Abstract
Autophagy has emerged as a critical innate immune mechanism for host elimination of intracellular pathogens, however, the role of the autophagy receptor Optineurin during mycobacterial infection is not fully understood. To address this lacuna, we infected bone marrow-derived macrophages (BMDMs) derived from Optn+/+ and Optn−/− mice with Mycobacterium smegmatis, and observed the infection outcome at sequential time points. While low multiplicity of infection (MOI) did not show any significant difference between BMDMs from the two groups, at high MOI Optn−/− mice-derived BMDMs showed significantly lower colony forming unit counts, as well as lower cell counts at 12 h and 24 h post-infection. Quantification of cell numbers and nuclear morphologies at various time points post-infection indicated a markedly higher cell death in the Optineurin-deficient macrophages. Optineurin-deficient BMDMs showed significantly lower levels of the autophagosomal protein LC3-II upon infection, indicating a potential role for Optineurin in regulating autophagy during mycobacterial infection. Moreover, when stimulated by bacterial LPS, Optineurin deficient macrophages, showed altered levels of the inflammatory cytokine pro-IL-1β. These observations taken together suggest a novel regulatory role for Optineurin during mycobacterial infection. Its deficiency leads to an impairment in macrophage responses, directly impacting the pathophysiology of infection.
Keywords: Optineurin, Mycobacteria, Macrophages, Autophagy, Cell death, Cytoprotectivity
Graphical abstract

Highlights
-
•
Mycobacterial infection induces increased cell death in Optineurin deficient BMDMs.
-
•
Infected Optineurin deficient macrophages display diminished autophagy.
-
•
Optineurin deficiency macrophages have disrupted inflammatory responses.
-
•
Its cytoprotective role makes Optineurin therapeutically relevant to TB infection.
1. Introduction
Optineurin is a multifunctional protein that is involved in vesicle trafficking, signal transduction, cell survival, innate immunity, inflammation, and autophagy [[1], [2], [3], [4]]. It functions by interacting with several proteins such as Myosin VI, Rab8, Transferrin receptor, LC3 (MAP1LC3B), TBK1 (Tank Binding Kinase 1), Transcription Factor IIIA, and Ubiquitin [[5], [6], [7], [8], [9], [10]]. Due to its interaction with various molecules that possess diverse functions, a deficiency or mutation affecting its interaction is likely to lead to alterations in cellular homeostasis. Optineurin is also essential for the maintenance of organelle structure and function [11], and its deficiency in cell lines leads to Golgi fragmentation, impaired vesicle trafficking and cell death [12,13]. The property of Optineurin to bind to LC3 and ubiquitinated molecules, simultaneously linking them to autophagosomal membranes, add to its function as an autophagy receptor [7]. Autophagy is an intracellular catabolic process that assists in the maintenance of homeostasis through lysosomal degradation [14]. Post-translational modifications such as phosphorylation of Optineurin by TBK1 regulate the autophagic activity of Optineurin [7,15]. Optineurin-mediated selective autophagy prevents neurodegeneration during herpes virus infection and prevents viral proliferation [16,17]. Intracellular bacterial pathogens such as Salmonella enterica which escape into the cytosol, are degraded by the cellular autophagy machinery, and this process is enhanced by phosphorylation of Optineurin at Serine-177 [7,18]. Optineurin is also known to play an important role in neutrophil recruitment and the associated inflammatory response, events critical to the control of bacterial infection [19].
Mycobacterium tuberculosis (M. tb) the causative agent of human tuberculosis, is one of the oldest coexisting pathogens of humans that still causes over a million deaths annually [20]. The ability of M. tb to establish infection is founded on its ability to evade host defence mechanisms [21,22]. This bacterium has evolved intricate mechanisms to manipulate the host's immune responses, which allow it to survive and replicate in the hostile macrophage environment. While the molecular details of the mycobacterium-macrophage chemistry continue to be elucidated, host autophagy seems to have a predominant role in the progress of the infection [23,24]. M. tb enters its human host via aerosols containing infectious bacilli, that are internalised by the lung's resident alveolar macrophages [20,25]. This initiates a cascade of events leading to the production of cytokines, induction of autophagy and other cellular homeostatic mechanisms, which coordinate to contain the infection [20,26]. However, the status of host immunity, and virulence of the infecting strain, are critical in determining the outcome of infection [27].
The ability to regulate cell death is critical during any microbial infection. Macrophages are the critical component of the innate immune system that serve as the first line of defence against infections [23,28]. However, M. tb has evolved sophisticated strategies that manipulate phagosome trafficking pathways, disrupting normal host cellular microbicidal activities [26]. Consequently, M. tb survives in the phagosome and limits its fusion with the lysosome, eventually leading to the death of macrophages and dissemination of the bacteria [[29], [30], [31]]. The three major consequences of a mycobacterial infection are necrosis, apoptosis, and survival of infected macrophages [22,32], with the kinetics of macrophage death being an important parameter that influences the outcome of infection. In this study, we have investigated the role of Optineurin during mycobacterial infection, using bone marrow-derived macrophages (BMDMs) derived from Optn+/+ and Optn−/− mice. Based on enumeration of cell numbers, classification of nuclear morphologies and quantifying markers of cell death post-infection, our data suggest that deficiency of Optineurin leads to an impairment in macrophage responses to mycobacterial infection, highlighting its novel cytoprotective role in this context.
2. Materials and methods
2.1. Isolation of bone marrow-derived macrophages
All mouse experiments were approved by the Institutional Animal Ethics Committee. Optineurin knockout mice were generated by replacing the Exon 2 of the Optineurin gene with a cassette containing a β-galactosidase reporter and neomycin resistance gene as described previously [33]. Genotyping of these mice was performed as described in Ref. [34]. Primary BMDMs were isolated from the femur, tibia, and humerus of littermate Optn+/+ and Optn−/− male mice. These long bones were sliced at both the ends and the bone marrow was flushed using sterile chilled PBS using a 23G 1.5-inch needle attached to a 10 ml syringe. The recovered bone marrow tissue was homogenized and cultured in 100 cm2 non-adherent dishes for 7 days in Macrophage culture media (80% DMEM +10% L929 conditioned media +10% FBS). After 7 days, the cells were scraped in chilled PBS using a sterile cell scraper, resuspended in macrophage culture media, counted, and plated in 6 well-plates for infection. To the best of our knowledge, the outcome of the experiments is unlikely to be affected by the sex of the mice from which BMDMs are derived.
2.2. Infection of BMDMs
All infection experiments were approved by the Institutional Biosafety Committee. M. smegmatis mc2155 was used for all experiments unless otherwise specified. The mycobacterial strains M. smegmatis mc2155 and M. tb H37Ra were grown in Middlebrook 7H9 broth and Middlebrook 7H10 agar (Difco) supplemented with albumin dextrose complex (5 g BSA, 2 g glucose and 0.85 g NaCl/L), 0.5% (v/v) glycerol and 0.05% (v/v) Tween 80, at 37 °C in a shaker incubator. Infections were carried out by resuspending exponentially growing M. smegmatis in DMEM +10% FBS +10% L929 conditioned medium without antibiotics, after proper declumping and dilution. For M. smegmatis, infections were performed for 4 h unless otherwise specified. Experiments that used M. tb H37Ra for infection allowed a 2 h infection time. Intracellular bacterial growth was measured by performing colony forming unit (CFU) counts, determined at the designated time points by lysing infected cells with 0.1% Triton X-100 followed by dilution plating on Middlebrook 7H10 agar. Cytokine and autophagy levels were assessed by western blotting, and quantification of cell death was performed using fluorescence microscopy.
2.3. Microscopy
Images of macrophage growth and microscopic assessment of cell death were performed with Ziess Axioimager Z1 fluorescence microscope at 10x, 20x and 40x magnification.
2.4. Western blotting
Western blotting was performed as described previously [34]. The signals were developed using the Vilber Lourmat Chemi Doc-5000 instrument and quantified using ImageJ. The antibodies used were as follows: Optineurin (Abcam, Ab23666), LC3B (Enzo life science, ALX80308), Actin (Millipore, MAB1501), Cleaved Caspase-3 (CST, #9664), IL-1β (Santa Cruz - sc-52012), and GAPDH (Millipore MAB374).
2.5. Statistical analysis
The Unpaired t-test was performed to determine levels of significance. A P-value less than 0.05 was considered significant. GraphPad Prism 5 and Microsoft Excel were used for data analysis and graph preparation followed by Adobe Photoshop for figure assembly.
3. Results
3.1. Fate of Mycobacteria upon infection of Optn+/+ & Optn−/− macrophages
To investigate the role of Optineurin in mycobacterial infection, BMDMs from Optn+/+ and Optn−/− mice were infected with Mycobacterium smegmatis (M. smegmatis), an avirulent saprophytic mycobacterial species, at low (10:1) and high (50:1) multiplicity of infection (MOI, Bacteria:Macrophages). This organism has been extensively used as a surrogate model to study the physiology and virulence mechanisms of pathogenic mycobacteria including M. tb [35]. At low MOI, we observed no significant differences in CFU counts in Optn+/+ vs Optn−/− BMDMs. At high MOI however, at 24 h post-infection, Optn−/− BMDMs showed lower mycobacterial counts than Optn+/+ BMDMs (Fig. 1). The reduction in CFU counts over time, signifies macrophage killing of M. smegmatis, a reflection of its non-pathogenic nature.
Fig. 1.
Intracellular viability of M. smegmatis in Optn−/−macrophages. CFU counts of M. smegmatis upon infection of Optn+/+ and Optn−/− BMDMs at an MOI of 10:1 (A) and 50:1 (B). (C) Optn+/+ and Optn−/− BMDM cell counts post 50:1 MOI M. smegmatis infection (n = 4 independent experiments, minimum 300 cells per experiment per genotype assessed) Bars represent mean ± SD. **p ≤ 0.01, ***p ≤ 0.001.
3.2. M. smegmatis induced higher cell death in optineurin-deficient macrophages at 50:1 MOI
Based on the above observations, we proceeded to investigate the effect of mycobacterial infection on Optn−/− BMDMs using an MOI of 50:1. We found that the loss of cell numbers due to M. smegmatis infection was higher in Optn−/− BMDMs compared to the Optn+/+ BMDMs. To quantify this effect, BMDMs were plated onto coverslips, and infected with M. smegmatis at an MOI of 50:1. After infection, coverslips with infected BMDMs were fixed and stained with DAPI at 4 h, 8 h, and 20 h time points, followed by assessment of total cell count per field. Random fields were selected and cells per field were counted under phase contrast at 400x magnification. On performing these counts, we observed a significantly lower number of Optn−/− BMDMs at 8 h and 20 h post infection (Fig. 1C). To gain a deeper insight into the loss of Optn/- BMDMs upon infection, we utilized DAPI-stained BMDMs for further microscopic examination. On further observation, we noticed alterations in the nuclear morphology of BMDMs post-infection, which were classified into three distinct types based on microscopic examination (Fig. 2A, B, C, D). - normal nuclei, pyknotic nuclei, and pale-staining/poorly staining nuclei. Uninfected BMDMs from Optn+/+ and Optn−/− mice showed similar gross cellular and nuclear morphology, and BMDMs from both Optn+/+ and Optn−/− mice showed aggregation and altered cellular morphology 8 h and 20 h post-infection. We observed a considerable reduction in the number of BMDMs 8 h post-infection, with altered nuclear morphology and an increase in pyknotic and pale staining nuclei. Notably, Optn−/− BMDMs showed a significantly higher percentage of pale staining nuclei compared to Optn+/+ BMDMs 20 h post-infection (Fig. 2E). We made similar observations with high MOI infection of BMDMs with M. tb H37Ra as well (Fig. S1). These findings suggest that mycobacterial infection alters the cellular and nuclear morphology of BMDMs, which may have implications in the immune response and pathogenesis of mycobacterial infections.
Fig. 2.
Microscopic observation of nuclear morphology changes in M. smegmatis infected BMDMs at 50:1 MOI: (A) Cellular and nuclear morphology of uninfected BMDMs from Optn+/+ and Optn−/− mice. Cellular and nuclear morphology of M. smegmatis infected BMDMs from Optn+/+ and Optn−/− mice, 0 h (B), 8 h (C), and 20 h (D) post infection. (E). Quantification of BMDMs based on nuclear morphology into normal, pyknotic and pale staining nucleus. (n = 4 independent experiments, minimum 300 cells per experiment per genotype assessed) Bars represent mean ± SD. ***p ≤ 0.001.
3.3. Optineurin-deficient macrophages infected with M. smegmatis at high MOI induce diminished LC3 II levels
The observed changes in nuclear morphology suggested that upon mycobacterial infection, BMDMs undergo time-dependent cellular changes that eventually lead to cell death. Since Optineurin is an autophagy receptor, we investigated changes in autophagy during mycobacterial infection. Induction of LC3II levels in BMDMs from both Optn+/+ and Optn−/− mice-derived BMDMs were observed post-infection with M. smegmatis (Fig. 3A), however, BMDMs derived from Optn−/− mice showed reduced induction of LC3II 20 h post infection (Fig. 3B). This finding is consistent with the observation of increased cell death in Optn−/− BMDMs and suggests that altered autophagy may be one of the reasons for this cell death phenomenon. We also quantified the levels of activated caspase-3 in BMDMs and found that the differences in these levels were not significant. This suggested that classical apoptosis did not play a significant role in the altered death of infected Optn−/− BMDMs.
Fig. 3.
Assessment of autophagy in M. smegmatis infected BMDMs from Optn+/+and Optn−/−mice: (A) Representative Western blot showing the levels of LC3, Cleaved Caspase 3, Optineurin, and β-Actin in M. smegmatis infected BMDMs isolated from Optn+/+ and Optn−/− mice. (B) Bar diagram showing quantified levels of LC3 II, at 20 h post-infection. (n = 4 independent experiments) Bars represent mean ± SD. ***p ≤ 0.001. ‘#’ indicates a non-specific band. Con. UI = Uninfected Control macrophages.
3.4. Optineurin-deficient macrophages show altered inflammatory responses
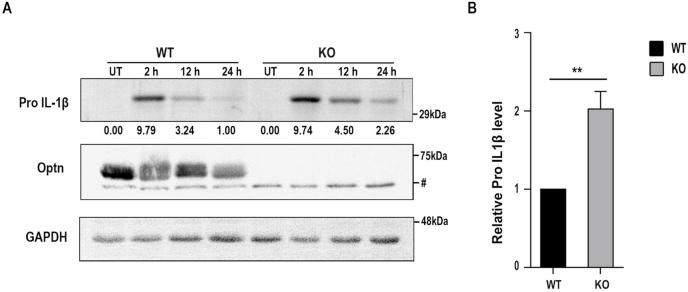
Interleukin-1β (IL-1β) is a crucial cytokine produced and secreted by macrophages in response to infection. The protein is synthesized as a 32 kDa pro-protein and processed by Caspase-1 into an active 17.5 kDa form, which is then secreted into the bloodstream. IL-1β functions as a lymphocyte activating factor and a leukocytic pyrogen, mediating inflammatory responses. To investigate the impact of Optineurin deficiency on the inflammatory response of BMDMs, we treated primary BMDMs derived from Optn+/+ and Optn−/− mice with 1 μg/ml LPS derived from E. coli and measured the induction of Pro IL-1β. Western blot analysis showed that Optn−/− BMDMs had higher levels of Pro IL-1β compared to Optn+/+ BMDMs after 24 h of LPS treatment (Fig. 4A). Statistical analysis revealed that the difference in Pro IL-1β levels was significant (Fig. 4B). These results suggest altered inflammatory response signalling in primary BMDMs under conditions of Optineurin deficiency.
Fig. 4.
Levels of Pro-IL-1β in BMDMs from Optn+/+and Optn−/−mice: (A) Levels of Pro IL-1β in BMDMs isolated from Optn+/+ and Optn−/− mice on treatment with 1 μg/ml LPS. B) Bar diagram showing quantified levels of Pro IL-1β at 24 h post LPS treatment (n = 3 independent experiments). Bars represent mean ± SD. **p ≤ 0.01. ‘#’ indicates a non-specific band.
4. Discussion
A report investigating the role of Optineurin in bacterial infection showed that optineurin restricts the growth of Salmonella in HeLa cells, consistent with its function as an autophagy receptor [7]. However, our results show that while Optineurin deficiency did not alter CFU counts under low MOI M. smegmatis infection of BMDMs, under high MOI conditions, CFU counts in infected BMDMs derived from Optn−/− mice were lower than BMDMs derived from Optn+/+ mice. It is probable that this results from the large increase in cell death in Optn−/− mice-derived BMDMs, which then become freely permeable to the gentamycin in the culture medium, thereby killing intracellular M. smegmatis. As a consequence, bacteria in dead BMDMs would not contribute to the CFU counts, leading to the lower CFU counts that we observe in infected Optn−/− BMDMs.
Xenophagy clears pathogenic intracellular organisms using autophagic receptors such as Optineurin, to recognise the ubiquitinated bacteria that are destined to be degraded [36,37]. The cargo destined for degradation is brought to LC3-positive phagophores called autophagosomes [38]. Optineurin appears to be important for pathogen clearance, as it was found to regulate the growth and replication of Salmonella, and Optineurin deficient mice were observed to be more susceptible to Salmonella enterica serovar typhimurium infection [7,19]. The association between Optineurin and mycobacterial infection has been a subject of recent investigations. Optineurin expression was observed to be upregulated upon M. marinum infection in macrophages. In addition, a deficiency of Optineurin leads to increased susceptibility to M. marinum infection [39,40]. Among various mycobacterial species M. smegmatis is recognized for its ability to induce the highest level of autophagy [41]. Since our focus was to investigate the role of the autophagy receptor Optineurin during mycobacterial infection, selecting this as the test organism for our studies was the logical choice. Optineurin deficiency in BMDMs causes reduced macrophage autophagy, as observed by reduced LC3II levels upon M. smegmatis infection. It is known from the literature that a deficiency of Optineurin leads to reduced autophagosome formation and reduces cargo selective autophagy [42,43]. In our experiments, infection with M. smegmatis resulted in increased pale staining nuclei in optineurin deficient BMDMs, which are likely to be dead macrophages. This cell death in infected BMDMs coinciding with diminished autophagy, could be the cause of increased cell death in Optn−/− mice derived BMDMs during M. smegmatis infection.
The observation that LPS treatment of Optineurin-deficient BMDMs leads to altered inflammatory stress response signalling, suggests that optineurin deficiency by itself predisposes cells to altered inflammatory signalling, and therefore leads to a poor infection response in BMDMs. Reduced expression of Optineurin is linked to impaired cytokine secretion by macrophages and altered inflammatory responses [44]. In addition, a reduction in Optineurin expression in mice led to diminished levels of pro-inflammatory TNFα in the serum, diminished cytokine secretion, and diminished neutrophil recruitment to sites of acute inflammation, resulting in greater mortality after bacterial stimulation [45]. Our findings show that Optineurin-deficient BMDMs are more sensitive to M. smegmatis induced cell death indicating that Optineurin-mediated modulation of the infection response has a cytoprotective function against M. smegmatis induced cell death.
Exposure of BMDMs to high MOI M. smegmatis leads to rapid and substantial bacterial uptake. In contrast to low MOI infection, this high bacterial load results in BMDMs becoming overwhelmed, leading to their death. Our results show that Optineurin deficient BMDMs have defective pro-inflammatory signalling. The excess of immune signalling molecules during high MOI infection can lead to cytotoxic effects on BMDMs themselves, contributing to increased cell death. BMDMs are known to generate reactive oxygen species as a defence mechanism against invading pathogens. Optineurin is expressed in bone and neuronal cells and has been reported to protect cells from ROS-induced cell damage [46,47]. However, in high MOI mycobacterial infection, the excessive production of ROS can cause cellular damage and contribute to death of BMDMs, with the absence of Optineurin intensifying these effects. The mechanistics of the causal relationships between cell death, autophagy and IL1β levels, presently remain unclear, and are part of our ongoing investigations.
5. Conclusion
In this study, Optineurin-deficient BMDMs exhibited an elevated cell death when infected with M. smegmatis compared to Optineurin-expressing BMDMs. Also, the deficiency of Optineurin was observed to impair autophagy, as demonstrated by the lower levels of autophagosomal protein LC3-II in Optineurin-deficient BMDMs. Stimulation with bacterial LPS altered pro-IL-1β levels in Optineurin-deficient BMDMs. These results underscore the pivotal role that Optineurin plays in modulating macrophage responses during mycobacterial infection, and its absence results in disrupted cytokine signals and autophagy, crucial events in the host infection response. Our observations emphasise the dual significance of Optineurin - as a cytoprotective factor and a potential therapeutic target in tuberculosis treatment.
Funding information
This work was supported by grant from the Council of Scientific and Industrial Research (MLP 0144), Government of India (CSIR) (to T. R. R.), and by a J.C. Bose National Fellowship grant (SR/S2/JCB-41/2010) from the Department of Science and Technology, Government of India (to G.S.). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript. G. R. was supported by a fellowship from the Indian Council of Medical Research, C.V.Y. was supported by a Research Associateship from the Department of Biotechnology, Government of India.
CRediT authorship contribution statement
Gopalakrishna Ramachandran: Writing – review & editing, Writing – original draft, Validation, Methodology, Investigation, Formal analysis, Data curation, Conceptualization. Chaitanya Veena Yeruva: Validation, Methodology, Investigation, Formal analysis. Ghanshyam Swarup: Writing – review & editing, Writing – original draft, Supervision, Resources, Project administration, Methodology, Funding acquisition, Conceptualization. Tirumalai R. Raghunand: Writing – review & editing, Writing – original draft, Supervision, Resources, Project administration, Methodology, Funding acquisition, Formal analysis, Conceptualization.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.bbrep.2024.101672.
Contributor Information
Gopalakrishna Ramachandran, Email: gopalakrishnaiyer@gmail.com.
Chaitanya Veena Yeruva, Email: chaitanyaveena@gmail.com.
Ghanshyam Swarup, Email: gshyam@ccmb.res.in.
Tirumalai R. Raghunand, Email: raghu@ccmb.res.in.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Toth R.P., Atkin J.D. Dysfunction of optineurin in amyotrophic lateral sclerosis and glaucoma. Front. Immunol. 2018;9:1017. doi: 10.3389/fimmu.2018.01017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Markovinovic A., Cimbro R., Ljutic T., Kriz J., Rogelj B., Munitic I. Optineurin in amyotrophic lateral sclerosis: multifunctional adaptor protein at the crossroads of different neuroprotective mechanisms. Prog. Neurobiol. 2017;154:1–20. doi: 10.1016/j.pneurobio.2017.04.005. [DOI] [PubMed] [Google Scholar]
- 3.Swarup G., Sayyad Z. Altered functions and interactions of glaucoma-associated mutants of optineurin. Front. Immunol. 2018;9:1287. doi: 10.3389/fimmu.2018.01287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ramachandran G., Moharir S.C., Raghunand T.R., Swarup G. Optineurin modulates ER stress-induced signaling pathways and cell death. Biochem. Biophys. Res. Commun. 2021;534:297–302. doi: 10.1016/j.bbrc.2020.11.091. [DOI] [PubMed] [Google Scholar]
- 5.Ying H., Shen X., Park B., Yue B.Y. Posttranslational modifications, localization, and protein interactions of optineurin, the product of a glaucoma gene. PLoS One. 2010;5 doi: 10.1371/journal.pone.0009168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chibalina M.V., Roberts R.C., Arden S.D., Kendrick-Jones J., Buss F. Rab8-optineurin-myosin VI: analysis of interactions and functions in the secretory pathway. Methods Enzymol. 2008;438:11–24. doi: 10.1016/S0076-6879(07)38002-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wild P., Farhan H., McEwan D.G., Wagner S., Rogov V.V., Brady N.R., Richter B., Korac J., Waidmann O., Choudhary C., Dotsch V., Bumann D., Dikic I. Phosphorylation of the autophagy receptor optineurin restricts Salmonella growth. Science (New York, N.Y.) 2011;333:228–233. doi: 10.1126/science.1205405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hattula K., Peranen J. FIP-2, a coiled-coil protein, links Huntingtin to Rab8 and modulates cellular morphogenesis. Curr. Biol. 2000;10:1603–1606. doi: 10.1016/s0960-9822(00)00864-2. [DOI] [PubMed] [Google Scholar]
- 9.Park B., Ying H., Shen X., Park J.S., Qiu Y., Shyam R., Yue B.Y. Impairment of protein trafficking upon overexpression and mutation of optineurin. PLoS One. 2010;5 doi: 10.1371/journal.pone.0011547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Moreland R.J., Dresser M.E., Rodgers J.S., Roe B.A., Conaway J.W., Conaway R.C., Hanas J.S. Identification of a transcription factor IIIA-interacting protein. Nucleic Acids Res. 2000;28:1986–1993. doi: 10.1093/nar/28.9.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ryan T.A., Tumbarello D.A. Optineurin: a coordinator of membrane-associated cargo trafficking and autophagy. Front. Immunol. 2018;9:1024. doi: 10.3389/fimmu.2018.01024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sahlender D.A., Roberts R.C., Arden S.D., Spudich G., Taylor M.J., Luzio J.P., Kendrick-Jones J., Buss F. Optineurin links myosin VI to the Golgi complex and is involved in Golgi organization and exocytosis. J. Cell Biol. 2005;169:285–295. doi: 10.1083/jcb.200501162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sippl C., Bosserhoff A.K., Fischer D., Tamm E.R. Depletion of optineurin in RGC-5 cells derived from retinal neurons causes apoptosis and reduces the secretion of neurotrophins. Exp. Eye Res. 2011;93:669–680. doi: 10.1016/j.exer.2011.08.011. [DOI] [PubMed] [Google Scholar]
- 14.Das G., Shravage B.V., Baehrecke E.H. Regulation and function of autophagy during cell survival and cell death. Cold Spring Harbor Perspect. Biol. 2012;4 doi: 10.1101/cshperspect.a008813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Richter B., Sliter D.A., Herhaus L., Stolz A., Wang C., Beli P., Zaffagnini G., Wild P., Martens S., Wagner S.A., Youle R.J., Dikic I. Phosphorylation of OPTN by TBK1 enhances its binding to Ub chains and promotes selective autophagy of damaged mitochondria. Proc. Natl. Acad. Sci. U. S. A. 2016;113:4039–4044. doi: 10.1073/pnas.1523926113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ames J., Yadavalli T., Suryawanshi R., Hopkins J., Agelidis A., Patil C., Fredericks B., Tseng H., Valyi-Nagy T., Shukla D. OPTN is a host intrinsic restriction factor against neuroinvasive HSV-1 infection. Nat. Commun. 2021;12:5401. doi: 10.1038/s41467-021-25642-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Patil C.D., Shukla D. OPTN (optineurin)-mediated selective autophagy prevents neurodegeneration due to herpesvirus infection. Autophagy. 2022;18:944–945. doi: 10.1080/15548627.2022.2037223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cemma M., Brumell J.H. Interactions of pathogenic bacteria with autophagy systems. Curr. Biol. 2012;22:R540–R545. doi: 10.1016/j.cub.2012.06.001. [DOI] [PubMed] [Google Scholar]
- 19.Slowicka K., Vereecke L., Mc Guire C., Sze M., Maelfait J., Kolpe A., Saelens X., Beyaert R., van Loo G. Optineurin deficiency in mice is associated with increased sensitivity to Salmonella but does not affect proinflammatory NF-κB signaling. Eur. J. Immunol. 2016;46:971–980. doi: 10.1002/eji.201545863. [DOI] [PubMed] [Google Scholar]
- 20.Smith I. Mycobacterium tuberculosis pathogenesis and molecular determinants of virulence. Clin. Microbiol. Rev. 2003;16:463–496. doi: 10.1128/CMR.16.3.463-496.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu C.H., Liu H., Ge B. Innate immunity in tuberculosis: host defense vs pathogen evasion. Cell. Mol. Immunol. 2017;14:963–975. doi: 10.1038/cmi.2017.88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Behar S.M., Divangahi M., Remold H.G. Evasion of innate immunity by Mycobacterium tuberculosis: is death an exit strategy? Nat. Rev. Microbiol. 2010;8:668–674. doi: 10.1038/nrmicro2387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Deretic V. Autophagy in tuberculosis. Cold Spring Harb Perspect Med. 2014;4 doi: 10.1101/cshperspect.a018481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bento C.F., Empadinhas N., Mendes V. Autophagy in the fight against tuberculosis. DNA Cell Biol. 2015;34:228–242. doi: 10.1089/dna.2014.2745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jones-Lopez E.C., Acuna-Villaorduna C., Ssebidandi M., Gaeddert M., Kubiak R.W., Ayakaka I., White L.F., Joloba M., Okwera A., Fennelly K.P. Cough aerosols of Mycobacterium tuberculosis in the prediction of incident tuberculosis disease in household contacts. Clin. Infect. Dis. 2016;63:10–20. doi: 10.1093/cid/ciw199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhai W., Wu F., Zhang Y., Fu Y., Liu Z. The immune escape mechanisms of Mycobacterium tuberculosis. Int. J. Mol. Sci. 2019;20 doi: 10.3390/ijms20020340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sia J.K., Rengarajan J. Immunology of Mycobacterium tuberculosis infections. Microbiol. Spectr. 2019;7 doi: 10.1128/microbiolspec.gpp3-0022-2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Weiss G., Schaible U.E. Macrophage defense mechanisms against intracellular bacteria. Immunol. Rev. 2015;264:182–203. doi: 10.1111/imr.12266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Armstrong J.A., Hart P.D. Response of cultured macrophages to Mycobacterium tuberculosis, with observations on fusion of lysosomes with phagosomes. J. Exp. Med. 1971;134:713–740. doi: 10.1084/jem.134.3.713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cambier C.J., Falkow S., Ramakrishnan L. Host evasion and exploitation schemes of Mycobacterium tuberculosis. Cell. 2014;159:1497–1509. doi: 10.1016/j.cell.2014.11.024. [DOI] [PubMed] [Google Scholar]
- 31.Pieters J. Mycobacterium tuberculosis and the macrophage: maintaining a balance. Cell Host Microbe. 2008;3:399–407. doi: 10.1016/j.chom.2008.05.006. [DOI] [PubMed] [Google Scholar]
- 32.Divangahi M., Behar S.M., Remold H. Dying to live: how the death modality of the infected macrophage affects immunity to tuberculosis. Adv. Exp. Med. Biol. 2013;783:103–120. doi: 10.1007/978-1-4614-6111-1_6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bansal M., Moharir S.C., Sailasree S.P., Sirohi K., Sudhakar C., Sarathi D.P., Lakshmi B.J., Buono M., Kumar S., Swarup G. Optineurin promotes autophagosome formation by recruiting the autophagy-related Atg12-5-16L1 complex to phagophores containing the Wipi2 protein. J. Biol. Chem. 2018;293:132–147. doi: 10.1074/jbc.M117.801944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Moharir S.C., Bansal M., Ramachandran G., Ramaswamy R., Rawat S., Raychaudhuri S., Swarup G. Identification of a splice variant of optineurin which is defective in autophagy and phosphorylation. Biochim. Biophys. Acta Mol. Cell Res. 2018;1865:1526–1538. doi: 10.1016/j.bbamcr.2018.08.009. [DOI] [PubMed] [Google Scholar]
- 35.Sparks I.L., Derbyshire K.M., Jacobs W.R., Jr., Morita Y.S. Mycobacterium smegmatis: the vanguard of mycobacterial Research. J. Bacteriol. 2023;205 doi: 10.1128/jb.00337-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mizushima N., Komatsu M. Autophagy: renovation of cells and tissues. Cell. 2011;147:728–741. doi: 10.1016/j.cell.2011.10.026. [DOI] [PubMed] [Google Scholar]
- 37.Gatica D., Lahiri V., Klionsky D.J. Cargo recognition and degradation by selective autophagy. Nat. Cell Biol. 2018;20:233–242. doi: 10.1038/s41556-018-0037-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Khaminets A., Behl C., Dikic I. Ubiquitin-dependent and independent signals in selective autophagy. Trends Cell Biol. 2016;26:6–16. doi: 10.1016/j.tcb.2015.08.010. [DOI] [PubMed] [Google Scholar]
- 39.Benard E.L., Rougeot J., Racz P.I., Spaink H.P., Meijer A.H. Transcriptomic approaches in the zebrafish model for tuberculosis-insights into host- and pathogen-specific determinants of the innate immune response. Adv. Genet. 2016;95:217–251. doi: 10.1016/bs.adgen.2016.04.004. [DOI] [PubMed] [Google Scholar]
- 40.Zhang R., Varela M., Vallentgoed W., Forn-Cuni G., van der Vaart M., Meijer A.H. The selective autophagy receptors Optineurin and p62 are both required for zebrafish host resistance to mycobacterial infection. PLoS Pathog. 2019;15 doi: 10.1371/journal.ppat.1007329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zullo A.J., Lee S. Mycobacterial induction of autophagy varies by species and occurs independently of mammalian target of rapamycin inhibition. J. Biol. Chem. 2012;287:12668–12678. doi: 10.1074/jbc.M111.320135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Qiu Y., Wang J., Li H., Yang B., Wang J., He Q., Weng Q. Emerging views of OPTN (optineurin) function in the autophagic process associated with disease. Autophagy. 2022;18:73–85. doi: 10.1080/15548627.2021.1908722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bansal M., Moharir S.C., Swarup G. Autophagy receptor optineurin promotes autophagosome formation by potentiating LC3-II production and phagophore maturation. Commun. Integr. Biol. 2018;11:1–4. doi: 10.1080/19420889.2018.1467189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Smith A.M., Sewell G.W., Levine A.P., Chew T.S., Dunne J., O'Shea N.R., Smith P.J., Harrison P.J., Macdonald C.M., Bloom S.L., Segal A.W. Disruption of macrophage pro-inflammatory cytokine release in Crohn's disease is associated with reduced optineurin expression in a subset of patients. Immunology. 2015;144:45–55. doi: 10.1111/imm.12338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chew T.S., O'Shea N.R., Sewell G.W., Oehlers S.H., Mulvey C.M., Crosier P.S., Godovac-Zimmermann J., Bloom S.L., Smith A.M., Segal A.W. Optineurin deficiency in mice contributes to impaired cytokine secretion and neutrophil recruitment in bacteria-driven colitis. Dis Model Mech. 2015;8:817–829. doi: 10.1242/dmm.020362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gao J., Ohtsubo M., Hotta Y., Minoshima S. Oligomerization of optineurin and its oxidative stress- or E50K mutation-driven covalent cross-linking: possible relationship with glaucoma pathology. PLoS One. 2014;9 doi: 10.1371/journal.pone.0101206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhu M., Li A., Chen J., Zhang S., Wu J. Effects of optineurin mutants on SH-SY5Y cell survival. Mol. Cell. Neurosci. 2016;74:18–24. doi: 10.1016/j.mcn.2016.03.003. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.