursors into R7 cells (4). As a result, the eyes of SevS11 flies are rough and disorganized due to irregular packing and frequent fusion of the abnormally formed ommatidia (Fig. 4B and F). Importantly, it has been reported that the rough-eye phenotype of SevS11 flies is suppressed by the inactivation of one allele of drk, Sos, Ras1, or dos (28, 44, 46). We found that dab mutations could also dominantly suppress the SevS11 phenotype as assayed either by the degree of eye roughness or by the average number of R7 cells per ommatidium (Fig. 4C and G; Table 1). While SevS11 flies have an average of 4.0 R7 cells per ommatidium, SevS11 flies heterozygous for a dab mutation have only 3.1. Furthermore, the effect of dab mutations on the SevS11 phenotype is reversed by the expression of a dab cDNA under sE transcriptional control (Fig. 4D and H). It is important to note that this assay tests whether the abundance of a component is a limiting factor during SEVS11 signaling. Loss-of-function mutations of many known components of the SEV pathway including CSW and Raf have no observable effect on SevS11 phenotype (51a). The fact that DAB mutations suppressed the rough eye phenotype of SevS11 flies demonstrates that the level of DAB function is critical for determining the efficiency of SEVS11 signaling and suggests that DAB may normally participate in SEV signal transduction.
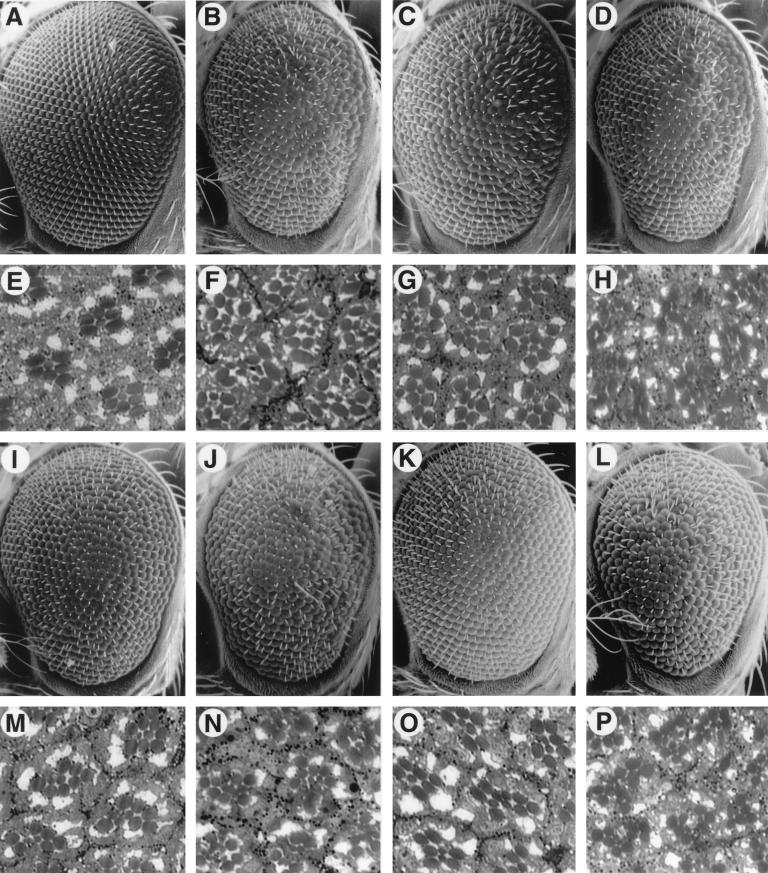
FIG. 4.
Effects of dab mutations on the phenotype of activated SEV. (A to D and I to L) SEM of adult eyes. (E to H and M to P) Photomicrographs of apical tangential sections of adult eyes. (A) SEM of a phenotypical wild-type eye of a w1118 fly. The eye is smooth in appearance, with uninterrupted ommatidial rows. The normal regular array of photoreceptor cells can be seen in panel E. The darkly stained structures are the rhabdomeres (light-sensing organelles) of each photoreceptor. The rhabdomere of R7 cell is smaller and more centrally located than the rhabdomeres of R1 to R6. The rhabdomere of R8 cell cannot be seen in this section because it is located underneath that of R7 cell. (B) SEM of an eye of a heterozygous P[sE-SevS11] fly. The external morphology of the eye is rough and disorganized due to the presence of multiple R7 cells per ommatidium and the frequent ommatidial fusions as seen in panel F. (C, D, G, and H) dab mutations attenuate SEVS11 signaling. (C) SEM of an eye of a fly heterozygous for both P[sE-SevS11] and dab mutation. Compared to the eye of the SevS11 fly, the surface of this eye is smoother, with the reappearance of organized ommatidial rows and very rare ommatidial fusion events. Sectioning of the eye revealed that the average number of R7 cells per ommatidium is reduced by the presence of the dab mutation (G). The flies represented in panels D and H are heterozygous for P[sE-SevS11], dab mutation, and P[sE-dab]. The suppression effect of dab mutations on SevS11 phenotype can be reversed by P[sE-dab], indicating that DAB is a limiting component during SEVS11 signaling. (I, J, M, and N) Mutation of a conserved residue in the PTB domain of DAB inhibits SEVS11 signaling. (I) SEM of a fly heterozygous for P[sE-SevS11] and P[sE-dabS119A]. Expression of DABS119A suppresses the rough-eye phenotype of SevS11 flies and reduces the average number of R7 cells per ommatidium (M). The dominant inhibitory effect that DABS119A exerts on SEVS11 signaling is relieved by an increased expression of wild-type DAB (J and N). (K, L, O, and P) DABΔPro enhances SEV signaling in vivo. (K) SEM of an eye of a SevS11-w fly. It is mildly roughened, with an average of 2.5 R7 cells per ommatidium (panel O and Table 1). The expression of DABΔPro in these flies enhances the roughness of the eye surface and increases the number of R7 to 3.6 cells per ommatidium (panel L and P and Table 1), while expression of wild-type DAB has no observable effect (data not shown).
TABLE 1.
dab mutations affect the average number of R7 cells per ommatidium in flies carrying a constitutively activated SEVa
| Genotype | Avg no. of R7 cells/ommatidium |
|---|---|
| P[sE-SevS11]/+; +/+ | 4.0 ± 0.2 |
| P[sE-SevS11]/+; dab/+ | 3.1 ± 0.3 |
| P[sE-SevS11]/+; dab/+; P[sE-dab]/+ | 4.1 ± 0.3 |
| P[sE-SevS11]/Y; P[sE-dabS119A]/+ | 2.5 ± 0.2 |
| P[sE-SevS11]/Y; P[sE-dabS119A]/+; P[sE-dab]/+ | 4.1 ± 0.3 |
| P[sE-SevS11-w]/+; +/+ | 2.5 ± 0.1 |
| P[sE-SevS11-w]/+; P[sE-dabΔPro]/+ | 3.6 ± 0.3 |
For each genotype, five to eight eyes were sectioned, and 70 to 100 ommatidia were analyzed in each eye. The average number of R7 cells per ommatidium and the standard of deviation were calculated for each eye and then used to tabulate the average number of R7 per ommatidium and the standard of deviation for each genotype.
DAB is tyrosine phosphorylated in response to SEV activation and can bind the SH2 domain of DRK.
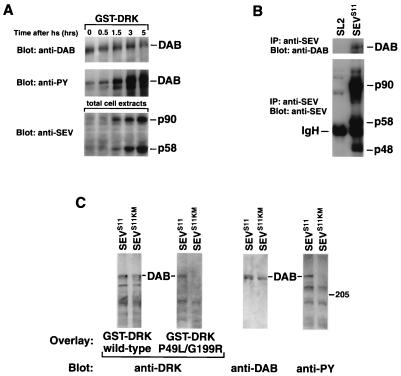
The crucial role of DAB during SEVS11 signaling led us to investigate the possibility that DAB becomes tyrosine phosphorylated in response to SEV activation. We expressed SEVS11 in SL2 cells under the control of a heat shock promoter and examined the tyrosine phosphorylation level of DAB. Following heat shock induction, SEVS11 autophosphorylation could be readily detected in these cells (data not shown). We found that upon induction of SEVS11, there was a significant increase in DAB phosphotyrosine content and a corresponding decrease in its electrophoretic mobility (Fig. 5A). In contrast, expression of a kinase-inactive SEVS11 in SL2 cells (generating SL2-SEVS11KM cells) had no effect. These results demonstrate that DAB tyrosine phosphorylation is responsive to SEV activation and imply that DAB is a direct or indirect target of SEV kinase activity.
FIG. 5.
Following induction of SEV kinase activity, DAB becomes tyrosine phosphorylated and then serves as a docking site for the SH2 domain of DRK. (A) DAB is tyrosine phosphorylated in response to induction of SEVS11. SL2-SEVS11 cells were placed at 37°C for 30 min and then allowed to recover at 23°C for 0, 0.5, 1.5, 3, or 5 h. Cell extracts were prepared in the presence of phosphatase inhibitors and used in a precipitation experiment with GST-DRK fusion protein coupled to glutathione-agarose beads. The precipitates were separated by SDS-PAGE, and immunoblotting was performed with rabbit anti-DAB antibodies (top). This blot was then stripped and reprobed with antiphosphotyrosine (anti-PY) antibody (middle). Total cell extracts were used in anti-SEV immunoblotting to verify the induction of SEVS11 expression (bottom). (B) DAB and SEVS11 coimmunoprecipitate. Extracts were prepared from either SL2 or SL2-SEVS11 cells and used in an immunoprecipitation (IP) experiment with anti-SEV monoclonal antibody. Immunoblotting was performed with either anti-DAB antibodies (top) or anti-SEV antibody (bottom). The positions of DAB and the three subunits of SEVS11 (the cytoplasmic domain precursor p90 and its processed forms p58 and p48) are indicated IgH, immunoglobulin heavy chain. (C) Tyrosine-phosphorylated DAB can bind to the SH2 domain of DRK. Cell extracts were prepared from SL2-SEVS11 or SL2-SEVS11KM cells, separated by SDS-PAGE on a 5 to 12% gradient gel, and transferred onto nitrocellulose filters. The filters were overlaid with GST fusion protein containing either wild-type DRK or DRK in which both SH3 domains had been inactivated (DRKP49L/G199R). The blots were then probed with anti-DRK antibodies. The level of DAB expression in each cell line and its tyrosine phosphorylation state were examined by immunoblotting with anti-DAB antibodies and antiphosphotyrosine antibody, respectively. The 205-kDa molecular mass marker is indicated; the 116-kDa marker is off the gel. DRKP49L/G199R can bind only to the tyrosine-phosphorylated DAB present in SL2-SEVS11 cell lysate.
The phosphorylation of DAB in response to SEV activation raises the possibility that DAB is also phosphorylated in response to the activation of other protein tyrosine kinases. We addressed this issue by examining DAB phosphorylation in SL2 cells either expressing an activated SRC64 or treated with insulin. While expression of activated SRC64 resulted in a marked overall increase in cellular tyrosine phosphorylation, it did not lead to DAB hyperphosphorylation (data not shown). Similarly, the activation of the insulin receptor had no effect on DAB tyrosine phosphorylation. These experiments demonstrate that the ability to induce tyrosine phosphorylation of DAB is not a property of all protein tyrosine kinases.
The tyrosine phosphorylation of DAB as well as the presence in DAB of potential consensus SH2 domain-binding sites suggest that phosphorylated DAB might function by binding the SH2 domains of signaling proteins. This possibility was examined for the DRK SH2 domain. To generate either phosphorylated or unphosphorylated DAB, we induced expression of either SEVS11 or SEVS11KM in SL2 cells. The resulting cell extracts were transferred onto nitrocellulose filters after SDS-PAGE. The filters were then probed with GST fusion proteins containing either wild-type DRK or DRK in which both SH3 domains were inactivated (DRKP49L/G199R). While wild-type DRK could bind to DAB from either extract, DRKP49L/G199R could bind to only the tyrosine-phosphorylated DAB present in the SL2-SEVS11 cell lysate (Fig. 5C). The dependence of the DAB-DRKP49L/G199R interaction on tyrosine phosphorylation suggests that it involves the SH2 domain of DRK. Thus, it appears that SEV-induced tyrosine phosphorylation of DAB can generate binding sites for the SH2 domain of DRK and perhaps for those of other signaling proteins as well.
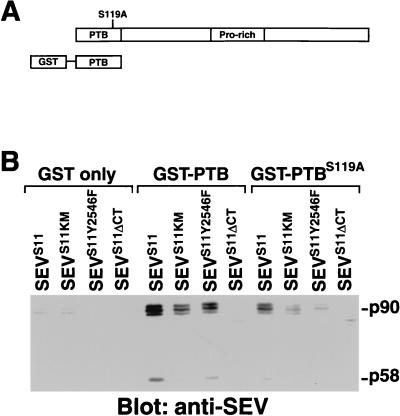
The PTB domain of DAB can bind SEV directly.
The PTB domains of insulin receptor substrate 1 (IRS-1) and Shc have been shown to bind to specific tyrosine-phosphorylated sequences of activated RTKs (17, 63). We examined whether the PTB domain of DAB could also bind to activated SEV. We generated a bacterially expressed fusion protein (GST-PTB) consisting of the PTB domain of DAB fused to GST (Fig. 6A) and tested whether it could precipitate SEVS11 from extracts of SL2-SEVS11 cells. While GST-PTB-coupled beads bound SEVS11, beads coupled to GST alone did not (Fig. 6B). Furthermore, since GST-PTB lacks the proline-rich region of DAB and does not bind to DRK (data not shown), the GST-PTB interaction with SEVS11 is likely due to a direct binding of the DAB PTB domain to SEVS11. Our experiments, however, do not exclude the possibility that the DAB PTB domain binds to other proteins that are associated with SEVS11.
FIG. 6.
DAB binds SEVS11 directly via its PTB domain, and mutation of a conserved residue in this domain reduces binding. (A) Schematic diagrams of the structures of DAB and the GST-PTB fusion protein. The relative position of the S119A point mutation is indicated. The GST-PTB fusion protein contains only the PTB domain of DAB fused to GST. It cannot bind to DRK due to the lack of the proline-rich core (data not shown). (B) The GST-PTB fusion protein can precipitate SEVS11. Expression of SEVS11 was induced by heat shock, and the cells were allowed to recover for 3 to 4 h. Cell extracts were prepared in the presence of phosphatase inhibitors and incubated with fusion proteins coupled to glutathione-agarose beads. Fusion proteins containing either GST only, GST-PTB, or GST-PTB with the S119A point mutation were used as indicated. The presence of SEVS11 in the complex was detected by immunoblotting with anti-SEV monoclonal antibody. GST-PTB fusion protein bound to the activated SEVS11 receptor. This ability was specific to the PTB portion of the fusion protein because glutathione-agarose beads coupled to GST alone did not precipitate SEVS11. Either inactivation of the kinase activity of SEVS11 (SEVS11KM) or mutation of the DRK-binding site on SEVS11 (SEVS11Y2546F) reduced binding. The C-terminal truncation of SEVS11 (SEVS11ΔCT), which removed the last 39 amino acids, including Y2546 but no other tyrosine residues, abolished binding. The S119A point mutation compromised binding of the DAB PTB domain to SEVS11.
Inactivation of the kinase activity of SEV (SEVS11KM) markedly reduced, but did not abolish, the ability of GST-PTB to associate with SEVS11 (Fig. 6B). The fact that the fusion protein still retained some binding activity for the kinase-inactive receptor is not unprecedented. It has been reported that the PTB domains of proteins such as Shc, X11, FE65, and Numb can bind to their targets independently of tyrosine phosphorylation (9, 14, 40).
We also found that mutation of the DRK-binding site on SEV (SEVS11Y2546F) (45) significantly decreased the amount of SEVS11 precipitated by the GST-PTB fusion protein (Fig. 6B). Since DRK is not directly involved in the interaction between the DAB PTB domain and SEVS11, this result suggests that the region of SEV containing this tyrosine residue might directly bind to the DAB PTB domain. This possibility is further supported by the result that the PTB domain of DAB could not bind a C-terminally truncated SEVS11 in which the last 39 amino acids, including Y2546 but no other tyrosine residues, were removed. However, we have not been able to demonstrate direct binding of the DAB PTB domain to peptides representing this region due to their poor solubility (data not shown).
The ability of SEVS11 to induce DAB tyrosine phosphorylation and the in vitro interaction of the DAB PTB domain with SEVS11 suggest that a complex of DAB and SEV might exist in vivo. To investigate this possibility, we examined anti-SEV immunoprecipitates from SL2-SEVS11 cells for the presence of DAB. We found that DAB and SEVS11 coimmunoprecipitated and thus provides evidence that SEV and DAB are present in a common complex in vivo (Fig. 5B).
A point mutation in the DAB PTB domain reduces SEV binding in vitro and inhibits SEV signaling in vivo.
The nuclear magnetic resonance structure of the PTB domain of human Shc (hShc) complexed to a tyrosine-phosphorylated peptide reveals that the phosphotyrosine of the peptide interacts with a positively charged site on hShc composed of Arg 67, Ser 151, Lys 169, and Arg 175 (63). Only one of these residues, Ser 151 in hShc, is highly conserved in PTB domain-containing proteins (10). We therefore mutated the corresponding Ser residue in DAB into Ala (S119A) (Fig. 6A) and tested its effect on DAB PTB domain binding. We found that SEVS11 could still bind to the mutated GST-PTB fusion protein but that the extent of binding was greatly reduced (Fig. 6B). Similarly, binding to either the kinase-inactive SEVS11KM or the DRK-binding-site-mutated SEVS11Y2546F was also compromised by the S119A mutation (Fig. 6B).
To investigate the role of the SEVS11-DAB PTB domain interaction in vivo, we examined the ability of DAB carrying the S119A mutation (DABS119A) to function during SEVS11 signaling in the developing eye. We found that expression of DABS119A under sE transcriptional control could not reverse the suppression of the SevS11 phenotype by dab mutations (data not shown). Moreover, DABS119A appeared to act as an inhibitor of wild-type DAB function. Expression of DABS119A in SevS11 flies that still carried two wild-type dab alleles strongly suppressed the SevS11 rough eye and reduced the average number of R7 cells per ommatidium from 4.0 to 2.5 (Fig. 4I and M; Table 1). The effects of DABS119A expression on SEVS11 signaling could be overcome by increased expression of wild-type DAB (Fig. 4J and N). Taken together, these results demonstrate that PTB domain function is important for DAB action during SEVS11 signaling and are consistent with the proposal that SEV and the DAB PTB domain interact during R7 development.
A deletion of the DAB proline-rich region enhances DAB function during SEV signaling.
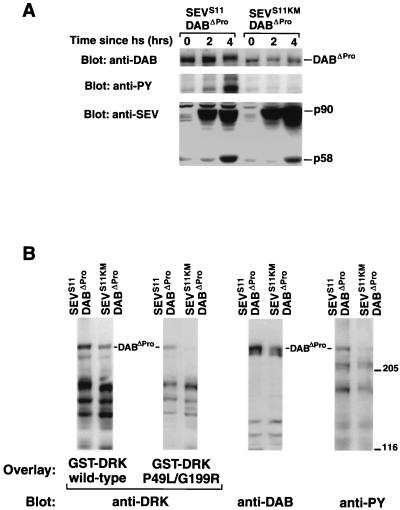
Our results have shown that the SH3 domains of DRK bind to the proline-rich region of DAB. To study the significance of this interaction on DAB function, we first examined the ability of DABΔPro to become phosphorylated following SEVS11 induction in SL2 cells. Although DABΔPro could not bind the DRK SH3 domains (Fig. 1C), it still became tyrosine phosphorylated and once phosphorylated could bind to the SH2 domain of DRK (Fig. 7). These results suggest that DAB tyrosine phosphorylation and its ability to act as docking sites for SH2 domains do not require interaction with the DRK SH3 domains.
FIG. 7.
DABΔPro still becomes tyrosine phosphorylated following SEV activation and can serve as a docking site for the SH2 domain of DRK. (A) The tyrosine phosphorylation of DABΔPro is responsive to SEVS11 induction. DABΔPro was coexpressed with either SEVS11 or the kinase-inactive SEVS11KM in SL2 cells. DABΔPro expression was under the control of the Actin5C promoter, while SEVS11 and SEVS11KM were under the control of a heat shock promoter. SEV expression was induced by heat shock (hs). Cells were collected either before heat shock (0 h, no induction) or 2 and 4 h after heat shock. The expression level of DABΔPro in each cell line was verified by immunoblotting with anti-DAB antibodies (top). The middle panel is an antiphosphotyrosine (anti-PY) stain of an identical blot. The tyrosine phosphorylation of DABΔPro was visibly increased at 2 h after heat shock, and became substantially enhanced at 4 h. Induction of the kinase-inactive receptor SEVS11KM had no effect on the tyrosine phosphorylation of DABΔPro. Both cell lines expressed comparable levels of SEV (bottom). (B) The tyrosine-phosphorylated DABΔPro can bind to the SH2 domain of DRK. Total cell extracts were prepared from the indicated cell lines, separated by SDS-PAGE on a 5 to 12% gradient gel, and transferred onto nitrocellulose filters. The filters were overlaid with GST fusion protein containing either wild-type DRK or DRK in which both SH3 domains had been inactivated (DRKP49L/G199R). The blots were then probed with anti-DRK antibodies. The level of DAB expression in each cell line and its tyrosine phosphorylation state were examined by immunoblotting with anti-DAB antibodies and antiphosphotyrosine antibody, respectively. The positions of the 205- and 116-kDa molecular mass markers are indicated.
We next tested whether DABΔPro could function during SEVS11 signaling in the developing eye. As for wild-type DAB, expression of DABΔPro had no apparent effect on the eye development of wild-type flies but did reverse the effects of dab mutations on the SevS11 phenotype. However, one difference between the effects of wild-type DAB and DABΔPro was that DABΔPro expression not only rescued the effects of insufficient dab function but also appeared to enhance the SevS11 rough-eye phenotype (data not shown). This finding suggests that expression of DABΔPro might actually increase the effectiveness of SEVS11 signaling. The enhancement of the SevS11 phenotype by DABΔPro was difficult to quantitate due to frequent fusions between ommatidial clusters. To overcome this problem, P-element transposition was used to generate a new strain of SevS11 flies (SevS11-w) that has a less severe phenotype (Fig. 4K and O). Expression of DABΔPro in SevS11-w flies enhanced the roughness of the eye surface and increased the average number of R7 cells from 2.5 to 3.6 per ommatidium (Fig. 4L and P; Table 1), while expression of wild-type DAB had no significant effect (data not shown). We also observed similar enhancement, although at a lesser extent, by expression of a DAB protein carrying point mutations in one of the proline-rich motifs (data not shown). The enhancement of the SevS11-w phenotype by DABΔPro expression suggests that the proline-rich region of DAB has an inhibitory effect on DAB function.
DISCUSSION
The primary function of RTKs is to activate specific intracellular biochemical pathways in response to binding of an extracellular ligand. Ligand binding leads to RTK activation and phosphorylation on tyrosine residues. The phosphotyrosine-containing regions in RTKs can then function as binding sites for cellular proteins containing either SH2 or PTB domains. These include the regulatory domain of phosphoinositide 3-kinase (PI3K), phospholipase C-γ, the Ras GTPase-activating protein (p120GAP), the phosphotyrosine phosphatase SHP2, Grb2/DRK, and Nck. In some cases, the associated proteins possess enzymatic activities that are regulated by their interaction with the activated RTK (36). In many other cases, the recruited proteins lack enzymatic activity and instead function by linking additional proteins to the activated receptor. Examples of these adaptor proteins include Grb2/DRK and p85, which serve to recruit SOS and the catalytic subunit of PI3K, respectively.
While many adaptor proteins are constitutively bound to their target molecules, the function of other adaptor proteins is dependent on their phosphorylation by the activated RTK. These adaptor proteins, once phosphorylated, may then function by recruiting proteins containing either SH2 or PTB domains into signaling complexes. This group of “switchable” adaptor proteins includes Shc, IRS-1, IRS-2, GAB1, Daughter of Sevenless (DOS), and fibroblast growth factor receptor substrate 2 (28, 30, 38, 46, 62). A structural property shared by all of these proteins is the presence of at least one domain that can direct localization either to the activated RTK (SH2 and PTB domains) or to the plasma membrane (pleckstrin homology domains and myristylation sites).
DAB as a putative adaptor protein during SEV signaling.
In this report, we have identified the PTB domain-containing protein DAB as a binding partner for the SH3 domains of DRK. We have shown that, as for drk, Sos, Ras1, and dos, inactivation of one allele of dab markedly attenuates signaling by the constitutively activated SEVS11 protein. Furthermore, expression of a PTB domain-mutated DAB suppresses the SevS11 phenotype. In contrast, the SevS11 phenotype is enhanced by the expression of a DAB lacking a proline-rich region. These results indicate that the level of DAB function can be critical for determining the strength of SEVS11 signaling and suggest that DAB is important for normal SEV signal transduction.
Several lines of evidence suggest that DAB may function as an adaptor protein whose function is regulated by phosphorylation. First, DAB contains a conserved N-terminal PTB domain that is capable of interacting with activated SEV. Second, SEV activation in Drosophila tissue culture cells leads to increased DAB tyrosine phosphorylation. Finally, we have evidence that this phosphorylation generates binding sites for the SH2 domain of DRK. These features are consistent with DAB functioning as an adaptor protein during SEV signaling. Further studies will be required to determine the importance of DRK SH2 domain binding and to identify additional SH2 domain-containing proteins that may bind to phosphorylated DAB.
Our analysis has provided evidence that the DAB PTB domain binds to the region including Y2546 of SEV. This conclusion is suggested by the effects on binding shown by SEVS11 proteins in which this site has been either mutated or deleted. One interesting feature of Y2546 is that it has previously been identified as a probable direct binding site on SEV for the DRK SH2 domain (45). The ability of both domains to interact with the same site on SEV suggests that a DRK-DAB complex, bound via the DRK SH3 domains and the proline-rich region of DAB, might interact with both subunits of an activated SEV dimer. Such an interaction might provide a mechanism for making DRK-DAB recruitment dependent on receptor dimerization. Alternatively, the DAB PTB and DRK SH2 domains might compete for SEV binding. This could provide a mechanism for the receptor to assemble different signaling complexes.
A potential role for DRK in regulating DAB function.
Our interest in the role of DAB during SEV signaling began with our identification of DAB as a DRK-binding partner. This in vitro interaction requires both the proline-rich region of DAB and the SH3 domains of DRK. Furthermore, complexes of DRK and DAB are present in Drosophila SL2 cells, and their formation depends on DRK SH3 domain function. To understand the significance of this interaction during SEV signaling, we assayed the ability of the proline-rich region truncated DABΔPro to participate in SEV signaling. Surprisingly, expression of DABΔPro enhanced the activated SevS11 phenotype. This result suggests that deletion of the proline-rich region leads to enhanced DAB function during SEV signaling.
Two models for the role of the DAB proline-rich region and its interaction with DRK are consistent with these results. One possibility is that the proline-rich region of DAB contains elements that act to inhibit DAB function. In this case, the role of DRK might be to relieve DAB inhibition through its bipartite binding to DAB proline-rich region via its SH3 domains and to the tyrosine-phosphorylated SEV via its SH2 domain. A similar role has been proposed for Grb2/DRK in regulating SOS activity (2, 35). A second possibility is that DRK binding to the DAB proline-rich region inhibits DAB function, and thus DRK acts as a negative regulator. In this model, the overall positive contribution of DRK to SEV signaling would likely result from its contribution to SOS function. It remains possible that the proline-rich region deletion has altered the function and/or localization of DAB. However, the fact that we observed similar enhancement by expression of a DAB protein carrying point mutations in one of the proline-rich motifs suggests that DABΔPro has an enhanced function due to its inability to interact with the SH3 domains.
The role of DAB in other RTK signaling pathways.
A murine DAB-related protein, mDAB1, has been identified as a tyrosine-phosphorylated protein that binds to the non-receptor protein tyrosine kinase Src (31). Recently, several reports have shown that mice lacking mDAB1 function have neuronal defects similar to those seen in reeler mice (32, 51, 61), including abnormal cortical lamination resulting from disruptions of neuronal migration processes. These results suggest that mDAB1 might participate in a signaling pathway triggered by REELIN, a secreted protein released near the targets of migrating neurons. In Drosophila, alleles of dab were originally identified due to their genetic interactions with mutations in abl. Homozygous abl mutant flies die either during pupation or shortly after eclosion. Inactivation of one allele of dab in homozygous abl mutant flies shifts the lethal phase from the pupal to the embryonic stage and results in severe disruptions of the axonal connections in the central nervous system (23). The neuronal defects associated with Drosophila and mouse dab mutations and our identification of DAB as a putative adaptor protein acting downstream of the receptor tyrosine kinase SEV suggest that DAB may function downstream of many RTKs, including ones required for proper development of the Drosophila central nervous system.
ACKNOWLEDGMENTS
We gratefully acknowledge F. M. Hoffmann, A. R. Comer, and S. M. Ahern for their generous discussion of unpublished results and provision of reagents. We thank A. Dingwall and M. Scott for the Drosophila embryonic λgt11 cDNA expression library and Carrie Steinberg for injection of the P-element constructs. We also thank M. Levin and members of our laboratory for critically reading the manuscript, useful advice, and general encouragement. We particularly thank R. Herbst for many invaluable suggestions and helpful discussions of this work.
This work was supported by a grant from the National Eye Institute (1RO1EY9845), a National Young Investigator Award from the NSF (MCB-9357009), and a Terman Fellowship Award to M.A.S. Further support was provided by National Institute of General Medical Sciences training grant GM07365 to N.L.
REFERENCES
- 1.Allard J D, Chang H C, Herbst R, McNeill H, Simon M A. The SH2-containing tyrosine phosphatase corkscrew is required during signaling by sevenless, Ras1 and Raf. Development. 1996;122:1137–1146. doi: 10.1242/dev.122.4.1137. [DOI] [PubMed] [Google Scholar]
- 2.Aronheim A, Engelberg D, Li N, al-Alawi N, Schlessinger J, Karin M. Membrane targeting of the nucleotide exchange factor Sos is sufficient for activating the Ras signaling pathway. Cell. 1994;78:949–961. doi: 10.1016/0092-8674(94)90271-2. [DOI] [PubMed] [Google Scholar]
- 3.Avruch J, Zhang X F, Kyriakis J M. Raf meets Ras: completing the framework of a signal transduction pathway. Trends Biochem Sci. 1994;19:279–283. doi: 10.1016/0968-0004(94)90005-1. [DOI] [PubMed] [Google Scholar]
- 4.Basler K, Christen B, Hafen E. Ligand-independent activation of the sevenless receptor tyrosine kinase changes the fate of cells in the developing Drosophila eye. Cell. 1991;64:1069–1081. doi: 10.1016/0092-8674(91)90262-w. [DOI] [PubMed] [Google Scholar]
- 5.Basler K, Siegrist P, Hafen E. The spatial and temporal expression pattern of sevenless is exclusively controlled by gene-internal elements. EMBO J. 1989;8:2381–2386. doi: 10.1002/j.1460-2075.1989.tb08367.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Biggs W H, Zavitz K H, Dickson B, van der Straten A, Brunner D, Hafen E, Zipursky S L. The Drosophila rolled locus encodes a MAP kinase required in the sevenless signal transduction pathway. EMBO J. 1994;13:1628–1635. doi: 10.1002/j.1460-2075.1994.tb06426.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Blanar M A, Rutter W J. Interaction cloning: identification of a helix-loop-helix zipper protein that interacts with c-Fos. Science. 1992;256:1014–1018. doi: 10.1126/science.1589769. [DOI] [PubMed] [Google Scholar]
- 8.Bonfini L, Karlovich C A, Dasgupta C, Banerjee U. The Son of sevenless gene product: a putative activator of Ras. Science. 1992;255:603–606. doi: 10.1126/science.1736363. [DOI] [PubMed] [Google Scholar]
- 9.Borg J P, Ooi J, Levy E, Margolis B. The phosphotyrosine interaction domains of X11 and FE65 bind to distinct sites on the YENPTY motif of amyloid precursor protein. Mol Cell Biol. 1996;16:6229–6241. doi: 10.1128/mcb.16.11.6229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bork P, Margolis B. A phosphotyrosine interaction domain. Cell. 1995;80:693–694. doi: 10.1016/0092-8674(95)90347-x. . (Letter.) [DOI] [PubMed] [Google Scholar]
- 11.Brunner D, Oellers N, Szabad J, Biggs W H, Zipursky S L, Hafen E. A gain of function mutation in Drosophila MAP kinase activates multiple receptor kinase signalling pathways. Cell. 1994;76:875–888. doi: 10.1016/0092-8674(94)90362-x. [DOI] [PubMed] [Google Scholar]
- 12.Cantley L L, Auger K, Carpenter C, Duckworth B, Graziani A, Kapeller R, Soltoff S. Oncogenes and signal transduction. Cell. 1991;64:281–302. doi: 10.1016/0092-8674(91)90639-g. [DOI] [PubMed] [Google Scholar]
- 13.Chardin P, Camonis J H, Gale N W, van Aelst L, Schlessinger J, Wigler M H, Bar-Sagi D. Human Sos1: a guanine nucleotide exchange factor for Ras that binds to GRB2. Science. 1993;260:1338–1343. doi: 10.1126/science.8493579. [DOI] [PubMed] [Google Scholar]
- 14.Charest A, Wagner J, Jacob S, McGlade C J, Tremblay M L. Phosphotyrosine-independent binding of SHC to the NPLH sequence of murine protein-tyrosine phosphatase-PEST. Evidence for extended phosphotyrosine binding/phosphotyrosine interaction domain recognition specificity. J Biol Chem. 1996;271:8424–8429. doi: 10.1074/jbc.271.14.8424. [DOI] [PubMed] [Google Scholar]
- 15.Clark S G, Stern M J, Horvitz H R. C. elegans cell-signalling gene sem-5 encodes a protein with SH2 and SH3 domains. Nature. 1992;356:340–344. doi: 10.1038/356340a0. [DOI] [PubMed] [Google Scholar]
- 16.Dickson B, Sprenger F, Morrison D, Hafen E. Raf functions downstream of Ras1 in the Sevenless signal transduction pathway. Nature. 1992;360:600–603. doi: 10.1038/360600a0. [DOI] [PubMed] [Google Scholar]
- 17.Eck M J, Dhe-Paganon S, Trub T, Nolte R T, Shoelson S E. Structure of the IRS-1 PTB domain bound to the juxtamembrane region of the insulin receptor. Cell. 1996;85:695–705. doi: 10.1016/s0092-8674(00)81236-2. [DOI] [PubMed] [Google Scholar]
- 18.Egan S E, Giddings B W, Brooks M W, Buday L, Sizeland A M, Weinberg R A. Association of Sos Ras exchange protein with Grb2 is implicated in tyrosine kinase signal transduction and transformation. Nature. 1993;363:45–51. doi: 10.1038/363045a0. [DOI] [PubMed] [Google Scholar]
- 19.Egan S E, Weinberg R A. The pathway to signal achievement. Nature. 1993;365:781–783. doi: 10.1038/365781a0. [DOI] [PubMed] [Google Scholar]
- 20.Fortini M E, Simon M A, Rubin G M. Signalling by the sevenless protein tyrosine kinase is mimicked by Ras1 activation. Nature. 1992;355:559–561. doi: 10.1038/355559a0. [DOI] [PubMed] [Google Scholar]
- 21.Fukazawa T, Reedquist K A, Trub T, Soltoff S, Panchamoorthy G, Druker B, Cantley L, Shoelson S E, Band H. The SH3 domain-binding T cell tyrosyl phosphoprotein p120. Demonstration of its identity with the c-cbl protooncogene product and in vivo complexes with Fyn, Grb2, and phosphatidylinositol 3-kinase. J Biol Chem. 1995;270:19141–19150. doi: 10.1074/jbc.270.32.19141. [DOI] [PubMed] [Google Scholar]
- 22.Gaul U, Mardon G, Rubin G M. A putative Ras GTPase activating protein acts as a negative regulator of signaling by the Sevenless receptor tyrosine kinase. Cell. 1992;68:1007–1019. doi: 10.1016/0092-8674(92)90073-l. [DOI] [PubMed] [Google Scholar]
- 23.Gertler F B, Bennett R L, Clark M J, Hoffmann F M. Drosophila abl tyrosine kinase in embryonic CNS axons: a role in axonogenesis is revealed through dosage-sensitive interactions with disabled. Cell. 1989;58:103–113. doi: 10.1016/0092-8674(89)90407-8. [DOI] [PubMed] [Google Scholar]
- 24.Gertler F B, Hill K K, Clark M J, Hoffmann F M. Dosage-sensitive modifiers of Drosophila abl tyrosine kinase function: prospero, a regulator of axonal outgrowth, and disabled, a novel tyrosine kinase substrate. Genes Dev. 1993;7:441–453. doi: 10.1101/gad.7.3.441. [DOI] [PubMed] [Google Scholar]
- 25.Guan K L, Dixon J E. Eukaryotic proteins expressed in Escherichia coli: an improved thrombin cleavage and purification procedure of fusion proteins with glutathione S-transferase. Anal Biochem. 1991;192:262–267. doi: 10.1016/0003-2697(91)90534-z. [DOI] [PubMed] [Google Scholar]
- 26.Hafen E, Dickson B, Brunner D, Raabe T. Genetic dissection of signal transduction mediated by the sevenless receptor tyrosine kinase in Drosophila. Prog Neurobiol. 1994;42:287–292. doi: 10.1016/0301-0082(94)90067-1. [DOI] [PubMed] [Google Scholar]
- 27.Henkemeyer M J, Gertler F B, Goodman W, Hoffmann F M. The Drosophila Abelson proto-oncogene homolog: identification of mutant alleles that have pleiotropic effects late in development. Cell. 1987;51:821–828. doi: 10.1016/0092-8674(87)90105-x. [DOI] [PubMed] [Google Scholar]
- 28.Herbst R, Carroll P M, Allard J D, Schilling J, Raabe T, Simon M A. Daughter of sevenless is a substrate of the phosphotyrosine phosphatase corkscrew and functions during sevenless signaling. Cell. 1996;85:899–909. doi: 10.1016/s0092-8674(00)81273-8. [DOI] [PubMed] [Google Scholar]
- 29.Herbst R, Shearman M S, Jallal B, Schlessinger J, Ullrich A. Formation of signal transfer complexes between stem cell and platelet-derived growth factor receptors and SH2 domain proteins in vitro. Biochemistry. 1995;34:5971–5979. doi: 10.1021/bi00017a026. [DOI] [PubMed] [Google Scholar]
- 30.Holgado-Madruga M, Emiet D R, Moscatello D K, Godwin A K, Wong A J. A Grb2-associated docking protein in EGF- and insulin-receptor signalling. Nature. 1996;379:560–564. doi: 10.1038/379560a0. [DOI] [PubMed] [Google Scholar]
- 31.Howell B W, Gertler F B, Cooper J A. Mouse disabled (mDab1): a Src binding protein implicated in neuronal development. EMBO J. 1997;16:121–132. doi: 10.1093/emboj/16.1.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Howell B W, Hawkes R, Soriano P, Cooper J A. Neuronal position in the developing brain is regulated by mouse disabled-1. Nature. 1997;389:733–737. doi: 10.1038/39607. [DOI] [PubMed] [Google Scholar]
- 33.Hsu J C, Perrimon N. A temperature-sensitive MEK mutation demonstrates the conservation of the signaling pathways activated by receptor tyrosine kinases. Genes Dev. 1994;8:2176–2187. doi: 10.1101/gad.8.18.2176. [DOI] [PubMed] [Google Scholar]
- 34.Kaelin W G, Jr, Krek W, Sellers W R, DeCaprio J A, Ajchenbaum F, Fuchs C S, Chittenden T, Li Y, Farnham P J, Blanar M A. Expression cloning of a cDNA encoding a retinoblastoma-binding protein with E2F-like properties. Cell. 1992;70:351–364. doi: 10.1016/0092-8674(92)90108-o. [DOI] [PubMed] [Google Scholar]
- 35.Karlovich C A, Bonfini L, McCollam L, Rogge R D, Daga A, Czech M P, Banerjee U. In vivo functional analysis of the Ras exchange factor son of sevenless. Science. 1995;268:576–579. doi: 10.1126/science.7725106. [DOI] [PubMed] [Google Scholar]
- 36.Kazlauskas A. Receptor tyrosine kinases and their targets. Curr Opin Genet Dev. 1994;4:5–14. doi: 10.1016/0959-437x(94)90085-x. [DOI] [PubMed] [Google Scholar]
- 37.Kimmel B E, Heberlein U, Rubin G M. The homeo domain protein rough is expressed in a subset of cells in the developing Drosophila eye where it can specify photoreceptor cell subtype. Genes Dev. 1990;4:712–727. doi: 10.1101/gad.4.5.712. [DOI] [PubMed] [Google Scholar]
- 38.Kouhara H, Hadari Y R, Spivak-Kroizman T, Schilling J, Bar-Sagi D, Lax I, Schlessinger J. A lipid-anchored Grb2-binding protein that links FGF-receptor activation to the Ras/MAPK signaling pathway. Cell. 1997;89:693–702. doi: 10.1016/s0092-8674(00)80252-4. [DOI] [PubMed] [Google Scholar]
- 39.Kramer H, Cagan R L, Zipursky S L. Interaction of bride of sevenless membrane-bound ligand and the sevenless tyrosine-kinase receptor. Nature. 1991;352:207–212. doi: 10.1038/352207a0. [DOI] [PubMed] [Google Scholar]
- 40.Li S C, Songyang Z, Vincent S J, Zwahlen C, Wiley S, Cantley L, Kay L E, Forman-Kay J, Pawson T. High-affinity binding of the Drosophila Numb phosphotyrosine-binding domain to peptides containing a Gly-Pro-(p)Tyr motif. Proc Natl Acad Sci USA. 1997;94:7204–7209. doi: 10.1073/pnas.94.14.7204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Marshall C J. MAP kinase kinase kinase, MAP kinase kinase, and MAP kinase. Curr Opin Genet Dev. 1997;4:82–89. doi: 10.1016/0959-437x(94)90095-7. [DOI] [PubMed] [Google Scholar]
- 42.McCormick F. Activators and effectors of ras p21 proteins. Curr Opin Genet Dev. 1994;4:71–76. doi: 10.1016/0959-437x(94)90093-0. [DOI] [PubMed] [Google Scholar]
- 43.Meisner H, Conway B R, Hartley D, Czech M P. Interactions of Cbl with Grb2 and phosphatidylinositol 3′-kinase in activated Jurkat cells. Mol Cell Biol. 1995;15:3571–3578. doi: 10.1128/mcb.15.7.3571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Olivier J P, Raabe T, Henkemeyer M, Dickson B, Mbamalu G, Margolis B, Schlessinger J, Hafen E, Pawson T. A Drosophila SH2-SH3 adaptor protein implicated in coupling the sevenless tyrosine kinase to an activator of Ras guanine nucleotide exchange, Sos. Cell. 1993;73:179–191. doi: 10.1016/0092-8674(93)90170-u. [DOI] [PubMed] [Google Scholar]
- 45.Raabe T, Olivier J P, Dickson B, Liu X, Gish G D, Pawson T, Hafen E. Biochemical and genetic analysis of the Drk SH2/SH3 adaptor protein of Drosophila. EMBO J. 1995;14:2509–2518. doi: 10.1002/j.1460-2075.1995.tb07248.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Raabe T, Riesgo-Escovar J, Liu X, Bausenwein B S, Deak P, Maroy P, Hafen E. DOS, a novel pleckstrin homology domain-containing protein required for signal transduction between sevenless and Ras1 in Drosophila. Cell. 1996;85:911–920. doi: 10.1016/s0092-8674(00)81274-x. [DOI] [PubMed] [Google Scholar]
- 47.Ren R, Mayer B J, Cicchetti P, Baltimore D. Identification of a ten-amino acid proline-rich SH3 binding site. Science. 1993;259:1157–1161. doi: 10.1126/science.8438166. [DOI] [PubMed] [Google Scholar]
- 48.Robinow S, White K. The locus elav of Drosophila melanogaster is expressed in neurons at all developmental stages. Dev Biol. 1988;126:294–303. doi: 10.1016/0012-1606(88)90139-x. [DOI] [PubMed] [Google Scholar]
- 49.Rozakis-Adcock M, Fernley R, Wade J, Pawson T, Bowtell D. The SH2 and SH3 domains of mammalian Grb2 couple the EGF receptor to the Ras activator mSos1. Nature. 1993;363:83–85. doi: 10.1038/363083a0. [DOI] [PubMed] [Google Scholar]
- 50.Schlessinger J, Ullrich A. Growth factor signaling by receptor tyrosine kinases. Neuron. 1992;9:383–391. doi: 10.1016/0896-6273(92)90177-f. [DOI] [PubMed] [Google Scholar]
- 51.Sheldon M, Rice D S, D’Arcangelo G, Yoneshima H, Nakajima K, Mikoshiba K, Howell B W, Cooper J A, Goldowitz D, Curran T. Scrambler and yotari disrupt the disabled gene and produce a reeler-like phenotype in mice. Nature. 1997;389:730–733. doi: 10.1038/39601. [DOI] [PubMed] [Google Scholar]
- 51a.Simon, M. Unpublished data.
- 52.Simon M A. Signal transduction during the development of the Drosophila R7 photoreceptor. Dev Biol. 1994;166:431–442. doi: 10.1006/dbio.1994.1327. [DOI] [PubMed] [Google Scholar]
- 53.Simon M A, Bowtell D D, Dodson G S, Laverty T R, Rubin G M. Ras1 and a putative guanine nucleotide exchange factor perform crucial steps in signaling by the sevenless protein tyrosine kinase. Cell. 1991;67:701–716. doi: 10.1016/0092-8674(91)90065-7. [DOI] [PubMed] [Google Scholar]
- 54.Simon M A, Bowtell D D, Rubin G M. Structure and activity of the sevenless protein: a protein tyrosine kinase receptor required for photoreceptor development in Drosophila. Proc Natl Acad Sci USA. 1989;86:8333–8337. doi: 10.1073/pnas.86.21.8333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Simon M A, Dodson G S, Rubin G M. An SH3-SH2-SH3 protein is required for p21Ras1 activation and binds to sevenless and Sos proteins in vitro. Cell. 1993;73:169–177. doi: 10.1016/0092-8674(93)90169-q. [DOI] [PubMed] [Google Scholar]
- 56.Singh H, Clerc R G, LeBowitz J H. Molecular cloning of sequence-specific DNA binding proteins using recognition site probes. BioTechniques. 1989;7:252–261. [PubMed] [Google Scholar]
- 57.Smith D B, Johnson K S. Single-step purification of polypeptides expressed in Escherichia coli as fusions with glutathione S-transferase. Gene. 1988;67:31–40. doi: 10.1016/0378-1119(88)90005-4. [DOI] [PubMed] [Google Scholar]
- 58.Songyang Z, Shoelson S E, Chaudhuri M, Gish G, Pawson T, Haser W G, King F, Roberts T, Ratnofsky S, Lechleider R J, Neel B G, Birge R B, Fajardo J E, Chou M M, Hanafusa H, Schaffhausen B, Cantley L C. SH2 domains recognize specific phosphopeptide sequences. Cell. 1993;72:767–778. doi: 10.1016/0092-8674(93)90404-e. [DOI] [PubMed] [Google Scholar]
- 59.Tomlinson A, Ready D F. Neuronal differentiation in the Drosophila ommatidium. Dev Biol. 1987;120:366–376. doi: 10.1016/0012-1606(87)90239-9. [DOI] [PubMed] [Google Scholar]
- 60.Tsuda L, Inoue Y H, Yoo M A, Mizuno M, Hata M, Lim Y M, Adachi-Yamada T, Ryo H, Masamune Y, Nishida Y. A protein kinase similar to MAP kinase activator acts downstream of the raf kinase in Drosophila. Cell. 1993;72:407–414. doi: 10.1016/0092-8674(93)90117-9. [DOI] [PubMed] [Google Scholar]
- 61.Ware M L, Fox J W, Gonzalez J L, Davis N M, Lambert de Rouvroit C, Russo C J, Chua S C, Jr, Goffinet A M, Walsh C A. Aberrant splicing of a mouse disabled homolog, mdab1, in the scrambler mouse. Neuron. 1997;19:239–249. doi: 10.1016/s0896-6273(00)80936-8. [DOI] [PubMed] [Google Scholar]
- 62.Yenush L, White M F. The IRS-signalling system during insulin and cytokine action. Bioessays. 1997;19:491–500. doi: 10.1002/bies.950190608. [DOI] [PubMed] [Google Scholar]
- 63.Zhou M M, Ravichandran K S, Olejniczak E F, Petros A M, Meadows R P, Sattler M, Harlan J E, Wade W S, Burakoff S J, Fesik S W. Structure and ligand recognition of the phosphotyrosine binding domain of Shc. Nature. 1995;378:584–592. doi: 10.1038/378584a0. [DOI] [PubMed] [Google Scholar]
- 64.Zipursky S L, Rubin G M. Determination of neuronal cell fate: lessons from the R7 neuron of Drosophila. Annu Rev Neurosci. 1994;17:373–397. doi: 10.1146/annurev.ne.17.030194.002105. [DOI] [PubMed] [Google Scholar]