Abstract
There are 78 loci associated with Parkinson’s disease in the most recent genome-wide association study (GWAS), yet the specific genes driving these associations are mostly unknown. Herein, we aimed to nominate the top candidate gene from each Parkinson’s disease locus and identify variants and pathways potentially involved in Parkinson’s disease. We trained a machine learning model to predict Parkinson’s disease-associated genes from GWAS loci using genomic, transcriptomic and epigenomic data from brain tissues and dopaminergic neurons. We nominated candidate genes in each locus and identified novel pathways potentially involved in Parkinson’s disease, such as the inositol phosphate biosynthetic pathway (INPP5F, IP6K2, ITPKB and PPIP5K2). Specific common coding variants in SPNS1 and MLX may be involved in Parkinson’s disease, and burden tests of rare variants further support that CNIP3, LSM7, NUCKS1 and the polyol/inositol phosphate biosynthetic pathway are associated with the disease. Functional studies are needed to further analyse the involvements of these genes and pathways in Parkinson’s disease.
Keywords: Parkinson’s disease, GWAS, machine learning, gene prioritization
Yu et al. train a machine learning model to predict Parkinson’s disease-associated genes from GWAS loci using genomic, transcriptomic and epigenomic data from brain tissues and dopaminergic neurons. They propose candidate genes for each locus, and identify novel pathways that may be involved in Parkinson’s disease.
See Lanore et al. (https://doi.org/10.1093/brain/awae043) for a scientific commentary on this article.
See Lanore et al. (https://doi.org/10.1093/brain/awae043) for a scientific commentary on this article.
Introduction
Genome-wide association studies (GWAS) have nominated many variants associated with complex traits. In Parkinson’s disease (PD), the most recent GWAS revealed 90 independent risk variants across 78 genomic loci.1 Although many single-nucleotide polymorphisms (SNPs) are in novel genomic loci, well-established PD genes discovered many years ago, such as LRRK2, PINK1, DJ-1, SNCA, GBA1, PRKN and MAPT still account for the vast majority of research on this disease.
Several disadvantages of GWAS limit additional functional analyses. First, over 90% of all GWAS significant SNPs are in non-coding regions.2 These SNPs are often passenger variants due to complex linkage disequilibrium (LD). Second, the causal gene associated with the causal SNPs remains unclear in most GWAS loci.3 To overcome these challenges, downstream GWAS analyses were established with the aim of identifying causal genes within GWAS loci. This involves techniques such as fine-mapping and co-localization methods to nominate causal SNPs, as well as transcriptome-wide association studies to nominate gene-trait associations.4-6 These models use LD structure, and gene expression panels to discover causal SNPs/genes. While these methods may propose causal variants and genes, additional biological evidence is generally required to pair causal variants with causal genes. Using multi-omic analyses, one can integrate a diverse range of comprehensive cellular and biological datasets such as genomic, transcriptomic and epigenetic datasets and use platforms such as Open Targets Genetics (https://genetics.opentargets.org/) to perform systematic analyses of gene prioritization across all publicly available GWASs.7 Although powerful, Open Targets Genetics lacks disease-specific tissues relevant to PD such as dopaminergic neurons and microglia. Using a similar approach, we may discover additional pathways and genetic targets involved in PD.
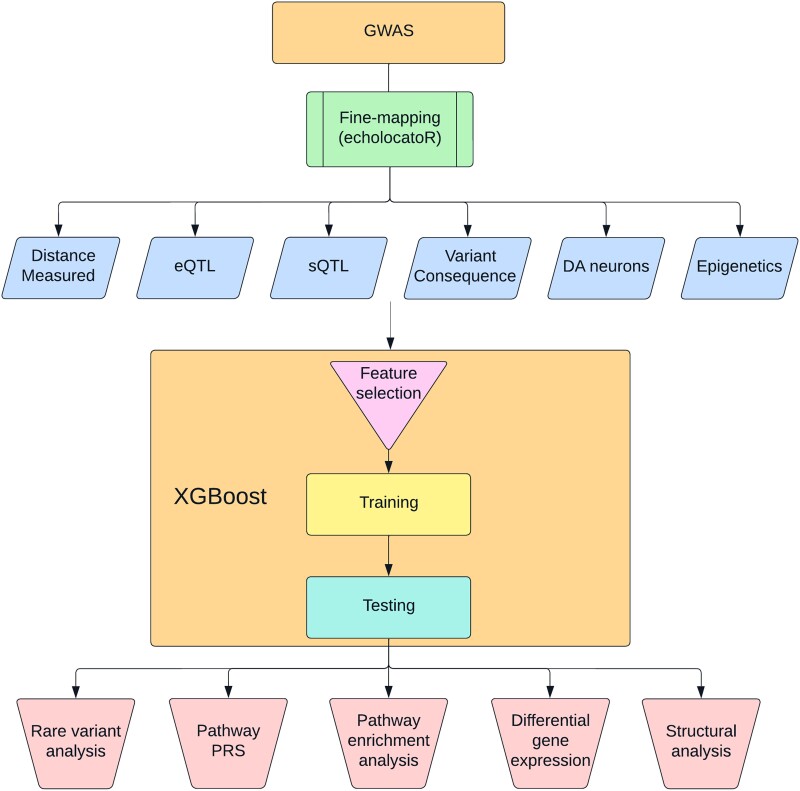
In this study, we leveraged PD-relevant transcriptomic, epigenomic and other datasets in our gradient boosting model (Fig. 1). We trained this model on well-established PD genes to nominate causal genes from PD GWAS loci.
Figure 1.
Workflow summary. This figure describes the analyses performed in this study. DA = dopaminergic; eQTL/sQTL = expression/splicing quantitative trait loci; GWAS = genome-wide association study; PRS = polygenic risk score.
Materials and methods
General design of the study
Our objective was to nominate the most probable genes to be involved in PD from each GWAS locus based on the most recent PD GWAS (see Fig. 1 for the study protocol).1 To do so, we first defined all the genes and SNPs that are within these loci (see later) and used to a machine learning approach to nominate the top genes in each locus. Based on the previous literature and consensus between authors, we identified seven genes from well-established loci associated with PD that can be considered the likeliest driving genes of their respective loci (GBA1, LRRK2, SNCA, GCH1, MAPT, TMEM175 and VPS13C). We then acquired data for multiple features, including different distance measures from top SNPs, different QTLs, expression in relevant tissues and cell types and predictions of variant consequences (78 features out of 284 were used after removal of redundant features; Supplementary Table 1). Using the seven well-established PD genes, which were labelled as positive, and 212 genes in the same loci that received negative labels (i.e. not likely to drive the association with PD, since the PD-driving gene is already well-established), we trained a machine learning model. This model enabled us to generate a prediction score for each gene within each locus, assessing their potential involvement in PD. The gene with the highest score in each locus is the nominated gene to be associated with PD. We then performed multiple post hoc analyses to further validate and explore our results: burden tests for rare variants in the top-scoring genes, pathway enrichment and pathway PRS analyses, differential expression analyses and structural analyses for candidate coding variants.
Definition of loci and genes within each locus
Following the definition by Nalls et al.,1 all loci were defined based on the 90 independent risk variants (Supplementary Table 2). Variants within 250 kb were merged into a single locus, which led to 78 loci. All protein coding genes within 1 Mb of the risk variants were included in the model. To exclude non-causal variants, echolocatoR was used as a comprehensive fine-mapping model.5 This method leverages Bayesian statistical and functional fine-mapping tools as well as epigenomic data to calculate the causal probability of SNPs in a locus.5 In our downstream analysis, we incorporated the SNPs nominated by echolocatoR into the credible gene sets generated by the same tool. Furthermore, we included the 90 independent SNPs obtained from the PD GWAS in our analysis.
Feature preprocessing
To leverage multi-omic data for the machine learning algorithm, we integrated a comprehensive list of datasets (Supplementary Table 1), which included SNP functional annotations, expression and splicing quantitative trait loci (eQTL/sQTL), single nuclear RNA sequencing (scRNA) and chromatin interaction. Since distance was previously shown to be the most predictive feature in about 60–70% of GWAS loci, the distances from each SNP to each gene in the locus and the distance to the transcription start site were included in the model.8 To predict the severity of variant consequences, we used VEP9 and PolyPhen-2.10 The SNP2GENE function on the FUMA platform was used to perform functional mapping of SNPs to eQTLs.11 In the FUMA settings, we chose the UK Biobank release2b 10k European reference panel, a maximum distance of 1000 kb from SNPs to gene, and included the major histocompatibility complex (MHC) region. All other FUMA settings were kept as default. Expressions QTL and 3D chromatin interaction mapping were performed using brain tissues, whole blood, Functional Annotation of the Mammalian genome (FANTOM) and Genotype-Tissue Expression (GTEx) datasets. Using scRNA datasets from Kamath et al.,12 we included gene expression from all ten subpopulations of dopaminergic neurons from post-mortem brains of seven PD and eight control donors. A complete list of all datasets can be found in Supplementary Table 3.
Neighbourhood scores
To integrate the concept of locus and LD in the model, we calculated the neighbourhood scores for each feature by transforming the data relative to the best-scoring gene within each locus,7 allowing the model to find the highest expressed genes across each locus. For example, if the feature is ‘maximum gene expression in blood’, the gene with the highest expression in each locus would have a score of 1 while the score of the remaining genes in the locus would be calculated following the expression of gene divided by the expression of highest expressed gene in the locus. Negative log transformation was applied so that the closest gene had the highest score.
Machine learning model to prioritize genes
We used XGBoost13 to train the machine learning model. We selected well-established genes from PD loci for the training dataset (GBA1, GCH1, LRRK2, MAPT, SNCA, TMEM175, VPS13C). These genes were labelled as positive labels, and the remaining genes from these same loci were labelled as negative labels. In total, the training set was composed of 212 genes (seven positive labelled and 205 negative labelled). The scale_pos_weight parameter in XGBoost was set to the ratio of negative to positive labels to control for the imbalance. The training process involved two steps. First, we performed feature selection to detect redundant features. This involved removing any variables from the dataset that were either redundant or uninformative. XGBoost was employed to transform the dataset into a subset containing the chosen features. To achieve this, we trained a model using the complete dataset and then retained the features present in the subset produced by XGBoost. In the second step, the final training model was created using the selected features. This two-step approach helps optimize the training process and ensures that the model focuses on relevant and informative features to make accurate predictions. We performed hyperparameter tuning and 5-fold cross-validation on both models. Mean average precision was used as an evaluation function to maximize the score of correct positive predictions made. Of the 284 features, 78 features passed feature selection for the final training model.
Functional enrichment analysis
To examine whether specific pathways may be involved in PD, based on the genes nominated in each locus, we performed an over-representation analysis using WebGestalt (Web-based Gene Set Analysis Toolkit) on 25 January 2023.14 We included the top candidate gene from each locus, and examined enrichment in terms of biological processes and cellular components from the Gene Ontology (GO) data. The genome protein-coding list was used as the reference list and pathways were considered to be associated with PD if significant after false discovery rate (FDR) correction.
Single-cell and bulk RNA sequencing analyses
To examine whether genes nominated by the machine learning model may be differentially expressed in PD relevant models, we used publicly available single-cell and bulk RNA sequencing (RNAseq) data from The Foundational Data Initiative for Parkinson’s disease (FOUNDIN-PD)15 and Kamath et al.12 FOUNDIN-PD scRNA data include 80 induced pluripotent stem cell (iPSC) lines collected after 65 days.15 We then performed differential gene expression analyses between PD cases and controls. For scRNA, we used the MAST16 package after adjusting for covariates, such as age, sex and batch. For bulk RNAseq, we used DESeq2,17 while adjusting for the same covariates.
Pathway polygenic risk score analyses
Pathway-specific polygenic risk score (PRS) analysis can further support a role for specific pathways in PD.18 Using PRSet,19 pathway-specific PRSs were calculated for pathways nominated by gene set analysis on 14 828 PD cases and 13 283 controls from seven cohorts [McGill, Parkinson’s Progression Markers Initiative (PPMI), Vance (dbGap phs000394), International Parkinson’s Disease Genomics Consortium (IPDGC) NeuroX dataset (dbGap phs000918.v1.p1), National Institute of Neurological Disorders and Stroke (NINDS) Genome-wide genotyping in Parkinson’s disease (dbGap phs000089.v4.p2), NeuroGenetics Research Consortium (NGRC) (dbGap phs000196.v3.p1) and UK Biobank]. The number of cases and controls for each cohort is described in Supplementary Table 4. Participants were unrelated individuals of European ancestry and were not gender matched. Rare SNPs (minor allele frequency < 0.01) with a P-value < 0.05 were excluded from the analysis. LD clumping was performed using r2 = 0.1 and 250 kb distance. Permutation testing was performed with 10 000 label permutations to generate an empirical P-value for each gene set after adjusting for a prevalence of 0.005, age at onset for cases, age at enrollment for control, sex and the top 10 principal components. The Vance cohort was excluded from the meta-analysis due to significant heterogeneity.
Rare variant burden analyses
To examine whether there is an association between rare variants in the genes nominated by the machine learning model and PD, we used MetaSKAT20 to perform a meta-analyses of rare variants. We used whole exome sequencing (WES) available for 602 PD patients, 6284 proxy patients and 140 207 controls from UK Biobank (n = 147 093) and 2600 PD patients, 3677 controls from Accelerating Medicines Partnership Parkinson’s Disease (AMP-PD)21 datasets (n = 6277). Additional selection criteria for UK Biobank and AMP PD were reported previously.22,23 We performed the analysis on several groups of rare variants (allele frequency < 0.01): loss of function variants; non-synonymous variants; potentially deleterious (CADD > 20) variants; and functional (including non-synonymous, frame-shift, stop-gain and splicing) variants. Pathway-specific rare variant analysis was performed by combining PD genes from the pathways nominated previously. All analyses were adjusted for age at onset for cases, age at sample for controls and sex.
Structural analysis
The atomic coordinates of SPNS1 (UniProt #H3BR82) were retrieved from the AlphaFold server (https://alphafold.ebi.ac.uk/). The structures of MLX-MAD1 and MLX-MLXIP were generated using AlphaFold-Multimer version 3, as implemented in ColabFold.24,25 The ternary complex with a DNA duplex was generated by superposing the heterodimers on the crystal structure of the MAD1-MAX-DNA complex (PDB 1NLW). The figures were generated using PyMol v.2.4.0.
Results
Machine learning model nominates PD-associated genes in each PD locus
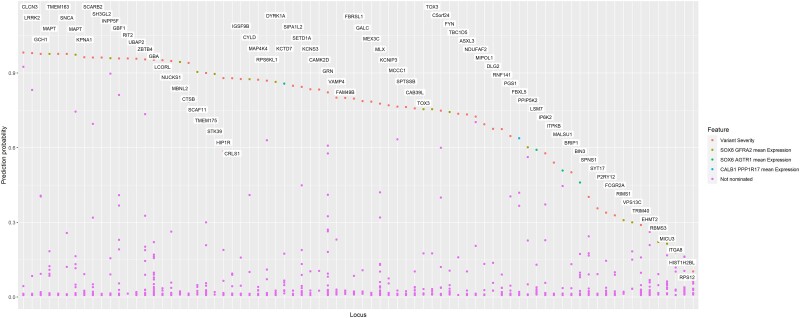
To train our machine learning model, we used seven well-established PD-associated genes from the PD GWAS (GBA1, LRRK2, SNCA, GCH1, MAPT, TMEM175 and VPS13C) as positive labels, and the remaining genes from the same loci (n = 205) were used as negative labels (i.e. genes that are unlikely to be involved in PD). We trained an XGBoost regression model to identify the best predictive features. Then, based the best predictive features, we assigned a probability score that indicated the likelihood that the gene was driving the association at each locus (Supplementary Table 2). We then nominated the top-scoring genes in each locus (Fig. 2 and Supplementary Table 2). Two genes, MAPT and TOX3, were nominated twice in neighbouring loci that harbour them, taking the total number of genes nominated in this model to 76 genes in 78 loci. A probability score higher than 0.75 was assigned to 48 of the 76 genes (63%). Of note, five genes (NEK1, FDFT1, PSD, BAG3 and SLC2A13) that were ranked second in their respective loci also had a probability score >0.75. However, the nominated genes in their loci (CLCN3, CTSB, GBF1, INPP5F and LRRK2, respectively) all had probability scores >0.94. In seven other loci, the top nominated genes had an especially low probability score (<0.3), including RBMS3, HIST1H2BL, TRIM40, EHMT2, RPS12, MICU3 and ITGA8.
Figure 2.
Probability score of the Parkinson’s disease genome-wide association study candidate genes. This figure shows the probability scores from the machine learning model for each locus in the Parkinson’s disease genome-wide association study loci sorted in descending order. For each gene, the top non-distance feature was used to colour the data.
Gene expression in subtypes of PD-associated dopaminergic neurons predicts PD-relevant genes
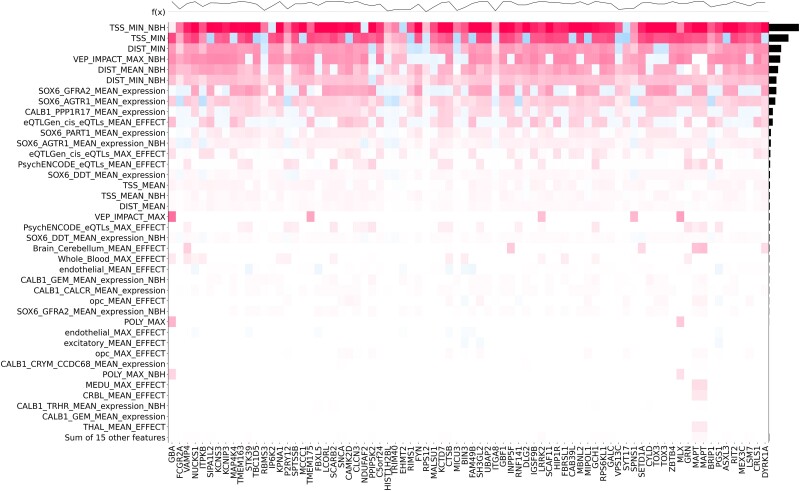
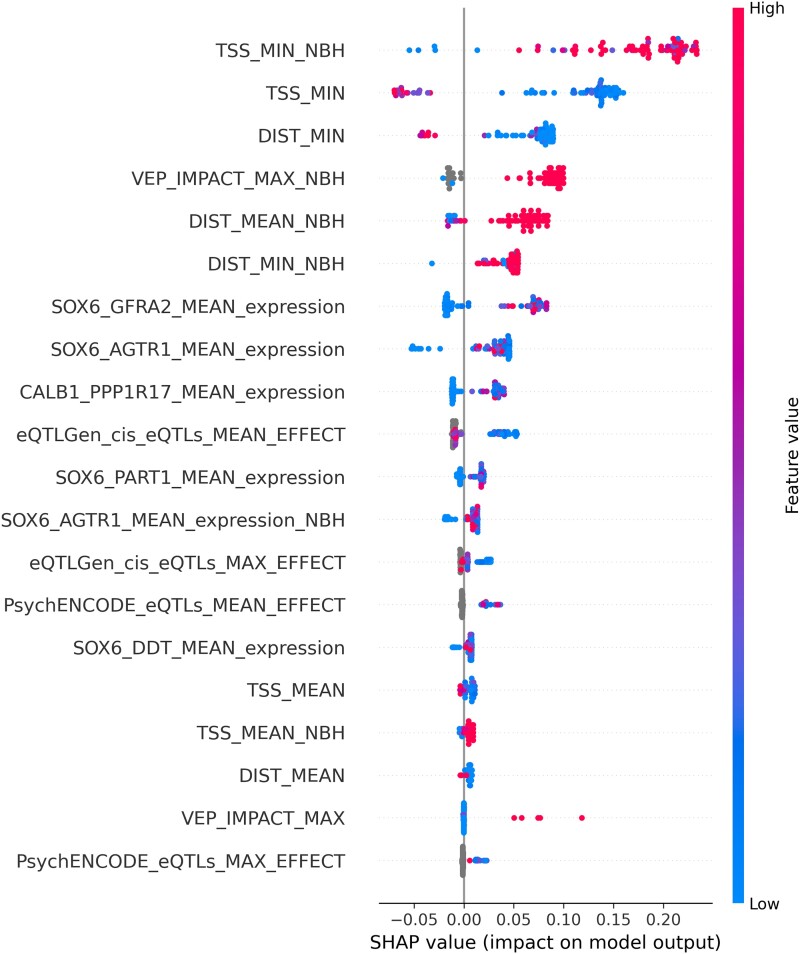
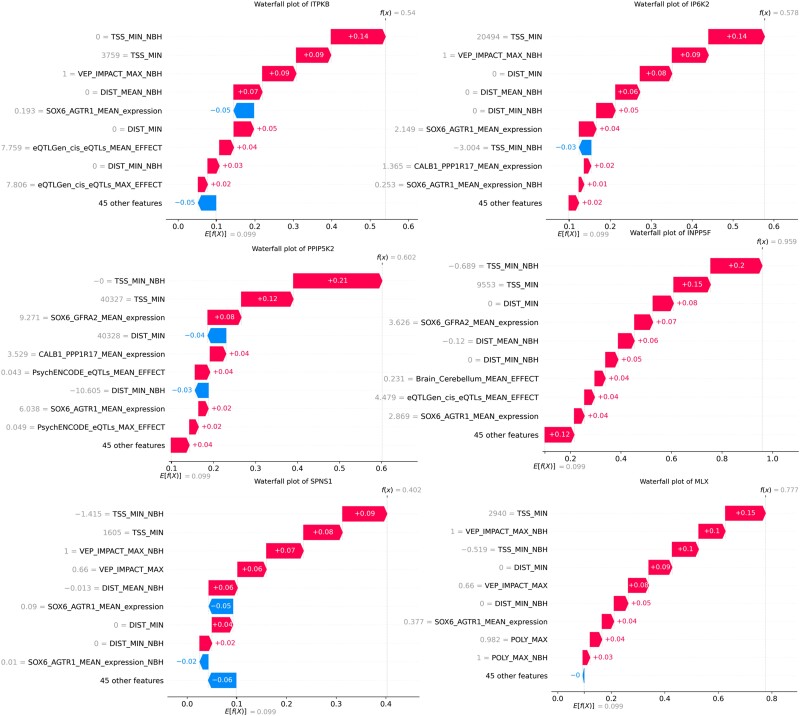
Next, we used Shapley Additive exPlanations (SHAP) values to determine which features of the model contributed most to the prediction.26,27 SHAP values provide, for each gene, the relative contribution of each feature to the selection of that gene. The most important features for the scoring of each gene are shown in Fig. 3. As expected, distance-related features, such as distance from the top-associated SNP in the locus to the transcription start site or distance to the beginning of the gene, were the most important features in our model.7 The next most important feature was the Variant Effect Predictor (VEP) value, followed by additional distance measures.9 Interestingly, the next most important features were mRNA expression values within specific dopaminergic neuron subtypes. These different dopaminergic neuron subtypes are defined by the expression of the genes GFRA2 and AGTR1 from single nuclear sequencing of post-mortem tissue. The latter is a specific subtype of dopaminergic neurons shown by Kamath et al.12 to be selectively degenerated in brains of PD patients.12 The remaining features include expression in other dopaminergic neuron subpopulations, eQTLs and other expression features. Epigenetic features were not predictive in our model. As shown in Fig. 4, all nominated genes had at least one of the distance features contributing to their selection. On top of the known contribution of missense variants in GBA1, LRRK2 and GCH1, we nominated missense SNPs that contributed to the score of two candidate genes: SPNS1 (p.L512M, rs7140) and MLX (p.Q139R, rs665268). In Europeans, both SNPs are in high LD with the candidate GWAS SNPs of their respective locus (SPNS1 D': 0. 88 r2: 0.74; MLX D': 1 r2: 1). In GTEx, rs7140 and rs665268 are also eQTLs/sQTLs for SPNS1 and MLX across several PD related tissues such as whole blood and anterior cingulate cortex. The eQTL and sQTL results from GTEx v8 are shown in Supplementary Table 5. SPNS1 and MLX have not previously been implicated in PD, and the important features identifying these genes as the top candidate for their respective GWAS loci are shown in Fig. 5.
Figure 3.
Feature importance for the Parkinson’s disease genome-wide association study gene prioritization model. Bee-swarm plot of feature importance using Shapley Additive exPlanations values along with the distribution of genes based on feature value.
Figure 4.
Heat map of feature importance. The heat map is generated using Shapley Additive exPlanations (SHAP) value for the top candidate gene in each locus. The plot at the top represents the probability score of each gene.
Figure 5.
Waterfall plots for Parkinson’s disease genome-wide association study candidate genes. Importance of the top 10 features using Shapley Additive exPlanations values for different selected candidate genes. E[f(x)] is the base score for each gene, which is calculated based on the average value of each features. f(x) is the final score after accounting for all features.
Differential expression of genes from the inositol phosphate biosynthetic pathway and MLX in PD
To further establish the importance of the nominated genes in PD, we examined whether they are differentially expressed in PD patients compared to controls, using expression data from single nuclear RNAseq (scRNA) from Kamath et al.12 and single nuclear and bulk RNAseq datasets from FOUNDIN-PD.15 Of the top nominated genes, INPP5F [average log fold-change (FC) = −7.22, P = 2.90 × 10−31] and MLX (average log FC = −1.80, P = 2.23 × 10−4) were associated with PD in the data published by Kamath et al.12 (Supplementary Table 6). In FOUNDIN-PD,15 after excluding prodromal cases, we found differential expression of many genes including INPP5F (average log FC = 0.070, P = 1.89 × 10−19) and IP6K2 (average log FC = −0.076, P = 1.35 × 10−35) in scRNA data (n = 80) from dopaminergic neurons by comparing PD and controls (Supplementary Table 7). Results from the bulk RNAseq analysis of FOUNDIN-PD (n = 92) can be found in Supplementary Table 8.
Structural analysis of SPNS1 and MLX
Since non-synonymous variants in SPNS1 and MLX were identified as major contributors to their selection as the nominated genes in their loci, we aimed to examine the potential consequences of these variants by performing in silico structural analyses of the protein encoded by these genes. SPNS1 encodes a transporter for phospholipids at the lysosome membrane.28 It mediates the efflux of lysophosphatidylcholine and lysophosphatidylethanolamine out of the lysosome. The SNP rs7140 is located in the 3′-untranslated region (UTR) of the canonical splice variant 1 transcript, which produces the 528 amino acid (aa) isoform that has been investigated functionally28 (UniProt #Q9H2V7). This canonical isoform has also been observed in numerous proteomics datasets in gpmDB (https://gpmdb.thegpm.org/index.html). However, six other potential isoforms generated by alternative splicing have been predicted, including a 538 aa fragment with an alternative C-terminus, whereas the rs7140 SNP is located within the coding region (UniProt #H3BR82). The rs7140 variant results in the p.L512M mutation in this isoform. To investigate the impact of this mutation on the function of this SNPS1 isoform, we inspected the 3D structure model generated by AlphaFold.29 Leu512 is located in the unstructured C-terminus of this membrane-bound protein, on the lumenal side of the lysosomal membrane (Supplementary Fig. 1A). The role of the C-terminus in this isoform of SPNS1 remains unclear, and thus the impact of the p.L512M mutation is unknown.
The Max-like protein (MLX) is at the heart of a transcriptional network pathway involved in energy metabolism and cell signalling.30,31 It interacts with at least six other related proteins including the MAD family of transcriptional repressors and the Mondo family of transcriptional activators. These proteins contain basic/helix-loop-helix/leucine zipper (bHLHZ) domains that form heterodimers and interact with DNA carrying the CACGTG E-box motif. To understand the impact of the p.Q223R MLX mutation (rs665268) on its activity, we modelled the structure of MLX heterodimers with both the MAD and Mondo families using AlphaFold. MLX dimerizes with MAD1,31 and thus we superposed its bHLHZ domain on the MAD1-MAX-DNA complex crystal structure32 to generate the ternary complex model. The model shows that Gln223 in MLX is at the end of the dimerization ‘zipper’ helix (Supplementary Fig. 1B). The mutation p.Q223R induces the formation of a salt bridge with Glu139 in MAD1, which could strengthen the interaction between MAD1 and MAX. This could then downregulate the interaction of MAD1 with MAX through competition, and thus affect the extent of the transcriptional repression. Glu139 is not conserved in other MAD-related proteins such as MXI1 and MAD3/4. Furthermore, the model of MLX interacting with MLXIP, a protein of the Mondo family also known as MondoA,33 shows that the mutation may negatively affect the formation of this heterodimer by introducing a charge next to a hydrophobic sidechain (Supplementary Fig. 1C). The nuclear localization of Mondo proteins is dependent on their interaction with MLX,30 and thus the mutation may down regulate activation by the Mondo family while strengthening repression via MAD1.
Gene enrichment analysis shows the inositol phosphate pathway as a novel pathway involved in PD
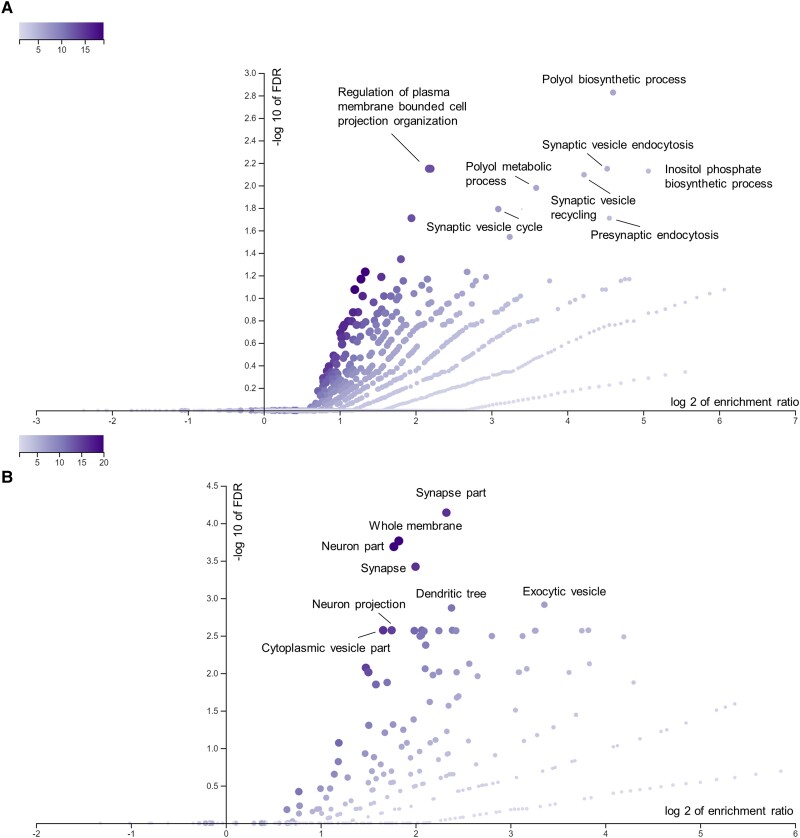
We further examined whether the nominated genes highlighted specific pathways and mechanisms associated with PD. We performed a pathway enrichment analysis by examining over-representation of the top nominated genes in biological processes and cellular components using the top genes in each locus. Among the biological processes passing the FDR correction, the inositol phosphate biosynthetic process (GO:0032958) and polyol biosynthetic process (GO:0046173) were strongly enriched (Fig. 6A). Inositol was associated with four candidate genes, namely ITPKB, IP6K2, PPIP5K2 and INPP5F. The features most important to the nomination of these genes as PD-associated by our ML model are shown in Fig. 5. Cellular components were also identified in the gene enrichment analysis (Fig. 6B).
Figure 6.
Volcano plots of gene ontology biological processes and cellular components. Volcano plots of gene-set enrichment analysis using WebGestalt showing the log of the false discovery rate (FDR) versus the enrichment ratio for biological processes (A) and cellular components (B). P-value are calculated using a hypergeometric test. All pathways that are significant after FDR correction were named.
Pathway-specific polygenic risk score of the inositol phosphate pathway is associated with PD
To further study the association between the putative novel PD pathways and PD status, pathway-specific PRSs were calculated for the above-mentioned gene sets. The association between these PRSs and PD was examined in six PD cohorts, followed by a meta-analysis as detailed in the ‘Materials and methods’ section. One outlier cohort was excluded due to heterogeneity. The pathway-specific PRSs were first calculated using all genes in that pathway. Then, to further validate that the specific pathway was indeed important in PD, we excluded the genes nominated by our machine learning pathway and recalculated the PRS. By removing these genes with GWAS significant signals, we could examine the residual effect of the remaining pathway. The inositol phosphate biosynthetic pathway was associated with PD even after excluding the genes nominated in our analysis [odds ratio (OR) 1.06, 95% confidence interval (CI) 1.03–1.09, P = 7.01 × 10−5], as well as other related pathways (Table 1). Forest plots of the all the pathway PRSs are shown in Supplementary Fig. 2.
Table 1.
Meta-analyses of pathway-specific polygenic risk scores
| Pathway-specific polygenic risk score | OR | 95% CI | P-value | Het P-value |
|---|---|---|---|---|
| POLYOL_BIOSYNTHETIC_PROCESS | 1.20 | 1.17–1.24 | 2.07 × 10−42 | 1.91 × 10−5 |
| INOSITOL_PHOSPHATE_BIOSYNTHETIC_PROCESS | 1.15 | 1.12–1.18 | 2.36 × 10−25 | 1.97 × 10−2 |
| POLYOL_BIOSYNTHETIC_PROCESS_filtered | 1.09 | 1.06–1.12 | 1.04 × 10−9 | 1.12 × 10−2 |
| INOSITOL_PHOSPHATE_BIOSYNTHETIC_PROCESS_filtered | 1.06 | 1.03–1.09 | 1.31 × 10−5 | 1.45 × 10−1 |
CI = confidence interval; Filtered = excluded Parkinson’s disease genome-wide association study top gene; GOBP_INOSITOL_PHOSPHATE_BIOSYNTHETIC_PROCESS = Gene Ontology inositol phosphate biosynthetic process (GO:0032958); GOBP_POLYOL_BIOSYNTHETIC_PROCESS = Gene Ontology polyol biosynthetic process (GO:0046173); Het = heterogeneity; OR = odds ratio.
Rare KCNIP3 and LSM7 variants and in the polyol/inositol biosynthetic pathway are involved in PD
To further establish the potential role of the nominated genes in PD, we performed rare variant burden tests in all the genes nominated by our model. As expected, genes that are known to harbour rare PD coding mutations including GBA1, LRRK2 and GCH1 were associated with PD (Table 2 and Supplementary Table 9). Three additional genes, including two genes that have not previously been implicated in PD (KCNIP3 and LSM7) showed a burden of rare variants after FDR correction for multiple comparisons. We then examined the genes from the pathway enrichment analysis and found that rare variants in the polyol/inositol biosynthetic pathway were also associated with PD (SKAT-O, P = 1.58 × 10−4), further supporting its role in PD.
Table 2.
Meta-analysis of rare variant analysis of putative causal genes
| Set | P-value | FDR P-value |
|---|---|---|
| GBA1 Rarefunctional | 2.04 × 10−12 | 6.22 × 10−10 |
| GBA1 Rarenonsyn | 3.38 × 10−11 | 5.15 × 10−9 |
| GBA1 RareLOF | 1.22 × 10−6 | 1.24 × 10−4 |
| GBA1 RareCADD | 2.32 × 10−6 | 1.77 × 10−4 |
| LSM7 RareLOF | 3.69 × 10−6 | 2.25 × 10−4 |
| KCNIP3 RareLOF | 1.12 × 10−5 | 5.69 × 10−4 |
| GCH1 RareLOF | 2.02 × 10−5 | 8.80 × 10−4 |
| LRRK2 RareCADD | 6.07 × 10−5 | 2.31 × 10−3 |
| Polyol Rarefunctional | 1.59 × 10−4 | 5.38 × 10−3 |
| Polyol Rarenonsyn | 2.86 × 10−4 | 8.74 × 10−3 |
| NUCKS1 RareCADD | 4.13 × 10−4 | 1.14 × 10−2 |
| Polyol RareLOF | 1.54 × 10−3 | 3.91 × 10−2 |
| SYT17 Rarenonsyn | 4.61 × 10−3 | 9.37 × 10−2 |
| P2RY12 RareLOF | 4.38 × 10−3 | 9.37 × 10−2 |
| CYLD RareLOF | 4.48 × 10−3 | 9.37 × 10−2 |
| SYT17 Rarefunctional | 7.39 × 10−3 | 1.38 × 10−1 |
| LCORL RareLOF | 7.66 × 10−3 | 1.38 × 10−1 |
| CAMK2D RareLOF | 8.62 × 10−3 | 1.46 × 10−1 |
| FBRSL1 RareLOF | 1.12 × 10−2 | 1.80 × 10−1 |
| CTSB RareLOF | 1.20 × 10−2 | 1.82 × 10−1 |
| KPNA1 RareCADD | 1.35 × 10−2 | 1.96 × 10−1 |
| ASXL3 RareLOF | 1.52 × 10−2 | 2.10 × 10−1 |
| KPNA1 RareLOF | 1.76 × 10−2 | 2.33 × 10−1 |
| LRRK2 Rarefunctional | 2.57 × 10−2 | 3.14 × 10−1 |
| MICU3 RareLOF | 2.56 × 10−2 | 3.14 × 10−1 |
| VAMP4 Rarenonsyn | 2.93 × 10−2 | 3.43 × 10−1 |
| MBNL2 RareCADD | 3.04 × 10−2 | 3.43 × 10−1 |
| LRRK2 Rarenonsyn | 3.28 × 10−2 | 3.57 × 10−1 |
| KPNA1 Rarefunctional | 3.46 × 10−2 | 3.64 × 10−1 |
| LSM7 Rarefunctional | 3.58 × 10−2 | 3.64 × 10−1 |
| HIP1R Rarenonsyn | 3.93 × 10−2 | 3.87 × 10−1 |
| KPNA1 Rarenonsyn | 4.23 × 10−2 | 3.91 × 10−1 |
| HIP1R Rarefunctional | 4.22 × 10−2 | 3.91 × 10−1 |
FDR = false discovery rate; Rarefunctional = rare functional variants; Rarenonsyn = rare non-synonymous variants; RareLOF = rare loss-of-function variants; RareCADD = rare variants with CADD score > 20; Set = variant set across genes/pathway.
Discussion
Using multi-omic data and machine learning, we nominated genes that potentially drive the associations with PD for each of the 78 PD GWAS loci. Our nominated genes included many not previously studied in the context of PD. Additionally, we identified two novel genes with rare variants (KCNIP3 and LSM7) as well as genes with GWAS significant coding variants such as SPNS1 and MLX that could be further studied. Furthermore, our gene enrichment, pathway-specific PRS and rare variant analyses suggested involvement of the inositol phosphate biosynthetic pathway in PD.
Four genes nominated by our machine learning model were associated with the inositol phosphate biosynthetic pathway, ITPKB, IP6K2, PPIP5K2 and SNCA,34 which showed strong enrichment of this pathway. In addition, INPP5F, also nominated by our analysis, is involved in inositol processing through a parallel pathway.35 Our results demonstrate that the inositol pathway-PRS, even when excluding the previously mentioned genes, is still associated with PD. Taken together, our findings support the importance of the inositol phosphate pathway in PD.
Based on the evidence from the candidate inositol genes and previous inositol studies, inositol could potentially be a therapeutic target for PD. In 1999, a clinical trial on inositol was conducted on nine PD patients.36 Treatment with inositol compared with placebo did not improve clinical outcomes; however, we cannot rule out inositol and inositol phosphates as potential therapeutic targets, as only nine patients were recruited for the trial.
ITPKB encodes for a ubiquitous kinase that phosphorylates inositol 14,5-trisphosphate (IP3) to inositol 1,3,4,5 tetrakisphosphate (IP4) using a Ca2+/calmodulin-dependent mechanism. IP3 is a secondary messenger that stimulates calcium release from the endoplasmic reticulum (ER). In primary neurons, ITPKB knockdown/overexpression was shown to increase/reduce levels of α-synuclein aggregation.37 Additionally, ITPKB knockdown in neurons leads to the accumulation of calcium in mitochondria. This accumulation can impair the process of autophagy, which is crucial for maintaining mitochondrial health. In neuroblastoma cells, ITPKB mRNA levels were also shown to be correlated with SNCA expression in the cortex and IPTKB protein levels were increased in wild-type α-synuclein, A53T and A30P mutants.38 Meanwhile, IP6K2 and PPIP5K2 interact with the same substrates. IP6K2 converts inositol hexakisphosphate (IP6) to 5-diphosphoinositol pentakisphosphate (5-IP7) or 1-diphosphoinositol pentakisphosphate (1-IP7) to bis-diphosphoinositol tetrakisphosphate (1,5-IP8), while PPIP5K2 convert 5-IP7 to 1,5-IP8 and IP6 to 1-IP7.39 In mice, IP6K2 has been implicated in cell death, apoptosis and neuroprotection.40 One study proposed that IP6K2 regulates mitophagy via the parkin/PINK1 pathway, but further evidence would be required to confirm this hypothesis.40PPIP5K2 has not previously been implicated in PD but is associated with hearing loss and colorectal carcinoma.41,42 Finally, INPP5F is involved with a different inositol pathway; it encodes SAC2, which converts phosphoinositides such as PI(4,5)P2 to phosphatidylinositol during endocytosis.35
Inositol phosphate has been suggested to be involved in obesity, insulin resistance and energy metabolism.43 In post-mortem brain tissues of PD patients, 3H-inositol 14,5-trisphosphate binding sites were found to be reduced in certain brain regions such as the caudate nucleus, putamen and pallidum.44 Additionally, IP6 was shown to be associated with PD. IP6 has a neuroprotective effect on dopaminergic cells by preventing 6-OHDA-induced apoptosis.45 IP6 inhibits the activity of β-secretase 1 (BACE1), an enzyme that cleaves amyloid-β precursor protein into toxic amyloid-β peptides.46 Paraquat-induced neurodegeneration in Drosophila was suggested to increase the levels of inositol phosphates metabolites.47 Previous studies have also suggested that different stereoisomers of inositol such as scyllo-inositol can inhibit the aggregation of α-synuclein48 or decrease the myoinositol concentration in patients with PD.49,50
Recent studies on inositol investigated the role of SYNJ1, an autosomal recessive form of early-onset parkinsonism.51 SYNJ1 is a lipid phosphatase of phosphatidylinositol-34,5-trisphosphate (PIP3).52 SYNJ1 knockout cell models were associated with an increase of α-synuclein and PIP3 levels. PIP3 dysregulation was suggested to promote α-synuclein aggregation, which increases the risk of PD. Together with our data, there is strong evidence for the involvement of the inositol phosphate biosynthetic pathway in PD, and this pathway should be further studied using both basic science and translational approaches.
Outside of the inositol pathway, SPNS1 and MLX were found to be the top causal gene in their respective loci with putative causal missense SNPs: rs7140 and rs665268. Rs7140 corresponds to p.Leu563Val on the SPNS1 transcript variant X1. We found that SPNS1 expression is lower in the SOX6_ATGR1 dopaminergic neuron subpopulation in PD compared with controls. This subcluster was previously highlighted to be the most susceptible to neurodegeneration in PD.12SPNS1 encodes a sphingolipid transmembrane transporter in the lysosome. The autophagy-lysosomal pathway has been well-established to be crucial in PD pathogenesis, especially the lysosomal sphingolipid metabolism pathway, which includes well established PD-associated genes including GBA1, GALC, SMPD1 and others.53,54 SPNS1 deficiency results in lipid accumulation in the lysosome and impaired lysosomal function.28
The second nominated gene in which we identified rare variants, MLX, encodes a Max-like protein X which belongs to a family of transcription factors regulating glucose metabolism. Rs665268 is a missense variant (p.Gln139Arg) that was found to be associated with Takayasu’s arteritis, an autoimmune systemic vasculitis.55MLX was also reported to be associated with age at onset of Alzheimer’s disease in females.56 This variant was suggested to affect two important PD pathways by increasing oxidative stress and suppressing autophagy in immune cells.55,56SPNS1 and MLX have not previously been implicated in PD. Both variants, rs7140 and rs665268, were found in high LD with the top candidate GWAS SNP. When examining missense SNPs in LD with the top GWAS SNPs, SPNS1, MLX and CD19 were the only genes with such features. CD19 was not nominated in our study, as it is located in the same locus as SPNS1, and it ranked lower than SPNS1. These findings indicate that these genes could play a role in PD and should be further studied.
Other studies have attempted to use machine learning to characterize genes involved in PD. Using machine learning, Ho et al.57 integrated tissue-specific eQTLs and the genotypes of PD patients and controls to identify PD-specific genes. They nominated the roles of two key variants in PD (rs7617877, rs6808178) and the potential role of heart atrial appendage tissue. Interestingly, AGTR1, a gene associated with many PD single-nuclei subpopulations included in our model, encodes for angiotensin II receptor type 1 protein.58 This protein is part of the renin-angiotensin system, which regulates blood pressure and the balance of fluids and salts in the body.58 Ho et al.57 also validated some of the top genes from our model such as INPP5F, P2RY12, HIP1R, STK39 and CTSB. Transcriptional changes to these genes could contribute to PD.
Interestingly, in certain regions of the genome, such as VPS13C, the top genes showed lower probability scores (Fig. 2). This could be due to complex LD structure, which weakens the effect size of eQTLS, as the variants in LD are associated with multiple genes. In such scenarios, the model might encounter challenges in precisely predicting the responsible gene. Additionally, the number of samples employed in statistically assessing attributes like eQTLs and enhancer-promoter interactions significantly impacts the model’s training. Features derived from studies with limited sample sizes may be less powered to detect eQTLs and more likely to be excluded from the model. For example, while data regarding enhancer-promoter interactions were incorporated into the training attributes, it might not have been important for the majority of variant-gene pairs. Overall, while VPS13C had a low probability score for a gene in the training set, it was still the top gene in its respective locus.
Although we identified candidate genes and new rare mutations, there were several limitations to this study. This study was based on a GWAS of European populations only. Therefore, our results are potentially restricted to Europeans. While there are some studies on the association of chromosome X in PD, the statistical power was limited compared with a PD GWAS of autosomes. As a result, no analysis was performed on chromosome X. In addition, the training set for the machine learning model was limited to a small set of known or highly likely PD genes with the assumption of one causal gene per locus. The study also lacked samples for a testing set due to the small number of well-established PD genes. Since these limitations may have introduced some bias, we used different strategies such as controlling for an imbalanced dataset and choosing balanced accuracy as an evaluation function to maximize the performance of the model. Although the distance between variants and genes holds significant predictive power in the model, it is crucial to acknowledge that not all top genes can accurately be predicted solely based on distance. Of the 78 genes analysed, 13 were not the closest genes in terms of distance from the gene to the top GWAS SNPs, and 25 were not the closest genes based on distance to the transcription start site. Additionally, when comparing the scRNA and bulk RNAseq results, most of the differentially expressed genes did not overlap across our datasets. For example, while INPP5F was nominated in scRNA of both datasets, it was not significant in the bulk RNAseq analysis. Lastly, the meta-analysis of rare variants can also be somewhat biased due to case/control imbalance. Larger GWAS and functional studies will be required to validate our findings.
Our results nominated multiple genes that have not been thoroughly studied in PD and provide a foundation for future functional studies of these genes. As larger PD GWASs will nominate more SNPs and loci, prioritizing causal genes will be crucial to understanding the underlying biological mechanisms and disease pathophysiology through additional studies. Future gene prioritization studies will be able to leverage larger datasets with more positive labels as new PD genes are discovered, increasing the accuracy of predictions.
Supplementary Material
Acknowledgements
E.Y. received a scholarship from the Canadian Institutes of Health Research (CIHR). S.R. received a scholarship from the Canadian Institutes of Health Research (CIHR). Z.G.O. is supported by the Fonds de recherche du Quebec Sante (FRQS) Chercheurs boursiers award and is a William Dawson and Killam Scholar. UK Biobank Resources were accessed under application number 45551. Data used in the preparation of this article were obtained from the AMP PD Knowledge Platform. For up-to-date information on the study, visit https://www.amp-pd.org. The AMP® PD program is a public-private partnership managed by the Foundation for the National Institutes of Health and funded by the National Institute of Neurological Disorders and Stroke (NINDS) in partnership with the Aligning Science Across Parkinson’s (ASAP) initiative; Celgene Corporation, a subsidiary of Bristol-Myers Squibb Company; GlaxoSmithKline plc (GSK); The Michael J. Fox Foundation for Parkinson's Research; Pfizer Inc.; Sanofi US Services Inc.; and Verily Life Sciences. Clinical data and biosamples used in preparation of this article were obtained from the (i) Michael J. Fox Foundation for Parkinson’s Research (MJFF) and National Institutes of Neurological Disorders and Stroke (NINDS) BioFIND study; (ii) Harvard Biomarkers Study (HBS); (iii) NINDS Parkinson’s Disease Biomarkers Program (PDBP); (iv) MJFF Parkinson’s Progression Markers Initiative (PPMI); and (vii) NINDS Study of Isradipine as a Disease-modifying Agent in Subjects With Early Parkinson Disease, phase 3 (STEADY-PD3). BioFIND is sponsored by MJFF with support from NINDS. The BioFIND Investigators have not participated in reviewing the data analysis or content of the manuscript. For up-to-date information on the study, visit michaeljfox.org/news/biofind. The HBS is a collaboration of HBS investigators (a full list of HBS investigators can be found at https://www.bwhparkinsoncenter.org/biobank/) funded through philanthropy and NIH and non-NIH funding sources. The HBS investigators have not participated in reviewing the data analysis or content of the manuscript. PPMI is sponsored by MJFF and supported by a consortium of scientific partners (full list of PPMI funding partners can be found at: https://www.ppmi-info.org/about-ppmi/who-we-are/study-sponsors). The PPMI investigators have not participated in reviewing the data analysis or content of the manuscript. For up-to-date information on the study, visit www.ppmi-info.org. The PDBP consortium is supported by NINDS at the National Institutes of Health. A full list of PDBP investigators can be found at https://pdbp.ninds.nih.gov/policy. The PDBP investigators have not participated in reviewing the data analysis or content of the manuscript. STEADY-PD3 is funded by NINDS at the National Institutes of Health with support from MJFF and the Parkinson Study Group. For additional study information, visit https://clinicaltrials.gov/ct2/show/study/NCT02168842. The STEADY-PD3 investigators have not participated in reviewing the data analysis or content of the manuscript.
Contributor Information
Eric Yu, Department of Human Genetics, McGill University, Montreal, Quebec H3A 0G4, Canada; The Neuro (Montreal Neurological Institute-Hospital), Montreal, Quebec H3A 2B4, Canada.
Roxanne Larivière, Department of Neurology and Neurosurgery, McGill University, Montreal, Quebec H3A 0G4, Canada.
Rhalena A Thomas, Department of Neurology and Neurosurgery, McGill University, Montreal, Quebec H3A 0G4, Canada; Early Drug Discovery Unit (EDDU), Montreal Neurological Institute-Hospital (The Neuro), Montreal, Quebec H3A 2B4, Canada.
Lang Liu, Department of Human Genetics, McGill University, Montreal, Quebec H3A 0G4, Canada; The Neuro (Montreal Neurological Institute-Hospital), Montreal, Quebec H3A 2B4, Canada.
Konstantin Senkevich, The Neuro (Montreal Neurological Institute-Hospital), Montreal, Quebec H3A 2B4, Canada; Department of Neurology and Neurosurgery, McGill University, Montreal, Quebec H3A 0G4, Canada.
Shady Rahayel, Centre for Advanced Research in Sleep Medicine, Hôpital du Sacré-Cœur de Montréal, Montreal, Quebec H4J 1C5, Canada; Department of Medicine, University of Montreal, Montreal, Quebec H3C 3J7, Canada.
Jean-François Trempe, Department of Pharmacology and Therapeutics and Centre de Recherche en Biologie Structurale, McGill University, Montreal, Quebec H3A 0G4, Canada.
Edward A Fon, Department of Neurology and Neurosurgery, McGill University, Montreal, Quebec H3A 0G4, Canada; Early Drug Discovery Unit (EDDU), Montreal Neurological Institute-Hospital (The Neuro), Montreal, Quebec H3A 2B4, Canada.
Ziv Gan-Or, Department of Human Genetics, McGill University, Montreal, Quebec H3A 0G4, Canada; The Neuro (Montreal Neurological Institute-Hospital), Montreal, Quebec H3A 2B4, Canada; Department of Neurology and Neurosurgery, McGill University, Montreal, Quebec H3A 0G4, Canada.
Data availability
The data used for this study can be accessed on: AMP-PD (https://amp-pd.org/); Bryois et al.59; Cuomo et al.60; FUMA https://fuma.ctglab.nl/; Kamath et al.12; PPMI ppmi-info.org; SMR https://yanglab.westlake.edu.cn/software/smr/and UK Biobank https://www.ukbiobank.ac.uk/. The scripts used for this study can be found on GitHub: github.com/gan-orlab/gene_prio.
Funding
This work was financially supported by the Michael J. Fox Foundation, Parkinson’s Society Canada, the Canadian Consortium on Neurodegeneration in Aging (CCNA), Canadian Institutes of Health Research (#476751) and the Canada First Research Excellence Fund (CFREF) awarded to McGill University for the Healthy Brains Healthy Lives (HBHL) program.
Competing interests
The authors report no competing interests.
Supplementary material
Supplementary material is available at Brain online.
References
- 1. Nalls MA, Blauwendraat C, Vallerga CL, et al. Identification of novel risk loci, causal insights, and heritable risk for Parkinson's disease: A meta-analysis of genome-wide association studies. Lancet Neurol. 2019;18:1091–1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Maurano MT, Humbert R, Rynes E, et al. Systematic localization of common disease-associated variation in regulatory DNA. Science. 2012;337:1190–1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Tam V, Patel N, Turcotte M, Bossé Y, Paré G, Meyre D. Benefits and limitations of genome-wide association studies. Nat Rev Genet. 2019;20:467–484. [DOI] [PubMed] [Google Scholar]
- 4. Li YI, Wong G, Humphrey J, Raj T. Prioritizing Parkinson’s disease genes using population-scale transcriptomic data. Nat Commun. 2019;10:994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Schilder BM, Raj T. Fine-mapping of Parkinson’s disease susceptibility loci identifies putative causal variants. Hum Mol Genet. 2022;31:888–900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Kia DA, Zhang D, Guelfi S, et al. Identification of candidate Parkinson disease genes by integrating genome-wide association study, expression, and epigenetic data sets. JAMA Neurol. 2021;78:464–472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Mountjoy E, Schmidt EM, Carmona M, et al. An open approach to systematically prioritize causal variants and genes at all published human GWAS trait-associated loci. Nat Genet. 2021;53:1527–1533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Lango Allen H, Estrada K, Lettre G, et al. Hundreds of variants clustered in genomic loci and biological pathways affect human height. Nature. 2010;467:832–838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. McLaren W, Gil L, Hunt SE, et al. The ensembl variant effect predictor. Genome Biol. 2016;17:122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Adzhubei IA, Schmidt S, Peshkin L, et al. A method and server for predicting damaging missense mutations. Nat Methods. 2010;7:248–249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Watanabe K, Taskesen E, van Bochoven A, Posthuma D. Functional mapping and annotation of genetic associations with FUMA. Nat Commun. 2017;8:1826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Kamath T, Abdulraouf A, Burris SJ, et al. Single-cell genomic profiling of human dopamine neurons identifies a population that selectively degenerates in Parkinson's disease. Nat Neurosci. 2022;25:588–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Chen T, Guestrin C. Xgboost: A scalable tree boosting system. In: Proceedings of the 22nd ACM SIGKDD international conference on knowledge discovery and data mining. Association for Computing Machinery. 2016:785-794.
- 14. Liao Y, Wang J, Jaehnig EJ, Shi Z, Zhang B. Webgestalt 2019: Gene set analysis toolkit with revamped UIs and APIs. Nucleic Acids Res. 2019;47(W1):W199–W205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Bressan E, Reed X, Bansal V, et al. The foundational data initiative for Parkinson disease: Enabling efficient translation from genetic maps to mechanism. Cell Genomics. 2023;3:100261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Finak G, McDavid A, Yajima M, et al. MAST: A flexible statistical framework for assessing transcriptional changes and characterizing heterogeneity in single-cell RNA sequencing data. Genome Biol. 2015;16:278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-Seq data with DESeq2. Genome Biol. 2014;15:550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Bandres-Ciga S, Saez-Atienzar S, Kim J, et al. Large-scale pathway specific polygenic risk and transcriptomic community network analysis identifies novel functional pathways in Parkinson disease. Acta Neuropathol. 2020;140:341–358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Choi SW, García-González J, Ruan Y, et al. PRSet: Pathway-based polygenic risk score analyses and software. PLoS Genet. 2023;19:e1010624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Lee S, Teslovich TM, Boehnke M, Lin X. General framework for meta-analysis of rare variants in sequencing association studies. Am J Hum Genet. 2013;93:42–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Iwaki H, Leonard HL, Makarious MB, et al. Accelerating medicines partnership: Parkinson's disease. Genetic resource. Mov Disord. 2021;36:1795–1804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Yu E, Ambati A, Andersen MS, et al. Fine mapping of the HLA locus in Parkinson's disease in Europeans. NPJ Parkinsons Dis. 2021;7:84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Senkevich K, Beletskaia M, Dworkind A, et al. Association of rare variants in ARSA with Parkinson’s disease. medRxiv. [Preprint] doi: 10.1101/2023.03.08.23286773 [DOI] [PMC free article] [PubMed]
- 24. Evans R, O’Neill M, Pritzel A, et al. Protein complex prediction with AlphaFold-Multimer. bioRxiv. [Preprint] doi: 10.1101/2021.10.04.463034 [DOI]
- 25. Mirdita M, Schütze K, Moriwaki Y, Heo L, Ovchinnikov S, Steinegger M. Colabfold: Making protein folding accessible to all. Nat Methods. 2022;19:679–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Lundberg SM, Erion G, Chen H, et al. From local explanations to global understanding with explainable AI for trees. Nat Mach Intell. 2020;2:56–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Lundberg SM, Lee S-I. A unified approach to interpreting model predictions. In: Advances in neural information processing systems 30 (NIPS 2017). NeuroIPS Proceedings. 2017.
- 28. He M, Kuk ACY, Ding M, et al. Spns1 is a lysophospholipid transporter mediating lysosomal phospholipid salvage. Proc Natl Acad Sci U S A. 2022;119:e2210353119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Tunyasuvunakool K, Adler J, Wu Z, et al. Highly accurate protein structure prediction for the human proteome. Nature. 2021;596:590–596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Billin AN, Ayer DE. The mlx network: Evidence for a parallel Max-like transcriptional network that regulates energy metabolism. Curr Top Microbiol Immunol. 2006;302:255–278. [DOI] [PubMed] [Google Scholar]
- 31. Billin AN, Eilers AL, Queva C, Ayer DE. Mlx, a novel Max-like BHLHZip protein that interacts with the Max network of transcription factors. J Biol Chem. 1999;274:36344–36350. [DOI] [PubMed] [Google Scholar]
- 32. Nair SK, Burley SK. X-ray structures of myc-Max and mad-Max recognizing DNA. Molecular bases of regulation by proto-oncogenic transcription factors. Cell. 2003;112:193–205. [DOI] [PubMed] [Google Scholar]
- 33. Billin AN, Eilers AL, Coulter KL, Logan JS, Ayer DE. Mondoa, a novel basic helix-loop-helix-leucine zipper transcriptional activator that constitutes a positive branch of a max-like network. Mol Cell Biol. 2000;20:8845–8854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Chakraborty A. The inositol pyrophosphate pathway in health and diseases. Biol Rev Camb Philos Soc. 2018;93:1203–1227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Nakatsu F, Messa M, Nández R, et al. Sac2/INPP5F is an inositol 4-phosphatase that functions in the endocytic pathway. J Cell Biol. 2015;209:85–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Mishori A, Levine J, Kahana E, Belmaker RH. Inositol is not therapeutic in Parkinson's disease. Hum Psychopharmacol. 1999;14:271–272. [Google Scholar]
- 37. Apicco DJ, Shlevkov E, Nezich CL, et al. The Parkinson’s disease-associated gene ITPKB protects against α-synuclein aggregation by regulating ER-to-mitochondria calcium release. Proc Natl Acad Sci U S A. 2021;118:e2006476118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Di Leva F, Filosi M, Oyston L, et al. Increased levels of the Parkinson’s disease-associated gene ITPKB correlate with higher expression levels of α-synuclein, independent of mutation Status. Int J Mol Sci. 2023;24:1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Chakraborty A, Kim S, Snyder SH. Inositol pyrophosphates as mammalian cell signals. Sci Signal. 2011;4:re1–re1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Nagpal L, Kornberg MD, Snyder SH. Inositol hexakisphosphate kinase-2 non-catalytically regulates mitophagy by attenuating PINK1 signaling. Proc Natl Acad Sci U S A. 2022;119:e2121946119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Cao C-H, Ling H, Han K, et al. PPIP5K2 Promotes colorectal carcinoma pathogenesis through facilitating DNA homologous recombination repair. Oncogene. 2021;40:6680–6691. [DOI] [PubMed] [Google Scholar]
- 42. Yousaf R, Gu C, Ahmed ZM, et al. Mutations in diphosphoinositol-pentakisphosphate kinase PPIP5K2 are associated with hearing loss in human and mouse. PLoS Genet. 2018;14:e1007297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Chatree S, Thongmaen N, Tantivejkul K, Sitticharoon C, Vucenik I. Role of inositols and inositol phosphates in energy metabolism. Molecules. 2020;25:5079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Kitamura N, Hashimoto T, Nishino N, Tanaka C. Inositol 1, 4, 5-trisphosphate binding sites in the brain: Regional distribution, characterization, and alterations in brains of patients with Parkinson's disease. J Mol Neurosci. 1989;1:181–187. [DOI] [PubMed] [Google Scholar]
- 45. Zhang Z, Hou L, Li X, et al. Neuroprotection of inositol hexaphosphate and changes of mitochondrion mediated apoptotic pathway and α-synuclein aggregation in 6-OHDA induced Parkinson׳ s disease cell model. Brain Res. 2016;1633:87–95. [DOI] [PubMed] [Google Scholar]
- 46. Abe TK, Taniguchi M. Identification of myo-inositol hexakisphosphate (IP6) as a β-secretase 1 (BACE1) inhibitory molecule in rice grain extract and digest. FEBS open bio. 2014;4:162–167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Shukla AK, Ratnasekhar C, Pragya P, et al. Metabolomic analysis provides insights on paraquat-induced Parkinson-like symptoms in Drosophila melanogaster. Mol Neurobiol. 2016;53:254–269. [DOI] [PubMed] [Google Scholar]
- 48. Ibrahim T, McLaurin J. α-Synuclein aggregation, seeding and inhibition by scyllo-inositol. Biochem Biophys Res Commun. 2016;469:529–534. [DOI] [PubMed] [Google Scholar]
- 49. Gröger A, Kolb R, Schäfer R, Klose U. Dopamine reduction in the substantia nigra of Parkinson's disease patients confirmed by in vivo magnetic resonance spectroscopic imaging. PloS one. 2014;9:e84081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Shah A, Han P, Wong M-Y, Chang R, Legido-Quigley C. Palmitate and stearate are increased in the plasma in a 6-OHDA model of Parkinson’s disease. Metabolites. 2019;9:31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Quadri M, Fang M, Picillo M, et al. Mutation in the SYNJ1 gene associated with autosomal recessive, early-onset parkinsonism. Hum Mutat. 2013;34:1208–1215. [DOI] [PubMed] [Google Scholar]
- 52. Choong C-J, Aguirre C, Kakuda K, et al. Phosphatidylinositol-3, 4, 5-trisphosphate interacts with alpha-synuclein and initiates its aggregation and formation of Parkinson’s disease-related fibril polymorphism. Acta Neuropathol. 2023;145:573–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Senkevich K, Gan-Or Z. Autophagy lysosomal pathway dysfunction in Parkinson's disease; evidence from human genetics. Parkinsonism Relat Disord. 2020;73:60–71. [DOI] [PubMed] [Google Scholar]
- 54. Senkevich K, Zorca CE, Dworkind A, et al. GALC Variants affect galactosylceramidase enzymatic activity and risk of Parkinson's disease. Brain. 2023;146:1859–1872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Tamura N, Maejima Y, Matsumura T, et al. Single-Nucleotide polymorphism of the MLX gene is associated with Takayasu arteritis. Circ Genom Precis Med. 2018;11:e002296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Li Y-J, Nuytemans K, La J, et al. Identification of novel genes for age-at-onset of Alzheimer's disease by combining quantitative and survival trait analyses. Alzheimers Dement. 2023;19:3148–3157. [DOI] [PubMed] [Google Scholar]
- 57. Ho D, Schierding W, Farrow SL, Cooper AA, Kempa-Liehr AW, O’Sullivan JM. Machine learning identifies six genetic variants and alterations in the heart atrial appendage as key contributors to PD risk predictivity. Front Genet. 2022;12:785436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Timmermans PBMWM. Angiotensin II receptor antagonists: An emerging new class of cardiovascular therapeutics. Hypertens Res. 1999;22:147–153. [DOI] [PubMed] [Google Scholar]
- 59. Bryois J, Calini D, Macnair W, et al. Cell-type-specific cis-eQTLs in eight human brain cell types identify novel risk genes for psychiatric and neurological disorders. Nat Neurosci. 2022;25:1104–1112. [DOI] [PubMed] [Google Scholar]
- 60. Cuomo ASE, Seaton DD, McCarthy DJ, et al. Single-cell RNA-Sequencing of differentiating iPS cells reveals dynamic genetic effects on gene expression. Nat Commun. 2020;11:810. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data used for this study can be accessed on: AMP-PD (https://amp-pd.org/); Bryois et al.59; Cuomo et al.60; FUMA https://fuma.ctglab.nl/; Kamath et al.12; PPMI ppmi-info.org; SMR https://yanglab.westlake.edu.cn/software/smr/and UK Biobank https://www.ukbiobank.ac.uk/. The scripts used for this study can be found on GitHub: github.com/gan-orlab/gene_prio.