Abstract
People living with human immunodeficiency virus (HIV) receiving integrase strand transfer inhibitors (INSTIs) have been reported to experience virological failure in the absence of resistance mutations in integrase. To elucidate INSTI resistance mechanisms, we propagated HIV-1 in the presence of escalating concentrations of the INSTI dolutegravir. HIV-1 became resistant to dolutegravir by sequentially acquiring mutations in the envelope glycoprotein (Env) and the nucleocapsid protein. The selected Env mutations enhance the ability of the virus to spread via cell-cell transfer, thereby increasing the multiplicity of infection (MOI). While the selected Env mutations confer broad resistance to multiple classes of antiretrovirals, the fold resistance is ~2 logs higher for INSTIs than for other classes of drugs. We demonstrate that INSTIs are more readily overwhelmed by high MOI than other classes of antiretrovirals. Our findings advance the understanding of how HIV-1 can evolve resistance to antiretrovirals, including the potent INSTIs, in the absence of drug-target gene mutations.
Mutations in the envelope glycoprotein and nucleocapsid confer HIV-1 resistance to integrase strand transfer inhibitors.
INTRODUCTION
Six classes of antiretrovirals (ARVs) have been approved for clinical use by the US Food and Drug Administration: nucleoside reverse transcriptase (RT) inhibitors (NRTIs), nonnucleoside RT inhibitors (NNRTIs), integrase strand transfer inhibitors (INSTIs), protease inhibitors (PIs), entry inhibitors, and a recently approved capsid inhibitor, lenacapavir (LEN) (1, 2). Combination antiretroviral therapy (cART) has markedly reduced human immunodeficiency virus (HIV)–associated morbidity and mortality. However, resistance to ARVs does arise in some people living with HIV (PLWH), often associated with poor adherence, use of suboptimal drug regimens, and/or lack of viral load monitoring, particularly in poorly resourced areas (3). In most cases, drug resistance is caused by mutations in the genes targeted by the drugs, often by interfering with the interaction between the drug and the viral target (3). Thus, in the clinical setting, drug resistance monitoring is largely focused on drug-target genes. Recently approved ARVs have been developed with the aim of overcoming resistant variants observed in the clinic. For example, second-generation INSTIs, such as dolutegravir (DTG) and bictegravir (BIC), show some efficacy against IN mutants that are resistant to first-generation INSTIs like raltegravir (RAL) (4). These second-generation INSTIs also exhibit higher genetic barriers to resistance compared to the first-generation INSTIs and RT inhibitors (5). At present, regimens containing DTG are therefore recommended as the preferred first-line regimen for most PLWH (6).
Retroviral integration requires two enzymatic reactions catalyzed by IN: 3′-end processing, during which the enzyme cleaves two nucleotides from the 3′ ends of the newly synthesized linear viral DNA, and DNA strand transfer, which entails the insertion of the viral DNA ends into host cell target DNA. The integration reaction takes place in a macromolecular complex known as the intasome, which comprises an IN multimer and the two viral DNA ends (4). INSTIs inhibit the strand transfer reaction by binding IN and the viral DNA ends in the intasome and chelating the Mg++ ions required for IN catalytic activity (4). Five INSTIs are currently approved for clinical use: two “first-generation” INSTIs, RAL and elvitegravir (EVG), and three “second-generation” INSTIs, DTG, BIC, and cabotegravir (CAB).
Despite the predominant role of drug-target gene mutations in HIV-1 drug resistance, mutations outside drug-target genes can contribute to drug resistance. Particularly in the case of PIs and INSTIs, some PLWH experience virological failure in the absence of mutations in the target genes (7–11). Mutations in Gag and the envelope glycoprotein (Env) have been implicated in PI resistance (12, 13). In vitro studies have reported that mutations in the 3′polypurine tract (3′PPT) reduce the susceptibility of HIV-1 to INSTIs (14–16). 3′PPT mutations may lead to the accumulation of unintegrated 1-LTR circles that can support the expression of viral proteins (14, 16) particularly in cell lines that express HTLV-1 Tax (14). Wijting et al. (11) reported a distinct set of mutations in the 3′PPT from a patient failing DTG monotherapy in the absence of INSTI resistance mutations in IN. However, in other studies, these in vivo–derived 3′PPT mutations were found not to confer resistance to INSTIs in vitro (17). It is therefore still unclear whether, or to what extent, 3′PPT mutations contribute to INSTI resistance in vivo. Nevertheless, as more potent inhibitors with higher genetic barriers to resistance are developed, unconventional drug resistance pathways will become important to consider.
The Env glycoproteins play a central role in HIV-1 entry and immune evasion. Env exists as a metastable trimer of three protomers comprising gp120 and gp41 heterodimers on the surface of the virion and the infected cell. The binding of gp120 to CD4 on the target cell triggers conformational rearrangement of the Env trimer that exposes coreceptor (CCR5 or CXCR4) binding sites in gp120. Subsequent binding of gp120 to coreceptor promotes insertion of the gp41 fusion peptide into the target cell membrane, and the refolding of gp41 heptad repeat 1 and 2 (HR1 and HR2) mediates the fusion of viral and cellular membranes, allowing viral entry into the cytosol of the target cell (18). Single-molecule Förster resonance energy transfer (smFRET) analysis has demonstrated that the Env trimer spontaneously transitions between at least three distinct prefusion conformations: state 1 (pretriggered, closed conformation), state 2 (necessary, intermediate conformation), and state 3 (fully CD4-bound, open conformation) (19–22). Env is heavily glycosylated with about half of its mass comprising N-linked glycans. The dense “shield” of glycans on gp120 protects the virus from host immune recognition. Moreover, these glycans contribute to correct Env glycoprotein folding, processing, antigenicity, and particle infectivity [reviewed in (23)]. The cytoplasmic tails (CTs) of lentiviral gp41 transmembrane glycoproteins also play important roles in a variety of aspects of Env function and contribute to viral pathogenesis (24–26).
HIV-1 spreads from infected donor cell to target cell via two different modes: cell-free virus infection and cell-cell (cell-associated) transfer. In the case of cell-free infection, virus particles are released from the donor cell, diffuse through the extracellular space, and then bind and infect the target cell. In the case of cell-cell transfer, virus particles accumulate at a point of close contact between producer and target cell, often referred to as a virological synapse (VS) (27). Interactions between Env on the producer cell and CD4 on the target cell promote the formation of the VS, stabilized by cellular adhesion molecules such as lymphocyte function–associated antigen 1 (LFA-1) and intercellular adhesion molecule 1 (ICAM-1) [reviewed in (27)]. Env expression can lead to cell-cell fusion (syncytium formation), a process that is tightly regulated by viral and cellular factors and often does not accompany VS formation and cell-cell viral transfer (28, 29).
A hallmark of cell-cell transmission is that HIV-1 can be efficiently transmitted at a higher multiplicity of infection (MOI) than with cell-free infection, resulting in the formation of multiple proviruses in target cells (multicopy infection) (30–32). This high MOI has been proposed to allow HIV-1 to overcome multiple barriers to infection, including those imposed by neutralizing antibodies (NAbs), ARVs, and restriction factors (27, 30, 33–35). Because lymphoid tissues, in which HIV-susceptible cells are tightly packed, are the dominant sites of viral replication, cell-cell transmission may play an important role in viral dissemination in vivo. Several animal studies have provided evidence for cell-cell transmission in vivo (36–40). However, the presence of multicopy infection in vivo is still debated. Some studies have shown that infected cells harboring multiple HIV-1 copies are present in the spleens of HIV-1–infected individuals (41–43), but other reports have demonstrated that most of the cells in the blood and lymph nodes of infected individuals carry a single provirus (44). While further analysis will be needed to clarify the contribution of cell-cell transfer to HIV-1 propagation in vivo, even in the absence of multiple proviruses per infected cell, it seems clear that highly efficient transfer of multiple virus particles to a target cell would be more likely to lead to a productive infection event than transfer of a single particle.
We previously reported that during in vitro propagation HIV-1 acquires mutations in the Env gp120-gp41 interface in response to ARVs including DTG (45, 46). These single–amino acid Env mutations confer broad but modest levels of resistance to multiple classes of approved ARVs and increase resistance to ARVs when coupled with drug-target gene mutations in the context of spreading infections. Most of the previously described Env mutations impair cell-free viral infectivity but increase cell-cell transfer capacity. Our single-genome sequence analysis of viruses from several study participants failing an INSTI-containing regimen despite a low frequency of resistance mutations in IN (7) revealed the presence of mutations at the gp120-gp41 interface analogous to those seen in our in vitro selections (7, 45). These observations raise the possibility that HIV-1 Env mutations may be involved in the acquisition by HIV-1 of high-level resistance to ARVs.
In the present study, we investigated the pathway used by HIV-1 to develop high-level INSTI resistance by long-term propagation of several clonal HIV-1 strains in the presence of DTG. We demonstrate that, over the course of nearly 1 year of continuous selection, HIV-1 became highly resistant to DTG by sequentially acquiring multiple mutations in the Env- and Gag-nucleocapsid (NC)–coding regions in the absence of resistance mutations in IN. The accumulated Env mutations led to high-level INSTI resistance by increasing the MOI via enhanced cell-cell transfer efficiency. To gain mechanistic insights into Env-mediated drug resistance, we characterized the effects of Env mutations on the conformation of the Env trimers and their sensitivity to a variety of Env-based inhibitors. We found that the combined Env mutations confer notably higher levels of resistance to INSTIs than to other classes of ARVs, leading to the finding that INSTIs are inherently more susceptible to loss of potency under high-MOI conditions relative to other classes of inhibitors.
RESULTS
HIV-1 acquires multiple mutations in the Env- and NC-coding regions during long-term propagation in DTG
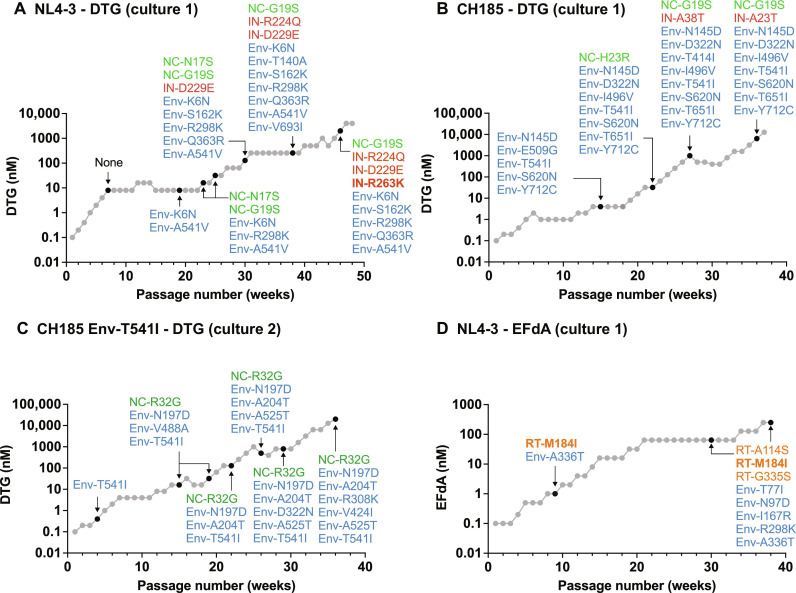
To investigate how HIV-1 develops high-level resistance to INSTIs, we infected the SupT1 T cell line with the HIV-1 NL4-3 clone (47) and serially propagated the virus in the presence of increasing concentrations of DTG (Fig. 1A). The passaging experiment was initiated at 0.1 nM DTG, which is less than the median inhibitory concentration (IC50) for wild-type (WT) NL4-3 in the context of spreading infection (46). After nearly 1 year of continuous passaging, we were able to increase the DTG concentration to 4.0 μM, which is >1000-fold the IC50. To identify the genetic changes in the viral genome responsible for DTG resistance, we extracted genomic DNA from the infected cells at a number of time points and performed sequencing analysis of gag, pol, env, and vpu. At passage 7 (8.0 nM DTG), we did not identify any mutations in these regions. However, upon continued propagation in 8.0 nM DTG (passage 19), we identified an Env-K6N mutation in the signal peptide (SP) and the Env-A541V mutation (HXB2 numbering) in gp41 HR1. We previously reported that the Env-A541V mutation confers an ~8-fold resistance to DTG in multicycle spreading infection (45, 46). After the emergence of Env-A541V, we could gradually increase the DTG concentration. At passage 23 (32 nM DTG), an additional Env mutation (Env-R298K in the gp120 V3 region) was acquired and two NC mutations (NC-N17S and G19S) were detected. Later, we identified two additional Env mutations (Env-S162K in the gp120 V2 region and Env-Q363R in the gp120 C3 region) and two IN mutations (IN-D229E and IN-R224Q). These IN mutations are not reported as INSTI resistance mutations in the Stanford HIV Drug Resistance Database (48). We determined that these IN mutations do not confer measurable resistance to DTG (fig. S1A). At passage 46 (2000 nM DTG), we identified the IN-R263K DTG resistance mutation at low frequency. Consistent with a previous report (49), IN-R263K conferred significant levels of resistance to DTG (fig. S1A). This analysis indicates that HIV-1NL4-3 sequentially acquires mutations in Env-, NC-, and IN-coding regions (Fig. 1A) during propagation in the SupT1 T cell line in the presence of escalating concentrations of DTG.
Fig. 1. In vitro selection of drug-resistant variants.
Long-term passaging of (A) WT NL4-3 (culture 1), (B) WT CH185 (culture 1), and (C) CH185 Env-T541I (culture 2) in the presence of the INSTI DTG. (D) Long-term passaging of WT NL4-3 (culture 1) in the presence of the NRTTI EFdA. SupT1 [(A) and (D)] or SupT1huR5 [(B) and (C)] T cell lines were infected with the indicated viruses to initiate the passaging experiments. At the time points indicated by the arrows, genomic DNA was extracted from the drug-treated cultures, and the Gag-, Pol-, and Env-coding regions were sequenced. Mutations detected at frequencies greater than 25% in the bulk sequencing are shown. Mutations highlighted in bold are established resistance mutations to DTG (A) or EFdA (D).
To verify these results, we performed repeated passaging experiments with WT NL4-3 in the presence of DTG (denoted as culture 2 and culture 3 in fig. S2). We also initiated long-term propagation experiments with the NL4-3 Env-A541V mutant, as this mutant is frequently detected early in selection experiments (Fig. 1A and fig. S2, C and D) (45, 46). We again identified multiple Env and NC mutations, but no resistance mutations in IN, in the presence of >1000 nM DTG. All of the NL4-3 cultures selected the Env-A541V mutation and Env mutations that disrupt N-linked glycosylation in the gp120 V1/V2 region (fig. S2, C and D).
The NL4-3 strain used in the experiments described above is a laboratory-adapted, subtype B, CXCR4-tropic HIV-1 isolate. To extend our investigation to CCR5-tropic and primary HIV-1 isolates, we performed multiple DTG selection experiments with the subtype C, transmitted founder, CCR5-tropic isolate CH185 (Fig. 1B and fig. S2) (50) and the subtype B, CCR5-tropic NL(AD8) clone (fig. S2) (51). Selections were performed in the SupT1huR5 T cell line, which expresses CCR5 (52). As observed in the passaging experiments using the NL4-3 isolate, both CH185 and NL(AD8) strains sequentially acquired mutations in Env-, NC-, and IN-coding regions. Despite the long-term passaging of NL(AD8) and CH185 in the presence of 500 to 20,000 nM DTG, we did not identify any reported INSTI resistance mutations in the IN-coding region (Fig. 1, B and C, and fig. S2B). NC-G19S was identified in all the viral strains tested, suggesting that mutations in the NC domain may contribute to INSTI resistance (Fig. 1, A and B, and fig. S2A). As shown in fig. S2E, the HIV-1 variants that we tested in this study frequently selected mutations in the outer domain of gp120 and at the gp120-gp41 interface. Mutations at amino acid positions Env-525, Env-541, Env-547, Env-558, and Env-561 (HXB2 numbering) in the gp120-gp41 interface were identified in multiple viral strains. We previously reported that mutations at Env-541 and Env-558 reduce sensitivity to DTG (45, 46). We also detected multiple mutations in the matrix (MA)-, capsid (CA)-, and RT-coding regions, but these mutations did not confer resistance (fig. S2, A and B). Overall, these data indicate that NC and Env mutations, rather than INSTI resistance mutations in IN, are predominantly selected independent of subtype and co-receptor usage upon HIV-1 propagation in the presence of DTG.
Next, we examined whether an INSTI-resistant IN mutant acquires NC and Env mutations during long-term passaging in the presence of high concentrations of DTG. The IN-G118R mutation is often seen in individuals experiencing virological failure on DTG-containing regimens (49). We propagated this mutant in the SupT1 T cell line at a starting concentration of 0.1 nM DTG and, over the course of nearly 1 year, escalated the DTG concentration to 2000 nM. The IN-G118R mutation was stable over the course of the selection experiment. The IN mutant first acquired multiple Env mutations, followed by two NC mutations (NC-N8S and M46I) (fig. S2A). The first Env mutation acquired was Env-A541V, which we previously reported can rescue defects imposed by drug resistance mutations (45). Env-A541V rescued replication defects imposed by IN-G118R and provided a replication advantage over WT in the presence of DTG (fig. S3). Together, these results suggest that mutations in NC, IN, and Env may cooperatively contribute to INSTI resistance in spreading infection.
HIV-1 more rapidly acquires resistance mutations in the drug-target gene in the presence of an RT inhibitor compared to the INSTI DTG
The data presented above indicate that HIV-1 predominantly acquires mutations in the Env- and NC-coding regions when propagated in the presence of the INSTI DTG. To investigate whether HIV-1 acquires mutations in these coding regions in the presence of another class of ARV, we propagated WT NL4-3 in the presence of increasing concentrations of the nucleoside RT translocation inhibitor (NRTTI), islatravir [4′-ethynyl-2-fluoro-2′-deoxyadenosine (EFdA)] (Fig. 1D). We chose EFdA because it is highly potent and active against HIV-1 strains resistant to existing NRTIs (53, 54). In contrast to our results with DTG, NL4-3 rapidly acquired a target-gene (RT) mutation in the presence of a low concentration of EFdA (Fig. 1D); at passage 8 (1.0 nM EFdA), the virus acquired the RT-M184I mutation, a primary RT mutation that confers modest levels of resistance to EFdA (fig. S1B) (53, 54). The virus also acquired Env-A336T at this time point. At a later time point (passage 30, 64 nM EFdA), the virus additionally acquired multiple Env mutations along with two accessory RT mutations (RT-A114S and G335S) (Fig. 1D and figs. S1B and S2). Two of the selected Env mutations (Env-T77I and N97D) are located at the gp120-gp41 interface, which we previously reported as the site of mutations that can rescue replication-defective mutants and confer broad ARV resistance (45, 46). In another culture of NL4-3 in the SupT1 T cell line, the RT-T165A mutation emerged after RT-M184I along with Env-S162R (fig. S2). The RT substitutions at residues 114 and 165 are accessory mutations that increase resistance to EFdA when coupled with RT-M184I/V (fig. S1B) (53, 54). No NC mutations were identified in any of the EFdA selection experiments (Fig. 1D and fig. S2A), suggesting that the emergence of NC mutations is specific to viral escape from INSTIs. These results demonstrate that, consistent with a large number of previous studies (3, 53, 54), HIV-1 can rapidly develop resistance to RT inhibitors by acquiring resistance mutations in RT. In contrast, we find that escape from INSTIs can be driven by mutations outside the drug-target gene (IN), specifically in Env and NC.
Accumulation of multiple Env mutations leads to high-level resistance to DTG
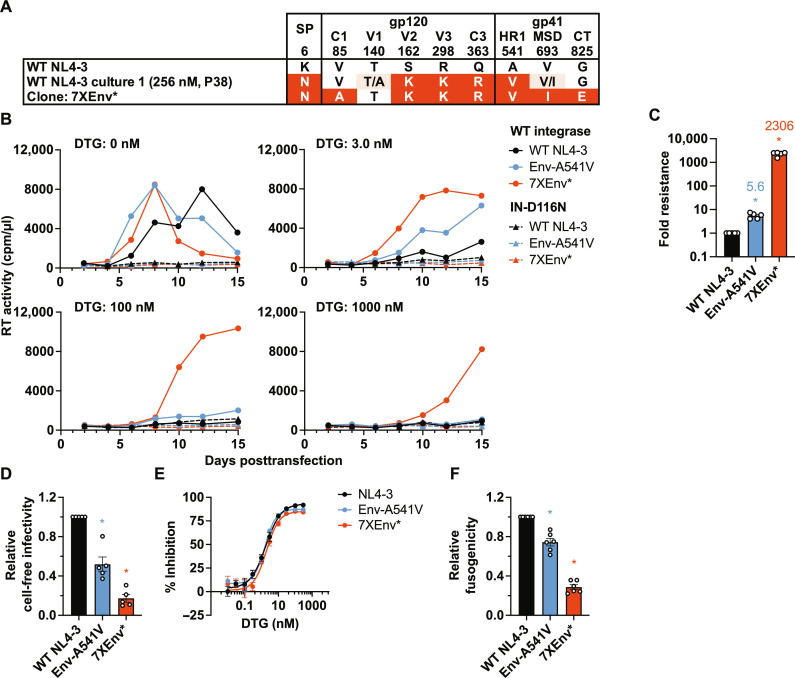
The passaging experiments presented in Fig. 1 and fig. S2 indicate the acquisition of high-level resistance to DTG. On the basis of our previous work with single Env mutations (45, 46), we hypothesized that the multiple Env mutations we identified are primarily responsible for the high-level DTG resistance achieved in our selection experiments. To test this hypothesis, we constructed an infectious NL4-3 molecular clone containing the heavily mutated Env derived from NL4-3 culture 1 in the presence of 256 nM DTG (Figs. 1A and 2A). The 7XEnv* clone contains seven Env mutations (V85A, S162K, R298K, and Q363R in gp120, and A541V, V693I, and G825E in gp41) and a mutation in the Vpu/Env-SP–coding region (Vpu-V60L/Env-SP-K6N, denoted by *). We examined the replication kinetics of this mutant in the presence or absence of a range of DTG concentrations (0.03 to 3000 nM) by transfecting SupT1 T cells with the WT NL4-3 and 7XEnv* molecular clones and monitoring virus replication by RT assay. Our previously reported Env-A541V mutant (45, 46) was included as a control (Fig. 2B). Consistent with our previous analyses (45, 46), Env-A541V exhibited faster-than-WT replication kinetics in the absence of DTG and a replication advantage over WT in the presence of 3.0 nM DTG (Fig. 2B). 7XEnv* exhibited similar replication kinetics as Env-A541V in the absence of DTG but displayed a replication advantage over both WT and Env-A541V in the presence of 3.0 nM DTG. Whereas replication of both WT and Env-A541V was blocked by 100 nM DTG, 7XEnv* could replicate in the presence of 100 and 1000 nM DTG. To determine whether the viruses acquired additional mutations in the presence of high concentrations of DTG, we performed sequencing analysis of the viruses at the peak of replication. We did not identify any changes in gag, pol, env, or nef compared to the starting viruses.
Fig. 2. The NL4-3 7XEnv* variant exhibits high-level DTG resistance in the context of spreading, multi-round infection but not in cell-free, single-round infection.
(A) An infectious molecular clone with seven Env mutations (7XEnv) and a Vpu mutation (denoted by the *) derived from the long-term passaging experiment was constructed by transferring the env amplicon from NL4-3 culture 1 (passage 38, 256 nM DTG; Fig. 1A) into pNL4-3. The locations of mutations in Env are indicated: C1 and C3, first and third conserved domains of gp120, respectively; V2 and V3, second and third variable domains of gp120, respectively; HR1, heptad repeat 1 of gp41; MSD, membrane-spanning domain; CT, cytoplasmic tail. (B) Replication kinetics of the NL4-3 Env variants in the SupT1 T cell line in the absence or presence of DTG. Replication curves obtained in the presence of 0, 3, 100, and 1000 nM DTG are shown. Data are representative of at least three independent experiments. (C) Fold changes in IC50 were calculated compared to that for the WT over a range of DTG concentrations (0.01 to 3000 nM). IC50 values were calculated on the basis of the AUC of the replication kinetics. (D) Single-round, cell-free viral infectivity of the Env variants. Relative infectivity is shown, normalized to 1 for WT NL4-3. (E) DTG sensitivity in the context of cell-free infection. TZM-bl cells were incubated with 100 TCID50 of WT virus or the Env mutants in the presence of various concentrations of DTG. (F) Cell-cell fusion activity of the NL4-3 Env variants. The transfected HEK293T cells were cocultured with TZM-bl cells in the presence of a cocktail of RPV and DTG to prevent productive infection of the TZM-bl cells. Data from at least three independent experiments are shown as means ± SEM. *P < 0.05, unpaired t test.
By calculating DTG IC50 based on the area under the curve (AUC) of the replication kinetics in the presence of 0.3 to 3000 nM DTG, we determined that 7XEnv* exhibited >2000-fold resistance to DTG (Fig. 2C). As reported previously (45, 46), A541V exhibited modest (~6-fold) resistance. These data indicate that accumulation of multiple Env mutations leads to high-level resistance to DTG. To examine the impact of the Vpu/Env-SP mutation in 7XEnv* on DTG sensitivity in a spreading infection, we also prepared the heavily mutated Env clone containing WT Vpu/Env-SP (denoted 7XEnv) (fig. S4). The Vpu/Env-SP mutation had only a subtle effect on the phenotype of 7XEnv* in spreading infection, indicating that the seven Env mutations present in 7XEnv play the dominant role in conferring high-level INSTI resistance.
Previous studies reported that, under certain conditions, unintegrated viral DNA can express viral proteins (55), leading to DTG resistance (14, 16). To examine whether integration is required for the high-level DTG resistance conferred by the Env mutations, we examined the replication of a catalytically inactive IN-D116N mutant (56) in the context of WT or the Env-A541V or 7XEnv* mutants in the presence of DTG. We found that the IN-D116N–containing mutants did not exhibit any measurable viral replication in the absence or presence of DTG (Fig. 2B). This finding indicates that unintegrated viral DNA does not contribute to viral replication and DTG resistance in our experiments.
We previously reported that single Env mutations at the gp120-gp41 interface (e.g., Env-P81S and Env-A558T) enhance virus replication capacity and confer ARV resistance in the context of a spreading infection but not in single-cycle infectivity assays (45, 46). These mutations promoted cell-cell virus transmission at the cost of cell-free viral infectivity and Env-mediated fusogenicity (45, 46). To examine the effect of accumulation of multiple Env mutations on infectivity and drug sensitivity in the context of cell-free viral infection, we measured single-round, cell-free viral infectivity of Env-A541V and 7XEnv* in the absence or presence of DTG in the TZM-bl indicator cell line (Fig. 2, D and E). 7XEnv* displayed impaired cell-free viral infectivity compared to WT NL4-3 (Fig. 2D). The Env mutations did not confer resistance to DTG in the context of single-round, cell-free viral infection (Fig. 2E). We also examined the fusogenicity of 7XEnv* and Env-A541V using the TZM-bl cell line. The fusogenicity of Env-A541V and 7XEnv* was significantly impaired relative to WT (Fig. 2F). These results indicate that the high-level resistance to INSTIs conferred by 7XEnv is not due to enhanced cell-free viral infection but rather is likely due to increased cell-cell transfer capacity (see below).
High-level Env-mediated INSTI resistance is independent of subtype and co-receptor usage
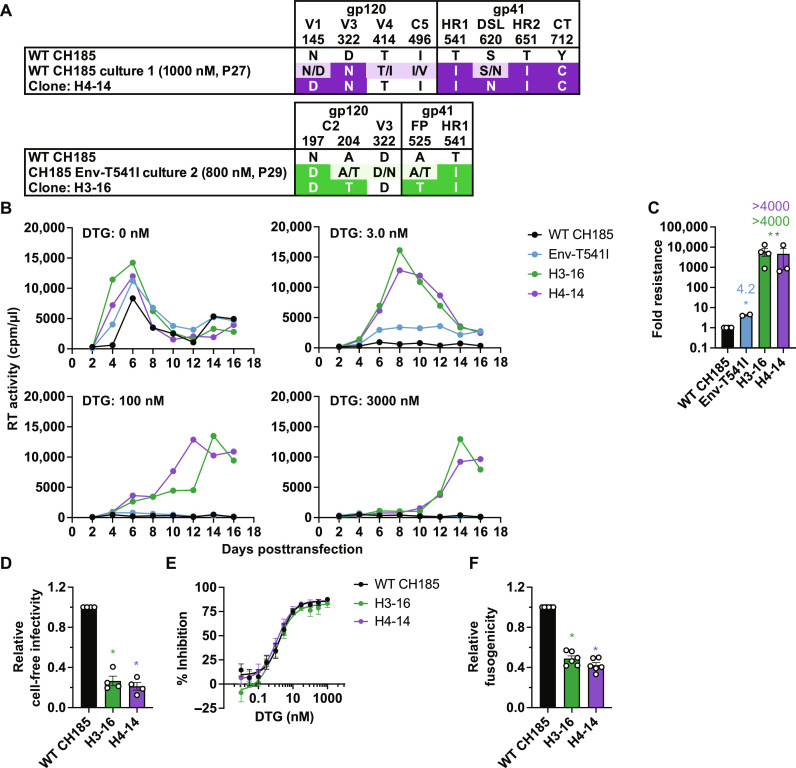
To examine whether the Env mutations selected in DTG in the context of a clinically relevant isolate (Fig. 1, B and C, and fig. S2, C and D) confer high-level resistance to INSTIs, we constructed infectious molecular clones containing the Env mutations selected in the context of the subtype C transmitted/founder strain CH185 (Fig. 3A). H4-14 is derived from the CH185 culture 1 in the presence of 4000 nM DTG (Fig. 1B). This clone has two mutations in gp120 (Env-N145D and D322N) and four in gp41 (T541I, S620N, T651I, and Y712C). H3-16 is derived from the CH185 Env-T541I culture 2 in the presence of 2000 nM DTG (Fig. 1C). This clone has two mutations in gp120 (Env-N197D and A204T) and two in the gp41 ectodomain (Env-A525T and T541I). Both clones harbor the Env-T541I mutation in gp41 and mutations in N-linked glycosylation motifs (Asn-X-Ser/Thr, where X is any amino acid) in the gp120 V1/V2 region (Env-N197D in H3-16 and Env-N145D in H4-14).
Fig. 3. The heavily mutated CH185 Env variants exhibit high-level DTG resistance in the context of spreading, multi-round infection but not in cell-free, single-round infection.
(A) Infectious molecular clones with the indicated Env mutations in the context of CH185. H3-16 and H4-14 clones were constructed by transferring the env amplicons from CH185 culture 1 (passage 27, 1000 nM DTG; Fig. 1B) and CH185 Env-T541I culture 2 (passage 29, 800 nM DTG; Fig. 1C), respectively, into CH185. V1 and V3, first and third variable domain of gp120, respectively; C2, second conserved domain; HR1 and HR2, heptad repeat 1 and 2 of gp41, respectively; FP, fusion peptide; DSL, disulfide loop; CT, cytoplasmic tail. (B) Replication kinetics of the CH185 Env variants in the SupT1 huR5 T cell line in the absence or presence of DTG. Replication curves obtained in the presence of 0, 3, 100, and 3000 nM DTG are shown. Data are representative of at least three independent experiments. (C) Fold changes in IC50 were calculated compared to that for the WT, based on the AUC of the replication kinetics. (D) Single-cycle, cell-free viral infectivity of the Env variants. RT-normalized virus stocks were used to infect TZM-bl cells. Luciferase activity was measured at 48 hours after infection. Relative infectivity is shown, normalized to 1 for WT CH185. (E) DTG sensitivity in the context of cell-free infection. TZM-bl cells were exposed to 100 TCID50 of WT or the Env mutants in the presence of various concentrations of DTG. (F) Cell-cell fusion activity of the CH185 Env variants. The transfected HEK293T cells were cocultured with TZM-bl cells in the presence of a cocktail of RPV and DTG to prevent productive infection of the TZM-bl cells. Data from at least three independent experiments are shown as means ± SEM. *P < 0.05, unpaired t test.
As observed with the NL4-3 7XEnv* and 7XEnv (Fig. 2 and fig. S4), in the absence of DTG, the Env-T541I single mutant and the H3-16 and H4-14 multiple mutants exhibit a replication advantage over WT CH185 (Fig. 3B). In the presence of 3 nM DTG, WT replication is blocked and that of Env-T541I is severely suppressed. In contrast, the H3-16 and H4-14 mutants can still replicate in 3000 nM DTG (Fig. 3B). Quantitative analyses over multiple experiments indicate that the H3-16 and H4-16 mutants exhibit a >4000-fold change in DTG IC50 relative to the WT in spreading infections (Fig. 3C). As seen with the NL4-3–derived mutants, the CH185-derived Env mutants display reduced single-round, cell-free infectivity (Fig. 3D), and DTG sensitivity in the context of cell-free viral infection is not affected by the Env mutations (Fig. 3E). The H3-16 and H4-16 mutants also exhibit reduced fusogenicity relative to WT (Fig. 3F). These results indicate that the ability of multiple Env mutations to confer high-level DTG resistance in multi-round, spreading infections is independent of subtype and co-receptor usage.
Heavily mutated Env clones that are highly resistant to DTG exhibit broad but modest reductions in sensitivity to other classes of ARVs
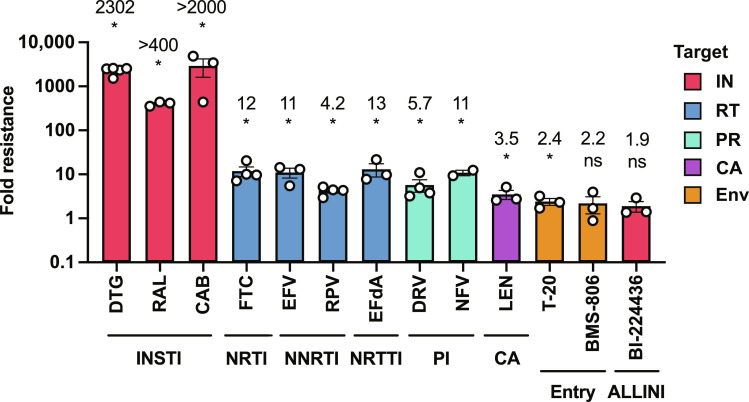
We previously reported that single Env mutations provide a modest replication advantage over WT virus in the presence of multiple classes of ARVs in spreading but not cell-free infections (45), and we show above that Env mutants with multiple Env substitutions exhibit high-level resistance to DTG. To determine whether 7XEnv* is resistant to other classes of ARVs, we compared viral replication of WT NL4-3 with 7XEnv* in multi-round, spreading infections in the presence of two other INSTIs (RAL and CAB), the NRTI emtricitabine (FTC), NNRTIs rilpivirine (RPV) and efavirenz (EFV), NRTTI EFdA, PIs darunavir (DRV) and nelfinavir (NFV), fusion inhibitor T-20, attachment inhibitor (conformational blocker) BMS-378806 (BMS-806), allosteric IN inhibitor (ALLINI) BI-224436 (57), and CA inhibitor LEN (Fig. 4). We found that 7XEnv* exhibits >400-fold and >2000-fold resistance to RAL and CAB, respectively, indicating that the heavily mutated Env confers high-level resistance to INSTIs other than DTG. 7XEnv* also exhibits broad resistance to the RT inhibitors FTC (12-fold), EFdA (13-fold), RPV (4.2-fold), EFV (11-fold), the PIs DRV (5.7-fold) and NFV (11-fold), and the capsid inhibitor LEN (3.5-fold). However, the fold resistance of 7XEnv* to these non-INSTI classes of ARVs is markedly lower than for the INSTIs. 7XEnv* does not exhibit resistance to the ALLINI BI-224436 despite the fact that this compound, like the INSTIs, targets IN (see Discussion).
Fig. 4. ARV sensitivity of the NL4-3 7XEnv* variant in the context of spreading infection.
Fold resistance of 7XEnv* to multiple classes of ARVs. The SupT1 T cell line was transfected with WT NL4-3 or 7XEnv* proviral clones in the absence or presence of a range of ARV concentrations. Virus replication kinetics were monitored by measuring RT activity. Fold changes in IC50 relative to WT were calculated. IC50 values were calculated on the basis of the AUC of the replication kinetics. IC50 values were calculated for INSTIs (DTG, RAL, and CAB), NRTI (FTC), NNRTI (EFV and RPV), NRTTI (EFdA), PI (DRV and NFV), entry inhibitors (T-20 and BMS-806), ALLINI (BI-224436), and CA inhibitor (LEN). Data from at least two independent experiments are shown as means ± SEM. *P < 0.05, unpaired t test.
Heavily mutated Env variants exhibit enhanced cell-cell transfer capacity
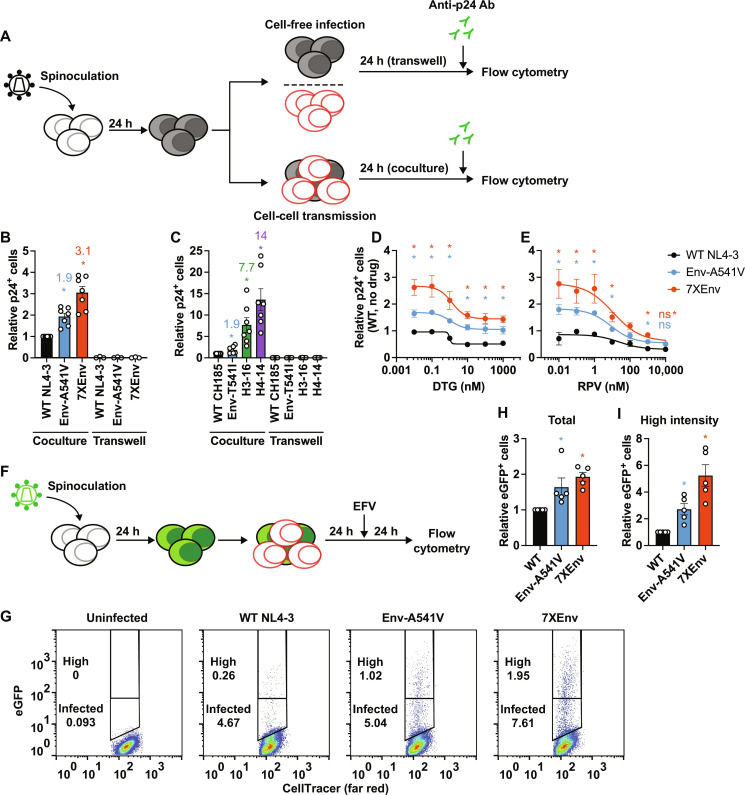
As shown above, the Env mutations selected in high concentrations of DTG confer high-level resistance to INSTIs in the context of spreading but not cell-free viral infection. This result suggests that the accumulation of Env mutations overcomes ARV inhibition by enhancing cell-cell transfer. To test this hypothesis, we compared cell-cell transfer of WT NL4-3, Env-A541V, and 7XEnv by coculturing virus-producing cells and uninfected target cells (Fig. 5A). Virus-producing SupT1 T cells were prepared by spinoculation, and target SupT1 cells were stained with a cell proliferation dye to distinguish them from producer cells. One day after coculture, cells were stained with fluorescein isothiocyanate (FITC)–conjugated anti-p24 Ab and infected target cells were enumerated by flow cytometry. As shown in Fig. 5B, this analysis demonstrated that productive infection of Env-A541V and 7XEnv through cell-cell contact is 1.9- and 3.1-fold more efficient than that of WT NL4-3, respectively. To monitor the contribution of cell-free viral infection in the coculture experiments, we also seeded the virus-producing cells in a transwell apparatus that prevents cell-cell contact and thus allows only cell-free infection to occur. During the 24-hour time frame, the contribution of cell-free infection was negligible (Fig. 5B). We also examined the efficiency of cell-cell transfer of the CH185 Env mutants under the same conditions. Similar to what we observed with the NL4-3–derived mutants, the CH185 mutants exhibited significantly enhanced cell-cell transfer, with increases of 1.9-, 7.7-, and 14-fold relative to WT for Env-T541I, H3-16, and H4-14, respectively (Fig. 5C). Again, the contribution of cell-free infection to viral transfer was negligible, as demonstrated by transwell experiments (Fig. 5C). Together with the results of Figs. 2D and 3D, these data indicate that the Env mutants selected to be resistant to DTG exhibit highly proficient cell-cell transfer capacity at the cost of reduced cell-free infectivity.
Fig. 5. The heavily mutated Env variants exhibit increased cell-cell transfer capacity.
(A) Experimental design of cell-cell transfer assay. Virus-producer SupT1 cells were spinoculated with the indicated viruses 24 hours before the assay. Target SupT1 cells were labeled with cell proliferation dye. Before the coculture, the number of infected donor cells was normalized by intracellular p24-FITC signals. The donor and the target SupT1 cells were cocultured at a 1:1 ratio in the absence or presence of ARVs. Twenty-four hours after coculture, intracellular p24 antigen was detected by staining with FITC-conjugated anti-p24 Ab. Virus-producing cells were also seeded in a transwell insert to monitor cell-free viral infection. (B) Cell-cell transfer of NL4-3 Env mutants. Fold change in the numbers of p24-positive target cells relative to WT. (C) Cell-cell transfer of CH185 variants. Fold change in the numbers of p24-positive target cells relative to WT. (D and E) Viral sensitivity to DTG and RPV, respectively, in the context of cell-cell transfer. Data are shown as the number of p24-positive cells relative to WT NL4-3 in the absence of drugs. (F) Experimental design of cell-cell transfer assay using pBR43IeG constructs (HIV eGFP). After 24 hours of coculture between infected and target cells, the second round of infection was blocked by adding 1 μM EFV, and then the cells were incubated for 24 hours. (G) Representative plots of eGFP expression after coculture. High–fluorescence intensity populations were defined as containing ~0.25% of the population showing a high-intensity eGFP signal in WT-infected target cells. (H) Relative numbers of eGFP-positive target cells in the absence of DTG. (I) Relative numbers of high–fluorescence intensity eGFP-positive target cells . Data from at least three independent experiments are shown as means ± SEM. *P < 0.05, unpaired t test.
Next, to examine whether the Env mutants can productively infect via the cell-cell route in the presence of ARVs, we performed coculture assays in the presence of DTG and RPV (Fig. 5, D and E). 7XEnv exhibited higher productive infection compared to WT and Env-A541V in the presence of low concentrations of DTG and RPV. Cell-cell transmission of 7XEnv was nearly completely inhibited by RPV in a dose-dependent manner (Fig. 5E), whereas even the highest concentration of DTG tested did not completely inhibit productive infection of 7XEnv (Fig. 5D). We noted that the inhibitory effect of DTG on viral infection reached a plateau at concentrations above 10 nM DTG, indicating that the maximum inhibitory effect of DTG is reduced in the context of cell-cell transfer.
To confirm that the p24 signals in the target cells are from de novo viral gene expression after viral DNA integration as opposed to p24 signal from incoming virus particles after viral transfer, we also performed coculture experiments using pBR43IeG constructs, which express enhanced green fluorescent protein (eGFP) under control of an internal ribosome entry site (IRES) after integration (Fig. 5F) (58). Consistent with the p24 staining results, the Env mutants produce more eGFP-expressing cells after coculture compared to WT (Fig. 5, G to I). The cells infected with the Env mutants also exhibit a higher eGFP signal intensity compared to WT (Fig. 5I), suggesting that the Env mutations increase the number of productive infection events per cell. Together, our data suggest that efficient cell-cell transfer conferred by the Env mutations results in higher levels of productive infection (a higher MOI), leading to high-level resistance to INSTIs.
The Env mutations alter the conformational landscape and stability of Env trimers
We previously reported that Env-A541V reduces the sensitivity of NL4-3 to NAbs recognizing the CD4-bound gp120 conformation, suggesting that this mutation stabilizes the closed Env conformation (45). To examine the impact of the accumulated Env mutations on Env conformation, we compared the sensitivity of WT and 7XEnv NL4-3 to NAbs that preferentially recognize the CD4-bound conformation (21, 59), specifically 17b [which binds a CD4-induced (CD4i) epitope], 447-52D (which binds the V3 loop), and F105 [which recognizes the CD4 binding site (CD4bs)] (Fig. 6A). 7XEnv was significantly more resistant to 17b, 447-52D, and F105 than WT (Fig. 6A) and was less sensitive than WT to inhibition by soluble CD4 (sCD4) (Fig. 6B). To measure the effect of the Env mutations on CD4 and NAb binding, we performed binding assays with Env on the surface of 293T cells (Fig. 6, C to E). Relative to WT, 7XEnv exhibited an increased binding to CD4–immunoglobulin (Ig) (60) but reduced binding to VRC03, suggesting that 7XEnv may sample a relatively open conformation in the unliganded state. We hypothesized that 7XEnv does not efficiently undergo CD4-induced conformational changes in the Env trimer, leading to less binding/neutralization by 17b. To test this hypothesis, we measured 17b binding in the presence of increasing concentrations of sCD4. We found that sCD4 could not sensitize 17b binding to 7XEnv compared to WT (Fig. 6E), indicating that sCD4 does not efficiently trigger conformational changes in the mutant Env trimer.
Fig. 6. Sensitivity of the heavily mutated Env variants to ligand binding and neutralization.
Sensitivity of NL4-3 7XEnv to (A) NAbs recognizing the CD4-bound conformation and (B) sCD4. TZM-bl cells were exposed to 100 TCID50 of viruses in the presence of various concentrations of NAbs or sCD4. Luciferase activity was measured at 48 hours after infection. (C to E) NAb/CD4-Ig binding to Env on the cell surface. 293T cells transfected with the indicated WT or 7XEnv pBR43IeG clones were preincubated with (C) CD4-Ig, (D) VRC03, and (E) 17b at 37°C for 30 min. The cells were washed, and Alexa Fluor 647–conjugated anti-human IgG was used to detect bound antibodies. For 17b binding, Env-expressing cells were treated with the indicated concentrations of sCD4. Alexa Fluor 647 signals were normalized by eGFP signals to calculate the Ab binding efficiency. (F) Effect of the 7XEnv mutations on cold sensitivity. RT-normalized viruses were incubated at 4°C for the indicated times and frozen at −80°C. The viral aliquots were quickly thawed, and infectivity was measured using TZM-bl cells. (G) CD4-Ig binding to CH185 Env on the cell surface. 293T cells transfected with the indicated Env mutants were preincubated with CD4-Ig at 37°C for 30 min. The cells were washed, and Alexa Fluor 647–conjugated anti-human IgG was used to detect bound antibodies. To detect p24 in the cells, the transfected cells were fixed and permeabilized and then stained with FITC-conjugated anti-p24 Ab. (H) Cold sensitivity of the CH185 Env mutants. RT-normalized viruses were incubated at 4°C for the indicated times and frozen at −80°C. The viral aliquots were quickly thawed, and infectivity was measured using TZM-bl cells. Data from at least three independent experiments are shown as means ± SEM. *P < 0.05, unpaired t test.
Several studies have reported that exposure to low temperatures causes gradual conformational changes in the Env trimer, leading to loss of function. Unstable Env trimers prone to open conformation are inactivated by exposure to cold (21, 61). We therefore examined the cold sensitivity of the 7XEnv mutant as a measure of their conformational stability. As shown in Fig. 6F, WT NL4-3 progressively lost infectivity after incubation at 4°C, whereas 7XEnv retained ~50% infectivity after 120 hours of incubation at 4°C.
We and others previously reported that Env mutations in primary isolates have different impacts on Env conformation and neutralization properties relative to laboratory-adapted strains (45, 62). We found that most of the Abs that recognize the CD4-bound conformation could not bind and neutralize subtype C CH185 Env. Therefore, we evaluated CD4-Ig binding and cold sensitivity. Similarly to NL4-3 7XEnv, we found that the H3-16 and H4-14 Env variants exhibit higher CD4-Ig binding (Fig. 6G) and are resistant to cold inactivation (Fig. 6H) relative to WT. These results suggest that the heavily mutated CH185 Env trimers have similar structural and functional phenotypes as 7XEnv.
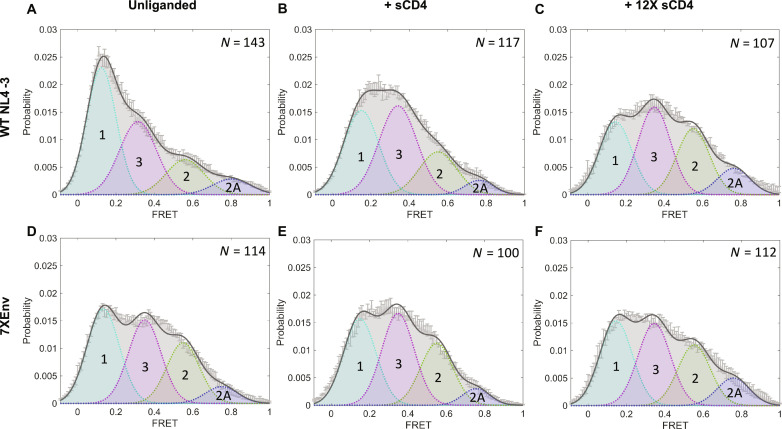
To examine further the conformational dynamics of 7XEnv, we performed a smFRET analysis of Env on the surface of virus particles (Fig. 7) using fluorescent probes in the gp120 V1 and V4 regions (20). We examined FRET states of unliganded Env, and Env treated with sCD4 or dodecameric CD4 (12XCD4) (63). smFRET analysis has previously shown that the HIV-1 Env trimer can sample at least three distinct conformational states: low, intermediate, and high FRET (denoted as states 1, 3, and 2/2A, respectively) (Fig. 7A), with state 1 representing a closed, ground-state configuration (19, 20, 22). Our analysis indicated that 7XEnv showed increased populations of intermediate- and high-FRET intensity in the unliganded state relative to WT (Fig. 7D), suggesting that 7XEnv more extensively samples partially open conformations in the unliganded state compared to WT. As reported previously (20, 63), treatment with sCD4 or 12XCD4 shifted the WT conformation to higher-FRET states, whereas 7XEnv remained relatively unaffected (Fig. 7, B and C to F). These results indicate that 7XEnv is relatively insensitive to conformational remodeling after interaction with CD4. Together, our biochemical and smFRET analyses demonstrate that the accumulation of Env mutations reduces CD4-induced conformational rearrangement of the Env trimer, which is necessary for viral entry in cell-free infection.
Fig. 7. 7XEnv displays an open conformation that is insensitive to soluble CD4.
HIV-1 NL4-3ΔRT virus particles containing WT (A to C) or 7XEnv (D to F) were produced, labeled, and analyzed by smFRET. Virus particles were incubated with soluble, monomeric CD4 [(B) and (E)] or dodecameric CD4 [(C) and (F)] at 100 μg/ml for 60 min before imaging. Histograms show the sum of N individual FRET trajectories ± SEM. Four-state Gaussian curve fitting was performed for each histogram, with FRET populations designated as follows: State 1, FRET ≈ 0.1 (pretriggered, closed conformation); state 2, FRET ≈ 0.6 (necessary, intermediate conformation); state 2A, FRET ≈ 0.8 (alternate intermediate conformation); state 3, FRET ≈ 0.3 (fully CD4-bound, open conformation).
The Env mutations stabilize gp120-gp41 interactions
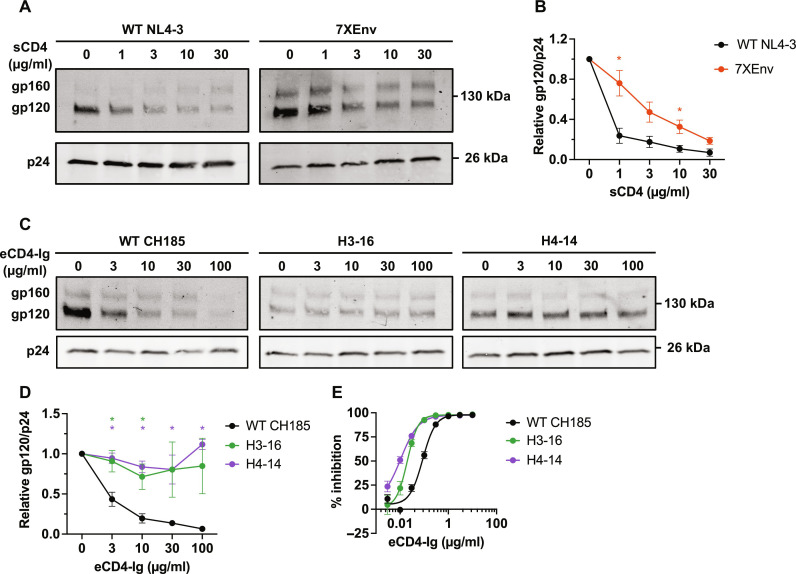
To gain insights into the mechanism of enhanced cell-cell transfer conferred by the accumulation of Env mutations, we incubated purified WT NL4-3 and 7XEnv virions with sCD4 at 37°C for 2 hours and then measured the amount of particle-associated gp120 and p24 by Western blotting (Fig. 8, A and B). As we previously reported (45), the levels of gp120 associated with WT NL4-3 virus particles decrease in a dose-dependent manner as the sCD4 concentration increases. 7XEnv exhibits a reduced rate of gp120 shedding relative to WT (Fig. 8, A and B), indicating a stabilized gp120-gp41 interaction.
Fig. 8. Effect of Env mutations on sCD4- or eCD4-Ig–induced gp120 shedding.
(A and B) The indicated viruses were incubated with a range of sCD4 concentrations at 37°C for 2 hours and purified through a 20% sucrose cushion, and viral proteins were detected by Western blotting. (A) Representative gel for sCD4-induced gp120 shedding assay. (B) The ratio of gp120 to p24 was quantified and plotted. (C and D) The indicated viruses were incubated with a range of eCD4-Ig concentrations at 37°C for 2 hours and purified through a 20% sucrose cushion, and viral proteins were detected by Western blotting. (E) Sensitivity of the CH185 Env mutants to eCD4-Ig. TZM-bl cells were exposed to 100 TCID50 of viruses in the presence of the indicated concentrations of eCD4-Ig. Luciferase activity was measured at 48 hours after infection. Data in the graphs from three independent experiments are shown as means ± SEM. *P < 0.05, unpaired t test.
It is known that Env glycoproteins from primary HIV-1 isolates tend to sample more closed Env conformations and are less prone to gp120 shedding compared to Env from laboratory-adapted strains (45, 64). It has been shown that eCD4-Ig, a fusion of CD4-Ig and a CCR5 coreceptor-mimetic peptide, can more potently neutralize viral infection and promote the shedding of gp120 of primary HIV-1 isolates relative to sCD4 (65). To examine the rate of gp120 shedding induced by eCD4-Ig, we incubated the CH185 Env variants with eCD4-Ig (0 to 100 μg/ml) at 37°C for 2 hours (Fig. 8, C and D). We found that eCD4-Ig strongly reduces the level of gp120 associated with WT CH185 particles in a dose-dependent manner. In contrast, the CH185 Env mutants exhibit minimal gp120 shedding in the presence of up to eCD4-Ig (100 μg/ml), indicating that the Env mutations stabilize the gp120-gp41 interaction. This is analogous to the stabilization of gp120/gp41 interactions seen above with NL4-3 7XEnv (Fig. 8, A and B). Despite this lack of gp120 shedding, eCD4-Ig efficiently neutralized the CH185 variants (Fig. 8E); the heavily mutated Env variants (H3-16 and H4-14) were more sensitive to eCD4-Ig–induced neutralization than WT CH185, suggesting that the heavily mutated Env may exhibit higher binding affinity with eCD4-Ig compared to WT CH185. These results indicate that enhanced cell-cell transmission capacity of the selected mutants from genetically distant viral strains (e.g., NL4-3 and CH185) is caused by analogous changes in conformational landscapes and Env trimer stability.
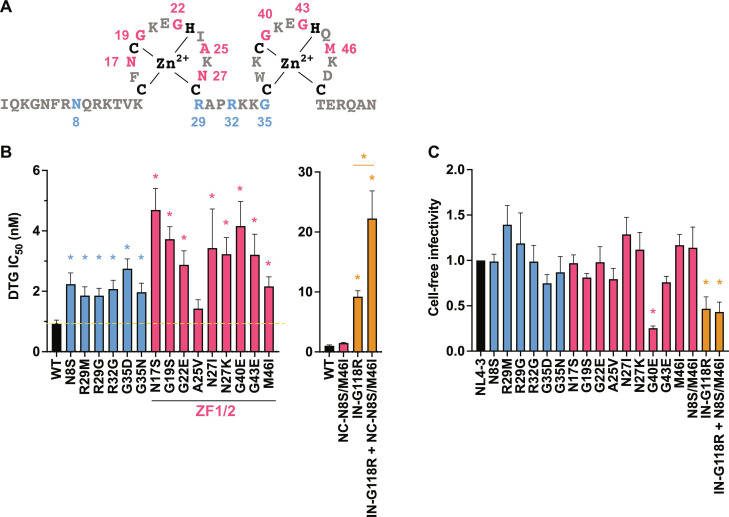
Mutations in the zinc-finger domains of NC confer modest levels of INSTI resistance
As demonstrated above, multiple independent passaging experiments using different viral strains in the presence of DTG led to the acquisition of mutations in NC (Figs. 1 and 9A and fig. S2A). To examine the impact of the NC mutations on DTG sensitivity, we introduced a panel of NC mutations obtained in our selection experiments into NL4-3 and measured their effect on susceptibility to INSTIs. We found that NC-N17S, G19S, G22E, N27I/K, G40E, and G43E mutations within the zinc-finger domains confer modest (three- to fivefold) but statistically significant levels of resistance not only to DTG (Fig. 9B) but also to other INSTIs, RAL and CAB (fig. S5, A and B). With the exception of NC-G40E, the single-round, cell-free infectivity of the NC mutants is comparable to that of WT (Fig. 9C). Although the fold resistance conferred by these NC mutations is lower than that conferred by the INSTI resistance mutation IN-G118R, INSTI resistance was increased when IN-G118R was coupled with the NC-N8S/M46I mutations, which we identified during propagation of IN-G118R in the presence of DTG (Fig. 9B and figs. S2A and S5, A and B). These data indicate that NC mutations can cooperatively increase INSTI resistance in combination with INSTI resistance mutations in IN. NC mutations selected with DTG did not affect viral sensitivity to the NRTTI EFdA or NRTI RPV (fig. S5, C and D), indicating that the observed resistance is specific to INSTIs.
Fig. 9. DTG sensitivity of the Gag-NC mutants.
(A) Sequence of the HIV-1 NC domain. The NC mutations selected in the presence of DTG are highlighted in red (within the zinc-finger domains) or blue (outside the zinc fingers). (B) Fold change in DTG IC50 of the NC mutants relative to WT. TZM-bl cells were exposed to 100 TCID50 of WT or the NC mutants in the presence of a range of concentrations of DTG (from 0.03 to 1000 nM DTG). Luciferase activity was measured at 48 hours after infection. (C) Cell-free viral infectivity of the NC mutants. RT-normalized virus stocks produced from 293T cells were used to infect TZM-bl cells. Luciferase activity was measured at 48 hours after infection. The infectivity of WT NL4-3 is normalized to 1.0. Data from at least three independent experiments are shown as means ± SEM. *P < 0.05, one-way analysis of variance (ANOVA) and Tukey’s multiple-comparison test or unpaired t test.
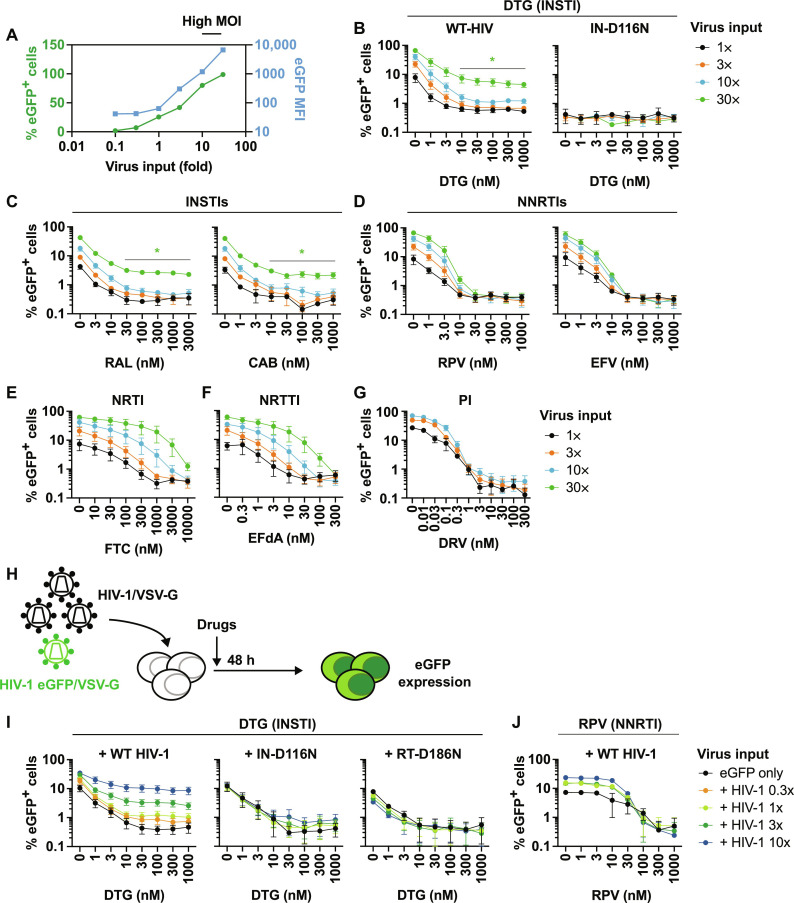
INSTI-mediated inhibition of HIV-1 infectivity can be overcome by high MOI
The data presented above indicate that heavily mutated Env clones display markedly higher resistance to INSTIs than to other classes of ARVs. This result, together with our observation that Env mutations confer drug resistance by increasing the MOI of the infection through enhanced cell-cell transfer capacity, raises the possibility that the antiviral activity of INSTIs is more readily overcome by high MOI than the antiviral activity of other classes of ARVs. To directly test this hypothesis, we used VSV-G–pseudotyped virus because such particles are highly infectious, making it possible to readily achieve high MOI in single-round infection (Fig. 10A). As expected, the number of eGFP+ cells increased in proportion to viral input; eGFP median fluorescence intensity (MFI) also increased sharply at high MOI, suggesting that the infected cells contain multiple copies of proviral DNA at high viral input (Fig. 10A). To compare the impact of high MOI on viral susceptibility to INSTIs versus other classes of ARVs, we titrated viral input of VSV-G–pseudotyped HIV-1 eGFP reporter virus in the absence or presence of the INSTIs DTG, RAL, and CAB; the NNRTIs RPV and EFV; the NRTI FTC; the NRTTI EFdA; and the PI DRV. In the case of DRV, the drug was used to treat the virus-producing cells, as PIs act at the maturation step of the virus replication cycle. Next, we titrated drug concentrations over a range of viral inputs and measured eGFP expression in the absence or presence of drugs. DTG potently inhibited viral infection under low-MOI conditions; however, the efficacy of DTG was substantially reduced in an MOI-dependent manner (Fig. 10B). Under these conditions, infection with virus encoding a catalytically inactive IN mutant (IN-D116N) did not result in any eGFP+ cells (Fig. 10B), indicating that eGFP expression from unintegrated DNA does not contribute to the loss of DTG susceptibility at high viral input. Similar results were obtained with the two other INSTIs, RAL and CAB (Fig. 10C). In contrast to the results obtained with INSTIs, the NNRTIs RPV and EFV could completely inhibit viral infection independent of MOI (Fig. 10, C and D). Consistent with previous studies (33, 34), the NRTI FTC and the NRTTI EFdA exhibited MOI-dependent viral inhibition (Fig. 10, E and F). However, unlike DTG, high concentrations of FTC and EFdA could block viral infection under high-MOI conditions. The PI DRV blocked infection at all MOIs tested (Fig. 10G). We also tested the CA inhibitor LEN and the ALLINI BI-224436. Infection was completely blocked by these inhibitors independent of MOI (fig. S6, A and B). These data indicate that INSTIs are more readily overwhelmed by high MOI than other classes of ARVs.
Fig. 10. Impact of MOI on the sensitivity of HIV-1 to ARVs.
(A) The SupT1 T cell line was exposed to a range of VSV-G–pseudotyped eGFP reporter virus inputs in the absence of drugs. eGFP MFI was markedly increased when SupT1 T cells were exposed to high input of VSV-G–pseudotyped viruses. Data representative of three independent experiments are shown. (B) DTG sensitivity of VSV-G–pseudotyped eGFP reporter virus harboring WT-IN or the catalytically inactive IN-D116N mutant. The SupT1 T cell line was infected with a 30-fold range of viral inputs in the presence of the indicated concentrations of DTG. The number of infected cells was enumerated by flow cytometry. Sensitivity of VSV-G–pseudotyped eGFP reporter viruses to (C) INSTIs RAL and CAB, (D) NNRTIs RPV and EFV, (E) NRTI FTC, and (F) NRTTI EFdA and (G) PI DRV. (H) Experimental design of abrogation experiments. The SupT1 T cell line was exposed to VSV-G–pseudotyped HIV-1 (dark virus) over a range of viral inputs together with a fixed amount of VSV-G–pseudotyped eGFP reporter virus. The amount of dark virus was 0.3, 1, 3, or 10 times the amount of eGFP reporter virus. Viral inputs were normalized by RT activity, except for the catalytically inactive RT-D186N mutant, in which case viral inputs were normalized by Gag Western blots. (I) DTG sensitivity of VSV-G–pseudotyped eGFP reporter virus in the presence of dark virus harboring WT-Pol, IN-D116N, or RT-D186N. (J) RPV sensitivity of VSV-G–pseudotyped eGFP reporter virus in the presence of WT dark virus. Data from at least three independent experiments are shown as means ± SEM. *P < 0.05, unpaired t test.
To examine further the ability of high viral input to reduce the sensitivity of HIV-1 to INSTIs, we performed an abrogation assay by coinfecting the SupT1 T cell line with VSV-G–pseudotyped eGFP reporter virus and VSV-G–pseudotyped HIV-1 lacking a reporter (denoted as “dark virus”) in the presence of ARVs. The amount of eGFP virus was held constant, while the amount of dark virus was increased (Fig. 10H). We observed that as the input of WT dark virus increased, the inhibitory effect of DTG on reporter virus infection decreased (Fig. 10I). Mutant dark viruses harboring catalytically inactive IN (IN-D116N) or RT (RT-D186N) mutations, which do not affect GagPol incorporation into the virion (56), did not affect the inhibitory activity of DTG (Fig. 10I), indicating that reverse transcription and intasome formation are required for the dark virus to abrogate DTG-mediated inhibition of eGFP reporter virus infection. In contrast to the ability of WT dark virus to overcome DTG inhibition of eGFP reporter virus infection, and consistent with the high-MOI infection data (Fig. 10D), the NNRTI RPV was able to completely block eGFP reporter virus infection even in the presence of a 10-fold excess of dark virus (Fig. 10J). Collectively, the data in Fig. 10 indicate that the inhibitory activity of INSTIs is more vulnerable to high-MOI conditions than that of other classes of ARVs.
DISCUSSION
In this study, we performed long-term propagation of HIV-1 in the presence of the INSTI DTG and the RT inhibitor EFdA. Independent of viral strain, in the presence of DTG, HIV-1 sequentially accumulated multiple mutations in Env, the NC domain of Gag, and, on rare occasions, the IN-coding region. In contrast, in the presence of the RT inhibitor EFdA, HIV-1 rapidly acquired resistance mutations in RT along with several mutations in Env. We found that the accumulation of Env mutations leads to broad resistance to multiple classes of ARVs. Notably, however, the fold resistance conferred by the heavily mutated Envs is markedly higher for INSTIs than for other classes of ARVs and is associated with highly efficient cell-cell transfer.
To gain insights into the mechanism by which the heavily mutated Envs selected at high concentrations of DTG enhance cell-cell transfer capacity, we analyzed the impact of the Env mutations on Env function and conformation. We found that 7XEnv exhibits resistance to NAbs that bind CD4-induced epitopes. Relative to WT, 7XEnv displays a higher binding affinity for CD4-Ig, but reduced affinities for 17b and 447-52D, which recognize CD4-induced epitopes, even in the presence of sCD4. smFRET analysis suggests that 7XEnv samples more open conformations in the unliganded state relative to WT but does not undergo the downstream conformational changes in response to CD4 binding. 7XEnv exhibits reduced sCD4-induced gp120 shedding, demonstrating that the gp120-gp41 interaction is stabilized by the Env mutations despite a higher affinity of 7XEnv for CD4. The CH185-derived Env mutants H3-16 and H4-14, like the NL4-3–derived 7XEnv, exhibit increased CD4 binding but were more resistant to CD4-induced conformational changes than 7XEnv. H3-16 and H4-14 showed reduced cold inactivation, and reduced eCD4-Ig–induced gp120 shedding relative to WT. We speculate that the Env mutations that provide high-level resistance to DTG in the SupT1 T cell line exhibit higher energy barriers for CD4-induced conformational activation, resulting in impaired cell-free viral infection and fusion. In turn, because the binding affinity of mutant Env for CD4 is high, and the propensity for CD4-induced conformational change in Env is low, mutant Env trimers on infected cells or budding virions may be able to stably interact with CD4 on neighboring target cells and more efficiently form VSs compared to WT. Cryo–electron tomography studies of Env trimers interacting with membrane-bound CD4 have revealed several stable CD4-bound conformational intermediates before activation (66). Stable Env-CD4 interactions would promote the formation of strong VSs, leading to enhanced cell-cell transfer observed with the heavily mutated Envs.
In addition to accumulating multiple Env mutations, we observed that in the presence of high concentrations of DTG, independent of viral strain, HIV-1 consistently acquired mutations in the NC domain of Gag. NC mutations were not identified during propagation of HIV-1 in the presence of EFdA. The selected NC mutations confer modest levels of resistance to INSTIs but not to RT inhibitors. The effects of the NC mutations on conferring INSTI resistance are additive with IN mutations. These results indicate that the selected NC mutations are INSTI-specific resistance mutations. Further study is needed to elucidate the mechanism by which mutations in NC reduce the susceptibility of HIV-1 to INSTIs.
We demonstrate that the Env variants selected in the presence of DTG exhibit multi-log resistance to INSTIs in spreading infections but more modest (~10-fold) resistance to RT and PR inhibitors and ~2- to 3-fold resistance to the entry inhibitor T-20 and the capsid inhibitor LEN. On the basis of our model that the mechanism of Env-mediated drug resistance is linked to increased MOI conferred by enhanced cell-cell transfer capacity, we investigated whether the antiviral activity of INSTIs is more readily overwhelmed by high MOI relative to that of other classes of ARVs. These experiments were performed using VSV-G–pseudotyped eGFP reporter virus. We found that RT inhibitors, a PR inhibitor, LEN, and an ALLINI could completely block virus infection even at high MOI. In contrast, we found that INSTIs, while being very potent inhibitors at low MOI, are not capable of completely blocking virus infection under high-MOI conditions. No eGFP signal was seen with the catalytically inactive IN mutant IN-D116N, indicating that the eGFP signal detected at high MOI in our assays is not produced by unintegrated viral DNA known to accumulate when integration is blocked (56, 67). Consistent with previous studies (33, 34), the NRTI FTC and NRTTI EFdA require higher concentrations to inhibit viral infection in the context of high MOI relative to an NNRTI, which is highly effective against high-MOI conditions even at relatively low drug concentrations (see slopes of the inhibition curves in Fig. 10, D to F).
To explore further the phenomenon of INSTI resistance conferred by high MOI, we devised an abrogation assay whereby cells were coinfected with a fixed amount of VSV-G–pseudotyped eGFP reporter virus and increasing amounts of virus not encoding a fluorescent reporter (so-called dark virus). This approach allowed us to confirm that the inhibitory activity of an INSTI, but not an RT inhibitor, could be overwhelmed by high MOI. Dark virus bearing inactivating mutations in RT or IN was unable to overcome INSTI antiviral activity, indicating that reverse transcription is required for the abrogating activity. Furthermore, because the catalytically inactive IN mutant (IN-D116N) may not be able to form an intasome capable of INSTI binding (68), the results suggest that formation of a functional intasome is required for abrogation. We speculate that under high-MOI conditions large numbers of functional intasomes in an infected cell can “swamp out” the ability of INSTIs to completely block all viral complexes. This phenomenon is illustrated in fig. S7 using the analogy of soldiers crossing a moat to capture a castle; while most soldiers (viruses) are blocked by the moat (antiviral drug), if enough soldiers attempt to cross the moat (high MOI), at least one will be successful and take the castle (establish a productive infection). In this study, high MOI is conferred by the increased ability of the selected heavily mutated Envs to mediate cell-cell transfer. In theory, however, mutations that greatly increase the efficiency of cell-free infection (infection by free virions) could enhance the efficiency of infection and also reduce the susceptibility of HIV-1 to INSTIs.
We speculate that the differential effect of MOI on RT inhibitors versus INSTIs is likely due to their different mechanisms of action. Reverse transcription involves thousands of catalytic events per infection cycle, providing a large number of opportunities for RT inhibitors to block productive infection; in contrast, each productive infection cycle requires only a single integration event. INSTIs exhibit very slow dissociation rates from the intasome (69), perhaps allowing intasomes to “soak up” INSTIs present in the cell, reducing their effective concentration. Even a single intasome not bound to drug could carry out successful integration, leading to a productive infection event. Further analysis will be needed to fully decipher the mechanism(s) by which high MOIs, whether conferred by highly efficient cell-free or cell-cell infection, reduce the susceptibility of HIV-1 to INSTIs. It is important to note that while ALLINIs, like INSTIs, target IN, these two classes of inhibitors have very different mechanisms of action. INSTIs target integration itself (specifically, strand transfer), whereas ALLINIs act primarily late in the replication cycle by inducing IN aggregation, thereby preventing IN from promoting the packaging of viral genomic RNA into the capsid during maturation (70).
The results presented here and in our previous studies (45, 46) raise the key question of whether Env and/or NC mutations contribute to virological failure in individuals treated with INSTI-containing regimens. A number of studies have reported INSTI failure in people living with HIV in the absence of drug resistance mutations in IN (7, 10, 11). However, systematic evaluation of Env and NC mutations in these individuals has not been conducted. Analysis of Env mutations in patient-derived material is particularly challenging given the high degree of sequence variation across Env (71) and the context dependence of Env mutations described here; i.e., different viral clones lead to distinct panels of mutations, making it difficult to draw conclusions based simply on viral genotyping. It will therefore be necessary to perform labor-intensive phenotypic analysis of env genes derived from individuals failing INSTI-containing regimens. Nevertheless, our data point to several features conserved across viral subtypes that are associated with INSTI resistance: mutations at the gp120/gp41 interface and the V1/V2 region, increased stability of the gp120/gp41 interaction, and enhanced cell-cell viral transmission. Although the role of cell-cell transfer and the prevalence of multicopy infection in PLWH remains a matter of debate, analyses thus far, to our knowledge, have not been performed in the context of INSTI failure. Given our finding that the antiviral potency of INSTIs can be overwhelmed by high MOI, it appears plausible that INSTI therapy may be more likely to drive the evolution of mutations that confer highly efficient cell-cell transfer relative to other drug regimens. On the basis of our observations that Env mutations are able to rescue the replication defects imposed by established drug resistance mutations in IN (fig. S3), we suggest that Env mutations, by facilitating more efficient virus replication, may increase the opportunity for the virus to acquire drug resistance mutations in IN. According to this hypothesis, Env mutations could serve as “stepping stones” on the path to achieving high-level resistance conferred by target-gene mutations. Thus, moving forward, analysis of patient samples should include both samples with, and without, resistance mutations in IN. We consider it unlikely that, in vivo, mutations in Env will confer the multi-log INSTI resistance observed here, as highly efficient cell-cell transfer would likely lead to virus-induced cytopathic effect and detection of infected cells by the host immune system (44, 72, 73). However, even in the absence of multicopy infection, mutations that increase the MOI would likely contribute to the acquisition of drug resistance (as illustrated in fig. S7) particularly in the context of INSTI-containing regimens.
It is important to note that individuals on PI-containing regimens have also been reported to experience virological failure in the absence of mutations in PR (8, 9, 13). Mutations in the CT of gp41 have been associated with PI resistance in vivo (12), and mutations in the gp41 HR region analogous to the gp41 HR mutations reported here and previously (45, 46) were also associated with PI failure (8).
In summary, the findings of this study demonstrate the importance of mutations outside drug-target genes in resistance to ARVs and demonstrate that INSTIs are particularly sensitive to high-MOI conditions. These data suggest that a full understanding of HIV-1 drug resistance will require exploring the contribution of mutations outside drug-target genes to virological failure. As more-potent drugs with higher genetic barriers to resistance are developed, it may become increasingly important to identify and elucidate such unconventional drug resistance pathways. We refer here to this phenomenon as drug epistasis, whereby a drug that targets the product of one gene leads to the acquisition of mutations in another gene.
MATERIALS AND METHODS
Cell lines
HeLa, human embryonic kidney (HEK) 293T, and TZM-bl (74) cells were maintained in Dulbecco’s modified Eagle’s medium supplemented with 10% fetal bovine serum (FBS) at 37°C in 5% CO2. The SupT1 and SupT1huR5 T cell lines (52) were cultured in RPMI 1640 medium supplemented with 10% FBS at 37°C in 5% CO2. The SupT1huR5 T cell line was a gift from J. Hoxie (Perelman School of Medicine, University of Pennsylvania, Philadelphia, PA).
Compounds and NAbs
DTG, BMS-378806, and LEN were purchased from MedChemExpress. EFdA, CAB, and RPV were purchased from Cayman Chemical. sCD4, RAL, FTC, EFV, DRV, NFV, T-20, 17b, VRC03, F105, 16H3, CD4-Ig, FITC-conjugated p24 monoclonal Ab (KC57), and HIV-Ig were obtained from the National Institutes of Health (NIH) HIV Reagent Program. eCD4-Ig (65) was a gift from M. Alpart (Emmune Inc., FL). BI-224436 (57) was a gift from A. Engelman (Dana Farber Cancer Institute, Boston, MA).
Plasmids
The full-length HIV-1 molecular clones pNL4-3 (47), pNL(AD8) (51), and CH185 (previously denoted K3016, a gift from C. Ochsenbauer and J. Kappes, University of Alabama) (50) were used in this study. pBR-NL43-IRES-eGFP-nef+ (pBR43IeG), a proviral clone that coexpresses Nef and eGFP from a single bicistronic RNA, was obtained from F. Kirchhoff (Ulm University Medical Center, Germany) through the NIH HIV Reagent Program (58). pNL4-3 Env-A541V (Env-A539V, NL4-3 numbering) was described previously (45, 46). The pBR43IeG/KFS clone, which does not express Env, was described previously (45, 46). The VSV-G–expressing plasmid, pHCMVG, was a gift from J. Burns (75). Mutations in Gag-, RT-, and IN-coding regions were introduced into pNL4-3–based plasmids by overlap polymerase chain reaction (PCR).
Preparation of virus stocks
The HEK293T and HeLa cell lines were transfected with HIV-1 proviral DNA using Lipofectamine 2000 (Invitrogen). VSV-G–pseudotyped viruses were prepared by cotransfecting HEK293T cells with pBR43IeG KFS harboring WT-IN or IN-D116N, and pHCMVG, at DNA ratios of 10:1 or 5:1. At 48 hours after transfection, virus-containing supernatants were passed through a 0.45-μm membrane filter (Merck Millipore). The amount of virus in the supernatant was quantified by RT assay. RT assays were performed as described previously (45) with minor modifications. Briefly, after incubation of the virus supernatants with RT reaction mixtures, which contained a template primer of poly(rA) (5 μg/ml) and oligo(dT)12–18 primers (1.57 μg/ml), in 50 mM tris (pH 7.8), 75 mM KCl, 2 mM dithiothreitol, 5 mM MgCl2, 0.05% NP-40, and 0.25 μCi of 32P-dTTP (3′-deoxythymidine 5′-triphosphate) at 37°C for 3 hours, the mixtures were spotted onto positively charged nylon membrane (MilliporeSigma). After washing the nylon membranes with 2× SSC buffer (300 mM NaCl and 30 mM sodium citrate), levels of bound 32P were measured on a Wallac MicroBeta2 microplate counter (PerkinElmer). The 50% tissue culture infectious dose (TCID50) of the virus stocks was determined using TZM-bl cells. Briefly, 1.0 × 104 TZM-bl cells were incubated with serially diluted virus stocks in the presence of diethylaminoethyl (DEAE)–dextran (30 μg/ml). At 48 hours after infection, luciferase activity was measured using the Britelite plus reporter gene assay system (PerkinElmer) and GloMax Navigator microplate luminometer (Promega). TCID50 was calculated according to the Reed-Muench method (76).
Long-term passaging of HIV-1 in the presence of ARVs
Selection of HIV-1 variants resistant to ARVs was performed by serial passaging of NL4-3 in the SupT1 (NL4-3 and derivatives) or SupT1huR5 [NL(AD8) and CH185] T cell lines with increasing concentrations of DTG or EFdA. The culture supernatant was harvested on day 7 and used to infect fresh cells for the next round of culture in the presence of the same or increasing concentrations of the compounds. Dose escalation was performed when a cytopathic effect was observed. At the indicated time points, genomic DNA was extracted from infected cells using the DNeasy Blood and Tissue Minikit (Qiagen), and the Gag-, Pol-, Env-, and Nef-coding regions were amplified by PrimeSTAR GXL DNA polymerase (Takara Bio) and sequenced (Psomagen or Poochon Scientific) using previously described primers (77, 78).
Construction of infectious molecular clones containing Env mutants
The Env mutants were constructed by the overlap PCR method (79). For 7XEnv*, the entire env region was amplified from genomic DNA from NL4-3–infected cells using the following primers (79): NL(6232)F (5′-AGCAGAAGACAGTGGCAATGAGAGTGAAG-3′) and EnvR (5′-TTTTGACCACTTGCCACCCATCTTATAGC-3′) (ref). A fragment encompassing the Eco RI restriction site in pNL4-3 was amplified using NL(5284)F (5′-GGTCAGGGAGTCTCCATAGAA-TGGAGG-3′) and NL(6232)Rv (5′-CTTCACTCTCATTGCCACT-GTCTTCTGCT-3′). Another fragment encompassing the Xho I restriction site in pNL4-3 was amplified using NL(8779)F (5′-GCTA-TAAGATGGGTGGCAAGTGGTCAAAA-3′) and NL(9045)R (5′-GATCTACAGCTGCCTTGTAAGTCATTGGTC-3′). Overlap PCR was performed using these three PCR products and primers NL(5284)F and NL(9045)R. The chimeric env amplicon was cloned into pNL4-3 using Eco RI and Xho I restriction sites. For 7XEnv, the Vpu-V60L/SP-K6N in 7XEnv* was mutated back to the WT sequence by overlap PCR. pBR43IeG 7XEnv was constructed using Age I and Xho I restriction sites in WT pNL4-3 and 7XEnv. For CH185 heavily mutated clones (H3-16 and H4-14), the entire env regions were amplified using K3016_env_startF (5′-AGTGGCAATGAGAGCGA-3′) and K3016_env_end R (5′-CATTTTATAGCAAAGCTGCT-3′). A fragment encompassing the Pac I restriction site in pK3016 was amplified using K3016_Pac I_F (5′-GGAGCCAGTAGATCCTAAC-CTAGAG-3′) and K3016_env_startR (5′-TCGCTCTCATTG-CCACT-3′). Another fragment encompassing the Xho I restriction site in pK3016 was amplified using K3016_env_end_F (5′-AGCAGCTTTGCTATAAAATG-3′) and K3016_Xho I_R (5′-CCCGGCGTGTCAATAATATCAC-3′). Overlap PCR was performed using these three PCR products and primers K3016_Pac I_F and K3016_Xho I_R. The chimeric env amplicon was cloned into pCH185 using Pac I and Xho I restriction sites. Constructed plasmids were verified by Sanger DNA sequencing (Psomagen).
Virus replication kinetic assays
Virus replication was monitored in SupT1 cells as previously described (45). Briefly, SupT1 cells were incubated with pNL4-3 clones (1.0 μg of DNA/1.0 × 106 cells) in the presence of DEAE-dextran (700 μg/ml) at 37°C for 15 min. Transfected cells (1.5 × 105 cells) were plated in 96-well flat-bottom plates and incubated at 37°C in the presence of various concentrations of ARVs. For CCR5-tropic viruses, SupT1huR5 cells were used. Aliquots of supernatants were collected to measure RT activity, and cells were split 1:3 every other day with fresh drug and medium. IC50 values were calculated on the basis of the AUC of the replication kinetics. IC50 was defined as the amount of inhibitor required to reduce the AUC by 50%.
Single-round, cell-free virus infectivity assays
Single-round infectivity assays were performed as previously described with minor modifications (45). TZM-bl cells (1.0 × 104 cells) in 96-well plates were incubated with RT-normalized virus stocks produced in HeLa cells. For drug sensitivity assays, 100 TCID50 of virus produced in 293T cells was incubated with TZM-bl cells (1.0 × 104 cells) in the presence of various concentrations of drugs or NAbs with DEAE-dextran (30 μg/ml). At 48 hours after infection, luciferase activity was measured using the Britelite plus reporter gene assay system (PerkinElmer) and GloMax Navigator microplate luminometer (Promega).
Cell-cell fusion assay
HEK293T cells were transfected with HIV-1 proviral DNA as described above. At 24 hours after transfection, serial dilutions of the transfected HEK293T cells were cocultured with 1.0 × 104 TZM-bl cells in the presence of a cocktail of RPV and DTG (1000 nM each) to prevent viral infection. At 24 hours after coculture, luciferase activity was measured as described above.
Cell-cell transfer assay
Virus-producing cells were prepared by spinoculation (2500g for 2.5 hours). Target cells were labeled with 1.0 μM dye from the CellTrace Far Red Cell Proliferation kit (Invitrogen). Forty-eight hours after spinoculation, virus-producing cells were cocultured with the same number of dye-labeled target cells in the absence or presence of DTG or RPV. The number of virus-producing cells was normalized by intracellular p24 antigen before coculture. In parallel with coculture experiments, virus-producing cells were seeded onto the insert of a transwell (Corning) with the labeled target cells in the lower chamber to measure cell-free viral infection. Twenty-four hours after coculture, the cells were fixed and permeabilized, and then incubated with KC57-FITC–conjugated Ab to stain intracellular p24 antigen. For cell-cell transfer assay using eGFP reporter virus, 1.0 μM EFV was added to the culture 24 hours after coculture of virus-producing cells with the target cells, and then the cultures were incubated for an additional 24 hours. Samples were analyzed with a FACSCalibur (BD Biosciences) flow cytometer.
Infection with VSV-G–pseudotyped eGFP reporter viruses
The SupT1 T cell line was incubated with VSV-G–pseudotyped eGFP reporter viruses over a range of viral inputs at 37°C for 2 hours. For competition experiments, the SupT1 T cell line was incubated with a fixed amount of VSV-G–pseudotyped eGFP reporter virus with varying amounts of VSV-G–pseudotyped HIV-1 (dark virus). Following incubation, the cells were washed and cultured in the presence or absence of drugs. Forty-eight after infection, eGFP expression was monitored by flow cytometry using a FACSCalibur flow cytometer (BD Biosciences). Data were analyzed by FCS Express Cytometry software 7 (De Novo Software) or FlowJo software (Tree Star Inc.).
Gp120 shedding assay
The gp120 shedding assay was performed as previously described (45). Briefly, concentrated viruses produced from HeLa cells were incubated with sCD4 or eCD4-Ig at 37°C for 2 hours. Following incubation, the viruses were purified by ultracentrifugation through 20% sucrose cushions (60,000g) for 45 min at 4°C. The levels of virion-associated gp160, gp120, and p24 were determined by Western blotting. Viral proteins were separated by SDS–polyacrylamide gel electrophoresis and transferred to polyvinylidene disulfide membranes (Merck Millipore). After blocking the membranes with Azure Fluorescent Blot Blocking Buffer (Azure Biosystems), the membranes were probed with 16H3 (1:10,000) and HIVIg (1:10,000) at 4°C overnight and then incubated for 1 hour with species-specific AzureSpectra Fluorescent Secondary Antibodies (Azure Biosystems). After the final washes, bands were detected by chemiluminescence with a Sapphire Biomolecular imager (Azure Biosystems). Quantification was performed using Image Studio Lite (LI-COR Biosciences) software.
Cold sensitivity assay
Cold sensitivity assay was performed as previously described with minor modifications (21). Virus aliquots (100 to 200 TCID50) were incubated at 4°C for different periods of time up to 120 hours. Following incubations, the viruses were stored at −80°C until infection. The frozen aliquots were quickly thawed at 37°C and incubated with 1.0 × 104 TZM-bl cells. Forty-eight hours after infection, luciferase activity was measured as described above.
Ab binding assay
HEK293T cells were transfected with pBR43IeG constructs as indicated above. At 48 hours after transfection, the cells were detached with 5 mM EDTA–phosphate-buffered saline (PBS) and washed with PBS. Cells (2.0 × 105) were incubated with the specified Abs at a final concentration of 1.0 to 3.0 μg/ml at 37°C. After 30 min of incubation, the cells were washed with PBS and incubated with Alexa Fluor 647–conjugated goat anti-human IgG Ab (Thermo Fisher Scientific). For CH185 Env mutants, intracellular p24 was stained with FITC-conjugated anti-p24 Ab after Env staining. The cells were washed with PBS and fixed with 4% paraformaldehyde (Boston Biosciences). Fixed cells were analyzed with a FACSCalibur (BD Biosciences) flow cytometer. Data were analyzed by FCS Express Cytometry Software 7 (De Novo Software) and FlowJo software (Tree Star Inc.). The Alexa Fluor 647 signal was normalized by eGFP or p24-FITC signals.
smFRET analysis of HIV-1 Env
HIV-1 NL4-3ΔRT virus particles containing WT or 7X Env were produced, labeled, and analyzed by smFRET as described previously (20). Briefly, HEK293T cells were plated in 10-cm dishes and transfected with NL4-3ΔRT plasmids encoding untagged and V1Q3/V4A1 peptide-tagged Env at a 40:1 ratio. Each dish was transfected with a total of 10 mg of DNA using PEI. Virus particles were harvested at 48 hours after transfection, passed through 0.45-mm syringe filters, and pelleted through 15% sucrose cushions. Viruses were then labeled overnight at ambient temperature with donor (Cy3) and acceptor (Cy5) fluorophores using recombinant acyl carrier protein synthase (ACPS) and transglutaminase enzymes. Viruses were then incubated with 1,2-distearoyl-sn-glycero-3-phosphoethanolamine-N-[biotinyl(polyethylene glycol)-2000] (ammonium salt) (DSPE)–biotin lipid and purified using continuous 6 to 18% iodixanol gradients. Purified virus particles were immobilized on streptavidin-passivated fused silica slides, placed in imaging buffer containing an enzymatic oxygen scavenging system and triplet-state quenchers, and imaged on a custom-built prism total internal reflection fluorescence microscope. For sCD4 experiments, virus particles were incubated with sCD4 proteins at 100 μg/ml for 60 min at ambient temperature before imaging. Particles were also imaged in the presence of sCD4 at the same concentration. smFRET data were analyzed in MATLAB using the SPARTAN analysis suite and custom MATLAB scripts (80).
Acknowledgments
We thank members of the Freed laboratory for discussions and critical review of the manuscript. We thank V. Pathak and W.-S. Hu for discussions and suggestions during manuscript preparation. We also thank S. Hughes, S. Smith, and T. Izumi for discussions. We thank J. Hoxie for providing the SupT1huR5 cell line. We also thank M. Alpart for providing eCD4-Ig. sCD4 protein was a gift from A. Finzi. 12XCD4 protein (sCD4-D1D2-Igatp) was a gift from J. Arthos and A. Fauci. We are also grateful to C. Ochsenbauer and J. Kappes for providing the pCH185 (pK3016) molecular clone. We thank J. Thomas and C. Mac Trubey for help with the flow cytometry analysis. The following reagents were obtained through the NIH HIV Reagent Program, NIAID, NIH: RAL, FTC, EFV, DRV, NFV, T-20, 17b, VRC03, F105, 16H3, CD4-Ig, FITC-conjugated p24–monoclonal antibody (KC57), and HIVIg.
Funding: Research in the Freed laboratory was supported by the Intramural Research Program of the Center for Cancer Research, National Cancer Institute, NIH. Y.H. was supported by a JSPS Research Fellowship for Japanese Biomedical and Behavioral Researchers at the NIH and an Intramural AIDS Research Fellowship. A.S. was supported by a Gruber Science Fellowship at Yale University. This work was also supported by federal grant R37AI150560 to W.M.
Author contributions: Conceptualization: Y.H., J.R.G., and E.O.F. Investigation: Y.H. performed the long-term passaging experiments, mutagenesis, infection, replication kinetics, drug sensitivity, Ab binding, and gp120 shedding assay; A.S. and J.R.G. performed smFRET analysis. Formal analysis: Y.H., J.R.G., and A.S. Methodology: Y.H., J.R.G., W.M., and E.O.F. Resources: Y.H. and J.R.G. Data curation: Y.H. and J.R.G. Validation: Y.H., J.R.G., and E.O.F. Software: J.R.G. Project administration: W.M. and E.O.F. Supervision: W.M. and E.O.F. Visualization: Y.H., J.R.G., and E.O.F. Funding: Y.H., A.S., W.M., and E.O.F. Writing—original draft: Y.H., J.R.G., and E.O.F. Writing—review and editing: Y.H., J.R.G., A.S., W.M., and E.O.F.
Competing interests: The authors declare that they have no competing interests.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. All materials will be made available upon request.
Supplementary Materials
This PDF file includes:
Figs. S1 to S7
REFERENCES AND NOTES
- 1.T. R. Kemnic, P. G. Gulick, HIV Antiretroviral Therapy (StatPearls Publishing, 2022). [PubMed] [Google Scholar]
- 2.Link J. O., Rhee M. S., Tse W. C., Zheng J., Somoza J. R., Rowe W., Begley R., Chiu A., Mulato A., Hansen D., Singer E., Tsai L. K., Bam R. A., Chou C. H., Canales E., Brizgy G., Zhang J. R., Li J., Graupe M., Morganelli P., Liu Q., Wu Q., Halcomb R. L., Saito R. D., Schroeder S. D., Lazerwith S. E., Bondy S., Jin D., Hung M., Novikov N., Liu X., Villasenor A. G., Cannizzaro C. E., Hu E. Y., Anderson R. L., Appleby T. C., Lu B., Mwangi J., Liclican A., Niedziela-Majka A., Papalia G. A., Wong M. H., Leavitt S. A., Xu Y., Koditek D., Stepan G. J., Yu H., Pagratis N., Clancy S., Ahmadyar S., Cai T. Z., Sellers S., Wolckenhauer S. A., Ling J., Callebaut C., Margot N., Ram R. R., Liu Y. P., Hyland R., Sinclair G. I., Ruane P. J., Crofoot G. E., McDonald C. K., Brainard D. M., Lad L., Swaminathan S., Sundquist W. I., Sakowicz R., Chester A. E., Lee W. E., Daar E. S., Yant S. R., Cihlar T., Clinical targeting of HIV capsid protein with a long-acting small molecule. Nature 584, 614–618 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Collier D. A., Monit C., Gupta R. K., The impact of HIV-1 drug escape on the global treatment landscape. Cell Host Microbe 26, 48–60 (2019). [DOI] [PubMed] [Google Scholar]
- 4.Jozwik I. K., Passos D. O., Lyumkis D., Structural biology of HIV integrase strand transfer inhibitors. Trends Pharmacol. Sci. 41, 611–626 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Clutter D. S., Jordan M. R., Bertagnolio S., Shafer R. W., HIV-1 drug resistance and resistance testing. Infect. Genet. Evol. 46, 292–307 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gandhi R. T., Bedimo R., Hoy J. F., Landovitz R. J., Smith D. M., Eaton E. F., Lehmann C., Springer S. A., Sax P. E., Thompson M. A., Benson C. A., Buchbinder S. P., Del Rio C., Eron J. J. Jr., Gunthard H. F., Molina J. M., Jacobsen D. M., Saag M. S., Antiretroviral Drugs for treatment and prevention of HIV infection in adults. JAMA 329, 63–84 (2023). [DOI] [PubMed] [Google Scholar]
- 7.La Rosa A. M., Harrison L. J., Taiwo B., Wallis C. L., Zheng L., Kim P., Kumarasamy N., Hosseinipour M. C., Jarocki B., Mellors J. W., Collier A. C., ACTG A5273 Study Group , Raltegravir in second-line antiretroviral therapy in resource-limited settings (SELECT): A randomised, phase 3, non-inferiority study. Lancet HIV 3, e247–e258 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Coetzer M., Ledingham L., Diero L., Kemboi E., Orido M., Kantor R., Gp41 and Gag amino acids linked to HIV-1 protease inhibitor-based second-line failure in HIV-1 subtype A from Western Kenya. J. Int. AIDS Soc. 20, e25024 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Castain L., Perrier M., Charpentier C., Palich R., Desire N., Wirden M., Descamps D., Sayon S., Landman R., Valantin M. A., Joly V., Peytavin G., Yazdanpanah Y., Katlama C., Calvez V., Marcelin A. G., Todesco E., New mechanisms of resistance in virological failure to protease inhibitors: Selection of non-described protease, Gag and Gp41 mutations. J. Antimicrob. Chemother. 74, 2019–2023 (2019). [DOI] [PubMed] [Google Scholar]
- 10.Hocqueloux L., Raffi F., Prazuck T., Bernard L., Sunder S., Esnault J. L., Rey D., Le Moal G., Roncato-Saberan M., Andre M., Billaud E., Valery A., Avettand-Fenoel V., Parienti J. J., Allavena C., MONCAY Study Group , Dolutegravir monotherapy versus dolutegravir/abacavir/lamivudine for virologically suppressed people living with chronic human immunodeficiency virus infection: The randomized noninferiority MONotherapy of TiviCAY trial. Clin. Infect. Dis. 69, 1498–1505 (2019). [DOI] [PubMed] [Google Scholar]
- 11.Wijting I. E. A., Lungu C., Rijnders B. J. A., van der Ende M. E., Pham H. T., Mesplede T., Pas S. D., Voermans J. J. C., Schuurman R., van de Vijver D. A. M. C., Boers P. H. M., Gruters R. A., Boucher C. A. B., van Kampen J. J. A., HIV-1 resistance dynamics in patients with virologic failure to dolutegravir maintenance monotherapy. J. Infect. Dis. 218, 688–697 (2018). [DOI] [PubMed] [Google Scholar]
- 12.Rabi S. A., Laird G. M., Durand C. M., Laskey S., Shan L., Bailey J. R., Chioma S., Moore R. D., Siliciano R. F., Multi-step inhibition explains HIV-1 protease inhibitor pharmacodynamics and resistance. J. Clin. Investig. 123, 3848–3860 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fun A., Wensing A. M., Verheyen J., Nijhuis M., Human immunodeficiency virus Gag and protease: Partners in resistance. Retrovirology 9, 63 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Richetta C., Subra F., Malet I., Leh H., Charpentier C., Corona A., Collin G., Descamps D., Deprez E., Parissi V., Calvez V., Tramontano E., Marcelin A. G., Delelis O., Mutations in the 3'-PPT lead to HIV-1 replication without integration. J. Virol. 96, e0067622 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Malet I., Subra F., Charpentier C., Collin G., Descamps D., Calvez V., Marcelin A. G., Delelis O., Mutations located outside the integrase gene can confer resistance to HIV-1 integrase strand transfer inhibitors. MBio 8, e00922-17 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dekker J. G., Klaver B., Berkhout B., Das A. T., HIV-1 3'-polypurine tract mutations confer dolutegravir resistance by switching to an integration-independent replication mechanism via 1-LTR circles. J. Virol. 97, e0036123 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Smith S. J., Ferris A., Zhao X., Pauly G., Schneider J. P., Burke T. R. Jr., Hughes S. H., INSTIs and NNRTIs potently inhibit HIV-1 polypurine tract mutants in a single round infection assay. Viruses 13, 2501 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen B., Molecular mechanism of HIV-1 entry. Trends Microbiol. 27, 878–891 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ma X., Lu M., Gorman J., Terry D. S., Hong X., Zhou Z., Zhao H., Altman R. B., Arthos J., Blanchard S. C., Kwong P. D., Munro J. B., Mothes W., HIV-1 Env trimer opens through an asymmetric intermediate in which individual protomers adopt distinct conformations. eLife 7, e34271 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Munro J. B., Gorman J., Ma X., Zhou Z., Arthos J., Burton D. R., Koff W. C., Courter J. R., Smith A. B. III, Kwong P. D., Blanchard S. C., Mothes W., Conformational dynamics of single HIV-1 envelope trimers on the surface of native virions. Science 346, 759–763 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Herschhorn A., Ma X., Gu C., Ventura J. D., Castillo-Menendez L., Melillo B., Terry D. S., Smith A. B. III, Blanchard S. C., Munro J. B., Mothes W., Finzi A., Sodroski J., Release of GP120 restraints leads to an entry-competent intermediate state of the HIV-1 envelope glycoproteins. MBio 7, e01598-16 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Alsahafi N., Bakouche N., Kazemi M., Richard J., Ding S., Bhattacharyya S., Das D., Anand S. P., Prevost J., Tolbert W. D., Lu H., Medjahed H., Gendron-Lepage G., Ortega Delgado G. G., Kirk S., Melillo B., Mothes W., Sodroski J., Smith A. B. III, Kaufmann D. E., Wu X., Pazgier M., Rouiller I., Finzi A., Munro J. B., An asymmetric opening of HIV-1 envelope mediates antibody-dependent cellular cytotoxicity. Cell Host Microbe 25, 578–587.e5 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhao C., Li H., Swartz T. H., Chen B. K., The HIV Env glycoprotein conformational states on cells and viruses. MBio 13, e0182521 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lawrence S. P., Elser S. E., Torben W., Blair R. V., Pahar B., Aye P. P., Schiro F., Szeltner D., Doyle-Meyers L. A., Haggarty B. S., Jordan A. P. O., Romano J., Leslie G. J., Alvarez X., O'Connor D. H., Wiseman R. W., Fennessey C. M., Li Y., Piatak M. Jr., Lifson J. D., LaBranche C. C., Lackner A. A., Keele B. F., Maness N. J., Marsh M., Hoxie J. A., A cellular trafficking signal in the SIV envelope protein cytoplasmic domain is strongly selected for in pathogenic infection. PLOS Pathog. 18, e1010507 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Breed M. W., Jordan A. P., Aye P. P., Lichtveld C. F., Midkiff C. C., Schiro F. R., Haggarty B. S., Sugimoto C., Alvarez X., Sandler N. G., Douek D. C., Kuroda M. J., Pahar B., Piatak M. Jr., Lifson J. D., Keele B. F., Hoxie J. A., Lackner A. A., Loss of a tyrosine-dependent trafficking motif in the simian immunodeficiency virus envelope cytoplasmic tail spares mucosal CD4 cells but does not prevent disease progression. J. Virol. 87, 1528–1543 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Checkley M. A., Luttge B. G., Freed E. O., HIV-1 envelope glycoprotein biosynthesis, trafficking, and incorporation. J. Mol. Biol. 410, 582–608 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dufloo J., Bruel T., Schwartz O., HIV-1 cell-to-cell transmission and broadly neutralizing antibodies. Retrovirology 15, 51 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Symeonides M., Murooka T. T., Bellfy L. N., Roy N. H., Mempel T. R., Thali M., HIV-1-induced small T cell syncytia can transfer virus particles to target cells through transient contacts. Viruses 7, 6590–6603 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Murakami T., Ablan S., Freed E. O., Tanaka Y., Regulation of human immunodeficiency virus type 1 Env-mediated membrane fusion by viral protease activity. J. Virol. 78, 1026–1031 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhong P., Agosto L. M., Ilinskaya A., Dorjbal B., Truong R., Derse D., Uchil P. D., Heidecker G., Mothes W., Cell-to-cell transmission can overcome multiple donor and target cell barriers imposed on cell-free HIV. PLOS ONE 8, e53138 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Russell R. A., Martin N., Mitar I., Jones E., Sattentau Q. J., Multiple proviral integration events after virological synapse-mediated HIV-1 spread. Virology 443, 143–149 (2013). [DOI] [PubMed] [Google Scholar]
- 32.Del Portillo A., Tripodi J., Najfeld V., Wodarz D., Levy D. N., Chen B. K., Multiploid Inheritance of HIV-1 during cell-to-cell infection. J. Virol. 85, 7169–7176 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Agosto L. M., Zhong P., Munro J., Mothes W., Highly active antiretroviral therapies are effective against HIV-1 cell-to-cell transmission. PLOS Pathog. 10, e1003982 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sigal A., Kim J. T., Balazs A. B., Dekel E., Mayo A., Milo R., Baltimore D., Cell-to-cell spread of HIV permits ongoing replication despite antiretroviral therapy. Nature 477, 95–98 (2011). [DOI] [PubMed] [Google Scholar]
- 35.Ikeda T., Symeonides M., Albin J. S., Li M., Thali M., Harris R. S., HIV-1 adaptation studies reveal a novel Env-mediated homeostasis mechanism for evading lethal hypermutation by APOBEC3G. PLOS Pathog. 14, e1007010 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kieffer C., Ladinsky M. S., Ninh A., Galimidi R. P., Bjorkman P. J., Longitudinal imaging of HIV-1 spread in humanized mice with parallel 3D immunofluorescence and electron tomography. eLife 6, e23282 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ladinsky M. S., Khamaikawin W., Jung Y., Lin S., Lam J., An D. S., Bjorkman P. J., Kieffer C., Mechanisms of virus dissemination in bone marrow of HIV-1-infected humanized BLT mice. eLife 8, e46916 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Law K. M., Komarova N. L., Yewdall A. W., Lee R. K., Herrera O. L., Wodarz D., Chen B. K., In vivo HIV-1 cell-to-cell transmission promotes multicopy micro-compartmentalized infection. Cell Rep. 15, 2771–2783 (2016). [DOI] [PubMed] [Google Scholar]
- 39.Ladinsky M. S., Kieffer C., Olson G., Deruaz M., Vrbanac V., Tager A. M., Kwon D. S., Bjorkman P. J., Electron tomography of HIV-1 infection in gut-associated lymphoid tissue. PLOS Pathog. 10, e1003899 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Murooka T. T., Deruaz M., Marangoni F., Vrbanac V. D., Seung E., von Andrian U. H., Tager A. M., Luster A. D., Mempel T. R., HIV-infected T cells are migratory vehicles for viral dissemination. Nature 490, 283–287 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Suspène R., Meyerhans A., Quantification of unintegrated HIV-1 DNA at the single cell level in vivo. PLOS ONE 7, e36246 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jung A., Maier R., Vartanian J. P., Bocharov G., Jung V., Fischer U., Meese E., Wain-Hobson S., Meyerhans A., Recombination: Multiply infected spleen cells in HIV patients. Nature 418, 144 (2002). [DOI] [PubMed] [Google Scholar]
- 43.Gratton S., Cheynier R., Dumaurier M. J., Oksenhendler E., Wain-Hobson S., Highly restricted spread of HIV-1 and multiply infected cells within splenic germinal centers. Proc. Natl. Acad. Sci. U.S.A. 97, 14566–14571 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Josefsson L., Palmer S., Faria N. R., Lemey P., Casazza J., Ambrozak D., Kearney M., Shao W., Kottilil S., Sneller M., Mellors J., Coffin J. M., Maldarelli F., Single cell analysis of lymph node tissue from HIV-1 infected patients reveals that the majority of CD4+ T-cells contain one HIV-1 DNA molecule. PLOS Pathog. 9, e1003432 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hikichi Y., Van Duyne R., Pham P., Groebner J. L., Wiegand A., Mellors J. W., Kearney M. F., Freed E. O., Mechanistic analysis of the broad antiretroviral resistance conferred by HIV-1 envelope glycoprotein mutations. MBio 12, e03134-20 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Van Duyne R., Kuo L. S., Pham P., Fujii K., Freed E. O., Mutations in the HIV-1 envelope glycoprotein can broadly rescue blocks at multiple steps in the virus replication cycle. Proc. Natl. Acad. Sci. U.S.A. 116, 9040–9049 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Adachi A., Gendelman H. E., Koenig S., Folks T., Willey R., Rabson A., Martin M. A., Production of acquired immunodeficiency syndrome-associated retrovirus in human and nonhuman cells transfected with an infectious molecular clone. J. Virol. 59, 284–291 (1986). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shafer R. W., Rationale and uses of a public HIV drug-resistance database. J. Infect. Dis. 194 (Suppl. 1), S51–S58 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rhee S. Y., Grant P. M., Tzou P. L., Barrow G., Harrigan P. R., Ioannidis J. P. A., Shafer R. W., A systematic review of the genetic mechanisms of dolutegravir resistance. J. Antimicrob. Chemother. 74, 3135–3149 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Parrish N. F., Gao F., Li H., Giorgi E. E., Barbian H. J., Parrish E. H., Zajic L., Iyer S. S., Decker J. M., Kumar A., Hora B., Berg A., Cai F., Hopper J., Denny T. N., Ding H., Ochsenbauer C., Kappes J. C., Galimidi R. P., West A. P. Jr., Bjorkman P. J., Wilen C. B., Doms R. W., O'Brien M., Bhardwaj N., Borrow P., Haynes B. F., Muldoon M., Theiler J. P., Korber B., Shaw G. M., Hahn B. H., Phenotypic properties of transmitted founder HIV-1. Proc. Natl. Acad. Sci. U.S.A. 110, 6626–6633 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Freed E. O., Englund G., Martin M. A., Role of the basic domain of human immunodeficiency virus type 1 matrix in macrophage infection. J. Virol. 69, 3949–3954 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Means R. E., Matthews T., Hoxie J. A., Malim M. H., Kodama T., Desrosiers R. C., Ability of the V3 loop of simian immunodeficiency virus to serve as a target for antibody-mediated neutralization: Correlation of neutralization sensitivity, growth in macrophages, and decreased dependence on CD4. J. Virol. 75, 3903–3915 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Diamond T. L., Ngo W., Xu M., Goh S. L., Rodriguez S., Lai M. T., Asante-Appiah E., Grobler J. A., Islatravir has a high barrier to resistance and exhibits a differentiated resistance profile from approved Nucleoside Reverse Transcriptase Inhibitors (NRTIs). Antimicrob. Agents Chemother. 66, e0013322 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Cilento M. E., Reeve A. B., Michailidis E., Ilina T. V., Nagy E., Mitsuya H., Parniak M. A., Tedbury P. R., Sarafianos S. G., Development of human immunodeficiency virus type 1 resistance to 4′-ethynyl-2-fluoro-2′-deoxyadenosine starting with wild-type or nucleoside reverse transcriptase inhibitor-resistant strains. Antimicrob. Agents Chemother. 65, e0116721 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Irwan I. D., Karnowski H. L., Bogerd H. P., Tsai K., Cullen B. R., Reversal of epigenetic silencing allows robust HIV-1 replication in the absence of integrase function. MBio 11, e01038-20 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Engelman A., Englund G., Orenstein J. M., Martin M. A., Craigie R., Multiple effects of mutations in human immunodeficiency virus type 1 integrase on viral replication. J. Virol. 69, 2729–2736 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Engelman A. N., Multifaceted HIV integrase functionalities and therapeutic strategies for their inhibition. J. Biol. Chem. 294, 15137–15157 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Schindler M., Würfl S., Benaroch P., Greenough T. C., Daniels R., Easterbrook P., Brenner M., Münch J., Kirchhoff F., Down-modulation of mature major histocompatibility complex class II and up-regulation of invariant chain cell surface expression are well-conserved functions of human and simian immunodeficiency virus nef alleles. J. Virol. 77, 10548–10556 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lu M., Ma X., Castillo-Menendez L. R., Gorman J., Alsahafi N., Ermel U., Terry D. S., Chambers M., Peng D., Zhang B., Zhou T., Reichard N., Wang K., Grover J. R., Carman B. P., Gardner M. R., Nikic-Spiegel I., Sugawara A., Arthos J., Lemke E. A., Smith A. B. III, Farzan M., Abrams C., Munro J. B., McDermott A. B., Finzi A., Kwong P. D., Blanchard S. C., Sodroski J. G., Mothes W., Associating HIV-1 envelope glycoprotein structures with states on the virus observed by smFRET. Nature 568, 415–419 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Jia M., Lu H., Markowitz M., Cheng-Mayer C., Wu X., Development of broadly neutralizing antibodies and their mapping by monomeric gp120 in human immunodeficiency virus type 1-infected humans and simian-human immunodeficiency virus SHIVSF162P3N-infected macaques. J. Virol. 90, 4017–4031 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Johnson J., Zhai Y., Salimi H., Espy N., Eichelberger N., DeLeon O., O'Malley Y., Courter J., Smith A. B. III, Madani N., Sodroski J., Haim H., Induction of a tier-1-like phenotype in diverse tier-2 isolates by agents that guide HIV-1 Env to perturbation-sensitive, nonnative states. J. Virol. 91, e00174-17 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Pacheco B., Alsahafi N., Debbeche O., Prevost J., Ding S., Chapleau J. P., Herschhorn A., Madani N., Princiotto A., Melillo B., Gu C., Zeng X., Mao Y., Smith A. B. III, Sodroski J., Finzi A., Residues in the gp41 ectodomain regulate HIV-1 envelope glycoprotein conformational transitions induced by gp120-directed inhibitors. J. Virol. 91, e02219-16 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Arthos J., Cicala C., Steenbeke T. D., Chun T. W., Dela Cruz C., Hanback D. B., Khazanie P., Nam D., Schuck P., Selig S. M., Van Ryk D., Chaikin M. A., Fauci A. S., Biochemical and biological characterization of a dodecameric CD4-Ig fusion protein: Implications for therapeutic and vaccine strategies. J. Biol. Chem. 277, 11456–11464 (2002). [DOI] [PubMed] [Google Scholar]
- 64.Moore J. P., McKeating J. A., Huang Y. X., Ashkenazi A., Ho D. D., Virions of primary human immunodeficiency virus type 1 isolates resistant to soluble CD4 (sCD4) neutralization differ in sCD4 binding and glycoprotein gp120 retention from sCD4-sensitive isolates. J. Virol. 66, 235–243 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Fetzer I., Gardner M. R., Davis-Gardner M. E., Prasad N. R., Alfant B., Weber J. A., Farzan M., ECD4-Ig variants that more potently neutralize HIV-1. J. Virol. 92, e02011-17 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Li W., Qin Z., Nand E., Grunst M. W., Grover J. R., Bess J. W. Jr., Lifson J. D., Zwick M. B., Tagare H. D., Uchil P. D., Mothes W., HIV-1 Env trimers asymmetrically engage CD4 receptors in membranes. Nature 623, 1026–1033 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Hazuda D. J., Felock P., Witmer M., Wolfe A., Stillmock K., Grobler J. A., Espeseth A., Gabryelski L., Schleif W., Blau C., Miller M. D., Inhibitors of strand transfer that prevent integration and inhibit HIV-1 replication in cells. Science 287, 646–650 (2000). [DOI] [PubMed] [Google Scholar]
- 68.Chen H., Wei S. Q., Engelman A., Multiple integrase functions are required to form the native structure of the human immunodeficiency virus type I intasome. J. Biol. Chem. 274, 17358–17364 (1999). [DOI] [PubMed] [Google Scholar]
- 69.Hightower K. E., Wang R., Deanda F., Johns B. A., Weaver K., Shen Y., Tomberlin G. H., Carter H. L. III, Broderick T., Sigethy S., Seki T., Kobayashi M., Underwood M. R., Dolutegravir (S/GSK1349572) exhibits significantly slower dissociation than raltegravir and elvitegravir from wild-type and integrase inhibitor-resistant HIV-1 integrase-DNA complexes. Antimicrob. Agents Chemother. 55, 4552–4559 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kleinpeter A. B., Freed E. O., HIV-1 maturation: Lessons learned from inhibitors. Viruses 12, 940 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Gaschen B., Taylor J., Yusim K., Foley B., Gao F., Lang D., Novitsky V., Haynes B., Hahn B. H., Bhattacharya T., Korber B., Diversity considerations in HIV-1 vaccine selection. Science 296, 2354–2360 (2002). [DOI] [PubMed] [Google Scholar]
- 72.Cooper A., Garcia M., Petrovas C., Yamamoto T., Koup R. A., Nabel G. J., HIV-1 causes CD4 cell death through DNA-dependent protein kinase during viral integration. Nature 498, 376–379 (2013). [DOI] [PubMed] [Google Scholar]
- 73.Galloway N. L., Doitsh G., Monroe K. M., Yang Z., Munoz-Arias I., Levy D. N., Greene W. C., Cell-to-cell transmission of HIV-1 is required to trigger pyroptotic death of lymphoid-tissue-derived CD4 T cells. Cell Rep. 12, 1555–1563 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Platt E. J., Wehrly K., Kuhmann S. E., Chesebro B., Kabat D., Effects of CCR5 and CD4 cell surface concentrations on infections by macrophagetropic isolates of human immunodeficiency virus type 1. J. Virol. 72, 2855–2864 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Yee J. K., Friedmann T., Burns J. C., Chapter 5 generation of high-titer pseudotyped retroviral vectors with very broad host range. Methods Cell Biol. 43, 99–112 (1994). [DOI] [PubMed] [Google Scholar]
- 76.Reed L. J., Muench H., A simple method of estimating fifty per cent endpoints12. Am. J. Epidemiol. 27, 493–497 (1938). [Google Scholar]
- 77.Mori M., Ode H., Kubota M., Nakata Y., Kasahara T., Shigemi U., Okazaki R., Matsuda M., Matsuoka K., Sugimoto A., Hachiya A., Imahashi M., Yokomaku Y., Iwatani Y., Nanopore sequencing for characterization of HIV-1 recombinant forms. Microbiol. Spectr. 10, e0150722 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Patro S. C., Brandt L. D., Bale M. J., Halvas E. K., Joseph K. W., Shao W., Wu X., Guo S., Murrell B., Wiegand A., Spindler J., Raley C., Hautman C., Sobolewski M., Fennessey C. M., Hu W. S., Luke B., Hasson J. M., Niyongabo A., Capoferri A. A., Keele B. F., Milush J., Hoh R., Deeks S. G., Maldarelli F., Hughes S. H., Coffin J. M., Rausch J. W., Mellors J. W., Kearney M. F., Combined HIV-1 sequence and integration site analysis informs viral dynamics and allows reconstruction of replicating viral ancestors. Proc. Natl. Acad. Sci. U.S.A. 116, 25891–25899 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Shibata J., Yoshimura K., Honda A., Koito A., Murakami T., Matsushita S., Impact of V2 mutations on escape from a potent neutralizing anti-V3 monoclonal antibody during in vitro selection of a primary human immunodeficiency virus type 1 isolate. J. Virol. 81, 3757–3768 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Juette M. F., Terry D. S., Wasserman M. R., Altman R. B., Zhou Z., Zhao H., Blanchard S. C., Single-molecule imaging of non-equilibrium molecular ensembles on the millisecond timescale. Nat. Methods 13, 341–344 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S7