Abstract
Qk1 is a member of the KH domain family of proteins that includes Sam68, GRP33, GLD-1, SF1, and Who/How. These family members are RNA binding proteins that contain an extended KH domain embedded in a larger domain called the GSG (for GRP33–Sam68–GLD-1) domain. An ethylnitrosourea-induced point mutation in the Qk1 GSG domain alters glutamic acid 48 to a glycine and is known to be embryonically lethal in mice. The function of Qk1 and the GSG domain as well as the reason for the lethality are unknown. Here we demonstrate that the Qk1 GSG domain mediates RNA binding and Qk1 self-association. By using in situ chemical cross-linking studies, we showed that the Qk1 proteins exist as homodimers in vivo. The Qk1 self-association region was mapped to amino acids 18 to 57, a region predicted to form coiled coils. Alteration of glutamic acid 48 to glycine (E➛G) in the Qk1 GSG domain (producing protein Qk1:E➛G) abolishes self-association but has no effect on the RNA binding activity. The expression of Qk1 or Qk1:E➛G in NIH 3T3 cells induces cell death by apoptosis. Approximately 90% of the remaining transfected cells are apoptotic 48 h after transfection. Qk1:E➛G was consistently more potent at inducing apoptosis than was wild-type Qk1. These results suggest that the mouse quaking lethality (E➛G) occurs due to the absence of Qk1 self-association mediated by the GSG domain.
The mouse quaking gene encodes the Qk1 RNA binding proteins (11). The type of RNA binding domain found in Qk1, known as a KH domain, was originally identified in the heterogeneous nuclear ribonucleoprotein K (hnRNP K [17, 35]). KH domains are evolutionary conserved domains that are thought to make direct protein-RNA contacts with a three-dimensional βααββα-fold (29). The Qk1 KH domain is embedded in a larger conserved domain of ∼200 amino acids called the GSG domain. The GSG domain was initially identified by aligning the first three family members (GRP33, Sam68, and GLD-1 [22]). The boundaries of this new protein module have become clearer with the identification of new family members (1, 11). This domain is also called STAR (for signal transduction and activator of RNA [39]) and the SGQ (Sam68, GLD-1, and Qk1 [25]) domain. GSG domain family members include Artemia salina GRP33 (9), human Sam68 (41), Caenorhabditis elegans GLD-1 (22), human SF1 (1), Drosophila Who/How (2, 16, 42), Xenopus Xqua (44), and mouse Qk1 (11). The features of the GSG domain include a single KH domain that is longer than most other KH domains (29). In addition to the KH domain, the GSG domain is composed of ∼75 amino acids N-terminal and ∼25 amino acids C-terminal of the KH domain (for a review, see reference 39). These regions in the Qk1 GSG domain are called QUA1 and QUA2, respectively (11).
GSG proteins share several properties, including RNA binding (1, 8, 25, 41, 44) and self-association (8, 45). With the exception of the human SF1 protein, which functions as a splicing factor (1), the roles of the GSG proteins in cellular processes are not known. Genetic studies with GSG domain proteins have demonstrated the roles of these proteins in development, differentiation, myelination, and tumorigenesis. In C. elegans, GLD-1 is required for germ cell differentiation (14, 15, 22). One class of alleles results in a recessive tumorous germ line phenotype, suggesting that GLD-1 functions as a tumor suppressor (22). The Qk1 proteins of Drosophila melanogaster, Xenopus laevis, and mice have been characterized. The Drosophila Who/How protein, a Qk1 homolog, has been shown to be critical for skeletal muscle development since weak alleles result in flies with “held-out” wings (2, 42). One such allele contains a point mutation in loop 4 of the Who/How KH domain (2). The Xenopus Xqua protein, another Qk1 homolog, has been shown to be necessary for notochord development (45). Mice that are homozygous for the quaking viable allele have a severe deficiency of myelin throughout their nervous systems and, as a consequence, develop a characteristic tremor (34). The genetic lesion in the quaking viable mouse has been mapped to the qk1 promoter-enhancer region (11). The defect in these mice is the absence of Qk1-6 and Qk1-7 protein expression from the myelin-forming oligodendrocytic cells (19). Another class of mouse quaking mutations is embryonic lethal (7, 23, 33). One such allele, qkkt4, was found to alter glutamic acid 48 to glycine in the QUA1 region of the Qk1 GSG domain (11); the cause for the lethality is unknown.
We have characterized the genetic point mutations identified in GLD-1 and Qk1 by using Sam68 (8). Substitution of the corresponding GLD-1 glycine 227 to aspartic acid in Sam68 abolishes RNA binding, suggesting that the mutation alters GLD-1 RNA binding in C. elegans. Substitution of the corresponding GLD-1 glycines 248 and 250 to arginines in Sam68 abolishes self-association, suggesting that some of the GLD-1 loss-of-function phenotypes observed in C. elegans may be due to the absence of protein-protein interactions. However, the replacement of Qk1 glutamic acid 48 by glycine in Sam68 had no effect on Sam68 RNA binding and oligomerization (8). Therefore, to better understand Qk1 and its lethal point mutation, we characterized the properties of these proteins in vitro and in vivo. Here we report that Qk1 self-associates into dimers via a GSG domain region predicted to form coiled coils. The introduction of the Qk1 lethal point mutation altering glutamic acid 48, located in the predicted coiled-coil region, to a glycine (E48G; resulting protein, Qk1:E➛G) abolished self-association. We also demonstrated that the expression of Qk1 and Qk1:E➛G in NIH 3T3 cells induces apoptosis. These data implicate GSG domain-mediated self-association in the normal function of Qk1.
MATERIALS AND METHODS
DNA constructions.
The deletion constructs encoding Qk1:1–205, Qk1:1–180, and Qk1:81–325 were generated by PCR with myc-Qk1 (8) as a DNA template. The sequences of the oligonucleotide pairs used are 5′-CTG GAA TTC GGT CGG GGA AAT GGA AAC GAA GG-3′ and 5′-ATG GAA TTC TAT CTG TAG GTG CCA TTC AG-3′ (for Qk1:1–205), 5′-CTG GAA TTC GGT CGG GGA AAT GGA AAC GAA GG-3′ and 5′-TCA GAA TTC TAT ACC AGT AAC TTC TTC AC-3′ (for Qk1:1–180), and 5′-ACC GAA TTC TCA GTT ACA AGA GAA ACT T-3′ and 5′-GCT GAA TTC TAG TCC TTC ATC CAG CAA GTC-3′ (for Qk1:81–325). The amplified DNA fragments were digested with EcoRI (the restriction sites are underlined) and subcloned into the EcoRI site of myc-Bluescript KS+ (32) and hemagglutinin (HA)-Bluescript KS (8). Myc-Qk1:E➛G was constructed by inverse PCR with myc-Qk1 as a DNA template and the following oligonucleotides as primers: 5′-GGA ATT AGC AGA GTA CGG AAA GAC-3′ and 5′-TTC GTC CAG CAG CCG CTC GAG-3′. HA-Qk1:E➛G was generated by subcloning the EcoRI fragment of myc-Qk1:E➛G into HA-Bluescript KS. The constructs encoding HA-Qk1, myc-GLD-1, myc-GRP33, and HA-GRP33 were previously described (8). The plasmids encoding glutathione S-transferase (GST)-Qk1 and GST-Qk1:E➛G fusion proteins were constructed by a two-step subcloning strategy. The BamHI-XhoI fragment of myc-Qk1 was first inserted in the corresponding sites of pGEX-KG, generating pGST-Qk1(BamHI-XhoI). The XhoI fragments of myc-Qk1 and myc-Qk1:E➛G were subcloned into the XhoI site of pGST-Qk1(BamHI-XhoI), resulting in pGST-Qk1 and pGST-Qk1:E➛G, respectively. The GST-Qk1 deletion constructs were generated by PCR amplification with myc-Qk1 as the DNA template. For Qk1:1–80, Qk1:1–57, and Qk1:1–37, the T7 primer was used as the forward primer and the following oligonucleotides were used as the reverse primers: 5′-CTC TCT AGA CTA AAC AAT GGG TCC CAC CGC-3′ (for Qk1:1–80), 5′-TAA TCT AGA CTA GTA CAT GTC TTT CCG TAC-3′ (for Qk1:1–57), and 5′-GAA TCT AGA TCA GAA GTT GGG CAG GCT GCT-3′ (for Qk1:1–37). The DNA fragments encoding Qk1:18–57 and Qk1:28–57 were generated by using the reverse primer employed for Qk1:1–57 and the following forward primers: 5′-CCA GGA TCC TTG ATG CAG CTG ATG AAC-3′ (for Qk1:18–57) and 5′-AAG GGA TCC ATG AGC AGC CTG CCC AAC-3′ (for Qk1:28–57). The oligonucleotides used to generate Qk1:38–80 were 5′-TGC GGA TCC TTC AAC CAC CTC GAG CGG-3′ and 5′-CTC TCT AGA CTA AAC AAT GGG TCC CAC CGC-3′. All of the amplified fragments were digested with BamHI and XbaI (the underlined nucleotide sequences denote the restriction sites) and subcloned into the corresponding sites of pGEX-KG. The GST proteins were purified by affinity chromatography with glutathione beads. The purified GST proteins were covalently coupled to Affi-Gel 10 (Bio-Rad) as described previously (32). The green fluorescence protein (GFP) fusion constructs GFP-Qk1 and GFP-Qk1:E➛G were generated by subcloning the EcoRI DNA fragments of myc-Qk1 and myc-Qk1:E➛G, respectively, into vector pEGFP-C1 (Clontech). pcDNA-Qk1, pcDNA-Qk1:E➛G, and pcDNA–GLD-1 were generated by subcloning the EcoRI fragments of myc-Qk1 and myc-Qk1:E➛G and the XhoI fragment of myc–GLD-1, respectively, into the corresponding sites of myc-pcDNA. myc-pcDNA was constructed by digesting myc-Bluescript KS+ (32) with BamHI and XhoI and subcloning the DNA fragment in the corresponding sites of pcDNA1 (Invitrogen). The identities of the plasmid constructs were verified by dideoxynucleotide sequencing with Sequenase (U.S. Biochemical).
Protein expression and analysis.
Proteins were expressed in HeLa cells, using the vaccinia virus T7 expression system as described previously (32). The HeLa cells were lysed in lysis buffer (1% Triton X-100, 150 mM NaCl, 20 mM Tris-HCl [pH 8.0], 50 mM NaF, 100 μM sodium vanadate, 0.01% phenylmethylsulfonyl fluoride, 1 μg of aprotinin per ml, 1 μg of leupeptin per ml), and the cellular debris and nuclei were removed by centrifugation. For immunoprecipitations, cell lysates were incubated on ice with the appropriate antibody for 1 h; then 20 μl of a 50% protein A-Sepharose slurry was added, and the mixture was incubated at 4°C for 30 min with constant end-over-end mixing. The beads were washed twice with lysis buffer and once with phosphate-buffered saline (PBS). The samples were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and transferred to nitrocellulose membranes. For GST pull-down assays, 20 μl of 50% slurry containing 2 mg of GST fusion protein, covalently coupled to beads, per ml was incubated with cell lysate expressing HA-Qk1 for 1 h with constant mixing. The samples were washed and analyzed as described previously (32). Immunoblotting was performed with anti-myc 9E10 (12), anti-HA, or anti-Qk1 rabbit polyclonal antibody. The anti-Qk1 antibody was generated by using GST-Qk1 (81–180) as an antigen. The designated primary antibody was followed by goat anti-mouse or goat anti-rabbit antibodies conjugated to horseradish peroxidase (Organon Teknika-Cappel), and chemiluminescence (Dupont) was used for protein detection.
In situ chemical cross-linking and analysis of Qk1 dimers.
HeLa cells transfected with Qk1 plasmids and rat C6 glioma cells (American Type Culture Collection) were treated in situ with 1 mM bis(maleimido)hexane (BMH) and analyzed as described previously (8). Rat astrocytes and oligodendrocytes were prepared by Guillermina Almazan (McGill University) as described elsewhere (31). Dimer formation was analyzed in HeLa cells transfected with myc-Qk1 and HA-Qk1. The transfected cells were treated with BMH in situ, and the cell lysates were immunoprecipitated with anti-myc antibody or mouse immunoglobulin G (IgG). The bound proteins as well as the cell lysates were separated by SDS-PAGE, transferred to nitrocellulose membranes, and immunoblotted with anti-HA antibody.
RNase treatment and RNA binding analysis.
RNase treatment was carried out by incubating cell lysates at 37°C for 1 h with RNase A (Boehringer Mannheim) at 1 mg/ml. Incubating the cell lysates at 37°C without RNase A was considered a mock treatment. RNA binding studies were carried out by incubating Qk1 immunoprecipitates with radiolabeled total cellular RNA. To obtain radiolabeled RNA, HeLa cells were incubated overnight with 50 μCi of [32P]orthophosphate (Dupont)/ml in phosphate-free Dulbecco’s modified Eagle’s medium (ICN). The cells were harvested, and RNA was extracted by using an RNeasy Mini Kit (Qiagen). myc-Qk1 expressed in HeLa cells was immunoprecipitated with anti-myc antibody or mouse IgG (control). The immunoprecipitates were incubated at 4°C for 30 min with 32P-labeled RNA (3 × 106 cpm) in the presence of 2 mg of heparin/ml. The beads were washed extensively, and the bound radioactivity was counted with a liquid scintillation counter. The bound proteins were then dissociated in 1× Laemmli sample buffer, separated by SDS-PAGE, transferred to nitrocellulose membranes, and analyzed by immunoblotting with anti-myc antibodies.
Apoptosis assays.
NIH 3T3 cells were plated 12 h before transfection, typically at a density of 105 cells/22-mm2 coverslip (Fisher Scientific Co.). Cells were transfected with DNA constructs encoding GFP alone, GFP-Qk1, GFP-Qk1:E➛G, pcDNA-Qk1, pcDNA-Qk1:E➛G, or pcDNA–GLD-1, using LipofectAMINE PLUS reagent (Gibco BRL). At 12, 24, 36, or 48 h after transfection, the cells were fixed with 4% paraformaldehyde in PBS for 15 min and permeabilized with 1% Triton X-100 in PBS for 5 min. For immunostaining, the fixed cells were incubated with the anti-myc 9E10 antibody (1:1,000) at room temperature for 1 h and subsequently with a rhodamine-conjugated goat anti-mouse secondary antibody (Jackson Laboratories; 1:300) for 30 min. The nuclei were stained with 3 μg of 4,6-diamidino-2-phenylindole (DAPI)/ml. The morphology of transfected cells was examined by fluorescence microscopy, and cells with morphological features such as nuclear condensation and fragmentation were considered apoptotic. Apoptosis was also examined by TUNEL (terminal deoxynucleotidyl transferase-mediated fluorescein-dUTP nick end labeling). The TUNEL reagents were obtained from Boehringer Mannheim, and the assay was performed as suggested by the manufacturer.
RESULTS
The mouse quaking gene products exist as homodimers in vivo.
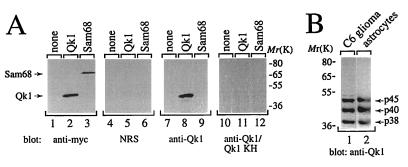
The quaking gene encodes three different alternatively spliced transcripts that generate the Qk1–5, Qk1–6, and Qk1–7 proteins (11). These Qk1 proteins differ in their C-terminal 30 amino acids and are predicted to migrate with apparent molecular masses of 45 to 38 kDa on SDS-polyacrylamide gels (11). To characterize the endogenous Qk1 proteins, a rabbit polyclonal antibody against Qk1 amino acids 81 to 180, encompassing the KH domain, was generated. This region is identical in all Qk1 splice variants, and therefore the antibody should recognize all three Qk1 isoforms. The specificity of the anti-Qk1 antibody was examined by using HeLa cells transfected with vector alone, myc epitope-tagged Qk1–7, or myc-Sam68. The cell lysates from the transfected cells were resolved by SDS-PAGE, transferred to nitrocellulose membranes, and immunoblotted with anti-myc (Fig. 1A, lanes 1 to 3), normal rabbit serum (lanes 4 to 6), anti-Qk1 (lanes 7 to 9), or anti-Qk1 antibodies preabsorbed with the GST-Qk1 antigen (lanes 10 to 12). The anti-Qk1 antibody recognized the transfected Qk1 protein but not Sam68 (lanes 8 and 9). Qk1 was not observed when using normal rabbit serum or anti-Qk1 antibodies preabsorbed with the antigen (lanes 4 to 6 and 10 to 12, respectively).
FIG. 1.
Characterization of the anti-Qk1 antibody. (A) Total cell lysates from HeLa cells transfected with vector (none), myc-Qk1 (Qk1), or myc-Sam68 (Sam68) were separated by SDS-PAGE. The proteins were transferred to nitrocellulose and immunoblotted with anti-myc, normal rabbit serum (NRS), anti-Qk1, or anti-Qk1 antibodies preabsorbed with 1 μg of GST-Qk1KH antigen/ml (anti-Qk1/Qk1 KH). The positions of Qk1 and Sam68 are shown on the left, and those of molecular mass markers (in kilodaltons) are on the right. (B) Total cell extracts from rat C6 glioma cells and rat astrocytes were immunoblotted with anti-Qk1. The presence of three Qk1-immunoreactive proteins with approximate molecular masses of 45, 40, and 38 kDa is shown.
To identify a cell line that contained all three Qk1 splice variants, a panel of neuronal cell lines was analyzed by immunoblotting with the anti-Qk1 antibody. Three major immunoreactive proteins in the 45- to 38-kDa range were detected in the rat C6 glioma cell line (Fig. 1B, lane 1) (5). Similar patterns of expression were observed for purified rat astrocytes (Fig. 1B, lane 2), purified rat oligodendrocytes, and whole-brain extracts from BALB/c, heterozygous, and homozygous quaking viable mice (data not shown). These data are consistent with the recent finding that Qk1–5, Qk1–6, and Qk1–7 are expressed in different brain cell types of normal and quaking viable mice (19), and therefore extracts from whole brains should contain all three Qk1 splice variants. Our results identify the C6 glioma cell line as a suitable cell system with which to investigate the properties of the endogenous Qk1 proteins.
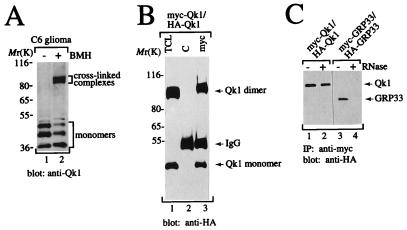
Since we have previously shown that the transfected Qk1–7 protein forms complexes in HeLa cells (8), we performed chemical cross-linking studies with C6 glioma cells to determine whether endogenous Qk1 proteins self-associated into similar complexes. C6 glioma cells were either left untreated or treated in situ with BMH, an irreversible chemical cross-linker, and the cell lysates were resolved by SDS-PAGE, transferred to nitrocellulose membranes, and immunoblotted with the anti-Qk1 antibody (Fig. 2A). In addition to the three monomeric Qk1 proteins (lanes 1 and 2), three distinct cross-linked Qk1 complexes with apparent molecular masses of 90 to 80 kDa were observed after cross-linking (lane 2). These data suggest that the Qk1 proteins exist as dimers.
FIG. 2.
Qk1 self-associates into homodimers in the absence of cellular RNA. (A) C6 glioma cells were treated in situ with (+) or without (−) BMH. The cells were lysed in sample buffer, and the proteins were separated by SDS-PAGE. The proteins were transferred to nitrocellulose and immunoblotted with rabbit anti-Qk1 antibody. The bands representing the three Qk1 isoforms are shown as monomers, and the cross-linked complexes are indicated. The positions of molecular mass markers (in kilodaltons) are indicated on the left. (B) HeLa cells cotransfected with myc- and HA-Qk1 were treated in situ with BMH. The cells were lysed, and an aliquot of the total cell lysate (TCL) as well as anti-myc (myc) and IgG (C) immunoprecipitates were separated by SDS-PAGE. The proteins were transferred to nitrocellulose and immunoblotted with anti-HA antibodies. (C) Qk1, unlike GRP33, does not require cellular RNA for self-association. HeLa cells expressing myc-Qk1, HA-Qk1, myc-GRP33, or HA-GRP33 were lysed separately, and each cell lysate was divided into two portions, either treated with RNase A (+) or not treated (−), mixed, and incubated with anti-myc antibodies. The anti-myc immunoprecipitates (IP) of the mixed lysates were separated by SDS-PAGE and immunoblotted with anti-HA antibodies.
To verify that Qk1 cross-linked complexes represented Qk1 dimers, HeLa cells coexpressing myc- and HA-Qk1 were treated in situ with BMH, and the cell lysates were immunoprecipitated with IgG (Fig. 2B, lane 2) or anti-myc antibody (lane 3). The immunoprecipitated proteins as well as the total cell lysate (lane 1) were analyzed by immunoblotting with anti-HA antibodies. BMH treatment of HeLa cells expressing Qk1 resulted in a 90-kDa complex, in addition to the 45-kDa Qk1 monomer (lane 1). This HA-Qk1-containing complex was present in anti-myc immunoprecipitates (lane 3), demonstrating that HA- and myc-Qk1 proteins dimerized. The presence of HA-Qk1 monomers in anti-myc immunoprecipitates indicated that not all complexes were chemically cross-linked (lane 3).
Since the mouse Qk1–7 protein is an RNA binding protein (8), we sought to investigate whether cellular RNA was required for Qk1 self-association. myc- and HA-Qk1 were expressed in HeLa cells separately, and the cell lysates were or were not treated with RNase A for 1 h at 37°C and then mixed and immunoprecipitated with anti-myc antibodies. The bound proteins were separated by SDS-PAGE, transferred to nitrocellulose, and immunoblotted with anti-HA antibodies. HA-Qk1 coprecipitated with myc-Qk1 regardless of whether the lysates were treated with RNase A (Fig. 2C, lanes 1 and 2), indicating that RNase treatment had no effect on the ability of Qk1 to self-associate. Under similar conditions, we also tested the ability of GRP33 to self-associate in the presence or absence of RNase. HA-GRP33 was observed in anti-myc immunoprecipitates when no RNase treatment was performed (lane 3), but it was not seen when RNase treatment was performed (lane 4). These findings show that unlike GRP33 and Sam68 (8), Qk1 does not require RNA for self-association.
The self-association and RNA binding properties of Qk1 map to the GSG domain.
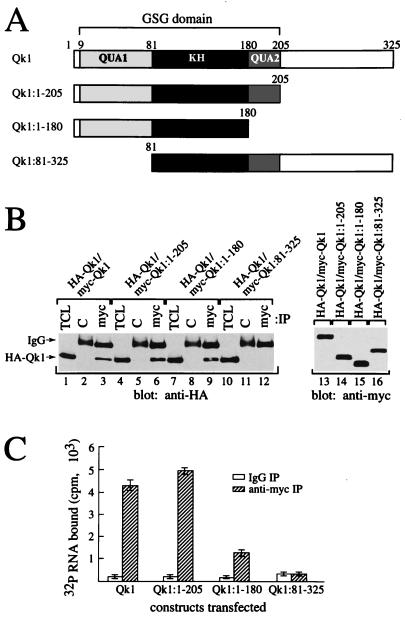
The Qk1 GSG domain spans amino acids 9 to 205, and the embedded KH domain spans amino acids 81 to 180 (Fig. 3A). A deletion analysis was performed to identify whether the GSG domain and the Qk1 C-terminal region were necessary and sufficient for Qk1 self-association and RNA binding. The truncated Qk1 proteins were tested for their ability to associate with HA-Qk1. HeLa cells were cotransfected with DNAs encoding HA-Qk1 and with wild-type myc-Qk1 or the truncated myc-Qk1 constructs (Fig. 3A). The cells were lysed and immunoprecipitated with control or anti-myc antibodies. The bound proteins were immunoblotted with anti-HA antibodies for detection of the presence of HA-Qk1. myc immunoprecipitates of wild-type myc-Qk1 (Fig. 3B, lane 3), myc-Qk1:1–205 (lane 6), and myc-Qk1:1–180 (lane 9) contained abundant levels of HA-Qk1. However, Qk1 lacking its N-terminal 80 amino acids, or the QUA1 region (myc-Qk1:81–325), did not coprecipitate HA-Qk1 (Fig. 3B, lane 12). These data demonstrate that the C-terminal 145 amino acids of Qk1 are dispensable and that the QUA1 region of the GSG domain is required for self-association. The levels of expression of the myc-Qk1 constructs were equivalent (Fig. 3B, lanes 13 to 16).
FIG. 3.
Mapping of the Qk1 self-association and RNA binding regions within the GSG domain. (A) Schematic diagrams of the Qk1 constructs utilized are shown. The black box denotes the KH domain, whereas the gray boxes represent the QUA1 and QUA2 regions as indicated. The entire region encompassing QUA1, KH, and QUA2 is the GSG domain. (B) Truncation of the N-terminal 80 amino acids of Qk1 or QUA1 abolishes self-association. HA-Qk1 was cotransfected in HeLa cells with various myc-Qk1 deletion constructs as indicated. Total cell lysates (TCL) as well as anti-myc (myc) and control IgG (C) immunoprecipitates were analyzed by immunoblotting with anti-HA antibodies. Total cell lysates of the myc-Qk1 proteins were immunoblotted with anti-myc antibodies (lanes 13 to 16). (C) The Qk1 GSG domain is required for RNA binding. HeLa cell lysates containing myc-tagged Qk1 or various truncated forms of Qk1 were immunoprecipitated with anti-myc antibody (hatched bars) or control IgG (white bars) and then incubated with 32P-labeled total cellular RNA. Each bar represents the mean ± standard deviation of data from more than six independent immunoprecipitations carried out during more than three separate experiments.
The RNA binding abilities of wild-type Qk1 and its truncated forms were compared. Wild-type and the various truncated myc-Qk1 proteins were expressed in HeLa cells, and the anti-myc and control IgG immunoprecipitates were incubated with 32P-labeled RNA in lysis buffer supplemented with 2 mg of heparin/ml. The radioactivity in the bound RNA was counted and expressed in counts per minute (Fig. 3C). Anti-myc immunoprecipitates of wild-type Qk1 bound 20 times more labeled RNA than did control immunoprecipitates (Fig. 3C, Qk1). The deletion of the C-terminal 120 amino acids had no effect on Qk1 RNA binding activity (Fig. 3C, Qk1:1–205). However, the deletion of the QUA2 region in the Qk1 GSG domain reduced the bound RNA by more than half (Fig. 3C, Qk1:1–180). Furthermore, the deletion of the QUA1 region of the Qk1 GSG domain completely abolished RNA binding (Qk1:81–325). These data suggest that the entire Qk1 GSG domain is required for optimal RNA binding.
The genetic mutation (E48G) identified in quaking lethal mice abolishes self-association but not RNA binding.
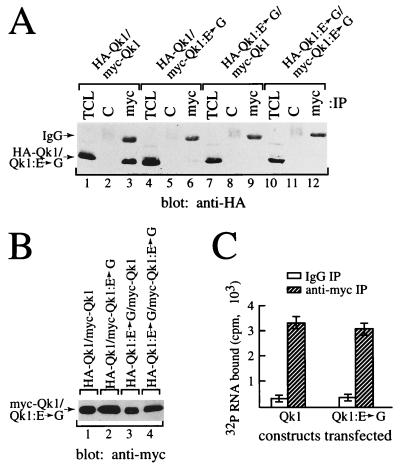
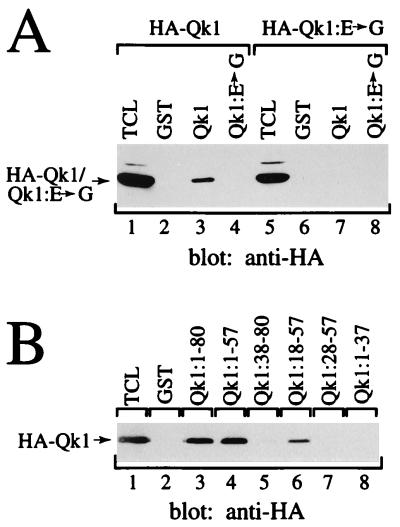
Our deletion studies indicated that the QUA1 region of the Qk1 GSG domain is required for Qk1 self-association and RNA binding. Interestingly, one ethylnitrosourea-induced point mutation that causes a mouse quaking lethal phenotype (23) has been identified in this region (11). This amino acid substitution, altering glutamic acid 48 to a glycine, was introduced in the mouse quaking protein (Qk1:E➛G) and tested for its effect on self-association and RNA binding. The abilities of Qk1:E➛G to associate with Qk1 and to self-associate were examined (Fig. 4A). HeLa cells were transfected with combinations of myc- and HA-Qk1 or Qk1:E➛G as indicated in Fig. 4A. The cells were lysed and immunoprecipitated with control or anti-myc antibodies. The proteins were separated by SDS-PAGE and analyzed by immunoblotting with anti-HA antibodies. HA-Qk1 coimmunoprecipitated with myc-Qk1 (Fig. 4A, lane 3) but not with myc-Qk1:E➛G (lane 6). Moreover, HA-Qk1:E➛G did not coprecipitate with myc-Qk1 (lane 9) or myc-Qk1:E➛G (lane 12), suggesting that the E48G mutation prevents Qk1 self-association. The levels of expression of the myc epitope-tagged constructs used were equivalent (Fig. 4B).
FIG. 4.
E48G substitution in Qk1 abolishes homodimerization but not RNA binding. (A) HeLa cells were transfected with HA- or myc-tagged Qk1 and/or Qk1:E➛G as indicated. The total cell lysate (TCL) as well as anti-myc (myc) and IgG (C) immunoprecipitates were immunoblotted with anti-HA antibody. The bands representing HA-Qk1 or HA-Qk1:E➛G are indicated on the left. The migration of the heavy chain of IgG is also indicated. (B) Total cell lysates corresponding to those of panel A were immunoblotted with anti-myc antibodies. (C) Immunoprecipitated Qk1 or Qk1:E➛G was incubated with labeled RNA as described in Materials and Methods.
The RNA binding properties of Qk1:E➛G and Qk1 were investigated. The Qk1:E➛G protein bound RNA to the same extent as the wild-type Qk1 protein (Fig. 4C), demonstrating that this point mutation has no effect on RNA binding. However, we cannot exclude the possibility that a difference in RNA binding will be observed once a high-affinity RNA target for Qk1 is identified.
We next examined the ability of wild-type and mutated Qk1 to self-associate in vitro. GST pull-down assays using bacterial fusion proteins were performed (Fig. 5A). HeLa cell lysates containing HA-Qk1 or HA-Qk1:E➛G were incubated with affinity matrices coupled with GST alone (lanes 2 and 6), GST-Qk1 (lanes 3 and 7), or GST-Qk1:E➛G (lanes 4 and 8). The proteins that bound the GST proteins were separated by SDS-PAGE and analyzed with anti-HA antibodies. HA-Qk1 bound the GST-Qk1 fusion protein (lane 3), indicating that Qk1 was able to self-associate in vitro. However, HA-Qk1 did not associate with GST-Qk1:E➛G (lane 4), and no interaction between HA-Qk1:E➛G and either GST-Qk1 (lane 7) or GST-Qk1:E➛G (lane 8) was observed. These results are consistent with our coimmunoprecipitation data, confirming that the quaking lethal point mutation abolishes Qk1 self-association. These observations demonstrate that Qk1 containing the quaking lethal point mutation is defective in GSG-mediated protein-protein interactions, such as the ability to self-associate.
FIG. 5.
Association of Qk1 in vitro and localization of the minimal region to amino acids 18 to 57. (A) HA-Qk1 or HA-Qk1:E➛G was transfected into HeLa cells. The cells were lysed, and GST pull-down assays were performed with full-length Qk1 or Qk1:E➛G expressed as a GST fusion protein. The bound proteins were separated by SDS-PAGE, transferred to nitrocellulose, and immunoblotted with anti-HA antibodies. (B) HA-Qk1 was expressed in HeLa cells and incubated with various Qk1 GST fusion proteins as indicated. The bound HA-Qk1 was analyzed as described for panel A. The migration of HA-Qk1 or HA-Qk1:E➛G is shown on the left. TCL, total cell lysate.
The QUA1 region of the GSG domain mediates Qk1 self-association.
The severe effect of the quaking point mutation on Qk1 self-association suggested that the QUA1 region of the Qk1 GSG domain is directly involved in Qk1 self-association. To further delineate the region responsible for self-association, a series of GST fusion proteins were generated. These fusion proteins were utilized in GST pull-down assays and tested for their ability to interact with HA-Qk1 produced in HeLa cells (Fig. 5B). The GST-Qk1:1–80 and GST-Qk1:1–57 fusion proteins bound HA-Qk1 (lane 3 and 4), but GST-Qk1:1–37 did not (lane 8). Amino-terminal deletions revealed that Qk1:18–57 was the minimum region capable of binding HA-Qk1 (lanes 5 to 7).
Qk1 and Qk1:E48➛G induce apoptosis in fibroblasts.
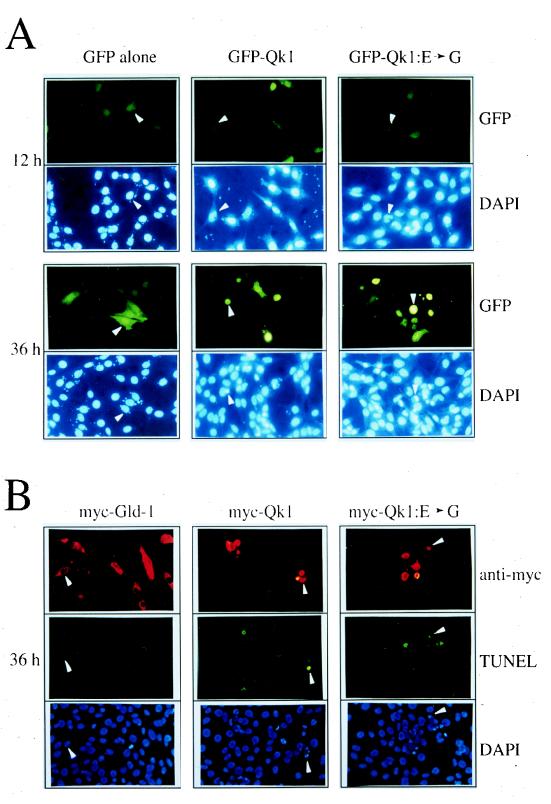
The phenotype of the quaking lethal mutation is arrested growth of an embryo with generalized abnormalities (23), suggesting that Qk1 may play a role in cell proliferation or differentiation. To determine the role of Qk1 and its lethal mutation, we expressed wild-type Qk1 and Qk1 with the E48G mutation in mouse fibroblast cells and examined their effects on cell growth. NIH 3T3 cells were transfected with expression vectors encoding GFP alone, GFP-Qk1, or GFP-Qk1:E➛G. Twelve hours after transfection, approximately 25% of the cells expressed GFP, GFP-Qk1, or GFP-Qk1:E➛G, as visualized by fluorescence microscopy. Interestingly, only 6 to 8% of the cells expressed GFP-Qk1 and GFP-Qk1:E➛G at 36 h, suggesting that the cells transfected with wild-type or mutant Qk1 were not surviving (data not shown). The cells expressing GFP alone appeared normal and healthy (Fig. 6A, left panels). Cells expressing GFP-Qk1 or Qk1:E➛G exhibited morphological changes characteristic of apoptosis, including cell shrinkage, cytoplasm condensation, and membrane blebbing (Fig. 6A, middle and right panels). GFP-transfected cells displayed normal nuclear morphology as visualized by DAPI staining (GFP panels, lower halves). GFP-Qk1- or GFP-Qk1:E➛G-transfected cells had irregular (GFP-Qk1 and GFP-Qk1:E➛G panels, 12 h, lower halves), condensed, or fragmented nuclei (36 h), consistent with apoptotic cell death. Similar data were obtained in HeLa cells (data not shown). To confirm that the morphological changes induced by GFP-Qk1 and GFP-Qk1:E➛G were indeed associated with apoptosis, we performed the TUNEL assay with fluoresceinated nucleotides, NIH 3T3 cells were transfected with myc-Qk1, myc-Qk1:E➛G, or myc-GLD-1 for 36 h. The cells were fixed, and the myc epitope-tagged proteins were detected by indirect immunofluorescence with a rhodamine-conjugated secondary antibody (Fig. 6B, top panels). As observed by TUNEL assay, most of the myc-Qk1- and myc-Qk1:E➛G-transfected cells fluoresced green, consistent with apoptotic cell death. The transfection of another cytoplasmic GSG protein, myc–GLD-1 (21), did not induce apoptosis and served as a negative control (Fig. 6B, left panels). All of the cells that stained in the TUNEL assay contained condensed or fragmented nuclei as visualized by DAPI staining (Fig. 6B). Therefore, the presence of nuclear condensation and fragmentation, as detected by DAPI staining, is a good indication of apoptotic cell death.
FIG. 6.
Qk1 and Qk1:E➛G induce apoptosis in NIH 3T3 cells. (A) NIH 3T3 cells were transfected with an expression vector encoding GFP alone, GFP-Qk1, or GFP-Qk1:E➛G. After 12, 24, 36, and 48 h, the cells were fixed and stained with DAPI to visualize the nuclei. The top photograph in each pair shows the fluoresceinated cells containing GFP, and the lower photograph shows the DAPI-stained nuclei. The white arrowheads were used to align the top and bottom photographs. (B) NIH 3T3 cells were transfected with expression vector encoding myc–GLD-1, myc-Qk1, or myc-Qk1:E➛G. The myc epitope-tagged proteins were visualized by indirect immunofluorescence with a rhodamine-conjugated secondary antibody (anti-myc). The apoptotic cells were visualized by TUNEL with fluorescein-containing nucleotides (TUNEL), and the nuclei were stained with DAPI (DAPI). The three photographs each for myc–GLD-1, myc-Qk1, and myc-Qk1:E➛G represent the same field of cells as visualized with different filters. The white arrowheads were used to orient the cells in the photographs.
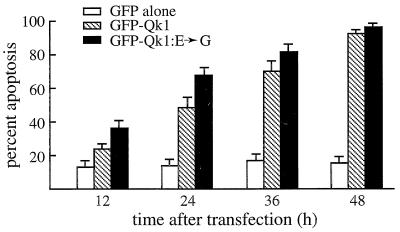
The levels of apoptosis induced by Qk1 and the Qk1:E➛G proteins were quantitated by randomly counting cells and expressing the number of apoptotic cells as a percentage of transfected (green) cells. NIH 3T3 cells were transfected with plasmids expressing GFP, GFP-Qk1, or GFP-Qk1:E➛G. A small fraction of GFP-expressing cells were apoptotic, and this fraction (∼15%) remained steady for up to 48 h (Fig. 7). The transfection of Qk1 or Qk1:E➛G resulted in a significant increase in the number of apoptotic cells with time. At 48 h posttransfection, ∼90% of the remaining cells were apoptotic (Fig. 7). Qk1:E➛G consistently resulted in a larger fraction of apoptotic cells upon transfection than did Qk1. This difference was more prominent at the early time points. At 12 and 24 h, 36.7 and 68.6% of the Qk1:E➛G-transfected cells were apoptotic. These values are in contrast to 24.4 and 49.0%, respectively, for Qk1-transfected cells, and these differences were statistically significant as calculated by the χ2 test (P < 0.01). Since the transfection efficiencies and levels of expression of GFP-Qk1 and GFP-Qk1:E➛G were similar (data not shown), these results suggested that Qk1:E➛G is more potent than Qk1. The majority, ∼70%, of the Qk1:E➛G-transfected cells were apoptotic at 24 h, whereas it took GFP-Qk1 36 h to reach a similar level of apoptosis. These data suggest that Qk1 induces apoptosis and that the E48G mutation in Qk1 contributes to aggravated apoptotic cell death.
FIG. 7.
Quantitation of the apoptosis induced by Qk1 and Qk1:E➛G. Eight hundred transfected NIH 3T3 cells, in three separate experiments, were counted and assessed for the presence of apoptotic nuclei. The presence of apoptotic nuclei was scored as cells undergoing apoptosis. The white, hatched, and black bars represent GFP, GFP-Qk1, and GFP-Qk1:E➛G, respectively.
DISCUSSION
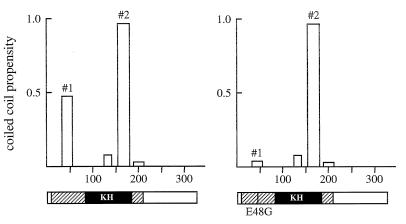
We have shown by coimmunoprecipitation and in situ chemical cross-linking studies that the Qk1 proteins exist as homodimers. The minimum region required for self-association consists of amino acids 18 to 57, which are located in the QUA1 region of the Qk1 GSG domain. This region contains several conserved residues, including glutamic acid at position 48. Alteration of glutamic acid 48 to glycine is thought to be the cause of the lethality in the qkkt4 mice (11). We demonstrated that the Qk1 E48G substitution abolishes self-association. Analysis of the Qk1 protein sequence with the computer program COILS (26) predicted that amino acids 38 to 57 have a high propensity to form coiled coils, which would be disrupted with the introduction of a glycine at position 48 (Fig. 8, coiled coil no. 1). Thus, it is likely that Qk1 dimerizes through coiled-coil interactions mediated by the GSG domain. These data suggest that the failure of Qk1 to dimerize causes embryonic lethality in the qkkt4 mice and implicate dimer formation in the normal function of Qk1 proteins. It is possible that Qk1 associates with other proteins via this region, and therefore we cannot exclude the possibility that the lethality results because Qk1 fails to mediate interactions with other proteins.
FIG. 8.
Coiled-coil motif predictions for Qk1 (left) and Qk1:E➛G (right). The Qk1 and Qk1:E➛G protein sequences were analyzed with the computer program COILS, and the putative coiled-coil motifs are shown (#1 and #2). The abscissa and ordinate represent amino acid numbers and the propensity to form coiled coils, respectively. The structures of the Qk1 proteins are shown below, with hatched and black boxes representing the GSG and KH domains, respectively. The vertical line in the GSG domain denotes the location of the quaking lethal point mutation E48G.
Mouse Qk1 amino acids 38 to 57, predicted to form coiled-coil no. 1, are conserved in Who/How and Xqua, with 13 of 20 and 19 of 20 identical residues, respectively (2, 11, 16, 42, 44). This region in Xqua has a coiled coil prediction similar to that of mouse Qk1 (data not shown), and since Xqua has been shown to self-associate in vitro (45), we predict that this region mediates the self-association. This region in Who/How is also predicted to form coiled coils, but with a lower propensity (0.15). Both Xqua and Who/How coiled coils in this region would be disrupted by the introduction of the corresponding mouse Qk1 E48G mutation. Based on our Qk1 data and the computer analyses, we predict that the point mutation identified in Who/How, altering arginine 185 to cysteine (2), should not alter self-association or RNA binding. Indeed, the introduction of R185C in Drosophila Who/How did not alter self-association or RNA binding (10).
The mouse Qk1 sequence is also predicted to form a second coiled coil, at the C terminus of the KH domain (Fig. 8, coiled coil no. 2). Although our data suggest that coiled coil no. 2 is not sufficient for Qk1 self-association (Qk1:81–325), it is most likely involved in protein-protein interactions other than self-association. Coiled-coil interactions have been predicted for KH domain proteins FMR1 and FXRs (36). These proteins have been observed to self-associate as well as to associate with ribosomes (24, 36). These interactions were demonstrated to occur outside the KH domains, by regions predicted to form coiled coils (36). Interestingly, the KH domains of the FMR1 protein are not predicted to form coiled coils like the KH domain of Qk1. Analysis of the sequences of GSG proteins SF1, Who/How, GLD-1, and Xqua demonstrated that they are all predicted to form coiled coil no. 2 at the C terminus of the KH domain (data not shown). This may represent one more difference between the KH domains of GSG proteins and other KH domains. Sam68 and GRP33 are not predicted to form such coiled coils and may represent a different subclass of GSG proteins. To this end, both GRP33 and Sam68 require RNA for self-association (Fig. 2C) (8). For Sam68, we have been unable to reconstitute the self-association in vitro with recombinant proteins (data not shown) as we have done with Qk1. Another difference is the minimum region required for Sam68 self-association, which is amino acids 103 to 269, the entire GSG domain, and within this region the KH domain loops 1 and 4 are essential (8).
We have mapped the Qk1 RNA binding domain to the N-terminal 205 amino acids encompassing the entire GSG domain. This demonstrates that the entire Qk1 GSG domain is sufficient for RNA binding, as has been previously demonstrated for Sam68 (8, 25). However, this is in contrast to what has been reported for Xqua by Zorn and Krieg; they demonstrated that the entire Xqua protein was required for optimal RNA binding (45). Deletion of the N-terminal GSG amino acids had only a slight reduction effect on RNA binding, whereas deletion of amino acids C terminal to the GSG domain (231 to 357) abolished RNA binding. Since the amino acid sequence identity between mouse, human, and Xenopus Qk1 proteins is 94% (44), Xqua and Qk1 are predicted to have similar RNA binding properties. Either the difference is intrinsic to the proteins or is due to the different stringencies of the RNA binding assays.
Although the exact function of the GSG domain is unknown, our observations with Qk1 and Sam68 indicate that the GSG domain is involved in oligomerization and RNA binding. It is presently unclear whether the GSG domain mediates protein-protein interactions with non-GSG family members. The presence of signaling protein motifs in GSG proteins suggests a role for these proteins in signal transduction (for a review, see reference 39). Interestingly, these potential SH2, SH3, and WW domain binding motifs lie outside the GSG domain, ruling out a direct role for the GSG domain in signal transduction. Nevertheless, the presence of signaling motifs and the presence of phosphorylation sites C terminal to the Sam68 GSG domain (32, 38, 41) suggest that signal transduction pathways may regulate GSG-mediated interactions. Indeed, we have demonstrated in previous studies that Sam68 RNA binding and oligomerization are abolished by the p59fyn tyrosine kinase (8, 40). Moreover, the binding of Sam68 to SH3 domains inhibits RNA binding in vitro (38). These data demonstrate that Sam68 has the potential to link signal transduction pathways with RNA metabolism. Similar data for the other GSG proteins has yet to be obtained. The absence of known signaling motifs in some GSG family members, such as GLD-1 (22), demonstrates that not all family members have the potential to act as signal transduction activators of RNA metabolism (39), but they may have other functions. It is possible that the only properties shared by all family members are GSG domain-mediated RNA binding and self-association.
The quaking genes from X. laevis, D. melanogaster, and mice are involved in a variety of processes, such as myelination, embryogenesis, muscle development, and notochord development (2, 7, 16, 20, 23, 33, 34, 42, 45). The pleiotropic roles and the high level of conservation of this gene suggest a general function for Qk1 in cellular processes. Our results suggest that a function of Qk1 might be to act as a regulator or effector of apoptosis. Since fibroblasts do not express Qk1 (Fig. 1, lanes 7 to 9), it is likely that the expression of Qk1 leads to perturbation of normal cellular processes. Qk1–7, the isoform used in our experiments, is predominantly cytoplasmic (Fig. 6 and data not shown) (19), and it may induce apoptosis by regulating or interfering with the translation and/or mRNA stability of apoptotic or survival proteins. There is a precedent for these mechanisms, since an increase in RNA degradation before the onset of apoptosis has been observed in T cells (28, 37) and hnRNP K has been shown to regulate translation (30).
The three mouse Qk1 splice variants have identical GSG domains and differ in their C termini (11). Qk1–5 is mainly expressed in the nucleus, whereas Qk1–6 and Qk1–7 are mainly expressed in the cytoplasm, suggesting that the last 30 amino acids determine the localization (19). The identicalness of the GSG domains suggests that all three Qk1 splice variants are able to associate with RNA, homodimerize, and heterodimerize. By performing cross-linking studies in C6 glioma cells, we have demonstrated the presence of at least three cross-linked Qk1 species that may represent homodimers and/or heterodimers. The presence of multiple Qk1 splice variants in glial cells and oligodendrocytes (19) suggests an interesting mechanism for the regulation of Qk1 cellular localization. Heterodimers of Qk1–5:Qk1–6 or Qk1–5:Qk1–7 may cause the retention of Qk1–5 in the cytoplasm. Alternatively, Qk1–6 and Qk1–7 might be dragged into the nucleus as Qk1–5 heterodimers. We speculate that formation and balance of such dimers are crucial for Qk1 function and are responsible for the phenotypes observed in the quaking viable and lethal mice.
The genetic lesion in the quaking viable mouse has been mapped to the qk1 promoter-enhancer region (11), and as a result, Qk1–6 and Qk1–7 are not expressed in oligodendrocytes (19). Oligodendrocytes still express nuclear Qk1–5. According to our data, Qk1–5 should be unable to form heterodimers with Qk1–6 or Qk1–7 in oligodendrocytes. This might interfere with Qk1 function and lead to the myelin dysregulation observed in the central nervous systems of these animals. The quaking viable mice have been extensively studied (20), and several defects in RNA metabolism have been observed. Alterations in the levels of alternatively spliced RNAs and in the processing and/or turnover of the mRNA transcripts encoding myelin-associated glycoprotein, myelin basic protein, and proteolipid protein have been demonstrated (4, 6, 13, 19). A defect in myelin basic protein mRNA transport has also been observed in the quaking viable mice (3). The challenge will be to determine whether Qk1 regulates splicing, RNA transport, mRNA stability, and/or translation. Since hnRNP K acts as a transcription factor (27) and Sam68 associates with double-stranded DNA (41), the possibility that nuclear Qk1 also functions as a transcription factor cannot be excluded.
The ethylnitrosourea-induced quaking alleles are known to be lethal at around day 9 or 10 of gestation (7, 23, 33). The only Qk1 isoform expressed in significant amounts at this early time is Qk1–5 (11). Our data suggest that this point mutation would be unable to homodimerize, thus possibly altering its function during embryogenesis. Interestingly, the Qk1:E➛G protein was significantly more potent than wild-type Qk1 at inducing apoptosis in NIH 3T3 cells. Since the Qk1 E48G point mutation is lethal in mice, it is tempting to speculate that unregulated apoptotic cell death occurs due to the absence of GSG-mediated dimerization. The apoptosis we observe with Qk1 in NIH 3T3 cells may be similar to the poisoning effects observed with certain GLD-1 point mutations in C. elegans (22).
By using a pan-Qk1 antibody, the C6 glioma cell line was identified to contain all three Qk1 isoforms. This cell line, which is of rat origin and is derived from glial cells (5), expresses several oligodendrocytic markers, such as myelin-associated glycoprotein, proteolipid protein, and 2′,3′-cyclic nucleotide 3′-phosphohydrolase (18, 43). Since the C6 glioma cell line expresses all three Qk1 isoforms, it should provide a cell system in which to study the properties of the Qk1 dimers and some of their biochemical functions. The expression of Qk1 and Qk1:E➛G in these cells did not readily induce apoptosis (data not shown), suggesting that either the endogenous Qk1 proteins provide a protective effect in these cells or C6 glioma cells are not a suitable system in which to study apoptosis.
In conclusion, we have defined the Qk1 GSG domain as the region required for dimerization and RNA binding. Replacement of glutamic acid 48 with a glycine, a mutation known to be lethal in mice, abolished Qk1 self-association but not RNA binding. The expression of Qk1 and Qk1:E➛G in fibroblast cells induced apoptotic cell death. Since Qk1 has signaling motifs (11), it will be essential to examine the potential role of signaling molecules in the regulation of Qk1 RNA binding, self-association, and apoptosis.
ACKNOWLEDGMENTS
We thank Guillermina Almazan for purified extracts of rat astrocytes and oligodendrocytes. We thank Janet Henderson and Antonis Koromilas for critically reading the manuscript and helpful comments. We are grateful to Rongtuan Lin, John Th’ng, and Hans Zingg for providing reagents and to Bassam Damaj for technical assistance with the rabbit polyclonal antibody.
T.C. is supported by a studentship from the Cancer Research Society of Canada and funds from Canderel. This work was supported by grants from the Medical Research Council of Canada, the Cancer Research Society of Canada, Fonds de la Recherche en Santé du Québec, and the Multiple Sclerosis Society of Canada. S.R. is a Scholar of the Medical Research Council of Canada.
REFERENCES
- 1.Arning S, Gruter P, Bilbe G, Kramer A. Mammalian splicing factor SF1 is encoded by variant cDNAs and binds to RNA. RNA. 1996;2:794–810. [PMC free article] [PubMed] [Google Scholar]
- 2.Baehrecke E H. who encodes a KH RNA binding protein that functions in muscle development. Development. 1997;124:1323–1332. doi: 10.1242/dev.124.7.1323. [DOI] [PubMed] [Google Scholar]
- 3.Barbarese E. Spatial distribution of myelin basic protein mRNA and polypeptide in Quaking oligodendrocytes in culture. J Neurosci Res. 1991;29:271–281. doi: 10.1002/jnr.490290302. [DOI] [PubMed] [Google Scholar]
- 4.Bartoszewicz Z P, Noronha A B, Fujita N, Sato S, Bo L, Trapp B D, Quarles R H. Abnormal expression and glycosylation of the large and small isoforms of myelin-associated glycoprotein in dysmyelinating quaking mutants. J Neurochem Res. 1995;41:27–38. doi: 10.1002/jnr.490410105. [DOI] [PubMed] [Google Scholar]
- 5.Benda P, Lightbody J, Sato G, Levine L, Sweet W. Differentiated rat glial cell strain in tissue culture. Science. 1968;161:370–371. doi: 10.1126/science.161.3839.370. [DOI] [PubMed] [Google Scholar]
- 6.Bo L, Quarles R H, Fujita N, Bartoszewiez Z, Sato S, Trapp B D. Endocytic depletion of L-MAG from CNS myelin in quaking mice. J Cell Biol. 1995;131:1811–1820. doi: 10.1083/jcb.131.6.1811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bode V C. Ethylnitrosourea mutagenesis and the isolation of mutant alleles for specific genes located in the t region of mouse chromosome 17. Genetics. 1984;108:457–470. doi: 10.1093/genetics/108.2.457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen T, Damaj B B, Herrera C, Lasko P, Richard S. Self-association of the single-KH-domain family members Sam68, GRP33, GLD-1, and Qk1: role of the KH domain. Mol Cell Biol. 1997;17:5707–5718. doi: 10.1128/mcb.17.10.5707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cruz-Alvarez M, Pellicer A. Cloning of a full-length complementary cDNA for an Artemia salina glycine-rich protein. J Biol Chem. 1987;262:13377–13380. [PubMed] [Google Scholar]
- 10.Di Fruscio, M., and S. Richard. Unpublished data.
- 11.Ebersole T A, Chen Q, Justice M J, Artzt K. The quaking gene unites signal transduction and RNA binding in the developing nervous system. Nat Genet. 1996;12:260–265. doi: 10.1038/ng0396-260. [DOI] [PubMed] [Google Scholar]
- 12.Evan G I, Lewis G K, Ramsay G, Bishop J M. Isolation of monoclonal antibodies specific for the human c-myc proto-oncogene product. Mol Cell Biol. 1985;5:3610–3616. doi: 10.1128/mcb.5.12.3610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Frail D E, Braun P E. Abnormal expression of the myelin-associated glycoprotein in the central nervous system of dysmyelinating mutant mice. J Neurochem. 1985;45:1071–1075. doi: 10.1111/j.1471-4159.1985.tb05525.x. [DOI] [PubMed] [Google Scholar]
- 14.Francis R, Barton M K, Kimbel J, Schedl T. Control of oogenesis, germline proliferation and sex determination by the C. elegans gene gld-1. Genetics. 1995;139:579–606. doi: 10.1093/genetics/139.2.579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Francis R, Maine E, Schedl T. gld-1, a cell-type specific tumor suppressor gene in C. elegans. Genetics. 1995;139:607–630. doi: 10.1093/genetics/139.2.607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fyrberg C, Becker J, Barthmaier P, Mahaffey J, Fyrberg E. A Drosophila muscle-specific gene related to the mouse quaking locus. Gene. 1997;197:315–323. doi: 10.1016/s0378-1119(97)00278-3. [DOI] [PubMed] [Google Scholar]
- 17.Gibson T J, Thompson J D, Heringa J. The KH domain occurs in a diverse set of RNA-binding proteins that include the antiterminator NusA and is probably involved in binding to nucleic acid. FEBS Lett. 1993;324:361–366. doi: 10.1016/0014-5793(93)80152-k. [DOI] [PubMed] [Google Scholar]
- 18.Goya L, Feng P T, Aliabadi S, Timiras P S. Effect of growth factors on the in vitro growth and differentiation of early and late passage of C6 glioma cells. Int J Dev Neurosci. 1996;14:409–417. [PubMed] [Google Scholar]
- 19.Hardy R J, Loushin C L, Friedrich V L, Jr, Chen Q, Ebersole T A, Lazzarini R A, Artzt K. Neural cell type-specific expression of QKI proteins is altered in the quaking viable mutant mice. J Neurosci. 1996;16:7941–7949. doi: 10.1523/JNEUROSCI.16-24-07941.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hogan E L, Greenfield S. Animal models of genetic disorders of myelin. In: Morrell P, editor. Myelin. 2nd ed. New York, N.Y: Plenum Press; 1984. pp. 489–534. [Google Scholar]
- 21.Jones A R, Francis R, Schedl T. GLD-1, a cytoplasmic protein essential for oocyte differentiation, shows stage- and sex-specific expression during Caenorhabditis elegans germline development. Dev Biol. 1996;180:165–183. doi: 10.1006/dbio.1996.0293. [DOI] [PubMed] [Google Scholar]
- 22.Jones A R, Schedl T. Mutations in GLD-1, a female germ cell-specific tumor suppressor gene in C. elegans, affect a conserved domain also found in Sam68. Genes Dev. 1995;9:1491–1504. doi: 10.1101/gad.9.12.1491. [DOI] [PubMed] [Google Scholar]
- 23.Justice M J, Bode V C. Three ENU-induced alleles of the murine quaking locus are recessive embryonic lethal mutations. Genet Res. 1988;51:95–102. doi: 10.1017/s0016672300024101. [DOI] [PubMed] [Google Scholar]
- 24.Khandjian E W, Corbin F, Woerly S, Rousseau F. The fragile X mental retardation protein is associated with ribosomes. Nat Genet. 1996;12:91–93. doi: 10.1038/ng0196-91. [DOI] [PubMed] [Google Scholar]
- 25.Lin Q, Taylor S J, Shalloway D. Specificity and determinants of Sam68 RNA binding. J Biol Chem. 1997;272:27274–27280. doi: 10.1074/jbc.272.43.27274. [DOI] [PubMed] [Google Scholar]
- 26.Lupas A, van Dyke M, Stock J. Predicting coiled coils from protein sequences. Science. 1991;252:1162–1164. doi: 10.1126/science.252.5009.1162. [DOI] [PubMed] [Google Scholar]
- 27.Michelotti E F, Michelotti G A, Aronsohn A I, Levens D. Heterogeneous nuclear ribonucleoprotein K is a transcription factor. Mol Cell Biol. 1996;16:2350–2360. doi: 10.1128/mcb.16.5.2350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mondino A, Jenkins M K. Accumulation of sequence-specific RNA-binding proteins in the cytosol of activated T cells undergoing RNA degradation and apoptosis. J Biol Chem. 1995;270:26593–26601. doi: 10.1074/jbc.270.44.26593. [DOI] [PubMed] [Google Scholar]
- 29.Musco G, Stier G, Joseph C, Morelli M A, Pastore A. Three-dimensional structure and stability of the KH domain: molecular insights into the fragile X syndrome. Cell. 1996;85:237–245. doi: 10.1016/s0092-8674(00)81100-9. [DOI] [PubMed] [Google Scholar]
- 30.Ostareck D H, Ostareck-Lederer A, Wilm M, Thiele B J, Mann M, Hentze M W. mRNA silencing in erythroid differentiation: hnRNP K and hnRNP E1 regulate 15-lipoxygenase translation from the 3′ end. Cell. 1997;89:597–606. doi: 10.1016/s0092-8674(00)80241-x. [DOI] [PubMed] [Google Scholar]
- 31.Radhakrichna M, Almazan G. Protein kinases mediate basic fibroblast growth factor’s stimulation of proliferation and c-fos induction in oligodendrocyte progenitors. Mol Brain Res. 1994;24:118–128. doi: 10.1016/0169-328x(94)90123-6. [DOI] [PubMed] [Google Scholar]
- 32.Richard S, Yu D, Blumer K J, Hausladen D, Olszowy M W, Connelly P A, Shaw A S. Association of p62, a multifunctional SH2- and SH3-domain-binding protein, with src family tyrosine kinases, Grb2, and phospholipase Cγ-1. Mol Cell Biol. 1995;15:186–197. doi: 10.1128/mcb.15.1.186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shedlovsky A, King T R, Dove W F. Saturation germ line mutagenesis of the murine t region including a lethal allele of the quaking locus. Proc Natl Acad Sci USA. 1988;85:180–184. doi: 10.1073/pnas.85.1.180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sidman R L, Dickie M M, Appel S H. Mutant mice (quaking and jimpy) with deficient myelination in the central nervous system. Science. 1964;144:309–311. doi: 10.1126/science.144.3616.309. [DOI] [PubMed] [Google Scholar]
- 35.Siomi H, Tunis M J M, Michael W M, Dreyfuss G. The pre-mRNA-binding K protein contains a novel evolutionarily conserved motif. Nucleic Acids Res. 1993;21:1193–1198. doi: 10.1093/nar/21.5.1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Siomi M C, Zhang Y, Siomi H, Dreyfuss G. Specific sequences in the fragile X syndrome protein FMR1 and the FXR proteins mediate their binding to 60S ribosomal subunits and the interaction among them. Mol Cell Biol. 1996;16:3825–3832. doi: 10.1128/mcb.16.7.3825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Taupin J-L, Tian Q, Kedersha N, Robertson M, Anderson P. The RNA-binding protein TIAR is translocated from the nucleus to the cytoplasm during Fas-mediated apoptotic cell death. Proc Natl Acad Sci USA. 1995;92:1629–1633. doi: 10.1073/pnas.92.5.1629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Taylor S J, Anafi M, Pawson T, Shalloway D. Functional interaction between c-src and its mitotic target, Sam68. J Biol Chem. 1995;270:10120–10124. doi: 10.1074/jbc.270.17.10120. [DOI] [PubMed] [Google Scholar]
- 39.Vernet C, Artzt K. STAR, a gene family involved in signal transduction and activation of RNA. Trends Genet. 1997;13:479–484. doi: 10.1016/s0168-9525(97)01269-9. [DOI] [PubMed] [Google Scholar]
- 40.Wang L L, Richard S, Shaw A S. p62 association with RNA is regulated by tyrosine phosphorylation. J Biol Chem. 1995;270:2010–2013. doi: 10.1074/jbc.270.5.2010. [DOI] [PubMed] [Google Scholar]
- 41.Wong G, Muller O, Clark R, Conroy L, Moran M F, Polakis P, McCormick F. Molecular cloning and nucleic acid binding properties of the GAP-associated tyrosine phosphoprotein p62. Cell. 1992;69:551–558. doi: 10.1016/0092-8674(92)90455-l. [DOI] [PubMed] [Google Scholar]
- 42.Zaffran S, Astier M, Gratecos D, Semeriva M. The held out wings (how) Drosophila gene encodes a putative RNA binding protein involved in the control of muscular and cardiac activity. Development. 1997;124:2087–2098. doi: 10.1242/dev.124.10.2087. [DOI] [PubMed] [Google Scholar]
- 43.Zhu W, Wiggins R C, Konat G W. Glucocorticoid-induced upregulation of proteolipid protein and myelin-associated glycoprotein genes in C6 cells. J Neurosci Res. 1994;37:208–212. doi: 10.1002/jnr.490370206. [DOI] [PubMed] [Google Scholar]
- 44.Zorn A M, Grow M, Patterson K D, Ebersole T A, Chen Q, Artzt K, Krieg P A. Remarkable sequence conservation of transcripts encoding amphibian and mammalian homologues of quaking, a KH domain RNA-binding protein. Gene. 1997;188:199–206. doi: 10.1016/s0378-1119(96)00795-0. [DOI] [PubMed] [Google Scholar]
- 45.Zorn A M, Krieg P A. The KH domain protein encoded by quaking functions as a dimer and is essential for notochord development in Xenopus embryos. Genes Dev. 1997;11:2176–2190. doi: 10.1101/gad.11.17.2176. [DOI] [PMC free article] [PubMed] [Google Scholar]