Abstract
Pancreatic ductal adenocarcinoma (PDAC) is characterized by its nutrient-scavenging ability, crucial for tumor progression. Here, we investigated the roles of caveolae-mediated endocytosis (CME) in PDAC progression. Analysis of patient data across diverse datasets revealed a strong association of high caveolin-1 (Cav-1) expression with higher histologic grade, the most aggressive PDAC molecular subtypes, and worse clinical outcomes. Cav-1 loss markedly promoted longer overall and tumor-free survival in a genetically engineered mouse model. Cav-1–deficient tumor cell lines exhibited significantly reduced proliferation, particularly under low nutrient conditions. Supplementing cells with albumin rescued the growth of Cav-1–proficient PDAC cells, but not in Cav-1–deficient PDAC cells under low glutamine conditions. In addition, Cav-1 depletion led to significant metabolic defects, including decreased glycolytic and mitochondrial metabolism, and downstream protein translation signaling pathways. These findings highlight the crucial role of Cav-1 and CME in fueling pancreatic tumorigenesis, sustaining tumor growth, and promoting survival through nutrient scavenging.
Pancreatic cancer cells survive in resource-limited environments, using caveolae-mediated endocytosis to scavenge vital nutrients.
INTRODUCTION
Pancreatic ductal adenocarcinoma (PDAC), which is derived from the pancreatic exocrine compartment, accounts for more than 90% of pancreatic cancers, is highly aggressive, and is characterized by poor vascularization, high interstitial pressure, and hypoxia (1). To overcome barriers to growth in this harsh tumor environment, tumor cells have developed strategies for the uptake of amino acids, glucose, and lipids that are required for growth and survival. These strategies include integrin-mediated nutrient scavenging, receptor-mediated nutrient uptake, macropinocytosis, and entosis for the uptake by entire cells (2). The presence of a KRAS oncogenic (activating) mutation circumvents the relatively poor nutrient environment through the up-regulation of several survival mechanisms such as increased macropinocytosis (3), autophagy (4), and metabolic rewiring (5). Therapeutic targeting of these pathways could lead to novel effective therapeutic strategies for both PDAC and other aggressive tumor types that use nutrient scavenging for tumor growth and progression.
Caveolae are 50- to 100-nm flask-shaped invaginations of the plasma membrane that are important for endocytosis and membrane trafficking in a process termed caveolae-mediated endocytosis (CME) (6). Caveolin-1 (Cav-1) is the principal structural 22-kDa protein component required for the formation of caveolae (7, 8), and overexpression of Cav-1 has been shown to predict poor prognosis in multiple cancers, including lung, kidney, esophageal, breast, and PDAC (9–15). Phenotypically, Cav-1 has been shown to promote aggressive PDAC cellular behaviors, including proliferation, migration, invasion, and resistance to radiation and chemotherapy (16–20). Furthermore, Cav-1 has been shown to have diverse roles in cellular processes that are important for cancer maintenance and growth, including membrane trafficking (21), vascular endothelial growth factor receptor signaling (22), metabolism (23), and focal adhesion dynamics (24). Because of the important role of CME in the uptake of extracellular albumin (25), a key protein that tumor cells use to obtain macromolecules for constructive metabolism (anabolism) and energy generation, we hypothesized that Cav-1 is up-regulated in the most aggressive forms of PDAC and mediates survival under low-nutrient conditions through nutrient scavenging to maintain the nutrient pool required for protein translation, cell metabolism, and cell growth.
In this study, we perform the most comprehensive study to date of the role of Cav-1 in PDAC, through analysis of large human PDAC datasets as well as our own institutional tumor tissue and blood sample dataset. We also developed a genetically engineered mouse model that allows conditional ablation of CAV1 in a tissue-specific fashion. Pancreas-specific ablation of CAV1 alters PDAC tumorigenesis using a canonical clinically relevant mouse model of PDAC. Last, we elucidate previously unidentified roles for Cav-1 in tumor metabolism and nutrient scavenging that provide a mechanistic rationale to support PDAC aggressiveness.
RESULTS
Elevated Cav-1 expression correlates with tumor aggressiveness and worse prognosis in PDAC
We aimed to comprehensively determine whether Cav-1 expression correlates with more aggressive pathological features and worse clinical outcomes in patients with PDAC by integrating genome and transcriptome sequencing data. Sequencing data from patients with PDAC were interrogated as part of a comprehensive CAV1 bioinformatics analysis. Genome and transcriptome sequencing data were integrated across three resectable PDAC cohorts: The Cancer Genome Atlas Program (TCGA) (n = 130 samples with at least one data type available), International Cancer Genome Consortium (ICGC) (n = 175) and Clinical Proteomic Tumor Analysis Consortium 3 (CPTAC-3) (n = 140) and two metastatic PDAC cohorts: Prospectively Defining Metastatic PDAC Subtypes by Comprehensive Genomic Analysis (PanGen) (n = 69) and Comprehensive Molecular Characterization of Advanced PDAC For Better Treatment Selection (COMPASS) (n = 195). The frequency of SNV/indels and copy number variants (CNVs) affecting CAV1 was assessed in each of the five cohorts. No mutations likely to infer loss of function (nonsense, missense, frameshift, and in-frame indels) were found in CAV1 in any of the cohorts. Across all samples with CNV data available (n = 542), the frequency of CNVs affecting CAV1 was limited, with 61 (11.2%) samples showing a heterozygous loss, 17 (3.1%) showing amplification, and 1 (0.18%) showing a homozygous loss. Heterozygous loss affecting CAV1 was more frequent in metastatic (42 of 264; 16%) compared to resectable (19 of 278; 6.8%) cohorts (P = 9.8 × 10−4). CAV1 expression levels were not substantially different between metastatic samples with and without heterozygous loss of CAV1 (P = 0.83).
In each of the resectable and metastatic cohorts, normalized CAV1 expression levels were compared in each of the molecular subgroups of the several existing PDAC classification schemes: Moffitt (basal-like and classical), Karasinska (quiescent, cholesterogenic, glycolytic, and mixed), Collisson (classical, exocrine-like, and quasi-mesenchymal), and Bailey (progenitor, immunogenic, ADEX, and squamous) subgroups (Fig. 1A) (26–29). Samples with clinical data available and overall survival (OS) greater than 1 month (TCGA n = 101, ICGC n = 147, CPTAC-3 n = 124, PanGen n = 63, COMPASS n = 187) were used for survival analysis. In each cohort, samples were stratified by low (<25% quantile), medium-low (25 to 50% quantile), medium-high (50 to 75% quantile), and high (>75% quantile) CAV1 expression levels, and OS was compared between CAV1 low and high groups in each cohort (Fig. 1B). High CAV1 expression was associated with shorter survival in all resectable and metastatic cohorts [TCGA: hazard ratio (HR) = 2.5, 95% confidence interval (CI) = [1.1 to 6.0], P = 0.029; ICGC: HR = 2.2, 95% CI = [1.3 to 3.6], P = 0.0035; CPTAC-3: HR = 2.5, 95% CI = [1.3 to 4.7], P = 0.0029; PanGen: HR = 3.4, 95% CI = [1.4 to 8.5], P = 0.0052; COMPASS: HR = 2.1, 95% CI = [1.3 to 3.4], P = 0.0019). Together, these data are consistent with the notion that heightened CAV1 transcriptional activity is associated with tumor aggressiveness in pancreatic cancer.
Fig. 1. CAV1 expression is associated with aggressive molecular subtypes and shorter survival in patients with resectable and metastatic PDAC.
(A) Boxplots comparing CAV1 expression levels between molecular subtype groupings for the Moffitt, Karasinska, Collisson, and Bailey subtyping methods (left to right) in each of the five patient cohorts (top to bottom). Box plots indicate the median (central line), 25 to 75% interquartile range (IQR; bounds of box), and whiskers extend from box bounds to the largest value no further than 1.5 times the IQR. Two-tailed Wilcoxon mean rank sum P values are shown. (B) Kaplan-Meier plots showing overall survival (OS) between groups stratified by CAV1 expression levels. Hazard ratio (HR), 95% confidence interval (CI), and log-rank P value are shown and represent the comparison between low and high CAV1 groups. Tables showing the number at risk are included below.
To assess Cav-1 at the protein level, we used two independent tissue microarrays (TMAs) to score Cav-1 expression levels and correlate them with tumor characteristics and clinical outcomes. We first developed and stained our institutional pancreatic TMA derived from resected tumor samples from 114 patients with PDAC. We observed little to no presence of Cav-1 expression in the normal pancreatic epithelium and the precancerous lesions known as pancreatic intraepithelial neoplasia (PanIN), which are known to progress into PDAC. This contrasted with the expected positive Cav-1 staining observed in blood vessel endothelial cells and other stromal cell types (30). However, there was a marked increase in Cav-1 expression as the differentiation of the tumor progressed from moderately differentiated (Mod Diff) to poorly differentiated (Poorly Diff) (Fig. 2A). As tumor grade increased, we found a statistically significant increase in the pathological Cav-1 H-score (Fig. 2B). Cav-1 expression in the commercially available pancreas Biomax TMA also demonstrated significantly increased levels of Cav-1 expression in poorly differentiated PDAC tumors compared to either Mod Diff or PanIN-1 or -2 samples (Fig. 2C). These results suggested that Cav-1 overexpression is a late event in PDAC tumorigenesis.
Fig. 2. Cav-1 expression in Human PDAC TMA.
(A) Representative immunohistochemistry (IHC) images of Cav-1 protein expression in normal pancreatic epithelium, pancreatic intraepithelial neoplasia (PanIN), moderately differentiated (Mod Diff), and poorly differentiated (Poorly Diff) pancreatic cancer tissue (original magnification, ×200) from samples in the OSU TMA. (B) Mean Cav-1 H-score correlated with tumor grade. (C) The BioMax TMA correlated with Cav-1 expression via IHC. (D and E) OS and distant metastasis–free survival (DMFS) in the patients with resected PDAC. The numbers below figure represent the number at risk for each time point. (F) Circulating Cav-1 in pancreatic cancer patients with metastasis (MP) compared to healthy volunteers (HV). (G) Cav-1 expression (H-score) compared between primary resected pancreatic tumors (RP) compared to sites of metastasis (MP) in those patients with developed metastatic disease and had metastatic tumors that were biopsied. *P < 0.05.
Clinical outcomes were retrospectively annotated per patient from our institutional TMA (OSU), and patients were grouped by either positive (+) or negative (−) Cav-1 expression (i.e., H-score > 0 versus 0). Patients with Cav-1(+) tumors had a significantly shorter time to death or metastasis than those with Cav-1(−) tumors. Median OS and distant metastasis–free survival (DMFS) were 22.7 and 19.1 months, respectively for Cav-1 (−) tumors, versus 15.2 and 10.0 months for Cav-1 (+) tumors [HR = 1.5 for Cav-1(+), log-rank P = 0.018 and P = 0.03 for OS and DMFS, respectively] (Fig. 2, D to E). Patient and clinical characteristics in terms of age, gender, CA-19-9 levels, and margin negative resection were not found to be statistically substantially different between Cav-1(+) and Cav-1(−) patients (table S1).
Cav-1 protein can be detected in the serum of patients with cancer and has been associated with a worse prognosis in other cancers, such as prostate cancer (31). While we did not have access to blood samples from patients included in the TMAs, we assessed Cav-1 protein levels in the serum of patients with metastatic PDAC (MP) using a prospective protocol and found significantly increased levels of Cav-1 compared to those in healthy volunteers (P = 0.0007; Fig. 2F). In addition, tumor expression of Cav-1 was significantly increased in metastatic biopsy specimens compared to that in patients with localized, resected pancreatic cancer (P < 0.0001; Fig 2G). Together, these robust human clinical and molecular data confirm the association of high Cav-1 expression, a marker of CME, with increased PDAC aggressiveness and poor clinical outcomes.
Genetic ablation of CAV1 attenuates pancreatic tumor growth and improves survival in mice
To examine the role of Cav-1 in PDAC tumorigenesis, we developed a CAV1 conditional knockout mouse model (exon 3 floxed; fig. S1). We then interbred this mouse with an autochthonous mouse model of PDAC (KPC; LSL-KrasG12D/+, LSL-p53R270H/+, and Pdx1-cre) (32, 33) to generate a pancreas-specific homozygous Cav-1–deficient mouse model (KPC-CAV1fl/fl) (Fig. 3A). CAV1 deletion and loss of Cav-1 protein expression were confirmed in KPC-CAV1fl/fl tumors by immunohistochemistry (IHC; Fig. 3A), Southern blotting, and polymerase chain reaction (PCR) (Fig. 3B). KPC-CAV1fl/fl mice had a significantly longer OS than heterozygous KPC-CAV1fl/+ and KPC mice (median OS 168 versus 128 versus 105 days, P < 0.0001; Fig. 3C). KPC-CAV1fl/fl mice also had significantly longer tumor-free survival than KPC mice (P < 0.0001, Fig. 3D), and the tumor burden measured as the pancreas/body weight ratio at 4 months of age was significantly lower in KPC-CAV1fl/fl mice than in KPC mice (1.7% versus 4.8%, P < 0.0001; Fig. 3E). Tumor incidence in the pancreas was also higher in KPC mice than in both KPC-CAV1fl/+ and KPC-CAV1fl/fl mice (76.7% versus 46.2% versus 52.4%, respectively, P < 0.05; Fig. 3F). The presence of low-grade and high-grade PanIN, as measured using quantitative digital pathology, did not substantially differ between KPC and KPC-CAV1fl/fl mice at 12 weeks of age, suggesting that CAV1 loss does not attenuate the development of precancerous lesions or tumor initiation. This finding corroborates the notion that up-regulation of CAV1 occurs later during tumor progression.
Fig. 3. CAV1 deletion reduces tumorigenesis in the KPC mouse model.
(A) The generation of the KrasLSL-G12D;p53LSL-R270H/þ;Pdx1-cre; CAV1flox/flox (KPC-CAV1fl/fl) mice (top). Representative IHC for Cav-1 protein in the KPC and KPC-CAV1fl/fl mice (bottom). (B) Southern blotting CAV1neo allele in tail DNA of F1 offspring (top left); Southern blotting for CAV1flox allele in tail DNA of mice after deletion of neo cassette (top right); genotyping of tail DNA from KPC, KPC-CAV1fl/+, and KPC-CAV1fl/fl mice (bottom left); genotyping of tail DNA from KPC and KPC-CAV1fl/fl mice or genotyping of pancreatic tumor DNA from KPC-CAV1fl/+ or KPC-CAV1fl/fl mice (bottom right). (C) Kaplan-Meier analysis of OS of KPC, KPC-CAV1fl/+, KPC-CAV1fl/fl, and CAV1fl/fl;Pdx1-cre mice. Numbers of animals per group are indicated. (D) Kaplan-Meier analysis of tumor-free survival of KPC, KPC-CAV1fl/+, KPC-CAV1fl/fl, and CAV1fl/fl; Pdx1-cre mice. (E) The tumor-to-body weight ratio (%) was calculated for each of the four groups of mice. *P < 0.0001. (F) Tumor incidence in the pancreata of mice in the KPC mice compared to either the KPC-CAV1fl/+ or KPC-CAV1fl/fl mice. *P < 0.05; ns, not significant.
Loss of Cav-1 impairs cell proliferation and growth during serum starvation
To better understand how Cav-1 affects PDAC tumorigenesis, we generated tumor-derived cell lines (TDCL) from KPC and KPC-CAV1fl/fl mice. Cav-1 protein expression was present in the KPC TDCLs but absent in the KPC-CAV1fl/fl TDCLs, as expected (Fig. 4A). In addition, we generated stable Cav-1 knockdown human PDAC cell lines in MIA PaCa-2 (MP2) and PANC-1 cells via transfection of short hairpin RNA (shRNA) targeting CAV1(Fig. 4, B and C). Under supraphysiological nutrient conditions [10% fetal bovine serum (FBS)], high Cav-1 cells proliferated at a rate similar to that of Cav-1–deficient cell lines (Fig. 4, D to F). Since PDAC is known to develop strategies for survival and progression in a low-nutrient environment (34), we posited that Cav-1 is critical for growth in nutrient-deprived conditions and that loss of Cav-1 would attenuate growth under such conditions. As expected, Cav-1–deficient TDCLs and Cav-1 knockdown human PDAC cell lines showed impaired growth under nutrient-deprived conditions (1% FBS) compared to Cav-1–proficient controls (Fig. 4, G to I). Similar results were demonstrated in colony-forming assays, where KPC and KPC-CAV1fl/fl TDCLs showed no differences in colony formation in 10% FBS growth media; however, when grown in serum deprivation conditions (1% FBS), KPC TDCL formed colonies, whereas KPC-CAV1fl/fl TDCL were unable to form colonies (Fig. 4J). Similarly, MP2 cells demonstrated no differences in colony formation in 10% FBS but showed significant differences in colony formation in 1% FBS (Fig. 4K). For PANC-1 cells, shCAV1 cells formed fewer colonies in both 10 and 1% FBS, with a more significant reduction in the colony formation fold change in the latter condition (Fig. 4L). Together, these results suggest that Cav-1 plays a critical role in the optimal growth of PDAC cells under serum-depleted conditions.
Fig. 4. Loss of Cav-1 renders cells with decreased proliferative capabilities in serum-deprived conditions.
(A) Western blot analysis of Cav-1 expression in cell lysates from TDCLs from the KPC and KPC-CAV1fl/fl tumors. Numbers refer to individual cell lines from different tumors. (B and C) Western blot analysis of (B) MiaPaca-2 (MP2) and (C) PANC-1 cells transfected with scrambled short hairpin RNA (shCtrl) or shRNA targeting CAV1 (shCAV1). (D to F) Proliferation assays measuring the change in confluence over time in regular media supplemented with 10% fetal bovine serum (FBS) in (D) KPC versus KPC-CAV1fl/fl, (E) MP2 shCtrl versus shCAV1, and (F) PANC-1 shCtrl versus shCAV1 cells. (G to I) Proliferation assays measuring the change in confluence over time in regular media supplemented with 1% FBS in (G) KPC versus KPC-CAV1fl/fl, (H) MP2 shCtrl versus shCAV1, and (I) PANC-1 shCtrl versus shCAV1 cells. (J to L) Cells were plated as single cells and incubated in either 10 or 1% FBS-supplemented media and colonies were assessed (>50 cells) after 2 weeks in (J) KPC versus KPC-CAV1fl/fl, (K) MP2 shCtrl versus shCAV1, and (L) PANC-1 shCtrl versus shCAV1 cells. *P < 0.05.
Loss of Cav-1 impairs glycolytic capacity and mitochondrial metabolism and reduces TCA cycle intermediates
Cav-1 has been implicated in metabolic alterations in cancer, such as glycolysis, mitochondrial bioenergetics, glutaminolysis, and fatty acid metabolism (5). However, the role of Cav-1 in regulating energy metabolism under nutrient stress in PDAC has not been elucidated. We first tested the impact of Cav-1 loss on glycolytic flux using the glycolysis stress test on the Seahorse XF analyzer. This assay measures the net proton production following the addition of glucose, oligomycin (mitochondrial proton pump inhibitor), and 2-deoxyglucose (2-DG; inhibits step 2 of glycolysis) to the cell system. Glycolytic capacity was significantly impaired in KPC-CAV1fl/fl compared to that in the wild-type KPC TDCL (Fig. 5A). These results were replicated in MP2 shControl (shCtrl) versus shCAV1 cell lines (Fig. 5B). We next tested mitochondrial respiration in TDCLs and MP2 cells using the mitochondrial stress test to measure oxygen consumption rates (OCR) in cells grown overnight in serum-repleted (10% FBS) or serum-starved (1% FBS) conditions. The OCR measurements of basal oxygen consumption, adenosine 5′-triphosphate (ATP) production, and maximal respiration were not statistically notable between KPC and KPC-CAV1fl/fl TDCLs grown in 10% FBS [including glucose (4.5 g/liter) and l-glutamine (584 mg/ml)] (Fig. 5, C and E). However, under serum-depleted conditions (1% FBS), KPC cells produced significantly higher OCR values than KPC-CAV1fl/fl cells (Fig. 5, C to E). MP2 control cells demonstrated higher OCR, basal oxygen consumption, ATP production, and maximal respiration under both serum-repleted and serum-starved conditions (Fig. 5, D and F). Next, we used liquid chromatography–tandem mass spectrometry to profile TCA cycle intermediates in the KPC and KPC-CAV1fl/fl TDCLs. After overnight incubation in serum-starved media, KPC cells demonstrated significantly increased levels of citric acid and isocitric acid compared with KPC-CAV1fl/fl cells (Fig. 5G). The levels of malic acid, succinic acid, and oxoglutaric acid were not notably different between the two cell lines (Fig. 5G). These results revealed the critical role of Cav-1 in maintaining cellular metabolism in PDAC cells, especially under serum-deprived conditions.
Fig. 5. Cav-1 regulates metabolic capacity in PDAC cells.
(A) The glycolysis stress test was performed in the Seahorse XFe96 Analyzer on KPC #1 and #4 as well as KPC-CAV1fl/fl #2 and #3 TDCLs. The extracellular acidification rate (ECAR) over time was plotted after treatment at set time points of glucose (10 mM), oligomycin (1 μg/ml), and 2-DG (at 50 mM, inhibition of glycolysis). The maximal glycolytic capacity was significantly higher in KPC versus KPC-CAV1fl/fl cells. (B) The glycolysis stress test was performed in MP2 shCtrl and shCAV1 cells as described above. The maximal glycolytic capacity was significantly higher in MP2 shCtrl versus shCAV1 cells. (C) The mitochondrial stress test was performed in the Seahorse XFe96 Analyzer KPC #1 and #4 TDCL and KPC-CAV1fl/fl #2 and #3 TDCL following either overnight seeding in 10% FBS (left) or 1% FBS (right). The oxygen consumption rate (OCR) over time was plotted after injection of oligomycin (0.5 μM), carbonyl cyanide p-trifluoromethoxyphenylhydrazone (FCCP) (1 μM), and rotenone/antimycin A (AA/Rot) (1 μM). Each data point represents an OCR measurement. (D) The mitochondrial stress test in MP2 shCtrl and shCAV1 cells following either overnight seeding in 10% FBS (left) or 1% FBS (right). (E) Basal consumption, adenosine 5′-triphosphate (ATP) production, and maximal respiration in 10% and 1% FBS media in the KPC cells compared to KPC-CAV1fl/fl cells. (F) Basal consumption, ATP production, and maximal respiration in both 10 and 1% FBS conditions in MP2 shCtrl and shCAV1 cells. (G) Liquid chromatography–tandem mass spectrometry chromatograms of TCA cycle intermediates extracted from KPC and KPC-CAV1fl/fl TDLC TDCLs after overnight incubation in 1% FBS. Data are expressed as means ± SEM, n = 2 independent experiments. (H) Diagram of the TCA cycle displaying the conversion of glutamine into alpha-ketoglutarate. *P < 0.05.
Cav-1 loss sensitizes tumor cells to glutamine deprivation through reduced albumin uptake
The two TCA metabolites, isocitrate and citrate, were found to be significantly reduced in the KPC-CAV1fl/fl cells, and metabolites immediately downstream of glutamine (Q) metabolism are used in the reductive carboxylation pathway, especially in hypoxic environments (35) (Fig. 5H). Therefore, we hypothesized that cells deficient in Cav-1 would require increased levels of glutamine for growth and would be more susceptible to glutamine deprivation. We found that KPC TDCLs maintained proliferation rates in glutamine concentrations as low as 0.4 mM but could not proliferate in glutamine concentrations of 0.2 mM and lower (Fig. 6A). On the other hand, KPC-CAV1fl/fl cells could only maintain proliferation at glutamine concentrations of 1 mM or higher, and proliferation was completely abrogated at threshold concentrations of 0.8 mM or lower (Fig. 6B). Thus, there was a 2.5-fold difference in glutamine requirement between the two TDCLs. Similar results were observed in the MP2 cells (Fig. 6, C and D).
Fig. 6. Cav-1 regulates growth under low glutamine environments and albumin uptake.
(A to D) KPC #4 TDCL, KPC-CAV1fl/fl #2 TDCL, MP2 shCtrl, and MP2 shCAV1 cells were grown in glutamine dilutions (0 to 4 mM) and proliferation was assessed by IncuCyte confluence measurements every 4 hours. (E to H) Cells were grown in the glutamine concentration just below the glutamine threshold of each individual cell line (KPC #4 = 0.2 mM, KPC-CAV1fl/fl = 0.8 mM, MP2 shCtrl = 0.4 mM, and MP2 shCAV1 = 1.0 mM) with or without supplementation with either mouse serum albumin (KPC) or human serum albumin (MP2) at 3% per volume and proliferation capacity measured by the IncuCyte. (I) Representative immunofluorescence images of 10 pancreatic cell lines after treatment with albumin for 30 min showing expression of Cav-1 (red), albumin (green), and 4′,6-diamidino-2-phenylindole (DAPI) (blue). Experiments were performed in triplicate and repeated three times with similar results. (J) The Cav-1 index was plotted against the albumin indexes to determine correlations.
Cav-1 plays an important role in albumin uptake in non-cancer and cancer cells as well as in susceptibility to albumin-bound chemotherapeutics (36). PDAC cells can use extracellular albumin for breakdown into constituent amino acids and central carbon metabolism, ultimately supporting growth under low glutamine conditions (34). We tested whether supplementation with physiological albumin concentrations under low glutamine conditions could rescue the growth of Cav-1–proficient cells. As predicted, mouse serum albumin (3%) was able to markedly rescue growth at 0.2 mM threshold glutamine concentration in KPC cells (Fig. 6E), while albumin supplementation could not rescue the growth of KPC-CAV1fl/fl cells at the threshold concentration of 0.8 mM glutamine (Fig. 6F). These results were replicated in the MP2 cells (Fig. 6, G and H).
Next, we tested whether Cav-1 expression correlated with albumin uptake. A panel of 10 human PDAC cell lines with varying levels of Cav-1 expression was serum-starved overnight, and then pulsed with human serum albumin (HSA) for 30 min before fixing cells for quantitative indirect immunofluorescence (Fig. 6I). The Cav-1 index in the 10 cell lines plotted against the albumin index showed a strong direct correlation (r2 = 0.657, P = 0.0005; Fig. 6J), indicating that elevated Cav-1 expression was correlated with elevated albumin uptake. These results revealed that Cav-1 is required for albumin uptake and that albumin can rescue proliferation in nutrient (glutamine)-deficient environments only in cells expressing Cav-1.
Cav-1 loss inhibits activation of ribosomal S6
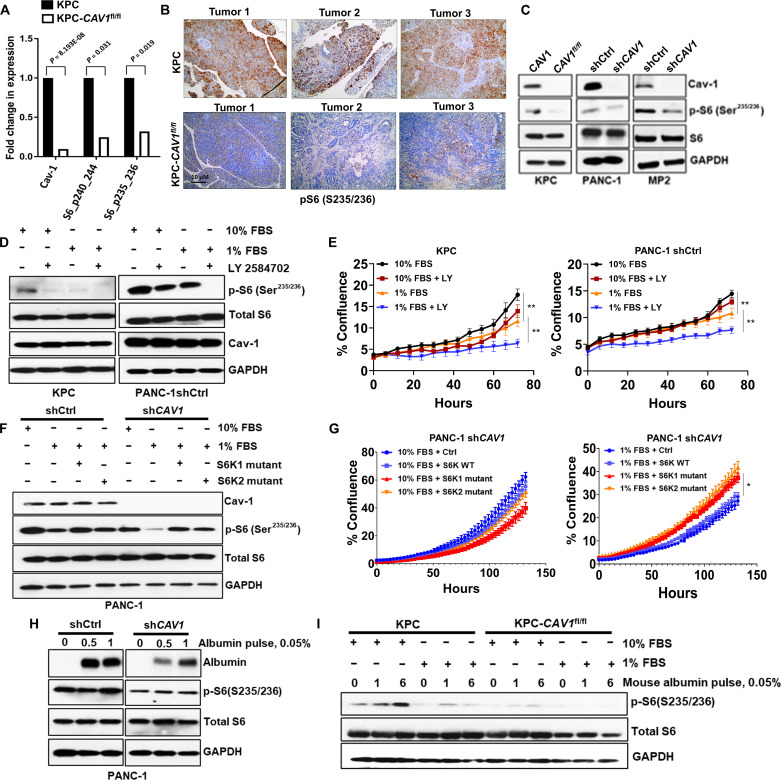
To determine the potential molecular mechanisms by which Cav-1 regulates growth under serum-depleted conditions, we performed reverse-phase protein microarray (RPPA) analyses on KPC and KPC-CAV1fl/fl TDCLs. The top three proteins found in RPPA with the largest fold change in levels between KPC and KPC-CAV1fl/fl cells were Cav-1, phospho-S6 (S240/244), and phospho-S6 (S235/236) (Fig. 7A). S6 is a component of ribosomal subunit 40S and is regulated by the mammalian target of rapamycin (mTOR) pathway. Following phosphorylation by S6 kinase (S6K) at the serine-235 or -236 residue, phosphorylated S6 increases cell proliferation, protein translation, and cell size (37). The RPPA data from the PAAD TCGA dataset also confirmed a direct correlation between phospho-S6 (S235/236) and Cav-1 protein levels (P = 0.0064; fig. S2). IHC using anti–phospho-S6 antibodies was performed on the KPC and KPC-CAV1fl/fl tumors and showed markedly higher levels of phospho-S6 (S235/236) in KPC tumors, compared to weak expression in KPC-CAV1fl/fl tumors (Fig. 7B). We also confirmed reduced levels of phospho-S6 in KPC-CAV1fl/fl, PANC-1 shCAV1, and MP2 shCAV1 cells compared to those in control cells (Fig. 7C), as well as other associated proteins in this pathway upstream of S6 in both PANC-1 and MP2 shCAV1 cells compared to those in shCtrl cells (fig. S3, A and B). Next, we tested whether serum-depleted conditions (1% FBS) could modify the phosphorylation of S6. The levels of phospho-S6 observed under serum deprivation (1% FBS) in KPC and PANC-1 shCtrl cells were significantly reduced, with attenuated cell proliferation compared to serum-repleted conditions (10% FBS) (Fig. 7, D and E). In addition, inhibition of upstream p70-S6K with LY-2584702 (LY) effectively abrogated S6 phosphorylation in both Cav-1–proficient and–deficient cells (Fig. 7D and fig. S4, A and B) and inhibited cell proliferation under nutrient-deprived conditions in KPC and PANC-1 shCtrl cells (Fig. 7E). p70-S6K inhibition had no additional negative impact on growth over Cav-1 deficiency in KPC-CAV1fl/fl or PANC-1 shCAV1 cells (fig. S4, C and D).
Fig. 7. Cav-1 regulates the activation of ribosomal S6 under nutrient stress.
(A) Mean comparisons of RPPA data for KPC versus KPC-CAV1fl/fl cells. Differential protein abundance was used using RPPA measures through limma to compare the two cell lines. P values are from a two-sided, unpaired t test comparing the mean of KPC cells versus KPC-CAV1fl/fl cells. Error bars denote SEM. (B) Representative IHC images of S6 (p235/236) protein expression KPC and KPC-CAV1fl/fl tumors (n = 3 each). (C) Immunoblots showing levels of phosphorylated S6 (Ser235/236), total S6, and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) in KPC/ KPC-CAV1fl/fl, MP2, and PANC-1 shCtrl/shCAV1 cells. (D) Immunoblots displaying levels of phosphorylated S6 (234/235), total S6, Cav-1, and GAPDH in KPC and PANC-1shCtrl cells in both 10 and 1% FBS ± LY-2584702 (5 μM, 2 hours). (E) Proliferation assays in the KPC #4 or PANC-1 shCtrl cells in 10 or 1% FBS ± 5 μM of LY-2584702 (**P < 0.01). (F) PANC-1 shCtrl and shCAV1 cells transfected with S6K mutant plasmids (S6K1 Mut or S6K2 Mut) for 48 hours followed by media exchange with 1% FBS overnight. Levels of phospho-S6 (S235/236), total S6, Cav-1, and GAPDH are shown. (G) Proliferation assays in the PANC-1 shCAV1 cells transfected with the S6K plasmids (S6K1 WT, S6K1 Mut, or S6K2 Mut) following 48-hour transfection with the S6K plasmids in either 10 or 1% FBS (*P < 0.05). (H) Immunoblots showing higher albumin uptake in PANC-1 shCtrl cells compared to shCAV1 cells, with induction of phospho-S6 activation at 1 hour. (I) Immunoblots indicating S6 activation in KPC cells compared to KPC-CAV1fl/fl cells. *P < 0.05 and **P < 0.01.
To validate that the Cav-1–S6K-S6 pathway is critical for cell survival and growth under low-nutrient conditions, we transiently expressed constitutively active p70S6K1 and p70S6K2 mutant plasmids (S6K1mut and S6K2mut, respectively) in PDAC cells (38, 39). First, as expected, growth in nutrient-depleted (1% FBS) conditions in the PANC-1 shCtrl and shCAV1 cells resulted in the reduction of phospho-S6 (Fig. 7F). We confirmed that transfection with these mutant S6K plasmids partially or fully restored the phosphorylation of S6 in PANC-1 shCtrl and shCAV1 cells (Fig. 7F), as well as in MP2 cells (fig. S5A). In terms of growth, we found no substantial changes in the proliferation rate of S6K1/2 mutant-expressing PANC-1 shCAV1 cells under nutrient-replete conditions (Fig. 7G, left). However, in nutrient-deficient conditions, S6K1/2 mutant-expressing PANC-1 shCAV1 cells proliferated at a substantially higher rate compared to either S6K wild-type or control (no vector) transfected cells (Fig. 7G, right; and fig. S5B). Last, albumin can be used as a fuel source in tumor cells, allowing for growth and survival in nutrient-deprived environments (40). To further investigate whether albumin contributes to the activation of S6 under nutrient-deprived conditions, we treated PANC-1 shCtrl and shCAV1 cells with albumin and found elevated albumin uptake and phospho-S6 levels in PANC-1 shCtrl cells compared to shCAV1 cells (Fig. 7H), which was also confirmed in KPC/ KPC-CAV1fl/fl cells (Fig. 7I). These findings suggest that Cav-1 is critical for protecting PDAC cells from nutrient deprivation by promoting the uptake of albumin and other nutrients, thereby stimulating S6K-S6 signaling to restart tumor cell growth and proliferation.
DISCUSSION
Cancer cells have a high nutrient requirement to support an accelerated rate of proliferation and have evolved mechanisms to identify and take in amino acids, lipids, glucose, macromolecules, and either live/dead cells to be repurposed into the building blocks for new growth, ATP production, and anabolism. This adaptation is often accomplished by transporting the material to lysosomes for degradation. Here, we identified that CME is critical for the survival of patients with pancreatic cancer. To our knowledge, this is the first study to develop a conditional (tissue-specific) knockout of CAV1 in a spontaneous pancreatic cancer mouse model, to clearly demonstrate that Cav-1 promotes tumor metabolism and nutrient scavenging in PDAC in a clinically relevant model. We found that mice with spontaneous pancreatic tumors lacking Cav-1 expression had longer survival and reduced tumor growth. We demonstrated that Cav-1 is highly associated with the most aggressive subtypes of PDAC and Cav-1 loss leads to reduced glycolysis, mitochondrial metabolism, and ribosomal signal transduction. Mechanistically, Cav-1 plays a critical role in cell growth and survival under low-nutrient conditions, in part through albumin uptake and S6K/S6 signaling, thereby reversing the metabolic and proliferative rewiring that occurs when Cav-1 is depleted.
Cav-1, the critical structural component of caveolae, is increasingly being studied for its role in tumorigenesis. Cav-1 is a small protein (~22 kDa), yet is necessary for caveolae formation, as genetic depletion of CAV1 ablates the development of caveolae (6). Cav-1 has diverse cellular roles depending on tissue type and environment (7, 41). Alterations in Cav-1 expression affect a diverse array of non-cancer disease phenotypes, such as diabetes, bladder dysfunction, and muscular dystrophy (8, 42–44). In cancer cells, Cav-1 has been shown to regulate a multitude of critical pro-tumorigenic functions including invasion (45), apoptosis (46), cell signaling (47), cholesterol metabolism (48), autophagy (49), metabolism (23), angiogenesis (22), and treatment resistance (19, 50). Specifically, in patients with PDAC, our work and that of others have shown that elevated Cav-1 expression correlates with worse clinical outcomes (15, 51, 52). We showed that genetic ablation of CAV1 in the pancreata of mice that spontaneously developed pancreatic tumors resulted in longer survival and reduced tumor burden compared to CAV1 wild-type mice, corroborating our patient data that showed elevated Cav-1 expression in pancreatic cancer occurs late in the disease course and predicts poor prognosis for cancer progression (Fig. 2).
Glutamine is an essential amino acid for cancer cells undergoing proliferation, and studies have found that PDAC cells are in effect, “glutamine-addicted” (53, 54). Glutamine is the most depleted amino acid in pancreatic tumor tissue compared to that in adjacent normal tissue (3). Oncogenic KRAS reprograms cells to convert glutamine into the reductive pathway while simultaneously suppressing the oxidative metabolism of glutamine (55). In contrast, non-KRAS-transformed cells are not affected by glutamine depletion (56). We found that Cav-1–depleted PDAC cells were more sensitive to glutamine restriction compared to Cav-1–expressing cells and that albumin could rescue proliferation only in Cav-1–expressing cells, not in Cav-1–deficient cells. These results suggested that Cav-1 plays a crucial role in maintaining glutamine supply for cancer cell growth. We found that Cav-1–depleted PDAC cells were more sensitive to glutamine restriction compared to Cav-1–expressing cells and that albumin could rescue proliferation only in Cav-1–expressing cells, but not in Cav-1–deficient cells. These results suggest that Cav-1 plays a crucial role in maintaining the glutamine supply for growth. In addition, in our metabolite analysis, we found lower levels of citrate in KPC-CAV1fl/fl cells than in KPC cells. One possible explanation for this finding could be related to reduced pyruvate availability due to decreased glycolysis in Cav-1–deficient cells, leading to a higher dependence on glutamine in TCA.
Cav-1 plays a pivotal role in maintaining the integrity of the lipid rafts within the cell membrane. Caveolae are enriched with a variety of transmembrane receptors, including insulin and growth factor receptors (57–59). The activation of these receptors triggers complex signaling pathways that influence various cellular activities such as energy production, protein biosynthesis, and cell proliferation, which aligns with the results of our study. Studies demonstrating that Cav-1 facilitates proper insulin and growth factor receptor signaling implicate that these receptors are actively regulated and modulated by Cav-1. Our study identified changes in metabolic responses based on relative levels of Cav-1. Therefore, it is plausible that the interaction between Cav-1 and these receptors may have a substantial impact on the observed outcomes and underscores the importance of considering these transmembrane receptors as potential key players in our findings. In the future, we will explore the molecular mechanisms by which Cav-1 and these receptors interact in relation to PDAC growth, survival, and progression, thereby enhancing our understanding of the biological processes underpinning our present findings.
Another finding from this study is the mechanistic association between Cav-1 expression and activation of ribosomal translation pathways (through phosphorylated rpS6 expression). The rpS6 protein is a substrate of S6K1 and S6K2 and plays important roles in regulating cell size, protein synthesis, glucose metabolism, and autophagy (60). We found that Cav-1–expressing cells were able to maintain the phosphorylation of rpS6 in nutrient-depleted concentrations in contrast to the Cav-1–deficient cell lines. Furthermore, we showed that inhibiting S6K-mediated phosphorylation of rpS6 resulted in decreased cell proliferation under serum deprivation, whereas induction of S6 phosphorylation (through S6K constitutively active mutants) could rescue growth in nutrient-challenged conditions. Our findings also support the notion that S6 may serve as a biomarker for tumor growth under nutrient-replete conditions. Together, these results indicate previously unknown roles of Cav-1 and CME in tumor metabolism, anabolism, and ribosomal translation to facilitate tumor growth and proliferation.
It is important to note several limitations of our study as well as future directions. First, the absence of blood samples from the 114 patients with PDAC was included in our TMA studies. Evaluation of systemic Cav-1 levels in correlation with metastatic outcomes could provide valuable insights into disease progression and warrant further investigation. Future prospective studies should explore the potential association between blood Cav-1 levels and the occurrence of metastasis. In addition, we were unable to perform a comprehensive pathologic analysis of metastases in our KPC mouse model to quantify the effects of Cav-1 on promoting metastasis in individual organs. In addition, while we have previously linked Cav-1 to cell death and apoptosis in PDAC cells (20), a more thorough understanding of the relationship between Cav-1, cell death pathways, and autophagy should be explored, given the central role of S6 in autophagy. Last, our findings that Cav-1 plays a role in optimal glycolytic capacity and mitochondrial energetics in PDAC also need further investigation, given that both alterations in glycolysis and mitochondrial function can occur in a complex relationship within tumor cells.
In summary, we demonstrated that PDAC is dependent on CME and Cav-1 in fueling pancreatic tumorigenesis and maintaining tumor growth and survival in nutrient-deprived conditions. These compelling results support the rationale for nominating this critical scavenging pathway as a target for therapeutic intervention, either as monotherapy or in combination with other therapies to attenuate tumor growth and progression.
MATERIALS AND METHODS
Sequencing data
Somatic mutation and RNA sequencing (RNA-seq) data for PanGen (NCT02869802) and COMPASS (NCT02750657) samples were generated and processed as described previously (27, 61). Publicly available TCGA, ICGC, and CPTAC-3 data were downloaded from their online data portals (https://portal.gdc.cancer.gov/, https://dcc.icgc.org/, and https://portal.gdc.cancer.gov/, respectively). All normalized gene expression values were log10-transformed before any analysis. RNA-seq batch correction was performed on all RNA-seq samples using ComBat v3.30.1, with default parameters. All somatic mutation data were based on human genome build GRCh37 (hg19).
PDAC molecular subtyping
Classification of samples into the Moffitt basal-like and classical groups was performed using the RNA-seq version of the Moffitt PurIST algorithm (26). PurIST scores (basal-like probability values) were used to stratify patients into basal-like (score > 0.75), classical (score < 0.25), and intermediate (score 0.25 to 0.75) subtype groups (26). Basal-like (n = 25) and classical (n = 25) genes from the original Moffitt subtyping manuscript (62) were used when directly investigating the expression values of Moffitt subtyping genes. Collisson and Bailey subtypes were determined separately using a consensus clustering approach, as previously described (27). Collisson classical (n = 22), exocrine-like (n = 20), and quasi-mesenchymal (n = 20) genes from the original Collisson subtyping paper (28) were used when directly investigating the expression values of Collisson subtyping genes. ADEX (n = 240), immunogenic (n = 370), squamous (n = 1061), and progenitor (n = 268) genes from the original Bailey subtyping paper (29) were used when directory comparing expression values of Bailey subtyping genes. Karasinska subtypes were determined on the basis of the relative expression of glycolytic (GAPDH, ALDOA, PKM, ENO1, TPI1, PGK1, GPI, PGAM1, PFKP, PFKFB3, ENO2, PPP2R5D, PFKM, and PFKFB4) and cholesterogenic (FDPS, FDFT1, DHCR24, EBP, IDI1, MVD, HMGCS1, SQLE, NSDHL, DHCR7, HMGCR, LSS, SC5D, MVK, and HSD1787) genes, as previously described (27).
Cell lines, antibodies, and drugs
The human PDAC cell lines AsPC1, BxPC3, CAPAN-1, CAPAN-2, CFPAC-1, G37, HPAF-II, MIAPaCa-2, PANC-1, and PL-45 were obtained from American Type Culture Collection (Manassas, VA). G37 PDX TDCL was obtained from Dr. J. Trevino. TDCLs were generated as follows (63, 64): The collected tumors were washed three times with phosphate-buffered saline (PBS) and transferred into a petri dish containing the complete Dulbecco’s modified Eagle’s medium (DMEM) media. The tumor was finely minced with a sterile scalpel and transferred into a sterile centrifuge tube with the complete medium containing collagenase P (10 mg/ml; Thermo Fisher Scientific, Waltham, MA). The mixture was incubated at 37°C for 30 min. The tubes were inverted every 3 min to ensure proper mixing. Following two washing cycles in a complete medium, the pellet obtained after centrifugation was suspended in the complete DMEM medium. Cell suspension without tissue debris was transferred into a new sterile 10-cm petri dish. After incubating overnight at 37°C in 5% CO2, the medium was replaced with fresh medium. Cells were trypsinized once confluent. Cells were maintained at 37°C in 5% CO2 in DMEM media supplemented with 10% or 1% FBS as indicated (Millipore-Sigma, St. Louis, MO) and penicillin/streptomycin (100 U/ml; Life Technologies, Grand Island, NY). Cells were cultured for no more than 3 months continuously. Cav-1 primary antibody (N-20) was purchased from Santa Cruz Biotechnology (Santa Cruz, CA). S6 ribosomal protein, phospho-S6 ribosomal protein (Ser235/236), p70 S6 kinase, phospho-p70 S6 kinase (Thr389), and GAPDH antibodies were purchased from Cell Signaling Technology (Danvers, MA). Anti-rabbit immunofluorescent secondary antibodies were purchased from LI-COR Biosciences (Lincoln, NE).
Cav-1 knockdown
For stable Cav-1 knockdown, MIAPaCa-2 and PANC1 cells were transduced with shRNA lentiviral particles (Millipore-Sigma) using hexadimethrine bromide (Sigma-Aldrich) as a transfection agent, according to the manufacturer’s protocol, and stable pools were selected with puromycin (1.0 mg/ml) for at least 7 days.
Expression analysis in RNA-based molecular subtype systems and TCGA dataset
CAV1 RNA expression was validated in three patient-derived sequencing cohorts, PanGen/POG (NCT 02869802 and NCT02155621, respectively), COMPASS (NCT02750657), and ICGC. For comparison of CAV1 expression between molecular subgroups of patient datasets, two-tailed Wilcoxon mean rank sum tests were used, and P values were subjected to Benjamini-Hochberg multiple test correction. For correlation analysis between Cav-1 and phosho-S6 (S235-236) at protein levels, the TCGA RPPA dataset was downloaded from www.cbioportal.org.
Tissue microarray
A TMA was created from 114 patients treated surgically for localized pancreatic cancer at The Ohio State University. The study was approved by The Ohio State University institutional review board (2014C0077), and informed consent was deemed not to be required. All hematoxylin and eosin (H&E) slides of PDAC were reviewed, and the most representative non-necrotic tumor areas were chosen by an experienced pancreas pathologist (W.C.). TMAs were constructed, and all tissue cores were obtained from diagnostic surgical samples (formalin-fixed paraffin-embedded tissue blocks from pancreatectomy specimens). Tissue cores (1.0 mm in diameter) were taken from spatially separate tumor areas in a single donor block from each case using a tissue microarrayer (Chemicon Advanced Tissue Arrayer, Temecula, CA, USA). The cores were arrayed into a recipient block at predetermined coordinates (two cores per patient). H&E sections from donor and recipient paraffin blocks were used to confirm the area of the tumor from which the cores were retrieved. Each case was duplicated using two tissue cores to ensure adequate tissue representation. After immunohistochemical staining, the expression of Cav-1 was scored using the H-score method (65) by an independent pathologist blinded to patient data (W.C.) by determining the average intensity score (0 = none, 1 = weak, 2 = intermediate, 3 = strong) and percent cell staining score (0 to 100%) and summing the scores for a range of 0 to 300. The scores from each of the two cores were averaged.
Histological analysis of pancreas tissues
Pancreatic tissues from the mice were collected at 1 to 4 months of age and fixed in 10% formalin buffer (Thermo Fisher Scientific). After 48 hours of fixation, tissues were processed for H&E staining. For histopathological analysis, H&E slides were scanned and converted into digital images, and the pathologist determined the pancreatic pathology as PanIN (low and high grade) and PDAC (well, moderately, and poorly differentiated).
Mouse models and care
Cav-1 conditional knockout (CAV1flox) mice were generated by Ozgene (Bentley, Australia) using standard homologous recombination cloning techniques (66). Briefly, a CAV1 targeting vector was designed to target exon 3 using the loxP/Cre system. A linearized targeting vector was used in standard homologous recombination techniques to introduce the targeted allele into mouse embryonic stem cells (ESC). The correctly recombined ESCs were injected into blastocytes and implanted into foster females to generate chimeric mice. The tail DNA from F1 offspring of chimeric mice (with Neo/TK) was confirmed by Southern blotting using ScaI digestion and EnP probes, as well as two probes that were located outside of 5′ or 3′ homology arms. The neomycin cassettes were removed using the FLP-FRT system in ESCs from the F1 offspring to generate the CAV1 conditional knockout mouse (CAV1flox). Southern blotting was performed using the ScaI restriction enzyme and EnP probe to confirm the floxed CAV1 mouse (deleted Neo/TK cassette). CAV1flox mice were bred with LSL-KrasG12D/+;LSL-p53R270H/+;Pdx-Cre (KPC) mice using the Targeted Validation Shared Resource at Ohio State University Comprehensive Cancer Center. CAV1flox/flox;KPC mice and CAV1+/+;KPC mice were on a mixed background (FVB/N, 129v/Sv, and C57BL/6). The use of mice was approved by the Institutional Animal Care and Use Committee. Mice were housed under standard husbandry conditions in a vivarium with a 12-hour light/dark cycle. Genotyping analysis was performed on the tails of 17-day-old mice or pancreatic tissues using standard PCR methods.
Generation of CAV1 conditional mice
CAV1flox mice were generated by Ozgene company (Bentley, Australia) using standard homologous recombination cloning techniques (66). Briefly, the CAV1 targeting vector was designed to target exon 3 using the loxP/Cre system. Linearized targeting vector was used for standard homologous recombination techniques to introduce the targeted allele into mouse ESCs. The correctly recombined ESCs were injected into blastocytes and implanted into foster females to generate chimeric mice. The tail DNA from F1 offspring of chimeric mice (with Neo/TK) was confirmed by Southern blots using ScaI digestion and EnP probes as well as two probes that were located outside of 5′ or 3′ homology arms. The neomycin cassettes were removed by the FLP-FRT system in ESCs from the F1 offspring to generate the CAV1 conditional knockout mouse (CAV1flox). Using the ScaI restriction enzyme and EnP probe, Southern blotting was performed to confirm the floxed CAV1 mouse (deleted Neo/TK cassette).
Immunoblotting
For assessment of Cav-1 expression, octyl-β-d-glucopyranoside (Millipore-Sigma; St. Louis, MO) was added at 60 mM final concentration to radioimmunoprecipitation assay buffer (Thermo Fisher Scientific, Waltham, MA) containing protease and phosphatase inhibitor cocktails (Roche, Basel, Switzerland). Protein concentration was determined with a DC Protein Assay Kit (Bio-Rad, Hercules, CA). Proteins were resolved by SDS–polyacrylamide gel electrophoresis and transferred to nitrocellulose membranes. Membranes were incubated in 5% bovine serum albumin (BSA) in a TBS-Tween blocking buffer for 1 hour at room temperature. Primary antibodies were allowed to bind overnight at 4°C and used at a dilution of 1:500 to 1:1000. Cav-1 primary antibody (N-20) was purchased from Santa Cruz Biotechnology (Santa Cruz, CA). S6 ribosomal protein, phospho-S6 ribosomal protein (Ser235/236), p70 S6 kinase, phospho-p70 S6 kinase (Thr389), phospho-AKT (T308), phospho-mTOR (Ser2448), mTOR, phospho-4Ebp1 (Thr37/46), and GAPDH antibodies were purchased from Cell Signaling Technology (Danvers, MA). Anti-rabbit immunofluorescent secondary antibodies were purchased from LI-COR Biosciences (Lincoln, NE). After washing in TBS-Tween three times for 10 min each, the membranes were incubated with immunofluorescent secondary antibodies at a 1:5000 dilution for 1 hour at room temperature. Membranes were washed with TBS-Tween before imaging via LI-COR Odyssey CLx Imaging System (Lincoln, NE).
Immunofluorescence
Cells were plated on coverslips overnight. Two hours before fixation, cells were pulsed with 0.05% of HSA. To remove membrane-bound albumin, two acid/salt washes were performed with 0.1 M glycine and 0.1 M NaCl (pH 3.02) on ice for 2 min each, followed by two washes with PBS to remove membrane-bound albumin. Cells were then fixed with 4% paraformaldehyde for 10 min at room temperature and washed with PBS two times for 5 min each. Cells were incubated in 1% Triton X-100 for 10 min to permeabilize, and then washed twice with PBS before blocking with 3% BSA in PBS overnight at 4°C. In a humidified chamber, primary antibodies diluted 1:50 in blocking buffer were added and incubated for 1 hour at 4°C, followed by three 10-min rinses with blocking buffer. A secondary antibody (conjugated to Alexa Fluor 488 or Alexa Flour 594, Invitrogen, Waltham, MA) was added along with DAPI for 1 hour at room temperature. Cells were then rinsed, and coverslips were mounted onto slides and then sealed. Cells were then imaged with a confocal microscope as previously described (64).
Cell proliferation using IncuCyte
Cells (500 to 2000 per well) were seeded in 96-well plates and treated according to schedule. Cell confluence as a measure of cell growth over time was monitored every 2 to 6 hours for up to 3 to 7 days using the IncuCyte Live-Cell Imaging System (Essen Biosciences).
Seahorse assay
Following procedures previously described in (67), OCR rates were measured with the Seahorse Biosciences Extracellular Flux Analyzer (XFe96). A total of 5 × 104 cells were seeded per well in normal growth media (RPMI 1640 with 10% FBS) and allowed to attach for 24 to 30 hours at 38.5°C and 8.5% CO2 before the study. Basal measurements were collected following 4 hours of incubation with either 10 or 1% FBS supplementation. SRC (maximal respiratory capacity-basal respiration rate) and total reserve capacity were measured by injecting the mitochondrial uncoupler FCCP (1.5 μM) and Complex III inhibitor antimycin (20 μM) from the XF24 ports as indicated. The extracellular acidification rate was monitored using a Seahorse XF96 Flux Analyzer (Seahorse Bioscience), according to the manufacturer’s instructions. Before the day of the assay [as previously described in (68)], the cartridge sensor was hydrated overnight with Seahorse Bioscience XF96 Calibration Buffer at 37°C without CO2. KPC and MP2 cells were seeded in a 96-well plate at a density of 1 × 104 cells per well, and the growth medium was replaced with serum-free DMEM/F12 medium lacking sodium bicarbonate. For assessment of the real-time glycolytic rate, an indicator of net proton loss during glycolysis, cells were incubated with unbuffered medium followed by a sequential injection of 10 mM glucose, 1 μM oligomycin (Agilent), and 80 mM 2-DG (Agilent).
Metabolic profiling by LC/ESI-MS
The cells (1 × 106) were grown in six-well plates for direct extraction from each well. Fresh media were added to the cells 24 hours before metabolite extraction. The six-well plates were placed on ice and washed with ice-cold PBS (pH 7.4). Next, 400 μl of ice-cold HPLC-grade methanol (Thermo Fisher Scientific, #A454-4) was added. Cells were scraped off the plate and transferred to a 1.5-ml Eppendorf tube. For the separation of polar metabolites from nonpolar metabolites, 400 μl of HPLC-grade chloroform was added (Thermo Fisher Scientific, C607-4). The samples were vortexed every 5 min for 15 to 30 min. The top aqueous layer was then transferred to a new tube containing glycolytic intermediates. The contents of both the tubes were dried using a vacuum concentrator. The samples were stored at −80°C until analysis. MS measurements were performed according to the methods described in the instrument documentation section. LC/ESI-MS analysis of the relative quantities of glycolytic intermediates was performed for KPC and KPC-CAV1 cells (69).
Statistical analysis
Fisher’s exact test was used to test differences in CNV frequency between resectable and metastatic patient groups. Log-rank tests were used to calculate P values in the Kaplan-Meier analysis. Wilcoxon mean rank sum tests were used for a two-group comparison of continuous variables. Gene set enrichment analysis was performed using two-tailed hypergeometric tests. Spearman’s correlation tests were used to correlate CAV1 expression versus other genes. All group comparison tests were two-tailed. All P values were subjected to Benjamini-Hochberg multiple test correction. All analyses were performed using R v3.6.3.
For cell and animal experiments, data are presented as means ± SEM for proliferation assays, mass spectrometry data, and tumor growth experiments. Group comparisons of the percentage change in tumor volume were performed at individual time points. Spearman’s rank correlation was used for correlation analysis. Kaplan-Meier survival analysis was performed. Statistical comparisons were made between the control and experimental conditions using the unpaired two-tailed Student’s t test, with significance assessed at P < 0.05. GraphPad Prism (GraphPad Software Inc.) was used for the statistical analyses.
Acknowledgments
We gratefully acknowledge the participation of patients and their families.
Funding: This work was supported by NIH grant R01CA246553 (to T.M.W.), NIH grant P30 CA016058 (to T.M.W.), NIH grant KL2 TR003108 (to A.W.), and RSNA Resident Research Grant (to A.W./T.M.W.). This research was supported through philanthropic donations received through the BC Cancer Foundation, as well as funding provided by the Terry Fox Research Institute (Project 1078), Ontario Institute for Cancer Research (PanCuRx Translational Research Initiative), Pancreatic Cancer Canada, Genome British Columbia (project B20POG), and VGH/UBC Hospital Foundation. RNA-seq data from the COMPASS trial were generated at the Ontario Institute for Cancer Research (PanCuRx Translational Research Initiative) through funding provided by the Government of Ontario, the Wallace McCain Centre for Pancreatic Cancer supported by the Princess Margaret Cancer Foundation, the Terry Fox Research Institute, the Canadian Cancer Society Research Institute, Pancreatic Cancer Canada, and the Canadian Friends of the Hebrew University (A. U. Soyka). The results published here are, in part, based on data generated by The Cancer Genome Atlas managed by the NCI and NHGRI (http://cancergenome.nih.gov), as well as data generated by the International Cancer Genome Consortium (https://icgc.org/) and the Clinical Proteomic Tumor Analysis Consortium (NCI/NIH).
Author contributions: Conceptualization: A.R.W., S.B., M.H., S.C.-G., R.R., D.-L.S., N.D., and T.M.W. Methodology: A.R.W., T.C., S.B., M.H., S.C.-G., R.R., P.F., J.T.T., and T.M.W. Investigation: A.R.W., T.C., S.B., S.C.-G., V.C.-A., M.H., R.R., R.P., D.-L.S., and T.M.W. Visualization: A.R.W., T.C., M.H., R.R., A.W., and T.M.W. Validation: A.R.W., T.C., S.B., M.H., S.C.-G., D.-L.S., V.C.-A., and T.M.W. Resources: A.R.W., T.C., S.B., M.H., D.F.S., W.C., J.M.K., M.L., P.F., D.J.R., and T.M.W. Data curation: A.R.W., T.C., S.B., and A.W. Formal analysis: A.R.W., T.C., S.B., M.H., R.R., A.W., J.T.T. and T.M.W. Project administration: A.R.W., M.H., and T.M.W. Software: T.C., J.T.T., and A.W. Writing—original draft: A.R.W., T.C., S.C.-G., J.T.T., and T.M.W. Writing—review and editing: A.R.W., T.C., V.C.-A., T.C.T., M.H., R.R., D.S., N.D., J.T.T. and T.M.W. Supervision: T.M.W. Funding acquisition: N.D. and T.M.W.
Competing interests: The other authors declare that they have no competing interests.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials.
Supplementary Materials
This PDF file includes:
Supplementary Text
Figs. S1 to S5
Table S1
REFERENCES AND NOTES
- 1.Koong A. C., Mehta V. K., Le Q. T., Fisher G. A., Terris D. J., Brown J. M., Bastidas A. J., Vierra M., Pancreatic tumors show high levels of hypoxia. Int. J. Radiat. Oncol. Biol. Phys. 48, 919–922 (2000). [DOI] [PubMed] [Google Scholar]
- 2.Finicle B. T., Jayashankar V., Edinger A. L., Nutrient scavenging in cancer. Nat. Rev. Cancer 18, 619–633 (2018). [DOI] [PubMed] [Google Scholar]
- 3.Commisso C., Davidson S. M., Soydaner-Azeloglu R. G., Parker S. J., Kamphorst J. J., Hackett S., Grabocka E., Nofal M., Drebin J. A., Thompson C. B., Rabinowitz J. D., Metallo C. M., Vander Heiden M. G., Bar-Sagi D., Macropinocytosis of protein is an amino acid supply route in Ras-transformed cells. Nature 497, 633–637 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yang S., Wang X., Contino G., Liesa M., Sahin E., Ying H., Bause A., Li Y., Stommel J. M., Dell'antonio G., Mautner J., Tonon G., Haigis M., Shirihai O. S., Doglioni C., Bardeesy N., Kimmelman A. C., Pancreatic cancers require autophagy for tumor growth. Genes Dev. 25, 717–729 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nwosu Z. C., Ebert M. P., Dooley S., Meyer C., Caveolin-1 in the regulation of cell metabolism: A cancer perspective. Mol. Cancer 15, 71 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Razani B., Woodman S. E., Lisanti M. P., Caveolae: From cell biology to animal physiology. Pharmacol. Rev. 54, 431–467 (2002). [DOI] [PubMed] [Google Scholar]
- 7.Williams T. M., Lisanti M. P., The Caveolin genes: From cell biology to medicine. Ann. Med. 36, 584–595 (2004). [DOI] [PubMed] [Google Scholar]
- 8.Cohen A. W., Hnasko R., Schubert W., Lisanti M. P., Role of caveolae and caveolins in health and disease. Physiol. Rev. 84, 1341–1379 (2004). [DOI] [PubMed] [Google Scholar]
- 9.Zhan P., Shen X. K., Qian Q., Wang Q., Zhu J. P., Zhang Y., Xie H. Y., Xu C. H., Hao K. K., Hu W., Xia N., Lu G. J., Yu L. K., Expression of caveolin-1 is correlated with disease stage and survival in lung adenocarcinomas. Oncol. Rep. 27, 1072–1078 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yoo S. H., Park Y. S., Kim H. R., Sung S. W., Kim J. H., Shim Y. S., Lee S. D., Choi Y. L., Kim M. K., Chung D. H., Expression of caveolin-1 is associated with poor prognosis of patients with squamous cell carcinoma of the lung. Lung Cancer 42, 195–202 (2003). [DOI] [PubMed] [Google Scholar]
- 11.Suzuoki M., Miyamoto M., Kato K., Hiraoka K., Oshikiri T., Nakakubo Y., Fukunaga A., Shichinohe T., Shinohara T., Itoh T., Kondo S., Katoh H., Impact of caveolin-1 expression on prognosis of pancreatic ductal adenocarcinoma. Br. J. Cancer 87, 1140–1144 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kato K., Hida Y., Miyamoto M., Hashida H., Shinohara T., Itoh T., Okushiba S., Kondo S., Katoh H., Overexpression of caveolin-1 in esophageal squamous cell carcinoma correlates with lymph node metastasis and pathologic stage. Cancer 94, 929–933 (2002). [PubMed] [Google Scholar]
- 13.Sloan E. K., Ciocca D. R., Pouliot N., Natoli A., Restall C., Henderson M. A., Fanelli M. A., Cuello-Carrion F. D., Gago F. E., Anderson R. L., Stromal cell expression of caveolin-1 predicts outcome in breast cancer. Am. J. Pathol. 174, 2035–2043 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Horiguchi A., Asano T., Asakuma J., Sumitomo M., Hayakawa M., Impact of caveolin-1 expression on clinicopathological parameters in renal cell carcinoma. J. Urol. 172, 718–722 (2004). [DOI] [PubMed] [Google Scholar]
- 15.Tanase C. P., Dima S., Mihai M., Raducan E., Nicolescu M. I., Albulescu L., Voiculescu B., Dumitrascu T., Cruceru L. M., Leabu M., Popescu I., Hinescu M. E., Caveolin-1 overexpression correlates with tumour progression markers in pancreatic ductal adenocarcinoma. J. Mol. Histol. 40, 23–29 (2009). [DOI] [PubMed] [Google Scholar]
- 16.Salem A. F., Bonuccelli G., Bevilacqua G., Arafat H., Pestell R. G., Sotgia F., Lisanti M. P., Caveolin-1 promotes pancreatic cancer cell differentiation and restores membranous E-cadherin via suppression of the epithelial-mesenchymal transition. Cell Cycle 10, 3692–3700 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Huang C., Qiu Z., Wang L., Peng Z., Jia Z., Logsdon C. D., Le X., Wei D., Huang S., Xie K., A novel FoxM1-caveolin signaling pathway promotes pancreatic cancer invasion and metastasis. Cancer Res. 72, 655–665 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hehlgans S., Eke I., Storch K., Haase M., Baretton G. B., Cordes N., Caveolin-1 mediated radioresistance of 3D grown pancreatic cancer cells. Radiother. Oncol. 92, 362–370 (2009). [DOI] [PubMed] [Google Scholar]
- 19.Cordes N., Frick S., Brunner T. B., Pilarsky C., Grutzmann R., Sipos B., Kloppel G., McKenna W. G., Bernhard E. J., Human pancreatic tumor cells are sensitized to ionizing radiation by knockdown of caveolin-1. Oncogene 26, 6851–6862 (2007). [DOI] [PubMed] [Google Scholar]
- 20.Chatterjee M., Ben-Josef E., Thomas D. G., Morgan M. A., Zalupski M. M., Khan G., Andrew Robinson C., Griffith K. A., Chen C.-S., Ludwig T., Bekaii-Saab T., Chakravarti A., Williams T. M., Caveolin-1 is associated with tumor progression and confers a multi-modality resistance phenotype in pancreatic cancer. Sci. Rep. 5, 10867 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Busija A. R., Patel H. H., Insel P. A., Caveolins and cavins in the trafficking, maturation, and degradation of caveolae: Implications for cell physiology. Am. J. Physiol. Cell Physiol. 312, C459–C477 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tahir S. A., Park S., Thompson T. C., Caveolin-1 regulates VEGF-stimulated angiogenic activities in prostate cancer and endothelial cells. Cancer Biol. Ther. 8, 2286–2296 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sotgia F., Martinez-Outschoorn U. E., Howell A., Pestell R. G., Pavlides S., Lisanti M. P., Caveolin-1 and cancer metabolism in the tumor microenvironment: Markers, models, and mechanisms. Annu. Rev. Pathol. 7, 423–467 (2012). [DOI] [PubMed] [Google Scholar]
- 24.Joshi B., Strugnell S. S., Goetz J. G., Kojic L. D., Cox M. E., Griffith O. L., Chan S. K., Jones S. J., Leung S.-P., Masoudi H., Leung S., Wiseman S. M., Nabi I. R., Phosphorylated caveolin-1 regulates Rho/ROCK-dependent focal adhesion dynamics and tumor cell migration and invasion. Cancer Res. 68, 8210–8220 (2008). [DOI] [PubMed] [Google Scholar]
- 25.Schubert W., Frank P. G., Razani B., Park D. S., Chow C. W., Lisanti M. P., Caveolae-deficient endothelial cells show defects in the uptake and transport of albumin in vivo. J. Biol. Chem. 276, 48619–48622 (2001). [DOI] [PubMed] [Google Scholar]
- 26.Rashid N. U., Peng X. L., Jin C., Moffitt R. A., Volmar K. E., Belt B. A., Panni R. Z., Nywening T. M., Herrera S. G., Moore K. J., Hennessey S. G., Morrison A. B., Kawalerski R., Nayyar A., Chang A. E., Schmidt B., Kim H. J., Linehan D. C., Yeh J. J., Purity independent subtyping of tumors (PurIST), A clinically robust, single-sample classifier for tumor subtyping in pancreatic cancer. Clin. Cancer Res. 26, 82–92 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Karasinska J. M., Topham J. T., Kalloger S. E., Jang G. H., Denroche R. E., Culibrk L., Williamson L. M., Wong H. L., Lee M. K. C., O'Kane G. M., Moore R. A., Mungall A. J., Moore M. J., Warren C., Metcalfe A., Notta F., Knox J. J., Gallinger S., Laskin J., Marra M. A., Jones S. J. M., Renouf D. J., Schaeffer D. F., Altered gene expression along the glycolysis-cholesterol synthesis axis is associated with outcome in pancreatic cancer. Clin. Cancer Res. 26, 135–146 (2020). [DOI] [PubMed] [Google Scholar]
- 28.Collisson E. A., Sadanandam A., Olson P., Gibb W. J., Truitt M., Gu S., Cooc J., Weinkle J., Kim G. E., Jakkula L., Feiler H. S., Ko A. H., Olshen A. B., Danenberg K. L., Tempero M. A., Spellman P. T., Hanahan D., Gray J. W., Subtypes of pancreatic ductal adenocarcinoma and their differing responses to therapy. Nat. Med. 17, 500–503 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bailey P., Chang D. K., Nones K., Johns A. L., Patch A.-M., Gingras M.-C., Miller D. K., Christ A. N., Bruxner T. J. C., Quinn M. C., Nourse C., Murtaugh L. C., Harliwong I., Idrisoglu S., Manning S., Nourbakhsh E., Wani S., Fink L., Holmes O., Chin V., Anderson M. J., Kazakoff S., Leonard C., Newell F., Waddell N., Wood S., Xu Q., Wilson P. J., Cloonan N., Kassahn K. S., Taylor D., Quek K., Robertson A., Pantano L., Mincarelli L., Sanchez L. N., Evers L., Wu J., Pinese M., Cowley M. J., Jones M. D., Colvin E. K., Nagrial A. M., Humphrey E. S., Chantrill L. A., Mawson A., Humphris J., Chou A., Pajic M., Scarlett C. J., Pinho A. V., Giry-Laterriere M., Rooman I., Samra J. S., Kench J. G., Lovell J. A., Merrett N. D., Toon C. W., Epari K., Nguyen N. Q., Barbour A., Zeps N., Moran-Jones K., Jamieson N. B., Graham J. S., Duthie F., Oien K., Hair J., Grützmann R., Maitra A., Iacobuzio-Donahue C. A., Wolfgang C. L., Morgan R. A., Lawlor R. T., Corbo V., Bassi C., Rusev B., Capelli P., Salvia R., Tortora G., Mukhopadhyay D., Petersen G. M.; Australian Pancreatic Cancer Genome Initiative, Munzy D. M., Fisher W. E., Karim S. A., Eshleman J. R., Hruban R. H., Pilarsky C., Morton J. P., Sansom O. J., Scarpa A., Musgrove E. A., Bailey U.-M. H., Hofmann O., Sutherland R. L., Wheeler D. A., Gill A. J., Gibbs R. A., Pearson J. V., Waddell N., Biankin A. V., Grimmond S. M., Genomic analyses identify molecular subtypes of pancreatic cancer. Nature 531, 47–52 (2016). [DOI] [PubMed] [Google Scholar]
- 30.Liu P., Rudick M., Anderson R. G., Multiple functions of caveolin-1. J. Biol. Chem. 277, 41295–41298 (2002). [DOI] [PubMed] [Google Scholar]
- 31.Tahir S. A., Frolov A., Hayes T. G., Mims M. P., Miles B. J., Lerner S. P., Wheeler T. M., Ayala G., Thompson T. C., Kadmon D., Preoperative serum caveolin-1 as a prognostic marker for recurrence in a radical prostatectomy cohort. Clin. Cancer Res. 12, 4872–4875 (2006). [DOI] [PubMed] [Google Scholar]
- 32.Hingorani S. R., Wang L., Multani A. S., Combs C., Deramaudt T. B., Hruban R. H., Rustgi A. K., Chang S., Tuveson D. A., Trp53R172H and KrasG12D cooperate to promote chromosomal instability and widely metastatic pancreatic ductal adenocarcinoma in mice. Cancer Cell 7, 469–483 (2005). [DOI] [PubMed] [Google Scholar]
- 33.Shakya R., Reid L. J., Reczek C. R., Cole F., Egli D., Lin C. S., deRooij D. G., Hirsch S., Ravi K., Hicks J. B., Szabolcs M., Jasin M., Baer R., Ludwig T., BRCA1 tumor suppression depends on BRCT phosphoprotein binding, but not its E3 ligase activity. Science 334, 525–528 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kamphorst J. J., Nofal M., Commisso C., Hackett S. R., Lu W., Grabocka E., Vander Heiden M. G., Miller G., Drebin J. A., Bar-Sagi D., Thompson C. B., Rabinowitz J. D., Human pancreatic cancer tumors are nutrient poor and tumor cells actively scavenge extracellular protein. Cancer Res. 75, 544–553 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fendt S.-M., Bell E. L., Keibler M. A., Olenchock B. A., Mayers J. R., Wasylenko T. M., Vokes N. I., Guarente L., Heiden M. G. V., Stephanopoulos G., Reductive glutamine metabolism is a function of the α-ketoglutarate to citrate ratio in cells. Nat. Commun. 4, 2236 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chatterjee M., Ben-Josef E., Robb R., Vedaie M., Seum S., Thirumoorthy K., Palanichamy K., Harbrecht M., Chakravarti A., Williams T. M., Caveolae-mediated endocytosis is critical for albumin cellular uptake and response to albumin-bound chemotherapy. Cancer Res. 77, 5925–5937 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Meyuhas O., Ribosomal protein S6 phosphorylation: Four decades of research. Int. Rev. Cell Mol. Biol. 320, 41–73 (2015). [DOI] [PubMed] [Google Scholar]
- 38.Schalm S. S., Blenis J., Identification of a conserved motif required for mTOR signaling. Curr. Biol. 12, 632–639 (2002). [DOI] [PubMed] [Google Scholar]
- 39.Lee-Fruman K. K., Kuo C. J., Lippincott J., Terada N., Blenis J., Characterization of S6K2, a novel kinase homologous to S6K1. Oncogene 18, 5108–5114 (1999). [DOI] [PubMed] [Google Scholar]
- 40.Davidson S. M., Jonas O., Keibler M. A., Hou H. W., Luengo A., Mayers J. R., Wyckoff J., del Rosario A. M., Whitman M., Chin C. R., Condon K. J., Lammers A., Kellersberger K. A., Stall B. K., Stephanopoulos G., Bar-Sagi D., Han J., Rabinowitz J. D., Cima M. J., Langer R., vander Heiden M. G., Direct evidence for cancer-cell-autonomous extracellular protein catabolism in pancreatic tumors. Nat. Med. 23, 235–241 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Williams T. M., Lisanti M. P., Caveolin-1 in oncogenic transformation, cancer, and metastasis. Am. J. Physiol. Cell Physiol. 288, C494–C506 (2005). [DOI] [PubMed] [Google Scholar]
- 42.Fielding C. J., Bist A., Fielding P. E., Caveolin mRNA levels are up-regulated by free cholesterol and down-regulated by oxysterols in fibroblast monolayers. Proc. Natl. Acad. Sci. U.S.A. 94, 3753–3758 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jung S., Kim S.-O., Cho K.-A., Song S. H., Kang T. W., Park K., Kwon D., Loss of Caveolin 1 is associated with the expression of Aquaporin 1 and bladder dysfunction in mice. Int. Neurourol. J. 19, 34–38 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Parker S., Peterkin H. S., Baylis H. A., Muscular dystrophy associated mutations in caveolin-1 induce neurotransmission and locomotion defects in Caenorhabditis elegans. Invert. Neurosci. 7, 157–164 (2007). [DOI] [PubMed] [Google Scholar]
- 45.Zhang C., Huang H., Zhang J., Wu Q., Chen X., Huang T., Li W., Liu Y., Zhang J., Caveolin-1 promotes invasion and metastasis by upregulating Pofut1 expression in mouse hepatocellular carcinoma. Cell Death Dis. 10, 477 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 46.Han F., Zhang L., Zhou Y., Yi X., Caveolin-1 regulates cell apoptosis and invasion ability in paclitaxel-induced multidrug-resistant A549 lung cancer cells. Int. J. Clin. Exp. Pathol. 8, 8937–8947 (2015). [PMC free article] [PubMed] [Google Scholar]
- 47.C. Boscher, I. R. Nabi, CAVEOLIN-1: Role in cell signaling, in Caveolins and Caveolae: Roles in Signaling and Disease Mechanisms, J.-F. Jasmin, P. G. Frank, M. P. Lisanti, Eds. (Springer US, 2012), pp. 29–50. [Google Scholar]
- 48.Frank P. G., Pavlides S., Cheung M. W.-C., Daumer K., Lisanti M. P., Role of caveolin-1 in the regulation of lipoprotein metabolism. Am. J. Physiol. Cell Physiol. 295, C242–C248 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nah J., Yoo S.-M., Jung S., Jeong E. I., Park M., Kaang B.-K., Jung Y.-K., Phosphorylated CAV1 activates autophagy through an interaction with BECN1 under oxidative stress. Cell Death Dis. 8, e2822 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tirado O. M., MacCarthy C. M., Fatima N., Villar J., Mateo-Lozano S., Notario V., Caveolin-1 promotes resistance to chemotherapy-induced apoptosis in Ewing's sarcoma cells by modulating PKCalpha phosphorylation. Int. J. Cancer 126, 426–436 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Witkiewicz A. K., Nguyen K. H., Dasgupta A., Kennedy E. P., Yeo C. J., Lisanti M. P., Brody J. R., Co-expression of fatty acid synthase and caveolin-1 in pancreatic ductal adenocarcinoma: Implications for tumor progression and clinical outcome. Cell Cycle 7, 3021–3025 (2008). [DOI] [PubMed] [Google Scholar]
- 52.Demirci N. S., Dogan M., Erdem G. U., Kacar S., Turhan T., Kilickap S., Cigirgan L. C., Kayacetin E., Bozkaya Y., Zengin N., Is plasma caveolin-1 level a prognostic biomarker in metastatic pancreatic cancer? Saudi J. Gastroenterol. 23, 183–189 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wise D. R., Thompson C. B., Glutamine addiction: A new therapeutic target in cancer. Trends Biochem. Sci. 35, 427–433 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Blum R., Kloog Y., Metabolism addiction in pancreatic cancer. Cell Death Dis. 5, e1065 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ying H., Kimmelman A. C., Lyssiotis C. A., Hua S., Chu G. C., Fletcher-Sananikone E., Locasale J. W., Son J., Zhang H., Coloff J. L., Yan H., Wang W., Chen S., Viale A., Zheng H., J.-h. Paik, Lim C., Guimaraes A. R., Martin E. S., Chang J., Hezel A. F., Perry S. R., Hu J., Gan B., Xiao Y., Asara J. M., Weissleder R., Wang Y. A., Chin L., Cantley L. C., DePinho R. A., Oncogenic Kras maintains pancreatic tumors through regulation of anabolic glucose metabolism. Cell 149, 656–670 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yang S., Hwang S., Kim M., Seo S. B., Lee J.-H., Jeong S. M., Mitochondrial glutamine metabolism via GOT2 supports pancreatic cancer growth through senescence inhibition. Cell Death Dis. 9, 55 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.de Laurentiis A., Donovan L., Arcaro A., Lipid rafts and caveolae in signaling by growth factor receptors. Open Biochem. J. 1, 12–32 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Méndez-Giménez L., Rodríguez A., Balaguer I., Frühbeck G., Role of aquaglyceroporins and caveolins in energy and metabolic homeostasis. Mol. Cell. Endocrinol. 397, 78–92 (2014). [DOI] [PubMed] [Google Scholar]
- 59.Palacios-Ortega S., Varela-Guruceaga M., Milagro F. I., Martinez J. A., de Miguel C., Expression of Caveolin 1 is enhanced by DNA demethylation during adipocyte differentiation. status of insulin signaling. PLOS ONE 9, e95100 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ruvinsky I., Meyuhas O., Ribosomal protein S6 phosphorylation: From protein synthesis to cell size. Trends Biochem. Sci. 31, 342–348 (2006). [DOI] [PubMed] [Google Scholar]
- 61.Aung K. L., Fischer S. E., Denroche R. E., Jang G. H., Dodd A., Creighton S., Southwood B., Liang S. B., Chadwick D., Zhang A., O'Kane G. M., Albaba H., Moura S., Grant R. C., Miller J. K., Mbabaali F., Pasternack D., Lungu I. M., Bartlett J. M. S., Ghai S., Lemire M., Holter S., Connor A. A., Moffitt R. A., Yeh J. J., Timms L., Krzyzanowski P. M., Dhani N., Hedley D., Notta F., Wilson J. M., Moore M. J., Gallinger S., Knox J. J., Genomics-driven precision medicine for advanced pancreatic Cancer: Early results from the COMPASS trial. Clin. Cancer Res. 24, 1344–1354 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Moffitt R. A., Marayati R., Flate E. L., Volmar K. E., Herrera Loeza S. G., Hoadley K. A., Rashid N. U., Williams L. A., Eaton S. C., Chung A. H., Smyla J. K., Anderson J. M., Kim H. J., Bentrem D. J., Talamonti M. S., Iacobuzio-Donahue C. A., Hollingsworth M. A., Yeh J. J., Virtual microdissection identifies distinct tumor- and stroma-specific subtypes of pancreatic ductal adenocarcinoma. Nat. Genet. 47, 1168–1178 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Torres M. P., Rachagani S., Souchek J. J., Mallya K., Johansson S. L., Batra S. K., Novel pancreatic cancer cell lines derived from genetically engineered mouse models of spontaneous pancreatic adenocarcinoma: Applications in diagnosis and therapy. PLOS ONE 8, e80580 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wolfe A. R., Robb R., Hegazi A., Abushahin L., Yang L., Shyu D. L., Trevino J. G., Cruz-Monserrate Z., Jacob J. R., Palanichamy K., Chakravarti A., Williams T. M., Altered gemcitabine and nab-paclitaxel scheduling improves therapeutic efficacy compared with standard concurrent treatment in preclinical models of pancreatic cancer. Clin. Cancer Res. 27, 554–565 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Fedchenko N., Reifenrath J., Different approaches for interpretation and reporting of immunohistochemistry analysis results in the bone tissue - a review. Diagn. Pathol. 9, 221 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Sharan S. K., Thomason L. C., Kuznetsov S. G., Court D. L., Recombineering: A homologous recombination-based method of genetic engineering. Nat. Protoc. 4, 206–223 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Strohecker A. M., Guo J. Y., Karsli-Uzunbas G., Price S. M., Chen G. J., Mathew R., McMahon M., White E., Autophagy sustains mitochondrial glutamine metabolism and growth of BrafV600E-driven lung tumors. Cancer Discov. 3, 1272–1285 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Jiang S. H., Dong F. Y., Da L. T., Yang X. M., Wang X. X., Weng J. Y., Feng L., Zhu L. L., Zhang Y. L., Zhang Z. G., Sun Y. W., Li J., Xu M. J., Ikarugamycin inhibits pancreatic cancer cell glycolysis by targeting hexokinase 2. FASEB J. 34, 3943–3955 (2020). [DOI] [PubMed] [Google Scholar]
- 69.TeSlaa T., Teitell M. A., Techniques to monitor glycolysis. Methods Enzymol. 542, 91–114 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Text
Figs. S1 to S5
Table S1